Abstract
Selfish genetic elements (SGEs) are DNA sequences that are transmitted to viable offspring in greater than Mendelian frequencies. Medea SGEs occur naturally in some populations of red flour beetle (Tribolium castaneum) and are expected to increase in frequency within populations and spread among populations. The large‐scale U.S. distributions of Medea‐4 (M4) had been mapped based on samples from 1993 to 1995. We sampled beetles in 2011–2014 and show that the distribution of M4 in the United States is dynamic and has shifted southward. By using a genetic marker of Medea‐1 (M1), we found five unique geographic clusters with high and low M1 frequencies in a pattern not predicted by microsatellite‐based analysis of population structure. Our results indicate the absence of rigid barriers to Medea spread in the United States, so assessment of what factors have limited its current distribution requires further investigation. There is great interest in using synthetic SGEs, including synthetic Medea, to alter or suppress pest populations, but there is concern about unpredicted spread of these SGEs and potential for populations to become resistant to them. The finding of patchy distributions of Medea elements suggests that released synthetic SGEs cannot always be expected to spread uniformly, especially in target species with limited dispersal.
Keywords: maternal effect, Medea, red flour beetle, selfish genetic element
This figure compares the Medea‐4 element distribution over time. The Medea‐4 element in the United States is widespread, but no longer in a latitudinal distribution.
1. INTRODUCTION
Selfish genetic elements (SGEs) have been found to occur naturally in a huge variety of taxa, but despite decades of study by evolutionary biologists, the origins, mechanisms, and population‐level impacts of these elements are still largely unknown (Burt & Trivers, 2006). Many populations of red flour beetle (Tribolium castaneum) harbor naturally occurring selfish Medea elements (Beeman & Friesen, 1999). Medea elements are genomic sequences which cause death of the non‐Medea offspring of Medea‐bearing mothers (Beeman, Friesen, & Denell, 1992). Currently, the most parsimonious model suggests Medea's action involves two tightly linked loci—one encoding a lethal, maternally expressed toxin in all eggs, and the other encoding a zygotic antidote that rescues only those progeny inheriting at least one Medea allele (Beeman & Friesen, 1999; Beeman et al., 1992). Because only non‐Medea (i.e., “wild‐type”) offspring produced by mothers that are heterozygous for the Medea element die, the Medea allele frequency is expected to increase within a population over time, provided that Medea introduction frequency is not extremely low, and the element does not carry a substantial fitness cost (Wade & Beeman, 1994). The influence of selfish genetic elements on populations and species can be substantial, from providing additional genetic variation that enables adaptation (e.g., Li, Schuler, & Berenbaum, 2007) to the lowering of population fitness (e.g., Carroll, Meagher, Morrision, Penn, & Potts, 2004). Beyond that, the basic population genetics and evolutionary history of elements such as Medea represent a fascinating yet understudied dimension in evolutionary biology (Burt & Trivers, 2006).
Beyond the importance of understanding natural SGEs for advancing basic science, knowledge of natural SGEs is relevant to the newly emerging technology of “gene drives” that aims at using synthetic SGEs to drive genes into pest populations that will act to suppress the populations or decrease vectorial capacity of the populations (e.g., Godfray, North, & Burt, 2017; Rode, Estoup, Bourguet, Courtier‐Orgogozo, & Debarre, 2019; Sinkins & Gould, 2006). Synthetic forms of Medea have been developed and tested in the laboratory (Buchman, Marshall, Ostrovski, Yang, & Akbari, 2018; Chen et al., 2007; Hay et al., 2010). Currently, there is concern among scientists and the public regarding predictability of spread of synthetic gene drives and the potential of pest populations to evolve resistance to the gene drive mechanisms or to linked sequences that impact traits and/or the viability of homozygote offspring (e.g., NASEM, 2016; Rode et al., 2019).
Two distinct Medea elements are known to be present in U.S. populations of red flour beetle: M1 and M4 (the M2 and M3 elements have each been identified only once, and in Asian populations) (Beeman & Friesen, 1999). Interestingly, M1 has only been found in wild beetles also harboring M4 (though the elements can be easily separated through crossing, and M1‐only strains are easily reared in the laboratory), while M4 is commonly found as the sole element present in wild populations (Beeman & Friesen, 1999). While both elements exhibit the same maternal‐effect lethality, they map to opposite ends of the same linkage group and do not cross‐rescue—for example, inheritance of an M4 allele cannot rescue the offspring of an M1‐bearing mother (Beeman & Friesen, 1999). M1 has been fully sequenced, and is associated with a transposon insertion, though the genetic mechanism involved in the maternal‐effect lethality remains a mystery (Lorenzen et al., 2008). A prior assessment of the distribution of the M4 element in the United States showed a striking latitudinal stepped cline (Beeman, 2003). All 26 sample locations above 33°N were fixed for the M4 element. In contrast, only two sampled locations below this latitude were fixed for the element, 21 lacked the element, and six had an intermediate frequency of M4. This delineation was so obvious that it spurred the hypothesis that distinct genetic races of T. castaneum might exist in the United States and that insufficient gene flow between these northern and southern races might create a barrier (Beeman, 2003). It is also possible that there is mating between the northern and southern populations, but that the southern populations are resistant to the action of Medea.
While they possess mechanisms allowing for their frequencies to increase, many naturally occurring SGEs appear to be maintained at low or intermediate frequency (reviewed in Hatcher, 2000). Despite their great potential for rapid spread, it is not uncommon for SGEs to be distributed in stable gradients. For example, the meiotic X‐chromosome driver SR in Drosophila pseudoobscura is distributed along a latitudinal gradient in North America, with the element more common in southern populations. This distribution appears to have been stable for at least the last half‐century potentially due to higher polyandry in northern populations (Price et al., 2014; Sturtevant & Dobzhansky, 1936). Drosophila melanogaster P elements show an east–west frequency gradient in Eurasia, with higher concentrations in western Europe fading out as sampling moves eastward (Anxolabéhère, Nouaud, Périquet, & Tchen, 1985). This gradient‐based distribution appears to be stable, likely due to the presence of “buffer populations” impervious to P elements (Bonnivard & Higuet, 1999).
Our goal was to determine whether the distribution of M4 in the United States is also stable, and if so, what genetic and/or environmental factors maintain the distribution. While M1 is also known to be present in U.S. populations, the only effort to assess its distribution involved few populations (Beeman & Friesen, 1999). Thus, it is unclear whether the same factors which shape the distribution of M4 also influence the distribution of M1.
Tribolium castaneum are thought to disperse primarily by human‐aided movement of infested stored grains and processed products. However, evidence for the importance of active dispersal via flight has been found in Australian populations of red flour beetle (Ridley et al., 2011). From traps spaced up to several dozen kilometers from the nearest food resources, roughly 88% of emigrating females were mated, showing the great potential for gene flow between populations. While the maximum extent of flight is unclear, it was evident that flight‐aided dispersal occurs at least on a scale of tens of kilometers (Ridley et al., 2011). Flight initiation peaks with warmer temperatures and increased daylight, so active dispersal will vary seasonally (Perez‐Mendoza, Campbell, & Throne, 2014).
Further, it is not yet known how successful long‐distance immigrants will be. Tribolium castaneum are not particularly attracted to undamaged or uninfested grains and flour, and success in locating a flour patch decreased as distance increased over a scale of only many centimeters (Romero, Campbell, Nechols, & With, 2010). The effectiveness of pheromones as attractants has been demonstrated on a scale of several meters (Boake & Wade, 1984; Obeng‐Ofori & Coaker, 1990), but may decay beyond this. If migrating beetles are effectively "flying blindly" until they luck upon a trail of aggregation pheromone or other food volatiles, the true impact of active dispersal on gene flow may be small.
An understanding of the factors which influence Medea spread is vital both for assessing Medea's potential as a synthetic gene drive mechanism and for garnering a better understanding of the element's evolutionary biology. A critical first step is to describe current Medea distributions and determine whether the distributions are changing. Here, we describe our analysis of the contemporary distribution of M4 in the United States relative to the distribution found in the earlier survey. We also present a description of the large‐scale distribution of M1 in the United States. Finally, we address the potential role of population structure in shaping these distributions.
2. MATERIALS AND METHODS
2.1. Sample collection
Red flour beetles used to assess the current distribution of Medea elements M1 and M4 were collected between November 2011 and May 2014. Resampling of two sites originally sampled in 2012 occurred in September 2013 to determine whether there were frequency changes over time within populations. Collection sites included rice and wheat flour mills, feed mills, farm supply stores, grain elevators, grain bins, and other on‐farm grain storage. When appropriate, as determined by facility layout and researcher safety, pheromone‐baited traps or probes (Trécé, Inc.) were employed to collect beetles from mills or larger stores of grain. Otherwise, beetles were collected by hand or sifted from the substrate. In cases where visiting the sampling site was not possible, beetles were collected and shipped directly from the site to our laboratory by an extension agent or facility employee.
Dead beetles were frozen as soon as possible after collection. Live beetles were permitted to mate and oviposit for up to 3 weeks on a mixture of organic whole wheat pastry flour and 5% (by volume) brewer's yeast. These beetles were reared at 22–23°C and approximately 60% relative humidity in a controlled quarantine facility in order to establish cultures from each location; after this period, the originally collected beetles were removed and frozen for later genotyping. Cultures were maintained at the same temperature and humidity in a quarantine facility via periodic subculturing and flour replenishment.
In an effort to make inferences about an earlier distribution of M1, red flour beetles were also obtained from samples collected between 2004 and 2007. Samples from 2007 were collected in traps containing oil. This oil was removed from the beetles prior to DNA extraction by rinsing the beetles for 5 min in CitriSolv (Fisher Scientific), followed by a wash in double‐distilled water.
2.2. M4 diagnosis
Because we lacked a reliable M4 molecular marker at the time of these experiments, the presence of M4 in selected populations was assessed via crosses. Females from the homozygous M4 pearl strain were crossed to the non‐Medea GA‐1 strain (Haliscak & Beeman, 1983) to generate heterozygous M4 females. These females were crossed to males from wild populations, and after 3 days, eggs were counted for each cross. Once the offspring completed development, the final number of surviving adult progeny was tallied. To minimize the impact of potentially unhealthy females on the survivorship data, crosses producing fewer than 10 eggs, or those which failed to produce any surviving adults, were not included in the analysis. Offspring survival frequency was used as an indicator of the presence/absence of the M4 element in the wild‐derived males. As in Beeman (2003), survival means from each location were compared to wild type and M4 means using the Mann–Whitney test in MATLAB (Version 8.0.0.783; Mathworks).
The expectation for offspring from crosses between known M4‐heterozygous females and wild‐derived males of unknown M4 status is close to 100% survivorship if the male is actually homozygous for M4, roughly 75% for crosses to males heterozygous for M4, and about 50% offspring survivorship in crosses to males lacking the M4 element. We used this information to categorize the probable genotypes of individual beetles.
2.3. M1 genotyping
Because the M1 element has been fully sequenced, we were able to design M1‐specific primers and genotype via PCR.
Genomic DNA was extracted for 25 individual beetles per location (or, if 25 individuals were not available, as many individuals as possible) using the Qiagen DNeasy Blood and Tissue Kit. Primers used for amplification were as follows:
Forward primer: 5′‐TGGCGATAGTCAAAATCCTTTGTCG‐3′
M1 reverse: 5′‐TGCCACCTTCACGTAGCCCG‐3′
Wild‐type reverse: 5′‐CAGGGCCCCGGAGTATTTTTCC‐3′
PCRs were performed in 25 µl volumes, each containing 1× PCR buffer, 3.5 mM MgCl2, 200 µM dNTPs, 2 µl DNA template, 1 μmol forward primer, 0.5 μmol of each reverse primer, 0.5 U Taq DNA polymerase (Genesee), and ddH2O to 25 µl. Thermal cycling consisted of initial denaturation of DNA template at 95°C for 4 min followed by 40 cycles of 95°C for 30 s, 58°C for 45 s, and 72°C for 75 s, and a final extension step of 72°C for 5 min. Alleles were separated on 2.5% agarose gels infused with ethidium bromide and visualized by ultraviolet (UV) illumination.
2.4. Confirmation of M1 lethality
Because our assessment of the distribution of M1 relied on genotyping via a molecular marker, crosses were performed in select M1‐fixed populations to diagnose whether the M1 sequences amplified during genotyping represented fully functional M1 elements. Females from populations that were genotyped as fixed for M1 were crossed to males from the GA‐1 strain, a laboratory strain devoid of Medea elements. Five presumably M1‐heterozygous females derived from each cross were then paired with GA‐1 males, and eggs from each cross were counted, with the proportion of surviving offspring used to determine whether maternal‐effect lethality had occurred. Survival rates near 100% would indicate the absence of Medea elements, while survival rates near 50% would imply the presence of a Medea element in the source population. Five additional heterozygous females per population were backcrossed to males from their population of origin, and the proportion of surviving offspring (presumably 100%, as sires were expected to be homozygous M1) was used as a control.
To determine whether any maternal‐effect lethality uncovered in these populations could be attributed to the M4 element, three heterozygous females from each population were also crossed to males from the homozygous M4 pearl strain. The absence of rescue in these crosses indicated the presence of a functional Medea element other than M4, presumably M1, given that M1 is the only other Medea element that has been found in the United States.
2.5. Distribution analysis
We used SaTscan v.7 to identify any regional clustering in our observed M1 distribution (Kulldorff & Information Management Services, Inc., 2009). This allowed us to find patterns that were significantly different from a random distribution of genotype frequencies. The geographic coordinates of each sampling location were entered, along with the total number of individuals genotyped at that location and the number of M1 individuals identified. The program scans across the overall distribution in an elliptical window that was varied in radius from zero to a size exceeding the total sampling area, where each window represents a potential cluster of M1 or wild‐type genotypes (Table S4). For each window, a likelihood ratio was found by comparing the observed and expected number of M1 genotypes under a Bernoulli probability model. The window with the maximum likelihood was assigned a p‐value, obtained through Monte Carlo simulation (Kulldorff, 1997).
2.6. Microsatellite genotyping
We selected populations for analysis based on the number of individuals available from each sampled population, as well as the geographic location of each population. Sites in this study were selected that represented the wide geographic range of our sampling effort, while others were selected because of their proximity to other sampling locations, representing a finer scale. Populations with at least 20 sampled individuals were preferred. Selected primers described in Demuth et al. (2007) were used for microsatellite amplification (Table S2).
For cost‐effective, high‐throughput genotyping, forward primers were designed with a 5' M13 (−29) sequence CACGACGTTGTAAAACGAC, and a universal M13 (−29) primer labeled with either IRDye 800 or IRDye 700 was added to the reaction (Schuelke, 2000). The fluorescent IRDye is integrated on to the end of the fragment containing the forward primer, allowing for fragment detection and size estimation.
PCR was performed in 10 µl reactions, with each containing 2 µl genomic DNA (approximately 50 ng), 1× PCR buffer, 2 mM MgCl2, 0.2 mM dNTPs, 0.016 µM unlabeled forward primer, 0.06 µM reverse primer, 0.06 µM IRDye‐labeled universal M13 primer (Integrated DNA Technologies), 0.5 U Taq polymerase (Genesee), and ddH2O to 10 µl. The cycling conditions consisted of an initial denaturation step at 94°C for 4 min, followed by 15 cycles of 94°C for 30 s, 65°C (−1°C/cycle) for 30 s, and 72°C for 1 min and 20 s, and ending with 30 cycles of 94°C for 15 s, 50°C for 15 s, and 72°C for 45 s.
The PCR products were diluted with 10 µl ddH2O, and 10 µl of a formamide stop solution (95% formamide, 20 mm EDTA, bromophenol blue) was added, followed by a denaturing step consisting of 95°C for 5 min. Fragments were separated by electrophoresis on a 6.5% Long Ranger 1× TBE polyacrylamide gel, run on a Li‐Cor 4300 automated DNA sequencer at a constant power of 40 W at 45°C for 1.5 hr. Fragments were sized using a 50–350 bp IRDye 800 or 700 standard (Li‐Cor, Inc). Allele sizes were scored using QuantarPro software (KeyGene). Individuals missing data at more than three loci were removed from further analyses. Because this left population SC‐2 with only 10 individuals, it was excluded from population‐level analyses.
2.7. Genetic diversity and differentiation
Observed and expected heterozygosity and deviation from Hardy–Weinberg equilibrium were assessed in Arlequin (Excoffier et al., 2005). Genepop (Raymond & Rousset, 1995) was used to carry out exact tests for linkage disequilibrium, employing a Markov chain with 10,000 dememorization steps, 250 batches, and 2,500 iterations per batch to estimate the exact p‐value. Significance levels of the tests were adjusted for multiple comparison following standard Bonferroni corrections (Rice, 1989). Pairwise F ST values were also estimated in Arlequin. Global R ST, allelic richness, and within‐population gene diversity were estimated in FSTAT (Goudet, 2001).
The program FreeNA (Chapuis & Estoup, 2007) was used to estimate null allele frequencies and to correct F ST values for bias from null allele presence. A global M1 F ST estimate was calculated using this same methodology, by assigning allele sizes to both the M1 and wild‐type alleles and constructing individual genotypes corresponding with our PCR genotyping results.
Potential population structure was investigated using the program STRUCTURE (Pritchard, Stephens, & Donnelly, 2000) to assess the most likely number of clusters (K), where both the "admixture" and "no admixture" models were run, with sampling location used as a prior, and allele frequencies correlated. Five replicates each from K = 1 to K = 15 were run with a burn‐in period of 200,000 steps followed by 200,000 MCMC iterations. The most likely value of K was determined in Structure Harvester (Earl & vonHoldt, 2012), which uses the delta K method described by Evanno, Regnaut, and Goudet (2005). For individual cluster assignments, STRUCTURE was run again for 20 replicates at the most likely K. The programs CLUMPP and DISTRUCT were used to visualize the raw data outputs from STRUCTURE (Jakobsson & Rosenberg, 2007).
The extent of genetic isolation due to geographic distance was assessed via a Mantel test with 10,000 permutations in Genepop's Isolde program (Raymond & Rousset, 1995), using two semimatrices: one consisting of F ST/(1−F ST) with ENA corrected F ST values and another semimatrix of natural log‐transformed kilometer linear distances between sample locations. These analyses were also performed with the original uncorrected F ST values, and the results obtained were compared to assess whether there were significant differences.
3. RESULTS
3.1. M4 genotyping and distribution
The overall result of testcrosses was that the M4 element was geographically widespread and found in nearly every sample population tested (Figure 1b). For most locations, all tested individuals were predicted to carry at least one M4 allele. This includes most locations south of 33°N, the previously described boundary of M4 fixation, indicating that M4 had moved southward. Only a single population, from southern Alabama, appeared to lack the M4 element. Two populations with intermediate predicted M4 frequencies were found in North Carolina, north of the 33rd parallel. North Carolina was not represented in the previous survey, and so it is unclear whether this represents a recent introduction of M4, or maintenance of an intermediate frequency.
Figure 1.
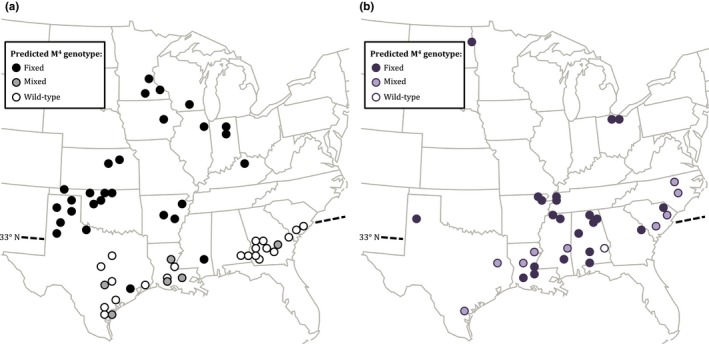
The M4 element in the United States is widespread, but no longer in a latitudinal distribution. (a) M4 distribution described by Beeman (2003), sampled 1993–1995. Figure adapted by authors from Beeman (2003) with permission. (b) M4 distribution of present study, sampled 2011–2014. Open circles indicate beetles genotyped were homozygous wild‐type, dark circles indicate beetles were homozygous M4, while light circles indicate both wild‐type and M4 beetles were present in the sample. The 33rd parallel (site of M4 delineation from Beeman, 2003) is indicated for reference
Out of a total of 176 productive diagnostic crosses, survival above 100% was found in two—in each case, this was the result of counting exactly one more surviving adult than the number of eggs originally tallied. This was likely the result of an egg eluding the initial census by sticking to the side of the vial, or of an unfortunate miscount. These errors indicate that there may have been undercounts in some of the other replicates (Table S3).
3.2. M1 genotyping and distribution analysis
M1 diagnostic crosses confirmed that our PCR‐based M1 genotyping was indeed amplifying genomic regions representing functional M1 sequence. For each of the test populations that genotyped as fixed for M1 with our PCR assay, maternal‐effect lethality was apparent in crosses to non‐Medea individuals (Figure S1). Further, this lethality could not be rescued by the M4 element (Figure S1). We did not find any M1 beetles present in AL‐9, the only population sample to have lacked M4 in our analyses (Table S3).
While we did not obtain enough archived samples from 2004 to 2007 to make a full, comprehensive assessment of the prior distribution of M1 (Table S1), the data we collected still reflect heterogeneity in the M1 distribution (Figure 2). In contrast with the distribution of M4 described by Beeman (2003), where the M4 element was fixed in northern regions and largely absent at the southern extreme of the sampled region, we see M1 at high frequency in the southern portion of the sampled area (with the exception of the relatively low frequency observed in the Louisiana locality), and largely absent at higher latitudes.
Figure 2.
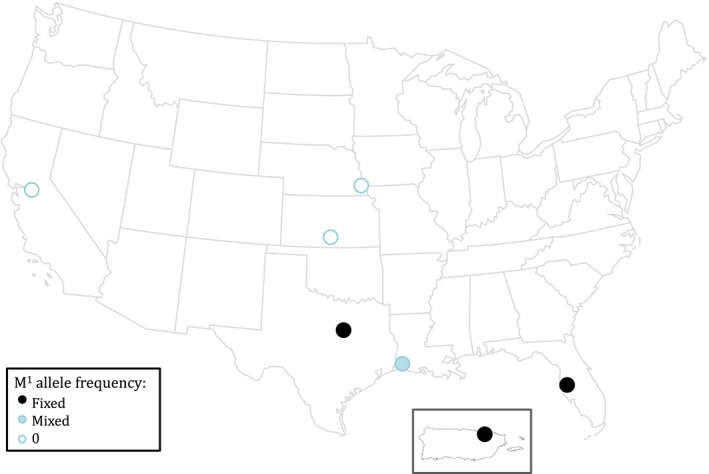
The M1 element was more prevalent in southern latitudes in wild beetles sampled 2004–2007. Open circles indicate beetles genotyped were homozygous wild‐type, dark circles indicate beetles were homozygous M1, while light circles indicate both wild‐type and M1 beetles were present in the sample. A sample from Puerto Rico is shown in an insert
The present‐day distribution of the M1 element in the United States (Figure 3) does not appear to show any latitudinal pattern, and certainly not one as distinct as the delineation found in prior M4 studies. There is, however, an interesting apparent clustering of high‐frequency Medea samples in the south‐central region, covering much of northern Alabama, northern Mississippi, western Tennessee, and eastern Arkansas.
Figure 3.
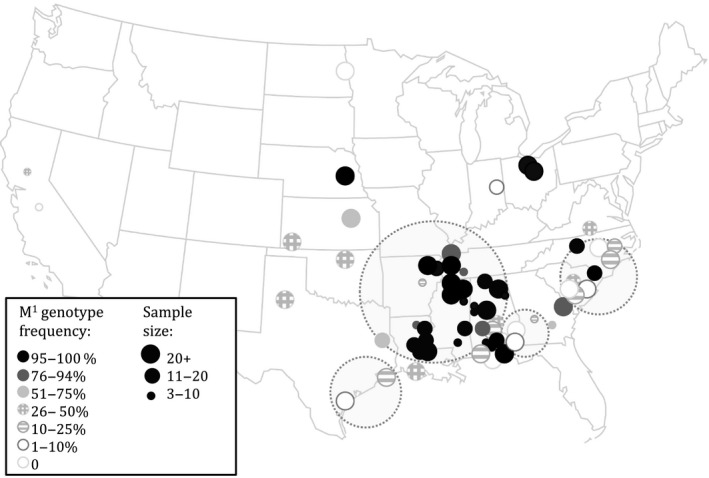
The present‐day distribution of the M1 element in the United States is patchy, with several significant clusters of high or low M1 frequency. Dotted circles indicate sites clustered by M1 allele frequency by SaTscan analysis. The color of the solid circle indicates the M1 genotype frequency of individuals sampled from that site, while the size of the circle is proportional to the number of individuals genotyped
Although the small number of sampled locations does not allow for a thorough comparison of the 2004–2007 distribution with the current‐day distribution, the pattern seen in the 2004–2007 samples is interesting, nonetheless. Our distribution analysis for the 2011–2014 samples revealed five nonoverlapping clusters (Figure 3). The largest cluster consisted of high‐frequency sites, incorporating 24 sampling locations stretching from Texas to eastern Alabama. Another high‐frequency cluster included three populations from Florida and southern Alabama. The remaining three clusters highlighted low‐frequency M1 regions: the eastern Carolinas (9 locations), eastern Alabama/western Georgia (4 locations), and the gulf coast (2 locations).
3.3. Genetic diversity and differentiation
The microsatellite markers were highly polymorphic, ranging between 6 and 31 alleles per locus across all samples (Table S5). Pairwise F ST across all loci and locations ranged from a high of 0.264 (between NE and SC) to a low of 0.002 (between MS and SC). Global per‐locus F ST values averaged 0.0669 (and ranged from 0.0365 to 0.1083, Table S7) and were comparable to another study of microsatellites in U.S. red flour beetle populations (0.018–0.149 in Semeao et al., 2012). In stark contrast, the F ST for the M1 locus was 0.640, roughly six times higher than our most differentiated microsatellite locus.
When looking at microsatellite loci at all sample locations, we find a trend toward isolation by distance (IBD) (using corrected F ST, r 2 = .012, p = .03); however, when excluding food production sites, a positive correlation between genetic distance and geographic distance remains, but the correlation is no longer statistically significant (p = .094) (Figure S3). Driving this trend toward IBD, these sites tended to have higher average pairwise F ST values than other locations (Table 1) and were also typically farther away from the next sampled location (the average pairwise geographic distance for these food production facilities was 1,094 km; the average pairwise distance excluding these sites was 861 km). Overall, samples from these sites also showed significantly lower allelic richness, lower observed heterozygosity, and lower within‐population genetic diversity. We tested to determine whether there was correlation between latitude and M1 frequency based on a sample of 31 populations. The correlation of 0.240 was not significant. We also tested for a correlation between M1 frequency and level of heterozygosity with the hypothesis that beetle populations with M1 would have lower heterozygosity due to linkage between the invading Medea element and alleles at other loci. We found no correlation (r = −.003).
Table 1.
Beetle samples from food production facilities showed lower metrics of diversity than beetles sampled in other locations
| Food production | Other | p‐Value | |
|---|---|---|---|
| Allelic richness | 4.353 | 5.094 | .004 |
| H O | 0.428 | 0.498 | .027 |
| H S | 0.576 | 0.670 | .001 |
| F IS | 0.258 | 0.257 | .982 |
| F ST | 0.152 | 0.043 | .001 |
Shown are the average allelic richness, observed heterozygosity (H O), within‐population gene diversity (H S), inbreeding coefficient (F IS), and F ST for the seven food production sites compared with the remaining 22 sampling locations. p‐Values were obtained after 1,000 permutations in FSTAT (Goudet, 2001).
No clear correlation was found between microsatellite and M1 frequency differences among populations (Figure S2b). We conclude from this that population structure does not appear to be a major factor shaping the Medea distributions. While the most likely number of clusters (K) from the structure analysis of microsatellite data was two, the delta K value (5.06) was quite small (Table S6). There was no clear geographic interpretation for the assignments, nor any clear relationship between clustering and Medea frequency.
4. DISCUSSION
Characterization of the spatial and temporal distribution of Medea elements is important for answering questions about Medea element evolution. Furthermore, from an applied perspective, there has been much attention paid to the potential use of Medea elements to spread beneficial genes into pest populations, but the only supporting data are from laboratory studies (Akbari et al., 2014; Buchman et al., 2018; Chen et al., 2007).
The current study is the first to assess the temporal dynamic nature of the distribution of Medea elements in a realistic landscape, and our data demonstrate that the M4 distribution has expanded in the past two decades (Beeman, 2003). Further, we present the first detailed descriptions of the distribution of M1 in the United States. We show that the M1 element is widespread but patchy, with some evidence of clusters of sampled areas with high and low frequencies. Finally, we show that there is little differentiation in frequencies of microsatellite alleles among the beetles collected in our geographically widespread samples, suggesting that Medea spread is not strongly restricted by gene flow. Other studies of red flour beetle, both in U.S. populations (Semeao et al., 2012) and in Australia (Ridley et al., 2011), have demonstrated similar levels of overall genetic differentiation among T. castaneum populations.
4.1. Medea‐4 has spread geographically
Our M4 genotyping demonstrates recent spread of the M4 element within southern regions of the United States. This increase in M4 frequency in the southern United States indicates that there are no rigid boundaries—genetic, environmental, or dispersal—preventing the spread of M4 in the United States. The spread of M4 in the southern United States as well as our analysis of population structure does not support the existence of distinct geographic T. castaneum races that had been hypothesized earlier (Beeman, 2003). Assessment of M4 status of each individual beetle required crosses and assessment of offspring survival, so sample size was small, and we could only determine whether M4 was likely absent, present, or fixed in a population.
4.2. Medea‐1 is widely but nonrandomly distributed within the United States
Because we have a molecular marker for active M1 elements, we were able to much more efficiently screen beetles for its presence/absence than was possible for M4, and we could roughly estimate frequency of M1 within populations. We were also able to check for the M1 marker in older, frozen beetles. Although M1 was present in most tested locations, the element was noticeably absent in some sampled regions, as revealed by our geographic clustering analysis. It is interesting to note that the lowest frequency clusters were in coastal regions. Because we lack a prior comprehensive, large‐scale M1 distribution for comparison, we do not yet know the temporal nature of these clusters.
Interestingly, M1 in our samples remains intertwined with M4. Even though the two elements can be easily separated in the laboratory with simple crosses, and with no apparent decrease in viability/fecundity of the M1‐only stocks, no M1 alleles were detected in beetles determined to lack the M4 genotype. One possible explanation is that M1 arose in an M4 background and has been tied to the M4 element's maternal‐effect lethality ever since. In the event of a dual introduction of M1 and M4 into a Medea‐naïve population (or an M4 population), we would expect both elements to persist, provided the introduction frequencies were not extremely low (Cash, 2016). Based on model predictions, in the case of a low‐frequency introduction, it is possible for just one of the two introduced Medea elements to be lost from the population based on recombination in heterozygous males.
4.3. Sampling locations and methodologies may impact differentiation metrics
A prior study of flour beetle populations in several U.S. commercial grain storage and processing facilities found evidence of population structure not associated with either geographic distance or the commodity type (rice or wheat) (Semeao et al., 2012). Beetle populations in these facilities are impacted by pest control treatments, resulting in population reduction or elimination. In a long‐term study of flour mills, the average number of beetles trapped postfumigation decreased by nearly 85% from prefumigation levels, indicating a significant population decrease (Campbell, Toews, Arthur, & Arbogast, 2010). Although these populations can rebound after treatment through propagation or immigration, the genetic makeup of the population is likely impacted by the bottleneck. In the present study, pairwise F ST of two samples taken at the same facility in Ohio roughly 16 months apart was 0.099, higher than the average pairwise F ST of 0.070 across all U.S. sites sampled. While pest management strategies are likely employed at other facilities, if storage or processing of products is not for human consumption, treatments may be less frequent or less stringent. An important caveat is that unlike most of the sampling locations, beetles from these sites were not collected directly by the researchers, who made an effort to collect from several locations within a site when possible. Instead, these beetles were sent by the facility, and may represent a nonrandom sample of the overall population, resulting in the lower levels of diversity seen.
4.4. Updating hypotheses about Medea history
While some other SGEs have stable regional distributions (Bonnivard & Higuet, 1999; Price, Hoskyns, Rapley, Evans, & Wedell, 2012), we have presented evidence for a dynamic Medea distribution in the United States. Medea distributions in other regions may be more stable, and could be influenced by ecological factors, or the presence of nonfunctional neutral Medea alleles or suppressors. Such regions could include the boundary between M4 and the H suppressor element in Asian populations (Thomson & Beeman, 1999; Thomson, Friesen, Denell, & Beeman, 1995), or low‐frequency regions such as Australia (Beeman & Friesen, 1999); however, the stability of these distributions has yet to be investigated.
The generation time of T. castaneum under optimal conditions is roughly 5 weeks at 30°C. However, development slows dramatically with temperature decreases and eggs fail to hatch below 17.5°C, halting reproduction in cooler months (Howe, 1956). With a Medea element that has no associated fitness cost, the time between immigration into a population at moderate frequency (10%) and fixation could take 2–5 years (Ward et al., 2010). If the initial frequency in a population is low (1%), fixation could take decades. Still, unless introductions to the United States were very recent, a Medea with no fitness cost would be expected to be spread more widely. Importation of T. castaneum‐infested grains and other stored commodities from regions where Medea is uncommon may reintroduce wild‐type beetles, suppressing the spread of Medea elements. There is a dramatic contrast between the lack of substantial microsatellite allelic differentiation found among our geographic populations and the strong differentiation in M1 allele frequencies among these same populations. If M1 had been in the United States for thousands of years, the clusters that we found would not be expected unless there was some physical, ecological, or genetic barrier to spread. In a companion study (Cash, Robert, Lorenzen, & Gould, in review), we tested to determine whether Medea spread is inhibited when it is introduced to laboratory colonies of beetles from populations that lack Medea.
While the current study reveals interesting information about the spatial and temporal dynamics of Medea elements, it is limited in scope. Future studies could examine distributions of Medea elements on a finer geographic scale and also could be expanded into other countries. Once there are molecular markers for multiple Medea elements, such studies would become less labor‐intensive.
Recent advances in molecular biology have enabled the development of synthetic Medea elements (Buchman et al., 2018; Chen et al., 2007; Hay et al., 2010) and other synthetic gene drive mechanisms (Macias, Ohm, & Rasgon, 2017) with the goal of suppressing pest populations or eliminating traits that cause their pest status (Gantz & Akbari, 2018; Piaggio et al., 2017). To date, these synthetic drives have only been tested in laboratory settings (e.g., Kyrou et al., 2018) and release in the field is complicated by the fact that the extent of spread and the potential for resistance to the drive is hard to predict (NASEM, 2016). A more detailed understanding of the temporal dynamics of Medea and other naturally occurring selfish genetic elements in species that differ in population structure could provide insights that would aid in the assessment and testing of synthetic gene drives. The finding in this study of patchy distributions of Medea elements suggests that releases of synthetic selfish genetic elements to alter pest population traits such as ability to transmit human diseases cannot be expected to spread uniformly, especially in target species such as Aedes aegypti, (the vector of dengue, Zika, yellow fever) that have limited dispersal.
CONFLICT OF INTEREST
None declared.
AUTHOR CONTRIBUTIONS
SAC, FG, and MDL designed the research; MDL provided reagents; SAC performed the research, analyzed the data, and wrote the original draft; SAC, FG, and MDL interpreted the data, and wrote, reviewed, and edited the manuscript.
Supporting information
ACKNOWLEDGMENTS
The author would like to thank J. Campbell for providing the 2004–2007 T. castaneum samples. We are also grateful to the many extension personnel and agricultural facilities that helped SAC collect beetle samples. This project was supported by the Agriculture and Food Research Initiative Competitive Grant No. 2013‐67011‐21111 to SAC from the USDA National Institute of Food and Agriculture. Additional funding was provided by the National Science Foundation Grant No. MCB‐1244772 to MDL and by W. M. Keck Foundation to FG.
Cash SA, Lorenzen MD, Gould F. The distribution and spread of naturally occurring Medea selfish genetic elements in the United States. Ecol Evol. 2019;9:14407–14416. 10.1002/ece3.5876
DATA AVAILABILITY STATEMENT
More data can be accessed in the following thesis: Cash, S. A. 2016. An Experimental and Theoretical Analysis of the Selfish Genetic Element Medea in Red Flour Beetle Populations. PhD thesis, North Carolina State University, Raleigh. Data have been archived on Dryad https://doi.org/10.5061/dryad.7sqv9s4p3.
REFERENCES
- Akbari, O. S. , Chen, C. H. , Marshall, J. M. , Huang, H. X. , Antoshechkin, I. , & Hay, B. A. (2014). Novel synthetic Medea selfish genetic elements drive population replacement in Drosophila; a theoretical exploration of Medea‐dependent population suppression. ACS Synthetic Biology, 3, 915–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anxolabéhère, D. , Nouaud, D. , Périquet, G. , & Tchen, P. (1985). P‐element distribution in Eurasian populations of Drosophila melanogaster: A genetic and molecular analysis. Proceedings of the National Academy of Sciences of the USA, 82, 5418–5422. 10.1073/pnas.82.16.5418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeman, R. W. (2003). Distribution of the Medea factor M4 in populations of Tribolium castaneum (Herbst) in the United States. Journal of Stored Products Research, 39, 45–51. 10.1016/S0022-474X(02)00016-4 [DOI] [Google Scholar]
- Beeman, R. W. , & Friesen, K. S. (1999). Properties and natural occurrence of maternal‐effect selfish genes (‘Medea’ factors in the red flour beetle, Tribolium castaneum). Heredity, 82, 529–534. 10.1038/sj.hdy.6885150 [DOI] [PubMed] [Google Scholar]
- Beeman, R. W. , Friesen, K. S. , & Denell, R. E. (1992). Maternal‐effect selfish genes in flour beetles. Science, 256, 89–92. 10.1126/science.1566060 [DOI] [PubMed] [Google Scholar]
- Boake, R. B. B. , & Wade, M. J. (1984). Populations of the red flour beetle Tribolium castaneum (Coleoptera: Tenebrionidae) differ in their sensitivity to aggregation pheromones. Environmental Entomology, 13, 1182–1185. [Google Scholar]
- Bonnivard, E. , & Higuet, D. (1999). Stability of European natural populations of Drosophila melanogaster with regard to the P‐M system: A buffer zone made up of Q populations. Journal of Evolutionary Biology, 12, 633–647. [Google Scholar]
- Buchman, A. , Marshall, J. M. , Ostrovski, D. , Yang, T. , & Akbari, O. S. (2018). Synthetically engineered Medea gene drive system in the worldwide crop pest Drosophila suzukii . Proceedings of the National Academy of Sciences of the USA, 115, 4725–4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burt, A. , & Trivers, R. (2006). Genes in conflict: The biology of selfish genetic elements. Cambridge, MA: Belknap Press. [Google Scholar]
- Campbell, J. F. , Toews, M. D. , Arthur, F. H. , & Arbogast, R. T. (2010). Long‐term monitoring of Tribolium castaneum in two flour mills: Seasonal patterns and impact of fumigation. Journal of Economic Entomology, 103, 991–1001. [DOI] [PubMed] [Google Scholar]
- Carroll, L. , Meagher, S. , Morrision, L. , Penn, D. , & Potts, W. (2004). Fitness effects of a selfish gene (the mus t complex) are revealed in an ecological context. Evolution, 58, 1318–1328. 10.1554/03-544 [DOI] [PubMed] [Google Scholar]
- Cash, S. A. (2016). An experimental and theoretical analysis of the selfish genetic element Medea in red flour beetle populations, PhD thesis, North Carolina State University, Raleigh. [Google Scholar]
- Cash, S. A. , Robert, M. A. , Lorenzen, M. D. , & Gould, F. (in review). The impact of local population genetic background on the spread of the selfish element Medea‐1 in red flour beetles. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapuis, M. , & Estoup, A. (2007). Microsatellite null alleles and estimation of population differentiation. Molecular Biology and Evolution, 24, 621–631. 10.1093/molbev/msl191 [DOI] [PubMed] [Google Scholar]
- Chen, C. , Huang, H. , Ward, C. M. , Su, J. T. , Schaeffer, L. V. , Guo, M. , & Hay, B. A. (2007). A synthetic maternal‐effect selfish genetic element drives population replacement in Drosophila . Science, 316, 597–600. 10.1126/science.1138595 [DOI] [PubMed] [Google Scholar]
- Demuth, J. P. , Drury, D. W. , Peters, M. L. , Van Dyken, J. D. , Priest, N. K. , & Wade, M. J. (2007). Genome‐wide survey of Tribolium castaneum microsatellites and description of 509 polymorphic markers. Molecular Ecology Notes, 7, 1189–1195. 10.1111/j.1471-8286.2007.01826.x [DOI] [Google Scholar]
- Earl, D. A. , & vonHoldt, B. M. (2012). STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources, 4, 359–361. 10.1007/s12686-011-9548-7 [DOI] [Google Scholar]
- Evanno, G. , Regnaut, S. , & Goudet, J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Molecular Ecology, 14, 2611–2620. 10.1111/j.1365-294X.2005.02553.x [DOI] [PubMed] [Google Scholar]
- Excoffier, L. , Laval, G. , & Schneider, S. (2005). Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evolutionary Bioinformatics, 1, 47–50. [PMC free article] [PubMed] [Google Scholar]
- Gantz, V. M. , & Akbari, O. S. (2018). Gene editing technologies and applications for insects. Current Opinion in Insect Science, 28, 66–72. 10.1016/j.cois.2018.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfray, H. C. J. , North, A. , & Burt, A. (2017). How driving endonuclease genes can be used to combat pests and disease vectors. BMC Biology, 15, 81 10.1186/s12915-017-0420-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudet, J. (2001). FSTAT, a program to estimate and test gene diversities and fixation indices (version 2.9.3). Retrieved from http://www.unil.ch/izea/softwares/fstat.html [Google Scholar]
- Haliscak, J. P. , & Beeman, R. W. (1983). Status of malathion resistance in five genera of beetles investing farm‐stored corn, wheat and oats in the United States. Journal of Economic Entomology, 76, 717–722. [Google Scholar]
- Hatcher, M. (2000). Persistence of selfish genetic elements: Population structure and conflict. Trends in Ecology & Evolution, 15, 271–277. 10.1016/S0169-5347(00)01875-9 [DOI] [PubMed] [Google Scholar]
- Hay, B. A. , Chen, C. , Ward, C. M. , Huang, H. , Su, J. T. , & Guo, M. (2010). Engineering the genomes of wild insect populations: Challenges and opportunities provided by synthetic Medea selfish genetic elements. Journal of Insect Physiology, 56, 1402–1413. 10.1016/j.jinsphys.2010.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe, R. W. (1956). The effect of temperature and humidity on the rate of development and mortality of Tribolium castaneum (Herbst) (Coleoptera, Tenebrionidae). The Annals of Applied Biology, 44, 356–368. 10.1111/j.1744-7348.1956.tb02128.x [DOI] [Google Scholar]
- Jakobsson, M. , & Rosenberg, N. A. (2007). CLUMPP: A cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics, 23, 1801–1806. 10.1093/bioinformatics/btm233 [DOI] [PubMed] [Google Scholar]
- Kulldorff, M. (1997). A spatial scan statistic. Communications in Statistics – Theory and Methods, 26, 1481–1496. 10.1080/03610929708831995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulldorff, M. , & Information Management Services, Inc. (2009). SaTScanTMv. 8.0: Software for the spatial and space‐time scan statistics. Retrieved from http://www.satscan.org/ [Google Scholar]
- Kyrou, K. , Hammond, A. M. , Galizi, R. , Kranjc, N. , Burt, A. , Beaghton, A. K. , … Crisanti, A. (2018). A CRISPR‐Cas9 gene drive targeting doublesex causes complete population suppression in caged Anopheles gambiae mosquitoes. Nature Biotechnology, 36, 1062–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X. , Schuler, M. A. , & Berenbaum, M. R. (2007). Molecular mechanisms of metabolic resistance to synthetic and natural xenobiotics. Annual Review of Entomology, 52, 231–253. 10.1146/annurev.ento.51.110104.151104 [DOI] [PubMed] [Google Scholar]
- Lorenzen, M. D. , Gnirke, A. , Margolis, J. , Garnes, J. , Campbell, M. , Stuart, J. J. , … Beeman, R. W. (2008). The maternal‐effect, selfish genetic element Medea is associated with a composite Tc1 transposon. Proceedings of the National Academy of Sciences of the USA, 105, 10085–10089. 10.1073/pnas.0800444105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macias, V. M. , Ohm, J. R. , & Rasgon, J. L. (2017). Gene drive for mosquito control: Where did it come from and where are we headed? International Journal of Environmental Research and Public Health, 14, E1006 10.3390/ijerph14091006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- NASEM (National Academies of Sciences, Engineering, and Medicine) (2016). Gene drives on the horizon; Advancing science, navigating uncertainty, and aligning research with public values. Washington, DC: The National Academies Press. [PubMed] [Google Scholar]
- Obeng‐Ofori, D. , & Coaker, T. H. (1990). Tribolium aggregation pheromone: Monitoring, range of attraction, and orientation behavior of T. castaneum (Coleoptera: Tenebrionidae). Bulletin of Entomological Research, 80, 441–451. [Google Scholar]
- Perez‐Mendoza, J. , Campbell, J. F. , & Throne, J. E. (2014). Effect of abiotic factors on initiation of red flour beetle (Coleoptera: Tenebrionidae) flight. Journal of Economic Entomology, 107, 469–472. 10.1603/EC13364 [DOI] [PubMed] [Google Scholar]
- Piaggio, A. J. , Segelbacher, G. , Seddon, P. J. , Alphey, L. , Bennett, E. L. , Carlson, R. H. , … Wheeler, K. (2017). Is it time for synthetic biodiversity conservation? Trends in Ecology & Evolution, 32, 97–107. 10.1016/j.tree.2016.10.016 [DOI] [PubMed] [Google Scholar]
- Price, T. A. R. , Bretman, A. , Gradilla, A. C. , Reger, J. , Taylor, M. L. , Giraldo‐Perez, P. , … Wedell, N. (2014). Does polyandry control population sex ratio via regulation of a selfish gene? Proceedings of the Royal Society B, 281, 20133259 10.1098/rspb.2013.3259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price, T. A. R. , Hoskyns, R. C. , Rapley, H. , Evans, J. C. , & Wedell, N. (2012). No evidence that temperature‐related fertility differences influence the distribution of a selfish genetic element. Functional Ecology, 26, 657–665. 10.1111/j.1365-2435.2012.01971.x [DOI] [Google Scholar]
- Pritchard, J. K. , Stephens, M. , & Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics, 155, 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond, M. , & Rousset, F. (1995). GENEPOP (version 1.2): Population genetics software for exact tests and ecumenicism. Journal of Heredity, 86, 248–249. [Google Scholar]
- Rice, W. R. (1989). Analyzing tables of statistical tests. Evolution, 43, 223–225. 10.1111/j.1558-5646.1989.tb04220.x [DOI] [PubMed] [Google Scholar]
- Ridley, A. W. , Hereward, J. P. , Daglish, G. J. , Raghu, S. , Collins, P. J. , & Walter, G. H. (2011). The spatiotemporal dynamics of Tribolium castaneum (Herbst): Adult flight and gene flow. Molecular Ecology, 20, 1635–1646. 10.1111/j.1365-294X.2011.05049.x [DOI] [PubMed] [Google Scholar]
- Rode, N. O. , Estoup, A. , Bourguet, D. , Courtier‐Orgogozo, V. , & Debarre, F. (2019). Population management using gene drive: Molecular design, models of spread dynamics and assessment of ecological risks. Conservation Genetics, 20, 671–690. [Google Scholar]
- Romero, S. A. , Campbell, J. F. , Nechols, J. R. , & With, K. A. (2010). Movement behavior of red flour beetle: Response to habitat cues and patch boundaries. Environmental Entomology, 39, 919–929. 10.1603/EN09324 [DOI] [PubMed] [Google Scholar]
- Schuelke, M. (2000). An economic method for the fluorescent labeling of PCR fragments. Nature Biotechnology, 18, 233–234. 10.1038/72708 [DOI] [PubMed] [Google Scholar]
- Semeao, A. A. , Campbell, J. F. , Beeman, R. W. , Lorenzen, M. D. , Whitworth, R. J. , & Sloderbeck, P. E. (2012). Genetic structure of Tribolium castaneum (Coleoptera: Tenebrionidae) populations in mills. Environmental Entomology, 41, 188–199. 10.1603/EN11207 [DOI] [PubMed] [Google Scholar]
- Sinkins, S. P. , & Gould, F. (2006). Gene drive systems for insect disease vectors. Nature Reviews Genetics, 7, 427–435. 10.1038/nrg1870 [DOI] [PubMed] [Google Scholar]
- Sturtevant, A. H. , & Dobzhansky, T. (1936). Geographical distribution and cytology of "Sex Ratio" in Drosophila pseudoobscura and related species. Genetics, 21, 473–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson, M. S. , & Beeman, R. W. (1999). Assisted suicide of a selfish gene. Journal of Heredity, 90, 191–194. 10.1093/jhered/90.1.191 [DOI] [PubMed] [Google Scholar]
- Thomson, M. S. , Friesen, K. S. , Denell, R. E. , & Beeman, R. W. (1995). A hybrid incompatibility factor in Tribolium castaneum . Journal of Heredity, 86, 6–11. 10.1093/oxfordjournals.jhered.a111527 [DOI] [Google Scholar]
- Wade, M. J. , & Beeman, R. W. (1994). The population dynamics of maternal‐effect selfish genes. Genetics, 138, 1309–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward, C. M. , Su, J. T. , Huang, Y. , Lloyd, A. L. , Gould, F. , & Hay, B. A. (2010). Medea selfish genetic elements as tools for altering traits of wild populations: A theoretical analysis. Evolution, 65, 1149–1162. 10.1111/j.1558-5646.2010.01186.x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
More data can be accessed in the following thesis: Cash, S. A. 2016. An Experimental and Theoretical Analysis of the Selfish Genetic Element Medea in Red Flour Beetle Populations. PhD thesis, North Carolina State University, Raleigh. Data have been archived on Dryad https://doi.org/10.5061/dryad.7sqv9s4p3.


