Abstract
Bacteriophage research has been instrumental to advancing many fields of biology, such as genetics, molecular biology, and synthetic biology. Many phage-derived technologies have been adapted for building gene circuits to program biological systems. Phages also exhibit significant medical potential as antibacterial agents and bacterial diagnostics due to their extreme specificity for their host, and our growing ability to engineer them further enhances this potential. Phages have also been used as scaffolds for genetically programmable biomaterials that have highly tunable properties. Furthermore, phages are central to powerful directed evolution platforms, which are being leveraged to enhance existing biological functions and even produce new ones. In this review, we discuss recent examples of how phage research is influencing these next-generation biotechnologies.
Keywords: synthetic biology, phage, phage therapy, phage display, phage-based materials, gene circuit, RNA polymerase, transcriptional regulator, integrase, recombinase, lysin, gene synthesis, diagnostics
INTRODUCTION
Over a century ago, the discovery of bacteriophages was met with excitement and controversy owing to their unknown biology and potential for treating bacterial infections. Bacteriophages have since been shown to be bacterial viruses that selectively infect specific bacterial strains. As such, bacteriophages have the potential to be used as biocontrol agents, but early attempts at developing phage-based therapies were largely foiled due to misunderstandings of phage biology (1). Fortunately, phage research endured. Because of their relatively small genome size [5 to~500 kb with a median around 50 kb (2)], fast growth rate, and ease of genetic manipulation, phages have become a major fixture in the field of genetics and have been used to establish some of the basic concepts of biology. The two most prominent examples are confirming DNA as the molecule carrying hereditary information (3) and establishing the triplet nature of codons (4). Phages were also instrumental in the discovery of enzymes that have made molecular biology and recombinant DNA possible (5–7). Most recently, phages have played a central role in deciphering the function of clustered regularly interspaced short palindromic repeat (CRISPR)-associated systems (Cas), the archaic immune systems that prokaryotes use to defend themselves against phages and other mobile genetic elements by expressing sequence specific nucleases (8). These nucleases are highly programmable, enable genetic manipulation with unprecedented ease and control, and are currently fueling a major biotechnological revolution (9).
Phage research continues to play a major role in biological research, as phage components are now being used for key developments in synthetic biology. Synthetic biology involves designing biological systems according to principles developed from engineering disciplines. In this review, we focus on the most recent contributions that phage research is making to the field of synthetic biology. Figure 1 summarizes these contributions and introduces the topics discussed.
Figure 1.
Summary schematic illustrating the various components of phages that have contributed to synthetic biology advancements. Various fields of biology have benefited greatly from phage research.
MINING SYNTHETIC BIOLOGY PARTS FROM PHAGE GENOMES
Synthetic biology’s goal of reprogramming biological systems depends upon having sets of functionally orthogonal parts, individual components that perform defined functions without interfering with one another. Phages offer a rich source of orthogonal parts for synthetic biology because they have evolved numerous ways to enact the same basic developmental program of taking over a bacterial host cell’s resources to produce phage progeny. This requires diverting bacterial resources to replicate the phage genome, producing an array of proteins that assemble into a capsid that packages the genome, and producing proteins that ultimately enable exiting the infected cell. The great diversity of phages lies, in part, in the multiplicity of genes involved in performing these functions.
Phage RNA Polymerases
Programming biology requires exerting precise control over gene expression. One common strategy is to build genetic circuits to perform logic functions by regulating mRNA production with promoter-specific RNA polymerases (RNAPs). Some phages produce their own single-unit RNAP that selectively recognizes promoter sequences not recognized by endogenous bacterial RNAPs. The most commonly studied of these enzymes originate from phages T7, SP6 (10), and N4 (11). However, many other phages possess a phage-specific RNAP that is selective for a corresponding promoter sequence. This specificity makes RNAPs inherently orthogonal to each other. In addition, phage RNAPs are largely host-independent and can work in most cellular contexts.
The diversity of phage RNAPs can be enhanced by engineering. For example, T7-like RNAP promoter specificity can be altered by replacement or mutation of a specificity-determining loop that is separate from the enzymatic core domain (12, 13). The specificity loop and core polymerase can be separated onto independent genes such that only coexpression of the two fragments reconstitutes activity. Using a split T7 RNAP, Segall-Shapiro et al. (14) built a resource allocator system in which the level of expression of the core fragment determines the combined expression level for all outputs, while the relative expression levels of the specificity loop fragments determine the relative expression for individual output genes (Figure 2). To ensure efficient complementation between the specificity loops and the core domain, leucine zippers were fused to each fragment to stabilize the assembled complex (14). Split RNAP fragments can also be fused to sensing domains that interact only under precisely defined conditions to yield synthetic sensor components. Pu et al. (15) constructed light-inducible and rapamycin-inducible split RNAPs and performed directed evolution to enhance RNAP sensor function. In addition, two other light-inducible T7 RNAPs were designed using different light-sensing domains, thus illustrating the versatility and flexibility of T7-like RNAP in synthetic circuit design (16, 17). This platform demonstrates how modular and orthogonal control over gene expression can be enabled by systematic engineering of phage components.
Figure 2.
Example of an application of a split T7 RNA polymerase (RNAP) in synthetic biology: a resource allocator. (a) Crystal structure (PBD ID 1CEZ) of the T7 RNAP highlighting the σ fragment (blue), which directs promoter specificity, and the catalytic core domain (gray). (b) Schematic illustrating the resource allocator strategy. A controller subunit encodes various σ fragments whose expression levels are determined by independent inputs. Once a σ fragment assembles with a core fragment, the reconstituted RNAP can recognize its cognate promoter and activate transcription of the downstream gene. Panel b modified from Reference 14. (c) Example of an output from the resource allocator. The expression level of each σ fragment is set to an arbitrary value defined by the genetic circuit (controller). As the level of expression of the core fragment is increased from an arbitrary value between 1 and 4, the output transcriptional expression level of all four actuators increases proportionally, leaving their relative signals unchanged but increasing the total combined signal.
Transcriptional Regulators
Phages can broadly be classified into two types: lytic phages, which are phages that infect cells and only reproduce and kill the cell, and temperate phages, which are phages that have developed the capacity to silence their genes, integrate their genome within their host’s genome and remain silent for many generations, and only become activated once DNA damage is detected. Temperate phages require many transcriptional control elements to integrate information and compute whether to grow lysogenically (gene silencing and integration into the genome) or lytically (immediate phage production). Despite a wide diversity of orthogonal phage transcriptional regulators, only the λ repressor, CI, has been widely used in synthetic biology, most likely because its function has been characterized over the course of more than 60 years (18). CI has been involved in many synthetic biology applications, such as the design of the first synthetic oscillator (19) and has been mutagenized to create artificial orthogonal repressor/promoter pairs (20), a memory recording device active in the mammalian gut (21), a kill switch (22), and a pulse-detecting circuit (23). Phage transcriptional regulators offer a rich source of parts for synthetic biology, but additional mechanistic studies of their DNA binding sites, orthogonality, and resource costs are necessary before they can be incorporated into the synthetic biologist’s arsenal.
Integrases
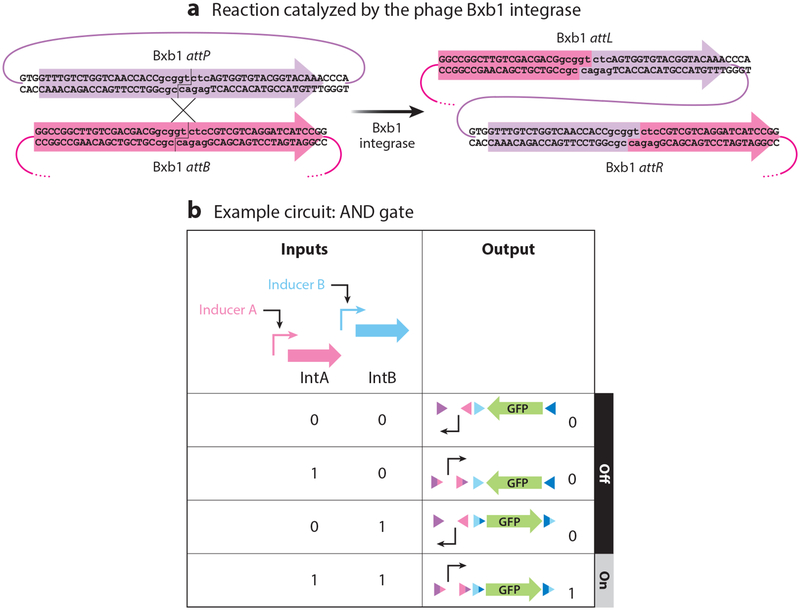
Most temperate phages rely on site-specific recombinases to integrate their genomes into their hosts’ DNA at a particular chromosomal site. These enzymes, known as integrases, recognize a specific phage attachment site, attP, and a specific bacterial site, attB, and catalyze the directional insertion of the phage genome into the bacterial chromosome. This process creates two new att sites at the termini of the phage genome: attR and attL, respectively (Figure 3a). The reverse reaction, excision, happens only in the presence of a recombination directionality factor. If the attP and attB sites are on the same DNA molecule, the outcome of recombination is dependent upon the relative orientations of the two sites. For example, if they are in parallel, the genetic region between the two sites is flipped, and if they are antiparallel, the intergenic region is excised. att sites and their cognate integrases can therefore be repurposed for rationally programming integration, displacement, and deletion events (Figure 3b). Moreover, integrase/att site pairs can be identified via genomic databases and have been shown to display a high degree of specificity toward their cognate recognition site (24).
Figure 3.
Example of integrase-based logic. (a) The Bxb1 integrase is used as an example to illustrate how integrases recombine attB (magenta) and attP (violet) sites to insert phage DNA into the bacterial chromosome and create two new hybrid sites, attL and attR. Lowercase letters indicate the essential core sequence of the attachment sites. (b) Schematic showing how recombinases can be used to build an AND logic gate as an example of genetic programming. Two different integrases, IntA and IntB, are induced by two different inducers or inputs. The circuit in the initial state has a promoter surrounded by attB (magenta) and attP (violet) sites specific for IntA while the reporter gene, gfp, is oriented such that it cannot be transcribed. The gfp reporter gene is surrounded by antiparallel attB (dark blue) and attP (light blue) sites specific for IntB. To induce GFP production, both inputs must be present long enough for both sets of att sites to be recombined by their respective integrases. In this particular system, there is no requirement for the order in which inputs are provided. Once the system is activated, it cannot be turned off, as integrases perform unidirectional recombination events.
The most common phage integrases used in synthetic biology are those of phages TP901–1, phiC31, and Bxb1, which belong to the large serine integrase family (25). These integrases do not require host factors for enzymatic catalysis, as opposed to the family of tyrosine integrases, which depend on integration host factor (IHF) (25). Both the host independence and the exquisite site specificity of large serine integrases have quickly led to their adoption in mammalian cell engineering projects (25). For example, landing pads containing one or more attB sites for different integrases have been created in dedicated cell lines to be used as recipients for integrative DNA constructs carrying desired gene circuits (26–28), thus speeding up genetic engineering tasks.
A major advantage of recombination-based genetic circuitry is its capacity to store information about past genetic events without requiring constant gene expression, even after cell death. By carefully intertwining parts surrounded by attB and attP sites in the appropriate orientations and cloning integrase genes under the control of inducible promoters, circuits can be built to perform complex logic and record molecular events. Using this design principle, it was possible to create all 16 two-input Boolean logic gates in Escherichia coli (29), build circuits that count pulses of a chemical with (30) or without (31) temporal integration, engineer robust sensors based on a combination of analog sensing components digitized by integrases (32), and assemble state machines with up to 3 inputs and 16 outputs (33). Moreover, many of these devices have been ported to mammalian systems, in which the digital response is particularly valuable (34, 35).
REFACTORING PHAGE GENOMES
As seen with the design of circuits that incorporate integrase attachment sites just described, synthetic biology is intimately linked to the capacity to assemble and re-arrange DNA sequences. Some phages, such as the filamentous phage M13 or RNA phage MS2, have genomes that are small enough (<10 kb) to be easily modified in vitro. However, most phages have genomes that are too large (>20 kb) for easy in vitro manipulation and are lethal to their bacterial hosts.
Temperate phages can be maintained stably in bacteria, enabling modification of their genomes using the same methodologies as those used for engineering bacterial genomes. However, genomes of strictly lytic phages cannot be cloned whole into bacteria for subsequent genetic manipulation. As a result, lytic phage genomes are traditionally edited via homologous recombination (allelic exchange), whereby the gene or genes to be modified are cloned into a plasmid and modified as needed before being introduced in the phage’s host. This strain is then infected with the wild-type phage, allowing recombination between the phage gene and the mutated gene copy on the plasmid (Figure 4a). Recombination requirements vary from phage to phage, but recombination often requires more than 1 kb of homology on each side of the target locus for recombination to occur at a frequency high enough for detection. Allelic exchange produces recombinants at frequencies that typically do not exceed ~1% and generally are below 10−5 recombinants/wild type (36). Such low recombination yields, compounded by the lack of selection markers for phage engineering, make screening for the desired phage mutants labor intensive and sometimes technically impossible. In addition to low recombination efficiency, many phage loci cannot be cloned because the activities they encode are detrimental to bacteria. Consequently, synthetic phage biology efforts have involved a strong push for developing new phage genome engineering methods.
Figure 4.
Approaches to phage genome engineering. (a) Phages are traditionally engineered by allelic exchange, whereby a modified gene (magenta) is provided with homology to the phage genome (gray and yellow boxes). Double crossovers will replace the normal phage gene with the mutated copy at low efficiency (<10−5). The wild-type phage is represented with a magenta DNA molecule in its head, and the mutant phage is represented with a blue DNA molecule in its head. (b) Recombineering uses linear DNA that has homology on each side of the target locus. Recombination between the linear DNA and the phage genome is enhanced by phage recombination functions, in this example λ gam, bet, and exo. Recombineering requires that the phage be stably maintained in the bacterial genome and is therefore applicable only for engineering temperate phages. (c) The CRISPR/Cas counterselection system increases the rate for isolating phage mutants when using the traditional allelic exchange engineering strategy. (d) Phage genomes can be reconstructed in full through the reassembly of polymerase chain reaction (PCR) products that carry overlapping ends that template the order in which the fragments are assembled. Assembly can be performed either in the yeast Saccharomyces cerevisiae by using yeast artificial chromosomes (YACs) or by isothermal assembly. The reconstructed genomes can then be rebooted to produce progeny through transformation of Escherichia coli or Listeria monocytogenes L-forms or by mixing with in vitro transcription/translation extracts.
Recombineering: Recombination with Linear DNAs Carrying Short Homologies
As a strategy to increase recombination efficiency, the recombination functions of some temperate phages have been co-opted to catalyze the recombination of linear DNA fragments (single-stranded or double-stranded DNA) into replicating DNA within bacteria. The most common system, known as recombineering, relies on the phage λ recombination genes exo, bet, and gam. The proteins encoded by these genes protect linear DNA from degradation while processing it for recombination with the programmed chromosomal target, which is templated by homologous regions ranging from ~30 to 1,000 bp (Figure 4b). Recombineering with oligonucleotides ~100 nt long reaches efficiencies up to a few percent and allows for direct screening for the desired mutations. However, recombineering with double-stranded DNA donors has a much lower recombination efficiency (in the range of 10−8 recombinants/surviving cell) and requires a strong selection marker to identify the recombinants. While recombineering has been transformative for bacterial genome manipulation and, by extension, for temperate phages that are part of bacterial chromosomes (37–40), it appears to be of limited value for engineering strictly lytic phages. The only documented success involves coliphage T4 (41). The authors exploited a peculiarity of the T4 life cycle, which stalls in the absence of a functional transcription termination factor, Rho, producing a window during which the T4 DNA is susceptible to recombineering. Rho dependence is a T4-specific feature; most other phage families are not Rho dependent, and therefore this approach is unlikely to be portable to other phage systems.
Recombineering has been expanded beyond engineering enterobacterial phages thanks to the discovery of orthologous recombination functions from phages infecting gram-positive bacteria leading to a technique termed bacteriophage recombineering of electroporated DNA (BRED) (37, 42). BRED can be used to introduce desired point mutations, insert genes, or create gene deletions, even in lytically growing phages in Mycobacterium smegmatis and E. coli hosts, and is efficient enough to eliminate the need for selection markers (37, 39).
CRISPR/Cas: The Most Versatile Counterselection System for Phage Engineering
Because phages are natural targets for CRISPR/Cas nucleases, they have been repurposed as a counterselection agent for phage engineering with exceptional versatility and are used to eliminate parental genotypes and enrich for rare mutants (Figure 4c).
For example, the cloned type IIA CRISPR/Cas system of Streptococcus thermophilus was used to select for deletions and replacements in the genome of Streptococcus thermophilus phage 2972 to assess the role of open reading frames of unknown function (43). Similarly, the type IE CRISPR/Cas system of E. coli was used to facilitate the selection of mutants of coliphage T7 (44). The CRISPR/Cas9 of Streptococcus pyogenes has been shown to efficiently alter the DNA of virulent lactococcal phages (45) and even the heavily glucosylated and hydroxymethylated DNA of E. coli phage T4, albeit with a drop in efficiency (46), thus demonstrating CRISPR/Cas9’s extraordinary versatility.
While CRISPR/Cas counterselection eliminates the need for direct selection markers, it does not address the problem of engineering regions of the phage genome that cannot be cloned due to their encoded toxicity for the bacterial host.
Complete Phage Genome Reconstruction in Nonhost Systems
To circumvent the toxicities associated with cloning phage DNA, hosts that are unable to express the toxic genes or are impervious to them can be used for cloning. For example, Jaschke et al. (47) reconstructed the ~5.5-kb-long genome of ϕX174 in yeast by recombineering PCR products of the phage genome and gene synthesis products that encode engineered genes. They also refactored the genome to eliminate gene overlaps and tuned ribosome binding sites and start codons to simplify the phage’s genetic makeup (47). Leveraging this principle, Ando et al. (48) showed that genomes of a wide variety of T7-like phages can be entirely reconstructed in Saccharomyces cerevisiae using sets of PCR products covering the entire phage genome, templated by short >30-bp-long homologies at each terminus. Desired mutations can be added anywhere in the genome during the PCR process, allowing an unprecedented level of flexibility for genome design. The resulting synthetic genomes are extracted from yeast and used to transform a highly competent E. coli strain so that it produces mutant phage particles (Figure 4d) (48). However, this method excludes phages that cannot use E. coli’s machinery to reboot, and it is unlikely that rebooting phages that infect gram-positive bacteria would be as efficient as rebooting gram-negative phages given the profound differences between the transcription and translation machineries of gram-negative and gram-positive organisms.
A recent and exciting report addressing this limitation describes using L-form bacteria as rebooting hosts. L-forms are bacteria that have been extensively engineered or selected to be devoid of a cell wall (49). The cells are very fragile but highly permeable to DNA in the presence of polyethylene glycol. Using Listeria monocytogenes L-forms, Kilcher et al. (50) managed to reboot a wide range of phages that normally infect gram-positive bacteria such as L. monocytogenes, Bacillus subtilis, or Staphylococcus aureus. The authors also rebooted synthetic phages reconstructed from overlapping PCR products generated using isothermal assembly (51). To demonstrate the versatility of this new rebooting host, variants of Listeria temperate phages were constructed (50) (Figure 4d).
Another promising strategy for phage engineering is to remove the cell completely and to produce phages using transcription/translation extracts (TXTL) from purified phage DNA. Phages MS2, φ174, and T7 have been produced using E. coli TXTL extracts (52) (Figure 4d).
PHAGE ENGINEERING FOR IMPROVED THERAPEUTIC ACTIVITY AND DIAGNOSTICS
The concept of phage therapy has gained considerable momentum because of the ever-increasing incidence of antibiotic resistance. Bacteriophages kill bacteria through mechanisms that are independent of those involved in antibiotic resistance, offering a complementary approach for treating antibiotic-resistant infections. In fact, phage therapy has been a staple medicine for several Eastern European countries and Russia for decades (53). However, its use has not been backed by rigorous clinical trials following modern standards of safety and efficacy. To be adopted as a standard treatment, regulatory strategies will have to be updated to account for the complexities of phage-based products (54). For example, it is of paramount importance to establish testing requirements for phage products. Empirical evidence suggests that phages are safe, but several studies show that phages can participate in the evolution of bacterial virulence (55) and elicit an immune response (56). In addition to regulatory hurdles, phages have other limitations causing barriers to entry, such as difficulties penetrating biofilms; limited host range, which necessitates the use of phage cocktails; complex pharmacokinetics; and the possibility of evolution of bacterial resistance. However, genetic engineering can potentially address these shortcomings.
Improving Phage Efficiency
As an illustration of this concept, phages engineered to express biofilm matrix-degrading enzymes penetrate and disperse biofilms more readily than their nonengineered counterpart (Figure 5). Coliphage T7 expressing dispersin B (DspB) cleared experimental E. coli biofilms approximately three times better than the non-DspB-expressing control, as judged by cell number counts (57). Biofilm matrices are complex, and other biofilm-dispersing cargos should be evaluated to expand beyond this proof-of-concept study, such as proteinaceous surfactants, antimicrobial peptides, and capsule depolymerases (58, 59).
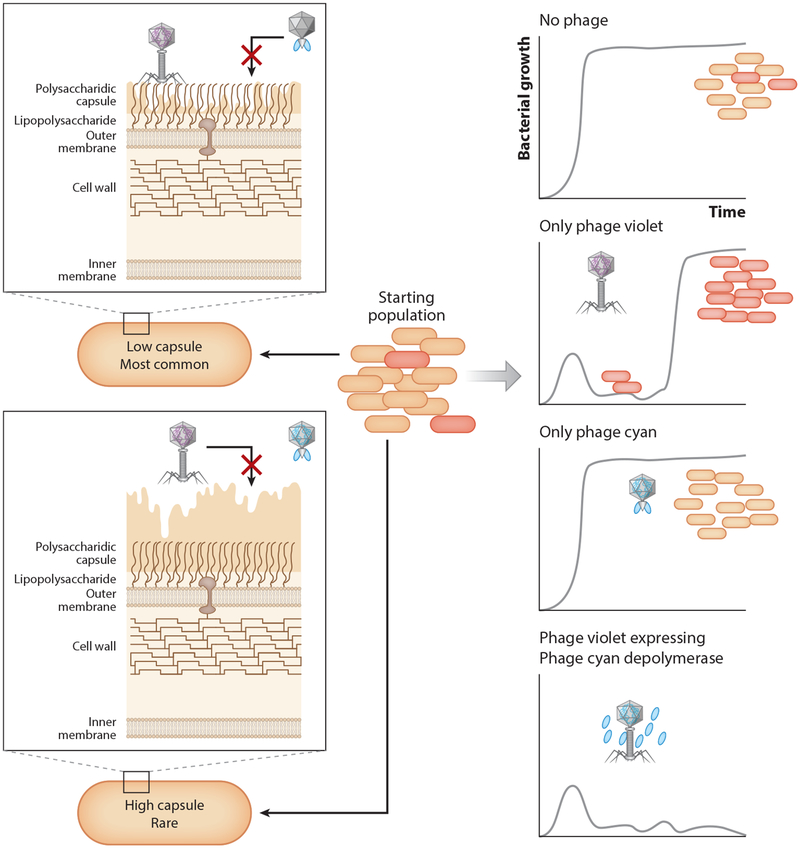
Figure 5.
Minimizing resistance to phage infection by expression of multiple host-recognition proteins. Many bacteria produce capsules that prevent phages from binding to their receptors. Capsules are metabolically expensive, and many bacteria only express them as a response to stress. In this example, inspired by Reference 60, phage violet recognizes and kills low-capsule bacteria, while phage cyan uses the capsule as a receptor and eliminates capsulated bacteria but not low-capsule ones. In this example, most bacteria would be low capsule. As a result, phage cyan would have no effect on bacterial growth, while phage violet would kill a large number of low-capsule bacteria. However, phage violet would also select for capsule-expressing mutants, which would eventually recolonize the environment. Because capsule-binding phages usually do so by using tail appendages that degrade capsules through depolymerization, engineering phage violet to express the phage cyan capsule depolymerase gene generates a phage that kills both the capsule-producing and the low-capsule population and minimizes resistance to phage infection.
Erwinia amylovora is a common plant pathogen, but few phages have proved effective for treating or protecting infected crops because Erwinia produces a capsule that is known to act as a barrier to phages. Cloning the capsule depolymerase gene of Erwinia phage L1 (dpoL1-C) into the genome of an unrelated phage Y2 resulted in a phage that produced larger and clearer plaques compared to Y2. Despite a lower burst size (number of phages produced per infected cell), the engineered phage killed Erwinia more efficiently in culture, resulting in an ~3-log reduction compared to wild type after 24 h of infection. These results were matched in experimental flower infections and illustrate how engineered phages that express depolymerases can help the phage perform better (Figure 5) (60). These two examples of engineering phage to encode a hydrolase or depolymerase gene demonstrate that synthetic biology can be used to greatly improve natural phages.
Altering Phage Host Range
Another way in which a phage can be altered to serve practical purposes is by narrowing or expanding its host range. Bacteriophage infection is initiated by the specific recognition of cell surface receptors, which may be surface polysaccharides, proteins, or even organelles such as pili or flagella (61). Phage receptors are highly variable, so most phages infect only a few strains and variants of a given pathogen. This specificity distinguishes phage from traditional broad-spectrum antibiotics, which kill bacteria with little discrimination between beneficial and pathogenic bacteria. However, cocktails consisting of more than one phage are likely to be required to achieve adequate clinical coverage. As pathogen populations continuously evolve during the course of an infection, these cocktails will need regular updating, which may present significant practical and regulatory challenges.
Methodologies have been developed for isolating phages that target many different strains of a given pathogen (62), but some phages are also capable of infecting bacteria of more than one species. For example, phage SH7 infects isolates of harmless laboratory E. coli, virulent O157:H7 E. coli, Shigella dysenteriae, Shigella flexneri, and Salmonella Paratyphi (63), and phage Mu infects E. coli, Citrobacter freundii, Shigella sonnei, Enterobacter spp., and Erwinia spp. (36). However, broadening phage host range may come with a trade-off involving decreased selectivity for pathogenic variants of the targeted bacterium. Another approach to remedy the host range obstacle is to develop pipelines that allow programming of host range in phage scaffolds that have already been approved as safe and effective. Striving toward this goal, Ando et al. (48) rationally constructed synthetic variants of T7-like phages that exhibited altered host range. In T7-like phages, host range is primarily controlled by tail appendages encoded by gene 17. While swapping gene 17 with analogous genes from several other related phages yielded predictable host range alterations, functional hybrids between the Klebsiella phage K11 and T7 required the replacement of all three tail genes of T7—genes 11, 12, and 17—with the corresponding genes from K11, a feat that would be extremely challenging for traditional allele exchange techniques. However, as this approach requires swapping tail components between known phages, it is inherently limited to producing phages active against bacteria for which phages have already been characterized and whose genomes have already been sequenced. Developing high-throughput methods for tuning phage host range would be extremely valuable to the future of phage therapy.
Phages as DNA Delivery Shuttles
A potential advantage of phage-based therapeutics is the ability of phages to replicate along with the bacterial cells that harbor them. However, there is concern that during an infection, uncontrolled self-amplification of a phage might trigger side effects caused by the sudden release of bacterial endotoxins upon lysis (64). To bypass these concerns, phages can be engineered to be nonreplicative and used as nanosyringes to inject gene circuits into specific bacterial populations in a process known as transduction. It is expected that phages can inject DNA into a broader range of hosts than those in which phage replication is observed macroscopically as plaques, because producing phage progeny requires many steps beyond DNA injection. Plaques are observed only if the phage has resisted bacterial defenses (restriction, CRISPR, superinfection exclusion); has expressed, processed, and assembled its components; and has lysed the host cell (65, 66).
Filamentous M13 phage and its relatives are commonly used as transducing vectors to deliver DNA payloads to bacteria. These genetic payloads are referred to as phagemids. M13 phagemids were used to screen for small antimicrobial peptides (AMPs) and peptide toxins with high potency against E. coli. The most potent AMPs identified were ceropin and apidaecin, while the most potent toxin was CcdB. These genes were subsequently cloned into a single phagemid and delivered to target bacteria. This construct decreased E. coli viability by four orders of magnitude after 6 h of treatment, thus producing an efficient therapeutic from a normally innocuous phage. This system also significantly protected mice from death in a peritonitis infection model using EMG2 E. coli as the pathogen (67). Another example of leveraging M13’s DNA delivery capabilities used the delivery of CRISPR/Cas nuclease constructs to specifically remove antibiotic resistance markers or virulence genes from microbiomes with great accuracy and efficacy (68). Although M13, like other filamentous phages, has a narrow host range (68), these examples showcase the potential phages have as delivery vehicles for synthetic gene circuits.
Developing efficient transduction platforms that deliver nonlethal genetic circuits to a wide range of bacterial species would be highly effective for microbiome engineering. Specifically, it would be useful to augment the metabolic capabilities of particular bacterial strains of the microbiome. T7-like phages can transduce plasmids if they harbor a specific DNA sequence, the pac sequence (69, 70). This ability was used to screen for T7-like tail components that enable efficient transduction into a variety of bacterial strains. Specifically, genes 11, 12, and 17 from 15 different T7-like phages that infect a variety of gram-negative bacteria were cloned into plasmids containing a pac sequence. Cells carrying these plasmids were infected by T7, and the capacity of the resulting lysates to transduce the plasmids into different bacteria was scored. A successful transduction indicated that the plasmid encodes a phage tail that can attach to the T7 head and catalyze injection of DNA into that particular bacterium. The successfully transduced plasmids were recovered and subjected to mutagenesis. The resulting mutagenized pools were then screened against their bacterial target to identify mutants with increased transduction efficiencies (71). This methodology takes advantage of the modularity of phages and demonstrates that engineering efforts can improve phage functions.
Repurposing Temperate Phages as Lytic Phages
Temperate phages are usually avoided for therapeutic applications because of the risk of lysogenizing their target strain, rendering it resistant to future phage infection, and potentially encoding virulence factors that worsen the pathogen’s virulence (72). However, some bacterial species appear to never produce strictly lytic phages (73). On the other hand, lysogeny is almost ubiquitous and could provide an almost unlimited source of therapeutic phages as long as strategies to eliminate their temperate nature can be developed. Technologies such as BRED can be used to engineer lytic derivatives of temperate phages in which the repressor is cleanly deleted (37). In addition, this same approach can be used to remove virulence factors and integrases, thus transforming temperate phages into therapeutically applicable lytic phages. This goal was one of the motivations for the L. monocytogenes L-form phage-engineering platform described above. Using the Listeria phage P335 as a scaffold, Kilcher et al. (50) built a virulent mutant that lacks the main repressor and further improved the phage’s lytic activity by having it express a second endolysin. The resulting phage did not lysogenize L. monocytogenes, killed L. monocytogenes efficiently, and was able to curtail the development of resistance because the additionally encoded endolysin targeted cells were not phage sensitive (50).
Killing Bacteria with Engineered Lysins
Phage parts can also be repurposed and engineered as therapeutics in addition to being used as biotechnology tools. The most common mechanism of phage-mediated bacterial cell lysis involves holin/antiholin pairs that insert into the cell membrane and produces pores, which serve as conduits for lysins (also called endolysin) to translocate across and degrade the host’s cell wall. Without an intact cell wall, bacterial cells lyse. Purified lysins applied exogenously are bactericidal against many gram-positive bacteria because their cell wall is exposed to the exterior medium and therefore directly accessible to the lysin (74). However, the cell wall of gram-negative bacteria is sandwiched between the inner and outer membranes and is thus inaccessible to most natural lysins. Lysins are particularly exciting antimicrobials because bacteria do not appear to mount effective resistance strategies against them. Strategies to produce lysins active against antibiotic-resistant gram-negative bacteria are being pursued.
Lukacik et al. (75) proposed an elegant solution to the problem of lysing gram-negative bacteria by engineering pesticin. Pesticin is a toxin produced by Yersinia pestis that is secreted in the milieu, killing nearby competing bacteria. Pesticin binds to the outer membrane protein FyuA of the target bacterium through its N-terminal domain. This binding triggers its translocation to the periplasm, where its C-terminal catalytic domain hydrolyses the cell wall, leading to cell lysis and death. Pesticin producers are protected by the expression of small immunity proteins that inhibit its catalytic activity in the periplasm. The N-terminal domain of pesticin is structurally similar to that of T4-lysozyme (T4’s lysin), which prompted the idea of fusing the C-terminal recognition domain of pesticin with T4-lysozyme, thus expanding the therapeutic scope to target pesticin-producing cells that have pesticin immunity. The resulting synthetic protein selectively killed FyuA-expressing cells and was insensitive to the immunity factors that inhibit pesticin. Since FyuA is a virulence factor for bacteria, mutants resistant to this engineered lysin are also attenuated (75). However, many pathogens do not express FyuA despite being fully virulent.
Briers et al. (76) developed synthetic phage lysins termed Artilysins, which incorporate peptides that permeate the outer membrane of gram-negative bacteria and enable uptake independent of any active transport. Their design is based on lipopolysaccharide-destabilizing peptides fused to the C terminus of select phage endolysins. The best of these Artilysins was bactericidal for Pseudomonas aeruginosa, Acinetobacter baumanii, E. coli, and Salmonella Typhimurium, decreasing their survival four to five orders of magnitude (76), and showed an extraordinary eight orders of magnitude killing efficacy on stationary phase A. baumannii (77).
Phage-Based Diagnostics
Phages have always been associated with bacterial diagnostics because of their exquisite specificity, exponential amplification, and simple readout, such as plaque formation. However, genetically engineered phages that express reporters offer a unique and simpler way to detect pathogenic bacteria. Fluorescent proteins are small and can be easily cloned into phage genomes. Fluorescence permits microscopic visualization of bacteria infected by phages and can be used as a signal for cell sorters. Starting from mycobacteriophage TM4, several generations of diagnostic phages having fluorescence as an output (fluorophages) have been developed for the detection of Mycobacterium tuberculosis and for the evaluation of the corresponding antibiotic resistance profiles (78–80). Nevertheless, fluorophage systems often suffer from poor signal-to-noise ratios because the natural fluorescence in biological samples contributes to background signal.
Bioluminescence, a process that produces light through enzymatic oxidation of a chemical substrate, does not suffer from the background problem associated with autofluorescence and is usually brighter than fluorescent systems. Luciferase is the bioluminescence enzyme responsible for producing light and has been used in a variety of phage systems to detect a wide range of pathogenic bacteria, often with sensitivities down to a single colony-forming unit: M. tuberculosis (81–84), Vibrio parahaemolyticus (85), Salmonella Typhimurium (86), Y. pestis (87), S. flexneri (88), Bacillus anthracis (89, 90), pathogenic E. coli (91–93), L. monocytogenes (94), and the plant pathogenE. amylovora (60).
A caveat to both fluorescence and bioluminescence signaling is that they require the phage to infect metabolically active bacteria to produce an output signal. Therefore, these systems may create false negatives when a significant fraction of the bacterial population resides in a persister (dormant) state. False negatives may also arise from post-infective bacterial defense mechanisms that inactivate or destroy laterally acquired DNA. To circumvent this limitation, Schmidt et al.(95) took advantage of the Salmonella O-antigen enzymatic specificities of the P22 and 9NA phage tailspikes to produce an ELISA-like diagnostic tool they termed ELISA-like tailspike adsorption assay (ELITA). ELITA can detect and differentiate Salmonella bacteria of the O4, O4, O5, and O9 serotypes. (95). This strategy can be easily adapted to detect other Salmonella serovars or even other bacteria by using tailspikes or other phage proteins that recognize different surface polysaccharides (96–100).
PHAGE-BASED NANOCARRIERS FOR DRUG DELIVERY AND VACCINE ADJUVANTS
Phage-Based Drug Delivery
Bacteriophage capsids possess remarkable three-dimensional quasi-crystalline structures that can be extensively tailored and repurposed for transporting drugs to specific tissues and preventing drug degradation. RNA phage MS2 is commonly used because its coat protein spontaneously assembles into noninfectious virus-like particles (VLPs) when expressed in E. coli, which are noninfectious because they do not contain genetic material. The MS2 coat protein can tolerate insertions at several locations, allowing cargo to be introduced within the VLP. Modifications include tags to attach therapeutic payloads (drugs, enzymes, nucleic acids, homing peptides), moieties for targeting to specific animal tissues, and chemical functionalization for improved resilience against degradation (101–108).
M13 is another commonly used drug delivery scaffold. Vaks & Benhar (109) loaded M13 particles with chloramphenicol by attaching the antibiotic to the particle surface via ester linkages, such that serum esterases triggered its slow release. When equipped with a Staphylococcus-recognizing antibody fragment, these bio-nanoparticles accumulated close to the pathogen and released a large local concentration of chloramphenicol that killed even chloramphenicol-resistant bacteria (109). Such targeted, phage-mediated delivery of a high local concentration of antibiotics may be generalizable to the treatment of other antibiotic-resistant infections and clinically useful because it minimizes the dosage required and associated toxicity of nontargeted antibiotics.
Phage particles can also be used for other types of drug delivery. The capsid of Salmonella phage P22 was modified to encapsulate the analgesic peptide ziconotide. The resulting VLPs were subsequently chemically altered to display cell-penetrating peptides from the human immunodeficiency virus Tat protein on the capsid surface. The engineered VLPs were able to cross the blood-brain barrier and release active peptide in various animal and human brain cell models (110). P22 VLPs have also been loaded with functional Cas9/single guide RNA complexes by genetically fusing Cas9 to a fragment of the phage scaffolding protein (111). These examples highlight the vast versatility of P22 VLPs for the delivery of protein therapeutics.
Phage-Based Vaccines
The size and composition of phages make them ideally suited to engage the immune system to elicit an immune response (112, 113). Phages are therefore being spotlighted as templates for vaccine design. Recently, filamentous phage f88 was engineered to display an immunizing epitope from influenza A virus, which could be produced continuously from a probiotic gut-colonizing E. coli strain. This E. coli strain was modified to express the Yersinia pseudotuberculosis invA gene, which promotes adhesion to the gut mucosa, enabling long-term colonization. This system yielded encouraging protection against influenza A virus infection (114). The benefits of such an approach include ease of oral immunization and the potential to eliminate booster inoculations due to the sustained colonization and delivery by the vaccine-producing probiotic, which make this approach well suited for low-resource settings. However, overstimulation of the immune system is a potential trade-off for sustained delivery.
Phages have undergone a selection process over hundreds of millions of years so that they are maximally productive when infecting bacteria but not necessarily amenable to human engineering. Thus, there is interest for biotechnology applications in developing fully synthetic phages. Inspired by RNA phages, such as MS2, Baker and colleagues (115) designed in silico a fully synthetic nucleocapsid that packages its own mRNA. This synthetic particle was successfully assembled and subsequently evolved in E. coli from random mutational libraries to select for capsids with increased packaging efficiency, improved residence time in the mouse circulatory system, and better modularity for cloning inserts (115), thus demonstrating that virus-like functions can be obtained from a fully synthetic template. This work is a seminal step toward developing more modular virus-based vaccines.
Phage-Based Nanomaterials
Filamentous phages are of great interest to materials scientists because of the ease of displaying peptides with specific functions on the phage surface. For example, screening of phage display libraries identified peptides capable of crystallizing metals onto filamentous phages to produce nanowires for bioelectronics (116–118). M13 can also be transformed to bind to carbon nanotubes through engineering of its coat protein (119). An asparagine-to-arginine mutation at position 381 of the M13 pVII protein allowed the phage to recognize and bind carbon nanotubes selectively, thus scaffolding the tubes into three-dimensional structures (120). Finally, M13 has been engineered to template gold nanoparticles, as well as nanoparticles made of other metals, illustrating its versatility in the creation of novel bionanomaterials (121, 122).
Bacteriophage capsids can be used to create structures that enhance enzymatic activities through spatial organization of enzymes in close proximity to protect unstable intermediates in enzymatic cascades, thus increasing metabolic flux. The self-assembling coat protein of phage MS2 was turned into a microreactor by genetically encoding a SpyTag on its inside surface and using it to attach two different enzymes, each fused to the complementary SpyCatcher peptide. SpyTag and SpyCatcher react together to form a covalent isopeptide bond that leads to the encapsulation and colocalization of both enzymes within the capsid core (123). As a proof of concept, the two enzymes that transform tryptophan into indigo, TnaA and FMO, were fused to SpyCatcher and incorporated into VLPs. The resulting VLP conjugate kept the enzymes active for at least 7 days and increased indigo production by 60% compared to production obtained with free enzymes. Because some substrates and products are more soluble in a lipid-rich environment, the same researchers developed a lipid-containing nanoreactor, using the lipid-containing phage φ6 as a scaffold (124).
PHAGE-BASED DISCOVERY TECHNOLOGIES
Phage Display
Phage display is a powerful technique to screen for variants of biological functions with improved characteristics (125). Originally, phage display fused the peptides to pIII, the coat protein of filamentous phage M13. Because M13 is maintained as a small, easily engineered plasmid in E. coli and can package its own genetic information, phenotypic selection can quickly be linked to genotypic variations. Phage display systems based on other phages (Fd, λ, T7, T4) have been described, and some afford the possibility of screening large proteins instead of small peptides. Phage display was quickly adopted for peptide/ligand studies, in particular for engineering antibody specificities and homing peptides. For an in-depth review of M13 phage display, we recommend the work of Wu et al. (126).
Phage-Assisted Continuous Evolution
Phages can also be used to evolve functions beyond binding. Phage-assisted continuous evolution (PACE) is a platform that links the growth of a phage to the evolution of a biological function by coupling the expression of an essential phage component to the desired biological function. As a result, only phages that solve the “problem” will reproduce, and the “solution” can easily be extracted by sequencing the genomes of the successful phages. The system can also increase the mutation rate and accelerate evolution through the use of an inducible mutagenic DNA polymerase.
In the pioneering paper by Esvelt et al. (127), the function under selection was the promoter specificity of T7 RNAP. To force the directed evolution of T7 RNAP, its gene was cloned into a defective M13 devoid of its coat protein gene pIII. The pIII gene was cloned in a separate plasmid under control of a T3 promoter, such that production of M13 was dependent upon T7 RNAP acquiring the capacity to initiate transcription from a T3 promoter. In such a system, only the M13 genomes acquiring a functional mutation in the T7 RNAP gene can produce progeny. This system was fully automated and generated a successful clone in less than 200 h of evolution (127). By creatively modulating the design of the promoter that drives pIII expression and accompanying selection pressures, many types of functions can be selected for or against (128). For example, PACE was applied for the evolution of novel amino-acyl tRNA synthetases (129), proteases with altered target motifs (130), transcription factors (20, 131), and even allowed the evolution of mutants of the Bacillus thuringiensis δ-endotoxin that are less prone to resistance by insects (132).
Recording Events by Rewriting DNA
Recombineering using phage λ recombination machinery allows efficient recombination between chromosomal DNA and single-stranded or double-stranded linear DNA. Farzadfard & Lu (133) creatively leveraged this function to record information into bacterial genomes at specific loci by building a synthetic programmable retron as a means of generating single-stranded DNA directly inside the target cells in an inducible fashion, which recombines into homologous regions in the bacterial genome when coexpressed with λ bet (133). This system yields revertants at a frequency of 10−5 to 10−7, but the authors were able to improve the efficiency by several orders of magnitude by deleting and xonA genes. Putting this system under the control of sensors sensitive to light or chemicals allowed analog recording of induction events through quantification of the reversion rates (133). Beyond recording sensing events, the ability to produce single-stranded DNA directly inside bacteria and the improved recombination efficiency make this system amenable to protein engineering–based applications.
CONCLUDING REMARKS
Phages have been indispensable players in the development of biology and have contributed immensely to biotechnology and to synthetic biology in particular. Phage genomes are important contributors of components for building genetic circuits, and based on the diversity of phages as assessed through genomics, many more useful functions hide within phage genomes waiting to be discovered. Particularly untapped are temperate phage antirepressors and termination factors, which could help build delicately regulated genetic circuits. Temperate phages are also a rich source of integrases, which will be invaluable as biological programming expands beyond the proof-of-concept stages. There is growing interest in using DNA for information storage, as demonstrated by the inception of companies such as Catalog, which raised ~$25 million to develop the use of DNA as archiving material, although achieving efficient read/write cycles on DNA is still complicated. Phages have contributed many of the enzymes used to clone or modify DNA and have evolved to replicate their genome quickly upon infection; therefore, we surmise that phage functions will supply part of the solution.
Another area of great interest for synthetic biologists is creating synthetic phages for therapeutic purposes. The biological nature of phages makes them quickly programmable and evolvable. We have presented recent examples of how researchers have increased the therapeutic potential of natural phages, and we foresee that with the ever-growing global threat of pan-antibiotic resistance, interest in clinically efficient phages will continue to grow as a complementary approach. This is illustrated by the record number of small biotech companies using phages in ever more clever ways to tackle afflictions that are otherwise difficult to treat. Of particular interest to the field will be the capacity to build biocontained phages, which are phages that replicate only under precise conditions, such as in the presence of a secondary drug. Biocontainment will be crucial for preventing the spread of phages built for maximum killing efficiency into environments where they may push bacterial populations toward resistance or disrupt natural bacterial communities. A better understanding of the biology of host recognition will also be imperative to control which bacterial populations are susceptible to therapeutic phages.
Isolated antibacterial phage components such as lysins are also witnessing a strong surge of interest, and natural or engineered phage lysins are being advanced into various stages of clinical or veterinary testing by companies such as ContraFect, GangaGen, and Lysando (134). This is an area of research that is sure to grow and one for which the capacity to predict, manipulate, and reprogram biological activities will be immensely useful.
The ease of engineering phages is a great inspiration for materials scientists, who can quickly create, select, and test functionalities displayed on phage particles. The examples discussed in this review highlight how these systems can improve the synthesis of a variety of organic and inorganic compounds with difficult-to-obtain geometries through chemistry. We expect that fully synthetic phage-inspired scaffolds will slowly replace naturally sourced ones as they can be built with improved modularity compared to natural phages and possibly with three-dimensional structures not achievable using natural phage capsids.
Finally, with an estimated 1031 bacteriophages on the planet and the recent discovery of an entirely new phage family that went unnoticed for decades despite being a major contributor to the phageome of oceans across the globe (135), we expect that exciting yet unknown possibilities will arise and quickly find their way to synthetic biologists, allowing for ever more precise programming of biological systems.
ACKNOWLEDGMENTS
We thank Karen Pepper for reading over the document and providing input. S.L., K.M.Y., and T.K.L. wrote the article. The Lu lab acknowledges financial support from the National Science Foundation (MCB-1350625, #1521925), the National Institutes of Health (5-P50-GM098792-05), the Office of Naval Research (N00014-13-1-0424), the Army Research Office (W911NF-ll-1-0281), the Broad Institute, the Koch Institute, the Defense Threat Reduction Agency (HDTRAl-14-1-0007), the Rainin Foundation (2016-3066), the Ellison Foundation (AG-NS-0948-12), the Wertheimer Fund, the Institute for Soldier Nanotechnologies (W9111NF-13-D-0001), J-WAFS, Novartis (14081955), the Desphande Center at the Massachusetts Institute of Technology (MIT), the Singapore-MIT Alliance for Research and Technology, and the Center for Microbiome Informatics and Technology (15127713).
Footnotes
DISCLOSURE STATEMENT
T.K.L. is a shareholder in phage-related companies, including AmpliPhi, BiomX, and Eligo Bioscience.
LITERATURE CITED
- 1.Summers WC, Stent G, Twort F. 2012. The strange history of phage therapy. Bacteriophage 2:130–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hatfull GF. 2008. Bacteriophage genomics. Curr. Opin. Microbiol 11:447–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hershey AD, Chase M. 1952. Independent functions of viral protein and nucleic acid in growth of bacteriophage. J. Gen. Physiol 36:39–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crick FH, Barnett L, Brenner S, Watts-Tobin RJ. 1961. General nature of the genetic code for proteins. Nature 192:1227–32 [DOI] [PubMed] [Google Scholar]
- 5.Bertani G, Weigle JJ. 1953. Host controlled variation in bacterial viruses. J. Bacteriol 65:113–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weiss B, Richardson CC. 1967. Enzymatic breakage and joining of deoxyribonucleic acid. I. Repair of single-strand breaks in DNA by an enzyme system from Escherichia coli infected with T4 bacteriophage. PNAS 57:1021–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chamberlin M, Ring J. 1973. Characterization of T7-specific ribonucleic acid polymerase. 1. General properties of the enzymatic reaction and the template specificity of the enzyme. J. Biol. Chem 248:2235–44 [PubMed] [Google Scholar]
- 8.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, et al. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709–12 [DOI] [PubMed] [Google Scholar]
- 9.Donohoue PD, Barrangou R, May AP. 2018. Advances in industrial biotechnology using CRISPR-Cas systems. Trends Biotechnol. 36:134–46 [DOI] [PubMed] [Google Scholar]
- 10.Sousa R, Mukherjee S. 2003. T7 RNA polymerase. Prog. Nucleic Acid Res. Mol. Biol 73:1–41 [DOI] [PubMed] [Google Scholar]
- 11.Lenneman B, Rothman-Denes L. 2015. Structural and biochemical investigation of bacteriophage N4-encoded RNA polymerases. Biomolecules 5:647–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Temme K, Hill R, Segall-Shapiro TH, Moser F, Voigt CA. 2012. Modular control of multiple pathways using engineered orthogonal T7 polymerases. Nucleic Acids Res. 40:8773–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meyer AJ, Ellefson JW, Ellington AD. 2015. Directed evolution of a panel of orthogonal T7 RNA polymerase variants for in vivo or in vitro synthetic circuitry. ACS Synth. Biol 4:1070–76 [DOI] [PubMed] [Google Scholar]
- 14.Segall-Shapiro TH, Meyer AJ, Ellington AD, Sontag ED, Voigt CA. 2014. A “resource allocator” for transcription based on a highly fragmented T7 RNA polymerase. Mol. Syst. Biol 10:742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pu J, Zinkus-Boltz J, Dickinson BC. 2017. Evolution of a split RNA polymerase as a versatile biosensor platform. Nat. Chem. Biol 13:432–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han T, Chen Q, Liu H. 2017. Engineered photoactivatable genetic switches based on the bacterium phage T7 RNA polymerase. ACS Synth. Biol 6:357–66 [DOI] [PubMed] [Google Scholar]
- 17.Baumschlager A, Aoki SK, Khammash M. 2017. Dynamic blue light-inducible T7 RNA polymerases (opto-T7RNAPs) for precise spatiotemporal gene expression control. ACS Synth. Biol 6:2157–67 [DOI] [PubMed] [Google Scholar]
- 18.Hochschild A, Lewis M. 2009. The bacteriophage λCI protein finds an asymmetric solution. Curr. Opin. Struct. Biol 19:79–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elowitz MB, Leibler S. 2000. A synthetic oscillatory network of transcriptional regulators. Nature 403:335–38 [DOI] [PubMed] [Google Scholar]
- 20.Brödel AK, Jaramillo A, Isalan M. 2016. Engineering orthogonal dual transcription factors for multi-input synthetic promoters. Nat. Commun 7:13858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kotula JW, Kerns SJ, Shaket LA, Siraj L, Collins JJ, et al. 2014. Programmable bacteria detect and record an environmental signal in the mammalian gut. PNAS 111:4838–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stirling F, Bitzan L, O’Keefe S, Redfield E, Oliver JWK, et al. 2017. Rational design of evolutionarily stable microbial kill switches. Mol. Cell 68:686–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Noman N, Inniss M, Iba H, Way JC. 2016. Pulse detecting genetic circuit—a new design approach. PLOS ONE 11:e0167162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang L, Nielsen AAK, Fernandez-Rodriguez J, McClune CJ, Laub MT, et al. 2014. Permanent genetic memory with >1-byte capacity. Nat. Methods 11:1261–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fogg PCM, Colloms S, Rosser S, Stark M, Smith MCM. 2014. New applications for phage integrases.J. Mol. Biol 426:2703–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Inniss MC, Bandara K, Jusiak B, Lu TK, Weiss R, et al. 2017. A novel Bxb1 integrase RMCE system for high fidelity site-specific integration of mAb expression cassette in CHO cells. Biotechnol. Bioeng 114:1837–46 [DOI] [PubMed] [Google Scholar]
- 27.Zhu F, Gamboa M, Farruggio AP, Hippenmeyer S, Tasic B, et al. 2014. DICE, an efficient system for iterative genomic editing in human pluripotent stem cells. Nucleic Acids Res. 42:e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matreyek KA, Stephany JJ, Fowler DM. 2017. A platform for functional assessment of large variant libraries in mammalian cells. Nucleic Acids Res. 45:e102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Siuti P, Yazbek J, Lu TK. 2013. Synthetic circuits integrating logic and memory in living cells. Nat. Biotechnol 31:448–52 [DOI] [PubMed] [Google Scholar]
- 30.Hsiao V, Hori Y, Rothemund PW, Murray RM. 2016. A population-based temporal logic gate for timing and recording chemical events. Mol. Syst. Biol 12:869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Siuti P, Yazbek J, Lu TK. 2014. Engineering genetic circuits that compute and remember. Nat. Protoc 9:1292–300 [DOI] [PubMed] [Google Scholar]
- 32.Daniel R, Rubens JR, Sarpeshkar R, Lu TK. 2013. Synthetic analog computation in living cells. Nature 497:619–23 [DOI] [PubMed] [Google Scholar]
- 33.Roquet N, Soleimany AP, Ferris AC, Aaronson S, Lu TK. 2016. Synthetic recombinase-based state machines in living cells. Science 353:aad8559. [DOI] [PubMed] [Google Scholar]
- 34.Weinberg BH, Pham NTH, Caraballo LD, Lozanoski T, Engel A, et al. 2017. Large-scale design of robust genetic circuits with multiple inputs and outputs for mammalian cells. Nat. Biotechnol 35:453–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fenno LE, Mattis J, Ramakrishnan C, Hyun M, Lee SY, et al. 2014. Targeting cells with single vectors using multiple-feature Boolean logic. Nat. Methods 11:763–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Calendar R, Abedon S. 2006. The Bacteriophages. Oxford, UK: Oxford Univ. Press [Google Scholar]
- 37.Marinelli LJ, Piuri M, Swigonová Z, Balachandran A, Oldfield LM, et al. 2008. BRED: a simple and powerful tool for constructing mutant and recombinant bacteriophage genomes. PLOS ONE 3:e3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oppenheim AB, Rattray AJ, Bubunenko M, Thomason LC, Court DL. 2004. In vivo recombineering of bacteriophage λ by PCR fragments and single-strand oligonucleotides. Virology 319:185–89 [DOI] [PubMed] [Google Scholar]
- 39.Fehér T, Karcagi I, Blattner FR, Pósfai G. 2012. Bacteriophage recombineering in the lytic state using the lambda red recombinases. Microb. Biotechnol 5:466–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thomason L, Court DL, Bubunenko M, Costantino N, Wilson H, et al. 2007. Recombineering: genetic engineering in bacteria using homologous recombination. Curr. Protoc. Mol. Biol 106:1.16.1–39 [DOI] [PubMed] [Google Scholar]
- 41.Pouillot F, Blois H, Iris F. 2010. Genetically engineered virulent phage banks in the detection and control of emergent pathogenic bacteria. Biosecur. Bioterrorism Biodefense Strateg. Pract. Sci 8:155–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Swingle B, Bao Z, Markel E, Chambers A, Cartinhour S. 2010. Recombineering using RecTE from Pseudomonas syringae. Appl. Environ. Microbiol 76:4960–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martel B, Moineau S. 2014. CRISPR-Cas: an efficient tool for genome engineering of virulent bacteriophages. Nucleic Acids Res. 42:9504–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kiro R, Shitrit D, Qimron U. 2014. Efficient engineering of a bacteriophage genome using the type I-E CRISPR-Cas system. RNA Biol. 11:42–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lemay ML, Tremblay DM, Moineau S. 2017. Genome engineering of virulent lactococcal phages using CRISPR-Cas9. ACS Synth. Biol 6:1351–58 [DOI] [PubMed] [Google Scholar]
- 46.Tao P, Wu X, Tang WC, Zhu J, Rao V. 2017. Engineering of bacteriophage T4 genome using CRISPR-Cas9. ACS Synth. Biol 6:1952–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jaschke PR, Lieberman EK, Rodriguez J, Sierra A, Endy D. 2012. A fully decompressed synthetic bacteriophage øX174 genome assembled and archived in yeast. Virology 434:278–84 [DOI] [PubMed] [Google Scholar]
- 48.Ando H, Lemire S, Pires DP, Lu TK. 2015. Engineering modular viral scaffolds for targeted bacterial population editing. Cell Syst. 1:187–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Allan EJ, Hoischen C, Gumpert J. 2009. Bacterial L-forms. Adv. Appl. Microbiol 68:1–39 [DOI] [PubMed] [Google Scholar]
- 50.Kilcher S, Studer P, Muessner C, Klumpp J, Loessner MJ. 2018. Cross-genus rebooting of custom-made, synthetic bacteriophage genomes in L-form bacteria. PNAS 115:567–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gibson DG, Young L, Chuang R, Venter JC, Hutchinson CA III, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6:343–45 [DOI] [PubMed] [Google Scholar]
- 52.Garamella J, Marshall R, Rustad M, Noireaux V. 2016. The all E. coli TX-TL toolbox 2.0: a platform for cell-free synthetic biology. ACS Synth. Biol 5:344–55 [DOI] [PubMed] [Google Scholar]
- 53.Kutter E, De Vos D, Gvasalia G, Alavidze Z, Gogokhia L, et al. 2010. Phage therapy in clinical practice: treatment of human infections. Curr. Pharm. Biotechnol 11:69–86 [DOI] [PubMed] [Google Scholar]
- 54.Cooper CJ, Khan Mirzaei M, Nilsson AS. 2016. Adapting drug approval pathways for bacteriophage-based therapeutics. Front. Microbiol 7:1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Canchaya C, Proux C, Fournous G, Bruttin A, Bru H. 2003. Prophage genomics. Microbiol. Mol. Biol. Rev 67:238–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Górski A, Międzybrodzki R, Borysowski J, Dąbrowska K, Wierzbicki P, et al. 2012. Phage as a modulator of immune responses. Adv. Virus Res. 83:41–71 [DOI] [PubMed] [Google Scholar]
- 57.Lu TK, Collins JJ. 2007. Dispersing biofilms with engineered enzymatic bacteriophage. PNAS 104:11197–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lin H, Paff ML, Molineux IJ, Bull JJ. 2017. Therapeutic application of phage capsule depolymerases against K1, K5, and K30 capsulated E. coli in mice. Front. Microbiol 8:2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fleming D, Rumbaugh KP. 2017. Approaches to dispersing medical biofilms. Microorganisms 5:E15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Born Y, Fieseler L, Thöny V, Leimer N, Duffy B, Loessner MJ. 2017. Engineering of bacteriophages Y2::dpoL1-C and Y2::luxAB for efficient control and rapid detection of the fire blight pathogen, Erwinia amylovora. Appl. Environ. Microbiol 83:e00341–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Silva JB, Storms Z, Sauvageau D. 2016. Host receptors for bacteriophage adsorption. FEMS Microbiol. Lett 363:fnw002. [DOI] [PubMed] [Google Scholar]
- 62.Yu P, Mathieu J, Li M, Dai Z, Alvarez PJJ. 2015. Isolation of polyvalent bacteriophages by sequential multiple-host approaches. Appl. Environ. Microbiol 82:808–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hamdi S, Rousseau GM, Labrie SJ, Tremblay DM, Kourda RS, et al. 2017. Characterization of two polyvalent phages infecting Enterobacteriaceae. Sci. Rep 7:40349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Matsuda T, Freeman TA, Hilbert DW, Duff M, Fuortes M, et al. 2005. Lysis-deficient bacteriophage therapy decreases endotoxin and inflammatory mediator release and improves survival in a murine peritonitis model. Surgery 137:639–46 [DOI] [PubMed] [Google Scholar]
- 65.Murooka Y, Takizawa N, Harada T. 1981. Introduction of bacteriophage Mu into bacteria of various genera and intergeneric gene transfer by RP4::Mu. J. Bacteriol 145:358–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Murooka Y, Harada T. 1979. Expansion of the host range of coliphage P1 and gene transfer from enteric bacteria to other gram-negative bacteria. Appl. Environ. Microbiol 38:754–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Krom RJ, Bhargava P, Lobritz MA, Collins JJ. 2015. Engineered phagemids for nonlytic, targeted antibacterial therapies. Nano Lett. 15:4808–13 [DOI] [PubMed] [Google Scholar]
- 68.Citorik RJ, Mimee M, Lu TK. 2014. Sequence-specific antimicrobials using efficiently delivered RNA-guided nucleases. Nat. Biotechnol 32:1141–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chung YB, Hinkle DC. 1990. Bacteriophage T7 DNA packaging. I. Plasmids containing a T7 replication origin and the T7 concatemer junction are packaged into transducing particles during phage infection.J. Mol. Biol 216:911–26 [DOI] [PubMed] [Google Scholar]
- 70.Hashimoto C 1988. Packaging and transduction of non-T3 DNA by bacteriophage T3. Virology 439:432–39 [DOI] [PubMed] [Google Scholar]
- 71.Yosef I, Goren MG, Globus R, Molshanski-Mor S, Qimron U. 2017. Extending the host range of bacteriophage particles for DNA transduction. Mol. Cell 66:721–28 [DOI] [PubMed] [Google Scholar]
- 72.Brüssow H 2012. What is needed for phage therapy to become a reality in Western medicine? Virology 434:138–42 [DOI] [PubMed] [Google Scholar]
- 73.Hargreaves KR, Clokie MRJ. 2014. Clostridium difficile phages: still difficult? Front. Microbiol 5:184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pastagia M, Schuch R, Fischetti VA, Huang DB. 2013. Lysins: the arrival of pathogen-directed anti-infectives. J. Med. Microbiol 62(Pt. 10):1506–16 [DOI] [PubMed] [Google Scholar]
- 75.Lukacik P, Barnard TJ, Keller PW, Chaturvedi KS, Seddiki N, et al. 2012. Structural engineering of a phage lysin that targets gram-negative pathogens. PNAS 109:9857–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Briers Y, Walmagh M, Van Puyenbroeck V, Cornelissen A, Cenens W, et al. 2014. Engineered endolysin-based “Artilysins” to combat multidrug-resistant gram-negative pathogens. mBio 5:e01379–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Defraine V, Schuermans J, Grymonprez B, Govers SK, Aertsen A, et al. 2016. Efficacy of Artilysin Art-175 against resistant and persistent Acinetobacter baumannii. Antimicrob. Agents Chemother. 60:3480–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Piuri M, Jacobs WR, Hatfull GF. 2009. Fluoromycobacteriophages for rapid, specific, and sensitive antibiotic susceptibility testing of Mycobacterium tuberculosis. PLOS ONE 4:e4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.O’Donnell MR, Pym A, Jain P, Munsamy V, Wolf A, et al. 2015. A novel reporter phage to detect tuberculosis and rifampin resistance in a high-HIV-burden population. J. Clin. Microbiol 53:2188–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jain P, Hartman TE, Eisenberg N, O’Donnell MR, Kriakov J, et al. 2012. ϕ2GFP10, a high-intensity fluorophage, enables detection and rapid drug susceptibility testing of Mycobacterium tuberculosis directly from sputum samples. J. Clin. Microbiol 50:1362–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jacobs WR, Barletta RG, Udani R, Chan J, Kalkut G, et al. 1993. Rapid assessment of drug susceptibilities of Mycobacterium tuberculosis by means of luciferase reporter phages. Science 260:819–22 [DOI] [PubMed] [Google Scholar]
- 82.Sarkis GJ, Jacobs WR, Hatfull GF. 1995. L5 luciferase reporter mycobacteriophages: a sensitive tool for the detection and assay of live mycobacteria. Mol. Microbiol 15:1055–67 [DOI] [PubMed] [Google Scholar]
- 83.Pearson RE, Jurgensen S, Sarkis GJ, Hatfull GF, Jacobs WR. 1996. Construction of D29 shuttle phasmids and luciferase reporter phages for detection of mycobacteria. Gene 183:129–36 [DOI] [PubMed] [Google Scholar]
- 84.Kumar V, Loganathan P, Sivaramakrishnan G, Kriakov J, Dusthakeer A, et al. 2008. Characterization of temperate phage Che12 and construction of a new tool for diagnosis of tuberculosis. Tuberculosis 88:616–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Peng Y, Jin Y, Lin H, Wang J, Khan MN. 2014. Application of the VPp1 bacteriophage combined with a coupled enzyme system in the rapid detection of Vibrio parahaemolyticus. J. Microbiol. Methods 98:99–104 [DOI] [PubMed] [Google Scholar]
- 86.Kim S, Kim M, Ryu S. 2014. Development of an engineered bioluminescent reporter phage for the sensitive detection of viable Salmonella Typhimurium. Anal. Chem 86:5858–64 [DOI] [PubMed] [Google Scholar]
- 87.Vandamm JP, Rajanna C, Sharp NJ, Molineux IJ, Schofield DA. 2014. Rapid detection and simultaneous antibiotic susceptibility analysis of Yersinia pestis directly from clinical specimens by use of reporter phage.J. Clin. Microbiol 52:2998–3003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Schofield DA, Wray DJ, Molineux IJ. 2015. Isolation and development of bioluminescent reporter phages for bacterial dysentery. Eur. J. Clin. Microbiol. Infect. Dis 34:395–403 [DOI] [PubMed] [Google Scholar]
- 89.Sharp NJ, Vandamm JP, Molineux IJ, Schofield DA. 2015. Rapid detection of Bacillus anthracis in complex food matrices using phage-mediated bioluminescence. J. Food Prot. 78:963–68 [DOI] [PubMed] [Google Scholar]
- 90.Sharp NJ, Molineux IJ, Page MA, Schofield DA. 2016. Rapid detection of viable Bacillus anthracis spores in environmental samples by using engineered reporter phages. Appl. Environ. Microbiol 82:2380–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhang D, Coronel-Aguilera CP, Romero PL, Perry L, Minocha U, et al. 2016. The use of a novel NanoLuc-based reporter phage for the detection of Escherichia coli O157:H7. Sci. Rep 6:33235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kim J, Kim M, Kim S, Ryu S. 2017. Sensitive detection of viable Escherichia coli O157:H7 from foods using a luciferase-reporter phage φV10 lux. Int. J. Food Microbiol. 254:11–17 [DOI] [PubMed] [Google Scholar]
- 93.Oosterik LH, Tuntufye HN, Tsonos J, Luyten T, Noppen S, et al. 2016. Bioluminescent avian pathogenic Escherichia coli for monitoring colibacillosis in experimentally infected chickens. Vet. J 216:87–92 [DOI] [PubMed] [Google Scholar]
- 94.Loessner MJ, Rudolf M, Scherer S. 1997. Evaluation of luciferase reporter bacteriophage A511::luxAB for detection of Listeria monocytogenes in contaminated foods. Appl. Environ. Microbiol 63:2961–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Schmidt A, Rabsch W, Broeker NK, Barbirz S. 2016. Bacteriophage tailspike protein based assay to monitor phase variable glucosylations in Salmonella O-antigens. BMC Microbiol. 16:207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Schmelcher M, Shabarova T, Eugster MR, Eichenseher F, Tchang VS, et al. 2010. Rapid multiplex detection and differentiation of Listeria cells by use of fluorescent phage endolysin cell wall binding domains. Appl. Environ. Microbiol 76:5745–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Na H, Kong M, Ryu S. 2016. Characterization of LysPBC4, a novel Bacillus cereus-specific endolysin of bacteriophage PBC4. FEMS Microbiol. Lett 363:fnw092. [DOI] [PubMed] [Google Scholar]
- 98.Gómez-Torres N, Dunne M, Garde S, Meijers R, Narbad A, et al. 2018. Development of a specific fluorescent phage endolysin for in situ detection of Clostridium species associated with cheese spoilage. Microb. Biotechnol 11:332–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Benešík M, Nováček J, Janda L, Dopitová R, Pernisová M, et al. 2018. Role of SH3b binding domain in a natural deletion mutant of Kayvirus endolysin LysF1 with a broad range of lytic activity. Virus Genes 54:130–39 [DOI] [PubMed] [Google Scholar]
- 100.Ugorcakova J, Medzova L, Solteszova B, Bukovska G. 2015. Characterization of a φBP endolysin encoded by the Paenibacillus polymyxa CCM 7400 phage. FEMS Microbiol. Lett 362:fnv098. [DOI] [PubMed] [Google Scholar]
- 101.Kovacs EW, Hooker JM, Romanini DW, Holder PG, Berry KE, Francis MB. 2007. Dual-surface-modified bacteriophage MS2 as an ideal scaffold for a viral capsid-based drug delivery system. Bioconjug. Chem 18:1140–47 [DOI] [PubMed] [Google Scholar]
- 102.Ashley CE, Carnes EC, Phillips GK, Durfee PN, Buley MD, et al. 2011. Cell-specific delivery of diverse cargos by bacteriophage MS2 virus-like particles. ACS Nano 5:5729–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rhee JK, Baksh M, Nycholat C, Paulson JC, Kitagishi H, Finn MG. 2012. Glycan-targeted virus-like nanoparticles for photodynamic therapy. Biomacromolecules 13:2333–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Galaway FA, Stockley PG. 2013. MS2 viruslike particles: a robust, semisynthetic targeted drug delivery platform. Mol. Pharm 10:59–68 [DOI] [PubMed] [Google Scholar]
- 105.Kato T, Yui M, Deo VK, Park EY. 2015. Development of rous sarcoma virus-like particles displaying hCC49 scFv for Specific targeted drug delivery to human colon carcinoma cells. Pharm. Res 32:3699–707 [DOI] [PubMed] [Google Scholar]
- 106.Deo VK, Kato T, Park EY. 2016. Virus-like particles displaying recombinant short-chain fragment region and interleukin 2 for targeting colon cancer tumors and attracting macrophages. J. Pharm. Sci 105:1614–22 [DOI] [PubMed] [Google Scholar]
- 107.Sánchez-Sánchez L, Tapia-Moreno A, Juarez-Moreno K, Patterson DP, Cadena-Nava RD, et al. 2015. Design of a VLP-nanovehicle for CYP450 enzymatic activity delivery. J. Nanobiotechnol 13:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lino CA, Caldeira JC, Peabody DS. 2017. Display of single-chain variable fragments on bacteriophage MS2 virus-like particles. J. Nanobiotechnol 15:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Vaks L, Benhar I. 2011. Antibacterial application of engineered bacteriophage nanomedicines: antibody-targeted, chloramphenicol prodrug loaded bacteriophages for inhibiting the growth of Staphylococcus aureus bacteria. Methods Mol. Biol 726:187–206 [DOI] [PubMed] [Google Scholar]
- 110.Anand P, O’Neil A, Lin E, Douglas T, Holford M. 2015. Tailored delivery of analgesic ziconotide across a blood brain barrier model using viral nanocontainers. Sci. Rep 5:12497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Qazi S, Miettinen HM, Wilkinson RA, McCoy K, Douglas T, Wiedenheft B. 2016. Programmed self-assembly of an active P22-Cas9 nanocarrier system. Mol. Pharm 13:1191–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jennings GT, Bachmann MF. 2008. The coming of age of virus-like particle vaccines. Biol. Chem 389:521–36 [DOI] [PubMed] [Google Scholar]
- 113.Lua LHL, Connors NK, Sainsbury F, Chuan YP, Wibowo N, Middelberg APJ. 2014. Bioengineering virus-like particles as vaccines. Biotechnol. Bioeng 111:425–40 [DOI] [PubMed] [Google Scholar]
- 114.Deng L, Roose K, Job ER, De Rycke R, Van Hamme E, et al. 2017. Oral delivery of Escherichia coli persistently infected with M2e-displaying bacteriophages partially protects against influenza A virus. J. Control. Release 264:55–65 [DOI] [PubMed] [Google Scholar]
- 115.Butterfield GL, Lajoie MJ, Gustafson HH, Sellers DL, Nattermann U, et al. 2017. Evolution of a designed protein assembly encapsulating its own RNA genome. Nature 552:415–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Han L, Shao C, Liang B, Liu A. 2016. Genetically engineered phage-templated MnO2 nanowires: synthesis and their application in electrochemical glucose biosensor operated at neutral pH condition. ACS Appl. Mater. Interfaces 8:13768–76 [DOI] [PubMed] [Google Scholar]
- 117.Li Y, Cao B, Yang M, Zhu Y, Suh J, Mao C. 2016. Identification of novel short BaTiO3-binding/nucleating peptides for phage-templated in situ synthesis of BaTiO3 polycrystalline nanowires at room temperature. ACS Appl. Mater. Interfaces 8:30714–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Manivannan S, Kang I, Seo Y, Jin HE, Lee SW, Kim K. 2017. M13 virus-incorporated biotemplates on electrode surfaces to nucleate metal nanostructures by electrodeposition. ACS Appl. Mater. Interfaces 9:32965–76 [DOI] [PubMed] [Google Scholar]
- 119.Yi H, Ghosh D, Ham MH, Qi J, Barone PW, et al. 2012. M13 phage-functionalized single-walled carbon nanotubes as nanoprobes for second near-infrared window fluorescence imaging of targeted tumors. Nano Lett. 12:1176–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Szot-Karpińska K, Golec P, Leśniewski A, Pałys B, Marken F, et al. 2016. Modified filamentous bacteriophage as a scaffold for carbon nanofiber. Bioconjug. Chem 27:2900–10 [DOI] [PubMed] [Google Scholar]
- 121.Golec P, Żelechowska K, Karczewska-Golec J, Karczewski J, Leśniewski A, et al. 2017. Bacteriophages as factories for Eu2O3 nanoparticle synthesis. Bioconjug. Chem 28:1834–41 [DOI] [PubMed] [Google Scholar]
- 122.Lee HK, Lee Y, Kim H, Lee HE, Chang H, et al. 2017. Screening of Pro-Asp sequences exposed on bacteriophage M13 as an ideal anchor for gold nanocubes. ACS Synth. Biol 6:1635–41 [DOI] [PubMed] [Google Scholar]
- 123.Giessen TW, Silver PA. 2016. A catalytic nanoreactor based on in vivo encapsulation of multiple enzymes in an engineered protein nanocompartment. ChemBioChem 17:1931–35 [DOI] [PubMed] [Google Scholar]
- 124.Myhrvold C, Polka JK, Silver PA. 2016. Synthetic lipid-containing scaffolds enhance production by colocalizing enzymes. ACS Synth. Biol 5:1396–403 [DOI] [PubMed] [Google Scholar]
- 125.Smith GP. 1985. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science 228:1315–17 [DOI] [PubMed] [Google Scholar]
- 126.Wu CH, Liu IJ, Lu RM, Wu HC. 2016. Advancement and applications of peptide phage display technology in biomedical science. J. Biomed. Sci 23:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Esvelt KM, Carlson JC, Liu DR. 2011. A system for the continuous directed evolution of biomolecules. Nature 472:499–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Carlson JC, Badran AH, Guggiana-Nilo DA, Liu DR. 2014. Negative selection and stringency modulation in phage-assisted continuous evolution. Nat. Chem. Biol 10:216–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Bryson DI, Fan C, Guo LT, Miller C, Söll D, Liu DR. 2017. Continuous directed evolution of aminoacyltRNA synthetases. Nat. Chem. Biol 13:1253–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Packer MS, Rees HA, Liu DR. 2017. Phage-assisted continuous evolution of proteases with altered substrate specificity. Nat. Commun 8:956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hubbard BP, Badran AH, Zuris JA, Guilinger JP, Davis KM, et al. 2015. Continuous directed evolution of DNA-binding proteins to improve TALEN specificity. Nat. Methods 12:939–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Badran AH, Guzov VM, Huai Q, Kemp MM, Vishwanath P, et al. 2016. Continuous evolution of Bacillus thuringiensis toxins overcomes insect resistance. Nature 533:58–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Farzadfard F, Lu TK. 2014. Genomically encoded analog memory with precise in vivo DNA writing in living cell populations. Science 346:1256272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sharma U, Paul VD. 2017. Bacteriophage lysins as antibacterials. Crit. Care 21:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kauffman KM, Hussain FA, Yang J, Arevalo P, Brown JM, et al. 2018. A major lineage of non-tailed dsDNA viruses as unrecognized killers of marine bacteria. Nature 554:118–22 [DOI] [PubMed] [Google Scholar]