Abstract
T-box riboregulators are a class of cis-regulatory RNAs that govern the bacterial response to amino acid starvation by binding, decoding, and reading the aminoacylation status of specific tRNAs. Here we provide a high-resolution crystal structure of a full-length T-box from Mycobacterium tuberculosis that explains tRNA decoding and aminoacylation sensing by this riboregulator . Overall, the T-box consists of decoding and aminoacylation sensing modules bridged by a rigid pseudoknot structure formed by the mid-region domains. Stem-I and the Stem-II S-turn assemble a claw-like decoding module, while the antiterminator, Stem-III, and the adjacent linker form a tightly interwoven aminoacylation sensing module. The uncharged tRNA is selectively recognized by an unexpected set of favorable contacts from the linker region in the aminoacylation sensing module. A complex structure with a charged tRNA mimic shows that the extra moiety dislodges the linker, indicative of the possible chain of events that lead to alternative base-pairing and altered expression output.
Keywords: T-box, tRNA, decoding, aminoacylation, riboswitch, RNA structure, gene regulation
A significant portion of bacterial genes are regulated at the mRNA level by cis-acting riboswitches in their 5’-UTRs. A typical riboswitch encodes an aptamer domain and an expression platform. Ligand binding to the aptamer domain triggers alternative base-pairing in the expression platform that alters transcription or translation levels of the associated mRNA1,2. The T-box riboregulator controls gene expression using the same conformational switching mechanism, however, instead of responding to small molecule ligands it recognizes a macromolecule, tRNA3. A typical T-box contains five structural domains: Stem-I, Stem-II, Stem IIA/B, Stem III, and Anti-Terminator (AntiT)/Anti-Sequestrator (AntiS) (Fig. 1a and inset). Two variant classes exist, glycine and atypical, which contain truncations outside the AntiT/S domain. Previous work has attributed distinct roles to structural domains: 1) A specifier motif in Stem-I decodes tRNA via codon-anticodon base-pairing3,4. 2) A conserved double T-loop structure in the distal half of Stem-I stacks against the D/T loops of the tRNA elbow and measures anticodon arm length in conjunction with the codon-anticodon pairing5-7. 3) The T-box sequence in AntiT/S specifies an uncharged tRNA 3’-NCCA tail through Watson-Crick (W-C) pairing8,9. Whereas the Stem-I-tRNA contacts have been defined by high-resolution structures6,7, the mechanism for aminoacylation sensing and the roles of the mid-region remain unclear.
Figure 1.
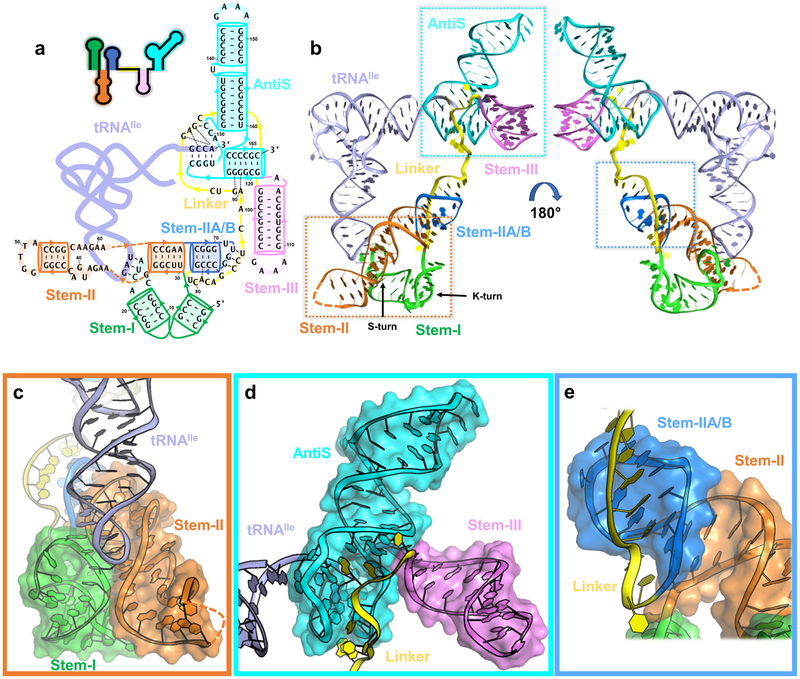
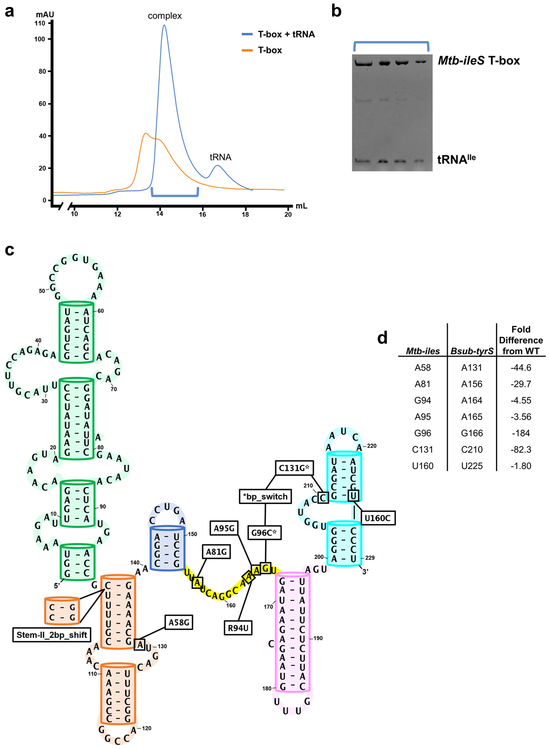
Overall structure of Mtb-ileS with details of domain arrangement. a, Secondary structure model with the sequence used for crystallization. Stem-I (green), Stem-II (orange), Stem-IIA/B (marine), linker (yellow), Stem-III (pink), AntiS (cyan), and tRNA (pale purple) are shown. Orange dashed lines link the sequence in Stem-II. Thin black dashes represent tertiary interactions. Arrows indicate direction 5’ to 3’. b, Front and back views of the Mtb-ileS 3D structure. c, Zoom-in of decoding module (dashed orange box in b). d, Zoom-in of aminoacylation sensing module (dashed cyan box in b). e, Zoom-in of Stem-IIA/B-linker pseudoknot (dashed marine box in b).
In this study, we present crystal structures of the full-length isoleucine T-box riboregulator from Mycobacterium tuberculosis (Mtb-ileS) in complex with its cognate uncharged tRNA or a charged tRNA mimic that contains a 2’−3’-cyclic phosphate at its terminus. Together they provide the structural basis for tRNA decoding and aminoacylation sensing by the T-box. We can now ascribe functions to the “variable” mid-region domains (Stem-II, Stem-IIA/B, and the linker), which play important roles in organizing the overall T-box structure. At the 5’-end, Stem-II assembles with the previously characterized Stem-I to form a decoding module. At the opposite end, Stem-IIA/B forms a rigid pseudoknot structure with the linker region. The mid-region domains combine to assemble an elongated structure that projects the aminoacylation sensing module near the tRNA 3’-NCCA tail. An unexpected finding of our work is that aminoacylation sensing not only requires the T-box antitermination domain as previously reported, but further involves Stem-III and the adjacent linker region. Together they form a tightly woven aminoacylation sensing module that co-axially stacks against the tRNA acceptor arm, guides the 3’-NCCA tail into a pocket through Watson-Crick (W-C) base-pairing, and cradles the uncharged tRNA terminus with favorable contacts. We further show that the aminoacylation sensing module is elastic enough to accommodate a tRNA with a 2’,3’-cyclic phosphate group at its 3’-end. However, the resulting steric hindrance tears the bottom of the binding pocket and increases overall conformational entropy in the T-box. This depicts a plausible scenario for how a charged tRNA could bind and actively initiate the transition to the alternative base-pairing scheme that switches the expression output. The T-box is the only other structured RNA capable of tRNA decoding and aminoacylation sensing besides the ribosome. A high-resolution understanding of this system opens discussions on its possible function in the primordial RNA world.
Results
Overall structure and domain organization in T-box-tRNA complex
Molecular dissection of the T-box-tRNA interaction has been difficult due to the lack of robust in vitro reconstitution. Speculating that this is because most T-boxes are transcriptional riboregulators whose folding is influenced by transcription speed and pausing elements10-12, we turned our attention to a class of translational T-boxes from actinobacteria reasoning that their folding is governed by thermodynamics13,14. In vitro reconstitution was indeed very robust (Extended Data Fig. 1a and b). We subsequently solved the co-crystal structure of a full-length isoleucine T-box from Mycobacterium tuberculosis bound to its uncharged cognate tRNA at 2.95 Å resolution (Fig. 1b, Table 1). The Mtb-ileS T-box includes all conserved structural features except the distal stacking module. It instead uses a degenerate Stem-I to decode tRNA (Fig 1a). Overall, the structure reveals that the Mtb-ileS T-box consists of two functional modules connected in the middle by a continuous structural unit assembled from the mid-region domains. On one side, the tail-to-tail stacked Stem-II and Stem-IIA/B further assemble with Stem-I to form the decoding module that latches onto the tRNA anticodon loop (Fig. 1c). On the opposite side, Stem-III and the nearby linker region weave around the AntiS domain to form the aminoacylation-sensing module that stacks to the end of the tRNA acceptor arm to interrogate the aminoacylation status of tRNA (Fig. 1d).
Table 1.
Data collection and refinement statistics
| Mtb-ileS_tRNA-OH_IrHex | Mtb-ileS_tRNA-OH_native (PDB 6UFG) |
Mtb-ileS_tRNA-cP_native (PDB 6UFH) |
|
|---|---|---|---|
| Data collectiona | |||
| Space group | P 41 | P 41 | P 1 21 1 |
| Cell dimensions | |||
| a, b, c (Å) | 76.769, 76.769, 170.857 | 77.209, 77.209, 172.754 | 76.276, 61.43, 122.52 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Peak | |||
| Wavelength | 1.105 | ||
| Resolution (Å)a | 76.77 - 3.12 (3.232 - 3.12)b | 77.21 - 2.929 (3.034 −2.929) | 62.68 - 3.104 (3.215 - 3.104) |
| Rmerge | 0.1096 (1.143) | 0.08105 (1.111) | 0.08573 (1.761) |
| I/σ(I) | 10.54 (1.37) | 12.77 (1.23) | 7.19 (0.83) |
| CC1/2 | 0.994 (0.813) | 0.994 (0.751) | 0.999 (0.626) |
| Completeness (%) | 99.84 (99.66) | 99.33 (96.16) | 96.34 (81.92) |
| Redundancy | 6.9 (7.0) | 6.8 (5.7) | 3.5 (3.3) |
| Refinement | |||
| Resolution (Å) | |||
| No. reflections | 21611 | 20047 | |
| Rwork / Rfree | 19.7/ 23.5 | 22.0/26.9 | |
| No. atoms | |||
| RNA | 5082 | 5172 | |
| MG | 11 | 6 | |
| K | 4 | - | |
| Water | - | - | |
| B factors | |||
| T-box | 112.65 | 171.16 | |
| tRNA | 126.34 | 147.07 | |
| MG | 91.5 | 97.5 | |
| K | 125.5 | - | |
| Water | - | - | |
| R.m.s deviations | |||
| Bond lengths (Å) | 0.003 | 0.003 | |
| Bond angles (°) | 0.69 | 0.76 |
Data in each column is from one crystal.
Values in parentheses are for highest-resolution shell.
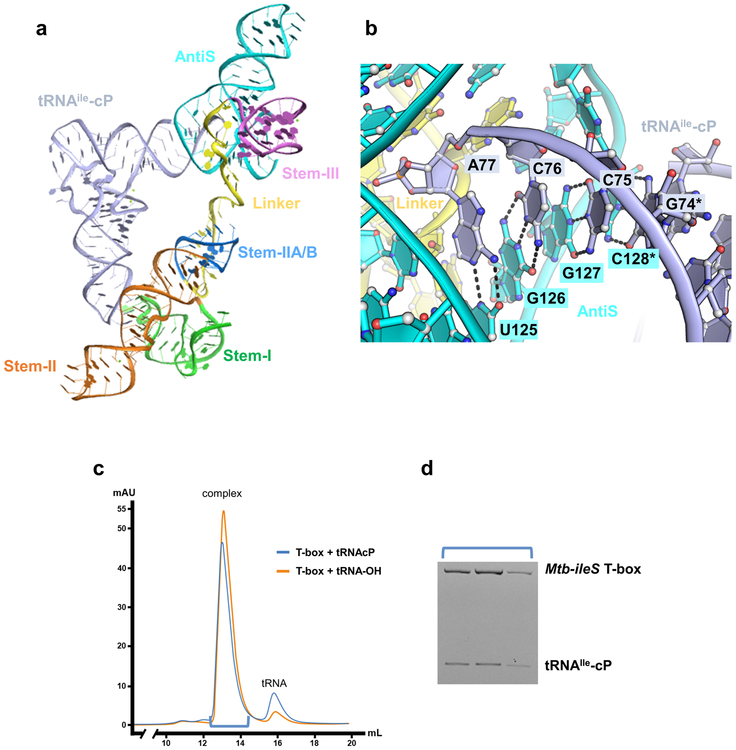
The long single-stranded linker between the two epicenters is largely absorbed through tertiary structure formation, leaving little flexibility therein (Fig. 1e). The 5’-half of the linker emerges from the stacking interface between Stem-II and IIA/B and travels along the minor groove of Stem-IIA/B, forming an extensive pseudoknot (Fig. 2a). Three W-C pairs14 and a ladder of stacked tertiary contacts to Stem-IIA/B are evident (Fig. 2a). The sharp bend required to redirect the linker from the stacking junction to the top of the pseudoknot is stabilized by a type-I A-minor interaction from A81 to the G68-C77 pair in Stem-IIA/B (Fig. 2a). The essentiality of this tertiary interaction explains the perplexing “F-box” sequence conservation in this region across T-boxes15. This and other structural features were evaluated by in vivo mutagenesis in the B. subtilis tyrS T-box, the model system where most of the previous genetic dissections were carried out3,15. For clarity, mutations will be referred to with Mtb-ileS notation throughout; the corresponding residue in tyrS and a secondary structure model showing the location of each mutation can be found in Extended Data Fig. 1c and d. Indeed, an F-box mutation (A81G) aimed at disrupting the A-minor contact reduced the B. subtilis tyrS T-box function to below the background level (Fig. 2b lime green). The pseudoknot interactions effectively shorten the distance between Stem-IIA/B and Stem-III. Thus, the pseudoknot contributes to T-box function not by making direct tRNA contact, but by introducing the necessary geometric constraints to place the aminoacylation sensing module close to the tRNA acceptor.
Figure 2.
tRNA decoding and pseudoknot formation. a, Detail of pseudoknot formation between Stem-IIA/B (marine) and the linker (yellow). Black arrows indicate backbone direction 5’ to 3’. b, Induction of tyrS T-box after tyrosine starvation. WT (purple) is the wildtype sequence in tyrosine(−) media. WT+ (orange) is wildtype sequence in tyrosine(+) media. Mutants are represented with dashed lines and use sequence notation from Mtb-ileS for clarity. A58G (red), Stem-II_2bp_shift (blue), and A81G (lime green) are shown. Error bars represent standard error of the mean with n = 3 biologically independent bacterial cultures. c, Zoom-in to the codon-anticodon pairing between Stem-I (green) and tRNAIle (pale purple) with Stem-II (orange) in the background. Hydrogen bonds are represented by black dashes. d, Stem-II tertiary contacts to each codon-anticodon pair arranged 5’ to 3’ relative to Stem-I. Weak electrostatic interaction between A16 and A60 (top) is shown as a grey dash. e, Detail of the Stem-II S-turn and its interaction with the codon-anticodon pair. Source data for panel b are available in Supplementary Table 3.
tRNA identity is specified by the Stem-I/II decoding module
With rare exceptions16, each T-box riboregulator selectively binds to one tRNA species to respond to the corresponding amino acid level. The key to this specificity is tRNA decoding via codon-anticodon pairing — a theme once thought to be unique to the ribosome. Previous structures of the glycine (glyQS) Stem-I-tRNA complex revealed an S-turn mediated codon-anticodon decoding mechanism and the further requirement of a second stacking contact to the tRNA D/T loop elbow6,7. The Mtb-ileS Stem-I lacks the stacking module and its decoding S-turn is replaced by a codon-embedded terminal specifier loop. It was puzzling how such T-boxes could use a simple stem-loop to efficiently decode tRNAIle. Here the structure reveals that decoding by Stem-I is supported by Stem-II; together they form a “claw” module that clamps onto the tRNA anticodon from both the major and minor groove sides (Fig. 1c and Fig. 2c).
Stem-II consists of a stem-loop with a conserved S-turn motif in the middle15 (Fig. 1b). It is one of the mid-region structural domains ubiquitous among typical and atypical T-boxes yet absent from glycine T-boxes. Thus, the novel tRNA contacts mediated by Stem-II that we observe are likely present among most T-box riboregulators. The Stem-I/II tertiary contact is enabled by the k-turn at the base of Stem-I, which orients the Stem-II S-turn motif to contact the codon-embedded specifier loop (Fig. 1b). The precise alignment of the Stem-I specifier loop and the Stem-II S-turn is important for T-box function. Increasing the Stem-II length by 2 base pairs (Stem-II_2bp_shift) projects the S-turn beyond the decoding center, causing T-box regulated induction to drop below the background level (Fig. 2b dark blue). At the interface of Stem-I and II, the S-turn sets the stage for tRNA binding by stapling the codon (A16, U17, and C18) into an optimal decoding conformation via A-minor contacts (A60•A16, G59•U17, and A58•C18) (Fig. 2d top, middle, and bottom). These are further supported from underneath by a ribose-zipper interaction (A39•A19) and a stacking interaction with specifier loop residue A19 (Fig. 2c). The network of interactions depends on the S-turn architecture, which is stabilized by the conserved G37•A58 Hoogsteen pair (Fig. 2e). Perturbing the S-turn motif with a mutation to A58 (A58G) in the tyrS T-box disrupts its ability to respond to tyrosine starvation in vivo, validating the importance of this structural feature (Fig. 2b red).
Upon tRNA recruitment, the geometry of the triplet W-C pairing between codon and anticodon allows additional stereospecific hydrogen (H-) bonds to form with the S-turn. These include a type-I-like A-minor contact from S-turn A38 to anticodon G34 (Fig. 2d bottom), a ribose zipper from S-turn G37 to anticodon A35 (Fig. 2d middle), and an N1-O2’ H-bond between S-turn A60 and anticodon U36 (Fig. 2d top). Modeling suggests that these contacts would not be disturbed by common tRNA anticodon loop modifications17. However, because position 34 in the anticodon is subject to a wide variety of modifications18, we cannot completely rule out the possibility that certain modifications may require the Stem-II S-turn to interact with the codon-anticodon pairing differently from what we observe here.
In summary, the structural features above enable the T-box to carry out A-minor motif assisted decoding to specify the cognate tRNA. Near-cognate tRNAs are rejected presumably because the non-W-C pairing would orient the ribose 2’-OH differently, preventing the strengthening minor groove contacts or introducing steric clashes14,19. This mechanism is conceptually similar to how the ribosome differentiates cognate and near-cognate tRNAs, although a different set of A-minor interactions are utilized. In the ribosome, the first two layers of codon-anticodon pairing are supported by type-I and type-II A-minor contacts, respectively20-22. Conversely, the A-minor contacts made by the T-box are focused on the second and wobble position pairs and are more extensive (Fig. 2d middle and bottom). Whereas the ribosome tolerates mismatches in the wobble position, the T-box is expected to have a much more stringent preference for tRNA isoacceptors19. In fact, a bias for a cytosine residue in the wobble position of codons found in T-box specifier loops has been noted23. The specific contacts in our structure between S-turn A38 and the sugar edge of tRNA G34 could explain the wobble cytosine preference from the complementary side (Fig. 2d bottom).
AntiS is insufficient to distinguish tRNA aminoacylation state
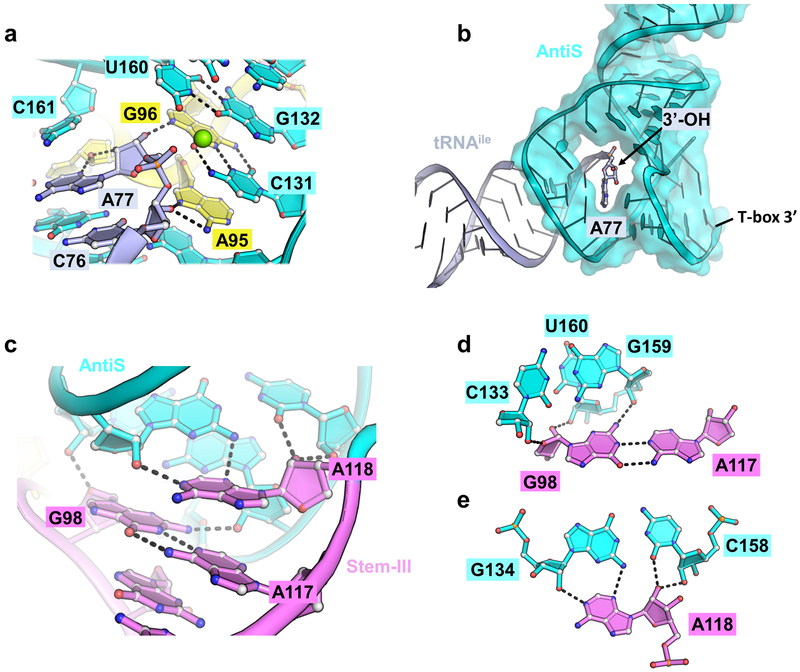
Whereas the tRNA decoding mechanism varies to some extent among the three T-box variants, the aminoacylation sensing mechanism is essentially identical. The AntiT or AntiS domains have long been established as the main contributors to aminoacylation sensing due to their characteristic “T-box” sequence in an asymmetric bulge that complements the tRNA NCCA tail8. In our structure, the AntiS helix is bent at the central asymmetric bulge to an L-shape (Fig. 3a). The “T-box” sequence inside the asymmetric bulge mediates four consecutive W-C base-pairs with the tRNA NCCA tail (Fig. 3b). The resulting four-base-pair helix is coaxially stacked on either side by the tRNA acceptor arm and the AntiS bottom helix, forming a continuous superhelix (Fig. 3a and Extended Data Fig. 2b). This arrangement ensures that only mature tRNA, but not precursors, are examined by T-boxes. An important finding from our structure is that the AntiS domain alone lacks specific contacts with the tRNA that interrogate its aminoacylation status. A residue (C161) from its bottom helix partially stacks with the tRNA terminal adenosine and a wobble pair (G132-U160) from the base of the AntiS top helix partially blocks the tRNA terminus (Extended Data Fig. 2a). However, these blockages are insufficient to fully distinguish a charged tRNA. For example, changing the wobble pair to a C-G pair (U160C) in the tyrS T-box should alleviate the partial blockage, but the mutation only decreases T-box induction by ~2-fold (Fig. 3f green). Overall, the AntiS forms a surprisingly incomplete binding pocket where the tRNA terminal adenosine is exposed in a large opening that could easily accommodate an aminoacylated tRNA terminus (Extended Data Fig. 2b).
Figure 3.
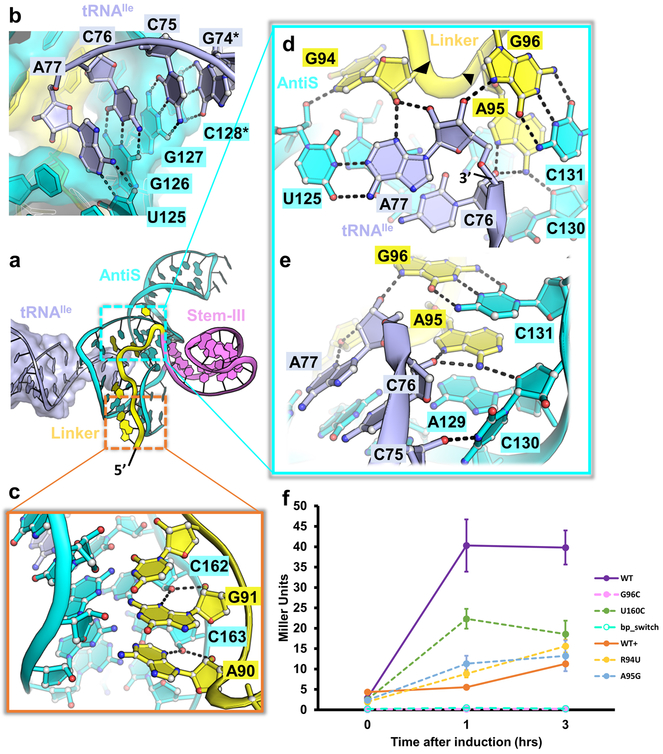
Details of the aminoacylation sensing module. a, Tertiary structure of the Mtb-ileS aminoacylation sensing module bound to tRNAIle (pale purple) with linker (yellow), Stem-III (pink), and AntiS (cyan) domains. b, Zoom-in of tRNAIle NCCA base pairing with AntiS. Hydrogen bonds are represented by black dashes. Asterisks denote residues mutated from wildtype sequence. c, Minor groove interactions between linker and AntiS. d, Aminoacylation sensing by the linker RAG (G94, A95, and G96) sequence. Black arrows indicate direction of backbone 5’−3’. e, Aminoacylation sensing pocket showing interaction between linker and conserved AntiS ACC (A129, C130, and C131) sequence. f, Induction of tyrS T-box after tyrosine starvation. WT (purple), WT+ (orange), G96C (pink), U160C (green), R94U (yellow), A95G (light blue), and bp_switch (teal) are shown. G96C and bp_switch overlap at bottom. Mutants are represented with dashed lines. Error bars represent standard error of the mean with n = 3 biologically independent bacterial cultures. WT and WT+ data are the same as plotted in Fig. 2b. Source data for panel f are available in Supplementary Table 3.
Aminoacylation sensing requires the nearby linker region
Formation of a functional aminoacylation sensing module necessarily involves Stem-III and the nearby linker region (Fig. 3a). This came as a surprise because neither the sequence nor the length of Stem-III is conserved23 and the linker sequence conservation was noted but not understood15. We can now ascribe three functions to the linker and Stem-III: they contribute key tertiary contacts to stabilize AntiS into the aminoacylation sensing conformation, are an integral component of the tRNA-binding pocket, and play pivotal roles in aminoacylation sensing. Beginning with a pair of type-II A-minor interactions from A90 and G91 (Fig. 3c), the linker meanders up the minor groove of the AntiS bottom helix to the opening where the tRNA terminus is pointed. Here, a GAG sequence (G94, A95, and G96) encompassing the previously noted AG motif15 specifies the aminoacylation status of the tRNA (Fig. 3d). Based on our structure and further inspection of T-box alignments24, we propose that a RAG sequence in the linker region adjacent to Stem-III is conserved across T-boxes and maintains this essential function.
In Mtb-ileS, this RAG sequence begins with G94, which makes a sequence-specific contact to the tRNA 3’-adenosine (A77) by contributing its 2’OH to a type-II A-minor interaction with the tRNA and its N7 to a H-bond with the AntiS ribose (U125) across the base pair (Fig. 3d). A transversion mutation (R94U) at this position in tyrS reduces induction to around background levels, illustrating the importance of the purine specific contacts we observe (Fig. 3f yellow). Following G94, A95 plays a pivotal role by mediating continuous stacking between AntiS A129 and C131 residues while H-bonding with the riboses of tRNA C76 and AntiS C130 using its Hoogsteen edge (Fig. 3e). The function of the tyrS T-box is especially sensitive to nucleotide identity at this position; an A95G mutation reduces T-box induction to near background (Fig. 3f light blue). The AntiS ACC (129–131) sequence that interacts with linker A95 (Fig. 3e) is almost universally found downstream of the NCCA-complementary region across T-boxes23,25. Our structure clearly shows that it serves as a platform to position the linker for aminoacylation sensing (Fig. 3e). Finally, G96 forms a long-range base pair with AntiS C131 (Fig. 3d) and positions its Hoogsteen edge to make an H-bond to the terminal 3’OH of the tRNA (Fig. 3d). This is the crux interaction for sensing uncharged tRNA because the 3’OH is strictly specified as a H-bond donor. As expected, a mutation to G96 (G96C) in the tyrS T-box precludes any induction (Fig. 3f pink). Importantly, T-box function could not be rescued with a purine-pyrimidine switch (bp_switch, G96C + C131G) that restores the linker-AntiS base pair between G96 and C131 (Fig. 3f teal). Together, the linker region interactions with the tRNA leave no room for an extra chemical moiety and are highly specific for an uncharged tRNA with a terminal adenosine containing intact 2’ and 3’ hydroxyls. Previous in vitro studies using cognate tRNAs without terminal hydroxyls or with a terminal 2-aminopurine decreased T-box response significantly9. We can now rationalize these results as being due to the modifications disrupting the observed linker contacts.
The bottom of Stem-III inserts into the AntiS minor groove with the rest of the stem-loop extending away (Fig. 3a). Its configuration in the aminoacylation sensing module suggests it likely functions as a twist-tie that maintains tension between the tRNA binding pocket and the AntiS bottom helix. All of Stem-III’s contacts with the AntiS are mediated by residues near the base of the stem, explaining the lack of sequence or length conservation in the body of the stem-loop23. The non-canonical G98•A117 pair expands the base of Stem-III, allowing it to span the minor groove of the AntiS top helix and place G98 to make cross-groove ribose contacts (Extended Data Fig. 2c and d). The following A118 residue connects Stem-III to the beginning of the AntiS bottom helix while anchoring to the third base pair of the AntiS top helix through a type-I A-minor interaction (Extended Data Fig. 2c and e). The close proximity of Stem-III and the linker allows a continuous belt of tertiary contacts, which act like a Velcro strip to patch up the tRNA binding pocket and stabilize the AntiS conformation.
Structure of T-box interacting with a charged tRNA mimic
There are two mechanistic possibilities for how T-boxes distinguish charged and uncharged tRNAs and thereby mediate switching between sequestrator and AntiS structures: 1) The aminoacylation sensing pocket may be rigid and sterically rejects the charged tRNA tail from binding. Or 2) the pocket may be elastic enough to accommodate the extra amino acid on the charged tRNA, but the deformation nucleates a cascade of conformational changes that facilitate switching to the sequestrator base-pairing scheme. To differentiate these two models, we determined the structure of another Mtb-ileS construct bound to tRNA with a terminal 2’,3’-cyclic phosphate (tRNAIle-cP) to mimic a charged tRNA. We hypothesized that the presence of a relatively small moiety at the tRNA terminus would not be enough to completely disrupt the T-box-tRNA complex, but instead would allow us to capture changes indicative of the actual mechanism. The resulting 3.1 Å co-crystal structure (Fig. 4a, Extended Data Fig. 3a, and Table 1) is consistent with the second mechanistic model – the gain from continuous W-C pairing with the tRNA tail offsets the penalty from steric hindrance induced disruption of tertiary contacts. Indeed, complex formation between Mtb-ileS and tRNAIle-cP is comparable to uncharged tRNA (Extended Data Fig. 3c and d).
Figure 4.
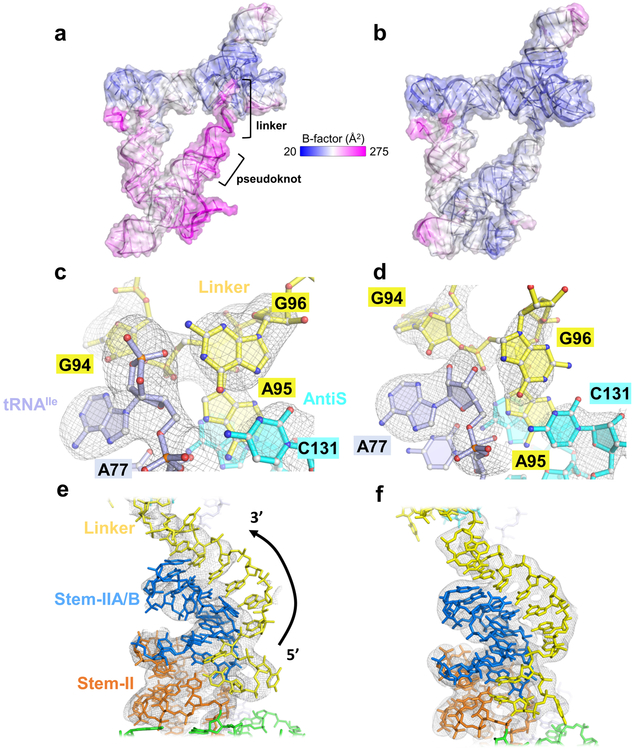
Comparison of Mtb-ileS complex structures bound to tRNAIle-cP or tRNAIle-OH. a, Overall structure of Mtb-ileS bound to tRNAIle-cP with b-factor coloring. b, Overall structure of Mtb-ileS bound to tRNAIle-OH with b-factor coloring. c, Omit map electron density (gray mesh) at 3.0 σ of the aminoacylation sensing pocket in the tRNAIle-cP structure. d, Omit map electron density at 3.0 σ of the aminoacylation sensing pocket in the tRNAIle-OH structure. e, 2Fo-Fc map electron density (gray mesh) at 1.0 σ of the pseudoknot region in the tRNAIle-cP structure. f, 2Fo-Fc map electron density at 1.0 σ of the pseudoknot region in the tRNAIle-OH structure.
In the structure, the entire tRNA NCCA tail is accommodated with continuous W-C pairing in the AntiS binding pocket (Extended Data Fig. 3b). Steric hindrance from the extra terminal moiety is absorbed locally by the linker region, leading to the disruption of its tertiary interaction with the sensing core and readjustments thereafter. As a result, the important long-range G96-C131 pairing is broken; G96 undergoes a syn-to-anti flip and makes alternative H-bonds (Fig. 4c and d). In fact, the entire linker appears to be peeling away: G94 is shifted out of its type-II A-minor interaction with the tRNA terminus (Fig. 4c and d) and B-factors for the linker at the start of its interaction with the AntiS are much higher in this structure (Fig. 4 a and b), indicating that it is no longer stably docked in its preferred conformation. Additionally, the weakened linker-AntiS interaction increases the conformational entropy of the nearby Stem-IIA/B pseudoknot (Fig. 4 a and b). Electron density corresponding to parts of the linker region and Stem-IIA/B diminishes, suggesting these residues are disordered upon binding to the cyclic-phosphate-containing tRNA (Fig. 4e and f). It is worth noting that even the smallest amino acid is bigger than a cyclic phosphate; a charged tRNA would introduce more severe steric clashing, particularly against G96, and would likely push the entire GAG motif out of the pocket. The complete ejection of the GAG sequence from the pocket could be the first domino to fall in a series of events that nucleate alternative base pairing. Overall, our data suggest that small changes to the linker in the aminoacylation sensing pocket can nucleate large global rearrangements.
Discussion
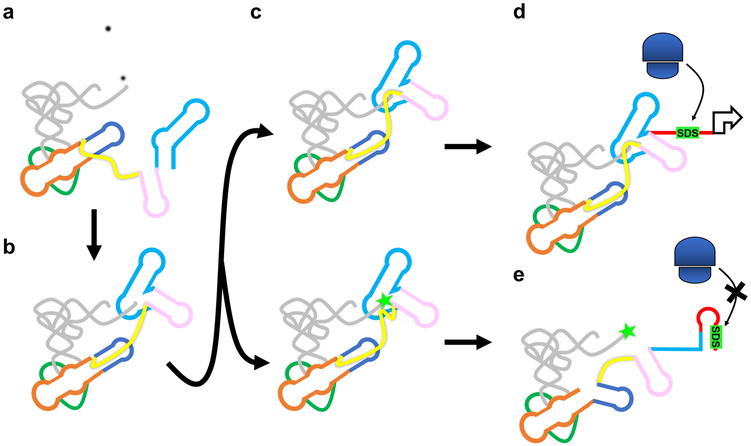
The discovery that T-box riboregulators are able to simultaneously decode and sense the aminoacylation status of tRNA3 has fascinated researchers for the past three decades. Such sophisticated tasks were thought to be reserved to the megadalton ribosome complex. Our T-box-tRNA structures finally provide a complete, high-resolution explanation of both processes. The structures best support a stepwise model for tRNA sensing. In the first step, Stem-I and Stem-II capture the tRNA and specify the anticodon irrespective of its aminoacylation status26 (Extended Data Fig. 4a). The next step is aminoacylation sensing, which our structures suggest proceeds through an intermediate where the AntiS interacts with the NCCA sequence of the tRNA while the linker samples the aminoacylation status (Extended Data Fig. 4b). An important new concept from our work is that the T-box may be capable of accommodating both versions of its ligand at this step, although they ultimately progress to different fates (Extended Data Fig. 4c). Interaction with the uncharged tRNA leads to stable complex formation, trapping the T-box in the AntiS conformation (Extended Data Fig. 4d). On the other hand, a charged tRNA is guided into the aminoacylation sensing pocket through W-C pairing to the tRNA 3’-tail, however, the extra moiety at the tail dislodges the linker region, which likely nucleates subsequent conformational changes (Extended Data Fig. 4e). We therefore conclude that charged tRNA binding leads to a short-lived intermediate and the aminoacylated tRNA terminus may kinetically facilitate the AntiS-to-sequestrator transition.
This model is in line with the recent single molecule studies that propose hierarchal tRNA recognition consisting of two distinct binding states11,12. These studies have also shown the emergence of a significantly longer-lived binding state for both charged and uncharged tRNA when a full-length T-box rather than the decoding-only fragment was used11. This is suggestive of the intermediate step we propose, where the tRNA terminus may be bound and examined by the aminoacylation sensing module. The uncharged tRNA was shown to induce even longer-lived binary complexes, which are likely stabilized by the linker contacts revealed in our structure. Due to limitations of the labeling scheme, the existing studies have not resolved the conformational differences and interconversion rates among different states. This limited our ability to draw more definitive conclusions. The improved spatial resolution from our structural study will surely stimulate new single molecule based mechanistic studies to probe deeper into the tRNA sensing and gene regulatory mechanisms of the T-box.
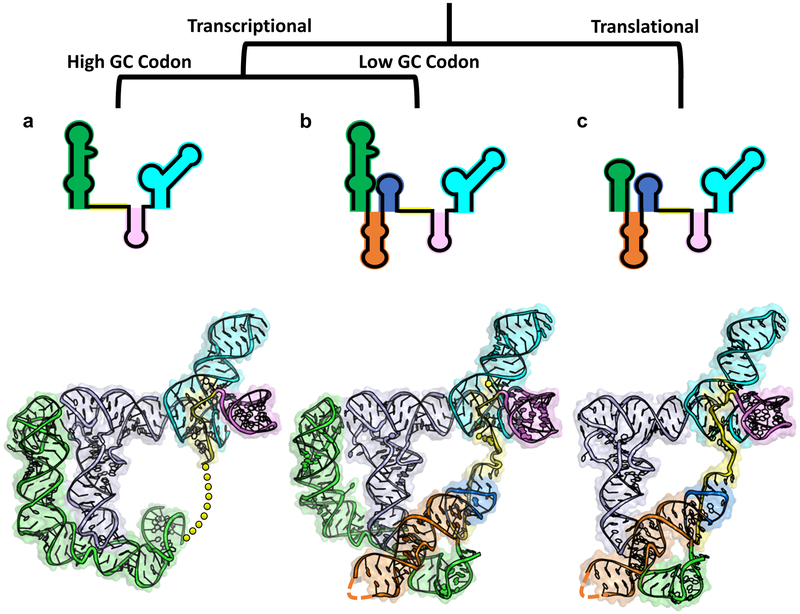
By synthesizing previous structural information with the current study, we can generate a family portrait depicting all three classes of T-boxes: glycine, typical, and atypical (Fig. 5a, b, and c). These snapshots allow us to begin rationalizing the differences among the three classes. Glycine T-boxes likely shed most of the mid-region because the GC-rich codon-anticodon interaction does not require further stabilization. Likewise, transcriptional T-boxes may rely on the distal stacking module in Stem-I to improve the kinetics of tRNA recruitment, whereas this is not as critical of a consideration for the thermodynamically driven translational T-boxes. As stated above, the T-box and the ribosome are the only RNA molecules capable of decoding and aminoacylation sensing. An intriguing hypothesis is that the ancestors of T-boxes may have played essential roles in translation in the primordial RNA world, possibly serving as a primitive tRNA synthetase27,28. The high-resolution information from this study opens the door to a more detailed investigation of these hypotheses.
Figure 5.
T-box family portrait. T-box classes are distinguished based on regulatory mechanism and characteristics of tRNA interaction. (a) Glycine T-box class. (top) Secondary structure representation. (bottom) Tertiary structure model combining glyQS Stem-I-tRNA structure (PDB 4LCK) with aminoacylation sensing module from Mtb-ileS. Linker represented with yellow dashes. (b) Typical T-box class. (top) Secondary structure representation. (bottom) Tertiary structure model combining glyQS Stem-I-tRNA (PDB 4MGN) and Mtb-ileS. (c) Atypical T-box class. (top) Secondary structure representation. (bottom) Overall structure of Mtb-ileS.
METHODS
Constructs, Plasmids, and Strains
Strains and plasmids are described in Supplementary Table 1. The sequences of the constructs used in the biochemical and crystallization experiments are documented in Supplementary Table 2.
RNA preparation
RNA constructs were cloned and produced as described previously5,29. T-box and tRNA sequences were cloned into the pUC19 plasmid downstream of a T7 RNA polymerase promoter and upstream of the hepatitis δ virus ribozyme (HDV) to produce homogeneous ends. The hammerhead ribozyme sequence was added upstream of tRNAIle sequences. Guanosine residues were added at the beginning of each construct to increase expression and non-conserved loops were mutated to GAAA tetraloops to facilitate crystallization as noted in Table S1. Plasmid templates for transcription of T-box constructs were prepared with Qiagen MegaPrep kits and linearized by restriction digestion after the HDV sequence. The transcription template for tRNAIle −3’-hydroxyl was produced by PCR with a 2’-O-methylated reverse primer. The plasmid transcription template for tRNAIle −3’-cyclic phosphate was produced as described for T-box constructs. 10 mL in vitro transcription reactions were performed as described previously29. RNA was gel purified by urea denaturing PAGE. The bands were excised from the gel, crushed, and eluted into MilliQ-purified water at 4 °C overnight. RNA was buffer-exchanged to 10 mM HEPES pH 7.0, 50 mM NaCl and refolded at 5 μM of the same buffer by heating at 95 °C for 2 min, cooling to 65 °C, adding 10 mM MgCl2 and equal volume of 5 μM unfolded tRNAIle, then cooling to 35 °C before putting on ice. Cooled samples were concentrated to 0.250 mM and then used for crystallization.
Crystallization and Data Collection
RNA constructs were screened for crystallization by sitting drop vapor diffusion at RNA concentrations of 0.125 and 0.250 mM, at 16 °C. Optimized conditions for Mtb-ileS tRNA-3’-hydroxyl were: 20 μM RNA in a mother liquor of 10% PEG 4000, 50 mM HEPES pH 7.0, 150 mM KCl, 40 mM MgCl2, at 16° C with a 1:1 (RNA: mother liquor) drop ratio. Optimized conditions for Mtb-ileS tRNA-3’-cyclic phosphate were: 20 μM RNA in a mother liquor of 10% PEG 4000, 50 mM sodium cacodylate pH 6.5, 200 mM KCl, 15 mM MgCl2, at 16° C with 1:1 (RNA: mother liquor) drop ratio. For phasing, 10 mM Iridium Hexamine (IrHex) and 20% ethylene glycol were added to the crystals overnight at 4° prior to flash freezing in liquid N2.
Data were collected at the Advanced Photon Source (APS) 24 ID-C Northeastern Collaborative Access Team (NE-CAT), as indicated in Table S2. Datasets were processed using HKL-200030 or XDS31 as part of NE-CAT’s RAPD pipeline. The Mtb-ileS tRNA-3’-hydroxyl structure was phased by the single-wavelength anomalous dispersion (SAD) method. The iridium hexamine sites were located using direct methods using SHELX32,33. These sites were fed to PHENIX AutoSol and further refined thererin34. A model was built into the experimental map using the program COOT35. The partial model was then used in MR-SAD to improve experimental map quality. Iterative rounds of such efforts eventually led to the completion of the T-box/tRNA model. Structural refinement was done in phenix.refine34, during which multiple rounds of simulated annealing refinement was carried out to remove possible model bias. The final tRNA-3’-hydroxyl model was built by molecular replacement of the SAD model into a higher resolution native dataset using PHENIX Phaser-MR followed by additional refinement. Simulated annealing omit maps were generated to ensure the correct interpretation of weaker electron densities. The tRNA-3’-cyclic phosphate structure was solved by molecular replacement using Phaser-MR and the model was built with subsequent rounds of rigidbody, positional, and temperature factor refinement.
T-box Induction Assay
The induction assay was adapted from Grundy and Henkin3. Briefly, the B. subtilis tyrS leader (native promoter and T-box) was cloned upstream of lacZ in the pDG1661 integration vector36. The reporter construct was transformed into a tyrosine auxotroph B. subtilis strain (BKK22610) from the Bacillus Genetic Stock Center and integrated into the amyE locus by homologous recombination. Mutant T-box sequences were generated using site directed mutagenesis of the WT tyrS reporter plasmid and transformed into the BKE22610 strain.
For T-box induction experiments, cells were grown overnight in Spizizen minimal media37 with 50 μg/mL tyrosine and the appropriate antibiotics at 37°C. Cells from the overnight cultures were subcultured into the same media and grown to mid-log phase. The cells were pelleted by centrifugation and resuspended in Spizizen minimal media with or without tyrosine supplementation. One mL of cells was harvested at 0, 1, and 3 hour timepoints after resuspension. Beta-galactosidase activity was measured as described previously38,39. All measurements were done in triplicate.
Data Availability
Atomic coordinates and structure factors for Mtb-ileS_tRNA-OH_native and Mtb-ileS_tRNA-cP_native have been deposited with the Protein Data Bank with the accession codes 6UFG and 6UFH, respectively. Source data for Figure 2b and 3f are available in Supplementary Table 3. Other data are available upon request.
Extended Data
Extended Data Fig. 1. Size exclusion chromatographic analysis and mutagenesis guide.
a, Size exclusion chromatogram of Mtb-ileS folded with (blue) and without (orange) tRNAIle. b, Urea denaturing PAGE gel of peak fractions from T-box + tRNA peak. c, Secondary structure model of tyrS T-box from Bacillus subtilis (Bsub-tyrS). Boxes highlight location and identity of each mutation. d, Conversion table for residue notation in Mtb-ileS and Bsub-tyrS. Fold difference from WT is difference between the average of three induction measurements at 1 hour for WT versus mutation of indicated residue.
Extended Data Fig. 2. Details of AntiS-tRNA and AntiS-Stem-III interactions.
a, Detail of G132-U160 wobble pair in the aminoacylation sensing pocket showing AntiS (cyan), tRNA (pale blue), and linker (yellow). Hydrogen bonds represented by black dashes. Magnesium ion represented by green sphere. b, View of AntiS bound to tRNA showing tRNA 3’-end exposed in a large opening. c, Stem-III (pink) and AntiS minor groove interactions. d, Details of G98 interactions with AntiS. e, Details of A118 type-I A-minor interaction with AntiS.
Extended Data Fig. 3. Structural details and size exclusion chromatography of Mtb-ileS with tRNAIle-cP.
a, Tertiary structure model of Mtb-ileS in complex with tRNAIle-cP. b, Detail of AntiS interaction with tRNAIle-cP NCCA sequence. Hydrogen bonds represented by black dashes. Asterisk indicates residues mutated from wildtype sequence. c, Size exclusion chromatograms of Mtb-ileS folded with tRNAIle-cP (blue) and with tRNAIle-OH (orange). d, Urea denaturing PAGE gel of peak fractions from T-box + tRNAIle-cP peak.
Extended Data Fig. 4. Mechanistic diagram of atypical T-box translational regulation.
a, tRNA recruitment and decoding. b, Pseudoknot formation positions AntiS to interact with tRNA NCCA sequence. c, Transient intermediate where aminoacylation sensing module interacts with uncharged (top) and charged (bottom) tRNA. d, Favorable interactions between uncharged tRNA and linker locks T-box into ON conformation. Exposed Shine-Dalgarno allows translation initiation. e, Steric clashing between charged tRNA and linker unravels the aminoacylation sensing module leading to alternative base pairing. Sequestrator formation prevents ribosome access to Shine-Dalgarno and prevents translation.
Supplementary Material
Acknowledgements
The authors would like to thank Ankita J. Sachla and John D. Helmann for advice on the β-galactosidase assay, the Bacillus Genetic Stock Center, the Nicholson Lab for use of their spectrophotometer, and the Fromme Lab for use of their microscope.
This work is supported by National Institutes of Health (NIH) grants GM118174 and GM116632 to A.K.
This work is based upon research conducted at the Northeastern Collaborative Access Team beamlines, which are funded by the National Institute of General Medical Sciences from the National Institutes of Health (P30 GM124165). The Pilatus 6M detector on 24-ID-C beam line is funded by a NIH-ORIP HEI grant (S10 RR029205). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357.
Footnotes
Competing Interests Statement
The authors declare no competing financial interests.
References
- 1.Mandal M & Breaker RR Gene regulation by riboswitches. Nat. Rev. Mol. Cell Biol 5, 451–63 (2004). [DOI] [PubMed] [Google Scholar]
- 2.Peselis A & Serganov A Themes and variations in riboswitch structure and function. Biochim. Biophys. Acta - Gene Regul. Mech 1839, 908–918 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grundy FJ & Henkin TM tRNA as a positive regulator of transcription antitermination in B. subtilis. Cell 74, 475–482 (1993). [DOI] [PubMed] [Google Scholar]
- 4.Grundy FJ, Winkler WC & Henkin TM tRNA-mediated transcription antitermination in vitro: Codon-anticodon pairing independent of the ribosome. Proc. Natl. Acad. Sci 99, 11121–11126 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grigg JC et al. T box RNA decodes both the information content and geometry of tRNA to affect gene expression. Proc. Natl. Acad. Sci. U. S. A 110, 7240–5 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang J & Ferré-D’Amaré AR Co-crystal structure of a T-box riboswitch stem I domain in complex with its cognate tRNA. Nature 500, 363–6 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grigg JC & Ke A Structural Determinants for Geometry and Information Decoding of tRNA by T Box Leader RNA. Structure 21, 2025–2032 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grundy FJ, Rollins SM & Henkin TM Interaction between the acceptor end of tRNA and the T box stimulates antitermination in the Bacillus subtilis tyrS gene: a new role for the discriminator base. J. Bacteriol 176, 4518–26 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang J & Ferré-D’Amaré AR Direct evaluation of tRNA aminoacylation status by the T-box riboswitch using tRNA-mRNA stacking and steric readout. Mol. Cell 55, 148–55 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grundy FJ & Henkin TM Kinetic analysis of tRNA-directed transcription antitermination of the Bacillus subtilis glyQS gene in vitro. J. Bacteriol 186, 5392–5399 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suddala KC et al. Hierarchical mechanism of amino acid sensing by the T-box riboswitch. Nat. Commun 1–14 doi: 10.1038/s41467-018-04305-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang J et al. Specific structural elements of the T-box riboswitch drive the two-step binding of the tRNA ligand. Elife 7, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seliverstov AV, Putzer H, Gelfand MS & Lyubetsky VA Comparative analysis of RNA regulatory elements of amino acid metabolism genes in Actinobacteria. BMC Microbiol. 5, 54 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sherwood AV, Grundy FJ & Henkin TM T box riboswitches in Actinobacteria: translational regulation via novel tRNA interactions. Proc. Natl. Acad. Sci. U. S. A 112, 1113–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rollins SM, Grundy FJ & Henkin TM Analysis of cis -acting sequence and structural elements required for antitermination of the Bacillus subtilis tyrS gene. Mol. Microbiol 25, 411–421 (1997). [DOI] [PubMed] [Google Scholar]
- 16.Saad NY et al. Two-codon T-box riboswitch binding two tRNAs. Proc. Natl. Acad. Sci 110, 12756–12761 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shepherd J & Ibba M Bacterial transfer RNAs. FEMS Microbiol. Rev 39, 280–300 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Agris PF, Vendeix FAP & Graham WD tRNA’s Wobble Decoding of the Genome: 40 Years of Modification. J. Mol. Biol 366, 1–13 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Putzer Harald, Condon Ciaran, Dominique Brechemier-Baey, R. B. and M. G.-M. C. Transfer RNA-mediated antitermination in vitro. Nucleic Acids Res. 30, 3026–3033 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ogle JM et al. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292, 897–902 (2001). [DOI] [PubMed] [Google Scholar]
- 21.Ogle JM, Murphy FV, Tarry MJ & Ramakrishnan V Selection of tRNA by the Ribosome Requires a Transition from an Open to a Closed Form. Cell 111, 721–732 (2002). [DOI] [PubMed] [Google Scholar]
- 22.Demeshkina N, Jenner L, Westhof E, Yusupov M & Yusupova G A new understanding of the decoding principle on the ribosome. Nature 484, 256–259 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Vitreschak AG, Mironov AA, Lyubetsky VA & Gelfand MS Comparative genomic analysis of T-box regulatory systems in bacteria. RNA 14, 717–735 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kalvari I et al. Non-Coding RNA Analysis Using the Rfam Database. Curr. Protoc. Bioinforma 62, e51 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henkin TM, Glass BL & Grundy FJ Analysis of the Bacillus subtilis tyrS gene: conservation of a regulatory sequence in multiple tRNA synthetase genes. J. Bacteriol 174, 1299–306 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grundy FJ, Yousef MR & Henkin TM Monitoring Uncharged tRNA During Transcription of the Bacillus subtilis glyQS Gene. J. Mol. Biol 346, 73–81 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Murakami H, Ohta A, Goto Y, Sako Y & Suga H Flexizyme as a versatile tRNA acylation catalyst and the application for translation. Nucleic Acids Symp. Ser 50, 35–36 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Golden BL, Chen J & Luptak A RIBOZYME WITH tRNA SYNTHETASE ACTIVITY AND METHODS OF MANUFACTURING AND USING THE SAME. US Pat. Appl (2016). [Google Scholar]
Methods References
- 29.Ke A & Doudna JA Crystallization of RNA and RNA–protein complexes. Methods 34, 408–414 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Otwinowski Z & Minor W Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997). [DOI] [PubMed] [Google Scholar]
- 31.Kabsch W Xds. Acta Crystallogr. Sect. D Biol. Crystallogr 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pape T et al. HKL2MAP : a graphical user interface for macromolecular phasing with SHELX programs. J. Appl. Crystallogr 37, 843–844 (2004). [Google Scholar]
- 33.Sheldrick GM et al. Experimental phasing with SHELXC / D / E : combining chain tracing with density modification. Acta Crystallogr. Sect. D Biol. Crystallogr 66, 479–485 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adams PD et al. PHENIX : a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Emsley P et al. Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guérout-Fleury A-M, Frandsen N & Stragier P Plasmids for ectopic integration in Bacillus subtilis. Gene 180, 57–61 (1996). [DOI] [PubMed] [Google Scholar]
- 37.Anagnostopoulos C & Spizizen J REQUIREMENTS FOR TRANSFORMATION IN BACILLUS SUBTILIS. J. Bacteriol 81, 741–6 (1961). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen L, James LP & Helmann JD Metalloregulation in Bacillus subtilis: isolation and characterization of two genes differentially repressed by metal ions. J. Bacteriol 175, 5428–37 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller JH Experiments in molecular genetics. (Cold Spring Harbor Laboratory, 1972) [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors for Mtb-ileS_tRNA-OH_native and Mtb-ileS_tRNA-cP_native have been deposited with the Protein Data Bank with the accession codes 6UFG and 6UFH, respectively. Source data for Figure 2b and 3f are available in Supplementary Table 3. Other data are available upon request.