Abstract
β-globin lentiviral vectors (β-LV) have faced challenges in clinical translation for gene therapy of sickle cell disease (SCD) due to low titer and sub-optimal gene transfer to hematopoietic stem and progenitor cells (HSPCs). To overcome the challenge of preserving efficacious expression while increasing vector performance, we used published genomic and epigenomic data available through ENCODE to redefine enhancer element boundaries of the β-globin locus control region (LCR) to construct novel ENCODE core sequences. These novel LCR elements were used to design a β-LV of reduced proviral length, termed CoreGA-AS3-FB, produced at higher titers and possessing superior gene transfer to HSPCs when compared to the full-length parental β-LV at equal MOI. At low vector copy number, vectors containing the ENCODE core sequences were capable of reversing the sickle phenotype in a mouse model of SCD. These studies provide a β-LV that will be beneficial for gene therapy of SCD by significantly reducing the cost of vector production and extending the vector supply.
Keywords: lentiviral vector, gene therapy, hematopoietic stem cell, transplantation, sickle cell disease, hemoglobinopathies, locus control region, ENCODE
Morgan and colleagues designed an improved globin expression vector capable of reversing the disease phenotype in a mouse model of sickle cell disease using epigenetic/genetic data available through ENCODE. This vector will be beneficial for gene therapy by significantly reducing the cost of vector production and extending vector supply.
Introduction
Sickle cell disease (SCD) is caused by a single nucleotide mutation in the β-globin gene, which results in the formation of abnormally charged hemoglobin (HbS) protein in red blood cells (RBCs).1 Vaso-occlusion is a major clinical feature of SCD and is caused by sickling of RBCs due to aggregation of HbS under low oxygen conditions.2 While two Food and Drug Administration approved drug treatments for SCD exist, both Hydroxyurea and L-Glutamine only modestly improve overall health of afflicted patients even when adherence is strictly maintained.3, 4
As an alternative therapy, allogenic hematopoietic stem cell transplantation (HSCT) has the potential to cure patients afflicted with SCD; however, identification of suitable histocompatibility leukocyte antigen (HLA)-matched donors remains a significant obstacle.5 Autologous HSCT combined with gene therapy is a more contemporary approach that entails the genetic modification of a patient’s own CD34+ hematopoietic stem and progenitor cells (HSPCs). By replacing a patient’s native HSPC population with genetically modified HSPCs harboring a normal or modified β-globin gene (such as Lenti/βAS3-FB6), SCD can be permanently cured without risks of graft versus host disease or graft rejection that accompanies the more traditional strategy of allogeneic HSCT.7 Preliminary data from clinical trials for hemoglobin disorders have indeed demonstrated a benefit from autologous HSCT with gene therapy.7, 8, 9, 10, 11
Low vector titer has represented a major barrier toward the effective advancement of therapeutic β-globin lentiviral vectors (β-LV) in clinical applications. Typically, β-LVs are produced at 10- to 100-fold lower titers when compared to LV with shorter genomes carrying small regulatory elements and cDNAs. In addition to the problem of low titer, clinical grade β-LV preparations (typically concentrated 100×–500×)12 showed reduced transduction efficiency for primary human HSPC and did not provide dose-dependent transduction of HSPCs with increasing vector concentrations (D.B.K. and R.A.M., unpublished data). The combinations of lower titer and subpar transduction efficiency mean that large volumes of vector need to be produced per patient dose, significantly adding to the clinical expense.
Poor gene transfer to HSPCs has also been a challenge in gene therapy trials and has limited the therapeutic benefit of gene therapy for patients with β-thalassemia and SCD.8, 13 Although the issues of gene transfer to HSPCs has been partially mitigated through the advent of transduction enhancers,14, 15, 16, 17 the purported use of transduction enhancers to achieve adequate vector copy numbers (VCNs) in clinic7, 9, 10 does not lower the cost of vector per patient dose when clinical vectors with inherently low infectivity are used.
One predominant factor that diminishes β-LV titer is the relatively large size of these vector genomes (e.g., 6–9 kb) due to the use of the human β-globin genomic sequences (promoter, exons, introns, 3′ UTR) and the β-globin Locus Control Region (LCR) DNase 1 hypersensitive site (HS) elements that augment expression. Another critical factor limiting vector performance is the inherent complexity of the human genomic elements; the β-globin genomic sequences and LCR are transcribed in reverse orientation relative to the vector genomic transcript (to retain intronic enhancers) and may contain problematic sequences that lead to non-functional vector genomes (e.g., cryptic polyadenylation signals).
The Globe-AS3-FB β-LV produced by Ferrari et al.18 is ∼2.7 kb smaller than the Lenti/βAS3-FB β-LV we have used19 (as well as other commonly used β-LVs)20, 21, 22, 23 and lacks the LCR HS4 sequences, the WPRE (woodchuck hepatitis virus post transcriptional regulatory element), and possesses a smaller 3′ UTR segment of the β-globin gene and a larger deletion of β-globin intron 2. It is unknown how sequence differences contribute to Globe-AS3-FB’s superior packaging and transduction efficiency when compared to Lenti/βAS3-FB24 or how these individual sequence differences influence β-globin expression.
Thus, we set out to determine whether the detrimental differences in titer and gene transfer of Lenti/βAS3-FB when compared to Globe-AS3-FB are due to proviral length or specific sequence elements. To reduce proviral size, we employed current genomic and epigenomic databases to identify and redefine functional elements present within the human genomic sequences of the LCR in Lenti/βAS3-FB to produce a composite LCR. A series of modified vectors were made with the aims of increasing titer and transduction efficiency while retaining high-level lineage-specific expression. Modified constructs were compared head-to-head and results were used to guide construction of a reduced length vector with superior titer and gene transfer and with sufficient expression of the anti-sickling βAS3-globin gene to induce hematologic correction of the sickle phenotype in the “Townes” mouse model of SCD.
Results
Proviral Length Influences Packaging and Transduction Efficiency
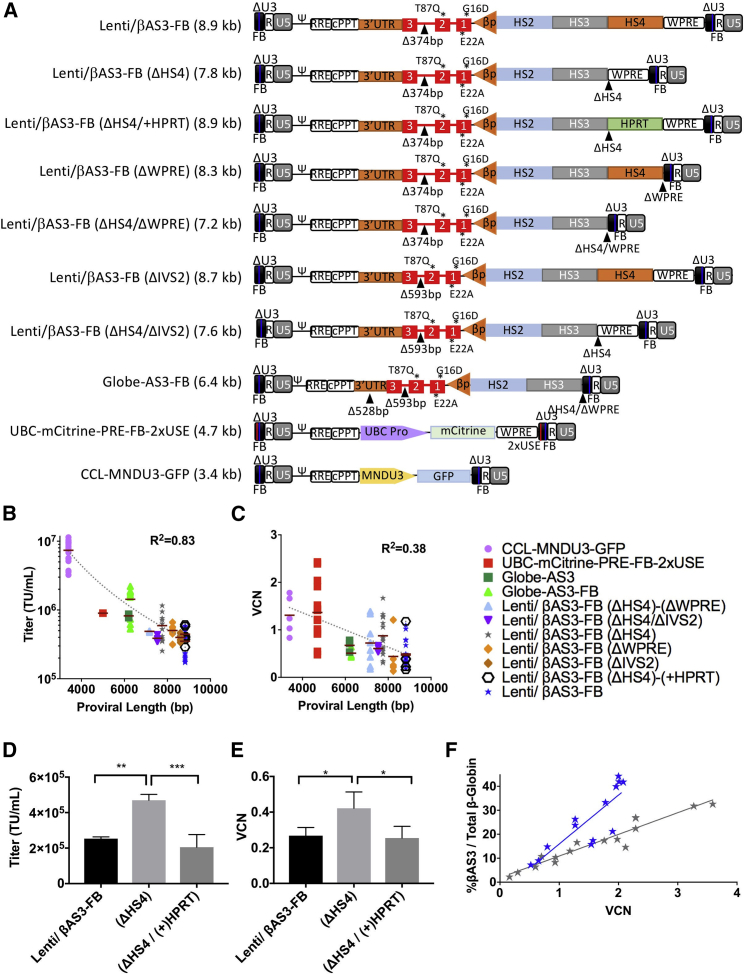
Previous reports have demonstrated that titer and transduction efficiency of lentiviral vectors inversely correlate with proviral length.25, 26 To determine whether the differences of performances between Lenti/βAS3-FB and Globe-AS3-FB were due to vector length (as opposed to the presence of specific adverse sequences), we cloned and tested a series of derivatives that reflect each difference between the two constructs (Figure 1A). When constructs were packaged and titered head-to-head, an inverse correlation between titer and vector genome length was observed (Figure 1B). When human CD34+ HSPCs were transduced at 2 × 107 transduction units (TU)/mL (MOI 20), cultured for 2 weeks under myeloid culture conditions and VCN plotted as a function of vector genome length, an inverse relationship was also observed between genome length and VCN (Figure 1C) with donor-to-donor variation in transduction efficiency diminishing the strength of the correlation. Transduced HSPCs were cultured under myeloid culture conditions (as opposed to erythroid culture conditions) as we have shown that short-term myeloid culture more stringently reflects the VCNs seen in bone marrow after xenotransplantation immune-deficient mouse hosts.19 Notably, the GLOBE-based constructs showed reduced transduction efficiency for their size, relative to the other vector.
Figure 1.
Reduction of Proviral Length Increases Titer and Infectivity
(A) Diagrams of 12 lentiviral vector constructs are shown with their proviral lengths (kb); the proviral length is the sequence length from start of 5′ LTR U3 through end of 3′ LTR U5. R, R region of the viral LTR; U5, U5 region of viral LTR; Ψ, packaging signal; RRE, Rev responsive element; cPPT, central polypurine tract; 3′ UTR, 3′ UTR; T87Q, G16D, and E22A, amino acid substitutions; βp, β-globin promoter; HS2, HS3, HS4, locus control region (LCR) DNase hypersensitive sites (HS) 2, 3, and 4; WPRE, woodchuck hepatitis virus post-transcriptional regulatory element; ΔU3, self-inactivating deletion in the U3 region; FB, FII-BEAD insulator. All major alterations introduced into each derivative are emphasized by triangle. (B) Constructs were packaged and titered and the quantity of infectious particles were plotted as a function of proviral length (bp). Each point in the plot represents an individual 10 cm plate of virus packaged and titered in parallel with CCL-MNDU3-GFP, Lenti/βAS3-FB, Globe-AS3-FB, and various derivatives. (C) Human CD34+ hematopoietic stem and progenitor cells (HSPCs) were transduced with constructs at 2 × 107 TU/mL and cultured under myeloid differentiation conditions to assess levels of infectivity. Each point in the plot represents an individual transduction, compared in parallel with CCL-MND-GFP (or CCL-UBC-mCitrine-PRE-FB-2xUSE), Lenti/βAS3-FB, Globe-AS3-FB, and various derivatives. (D) Quantity of infectious particles of constructs when packaged and titered in parallel. n = 6. **p < 0.01; ***p < 0.001. (E) Vector copy number (VCN) was determined by ddPCR 14 days after human CD34+ HSPCs were transduced at 2 × 107 TU/mL and cultured under myeloid differentiation conditions to assess infectivity. n = 3. *p < 0.05. (F) Percent βAS3-globin RNA expression was determined by RT-ddPCR and shown in relation to VCN for Lenti/βAS3-FB (blue [n = 13]) and Lenti/βAS3-FB-(ΔHS4) (gray [n = 16]). Each point represents an individual transduction done in parallel with Lenti/βAS3-FB. Difference between slopes p = 0.0010. Error bars represent mean ± SD.
Direct evidence that vector genome length was the major factor impacting vector performance was obtained through simply deleting the HS4 region from Lenti/βAS3-FB, as it was a major difference between Lenti/βAS3-FB and the higher titer vector Globe-AS3-FB. A 2-fold increase in both titer (p < 0.01) and gene transduction efficiency to CD34+ HSPCs (p < 0.05) was observed when the 1.1 kb HS4 element was removed from Lenti/βAS3-FB. These gains in performance were not sequence-related, as replacing HS4 with a similarly sized 1.1 kb stuffer sequence from the human HPRT gene intron 1 similarly reduced titer and infectivity (Figures 1D and 1E). As expected, the removal of HS4 predictably decreased expression of βAS3-globin (Figure 1F).27, 28
Additional sequence deletions (>1.1 kb) offered increasing gains in titer and infectivity suggesting that length influenced vector performance more strongly than the presence of discreet sequences (additional analyses examining derivatives of Lenti/βAS3-FB are provided in the Supplemental Results). Taken together, these data suggested that the titer and infectivity of Lenti/βAS3-FB could be improved through shortening vector length. We hypothesized that a decrease in vector length of 2–3 kb would be required to generate a construct produced at 3- to 4-fold higher raw titer that would have superior gene transfer to HSPCs when compared to Lenti/βAS3-FB at equivalent MOI. None of the smaller vectors were studied moving forward; we decided to focus on reducing the size of the LCR.
Redefining the Putative Boundaries of HS Elements
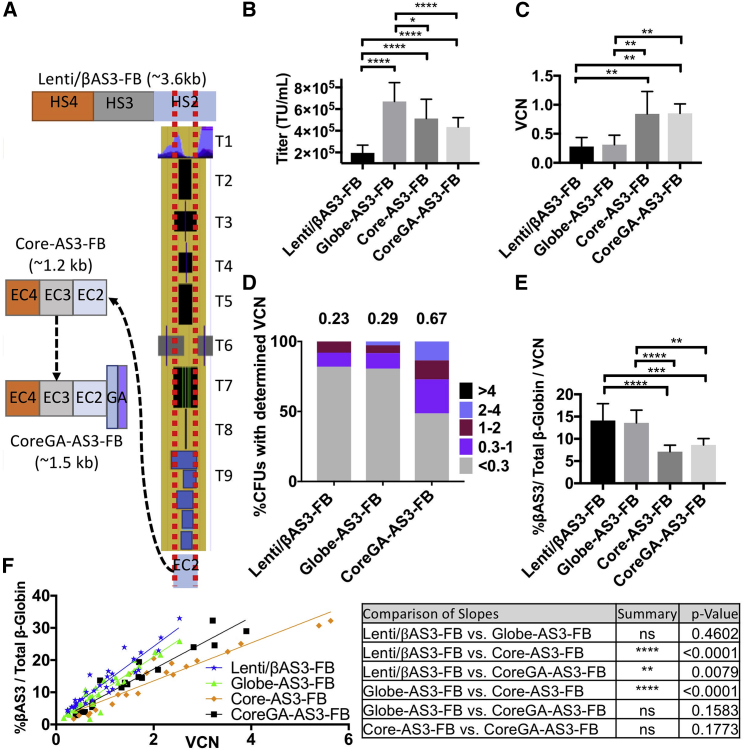
Vector length reduction was achieved by redefining the boundaries of the LCR HS (2, 3, and 4) elements of Lenti/βAS3-FB by using published genomic and epigenomic data available through ENCODE (accessible via the UCSC Genome Browser).29 To generate new boundaries for the HS sequences of Lenti/βAS3-FB, we looked specifically for clues to identify the Core enhancer sequences of HS 2, 3, and 4, such as histone markers of active chromatin (H3k27ac) and characteristics of open chromatin (DNase I hypersensitive and formaldehyde-assisted isolation of regulatory elements [FAIRE]), as well as TF binding (transcription factor chromatin immunoprecipitation sequencing [ChiP-seq]) and conserved TF motifs. These novel defined HS sequences, termed “ENCODE core sequences” (EC2, ∼420 base pairs [bp]; EC3, ∼340 bp; EC4, ∼410 bp), were used to replace the HS sequences present within Lenti/βAS3-FB (∼3.6 kb reduced to ∼1.2 kb) to generate a reduced length construct termed “Core-AS3-FB.” We then added a 360 bp fusion element to facilitate position-independent expression of βAS3-globin by combining a 202 bp murine Gata1 gene HS element with a 143 bp Human Ankyrin gene element30 to create CoreGA-AS3-FB (Figure 2A).
Figure 2.
Approach for Length Reduction and In Vitro Characterization of Reduced Length Vectors
(A) Published genomic data available through ENCODE were used to redefine the putative HS boundaries. The track sets (T) 1–8 were used to generate new boundaries for the LCR’s HS sequences. T1, H3k27ac; T2, DNase I (Encode); T3, DNase I (University Wisconsin); T4, DNase I (UNC Chapel Hill); T5, Formaldehyde-Assisted Isolation of Regulatory Elements, FAIRE (UNC Chapel Hill); T6, DNase I/FAIRE/ChiP-seq (Duke/UNC Chapel Hill/UT-Austin); T7, TF ChiP-seq; T8, conserved TF motifs; and T9, regulatory elements (Open Regulatory Annotation database). These new sequences were termed “ENCODE Cores” (EC) 2, 3, and 4 and combined to create Core β-LV. The 143 bp human Ankyrin Element was then combined with the 218 bp murine Gata1 HS4 element and added to EC 2, 3, and 4 to create CoreGA-AS3-FB β-LV. (B) Quantity of infectious particles of constructs when packaged and titered in parallel. n = 5. *p < 0.05; ****p < 0.0001. (C) Human CD34+ HSPCs were transduced with constructs at 1 × 107 TU/mL and cultured under myeloid culture conditions for 14 days. VCN was determined by ddPCR. n = 3. **p < 0.01. (D) Human CD34+ HSPCs were transduced in parallel at 1 × 106 TU/mL and used to seed a methylcellulose-based colony forming unit assay. Shown are stacked bar graphs representing percentages of colonies with a determined VCN range. n = 50 colonies, Lenti/βAS3-FB; n = 36 colonies, Globe-AS3-FB; n = 37 colonies, CoreGA-AS3-FB. (E) Human CD34+ HSPCs were transduced in parallel at 1 × 107 TU/mL and cultured under erythroid culture conditions for 14 days. Expression of different constructs is presented as percentage of βAS3-globin normalized to VCN. n = 9. ***p < 0.001 (F) Human CD34+ HSPCs were transduced at various MOI and differentiated under erythroid culture conditions for 14 days. Percent βAS3-globin RNA expression was determined by RT-ddPCR and is shown in relation to VCN. Each point in the plot represents an individual transduction done in parallel with Lenti/βAS3-FB and Globe-AS3-FB. n = 33, Lenti/βAS3-FB; n = 36, Globe-AS3-FB; n = 21, Core; n = 18, CoreGA-AS3-FB. Error bars represent mean ± SD.
The β-LV constructs were packaged in parallel using HEK293T cells and titered head-to-head on HT-29 cells.12 From five independent packaging experiments, titers of the Globe-AS3-FB, Core-AS3-FB, and CoreGA-AS3-FB β-LVs were on average about 3-fold higher than that of Lenti/βAS3-FB (p < 0.0001), presumably due to their reduced proviral lengths in comparison to Lenti/βAS3-FB (Figure 2B).
To evaluate the efficiency of transduction of Core-AS3-FB and CoreGA-AS3-FB in comparison to Lenti/βAS3-FB and GLOBE-AS3-FB, we transduced human CD34+ HSPCs isolated from bone marrow (BM) of healthy donors at 1 × 107 TU/mL (MOI 10) and cultured them for 14 days under myeloid differentiation conditions. From three independent experiments, the gene transfer efficiency of Core-AS3-FB was on average about 3-fold higher when compared to Lenti/βAS3-FB and Globe-AS3-FB (p < 0.01), and the further addition of the fusion element to produce CoreGA-AS3-FB did not negate gains in gene transfer efficiency achieved by Core-AS3 (p < 0.01) (Figure 2C).
To confirm that CoreGA-AS3-FB retained superior transduction efficiency when compared to Lenti/βAS3-FB and GLOBE-AS3-FB at equal MOI (1 × 106 TU/mL), we assessed infectivity at the clonal level. Transduced CD34+ HSPCs were plated in a methylcellulose-based medium to allow growth of colony-forming units (CFUs). After 14 days of culture, colonies were scored manually by morphologic criteria to determine lineage differentiation (blast forming unit-erythroid [BFU-E], CFU-granulocyte-monocyte, CFU-granulocyte-erythroid-monocyte-megakaryocyte). The VCNs of bulk transduced cells used to seed the CFU assay were 0.23, 0.29, and 0.67 for Lenti/βAS3-FB, Globe-AS3-FB, and CoreGA-AS3-FB, respectively, reflecting the higher infectivity of the CoreGA-AS3-FB vector. Colonies were grouped by their VCN value as determined by droplet digital PCR (ddPCR). Colonies with VCN > 0.3 were considered PCR positive for the presence of vector and further classified by VCN (0.3–1, 1–2, 2–4, or >4). The percentages of PCR-positive colonies seen for CoreGA-AS3-FB were significantly higher than for Lenti/βAS3-FB and Globe-AS3-FB (p < 0.004; chi-square test) reaching 51%, 18%, and 19%, respectively (Figure 2D). Among the positive colonies, CoreGA-AS3-FB achieved greater numbers of integrations per colony when compared to Lenti/βAS3-FB (0.3–1 VCN; p < 0.01, 1–2 VCN, p < 0.05; 2–4 VCN, p < 0.005; chi-square test). These results demonstrate that when assessed at comparable MOI, CoreGA-AS3-FB was able to transduce greater numbers of CD34+ HSPCs with more integrant per cells when compared to current clinical vectors.
To determine whether the newly designed “Core” constructs retained the ability to achieve therapeutic levels of βAS3-globin expression, we performed in vitro erythroid differentiation studies using BM CD34+ HSPCs from healthy donors. After HSPCs were transduced at equal MOI, βAS3-globin and total β-globin RNA transcript levels were measured by reverse transcription (RT)-ddPCR 14 days after transduction and in vitro differentiation. The percentages of βAS3-globin RNA transcripts over total β-globin RNA transcript were normalized to VCN for nine independent experiments. Expression levels normalized by VCN of Core-AS3-FB and CoreGA-AS3-FB were 0.5-fold (p < 0.0001), and 0.4-fold (p < 0.001) lower than that of Lenti/βAS3-FB, respectively (Figure 2E).
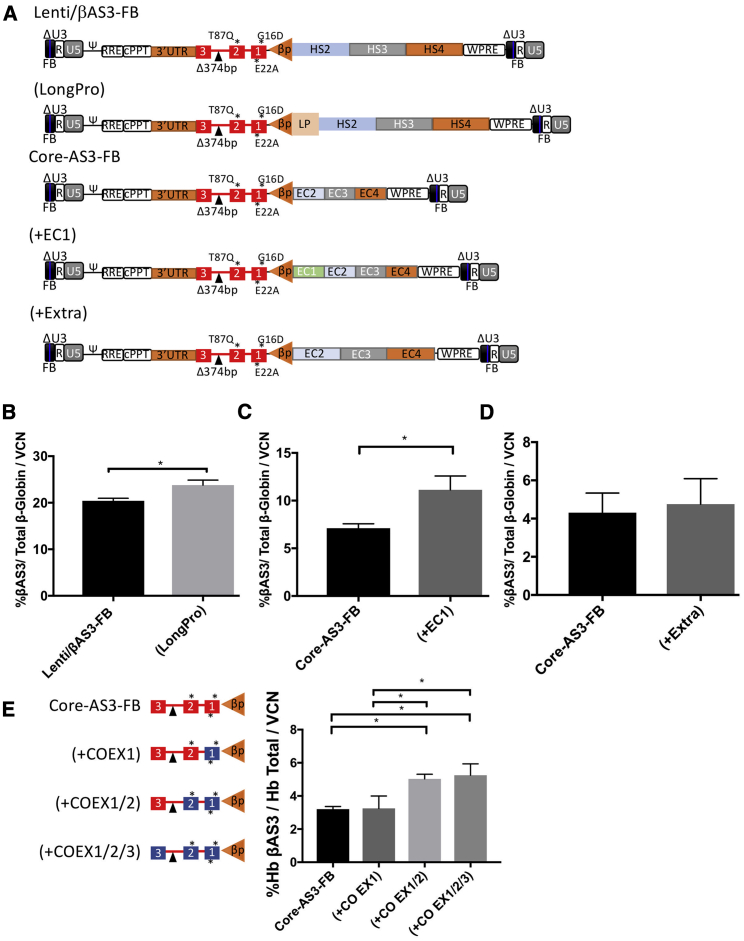
To examine the linear relationship between VCN and %βAS3-globin transcript levels, we transduced HSPCs derived from multiple healthy donors at different MOIs and subjected them to in vitro erythroid differentiation for 14 days, followed by measurement of expression of βAS3-globin and VCN. Globe-AS3-FB had slightly higher levels of expression than did CoreGA-AS3-FB, (as shown by the increased slope of the fitted line), but this difference failed to reach significance. As Core-AS3-FB and CoreGA-AS3-FB were able to transduce HSPCs to higher VCN than the other vectors, total expression levels also increased in a linear fashion to compensate for the decreased expression per vector genome (Figure 2F). Several efforts were made to increase expression per VCN, including codon optimization, using a larger promoter sequence or adding Core HS1 sequence to the enhancer. Although increments in expression were seen (Figure 3), gene transfer was diminished (data not shown). We also tried arbitrarily increasing the Core sequences by 100 bp in both directions to see whether that would increase expression, but this did not improve expression from the vector (Figure 3D). As a result, CoreGA-AS3-FB was deemed to be the best candidate vector to test in vivo.
Figure 3.
Modifications Increase Expression per Vector Genome
(A) Diagrams of lentiviral vector constructs. LP, longer β-globin promoter; EC1, ENCODE core 1; Extra, ECs extended 100 bp in each direction; all other annotations similar to those shown in Figure 1A. (B) Human CD34+ HSPCs were transduced in parallel at 2 × 107 TU/mL and cultured under erythroid culture conditions for 14 days. Expression of different constructs is presented as percentage of βAS3-globin normalized to VCN. n = 5. *p < 0.05. (C) Human CD34+ HSPCs were transduced in parallel at 1 × 107 TU/mL and cultured under erythroid culture conditions for 14 days. Presentation of results the same as (B). n = 3. (D) Human CD34+ HSPCs were transduced and cultured under conditions same as (B) with results presented the same. n = 3. (E) Diagrams showing modified exons (in blue) of βAS3 transgene. All modifications were assessed in Core-AS3-FB backbone. Human CD34+ HSPCs were transduced in parallel at 2 × 107 TU/mL and cultured under erythroid culture conditions for 21 days. Expression of different constructs is presented as percentages of Hb βAS3 tetramers from cell lysates normalized VCN. n = 3. Error bars represent ± SD.
Thus, in vivo studies comparing CoreGA-AS3-FB to Lenti/βAS3-FB and Globe-AS3-FB were conducted in the “Townes” mouse model of SCD31, 32, 33 to evaluate long-term expression of βAS3-globin and to determine whether CoreGA-AS3-FB could ameliorate the sequelae related to SCD.
In Vivo Analysis of Peripheral Blood from SCD Mouse Model
The “Townes” mouse model of SCD was used to evaluate the hematologic correction potential of CoreGA-AS3-FB. While we hypothesized that the superior transduction efficiency of CoreGA-AS3-FB would compensate for its decreased expression per vector genome when compared to Lenti/βAS3-FB at equal MOI, we chose to perform a direct comparison of CoreGA-AS3-FB to Lenti/βAS3-FB and Globe-AS3-FB (Figure S3A) at equivalent VCN. Using constructs under conditions where transduction efficiencies were comparable (achieved by using vectors at different MOI) enabled a more appropriate evaluation for hematologic correction at low copy number. An outline of the experiment is provided in Figure S3B.
Lineage-depleted BM cells were obtained from homozygous βS/βS donor mice and pre-stimulated for 24 h. Cells were transduced with different amounts of vector to achieve equivalent copy numbers and delivered by retro-orbital injection into lethally irradiated recipients (Pep Boy/J [CD45.1 congenic]) 24 h after transduction. Table 1 provides the concentrations of vector used to transduce murine lineage depleted (Lin−) BM cells. Two independent experiments were conducted and in vitro VCN was determined from the transduced cell product 14 days after transduction (shown in Table 1).
Table 1.
In Vitro VCN of Gene Modified βS/βS Lin− BM Cells Before Transplant
| Lenti/βAS3-FB | Globe-AS3-FB | CoreGA-AS3-FB | |
|---|---|---|---|
| Transduction Condition (TU/mL) | 6.6 × 106 | 1 × 107 | 3.3 × 106 |
| In vitro VCN experiment 1 | 2 | 2 | 2 |
| In vitro VCN experiment 2 | 2 | 2 | 2 |
TU, transduction units; VCN, vector copy number.
Engraftment by flow cytometry for CD45.2+ donor cells, transduction efficiency (VCN by ddPCR), βAS3-globin expression (by RT-ddPCR), blood hemoglobin concentration, and RBC counts and hemoglobin composition by high-performance liquid chromatography (HPLC) were measured on peripheral blood (PB) samples acquired 4, 16, and 20 weeks post-transplantation. At week 20, mice were euthanized, and BM cells collected to measure engraftment, transduction efficiency, and βAS3-globin expression. Mice with BM donor engraftment <97% at week 20 were excluded from analyses to prevent inclusion of artifactually corrected mice (as ≥4% residual WT recipient RBCs would mask the adverse pathophysiology induced by βS/βS donor cells.32 At the time of euthanasia, BM cells taken from two mice from each arm were used to seed a methylcellulose-based CFU assay; after 2 weeks growth, individual primitive erythroid progenitor cell colonies (BFU-E) were isolated from the CFU dishes and assayed to measure VCN and βAS3-globin expression. Peripheral blood cells were also collected at time of euthanasia and used to quantify the percentages of sickled erythrocytes present within the peripheral circulation.
As intended, the average gene transfer efficiency seen in PB cells did not significantly differ among experimental arms and the average PB VCNs were similar to those of in vitro cultured cells before transplantation. Moreover, gene marking of cells remained stable across all three time points, indicating that cells with long-term engraftment potential were stably transduced and able to contribute to hematopoiesis (Figure 4A).
Figure 4.
In Vivo Analysis of Hematologic Correction of SCD Mouse Model
Mice were bled at weeks 4, 16, and 20 and peripheral blood (PB) analyzed. Only mice with >97% BM donor engraftment were included in analysis. n = 9, Mock; n = 8, Lenti/βAS3-FB; n = 9, Globe-AS3-FB; n = 8, CoreGA-AS3-FB. (A) Peripheral blood VCN by ddPCR. ****p < 0.0001. (B) Percentages of Hb βAS3 tetramers in PB lysates measured by HPLC. *p < 0.05. (C) Percentages of Hb βAS3 tetramers normalized to PB VCN. (D) Hemoglobin (HB [g/dL]) levels. **p < 0.01. (E) Red blood cell (RBC) count (×106). ***p < 0.001. (F) Percentage of sickled red blood cells in PB quantified by ImageStream at 20 weeks post transplantion. "Townes" SCD (homozygous SCD mice) were included as negative control; WT C57BL/6J (Pep Boy/J [CD45.1 congenic]) mice were included as postitive control. Only mice from experiment two with >97% BM donor engraftment were included in analysis. n = 2, WT C57BL/6J; n = 2, "Townes" SCD; n = 4, Mock; n = 4, Lenti/βAS3-FB; n = 5, Globe-AS3-FB; n=3, CoreGA-AS3-FB. Error bars represent mean ± SD.
Quantification of HbβAS3 tetramers in peripheral blood lysates was accomplished using HPLC. Figure S4 provides representative HPLC chromatograms of globin tetramers in PB obtained at time of euthanasia from representative mice. At 20 weeks post transplantation, the average levels of HbβAS3/total hemoglobin tetramers were 37.4%, 28.8%, and 19.1% for Lenti/βAS3-FB, Globe-AS3-FB, and CoreGA-AS3-FB, respectively (Figure 4B).
To compare the difference in normalized expression between each experimental arm, we normalized and plotted %HbβAS3 to PB VCN for each mouse. An average of 18.3, 13.4, and 8.8%HbβAS3/VCN was seen for Lenti/βAS3-FB, Globe-AS3-FB, and CoreGA-AS3-FB, respectively (Figure 4C). Table S1 provides additional characterizations of globin tetramers in PB lysates generated by HPLC for weeks 4, 16, and 20.
The Hb levels seen at time of euthanasia were significantly higher when comparing recipients of BM transduced with Lenti/βAS3-FB (p < 0.0001), Globe-AS3-FB (p < 0.05), and CoreGA-AS3-FB (p < 0.05) to recipients of mock-transduced BM cells. The Hb levels of mice that received mock-transduced cells were 8.5 g/dL on average while the Hb levels of mice that received BM transduced with Lenti/βAS3-FB, Globe-AS3-FB, or CoreGA-AS3-FB were 10.7, 10.2, and 9.9 g/dL on average, respectively (Figure 4D).
RBC counts were also significantly higher for recipients of Lenti/βAS3-FB (p < 0.001), Globe-AS3-FB (p < 0.01), and CoreGA-AS3-FB (p < 0.001) transduced BM cells compared to recipients of mock-transduced BM cells. The RBC counts of mice that received mock-transduced cells were 6.0 × 106 cells/μL on average, while the RBC counts of mice that received Lenti/βAS3-FB, Globe-AS3-FB, or CoreGA-AS3-FB were 8.4, 8.5, and 8.3 × 106 cells/μL on average, respectively (Figure 4E). Notably, mice that received mock transduced BM cells developed progressive anemia as noted by decreasing Hb levels and RBC counts observed in PB over time.
Lastly, the quantity of sickled RBCs present within PB at time of euthanasia from a subset of representative mice was measured using the ImageStream X Mark II Imaging Flow Cytometer (ISX). Equal volumes of peripheral blood and 2% sodium metabisulfite solution were mixed together and incubated under hypoxic conditions (2% oxygen) at 37°C for 10 minutes, diluted in 2% glutaraldehyde solution, and then analyzed by ISX. Peripheral blood taken from “Townes” homozygous βS/βS SCD mice or mice transplanted with mock-transduced HSPCs displayed averages of 41.0% and 34.3% RBC sickling, respectively. Mice transplanted with Lenti/βAS3-FB, Globe-AS3-FB, or CoreGA-AS3-FB transduced HSPCs displayed a significant decrease in the percentages of sickled cells present within peripheral blood (p < 0.0001). The average amount of sickling seen for Lenti/βAS3-FB, Globe-AS3-FB, and CoreGA-AS3-FB were 7.5%, 5.76%, and 11.5%, respectively (Figure 4F).
In Vivo Analysis of Bone Marrow βAS3-Globin Expression from an SCD Mouse Model
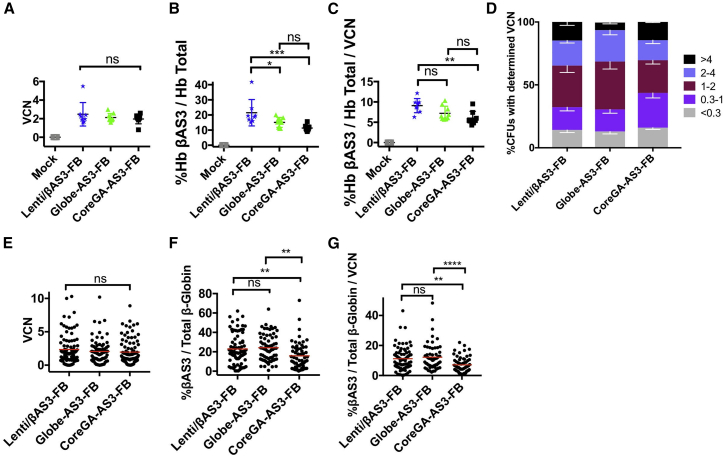
BM cells were collected at time of euthanasia and used to measure engraftment, transduction efficiency, and βAS3-globin expression. As expected, average BM VCN was similar among all experimental arms and matched the average PB VCN seen at euthanasia (Figure 5A).
Figure 5.
In Vivo Analysis of Bone Marrow βAS3-Globin Expression from SCD Mouse Model
Whole BM was taken from each mouse at time of euthanasia and mice with >97% donor engraftment were analyzed. n = 9, Mock; n = 8, Lenti/βAS3-FB; n = 9, Globe-AS3-FB; n = 8, CoreGA-AS3-FB. (A) BM VCN by ddPCR. ns = not significant. (B) Percentages βAS3-globin RNA expression of BM determined by RT-ddPCR. *p < 0.05; **p < 0.01. (C) Percentage of BM βAS3-globin RNA expression normalized to BM VCN. Error bars represent mean ± SD. (D) BM obtained at time of euthanasia was used to seed a methylcellulose based CFU assay. Individual colonies were plucked 2 weeks after plating and gDNA and RNA extracted. Shown are stacked bar graphs representing percentages of colonies with a determined VCN range. Shown are mean ± SEM from two independent experiments (n = 2 donors/arm). n = 189 colonies, Lenti/βAS3-FB; n = 129 colonies, Globe-AS3-FB; n = 156 colonies, CoreGA-AS3-FB. (E) VCN of CFUs from two independent experiments (n = 2 donors/arm); same data as (D). Only PCR-positive CFUs with detectable endogenous globin expression were included. n = 93 colonies, Lenti/βAS3-FB; n = 84 colonies, Globe-AS3-FB; n = 94 colonies, CoreGA-AS3-FB. Each dot represents a single colony. Bar represents grand average of VCN from two independent experiments. (F) Percent βAS3-globin RNA expression was determined by RT-ddPCR. ****p < 0.0001. (G) Percent βAS3-globin RNA expression normalized to VCN for matched colony. ***p < 0.001. (H) Percentage of sickled RBCs in PB quantified by ImageStream at 20 weeks post transplantation. “Townes” SCD (homozygous SCD mice), included as negative control; WT C57BL/6J (Pep Boy/J [CD45.1 congenic]) mice are included as a positive control. Only mice from experiment two with >97% BM donor engraftment were included in analysis. n = 2, WT C57BL/6J; n = 1, “Townes” SCD; n = 4, Mock; n = 4, Lenti/βAS3-FB; n = 5, Globe-AS3-FB; n = 3, CoreGA-AS3-FB.
To determine whether CoreGA-AS3-FB retained the ability to achieve therapeutic levels of expression in BM cells, we measured βAS3-globin and total β-globin RNA transcript levels by RT-ddPCR. Average levels of %βAS3-globin were 21.5%, 15.1%, and 11.4% for Lenti/βAS3-FB, Globe-AS3-FB, and CoreGA-AS3-FB, respectively (Figure 5B). To compare the difference in expression per vector genome between all treatment arms, we normalized %βAS3-globin expression to BM VCN for each mouse and plotted in Figure 5C. An average of 9.1%, 7.2%, and 6.1% βAS3-globin/total β-globin/VCN was seen for Lenti/βAS3-FB, Globe-AS3-FB, and CoreGA-AS3-FB, respectively.
Persistence of long-term βAS3-globin expression was determined using a methylcellulose-based CFU assay. BM was taken from two mice from each treatment arm at time of euthanasia and used to seed methylcellulose supplemented with recombinant cytokines that promoted primitive erythroid progenitor cell growth. Two independent experiments were performed. The VCNs of the bulk BM cells used to seed methylcellulose at the time of euthanasia are provided in Table 2. Colonies recognized as BFU-E were plucked 14 days after seeding and both DNA and RNA were obtained from the same colony. The VCN of each colony was determined by ddPCR and colonies were then grouped by their VCN value. The percentages of PCR-positive colonies seen for Lenti/βAS3-FB, Globe-AS3-FB, and CoreGA-AS3-FB were 86%, 87%, and 84%, respectively (Figure 5D).
Table 2.
VCNs of Bulk BM Cells Used to Seed Methylcellulose Based CFU Assay
| Lenti/βAS3-FB | Globe-AS3-FB | CoreGA-AS3-FB | |
|---|---|---|---|
| In vitro CFU experiment 1 | 1.6, 1.9 | 1.9, 1.36 | 1.9, 2.1 |
| In vitro CFU experiment 2 | 2.4, 2.6 | 2.3, 1.9 | 2.0, 2.6 |
Persistence of long-term βAS3-globin expression was determined by measuring %βAS3-globin for each BFU-E. The VCNs of PCR-positive BFU-Es with detectable endogenous β-globin expression were plotted in Figure 5E. While all experimental arms displayed populations of BFU-Es with >20%βAS3-globin, average levels of %βAS3-globin/total β-globin mRNA were 22.9%, 24.2%, and 15.8% for Lenti/βAS3-FB, Globe-AS3-FB, and CoreGA-AS3-FB, respectively (Figure 5F). When %βAS3-globin expression values of individual CFUs were normalized to their corresponding VCNs, averages of 11.2%, 12.2%, and 7.1% βAS3-globin/VCN were observed for Lenti/βAS3-FB, Globe-AS3-FB, and CoreGA-AS3-FB, respectively (Figure 5G). Taken together, these data demonstrate that long-term and persistent expression was observed in each experimental arm and differences in expression observed in bulk BM cells were reflected at the clonal level.
Incorporating ENCODE Core Sequences into a Shorter β-LV Expression Cassette
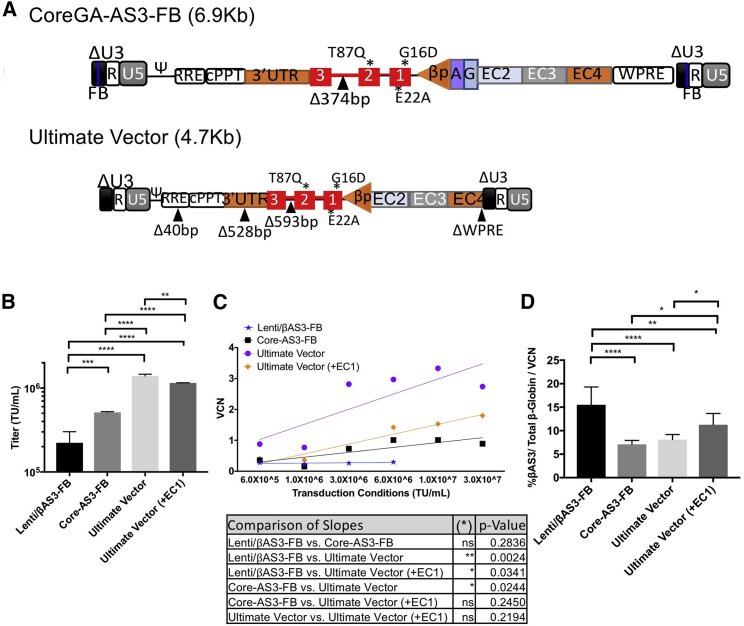
The expression studies of CoreGA-AS3-FB were performed to ensure that exchanging the original LCR sequence for the ENCODE core sequences could still provide sufficient expression to ameliorate disease sequelae in the Towne’s mouse model of SCD. Because reduction in vector size was determined to be the largest factor influencing titer and gene transfer, we made further modifications to the CoreGA-AS3-FB to remove approximately 2 kb of sequence from the provirus. The main deletions were of the WPRE sequence, β-globin 3′ UTR and IVS2 (matching those found in the Globe-AS3-FB). The resulting ∼4.7 kb UV-AS3-FB “Ultimate Vector” (shown in Figure 6A) has a titer that is 3-fold higher than CoreGA-AS3-FB (Figure 6B). Gene transfer is also improved 3-fold compared to CoreGA-AS3-FB (Figure 6C) and the expression remains identical (Figure 6D).
Figure 6.
Size Reduction Improves Vector Performance
(A) Diagrams of lentiviral vector constructs. All annotations are similar to those shown in Figure 1A. (B) Quantity of infectious particles of constructs when packaged and titered in parallel. n = 3. *p < 0.05; **p < 0.01, ***p < 0.001, ****p < 0.0001. (C) Human CD34+ HSPCs were at various MOIs and cultured under myeloid culture conditions for 14 days. VCN was determined by ddPCR. (D) Human CD34+ HSPCs were transduced in parallel at 2.0 × 107 TU/mL and cultured under erythroid culture conditions for 14 days. Expression of different constructs is presented as percentage of βAS3-globin normalized to VCN. n = 12, Lenti/βAS3-FB; n = 3, Core-AS3-FB; n = 9, Ultimate Vector; n = 9, Ultimate Vector (+EC1). Error bars represent mean ± SD.
Discussion
The requirements to be met for a gene therapy vector to be considered as a candidate for clinical translation include high-titer, efficient, and stable proviral transmission, high-level, lineage-specific, and persistent expression, and safe integration profile. The results presented herein demonstrate that some of the major hurdles that have historically restricted clinical translation of β-globin gene therapy vectors for treatment of SCD can be surmounted through vector engineering.
One predominant factor that diminishes β-LV titer is suspected to be the relatively large size of these vectors due to the use of the human β-globin genomic sequences and LCR elements. Both Kumar et al.25 and Barrett et al.26 demonstrated that titers of LVs decrease proportionally to the length of insert irrespective of sequence. We confirmed that vector length was the major factor contributing to the difference in titer observed between Lenti/βAS3-FB and Globe-AS3-FB. Moreover, introduction of additional sequence deletions to Lenti/βAS3-FB offered increasing gains in titer and infectivity confirming that vector length strongly influenced titer and infectivity. Although the dissection revealed that vector length could be reduced by ∼1 kb by removing redundant sequences (see Supplemental Information), that level of size reduction fell short of our 2–3 kb target.
The critical minimal segments of the ∼20 kb human β-globin LCR needed for high-level, position-independent, erythroid expression of β-globin were elucidated using transgenic mouse models that, at the time, relied on a limited repertoire of restriction enzymes to introduce sequence deletions to the hypersensitive sites.34, 35, 36, 37, 38, 39 The findings of these studies informed the development of current first generation β-globin lentiviral vectors, which were engineered to contain specific LCR fragments that confer erythroid-specific expression of β-globin.40, 41, 42, 43, 44 Given the methods that were deployed to define the boundaries of the LCR HS elements, we hypothesized that these fragments included core sequences that bestow erythroid specific expression, but also some extraneous sequences that could be eliminated with minimal decrease in enhancer activity. Therefore, emphasis was given to develop a strategy for reducing the length of the LCR HS elements recognizing potential risks for reducing overall expression per vector genome.
When designing the new core HS elements, several genetic and epigenetic factors were considered, including the sites binding histones associated with active genes, DNase1 hypersensitivity in erythroid lineages, and sequence conservation. Using this design approach, the overall lengths of combined HS elements 2, 3, and 4 were reduced from 3.6 kb to 1.2 kb. Regulatory elements from the erythroid-specific murine Gata1 and human ANK1 genes were added to aid in position-independent expression.
When compared to the standard Lenti/βAS3-FB design, we found that the CoreGA-AS3-FB was produced at nearly 3-fold higher titer (p < 0.0001), possessed nearly 3-fold higher gene transfer to CD34+ HSPCs at the same MOI as for standard vectors (p < 0.01), and expressed nearly 60% of the amount of βAS3-globin transcripts per vector genome in CD34+ HSPCs cultured under erythroid conditions (p < 0.001). These observations of improved titer and transduction efficiency taken together spurred the continued study of CoreGA-AS3-FB to determine whether the observed difference in expression when compared to Lenti/βAS3-FB was relevant to a clinical scenario.
Previous reports examining SCD patients who co-inherit the condition of hereditary persistence of fetal hemoglobin (where the distribution of HbF is thought to be pancellular) indicate that anti-sickling globin protein levels greater than 10%–15% can ameliorate the clinical sequelae of SCD.45, 46, 47, 48, 49 When CoreGA-AS3-FB was evaluated in a mouse model of SCD at an average BM VCN = 2, βAS3-globin RNA comprised an average of 12% of the total β-globin transcripts and HbβAS3 tetramers made-up 18.5% of the total hemoglobin tetramers. The finding that the percentage of HbβAS3 tetramers exceeded the relative abundance of βAS3-globin mRNA is consistent with the increased affinity for α-globin of the βAS3-globin protein due to the G16D amino acid substitution.32, 50 Thus, the in vivo percentages of βAS3-globin RNA transcripts and protein seen for CoreGA-AS3-FB are at levels expected to be therapeutic, as the βAS3-globin gene has been shown to have equivalent anti-sickling activity as γ-globin.33
A methylcellulose-based CFU assay using BM cells obtained from mice 20 weeks after transplantation also revealed the presence of BFU-Es with >20% βAS3-globin in all experimental arms. This finding suggested that populations of corrected HSPCs capable of contributing to long-term erythropoiesis were established by each vector. More importantly, when murine HSPCs were transduced with CoreGA-AS3-FB at lower MOI (in comparison to those MOIs used to transduce cells with Lenti/βAS3-FB or Globe-AS3-FB), CoreGA-AS3-FB transduced similar percentages of colony-forming progenitors resulting in similar bulk VCNs. These data suggest that CoreGA-AS3-FB has superior infectivity in comparison to Lenti/βAS3-FB and Globe-AS3-FB, exemplified by its transduction efficiency even when constrained by using less vector.
The most critical pre-clinical test of any novel β-LV design being considered as a candidate for clinical translation is amelioration of hematologic parameters defining the pathological phenotype of a SCD mouse model. While the amounts of βAS3-globin expressed per VCN by CoreGA-AS3-FB were lower when compared to the other vectors, the levels were sufficient to result in physiologic improvements in the signs of SCD in the mice, with significant increases in both RBC count and hemoglobin content in comparison to recipients of mock-transduced BM.
When the quantity of sickled RBCs were measured in PB at time of euthanasia from a subset of representative mice, a higher amount of sickling was seen in PB of CoreGA-AS3-FB treated mice. This observation may have resulted from transplantation of a larger number of non-transduced cells into recipient mice; a result of using less vector.
Incorporation of the ENCODE core sequences from CoreGA-AS3-FB into the approximately 2 kb shorter UV-AS3-FB resulted in a superior titer and gene transfer. The UV-AS3-FB was designed after the in vivo studies were complete and will be the subject of a future submission. This new lentiviral vector design with reduced sizes of the LCR components and expression cassette should have significant advantages for clinical-scale production by providing far higher level of gene transfer to HSPC from lower amounts of vector. It may extend the benefits of autologous gene therapy for SCD by significantly reducing the cost of vector production and extending the vector supply.
Materials and Methods
Cloning and Vector Production
The Lenti/BAS3-FB vector has been described previously.19 To introduce desired sequence deletions, we used sets of reverse-oriented primers to PCR amplify the Lenti/BAS3-FB plasmid backbone and then phosphorylated and ligated the resultant linearized plasmids. To seamlessly introduce sequences in place of deletions, we ordered gBlock gene fragments (Integrated DNA Technologies, Skokie, IL) containing homology to the plasmid backbone and joined them to linearized plasmids using the NEBuilder HiFi DNA Assembly kit (New England Biolabs, Ipswich, MA). All plasmids were sequence verified by Sanger sequencing (Laragen, Culver City, CA). The murine Gata1 element was from bp 438–639 (GenBank: U89137.1) and the human ANK1 element is from bp 103987–104129 (NCBI: NG_012820.2). The UV-AS3-FB vector was created by using NEBuilder HiFi DNA Assembly kit to fuse together PCR amplified EC2 from CoreGA-AS3-FB to gBlock gene fragments for the: cPPT, β-globin expression cassette, EC3, EC4, 3′ long terminal repeat (LTR) with polyadenylation enhancements (BGH polyA), and SV40 origin of replication.
Transient transfection of 293T cells using the third-generation packaging system51 provided packaged virus particles. Viral supernatants were then directly used for titer determination or concentrated by tangential flow filtration, as described by Cooper et al.12 Briefly, the HT-29 human colorectal carcinoma cell line was transduced with different dilutions of both raw and concentrated vectors. To calculate titers, we harvested cells and determined VCNs by ddPCR approximately 60 h post transduction.
BM CD34+ Cell Culture and Transduction
All BM aspirates were obtained from voluntary healthy donors supplied by AllCells (Alameda, CA), which obviated the need for Instituional Review Board review. BM mononuclear cells were isolated by Ficoll-Hypaque density gradient centrifugation. CD34+ HSPCs were enriched using CD34+ MicroBead Kit (Miltenyi Biotec, Bergisch Gladbach, Germany). Enriched CD34+ HSPCs were cryopreserved in fetal bovine serum supplemented with 10% DMSO (Sigma-Aldrich, St. Louis, MO) in liquid nitrogen. Cells were thawed and plated on non-tissue culture-treated six-well plates pre-coated with RectroNectin (20 μg/mL, Takara Shuzo, Otsu Japan) at 1 × 106 cells/mL. Cells were pre-stimulated for 16–24 h in X-Vivo 15 medium (Lonza, Basel, Switzerland) supplemented with 1× glutamine, penicillin, and streptomycin (Gemini Bio-Products, Sacramento, CA), human stem cell factor (50 ng/mL), human Flt-3 ligand (50 ng/mL), human thrombopoietin (50 ng/mL), and human interleukin-3 (20 ng/mL; all cytokines were acquired from PeproTech, Rocky Hill, NJ). Concentrated viral supernatants were used at various MOI to transduce CD34+ HSPCs for 24 h. These cells were washed, re-plated, and cultured under myeloid or erythroid culture conditions, as described by Romero et al.19 On day 14 of culture, genomic DNA and/or mRNA was extracted from transduced cells.
ddPCR for VCN and %βAS3 mRNA Quantification
Genomic DNA was extracted using PureLink Genomic DNA Mini Kit (Invitrogen, Waltham, MA). VCN was calculated by using probes SCD4 (Human Syndecan 4) as a reference and HIV-1 PSI as a target. ddPCR was carried out as described in Urbinati et al.6 RNeasy Plus Mini Kit (QIAGEN, Valencia, CA) was used for RNA extraction followed by reverse transcription as described by Urbinati et al.6 Probes HBBTOTAL as a reference and HbβAS3 as a target were used to generate droplets for ddPCR, as described by Hindson et al.52 Droplets were analyzed for absolute quantification of the βAS3 gene expression normalized to the total B-globin gene expression.
In Vivo Experiment in SCD Mouse Model
All mouse work was performed under protocols approved by the UCLA Animal Care Committee. BM from 8- to 12-week-old homozygous βS/βS “Townes” mice (JAX stock #013071) were lineage-depleted using the lineage cell depletion kit from Miltenyi Biotec. Lin− cells were pre-stimulated for 24 h in StemSpan (Stem Cell Technologies, Vancouver, Canada) supplemented with murine stem cell factor (100 ng/mL), human interleukin-11 (100 ng/mL), murine interleukin-3 (20 ng/mL), and human FLT-3 ligand (100 ng/mL). Pre-stimulated Lin− cells were then transduced at various MOIs to obtain similar VCNs in the bulk cell product or mock transduced. Twenty-four h later, one to two million transduced cells were delivered by retro-orbital injection after recipient mice (Pep Boy/J [CD45.1 congenic]) were lethally irradiated (1,075 cGy, split in two fractions). A portion of the transduced cells was cultured for 2 weeks in vitro under myeloid differentiation conditions to determine VCN in the cell product.
Peripheral blood samples were collected at weeks 4, 16, and 20 to measure VCN of engrafted cells by ddPCR, expression of HbβAS3 hemoglobin by HPLC, and to determine RBC indices. At week 20, mice were euthanized and BM cells were used to measure engraftment by flow cytometry (CD45.2/CD45.1), VCN, and expression and to seed a methylcellulose based CFU assay. The percentages of sickled erythrocytes present within peripheral blood were also quantified using the Amnis ImageStream Mark II Imaging Flow Cytometer (Luminex, Austin, TX) (as described below).
HPLC
To characterize and quantify hemoglobin tetramers, including human HbS and HbBAS3, and murine HbA and HbF, we lysed 1 μL of murine peripheral blood in 25 μL hemolysate and incubated at room temperature. Hemolysates were then centrifuged at 500 g for 10 minutes at 4°C to remove RBC ghost. The lysates were then stored frozen at −80°C and later thawed and processed as described by Urbinati et al.6
CFU Progenitor Assay
To quantify VCN range and production of BAS3-globin transcripts in individual BM progenitor cells, we acquired total murine BM from transduced or mock-transduced βS/βS mice and plated it in a methylcellulose medium that supports murine BFU-E growth (Methocult SF M3436; Stem Cell Technologies), as described by Urbinati et al., 2017.6 Cells were plated at three densities from a serial dilution: 20,000, 60,000, and 120,000 cells per 35 mm gridded plate with two plates per each density. After 14 days of culture at 37°C supplemented with 5% CO2, individual BFU-E colonies were enumerated, plucked, and separated into two portions for DNA and mRNA isolation (NucleoSpin Tissue XS; Clontech Laboratories, Mountain View, CA).
Cell Morphology Determination and Quantification
The Amnis ImageStream Mark II Imaging Flow Cytometer was used to determine and quantify the morphologies of RBCs isolated from experimental mice at the time of euthanasia. A 2% solution of sodium metabisulfite was made fresh using ultra-distilled water (Invitrogen, Carlsbad, California). 5 μL of blood was mixed with 5 μL of 2% sodium metabisulfite solution (MBS) and incubated under hypoxic conditions (2% oxygen) for 10 min at 37°C. After incubation, 5 μL of the 1:1 blood and MBS solution was resuspended in 95 μL of a 2% glutaraldehyde solution. The 2% glutaraldehyde solution was made by diluting 25% stock glutaraldehyde solution (Sigma-Aldrich, St. Louis, MO) in PBS. Cell images were collected on the ImageStream Flow Cytometer at 60× magnification with bright-field set to channel four and all lasers off. About 40,000 images were collected for each sample and then analyzed using the IDEAS software (Amnis Corporation, Seattle, WA) using a custom template designed with the assistance of Amnis Corporation.
Statistical Analysis
All data are reported as mean ± SD unless otherwise stated. Statistical analyses were performed using GraphPad Prism version 7.0 (GraphPad Software, San Diego, California, USA). The statistical significance between two averages was established using unpaired t test. When the statistical significance between three or more averages were evaluated, a one-way ANOVA was applied followed by multiple paired comparisons for normally distributed data (Tukey test). When normality assumption was violated, Mann-Whitney U test was performed for group-wise comparison instead. Chi-square tests were used to compare the frequency of transduced CFUs. Linear regression analyses were used to determine the correlation between VCN and βAS3-globin RNA transcripts quantities. All statistical tests were two-tailed and a p value of < 0.05 was deemed significant.
Author Contributions
R.A.M., R.P.H. and D.B.K. conceived and designed all experiments. R.A.M. executed and analyzed all experiments. M.J.U., B.A., R.O.’B., K.S.O., C.K., O.B.S., S.S, B.A., D.B., M.R, J.P.Q., P.G.A., and E.M. helped execute portions of experiments. R.A.M and R.P.H. provided research materials. R.P.H and D.B.K advised experiments. R.A.M and D.B.K. provided financial and administrative support. R.A.M., R.P.H., and D.B.K. wrote the manuscript. R.A.M. and D.B.K. approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by a Sponsored Research Agreement from Biomarin Pharmaceutical, Inc. Training grants provided support to R.A.M. (NIH/NHLBI F31 HL134313; PhRMA Foundation Paul Calabresi Predoctoral Fellowship 20174717; and UCLA MSTP T32 GM008042). The Flow Cytometry Core and the DNA Sequencing Core of the UCLA Eli & Edythe Broad Center of Regenerative Medicine and Stem Cell Research and the Jonsson Comprehensive Cancer Center were used to support studies.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.ymthe.2019.09.020.
Supplemental Information
References
- 1.Ingram V.M. Gene mutations in human haemoglobin: the chemical difference between normal and sickle cell haemoglobin. Nature. 1957;180:326–328. doi: 10.1038/180326a0. [DOI] [PubMed] [Google Scholar]
- 2.Francis R.B., Jr., Johnson C.S. Vascular occlusion in sickle cell disease: current concepts and unanswered questions. Blood. 1991;77:1405–1414. [PubMed] [Google Scholar]
- 3.Charache S., Terrin M.L., Moore R.D., Dover G.J., Barton F.B., Eckert S.V., McMahon R.P., Bonds D.R., Investigators of the Multicenter Study of Hydroxyurea in Sickle Cell Anemia Effect of hydroxyurea on the frequency of painful crises in sickle cell anemia. N. Engl. J. Med. 1995;332:1317–1322. doi: 10.1056/NEJM199505183322001. [DOI] [PubMed] [Google Scholar]
- 4.Niihara Y., Miller S.T., Kanter J., Lanzkron S., Smith W.R., Hsu L.L., Gordeuk V.R., Viswanathan K., Sarnaik S., Osunkwo I., Investigators of the Phase 3 Trial of l-Glutamine in Sickle Cell Disease A Phase 3 Trial of l-Glutamine in Sickle Cell Disease. N. Engl. J. Med. 2018;379:226–235. doi: 10.1056/NEJMoa1715971. [DOI] [PubMed] [Google Scholar]
- 5.Justus D., Perez-Albuerne E., Dioguardi J., Jacobsohn D., Abraham A. Allogeneic donor availability for hematopoietic stem cell transplantation in children with sickle cell disease. Pediatr. Blood Cancer. 2015;62:1285–1287. doi: 10.1002/pbc.25439. [DOI] [PubMed] [Google Scholar]
- 6.Urbinati F., Wherley J., Geiger S., Fernandez B.C., Kaufman M.L., Cooper A., Romero Z., Marchioni F., Reeves L., Read E. Preclinical studies for a phase 1 clinical trial of autologous hematopoietic stem cell gene therapy for sickle cell disease. Cytotherapy. 2017;19:1096–1112. doi: 10.1016/j.jcyt.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 7.Ribeil J.A., Hacein-Bey-Abina S., Payen E., Magnani A., Semeraro M., Magrin E., Caccavelli L., Neven B., Bourget P., El Nemer W. Gene Therapy in a Patient with Sickle Cell Disease. N. Engl. J. Med. 2017;376:848–855. doi: 10.1056/NEJMoa1609677. [DOI] [PubMed] [Google Scholar]
- 8.Thompson A.A., Kwiatkowski J., Rasko J., Hongeng S., Schiller G.J., Anurathapan U., Cavazzana M., Joy Ho P., von Kalle C., Kletzel M. Lentiglobin Gene Therapy for Transfusion-Dependent β-Thalassemia: Update from the Northstar Hgb-204 Phase 1/2 Clinical Study. Blood. 2016;128:1175. [Google Scholar]
- 9.Kanter J., Walters M.C., Hsieh M., Krishnamurti L., Kwiatkowski J.L., Kamble R., von Kalle C., Joseney-Antoine M., Pierciey F.J., Jr., Shi W. Interim Results from a Phase 1/2 Clinical Study of Lentiglobin Gene Therapy for Severe Sickle Cell Disease. Blood. 2017;130:527. [Google Scholar]
- 10.Walters M.C., Hongeng S., Kwiatkowski J.L., Locatelli F., Porter J.B., Sauer M.G. Results from the Hgb-207 (Northstar-2) Trial: A Phase 3 Study to Evaluate Safety and Efficacy of Lentiglobin Gene Therapy for Transfusion-Dependent β-Thalassemia (TDT) in Patients with Non-β0/β0 Genotypes. Blood. 2017;130:526. [Google Scholar]
- 11.Thompson A.A., Walters M.C., Kwiatkowski J., Rasko J.E.J., Ribeil J.A., Hongeng S., Magrin E., Schiller G.J., Payen E., Semeraro M. Gene Therapy in Patients with Transfusion-Dependent β-Thalassemia. N. Engl. J. Med. 2018;378:1479–1493. doi: 10.1056/NEJMoa1705342. [DOI] [PubMed] [Google Scholar]
- 12.Cooper A.R., Patel S., Senadheera S., Plath K., Kohn D.B., Hollis R.P. Highly efficient large-scale lentiviral vector concentration by tandem tangential flow filtration. J. Virol. Methods. 2011;177:1–9. doi: 10.1016/j.jviromet.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cavazzana-Calvo M., Payen E., Negre O., Wang G., Hehir K., Fusil F., Down J., Denaro M., Brady T., Westerman K. Transfusion independence and HMGA2 activation after gene therapy of human β-thalassaemia. Nature. 2010;467:318–322. doi: 10.1038/nature09328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hauber I., Beschorner N., Schrödel S., Chemnitz J., Kröger N., Hauber J., Thirion C. Improving Lentiviral Transduction of CD34+ Hematopoietic Stem and Progenitor Cells. Hum. Gene Ther. Methods. 2018;29:104–113. doi: 10.1089/hgtb.2017.085. [DOI] [PubMed] [Google Scholar]
- 15.Federico M., editor. Lentiviral Vectors and Exosomes as Gene and Protein Delivery Tools. Humana Press; New York, NY: 2016. pp. 49–61. [Google Scholar]
- 16.Delville M., Soheili T., Bellier F., Durand A., Denis A., Lagresle-Peyrou C., Cavazzana M., Andre-Schmutz I., Six E. A Nontoxic Transduction Enhancer Enables Highly Efficient Lentiviral Transduction of Primary Murine T Cells and Hematopoietic Stem Cells. Mol. Ther. Methods Clin. Dev. 2018;10:341–347. doi: 10.1016/j.omtm.2018.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Petrillo C., Thorne L.G., Unali G., Schiroli G., Giordano A.M.S., Piras F., Cuccovillo I., Petit S.J., Ahsan F., Noursadeghi M. Cyclosporine H Overcomes Innate Immune Restrictions to Improve Lentiviral Transduction and Gene Editing In Human Hematopoietic Stem Cells. Cell Stem Cell. 2018;23:820–832.e9. doi: 10.1016/j.stem.2018.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miccio A., Cesari R., Lotti F., Rossi C., Sanvito F., Ponzoni M., Routledge S.J., Chow C.M., Antoniou M.N., Ferrari G. In vivo selection of genetically modified erythroblastic progenitors leads to long-term correction of beta-thalassemia. Proc. Natl. Acad. Sci. USA. 2008;105:10547–10552. doi: 10.1073/pnas.0711666105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Romero Z., Urbinati F., Geiger S., Cooper A.R., Wherley J., Kaufman M.L., Hollis R.P., de Assin R.R., Senadheera S., Sahagian A. β-globin gene transfer to human bone marrow for sickle cell disease. J. Clin. Invest. 2013;123:3317–3330. doi: 10.1172/JCI67930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pawliuk R., Westerman K.A., Fabry M.E., Payen E., Tighe R., Bouhassira E.E., Acharya S.A., Ellis J., London I.M., Eaves C.J. Correction of sickle cell disease in transgenic mouse models by gene therapy. Science. 2001;294:2368–2371. doi: 10.1126/science.1065806. [DOI] [PubMed] [Google Scholar]
- 21.May C., Rivella S., Chadburn A., Sadelain M. Successful treatment of murine beta-thalassemia intermedia by transfer of the human beta-globin gene. Blood. 2002;99:1902–1908. doi: 10.1182/blood.v99.6.1902. [DOI] [PubMed] [Google Scholar]
- 22.Negre O., Bartholomae C., Beuzard Y., Cavazzana M., Christiansen L., Courne C., Deichmann A., Denaro M., de Dreuzy E., Finer M. Preclinical evaluation of efficacy and safety of an improved lentiviral vector for the treatment of β-thalassemia and sickle cell disease. Curr. Gene Ther. 2015;15:64–81. doi: 10.2174/1566523214666141127095336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perumbeti A., Higashimoto T., Urbinati F., Franco R., Meiselman H.J., Witte D., Malik P. A novel human gamma-globin gene vector for genetic correction of sickle cell anemia in a humanized sickle mouse model: critical determinants for successful correction. Blood. 2009;114:1174–1185. doi: 10.1182/blood-2009-01-201863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Urbinati F., Campo Fernandez B., Masiuk K.E., Poletti V., Hollis R.P., Koziol C., Kaufman M.L., Brown D., Mavilio F., Kohn D.B. Gene Therapy for Sickle Cell Disease: A Lentiviral Vector Comparison Study. Hum. Gene Ther. 2018;29:1153–1166. doi: 10.1089/hum.2018.061. [DOI] [PubMed] [Google Scholar]
- 25.Kumar M., Keller B., Makalou N., Sutton R.E. Systematic determination of the packaging limit of lentiviral vectors. Hum. Gene Ther. 2001;12:1893–1905. doi: 10.1089/104303401753153947. [DOI] [PubMed] [Google Scholar]
- 26.Canté-Barrett K., Mendes R.D., Smits W.K., van Helsdingen-van Wijk Y.M., Pieters R., Meijerink J.P. Lentiviral gene transfer into human and murine hematopoietic stem cells: size matters. BMC Res. Notes. 2016;9:312. doi: 10.1186/s13104-016-2118-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Navas P.A., Peterson K.R., Li Q., McArthur M., Stamatoyannopoulos G. The 5'HS4 core element of the human beta-globin locus control region is required for high-level globin gene expression in definitive but not in primitive erythropoiesis. J. Mol. Biol. 2001;312:17–26. doi: 10.1006/jmbi.2001.4939. [DOI] [PubMed] [Google Scholar]
- 28.Lisowski L., Sadelain M. Locus control region elements HS1 and HS4 enhance the therapeutic efficacy of globin gene transfer in beta-thalassemic mice. Blood. 2007;110:4175–4178. doi: 10.1182/blood-2007-08-108647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Romero Z., Campo-Fernandez B., Wherley J., Kaufman M.L., Urbinati F., Cooper A.R., Hoban M.D. The human ankyrin 1 promoter insulator sustains gene expression in a β-globin lentiviral vector in hematopoietic stem cells. Mol. Ther. Methods Clin. Dev. 2015;2:15012. doi: 10.1038/mtm.2015.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ryan T.M., Ciavatta D.J., Townes T.M. Knockout-transgenic mouse model of sickle cell disease. Science. 1997;278:873–876. doi: 10.1126/science.278.5339.873. [DOI] [PubMed] [Google Scholar]
- 32.Levasseur D.N., Ryan T.M., Pawlik K.M., Townes T.M. Correction of a mouse model of sickle cell disease: lentiviral/antisickling beta-globin gene transduction of unmobilized, purified hematopoietic stem cells. Blood. 2003;102:4312–4319. doi: 10.1182/blood-2003-04-1251. [DOI] [PubMed] [Google Scholar]
- 33.Levasseur D.N., Ryan T.M., Reilly M.P., McCune S.L., Asakura T., Townes T.M. A recombinant human hemoglobin with anti-sickling properties greater than fetal hemoglobin. J. Biol. Chem. 2004;279:27518–27524. doi: 10.1074/jbc.M402578200. [DOI] [PubMed] [Google Scholar]
- 34.Grosveld F., van Assendelft G.B., Greaves D.R., Kollias G. Position-independent, high-level expression of the human beta-globin gene in transgenic mice. Cell. 1987;51:975–985. doi: 10.1016/0092-8674(87)90584-8. [DOI] [PubMed] [Google Scholar]
- 35.Ryan T.M., Behringer R.R., Martin N.C., Townes T.M., Palmiter R.D., Brinster R.L. A single erythroid-specific DNase I super-hypersensitive site activates high levels of human beta-globin gene expression in transgenic mice. Genes Dev. 1989;3:314–323. doi: 10.1101/gad.3.3.314. [DOI] [PubMed] [Google Scholar]
- 36.Peterson K.R., Clegg C.H., Navas P.A., Norton E.J., Kimbrough T.G., Stamatoyannopoulos G. Effect of deletion of 5′HS3 or 5′HS2 of the human beta-globin locus control region on the developmental regulation of globin gene expression in beta-globin locus yeast artificial chromosome transgenic mice. Proc. Natl. Acad. Sci. 1996;93:6605–6609. doi: 10.1073/pnas.93.13.6605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tanimoto K., Sugiura A., Omori A., Felsenfeld G., Engel J.D., Fukamizu A. Human beta-globin locus control region HS5 contains CTCF- and developmental stage-dependent enhancer-blocking activity in erythroid cells. Mol. Cell. Biol. 2003;23:8946–8952. doi: 10.1128/MCB.23.24.8946-8952.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fedosyuk H., Peterson K.R. Deletion of the human beta-globin LCR 5‘HS4 or 5’HS1 differentially affects beta-like globin gene expression in beta-YAC transgenic mice. Blood Cells Mol. Dis. 2007;39:44–55. doi: 10.1016/j.bcmd.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peterson K.R., Fedosyuk H., Harju-Baker S. LCR 5′ hypersensitive site specificity for globin gene activation within the active chromatin hub. Nucleic Acids Res. 2012;40:11256–11269. doi: 10.1093/nar/gks900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Forrester W.C., Novak U., Gelinas R., Groudine M. Molecular analysis of the human beta-globin locus activation region. Proc. Natl. Acad. Sci. USA. 1989;86:5439–5443. doi: 10.1073/pnas.86.14.5439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Collis P., Antoniou M., Grosveld F. Definition of the minimal requirements within the human beta-globin gene and the dominant control region for high level expression. EMBO J. 1990;9:233–240. doi: 10.1002/j.1460-2075.1990.tb08100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fraser P., Hurst J., Collis P., Grosveld F. DNaseI hypersensitive sites 1, 2 and 3 of the human beta-globin dominant control region direct position-independent expression. Nucleic Acids Res. 1990;18:3503–3508. doi: 10.1093/nar/18.12.3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fraser P., Pruzina S., Antoniou M., Grosveld F. Each hypersensitive site of the human beta-globin locus control region confers a different developmental pattern of expression on the globin genes. Genes Dev. 1993;7:106–113. doi: 10.1101/gad.7.1.106. [DOI] [PubMed] [Google Scholar]
- 44.May C., Rivella S., Callegari J., Heller G., Gaensler K.M., Luzzatto L., Sadelain M. Therapeutic haemoglobin synthesis in beta-thalassaemic mice expressing lentivirus-encoded human beta-globin. Nature. 2000;406:82–86. doi: 10.1038/35017565. [DOI] [PubMed] [Google Scholar]
- 45.Pembrey M.E., Wood W.G., Weatherall D.J., Perrine R.P. Fetal haemoglobin production and the sickle gene in the oases of Eastern Saudi Arabia. Br. J. Haematol. 1978;40:415–429. doi: 10.1111/j.1365-2141.1978.tb05813.x. [DOI] [PubMed] [Google Scholar]
- 46.Platt O.S., Brambilla D.J., Rosse W.F., Milner P.F., Castro O., Steinberg M.H., Klug P.P. Mortality in sickle cell disease. Life expectancy and risk factors for early death. N. Engl. J. Med. 1994;330:1639–1644. doi: 10.1056/NEJM199406093302303. [DOI] [PubMed] [Google Scholar]
- 47.Forget B.G. Molecular basis of hereditary persistence of fetal hemoglobin. Ann. N. Y. Acad. Sci. 1998;850:38–44. doi: 10.1111/j.1749-6632.1998.tb10460.x. [DOI] [PubMed] [Google Scholar]
- 48.Akinsheye I., Alsultan A., Solovieff N., Ngo D., Baldwin C.T., Sebastiani P., Chui D.H.K., Steinberg M.H. Fetal hemoglobin in sickle cell anemia. Blood. 2011;18:19–27. doi: 10.1182/blood-2011-03-325258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bao E.L., Lareau C.A., Brugnara C., Fulcher I.R., Barau C., Moutereau S., Habibi A., Badaoui B., Berkenou J., Bartolucci P. Heritability of fetal hemoglobin, white cell count, and other clinical traits from a sickle cell disease family cohort. Am. J. Hematol. 2019;94:522–527. doi: 10.1002/ajh.25421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baglioni C., Weatherall D.J. ABNORMAL HUMAN HEMOGLOBINS. IX. CHEMISTRY OF HEMOGLOBIN J-BALTIMORE. Biochim. Biophys. Acta. 1963;78:637–643. doi: 10.1016/0006-3002(63)91029-1. [DOI] [PubMed] [Google Scholar]
- 51.Dull T., Zufferey R., Kelly M., Mandel R.J., Nguyen M., Trono D., Naldini L. A third-generation lentivirus vector with a conditional packaging system. J. Virol. 1998;72:8463–8471. doi: 10.1128/jvi.72.11.8463-8471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hindson B.J., Ness K.D., Masquelier D.A., Belgrader P., Heredia N.J., Makarewicz A.J., Bright I.J., Lucero M.Y., Hiddessen A.L., Legler T.C. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal. Chem. 2011;83:8604–8610. doi: 10.1021/ac202028g. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.