Abstract
Development of new, selective inhibitors of nicotinamide adenine dinucleotide phosphate oxidase (NOX) isoforms is important both for basic studies on the role of these enzymes in cellular redox signaling, cell physiology, and proliferation and for development of new drugs for diseases carrying a component of increased NOX activity, such as several types of cancer and cardiovascular and neurodegenerative diseases. High-throughput screening (HTS) of large libraries of compounds remains the major approach for development of new NOX inhibitors. Here, we describe the protocol for the HTS campaign for NOX inhibitors using rigorous assays for superoxide radical anion and hydrogen peroxide, based on oxidation of hydropropidine, coumarin boronic acid, and Amplex Red. We propose using these three probes to screen for and identify new inhibitors, by selecting positive hits that show inhibitory effects in all three assays. Protocols for the synthesis of hydropropidine and for confirmatory assays, including oxygen consumption measurements, electron paramagnetic resonance spin trapping of superoxide, and simultaneous monitoring of superoxide and hydrogen peroxide, are also provided.
Keywords: High-throughput screening, Superoxide radical anion, Hydrogen peroxide, Hydropropidine, Coumarin boronic acid, Amplex Red, Fluorescence, EPR spin trapping, Seahorse extracellular flux analyzer
1. Introduction
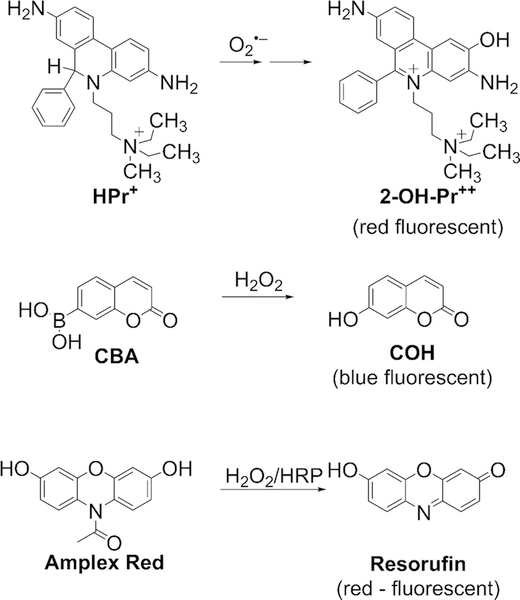
The members of the family of nicotinamide adenine dinucleotide phosphate (NADPH) oxidase enzymes (NOX1–5, DUOX1–2) are specialized multicomponent enzymes, activatable or constitutively active, for which a sole function is to transfer electrons from the NADPH coenzyme to molecular oxygen, producing superoxide radical anion (O2•−) and hydrogen peroxide (H2O2), a product of dismutation of O2•− (Fig. 1) [1, 2]. Different isoforms of NADPH oxidases have been implicated in cellular redox signaling, immune response to pathogens, and pathological conditions, including cancer and cardiovascular and neurodegenerative diseases [3–5]. The availability of specific pharmacological inhibitors, therefore, provides both basic scientific and translational potential. However, the development of specific NOX enzyme inhibitors is not trivial because no crystal structure of any member of the NOX family of enzymes is available, limiting the ability to use computational tools or rational design of new inhibitors. Thus, new approaches to discovering inhibitors of NADPH oxidases are mostly based on “untargeted” screening of the libraries of chemical compounds, including large libraries available in the specialized screening centers [6–15]. Typically, high-throughput screening (HTS) assays are based on the detection of oxidants generated by NOX enzymes, using chemiluminescent or fluorescent probes [16–18]. Many of those probes, however, do not react directly with O2•− or H2O2 and require “activation” via electron transfer mechanisms or a catalyst. Thus, many compounds inhibiting the luminescent (chemiluminescence, fluorescence) signal may act by blocking the activation step or competing for the catalyst with the probe rather than by blocking the formation of O2•− or H2O2. This leads to a high rate of false-positive hits, necessitating cost-and time-consuming confirmatory assays. In fact, a recent report on myeloperoxidase inhibitors was based on the initial screening campaign for NOX inhibitors [19]. The results of confirmatory assays were critical to accurately assign the mode of action of the identified positive hits. In several cases, the claims of NOX-inhibitory activity have not been confirmed using more rigorous assays and/or monitoring NOX activity in cell-free systems in independent laboratories [11, 13]. Recent development in fluorescent probes for O2•− and H2O2 (Fig. 2) enables the use of probes reacting directly with O2•− (hydropropidine, HPr+) or H2O2 (coumarin boronic acid, CBA), avoiding many limitations of the previously used probes [14, 20–22]. Upon the reaction between HPr+ and O2•−, a highly specific product, 2-hydroxypropidium (2-OH-Pr++), is formed. The fluorescence yield of this product is further increased upon binding to DNA [20]. In the presence of H2O2, CBA is oxidized to 7-hydroxycoumarin (COH, also known as umbelliferone). In addition, Amplex Red (10-acetyl-3,7-dihydroxyphenoxazine) undergoes oxidation by H2O2 to resorufin in a peroxidase-catalyzed reaction. Understanding the chemistry behind the detection of O2•− and H2O2 using these probes opened the possibility of rigorous monitoring of NOX activity in a high throughput manner [17, 23–27]. Correlation of the results obtained with three probes with different probing mechanisms will enable identification of NOX inhibitors with higher confidence, as compared with previously used probes.
Fig. 1.
Scheme showing the enzymatic activity of NADPH oxidases (NOX)
Fig. 2.
Chemical structures of probes and their oxidation products used for HTS of NOX inhibitors
Here, we describe the protocol for HTS of NOX inhibitors in intact cells using three probes of different ROS sensing mechanisms, including HPr+ for monitoring O2•− generation and CBA for monitoring H2O2 production. The third probe, Amplex Red, is used to detect H2O2 in the presence of horseradish peroxidase (HRP). We also provide protocols for cytotoxicity measurement, electron paramagnetic resonance (EPR) spin trapping, oximetry, and simultaneous monitoring of O2•− and H2O2 for confirmatory purposes for potential NOX inhibitors identified in the HTS campaign.
2. Materials
2.1. Components for the Synthesis of Hydropropidine
0.1 g (0.15 mmol) of propidium iodide.
20 mL of methanol (MeOH).
6 mg (0.16 mmol) of sodium borohydride.
20 mL of dichloromethane (DCM).
Argon gas.
1 g of anhydrous sodium sulfate.
2.2. Cell Incubation and Differentiation Components
Cell growth medium: Roswell Park Memorial Institute (RPMI) 1640 medium containing 10% fetal bovine serum (FBS), 100 units/mL penicillin, and 100 μg/mL streptomycin (see Note 1).
0.1 M all-trans retinoic acid (ATRA) in ethanol. The solution may be stored at −80 °C for at least 1 year.
2.3. HTS Assay Components
HBSS containing 25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), pH 7.4, and 0.1 mM diethylenetriaminepentaacetic acid (DTPA). Adjust pH to 7.4 using concentrated HCl or NaOH if necessary.
0.25mMsolution of hydropropidine (HPr+) in HBSS containing 0.5 mg/mL DNA, 25 mM HEPES, pH 7.4, 0.1 mM DTPA, and 0.1 mg/mL bovine serum albumin (BSA) (see Note 2).
0.5 mM solution of coumarin boronic acid (CBA) in HBSS containing 25 mM HEPES, pH 7.4, 0.1 mM DTPA, and 0.1 mg/mL BSA (see Note 2).
0.25 mM solution of Amplex Red in HBSS containing 0.5 U/mL HRP, 25 mM HEPES, pH 7.4, 0.1 mM DTPA, and 0.1 mg/mL BSA (see Note 2).
5 μM solution of phorbol 12-myristate 13-acetate (PMA) in HBSS containing 25 mM HEPES, pH 7.4, 0.1 mM DTPA, and 0.1 mg/mL BSA (see Note 2).
2.4. ATP Measurements (See Note 3)
RPMI medium without phenol red and bicarbonate.
Adenosine triphosphate (ATP) releasing reagent (lysis buffer, component of a kit for bioluminescence-based ATP measurements).
ATP bioluminescence reagent (mix of luciferase and cofactors, component of a kit for bioluminescence-based ATP measurements).
2.5. Seahorse XF96-Based Oximetry
RPMI medium without phenol red and bicarbonate.
10 μM PMA solution in RPMI medium without phenol red and bicarbonate, containing 0.1 mg/mL BSA.
2.6. EPR Spin Trapping of O2•−
2.7. Simultaneous Measurements of O2•− and H2O2
HBSS containing 25 mM HEPES, pH 7.4, and 0.1 mM DTPA.
Mixture of hydroethidine (HE, 6.7 mM), CBA (33 mM), and PMA (0.33 mM) in dimethyl sulfoxide (DMSO) (see Note 2).
A mixture of superoxide dismutase (SOD, 50×) and catalase (CAT, 50×): prepare a solution of SOD (5 mg/mL) and CAT (5 kU/mL) in water (see Note 4).
3. Methods
3.1. Preparation of HPr+
Prepare HPr+ (steps 2–12) inside a well-ventilated hood.
Deoxygenate 20 mL of MeOH by slowly passing argon gas through it for 10 min.
Deoxygenate 20 mL of DCM by slowly passing argon gas through it for 10 min.
Dissolve sodium borohydride (6 mg) in 1 mL of deoxygenated MeOH.
Dissolve propidium iodide (0.1 g) in 5 mL of deoxygenated MeOH.
Add sodium borohydride solution dropwise to the solution of propidium iodide, stirring constantly.
After 10 min of stirring under Ar atmosphere, add 1 mL of deoxygenated water and 5 mL of deoxygenated DCM, and vigorously shake the mixture for 1 min.
Allow the two phases to separate and transfer the DCM phase (lower) into an empty 20 mL vial.
Repeat steps 7–8 five times and combine the DCM extracts.
Add 1 g of anhydrous sodium sulfate to the DCM extract, and stir the suspension for 10 min.
Filter the suspension and place the filtrate in an empty 20 mL glass vial.
Remove the solvent under vacuum (using a rotary evaporator) or by flushing the solution with argon or nitrogen gas.
Store the solid at −20 °C or lower.
Dissolve in deoxygenated aqueous solution of 1 mM HCl to a final concentration of 1 mM, freeze in liquid nitrogen, and store at −80 °C (see Note 5).
3.2. Preparation of Differentiated HL60 Cells Expressing NOX2 Isoform (See Note 1)
Grow HL60 cells in RPMI 1640 medium supplemented with FBS and antibiotics.
Count cells in suspension and prepare a suspension of 105 cells/mL.
Add ATRA to cells to a final concentration of 1 μM.
Incubate cells for 4 days.
Inspect cell morphology to confirm differentiation, as described elsewhere [28, 29]. Collect and save some cells to test for NOX2 expression (see Note 6).
3.3. HTS of NOX Inhibitors
Place the suspension of differentiated HL60 (dHL60) cells in 50 mL centrifuge tube(s), and spin down the cells (see Note 7).
Remove the supernatant and resuspend the cells in pre-warmed HBSS containing 25 mM HEPES, pH 7.4, and 0.1 mM DTPA.
Determine the cell density by cell counting, and dilute the cells to the density of 105 cells/mL.
Transfer the cell suspension into black 384-well plates; use 30 μL/well (see Note 8).
Add ~100 nL of stock solutions of the compounds to be tested (the chemical library) (see Notes 9 and 10).
Incubate the cells with compounds for 30 min at 37 °C in a carbon dioxide (CO2)-free incubator under ambient oxygen (21% O2).
After incubation, add 10 μL/well of freshly prepared solution of the probe (5× solution) in HBSS containing 25 mM HEPES, pH 7.4, 0.1 mM DTPA, and 0.1 mg/mL BSA (see Note 8).
Add 10 μL/well of 5 μM solution of PMA in HBSS containing 25mMHEPES, pH 7.4, 0.1mMDTPA, and 0.1 mg/mL BSA (see Note 8).
Incubate the cells with reactive oxygen species (ROS) probes for 60 min at 37° C in a CO2-free incubator (see Note 11).
Measure fluorescence intensity using a plate reader. The parameters for excitation and emission filters for each probe are shown in Table 1 (see Note 12).
Repeat steps 1–10 for each ROS probe.
Table 1.
Fluorescence parameters used for monitoring the oxidation products of HPr+, CBA, and Amplex Red probes
| Probe | Excitation wavelength (nm) | Emission wavelength (nm) |
|---|---|---|
| HPr+ | 485 | 574 |
| CBA | 355 | 460 |
| Amplex Red | 535 | 595 |
3.4. HTS Data Analysis
-
For each plate, determine the Z’ value using control positive (PMA, no inhibitor, “control+”) and control negative (PMA + known inhibitor, “control−”) samples in columns 1, 2, 23, and 24 (see Note 9). Use the following formula:
-
Normalize the fluorescence intensity data for each sample (Ix) so that the mean fluorescence intensity of sample control+ (Ic+) is set to 0% and the mean fluorescence intensity of sample control −(Ic−) is set to −100%, using the following formula:
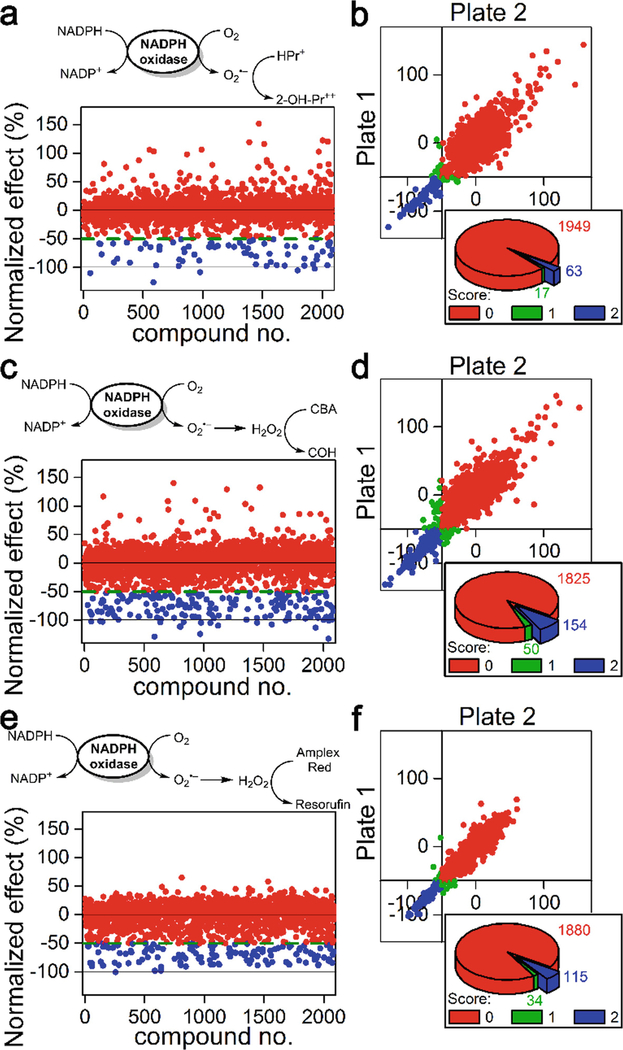
Identify the positive hits, exhibiting normalized fluorescence (NIx) values (Fig. 3) below a set threshold (see Note 13).
Perform steps 1–3 for each probe used.
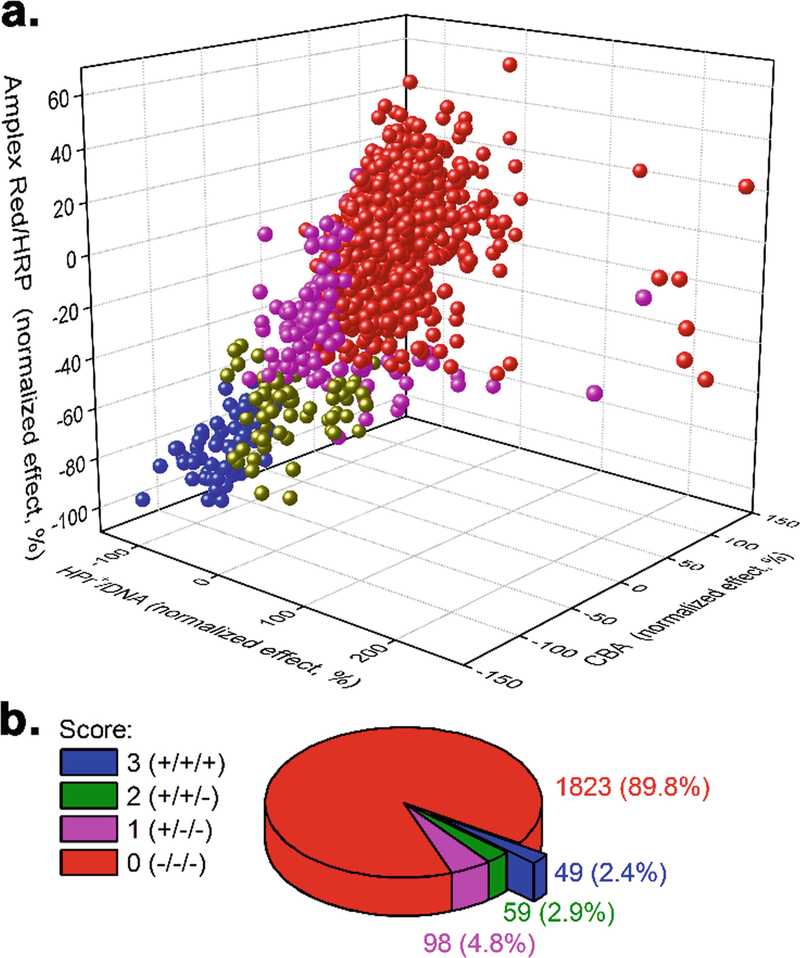
Identify the compounds that are positive hits in all three assays (Fig. 4). These compounds are most likely to inhibit NADPH oxidase activity in the cellular model tested.
Evaluate the positive hits for the presence of pan assay interference compounds (PAINS) [30, 31]. Remove those compounds from the list of positive hits (see Note 14).
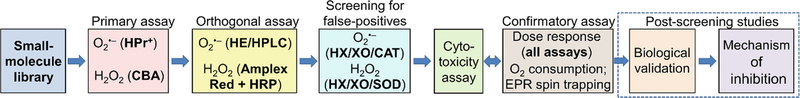
The remaining positive hits are potential inhibitors of NADPH oxidases and should be further evaluated using an array of confirmatory assays. See Fig. 5 for the complete workflow (see Note 15).
Fig. 3.
Results of screening of a library of bioactive compounds (~2000) using three probes: HPr+ (50 μM) in the presence of DNA (0.1 mg/mL) as a probe for O2Superscript>/Superscript> (a and b) and CBA (100 μM, c and d) or Amplex Red (50 μM) in the presence of HRP (0.1 U/mL, e and f) as probes for H2O2. dHL60 cells were stimulated with PMA (1 μM) to induce NOX2 activity (a, c, and e). Schemes of oxidation of the probes and the results of screening after normalization (b, d, and f). Plate-to-plate reproducibility data and number of negative/inconclusive/positive hits for each assay. Score “0” (red color) corresponds to negative, “1” (green color) to inconclusive, and “2” (blue color) to positive hits. (This research was originally published in Journal of Biological Chemistry. Zielonka, J., Zielonka, M., VerPlank, L., Cheng, G., Hardy, M., Ouari, O., Ayhan, M. M., Podsiadly, R., Sikora, A., Lambeth, J. D., & Kalyanaraman, B. Mitigation of NADPH oxidase 2 activity as a strategy to inhibit peroxynitrite formation. J. Biol. Chem. 2016; 291:7029–7044. © the American Society for Biochemistry and Molecular Biology)
Fig. 4.
Results of the screening of a library of bioactive compounds (~2000) using three probes: HPr+ in the presence of DNA as a probe for O2Superscript>/Superscript> and CBA or Amplex Red in the presence of HRP as probes for H2O2. (a) Correlation of the results of the three assays for NOX2 activity. (b) Results of screening as a percentage of positive hits in one, two, or all three assays. (This research was originally published in Journal of Biological Chemistry. Zielonka, J., Zielonka, M., VerPlank, L., Cheng, G., Hardy, M., Ouari, O., Ayhan, M. M., Podsiadly, R., Sikora, A., Lambeth, J. D., & Kalyanaraman, B. Mitigation of NADPH oxidase 2 activity as a strategy to inhibit peroxynitrite formation. J. Biol. Chem. 2016; 291:7029–7044. © the American Society for Biochemistry and Molecular Biology)
Fig. 5.
The workflow scheme for the screening of NOX inhibitors. (This research was originally published in Journal of Biological Chemistry. Zielonka, J., Cheng, G., Zielonka, M., Ganesh, T., Sun, A., Joseph, J., Michalski, R., O’Brien, W. J., Lambeth, J. D., & Kalyanaraman, B. High-throughput assays for superoxide and hydrogen peroxide: design of a screening workflow to identify inhibitors of NADPH oxidases. J. Biol. Chem. 2014; 289:16176–16189. © the American Society for Biochemistry and Molecular Biology)
3.5. Cytotoxicity Measurements by Monitoring Cellular ATP
Load an empty 96-deep well plate (2 mL well volume) with potential NOX inhibitors (1.0 μL of 1000 × solution in DMSO or other solvent), identified during the HTS campaign.
Place the suspension of dHL60 cells in 50 mL centrifuge tube (s), and spin down the cells (see Note 1).
Remove the supernatant, and resuspend the cells in pre-warmed bicarbonate-and phenol red-free RPMI.
Determine the cell density by cell counting, and dilute the cells in RMPI medium to the density of 1 × 105 cells/mL.
Transfer the cell suspension into the 96-deep well plate preloaded with potential inhibitors (1.0 mL/well). Mix the cell suspension with inhibitors by three aspirating/dispensing cycles.
Place the plate in CO2-free incubator (37 C) for 120 min.
Spin down the cells by centrifuging the plate for 1 min at 300 × g.
Remove supernatant using a 12-channel pipettor.
Add ATP-releasing reagent to cell pellets (40 μL/well) using a 12-channel pipettor. Shake the plate for 3 min.
Transfer 20 μL of the lysate to 96-well solid bottom white plate (for luminescence measurements) using a 12-channel pipettor.
Add the ATP reagent containing luciferase and cofactors using a 12-channel pipettor, and immediately measure luminescence intensity (see Note 16).
3.6. Oxygen Consumption Measurements Using the Seahorse XF96 Extracellular Flux Analyzer
Hydrate the Seahorse XF96 cartridge overnight according to the manufacturer’s instructions.
Place the suspension of dHL60 cells in 50 mL centrifuge tube (s), and spin down the cells (see Note 1).
Remove the supernatant, and resuspend the cells in pre-warmed bicarbonate-and phenol red-free RPMI (assay medium).
Determine the cell density by cell counting, and dilute the cells in the assay medium to the density of 2.5×105 cells/mL.
Transfer the cell suspension into the Seahorse XF96 well plate, 80 μL/well, to a total of 2×104 cells/well (see Note 17).
Spin down the cells by centrifuging the plate for 3 min at 100 × g.
Gently add 100 μL of the assay medium, without disturbing the cells at the bottom. Keep the plate at 37 °C in a CO2-free incubator.
Load the Seahorse XF96 cartridge port A with solutions of potential NOX inhibitors (20 μL of 10 ×solution), identified during the HTS campaign. Use RPMI containing 0.1% BSA as the solvent. Additional wells containing the same amounts of the organic solvent (typically DMSO) as introduced with the inhibitors also should be included. In case of limited solubility of the inhibitors in the medium, preincubation of cells with the inhibitors (1 × concentration in the medium) before starting the measurements of oxygen consumption is also possible (see Note 17).
Load port B with 22.2 μL of 10 μM PMA solution in RPMI containing 0.1% BSA.
Place the cartridge in the Seahorse XF96 instrument, and start the calibration process.
Set up the oxygen consumption rate (OCR) measurements over 240 min including basal OCR monitoring, followed by injection of potential NOX inhibitors (port A) at the 60 min time point and injection of PMA solution (port B) at the 120 min time point (see Note 18).
Insert the plate with cells when prompted and start measurements.
3.7. EPR Spin Trapping of O2•−
Place the suspension of dHL60 cells in 50 mL centrifuge tube (s), and spin down the cells (see Note 1).
Remove the supernatant, and resuspend the cells in pre-warmed HBSS containing 25 mM HEPES, pH 7.4, 0.1 mM DTPA.
Determine the cell density by cell counting, and dilute the cells in RMPI medium to the density of 1.25 × 105 cells/mL.
To a 1.5 mL microcentrifuge tube, add 10 μL of potential NOX inhibitor (10 μL of 10 × solution), identified during the HTS campaign.
Add 80 μL of cell suspension (1.25 × 105 cells/mL), and mix by three aspirating/dispensing cycles.
Incubate cells with inhibitor for 30 min.
Add 5 μL of 1 M DEPMPO spin trap in water.
Add 5 μL of 20 μM PMA in HBSS containing 25 mM HEPES, pH 7.4, 0.1 mM DTPA, and 0.1 mg/mL BSA, and mix by three aspirating/dispensing cycles.
Incubate the mixture for 1 h.
Transfer the solution into the EPR capillary (50 or 75 μL), seal the capillary, and place in the EPR instrument.
Measure EPR spectra. Average multiple scans if necessary to obtain an acceptable signal-to-noise ratio.
Repeat steps 4–11 for each inhibitor to be tested, and compare to a control sample without inhibitors. All samples should be run using the same number of averaged spectra.
Quantify the spin adduct by double integration of the EPR peaks, and compare the integrated peak areas of the PMA-stimulated superoxide spin adduct between control and inhibitor-treated cells.
3.8. Rapid HPLC-Based Simultaneous Measurements of O2•− and H2O2
Load the empty 96-deep well plate (2 mL well volume) with potential NOX inhibitors (1.2 μL of 1000 × solution in DMSO or other solvent), identified during the HTS campaign.
Place the suspension of dHL60 cells in 50 mL centrifuge tube (s), and spin down the cells (see Note 1).
Remove the supernatant, and resuspend the cells in pre-warmed HBSS containing 25 mM HEPES, pH 7.4, 0.1 mM DTPA.
Determine the cell density by cell counting, and dilute the cells in HBSS containing 25 mM HEPES, pH 7.4, and 0.1 mM DTPA to the density of 1 × 105 cells/mL.
Transfer the cell suspension into the 96-deep well plate preloaded with potential inhibitors (1.2 mL/well). Mix the cell suspension with inhibitors by three aspirating/dispensing cycles.
Incubate the cells with inhibitors for 30 min.
Preload a second 96-deep well plate with a mix of HE (6.7 mM), CBA (33 mM), and PMA (0.33 mM) in DMSO (3 μL/well).
Transfer cells and inhibitors suspensions (1 mL/well) to a plate preloaded with HE, CBA, and PMA. Mix the cell suspension with inhibitors by three aspirating/dispensing cycles.
Incubate the cells for 1 h at 37 °C, protected from light.
Add SOD + CAT solution (50 × 20 μL/well) to stop oxidation of the probes (HE, CBA).
Spin down the cells by centrifuging the plate for 1 min at 300 × g.
Transfer the supernatants into high-performance liquid chromatography (HPLC) vials (400 μL/vial), a 96-well plate (250 μL/well), or a 384-well plate (80 μL/well), depending on the HPLC autosampler compatibility.
Run rapid HPLC analyses of HE, 2-OH-E+, CBA, and COH, as described elsewhere [14, 15] (see Note 19). The levels of 2-OH-E+ and COH reflect the production of O2•− and H2O2, respectively.
4. Notes
The cell culture medium depends on the cell type and should be based on the requirements for normal cell growth, unless intended otherwise. In the protocol described here, the cell culture and differentiation conditions are provided for HL60 promyelocytic leukemia cells, differentiated into neutrophil-like cells to study the NOX2 isoform. These cells are grown and assayed in suspension. Some modifications of the protocols may be needed to study cellular models of NOX isoforms using adherent cells.
The solutions should be prepared immediately before being added to the cells and should be shielded from light. Any leftover solutions should be discarded and not reused.
For ATP measurements, any commercially available bioluminescence kit based on ATP-dependent luciferase-catalyzed oxidation of luciferin should be appropriate. Final solutions should be prepared immediately before measurements and shielded from light.
From our experience, different preparations of commercial catalase exhibit peroxidatic activity to different extents. To minimize the peroxidatic activity, we recommend using catalase from Corynebacterium glutamicum (Sigma, cat. no. 02071).
The purity of the prepared HPr+ should be confirmed by HPLC to ensure the amount of the oxidation product, propidium dication, does not exceed 1%. If necessary, HPLC-based purification of the probe can be carried out, as described elsewhere [20]. HPLC purification using mobile phase containing trifluoroacetic acid (TFA) will result in anion exchange into TFA, which should be considered when calculating the molecular weight of the product. The concentration of the stock solution of HPr+ should be determined by UV-Vis spectroscopy at 350 nm, using the extinction coefficient of 1.15 × 104 M −1 cm −1 [20]. No absorption band above 400 nm, indicative of the presence of HPr+ oxidation product (s), should be observed.
The expression of the NOX isoform of interest should be confirmed by immunoblotting and/or other technique. This also applies to cytosolic components, when appropriate.
As mentioned in Note 1, the protocol described here is for cells grown in suspension. Adherent cells could be grown directly in a 384-well plate and assayed while attached to the plate bottom. Alternatively, cells could be grown in a cell culture flask, harvested, and assayed in suspension, similar to the protocol for dHL60 cells. The choice of experimental protocol should be based on the optimal conditions to measure the activity of NADPH oxidase, as determined by the calculated Z’ value.
When screening hundreds or thousands of compounds, an automatic dispensing/pipetting instrument should be used for fast and accurate pipetting of the assay components.
For each assay plate, columns 1 and 23 are reserved for DMSO (or other solvent used in the solutions of inhibitors), and these wells will be used as control+ samples. Columns 2 and 24 are reserved for known inhibitors of NADPH oxidase (diphenyleneiodonium or phenylarsine oxide for the NOX2 isoform), and these wells will be used as control-samples. From these samples, the Z’ parameter should be determined for each plate, as should its reproducibility between plates and the days of measurements for each probe/experimental condition.
Typically, the chemical libraries of compounds contain DMSO solutions, with concentrations in the millimolar range (5–10 mM). To minimize the final concentration of DMSO, small volumes of the solutions should be added to the cell medium to keep the total DMSO concentration below 0.5% (v/v). The maximum DMSO content, which does not significantly affect the activity of NADPH oxidase in the studied cellular model, should be experimentally determined. Low volumes are typically added using specialized instruments (e.g., equipped with pin tools) available in HTS centers. For most labs, more cost-effective solutions, including disposable pin tools, delivering 100–130 nL/well may be used.
The incubation time as well as optimal density should be determined experimentally, by determining Z’ values and choosing the conditions yielding the maximal Z’ value. The plate should contain control+ and control − samples (see Note 9) in at least triplicates for each experimental condition. For the HTS campaign, the Z’ value should be equal to or higher than 0.5. The best approach is to run the initial experiment, using different cell densities and varying other conditions (concentrations of probes, activators, etc.), in the real-time (kinetic) mode and calculate Z’ values for all time points before selecting the optimal time point. It may be best to choose the time at which the signal is close to reaching plateau, so that no large changes are expected due to small shifts in the total time of incubation (from starting the incubation with the probes through signal measurement) between all samples within and between the plates.
The fluorescence parameters used in our lab are listed in Table 1. However, small shifts in the wavelengths used may be acceptable. The excitation/emission maxima for each probe are as follows: 2-OH-Pr++/DNA, 508 nm/575 nm [20]; COH, 332 nm/450 nm [21]; and resorufin, 570 nm/583 nm [25].
Typically, the threshold is set at –50% of the normalized intensity (NIx). Minimum inhibitory activity for the threshold should be equal to three standard deviations of the positive control (SDcontrol+). The choice of threshold will affect the number of positive hits to be evaluated in the confirmatory assays, possibly with higher rates of false-positive hits. However, this will also enable identification of weaker, but potentially more selective, inhibitors, the potency of which could be later improved using medicinal chemistry approaches.
PAINS are compounds that are frequent hitters in most screening campaigns [30] and are deemed unsuitable for probe/drug development due to their promiscuity. PAINS could be eliminated from the identified positive hits using an online filter: http://www.cbligand.org/PAINS/search_struct.php.
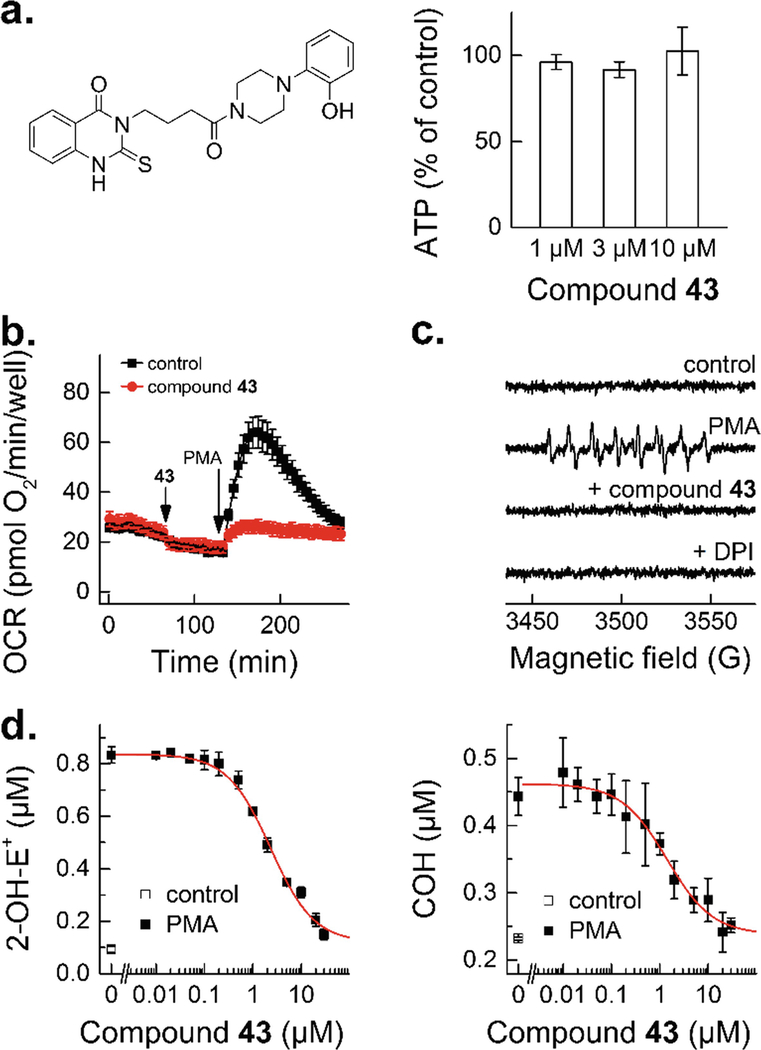
The positive hits identified during the HTS campaign may work by direct interaction with NOX isoform or show the activity due to its cytotoxicity or the interference with the upstream events affecting NOX activity. Thus, additional (confirmatory) assays need to be performed to understand the mechanism of the inhibitory effects observed, as shown in Fig. 5. These include cytotoxicity screening, oxygen consumption measurements, EPR spin trapping of O2 −, and HPLC-based monitoring of the products from HPr+ and CBA oxidation. The dose response should be also tested. See Fig. 6 for an example of the confirmatory assays used for an identified inhibitor of NOX2 activity in dHL60 cells [14]. Further studies establishing the mechanisms of inhibition using cell-free assays and NOX isoform selectivity are also required for complete characterization of the mechanism of action and selectivity of the identified inhibitor [11, 13, 32].
A decrease in the total ATP level may indicate the cytotoxic activity of the positive hits. It is also possible that the compound exhibits cytotoxicity only in the presence of a NOX activator (e.g., PMA), and therefore this assay should be performed in duplicate plates, one with non-stimulated and one with stimulated cells, if appropriate.
Alternatively, cells (at the density of 2.5 ×105 cells/mL) may be preincubated with inhibitors before being loaded onto the plate. In such case, only the PMA solution needs to be loaded into the cartridge. The advantage of this approach is that preparation of 10 × solutions of the inhibitors in RPMI could be avoided, which may be necessary in the case of limited solubility of the potential inhibitors. The advantage of direct injection of 10 × solutions during OCR measurements is that each sample is its own control, when determining the effect of the potential inhibitor on basal OCR (mitochondrial respiration).
The basal OCR value is, in most cases, due largely to mitochondrial respiration, as it is inhibitable by injection of rotenone (complex I inhibitor) and antimycin A (complex III inhibitor) [14]. Injection of PMA induces NOX-dependent oxygen consumption and reflects NOX activity. Injection of the compound may affect both mitochondrial respiration (basal OCR) and NOX-dependent oxygen consumption (PMA-stimulated OCR), as shown for the VAS2870 compound [14]. Notably, inhibition of mitochondrial respiration by rotenone does not block the PMA-induced oxidative burst, indicating that both processes are independent in the cellular model tested.
Rapid HPLC analyses of HE, CBA, 2-OH-E+, and COH are performed using HPLC equipped with UV-Vis absorption, fluorescence detectors, and a column Ascentis Express Phenyl-Hexyl 50 mm × 4.6 mm, 2.7 μm (Supelco), as described elsewhere [14, 15]. The HE and CBA levels are monitored using a UV-Vis absorption detector set at 370 and 290 nm, respectively, and the 2-OH-E+ and COH levels are monitored using a fluorescence detector with excitation set at 370 nm and emission set at 565 nm. Retention times are 19 and 36 s for HE and 2-OH-E+, and 21 and 25 s for CBA and COH, respectively.
Fig. 6.
Examples of confirmatory assays used for characterization of the positive hits from the HTS campaign. (a) Structure of the identified hit (compound 43 from ref. 14) and its effect on total cellular ATP level. (b and c) Effect of the identified hit on the PMA-stimulated oxygen consumption rates (b) and formation of DEPMPO superoxide spin adduct (c). (d) Concentration dependence of the effect of compound 43 on PMA-stimulated probe oxidation by dHL60 cells in the HPLC-based assays for simultaneous monitoring of O2Superscript>/Superscript> and H2O2. (This research was originally published in Journal of Biological Chemistry. Zielonka, J., Cheng, G., Zielonka, M., Ganesh, T., Sun, A., Joseph, J., Michalski, R., O’Brien, W. J., Lambeth, J. D., & Kalyanaraman, B. High-throughput assays for superoxide and hydrogen peroxide: design of a screening workflow to identify inhibitors of NADPH oxidases. J. Biol. Chem. 2014; 289:16176–16189. © the American Society for Biochemistry and Molecular Biology)
Acknowledgment
This work was supported by NIH grants NCI U01 CA178960 and R01 AA022986 to B.K.
References
- 1.Lambeth JD (2004) NOX enzymes and the biology of reactive oxygen. Nat Rev Immunol 4:181–189 [DOI] [PubMed] [Google Scholar]
- 2.Bedard K, Krause KH (2007) The NOX family of ROS-generating NADPH oxidases: physiology and pathophysiology. Physiol Rev 87:245–313 [DOI] [PubMed] [Google Scholar]
- 3.Cifuentes-Pagano E, Meijles DN, Pagano PJ (2014) The quest for selective nox inhibitors and therapeutics: challenges, triumphs and pit-falls. Antioxid Redox Signal 20:2741–2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bedard K, Whitehouse S, Jaquet V (2015) Challenges, Progresses, and Promises for Developing Future NADPH Oxidase Therapeutics. Antioxid Redox Signal 23:355–357 [DOI] [PubMed] [Google Scholar]
- 5.Diebold BA, Smith SM, Li Y, Lambeth JD (2015) NOX2 as a target for drug development: indications, possible complications, and progress. Antioxid Redox Signal 23:375–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cifuentes-Pagano E, Csanyi G, Pagano PJ (2012) NADPH oxidase inhibitors: a decade of discovery from Nox2ds to HTS. Cell Mol Life Sci 69:2315–2325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith SM et al. (2012) Ebselen and congeners inhibit NADPH oxidase 2-dependent superoxide generation by interrupting the binding of regulatory subunits. Chem Biol 19:752–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borbely G et al. (2010) Small-molecule inhibitors of NADPH oxidase 4. J Med Chem 53:6758–6762 [DOI] [PubMed] [Google Scholar]
- 9.Seitz PM et al. (2010) Development of a high-throughput cell-based assay for superoxide production in HL-60 cells. J Biomol Screen 15:388–397 [DOI] [PubMed] [Google Scholar]
- 10.Cifuentes-Pagano E et al. (2013) Bridged tetra-hydroisoquinolines as selective NADPH oxidase 2 (Nox2) inhibitors. Medchemcomm 4:1085–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seredenina T et al. (2015) A subset of N-substituted phenothiazines inhibits NADPH oxidases. Free Radic Biol Med 86:239–249 [DOI] [PubMed] [Google Scholar]
- 12.Gianni D et al. (2010) A novel and specific NADPH oxidase-1 (Nox1) small-molecule inhibitor blocks the formation of functional invadopodia in human colon cancer cells. ACS Chem Biol 5:981–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirano K et al. (2015) Discovery of GSK2795039, a novel small molecule NADPH oxidase 2 inhibitor. Antioxid Redox Signal 23:358–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zielonka J et al. (2014) High-throughput assays for superoxide and hydrogen peroxide design of a screening workflow to identify inhibitors of NADPH oxidases. J Biol Chem 289:16176–16189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zielonka J et al. (2016) Mitigation of NADPH oxidase 2 activity as a strategy to inhibit peroxynitrite formation. J Biol Chem 291:7029–7044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maghzal GJ, Krause KH, Stocker R, Jaquet V (2012) Detection of reactive oxygen species derived from the family of NOX NADPH oxidases. Free Radic Biol Med 53:1903–1918 [DOI] [PubMed] [Google Scholar]
- 17.Zielonka J et al. (2017) Recent developments in the probes and assays for measurement of the activity of NADPH oxidases. Cell Biochem Biophys 75:335–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wardman P (2007) Fluorescent and luminescent probes for measurement of oxidative and nitrosative species in cells and tissues: progress, pitfalls, and prospects. Free Radic Biol Med 43:995–1022 [DOI] [PubMed] [Google Scholar]
- 19.Li Y et al. (2015) Thioxo-dihydroquinazolin-one compounds as novel inhibitors of myeloperoxidase. ACS Med Chem Lett 6:1047–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Michalski R, Zielonka J, Hardy M, Joseph J, Kalyanaraman B (2013) Hydropropidine: a novel, cell-impermeant fluorogenic probe for detecting extracellular superoxide. Free Radic Biol Med 54:135–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zielonka J, Sikora A, Joseph J, Kalyanaraman B (2010) Peroxynitrite is the major species formed from different flux ratios of co-generated nitric oxide and superoxide: direct reaction with boronate-based fluorescent probe. J Biol Chem 285:14210–14216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kalyanaraman B, Hardy M, Zielonka J (2016) A critical review of methodologies to detect reactive oxygen and nitrogen species stimulated by NADPH oxidase enzymes: implications in pesticide toxicity. Curr Pharmacol Rep 2:193–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zielonka J, Kalyanaraman B (2010) Hydroethidine-and MitoSOX-derived red fluorescence is not a reliable indicator of intracellular superoxide formation: another inconvenient truth. Free Radic Biol Med 48:983–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zielonka J, Sikora A, Hardy M, Joseph J, Dranka BP, Kalyanaraman B (2012) Boronate probes as diagnostic tools for real time monitoring of peroxynitrite and hydroperoxides. Chem Res Toxicol 25:1793–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zielonka J et al. (2012) Global profiling of reactive oxygen and nitrogen species in biological systems: high-throughput real-time analyses. J Biol Chem 287:2984–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Michalski R, Michalowski B, Sikora A, Zielonka J, Kalyanaraman B (2014) On the use of fluorescence lifetime imaging and dihydroethidium to detect superoxide in intact animals and ex vivo tissues: a reassessment. Free Radic Biol Med 67:278–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Debski D et al. (2016) Mechanism of oxidative conversion of Amplex(R) Red to resorufin: pulse radiolysis and enzymatic studies. Free Radic Biol Med 95:323–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin SJ, Bradley JG, Cotter TG (1990) HL-60 cells induced to differentiate towards neutrophils subsequently die via apoptosis. Clin Exp Immunol 79:448–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dufer J, Biakou D, Joly P, Benoist H, Carpentier Y, Desplaces A (1989) Quantitative morphological aspects of granulocytic differentiation induced in HL-60 cells by dimethylsulf-oxide and retinoic acid. Leuk Res 13:621–627 [DOI] [PubMed] [Google Scholar]
- 30.Baell JB, Holloway GA (2010) New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J Med Chem 53:2719–2740 [DOI] [PubMed] [Google Scholar]
- 31.Baell J, Walters MA (2014) Chemistry: chemical con artists foil drug discovery. Nature 513:481–483 [DOI] [PubMed] [Google Scholar]
- 32.Pick E (2014) Cell-free NADPH oxidase activation assays: “in vitro veritas”. Methods Mol Biol 1124:339–403 [DOI] [PubMed] [Google Scholar]