Abstract
Monocytes are derived from hematopoietic stem cells through a series of intermediate progenitor stages, but the factors that regulate this process are incompletely defined. Using a Ccr2/Cx3cr1 dual-reporter system to model murine monocyte ontogeny, we conducted a small molecule screen that identified an essential role of mechanistic target of rapamycin complex 1 (mTORC1) in the development of monocytes and other myeloid cells. Confirmatory studies using mice with inducible deletion of the mTORC1 component Raptor demonstrated absence of mature circulating monocytes, as well as disruption in neutrophil and dendritic cell development, reflecting arrest of terminal differentiation at the granulocyte-monocyte progenitor (GMP) stage. Conversely, excess activation of mTORC1 through deletion of the mTORC1 inhibitor tuberous sclerosis complex 2 (Tsc2) promoted spontaneous myeloid cell development and maturation. Inhibitor studies and stage-specific expression profiling identified failure to downregulate the transcription factor Myc by the mTORC1 target ribosomal S6 kinase 1 (S6K1) as the mechanistic basis for disrupted myelopoiesis. Together, these findings define the mTORC1-S6K1-Myc pathway as a key checkpoint in terminal myeloid development.
One Sentence Summary
The mTORC1-S6K1 pathway controls terminal differentiation of myeloid progenitors via regulation of Myc.
Introduction
Monocytes are innate immune cells with pleiotropic functions including cytokine production, pathogen clearance, antigen presentation and wound healing (1–3). Studies over the past two decades have delineated two major subsets of monocytes in mice and humans. Inflammatory monocytes, characterized by high expression of the chemokine receptor Ccr2, are continuously released from the bone marrow into the circulation. These cells, alternately known as classical monocytes, are Ly6Chi in mice and correspond to the CD14hiCD16lo monocyte subset in humans; they migrate to inflamed tissues where they may reside as tissue monocytes or differentiate into macrophages (3–5). In contrast, patrolling monocytes represent a more differentiated subset and are marked by higher surface expression of Cx3cr1. Patrolling monocytes express low levels of Ly6C in mice and are CD14loCD16hi in humans; they survey the endothelium for evidence of inflammation or injury (6). Adoptive transfer and fate-mapping studies support the hypothesis that monocytes develop along a differentiation continuum in which inflammatory monocytes give rise to the patrolling subset in the circulation (7, 8).
Development of monocytes from bone marrow progenitors is a complex process that combines the regulated expression of numerous transcription factors with influence by growth factors and cytokines (9). Monocytes and neutrophils are both derived from hematopoietic stem cells (HSC) via common myeloid progenitors (CMP), but their differentiation bifurcates after the granulocyte-monocyte progenitor (GMP) stage (10). GMP give rise to monocyte-dendritic cell progenitors (MDP) and the downstream common monocyte progenitors (CMoP) that ultimately differentiate into Ly6Chi monocytes (11, 12). The transcription factors Spi-1 / PU.1, interferon regulatory factor-8 (Irf8) and the downstream Kruppel-like factor-4 (Klf4) are critical for the production of Ly6Chi monocytes in mice (13–15), while the orphan receptor Nr4a1 and macrophage colony stimulating factor (M-CSF) signaling are essential for the differentiation of Ly6Clo monocytes (16).
Evaluation of monocyte differentiation from myeloid progenitors is challenging due to the paucity of precursor cells and the potential complications introduced by ex vivo antibody-mediated cell sorting strategies. To overcome this limitation, we employed murine myeloid progenitors cells immortalized via conditional overexpression of Hoxb8, a homeobox gene that functions to suppress the myeloid differentiation program (17). We utilized Hoxb8 progenitors bearing fluorescent protein reporters for Ccr2 and Cx3cr1 expression as an experimental model to recapitulate the monocyte development continuum. Using this model to screen a small molecule library, followed by confirmatory in vivo validation strategies, we identified a previously unrecognized role for the mTORC1-S6K-Myc pathway in the regulation of terminal myeloid differentiation, thereby delineating a new pathway through which metabolism modulates immunity.
Results
Dual-reporter ER-Hoxb8 myeloid progenitor cells model normal monocyte differentiation
Transduction of bone marrow progenitors with murine stem cell virus (MSCV) encoding an estrogen-receptor–Hoxb8 fusion protein (ER-Hoxb8) results in the conditional expansion of myeloid progenitors capable of generating mature neutrophils and macrophages (18). In the presence of β-estradiol and stem cell factor (SCF), Hoxb8 functions to suppress myeloid differentiation and these cells proliferate in the myeloid progenitor state. We evaluated the utility of this system to model normal monocyte development. ER–Hoxb8 progenitors are Lin− Sca-1− c-kit+ FcγRII/III+ CD34+ at baseline (Fig. S1A), resembling the profile of CMP. Upon β-estradiol withdrawal, ER-Hoxb8 cells spontaneously differentiate into monocytes and neutrophils, with up-regulation of CD11b and Ly6C (Fig. S1B). Monocytes are distinguished by expression of M-CSF receptor (CD115). Undifferentiated ER–Hoxb8 cells resemble progenitor cells with a high nucleus-to-cytoplasm ratio and acquired monocyte and macrophage-like morphology by day 4 and 8 after estrogen withdrawal, respectively (Fig. 1A).
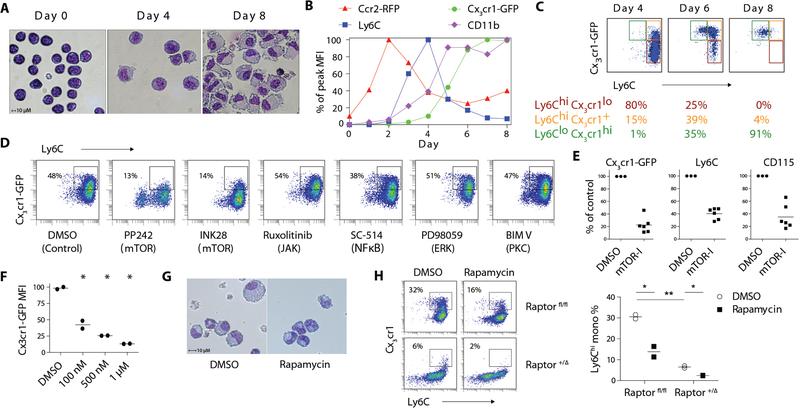
Figure 1. Small molecule library screen identifies an essential role of mTORC1 signaling in monocyte development.
A) Giemsa-Wright staining of ER-Hoxb8 cells before and after differentiation. B) Longitudinal FACS quantification of Cx3cr1-GFP, Ccr2-RFP, CD11b and Ly6C expression in DR-ER-Hoxb8 cells. MFI of each marker was normalized to the peak expression during the time course. C) FACS analysis of CD115+ DR-ER-Hoxb8 cells from day 4 to day 8 of differentiation. D) FACS plot of DR-ER-Hoxb8 cells differentiated in the presence of small molecule inhibitors. Boxes depict the percentage of Ly6Chi monocytes. E) MFI quantification of monocyte marker expression in DR-ER-Hoxb8 cells treated with mTOR inhibitors (mTOR-I) normalized to vehicle control (DMSO). Each data point for inhibitors (n = 6) represents a distinct compound. F) Dose-response curve of Cx3cr1-GFP expression in DR–ER–Hoxb8 cells treated with rapamycin (n = 2 per condition). G) Giemsa-Wright stain of DR-ER-Hoxb8 cells differentiated for 6 days in the presence of rapamycin (500 nM) or DMSO. H) FACS plots and quantification of monocyte production in Raptor fl/fl and +/Δ ER–Hoxb8 cells with or without rapamycin (500 nM; n = 2 per condition). Boxes indicate percentage of Ly6Chi monocytes. * p < 0.05
To study the dynamics of monocyte development and maturation, we generated dual-reporter ER–Hoxb8 (DR–ER–Hoxb8) cells from Ccr2RFP/+ Cx3cr1GFP/+ knock-in mice (19). DR–ER–Hoxb8 cells displayed low baseline red and green fluorescence (Fig. S1C). Longitudinal differentiation analysis revealed the coordinated, sequential expression of Ccr2-RFP, Ly6C and Cx3cr1-GFP following the removal of β-estradiol (Fig. 1B and Fig. S1D). By day 4, monocytes acquired a Ly6Chi phenotype with gradual up-regulation of Cx3cr1-GFP. A rapid increase in Cx3cr1-GFP expression by day 6 was accompanied by declining Ly6C expression, illustrating a maturation continuum from Ly6Chi to Ly6Clo monocytes (Fig. 1B–C).
In addition to their normal morphology and surface-marker expression, monocytes derived from ER-Hoxb8 cells also behaved as functional monocytes, as assayed by the production of proinflammatory cytokines and phagocytosis of E. coli (Fig. S1E). Neutralization of endogenous M-CSF showed minimal effects on the development of Ly6Chi monocytes but strongly inhibited the yield of Ly6Clo monocytes (Fig. S1F), consistent with the requirement of M-CSF for monocyte maturation (20). Correspondingly, the addition of exogenous M-CSF augmented the conversion of Ly6Chi to Ly6Clo monocytes (Fig. S1G). These features mimicked the biology of sorted bone marrow CMP that differentiated into monocytes with sequential expression of Ly6C and Cx3cr1, a process accelerated by M-CSF (Fig. S1H).
An advantage of ER-Hoxb8 cells is that they are amenable to genome editing at the CMP stage using techniques such as CRISPR (clustered regularly interspaced short palindromic repeats) / Cas. We generated ER-Hoxb8 cells from Cas9-EGFP transgenic mice (21) and delivered guide RNAs targeting EGFP or Ly6C via lentivirus. Protein expression of the targeted genes was markedly abrogated as assessed by flow cytometry (Fig. S1I), demonstrating effective genetic modification in ER-Hoxb8 cells using CRISPR/Cas technology. Taken together, these findings show that ER–Hoxb8 cells model the normal pattern of monocyte development, can easily be generated and expanded, and represent a convenient system in which to study murine monocyte ontogeny, in particular in dual-reporter cells expressing monocyte differentiation markers.
A kinase inhibitor screen in DR-ER-Hoxb8 cells identifies modulators of monocyte development
Given the predictable kinetics of DR–ER–Hoxb8 cell differentiation, we employed this system to identify modulators of monocyte development using a small molecule library of 152 kinase inhibitors (Table S1). Inhibitors of mechanistic target of rapamycin (mTOR) potently suppressed the expression of Ly6C, CD115 and Cx3cr1-GFP (Fig. 1D–E and Table S2). Inhibition of other kinases involved in common signaling pathways (e.g. Janus kinases, nuclear factor kappa b, c-Jun N-terminal kinases, protein kinase C and extracellular signal-regulated kinases) had minimal effect on monocyte development (Fig. 1D).
Two distinct mTOR complexes have been characterized, with distinct biological functions (22). Treatment of DR–ER–Hoxb8 cells with rapamycin, a prototypical partial inhibitor of mTORC1, delayed up-regulation of Ly6C and potently suppressed the expression of Cx3cr1 and CD115 (Fig. 1F and S1J). DR–ER–Hoxb8 cells differentiated in the presence of rapamycin were smaller and appeared less morphologically mature (Fig. 1G). Consistent with the findings from DR–ER–Hoxb8 cells, rapamycin reduced spontaneous monocyte differentiation from sorted CMP (Fig. S1K).
To further investigate the role of mTORC1 in monocyte development, we generated ER–Hoxb8 cells with hemizygous deficiency of regulatory-associated protein of mTOR (Raptor), a critical component of mTORC1 but not mTORC2 (Fig. S1L) (23). Monocytic differentiation towards the Ly6Chi subset from Raptor +/Δ ER–Hoxb8 cells was profoundly impaired and the effects were further amplified by blocking the residual mTORC1 activity with rapamycin (Fig. 1H).
mTORC1 is required for myeloid differentiation in vivo
To study the effects of mTORC1 ablation, we generated Raptor inducible KO mice (iKO; UBC-Cre/ERT2 Raptor fl/fl). Baseline distribution of monocyte subsets was comparable in the bone marrow, spleen and peripheral blood of untreated Raptor iKO and wild-type C57BL/6 mice (Fig S2A–B). Five days after tamoxifen-induced deletion of Raptor alleles, a profound reduction of Ly6Chi monocytes, but not Ly6Clo monocytes, was seen in the peripheral blood of iKO mice (Fig. 2A). Both monocyte subsets were substantially depleted in Raptor iKO mice 14 days post-tamoxifen, consistent with the expected development of patrolling from inflammatory monocytes. Neutrophils were also profoundly depleted during this time course (Fig 2B).
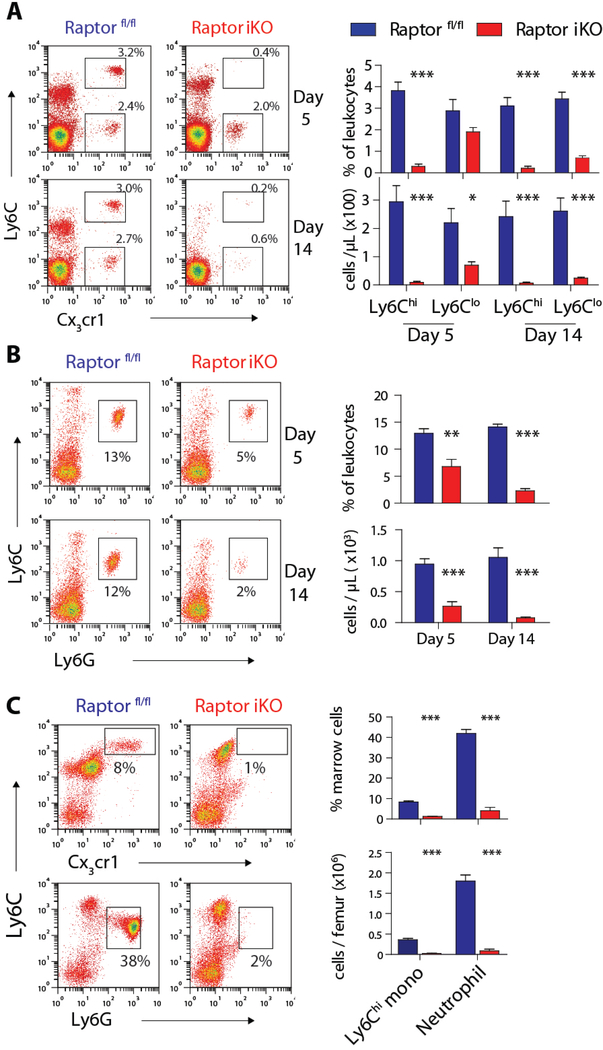
Figure 2. mTORC1 is required for the generation of monocytes and neutrophils in vivo.
A) FACS plots and quantification of peripheral blood monocyte subsets in Raptor fl/fl and Raptor iKO mice on day 5 and 14 after tamoxifen treatment. Boxes indicate Ly6Chi and Ly6Clo monocyte subsets (n = 5 per group). B) FACS plots and quantification of peripheral blood neutrophils in Raptor fl/fl and Raptor iKO mice after tamoxifen treatment. Boxes indicate neutrophils (n = 4–6 per group). C) FACS plot and quantification of bone marrow Ly6Chi monocytes and neutrophils in Raptor fl/fl and Raptor iKO mice (n = 5 per group). * p < 0.05, ** p < 0.01, *** p< 0.001
The depletion of peripheral blood myeloid cells in Raptor iKO mice might be explained by increased cell death or defective myeloid development. In contrast to the conditional deletion of the scaffold protein Mtor (25), which results in the loss of both mTORC1 and mTORC2 signaling, increased apoptosis was not seen in the bone marrow, peripheral blood or lymph nodes following the deletion of Raptor (Fig. S2C). These results implicate disrupted myeloid development as the explanation for the marked reduction of peripheral myeloid cells in Raptor iKO mice. Indeed, Raptor was effectively deleted in the majority of bone marrow cells (Fig. S2D) and the marrow pool of Ly6Chi monocytes and neutrophils were profoundly reduced in Raptor iKO mice (Fig. 2C).
Next, we analyzed bone marrow progenitor populations to study the defect in myelopoiesis. Consistent with previous studies (24, 26), c-kit+ progenitors were increased in Raptor iKO mice despite a reduction of total bone marrow cellularity by ~50%, with the largest difference seen in GMP and Lin− Sca-1+ c-kit+ (LSK) cells, which contains HSC and multipotent progenitors (Fig. 3A–C and S3A). GMP from Raptor iKO mice failed to generate MDP and CMoP based on the established markers for these progenitor populations in the absence of significant apoptosis, consistent with a developmental block at the GMP stage (Fig. 3C and S3B). The proportion of common lymphoid progenitors (CLP) and megakaryocyte-erythrocyte progenitors (MEP) were minimally affected in Raptor iKO mice Fig. 3C).
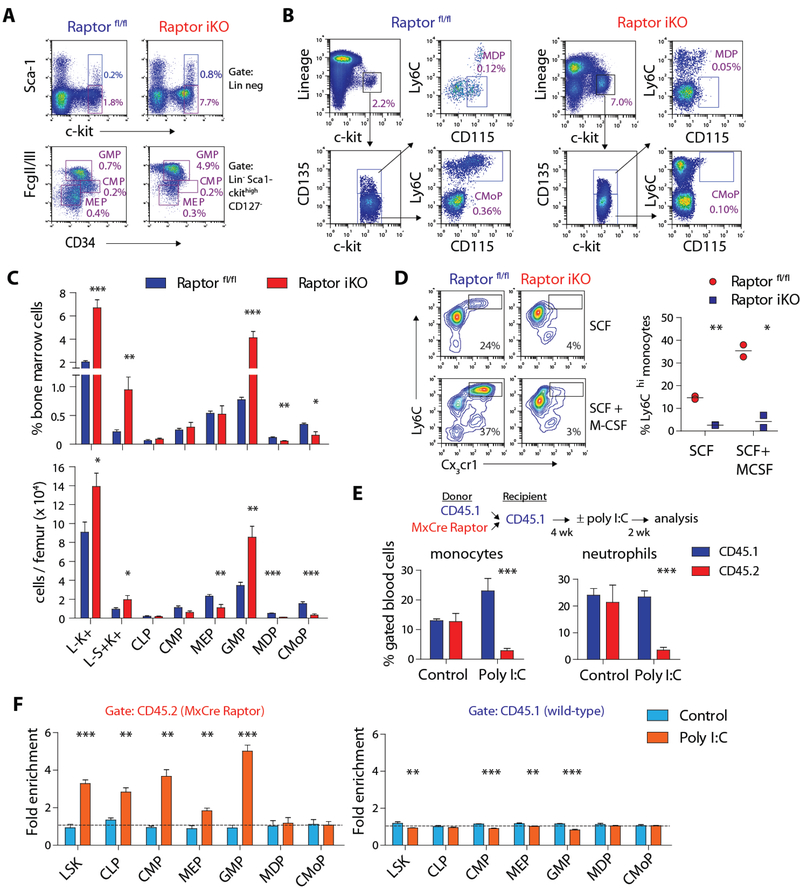
Figure 3. mTORC1 is critical for proper differentiation of myeloid progenitors.
A) FACS plots illustrating the analysis of LSK cells (Lin− Sca-1+ c-kit+), CMP (L-S-K+ CD127− FcγR II/IIImid CD34+), MEP (L-S-K+ FcγR II/IIIlo CD34-) and GMP (L-S-K+ FcγR II/IIIhi CD34lo). B) FACS plots depicting the analysis of MDP (L-S-K+ CD135+ Ly6Clo CD115+) and CMoP (L-S-K+ CD135− Ly6Chi CD115+). Boxes indicate the percentage of specified progenitor populations relative to the total bone marrow cell count. C) Relative frequency and absolute count of bone marrow progenitor populations in Raptor fl/fl and Raptor iKO mice (n = 5–6 per group). D) FACS plots and quantification of in vitro monocyte differentiation by sorted GMP from Raptor fl/fl and Raptor iKO mice in the presence SCF ± M-CSF (10 ng/mL) for 4 days (n = 2 per condition). Boxes indicate the percentage of Ly6Chi monocytes. E) Schematic of mixed chimera bone marrow transplant and quantification of peripheral blood monocytes (CD11b+Cx3cr1+) and neutrophils (Ly6G+) in transplanted mice (n = 3–6 per group). F) Quantification of bone marrow progenitor populations in transplanted mice with or without poly I:C treatment (n = 3–6 per group; see Materials and Methods for fold enrichment calculation). For all panels, mice were treated with tamoxifen or poly I:C two weeks before analysis. * p < 0.05, ** p < 0.01, *** p< 0.001
To assess whether the defect was intrinsic to progenitor cells, we examined in vitro differentiation of sorted bone marrow GMP from Raptor fl/fl and iKO mice treated with tamoxifen. GMP expressing Raptor spontaneously gave rise to Ly6Chi Cx3cr1+ monocytes when maintained in medium containing low levels of SCF, and the process was enhanced by addition of M-CSF (Fig. 3D). In contrast, Raptor-deficient GMP were unable to undergo normal monocyte differentiation under either condition.
Because GMP and MDP are also intermediate stages of dendritic cell (DC) development, we tested whether DC populations are affected in in Raptor iKO mice. Previous studies have demonstrated inhibitory effects of rapamycin on DC development and function (27, 28). Consistent with these findings, splenic CD8+, CD8−, and plasmacytoid DC subsets were all significantly reduced in Raptor iKO mice (Fig. S3C).
As expected from previous reports (24), Raptor iKO mice developed significant weight loss and ~50% mortality two weeks after tamoxifen treatment. To rule out the potential influence of poor health of Raptor iKO mice and chronic peritonitis from tamoxifen treatment, we generated Mx-Cre Raptor fl/fl mice as a second model of inducible Raptor deletion. Mx-Cre Raptor mice showed near complete gene deletion in hematopoietic cells after poly I:C (polyinosinic : polycytidylic acid) administration but did not display weight loss or early mortality (26), possibly due to more limited activation of Cre-recombinase in non-hematopoietic tissues (29). Importantly, the key findings in Raptor iKO mice including depletion of monocytes and neutrophils in the peripheral blood and bone marrow as well as the changes in progenitor cell populations were all replicated in Mx-Cre Raptor mice (Fig. S3D–F). Consistent with the observation by Kalaitzidis et al., an accumulation of CD11b+ Ly6Clo macrophages was seen in the bone marrow of Mx-Cre Raptor mice (Fig. S3F).
To determine whether these findings are intrinsic to Raptor-deleted cells, we created mixed chimeras by transplanting wild-type CD45.1 and Mx-Cre Raptor (CD45.2) bone marrow cells into CD45.1 recipients. The proportion of peripheral blood monocytes and neutrophils in CD45.1 and CD45.2 fractions were equivalent after engraftment (Fig 3E). Consistent with the effects of Raptor deletion, administration of poly I:C depleted CD45.2+ myeloid (Fig 3E and S3F). Assessment of bone marrow cells confirmed the expansion of early progenitor cells after Raptor deletion and but not downstream MDP and CMoP despite the accumulation of GMP (Fig 3F). Cell extrinsic effects were unlikely as wild-type (CD45.1+) progenitor cells were not enriched after poly I:C treatment.
Myeloid-specific deletion of Raptor does not affect homeostasis of differentiated myeloid cells
To quantify mTORC1 activity during myelopoiesis, we measured the phosphorylation of target substrates 4E-BP1 (eukaryotic initiation factor 4E-binding protein 1) and S6K1 (ribosomal protein S6 kinase-1) in various progenitor stages of monocyte development. The most prominent activity was found in the LSK fraction and decreased over the course of myeloid differentiation, reaching a nadir in Ly6Clo monocytes (Fig. 4A). In contrast, mTORC2 activity reflected by phosphorylation of Akt at Ser473 was relatively stable among progenitor cells.
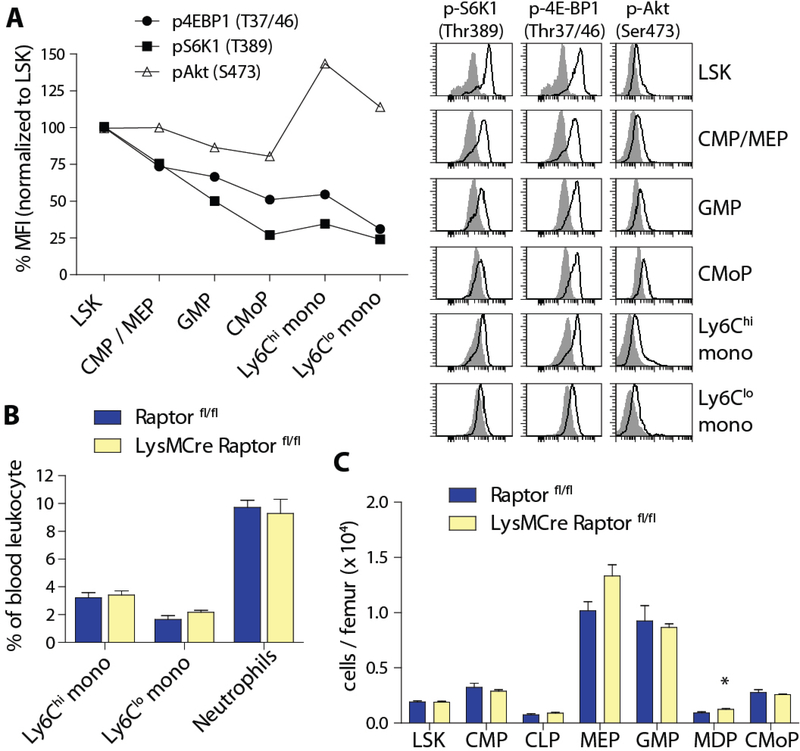
Figure 4. Raptor is dispensable in differentiated myeloid cells.
A) Histograms and quantification of phospho-4E-BP1 (Thr37/46), phospho-S6K1 (Thr389) and phospho-Akt (Ser473) in bone marrow progenitors and monocytes by intracellular FACS. Values represent mean MFI normalized to LSK cells after subtraction of background from isotype control staining (n = 2). B) Quantification of peripheral blood myeloid cells and C) bone marrow progenitor cells in LysMCre Raptor fl/fl mice and controls (n = 4–8 per group). * p< 0.05.
We next asked whether mTORC1 signaling is required for the homeostasis of myeloid populations after lineage specification. To address this question, we sorted bone marrow GMP and Ly6Chi monocytes from uninduced Raptor iKO mice and added tamoxifen to drive cre-mediated deletion in culture. Analysis after 4 days showed decreased expression of monocyte markers in tamoxifen-treated GMP, but not in Ly6Chi monocytes (Fig S4A).
In addition, we generated mice with myeloid-specific deletion of Raptor (LysMCre Raptor fl/fl) and confirmed effective gene deletion in bone marrow monocytes and neutrophils (Fig. S4B). Unlike the two models of inducible KO, Raptor fl/fl mice with heterozygous or homozygous LysMCre alleles did not show quantitative differences in monocyte and neutrophil populations in the peripheral blood, bone marrow or spleen (Fig 4B and Fig. S4C) compared to controls. The distribution of bone marrow myeloid progenitor subsets was also largely comparable to controls (Fig. 4C and S4D). Therefore, mTORC1 signaling is required for myeloid development but not essential for the survival or distribution of differentiated myeloid cell subsets.
Enhanced mTORC1 activity promotes myeloid development
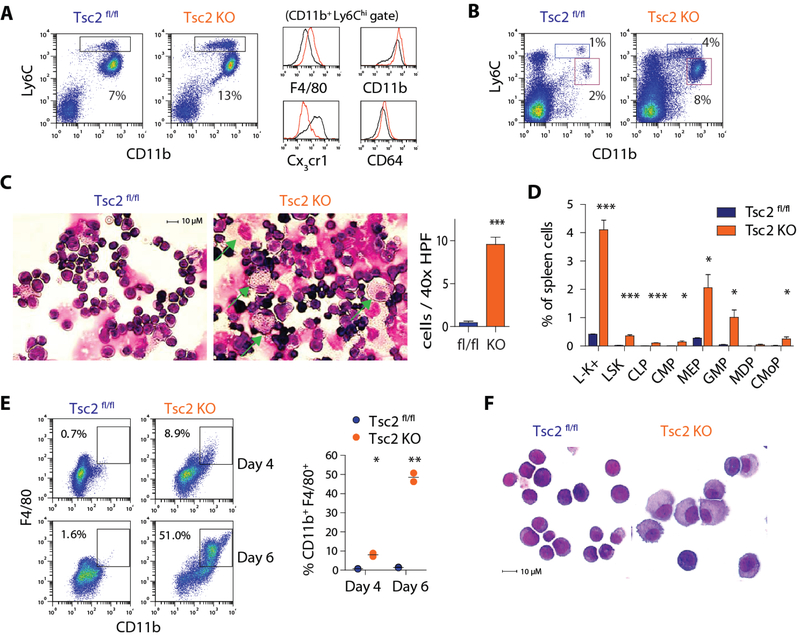
The tuberous sclerosis complex (Tsc) formed by the heterodimer of Tsc1 and Tsc2 proteins functions as a negative regulator of mTORC1 activity. We generated Mx-Cre Tsc2 fl/fl (Tsc2 KO) mice to ask whether mTORC1 over-activation promotes myelopoiesis. As expected, poly I:C-treated Tsc2 KO mice showed gene deletion in bone marrow cells with heightened mTORC1 signaling as illustrated by enhanced phosphorylation of ribosomal protein S6 and 4E-BP1 (Fig. S5A–B). Interestingly, the proportion of Ly6Chi monocytes, but not other lineages, was increased in the bone marrow of Tsc2 KO mice (Fig. 5A and S5C). These monocytes displayed greater surface expression of F4/80, CD11b, and FcγRI (CD64) but lower expression of Cx3cr1 (Fig. 5A), indicative of a macrophage-like phenotype. Examination of bone marrow progenitor populations revealed that LSK cells and GMP were expanded after Tsc2 deletion while the proportion of MDP and CMoP were unchanged (Fig. S5C).
Figure 5. Deletion of Tsc2 enhances mTORC1 signaling and augments myelopoiesis.
A) FACS plots of bone marrow Ly6Chi monocytes (box) in Tsc2 KO and controls. Histograms depict myeloid marker expression in Ly6Chi monocytes. B) FACS plots of isolated spleen cells from Tsc2 KO and controls. Boxes indicate Ly6Chi monocytes (blue) and neutrophils (red). C) Giemsa-Wright staining of isolated spleen cells and quantification of large macrophages (arrow; n = 9 HPF counted per group from 3 animals). D) Quantification of splenic progenitor cell populations in Tsc2 KO and controls (n = 3 per group). E) FACS analysis and F) Giemsa-Wright staining of macrophages differentiated from sorted LSK cells from control or Tsc2 KO mice maintained in SCF-containing medium (10 ng/mL; n = 2 per condition). * p < 0.05, ** p < 0.01, *** p< 0.001
While the absence of Tsc2 did not affect the distribution of monocyte subsets in the peripheral blood (Fig. S5D), Tsc2 KO mice developed splenomegaly with a striking expansion of myeloid cells (Fig. 5B and Fig. S5E–F). Ly6Chi monocytes and neutrophils were both increased by four-fold while lymphocytes were reduced. Cytospin of spleen cells from Tsc2 KO mice further revealed large macrophages that were not evident in controls (Fig. 5C). We assessed whether extramedullary myelopoiesis was responsible for the myeloid skew in the spleen of Tsc2 KO mice. In control animals, c-kit+ progenitor cells were rare (~0.3%) and most resembled megakaryocyte-erythrocyte progenitors (MEP) by surface marker expression (Fig. S5F–G). Tsc2 KO mice displayed a 10-fold expansion of splenic Lin− c-kit+ progenitor cells (~3%) with a prominent increase of GMP (Fig. 5D). Importantly, downstream monocyte progenitors including MDP and CMoP were found in the spleen of Tsc2 KO, but not controls, illustrating the full continuum of monocyte progenitors normally seen in the bone marrow.
To understand how excess mTORC1 signaling in progenitor cells directly influences myelopoiesis, we studied the differentiation of sorted LSK cells in vitro. When cultured in the presence of SCF alone to support stem cell proliferation, LSK cells from Tsc2fl/fl control did not generate mature myeloid cells after 6 days, while those from Tsc2 KO spontaneously gave rise to CD11b+ F4/80+ cells in the absence of additional growth factors (Fig. 5E). Similarly, cytospin analysis revealed mostly immature cells expanded from control LSK cells and large macrophages from Tsc2 KO counterparts (Fig. 5F). The accelerated myeloid differentiation in Tsc2-deficient LSK cells was entirely dependent on the mTORC1 pathway and effectively reversed by rapamycin treatment (Fig. S5H). In contrast, no difference was seen with inhibition of JNK, which is known to mediate mTORC1-independent Tsc1 signaling (30). The development of macrophages from Tsc2 KO LSK cells was not affected by neutralizing antibodies to M-CSF or GM-CSF (Fig. S5I), and co-culture with Tsc2 KO LSK did not affect the differentiation of wild-type LSK (Fig. S5J), consistent with a cell-intrinsic mechanism.
The effect of mTORC1 on myelopoiesis is mediated by the S6 kinase pathway
Functionally, mTORC1 is a serine / threonine kinase that phosphorylates effector molecules involved in the regulation of protein synthesis, lipid synthesis, mitochondrial metabolism and autophagy (22). Phosphorylation of 4E-BP1 by mTORC1 leads to its dissociation from eIF4E to allow cap-dependent translation, while S6K1 is itself a serine / threonine kinase with critical roles in transcription, translation and ribosomal protein synthesis (31).
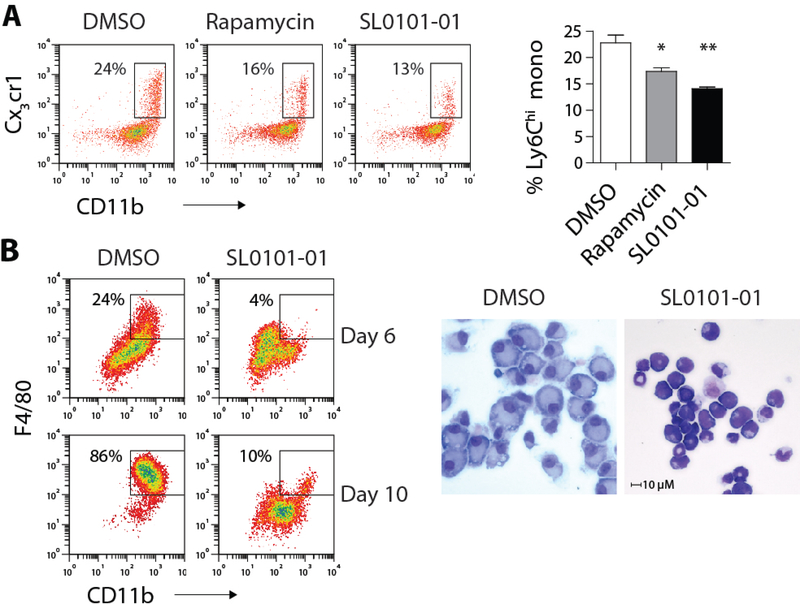
Rapamycin is a partial inhibitor of mTOR with differential effects on S6K1 and 4E-BP1 phosphorylation depending on the cell type (32). ER-Hoxb8 cells treated with rapamycin showed lasting reduction in S6 phosphorylation, a functional read-out of S6K1 activity, while phospho-4E-BP1 level was unaffected (Fig. S6A), suggesting that the S6K1 pathway is the likely target of mTORC1 that licenses myeloid development. To isolate the effects of S6K1 downstream of mTORC1, we utilized the ribosomal S6K inhibitor SL0101–01, which inhibited S6 phosphorylation without affecting 4E-BP1 phosphorylation in ER-Hoxb8 cells (Fig. S6B). Indeed, SL0101–01 was nearly equipotent to rapamycin in blocking monocyte development from isolated CMP and ER-Hoxb8 cells (Fig. 6A and S6C). The same pathway was responsible for the augmented myeloid development in Tsc2 KO mice, as SL0101–01 prevented the development of macrophages from Tsc2-deficient LSK cells (Fig. 6B). The findings collectively show that steady-state myelopoiesis is controlled by mTORC1 via activation of the S6K1 pathway.
Figure 6. mTORC1 activates S6K1 to promote myelopoiesis.
A) FACS plots and B) quantification of sorted CMP differentiated in the presence of DMSO, rapamycin (100 nM) or SL0101–01 (10 nM) for 4 days (n = 3 per condition). Box indicates the percentage of Ly6Chi monocytes. B) FACS plots and Giemsa-Wright stain of Tsc2 KO LSK cells differentiated in the presence of DMSO or SL0101–01 for 6 days. * p < 0.05
mTORC1 modulates myeloid development via regulation of c-Myc expression
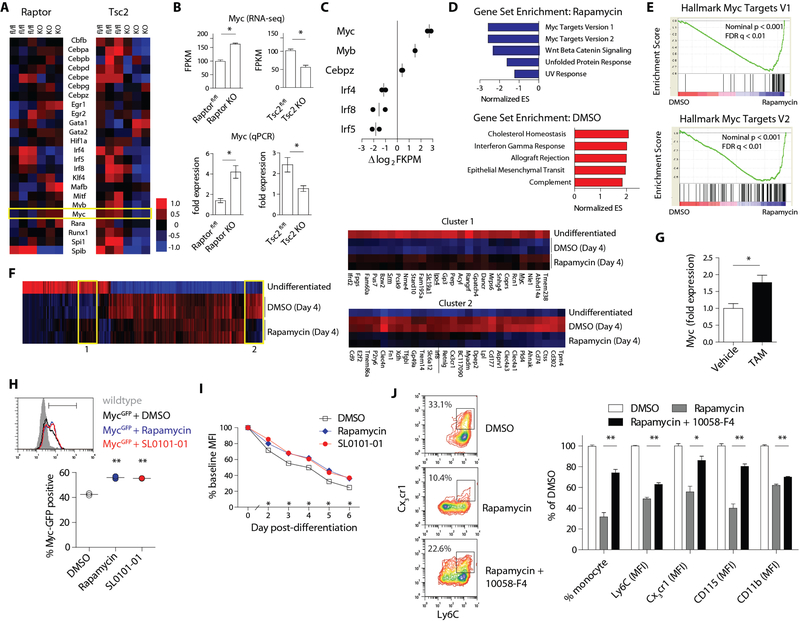
To understand how mTORC1-S6K1 signaling modulates myelopoiesis, we performed transcriptomic analysis of GMPs from Raptor-deficient, Tsc2-deficient and controls mice using RNA sequencing (RNA-seq). We focused the analysis on transcription factors that regulate myeloid cell development (Fig. 7A) and statistically significant comparisons are provided in Fig S7A. Interestingly, Myc (c-Myc) was the only transcription factor reciprocally regulated by the deletion of Raptor and Tsc2. Increased Myc expression in Raptor-deficient GMP and downregulation in Tsc2 KO were confirmed by quantitative PCR (Fig. 7B).
Figure 7. mTORC1 modulates Myc expression during myeloid development.
A) Heat-map display of myeloid transcription factor expression by RNA-seq in GMP from MxCre Raptor, MxCre Tsc2 and control animals (n = 3 per strain). B) Confirmation of Myc expression data from RNA-seq by qPCR (n = 3 per strain). C) Differentially regulated myeloid transcription factors in ER-Hoxb8 cells differentiated for 4 days in the presence of DMSO vs. rapamycin (100 nM; n = 2 per condition). D) Top 5 enriched gene sets in DMSO vs. rapamycin-treated ER-Hoxb8 cells (day 4 of differentiation) based on normalized enrichment score (NES) from GSEA. E) Enrichment plot of Myc gene sets from GSEA. F) Hierarchical cluster analysis of gene expression in undifferentiated vs. differentiated ER-Hoxb8 cells (DMSO or Rapamycin-treated; n = 2 per group). Left panel: heat map of 385 genes differentially expressed (≥ 2 fold) among the groups. Two clusters were selected for detailed analysis (right panels) and Myc and its target genes are underlined in Cluster 1. G) qPCR analysis of Myc expression in sorted CMP from Raptor iKO mice treated with vehicle (methanol) or 4OH-tamoxifen (1 μM) for 3 days (n = 3 per group). Quantification of GFP expression in H) sorted CMP from MycGFP mice and I) MycGFP ER-Hoxb8 cells cultured in the presence of DMSO, rapamycin (100 nM) or SL0101–01 (10 nM) (n = 2 per condition). J) FACS analysis and quantification of surface marker expression in ER-Hoxb8 cells treated with DMSO, rapamycin (100 nM), or rapamycin + 10058-F4 (50 nM). Box in FACS plots indicate Ly6Chi monocytes (n = 3 per condition). * p < 0.05, ** p < 0.01
Myc is an oncogene with well-established role in controlling stem cell proliferation and differentiation through transcriptional regulation (33). Overexpression of Myc is a hallmark of tumorigenesis (34) and downregulation of Myc is required for terminal myeloid differentiation (35). To study whether mTORC1 signaling directly modulates Myc expression, we performed RNA-seq in ER-Hoxb8 cells differentiated in the presence of rapamycin or vehicle control. Supporting the data from Raptor-deficient GMPs, pharmacologic inhibition of mTORC1 resulted in upregulation of Myc and decreased expression of Irf4, Irf5 and Irf8 (Fig. 7C). Moreover, gene set enrichment analysis (GSEA) using the Hallmark signatures (36) showed significant enrichment of Myc target genes in rapamycin-treated cells, confirming enhanced Myc transcriptional activity (Fig. 7D–E and Fig. S7B). In line with the downstream role of S6K1, treatment with SL0101–01 yielded similar upregulation of Myc and enrichment of Myc target gene transcripts (Fig. S7C–F)
To study the expression kinetics of Myc and its gene targets during myelopoiesis, we incorporated undifferentiated and differentiated ER-Hoxb8 cells in hierarchical clustering analysis (Fig. 7F). Myc and several of its target genes (Slc19a1, Ipo4, Srm) clustered with transcripts that are normally diminished during differentiation but the downregulation is partially reversed by rapamycin (Cluster 1). This pattern suggests that intact mTORC1 signaling is required for proper downregulation of the Myc transcription program during myelopoiesis. In contrast, Cluster 2 contains genes that are normally upregulated during myeloid development but inhibited by rapamycin. Not surprisingly, this group was highlighted by markers of myeloid maturation including Cx3cr1, Irf8, Treml4, Mmp9 and members of the clec4 family (Fig. 7F).
Consistent with mTORC1-mediated suppression of Myc expression, tamoxifen treatment of CMP from untreated Raptor iKO mice showed upregulated expression of Myc (Fig. 7G). Moreover, CMP expanded from MycGFP reporter mice (37) showed enhanced GFP expression in the presence of mTORC1 or S6K1 inhibitor (Fig. 7H and S7G). ER-Hoxb8 cells generated from MycGFP mice similarly displayed enhanced GFP expression under these conditions (Fig. 7I and S7H).
Finally, we investigated whether concomitant inhibition of Myc can overcome the effects of mTORC1 blockade on myeloid development. ER-Hoxb8 cells differentiated in the presence of rapamycin were treated with 10058-F4, a small molecule inhibitor of Myc (38). Indeed, the potent inhibitory effects of rapamycin on monocyte development were markedly inhibited by the addition of 10058-F4 (Fig. 7J). Myc blockade improved the yield of Ly6Chi monocytes and enhanced the surface expression of monocyte markers. Importantly, treatment with 10058-F4 did not alter mTOR-S6K1 signaling (Fig. S7I). These data collectively show that mTORC1 modulates myeloid development, and demonstrates an integral role for S6K1-mediated regulation of Myc expression in this process.
Discussion
mTOR complexes regulate a spectrum of essential biological functions including energy metabolism, growth factor signaling, immune cell activation and cell proliferation (22, 39). A balance of mTOR signaling appears to be essential for hematopoiesis, as either over-activation or disruption of this pathway can result in HSC exhaustion and bone marrow failure (25, 40). We now identify a specific requirement for mTORC1 signaling in terminal myeloid development.
Our data indicate that mTORC1 signaling is critical for proper myeloid differentiation at or prior to the GMP stage, as downstream differentiation of inflammatory monocytes is profoundly impaired in Raptor iKO mice. Disruption of mTORC1 signaling via conditional deletion of Raptor resulted in an accumulation of GMP that fail to generate MDP and CMoP. On the other hand, overactivation of mTORC1 via Tsc2 deletion augmented myeloid development with prominent extramedullary myelopoiesis. An expansion of HSC in the bone marrow and spleen was previously described in Tsc1-deficient mice (41), but myeloid cells appear to be reduced and monocytes were not studied in detail.
While our study is focused on monocyte development, a myeloid differentiation defect at the GMP stage would predict defective development of neutrophils and DCs since they are also derived from GMP. Consistent with previous studies using Raptor iKO and Mtor hypomorphic mice (24, 42), we found marked reduction in bone marrow and peripheral blood neutrophil count in both models of inducible Raptor KO mice. DC development in vivo and in vitro was also substantially impaired in these mice. A differentiation defect at the progenitor level likely explains the negative effects of rapamycin on neutrophil and dendritic cell development in vitro (27, 28, 43).
In vitro deletion of Raptor also led to impaired differentiation at the GMP level but had no detectable phenotype in sorted Ly6Chi monocytes. Correspondingly, mice with myeloid-specific deletion of Raptor (LysMCre) exhibited normal numbers of monocytes in the bone marrow and periphery, delimiting the requirement for mTORC1 to early myeloid development. Correspondingly, DC-specific deletion of Raptor (via CD11c-Cre) was shown not to deplete this lineage but rather to alter DC subset distribution (44). These studies, together with the sequential depletion of Ly6Chi and Ly6Clo monocyte subsets we observe with Raptor iKO, implicate mTORC1 in development but not survival of myeloid cells under the conditions studied.
Our data further demonstrate a role of mTORC1-S6K1 signaling in the physiologic downregulation of Myc transcription during myelopoiesis. This view contradicts an earlier study that showed accelerated retinoid-induced granulocyte development by mTOR inhibition due to suppression of Myc (45). While the in vitro systems cannot be directly compared, our in vivo findings clearly support a critical role of mTORC1 in myeloid development, and Myc expression was reciprocally upregulated Raptor iKO GMPs and decreased in Tsc2 KO GMPs. Furthermore, mTORC1 or S6K inhibitors both enhanced the expression of Myc and its target genes and simultaneous inhibition of Myc ameliorated blockade of myeloid development.
Additional roles of mTORC1 in myelopoiesis remain possible, as mTORC1 may serve as a checkpoint for the availability of nutrients to support downstream myeloid differentiation and effector function. Regulation of metabolic enzymes at both transcriptional and posttranscriptional levels has been observed during early stages of myeloid commitment (46, 47). While defects in glucose metabolism have been associated with congenital neutropenia (48), such association with monocyte development has not been reported. Beyond a role in myeloid development, the mTORC1-S6K1 axis regulates many immune functions of differentiated myeloid cells (39). Metabolic reprogramming by mTOR is required for the establishment of trained immunity in monocytes (49). A regulatory role of mTOR and S6K1 in macrophages has been described in the context of Toll-like receptor signaling (50, 51). Proinflammatory features associated with mTORC1 activation are also well-described (39) as this pathway mediates the production and/or downstream effects of inflammatory cytokines including IL-6, tumor necrosis factor alpha and type I interferons (52, 53).
Our studies did not specifically address a role for mTORC2 in monocyte development. Conditional deletion of the mTORC2-specific component Rictor (rapamycin insensitive companion of TOR) results in negligible effects in the setting of steady-state myelopoiesis (26, 54). Unlike mTORC1, which is most active in early progenitor stages, mTORC2 activity as measured by phosphorylation of Akt (Ser473) appears to be more abundant in differentiated monocytes. Nevertheless, Rictor / mTORC2 has a role in macrophage polarization, as myeloid-specific Rictor deficiency favors M1 skewing and enhances the inflammatory response to LPS (55).
As illustrated here, the ER–Hoxb8 system can be adapted to address a variety of questions surrounding early myelopoiesis and immune cell function. Immortalized murine myeloid progenitors can be created from the bone marrow, fetal liver, or embryonic stem cell populations and large number of cells can be generated and maintained with ease (18, 43). Our work employed the ER–Hoxb8 technology to study monocyte development via fluorescent reporters for the expression of Ccr2 and Cx3cr1, in a system amenable to small molecule screening studies, conditional gene deletion and genomic manipulations via CRISPR/Cas. Our findings confirm that results obtained using this system reliably predict the consequences of pathway disruption in vivo, thereby representing a powerful and flexible tool to understand myeloid biology.
Taken together, these results identify a previously unrecognized role for the mTORC1-S6K1-Myc axis in myeloid development. Disruption of this pathway abrogates development beyond the GMP stage, resulting in sequential depletion of circulating inflammatory and then patrolling monocytes, as well as neutrophils and dendritic cells. By contrast, mTORC1 has no obligatory role in the survival of more mature cells, at least under basal conditions. These finding therefore establish the mTORC1-S6K1-Myc pathway as a critical checkpoint in terminal myeloid development.
Material and Methods
Study Design
The goal of this study is to understand the role of mTORC1 in myeloid cell development. These studies were approved by the IACUC of the Dana Farber Cancer Institute and Brigham and Women’s Hospital. Pilot experiments were performed to determine the number of animals needed to ensure statistical power. For all animal studies, each experimental group contained three to eight biological replicates. At least two biological replicates were performed for all in vitro experiments. No randomization or blinding was performed. All results were confirmed by two or more independent experiments.
Reagents
The Kinase Screening Library, other chemical inhibitors and tamoxifen were purchased from Cayman Biochem. Recombinant IL-3 and SCF were purchased from Peprotech. Recombinant M-CSF was obtained from Biolegend. β-estradiol, 4-OH tamoxifen, Tween 80 and PEG-400 were purchased from Sigma-Aldrich. Poly I:C was purchased from GE Healthcare Life Sciences. All antibodies used in this study and their sources are listed in Table S3.
Animals
Wild-type C57BL/6, Ccr2RFP reporter (B6.129(Cg)-Ccr2tm2.1Ifc/J), Cx3cr1GFP reporter (B6.129P-Cx3cr1tm1Litt/J), CD45.1 (B6.SJL-Ptprca Pepcb/BoyJ), Raptor fl/fl (B6.Cg-Rptortm1.1Dmsa/J), ubiquitin-ERT2-cre (B6.Cg-Tg(UBC-cre/ERT2)1Ejb/1J), Mx1-Cre (B6.Cg-Tg(Mx1-cre)1Cgn/J), and LysM-Cre (B6.129P2-Lyz2tm1(cre)Ifo/J) mice were obtained from Jackson Laboratories. Femurs from MycGFP mice (B6;129-Myctm1Slek/J) were also obtained from Jackson Laboratories. Ccr2RFP/+ Cx3cr1GFP/+ (dual reporter) mice, Cre/ERT2 Raptor fl/fl mice (Raptor iKO), Mx1-Cre Raptor fl/fl mice and LysMCre Raptor fl/fl mice were generated as described (19, 26, 56). Tsc2 fl/fl (Tsc2tm1.1Mjg/J) mice were kindly provided by Dr. Michael Gambello (57) and bred with Mx1-Cre mice to generate Tsc2 KO mice. Male and female mice age between 6 to 8 weeks were used with matched littermate controls.
Conditional gene deletion
To induce deletion of Raptor, 6 week old Raptor iKO mice were treated with tamoxifen (75mg/kg/day; solubilized in corn oil) i.p. daily for three consecutive days. To induce gene deletion in Mx1-Cre Raptor fl/fl and Mx1-Cre Tsc2 fl/fl mice, 6 week-old mice were given poly I:C (250 μg i.p.) every other day for 3 doses. Wild type, floxed, and deleted alleles of Raptor were determined by conventional PCR using the primers: GCGCAGTGAGTACTGTTACCACCATG and GGGTACAGTATGTCAGCACAG. PCR Analysis of Tsc2 alleles were performed using the primers: AAGATTCCGGCTTGAAGGAG, CACTAGTCTAGCCTGACTCT, and GAGGACAAGCCAACATCCAT.
Bone marrow transplant
Recipient CD45.1 mice were irradiated using two split doses of 500 rads and given 2 × 106 bone marrow mononuclear cells from CD45.1 mice and CD45.2 Mx-Cre Raptor fl/fl donors via intravenous injection. Engraftment was confirmed by FACS analysis of PBMC after 4 weeks followed by induction of gene deletion by poly I:C. For analysis of bone marrow progenitor cells, fold enrichment of CD45.2 (or CD45.1) progenitor subsets was determined by dividing the CD45.2+ (or CD45.1+) percentage of each progenitor subset by the CD45.2+ (or CD45.1+) percentage of all bone marrow cells.
ER–HoxB8 cell lines
Detailed protocols for construction of the retroviral vector encoding ER–Hoxb8, virus production, and transduction of bone marrow myeloid progenitor cells were described previously (18). In brief, bone marrow mononuclear cells were isolated and expanded in medium containing SCF and IL-3 for 48 h. Cells were placed in 12-well plates pre-coated with human fibronectin (BD Biosciences) and spin-infected with murine stem cell virus encoding ER–HoxB8. Infected cells were cultured in RPMI medium containing SCF (20 ng/mL) and β-estradiol (500 nM) for two days prior to addition of G418 for selection. The cells were passaged to new plates with fresh medium every 3–4 days, and non-adherent immortalized cells grew out in ~3 weeks. Raptor +/Δ ER-Hoxb8 cells were generated using bone marrow cells from Ubc-Cre-ERT2 Raptor +/fl mice and Cre-recombinase was activated by the presence of estrogen in culture medium.
Genome editing by CRISPR / Cas
Hoxb8 cells with constitutive expression of Cas9-EGFP were derived from bone marrow cells of Cas9-EGFP transgenic mice (21). Lentivirus encoding gRNA for EGFP and Ly6C were generated by the Broad Institute Genetic Perturbation Platform using the pXPR_003 vector. Sequences of gRNA are EGFP #1: GAAGTTCGAGGGCGACACCC; EGFP #2: GGTGAACCGCATCGAGCTGA; Ly6C #1: GCTGGGCAGGAAGTCTCAAT; and Ly6C #2: AGCAATGCAGAATCCATCAG. Cas9-EGFP Hoxb8 cells were spin-infected with gRNA lentivirus (multiplicity of infection 10) and selected in puromycin for 1 week prior to differentiation and evaluation of gene expression.
Cell culture
Immortalized ER–Hoxb8 cells were cultured in “maintenance medium” with 10% FCS, penicillin-streptomycin, 4% SCF conditioned medium from stably-transfected CHO cells (~ 20 ng/mL of SCF) and β-estradiol (500 nM) in RPMI 1640. Prior to differentiation, cells were pelleted by centrifugation and washed with PBS twice before resuspension in “differentiation medium” lacking β-estradiol and containing 10% FCS, penicillin-streptomycin and 0.4% SCF conditioned medium (~ 2 ng/mL) in RPMI. For all experiments, day 0 was defined as the day of estrogen withdrawal. For Cre-recombinase activation in vitro, sorted bone marrow cells were cultured in complete RPMI containing 4OH-tamoxifen (1 μM) and SCF (10 ng/mL) for 3 days prior to differentiation with or without M-CSF (10–20 ng/mL) to generate monocytes. For measurement of intracellular cytokine production, LPS (1 μg/mL) was added to ER–Hoxb8 cells on day 4 of differentiation in the presence of monensin (Biolegend, 1:1000 dilution). Cells were collected after 16 hours for intracellular flow cytometry analysis. Phagocytosis assay was performed by co-culturing ER-Hoxb8 cells (day 4 of differentiation) with fluorescein-conjugated Escherichia coli (K-12 strain; Invitrogen) at a cell to target ratio of 1:100 for 30 minutes in RPMI medium. Cells were washed with PBS before FACS analysis.
Small molecule screening and cytokine stimulation
DR–ER–HoxB8 cells were placed in 96 well plates (5×104 cells/well) in differentiation medium after estrogen removal. For small molecule screening, individual compound from the Cayman Kinase Screening Library were added to final concentrations of 10 μm and 1 μM on day 0. For compounds that induced > 50% cell death at the initial concentrations, further dilutions to 100 nM, 10 nM and 1 nM were analyzed on repeat experiments. The expression of Ccr2-RFP, Cx3cr1-GFP, Ly6C, CD11b and CD115 were measured by flow cytometry and expressed as percentage of MFI relative to DMSO-treated controls.
Flow cytometry
Before surface staining, cells were incubated with anti–mouse CD16/32 (Fc block) for 10 min. Cells were then stained with an optimized amount of primary antibody or the appropriate isotype control for 15 min at room temperature before washing and resuspending in PBS supplemented with 0.1% BSA. Intracellular staining was performed after surface staining using fixation and permeabilization buffer (Ebioscience). Antibodies were added to cells after the permeabilization step and samples were incubated for 1 hour on ice before analysis. Samples were acquired using a Becton-Dickinson FACS Canto II flow cytometer and analyzed with FCS Express 5 software (De Novo Software). In all FACS experiments, live cells were identified by size and singlets were gated for analysis. Isotype control antibodies were used to establish gates for specific cell populations. Apoptosis assays were performed using the FITC Annexin V Apoptosis Detection Kit with PI (Biolegend) following manufacturer’s instructions.
FACS sorting
Bone marrow cells were isolated and stained with antibodies as described above. After lysis of erythrocytes in ACK buffer, cells were washed in PBS supplemented with 0.1% BSA and resuspended in complete RPMI. Bone marrow progenitors and myeloid cells were sorted using a BD FACS Arias III cell sorter using defined gating strategies (11, 12) (Fig 4 and S2). For RNA sequencing, cells are sorted directly into RLT buffer. To culture stem / progenitor cells, sorted cells were cultured in RPMI with 10% FCS, penicillin-streptomycin, 4% SCF conditioned medium from stably-transfected CHO cells.
RNA sequencing
RNA was extracted from sorted cells (1 × 104) using Qiagen RNAeasy micro kit. RNA-seq was performed using the Smart-seq2 platform (58, 59). Smart-Seq2 libraries were prepared by the Broad Technology Labs and sequenced by the Broad Genomics Platform. Transcripts were quantified by the BTL computational pipeline using Cuffquant version 2.2.1 (60). Transcriptions factors with known roles in myeloid development were selected for focused analysis (9, 61). Gene set enrichment analysis was performed using software v3.0 (Broad Institute) with the Molecular Signatures Database (MSigDB) hallmark gene set collection (36, 62). Hierarchical clustering analysis was performed using Cluster 3.0 (63) and heat maps were was constructed using Java Treeview (64).
Quantitative PCR
Total RNA was extracted from sorted cells using RNAeasy Micro kit, and cDNA was synthesized using the Superscript III First-Strand Synthesis Kit (Thermo Fisher). Q-PCR was performed using RT2 SYBR green master mix (Qiagen) with a StepOne thermocycler (Thermo Fisher). Amplification conditions were: 95°C for 10 min, followed by 45 cycles of 94°C for 15 seconds and 60°C for 1 minute. After the final extension (72°C for 10 min), a melting-curve analysis was performed to ensure specificity of the products. Gene expression was normalized to 18S RNA, and expression relative to the sample with the lowest expression was calculated using the 2−ΔΔCt method. Primers used in this study were all described previously (65) with the exception of Myc (forward: TTGAGGAAACGACGAGAACAG; reverse: AGCCAAGGTTGTGAGGTTAG).
Statistical analyses
For all bar graphs, bars represent mean ± SEM. For quantitative variables, differences between two groups were analyzed by the unpaired Student’s t test and one-way ANOVA was used for comparison of multiple groups. All tests were two-sided, and p < 0.05 was considered significant. Statistical analyses were performed using Prism 5.0 software (GraphPad Software).
Supplementary Material
Fig. S1. Phenotypic and functional analysis of DR–ER–Hoxb8 cells and their response to mTOR inhibition.
Fig. S2. Raptor deletion impairs monocyte development without causing apoptosis
Fig. S3. Impaired development of myeloid progenitors in Raptor-deficient mice
Fig. S4. Myeloid-specific deletion of Raptor does not affect monocyte or neutophil homeostasis
Fig. S5. Deletion of Tsc2 augments myelopoiesis via mTORC1 activation
Fig. S6. mTORC1 modulates myeloid development via S6K1
Fig. S7. mTORC1-S6K1 signaling modulates the expression of Myc and its target genes
Fig. S8. Complete images from gel electrophoresis
Table S1. Results from small molecule library screen using DR-ER-Hoxb8 cells
Table S2. Effects of mTOR inhibitors on monocyte marker expression in DR-ER-Hoxb8 cells
Table S3. Table of antibodies
Table S4. Compilation of raw data and analysis
Acknowledgements
We thank Dr. Mark Kamps (University of California San Diego) for providing Hoxb8 retroviral constructs, Dr. Randall Platt and Dr. Feng Zhang (Broad Institute) for providing bone marrow cells from Cas9-EGFP transgenic mice, Drs. Michael Gambello (Emory University) and Elisabeth Henske for providing Tsc2 fl/fl mice, and Drs. Victor Hsu and Tiffany Horng for helpful discussions.
Funding: This work was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases R01AR065538 (PAN), P30AR070253 (PAN), The Broad Institute-Boston Children’s Hospital Collaborative Grant (PAN), K08AR062590 (JFC), the Rheumatology Research Foundation Scientist Development Award (PYL), the Frederick Lovejoy Research Award of Boston Children’s Hospital (PYL), and National Institute of General Medical Sciences SC1GM095426 (AEC). PYL and KW are National Institutes of Health T32 trainees (T32AI007512 and T32AR007530, respectively). DK was supported by NIH NIDDK award (K01DK092300). PAN was supported by the Cogan Family Foundation and the Fundación Bechara.
Footnotes
Competing Interests: The authors declare no competing financial interests. The ER-HoxB8 system was obtained under a material transfer agreement with Dr. Mark Kamps and UCSD.
References
- 1.Auffray C, Sieweke MH, Geissmann F, Blood monocytes: development, heterogeneity, and relationship with dendritic cells. Annu Rev Immunol 27, 669 (2009). [DOI] [PubMed] [Google Scholar]
- 2.Gordon S, Taylor PR, Monocyte and macrophage heterogeneity. Nat Rev Immunol 5, 953 (December, 2005). [DOI] [PubMed] [Google Scholar]
- 3.Ginhoux F, Jung S, Monocytes and macrophages: developmental pathways and tissue homeostasis. Nat Rev Immunol 14, 392 (June, 2014). [DOI] [PubMed] [Google Scholar]
- 4.Geissmann F, Jung S, Littman DR, Blood monocytes consist of two principal subsets with distinct migratory properties. Immunity 19, 71 (July, 2003). [DOI] [PubMed] [Google Scholar]
- 5.Tacke F, Randolph GJ, Migratory fate and differentiation of blood monocyte subsets. Immunobiology 211, 609 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Auffray C, Fogg D, Garfa M, Elain G, Join-Lambert O, Kayal S, Sarnacki S, Cumano A, Lauvau G, Geissmann F, Monitoring of blood vessels and tissues by a population of monocytes with patrolling behavior. Science 317, 666 (August 3, 2007). [DOI] [PubMed] [Google Scholar]
- 7.Sunderkotter C, Nikolic T, Dillon MJ, Van Rooijen N, Stehling M, Drevets DA, Leenen PJ, Subpopulations of mouse blood monocytes differ in maturation stage and inflammatory response. J Immunol 172, 4410 (April 1, 2004). [DOI] [PubMed] [Google Scholar]
- 8.Yona S, Kim KW, Wolf Y, Mildner A, Varol D, Breker M, Strauss-Ayali D, Viukov S, Guilliams M, Misharin A, Hume DA, Perlman H, Malissen B, Zelzer E, Jung S, Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity 38, 79 (January 24, 2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Friedman AD, Transcriptional control of granulocyte and monocyte development. Oncogene 26, 6816 (October 15, 2007). [DOI] [PubMed] [Google Scholar]
- 10.Akashi K, Traver D, Miyamoto T, Weissman IL, A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature 404, 193 (March 9, 2000). [DOI] [PubMed] [Google Scholar]
- 11.Fogg DK, Sibon C, Miled C, Jung S, Aucouturier P, Littman DR, Cumano A, Geissmann F, A clonogenic bone marrow progenitor specific for macrophages and dendritic cells. Science 311, 83 (January 6, 2006). [DOI] [PubMed] [Google Scholar]
- 12.Hettinger J, Richards DM, Hansson J, Barra MM, Joschko AC, Krijgsveld J, Feuerer M, Origin of monocytes and macrophages in a committed progenitor. Nature immunology 14, 821 (August, 2013). [DOI] [PubMed] [Google Scholar]
- 13.DeKoter RP, Walsh JC, Singh H, PU.1 regulates both cytokine-dependent proliferation and differentiation of granulocyte/macrophage progenitors. The EMBO journal 17, 4456 (August 3, 1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feinberg MW, Wara AK, Cao Z, Lebedeva MA, Rosenbauer F, Iwasaki H, Hirai H, Katz JP, Haspel RL, Gray S, Akashi K, Segre J, Kaestner KH, Tenen DG, Jain MK, The Kruppel-like factor KLF4 is a critical regulator of monocyte differentiation. The EMBO journal 26, 4138 (September 19, 2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tamura T, Nagamura-Inoue T, Shmeltzer Z, Kuwata T, Ozato K, ICSBP directs bipotential myeloid progenitor cells to differentiate into mature macrophages. Immunity 13, 155 (August, 2000). [DOI] [PubMed] [Google Scholar]
- 16.Hanna RN, Carlin LM, Hubbeling HG, Nackiewicz D, Green AM, Punt JA, Geissmann F, Hedrick CC, The transcription factor NR4A1 (Nur77) controls bone marrow differentiation and the survival of Ly6C-monocytes. Nature immunology 12, 778 (August, 2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blatt C, Lotem J, Sachs L, Inhibition of specific pathways of myeloid cell differentiation by an activated Hox-2.4 homeobox gene. Cell growth & differentiation : the molecular biology journal of the American Association for Cancer Research 3, 671 (October, 1992). [PubMed] [Google Scholar]
- 18.Wang GG, Calvo KR, Pasillas MP, Sykes DB, Hacker H, Kamps MP, Quantitative production of macrophages or neutrophils ex vivo using conditional Hoxb8. Nat Methods 3, 287 (April, 2006). [DOI] [PubMed] [Google Scholar]
- 19.Saederup N, Cardona AE, Croft K, Mizutani M, Cotleur AC, Tsou CL, Ransohoff RM, Charo IF, Selective chemokine receptor usage by central nervous system myeloid cells in CCR2-red fluorescent protein knock-in mice. PLoS One 5, e13693 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.MacDonald KP, Palmer JS, Cronau S, Seppanen E, Olver S, Raffelt NC, Kuns R, Pettit AR, Clouston A, Wainwright B, Branstetter D, Smith J, Paxton RJ, Cerretti DP, Bonham L, Hill GR, Hume DA, An antibody against the colony-stimulating factor 1 receptor depletes the resident subset of monocytes and tissue- and tumor-associated macrophages but does not inhibit inflammation. Blood 116, 3955 (November 11, 2010). [DOI] [PubMed] [Google Scholar]
- 21.Platt RJ, Chen S, Zhou Y, Yim MJ, Swiech L, Kempton HR, Dahlman JE, Parnas O, Eisenhaure TM, Jovanovic M, Graham DB, Jhunjhunwala S, Heidenreich M, Xavier RJ, Langer R, Anderson DG, Hacohen N, Regev A, Feng G, Sharp PA, Zhang F, CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 159, 440 (October 9, 2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laplante M, Sabatini DM, mTOR signaling in growth control and disease. Cell 149, 274 (April 13, 2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM, mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 110, 163 (July 26, 2002). [DOI] [PubMed] [Google Scholar]
- 24.Hoshii T, Tadokoro Y, Naka K, Ooshio T, Muraguchi T, Sugiyama N, Soga T, Araki K, Yamamura K, Hirao A, mTORC1 is essential for leukemia propagation but not stem cell self-renewal. The Journal of clinical investigation 122, 2114 (June, 2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo F, Zhang S, Grogg M, Cancelas JA, Varney ME, Starczynowski DT, Du W, Yang JQ, Liu W, Thomas G, Kozma S, Pang Q, Zheng Y, Mouse gene targeting reveals an essential role of mTOR in hematopoietic stem cell engraftment and hematopoiesis. Haematologica 98, 1353 (September, 2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kalaitzidis D, Sykes SM, Wang Z, Punt N, Tang Y, Ragu C, Sinha AU, Lane SW, Souza AL, Clish CB, Anastasiou D, Gilliland DG, Scadden DT, Guertin DA, Armstrong SA, mTOR complex 1 plays critical roles in hematopoiesis and Pten-loss-evoked leukemogenesis. Cell stem cell 11, 429 (September 7, 2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hackstein H, Taner T, Zahorchak AF, Morelli AE, Logar AJ, Gessner A, Thomson AW, Rapamycin inhibits IL-4--induced dendritic cell maturation in vitro and dendritic cell mobilization and function in vivo. Blood 101, 4457 (June 1, 2003). [DOI] [PubMed] [Google Scholar]
- 28.Sathaliyawala T, O’Gorman WE, Greter M, Bogunovic M, Konjufca V, Hou ZE, Nolan GP, Miller MJ, Merad M, Reizis B, Mammalian target of rapamycin controls dendritic cell development downstream of Flt3 ligand signaling. Immunity 33, 597 (October 29, 2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuhn R, Schwenk F, Aguet M, Rajewsky K, Inducible gene targeting in mice. Science 269, 1427 (September 8, 1995). [DOI] [PubMed] [Google Scholar]
- 30.Zhu L, Yang T, Li L, Sun L, Hou Y, Hu X, Zhang L, Tian H, Zhao Q, Peng J, Zhang H, Wang R, Yang Z, Zhao Y, TSC1 controls macrophage polarization to prevent inflammatory disease. Nat Commun 5, 4696 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Burnett PE, Barrow RK, Cohen NA, Snyder SH, Sabatini DM, RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc Natl Acad Sci U S A 95, 1432 (February 17, 1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choo AY, Yoon SO, Kim SG, Roux PP, Blenis J, Rapamycin differentially inhibits S6Ks and 4E-BP1 to mediate cell-type-specific repression of mRNA translation. Proc Natl Acad Sci U S A 105, 17414 (November 11, 2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Delgado MD, Leon J, Myc roles in hematopoiesis and leukemia. Genes & cancer 1, 605 (June, 2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dang CV, MYC on the path to cancer. Cell 149, 22 (March 30, 2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Selvakumaran M, Liebermann D, Hoffman B, The proto-oncogene c-myc blocks myeloid differentiation independently of its target gene ornithine decarboxylase. Blood 88, 1248 (August 15, 1996). [PubMed] [Google Scholar]
- 36.Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P, The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell systems 1, 417 (December 23, 2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang CY, Bredemeyer AL, Walker LM, Bassing CH, Sleckman BP, Dynamic regulation of c-Myc proto-oncogene expression during lymphocyte development revealed by a GFP-c-Myc knock-in mouse. European journal of immunology 38, 342 (February, 2008). [DOI] [PubMed] [Google Scholar]
- 38.Wang H, Hammoudeh DI, Follis AV, Reese BE, Lazo JS, Metallo SJ, Prochownik EV, Improved low molecular weight Myc-Max inhibitors. Molecular cancer therapeutics 6, 2399 (September, 2007). [DOI] [PubMed] [Google Scholar]
- 39.Weichhart T, Hengstschlager M, Linke M, Regulation of innate immune cell function by mTOR. Nat Rev Immunol 15, 599 (September 25, 2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gan B, Sahin E, Jiang S, Sanchez-Aguilera A, Scott KL, Chin L, Williams DA, Kwiatkowski DJ, DePinho RA, mTORC1-dependent and -independent regulation of stem cell renewal, differentiation, and mobilization. Proceedings of the National Academy of Sciences of the United States of America 105, 19384 (December 9, 2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen C, Liu Y, Liu R, Ikenoue T, Guan KL, Zheng P, TSC-mTOR maintains quiescence and function of hematopoietic stem cells by repressing mitochondrial biogenesis and reactive oxygen species. The Journal of experimental medicine 205, 2397 (September 29, 2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang S, Readinger JA, DuBois W, Janka-Junttila M, Robinson R, Pruitt M, Bliskovsky V, Wu JZ, Sakakibara K, Patel J, Parent CA, Tessarollo L, Schwartzberg PL, Mock BA, Constitutive reductions in mTOR alter cell size, immune cell development, and antibody production. Blood 117, 1228 (January 27, 2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li D, Yang H, Nan H, Liu P, Pang S, Zhao Q, Karni R, Kamps MP, Xu Y, Zhou J, Wiedmer T, Sims PJ, Wang F, Identification of key regulatory pathways of myeloid differentiation using an mESC-based karyotypically normal cell model. Blood 120, 4712 (December 6, 2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ohtani M, Hoshii T, Fujii H, Koyasu S, Hirao A, Matsuda S, Cutting edge: mTORC1 in intestinal CD11c+ CD11b+ dendritic cells regulates intestinal homeostasis by promoting IL-10 production. J Immunol 188, 4736 (May 15, 2012). [DOI] [PubMed] [Google Scholar]
- 45.Wall M, Poortinga G, Hannan KM, Pearson RB, Hannan RD, McArthur GA, Translational control of c-MYC by rapamycin promotes terminal myeloid differentiation. Blood 112, 2305 (September 15, 2008). [DOI] [PubMed] [Google Scholar]
- 46.Klimmeck D, Cabezas-Wallscheid N, Reyes A, von Paleske L, Renders S, Hansson J, Krijgsveld J, Huber W, Trumpp A, Transcriptome-wide profiling and posttranscriptional analysis of hematopoietic stem/progenitor cell differentiation toward myeloid commitment. Stem cell reports 3, 858 (November 11, 2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Klimmeck D, Hansson J, Raffel S, Vakhrushev SY, Trumpp A, Krijgsveld J, Proteomic cornerstones of hematopoietic stem cell differentiation: distinct signatures of multipotent progenitors and myeloid committed cells. Molecular & cellular proteomics : MCP 11, 286 (August, 2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jun HS, Lee YM, Cheung YY, McDermott DH, Murphy PM, De Ravin SS, Mansfield BC, Chou JY, Lack of glucose recycling between endoplasmic reticulum and cytoplasm underlies cellular dysfunction in glucose-6-phosphatase-beta-deficient neutrophils in a congenital neutropenia syndrome. Blood 116, 2783 (October 14, 2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cheng SC, Quintin J, Cramer RA, Shepardson KM, Saeed S, Kumar V, Giamarellos-Bourboulis EJ, Martens JH, Rao NA, Aghajanirefah A, Manjeri GR, Li Y, Ifrim DC, Arts RJ, van der Veer BM, Deen PM, Logie C, O’Neill LA, Willems P, van de Veerdonk FL, van der Meer JW, Ng A, Joosten LA, Wijmenga C, Stunnenberg HG, Xavier RJ, Netea MG, mTOR- and HIF1alpha-mediated aerobic glycolysis as metabolic basis for trained immunity. Science 345, 1250684 (September 26, 2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim SY, Baik KH, Baek KH, Chah KH, Kim KA, Moon G, Jung E, Kim ST, Shim JH, Greenblatt MB, Chun E, Lee KY, S6K1 negatively regulates TAK1 activity in the toll-like receptor signaling pathway. Molecular and cellular biology 34, 510 (February, 2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weichhart T, Costantino G, Poglitsch M, Rosner M, Zeyda M, Stuhlmeier KM, Kolbe T, Stulnig TM, Horl WH, Hengstschlager M, Muller M, Saemann MD, The TSC-mTOR signaling pathway regulates the innate inflammatory response. Immunity 29, 565 (October 17, 2008). [DOI] [PubMed] [Google Scholar]
- 52.Cao W, Manicassamy S, Tang H, Kasturi SP, Pirani A, Murthy N, Pulendran B, Toll-like receptor-mediated induction of type I interferon in plasmacytoid dendritic cells requires the rapamycin-sensitive PI(3)K-mTOR-p70S6K pathway. Nature immunology 9, 1157 (October, 2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen C, Liu Y, Zheng P, Mammalian target of rapamycin activation underlies HSC defects in autoimmune disease and inflammation in mice. The Journal of clinical investigation 120, 4091 (November, 2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Magee JA, Ikenoue T, Nakada D, Lee JY, Guan KL, Morrison SJ, Temporal changes in PTEN and mTORC2 regulation of hematopoietic stem cell self-renewal and leukemia suppression. Cell stem cell 11, 415 (September 7, 2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Festuccia WT, Pouliot P, Bakan I, Sabatini DM, Laplante M, Myeloid-specific Rictor deletion induces M1 macrophage polarization and potentiates in vivo pro-inflammatory response to lipopolysaccharide. PLoS One 9, e95432 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ai D, Jiang H, Westerterp M, Murphy AJ, Wang M, Ganda A, Abramowicz S, Welch C, Almazan F, Zhu Y, Miller YI, Tall AR, Disruption of mammalian target of rapamycin complex 1 in macrophages decreases chemokine gene expression and atherosclerosis. Circulation research 114, 1576 (May 9, 2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hernandez O, Way S, McKenna J 3rd, Gambello MJ, Generation of a conditional disruption of the Tsc2 gene. Genesis 45, 101 (February, 2007). [DOI] [PubMed] [Google Scholar]
- 58.Picelli S, Bjorklund AK, Faridani OR, Sagasser S, Winberg G, Sandberg R, Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Methods 10, 1096 (November, 2013). [DOI] [PubMed] [Google Scholar]
- 59.Trombetta JJ, Gennert D, Lu D, Satija R, Shalek AK, Regev A, Preparation of Single-Cell RNA-Seq Libraries for Next Generation Sequencing. Current protocols in molecular biology 107, 4 22 1 (July 01, 2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L, Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature protocols 7, 562 (March 01, 2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rosenbauer F, Tenen DG, Transcription factors in myeloid development: balancing differentiation with transformation. Nat Rev Immunol 7, 105 (February, 2007). [DOI] [PubMed] [Google Scholar]
- 62.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP, Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545 (October 25, 2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.de Hoon MJ, Imoto S, Nolan J, Miyano S, Open source clustering software. Bioinformatics 20, 1453 (June 12, 2004). [DOI] [PubMed] [Google Scholar]
- 64.Saldanha AJ, Java Treeview--extensible visualization of microarray data. Bioinformatics 20, 3246 (November 22, 2004). [DOI] [PubMed] [Google Scholar]
- 65.Lee PY, Kumagai Y, Li Y, Takeuchi O, Yoshida H, Weinstein J, Kellner ES, Nacionales D, Barker T, Kelly-Scumpia K, van Rooijen N, Kumar H, Kawai T, Satoh M, Akira S, Reeves WH, TLR7-dependent and FcgammaR-independent production of type I interferon in experimental mouse lupus. The Journal of experimental medicine 205, 2995 (December 22, 2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Edgar R, Domrachev M, Lash AE, Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic acids research 30, 207 (January 01, 2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Phenotypic and functional analysis of DR–ER–Hoxb8 cells and their response to mTOR inhibition.
Fig. S2. Raptor deletion impairs monocyte development without causing apoptosis
Fig. S3. Impaired development of myeloid progenitors in Raptor-deficient mice
Fig. S4. Myeloid-specific deletion of Raptor does not affect monocyte or neutophil homeostasis
Fig. S5. Deletion of Tsc2 augments myelopoiesis via mTORC1 activation
Fig. S6. mTORC1 modulates myeloid development via S6K1
Fig. S7. mTORC1-S6K1 signaling modulates the expression of Myc and its target genes
Fig. S8. Complete images from gel electrophoresis
Table S1. Results from small molecule library screen using DR-ER-Hoxb8 cells
Table S2. Effects of mTOR inhibitors on monocyte marker expression in DR-ER-Hoxb8 cells
Table S3. Table of antibodies
Table S4. Compilation of raw data and analysis