Abstract
Hydroethidine is a fluorogenic probe that in the presence of the superoxide radical anion is oxidized to a red fluorescent product, 2-hydroxyethidium. In cells, hydroethidine is also oxidized to other products, including red fluorescent ethidium. Thus, selective monitoring of 2-hydroxyethidium is required for specific detection of the superoxide radical anion. Here, we provide protocols for HPLC- and LC-MS-based quantitation of 2-hydroxyethidium, among other oxidation products. Also, a protocol for continuous sampling for real-time monitoring of superoxide production using rapid HPLC measurements of 2-hydroxyethidium is described.
Keywords: Superoxide radical anion, Hydroethidine, 2-hydroxyethidium, HPLC, LC-MS
1. Introduction
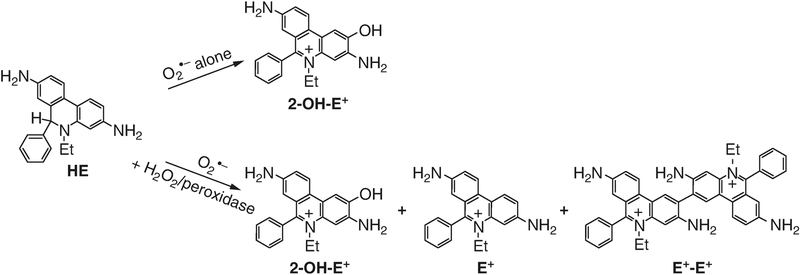
Superoxide radical anion (O2•−) is a primary species formed upon one-electron reduction of molecular oxygen, a key step in the enzymatic function of nicotinamide adenine dinucleotide phosphate (NADPH) oxidases (NOX enzymes) [1, 2]. Because production of O2•− and hydrogen peroxide (H2O2, a product of dismutation of O2•−) is regarded as a primary function of the members of the family of NADPH oxidases [1, 3], their detection is often used to monitor the activity of NOX enzymes in cell-free and cell-based assays [4]. Luminescent (chemiluminescent and fluorescent) probes are typically used for this purpose, as they allow for sensitive, nondestructive, and real-time measurement of O2•− or H2O2 production [5, 6]. The applicability of currently available chemiluminescent probes for specific detection of O2•− is rather limited, due to their inherent chemical reactivity and the reactivity of reaction intermediates on the pathways to chemiluminescence [5–7]. Among fluorescent probes for O2•−, hydroethidine (HE, also known as dihydroethidium) has proven to be most useful [8]. In the presence of O2•−, HE is oxidized to a red fluorescent product, 2-hydroxyethidium (2-OH-E+, Fig. 1). Because this product was demonstrated to be formed only by O2•−, but not other biologically relevant oxidizing species, assays based on the conversion of HE into 2-OH-E+ are currently regarded as a gold standard for O2•− detection. In cells, HE is also oxidized to other products, including ethidium and dimers. Because ethidium is also red fluorescent and is formed intracellularly typically at more than tenfold excess of 2-OH-E+, selective detection of 2-OH-E+ requires separation of the products by high-performance liquid chromatography (HPLC) or other techniques. In addition, one-electron oxidants (e.g., peroxidase + H2O2) have been shown to increase the yield of 2-OH-E+ in the presence of a steady flux of O2•−, as well as to produce both E+ and E+-E+ (Fig. 1). Thus, profiling all oxidation products of HE, including dimers, is crucial for accurate interpretation of the changes in 2-OH-E+ formation. Several papers reported the use of HPLC-based detection of 2-OH-E+ for monitoring the activity of NADPH oxidases in cellular systems [9–15].
Fig. 1.
HE oxidation scheme
Here, we describe how profiling the oxidation products of the HE probe can be utilized to selectively monitor superoxide produced by NADPH oxidases, both intracellularly and in the extracellular medium. The method is based on HPLC or liquid chromatography-mass spectrometry (LC-MS)-based separation and quantification of the probe and the oxidation products, including 2-OH-E+, E+, and E+-E+. We describe the protocols for cell sample collection, processing, and HPLC and LC-MS-based analyses of the products formed.
2. Materials
Prepare all solutions using ultrapure water, endotoxin-free dimethyl sulfoxide (DMSO), and analytical-grade reagents. Samples and solutions should be protected from light and stored on ice (unless indicated otherwise) (see Note 1).
2.1. Probes and Standards of the Oxidation Products
Hydroethidine: prepare stock solution (20 mM) in DMSO, aliquot in brown or black tubes (20 μL per tube), and store at −80 °C. Discard any leftovers; do not reuse (see Note 2).
2-Hydroxyethidium: prepare using a published protocol [16, 17]. Prepare a stock solution of 20 mM in DMSO and store at −80 °C.
Ethidium bromide: prepare a stock solution of 20 mM in DMSO and store at −80 °C.
Diethidium: prepare using a published protocol [16, 17]. Prepare a stock solution of 20 mM in DMSO and store at −80 °C.
Internal standard: 3,8-diamino-6-phenylphenanthridine (DAPP). Prepare a stock solution of 1 mM in DMSO and store at −20 °C.
2.2. Cell Incubation Components
2.3. Cell Extraction Components
Cell lysis buffer: 4 mL of DPBS containing 0.1% Triton X-100, and 1 μM DAPP (internal standard), and place on ice.
10 mL of ice-cold LC-MS-grade acetonitrile (MeCN), containing 0.1% (v/v) formic acid (FA).
10 mL of ice-cold LC-MS-grade water, containing 0.1% (v/v) FA.
Protein assay reagent (e.g., Bradford reagent).
Bovine serum albumin (BSA) in the lysis buffer: 20 mg/mL BSA. Prepare a series of BSA solutions for a calibration curve for protein assay. Dilute the 20 mg/mL BSA solution in the lysis buffer to final concentrations of 0.5, 1, 1.5, 2, 3, 4, 5, 7, and 10 mg/mL. Keep all solutions on ice or refrigerated.
2.4. Medium Extraction Components
10 mL of ice-cold LC-MS-grade MeCN, containing 0.1% (v/v) FA and 1 μM DAPP (internal standard).
10 mL of ice-cold LC-MS-grade water, containing 0.1% (v/v) FA.
2.5. Stopping Solution for “Real-Time” Analyses
1 mL of a mixture of superoxide dismutase (SOD, 50×) and catalase (CAT, 50×): prepare a solution of SOD (5 mg/mL) and catalase (5 kU/mL) in water (see Note 6).
2.6. HPLC and LC-MS Analysis Components
HPLC mobile phase: 0.1% trifluoroacetic acid in water (mobile phase A) and 0.1% trifluoroacetic acid in MeCN (mobile phase B). LC-MS mobile phase: 0.1% FA in water (mobile phase A) and 0.1% FA in MeCN (mobile phase B) (see Note 7).
Solvent for standards: water-to-MeCN (3:1, v/v) mixture, containing 0.1% FA and 1 μM DAPP (internal standard).
Mixture of the HE oxidation products standards (2-OH-E+, E+, and E+-E+; 100 μM each) in the solvent for standards (prepared in the previous step). Prepare serial dilutions from 1 μM down to 1 nM.
Prepare serial dilutions of HE probe from 10 μM down to 10 nM. Use the solvent for standards prepared in step 2.
3. Methods
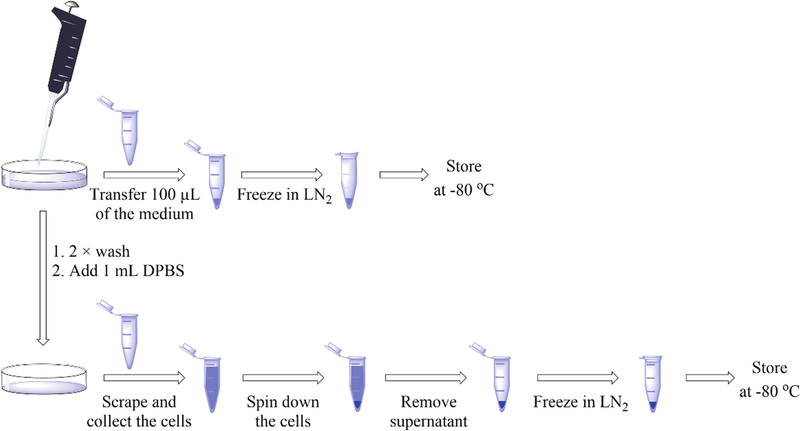
3.1. Cell Incubation with the Probe (Fig. 2)
Fig. 2.
Scheme showing collection of cells and media from cell experiment
Prepare the cells according to the required experimental conditions.
Add HE (from the 20 mM stock solution in DMSO) to obtain a final concentration in the medium of 10 μM (see Note 8).
Incubate the cells for 1 h at 37 °C.
Collect an aliquot of the medium (100 μL) in a 1.5 mL microcentrifuge tube and freeze immediately in liquid nitrogen.
Remove the rest of medium and wash the cells twice with ice-cold DPBS (2 × 10 mL in 15 mL conical tubes).
Add 1 mL of ice-cold DPBS and harvest the cells, transfer the cells into a 1.5 mL microcentrifuge tube, and spin down the cells by quick (30 s, 2000 × g) centrifugation. Discard the supernatant and freeze the cell pellet in liquid nitrogen.
Repeat steps 4–6 for each sample (see Note 8).
Frozen cells and pellets can be stored at −80 °C for up to 1 week before analysis.
3.2. Extraction of the Products
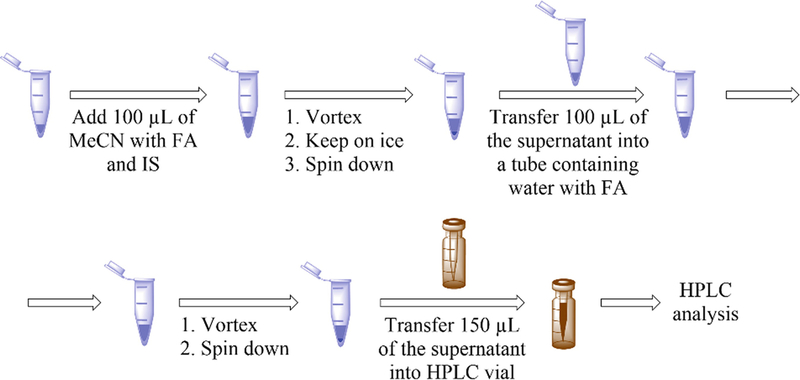
3.2.1. Cell Pellets (Fig. 3)
Fig. 3.
Scheme showing preparation of the cell samples for HPLC analysis
Preload one set of 1.5 mL microcentrifuge tubes with 100 μL of MeCN containing 0.1% FA and place on ice.
Preload a second set of 1.5 mL microcentrifuge tubes with 100 μL of water containing 0.1% FA and place on ice.
Prepare a clear-bottom 96-well plate for protein assay and place on ice.
Place the tubes with frozen cell pellets on ice.
Add 150 μL of the lysis buffer containing DAPP (1 μM), and lyse the cells by 10 syringe strokes using a 0.5 mL insulin syringe with needle 28 G × 0.5 in (0.36 mm × 13 mm).
Immediately after lysing, transfer 100 μL of the cell lysate into a tube containing 100 μL MeCN with 0.1% FA, vortex for 10 s, and put back on ice. Transfer 3 × 2 μL aliquots of the cell lysate into three wells on a 96-well plate for the protein assay.
Repeat steps 5 and 6 for each sample (see Note 9).
Incubate the mixtures of cell lysates with MeCN for 30 min on ice.
During incubation, measure protein concentration in the lysates using the Bradford assay and a plate reader with absorption detection.
Vortex the tubes for 5 s and centrifuge for 30 min at 20,000 × g at 4 °C.
Place the tubes back on ice and transfer 100 μL of the supernatants into the second set of tubes containing 0.1% FA in water.
Vortex the tubes for 5 s and centrifuge for 15 min at 20,000 × g at 4 °C.
Transfer 150 μL of the supernatants into HPLC vials preloaded with conical inserts, seal the vials, and place on ice. After all solutions have been transferred, place the vials in an HPLC autosampler precooled to 4 °C.
3.2.2. Media (Fig. 4)
Fig. 4.
Scheme showing preparation of the media samples for HPLC analysis
Preload one set of 1.5 mL microcentrifuge tubes with 100 μL of water containing 0.1% FA and place on ice.
Place the tubes with frozen media on ice.
Add 100 μL of the ice-cold MeCN, containing 1 μM DAPP (internal standard, IS), vortex for 10 s, and place back on ice.
Incubate the mixtures of media with MeCN for 30 min on ice.
Vortex the tubes for 5 s and centrifuge for 30 min at 20,000 × g at 4 °C.
Place the tubes back on ice and transfer 100 μL of the supernatants into the second set of tubes containing 0.1% FA in water.
Vortex the tubes for 5 s and centrifuge for 15 min at 20,000 × g at 4 °C.
Transfer 150 μL of the supernatants into HPLC vials preloaded with conical inserts, seal the vials, and place on ice. After all solutions have been transferred, place the vials in an HPLC autosampler precooled to 4 °C.
3.3. HPLC Analyses of the Extracts
Install the column Kinetex C18 100 mm × 4.6 mm, 2.6 μm (Phenomenex), in the HPLC system equipped with UV-Vis absorption and fluorescence detectors (HPLC-Abs/Fl). The column should be equipped with a UHPLC column filter or guard column to protect the column and extend its lifetime.
Equilibrate the column with the mobile phase (80% of mobile phase A and 20% of mobile phase B).
Set up the HPLC method and detection parameters according to Tables 1 and 2, respectively.
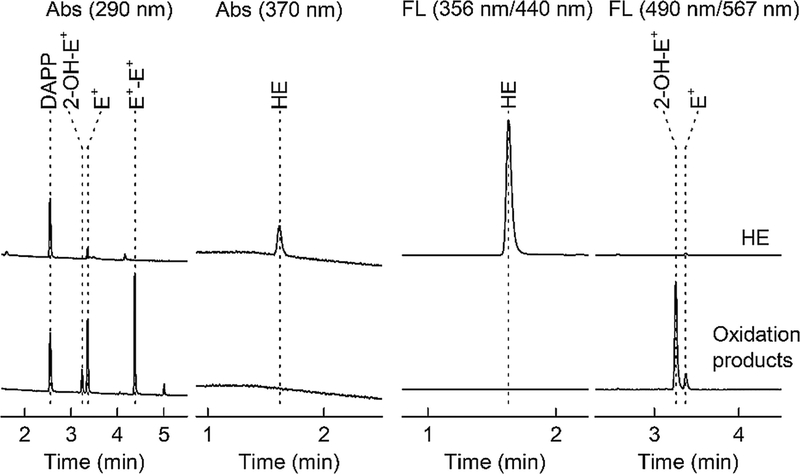
Test the system by three injections of a mix of standards (HE, 2-OH-E+, E+, E+-E+, and DAPP; 1 μM each) for the reproducibility of the retention times and peak intensities for all analytes and the internal standard, as shown in Fig. 5.
Set up the batch table including the calibration standards.
Run the analysis of the batch of samples.
Include the system and column wash with the water-to-methanol mixture (1:1) at the end of batch.
Quantify each analyte based on the detection parameters shown in Table 2 and calibration curves constructed in the concentration range relevant to levels detected in the samples analyzed, using an internal standard-based method.
When appropriate, normalize the concentrations of analytes to the protein levels in cell lysates, as determined by the Bradford method.
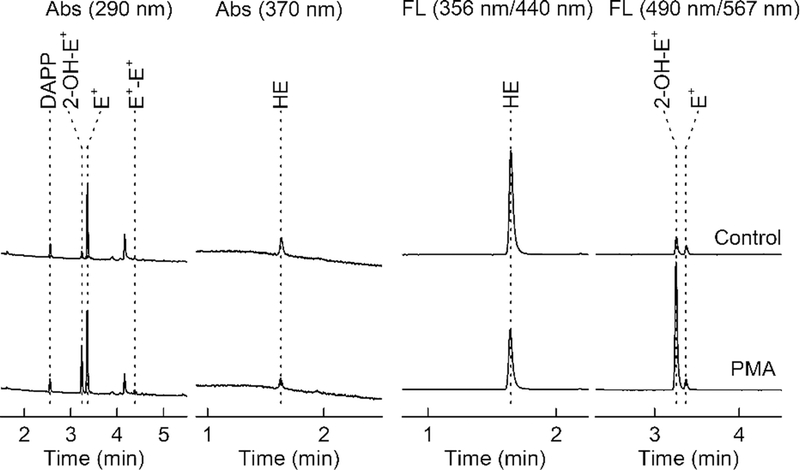
An increase in the peak intensity of 2-OH-E+ but not E+-E+ indicates increased superoxide formation (see Fig. 6 as an example). A concomitant increase in 2-OH-E+ and E+-E+ suggests an increase in peroxidatic activity.
Table 1.
Gradient HPLC method parameters
| Flow rate | 1.5 mL/min | ||
| Gradient | 0 min | 80% A | 20% B |
| 4.5 min | 44% A | 56% B | |
| 5.0 min | 0% A | 100% B | |
| 6.5 min | 0% A | 100% B | |
| 7.0 min | 80% A | 20% B | |
| 9.0 min | 80% A | 20% B | |
| ABS detector | 290 nm | 370 nm | |
| FL detector | 0 min | Excitation at 358 nm Emission at 440 nm |
|
| 2.3 min | Excitation at 490 nm Emission at 567 nm |
Table 2.
UV-Vis absorption and fluorescence detection parameters
| Analyte | Detector | Wavelength | Retention time (min) |
|---|---|---|---|
| DAPP | ABS | 290 nm | 2.5 |
| HE | ABS FL |
370 nm Exc. 358 nm Emi. 440 nm |
1.6 |
| 2-OH-E+ | FL | Exc. 490 nm Emi. 567 nm |
3.2 |
| E+ | FL | Exc. 490 nm Emi. 567 nm |
3.3 |
| E+-E+ | ABS | 290 nm | 4.3 |
Fig. 5.
HPLC-Abs/Fl chromatograms of standards of HE (2 μM, upper traces) and its oxidation products (2-OH-E+, E+, and E+-E+; 1 μM each, lower traces). Both samples contained DAPP (1 μM) as the internal standard. Samples were analyzed using the gradient elution method, as described in Subheading 3.3
Fig. 6.
HPLC-Abs/Fl chromatograms of the extracts from RAW 264.7 cells. The upper traces correspond to untreated (control) cells, and the lower traces correspond to cells stimulated with a phorbol ester (PMA) to activate superoxide production. Samples were analyzed using the gradient elution method, as described in Subheading 3.3
3.4. LC-MS Analyses of the Extracts
Install the column Titan C18 100 mm × 2.1 mm, 1.9 μm (Supelco), in the LC-MS system equipped with a triple quadrupole (MS/MS) detector. The column should be equipped with an ultrahigh-performance liquid chromatography (UHPLC) column filter or guard column to protect the column and extend its lifetime.
Equilibrate the column with the mobile phase (80% of mobile phase A and 20% of mobile phase B).
Set up the HPLC-MS/MS method and detection parameters according to Tables 3 and 4, respectively.
Test the system by three injections of a mix of standards (HE, 2-OH-E+, E+, E+-E+, and DAPP; 1 μM each) for the reproducibility of the retention times and peak intensities for all analytes and the internal standard, as shown in Fig. 7.
Set up the batch table including the calibration standards.
Run the analysis of the batch of samples.
Include the system and column wash with the water-to-methanol mixture (1:1) at the end of batch.
Quantify each analyte based on the specific multiple reaction monitoring (MRM) transitions (Table 4) and calibration curves constructed in the concentration range relevant to the levels detected in the samples analyzed using an internal standard-based method.
When appropriate, normalize the concentrations of analytes to the protein levels in cell lysates, as determined by the Bradford method.
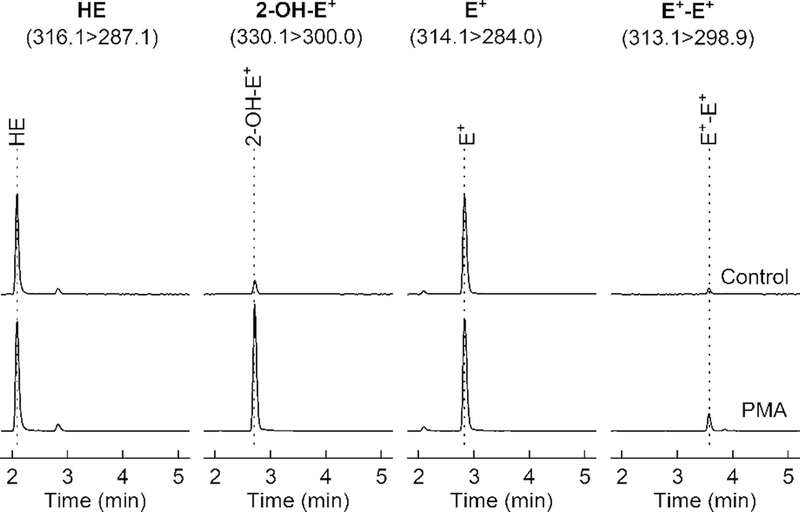
An increase in the peak intensity of 2-OH-E+ but not E+-E+ indicates increased superoxide formation (Fig. 8). A concomitant increase in 2-OH-E+ and E+-E+ suggests an increase in the peroxidatic activity.
Table 3.
LC-MS method parameters
| Flow rate | 0.5 mL/min | ||
| Gradient | 0 min | 80% A | 20% B |
| 7.0 min | 50% A | 50% B | |
| 7.25 min | 0% A | 100% B | |
| 7.75 min | 0% A | 100% B | |
| 8.0 min | 80% A | 20% B | |
| 9.0 min | 80% A | 20% B | |
| Diverter valve | 0 min | Waste | |
| 1.8 min | Detector | ||
| 7.0 min | Waste |
Table 4.
MS/MS detection parameters
| Analyte | MRM transition | Retention time (min) |
|---|---|---|
| DAPP | 286.0 > 208.0 | 2.2 |
| HE | 316.1 > 287.1 | 2.1 |
| 2-OH-E+ | 330.1 > 300.0 | 2.7 |
| E+ | 314.1 > 284.0 | 2.8 |
| E+-E+ | 313.1 > 298.9 | 3.5 |
Fig. 7.
LC-MS/MS chromatograms of standards of HE (2 μM, upper traces) and its oxidation products (2-OH-E+, E+, and E+-E+; 1 μM each, lower traces). Samples were analyzed using the gradient elution method, as described in Subheading 3.4
Fig. 8.
LC-MS/MS chromatograms of the extracts from RAW 264.7 cells. The upper traces correspond to untreated (control) cells, and the lower traces correspond to cells stimulated with a phorbol ester (PMA) so as to induce superoxide production. Samples were analyzed using the gradient elution method, as described in Subheading 3.4
3.5. Cell Incubation with the Probe Combined with Media Sampling for Real-Time Monitoring of Superoxide Released into Extracellular Medium (Fig. 9)
Fig. 9.
Scheme showing sampling of the cell media samples for real-time monitoring of O2•−formation
Prepare the cells according to the required experimental conditions (see Note 10).
Remove the medium and wash the cells with Hank’s Balanced Salt Solution (HBSS) supplemented with HEPES buffer (25 mM) and DTPA (0.1 mM) (see Note 4).
Add the assay medium containing 10 μM HE and all necessary supplements for cell function (see Notes 4 and 11).
Incubate the cells at 37 °C.
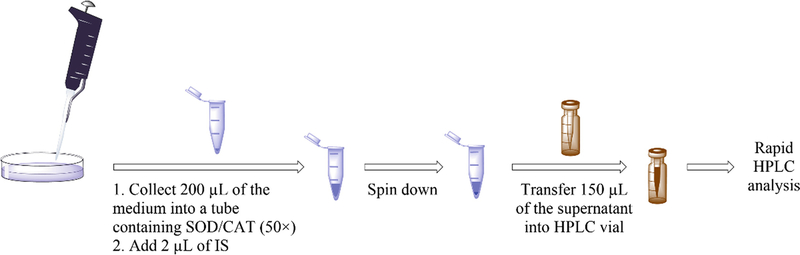
Collect an aliquot of the medium (200 μL) into a 1.5 mL microcentrifuge tube containing 4 μL of SOD and CAT solution (50×). Add 2 μL of 100 μM DAPP (internal standard). Invert the tube three times for complete mixing.
Spin down any floating cells by quick (1 min, 2000 × g) centrifugation.
Transfer 150 μL of the supernatant into an HPLC vial preloaded with conical inserts, seal the vial, place it in the HPLC autosampler, and start the analysis.
Repeat steps 5–7 at the intended time intervals.
3.6. Rapid HPLC Analyses
Install the column Ascentis Express Phenyl-Hexyl 50 mm × 4.6 mm, 2.7 μm (Supelco), in the HPLC system equipped with UV-Vis absorption and fluorescence detectors. The column should be equipped with a UHPLC column filter or guard column to protect the column and extend its lifetime (see Note 12).
Equilibrate the column with the mobile phase (65% of mobile phase A and 35% of mobile phase B) (see Note 13).
Set up the HPLC method and detection parameters according to Tables 5 and 6, respectively (see Note 14).
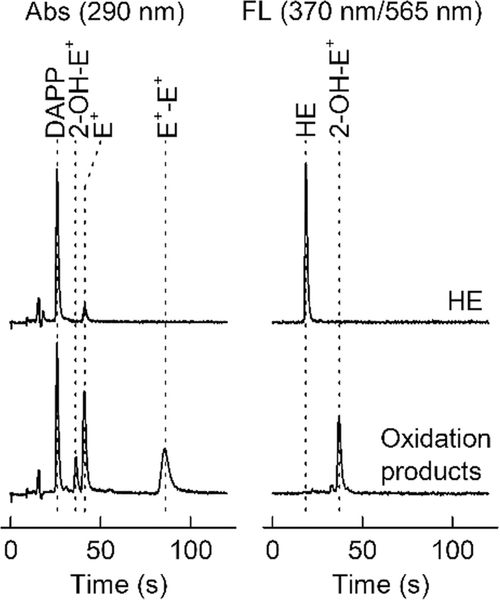
Test the system by three injections of a mix of standards (HE, 2-OH-E+, E+, E+-E+, and DAPP; 1 each) for the reproducibility of the retention times and peak intensities for all analytes and the internal standard, as shown in Fig. 10.
Run analyses of standards for calibration purposes.
Run the analysis of the batch of samples.
Include the system and column wash with the water-to-methanol mixture (1:1) at the end of batch.
Quantify each analyte based on the detection parameters shown in Table 6 and calibration curves constructed in the concentration range relevant to the levels detected in the samples analyzed, using an internal standard-based method.
When appropriate, normalize the concentrations of analytes to the number of cells.
An increase in the peak intensity of 2-OH-E+ indicates increased superoxide formation and release into medium (See Fig. 11 for an example).
Table 5.
Rapid HPLC method parameters
| Flow rate | 2.0 mL/min | ||
| Mobile phase | 65% A | 35% B | |
| ABS detector | 290 nm | 370 nm | |
| FL detector | Excitation at 370 nm Emission at 565 nm |
Table 6.
UV-Vis absorption and fluorescence detection parameters for the rapid HPLC method
| Analyte | Detector | Wavelength | Retention time (s) |
|---|---|---|---|
| DAPP | ABS | 290 nm | 26 |
| HE | FL | Exc. 370 nm Emi. 565 nm |
19 |
| 2-OH-E+ | FL | Exc. 370 nm Emi. 565 nm |
36 |
| E+ | ABS | 290 nm | 41 |
| E+- E+ | ABS | 290 nm | 86 |
Fig. 10.
HPLC-Abs/Fl chromatograms of standards of HE (1 μM) and its oxidation products (2-OH-E+, E+, and E+-E+; 1 μM each, lower traces). Both samples contained DAPP (1 μM) as the internal standard. Samples were analyzed using the rapid HPLC method with isocratic elution, as described in Subheading 3.6
Fig. 11.
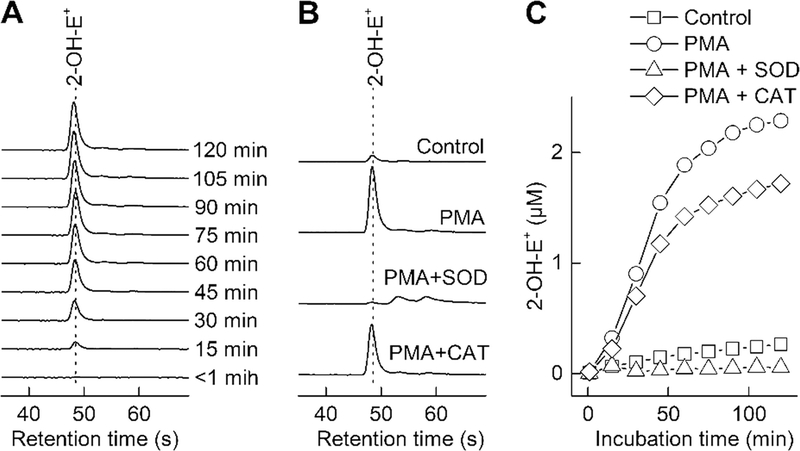
Real-time monitoring of superoxide production by differentiated HL60 (dHL60) cells stimulated with PMA. (A) HPLC peak of 2-OH-E+ as a function of incubation time of dHL60 cells with the HE probe. (B) Effect of PMA, SOD, and CAT on the HPLC peak of 2-OH-E+ detected at 60 min of incubation time of dHL60 cells with the HE probe. (C) Effect of PMA, SOD, and CAT on the dynamics of 2-OH-E+ formation during incubation of dHL60 cells with the HE probe. Samples were analyzed using the isocratic elution method, as described in Subheading 3.6, but using the column and mobile phase composition as detailed in Note 12
4. Notes
HE has been shown to be prone to photooxidation [18]. Also, 2-OH-E+ may act as a photosensitizer, catalyzing the photooxidation process. Thus, it is important to minimize the light exposure of the samples at all stages of the experiment.
If the stock solution of HE is prepared and aliquoted under anaerobic conditions, the aliquots can be stored at −80 °C for at least 6 months without probe degradation.
The cell culture medium depends on the cell type and should be based on the requirements for normal cell growth, unless intended otherwise.
For most studies, the regular cell culture medium can be used during incubation of the cells with the HE probe. For real-time sampling of media combined with the rapid HPLC method (isocratic elution), DPBS or HBSS with necessary supplements (glucose, pyruvate, etc.) and buffer + metal chelator (e.g., 25 mM HEPES + 0.1 mM DTPA) are recommended. This is to avoid clogging the column with hydrophobic components of the regular cell growth medium while providing the bioenergetic substrates and preventing metal-catalyzed oxidation of the probe.
The 15 mL centrifuge tubes should be prefilled with DPBS and placed on ice before the experiment. That way, cells can be washed twice with ice-cold DPBS quickly (<30 s). DPBS should be poured on the dish gently to avoid cell detachment.
From our experience, different preparations of commercial catalase exhibit peroxidatic activity to different extents. To minimize the peroxidatic activity, we recommend using catalase from Corynebacterium glutamicum (Sigma, cat. no. 02071).
HPLC-grade solvents should be used for HPLC analyses. For LC-MS analyses, UHPLC/MS-grade solvents should be used. After preparation, the mobile phase should be passed through a 0.2 μm filter. The mobile phase should be used within 1 week after preparation and stored at 4 °C in between analyses.
The probe in DMSO should be added slowly, with a gentle shaking of the dish, to avoid exposing the cells to a local high concentration of DMSO. Alternatively, a large volume of the medium containing the probe can be prepared immediately before starting the experiment. For all samples, incubation with the probe should be started in time intervals of 3–5 min, depending on the predetermined time needed to harvest cells per a single sample. That way, cells and media are placed in liquid nitrogen as quickly as possible after stopping the incubation, and the time between stopping the incubation and freezing the pellet should be consistent for all samples. To decrease the time, two persons may be needed for sample collection.
Steps 5–7 should be done for one sample at a time; i.e., after finishing step 7 for one sample, start lysing another sample (step 5). To decrease the time, two persons may be needed for cell lysis and extraction.
The protocol described is for adherent cells, but it also can be used for cells in suspension (e.g., in differentiated HL60 cells [13, 14], see Fig. 11).
The assay medium volume should be large enough to not significantly change upon sampling. The combined sampling volume per sample over whole experiment should be less than 10% of the initial volume.
Alternatively, the Kinetex Phenyl-Hexyl column (Phenomenex, 50 mm × 4.6 mm, 2.6 μm) can be used with isocratic elution at the mobile phase composition of 70% mobile phase A and 30% mobile phase B [14]. All other conditions are the same as described in Subheading 3.6. An example of data obtained with this column is shown in Fig. 11.
The same mobile phase (with trifluoroacetic acid) should be used as is used for a gradient method (described in Subheading 3.3).
The same HPLC method and detection parameters may also be used for simultaneous monitoring of superoxide and H2O2 production if hydroethidine and coumarin boronic acid probes are combined in the assay medium and both 2-OH-E+ and 7-hydroxycoumarin products are monitored [13, 14].
Acknowledgment
This work was supported by NIH grants NCI U01 CA178960 and R01 AA022986 to B.K.
References
- 1.Lambeth JD (2004) NOX enzymes and the biology of reactive oxygen. Nat Rev Immunol 4:181–189 [DOI] [PubMed] [Google Scholar]
- 2.Winterbourn CC (2008) Reconciling the chemistry and biology of reactive oxygen species. Nat Chem Biol 4:278–286 [DOI] [PubMed] [Google Scholar]
- 3.Bedard K, Krause KH (2007) The NOX family of ROS-generating NADPH oxidases: physiology and pathophysiology. Physiol Rev 87:245–313 [DOI] [PubMed] [Google Scholar]
- 4.Maghzal GJ, Krause KH, Stocker R, Jaquet V (2012) Detection of reactive oxygen species derived from the family of NOX NADPH oxidases. Free Radic Biol Med 53:1903–1918 [DOI] [PubMed] [Google Scholar]
- 5.Wardman P (2007) Fluorescent and luminescent probes for measurement of oxidative and nitrosative species in cells and tissues: progress, pitfalls, and prospects. Free Radic Biol Med 43:995–1022 [DOI] [PubMed] [Google Scholar]
- 6.Zielonka J, Kalyanaraman B (2018) Small-molecule luminescent probes for the detection of cellular oxidizing and nitrating species. Free Radic Biol Med 128:3–22. 10.1016/j.freeradbiomed.2018.03.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zielonka J, Lambeth JD, Kalyanaraman B (2013) On the use of L-012, a luminol-based chemiluminescent probe, for detecting superoxide and identifying inhibitors of NADPH oxidase: a reevaluation. Free Radic Biol Med 65:1310–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zielonka J, Kalyanaraman B (2010) Hydroethidine- and MitoSOX-derived red fluorescence is not a reliable indicator of intracellular superoxide formation: another inconvenient truth. Free Radic Biol Med 48:983–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fernandes DC, Goncalves RC, Laurindo FR (2017) Measurement of superoxide production and NADPH oxidase activity by HPLC analysis of dihydroethidium oxidation. Methods Mol Biol (Clifton, NJ) 1527:233–249 [DOI] [PubMed] [Google Scholar]
- 10.Laurindo FR, Fernandes DC, Santos CX (2008) Assessment of superoxide production and NADPH oxidase activity by HPLC analysis of dihydroethidium oxidation products. Methods Enzymol 441:237–260 [DOI] [PubMed] [Google Scholar]
- 11.Fernandes DC, Wosniak J Jr, Pescatore LA, Bertoline MA, Liberman M, Laurindo FR, Santos CX (2007) Analysis of DHE-derived oxidation products by HPLC in the assessment of superoxide production and NADPH oxidase activity in vascular systems. Am J Physiol Cell Physiol 292:C413–C422 [DOI] [PubMed] [Google Scholar]
- 12.Zielonka J et al. (2017) Recent developments in the probes and assays for measurement of the activity of NADPH oxidases. Cell Biochem Biophys 75:335–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zielonka J et al. (2016) Mitigation of NADPH oxidase 2 activity as a strategy to inhibit peroxynitrite formation. J Biol Chem 291:7029–7044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zielonka J et al. (2014) High-throughput assays for superoxide and hydrogen peroxide design of a screening workflow to identify inhibitors of NADPH oxidases. J Biol Chem 289:16176–16189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zielonka J et al. (2012) Global profiling of reactive oxygen and nitrogen species in biological systems: high-throughput real-time analyses. J Biol Chem 287:2984–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zielonka J, Hardy M, Kalyanaraman B (2009) HPLC study of oxidation products of hydroethidine in chemical and biological systems: ramifications in superoxide measurements. Free Radic Biol Med 46:329–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zielonka J, Vasquez-Vivar J, Kalyanaraman B (2008) Detection of 2-hydroxyethidium in cellular systems: a unique marker product of superoxide and hydroethidine. Nat Protoc 3:8–21 [DOI] [PubMed] [Google Scholar]
- 18.Zielonka J, Vasquez-Vivar J, Kalyanaraman B (2006) The confounding effects of light, sonication, and Mn(III)TBAP on quantitation of superoxide using hydroethidine. Free Radic Biol Med 41:1050–1057 [DOI] [PubMed] [Google Scholar]