Abstract
DNA molecules are known as the genetic information carriers. Recently, they are being explored as a new generation of biocatalysts or chiral scaffolds for metal catalysts. There is also a growing interest of finding alternative solvents for DNA preservation and stabilization, including two unique types of solvents: ionic liquids (ILs) and deep eutectic solvents (DES). Therefore, it is important to understand how DNA molecules interact with these novel ionic solvent systems (i.e. ILs and DES). It is well known that inorganic di- and monovalent ions preferentially bind with major and minor grooves of DNA structures. However, in the case of ILs and DES, organic cation may intrude into the DNA minor grooves; more importantly, electrostatic attraction between organic cations and the DNA phosphate backbone becomes a predominant interaction, accompanying by hydrophobic and polar interactions between ILs and DNA major and minor grooves. In addition, anions may form hydrogen-bonds with cytosine, adenine and guanine bases. Despites these strong interactions, DNA molecules maintain double helical structure in most ionic solvent systems, especially in aqueous IL solutions. Furthermore, the exciting advances of G-quadruplexe DNA structures in ILs and DES are discussed.
Keywords: DNA, groove, ionic liquid, deep eutectic solvent, DNA-ion interaction
Introduction
Deoxyribonucleic acid (DNA) is a macromolecule that consists of repeating units called nucleotides and carries genetic information for the growth and functioning of living organisms and many viruses. In the laboratories, some DNA molecules have been found to have catalytic capability through a systematic evolution of ligands by exponential enrichment (SELEX) process.1, 2 These DNAs are known as the DNAzymes (or deoxyribozymes or DNA enzymes). They are commonly used as catalysts to cleave or ligate two RNA substrates, and also as photolyase (to photocleave thymine cyclobutane dimers), or peroxidase (for DNA detection), etc.2, 3 Recently, the double-stranded DNA from herring sperm was found capable of catalyzing the dithioacetalization of different aldehydes in water.4 Also, G-quadruplex DNA have been demonstrated as promising catalysts for enantioselective Diels–Alder5–7 and Friedel–Crafts8 reactions.
In addition, due to its unique chiral structure, DNA is being evaluated as a chiral scaffold for metal complexes resulting in ‘DNA-based hybrid catalysts’. These new hybrid catalysts are becoming very promising for asymmetric synthesis,9–11 and have exhibited high activities and enantioselectivities in a number of reactions including Michael addition,12 Diels-Alder reaction,13–15 Friedel-Crafts alkylation,16 intramolecular cyclopropanation,17 electrophilic fluorination of β-keto esters,18 and asymmetric hydration,14 etc. Many of these asymmetric reactions are very valuable to the preparation of natural products and the synthesis of chiral intermediates for pharmaceuticals and agrochemicals. The Roelfes group19 indicated that the addition of organic solvents (10–33%, v/v) including acetonitrile, THF and alcohols usually has no negative impact on the ee values of DNA-based asymmetric catalytic Diels–Alder, Michael addition and Friedel–Crafts alkylation reactions, and could even accelerate the Michael addition and Friedel–Crafts alkylation reaction. However, most of these reactions were carried out in aqueous buffers which do not typically dissolve the organic substrates; this leads to long reaction times and low catalytic efficiencies. Therefore, there is a great need to introduce non-aqueous solvents in these DNA-based asymmetric reactions.
To better utilize DNA in non-aqueous solvents, it is of great interest to understand the impact of different solvent media on the DNA structural changes and binding properties. The canonical DNA structure is a B-form duplex comprising Watson–Crick base pairs. DNA loses its double-helical structure in 99% dimethyl sulfoxide, formamide, or methanol, whereas the structure is retained in glycerol and to some extent in ethylene glycol (although the thermal stability in these two solvents is much lower than that in aqueous buffer).20 The DNA melting transition temperature in aqueous ethylene glycol was found to vary linearly with the solvent fraction of ethylene glycol.21 Recently, ionic liquids (ILs) and deep eutectic solvents (DES) have been actively investigated as non-conventional replacements to volatile organic solvents in biocatalysis.
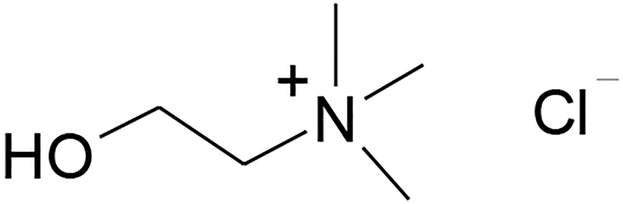
Ionic liquids (ILs) consist of ions and remain liquid at temperatures lower than 100 °C. As a new generation of non-aqueous solvents, ILs have many favorable properties such as near-zero vapor pressure, a wide liquid range, low flammability, high ionic conductivity, high thermal conductivity, high dissolution capability towards many substrates, high thermal and chemical stability, and a wide electrochemical potential window.22 More importantly, the physical properties of ILs can be finely tuned through the judicious selection of cations and anions. All these adjustable properties are very important to biomolecule stabilization and activation; therefore, many enzymatic reactions have been reported in different types of ILs.23–25 Alternatively, a new type of solvents, so called ‘deep eutectic solvents’ (DES), can be produced by mixing a solid organic salt (such as choline chloride, Scheme 1) and a suitable amount of complexing agent (such as urea or glycerol) to form a liquid with lowest possible freezing point (Tf). A representative example can be illustrated by mixing choline chloride (m.p. = 302 °C, Scheme 1) and urea (m.p. = 133 °C) at a 1:2 molar ratio yielding a room-temperature liquid (Tf = 12 °C).26 The reason of forming the deep eutectic system is that the complexing agent (typically a H-bond donor) interacts with the anion and increases its effective size, which consecutively reduces the anion interaction with the cation and thus decreases the freezing point (Tf) of the mixture. Although DES typically contain a substantial portion of molecular component, they share many attractive solvent properties with regular ILs. In addition, many DES systems have other advantages such as low-cost, high biodegradability and low toxicity. For these reasons, these new solvents have recently been actively studied in a number of applications such as solvents for electrodeposition and electropolishing of metals, catalyst or solvents for chemical and enzymatic reactions, and solvents for extractions.26–29 Several studies have reported high enzyme activity and stability in DES, including Candida antarctica lipase B (CALB), Candida antarctica lipase A (CALA), and epoxide hydrolase in DES,30 potato epoxide hydrolase StEH1 in solutions of eutectic mixtures,31 and α-chymotrypsin in aqueous mixtures of triethylammonium acetate and urea.32 Recently, our group developed new DES (such as choline chloride/glycerol at 1:2 molar ratio and choline acetate/glycerol at 1:1.5) that are also biocompatible with immobilized CALB,33, 34 and cross-linked proteases (subtilisin and α-chymotrypsin) immobilized on chitosan.35
Scheme 1.
Structure of choline chloride.
Our recent finding36 suggested that different ILs and DES have significant impact on the enantioselectivity and catalytic efficiency of DNA-based asymmetric Michael addition. Although current studies on the DNA-based catalysis in ILs and DES are very limited, it is very valuable to visualize how DNA molecules interact with neat ILs and DES or their aqueous solutions. This would enable us to demystify the compatibility of ionic solvents with DNA, and to rationally design ILs and DES for DNA-based applications. This concise review intends to provide a quick overview of the DNA stability in ionic environments, i.e. solutions of inorganic salts, ILs and DES, and to stimulate the further research on DNA-based applications in ionic media.
DNA-Inorganic Ion Interactions
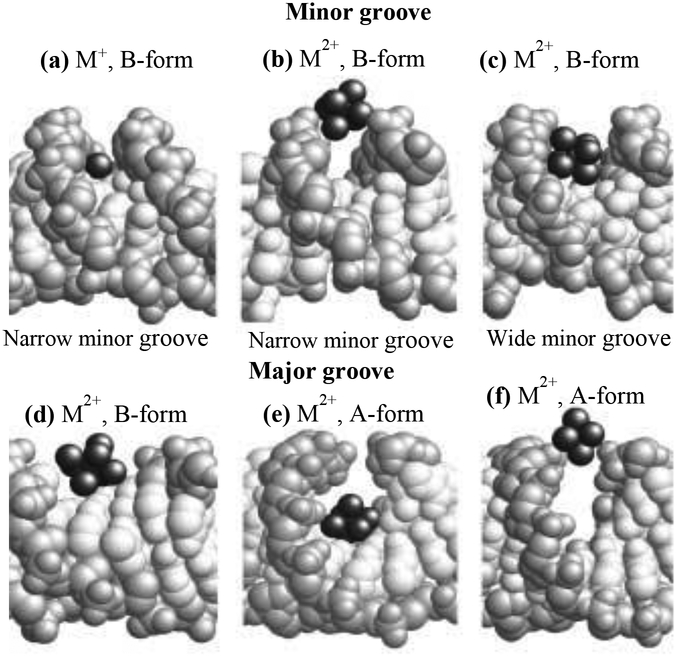
Previous studies have focused on the understanding of the interactions of DNA molecules with mono- and divalent inorganic ions. Experimental results from X-ray crystallography, NMR spectroscopy and capillary electrophoresis, as well as theoretical studies from molecular dynamic simulations, have led to some basic understandings of how di- and monovalent metal cations preferentially interact with the major and minor grooves of DNA duplexes (see Fig 1):37–39 (1) Preferential cation binding with major and minor grooves depends on both the DNA sequence (electrostatic property of grooves and the specific position of electronegative groups in grooves) and the type of cations (di- and monovalent); (2) AT-rich sequences drive the binding of cations in the minor groove, whilst GC-rich sequences promote the preferential binding of cations in the major groove; (3) Divalent Mg2+ and Ca2+ bind at GpN steps in the major grooves of A- and B-DNA, and bind to G-tracts or at the G/A tract interface in the B-DNA minor groove; (4) Monovalent cations (e.g. Na+, K+, Cs+, Rb+, and Tl+) bind to G-tracts in the major groove of B-DNA or bind at GpT steps in A-DNA, and bind to thymines from opposite strands at the central ApT step of A tracts in the B-DNA minor groove. In particular, Hud et al.40 confirmed that ammonium cations bind with DNA in the minor groove of A-tract sequences based on the high resolution NMR spectroscopy experiments. The sequence-specific binding of monovalent cations with A-tracks suggests an elucidation for the origin of sequence-directed bending. Based on molecular dynamics (MD) simulations, Hamelberg et al.41 suggested that cations (such as Na+) could cause the minor groove in the AT sequence of DNA to become narrow, primarily due to sequence-dependent Na+‒phosphates and base‒water interactions.
Fig. 1.
Space-filling models of X-ray crystal structures with cations bound in the minor and major grooves illustrating the correlation between DNA groove width and cation coordination sites. DNA base atoms are white and backbone atoms are light gray; cations and water oxygen atoms are dark gray (Reprinted from Ref,38 Copyright © 2001, with permission from Elsevier).
In addition, multivalent cations (such as spermidine3+ and cobalt(III) hexaammine3+) can provoke the condensation of high molecular weight DNA (at least several hundred base pairs) to form a compact and highly ordered toroidal or rodlike structure.42 The DNA condensation can be attributed to various intermolecular interactions, which can be further credited to the contributions of ions and water. Furthermore, the DNA condensing ions may cause subtle changes of local structures of the double helix. Small multivalent cations can induce DNA bending since these cations binds at the entrance to the B-DNA major groove electrostatically repelling Na+ cations from the adjacent phosphate strands.43 The phosphate groups strongly interact with the groove-bound cation, resulting in groove collapse and subsequent DNA bending toward the cation. The bending is expected to be greater in GC-rich than in AT-rich DNA. The cation is at least divalent with high charge density (such as Mg2+, putrescine2+, spermidine3+, spermine4+, and cobalt(III) hexaammine3+). It is important to realize that a cationic ligand with high binding specificity does not induce considerable DNA bending, unless its binding site is located in the DNA major groove. Based on the DNA melting temperatures in metal solutions and other studies, Eichhorn and Shin44 suggested an increasing order of apparent relative affinity of metal ions toward phosphate and base sites on DNA as Mg(II) < Co(II), Ni(II) < Mn(II) < Zn(II) < Cd(II) < Cu(II) < Ag(I) < Hg(II). Kasyanenko et al.45 suggested that trivalent ions in solutions of [Co(NH3)6]Cl3, [Co(NO2)6]Na3 or FeCl3 bind to DNA molecules causing DNA shrinkage, which is concluded from the results of decreasing DNA intrinsic viscosity and increasing optical anisotropy with the salt concentration. Through examining 1 Å resolution X-ray crystal structures of Mg2+ and Ca2+ salts of the B-DNA decamers CCAACGTTGG and CCAGCGCTGG, Chiu and Dickerson46 suggested that DNA bending by base-roll compression towards the major groove is induced by sequence-specific and strand-specific binding of Mg2+ and Ca2+ to the major groove, while a minimum impact on the helical structure is caused by the sequence-specific binding of Mg2+ and Ca2+ in the minor groove.
DNA Stability in Neat ILs
Several studies have evaluated the DNA stability in neat ILs. Zhang et al.47 found that poly[3-butyl-1-vinylimidazolium l-proline salt] could condense plasmid DNA to form stable complexes against the enzymatic degradation by deoxyribonuclease I, as suggested by the agarose gel electrophoresis. The DNA could be released from the complexes by competitive exchange with sodium polyacrylate without losing its structures. Therefore, this poly(IL) has a potential application as DNA vectors. By laser light scattering, Chen48 studied the solubility and chain conformation of DNA (from salmon testes) in [AMIM]Cl and [BMIM][HCOO]; they suggested that the DNA chain is in random conformation in [AMIM]Cl and DNA is denatured and condensed in [BMIM][HCOO].
The Hud group49 indicated that the oligonucleotide d(CG)8 exhibits secondary-structure formation in [HMIM][BF4], however, the 32 bp DNA duplex seems partially denatured in this IL. Cardoso and Micaelo50 examined the molecular solvation of double-stranded DNA (dsDNA) and single-stranded DNA (ssDNA) in neat room-temperature ILs based on imidazolium, oxazolium, pyrrolidinium, pyrimidinium, quaternary ammonium and cholinium cations, carrying BF4− and PF6− anions. They suggested that cations tend to move close to the DNA main chain due to strong electrostatic interactions with the phosphate groups as well as hydrogen-bonding and edge-to-face NH···π interactions with the DNA bases, while anions mainly form hydrogen-bonds with cytosine, adenine and guanine bases. In addition, ssDNA bases are preferentially solvated by the anions via hydrogen bonding with the fluorine atoms. Their modeling and molecular dynamics simulation studies indicate that the DNA structure is highly stable in these ILs, retaining the native B-form dsDNA crystallographic structure. In term of stabilizing DNA secondary structures, the best ILs are based on cholinium and pyridinium cations and PF6− anions; the BF4− anions tend to form more hydrogen bonds with the dsDNA bases than the PF6− anions. On the other hand, ssDNA bases are more accessible by the solvent, especially the BF4− anions. Furthermore, the radial distributions (RDF) of the cations suggest that the cation “head” charge group is preferentially located at 0.5 nm from the DNA phosphate groups while the anions are completely excluded from this region. The Prasad group51 dissolved up to 3.5 wt% DNA (from salmon testes) in two bio-based ILs, namely choline indole-3-acetate and choline indole-3-butyrate. DNA maintains B-form structures and a long-term stability (up to 6 months) in the first IL but is denatured by the second one.
DNA Stability in Hydrated or Aqueous ILs
More studies focus on the interactions of DNA with aqueous ILs. First of all, hydration itself has an impact on the DNA structure. Poater et al.52 theoretically computed a series of 51 Watson-Crick and mismatched DNA base pairs using dispersion-corrected DFT, and indicated that the DNA solvation in water could lead to a weakening and lengthening of hydrogen bonds between the DNA bases. The existing of ILs in water complicates the DNA interactions and stability. The Wang group53 found that [BMIM][PF6] was effective for extraction of salmon testes DNA from aqueous solutions. Their 31P NMR and FT-IR results suggest that [BMIM]+ cations displace Na+ ions and interact with P-O bonds of phosphate groups in the DNA strands. The same group54 further observed that in [BMIM][PF6], the resonance light scattering (RSL) signals of ethidium bromide decreased with the addition of double-stranded DNA (dsDNA), which is the opposite trend of that in aqueous solutions. They explained that the cations intercalate into the DNA helix structure and interact with the P‒O bonds of DNA phosphate groups, which results in a decrease of the base-pair interstice and the transformation of DNA conformations, as well as a reduction in the intercalation between ethidium bromide and DNA. On the contrary, the Liu group55 reported that hydrophobic ILs ([BMIM][PF6] and [HMIM][PF6]) are not effective for extracting DNA from aqueous solutions, but are efficient for extraction DNA staining dyes.
Xie et al.56 confirmed the electrostatic interaction between calf thymus DNA and [BMIM][BF4] using cyclic voltammograms of Co(bpy)33+/2+ (bpy = 2,2′-bipyripyl) at a DNA-modified gold electrode in phosphate buffer solution containing [BMIM][BF4]. They calculated the binding constant as 4.26 × 104 M−1 for [BMIM][BF4] bound to DNA, and the Gibbs energy of surface binding ‒26.4 kJ mol−1 at 298 K. They also noticed that the dissociation rate constant of Co(bpy)33+/2+ from the electrode surface in the presence of 1.5% [BMIM][BF4] is over 40% greater than that without the IL, suggesting a competitive interaction between [BMIM][BF4] and DNA.
The Guo group57 studied the binding characteristics and molecular mechanism of interactions between DNA and [BMIM]Cl in aqueous solutions. They observed a decreased critical aggregation concentration of [BMIM]Cl in the presence of DNA, and a continuous fluorescence quenching of the intercalated probe ethidium bromide with the addition of [bmim]Cl, which implies the interaction between the IL and DNA. Dynamic light scattering (DLS) results suggest that a low concentration of [BMIM]Cl could induce a coil-to-globule transition of DNA, which was confirmed by the cryo-TEM images of DNA-IL complexes. During the binding process, DNA retains the B-form helix; however, the base packing and helical structure of DNA are altered to a certain degree. Based on the experimental data and quantum chemical calculations, this group proposed a DNA-IL interaction mechanism: when [BMIM]Cl concentration is lower than 0.06 M, the cationic headgroups are within several angstroms from DNA phosphates, and the hydrophobic chains are organized parallel to the DNA surface; when [BMIM]Cl concentration is greater than 0.06 M, the cationic headgroups are located near DNA phosphates while the hydrocarbon chains are perpendicularly attached to the DNA surface. This group further emphasized that although the electrostatic interaction is critical to the binding of [bmim]Cl to DNA, the hydrophobic interaction between the hydrocarbon chains of [bmim]Cl and the bases of DNA also plays a crucial role.
Singh et al.58 probed the DNA-IL interaction by studying the binding between calf thymus DNA and Thioflavin-T via UV-Vis emission and femtosecond fluorescence spectra. The addition of higher concentrations of ILs ([EMIM][BF4], [BMIM][BF4] and [HMIM][BF4]) and more hydrophobic ILs (e.g. [HMIM][BF4]) reduces the emission intensity, which suggests a weaker binding between DNA and Thioflavin-T but a stronger binding between DNA and ILs. These results further confirm the electrostatic interaction is essential to the DNA-IL interaction; however, the hydrophobic interaction between the cation’s alkyl chain and DNA is also important. He et al.59 studied the interaction between [C12MIM]Br and DNA in dilute brine by various techniques including isothermal titration calorimetry, micropolarity, dynamic light scattering, UV–Vis transmittance, atomic force microscopy (AFM), circular dichroism and molecular dynamics simulation. They noticed the strong electrostatic attraction between DNA phosphate groups and IL headgroups, and the hydrophobic interactions between the [C12MIM]Br alkyl chains. They also observed the size transition and conformational change of DNA in the presence of [C12MIM]Br.
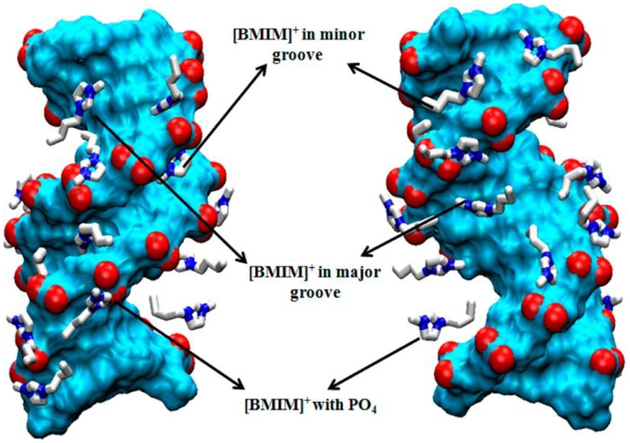
The MacFarlane group60 observed the long-term (up to 6 months) stability of salmon testes DNA in [choline][lactate], [choline][H2PO4] (containing 20–50 wt% water) and choline nitrate (containing 20% water), as confirmed by the circular dichroism and fluorescence spectra as well as the gel electrophoresis. In addition, DNA is thermally stable (up to 100 °C) in [choline][H2PO4] (with 50 wt% water) as confirmed by fluorescence spectroscopy and differential scanning calorimetry. It was explained that the mild hydrogen-bonding and low-water environment of these IL systems minimize the hydrolytic reactions depolymerizing the DNA molecules. Following these encouraging results, Chandran et al.61 carried out molecular dynamics simulations and spectroscopic experiments (CD, UV-Vis and fluorescence) to understand the mechanism of high DNA stability in hydrated ILs. They confirmed the model DNA (Dickerson−Drew dodecamer) maintained the B-conformation in hydrated (up to 80 wt%) ILs ([BMIM]Cl, [BMIM][NO3], [BMIM][lactate], [choline][NO3] and [choline][lactate]); however, [BMIM]Cl and [BMIM][NO3] bind to DNA more strongly than [BMIM][lactate]. In the presence of 80 wt% IL, 55−75% of the water molecules could be stripped from the solvation shell of DNA; [BMIM]+ cations seem to disrupt the spine of hydration by intruding into the DNA minor groove and; [BMIM]+ cations bind to DNA minor grooves more effectively than [choline]+ cations (Fig 2). They also suggested that the electrostatic interactions between ILs and DNA backbone, as well as hydrophobic and polar interactions between ILs and DNA major and minor grooves lead to the DNA dehydration and high stability. Furthermore, Tateishi-Karimata and Sugimoto62 evaluated the melting temperatures of the formation of DNA duplexes consisting of different A-T and G-C base pairs, and suggested that A-T base pairs are more stable than G-C base pairs in the hydrated [choline][H2PO4] (4 M or 80 wt%). The molecular dynamic calculations reveal that choline cations bind strongly in the major groove of A–T regions but weakly to the G–C rich regions; however, choline cations bind preferentially to G–C rich single-stranded DNA vs to A or T rich strands; the binding of choline cation to guanines destabilizes G–C rich DNA duplexes.
Fig. 2.
Typical spatial distribution map of [BMIM]+ cations illustrating their association with the DNA backbone, major groove, and minor groove (color scheme: cyan, DNA; red, DNA phosphates; white, IL cations; blue, ring nitrogens of the [BMIM]+ cations) (Reprinted with permission from Ref,61 Copyright © 2012 American Chemical Society).
Wang et al.63 examined the binding of calf thymus DNA to [CnMIM]Br (n = 4, 6, 8, 10, and 12) in aqueous solutons using pyrene as the fluorescence probe, and reported that the binding Gibbs energy decreases linearly with the increase of alkyl chain length of cations and the binding constants increase with the alkyl chain length. They further suggested that the main contributor to the DNA-IL interaction is the electrostatic attraction although the hydrophobic interaction increases with the alkyl chain length.
DNA Stability in Deep Eutectic Solvents (DES)
Since DES share many common properties with ILs, a number of groups also evaluated the DNA stability in DES. The Hud group49 investigated the secondary structures of DNA and RNA in a neat DES known as choline chloride/urea (1:2) using circular dichroism (CD) spectroscopy. They suggested that: (1) the 32 bp mixed-sequence DNA exhibits an A-form duplex in the neat DES but B-form helix in aqueous salt solutions (such as 3.7 m NaCl); (2) the oligonucleotide [d(CG)8]2 shows a left-handed Z-form helix in the neat DES and aqueous solutions of choline chloride (3.7 m) and NaCl (3.7 m); (3) the oligonucleotide [d(AT)16]2 has a similar B-form helix in the neat DES as in aqueous solutions; (4) the sequence [d(A4T4)4]2 adopts an altered B-form helical structure (B* form). However, the melting transition midpoints (TM) of these DNAs in the DES are lower than those in aqueous solutions, suggesting a lower duplex stability in dehydrating and high ionic strength conditions. In addition, DNA can maintain certain triplex and G-quadruplex structures in the DES although these structures are different from those in aqueous solutions. The Prasad group64 has demonstrated that two DES (choline chloride/ethylene glycol at 1:2 and choline chloride/glycerol at 1:2) could dissolve up to 5.5 wt% and 2.5 wt% DNA from salmon testes. They observed that DNA maintains B-form helical structures in DES and the regenerated DNA shows a high thermal and pH stability. They also proposed a likely DNA solvation mechanism as the interaction of choline cations with DNA phosphate.
G-quadruplex DNA in ionic solvents
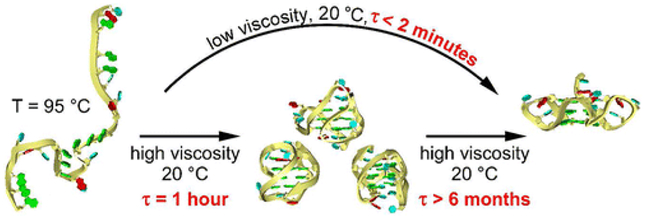
G-quadruplexe DNA has unique four-stranded helices, and is folded from guanine-rich DNA sequences by the stacking of planar quartets composed of four guanines that interact by Hoogsteen hydrogen bonding.65, 66 G-quadruplex structures are highly polymorphic; for example, human telomeric sequences can adopt at least five different intramolecular G-quadruplexes.67 Therefore, G-quadruplex DNA molecules have shown potential applications in catalysts, biosensors, and DNA-based architectures. G-quadruplex DNA structures are usually maintained in aqueous solutions and thus most related applications are explored in aqueous phases. Fujita and Ohno68 found that dG3(T2AG3)3 DNA maintains the G-quadruplex structure in aqueous [choline][H2PO4] regardless of its concentration, but not in most other salt solutions. They suggested that the G-quadruplex structure is maintained in kosmotropic salt solutions since the DNA hydration state is strongly influenced by the ion kosmotropicity. Lannan et al.69 found that human telomere sequence (HTS) DNA exhibits a G-quadruplex with the parallel-stranded fold in choline chloride/urea (1:2). This is in agreement with the observation that a reduced water activity leads to the parallel fold whilst a high water activity prefers alternative folds. More fascinatingly, after HTS DNA is thermally denatured and then quickly cooled to room temperature, the refolding of DNA back to the parallel structure in the DES requires several months (vs < 2 min in an aqueous solution) (Fig 3).
Fig. 3.
DNA refolding in viscous choline chloride/urea (1:2) (Reprinted with permission from Ref,69 Copyright © 2012 American Chemical Society).
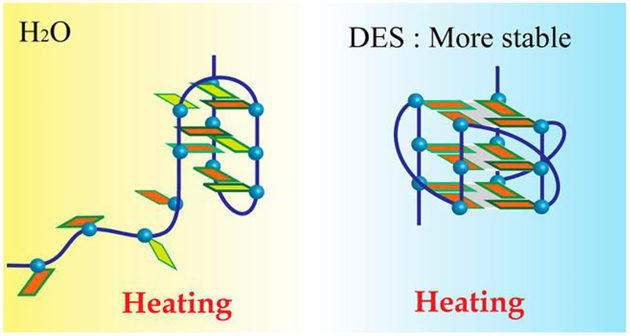
Zhao et al.70 examined the structures of ten G-quadruplexes in neat DES (choline chloride/urea, 1:2 molar ratio) via UV melting, CD and fluorescence spectroscopy, and observed various intramolecular, intermolecular, and higher-order G-quadruplex structures in DES, particularly the parallel structure (Fig 4). G-quadruplexes in neat DES become more stable than in aqueous media; some G-quadruplex DNA molecules are even thermally stable at over 110 °C.
Fig. 4.
Human telomeric DNA, Tel22, adopting a more stable parallel G-quadruplex structure in water-free DES (containing K+) (Reprinted with permission from Ref,70 Copyright © 2013 American Chemical Society).
Summary
It is important to realize that the ion interaction with DNA is not the determining factor of DNA structure, but contribute to the heterogeneity of DNA structure. The major interaction between ionic solvents and DNA is the electrostatic attraction of organic cations and DNA phosphate backbone, followed by the hydrophobic and polar interactions between ILs and DNA major and minor grooves. Anions seem to play a less intense role on the DNA stability by interacting with the bases through hydrogen bonds. The existence of G-quadruplexe DNA structures in ILs and DES could have a profound impact on the future advances of the DNA-based catalysis and other applications.
Acknowledgements
The author acknowledges the supports by the Henry Dreyfus Teacher-Scholar Award (2012), NIH MBRS-RISE grant (1R25GM096956), NIH NIBIB contract award (HHSN268201200011C), and the National Natural Science Foundation of China (21328601).
References
- 1.Breaker RR, and Joyce GF, A DNA enzyme that cleaves RNA. Chem. Biol 1: 223–229 (1994). [DOI] [PubMed] [Google Scholar]
- 2.Sen D, and Geyer CR, DNA enzymes. Curr. Opin. Chem. Biol 2: 680–687 (1998). [DOI] [PubMed] [Google Scholar]
- 3.Höbartner C, and Silverman SK, Recent advances in DNA catalysis. Biopolymers 87: 279–292 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Wang C, Li Y, Jia G, Lu S, Liu Y, and Li C, DNA catalyzed dithioacetalization in water. Acta Chim. Sinica 71: 36–39 (2013). [Google Scholar]
- 5.Roe S, Ritson DJ, Garner T, Searle M, and Moses JE, Tuneable DNA-based asymmetric catalysis using a G-quadruplex supramolecular assembly. Chem. Commun 46: 4309–4311 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Wang C, Jia G, Zhou J, Li Y, Liu Y, Lu S, and Li C, Enantioselective Diels–Alder reactions with G-quadruplex DNA-based catalysts. Angew. Chem. Int. Ed 51: 9352–9355 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Wilking M, and Hennecke U, The influence of G-quadruplex structure on DNA-based asymmetric catalysis using the G-quadruplex-bound cationic porphyrin TMPyP4·Cu. Org. Biomol. Chem 11: 6940–6945 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Wang C, Li Y, Jia G, Liu Y, Lu S, and Li C, Enantioselective Friedel–Crafts reactions in water catalyzed by a human telomeric G-quadruplex DNA metalloenzyme. Chem. Commun 48: 6232–6234 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Boersma AJ, Megens RP, Feringa BL, and Roelfes G, DNA-based asymmetric catalysis. Chem. Soc. Rev 39: 2083–2092 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Park S, and Sugiyama H, DNA as a chiral scaffold for asymmetric synthesis. Molecules 17: 12792–12803 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park S, and Sugiyama H, DNA-based hybrid catalysts for asymmetric organic synthesis. Angew. Chem. Int. Ed 49: 3870–3878 (2010). [DOI] [PubMed] [Google Scholar]
- 12.Coquière D, Feringa BL, and Roelfes G, DNA-based catalytic enantioselective Michael reactions in water. Angew. Chem. Int. Ed 46: 9308–9311 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Roelfes G, Boersma AJ, and Feringa BL, Highly enantioselective DNA-based catalysis. Chem. Commun: 635–637 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Rosati F, and Roelfes G, A ligand structure–activity study of DNA-based catalytic asymmetric hydration and Diels–Alder reactions. ChemCatChem 3: 973–977 (2011). [Google Scholar]
- 15.Boersma AJ, Feringa BL, and Roelfes G, α,β-Unsaturated 2-acyl imidazoles as a practical class of dienophiles for the DNA-based catalytic asymmetric Diels−Alder reaction in water. Org. Lett 9: 3647–3650 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Boersma AJ, Feringa BL, and Roelfes G, Enantioselective Friedel–Crafts reactions in water using a DNA-based catalyst. Angew. Chem. Int. Ed 48: 3346–3348 (2009). [DOI] [PubMed] [Google Scholar]
- 17.Oelerich J, and Roelfes G, DNA-based asymmetric organometallic catalysis in water. Chem. Sci 4: (2013). [Google Scholar]
- 18.Shibata N, Yasui H, Nakamura S, and Toru T, DNA-Mediated enantioselective carbon–fluorine bond formation. Synlett 7: 1153–1157 (2007). [Google Scholar]
- 19.Megens RP, and Roelfes G, Organic co-solvents in aqueous DNA-based asymmetric catalysis. Org. Biomol. Chem 8: 1387–1393 (2010). [DOI] [PubMed] [Google Scholar]
- 20.Bonner G, and Klibanov AM, Structural stability of DNA in nonaqueous solvents. Biotechnol. Bioeng 68: 339–344 (2000). [DOI] [PubMed] [Google Scholar]
- 21.Hammouda B, and Worcester D, The denaturation transition of DNA in mixed solvents. Biophys. J 91: 2237–2242 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wasserscheid P, and Welton T, Ionic Liquids in Synthesis Wiley-VCH, Weinheim: (2008). [Google Scholar]
- 23.van Rantwijk F, and Sheldon RA, Biocatalysis in ionic liquids. Chem. Rev 107: 2757–2785 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Moniruzzaman M, Nakashima K, Kamiya N, and Goto M, Recent advances of enzymatic reactions in ionic liquids. Biochem. Eng. J 48: 295–314 (2010). [Google Scholar]
- 25.Zhao H, Methods for stabilizing and activating enzymes in ionic liquids — A review. J. Chem. Tech. Biotechnol 85: 891–907 (2010). [Google Scholar]
- 26.Abbott AP, Capper G, Davies DL, Rasheed RK, and Tambyrajah V, Novel solvent properties of choline chloride/urea mixtures. Chem. Commun: 70–71 (2003). [DOI] [PubMed] [Google Scholar]
- 27.Abbott AP, Boothby D, Capper G, Davies DL, and Rasheed RK, Deep eutectic solvents formed between choline chloride and carboxylic acids: Versatile alternatives to ionic liquids. J. Am. Chem. Soc 126: 9142–9147 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Abbott AP, Capper G, and Gray S, Design of improved deep eutectic solvents using hole theory. ChemPhysChem 7: 803–806 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Tang S, Baker GA, and Zhao H, Ether- and alcohol-functionalized task-specific ionic liquids: attractive properties and applications. Chem. Soc. Rev 41: 4030–4066 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gorke JT, Srienc F, and Kazlauskas RJ, Hydrolase-catalyzed biotransformations in deep eutectic solvents. Chem. Commun: 1235–1237 (2008). [DOI] [PubMed] [Google Scholar]
- 31.Lindberg D, de la Fuente Revenga M, and Widersten M, Deep eutectic solvents (DESs) are viable cosolvents for enzyme-catalyzed epoxide hydrolysis. J. Biotechnol 147: 169–171 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Attri P, Venkatesu P, Kumar A, and Byrne N, A protic ionic liquid attenuates the deleterious actions of urea on α-chymotrypsin. Phys. Chem. Chem. Phys 13: 17023–17026 (2011). [DOI] [PubMed] [Google Scholar]
- 33.Zhao H, Baker GA, and Holmes S, New eutectic ionic liquids for lipase activation and enzymatic preparation of biodiesel. Org. Biomol. Chem 9: 1908–1916 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao H, Zhang C, and Crittle TD, Choline-based deep eutectic solvents for enzymatic preparation of biodiesel from soybean oil. J. Mol. Catal. B: Enzym 85–86: 243–247 (2013). [Google Scholar]
- 35.Zhao H, Baker GA, and Holmes S, Protease activation in glycerol-based deep eutectic solvents. J. Mol. Catal. B: Enzym 72: 163–167 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao H, and Shen K, DNA-based asymmetric catalysis: Role of ionic solvents and glymes. RSC Adv: submitted (2014). [DOI] [PMC free article] [PubMed]
- 37.Egli M DNA-cation interactions: quo vadis? Chem Biol 9: 277–86 (2002). [DOI] [PubMed] [Google Scholar]
- 38.Hud NV, and Polak M, DNA–cation interactions: the major and minor grooves are flexible ionophores. Curr. Opin. Struct. Bio 11: 293–301 (2001). [DOI] [PubMed] [Google Scholar]
- 39.McFail-Isom L, Sines CC, and Williams LD, DNA structure: cations in charge? Curr. Opin. Struct. Bio 9: 298–304 (1999). [DOI] [PubMed] [Google Scholar]
- 40.Hud NV, Sklenář V, and Feigon J, Localization of ammonium ions in the minor groove of DNA duplexes in solution and the origin of DNA A-tract bending. J. Mol. Biol 286: 651–660 (1999). [DOI] [PubMed] [Google Scholar]
- 41.Hamelberg D, McFail-Isom L, Williams LD, and Wilson WD, Flexible structure of DNA: Ion dependence of minor-groove structure and dynamics. J. Am. Chem. Soc 122: 10513–10520 (2000). [Google Scholar]
- 42.Bloomfield VA DNA condensation by multivalent cations. Biopolymers 44: 269–282 (1997). [DOI] [PubMed] [Google Scholar]
- 43.Rouzina I, and Bloomfield VA, DNA bending by small, mobile multivalent cations. Biophys. J 74: 3152–3164 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eichhorn GL, and Shin YA, Interaction of metal ions with polynucleotides and related compounds. XII. The relative effect of various metal ions on DNA helicity. J. Am. Chem. Soc 90: 7323–7328 (1968). [DOI] [PubMed] [Google Scholar]
- 45.Kasyanenko NA, Zanina AV, Nazarova OV, and Panarin EF, DNA interaction with complex ions in solution. Langmuir 15: 7912–7917 (1999). [Google Scholar]
- 46.Chiu TK, and Dickerson RE, 1 Å crystal structures of B-DNA reveal sequence-specific binding and groove-specific bending of DNA by magnesium and calcium. J. Mol. Bio 301: 915–945 (2000). [DOI] [PubMed] [Google Scholar]
- 47.Zhang Y, Chen X, Lan J, You J, and Chen L, Synthesis and biological applications of imidazolium-based polymerized ionic liquid as a gene delivery vector. Chem. Biol. Drug Des 74: 282–288 (2009). [DOI] [PubMed] [Google Scholar]
- 48.Chen Y, Zhang Y, Ke F, Zhou J, Wang H, and Liang D, Solubility of neutral and charged polymers in ionic liquids studied by laser light scattering. Polymer 52: 481–488 (2011). [Google Scholar]
- 49.Mamajanov I, Engelhart AE, Bean HD, and Hud NV, DNA and RNA in anhydrous media: Duplex, triplex, and G-quadruplex secondary structures in a deep eutectic solvent. Angew. Chem. Int. Ed 49: 6310–6314 (2010). [DOI] [PubMed] [Google Scholar]
- 50.Cardoso L, and Micaelo NM, DNA molecular solvation in neat ionic liquids. ChemPhysChem 12: 275–277 (2011). [DOI] [PubMed] [Google Scholar]
- 51.Mukesh C, Mondal D, Sharma M, and Prasad K, Rapid dissolution of DNA in a novel bio-based ionic liquid with long-term structural and chemical stability: successful recycling of the ionic liquid for reuse in the process. Chem. Commun 49: 6849–6851 (2013). [DOI] [PubMed] [Google Scholar]
- 52.Poater J, Swart M, Guerra CF, and Bickelhaupt FM, Solvent effects on hydrogen bonds in Watson–Crick, mismatched, and modified DNA base pairs. Comput. Theor. Chem 998: 57–63 (2012). [Google Scholar]
- 53.Wang J-H, Cheng D-H, Chen X-W, Du Z, and Fang Z-L, Direct extraction of double-stranded DNA Into ionic liquid 1-butyl-3-methylimidazolium hexafluorophosphate and its quantification. Anal. Chem 79: 620–625 (2007). [DOI] [PubMed] [Google Scholar]
- 54.Cheng D-H, Chen X-W, Wang J-H, and Fang Z-L, An abnormal resonance light scattering arising from ionic-liquid/DNA/ethidium interactions. Chem. Eur. J 13: 4833–4839 (2007). [DOI] [PubMed] [Google Scholar]
- 55.Khimji I, Doan K, Bruggeman K, Huang P-JJ, Vajha P, and Liu J, Extraction of DNA staining dyes from DNA using hydrophobic ionic liquids. Chem. Commun 49: 4537–4539 (2013). [DOI] [PubMed] [Google Scholar]
- 56.Xie Y-N, Wang S-F, Zhang Z-L, and Pang D-W, Interaction between room temperature ionic liquid [bmim]BF4 and DNA investigated by electrochemical micromethod. J. Phys. Chem. B 112: 9864–9868 (2008). [DOI] [PubMed] [Google Scholar]
- 57.Ding Y, Zhang L, Xie J, and Guo R, Binding characteristics and molecular mechanism of interaction between ionic liquid and DNA. J. Phys. Chem. B 114: 2033–2043 (2010). [DOI] [PubMed] [Google Scholar]
- 58.Singh PK, Sujana J, Mora AK, and Nath S, Probing the DNA–ionic liquid interaction using an ultrafast molecular rotor. J. Photochem. Photobiol. A: Chem 246: 16–22 (2012). [Google Scholar]
- 59.He Y, Shang Y, Liu Z, Shao S, Liu H, and Hu Y, Interactions between ionic liquid surfactant [C12mim]Br and DNA in dilute brine. Colloids Surf. B 101: 398–404 (2013). [DOI] [PubMed] [Google Scholar]
- 60.Vijayaraghavan R, Izgorodin A, Ganesh V, Surianarayanan M, and MacFarlane DR, Long-term structural and chemical stability of DNA in hydrated ionic liquids. Angew. Chem. Int. Ed 49: 1631–1633 (2010). [DOI] [PubMed] [Google Scholar]
- 61.Chandran A, Ghoshdastidar D, and Senapati S, Groove binding mechanism of ionic liquids: A key factor in long-term stability of DNA in hydrated ionic liquids? J. Am. Chem. Soc 134: 20330–20339 (2012). [DOI] [PubMed] [Google Scholar]
- 62.Tateishi-Karimata H, and Sugimoto N, A–T base pairs are more stable than G–C base pairs in a hydrated ionic liquid. Angew. Chem. Int. Ed 51: 1416–1419 (2012). [DOI] [PubMed] [Google Scholar]
- 63.Wang H, Wang J, and Zhang S, Binding Gibbs energy of ionic liquids to calf thymus DNA: a fluorescence spectroscopy study. Phys. Chem. Chem. Phys 13: 3906–3910 (2011). [DOI] [PubMed] [Google Scholar]
- 64.Mondal D, Sharma M, Mukesh C, Gupta V, and Prasad K, Improved solubility of DNA in recyclable and reusable bio-based deep eutectic solvents with long-term structural and chemical stability. Chem. Commun 49: 9606–9608 (2013). [DOI] [PubMed] [Google Scholar]
- 65.Parkinson GN, Lee MPH, and Neidle S, Crystal structure of parallel quadruplexes from human telomeric DNA. Nature 417: 876–880 (2002). [DOI] [PubMed] [Google Scholar]
- 66.Huppert JL, Four-stranded nucleic acids: Structure, function and targeting of G-quadruplexes. Chem. Soc. Rev 37: 1375–1384 (2008). [DOI] [PubMed] [Google Scholar]
- 67.Mashimo T, Yagi H, Sannohe Y, Rajendran A, and Sugiyama H, Folding pathways of human telomeric type-1 and type-2 G-quadruplex structures. J. Am. Chem. Soc 132: 14910–14918 (2010). [DOI] [PubMed] [Google Scholar]
- 68.Fujita K, and Ohno H, Stable G-quadruplex structure in a hydrated ion pair: cholinium cation and dihydrogen phosphate anion. Chem. Commun 48: 5751–5753 (2012). [DOI] [PubMed] [Google Scholar]
- 69.Lannan FM, Mamajanov I, and Hud NV, Human telomere sequence DNA in water-free and high-viscosity solvents: G-Quadruplex folding governed by Kramers rate theory. J. Am. Chem. Soc 134: 15324–15330 (2012). [DOI] [PubMed] [Google Scholar]
- 70.Zhao C, Ren J, and Qu X, G-quadruplexes form ultrastable parallel structures in deep eutectic solvent. Langmuir 29: 1183–1191 (2013). [DOI] [PubMed] [Google Scholar]