Abstract
4-cyanoindole-2ʹ-deoxyribonucleoside (4CIN) is a fluorescent isomorphic nucleoside analogue with superior spectroscopic properties in terms of Stokes shift and quantum yield in comparison to the widely utilized isomorphic nucleoside analogue, 2-aminopurine-2ʹ-deoxyribonucleoside (2APN). Notably, when inserted into single- or double-stranded DNA, 4CIN experiences substantially less in-strand fluorescence quenching compared to 2APN. Given the utility of these properties for a spectrum of research applications involving oligonucleotides and oligonucleotide-protein interactions (e.g., enzymatic processes, DNA hybridization, DNA damage), we envision that additional reagents based on 4-cyanoindole-nucleosides may be widely utilized. This protocol expands on the previously published synthesis of 4CIN to include synthetic routes to both 4-cyanoindole-ribonucleoside (4CINr) and 4-cyanoindole-2ʹ-deoxyribonucleoside-5ʹ-triphosphate (4CIN-TP), as well as a method for the enzymatic incorporation of 4CIN-TP into DNA by a polymerase. These methods are anticipated to further enable the utilization of 4CIN in diverse applications involving DNA and RNA oligonucleotides.
BASIC PROTOCOL 1: Synthesis of 4-cyanoindole-2ʹ-deoxyribonucleoside (4CIN) and 4CIN phosphoramidite (4)
BASIC PROTOCOL 2: Synthesis of 4-cyanoindole-ribonucleoside (4CINr)
BASIC PROTOCOL 3: Synthesis of 4-cyanoindole-2ʹ-deoxyribonucleoside-5ʹ-triphosphate (4CIN-TP)
BASIC PROTOCOL 4: Steady State Incorporation Kinetics of 2AP-TP and 4CIN-TP by DNA Polymerases
Keywords: fluorescent nucleosides, 4-cyanoindole, polymerase incorporation
INTRODUCTION
Fluorescent nucleosides are an ever-growing class of molecules with diverse biological applications (Sinkeldam et al., 2010; Xu et al., 2017). By exploiting the unique spectral properties and environmental sensitivities of fluorescent nucleosides, a variety of fundamental studies involving polymerases (Joyce et al., 2008), base-flipping enzymes (Kilin et al., 2017), and processes associated with DNA damage (Wilson et al., 2018) have been achieved. This unit provides detailed synthesis procedures for four different nucleoside and nucleotide molecules based on the fluorophore 4-cyanoindole (4CI), including the recently published 4-cyanoindole-2ʹ-deoxyribonucleoside (4CIN) and its phosphoramidite (Passow & Harki, 2018), as well as new analogues 4-cyanoindole-ribonucleoside (4CINr) and the 5ʹ-triphosphate of 4CIN, 4CIN-TP. The unit ends with assay protocols to evaluate the enzymatic incorporation of 4CIN-TP into DNA using DNA polymerases.
Basic Protocol 1 provides additional experimental insight beyond the published synthesis of 4CIN and its phosphoramidite 4, which can be incorporated into DNA using phosphoramidite chemistry. Basic Protocol 2 details the synthesis of the ribonucleoside analogue, 4CINr, which depends on a TDA-1 and KOH mediated bond formation between a chlorinated ribose and the indole base. Basic Protocol 3 describes the synthesis of 4CIN-TP starting from 4CIN that proceeds through a phosphonate precursor (12), which delivers the final compound in approximately 2% overall yield. 4CIN-TP is used in turn in Basic Protocol 4 alongside commercially available 2-aminopurine-2ʹ-deoxyribonucleoside-5ʹ-triphosphate (2AP-TP) to establish general kinetics of incorporation by a DNA polymerase.
BASIC PROTOCOL 1
Synthesis of 4-cyanoindole-2ʹ-deoxyribonucleoside (4CIN) and 4CIN phosphoramidite (4)
Basic Protocol 1 reiterates the published synthesis of 4CIN and its corresponding phosphoramidite (4) for use in automated DNA synthesis (Passow & Harki, 2018). The synthesis route is shown in Figure 1 and starts from commercially available 4-cyanoindole (4CI) and couples this nucleobase to commercial 3,5-di-O-toluoyl-α−1-chloro-2-deoxy-D-ribofuranose (1) to yield 2. Base-mediated deprotection of the toluoyl groups yields 4CIN and subsequent 5ʹ-O-DMT protection (3) and 3ʹ-O-phosphoramidite coupling yields 4.
Figure 1.
Synthesis of 4CIN and phosphoramidite 4.
Materials
Sodium Hydride (NaH) (Sigma Aldrich cat. no. 452912–100G, 60% dispersion in mineral oil)
4-cyanoindole (4CI) (Sigma Aldrich cat. no. 645532–5G)
3,5-di-O-toluoyl-α−1-chloro-2-deoxy-D-ribofuranose (1) (Carbosynth cat. no. MC02788)
Acetonitrile (MeCN) (Fisher Chemical cat. no. A998–4, HPLC grade, Anhydrous)
Anhydrous nitrogen (N2) gas
Dichloromethane (DCM) (Fisher Chemical cat. no. D37–20, ACS grade)
Celite (Sigma Aldrich cat. No. 419931)
Hexanes (Fisher Chemical cat. no. H292–20, ACS grade)
Ethyl acetate (EtOAc) (J.T. Baker cat. no. 9290–07, ACS grade)
Silica gel (40 – 64 μm, Silicycle cat. no. R12030B or Teledyne ISCO column sizes 4g, 40g cat. no. 69–2203-304 and 69–2203-340, respectively)
Deuterated chloroform (CDCl3) (Cambridge Isotopes, cat. no. DLM-7–100 or DLM-7TB-100)
Sodium Hydroxide (NaOH) (Sigma Aldrich cat. no. 221465–1KG, >97% pellets)
Methanol (MeOH) (Fisher Chemical cat. no. A412–4, ACS grade)
Deuterated dimethylsulfoxide (DMSO-d6) (Cambridge Isotopes, cat. no. DLM-10–10 or DLM-10TB-10)
Pyridine (Sigma Aldrich cat. no. 270970–1L, anhydrous, 99.8%)
4,4ʹ-dimethoxytrityl chloride (DMTrCl) (Alfa Aesar cat. no. A11626, 98%)
Sodium sulfate (Na2SO4) (Beantown Chemical cat. no. 136810–10KG)
Diisopropylethylamine (DIPEA) (Sigma Aldrich cat. no. D125806–500ML, reagent grade)
Calcium hydride (CaH2) (Sigma Aldrich cat. no. 208027–500G, reagent grade)
Triethylamine (TEA) (Fisher Chemical cat. no. 04885–1, reagent grade)
2-cyanoethyl N,N’-diisopropylchlorophosphoramidite (Sigma Aldrich, cat. no. 302309)
Cyclohexane (Fisher Chemical cat. no. C556–4, ACS grade)
10-, 25-, 250-, and 500-mL round-bottom flasks
250-and 500- mL beaker
Drying oven or propane torch
Rubber septa
Magnetic stir bars
Magnetic stir plate/oil bath heater
High vacuum pump
1-, 3-, and 5- mL plastic syringe (luer lock desirable)
18-, 20-, 21-G needles (0.8 × 40 mm)
10-, 25-, 50-, 100-, and 500- mL graduated cylinder
Erlenmeyer flasks (100 mL, 250 mL, and 500 mL)
60 mL Fritted funnel
Buchner funnel
500 mL vacuum flask
Filter paper
Rotary evaporator
Short path distillation apparatus
4 Å molecular sieves (dried in an oven at 135°C for at least 24 hours)
UV lamp, 254 nm
Thin-layer chromatography (TLC) plate (EMD Chemicals Silica gel 60 F254, 250 μm thickness)
TLC Chamber
Combiflash Rf (Teledyne ISCO or comparable normal phase chromatography system) or standard flash column chromatography setup
NMR (e.g. Brucker Advance 500 MHz)
Protocol Steps
Synthesis of 3′,5′-di-O-toluoyl-4-cyanoindole-2′-deoxyribonucleoside (2)
-
1
To a flame-dried 500 mL round bottom flask equipped with a magnetic stir bar, add 4-cyanoindole (4CI, 1.0 g, 7.0 mmol, 1.0 equiv).
-
2To this flask add anhydrous MeCN (150 mL).Unless otherwise stated, all anhydrous MeCN, DCM, and THF used in these protocols were made anhydrous by running purchased solvents over a column of activated alumina or molecular sieves (for anhydrous DMF only) using a solvent purification system, e.g. the MBraun Solvent Purification System (SPS) line of products. Other anhydrous solvents, such as pyridine, were purchased in sealed anhydrous bottles. All other solvents were used as purchased.
-
3
Following dissolution of 4CI, add NaH (60% mineral oil dispersion, 0.34 g, 8.4 mmol, 1.2 equiv) portion-wise to the solution over 1 minute. Stir this mixture for 30 minutes at room temperature.
-
4
After the 30 minute reaction, add 3,5-di-O-toluoyl-α−1-chloro-2-deoxy-D-ribofuranose (1, 3.3 g, 8.4 mmol, 1.2 equiv) in small portions over 5 minutes. Place the mixture under a N2 atmosphere. Stir this mixture for 18 hours.
-
5
Dilute the mixture with DCM (200 mL). Filter the mixture through a pad of celite on a fritted funnel to remove any insoluble materials.
-
6
Evaporate this mixture in vacuo using a rotary evaporator onto approximately 2 g of SiO2.
-
7Purify by SiO2 chromatography using a 40 g silica gel cartridge (Teledyne ISCO) and run at 40 mL/min flowrate while monitoring at 254 / 300 nm using a gradient of EtOAc (0 to 60%) in hexanes collecting approximately 40 × 18 mL fractions.This step and all subsequent silica gel chromatography steps in this paper can be completed using whatever means available to the user including other automated systems or standard flash chromatography.
-
8Collect fractions containing the desired product and evaporate in vacuo using a rotary evaporator to yield 3′,5′-di-O-toluoyl-4-cyanoindole-2′-deoxyribonucleoside (2) as a pale yellow foam (2.5 g, 5.1 mmol, 72% yield).Given the fluorescent nature of the molecule, all 4-cyanoindole containing molecules in this protocol stand out on TLC plates with a bright-blue appearance under a 254 nm lamp. This will easily distinguish 4CI-related materials found in this protocol from other common aromatic compounds that appear purple.
-
9
Characterize the product by TLC (on silica gel), 1H NMR, 13C NMR, and HR-MS.
3′,5′-di-O-toluoyl-4-cyanoindole-2′-deoxyribonucleoside (2)
Rf = 0.30 (4/1, v/v hexanes/EtOAc)
Chemical shifts referenced to TMS (0.05% v/v; δ = 0.0 ppm) for 1H NMR and solvent signal for 13C (δ = 77.0 ppm for CDCl3).
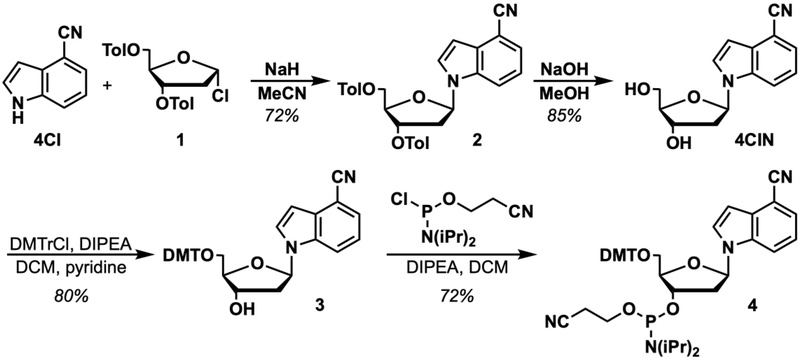
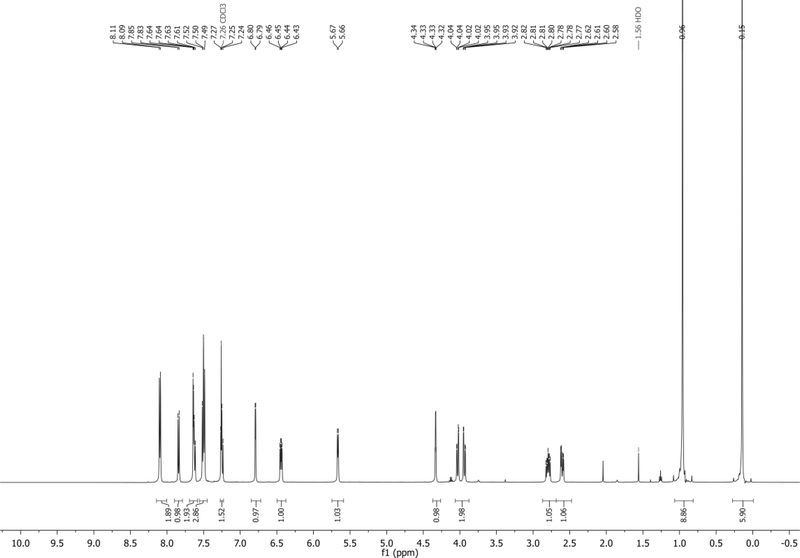
1H NMR (500 MHz, CDCl3): δ 7.98 (d, J = 8.0 Hz, 2H), 7.89 (d, J = 8.0 Hz, 2H), 7.75 (d, J = 8.4 Hz, 1H), 7.48 – 7.42 (m, 2H), 7.29 (d, J = 7.9 Hz, 2H), 7.24 (d, J = 8.0 Hz, 2H), 7.15 (app t, J = 7.9 Hz, 1H), 6.72 (d, J = 3.3 Hz, 1H), 6.45 (dd, J = 8.3, 5.6 Hz, 1H), 5.77 – 5.70 (m, 1H), 4.69 (dd, J = 12.0, 3.6 Hz, 1H), 4.62 (dd, J = 12.0, 3.7 Hz, 1H), 4.60 – 4.56 (m, 1H), 2.91 – 2.83 (m, 1H), 2.70 (ddd, J = 14.3, 5.8, 2.1 Hz, 1H), 2.44 (s, 3H), 2.43 (s, 3H) ppm. Spectrum shown in Figure 2.
13C NMR (126 MHz, CDCl3): δ 166.1, 166.0, 144.6, 144.2, 135.5, 130.6, 129.8, 129.6, 129.3, 129.3, 126.7, 126.7, 126.43, 125.7, 121.8, 118.4, 114.9, 103.5, 102.4, 85.7, 82.1, 74.7, 63.9, 38.0, 21.74, 21.70 ppm.
HRMS-ESI+ (m/z) calc’d [M+Na]+ for C30H26N2O5Na: 517.1739, found: 517.1725.
Figure 2.
1H NMR of 2 in CDCl3 with tetramethylsilane (TMS).
Synthesis of 4-cyanoindole-2′-deoxyribonucleoside (4CIN)
-
10
To a flame-dried 250 mL round bottom flask equipped with a magnetic stir bar, add 2 (2.3 g, 4.6 mmol, 1.0 equiv).
-
11
Prepare 100 mL of 1% wt/v solution of NaOH in MeOH by dissolving NaOH (1.0 g, 25 mmol) in 100 mL of MeOH.
-
12Add this NaOH solution to the 250 mL round bottom flask and stir the mixture for 30 minutes.At first the material will not be soluble in methanol and will slowly dissolve over 30 minutes.
-
13
Evaporate the resulting solution in vacuo using a rotary evaporator onto approximately 3 g of silica gel.
-
14
Purify by chromatography on silica gel using a 40 g silica gel cartridge (Teledyne ISCO) and run at 40 mL/min flowrate while monitoring at 254 / 300 nm using a gradient of MeOH (0 to 10%) in DCM collecting approximately 40 × 18 mL fractions.
-
15
Collect fractions containing the desired product and evaporate in vacuo using a rotary evaporator to yield 4-cyanoindole-2′-deoxyribonucleoside (4CIN) as a glassy colorless solid (1.0 g, 3.9 mmol, 85% yield).
-
16
Characterize the product by TLC (on silica gel), 1H NMR, 13C NMR, and HR-MS.
4-cyanoindole-2′-deoxyribonucleoside (4CIN)
Rf = 0.52 (9/1, v/v DCM/MeOH)
Chemical shifts referenced to TMS (0.05% v/v; δ = 0.0 ppm) for 1H NMR and solvent signal for 13C (δ = 39.7 ppm for DMSO-d6).
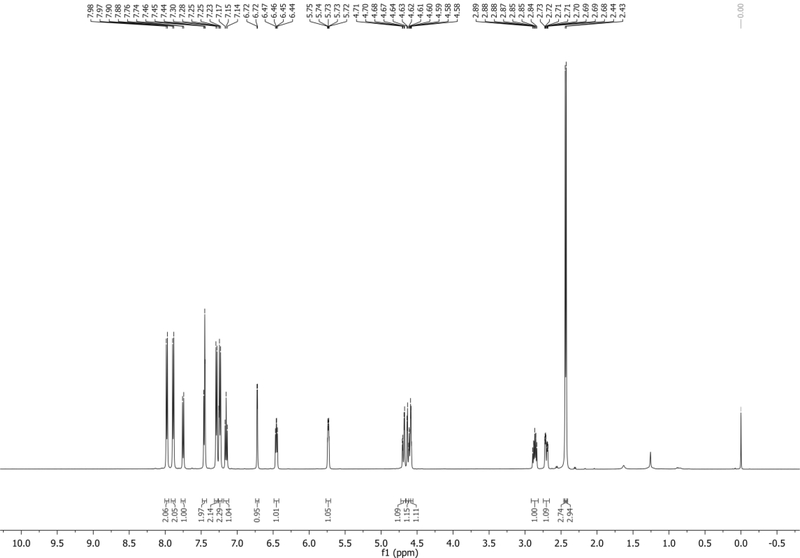
1H NMR (500 MHz, DMSO-d6): δ 8.02 (d, J = 8.4 Hz, 1H), 7.91 (d, J = 3.3 Hz, 1H), 7.59 (d, J = 7.3 Hz, 1H), 7.30 (app t, J = 7.9 Hz, 1H), 6.67 (m, 1H), 6.46 (app t, J = 6.9 Hz, 1H), 5.31 (d, J = 3.9 Hz, 1H), 4.93 (app t, J = 5.4 Hz, 1H), 4.40 – 4.34 (m, 1H), 3.87 – 3.81 (m, 1H), 3.60 – 3.47 (m, 2H), ~2.5 (obstructed by DMSO, 1H; Visible in D2O), 2.31 – 2.23 (m, 1H) ppm. Spectrum shown in Figure 3.
13C NMR (126 MHz, DMSO-d6): δ 135.2, 129.3, 128.9, 125.1, 121.4, 118.3, 115.8, 101.5, 100.3, 87.2, 84.7, 70.5, 61.6, 40.3 (CH2 obstructed by DMSO, visible in DEPT135) ppm.
HRMS-ESI+ (m/z) calc’d [M + Na]+ for C14H14N2O3Na: 281.0902, found: 281.0897.
Figure 3.
1H NMR of 4CIN in DMSO-d6 with TMS.
Synthesis of 5′-O-dimethoxytrityl-4-cyanoindole-2′-deoxyribonucleoside (3)
-
17
Prepare anhydrous diisopropylethylamine (DIPEA) by short path distillation over CaH2 (Armarego & Chai, 2009). After distilling, add oven-dried 4 Å molecular sieves and store the resulting anhydrous DIPEA under Ar gas. Store until needed at room temperature (up to a week wrapped in aluminum foil).
-
18
To a flame-dried 25 mL round bottom flask equipped with a magnetic stir bar, add 4CIN (85.0 mg, 0.329 mmol, 1.0 equiv).
-
19
To this flask add anhydrous pyridine (2 mL). Evaporate the mixture in vacuo using a rotary evaporator. Repeat this step twice more.
-
20
To the flask add an anhydrous mixture of pyridine:DCM (1:1, 9 mL).
-
21
Add freshly distilled DIPEA (86.0 μL, 0.494 mmol, 1.5 equiv) and initiate stirring.
-
22
To this mixture add 4,4ʹ-dimethoxytrityl chloride (DMTrCl, 197 mg, 0.581 mmol, 1.75 equiv) slowly in small portions to the mixture over 1 minute. Place the reaction mixture under a N2 atmosphere and stir the reaction for 3 hours.
-
23
Quench the reaction by adding MeOH (1.0 mL) and then concentrate the mixture in vacuo using a rotary evaporator onto approximately 200 mg of silica gel.
-
24
Purify by chromatography on silica gel using a 4g silica gel cartridge (Teledyne ISCO) and run at 18 mL/min flowrate while monitoring at 254 nm / 300 nm using a gradient of EtOAc (0 to 70%) in hexanes collecting approximately 30 × 8 mL fractions.
-
25
Collect fractions containing the desired product and evaporate in vacuo using a rotary evaporator to yield 5′-O-dimethoxytrityl-4-cyanoindole-2′-deoxyribonucleoside (3) as a yellow foam (148 mg, 0.264 mmol, 80% yield).
-
26
Characterize the product by TLC (on silica gel), 1H NMR, 13C NMR, and HR-MS.
5′-O-dimethoxytrityl-4-cyanoindole-2′-deoxyribonucleoside (3)
Rf = 0.31 (1/1, v/v hexanes/EtOAc)
Chemical shifts referenced to TMS (0.05% v/v; δ = 0.0 ppm) for 1H NMR and solvent signal for 13C (δ = 77.0 ppm for CDCl3).
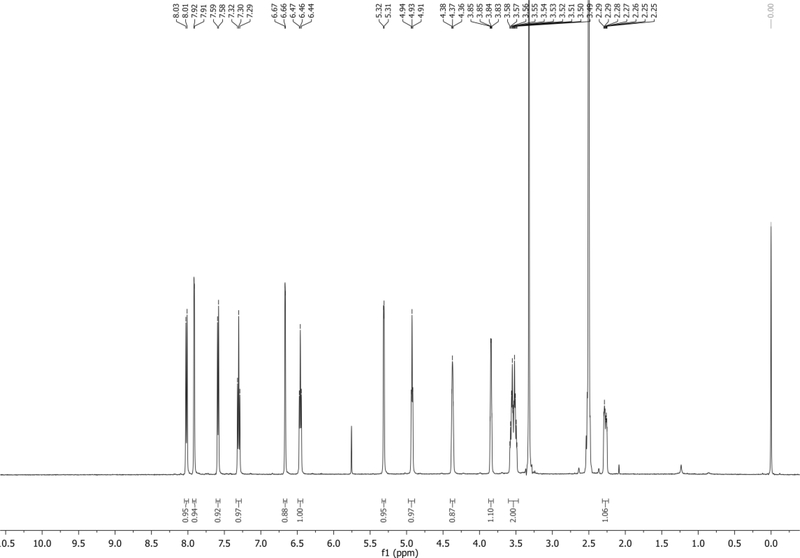
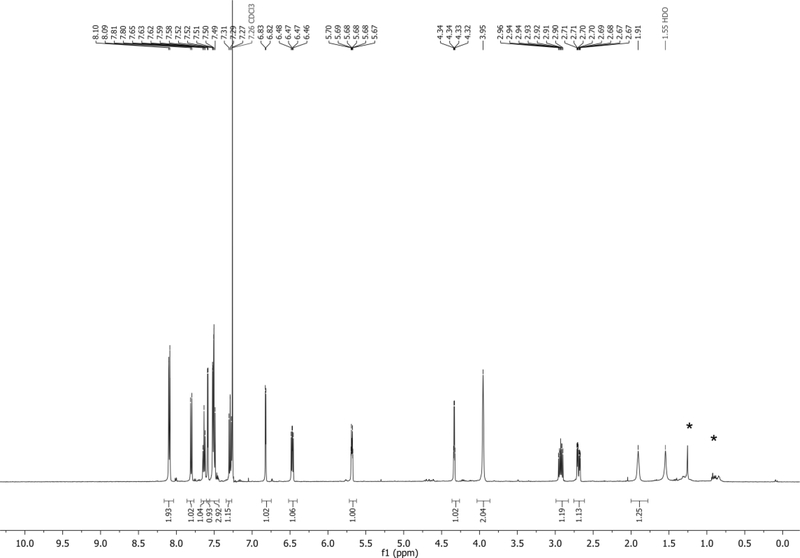
1H NMR (500 MHz,CDCl3): δ 7.74 (d, J = 8.4 Hz, 1H), 7.45 (d, J = 7.3 Hz, 1H), 7.42 – 7.36 (m, 3H), 7.35 – 7.18 (m, 8H), 7.14 (app t, J = 7.9 Hz, 1H), 6.88 – 6.74 (m, 4H), 6.72 – 6.66 (m, 1H), 6.38 (app t, J = 6.7 Hz, 1H), 4.66 – 4.59 (m, 1H), 4.14 – 4.06 (m, 1H), 3.76 (s, 6H), 3.36 (dd, J = 10.3, 4.1 Hz, 1H), 3.32 (dd, J = 10.2, 4.6 Hz, 1H), 2.67 – 2.57 (m, 1H), 2.46 – 2.38 (m, 1H) ppm. Spectrum shown in Figure 4.
13C NMR (126 MHz,CDCl3): δ 158.6, 144.5, 135.62, 135.58, 135.53, 130.4, 130.1, 128.1, 127.9, 127.1, 127.0, 125.5, 121.6, 118.6, 115.1, 113.2, 103.1, 101.8, 86.6, 85.5, 85.2, 72.5, 63.8, 55.2, 40.3 ppm.
HRMS-ESI+ (m/z) calc’d [M + Na]+ for C35H32N2O5Na: 583.2209, found: 583.2195.
Figure 4.
1H NMR of 3 in CDCl3 with TMS.
Synthesis of 5′-O-dimethoxytrityl-4-cyanoindole-2′-deoxyribonucleoside phosphoramidite (4)
-
27
To a flame-dried 10 mL round bottom flask equipped with a magnetic stir bar, add 3 (148 mg, 0.264 mmol, 1.0 equiv). Dry this flask under high vacuum for 24 hours.
-
28
Preparing triethylamine (TEA)-deactivated silica gel: In a 500 mL beaker containing a large magnetic stir bar, add 200 g silica gel. To this beaker add 200 mL of a 1% v/v triethylamine in hexanes solution. Stir the resulting slurry for 1 hour. Pour the suspension into a prepared Büchner funnel equipped with filter paper and placed on a vacuum flask setup to collect the silica gel by vacuum filtration. Allow the silica gel to dry in the funnel before collecting in a clean beaker. This deactivated silica gel can be stored and used as needed.
-
29
Backfill the prepared flask with N2 and add anhydrous DCM (3 mL).
-
30
To this flask add freshly distilled DIPEA (0.138 mL, 0.793 mmol, 3.0 equiv).
-
31To the flask add 2-cyanoethyl N,Nʹ-diisopropylchlorophosphoramidite (88.3 μL, 0.396 mmol, 1.5 equiv) dropwise to the mixture. Stir the mixture under N2 for one hour.To ensure reaction success, proper anhydrous technique must be observed. The chlorophosphoramidite reagent is shipped in a screw-top vial (no septa) and should ideally be used once and then disposed. We suggest buying the reagent in small aliquots.
-
32Directly evaporate the mixture onto 300 mg of pre-prepared TEA-deactivated silica gel.The phosphoramidite product is sensitive to acid, therefore trimethylamine-treated silica gel is used to temper silica acidity to prevent product degradation.
-
33
Prior to chromatography, wash a 4 g silica gel cartridge with a 1% v/v solution of TEA in cyclohexane (3 column volumes) and then elute excess TEA with cyclohexane. Purify by chromatography on silica gel using this deactivated 4 g silica gel cartridge (Teledyne ISCO) and run at 18 mL/min flowrate while monitoring at 254 nm / 300 nm using a gradient of EtOAc (0 to 30%) in cyclohexane collecting approximately 30 × 8 mL fractions.
-
34Collect fractions containing the desired product and evaporate in vacuo using a rotary evaporator to yield 5′-O-dimethoxytrityl protected 4CIN phosphoramidite (4) as a white foam (144 mg, 0.189 mmol, 72% yield).Molecule 4 can be stored under inert gas for >1 year at 0 °C without any observed degradation.
-
35
Characterize the product by TLC (on silica gel), 1H NMR, 13C NMR, and HR-MS.
5′-O-dimethoxytrityl-4-cyanoindole-2′-deoxyribonucleoside phosphoramidite (4)
Rf = 0.30 (7/3, v/v hexanes/EtOAc)
Chemical shifts referenced to TMS (0.05% v/v; δ = 0.0 ppm) for 1H NMR and solvent signal for 13C (δ = 77.0 ppm for CDCl3).
1H NMR (500 MHz, CDCl3): δ 7.82 (d, J = 3.3 Hz, resolved diastereomer, 0.5H), 7.80 (d, J = 3.3 Hz, resolved diastereomer, 0.5H), 7.47 (d, J = 7.4 Hz, 1H), 7.44 (app t, J = 3.8 Hz, 1H), 7.39 (app t, J = 6.6 Hz, 2H), 7.31 – 7.18 (m, 7H), 7.14 (app t, J = 7.7 Hz, 1H), 6.81 – 6.73 (m, 4H), 6.70 (app t, J = 4.0 Hz, 1H), 6.42 – 6.34 (m, 1H), 4.78 – 4.69 (m, 1H), 4.29 – 4.21 (m, 1H), 3.95 – 3.81 (m, 1H), 3.80 – 3.75 (m, 6H), 3.74 – 3.65 (m, 1H), 3.65 – 3.54 (m, 2H), 3.42 – 3.24 (m, 2H), 2.79 – 2.43 (m, 4H), 1.21 – 1.16 (m, 9H), 1.11 (d, J = 6.8 Hz, 3H) ppm. Spectrum shown in Figure 5.
13C NMR (126 MHz, CDCl3): δ 158.54, 158.51, 144.51 (br), 135.67, 135.61, 135.59, 135.57, 135.56, 135.53, 130.51, 130.49, 130.15, 130.13, 130.09, 128.24, 128.18, 127.82, 127.80, 127.25, 127.21, 126.91, 126.87, 125.47, 125.46, 121.56, 121.53, 118.61, 118.58, 117.55, 117.43, 115.35, 115.26, 113.09 (br), 103.24, 103.18, 101.75, 101.69, 86.48, 86.46, 85.56, 85.28, 85.25, 85.02, 84.97, 74.0 (JC-P = 17.6 Hz, one diastereomer), 73.5 (JC-P = 17.6 Hz, one diastereomer) 63.45, 63.34, 58.52, 58.35 (JC-P = 12.6 Hz, one diastereomer), 58.19 (JC-P = 12.6 Hz, one diastereomer), 58.37, 55.21, 55.19, 43.30 (JC-P = 5.0 Hz, one diastereomer), 43.20 (JC-P = 5.0 Hz, one diastereomer), 39.70, 39.67, 26.89, 24.6–24.5 (complex), 24.49, 20.42, 20.41 (JC-P = 7.6 Hz, one diastereomer) 20.37, 20.23 (JC-P = 7.6 Hz, one diastereomer) ppm.
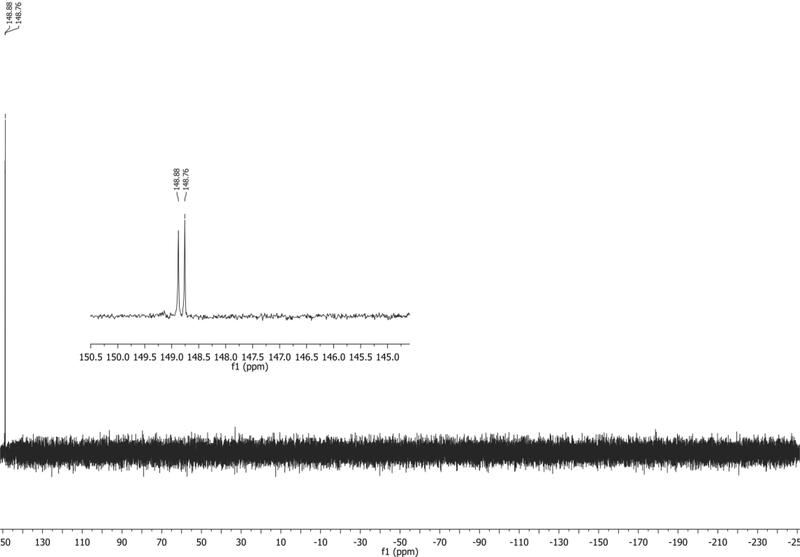
31P NMR (202 MHz, CDCl3): δ 148.88, 148.76 ppm. Spectrum shown in Figure 6.
HRMS-ESI+ (m/z) calc’d [M + Na]+ for C44H49N4O6PNa: 783.3287, found:783.3273.
Figure 5.
1H NMR of 4 in CDCl3 with TMS.
Figure 6.
31P NMR of 4 in CDCl3 with TMS.
BASIC PROTOCOL 2
Synthesis of 4-cyanoindole-ribonucleoside (4CINr)
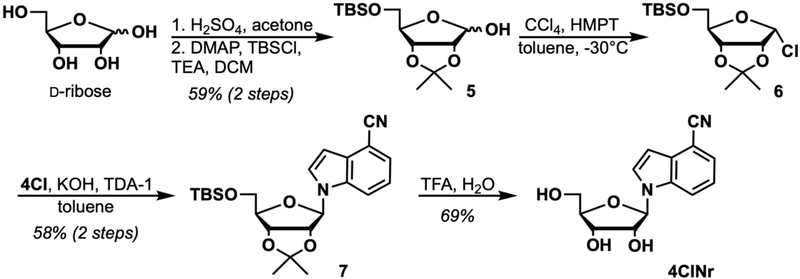
Basic Protocol 2 describes the synthesis of 4CINr starting from D-ribose and 4-cyanoindole (4CI). The basic step outline shown in Figure 7 involves 5 synthetic steps, first protecting the 2- and 3- alcohols to form an acetonide and a subsequent 5-alcohol protection with TBS yielding 5. Next, 5 is α-chlorinated using the method of Wilcox and Otoski (1986) yielding 6, which is then immediately coupled with 4CI to afford protected ribonucleoside 7. Global deprotection with TFA yields the final compound, 4CINr.
Figure 7.
Synthesis of 4CINr.
Materials
Acetone (Fisher Chemical cat. no. A18P-4, ACS grade)
Sulfuric Acid (H2SO4) (Fisher Chemical cat. no. A300–212, ACS plus)
D-ribose (SAFC cat. No. W379301, >98%)
Triethylamine (TEA) (Fisher Chemical cat. no. 04885–1, reagent grade)
Celite (Sigma Aldrich cat. No. 419931)
Dichloromethane (DCM) (Fisher Chemical cat. no. D37–20, ACS grade)
Deuterated chloroform (CDCl3) (Cambridge Isotopes, cat. no. DLM-7–100 or DLM-7TB-100)
N,N-dimethylaminopyridine (DMAP) (Oakwood Chemical cat. no. 001704)
Tert-butyldimethylsilyl chloride (TBSCl) (Oakwood Chemical cat. no. 003869)
Anhydrous nitrogen (N2) gas
Anhydrous Argon (Ar) gas
Dry ice
Brine (saturated aqueous sodium chloride solution, NaCl)
Sodium sulfate (Na2SO4) (Beantown Chemical cat. no. 136810–10KG)
Magnesium sulfate (MgSO4) (Acros cat. no. 41348–5000, 97%)
Silica gel (40 – 64 μm, Silicycle or Teledyne ISCO column sizes 24 g, 40 g)
Ammonium molybdate (Alfa Aesar cat. no. A13766, 99%)
Ceric ammonium molybdate (CAM) (Sigma Aldrich cat. no. 431346, ACS, 99.98%)
Hexanes (Fisher Chemical cat. no. H292–20, ACS grade)
Ethyl acetate (EtOAc) (J.T. Baker cat. No. 9290–07, ACS grade)
Toluene (Fisher Chemical cat. no. T290–4, ACS grade)
Hexamethylphosphorous triamide (HMPT) (Sigma Aldrich cat. no. 393290–25ML, 97%)
Carbon tetrachloride (Sigma Aldrich, cat. no. 270652, anhydrous, 99%)
4-cyanoindole (4CI) (Sigma Aldrich cat. no. 645532–5G)
Potassium Hydroxide (KOH) (Fisher Chemical cat. no. P250–3, ACS grade)
Tris[2-(2-methoxyethoxy)ethyl]amine (TDA-1) (TCI cat. no. T1231, >90%)
Ammonium chloride (saturated aqueous solution, NH4Cl) (Chem. Impex. Int. cat. no. 01207, ACS grade)
Trifluoroacetic acid (TFA) (Acros cat. no. 139720010, 99%)
Methanol (MeOH) (Fisher Chemical cat. no. A412–4, ACS grade)
Deuterated Methanol (MeOD) (Cambridge Isotopes cat. no. DLM-24–10)
500 mL jar (for TLC stain)
25-, 50-, and 500- mL round-bottom flasks
Drying oven or propane torch
Heat gun
Rubber septa
Magnetic stir bars
Magnetic stir plate/oil bath heater
High vacuum pump
1-, 3-, and 5- mL plastic syringe (luer lock desirable)
18-, 20-, 21-G needles (0.8 × 40 mm)
60 and 2000 mL separatory funnel
25-, 50-, 100-, and 500- mL graduated cylinder
Low temperature thermometer
Filter paper
Buchner funnel
60 mL Fritted funnel
Rotary evaporator
Cannula
Mortar and pestle
Cold water bath
Vacuum dewar (for dry ice bath)
Short path distillation apparatus
Thermometer
Polycarbonate splash safety shield
Sealed tube reaction vessel (ChemGlass Part no. CG-1880–02, 35 mL) and accompanying seal and o-ring
UV lamp, 254 nm
Thin-layer chromatography (TLC) plate (EMD Chemicals Silica gel 60 F254, 250 μm thickness)
TLC Chamber
Combiflash Rf (Teledyne ISCO, or comparable normal phase chromatography system, or standard flash chromatography setup
NMR (e.g. Brucker Advance 500 MHz)
Protocol steps
Synthesis of 2,3-O-isopropylidene-5-O-TBS-D-ribose (5). Original procedure adapted from Stewart and Williams (1985) and Kane and Mann (1984).
-
1
Prepare cerium ammonium molybdate (CAM) stain by mixing ammonium molybdate (12 g), ceric ammonium molybdate (0.5 g), and sulfuric acid (25 mL) to distilled water (235 mL).
-
2
To a flame-dried 500 mL round bottom flask equipped with a magnetic stir bar, add D-ribose (15.0 g, 99.9 mmol, 1.0 equiv).
-
3
Add acetone (200 mL) to the flask and initiate stirring to suspend the solid D-ribose.
-
4Add H2SO4 (426 μL, 7.99 mmol, 0.08 equiv), place the flask under a N2 atmosphere, and stir rapidly for 3 hours.The solution will slowly become translucent as the product forms and the mixture solubilizes.
-
5
After 3 hours, add TEA (approximately 1 mL) to neutralize the acid and then vacuum filter the mixture through a pad of celite on a fritted funnel into a vacuum flask. Rinse the celite pad with 50 mL of DCM.
-
6
Evaporate the crude mixture and then resuspend in anhydrous DCM (240 mL) along with a clean magnetic stir bar.
-
7
To this new mixture add TEA (27.9 mL, 199 mmol, 2.0 equiv) and DMAP (1.83 g, 14.9 mmol, 0.15 equiv).
-
8Lastly, add TBSCl (18.1 g, 120. mmol, 1.2 equiv) portion-wise to the rapidly stirring mixture and stir for 24 hours.As the reaction proceeds, the mixture changes from a yellow solution to a cloudy white suspension.
-
9
Dilute the mixture with DCM (200 mL) and quench with H2O (300 mL) in a 2L separatory funnel. Wash the organic layer further with 300 mL of brine.
-
10
Dry the organic layer over Na2SO4, filter, and concentrate onto approximately 30 g of silica gel in vacuo using a rotary evaporator.
-
11
Purify by flash column chromatography on silica gel using a fritted glass column using a gradient of EtOAc (0 to 5%) in hexanes collecting approximately 20 × 100 mL fractions.
-
12
Monitor TLC fractions for product by staining using prepared CAM stain from Step 1. Dip the TLC plate into the stain and then heat the plate with a heat gun or hot plate to develop. Stained molecules will appear blue on the plate.
-
13Collect fractions containing the desired product and evaporate in vacuo using a rotary evaporator to yield 2,3-O-isopropylidene-5-O-TBS-D-ribose (5) as a mixture of diastereomers (18.1 g, 59.3 mmol, 59% yield).Depending on product purity and dryness, the product may appear as a colorless liquid or white solid. Cooling the liquid-form product can facilitate solidification. Otherwise, the liquid product can still be used without issue if the purity is sufficient by NMR.
-
14
Characterize the product by TLC (on silica gel). 1H NMR matches literature known spectra (Kane & Mann, 1984).
2,3-O-isopropylidene-5-O-TBS-D-ribose (5)
Rf = 0.17 (9/1, v/v hexanes/EtOAc)
Chemical shifts referenced to solvent (δ = 7.26 ppm for CDCl3) for 1H NMR.
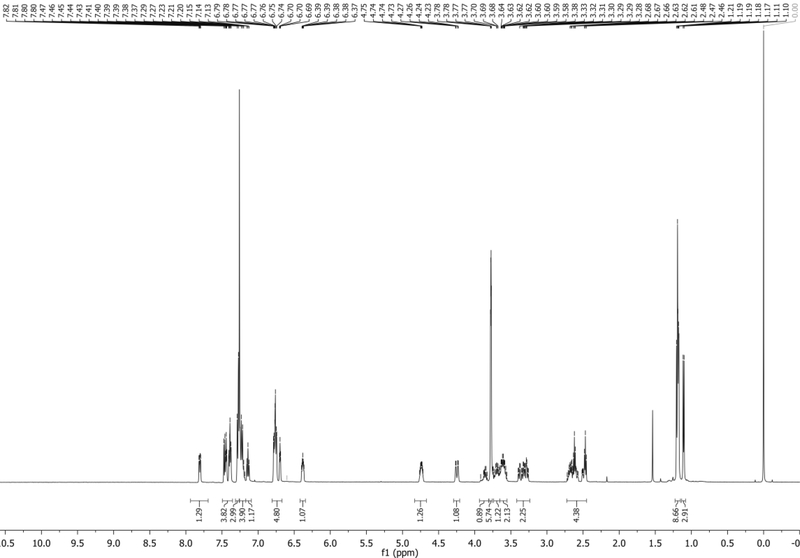
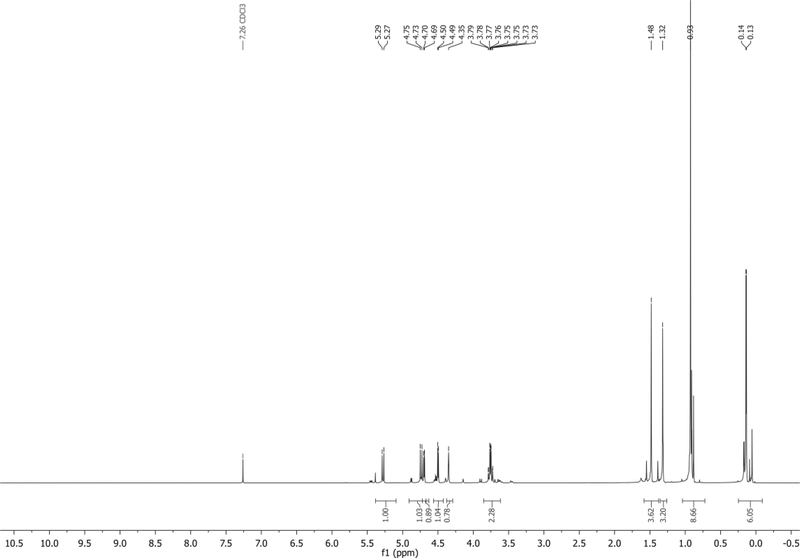
The NMR of the major β-anomer is reported here: 1H NMR (500 MHz, CDCl3): δ 5.28 (d, J = 11.8 Hz, 1H), 4.74 (d, J = 11.9 Hz, 1H), 4.70 (d, J = 5.7 Hz, 1H), 4.50 (d, J = 6.0 Hz, 1H), 4.35 (s, 1H), 3.85 – 3.62 (m, 2H), 1.48 (s, 3H), 1.32 (s, 3H), 0.93 (s, 9H), 0.14 (d, J = 3.4 Hz, 6H) ppm. Spectrum shown in Figure 8.
Figure 8.
1H NMR of 5 in CDCl3.
Synthesis of 2,3-O-isopropylidene-5-O-TBS-1-α-Cl-D-ribose (6) and 4-cyanoindole-2ʹ,3ʹ-O-isopropylidene-5ʹ-O-TBS-ribonucleoside (7). Original chlorination procedure by Wilcox and Otoski (1986) and Rosemeyer and Seela (1988). Our procedure is adapted from Ugarkar et al. (2000).
-
15
Place a solution of brine in a refrigerator or freezer to cool.
-
16
Prepare Mixture A: To a flame-dried 25 mL round bottom flask equipped with a magnetic stir bar, add 5 (1.87 g, 6.14 mmol, 2.0 equiv), anhydrous toluene (12.5 mL), and CCl4 (889 μL, 9.21 mmol, 3.0 equiv) and place under a N2 atmosphere.
-
17
Cool the mixture to −30°C by making an acetone and dry ice bath in a dewar, monitoring the temperature using a low temperature thermometer.
-
18
Add HMPT (1.45 mL, 7.98 mmol, 2.6 equiv) dropwise to the mixture slowly over 30 minutes while ensuring the bath temperature does not exceed −30°C.
-
19
When addition is complete, allow the mixture to warm to 0°C and allow to react for an additional 1 hour at this temperature by placing the flask in an ice water bath.
-
20After 30 minutes of the reaction for Mixture A, prepare Mixture B: To a flame-dried 50 mL round bottom flask equipped with a stir bar, add 4CI (439 mg, 3.07 mmol, 1.0 equiv), powdered KOH (379 mg, 6.75 mmol, 2.2 equiv), and TDA-1 (493 μL, 1.54 mmol, 0.5 equiv).A mortar and pestle can be used to crush KOH pellets into a powder to facilitate dissolution. Alternatively, powdered KOH can be purchased.
-
21
Suspend Mixture B in anhydrous toluene (12.5 mL) and stir rapidly under a N2 atmosphere until Step 25.
-
22Work up Mixture A: After one hour has passed, transfer Mixture A to a 60 mL separatory funnel and wash with 12.5 mL of cold brine from Step 15.Note: Chlorination using HMPT produces hexamethylphosphoric triamide (HMPA), the oxidized product. HMPA is known to be toxic in animals and believed to be harmful to humans. Therefore, users are urged to use appropriate personal protective equipment (PPE) and to properly dispose of the all waste from this reaction. See this report on HMPA from the United States Centers for Disease Control and Prevention at: https://www.cdc.gov/niosh/docs/78–127/78127_6.html
-
23Dry the organic layer over MgSO4 and filter off the MgSO4 to yield a crude solution of 6 which can be used without further purification.Na2SO4 is not satisfactory in this case and thus MgSO4 should be used. This crude solution should be used as quickly as possible.
-
24
Cannulate the dried crude toluene mixture of 6 from Mixture A into Mixture B. Stir for 24 hours under N2 at room temperature.
-
25
Transfer the mixture to a 60 mL separatory funnel and wash the organic layer with 15 mL each of saturated NH4Cl and brine.
-
26
Dry the organic layer over Na2SO4, filter, and concentrate onto approximately 1 g of silica gel in vacuo using a rotary evaporator.
-
27
Purify by chromatography on silica gel using a 40 g silica gel cartridge (Teledyne ISCO) and run at 40 mL/min flowrate while monitoring at 254 nm/ 300 nm using a gradient of EtOAc (0 to 100%) in hexanes collecting approximately 40 × 18 mL fractions.
-
28
Collect fractions containing the desired product and evaporate in vacuo using a rotary evaporator to yield 4-cyanoindole-2ʹ,3ʹ-O-isopropylidene-5ʹ-O-TBS-ribonucleoside (7) as a colorless liquid (768 mg, 1.79 mmol, 58% yield).
-
29
Characterize the product by TLC (on silica gel), 1H NMR, 13C NMR, and HR-MS.
4-cyanoindole-2ʹ,3ʹ-O-isopropylidene-5ʹ-O-TBS-ribonucleoside (7)
Rf = 0.23 (9/1, v/v hexanes/EtOAc)
Chemical shifts referenced to solvent (δ = 7.26 ppm for CDCl3) for 1H NMR and solvent signal for 13C (δ = 77.0 ppm for CDCl3).
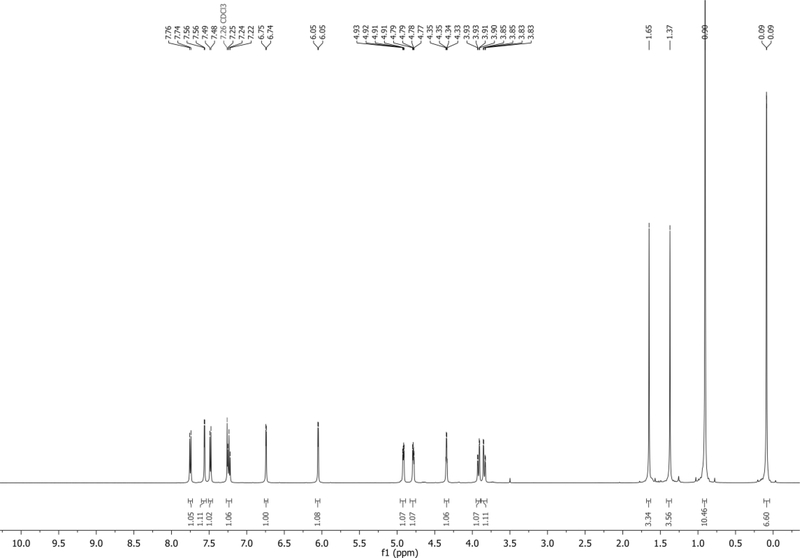
1H NMR (500 MHz, CDCl3): δ 7.75 (d, J = 8.3 Hz, 1H), 7.56 (d, J = 3.4 Hz, 1H), 7.48 (d, J = 7.3 Hz, 1H), 7.24 (app t, J = 7.9 Hz, 1H), 6.74 (d, J = 3.3 Hz, 1H), 6.05 (d, J = 3.5 Hz, 1H), 4.92 (dd, J = 6.4, 3.1 Hz, 1H), 4.78 (dd, J = 6.4, 3.5 Hz, 1H), 4.34 (q, J = 3.0 Hz, 1H), 3.92 (dd, J = 11.4, 3.0 Hz, 1H), 3.84 (dd, J = 11.3, 3.1 Hz, 1H), 1.65 (s, 3H), 1.37 (s, 3H), 0.90 (s, 9H), 0.09 (m, 6H) ppm. Spectrum shown in Figure 9.
13C NMR (126 MHz, CDCl3): δ 134.9, 130.7, 127.6, 125.5, 121.6, 118.6, 115.2, 114.5, 103.3, 101.6, 92.5, 85.3, 85.0, 80.5, 63.1, 27.4, 25.9, 25.4, 18.4, −5.4, −5.5 ppm.
HRMS-ESI+ (m/z) calc’d [M + H]+ for C23H33N2O4Si: 429.2165, found: 429.2188.
Figure 9.
1H NMR of 7 in CDCl3.
Synthesis of 4-cyanoindole-ribonucleoside (4CINr)
-
30
To a flame-dried 50 mL round bottom flask equipped with a magnetic stir bar, transfer 7 (631 mg, 1.47 mmol, 1.0 equiv).
-
31
To this flask add a mixture of TFA and H2O (1:1 v/v, 17 mL). Stir for 40 minutes.
-
32
Evaporate the mixture in vacuo using a rotary evaporator and then co-evaporate 3x with 10 mL aliquots of MeOH to remove all residual TFA.
-
33
Evaporate this crude product onto approximately 1 g of silica gel.
-
34
Purify by chromatography on silica gel using a 24g silica gel cartridge (Teledyne ISCO) and run at 35 mL/min flowrate while monitoring at 254 nm / 300 nm using a gradient of MeOH (0 to 10%) in DCM collecting approximately 30 × 18 mL fractions.
-
35
Collect fractions containing the desired product and evaporate in vacuo using a rotary evaporator to yield 4-cyanoindole-ribonucleoside (4CINr) as a light brown solid (278 mg, 1.01 mmol, 69% yield).
-
36
Characterize the product by TLC (on silica gel), 1H NMR, 13C NMR, and HR-MS.
4-cyanoindole-ribonucleoside (4CINr)
Rf = 0.45 (9/1, v/v DCM/MeOH)
Chemical shifts referenced to solvent (δ = 3.31 ppm for MeOD) for 1H NMR and solvent signal for 13C (δ = 49.2 ppm for MeOD).
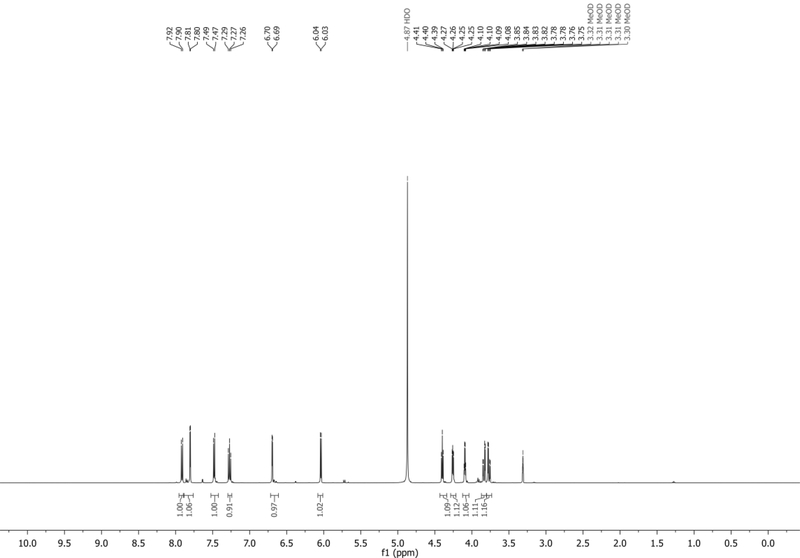
1H NMR (500 MHz, MeOD): δ 7.91 (d, J = 8.4 Hz, 1H), 7.80 (d, J = 3.4 Hz, 1H), 7.48 (d, J = 7.3 Hz, 1H), 7.27 (t, J = 8.0 Hz, 1H), 6.69 (d, J = 3.3 Hz, 1H), 6.04 (d, J = 6.0 Hz, 1H), 4.40 (t, J = 5.7 Hz, 1H), 4.26 (dd, J = 5.5, 3.7 Hz, 1H), 4.09 (app q, J = 3.6 Hz, 1H), 3.83 (dd, J = 12.1, 3.3 Hz, 1H), 3.77 (dd, J = 12.1, 3.9 Hz, 1H) ppm. Spectrum shown in Figure 10.
13C NMR (126 MHz, MeOD): δ 136.1, 130.2, 128.0, 125.0, 121.2, 118.0, 115.2, 102.2, 100.6, 89.3, 85.1, 74.7, 70.6, 61.6 ppm.
HRMS-ESI+ (m/z) calc’d [M + H]+ for C14H15N2O4: 275.0987, found: 275.1021.
Figure 10.
1H NMR of 4CINr in MeOD.
BASIC PROTOCOL 3
Synthesis of 4-cyanoindole-2ʹ-deoxyribonucleoside-5ʹ-triphosphate (4CIN-TP)
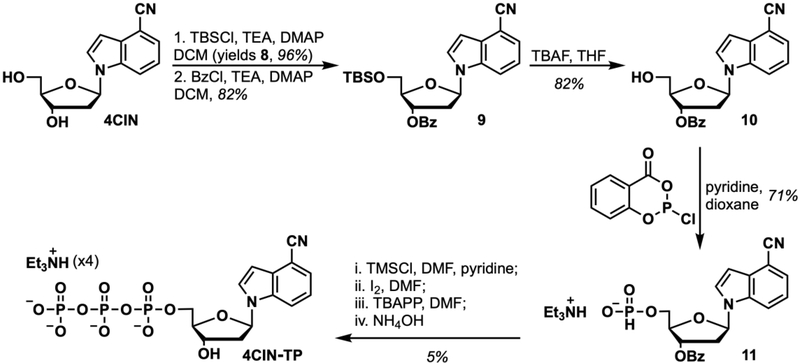
Basic Protocol 3 reports the synthesis of 4CIN-TP starting from 4CIN (Basic Protocol 1). The basic step outline follows 5 synthetic steps shown in Figure 11, starting with protecting the 5ʹ-OH of 4CIN with a silyl protecting group, yielding 9, subsequent benzoyl protection of the 3ʹ-OH, yielding 10, and then deprotection removal of the 5ʹ-OTBS, affording 11. Nucleoside 11 is then converted to phosphonate salt 12, followed by elaboration to the triphosphate, 4CIN-TP, in a four-step, one-pot procedure adapted from the method described by Sun et al. (2008).
Figure 11.
Synthesis of 4CIN-TP.
Materials
Dichloromethane (DCM) (Fisher Chemical cat. no. D37–20, ACS grade)
Triethylamine (TEA) (Fisher Chemical cat. no. 04885–1, reagent grade)
N,N-dimethylaminopyridine (DMAP) (Oakwood Chemical cat. no. 001704)
Tert-butyldimethylsilyl chloride (TBSCl) (Oakwood Chemical cat. no. 003869)
Anhydrous N2 gas
Brine (saturated aqueous sodium chloride solution, NaCl) (Fisher Chemical cat. no. S271–3, ACS grade)
Sodium sulfate (Na2SO4) (Beantown Chemical cat. no. 136810–10KG)
Silica gel (40 – 64 μm, Silicycle or Teledyne ISCO sizes 12 g, 24 g, 40 g)
Hexanes (Fisher Chemical cat. no. H292–20, ACS grade)
Ethyl acetate (EtOAc) (J.T. Baker cat. No. 9290–07, ACS grade)
Deuterated chloroform (CDCl3) (Cambridge Isotopes, cat. no. DLM-7–100 or DLM-7TB-100)
Benzoyl chloride (BzCl) (Sigma Aldrich cat. no. 259950, ACS grade, 99%)
Tetrahydrofuran (THF) (Fisher Chemical cat. no. T397–4, Certified grade)
Sodium Bicarbonate (NaHCO3 solution, saturated) (Fisher Chemical cat. no. S233–3, ACS grade)
Tetrabutylammonium fluoride (TBAF) (Sigma Aldrich cat. no. 216143, 1.0 M THF solution)
Ammonium chloride (saturated aqueous solution, NH4Cl) (Chem. Impex. Int. cat. no. 01207, ACS grade)
2-chloro-1,3,2-benzodioxaphosphorin-4-one (Sigma Aldrich cat. no. 324124, 95%)
Pyridine (Sigma Aldrich cat. No. 270970–1L, anhydrous, 99.8%)
Dioxane (Sigma Aldrich cat. no. 360481, ACS grade, 99%)
Methanol (MeOH) (Fisher Chemical cat. no. A412–4, ACS grade)
Deuterated Methanol (MeOD) (Cambridge Isotopes cat. no. DLM-24–10)
N,N-dimethylformamide (DMF) (Fisher Biology Reagents cat. no. BP1160–4, sequencing grade > 99.5%)
Trimethylsilyl chloride (TMSCl) (Acros, cat. no. 110122500, 98%)
Calcium hydride (CaH2) (Sigma Aldrich cat. no. 208027–500G, reagent grade)
Anhydrous argon (Ar) gas
Tris(tetrabutylammonium) hydrogen pyrophosphate (TBAPP) (Sigma Aldrich cat no. 93397, >97%)
Iodine (I2) (Alfa Aesar, cat. no. 41955, 99.8%)
Ammonium hydroxide (NH4OH) (Millipore Sigma cat. no. AX1303–75, aqueous, concentrated)
Triethylammonium acetate, 1.0 M solution in water, pH 7 (TEAA) (Sigma Aldrich cat. no. 90358)
Acetonitrile (MeCN) (Fisher Chemical cat. no. A998–4, HPLC grade, Anhydrous)
Acetic Acid (AcOH) (Fisher Chemical cat. no. A38–212, ACS grade, glacial)
Sephadex LH-20 Resin (GE Healthcare Life Sciences cat. no. 17009001)
Deuterated Water (D2O) (Cambridge Isotopes cat. no. DLM-4–100)
10-, 25-, and 50- mL round-bottom flasks
Drying oven or propane torch
Rubber septa
Magnetic stir bars
Magnetic stir plate/oil bath heater
High vacuum pump
1-, 3-, and 5- mL plastic syringe (luer lock desirable)
18-, 20-, 21-G needles (0.8 × 40 mm)
4” non-disposable needles and glass gas-tight syringes (100 μl to 1 mL, Hamilton)
125-and 250- mL separatory funnel
10-, 25-, 50-, 100- mL graduated cylinder
Erlenmeyer flasks
Filter paper
Buchner funnel
Fritted funnel
Rotary evaporator
Cold water bath
Balloons
Short path distillation apparatus
Thermometer
Polycarbonate splash safety shield
Sealed tube reaction vessel (ChemGlass Part no. CG-1880–02, 35 mL) and accompanying seal and o-ring
UV lamp, 254 nm
Thin-layer chromatography (TLC) plate (EMD Chemicals Silica gel 60 F254, 250 μm thickness)
TLC Chamber
Combiflash Rf (Teledyne ISCO, or comparable normal phase chromatography system, or standard flash chromatography setup
Electronic pH meter
Lyophilizer and bucket attachments
Preparative HPLC column (Agilent Technologies Zorbax SB-C18, 21.2 × 250 mm, 7 μm pore, cat. no. 877250–102)
Preparative HPLC System comprising:
Autosampler
Multiple Wavelength Detector (MWD)
Fraction Collector
Preparative pumps x2
NMR (e.g. Brucker Advance 500 MHz)
Protocol steps—Step annotation
Synthesis of 5ʹ-O-TBS-4-cyanoindole-2ʹ-deoxyribonucleoside (8)
-
1
Transfer a sample of 4CIN (464 mg, 1.80 mmol, 1 equiv) into a flame-dried 50 mL round-bottom flask equipped with a magnetic stir bar.
-
2
To the flask add anhydrous DCM (15 mL).
-
3
To the flask add TEA (301 μL, 2.16 mmol, 1.2 equiv).
-
4
Add DMAP (22.0 mg, 0.180, 0.1 equiv) to the mixture.
-
5
Add TBSCl (298 mg, 1.98 mmol, 1.1 equiv) to the mixture. Place the flask under a N2 atmosphere. Stir the mixture 24 hours under a N2 atmosphere.
-
6
Dilute the mixture with DCM (50 mL) and wash the organic layer with H2O (50 mL) and brine (50 mL) in a 250 mL separatory funnel.
-
7
Dry the organic layer over Na2SO4, filter, and concentrate onto approximately 1g of silica gel in vacuo using a rotary evaporator.
-
8
Purify by chromatography on silica gel using a 24g silica gel cartridge (Teledyne ISCO) and run 35 mL/min flowrate while monitoring at 254 nm / 300 nm using a gradient of EtOAc (0 to 80%) in hexanes.
-
9
Collect the fractions containing the desired product and evaporate in vacuo using a rotary evaporator to yield 5ʹ-O-TBS-4-cyanoindole-2ʹ-deoxyribonucleoside (8) as a pale yellow foam (643 mg, 0.264 mmol, 96% yield) collecting approximately 30 × 18 mL fractions.
-
10
Characterize the product by TLC (on silica gel), 1H NMR, 13C NMR, and HR-MS.
5ʹ-O-TBS-4-cyanoindole-2ʹ-deoxyribonucleoside (8)
Rf = 0.5 (1/1, v/v hexanes/EtOAc)
Chemical shifts referenced to solvent (δ = 7.26 ppm for CDCl3) for 1H NMR and solvent signal for 13C (δ = 77.0 ppm for CDCl3).
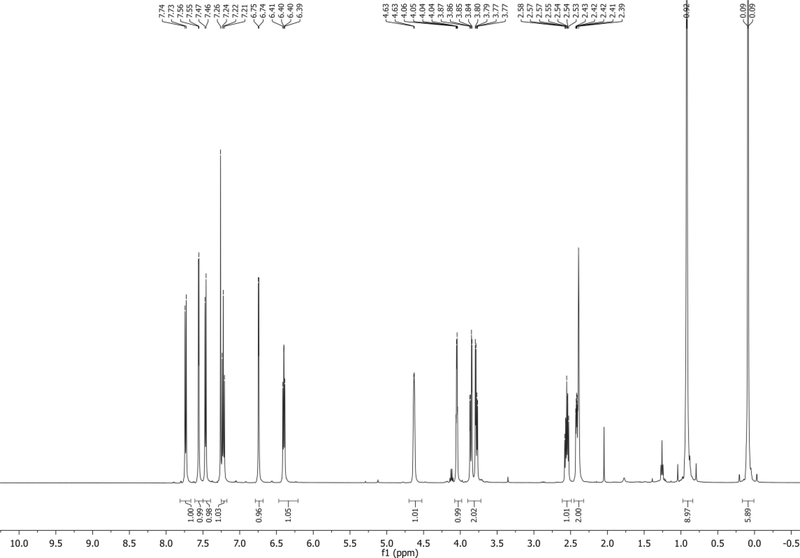
1H NMR (500 MHz, CDCl3): δ 7.73 (d, J = 8.4 Hz, 1H), 7.56 (d, J = 3.4 Hz, 1H), 7.46 (d, J = 7.4 Hz, 1H), 7.22 (app t, J = 7.9 Hz, 1H), 6.74 (d, J = 3.4 Hz, 1H), 6.40 (app t, J = 6.7 Hz, 1H), 4.63 (s, 1H), 4.05 (app q, J = 3.5 Hz, 1H), 3.86 (dd, J = 11.0, 3.2 Hz, 1H), 3.78 (dd, J = 10.9, 4.5 Hz, 1H), 2.55 (m, 1H), 2.48 – 2.33 (m, 2H), 0.92 (s, 9H), 0.09 (d, J = 2.1 Hz, 6H) δ ppm. Spectrum shown in Figure 12.
13C NMR (126 MHz, CDCl3): δ 135.7, 130.5, 127.2, 125.5, 121.5, 118.7, 114.9, 103.1, 101.7, 86.5, 85.3, 72.6, 63.7, 40.8, 26.0, 18.4, −5.4, −5.5 ppm.
HRMS-ESI+ (m/z) calc’d [M + H]+ for C20H29N2O3Si: 373.1903, found: 373.1930
Figure 12.
1H NMR of 8 in CDCl3.
Synthesis of 5ʹ-O-TBS-3ʹ-O-Bz-4-cyanoindole-2ʹ-deoxyribonucleoside (9)
-
11
Transfer a sample of 8 (1.42 g, 3.81 mmol, 1.0 equiv) to a flame-dried 50 mL round-bottom flask equipped with a magnetic stir bar.
-
12
To the flask add anhydrous DCM (20 mL).
-
13
To the flask add TEA (1.59 mL, 11.4 mmol, 3.0 equiv).
-
14
To the flask add DMAP (93.1 mg, 0.762 mmol, 0.2 equiv).
-
15
Prepare an ice water bath. Place the flask into the ice bath to cool the mixture to 0°C.
-
16
Connect the mixture to a N2 atmosphere.
-
17
Add BzCl (885 μL, 7.62 mmol, 2.0 equiv) to the cooled mixture dropwise over 1 minute.
-
18
Stir the mixture under N2 18 hours, allowing it to warm to room temperature gradually.
-
19Quench the reaction using 10 mL of saturated NaHCO3 followed by rapid stirring.Perform this step with care. Evolution of CO2 will occur.
-
20
Dilute the mixture with an additional 20 mL of DCM and wash the organic layer with 50 mL of brine solution.
-
21
Dry the organic layer over Na2SO4, filter, and concentrate onto approximately 3 g of silica gel in vacuo using a rotary evaporator.
-
22
Purify by chromatography on silica gel using a 40g silica gel cartridge (Teledyne ISCO) and run at 40 mL/min flowrate while monitoring at 300 nm/ 254 nm using a gradient of EtOAc (0 to 40%) in hexanes collecting approximately 40 × 18 mL fractions.
-
23
Collect fractions containing the desired product and evaporate in vacuo using a rotary evaporator to yield 5ʹ-O-TBS-3ʹ-O-Bz-4-cyanoindole-2ʹ-deoxyribonucleoside (9) as a white foam (1.49 g, 3.12 mmol, 82% yield).
-
24
Characterize the product by TLC (on silica gel), 1H NMR, 13C NMR, and HR-MS.
5ʹ-O-TBS-3ʹ-O-Bz-4-cyanoindole-2ʹ-deoxyribonucleoside (9)
Rf = 0.6 (3/1, v/v hexanes/EtOAc)
Chemical shifts referenced to solvent (δ = 7.26 ppm for CDCl3) for 1H NMR and solvent signal for 13C (δ = 77.0 ppm for CDCl3).
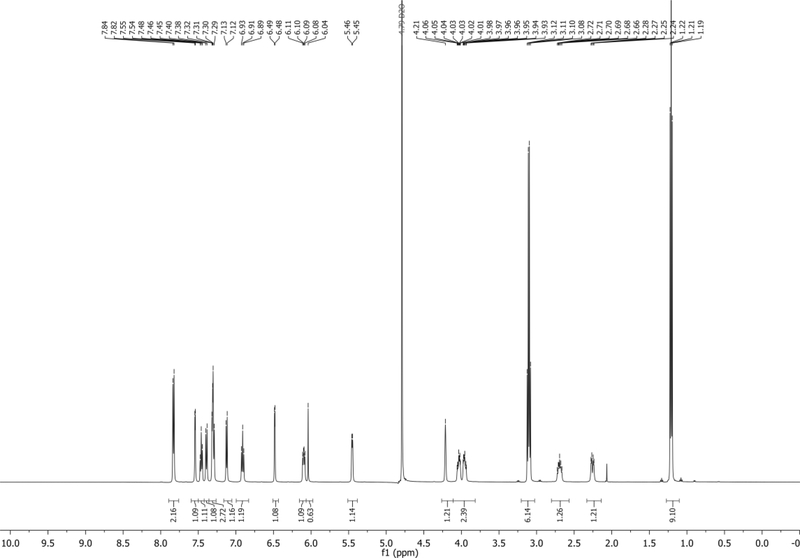
1H NMR (500 MHz, CDCl3): δ 8.10 (d, J = 7.8 Hz, 2H), 7.84 (d, J = 8.3 Hz, 1H), 7.66 – 7.59 (m, 2H), 7.54 – 7.47 (m, 3H), 7.29 – 7.22 (m, 1H), 6.79 (d, J = 3.4 Hz, 1H), 6.44 (dd, J = 9.2, 5.3 Hz, 1H), 5.67 (d, J = 5.9 Hz, 1H), 4.33 (app q, J = 2.4 Hz, 1H), 4.03 (dd, J = 11.3, 2.6 Hz, 1H), 3.94 (dd, J = 11.3, 2.6 Hz, 1H), 2.84 – 2.75 (m, 1H), 2.60 (dd, J = 13.9, 5.2 Hz, 1H), 0.96 (s, 9H), 0.15 (s, 6H) ppm. Spectrum shown in Figure 13.
13C NMR (126 MHz, CDCl3): δ 166.1, 135.5, 133.5, 130.6, 129.7, 129.5, 128.6, 127.4, 125.6, 121.6, 118.6, 115.0, 103.5, 102.0, 86.1, 85.1, 75.9, 63.7, 38.6, 26.0, 18.5, −5.3, −5.5 ppm.
HRMS-ESI+ (m/z) calc’d [M + H]+ for C27H33N2O4Si: 477.2165, found: 477.2185.
Figure 13.
1H NMR of 9 in CDCl3.
Synthesis of 3ʹ-O-Bz-4-cyanoindole-2ʹ-deoxyribonucleoside (10)
-
25
Transfer a sample of 9 (172 mg, 0.361 mmol, 1.0 equiv) to a flame-dried 25 mL round-bottom flask equipped with a magnetic stir bar.
-
26
To the flask add anhydrous THF (5 mL) and cool the mixture to 0°C using an ice water bath.
-
27To this flask add TBAF (1.0M solution in THF, 433 μL, 0.433 mmol, 1.2 equiv) slowly and dropwise (approximately 1 drop every 2 seconds).The reaction should near completion after dropwise addition is complete. Follow TLC until completion.
-
28
Quench the reaction with 10 mL of saturated NH4Cl and extract with 20 mL of DCM. Wash the organic layer with 10 mL of brine.
-
29
Dry the organic layer over Na2SO4, filter, and concentrate onto approximately 0.4 g of silica gel in vacuo using a rotary evaporator.
-
30
Purify by chromatography on silica gel using a 12g silica gel cartridge (Teledyne ISCO) and run at 30 mL/min flowrate while monitoring at 254 nm/ 300 nm using a gradient of EtOAc (0 to 70%) in hexanes collecting approximately 25 × 18 mL fractions.
-
31
Collect fractions containing the desired product and evaporate in vacuo using a rotary evaporator to yield 3ʹ-O-Bz-4CIN (10) as a white foam (107 mg, 0.295 mmol, 82% yield).
-
32
Characterize the product by TLC (on silica gel), 1H NMR, 13C NMR, and HR-MS.
3ʹ-O-Bz-4-cyanoindole-2ʹ-deoxyribonucleoside (10)
Rf = 0.55 (1/1, v/v hexanes/EtOAc)
Chemical shifts referenced to solvent (δ = 7.26 ppm for CDCl3) for 1H NMR and solvent signal for 13C (δ = 77.0 ppm for CDCl3).
1H NMR (500 MHz, CDCl3): δ 8.09 (d, J = 7.0 Hz, 2H), 7.80 (d, J = 8.3 Hz, 1H), 7.63 (t, J = 7.5 Hz, 1H), 7.59 (d, J = 3.4 Hz, 1H), 7.56 – 7.42 (m, 3H), 7.29 (app t, J = 7.8 Hz, 1H), 6.82 (d, J = 3.4 Hz, 1H), 6.47 (dd, J = 8.5, 5.6 Hz, 1H), 5.68 (dt, J = 6.6, 2.6 Hz, 1H), 4.33 (q, J = 3.2 Hz, 1H), 3.95 (s, 2H), 2.93 (ddd, J = 14.3, 8.5, 6.9 Hz, 1H), 2.69 (ddd, J = 14.1, 5.7, 2.2 Hz, 1H), 1.91 (s, 1H) ppm. Spectrum shown in Figure 14.
13C NMR (126 MHz, CDCl3): δ 166.2, 135.9, 133.7, 130.6, 129.7, 129.3, 128.6, 126.8, 125.8, 121.9, 118.4, 114.8, 103.6, 102.6, 85.3, 84.6, 75.2, 62.8, 37.9 ppm.
HRMS-ESI+ (m/z) calc’d [M + H]+ for C21H19N2O4: 363.1300, found: 363.1328
Figure 14.
1H NMR of 10 in CDCl3. * = Hexanes/grease impurity.
Synthesis of 5ʹ-H-phosphonate-3ʹ-OBz-4-cyanoindole-2ʹ-deoxyribonucleoside (11)
-
33
Transfer a sample of 10 (370. mg, 1.02 mmol, 1.0 equiv) to a flame-dried 25 mL round-bottom flask. Place this flask under vacuum for 24 hours.
-
34
In a second flame-dried 25 mL round-bottom flask equipped with a stir bar, add 2-chloro-1,3,2-benzodioxaphosphorin-4-one (414 mg, 2.04 mmol, 2.0 equiv). Similarly, place this flask under vacuum for 24 hours.
-
35
On the day of the reaction, backfill each flask with N2.
-
36
To the flask containing 2-chloro-1,3,2-benzodioxaphosphorin-4-one add a mixture of anhydrous pyridine and anhydrous dioxane (3 mL and 3 mL).
-
37
Similarly, dissolve 10 in a mixture of anhydrous pyridine and anhydrous dioxane (2 mL and 2 mL) and then using a syringe equipped with a needle remove the solution of 10 under N2.
-
38Add the solution of 10 dropwise to the stirring solution of 2-chloro-1,3,2-benzodioxaphosphorin-4-one. Stir this new mixture for two hours under N2 at room temperature.Typically, the reaction mixture is colorless to brown and translucent, but as the reaction progresses, the mixture will begin to cloud and become dark.
-
39Add H2O (1 mL) to quench the reaction and stir for two minutes, and then concentrate onto approximately 1g of deactivated silica gel (as prepared on Basic Protocol 1 Step 28).After the addition of water, the mixture will turn translucent again.
-
40Prior to chromatography, wash a 12g silica gel cartridge with 1% v/v TEA in DCM (3 column volumes) and then elute excess TEA with excess DCM. Purify by chromatography on silica gel using this deactivated 12g silica gel cartridge (Teledyne ISCO) and run at 30 mL/min flowrate while monitoring at 254 nm/ 300 nm using a gradient of MeOH (0 to 20%) in DCM collecting approximately 50 × 8 mL fractions.Typically, two column purifications were required for adequate purification and removal of excess TEA.
-
41
Collect fractions containing the desired product and evaporate in vacuo using a rotary evaporator to yield 5ʹ-H-phosphonate-3ʹ-OBz-4-cyanoindole-2ʹ-deoxyribonucleoside (11)-triethylamine salt as a colorless glassy solid (380. mg, 0.720 mmol, 71% yield).
-
42
Characterize the product by TLC (on silica gel), 1H NMR, 13C NMR, 31P NMR, and HR-MS.
5ʹ-H-phosphonate-3ʹ-OBz-4-cyanoindole-2ʹ-deoxyribonucleoside triethylamine salt (11)
Rf = 0.42 (4/1, v/v DCM/MeOH with one drop of triethylamine)
This product can smear on TLC without triethylamine.
Chemical shifts referenced to solvent (δ = 4.79 ppm for D2O) for 1H NMR.
1H NMR (500 MHz, D2O): δ 7.83 (d, J = 7.7 Hz, 2H), 7.54 (d, J = 3.4 Hz, 1H), 7.46 (t, J = 7.5 Hz, 1H), 7.39 (d, J = 8.3 Hz, 1H), 7.34 – 7.27 (m, 3H), 7.12 (d, J = 7.3 Hz, 1H), 6.91 (t, J = 7.8 Hz, 1H), 6.48 (d, J = 3.2 Hz, 1H), 6.10 (dd, J = 9.3, 5.5 Hz, 1H), 6.04 (s, 1H), 5.45 (d, J = 5.7 Hz, 1H), 4.21 (s, 1H), 4.08 – 3.91 (m, 2H), 3.10 (q, J = 7.3 Hz, 6H, TEA salt), 2.74 – 2.64 (m, 1H), 2.26 (dd, J = 14.1, 5.2 Hz, 1H), 1.21 (t, J = 7.3 Hz, 9H, TEA salt) ppm.
13C NMR (126 MHz, D2O): δ 167.1, 135.4, 133.8, 129.8, 129.3, 128.7, 128.5, 127.8, 125.6, 121.5, 118.8, 115.0, 101.7, 101.5, 85.0, 82.7 (d, JC-P = 8.2 Hz), 76.0, 63.5 (d, JC-P = 3.4 Hz), 46.5, 36.4, 8.2 ppm. Spectrum shown in Figure 15.
31P NMR (202 MHz, D2O): δ 6.40 ppm.
HRMS-ESI+ (m/z) calc’d [M + Et3N + H]+ for C27H35N3O6P: 528.2219, found:528.2234
Figure 15.
1H NMR of 11 in D2O.
Synthesis of 4-cyanoindole-2ʹ-deoxyribonucleoside-5ʹ-triphosphate (4CIN-TP)
-
43
Transfer a sample of 11 (92.0 mg, 0.174 mmol, 1.0 equiv) to a flame-dried 50 mL round-bottom flask equipped with a magnetic stir bar. Place the flask under vacuum for 24 hours.
-
44
Fill two flame-dried 25 mL round-bottom flasks with 15 mL of anhydrous DMF and 15 mL of anhydrous pyridine, respectively. To each add oven-dried 4Å molecular sieves and put under an argon atmosphere. Store these solvents for 24 hours before using.
-
45
Perform a short path distillation of TMSCl over CaH2 (Armarego & Chai, 2009) and place it under an Ar atmosphere.
-
46
To a flame-dried 5 mL round-bottom flask add TBAPP (315 mg, 0.349 mmol, 2.0 equiv), and then place under vacuum for 24 hours.
-
47
On the day of the reaction: To the flask containing 11 add anhydrous DMF (1.5 mL) and anhydrous pyridine (350 μL, 4.35 mmol, 25 equiv) from the prepared stocks, and then stir to solubilize the material.
-
48
Add TMSCl (110 μL, 0.870 mmol, 5 equiv) from the prepared stock to the solution of 11. Stir this mixture for approximately 5 minutes.
-
49During this 5 minutes, weigh solid I2 (66.2 mg, 0.261 mmol, 1.5 equiv) and dissolve in 1 mL of anhydrous DMF. Add this solution dropwise to the mixture until the orange-brown color persists. Stir the now brown mixture for approximately 2 minutes.The whole I2 solution is usually not necessary to achieve the desired permanent orange-brown color.
-
50To the flask containing TBAPP add 2 mL of anhydrous DMF to dissolve the solid.Sonication is often helpful to dissolve the solid. Default settings on a bath sonicator were used on our sample without any additional temperature control. Dissolution took approximately 2–3 minutes.
-
51
Add the TBAPP solution rapidly to the brown reaction mixture and stir for 30 minutes.
-
52Transfer the entire mixture and stir bar into a sealed tube reaction vessel (35 mL capacity).Rinse the original flask with 5 mL of water and transfer to ensure no material is lost. Do not fill the new reaction vessel more than half full.
-
53Add 5 mL of NH4OH to the vessel, attach the screw cap to seal and place the mixture in a 50°C oil bath for 18 hours.Put a polycarbonate splash safety shield in front of the flask for safety purposes.
-
54
Leaving the vessel sealed, cool the mixture back to room temperature. Leaving the safety shield in place, reach around the shield and unseal the vessel slowly to allow venting of volatile gases. Once properly vented, remove the shield and transfer this solution into a 50 mL round bottom flask.
-
55
Evaporate the mixture in vacuo using a rotary evaporator until most of the solvent is removed. Next, attach the flask to a high vacuum pump equipped with a cold trap for 18 hours to remove the remaining high boiling solvent.
-
56Prepare a 10 mM solution of triethylammonium acetate (TEAA, pH 8.0, adjusted with either glacial acetic acid or triethylamine).We suggest using a pH meter instead of pH papers.
-
57
Prepare a glass fritted column with Sephadex LH-20 resin (a bed approximately 2.5 × 40 cm in size) in a chemical hood. Equilibrate this column with the prepared TEAA buffer.
-
58Dissolve the crude triphosphate sample in this buffer and load onto a pre-equilibrated Sephadex LH-20 column (2.5 cm x 40 cm) with the aforementioned TEAA buffer. Elute by running the column at 2.0 mL/min (approximately gravity filtration rate) and collect 200 mL total with 10 mL fractions up to 100 mL and then two 50 mL fractions.We found that trisphosphate-containing fractions typically elute between 30–50 mL.
-
59
Freeze and lyophilize the samples to dryness. Identify fractions containing the three prototypical 31P shifts of a triphosphate by NMR (approx. 8 ppm, 10 ppm, and 20 ppm see Figure 14) and combine.
-
60
Dissolve identified triphosphate fractions in 10 mM TEAA pH 7.5 buffer and repurify as needed using preparative reverse phase HPLC:
Column: Agilent Technologies Zorbax SB-C18, 21.2 × 250 mm size, 7 μm pore
Method: Solvent A: 10 mM TEAA (pH 7.5); Solvent B: 1:1, 10 mM TEAA (pH 7.5) and MeCN. Monitor wavelengths 300 nm and 215 nm.
Gradient: A:B 98:2 from 0–5 minutes, followed by a linear gradient to A:B 60:40 from 5–30 minutes, and from 30–35 minutes an isocratic A:B 60:40.A typical run would have product elute between 15–20 minutes on our system. -
61
Collect fractions from the HPLC absorbing at 300 nm, freeze, and lyophilize. Check for product using NMR spectroscopy or mass spectrometry.
-
62Repeat HPLC procedure until pure samples were achieved.Between 1 and 2 HPLC runs after the Sephadex column are usually necessary.
-
63Lyophilize the product-containing fractions multiple times with ddH2O to yield the tetra(triethylammonium) 4CIN-TP salt form as a colorless to yellowed glassy solid (7.93 μmol, 5% yield).Calculate the yield of the reaction by dissolving the final product in ddH2O. Using Beer’s law and the published 4CIN ε300 = 7790 M−1cm−1, calculate a molarity and then moles of product (Passow & Harki, 2018).
-
64
Characterize the product by 1H NMR, 31P NMR, LR-MS, and analytical HPLC.
Chemical shifts referenced to solvent (δ = 4.79 ppm for D2O) for 1H NMR.
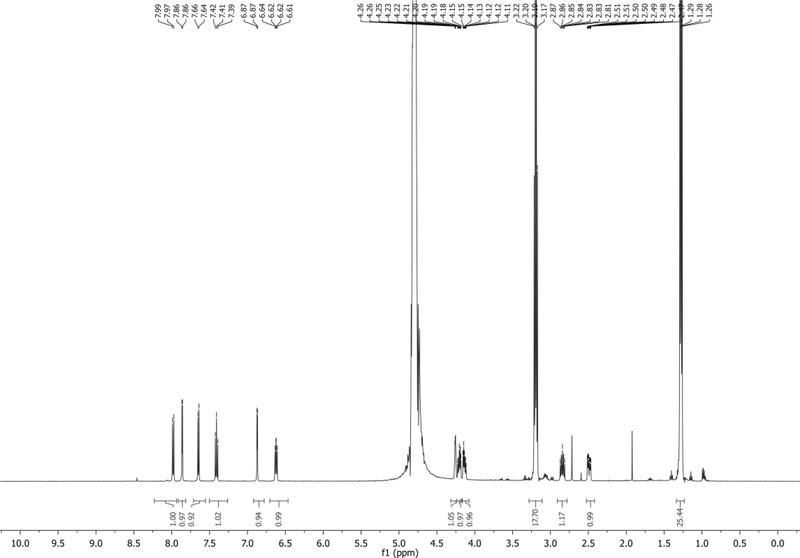
1H NMR (500 MHz, D2O): δ 7.98 (d, J = 8.4 Hz, 1H), 7.86 (d, J = 3.5 Hz, 1H), 7.65 (d, J = 7.4 Hz, 1H), 7.41 (t, J = 7.9 Hz, 1H), 6.87 (d, J = 3.4 Hz, 1H), 6.62 (dd, J = 8.3, 6.1 Hz, 1H), 4.28 – 4.24 (m, 1H), 4.23 – 4.09 (m, 2H), 3.19 (q, J = 7.3 Hz, 18H), 2.84 (ddd, J = 14.3, 8.3, 6.3 Hz, 1H), 2.49 (ddd, J = 14.0, 6.1, 2.9 Hz, 1H), 1.28 (t, J = 7.3 Hz, 26H) ppm. Spectrum shown in Figure 16.
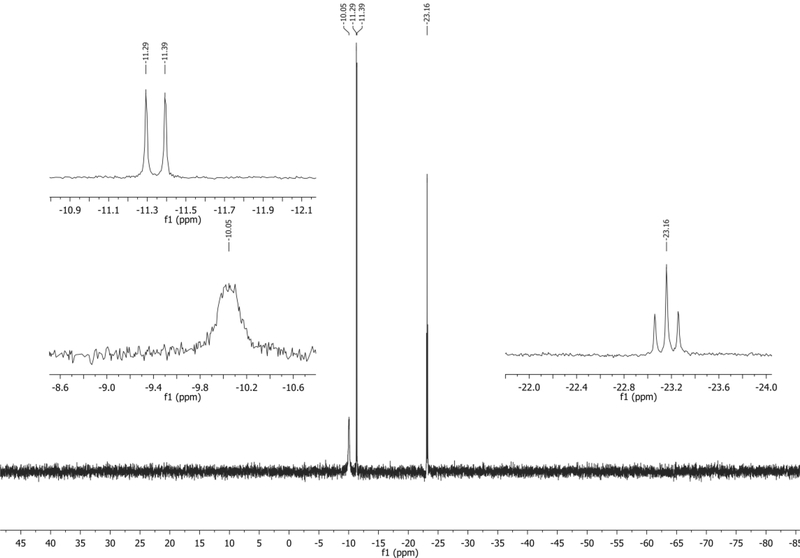
31P NMR (202 MHz, D2O): δ −10.05 (s, broad), −11.34 (d, J = 20.0 Hz), −23.16 (t, J = 19.9 Hz) ppm. Spectrum shown in Figure 17.
LRMS-ESI+ (m/z) calc’d [M + H, no salt]+ for C14H18N2O12P3: 499.0, found:498.8
LRMS-ESI+ (m/z) calc’d [M – H, no salt]- for C14H16N2O12P3: 497.0, found: 497.1
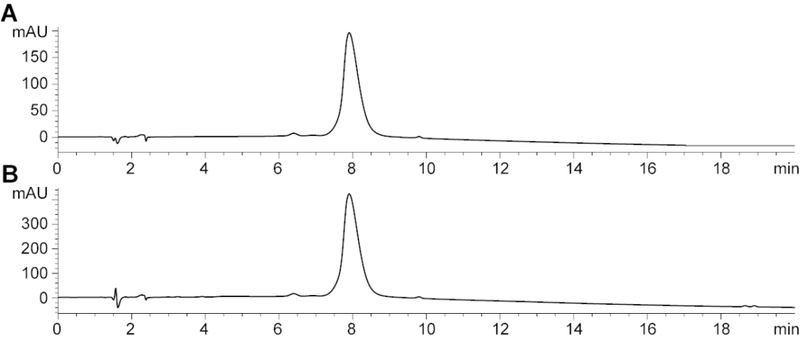
Analytical traces of purified 4CIN-TP shown in Figure 18.
Figure 16.
1H NMR of 4CIN-TP in D2O.
Figure 17.
31P NMR of 4CIN-TP in D2O.
Figure 18.
Analytical trace of 4CIN-TP collected using a previously described analytical method (Harki et al., 2002). Traces collected at 300 nm (A) and 215 nm (B).
BASIC PROTOCOL 4
Steady State Incorporation Kinetics of 2AP-TP and 4CIN-TP by DNA Polymerases
Basic Protocol 4 details the steps necessary to measure the incorporation kinetics of 2AP-TP and 4CIN-TP by DNA Polymerase I Klenow exo- fragment into a fluorescently labeled oligonucleotide primer. Analysis by gel electrophoresis imaging allows the calculation of KM, Vmax, kcat, and catalytic efficiency (Cat eff, kcat/KM).
Materials
Magnesium Acetate (Mg(OAc)2) (Sigma-Aldrich, cat. no. M5661–250G, for molecular biology, >99%)
HEPES (1 M pH 7.5 at 25 °C solution Sigma-Aldrich, cat. no. 83264–100ML)
DNase/RNase free water (Sigma-Aldrich, cat. no. W4502–1L)
Sodium Chloride (NaCl) (Fisher, cat. no. S316060, analytical grade)
Magnesium Chloride (MgCl2) (Mallinckrodt, cat. no. 5958, ACS grade)
dl-Dithiothreitol (DTT) (Sigma-Aldrich cat. no. 43815, Bioultra, >99.5%)
Formamide (Sigma-Aldrich, cat. no. 47670–250ML, ACS reagent)
EDTA (Biorad, cat. no. 161–0729, electrophoresis purity)
Bromophenol Blue sodium salt (Fluka, cat. no. 18040)
Boric Acid (Sigma-Aldrich, cat. no. B16768–1KG, Bioreagent)
Tris base (Roche, cat. no. 10708976001)
Urea (VWR International, ACS grade)
40 % acryl-bisacrylamide, 19:1 (Serva (DE), cat. no. 10679.1)
Acetone
Sigmacote (Sigma-Aldrich, cat. no. SL2)
N,N,Ń,Ń-Tetramethylethylenediamine (TEMED) (Sigma-Aldrich, cat. no. T9821–50ML, Bioreagent, for electrophoresis, >99%)
Ammonium persulfate (APS) (Sigma-Aldrich, cat. no. A3678–25G, for molecular biology, for electrophoresis, >98%)
2-aminopurine-2ʹ-deoxyriboside-5ʹ-triphosphate (2AP-TP, Trilink Biotechnologies, Part no. N-2004–5)
4CIN-TP (from Basic Protocol 3, 1–3 μmol will be required)
10x Annealing buffer (50 mM Mg(OAc)2, 200 mM HEPES, pH 7.5, DNase/RNase free water)
5x Assay buffer (250 mM NaCl, 50 mM TrisHCl (pH 7.9 at 25 °C), 50 mM MgCl2, 5 mM DTT, DNase/RNase free water. Combine everything but DTT, add DTT to fresh aliquot prior to use.)
Quench Buffer (80 % (v/v) Formamide, 0.1 M EDTA, 0.01 % (w/v) Bromophenol Blue)
10x Tris Borate EDTA Buffer (10x TBE, 1M Tris base, 0.9M boric acid, 0.1M EDTA)
5ʹ-Cy5 labeled DNA primer (Eurogentec):
Sequence: 5ʹ- Cy5 - GGTGAGGATGGGCCTC
Mass 5511.9 g mol−1
ε260 164,000 L mol−1 cm−1
Tm (calculated) = 54 °C
DNA templates (unlabeled) (Eurogentec):
TempA 5ʹ GCATGAACCAGAGGCCCATCCTCACC
Mass 7870.1 g mol−1
ε260 243,100 L mol−1 cm−1
Tm (calculated) = 59.2 °C
TempC 5ʹ GCATGAACCCGAGGCCCATCCTCACC
Mass 7846.1 g mol−1
ε260 237,500 L mol−1 cm−1
Tm (calculated) = 60.8 °C
TempG 5ʹ GCATGAACCCGAGGCCCATCCTCACC
Mass 7886.1.9 g mol−1
ε260 240,400 L mol−1 cm−1
Tm (calculated) = 60.8 °C
TempT 5ʹ GCATGAACCCTAGGCCCATCCTCACC
Mass 7861.1 g mol−1
ε260 237,800 L mol−1 cm−1
Tm (calculated) = 59.2 °C
Glycerol (Sigma-Aldrich, cat. no. G5516, for molecular biology, >99%)
Klenow fragment (3’→5’ exo- ) (NEB, M0212L)
Microcentrifuge tubes (500 μL)
Adjustable Slab Gel Apparatus (C. B. S. Scientific, cat. no. DASG-250)
Bio-Rad ChemiDoc MP Imager
Bio-Rad Image Lab software
Clamps (C. B. S. Scientific)
Digital Timer
Glass plates – 16.5 × 22 cm (C. B. S. Scientific, cat. no. NGP-200N)
GraphPad Prism software (v. 8.2.0)
Heat blocks (Labnet, Model D1200)
High voltage power supply
Magnetic stir plate and stir bar
Microsoft Excel
Spacers – 1 mm thick (C. B. S. Scientific, cat. no. VGS-1020)
Vertical square tooth comb – 1 mm thick, 30 wells (C. B. S. Scientific, cat. no. VGC-1030)
Gel wrap gaskets - 1 mm thick (C.B.S. Scientific, cat. no. VGE-1020)
Protocol steps—Step annotations
Annealing of the primer-template.
-
1To make 60 μL of primer-template for annealing, combine 3 μL of 10 μM template (TempA, TempC, TempG, or TempT) and 3 μL of 10 μM Cy5-labeled primer with 6 μL of 10x annealing buffer and DNase/RNase-free water in a microcentrifuge tube to reach a final concentration of 500 nM DNA duplex and 1x annealing buffer.Typically, 60 μL of the 500 nM primer-template solution is enough for one replicate of steady-state kinetics. Other fluorescent labels can be used as well, such as Cy3 or fluorescein. Repeat this step as necessary to build each primer-template duplex.
-
2
Heat the primer-template DNA solution at 95 °C for 10 min in a heat block and then turn off heat block and allow to slowly cool to room temperature overnight.
Single Nucleotide Incorporation and Steady State Kinetics Assays
-
3
Prepare 150 μL of 5x assay buffer by adding 2.5 μL fresh 500 mM DTT stock to 147.5 μL of 5x assay buffer (see materials section for Basic Protocol 4).
-
4Set heat block containing the primer-template DNA solution to 25 °C. Set a second heat block to 75 °C for heat inactivation of the Klenow fragment (3’→5’ exo-) (Kf -) which occurs in Step 11.Some literature shows enzymatic assays using Kf - as the DNA polymerase completed at 25 °C due to a higher binding affinity for DNA (Datta et al., 2006). For other polymerases, 37 °C is the preferred reaction temperature.
-
5
Calculate the concentration of purchased 2AP-TP stock solution using its molar extinction coefficient at 303 nm, ε303 = 6422 M−1 cm−1, using Beer’s Law.
The molar extinction coefficient at 303 nm was taken from the reagent vendor Trilink (Part no. N-2004–5).
-
6To determine kinetic parameters for 2AP-TP incorporation onto a position templated by T at 0.8 μM, first prepare a 2AP-TP 5x stock at a concentration 4.0 μM.The 5x is ultimately diluted as described in Step 8. You will need approximately 6 μL of this 5x stock per assay.Choice of dNTP concentrations is discussed in Step 13.
-
7
Prepare 5 microcentrifuge tubes for time points 0 s, 30 s, 1 min, 1 min 30 s, and 2 min. Label the tubes appropriately and add 15 μL of quench buffer to each tube.
-
8Prepare the reaction stock solution in a single, appropriately labeled microcentrifuge tube by combining RNase/DNase-free water, 5x assay buffer, annealed primer/template, and DNA polymerase (Kf -) as shown in Table 1. Do not add the dNTP solution at this step.The setup shown in Table 1 is for only one dNTP concentration. For each additional component added to the mixture, ensure mixing by pipetting up and down several times. Once Kf - is added do not mix vigorously with a vortex device.
-
9
Add 4 μL of solution prepared in Step 8 to the tube designated for 0 s.
-
10Start the timer and in 15 s add 6 μL of the 4.0 μM 2AP-TP.Because of the 15 s delay, time points will be taken at 15s + the selected time point as displayed on the digital timer. For example, our 30 s interval time points were therefore taken at 45 s, 1 min 15 s, etc.This time delay can be used to support a higher throughput experimental setup. Depending on the time point intervals chosen, users can choose to run several reactions at the same time as 4 reactions can be started within 1 minute. For 2 min time points, 8 reactions can be done. For 30 s time points, only two reactions can be done. See the following reference for a similar experimental set up (O’Flaherty & Guengerich, 2014).
-
11At the desired time points, remove 5 μL from the reaction tube and dispense it in the correctly labeled microcentrifuge tube for the given timepoint containing the quench buffer. Take the microfuge tube and put it in the second heat block set at 75 °C for 20 min to heat inactivate the Kf −.Use precise timing when removing the aliquots as exact time values will be vital for accurate data analysis.Quench buffer containing xylene cyanol is commonly used, however, it fluoresces at the same wavelength as Cy5 so it should be avoided in this case.The quench buffer contains a dye to assist in gel electrophoresis experiments in the next section and thus combines a quenching step and dye loading step into one.
-
12Store the quenched samples at −20 °C until ready to analyze by gel electrophoreses in Steps 16–28.Steps 6–12 should be repeated 3 times for a total of 3 replicate experiments for incorporation at this concentration.
-
13To test the relationship of 2AP-TP incorporation efficiency with respect to concentration, repeat steps 6–12 as follows. Start by preparing 5x concentration stocks of 2AP-TP of 5, 10, 25, 37.5 50, 75, and 125 μM (to measure 2AP-TP kinetics at 1.0, 2.0, 5.0, 7.5, 10, 15, and 25 μM in addition to the 0.8 μM measurements performed in steps 6–12). Three replicates should be completed for each 2AP-TP concentration for incorporation opposite each template base (three replicates each for incorporation opposite template A, C, G, and T).In the end, 24 total experiments (3 replicates for each of the 8 concentrations of 2AP-TP) should be conducted for one primer-template duplex (i.e. primer vs TempA), resulting in a total of 96 total experiments to validate the incorporation kinetics of 2AP-TP against each of the four templates.No incorporation kinetic values have been reported previously for 4CIN-TP, therefore values available for 2AP-TP were used as an approximation. The KM of incorporation of 2AP-TP opposite a templating T by Klenow exo- was reported to be in the range of 0.77 to 4.6 μM using a DNA trap assay (Bloom et al., 1993). Incorporation, even at longer time points, was not seen at 2AP-TP concentrations below 0.8 μM; therefore, we predicted the KM to be closer to the 4.6 μM value and used values approximately 5-fold above and below the 4.6 μM concentration.
-
14
Resuspend fresh lyophilized pellet of 4CIN-TP in DNase/RNase-free water and calculate the concentration using its molar extinction coefficient at 300 nm, ε300 = 7790 M−1 cm−1 using Beer’s Law (Passow & Harki, 2018).
-
15
Repeat Steps 6–13 for the evaluation of 4CIN-TP as was done for 2AP-TP.
See Supplementary Table 1 for a summary of concentrations of 2AP-TP and 4CIN-TP used for steady state kinetics experiments and their calculated KM values.
Table 1.
Components for one reaction with a single nucleotide triphosphate concentration.
| Initial Concentration | Final concentration* | Volume (μL) | |
|---|---|---|---|
| Primer/Template | 500 nM | 100 nM | 7 |
| DNA polymerase | 5 nM | 1 nM | 7 |
| 5× assay buffer | 5X | 1X | 7 |
| Water | 7 | ||
| Total | 28 | ||
| Take out for zero point | −4 | ||
| Total | 24 | ||
| Initiate with 2AP-TP or 4CIN-TP | 6 | ||
| 30 | |||
The final concentration column in Table 1 is calculated assuming the nucleotide has been added as in Step 10, i.e. the final 30 μL volume solution has been mixed and the assay has started.
Gel Electrophoresis and Imaging analysis of Enzymatic Reactions
-
16Prepare denaturing 7 M urea, 20 % acrylamide gel in a small flask or plastic tube.For 40 mL of urea-acrylamide solution combine 16.8 g urea, 20 mL of 40% acryl-bisacrylamide, 19:1, 4 mL of 10x TBE, and 4 mL of water. Add a magnetic stir bar and stir until urea dissolves completely.
-
17
Clean 16.5 × 22 cm glass plates with acetone or ethanol, add sigmacote to one plate.
-
18Setup the glass plates and spacers for pouring the gel as per the manufacturer’s specifications.Prop the gel at an angle to help with pouring and polymerization.
-
19
To initiate the polymerization of the gel add 400 μL of 10% (w/v) APS (1 g in 10 mL of water) and 40 μL of TEMED to the acrylamide/urea solution while stirring.
-
20Then quickly pour the gel solution in between the glass plates, keeping the glass plate setup at an angle for easier pouring.Polymerization should be completed in 20 to 30 min. This can be monitored using the unused gel solution remaining in the flask.
-
21Once the gel is poured, insert the comb carefully without introducing bubbles.While the gel is polymerizing take out the DNA samples from Step 12 to allow to warm to room temperature.
-
22Once polymerization is complete, remove the gasket or tape seal and place the gel into the gel apparatus. Dilute 10X TBE (see Materials section) to 1x and add to the buffer reservoirs and attach the apparatus to a power supply. Pre-run the gel for 20 min to equilibrate the gel.For the 16.5 × 22 cm size gels a voltage of 500 V is sufficient. Depending on the gel system the comb may need to be removed prior to equilibration, if that is the case, clean out the wells with the 1x TBE using a syringe and needle prior to equilibration.
-
23
While pre-running the gel, heat the thawed DNA samples at 95 °C for 2 min in a heat block and then centrifuge the samples at 9000xg for 2 minutes.
-
24
After 20 minutes of pre-run, clean out the gel wells with a syringe and needle prior to loading the samples by syringing up and down multiple times.
-
25Load the DNA samples (3 μL; they already contain a gel dye from the quench buffer recipe), starting 4–5 wells from the end, and changing the tips for each sample.Be sure to load the samples gently as vigorous dispensing of the sample could result in loss of sample or dispersion, which can affect the analysis. Loading samples 4–5 wells in from the end of the gel is helpful to avoid any smiling of bands and make the gel easier to analyze as the gel tends to run hotter in the middle than towards the edges.
-
26Once loaded, continue the electrophoresis at 500 V for 3–3.5 hours to obtain single-nucleotide extension of the 17mer oligomer.In this case, the bromophenol blue from the quench buffer in the DNA sample should be at the very bottom of the gel nearly running off the bottom. The electrophoresis time depends on the size of the oligomers as well as the acrylamide/bis-acrylamide percentage.
-
27Disassemble the gel from the gel apparatus and slowly separate the glass plates to avoid disrupting the gel.Remove the smaller, “back” glass plate so that when the gel is placed on the imaging platform the samples will be in the same order as they were loaded.
-
28Place the gel on the gel imager platform and image using the Cy5 excitation wavelength at 633 or 647 nm and emission wavelength at 666 nm (default instrument settings may include a setup for “red fluorophores”).Flip the front glass plate with the gel still on the plate on top of the imaging platform to minimize handling of the gel itself.
Gel Analysis using Biorad Image Lab 6 and GraphPad Prism
-
29
Band quantification can be done using a variety of software programs that use density analysis, such as Biorad Image Lab. Create a box around a band of interest and copy the box to cover all bands of interest so that the boxes are all the same size.
-
30Export density measurements to spreadsheet to determine the ratio of extension product band to primer substrate band using Equation 1. Where P0 is the “extended” band for the zero timepoint, i.e. background. P1E is the extended band at one of the four non-zero timepoints and P1 is the primer band at non-zero timepoints (see Figure 19).
Equation 1: -
31
Plot the ratio of extension product to background vs. time for each dNTP concentration experiment (one round through Steps 6–12) and calculate the slope of each line. This represents the initial velocity, V0. Repeat this process for each dNTP concentration assayed.
-
32Using software, such as GraphPad Prism, perform nonlinear regression analysis. This can be done by plotting the calculated V0 slopes versus the concentration of dNTP at which the V0 was calculated (V0 vs [dNTP]). The resulting plot should be a hyperbola, as shown in Figure 20.In GraphPad Prism, select ‘Analysis’,’fit with nonlinear regression,’ and select within the equation list Enzyme Kinetics – velocity as a function of substrate, Michaelis-Menten. This will calculate the KM and Vmax values for the fitted curve.
-
33Once the Vmax has be obtained, the kcat (turnover number) for polymerase can be calculated using Vmax and total enzyme concentration ([ET]) as shown in Equation 2. The kcat represents the number of dNTP molecules added to the primer per unit time.
Equation 2: The enzyme concentration [ET] = 1 nM as per Table 1 for both 2AP-TP and 4CIN-TP measurements.
-
34
The entire protocol steps 1–33 must be repeated for at least 3 replicates of 2AP-TP incorporation opposite each template base. In this case, triplicate data for incorporation opposite TempA, TempC, TempG, and TempT. This was then repeated for incorporation of 4CIN-TP opposite each template base.
-
35
Final KM, Vmax values between triplicate measurements can then be averaged and the standard deviation measured. All final kinetic parameter calculations for 2AP-TP (our KM measurements align with results reported by Bloom et al., 1993) and 4CIN-TP are listed in Table 2.
Figure 19.
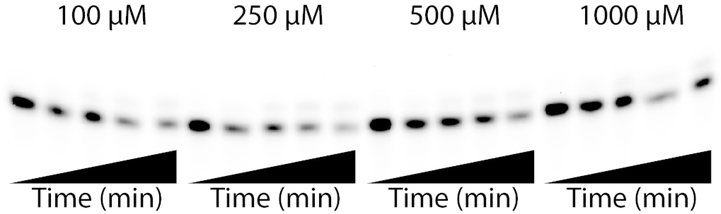
Gel (20% acrylamide, 7 M urea) image of a representative kinetics experiment with Klenow fragment exo-, 4CIN-TP (incorporating opposite template T), Cy5 labeled primer. The data show four increasing 4CIN-TP concentrations (right to left, 100, 250, 500, and 1000 μM) and time course for each concentration (5 points per concentration). Where the first (lane 1, left) is the reaction prior to initiation with 4CIN-TP followed by 4 time points of 30 s each for final timepoint at 2 min. The primer substrate band is the lower band, and the “extended” band is the higher band, indicating a larger DNA has been synthesized which incorporated 4CIN-TP.
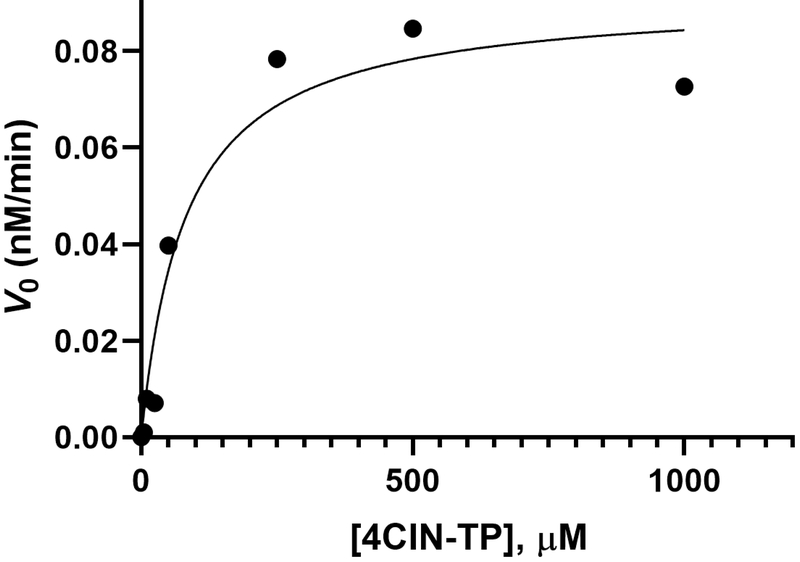
Figure 20.
Representative Michaelis-Menten analysis steady state kinetics of the incorporation of 4CIN-TP opposite TempT by Klenow fragment exo – (Kf -) using GraphPad Prism.
Table 2.
Steady State kinetics incorporation of 2APTP and 4CIN-TP by Kf− opposite the four natural DNA bases.
| Nucleotide/Template base | KM (μM) | Vmax (nM min−1) | kcat (min−1) | Catalytic efficiency (kcat/KM) (μM−1 min−1) |
|---|---|---|---|---|
| 2AP-TP | ||||
| vs TempA | 175.3 ± 27.4 | 0.0127 ± 0.0060 | 0.0127 | 7.23×10−5 |
| vs TempC | 195.3 ± 10.8 | 0.182 ± 0.053 | 0.182 | 9.44×10−4 |
| vs TempG | 98.3 ± 63.8 | 0.0167 ± 0.008 | 0.0167 | 1.82×10−4 |
| vs TempT | 3.67 ± 0.54 | 1.16 ± 0.59 | 1.16 | 3.19×10−1 |
| 4CIN-TP | ||||
| vs TempA | 42.1 ± 12.6 | 0.0952 ± 0.0391 | 0.0952 | 2.49×10−3 |
| vs TempC | 34.6 ± 8.9 | 0.0321 ± 0.0238 | 0.0321 | 9.47×10−4 |
| vs TempG | 40.7 ± 9.7 | 0.0185 ± 0.0151 | 0.0185 | 4.33×10−4 |
| vs TempT | 78.7 ± 15.4 | 0.0858 ± 0.0051 | 0.0858 | 1.11×10−3 |
These kinetics were done in triplicate at 25 °C with 1 nM Kf−, and 100 nM Cy5-labelled primer/template.
COMMENTARY
Background Information
Fluorescent nucleoside analogues can be classified by their general structure as chromophoric analogues, pteridines, extended nucleosides, nucleobase expansions, or isomorphic analogues (Sinkeldam et al., 2010). While each class has unique uses and properties, our laboratories are most interested in new isomorphic analogues, which are analogues that remain faithful to the size and shape of a naturally occurring nucleobase. In a recent publication, we described the synthesis and extensive characterization of 4CIN, which exhibits strong fluorescence not only as a monomer, but also in a DNA context (Passow & Harki, 2018). This molecule was subsequently used by Gai and coworkers in a proof of concept study showing that 4CIN could be used to report DNA-protein binding including the calculation of a binding affinity (Ahmed et al., 2019). This, in turn, exemplifies the utilities of 4CI-based nucleosides and nucleotides as biological reporters.
In describing the syntheses of the nucleosides and nucleotides in Basic Protocols 1–3, we often used established nucleoside chemistry. In the synthesis of 4CIN, previously reported synthesis procedures for indole-containing 2ʹ-deoxyribonucleosides were employed and we found these methods to be quite robust (Girgis et al., 1988; Struntz & Harki, 2016). Ribonucleoside syntheses are similarly well established, where typical methods utilize either a Lewis acid-mediated (TiCl4) chlorination of 1-O-acetyl-2,3,5-tri-O-benzoyl-β-D-ribofuranose (Harki et al., 2007) or utilize intermediates 5 (Kane & Mann, 1984; Stewart & Williams, 1985) and 6 (Rosemeyer & Seela, 1988; Ugarkar et al., 2000; Wilcox & Otoski, 1986). We found that displacement of 1-chloro-2,3,5-tri-O-benzoyl-β-D-ribofuranose with the sodium salt of 4CI yielded what is likely a trapped amide acetal intermediate indicating a lack of nucleophilicity and thus did not produce the desired coupling product (see Kobayashi et al., 2018 and Sokolova et al., 1980 for examples of such structures). We therefore chose to use the 5/6 sequence.
After considering extensive reviews on nucleoside triphosphate synthesis (Burgess & Cook, 2000; Roy et al., 2016), we decided to forego the use of the popular “one pot, three step” chemistry (Ludwig, 1981; Yoshikawa et al., 1967) and instead utilized a more recent methodology that employs a phosphonate precursor before elaborating to the full triphosphate (Sun et al., 2008), which we found to be easier to purify and would also allow for simple adaption to synthesize mono- and diphosphate derivatives if desired.
Polymerase extension assays are a common way to evaluate the suitability of novel nucleoside triphosphates for biological applications. These assays have been performed previously with similarly fluorescent molecules like 2-aminopurine-2ʹ-deoxyribonucleoside-5ʹ-triphosphate (2AP-TP) (Lee & Berdis, 2006; Mcclure & Scheit, 1973) and monitoring reaction kinetics is commonly used to evaluate the proficiency and selectivity of the process (Gahlon & Sturla, 2019). 2-aminopurine is a well-established isomorphic nucleoside fluorophore (Ward et al., 1969) and was therefore used as a benchmark comparison in our previous study (Passow & Harki, 2018) and this current protocol. Taken together, these studies provide important experimental details concerning the synthesis and utilization of 4-cyanoindole-nucleoside-based reagents, which is expected to facilitate their utilization in a variety of new biomedical research applications.
2AP-TP has been shown to be efficiently incorporated opposite a template T (Sowers et al., 1986; Watanabe & Goodman, 1981) and this is reflected in the data shown in Table 2, with KM values 33–66-fold weaker for template bases A, C, and G. Here, we show with polymerase primer extension assays that 4CIN-TP has a similar KM for all template bases (Table 2). This correlates well with the previous finding that 4CIN hybridizes non-discriminately with all bases in a similar thermal range in duplex DNA (Passow & Harki, 2018). Therefore, we conclude that 4CIN-TP is a fluorescent nucleotide that can be enzymatically incorporated into DNA across all of the natural templating nucleobases with similar efficiencies.
Critical Parameters & Troubleshooting
There are three transformations that require particular attention for success. For the synthesis of 5, the di-TBS protected byproduct must be separated from product 5. This byproduct is less polar and elutes first by SiO2 chromatography. For the preparation of 7, the molar ratio of 6 to 4-cyanindole is critical since excess equivalents of 4-cyanoindole improves yield and the unreacted 4-cyanindole can be recovered during chromatography. Lastly, for the synthesis of 4CIN-TP, controlling moisture in critical since TBAPP and TMSCl are both water sensitive and vital to the success of the reaction. A major side-product observed in this reaction sequence is 4-cyanoindole-2ʹ-deoxyribonucleoside-5ʹ-monophosphate that results from a failed substitution by the pyrophosphate reagent after oxidation by iodine. If desired, this material can be isolated. The multiple chromatography steps required for the preparation of pure 4CIN-TP (purity is more essential than yield for our applications) contributes to the low overall yield.
Repeated freezing and thawing of the polymerase can affect the activity over time. It is best to aliquot the polymerase into smaller aliquots upon receiving to avoid several freeze-thaw cycles.
Most intermediates synthesized in this protocol are stable and can be stored at room temperature, with a few exceptions as mentioned in the protocol. Nucleoside phosphoramidite 4 should be stored between −20°C - 0°C. Triphosphates 4CIN-TP and 2AP-TP should be stored at −20°C or colder.
Understanding Results
Basic Protocol 1 has an overall yield of 35% over 4 steps. We have scaled Protocol 1 to produce almost 2 g of 4 in other work.
Basic Protocol 2 has an overall yield of 24% yield over 4 steps. Although users may wish to start with smaller test reactions, the synthesis of 5 is quite amenable to multi-gram preparations and that material can be stored for future applications. Overall, 1 g of 5 along with ~500 mg of 4-cyanoindole can produce hundreds of milligrams of 4CINr.
Basic Protocol 3 has an overall yield of 46% up to the preparation of 11 (assuming 4CIN has already been synthesized). Intermediate 11 can be prepared on the hundreds of milligrams scale. The final transformations and extensive purifications required for the production of 4CIN-TP were performed in a 5% overall yield (from 11). Since only small amounts of 4CIN-TP are needed for enzymatic assays, significant yield optimization efforts were not undertaken. 4CIN-TP is stable upon storage at −20 °C.
Time Considerations
With a reasonable organic chemistry skill set a typical user can complete each step of the synthesis within 1–2 days including purification and characterization. Exceptions include steps that require extensive drying, which includes the synthesis of phosphoramidite 4, phosphonate 11, and 4CIN-TP. The synthesis and purification of 4CIN-TP in particular can take over a week. In our case, each purification step in the triphosphate synthesis step requires approximately 2 days to complete, including the chromatography purification, fraction collecting, lyophilizing, and analysis.
For the steady state kinetics assays, with familiarity in performing enzymatic assays, especially with polymerases, a user could accomplish one experiment in two days. The limiting time factor is any adjustments that may need to be made to the nucleotide triphosphate concentrations to obtain a Michaelis-Menten curve. Depending on what can be found in literature, the determination of nucleotide triphosphate concentrations can take several trial-and-error attempts to find a workable range.
Supplementary Material
Statement of Significance.
Fluorescent nucleosides are utilized in a variety of research applications and have yielded critical insights into oligonucleotide and oligonucleotide-protein interactions. These utilities have spurred the continual development of structurally and spectroscopically diverse analogues. We recently developed a novel fluorescent nucleoside, 4-cyanoindole-2ʹ-deoxyribonucleoside (4CIN), which exhibits a high quantum yield as both the nucleoside and as a component of DNA. Here, we expand the utility of 4CIN by reporting syntheses of the ribonucleoside analogue, 4CINr, and 5’-triphosphate of 4CIN, 4CIN-TP, and demonstrate the first enzymatic incorporation of 4CIN-TP into DNA by a polymerase. These new reagents and protocols are expected to facilitate the utilization of 4CIN-based fluorescent nucleotides in a variety of applications.
ACKNOWLEDGEMENT
This work was supported by the National Institutes of Health (R01-GM110129 to DAH), the Swiss National Science Foundation (310030_185020), and a 3M Graduate Research Fellowship to KTP. Mass spectrometry was performed at the Analytical Biochemistry Core Facility of the University of Minnesota Masonic Cancer Center, which is supported by the NIH (P30-CA77598)
LITERATURE CITED
- Ahmed IA, Acharyya A, Eng CM, Rodgers JM, DeGrado WF, Jo H, and Gai F 2019. 4-Cyanoindole-2’-deoxyribonucleoside as a Dual Fluorescence and Infrared Probe of DNA Structure and Dynamics. Molecules 24:602. doi: 10.3390/molecules24030602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armarego WLF, and Chai CLL 2009. Purification of Laboratory Chemicals, 6th Edition. Purification of Laboratory Chemicals, 6th Edition 1–743. [Google Scholar]
- Bloom LB, Otto MR, Beechem JM, and Goodman MF 1993. Influence of 5’-nearest neighbors on the insertion kinetics of the fluorescent nucleotide analog 2-aminopurine by Klenow fragment. Biochemistry 32:11247–11258. doi: 10.1021/bi00092a039. [DOI] [PubMed] [Google Scholar]
- Burgess K, and Cook D 2000. Syntheses of nucleoside triphosphates. Chem. Rev 100:2047–2060. doi: 10.1021/cr990045m. [DOI] [PubMed] [Google Scholar]
- Datta K, Wowor AJ, Richard AJ, and LiCata VJ 2006. Temperature dependence and thermodynamics of Klenow polymerase binding to primed-template DNA. Biophys J. 90:1739–1751. doi: 10.1529/biophysj.105.071837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahlon HL, and Sturla SJ 2019. Determining Steady-State Kinetics of DNA Polymerase Nucleotide Incorporation. Methods Mol. Biol 1973:299–311. doi: 10.1007/978-1-4939-9216-4_19. [DOI] [PubMed] [Google Scholar]
- Girgis NS, Cottam HB, and Robins RK 1988. Synthesis of 2’-Deoxyribofuranosyl Indole Nucleosides Related to the Antibiotics Sf-2140 and Neosidomycin. J. Heterocycl. Chem 25:361–366. doi: 10.1002/jhet.5570250202. [DOI] [Google Scholar]
- Harki DA, Graci JD, Edathil JP, Castro C, Cameron CE, and Peterson BR 2007. Synthesis of a universal 5-nitroindole ribonucleotide and incorporation into RNA by a viral RNA-dependent RNA polymerase. ChemBioChem 8:1359–1362. doi: 10.1002/cbic.200700160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harki DA, Graci JD, Korneeva VS, Ghosh SK, Hong Z, Cameron CE, and Peterson BR 2002. Synthesis and antiviral evaluation of a mutagenic and non-hydrogen bonding ribonucleoside analogue: 1-beta-D-Ribofuranosyl-3-nitropyrrole. Biochemistry 41:9026–9033. doi: 10.1021/bi026120w. [DOI] [PubMed] [Google Scholar]
- Joyce CM, Potapova O, Delucia AM, Huang X, Basu VP, and Grindley ND 2008. Fingers-closing and other rapid conformational changes in DNA polymerase I (Klenow fragment) and their role in nucleotide selectivity. Biochemistry 47:6103–6116. doi: 10.1021/bi7021848. [DOI] [PubMed] [Google Scholar]
- Kane PD, and Mann J 1984. The Preparation and Utility of Ethyl 2-(5’-O-Tert-Butyldimethylsilyl-2’,3’-O-Isopropylidene-Beta-D-Ribofuranosyl)Propenoate as a Key Intermediate for C-Nucleoside Synthesis. J. Chem. Soc. Perkin Trans 1 657–660. doi: 10.1039/p19840000657. [DOI] [Google Scholar]
- Kilin V, Gavvala K, Barthes NP, Michel BY, Shin D, Boudier C, … Mely Y 2017. Dynamics of Methylated Cytosine Flipping by UHRF1. J. Am. Chem. Soc 139:2520–2528. doi: 10.1021/jacs.7b00154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Nakatsuji Y, Li S, Tsuzuki S, and Takemoto Y 2018. Direct N-Glycofunctionalization of Amides with Glycosyl Trichloroacetimidate by Thiourea/Halogen Bond Donor Co-Catalysis. Angew. Chem. Int. Ed 57:3646–3650. doi: 10.1002/anie.201712726. [DOI] [PubMed] [Google Scholar]
- Lee I, and Berdis A 2006. Fluorescent analysis of translesion DNA synthesis by using a novel, non-natural nucleotide analogue. ChemBioChem 7:1990–1997. doi: 10.1002/cbic.200600128. [DOI] [PubMed] [Google Scholar]
- Ludwig J 1981. A new route to nucleoside 5’-triphosphates. Acta Biochim. Biophys. Acad. Sci. Hung 16:131–133. [PubMed] [Google Scholar]
- Mcclure WR, and Scheit KH 1973. Enzyme Kinetic Parameters of Fluorescent Atp Analog, 2-Aminopurine Triphosphate. Febs Letters 32:267–269. doi: 10.1016/0014-5793(73)80849-X. [DOI] [PubMed] [Google Scholar]
- O’Flaherty DK, and Guengerich FP 2014. Steady-state kinetic analysis of DNA polymerase single-nucleotide incorporation products. Curr. Protoc. Nucleic Acid Chem 59:7.21.1–7.21.13. doi: 10.1002/0471142700.nc0721s59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passow KT, and Harki DA 2018. 4-Cyanoindole-2’-deoxyribonucleoside (4CIN): A Universal Fluorescent Nucleoside Analogue. Org. Lett 20:4310–4313. doi: 10.1021/acs.orglett.8b01746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosemeyer H, and Seela F 1988. Stereoselective Synthesis of Pyrrolo[2,3-D]Pyrimidine Alpha-D-Ribonucleosides and Beta-D-Ribonucleosides from Anomerically Pure D-Ribofuranosyl Chlorides - Solid-Liquid Phase-Transfer Glycosylation and N-15-Nmr Spectra. Helvetica Chimica Acta 71:1573–1585. doi: 10.1002/hlca.19880710623. [DOI] [Google Scholar]
- Roy B, Depaix A, Perigaud C, and Peyrottes S 2016. Recent Trends in Nucleotide Synthesis. Chem. Rev 116:7854–7897. doi: 10.1021/acs.chemrev.6b00174. [DOI] [PubMed] [Google Scholar]
- Sinkeldam RW, Greco NJ, and Tor Y 2010. Fluorescent analogs of biomolecular building blocks: design, properties, and applications. Chem. Rev 110:2579–2619. doi: 10.1021/cr900301e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolova TN, Shevchenko VE, and Preobrazhenskaya MN 1980. Interaction of Indoles with Glycosyl Halides in the Presence of Silver-Oxide. Carbohydrate Research 83:249–261. doi: 10.1016/S0008-6215(00)84538-3. [DOI] [Google Scholar]
- Sowers LC, Fazakerley GV, Eritja R, Kaplan BE, and Goodman MF 1986. Base pairing and mutagenesis: observation of a protonated base pair between 2-aminopurine and cytosine in an oligonucleotide by proton NMR. Proc. Natl. Acad. Sci. U.S.A 83:5434–5438. doi: 10.1073/pnas.83.15.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart AO, and Williams RM 1985. C-Glycosidation of Pyridyl Thioglycosides. J. Am. Chem. Soc 107:4289–4296. doi: 10.1021/ja00300a036. [DOI] [Google Scholar]
- Struntz NB, and Harki DA 2016. Catch and Release DNA Decoys: Capture and Photochemical Dissociation of NF-kappa B Transcription Factors. ACS Chem. Biol 11:1631–1638. doi: 10.1021/acschembio.6b00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Edathil JP, Wu R, Smidansky ED, Cameron CE, and Peterson BR 2008. One-pot synthesis of nucleoside 5’-triphosphates from nucleoside 5’-H-phosphonates. Org. Lett 10:1703–1706. doi: 10.1021/ol8003029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugarkar BG, Castellino AJ, DaRe JM, Kopcho JJ, Wiesner JB, Schanzer JM, and Erion MD 2000. Adenosine kinase inhibitors. 2. Synthesis, enzyme inhibition, and antiseizure activity of diaryltubercidin analogues. J. Med. Chem 43:2894–2905. doi: 10.1021/jm0000259. [DOI] [PubMed] [Google Scholar]
- Ward DC, Reich E, and Stryer L 1969. Fluorescence studies of nucleotides and polynucleotides. I. Formycin, 2-aminopurine riboside, 2,6-diaminopurine riboside, and their derivatives. J. Biol. Chem 244:1228–1237. [PubMed] [Google Scholar]
- Watanabe SM, and Goodman MF 1981. On the molecular basis of transition mutations: frequencies of forming 2-aminopurine.cytosine and adenine.cytosine base mispairs in vitro. Proc. Natl. Acad. Sci. U.S.A 78:2864–2868. doi: 10.1073/pnas.78.5.2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox CS, and Otoski RM 1986. Stereoselective Preparations of Ribofuranosyl Chlorides and Ribofuranosyl Acetates - Solvent Effects and Stereoselectivity in the Reaction of Ribofuranosyl Acetates with Trimethylallylsilane. Tetrahedron Lett. 27:1011–1014. doi: 10.1016/S0040-4039(86)80035-1. [DOI] [Google Scholar]
- Wilson DL, Beharry AA, Srivastava A, O’Connor TR, and Kool ET 2018. Fluorescence Probes for ALKBH2 Allow the Measurement of DNA Alkylation Repair and Drug Resistance Responses. Angew. Chem. Int. Ed 57:12896–12900. doi: 10.1002/anie.201807593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Chan KM, and Kool ET 2017. Fluorescent nucleobases as tools for studying DNA and RNA. Nat. Chem 9:1043–1055. doi: 10.1038/nchem.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa M, Kato T, and Takenishi T 1967. A novel method for phosphorylation of nucleosides to 5’-nucleotides. Tetrahedron Lett. 50:5065–5068. doi: 10.1016/S0040-4039(01)89915-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.