Abstract
Posttraumatic stress disorder (PTSD) is a major problem among military veterans and civilians alike, yet its pathophysiology remains poorly understood. We used genomewide association study (GWAS) and bioinformatic analyses, including 146,660 European-Americans (EAs) and 19,983 African-Americans (AAs) in the US Million Veteran Program, to identify genetic risk factors relevant to Intrusive reexperiencing of trauma -- the most characteristic symptom cluster of PTSD. In EAs, 8 distinct significant regions were identified. Three regions had P<5×10−10 -- CAMKV; chromosome 17 closest to KANSL1 but within a large high-LD region that also includes CRHR1; and TCF4. Associations were enriched with respect to the transcriptomic profiles of striatal medium spiny neurons. No significant associations were observed in the AA part of the sample. Results in EAs were replicated in UK Biobank. These results provide new insights into the biology of PTSD in a well--powered GWAS.
Introduction
Posttraumatic stress disorder (PTSD) is a worldwide public health problem and of particular concern to the US military. While PTSD can be attributable to a range of traumatic events, the nature and intensity of traumatic events experienced by some military personnel result in particularly high prevalence among US military veterans1. Published PTSD genome-wide association studies (GWAS) have identified genomewide-significant risk loci2–4; however these studies have tended to be small and underpowered for a trait as complex as PTSD. The largest to date was a meta-analysis undertaken by the Psychiatric Genomics Consortium (PGC)5. While there have been promising possible gene identifications, there are none yet that are both significant and replicated.
The PTSD Checklist (PCL) is a widely used 17-item self-report measure of past-month PTSD symptoms6. It collects information relevant to the three (according to DSM-IV) PTSD symptom clusters: reexperiencing, avoidance, and hyperarousal. Of these, hyperarousal and avoidance are common to many anxiety-related disorders, but re-experiencing symptoms (e.g., nightmares and flashbacks about a traumatic event) are largely unique to PTSD and are core to the diagnosis.
To increase power for risk variant identification, the two main strategies are to increase sample size, and to increase the informativity of the phenotype. In the present study, we did both: we included over 165,000 US Million Veteran Program (MVP) participants, and we used a quantitative phenotype for reexperiencing.
Results
In the EAs, (137,044 males and 9,616 females), SNP-h2=0.067±0.005 (SE) (p = 3.02×10−41). Estimated heritability was similar in males only (h2 = 0.066±0.005, p=4.38×10−40). In the AAs, SNP-h2=0.048±0.039 (p=0.109).
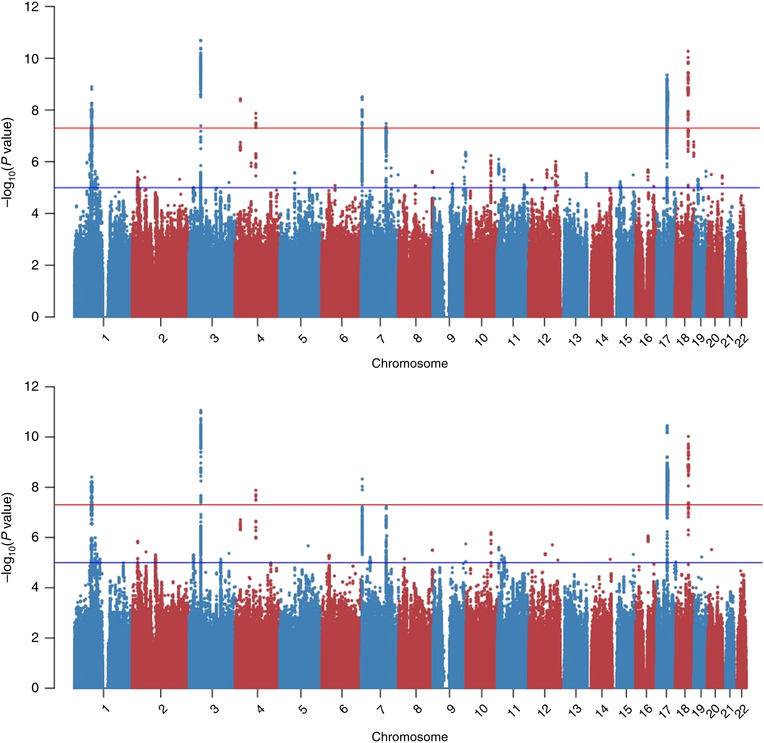
Due to the large sample size of the EA cohort investigated (N=146,660), we observed a consistent polygenic signal in the summary association data (λ=1.20), with negligible inflation observed (LD score regression intercept=1.05±0.01) due to possible confounding factors, including population stratification and the distribution of the reexperiencing items in the cohort investigated (Table S1).. Eight distinct common-variant GWS regions were identified in this ancestry group (Table 1 and Figure 1), three of these with significance p<5×1010. These latter three regions map to chromosome 3, lead SNP rs2777888 (beta=0.111, p=2.1×10−11), gene CAMKV (CaM Kinase Like Vesicle Associated); chromosome 17, lead SNP rs2532252 (beta=0.122, p=4.5×10−10), closest to KANSL1 but within a long high-LD region that also includes CRHR1 (corticotropin releasing hormone receptor 1); and chromosome 18, lead SNP rs2123392 (beta=0.113, p=5.4×10−11), at TCF4 (Transcription Factor 4). Other significant associations were observed at KCNIP4 (Potassium Voltage-Gated Channel Interacting Protein 4, rs4697248: beta=0.105, p=3.73×10−9), HSD17B11 (Hydroxysteroid 17-Beta Dehydrogenase 11, rs7688962: beta=−0.126, p=1.34×10−8), MAD1L1 (MAD1 Mitotic Arrest Deficient Like 1, rs10235664: beta=−0.113, p = 3.09×10−9), SRPK2 (SRSF Protein Kinase 2, rs67529088: beta=−0.09, p=3.32×10−8), and LINC01360 (Long Intergenic Non-Protein Coding RNA 1360, an RNA Gene, rs7519147: beta=−0.101, p=1.29×10−9). The 10,000 statistically strongest associations for EAs and AAs are presented in Tables S3 and S4.
Table 1:
Top markers in EA GWAS of REX in MVP cohort and replication in UKB sample.
| rsID | CHR | BP | Closest Gene (location) | effect allele | other allele | MVP Re-experiencing | UKB REX | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Beta | SE | P | Beta | SE | P | ||||||
| rs7519147 | 1 | 73994416 | LINC01360 (downstream) | T | C | −0.101 | 0.017 | 1.29E-09 | −0.001 | 0.003 | 0.740 |
| rs2777888 | 3 | 49898000 | CAMKV (intron) | G | A | 0.111 | 0.017 | 2.07E-11 | 0.007 | 0.003 | 0.034 |
| rs4697248 | 4 | 21931195 | KCNIP4 (intron) | C | T | 0.105 | 0.018 | 3.73E-09 | 0.006 | 0.003 | 0.078 |
| rs7688962 | 4 | 88281182 | HSD17B11 (intron) | A | G | −0.126 | 0.022 | 1.34E-08 | −0.011 | 0.004 | 0.012 |
| rs10235664 | 7 | 2086814 | MAD1L1 (intron) | C | T | −0.113 | 0.019 | 3.09E-09 | −0.0003 | 0.004 | 0.940 |
| rs67529088 | 7 | 104907066 | SRPK2 (intron) | C | G | −0.09 | 0.016 | 3.32E-08 | −0.007 | 0.003 | 0.030 |
| rs2532252 | 17 | 44257783 | KANSL1 (intron) | G | A | 0.122 | 0.019 | 4.50E-10 | 0.010 | 0.004 | 0.007 |
| rs2123392 | 18 | 53214865 | TCF4 (intron) | C | T | 0.113 | 0.017 | 5.38E-11 | 0.011 | 0.003 | 3.62E-04 |
Figure 1:
Manhattan plots – European-Americans (top; N=146,660), and meta-analyzed European- and African-Americans (bottom; AA N=19,983). Negative logarithm of the association two-sided p value (from a linear regression model) for each variant tested is reported on the y-axis. Red and blue line indicates the Bonferroni multiple testing correction and suggestive significances, respectively.
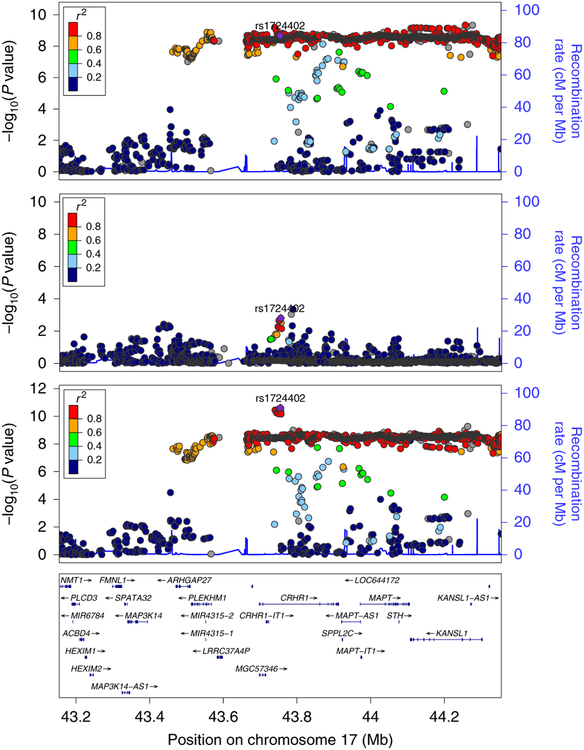
There were no GWS associations in the smaller AA sample (Figure S2). The meta-analysis of the EA and AA results showed mostly similar results, with the exception of regions on chromosome 1 and chromosome 17, which improved in significance. In other regions, significance was similar to what was seen for EAs-only, but the position of the lead SNP sometimes shifted, and two regions were not GWS in the transpopulation meta-analysis (SPRK2, KCNIP4) (Table S5). The most noteworthy difference in the transpopulation meta-analysis was seen on chromosome 17 (Figure 2), where the association signal improved by over an order of magnitude and shifted from KANSL1 rs2532252 (p=4.5×10−10) to CRHR1 rs1724409 (p=3.55×10−11). Regional Manhattan plots for significant regions in EAs, AAs, and meta-analysis are presented in Figure S3.
Figure 2:
Regional Manhattan plots, associated region on chromosome 17, European-Americans, African-Americans, and European- and African-Americans, meta-analyzed – showing that associated region localizes to gene CRHR1 only on trans-population meta-analysis. Details regarding sample size and statistical test are reported in the Figure 1 legend.
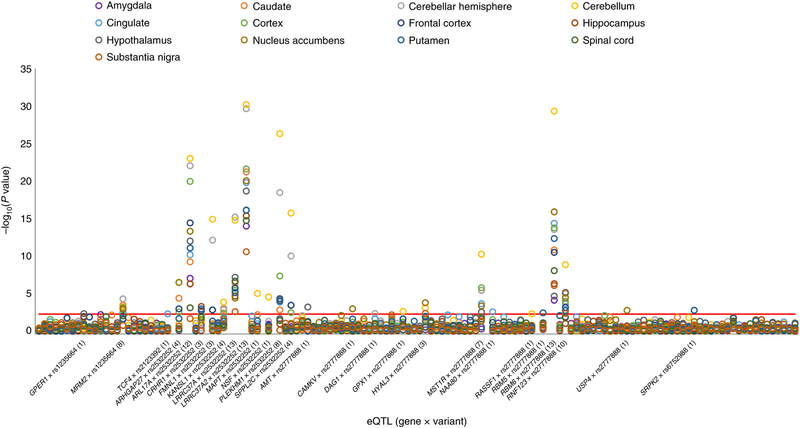
To explore further the functional role of the GWS variants identified, we tested association of these loci with gene expression (cis-eQTL: variants located ±1Mb from the transcription site start of the gene tested) considering the 13 tissues related to the central nervous system (CNS) available in GTEx V77. After applying a FDR 5% correction accounting for the number of variants, genes, and tissues tested, we observed 257 significant eQTLs (of 2,514 tests) conducted with respect to the GWS loci observed in the EUR samples (Supplemental Table 6). The eight GWS variants observed in EUR are associated with the expression of 52 genes, of which 27 are protein-coding. However, most of the eQTLs relate to two variants: rs2532252 on chromosome 17 is associated with the expression of 30 different genes; rs2777888 on chromosome 3 is associated with the expression of 14 different genes. Figure 3 shows the strength of the eQTLs identified with respect to protein-coding genes across the 13 brain tissues investigated. Due to the strong LD with the loci identified in the EUR analysis, similar eQTL results were observed with respect to the variants identified in the trans-ancestry meta-analysis.
Figure 3:
eQTLs observed with respect to the eight GWS variants identified in EUR meta-analysis (N=146,660) considering protein-coding genes and 13 brain tissues. The eQTL IDs (x-axis, gene*variant) are reported only for those with at least one result surviving FDR multiple testing correction. Negative logarithm of the association two-sided p value for each eQTL tested is reported on the y-axis. The number reported between parentheses in each eQTL corresponds to the significant results observed with respect to the 13 brain tissues tested. The red line corresponds to the FDR multiple testing correction (q<5%).
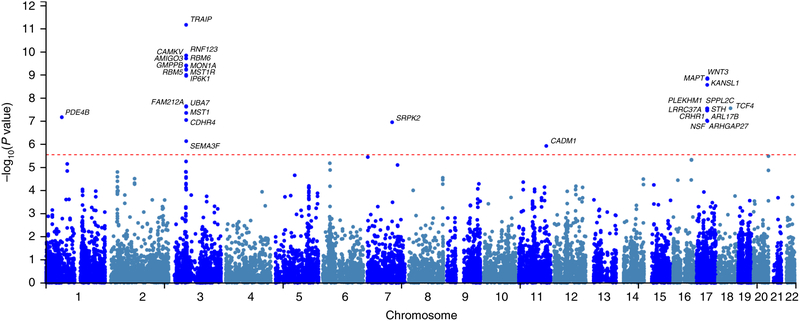
Gene-based analysis
There were 30 GWS gene-based associations detected using MAGMA (Figure 4; Supplemental Table S7). The strongest significance was observed for TRAIP (p = 6.64×10−12) on chromosome 3 (there was no significant SNP association mapped to this locus). An additional 14 genes reached GWS in the same chromosomal region. A second large cluster of significant gene-based associations was observed on chromosome 17, where CRHR1 reached GWS (p = 3.44×10−8) together with another 10 genes located in the same chromosomal region.
Figure 4:
Manhattan plot reporting gene-based association results generated from the EA cohort (N=146,660). Details are reported in Supplemental Table S7. Negative logarithm of the association p value for each gene tested is reported on the y-axis. The red dotted line represents the Bonferroni threshold for gene-based association (0.05/18120).
Genetic correlations
LDSC revealed significant genetic correlations (FDR q<0.05) with 400 of nearly 1800 traits (Supplementary Table S8). The most significant of these were poor overall-health rating (UKB, rg=0.55, p=9.71×10−57; 4-category scale, from 1-Excellent to 4-Poor) and mood swings (UKB, rg =0.66, p=1.07×10−56). Among the other most interesting statistically significant genetic correlations were with neuroticism (UKB, rg=0.56, p=1.28×−10−44), having seen a doctor (GP) for nerves, anxiety, tension or depression (UKB, rg = 0.58, p=1.54×10−41), years of education (LDHub, r =−0.42, p=9.32×10−32), “loneliness, isolation” (UKB, rg =0.56, p=6.63×10−31), depressive symptoms (LDHub, rg =0.64, p=1.00×10−29), current smoker (UKB, rg =0.43, p=3.49×10−29), intelligence (LDHub, rg=−0.53, p=1.20×10−28), alcohol intake frequency (UKB, rg =0.37, p=2.70×10−22), schizophrenia (LD Hub – PGC, rg=0.25, p=5.17×10−12), insomnia (LDHub, rg=0.38, p=9.49×10−10), risk taking (UKB, rg=0.25, p=2.59×10−9), and hypertension (UKB, rg=0.21, p=3.17×10−09).
Tissue and Cell Type Enrichment
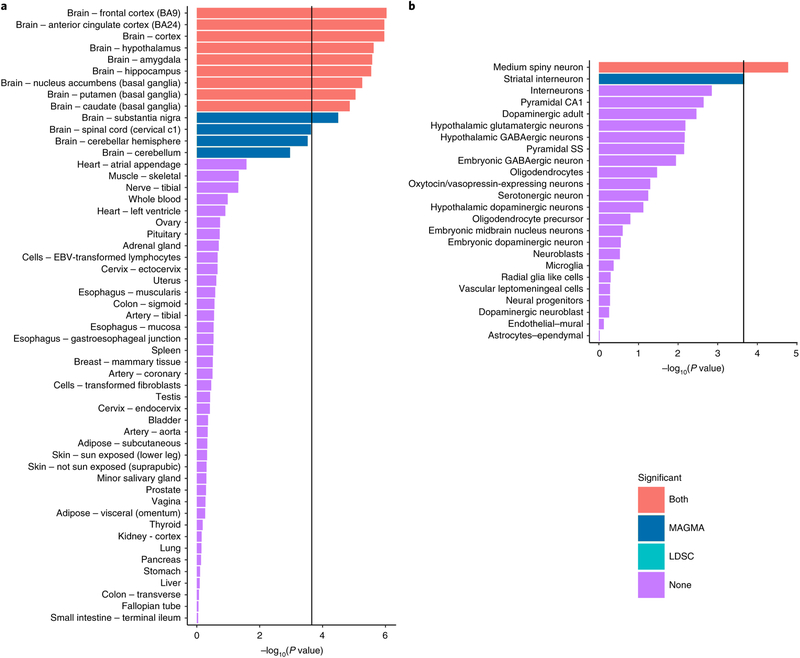
We tested whether genetic associations with REX were enriched with respect to the transcriptomic profiles of human tissues8 and mouse brain cell types9. At the tissue level, we found the transcriptomic profiles related to brain tissues were associated with REX (Figure 5a). The most significant association was observed with respect to the brain cortex, followed by hypothalamus, amygdala, hippocampus and the basal ganglia. At the cell type level, we found the transcriptomic profile of medium spiny neurons (located in the striatum) to be associated with REX using two different methods (Figure 5b), MAGMA and LDSC, while striatal interneurons were implicated only by MAGMA. A more detailed analysis using narrowly-defined cell types found gene expression of medium spiny neurons and one type of cortical interneuron associated with REX LDSC, while MAGMA implicated two types of hypothalamic neurons, dopaminergic neurons from the ventral tegmental area and striatal interneurons (Figure S4). We observed little association evidence with respect to non-neuronal cell types with REX. Pathway analysis are reported in the Supplement - Table S9.
Figure 5:
a. Enrichment analysis between human tissue specific expression and PTSD re-experiencing genetic associations obtained from the EUR sample (N=146,660). Each bar represents the negative logarithm of the enrichment p value for each tissue tested obtained from two different statistical methods (LDSC and MAGMA, see details in the method section). The black bar represents the Bonferroni threshold (0.05/(53+24+149)). b. Enrichment analysis between mouse brain cell type specific expression (broadly defined) and PTSD re-experiencing genetic associations obtained from the EUR sample (N=146,660). Each bar represents the negative logarithm of the enrichment p value for each cell type tested obtained from two different statistical methods (LDSC and MAGMA, see details in the method section). The black bar represents the Bonferroni threshold (0.05/(53+24+149)).
Replication
Although the UKB data are derived from a single item, we observed very strong genetic correlation between MVP EA and UK Biobank REX phenotypes (rg=0.88, SE=0.07, p=8.47 ×10−34). Considering our eight GWS results in EA subjects, we observed nominal replication for five loci – TCF4 rs2123392 (beta=0.011, p=3.62×10−4), KANSL1 rs2532252 (beta=0.010, p=0.007), HSD17B11 rs7688962 (beta=−0.011, p=0.012), SRPK2 rs67529088 (beta=−0.007, p=0.03), and CAMKV rs2777888 (beta=0.007, p=0.034). Applying a more stringent Bonferroni adjustment (p<0.006), TCF4 rs2123392 survived the multiple testing correction. However, all GWS variants signals have the same direction of effect in MVP and UKB analyses (Table 1). The probability to observe concordant direction of all eight MVP-identified loci in UKB GWAS by chance is 0.4%. The whole-genome polygenic risk score (PRS) based on the MVP REX data can explain up to 0.36% (p=7.53×10−95; Figure S5) of the variance in the UKB REX item. The relatively low REX variance explained by MVP PRS in the UK Biobank cohort is consistent with the limited SNP heritability estimates for MVP and UKB REX phenotypes, 6.7% and 4.3% respectively. Since the MVP trans-ancestry meta-analysis is mainly driven by the EA sample (N=146,660) with a small contribution by AAs (N=19,983), we also checked the replication of the trans-ancestry meta-analysis in the UKB sample, which includes only individuals of European descent. Among the six variants that reached genome-wide significance in the trans-ancestry meta-analysis (Table S5), four were replicated in the UKB cohort: TCF4 rs12458015 (MVP p=9.6 ×10−11; UKB p=9.91×10−5), CRHR1 rs1724409 (MVP p=3.55×10−11; UKB p=2.42 ×10−3), HSD17B11 rs12649023 (MVP p=1.32×10−8; UKB p=8.78 ×10−3), and CAMVK rs2681780 (MVP p=8.82×10−12; UKB p=2.42 ×10−3).
Discussion
PTSD is a commonly occurring psychiatric consequence of exposure to extreme, life threatening stress. We report herein the results from MVP of the first GWAS of the PTSD re-experiencing (REX) symptom cluster, which reflects the most characteristic set of PTSD symptoms. Our approach of using severity of the core PTSD phenotype – REX symptoms – for GWAS obviates the reliance on a categorical phenotype such as DSM or ICD-defined PTSD; these have changed with various iterations to the diagnostic criteria and as a result have proven to identify individuals with varying levels of symptom severity.10 Our SNP results support eight separate GWS regions; our gene-based results point to CRHR1 as well as several other genes expressed in brain; and our LD score regression results support genetic overlap with multiple psychiatric, behavioral and other medical phenotypes. While these data identify numerous biological mechanisms that may be associated with REX scores, two patterns stand out. The first pattern relates to steroid response or metabolism. We identified a GWS association mapped to a well-known 900 kb inversion region (MAF ~0.2 in Europeans)11 that includes CRHR1, although in EAs the lead SNP mapped at KANSL1. In general, fine-mapping risk loci by statistical methods is difficult in regions of high LD. The inversion is much less common in Africans11,12 so meta-analysis between EAs and AAs would have the potential to narrow the associated region greatly, assuming a meaningful association is present in the latter population also. And indeed, on transpopulation meta-analysis, the statistical significance increased by about an order of magnitude (to rs174409 p=3.6×10−11) with the lead SNP now mapped to CHRH1, a locus with considerable prior support on the basis of known biology.13 Thus, even though our AA sample was too small for locus identification when taken individually, transpopulation meta-analysis was very valuable because of the differences not just in local LD but in larger-scale genomic structure (Figure S6). CRHR1 also emerged as one of top genes in the gene-based genomewide analysis using MAGMA; and it was also replicated in the UKB cohort. CRHR1 is involved in steroid signaling and response. This is a stress-response pathway long believed to be important in PTSD pathophysiology, and there has been considerable prior interest in CRHR114,15 for PTSD and related phenotypes (notably neuroticism,16 with which we find PTSD to be genetically correlated). The two lines of evidence presented herein – SNP-based and gene-based – together with the prior biological support, make a strong case for the validity and importance of this discovery.
Another significant locus, HSD17B11, encoding hydroxysteroid 17-beta dehydrogenase 11, shares some biological features with CHRH1. Its protein product functions in steroid hormone metabolism and synthesis. Two of the eight GWS loci (one also significant on the gene level) thus fit closely with our current understanding of stress response and its relationship to PTSD. HSD17B11 was replicated in the UKB sample.
The second pattern involves another two loci previously associated with schizophrenia and gene-level pleiotropy with other psychiatric traits. TCF4 (encoding transcription factor 4; lead SNP associated at p=5.4×10−11 in the present study), important in CNS development, has long been known to be associated to schizophrenia17, and was one of the first loci implicated via well-powered cross disorder analysis in the Psychiatric Genomics Consortium18. An additional significant locus, MAD1L1, encodes MAD1 Mitotic Arrest Deficient Like 1, and has previously been significantly associated with schizophrenia and with bipolar disorder.19
This second pattern gives rise to additional pathophysiological hypotheses. We found both single-SNP- and gene-level genetic evidence for overlap of REX with schizophrenia and other psychiatric traits based on LD score regression. Genetic correlation with schizophrenia –also observed in PGC-PTSD5 – is of particular interest, owing to the phenomenological similarities between REX and hallucinating. We might consider the main difference between these two experiences to be thought alienation in the case of hallucinations – the subject, in the case of a hallucination, cannot recognize the re-experienced thought or event as his or her own. Psychotic symptoms, per se, occur in some PTSD patients and, even more broadly, among individuals exposed to traumatic events.20 Auditory hallucinations occur not only in schizophrenia but in trauma-related disorders such as borderline personality disorder and, notably, PTSD.21 In context of these observed epidemiological associations between trauma exposure, PTSD, and psychosis, our findings of shared genetic risk for PTSD and schizophrenia should be further interrogated with the aim of revealing shared pathophysiological mechanisms and, possibly, new treatments. Regarding treatment, atypical antipsychotics have been used to treat PTSD, though the supporting evidence is controversial.22 In the largest single trial of an adjunctive atypical antipsychotic, risperidone, for PTSD, the drug was superior to placebo for REX (and hyperarousal) symptoms.23 The possibility that a subset of patients with PTSD – possibly those with the greatest shared risk for schizophrenia (or the most prominent REX symptoms) – may be more likely to benefit from antipsychotic medications, should be tested in the drive toward precision medicine in psychiatry.
A fifth significant locus, KCNIP4, Potassium Voltage-Gated Channel Interacting Protein 4, relates to response to intracellular calcium, and interacts with presenilin. Calcium signaling is important in an extensive range of behavioral traits.18,24 A sixth locus, CAMKV (CaM Kinase Like Vesicle Associated), was associated with educational attainment in the UK Biobank sample25. The other two GWS associations are at LINC01360 and SRPK2, both less well understood in terms of their function. One gene significantly associated with REX in the gene-based analysis, RAB27B, has been implicated in exosome production necessary for appropriate responsiveness to inflammatory stimuli.26 Given recent hypotheses about immune dysfunction in PTSD,3,27 the involvement of this locus deserves further scrutiny.
In our investigation of tissue and cell type enrichment9, we found that brain tissues from specific regions were associated with REX: cortex, hypothalamus, amygdala, hippocampus, and basal ganglia. We also found striatal medium spiny neurons associated with REX using MAGMA and LDSC. Other associations observed were congruent with these findings and there was little association evidence for non-neuronal cell types. These results suggest that different types of neurons located in different brain regions (striatum, cortex, hypothalamus, ventral tegmental area) may play roles in REX symptoms and PTSD. The finding of association with striatal medium spiny neurons, coupled with the aforementioned genetic correlation between REX and schizophrenia, should spur further investigation into a role for striatal dysfunction in PTSD, and conceivably may point to new avenues for pharmacotherapy.
We found replication evidence for GWS loci and a polygenic signal identified in the MVP cohort in an independent sample from UKB. Beyond the consistency of the results obtained, the comparison of MVP and UKB results strongly indicated that, although there is a large difference in combat-exposure between MVP and UKB (41.3% vs. 3.6%), there is still a large genetic overlap between REX phenotypes assessed in these two cohorts. Modest heritability (comparable to that observed for other psychiatric traits, e.g., major depression28) was observed in both samples (MVP h2 = 6.7%; UKB h2 = 4.3%).
Replication in UKB is particularly reassuring considering the differences between the UKB and MVP cohorts. Presently the UKB has more analyzable subjects (~500,000) than the MVP, but the MVP is still recruiting towards a final sample size of at least 1,000,000. MVP, being a sample of US veterans, includes disproportionately many subjects with military-related illnesses (like PTSD) and therefore is highly informative for such traits; its subjects tend to be lower in socioeconomic status than the population-based UKB (a means test is applied to determine eligibility for VA care). Owing to its military origin, MVP has a strong male predominance. The MVP moreover has strong representation of non-European ancestry populations and therefore is a major resource for mapping in, especially, African-Americans.
In addition to strong genetic correlations with multiple mood and anxiety-related phenotypes (e.g., neuroticism, treatment for depression or anxiety, depressive symptoms, insomnia), REX symptoms were genetically correlated with hypertension. PTSD has been shown in clinical epidemiological studies to be strongly associated with hypertension (and other cardiovascular disorders),29,30 and these data suggest a shared genetic basis for this comorbidity. Moreover, PTSD patients with clinical or subclinical hypertension might be particularly likely to respond to anti-adrenergic agents such as prazosin, given that this drug does not seem to be broadly effective for all patients with PTSD.31 The present findings suggest that this might be understood in terms of shared genetic risk. Stratifying those patients with PTSD with increased polygenic risk for hypertension might identify a subgroup especially responsive to the anti-PTSD effects of prazosin -- worth testing as the field moves toward a personalized approach to treatment, involving medication repurposing at the forefront of this movement.
In summary, we have presented results from the first large GWAS of reexperiencing, a quantitative trait highly characteristic of PTSD. Significant markers identified correspond to genes that influence corticosteroid and steroid function (e.g. CRHR1 and HSD17B11), and have also been associated with schizophrenia (TCF4 and MAD1L1), and other psychiatric traits (the latter two having been previously implicated for pleiotropic effects on psychiatric traits). Gene-based analyses identified numerous additional associated loci. At the cell type level, we found the transcriptomic profile of medium spiny neurons (located in the striatum) to be associated with reexperiencing symptoms. There were highly significant genetic correlation results with numerous mental and physical health traits. Our results confirm prior biological knowledge regarding relationships with steroid response, and suggest new biological relationships with implications for treatment response and precision medicine. They also demonstrate the immediate utility of the MVP sample for disorders prevalent in US veterans.
Methods
Subjects
All subjects are enrollees in the MVP32. Active users of the Veterans Health Administration healthcare system (>8 million veterans) learn of MVP via an invitational mailing and/or through MVP staff while receiving clinical care with informed consent and HIPAA authorization as the only inclusion criteria. As of August 2018, more than 670,000 veterans have enrolled in the program; for the current analyses, genotyping data were available from approximately 350,000 participants. Research involving MVP in general is approved by the VA Central IRB; the current project was also approved by IRBs in Boston, San Diego, and West Haven.
Enrollment involves providing a blood sample for genomic analyses, allowing ongoing access to medical records and other administrative health data by authorized MVP staff, and completing questionnaires. Two surveys are provided to MVP participants: the MVP Baseline Survey, for demographic factors, family pedigree, health status, lifestyle habits, military experiences, medical history, family history of specific illnesses, and physical features; and the optional MVP Lifestyle Survey, which includes the PCL (DSM-IV version)33. This PCL version resembles the civilian version (PCL-C) in that it asks respondents to report how much they have been bothered in the past 30 days by symptoms in response to “stressful experiences” (i.e., not just military experiences). We scrutinized the symptom cluster most distinctive for PTSD, re-experiencing (REX) symptoms (range 5–25). REX items and their distributions in EAs and AAs are shown in Table S1 and Figure S1. Chronbach’s alpha for REX was calculated with the alpha function in the psych R package to test the internal consistency of the 5 items which make up the re-experiencing cluster. Chronbach’s alpha for the 5 reexperiencing items was 0.94 in EAs and 0.95 in AAs. Based on the population definition results, there were initially ~247,000 EA and ~60,000 AA subjects. After accounting for missing phenotype data as only subjects who completed the voluntary surveys could be included, the final sample sizes were 146,660 EAs and 19,983 AAs. Of 146,660 EA subjects, 41.3% were combat-exposed; and of 19,983 AA subjects, 36.2% were combat exposed (extended demographic information, Table S2).
Data Analysis
Genotyping was accomplished via a 723,305-SNP Affymetrix Axiom biobank array, customized for the MVP32. The first MVP data release (used here) includes genotyping data from 360,062 individuals. After quality control, there were 353,948 samples and 657,459 SNPs remaining. Of these, 187,644 subjects had available PCL scores. Population group assignment is discussed below. Due to missing PCL scores or covariate information, GWAS analysis considered 146,660 EA samples and 19,983 AA samples. The MVP genotype data were imputed through Minimac334 using data from the 1000 Genomes Project reference panel (version 3), resulting in imputed genotypes for 49,134,253 SNPs. QC thresholds for post-imputation data were: info score R2 >0.9, MAF>0.01, and Hardy-Weinberg equilibrium test p-value >1 × 10−6. Samples with call rate >98.5% and SNPs with minor allele frequency (MAF) >2×10−6 were retained.
Population group assignment:
Ethnicity self-definitions for the MVP samples include AFR, EUR, Asian, American Indian or Alaska native, native Hawaiian or other Pacific Islander, and “other.” The MVP admixture analysis results from the first data release provided the probability for each sample to be “GBR (British in England and Scotland), PEL (Peruvians from Lima, Peru), YRI (Yoruba in Ibadan, Nigeria), CHB (Han Chinese in Beijing), and LWK (Luhya in Webuye, Kenya)”. A kinship coefficient cutoff of ≥0.0884 was used to remove 10,662 related individuals. We selected 26,497 SNPs through linkage disequilibrium (LD) clumping using PLINK35, and then applied flashpca36 to 343,286 MVP samples and 2,504 1000 Genomes samples37. The annotated population groups (EUR, EAS, AFR, AMR, or SAS) in the 1000 Genomes samples, in addition to the admixture analysis results with a probability cut-off of 0.9, were used to define the final EUR (n=245,123) and AFR (n=62,034) populations (from which study subjects were drawn based on availability of phenotype information). Specifically, the subjects with a probability cut off of 0.9 relative to the genetic component observed in YRI (Yoruba in Ibadan, Nigeria; West Africa) or LWK (Luhya in Webuye, Kenya; East Africa) in the admixture analysis and the AFR samples of the 1,000 Genomes Project indicated the genetic diversity cluster for African-Americans on the pairwise PC-based scatter plots. The MVP subjects enclosed in the AFR cluster on the scatter plots of PC1 to PC4 are identified as the African-Americans, whose probabilities of being YRI or LWK from the admixture analysis are >0.6. Similarly, the EUR cluster in the pairwise PC-based scatter plots is defined by the subjects with a probability cut off of 0.9 relative to the genetic component observed in GBR (British in England and Scotland) and the EUR samples of the 1000 Genomes Project; the MVP samples within the EUR cluster in the PC-based plots are identified as European-Americans, whose probabilities of being GBR from the admixture analysis are >0.8.
For each GWAS analysis, we performed single variant tests using RVTEST38 software, which implements association tests that can be used for common and rare variants using regression models also implemented in the better-known PLINK35. We included the first 10 principal components, age, and sex as covariates in the linear regression association analyses. The results were filtered with imputation quality score R2->0.9, minor allele frequency (MAF) >0.01, and Hardy-Weinberg equilibrium test p-value >1×10−06. A threshold of GWAS p-value of 5×10−8 was adopted for genome wide significant (GWS) association results. Meta-analysis was performed for EA and AA results using PLINK.
Gene-based association analysis was conducted using MAGMA39 implemented in the FUMA platform40. Genome-wide significance was defined as 2.77×10−6 (Bonferroni correction based on 18,120 genes tested).
LD score regression (LDSC):
To investigate shared molecular mechanisms, we tested the genetic overlap (i.e., shared risk alleles) of PTSD REX symptoms with respect to a wide range of phenotypic characteristics, including pathological and physiological traits. Genetic correlations were calculated using the LDSC method (available at https://github.com/bulik/ldsc)41. LDSC results regarding 232 traits were extracted from the data available at LDHub v2.0 (http://ldsc.broadinstitute.org/ldhub/)42,43,44. Genetic correlations for an additional 1,547 traits were calculated using the GWAS summary association results available at https://sites.google.com/broadinstitute.org/ukbbgwasresults. These GWAS used data regarding ~337,000 unrelated individuals of British descent from the UK Biobank (UKB)45. False Discovery Rate (FDR) was applied to correct the genetic-correlation results for multiple testing and q values < 0.05 were considered to be significant. Individuals with admixed ancestral background such as AAs are a mosaic of haplotypes from different ancestral origins. As LDSC requires a LD reference panel, we did not perform these analyses for AA samples because LD reference is not considered reliable with respect to admixed populations, as also highlighted by the LDSC developers42.
Heritability estimation:
We applied LDSC to perform heritability estimation and enrichment analysis for PCL REX scores.
Tissue and Cell Type Enrichment:
At the tissue level, we used data from 53 human tissues (Genotype-Tissue Expression (GTEx) project version 7)8; at the cell type level, we used mouse data from 5 different brain regions broadly-defined (24 cell types) and narrowly-defined (149 cell types)9. Analyses were performed using MAGMA and partitioned LD score regression as previously described9 with minor modifications. Briefly, for the single cell data set9, gene expression for each cell type was scaled to 1,000,000 unique molecular identifiers (UMI) prior to computing specificity scores (defined as the proportion of total expression of specific genes performed in a given cell type). Median transcripts per million (TPM) across individuals were downloaded from the GTEx portal website (https://gtexportal.org/) and used to compute specificity scores. For each tissue or cell-type, the specificity scores were then rank-transformed to a standard normal distribution usingthe rntranform function from the GenABEL R package46. Using MAGMA, the standard normalized specificity scores were then regressed on gene-wise association, defined as the mean p-value across all SNPs assigned to the gene (+35kb upstream to −10kb downstream). Using partitioned LD score regression47, we selected the top 10% most specific genes for each tissue/cell type, extended the coordinates of each gene by 100kb upstream and downstream, and tested for heritability enrichment of these regions48. Results from both methods were integrated by averaging the -log10 p-values obtained for each tissue/cell type. Bonferroni multiple-testing correction was applied for all tests performed in this analysis (0.05/(53+24+149)).
Pathway analysis:
A list of 231 genes was created by using a 0.10 FDR cutoff from the MAGMA GWGAS output. This list was imported into Ingenuity IPA (Ingenuity Systems, Redwood City, CA, USA) to identify the most enriched canonical pathways. This same list of 231 genes was also input into STRING49 to identify associations between genes and protein products.
Replication:
To verify the consistency of our results in an independent sample, we used UKB summary association data (117,900 subjects of European descent) for an item from the traumatic events assessment which is one of the five REX items in MVP: “Repeated disturbing thoughts of stressful experience in past month” (UKB Field ID: 20497). A detailed description of the quality control and the association analysis used to generate these data is available at https://github.com/Nealelab/UK_Biobank_GWAS.
Polygenic risk scoring:
The PRS analysis was conducted on the basis of the GWAS summary association data for both training and target datasets using the gtx R package incorporated in PRSice software50. Specifically, we calculated estimates of the explained variance from a multivariate regression model.
Supplementary Material
Acknowledgments
This research is based on data from the Million Veteran Program (MVP), Office of Research and Development, Veterans Health Administration, and was supported by MVP and the VA Cooperative Studies Program (CSP) study #575B.
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
Publisher's Disclaimer: Disclaimer
The views expressed in this article are those of the authors and do not necessarily reflect the position or policy of the Department of Veterans Affairs or the United States government.
Disclosures
Dr. Gelernter is named as co-inventor on PCT patent application #15/878,640 entitled: “Genotype-guided dosing of opioid agonists,” filed January 24, 2018.
Dr. Stein has in the past three years been a consultant for Actelion, Aptinyx, Bionomics, Dart Neuroscience, Healthcare Management Technologies, Janssen, Jazz Pharmaceuticals, Neurocrine Biosciences, Oxeia Biopharmaceuticals, Pfizer, and Resilience Therapeutics. Dr. Stein owns founders shares and stock options in Resilience Therapeutics and has stock options in Oxeia Biopharmaceuticals.
Data Availability
The GWAS summary statistics generated during and/or analyzed during the current study are available via dbGAP; the dbGaP accession assigned to the Million Veteran Program is phs001672.v1.p. The website is: https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001672.v1.p1 Additionally, the data that support the findings of this study are available from the corresponding authors upon request.
REFERENCES
- 1.Fulton JJ et al. The prevalence of posttraumatic stress disorder in Operation Enduring Freedom/Operation Iraqi Freedom (OEF/OIF) Veterans: a meta-analysis. J Anxiety Disord 31, 98–107 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Logue MW et al. A genome-wide association study of post-traumatic stress disorder identifies the retinoid-related orphan receptor alpha (RORA) gene as a significant risk locus. Mol Psychiatry 18, 937–42 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stein MB et al. Genome-wide Association Studies of Posttraumatic Stress Disorder in 2 Cohorts of US Army Soldiers. JAMA Psychiatry 73, 695–704 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xie P et al. Genome-wide association study identifies new susceptibility loci for posttraumatic stress disorder. Biological psychiatry 74, 656–63 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duncan LE et al. Largest GWAS of PTSD (N=20 070) yields genetic overlap with schizophrenia and sex differences in heritability. Mol Psychiatry 23, 666–673 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blanchard EB, Jones-Alexander J, Buckley TC & Forneris CA Psychometric properties of the PTSD Checklist (PCL). Behav Res Ther 34, 669–73 (1996). [DOI] [PubMed] [Google Scholar]
- 7.Battle A, Brown CD, Engelhardt BE & Montgomery SB Genetic effects on gene expression across human tissues. Nature 550, 204–213 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Consortium GT Genetic effects on gene expression across human tissues. Nature 550, 204 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Skene NG et al. Genetic identification of brain cell types underlying schizophrenia. Nature Genetics 50, 825–833 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barbano AC et al. Clinical implications of the proposed ICD-11 PTSD diagnostic criteria. Psychol Med, 1–8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stefansson H et al. A common inversion under selection in Europeans. Nature Genetics 37, 129 (2005). [DOI] [PubMed] [Google Scholar]
- 12.Cáceres A, Sindi SS, Raphael BJ, Cáceres M & González JR Identification of polymorphic inversions from genotypes. BMC Bioinformatics 13, 28 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amstadter AB et al. Corticotrophin-releasing hormone type 1 receptor gene (CRHR1) variants predict posttraumatic stress disorder onset and course in pediatric injury patients. Disease markers 30, 89–99 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kasckow JW, Baker D & Geracioti TD Jr. Corticotropin-releasing hormone in depression and post-traumatic stress disorder. Peptides 22, 845–51 (2001). [DOI] [PubMed] [Google Scholar]
- 15.McFarlane AC, Barton CA, Yehuda R & Wittert G Cortisol response to acute trauma and risk of posttraumatic stress disorder. Psychoneuroendocrinology 36, 720–7 (2011). [DOI] [PubMed] [Google Scholar]
- 16.Smith DJ et al. Genome-wide analysis of over 106 000 individuals identifies 9 neuroticism-associated loci. Mol Psychiatry 21, 749–57 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stefansson H et al. Common variants conferring risk of schizophrenia. Nature 460, 744–7 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cross-Disorder Group of the Psychiatric Genomics, C. et al. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet 381, 1371–9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruderfer DM et al. Polygenic dissection of diagnosis and clinical dimensions of bipolar disorder and schizophrenia. Mol Psychiatry 19, 1017–1024 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McGrath JJ et al. Trauma and psychotic experiences: transnational data from the World Mental Health Survey. Br J Psychiatry 211, 373–380 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Waters F, Blom JD, Jardri R, Hugdahl K & Sommer IEC Auditory hallucinations, not necessarily a hallmark of psychotic disorder. Psychol Med 48, 529–536 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Ravindran LN & Stein MB The pharmacologic treatment of anxiety disorders: a review of progress. The Journal of clinical psychiatry 71, 839–54 (2010). [DOI] [PubMed] [Google Scholar]
- 23.Krystal JH et al. Adjunctive risperidone treatment for antidepressant-resistant symptoms of chronic military service-related PTSD: a randomized trial. JAMA 306, 493–502 (2011). [DOI] [PubMed] [Google Scholar]
- 24.Gelernter J et al. Genome-wide association study of opioid dependence: multiple associations mapped to calcium and potassium pathways. Biol Psychiatry 76, 66–74 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okbay A et al. Genome-wide association study identifies 74 loci associated with educational attainment. Nature 533, 539–42 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alexander M et al. Rab27-Dependent Exosome Production Inhibits Chronic Inflammation and Enables Acute Responses to Inflammatory Stimuli. J Immunol 199, 3559–3570 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Michopoulos V, Powers A, Gillespie CF, Ressler KJ & Jovanovic T Inflammation in Fear- and Anxiety-Based Disorders: PTSD, GAD, and Beyond. Neuropsychopharmacology 42, 254–270 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Howard DM et al. Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat Neurosci (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sumner JA et al. Post-traumatic stress disorder symptoms and risk of hypertension over 22 years in a large cohort of younger and middle-aged women. Psychol Med 46, 3105–3116 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roy SS, Foraker RE, Girton RA & Mansfield AJ Posttraumatic Stress Disorder and Incident Heart Failure Among a Community-Based Sample of US Veterans. Am J Public Health 105, 757–63 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raskind MA et al. Trial of Prazosin for Post-Traumatic Stress Disorder in Military Veterans. N Engl J Med 378, 507–517 (2018). [DOI] [PubMed] [Google Scholar]
Methods References
- 32.Gaziano JM et al. Million Veteran Program: A mega-biobank to study genetic influences on health and disease. J Clin Epidemiol 70, 214–23 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Wilkins KC, Lang AJ & Norman SB Synthesis of the psychometric properties of the PTSD checklist (PCL) military, civilian, and specific versions. Depress Anxiety 28, 596–606 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Das S et al. Next-generation genotype imputation service and methods. Nat Genet 48, 1284–7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Purcell S et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. The American Journal of Human Genetics 81, 559–575 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abraham G & Inouye M Fast principal component analysis of large-scale genome-wide data. PLoS One 9, e93766 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Auton A et al. A global reference for human genetic variation. Nature 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhan X, Hu Y, Li B, Abecasis GR & Liu DJ RVTESTS: an efficient and comprehensive tool for rare variant association analysis using sequence data. Bioinformatics 32, 1423–6 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Leeuw CA, Mooij JM, Heskes T & Posthuma D MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol 11, e1004219 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Watanabe K, Taskesen E, van Bochoven A & Posthuma D FUMA: Functional mapping and annotation of genetic associations. bioRxiv (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bulik-Sullivan B et al. An atlas of genetic correlations across human diseases and traits. Nat Genet 47, 1236–41 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bulik-Sullivan BK, Loh PR, Finucane HK, Ripke S & Yang J LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. 47, 291–5 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zheng J et al. HAPRAP: a haplotype-based iterative method for statistical fine mapping using GWAS summary statistics. Bioinformatics 33, 79–86 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng J et al. LD Hub: a centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics 33, 272–279 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bycroft C et al. Genome-wide genetic data on ~500,000 UK Biobank participants. bioRxiv (2017). [Google Scholar]
- 46.Aulchenko YS, Ripke S, Isaacs A & van Duijn CM GenABEL: an R library for genome-wide association analysis. Bioinformatics 23, 1294–1296 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Finucane HK et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet 47, 1228–35 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Finucane HK et al. Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. Nat Genet 50, 621–629 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Szklarczyk D et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res 45, D362–D368 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Euesden J, Lewis CM & O’Reilly PF PRSice: Polygenic Risk Score software. Bioinformatics 31, 1466–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.