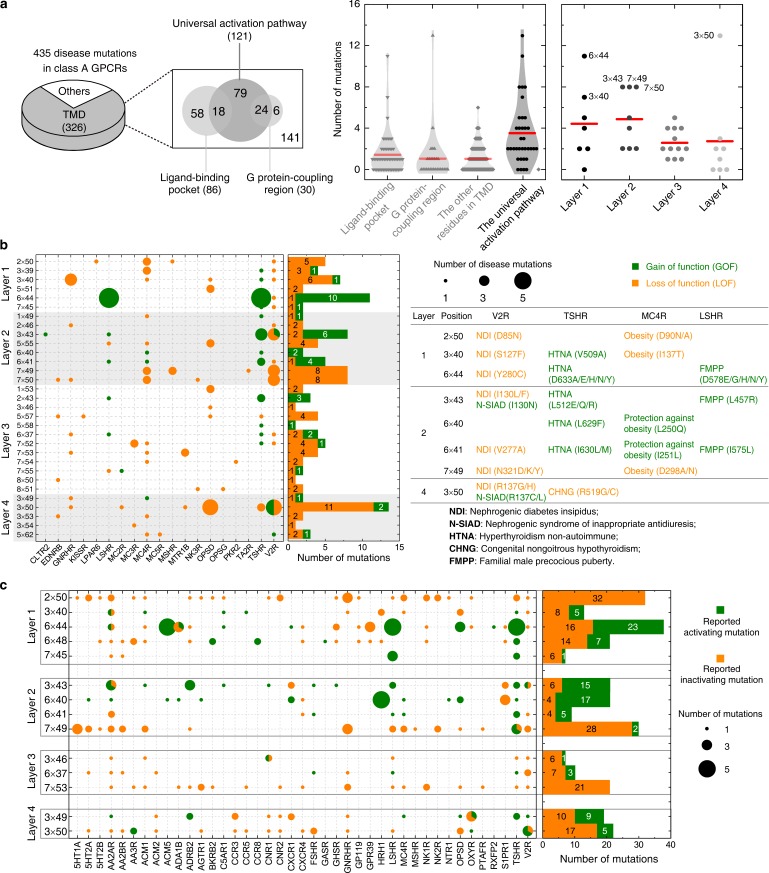
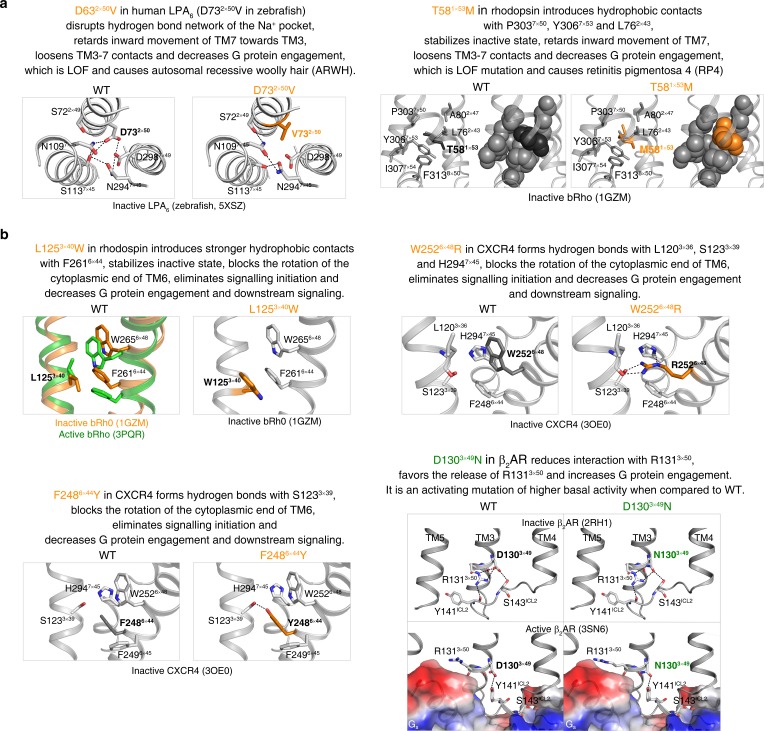
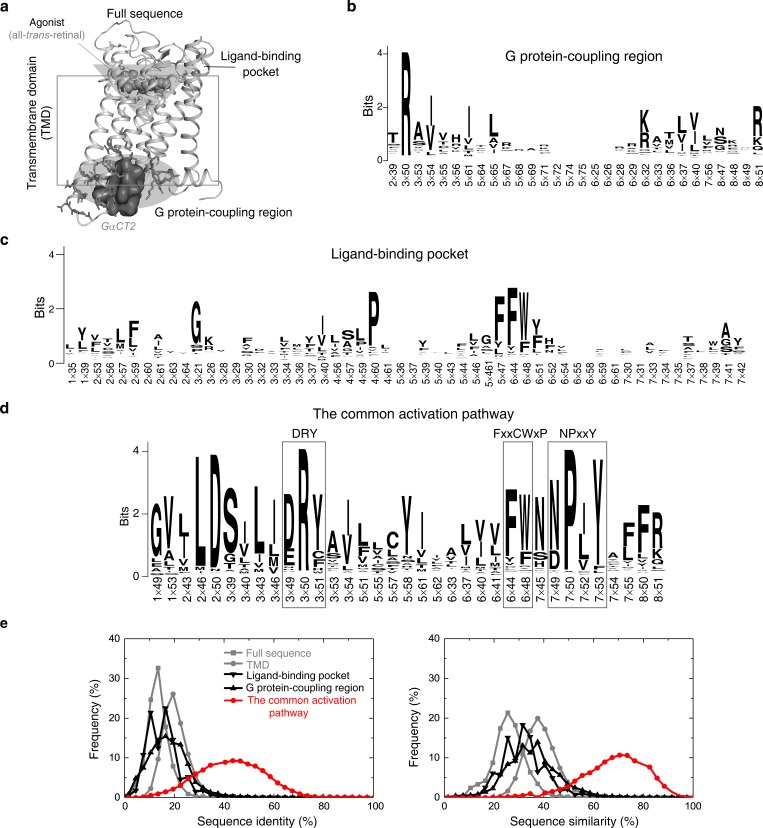
Figure 7. Importance of the common activation pathway in pathophysiological and biological contexts.
(a) Comparison of disease-associated mutations in the common activation pathway (further decomposed into layers 1–4), ligand-binding pocket, G-protein-coupling region and other regions. Red line denotes the mean value. (b) Mapping of disease-associated mutations in class A GPCRs to the common activation pathway. (c) Key roles of the residues constituting the common activation pathway have been reported in numerous experimental studies on class A GPCRs. Two hundred seventy two (272) CAMs/CIMs from 41 receptors were mined from the literature for the 14 hub residues (i.e. residues that have more than one edges in the pathway).