Abstract
Observational studies have consistently reported elevated plasma homocysteine as a risk factor for Alzheimer’s disease (AD). However, results from clinical trials of homocysteine-lowering treatments are inconsistent. This discrepancy may be explained by a lack of causal association between homocysteine and AD. Mendelian randomization studies have the potential to provide insight into the causality of this association through studying the effect of genetic predisposition to high homocysteine on AD. Our analyses using summarized (n = 54,162) and individual participant (n = 6987) data from Caucasian participants did not show an effect of plasma homocysteine genetic risk on susceptibility to AD. Although with smaller sample sizes, further subanalyses also did not support an effect of genetically determined plasma homocysteine on cognitive impairment and decline, beta-amyloid and tau pathology and gray matter atrophy in AD. However, we found associations with tau tangle burden (n = 251) and gray matter atrophy (n = 605) in cognitively normal elderly. Our results do not support a causal association between elevated homocysteine and risk, severity, and progression of AD. However, the relationship between genetically determined homocysteine and brain pathology in cognitively normal elderly requires further exploration.
Keywords: Mendelian randomization, Alzheimer’s disease, Aging, Plasma homocysteine, Causal association, Polygenic score
1. Introduction
Late-onset Alzheimer’s disease (AD) is a complex disorder with several genetic and environmental risk factors. Despite its burden, there are no definitive or disease-modifying treatments yet available for AD. Therefore, modifiable risk factors make accessible targets for its treatment and prevention. Elevated circulating homocysteine (Hcy) level has been implicated in some studies as a modifiable risk factor in AD and age-related cognitive decline.
Several cross-sectional and prospective longitudinal epidemiological studies have demonstrated that higher Hcy level is associated with both an increased risk of developing AD (Annerbo et al., 2006; Seshadri et al., 2002; Zylberstein et al., 2011) and accelerated cognitive aging (Dufouil et al., 2003; Tucker et al., 2005). Evidence from cell biology and animal studies have further demonstrated the effects of high Hcy levels on neuronal endoplasmic reticulum stress (Kim et al., 2008), oxidative stress (Streck et al., 2003), and excitotoxicity (Abushik et al., 2014), and even directly through induction of amyloid beta (Aβ) deposition (Zhang et al., 2009) and tau phosphorylation (Luo et al., 2007). Hcy can be recycled into methionine with the aid of vitamin B 12, folic acid, or trimethylglycine or converted into cysteine with vitamin B 6 as the cofactor. Therefore, elevated Hcy levels can be reduced by simple drug-based interventions. However, the causality of the effect of Hcy in AD has remained controversial (Zhuo et al., 2011). While some clinical trials have reported benefit from Hcy-lowering interventions (Douaud et al., 2013; Durga et al., 2007; Jager et al., 2012), others have shown no improvement or even detrimental effects (Eussen et al., 2006; McMahon et al., 2006). Given that such vitamin supplementation therapies may affect various other metabolic pathways, it is possible that observed effects in these trials are not purely a result of lowering Hcy levels. It is also possible that findings from observational studies are influenced by confounding factors or reverse causality.
In the absence of definitive results from randomized clinical trials, Mendelian randomization (Lawlor et al., 2008) studies can help determine the causality of observed associations between exposure and disease. This approach exploits the random inheritance of genotypes with known associations with a disease risk factor. The risk factor’s causal effect on disease can then be deciphered independent of confounding factors and reverse causality. Here, using data from large- and medium-scale publicly available AD data sets, we examined whether genetic predisposition to elevated circulating Hcy influences the risk of developing AD. Moreover, we assessed its effects on a number of AD-related outcomes including cognitive decline, brain and cerebrospinal fluid (CSF) pathologic biomarkers of AD, and brain atrophy, separately in AD and cognitively normal (CN) elderly, where data were available.
2. Method
2.1. Mendelian randomization study using summarized data
Top polymorphisms from 13 independent genome-wide significant loci (p < 5 × 10–8) influencing plasma Hcy level and their effect size (amount of standard deviation change in ln(Hcy) per the effect allele) were obtained from the largest available meta-analysis of genome-wide association studies on Hcy (van Meurs et al., 2013) (n = 44,147 individuals of European descent). Summarized data on the effect of each single nucleotide polymorphism (SNP) on AD risk were extracted from the largest available meta-analysis of genome-wide association studies on AD (Lambert et al., 2013) (International Genomics of Alzheimer’s Project [IGAP] stage 1; 17,008 AD patients and 37,154 controls of European descent). Mendelian randomization analysis was performed using an inverse-variance weighted fixed-effect meta-analysis approach (Burgess et al., 2013) to estimate the effect of genetically determined 1-standard deviation change in ln(Hcy) on the risk of AD.
2.2. Mendelian randomization studies using individual participant data (polygenic score association studies)
Polygenic score (PGS) association studies were performed using available individual-level data from AD patients and CN elderly from 4 study groups: the National Institute on Aging Alzheimer’s Disease Centers (ADCs) Cohort (Naj et al., 2011), Multi-Site Collaborative Study for Genotype-Phenotype Associations in Alzheimer’s Disease (GenADA) (Li et al., 2008), Alzheimer’s Disease Neuroimaging Initiative (ADNI) (Weiner et al., 2010), and Rush Alzheimer’s Disease Center Religious Orders Study and Memory and Aging Project (ROS/MAP) (Bennett et al., 2012a, Bennett et al., 2012b). The appropriate ethics committee or institutional review board of each study/recruitment site approved study protocols. Written informed consent was obtained from all study participants or in case of substantial cognitive impairment, from a caregiver, legal guardian, or other proxy.
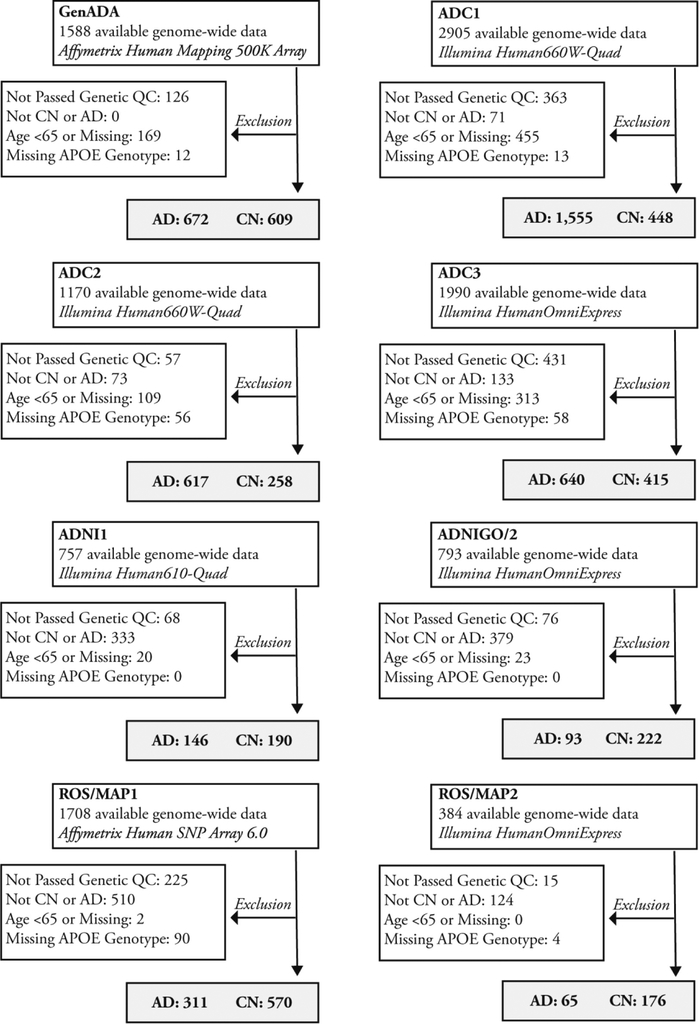
Genome-wide genotyping data from ADC, GenADA, ADNI, and ROS/MAP were available in 8 batches (Fig. 1). Genetic quality control, multidimensional scaling, and imputation were performed for each batch separately. Imputed data for the 13 SNPs were extracted for participants with European ancestry (eTable 1). Hcy-PGSs were calculated as the sum of the expected number of alleles associated with higher Hcy weighted by their effect size (extracted from the Hcy meta-analysis [van Meurs et al., 2013]).
Fig. 1.
Quality control of available individual participant data. Abbreviations: AD, Alzheimer’s disease; CN, cognitively normal.
Cross-sectional and longitudinal Mini–Mental State Examination (MMSE) scores indexing cognitive performance in AD patients were available from GenADA, ADNI, and ROS/MAP. Cross-sectional and longitudinal measurements of global cognitive function (average of normalized scores of 17 tasks in 5 cognitive domains) assessing cognitive performance in CN participants were available from ROS/MAP.
Individual postmortem neuropathology composite scores for overall brain Aβ burden and tau tangle density were available for analysis from ROS/MAP. Cross-sectional in vivo cortical Aβ deposition burden assessed using [18F]Florbetapir positron emission tomography imaging, and CSF Aβ1–42, total tau, and phosphorylated tau (p-tau181p) levels were obtained from ADNI.
Gray matter (GM) atrophy was assessed using cross-sectional T1-weighted brain magnetic resonance imaging available from ADNI and ROS/MAP. Global GM atrophy was assessed using total GM volumes segmented from the T1-weighted images. Regional GM atrophy was assessed using voxel-based morphometry (VBM) performed on the smoothed modulated GM probability maps. ADNI cortical thickness data were used for post hoc region of interest analysis.
Linear mixed models were used to assess the effect of Hcy-PGS on diagnosis, cognitive performance, neuropathological biomarkers, and GM atrophy. Unless otherwise specified, all analyses were performed while adjusting for the fixed effects of age, sex, and the number of APOE ε4 alleles, and the random effect of the study/imputation batch. Total GM volume analyses were also adjusted for intracranial volume. Studies/imputation batches with data available from less than 20 participants were excluded from the analyses. Longitudinal analyses were performed using linear mixed models accounting for the aforementioned covariates in addition to the fixed effect of follow-up year and assuming a random intercept and slope for the baseline measure of outcome variable and its change over time per individual. Neuropathology data underwent square-root transformation whenever required before statistical analysis in order to meet the normality assumptions. Voxelwise analyses were performed while adjusting for the effects of age, sex, handedness, number of APOE ε4 alleles, and the study/imputation batch.
The methods are described in detail in eMethods.
3. Results
3.1. Genetic influence of Hcy on AD risk
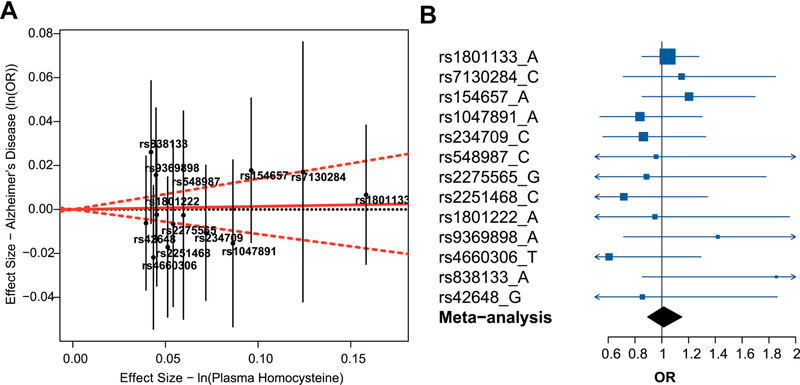
Using IGAP summarized data, none of the 13 SNPs affecting Hcy were associated with AD (all p > 0·05), and no significant heterogeneity was observed in their effects on AD risk (heterogeneity p = 0·69) (Fig. 2A). The Mendelian randomization meta-analysis of the summarized data from the 13 SNPs did not support a causal effect of genetically determined Hcy on AD risk (odds ratio [OR] [95% confidence interval {CI}] = 1.01 [0.89–1.15], p = 0.84). Further Mendelian randomization subanalyses using data from individual SNPs also did not show any significant association (Fig. 2B).
Fig. 2.
Mendelian randomization analysis of the effect of plasma homocysteine on AD risk using summarized data. (A) The effect sizes of polymorphisms affecting plasma homocysteine on natural log-transformed homocysteine levels and risk for AD. Effect sizes for plasma homocysteine are extracted from van Meurs et al. (2013) and for AD risk are obtained from summary statistics from IGAP (Lambert et al., 2013). The effect estimate of plasma homocysteine on AD risk is represented by the red solid line. The 95% CI of this estimate is represented by the red dashed lines. The plot is created using the “gtx” package in R. (B) Mendelian randomization estimates for the effect of 1-standard deviation increase in genetically determined plasma homocysteine on AD risk (OR and 95% CI) are demonstrated for each plasma homocysteine risk allele (dark blue) and the overall fixed-effect meta-analysis of all alleles (black). Abbreviations: AD, Alzheimer’s disease; CI, confidence interval; IGAP, International Genomics of Alzheimer’s Project; OR, odds ratio.
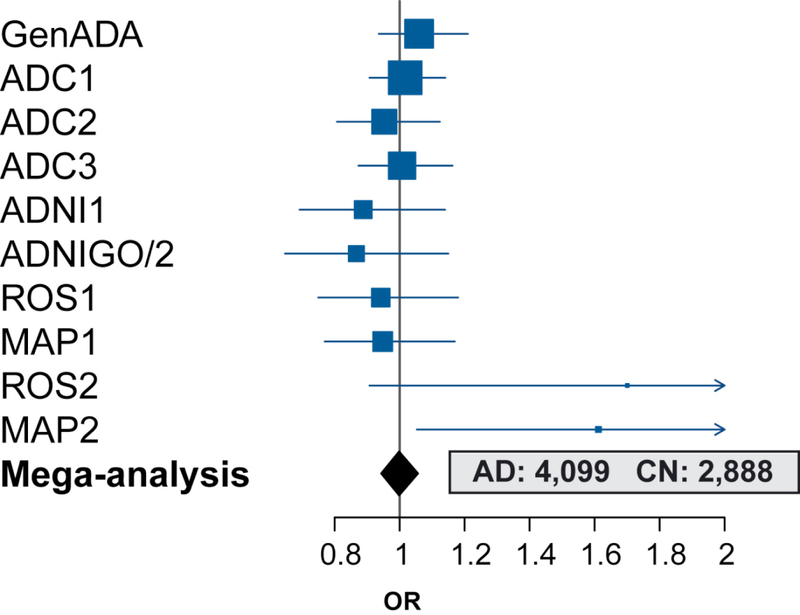
Characteristics of the participants from each of the 10 study/imputation batches included in the PGS association analyses are shown in Table 1. Hcy-PGS distributions are demonstrated in eFig. 1. Performing analysis on individual participant data (n = 6987: AD = 4,099, CN = 2888), we observed no evidence for a significant association between Hcy-PGS and risk for AD (OR [95% CI] = 1.0 [0.94–1.06], p = 0.96; Fig. 3). Further analyses demonstrated no significant association between Hcy-PGS and AD after stratification for sex (female: n = 3953, %AD = 55, OR [95% CI] = 0.98 [0.91–1.06], p = 0.57; male: n = 3034, %AD = 64, OR [95% CI] = 1.03 [0.95–1.13], p = 0.45), and APOE ε4 allele carrier status (noncarriers: n = 3648, %AD = 40, OR [95% CI] = 1.01 [0.94–1.08], p = 0.85; carriers: n = 3339, %AD = 79, OR [95% CI] = 0.98 [0.90–1.08], p = 0.71). Also, no interaction effect was observed between age and Hcy-PGS in relation to AD risk (p = 0.14).
Table 1.
Characteristics of participants included in the Mendelian randomization study of the effect of plasma homocysteine on AD risk using individual participant data
| Study | Diagnosis | |
|---|---|---|
| GenADA | CN (n = 609) | AD (n = 672) |
| Age (y) (mean ± SD) | 75.7 ± 6.2 | 79.7 ± 6.6 |
| Sex (% female) | 64% | 59% |
| APOE ε4 (% 0/1/2 alleles) | 75/23/2% | 35/51/14% |
| MMSE score, mean ± SD (range) | 29.1 ± 1.1 (20–30) | 17.8 ± 8.6 (0–30) |
| ADC1 | CN (n = 448) | AD (n = 1555) |
| Age (y) (mean ± SD) | 77.0 ± 6.5 | 78.7 ± 6.2 |
| Sex (% female) | 57% | 50% |
| APOE ε4 (% 0/1/2 alleles) | 71/27/2% | 32/50/18% |
| MMSE score, mean ± SD (range) | - | - |
| ADC2 | CN (n = 258) | AD (n = 617) |
| Age (y) (mean ± SD) | 76.6 ± 6.1 | 79.4 ± 6.0 |
| Sex (% female) | 71% | 49% |
| APOE ε4 (% 0/1/2 alleles) | 70/27/3% | 33/50/17% |
| MMSE score, mean ± SD (range) | - | - |
| ADC3 | CN (n = 415) | AD (n = 640) |
| Age (y) (mean ± SD) | 75.6 ± 7.2 | 78.8 ± 6.6 |
| Sex (% female) | 63% | 51% |
| APOE ε4 (% 0/1/2 alleles) | 76/22/2% | 32/52/16% |
| MMSE score, mean ± SD (range) | - | - |
| ADNI1 | CN (n = 190) | AD (n = 146) |
| Age (y) (mean ± SD) | 76.1 ± 4.6 | 77.3 ± 5.7 |
| Sex (% female) | 44% | 45% |
| APOE ε4 (% 0/1/2 alleles) | 73/25/2% | 33/51/16% |
| MMSE score, mean ± SD (range) | 29.1 ± 1.1 (25–30) | 23.4 ± 2.1 (18–28) |
| ADNIGO/2 | CN (n = 222) | AD (n = 93) |
| Age (y) (mean ± SD) | 73.8 ± 5.7 | 77.4 ± 5.9 |
| Sex (% female) | 52% | 35% |
| APOE ε4 (% 0/1/2 alleles) | 71/27/2% | 33/53/14% |
| MMSE score, mean ± SD (range) | 29.0 ± 1.3 (24–30) | 23.2 ± 2.1 (19–26) |
| ROS1 | CN (n = 264) | AD (n = 151) |
| Age (y) (mean ± SD) | 84.2 ±6.3 | 89.6 ± 6.2 |
| Sex (% female) | 61% | 64% |
| APOE ε4 (% 0/1/2 alleles) | 82/17/1% | 70/27/3% |
| MMSE score, mean ± SD (range) | 28.2 ± 1.7 (20–30) | 12.8 ± 8.5 (0–27) |
| MAP1 | CN (n = 306) | AD (n = 160) |
| Age (y) (mean ± SD) | 86.5 ± 6.8 | 90.9 ± 5.3 |
| Sex (% female) | 74% | 68% |
| APOE ε4 (% 0/1/2 alleles) | 84/14/2% | 64/35/1% |
| MMSE score, mean ± SD (range) | 28.0 ± 1.8 (17–30) | 13.5 ± 8.0 (0–27) |
| ROS2 | CN (n = 50) | AD (n = 22) |
| Age (y) (mean ± SD) | 81.4 ± 7.0 | 87.5 ± 7.9 |
| Sex (% female) | 74% | 73% |
| APOE ε4 (% 0/1/2 alleles) | 78/20/2% | 55/41/4% |
| MMSE score, mean ± SD (range) | 28.3 ± 1.4 (24–30) | 14.6 ± 7.8 (0–28) |
| MAP2 | CN (n = 126) | AD (n = 43) |
| Age (y) (mean ± SD) | 84.0 ± 7.1 | 91.0 ± 5.6 |
| Sex (% female) | 70% | 74% |
| APOE ε4 (% 0/1/2 alleles) | 82/17/1% | 65/35/0% |
| MMSE score, mean ± SD (range) | 28.0 ± 1.6 (22–30) | 14.5 ± 7.9 (0–28) |
Age and MMSE data from GenADA and ADNI baseline and ROS/MAP last follow-up visits are used in the cross-sectional analyses and reported in the Table. MMSE data for cognitively normal participants are presented for the purpose of comparison and are not used in the analyses.
Key: AD, Alzheimer’s disease; ADC, Alzheimer’s Disease Centers; ADNI, Alzheimer’s Disease Neuroimaging Initiative; CN, cognitively normal elderly; GenADA, Genotype-Phenotype Associations in Alzheimer’s Disease; MAP, Memory and Aging Project; MMSE, Mini–Mental State Examination; ROS, Religious Orders Study; SD, standard deviation.
Fig. 3.
Mendelian randomization analysis of the effect of plasma homocysteine on AD risk using individual participant data (plasma homocysteine polygenic score association study). Estimates for the effect of 1-standard deviation increase in plasma homocysteine polygenic score on AD risk (OR and 95% CI) while accounting for the effects of age, sex, and APOE ε4 allele status are demonstrated for each study/imputation batch (dark blue). The overall result from the mega-analysis is shown in black. Abbreviations: AD, Alzheimer’s disease; CI, confidence interval; CN, cognitively normal; OR, odds ratio.
3.2. Hcy polygenic score and cognitive impairment
Hcy-PGS was not associated with cross-sectional MMSE score (n = 1262 from GenADA, ROS/MAP, and ADNI AD participants; t = −1.1, p = 0.29) or longitudinal MMSE performance (n = 1473 observations from 478 AD patients, median follow-up = 2 years; interaction between Hcy-PGS and year of follow-up: t = 1.47, p = 0.14).
Using global cognitive function assessments from ROS/MAP participants who were CN at baseline, we did not observe any significant association between Hcy-PGS and either cross-sectional (n = 1,194, age at baseline [mean ± standard deviation] = 78 ± 7; t = −1.60 p = 0.11) or longitudinal (n = 11,344 observations from 1146 participants, follow-up period = 1–21 years, median follow-up = 9 years; t = 0.46, p = 0.64) cognitive performance. Excluding participants who likely developed mild cognitive impairment or dementia due to mixed and non-AD causes during the course of follow-up also yielded similar findings (n = 10,613 observations on 1077 individuals: t = −0.71, p = 0.48).
3.3. Hcy polygenic score and brain and CSF pathologic biomarkers of AD
Using ROS/MAP postmortem neuropathology and last antemortem clinical assessment data, we found no evidence for association between Hcy-PGS and brain amyloid burden in AD (n = 260, p = 0.68) and CN (n = 255, p = 0.16) participants. These results were further supported by in vivo ADNI [18F]Florbetapir positron emission tomography cortical amyloid (AD: n = 130, p = 0.39; CN: n = 247, p = 0.87) and CSF Aβ1–42 data (AD: n = 167, p = 0.68; CN: n = 294, p = 0.85).
Although no association was found between Hcy-PGS and postmortem tau neurofibrillary tangle burden in AD in ROS/MAP (n = 256, p = 0.35), a positive association was observed in CN individuals (n = 251, t = 2.4, p = 0.017). However, we did not observe any significant association between Hcy-PGS and ADNI CSF tau (AD: n = 164, p = 0.69; CN: n = 293, p = 0.58) or p-tau181p (AD: n = 167, p = 0.91; CN: n = 294, p = 0.46).
3.4. Hcy polygenic score and gray matter atrophy
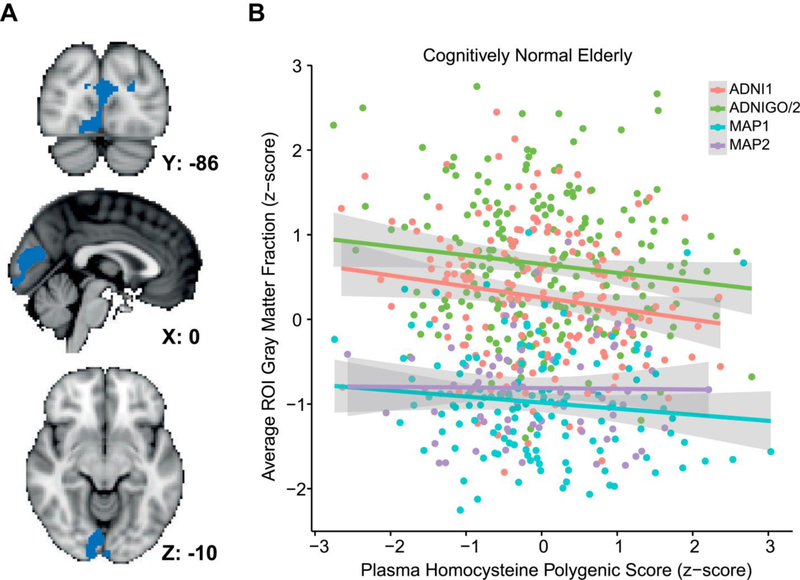
No association was found between Hcy-PGS and total GM volume in AD (n = 269 participants from ADNI, t = −0.56, p = 0.57). However, higher Hcy-PGS was associated with lower total GM volume in CN elderly (n = 605 participants from ADNI and MAP, t = −2.32, p = 0.02). VBM revealed significantly greater regional GM atrophy in the occipital cortex of CN participants (pericalcarine, cuneus, and lingual areas; peak family-wise error corrected p = 0.04) associated with higher Hcy-PGS (Fig. 4). ADNI FreeSurfer cortical thickness data that had passed quality control for the occipital lobe were used for post hoc region of interest analysis. Results demonstrated an interaction effect between Hcy-PGS and diagnosis (AD vs. CN) on the average thickness of right and left pericalcarine, cuneus, and lingual cortices (interaction: n = 610, p = 0.04; after stratifying for diagnosis: CN: n = 377, β ± standard error = −0.11 ± 0.04, p = 0.004; AD: n = 233, β ± standard error = 0.02 ± 0.05, p = 0.71).
Fig. 4.
Effect of plasma homocysteine polygenic score on gray matter atrophy in cognitively normal elderly. (A) Results for whole-brain voxel-based morphometry of the effect of plasma homocysteine polygenic score on gray matter atrophy in 605 cognitively normal elderly depicted in the Montreal Neurological Institute (MNI) space. Significant voxels are shown in blue (threshold-free cluster enhancement, family-wise error corrected p < 0.05). (B) Bivariate correlations between average gray matter volume fraction in the significant voxels shown in (A) and plasma homocysteine polygenic score in each individual. Abbreviation: ROI, region of interest.
4. Discussion
Using Mendelian randomization approach, we did not find evidence supporting a causal association between plasma Hcy level and susceptibility to AD in participants with European ancestry. Furthermore, we did not find evidence for significant associations between Hcy-PGS and cognitive impairment and decline in AD and CN elderly, and brain and CSF beta-amyloid and tau pathology and GM atrophy in AD. Although no association was observed between Hcy-PGS and beta-amyloid pathology in CN participants, we found associations with brain tangle burden and GM atrophy in these individuals.
Using summarized and individual participant data comparing AD and CN individuals, we found no association between genetically determined Hcy and AD risk. Similar finding from both approaches (OR [95% CI] = 1.01 [0.89–1.15] and 1.0 [0.94–1.06], respectively) increases confidence in the robustness of our results. The sample sizes used for assessing the association with AD risk were from multiple sources and fairly large. Therefore, our results can be considered generalizable to people from European ancestry. Our analysis on longitudinal quantitative cognitive data from CN elderly (with some individuals developing mild cognitive impairment or dementia during the course of follow-up) further supports our main finding on cross-sectional binary diagnostic data.
Previous Mendelian randomization meta-analyses (Hu et al., 2016, Hua et al., 2011, Peng et al., 2015, Rai, 2016, Zhang et al., 2010) on association between Hcy and AD risk have used summarized data from a highly overlapping number of previously published case-control studies on a single polymorphism (rs1801133 [MTHFR C677T]). The included studies are from participants with both European and non-European ancestries, with a maximum total Caucasian subsample of ~2000 AD and ~2000 controls. Although all these meta-analyses have reported significant causal associations between Hcy and AD risk in their overall sample, subanalyses in Caucasians were nonsignificant in all, except for 1 (Rai, 2016). There are also some discrepancies between these meta-analyses in terms of classifying publications from countries with specific ethnicities (e.g., Israel, Egypt, and Tunisia) or with mixed populations (e.g., USA) as Caucasian or non-Caucasian. In line with our findings and the majority of the Caucasian case-control meta-analyses, IGAP summarized data from ~17,000 AD to ~37,000 controls with European ancestry do not support a significant association between rs1801133 and AD risk (p = 0·68) (Lambert et al., 2013).
Our study has overcome several limitations in the aforementioned studies (Hu et al., 2016, Hua et al., 2011, Peng et al., 2015, Rai, 2016, Zhang et al., 2010). By using a polygenic rather than a single polymorphism approach, we were able to explain a greater proportion of the variance of Hcy levels (van Meurs et al., 2013), and in turn, provide a more accurate estimation of the effect of genetically determined Hcy. Extracting whole-genome data could overcome potential publication biases of single-gene case-control association studies, which might have shifted the meta-analyses results towards significant findings. Moreover, genomic data provided the means for performing multidimensional scaling, which helped objectively determine participants’ ancestries and also detect ethnic outliers. Statistical models for analyses on individual participant data could account for confounding variables and covariates such as age, sex, and APOE ε4 allele status. Finally, the majority of our individual participant data came from studies that are less likely affected by selection bias (i.e., not including many of the individuals with the disease risk factor in the study due to death or ill health). The ADC cohort did not depend on participants necessarily being alive and included living and also autopsy-confirmed AD and CN participants; and ROS/MAP is a longitudinal community cohort study of participants who are mainly dementia free at enrollment and are naturally followed so that a number of them develop cognitive impairment over the course of follow-up.
A limited number of studies have assessed the causal relationship between Hcy and the severity and progression of AD (Gorgone et al., 2009, Seripa et al., 2003). Our study on cross-sectional and longitudinal MMSE did not support a causal association between Hcy and AD severity (n > 1200) and progression (n ~500 followed-up up to 4 years). Although with more limited sample sizes, our nonsignificant results from other studied AD-related outcomes (brain and CSF beta-amyloid and tau burden and GM atrophy) in AD patients were in line with these findings.
In support of our nonsignificant findings on the effect of Hcy-PGS on cognitive performance and decline and beta-amyloid pathology in CN elderly, previous studies have demonstrated a lack of association between rs1801133 and cognitive performance (Almeida et al., 2005, de Lau et al., 2010, Durga et al., 2006), cognitive decline (Bathum et al., 2007, Schiepers et al., 2011a, Schiepers et al., 2011b), and conversion to AD (Gussekloo et al., 1999), except for 2 studies reporting weak associations (Elkins et al., 2007, Ford et al., 2012). However, we found a positive association with tau tangle burden and a negative association with GM atrophy (prominently in the occipital lobe) in CN individuals.
We cannot rule out the possibility of tau and magnetic resonance imaging false-positive findings in CN (due to multiple comparisons) and false-negative findings in AD (due to limited sample size). On the other hand, it is also possible that our results reflect associations between Hcy-PGS and non-AD pathologies (such as e.g., primary age-related tauopathy or age-related macular degeneration). In support of our tau tangle finding, MTHFR knockout mice have increased brain phosphorylated tau in comparison to wild-type controls, with enhanced effects notably in aged compared to young mice (Sontag et al., 2014). Although our postmortem brain tangle finding in ROS/MAP CN individuals was not supported by ADNI CSF data, this can be explained by the imperfect correlation between brain and CSF tau (Chhatwal et al., 2016). Our CN GM findings were present in almost all subsamples included in the study (Fig. 4B), and our results were not dependent on the method of analysis (VBM vs. FreeSurfer cortical thickness data), both of which increase confidence in our finding. Hence, we believe that the causal association between Hcy and tau burden and GM atrophy in CN elderly deserves further exploration.
While results from Hcy-lowering randomized clinical trials are inconsistent and Mendelian randomization results are nonsignificant, various mechanisms may explain previously observed significant associations between Hcy and AD-related outcomes in cross-sectional and longitudinal observational studies. One possible explanation is reverse causality, as it could be argued that Hcy levels may increase secondary to AD pathological processes, and are the result, not the cause of AD pathology. Other than pathological processes inherent to the disease, it is also possible that changes in lifestyle and diet caused by AD dementia-related behavioral alterations influence Hcy metabolism. Another possible explanation could be confounding factors that are associated with both AD risk and elevated Hcy levels. Potential confounding mechanisms can include vascular, metabolic, lifestyle, and dietary risk factors. Moreover, as it has been suggested that a causal relationship is present between the MTHFR C677T polymorphism and AD risk in individuals with non-European ancestry (Hu et al., 2016, Hua et al., 2011, Peng et al., 2015, Rai, 2016, Zhang et al., 2010), ethnicity might also modulate the effect of genetically determined Hcy on AD, either via genetic interactions or environmental factors such as differences in lifestyle.
Our study was limited to individuals of European descent. However, because of the suggested differences in the effect of Hcy-related polymorphisms in populations of different ancestries, it is important to perform separate large-scale and less-biased analyses in individuals from each ancestry. Another limitation to our study is the limited proportion of variation of Hcy levels explained by the polygenetic score (van Meurs et al., 2013). However, our study was well powered to detect a significant association between the Hcy-PGS and AD risk using a sample size of ~54,000 people (reported OR [Seshadri et al., 2002] for 1-standard deviation increase in log-transformed Hcy on AD risk: OR [95% CI] = 1.4[1.2–1.7], variance explained by Hcy-PGS: ~5% [van Meurs et al., 2013], power: >80% assuming an odds ratio as low as 1.12 for the effect of Hcy on AD risk [Brion et al., 2013]). Using PGS has the advantage of providing evidence for the effect of lifetime exposure to higher Hcy levels, rather than the short-term effects that are assessed using Hcy-lowering randomized clinical trials. However, we cannot rule out the effect of canalization, that is, the compensatory feedback interactions that may buffer the effect of genetically determined Hcy.
Taken together, our integrative genetic analyses of multiple data sources, data types, outcome variables, and methods of analysis suggest that there is no causal association between elevated Hcy and AD in people of European ancestry. Accordingly, our results suggest that treating elevated Hcy levels would not necessarily protect against the development of AD. High Hcy has previously been proposed as a risk factor for coronary artery disease and ischemic stroke, both of which have higher prevalence in the aged population similar to AD. To date, there is also no evidence supporting a causal relationship between genetically determined Hcy and these diseases (Cotlarciuc et al., 2014, van Meurs et al., 2013).
Supplementary Material
Acknowledgements
The work was supported by the Isabel Johnson Biomedical Postdoctoral Award, Alzheimer Society of Canada Research Program (awarded to TR). The funding source had no involvement in the study design; collection, analysis, and interpretation of data; writing of the report; and the decision to submit the article for publication.
DF is supported by a CIHR Postdoctoral Fellowship. AN was funded by the Centre for Addiction and Mental Health and Canadian Institutes of Health Research Fellowship awards. ANV is funded by the Canadian Institutes of Health Research, Ontario Mental Health Foundation, Brain and Behavior Research Foundation, and the National Institute of Mental Health (R01MH099167 and R01MH102324).
The authors thank the International Genomics of Alzheimer’s Project (IGAP), National Institute on Aging Alzheimer’s Disease Centers (ADCs) Cohort, Multi-Site Collaborative Study for Genotype-Phenotype Associations in Alzheimer’s Disease (GenADA), Alzheimer’s Disease Neuroimaging Initiative (ADNI), and Rush Alzheimer’s Disease Center Religious Orders Study and Memory and Aging Project (ROS/MAP) for providing data for these analyses. These studies were made possible by the generous participation of the control subjects, the patients, and their families.
The i-Select chips used in IGAP was funded by the French National Foundation on Alzheimer’s disease and related disorders. EADI was supported by the LABEX (laboratory of excellence program investment for the future) DISTALZ grant, Inserm, Institut Pasteur de Lille, Université de Lille 2, and the Lille University Hospital. GERAD was supported by the Medical Research Council (Grant no. 503480), Alzheimer’s Research UK (Grant no. 503176), the Wellcome Trust (Grant no. 082604/2/07/Z), and German Federal Ministry of Education and Research (BMBF): Competence Network Dementia (CND) grant no. 01GI0102, 01GI0711, 01GI0420. CHARGE was partly supported by the NIH/NIA grant R01 AG033193 and the NIA AG081220 and AGES contract N01-AG-12100, the NHLBI grant R01 HL105756, the Icelandic Heart Association, and the Erasmus Medical Center and Erasmus University. ADGC was supported by the NIH/NIA grants: U01 AG032984, U24 AG021886, U01 AG016976, and the Alzheimer’s Association grant ADGC-10-196728. The genotypic and associated phenotypic data used in GenADA were provided by the GlaxoSmithKline, R&D Limited. Data collection and sharing for ADNI was funded by ADNI National Institutes of Health Grant U01 AG024904 and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc; Eisai Inc; Elan Pharmaceuticals, Inc; Eli Lilly and Company; Euroimmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc; Fujirebio; GE Healthcare; IXICO Ltd; Janssen Alzheimer Immunotherapy Research & Development, LLC; Johnson & Johnson Pharmaceutical Research & Development LLC; Lumosity; Lundbeck; Merck & Co, Inc; Meso Scale Diagnostics, LLC; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California. ROS/MAP is funded by NIA grants P30AG10161 and RF1AG15819 for the Religious Orders Study and R01AG17917 for the Memory and Aging Project.
Footnotes
Disclosure statement
The authors have no actual or potential conflicts of interest.
References
- Abushik PA, Niittykoski M, Giniatullina R, Shakirzyanova A, Bart G, Fayuk D, Sibarov DA, Antonov SM, Giniatullin R, 2014. The role of NMDA and mGluR5 receptors in calcium mobilization and neurotoxicity of homocysteine in trigeminal and cortical neurons and glial cells. J. Neurochem 129, 264e274. [DOI] [PubMed] [Google Scholar]
- Almeida OP, Flicker L, Lautenschlager NT, Leedman P, Vasikaran S, van Bockxmeer FM, 2005. Contribution of the MTHFR gene to the causal pathway for depression, anxiety and cognitive impairment in later life. Neurobiol. Aging 26, 251e257. [DOI] [PubMed] [Google Scholar]
- Annerbo S, Wahlund L-O, Lökk J, 2006. The significance of thyroid-stimulating hormone and homocysteine in the development of Alzheimer’s disease in mild cognitive impairment a 6-year follow-up study. Am. J. Alzheimers Dis. Other Demen. 21, 182e188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bathum L, von Bornemann Hjelmborg J, Christiansen L, McGue M, Jeune B, Christensen K, 2007. Methylenetetrahydrofolate reductase 677C>T and methionine synthase 2756A>G mutations: no impact on survival, cognitive functioning, or cognitive decline in nonagenarians. J. Gerontol. A. Biol. Sci. Med. Sci 62, 196e201. [DOI] [PubMed] [Google Scholar]
- Bennett DA, Schneider JA, Arvanitakis Z, Wilson RS, 2012a. Overview and findings from the religious orders study. Curr. Alzheimer Res. 9, 628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett DA, Schneider JA, Buchman AS, Barnes LL, Boyle PA, Wilson RS, 2012b. Overview and findings from the rush memory and aging Project. Curr Alzheimer Res. 9, 646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brion MJ, Shakhbazov K, Visscher PM, 2013. Calculating statistical power in Mendelian randomization studies. Int. J. Epidemiol 42, 1497e1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess S, Butterworth A, Thompson SG, 2013. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet. Epidemiol 37, 658e665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhatwal JP, Schultz AP, Marshall GA, Boot B, Gomez-Isla T, Dumurgier J,LaPoint M, Scherzer C, Roe AD, Hyman BT, Sperling RA, Johnson KA,2016. Temporal T807 binding correlates with CSF tau and phospho-tau innormal elderly. Neurology 87, 920e926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotlarciuc I, Malik R, Holliday EG, Ahmadi KR, Pare G, Psaty BM, Fornage M,Hasan N, Rinne PE, Ikram MA, Markus HS, Rosand J, Mitchell BD,Kittner SJ, Meschia JF, van Meurs JB, Uitterlinden AG, Worrall BB,Dichgans M, Sharma P, 2014. Effect of genetic variants associated with plasma homocysteine levels on stroke risk. Stroke 45, 1920e1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lau LM, van Meurs JB, Uitterlinden AG, Smith AD, Refsum H, Johnston C, Breteler MM, 2010. Genetic variation in homocysteine metabolism, cognition, and white matter lesions. Neurobiol. Aging 31, 2020e2022. [DOI] [PubMed] [Google Scholar]
- Douaud G, Refsum H, de Jager CA, Jacoby R, Nichols TE, Smith SM, Smith AD, 2013. Preventing Alzheimer’s disease-related gray matter atrophy by B-vitamin treatment. Proc. Natl. Acad. Sci. U. S. A 110, 9523e9528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufouil C, Alpérovitch A, Ducros V, Tzourio C, 2003. Homocysteine, white matter hyperintensities, and cognition in healthy elderly people. Ann. Neurol 53, 214e221. [DOI] [PubMed] [Google Scholar]
- Durga J, van Boxtel MP, Schouten EG, Bots ML, Kok FJ, Verhoef P, 2006. Folate and the methylenetetrahydrofolate reductase 677C–>T mutation correlate with cognitive performance. Neurobiol. Aging 27, 334e343. [DOI] [PubMed] [Google Scholar]
- Durga J, Van Boxtel MPJ, Schouten EG, Kok FJ, Jolles J, Katan MB, Verhoef P, 2007. Effect of 3-year folic acid supplementation on cognitive function in older adults in the FACIT trial: a randomised, double blind, controlled trial. The Lancet 369, 208e216. [DOI] [PubMed] [Google Scholar]
- Elkins JS, Johnston SC, Ziv E, Kado D, Cauley JA, Yaffe K, 2007. Methylenetetrahydrofolate reductase C677T polymorphism and cognitive function in older women. Am. J. Epidemiol 166, 672e678. [DOI] [PubMed] [Google Scholar]
- Eussen SJ, de Groot LC, Joosten LW, Bloo RJ, Clarke R, Ueland PM, Schneede J, Blom HJ, Hoefnagels WH, van Staveren WA, 2006. Effect of oral vitamin B-12 with or without folic acid on cognitive function in older people with mild vitamin B-12 deficiency: a randomized, placebo-controlled trial. Am.J. Clin. Nutr 84, 361e370. [DOI] [PubMed] [Google Scholar]
- Ford AH, Flicker L, Hankey GJ, Norman P, van Bockxmeer FM, Almeida OP, 2012. Homocysteine, methylenetetrahydrofolate reductase C677T polymorphism and cognitive impairment: the health in men study. Mol. Psychiatry 17, 559e566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgone G, Ursini F, Altamura C, Bressi F, Tombini M, Curcio G, Chiovenda P, Squitti R, Silvestrini M, Ientile R, Pisani F, Rossini PM, Vernieri F, 2009. Hyperhomocysteinemia, intima-media thickness and C677T MTHFR gene polymorphism: a correlation study in patients with cognitive impairment. Atherosclerosis 206, 309e313. [DOI] [PubMed] [Google Scholar]
- Gussekloo J, Heijmans BT, Slagboom PE, Lagaay AM, Knook DL, Westendorp RG, 1999. Thermolabile methylenetetrahydrofolate reductase gene and the risk of cognitive impairment in those over 85. J. Neurol. Neurosurg. Psychiatry 67, 535e538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q, Teng W, Li J, Hao F, Wang N, 2016. Homocysteine and Alzheimer’s disease: evidence for a causal link from Mendelian randomization. J. Alzheimers Dis. 52, 747e756. [DOI] [PubMed] [Google Scholar]
- Hua Y, Zhao H, Kong Y, Ye M, 2011. Association between the MTHFR gene and Alzheimer’s disease: a meta-analysis. Int. J. Neurosci 121, 462e471. [DOI] [PubMed] [Google Scholar]
- Jager CA, Oulhaj A, Jacoby R, Refsum H, Smith AD, 2012. Cognitive and clinical outcomes of homocysteine-lowering B-vitamin treatment in mild cognitive impairment: a randomized controlled trial. Int. J. Geriatr. Psychiatry 27, 592e600. [DOI] [PubMed] [Google Scholar]
- Kim H-J, Cho H-K, Kwon YH, 2008. Synergistic induction of ER stress by homocysteine and b-amyloid in SH-SY5Y cells. J. Nutr. Biochem 19, 754e761. [DOI] [PubMed] [Google Scholar]
- Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, DeStafano AL, Bis JC, Beecham GW, Grenier-Boley B, Russo G, Thorton- Wells TA, Jones N, Smith AV, Chouraki V, Thomas C, Ikram MA, Zelenika D, Vardarajan BN, Kamatani Y, Lin CF, Gerrish A, Schmidt H, Kunkle B, Dunstan ML, Ruiz A, Bihoreau MT, Choi SH, Reitz C, Pasquier F, Cruchaga C, Craig D, Amin N, Berr C, Lopez OL, De Jager PL, Deramecourt V, Johnston JA, Evans D, Lovestone S, Letenneur L, Moron FJ, Rubinsztein DC, Eiriksdottir G, Sleegers K, Goate AM, Fievet N, Huentelman MW, Gill M, Brown K, Kamboh MI, Keller L, Barberger-Gateau P, McGuiness B, Larson EB, Green R, Myers AJ, Dufouil C, Todd S, Wallon D, Love S, Rogaeva E, Gallacher J, St George-Hyslop P, Clarimon J, Lleo A, Bayer A, Tsuang DW, Yu L, Tsolaki M, Bossu P, Spalletta G, Proitsi P, Collinge J, Sorbi S, Sanchez-Garcia F, Fox NC, Hardy J, Deniz Naranjo MC, Bosco P, Clarke R, Brayne C, Galimberti D, Mancuso M, Matthews F, Moebus S, Mecocci P, Del Zompo M, Maier W, Hampel H, Pilotto A, Bullido M, Panza F, Caffarra P, Nacmias B, Gilbert JR, Mayhaus M, Lannefelt L, Hakonarson H, Pichler S, Carrasquillo MM, Ingelsson M, Beekly D, Alvarez V, Zou F, Valladares O, Younkin SG, Coto E, Hamilton-Nelson KL, Gu W, Razquin C, Pastor P, Mateo I, Owen MJ, Faber KM, Jonsson PV, Combarros O, O’Donovan MC, Cantwell LB, Soininen H, Blacker D, Mead S, Mosley TH Jr., Bennett DA, Harris TB, Fratiglioni L, Holmes C, de Bruijn RF, Passmore P, Montine TJ, Bettens K, Rotter JI, Brice A, Morgan K, Foroud TM, Kukull WA, Hannequin D, Powell JF, Nalls MA, Ritchie K, Lunetta KL, Kauwe JS, Boerwinkle E, Riemenschneider M, Boada M, Hiltuenen M, Martin ER, Schmidt R, Rujescu D, Wang LS, Dartigues JF, Mayeux R, Tzourio C, Hofman A, Nothen MM, Graff C, Psaty BM, Jones L, Haines JL, Holmans PA, Lathrop M, Pericak-Vance MA, Launer LJ, Farrer LA, van Duijn CM, Van Broeckhoven C, Moskvina V, Seshadri S, Williams J, Schellenberg GD, Amouyel P, 2013. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat. Genet 45, 1452e1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawlor DA, Harbord RM, Sterne JA, Timpson N, Davey Smith G, 2008. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat. Med 27, 1133e1163. [DOI] [PubMed] [Google Scholar]
- Li H, Wetten S, Li L, St Jean PL, Upmanyu R, Surh L, Hosford D, Barnes MR, Briley JD, Borrie M, Coletta N, Delisle R, Dhalla D, Ehm MG, Feldman HH, Fornazzari L, Gauthier S, Goodgame N, Guzman D, Hammond S, Hollingworth P, Hsiung GY, Johnson J, Kelly DD, Keren R, Kertesz A, King KS, Lovestone S, Loy-English I, Matthews PM, Owen MJ, Plumpton M, Pryse-Phillips W, Prinjha RK, Richardson JC, Saunders A, Slater AJ, St George-Hyslop PH, Stinnett SW, Swartz JE, Taylor RL, Wherrett J, Williams J, Yarnall DP, Gibson RA, Irizarry MC, Middleton LT, Roses AD, 2008. Candidate single-nucleotide polymorphisms from a genomewide association study of Alzheimer disease. Arch. Neurol 65, 45e53. [DOI] [PubMed] [Google Scholar]
- Luo Y, Zhou X, Yang X, Wang J, 2007. Homocysteine induces s hyperphosphorylation in rats. Neuroreport 18, 2005e2008. [DOI] [PubMed] [Google Scholar]
- McMahon JA, Green TJ, Skeaff CM, Knight RG, Mann JI, Williams SM, 2006. A controlled trial of homocysteine lowering and cognitive performance. N. Engl. J. Med 354, 2764e2772. [DOI] [PubMed] [Google Scholar]
- Naj AC, Jun G, Beecham GW, Wang LS, Vardarajan BN, Buros J, Gallins PJ, Buxbaum JD, Jarvik GP, Crane PK, Larson EB, Bird TD, Boeve BF, Graff- Radford NR, De Jager PL, Evans D, Schneider JA, Carrasquillo MM, Ertekin-Taner N, Younkin SG, Cruchaga C, Kauwe JS, Nowotny P, Kramer P, Hardy J, Huentelman MJ, Myers AJ, Barmada MM, Demirci FY, Baldwin CT, Green RC, Rogaeva E, St George-Hyslop P, Arnold SE, Barber R, Beach T, Bigio EH, Bowen JD, Boxer A, Burke JR, Cairns NJ, Carlson CS, Carney RM, Carroll SL, Chui HC, Clark DG, Corneveaux J, Cotman CW, Cummings JL, DeCarli C, DeKosky ST, Diaz-Arrastia R, Dick M, Dickson DW, Ellis WG, Faber KM, Fallon KB, Farlow MR, Ferris S, Frosch MP, Galasko DR, Ganguli M, Gearing M, Geschwind DH, Ghetti B, Gilbert JR, Gilman S, Giordani B, Glass JD, Growdon JH, Hamilton RL, Harrell LE, Head E, Honig LS, Hulette CM, Hyman BT, Jicha GA, Jin LW, Johnson N, Karlawish J, Karydas A, Kaye JA, Kim R, Koo EH, Kowall NW, Lah JJ, Levey AI, Lieberman AP, Lopez OL, Mack WJ, Marson DC, Martiniuk F, Mash DC, Masliah E, McCormick WC, McCurry SM, McDavid AN, McKee AC, Mesulam M, Miller BL, Miller CA, Miller JW, Parisi JE, Perl DP, Peskind E, Petersen RC, Poon WW, Quinn JF, Rajbhandary RA, Raskind M, Reisberg B, Ringman JM, Roberson ED, Rosenberg RN, Sano M, Schneider LS, Seeley W, Shelanski ML, Slifer MA, Smith CD, Sonnen JA, Spina S, Stern RA, Tanzi RE, Trojanowski JQ, Troncoso JC, Van Deerlin VM, Vinters HV, Vonsattel JP, Weintraub S, Welsh-Bohmer KA,Williamson J,Woltjer RL, Cantwell LB, Dombroski BA, Beekly D, Lunetta KL, Martin ER, Kamboh MI, Saykin AJ, Reiman EM, Bennett DA, Morris JC, Montine TJ, Goate AM, Blacker D, Tsuang DW, Hakonarson H, Kukull WA, Foroud TM, Haines JL, Mayeux R, Pericak- Vance MA, Farrer LA, Schellenberg GD, 2011. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat. Genet 43, 436e441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Q, Lao X, Huang X, Qin X, Li S, Zeng Z, 2015. The MTHFR C677T polymorphism contributes to increased risk of Alzheimer’s disease: evidence based on 40 case-control studies. Neurosci. Lett 586, 36e42. [DOI] [PubMed] [Google Scholar]
- Rai V, 2016. Methylenetetrahydrofolate reductase (MTHFR) C677T polymorphism and Alzheimer disease risk: a meta-analysis. Mol. Neurobiol 54, 1173e1186. [DOI] [PubMed] [Google Scholar]
- Schiepers OJ, van Boxtel MP, de Groot RH, Jolles J, Bekers O, Kok FJ, Verhoef P, Durga J, 2011a. Genetic variation in folate metabolism is not associated with cognitive functioning or mood in healthy adults. Prog. Neuropsychopharmacol. Biol. Psychiatry 35, 1682e1688. [DOI] [PubMed] [Google Scholar]
- Schiepers OJ, van Boxtel MP, Harris SE, Gow AJ, Pattie A, Brett CE, de Groot RH, Jolles J, Starr JM, Deary IJ, 2011b. MTHFR polymorphisms and cognitive ageing in the ninth decade: the Lothian Birth Cohort 1921. Genes Brain Behav. 10, 354e364. [DOI] [PubMed] [Google Scholar]
- Seripa D, Forno GD, Matera MG, Gravina C, Margaglione M, Palermo MT, Wekstein DR, Antuono P, Davis DG, Daniele A, Masullo C, Bizzarro A, Gennarelli M, Fazio VM, 2003. Methylenetetrahydrofolate reductase and angiotensin converting enzyme gene polymorphisms in two genetically and diagnostically distinct cohortof Alzheimer patients.Neurobiol.Aging 24, 933e939. [DOI] [PubMed] [Google Scholar]
- Seshadri S, Beiser A, Selhub J, Jacques PF, Rosenberg IH, D’Agostino RB, Wilson PWF, Wolf PA, 2002. Plasma homocysteine as a risk factor for dementia and Alzheimer’s disease. N. Engl. J. Med 346, 476e483. [DOI] [PubMed] [Google Scholar]
- Sontag JM,Wasek B, Taleski G, Smith J, Arning E, Sontag E, Bottiglieri T, 2014. Altered protein phosphatase 2A methylation and Tau phosphorylation in the young and aged brain of methylenetetrahydrofolate reductase (MTHFR) deficient mice. Front. Aging Neurosci. 6, 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streck EL, Vieira PS, Wannmacher CMD, Dutra-Filho CS, Wajner M, Wyse ATS, 2003. In vitro effect of homocysteine on some parameters of oxidative stress in rat hippocampus. Metab. Brain Dis. 18, 147e154. [DOI] [PubMed] [Google Scholar]
- Tucker KL, Qiao N, Scott T, Rosenberg I, Spiro A, 2005. High homocysteine and low B vitamins predict cognitive decline in aging men: the Veterans Affairs Normative Aging Study. Am. J. Clin. Nutr 82, 627e635. [DOI] [PubMed] [Google Scholar]
- van Meurs JB, Pare G, Schwartz SM, Hazra A, Tanaka T, Vermeulen SH, Cotlarciuc I, Yuan X, Malarstig A, Bandinelli S, Bis JC, Blom H, Brown MJ, Chen C, Chen YD, Clarke RJ, Dehghan A, Erdmann J, Ferrucci L, Hamsten A, Hofman A, Hunter DJ, Goel A, Johnson AD, Kathiresan S, Kampman E, Kiel DP, Kiemeney LA, Chambers JC, Kraft P, Lindemans J, McKnight B, Nelson CP, O’Donnell CJ, Psaty BM, Ridker PM, Rivadeneira F, Rose LM, Seedorf U, Siscovick DS, Schunkert H, Selhub J, Ueland PM, Vollenweider P, Waeber G, Waterworth DM, Watkins H, Witteman JC, den Heijer M, Jacques P, Uitterlinden AG, Kooner JS, Rader DJ, Reilly MP, Mooser V, Chasman DI, Samani NJ, Ahmadi KR, 2013. Common genetic loci influencing plasma homocysteine concentrations and their effect on risk of coronary artery disease. Am. J. Clin. Nutr 98, 668e676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner MW, Aisen PS, Jack CR Jr., Jagust WJ, Trojanowski JQ, Shaw L, Saykin AJ, Morris JC, Cairns N, Beckett LA, Toga A, Green R, Walter S, Soares H, Snyder P, Siemers E, Potter W, Cole PE, Schmidt M, 2010. The Alzheimer’s disease neuroimaging initiative: progress report and future plans. Alzheimers Dement. 6, 202e211.e207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C-E, Wei W, Liu Y-H, Peng J-H, Tian Q, Liu G-P, Zhang Y, Wang J-Z, 2009. Hyperhomocysteinemia increases b-amyloid by enhancing expression of g-secretase and phosphorylation of amyloid precursor protein in rat brain. Am. J. Pathol 174, 1481e1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang MY, Miao L, Li YS, Hu GY, 2010. Meta-analysis of the methylenetetrahydrofolate reductase C677T polymorphism and susceptibility to Alzheimer’s disease. Neurosci. Res 68, 142e150. [DOI] [PubMed] [Google Scholar]
- Zhuo JM, Wang H, Pratico D, 2011. Is hyperhomocysteinemia an Alzheimer’s disease (AD) risk factor, an AD marker, or neither? Trends Pharmacol. Sci. 32, 562e571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zylberstein DE, Lissner L, Björkelund C, Mehlig K, Thelle DS, Gustafson D, Östling S,Waern M, Guo X, Skoog I, 2011. Midlife homocysteine and late-life dementia in women. A prospective population study. Neurobiol. Aging 32,380e386. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.