Abstract
Background
Bacterial resistance to antibiotics has become a major public health concern. This study aimed to determine the resistance mechanisms to carbapenem in clinical isolates of Pseudomonas aeruginosa.
Methods
A total of 62 clinical isolates of carbapenem-resistant P. aeruginosa (CRPA) were collected from 2015 to 2017. Imipenem (IPM)–EDTA disk synergy test was used to screen strains that produced metallo-β-lactamase. In addition, the genes for outer membrane protein OprD2, metallo-β-lactamase and mexR gene were amplified and sequenced. Expression of mexA was detected by real-time PCR.
Results
Disk synergy test showed that 51.6% (32/62) of the strains were positive for metallo-β-lactamase. PCR showed that 84.4% of the strains were SIM-positive (27/32), 15.6% of the strains were IMP-positive (5/32), and 12.5% of the strains were VIM-positive (4/32). SPM-positive and GIM-positive strains were not detected. In addition, 5 of the 62 strains had small deletions and/or point mutations in OprD2. Three strains had a high expression of mexA, while eight strains were positive for the regulatory gene mexR with no mutations detected by DNA sequencing.
Conclusion
Expression of metallo-β-lactamase is the main resistance mechanism of P. aeruginosa to carbapenem. Mutations in OprD2 and/or the overexpression of efflux pump MexAB-OprM may contribute to P. aeruginosa resistance to carbapenem.
Keywords: Pseudomonas aeruginosa, carbapenem, metallo-β-lactamase, outer membrane protein OprD2, efflux pump MexAB-OprM
Introduction
Bacterial resistance has attracted worldwide attention. ESKAPE bacteria as defined by the American Society of Infectious Diseases indicate a class of bacteria including Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species, which can evade antimicrobial activity and are highly resistant to antibiotics.1
P. aeruginosa is a common gram-negative bacterium and is a conditional pathogen that causes severe nosocomial-acquired infections. Carbapenem-resistant Pseudomonas isolates have been occasionally reported in severely ill or immunocompromised patients. The majority of P. aeruginosa infections have been observed in neurosurgery intensive care units (NICU) and respiratory medicine departments.2 P. aeruginosa is often isolated from bronchiectasis or cystic fibrosis (CF) patients.
Carbapenem is a type of β-lactam antibiotic with a broad antimicrobial spectrum and strong antimicrobial activity and is widely used for the treatment of P. aeruginosa infections. However, due to antibiotic overuse, carbapenem-resistant P. aeruginosa (CRPA) strains emerge and cause severe and fatal infections and increased cost of therapy. An epidemiological report showed that South America, Russia and South-West Asia are the areas with the highest incidences of CRPA infections. In Russia, the resistance to carbapenem antibiotics is as high as 75.3%.3 Therefore, it is urgent to elucidate the mechanism of resistance to carbapenem in order to develop therapeutics to combat drug resistance strains.
Resistance to carbapenem in P. aeruginosa strains is often induced by the production of metallo‑β‑lactamases (MBLs), loss of the outer membrane protein OprD2 or the overexpression of efflux pumps, particularly MexAB-OprM. The majority of MBLs‑encoding genes are located in plasmids and are spread to other strains, ultimately leading to antibiotic resistance. Various types of MBLs such as IMP, VIM, GIM, SPM, SIM, NDM-1, FIM-1 and HMB-1 have been reported in P. aeruginosa. MBLs can hydrolyze the majority of β-lactam antibiotics. Metallo-beta-lactamase-producing P. aeruginosa (MPPA) is resistant to a variety of antibiotics and causes serious nosocomial infection outbreaks.4
OprD2 is an outer membrane pore protein that plays a major role in antibiotic permeability in P. aeruginosa. Imipenem (IPM) binds to a specific site on OprD2. Several studies have demonstrated that absence or low expression of OprD2 affected the permeability of P. aeruginosa to carbapenem, thereby reducing the minimum inhibitory concentration of carbapenem. Moreover, the insertion of OprD2 sequence (IS) elements can lead to the inactivation of OprD2 gene.5 In addition, the overexpression of efflux pumps is closely associated with the resistance of P. aeruginosa to carbapenems.6–8 The multi-drug efflux pump MexAB-OprM was the first efflux pump discovered in P. aeruginosa, composed of three-component membrane fusion protein MexA, transporter MexB and outer membrane protein OprM.9 It was reported that MexAB-OprM genes were overexpressed in 62% of CRPA strains.10 On the other hand, mexR, nalD and nalC negatively regulate the expression of MexAB-OprM efflux pump, and mutations in these genes can lead to increased expression of MexAB-OprM.
In this study, we aimed to determine the resistance mechanisms to carbapenem in clinical isolates of P. aeruginosa. First, phenotypic assay for MBLs was performed using the imipenem (IPM)–EDTA disk method. Next, blaIMP, blaVIM, blaSIM, blaGIM and blaSPM genes were amplified by PCR. PAβN (MC207110), an efflux pump inhibitor, was used to screen the phenotypes of efflux pumps. The relative expression of mexA and MexAB-OprM was detected by real-time PCR. Mutations in mexR were detected by sequencing after PCR amplification.
Materials and Methods
Bacterial Isolates
This study was approved by the Ethics Committee of the First Affiliated Hospital of Baotou Medical College and carried out in accordance with the principles of the Declaration of Helsinki. All patients provided written informed consent for their data to be used in the study and their data kept confidential. A total of 62 non-duplicate carbapenem‑resistant clinical isolates of P. aeruginosa were collected from January 2015 to December 2017. Isolates were obtained from different clinical specimens, including sputum, urine, and wounds, from patients admitted to respiratory, neurology, and neurosurgery department of the First Affiliated Hospital of Baotou Medical College. P. aeruginosa ATCC27853 strain was used as positive quality control.
Antibiotic Susceptibility Tests
Antibiotic sensitivity of the isolates was examined by Kirby Bauer’s Method. Antibiotics used in this study were as follows: ceftazidime (30 µg) (CAZ), amikacin (30 µg) (AMK), piperacillin/tazobactam (100/10 µg) (TZP), gentamicin (10 µg) (GEN), imipenem (10 µg) (IPM), cefepime (30 µg) (FEP), meropenem (10 µg) (MEM), ciprofloxacin (5 µg) (CIP), tobramycin (10 µg) (TOB), piperacillin (100 µg) (PIP) and levofloxacin (5µg) (LVX). All antibiotic disks were purchased from OXOID (UK). Results were interpreted based on the 2013 CLSI guidelines.
IPM-EDTA Disk Method
Experimental strains were diluted to 0.5 M bacterial suspension using sterilized saline water and then evenly coated onto Müller-Hinton (M-H) agar plates (OXOID, UK). Two 10 µg IPM drug sensitivity test papers (OXOID, UK) were pasted. One of the test papers had 10 µL EDTA at a concentration of 0.5 mmol/l (pH 8.0). The M-H plates with pasted drug sensitivity test papers were incubated at 35°C for 18 h. Next, the diameter of the inhibition zone of IPM drug sensitivity test paper with EDTA was compared to that with IPM.
PCR Amplification of Genes for MBLs and OprD2
DNA was extracted using the DNA extraction kit (Tiangen, Beijing, China) according to the manufacturer’s instructions. The primers are shown in Table 1. The PCR mixture consisted of 1 µL of each primer, 12.5 μL 2×Tap PCR Master Mix, 1 μL of DNA template and 9.5 ul double-distilled water (total volume of 25 μL). The cycling conditions were as follows: initial denaturation at 94°C for 5 min followed by 30 cycles of denaturation at 94°C for 60 sec, annealing at 53–60°C (45 sec), extension at 72°C (1 min) and a final extension at 72°C for 7 min. PCR products were electrophoresed on a 2.0% agarose gel. PCR bands were imaged under an ultraviolet transilluminator.
Table 1.
Primers Used in This Study
| Gene | Primer Sequence (5ʹ→3ʹ) | Amplicon Size (bp) | Annealing Temperature (°C) |
|---|---|---|---|
| SIM | F- TACAAGGGATTCGGCATCG R-TAATGGCCTGTTCCCATGTG |
741 | 60 |
| IMP | F-GAAGGCGTTTATGTTCATAC R-GTATGTTTCAAGAGTGATGC |
587 | 53 |
| GIM | F- CCTGTAGCGTTGCCAGCTTTA R- CAGCCCAAGAGCTAATTGAGG |
562 | 55 |
| VIM | F-GTTTGGTCGCATATCGCAAC R-AATGCGCAGCACCAGGATAG |
382 | 55 |
| SPM | F- CTGCTTGGATTCATGGGCGC R- CCTTTTCCGCGACCTTGATCG |
784 | 55 |
| OprD2 | F-CGCCGACAAGAAGAACTAGC R-GTCGATTACAGGATCGACAG |
1332 | 55 |
Phenotypic Detection of Efflux Pump Activity
Efflux pump activity of the strains was phenotypically detected using meropenem (MEM, 10 μg) with or without PaβN (MC207110). Minimum inhibitory concentration (MIC) assay was performed using meropenem alone or in combination with PaβN at 20 mg/l. An efflux pump overexpressed phenotype was defined as any strain exhibiting at least a four-fold decrease in MIC when tested in the presence of PAβN.
Real-Time PCR
Total RNA was extracted using the RNA extraction kit (Tiangen, Beijing, China), and cDNA was synthesized according to the manufacturer’s instructions (Tiangen, Beijing, China). Real-time PCR was used to determine the expression level of mexA using the SYBR green PCR master mix, primers and template cDNA. Relative expression levels of mexA were calculated using the 2−ΔΔCT method and normalized to rpsL gene which was used as an internal control. The standard P. aeruginosa ATCC27853 strain was used as control.
Sequencing of mexR Gene
PCR was performed for all isolates identified as MexAB-OprM overproducing strains. DNA was extracted using the DNA extraction kit (Tiangen). Primers for mexR were F-ACCAATGAACTACCCCGTGA and R-AATGTTCTTAAATATCCTCAA. PCR was performed with the following conditions: denaturation at 94°C for 5 min followed by 30 cycles of denaturation at 94°C for 60 sec, annealing at 58°C (45 sec), extension at 72°C (1 min) and a final extension at 72°C for 7 min. PCR products were electrophoresed on a 2.0% agarose gel, then imaged using an ultraviolet transilluminator. The purified PCR products were then sequenced and analyzed using the Basic Local Alignment Search Tool (BLAST) online program from the National Center for Biotechnology Information.
Statistical Analysis
Data were analyzed using SPSS 21.0 statistical software for Windows. Differences between proportions were analyzed using the χ2 test. p < 0.05 was considered significantly different.
Results
Basic Information and Drug Resistance Patterns of the Isolates
A total of 62 isolates of CRPA were collected from different clinical specimens including sputum (52 isolates; 83.9%), urine (2 isolates; 3.2%), wounds (5 isolates; 8.1%), and drainage (3 isolates; 4.8%) (Table 2). The isolates were obtained from patients admitted to different departments as shown in Table 3.
Table 2.
Distribution of CRPA Isolates in Clinical Specimens
| Sources of Specimen | Strains | Ratio (%) |
|---|---|---|
| Sputum | 52 | 83.9 |
| Wound | 5 | 8.1 |
| Drainage | 3 | 4.8 |
| Urine | 2 | 3.2 |
| Total | 62 | 100 |
Table 3.
Distribution of CRPA Isolates from Various Clinical Departments
| Department | Strains | Ratio (%) |
|---|---|---|
| Respiratory | 22 | 35.6 |
| Neurosurgery | 13 | 21.0 |
| Neurology | 6 | 9.7 |
| Orthopaedics | 3 | 4.8 |
| General surgery | 3 | 4.8 |
| Intensive care unit | 3 | 4.8 |
| Health Centers | 3 | 4.8 |
| Urology | 2 | 3.2 |
| Others | 7 | 11.3 |
| Total | 62 | 100 |
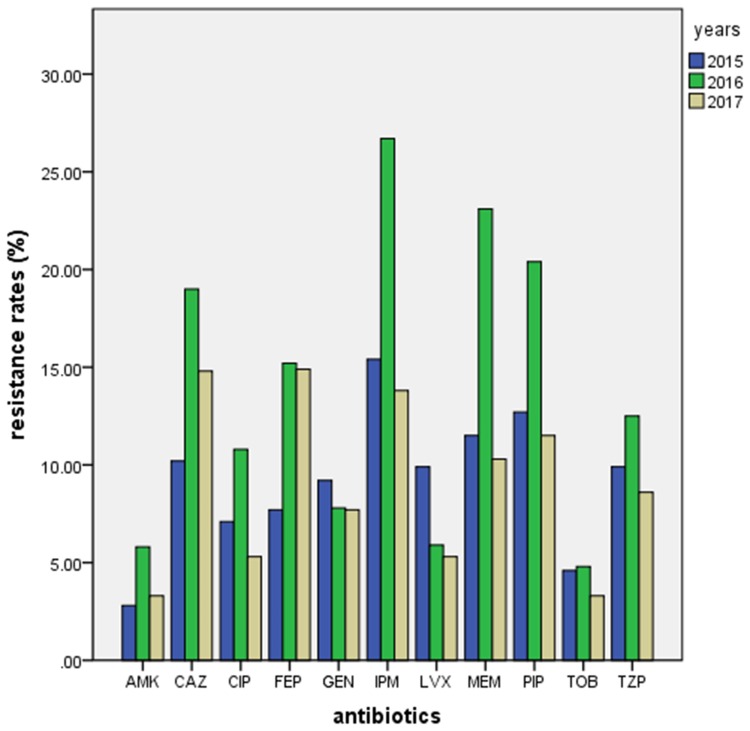
Antimicrobial tests showed high resistance to carbapenem of the isolates obtained at different time periods. The resistance rate for imipenem was 15.4%, 26.7% and 13.8% in 2015, 2016 and 2017, respectively, and the resistance rate for meropenem was 11.5%, 23.1% and 10.3% in 2015, 2016 and 2017, respectively. The resistance rates for imipenem and meropenem in 2016 were significantly higher than in 2015 and 2017 (p < 0.05). In addition, the resistance rates for amikacin and tobramycin were very low for all 3 years (Table 4, Figure 1).
Table 4.
Drug Resistance Rates of P. aeruginosa Isolates from 2015 to 2017 (%)
| Antimicrobial Agents | 2015 (n=143) | 2016 (n=105) | 2017 (n=95) |
|---|---|---|---|
| Imipenem (IPM) (MIC: 8 µg/mL) | 15.4* | 26.7 | 13.8* |
| Meropenem (MEM) (MIC: 8 µg/mL) | 11.5* | 23.1 | 10.3* |
| Piperacillin (PIP) (MIC: 128 µg/mL) | 12.7 | 20.4 | 11.5 |
| Ciprofloxacin (CIP) (MIC: 4 µg/mL) | 7.1 | 10.8 | 5.3 |
| Levofloxacin (LVX) (MIC: 8 µg/mL) | 9.9 | 5.9 | 5.3 |
| Gentamicin (GEN) (MIC: 16 µg/mL) | 9.2 | 7.8 | 7.7 |
| Ceftazidime (CAZ) (MIC: 32 µg/mL) | 10.2 | 19.0 | 14.8 |
| Piperacillin/Tazobactam (TZP) (MIC: 128 µg/mL) | 9.9 | 12.5 | 8.6 |
| Cefepime (FEP) (MIC: 32 µg/mL) | 7.7 | 15.2 | 14.9 |
| Tobramycin (TOB) (MIC: 4 µg/mL) | 4.6 | 4.8 | 3.3 |
| Amikacin (AMK) (MIC: 64 µg/mL) | 2.8 | 5.8 | 3.3 |
Notes: Differences between proportions were analyzed using the χ2 test. *p<0.05 compared to resistance rates in 2016.
Figure 1.
Drug resistance rates of P. aeruginosa isolates from 2015 to 2017.
We found that 29 isolates (46.8%) had multiple drug resistance (MDR) patterns. Among them, four strains were resistant to three antibiotics concurrently, 9 strains were resistant to four antibiotics concurrently, three strains were resistant to five antibiotics concurrently, six strains were resistant to six antibiotics concurrently, and four strains were resistant to seven antibiotics concurrently.
Detection of Drug Resistance Related Genes in the Isolates
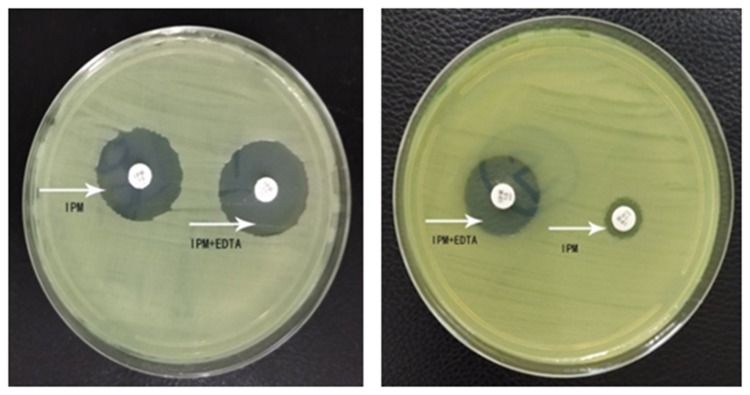
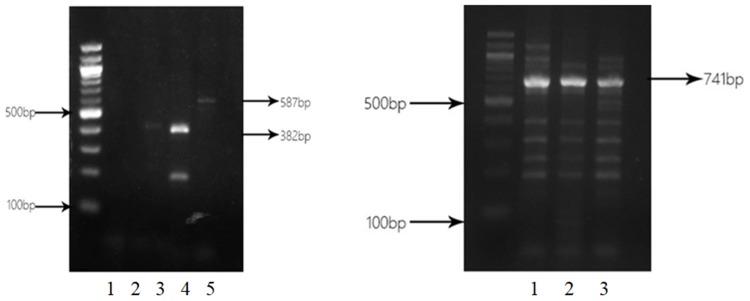
Among 62 isolates of CRPA, 32 (51.6%) isolates were identified to produce MBL. Bla SIM (27 strains, 84.4%) was the most common, followed by bla IMP (5 strains, 15.6%) and bla VIM (4 strains, 12.5%). SPM-positive and GIM-positive strains were not detected. Two strains were positive for IMP, VIM and SIM simultaneously. IPM-EDTA disk synergy assay showed the production of MBLs in clinical isolates (Figure 2). PCR analysis of blaIMP, blaVIM, blaSIM, blaSPM and blaGIM is shown in Figure 3.
Figure 2.
IPM-EDTA disk synergy assay for P. aeruginosa standard strain ATCC27853 (left) and P. aeruginosa clinical strain MBL (+) (right). The arrows indicated test papers loaded with IPM or IPM+EDTA, respectively.
Figure 3.
Electrophoresis of PCR products of blaIMP, blaVIM, and blaSIM. Left panel: lane 1. CRPA38 isolate without blagGIM PCR product (562 bp); lane 2: CRPA38 isolate without blaSPM PCR product (784 bp); lanes 3 and 4: CRPA38 and CRPA48 isolates with blaVIM PCR product (382 bp); lane 5: CRPA42 isolate with blaIMP PCR product (587 bp). Right panel: lane 1: CRPA30 isolate with blaSIM PCR product (741 bp); lane 2: CRPA35 isolate with blaSIM PCR product (741 bp); lane 3: CRPA36 isolate with blaSIM PCR product (741 bp).
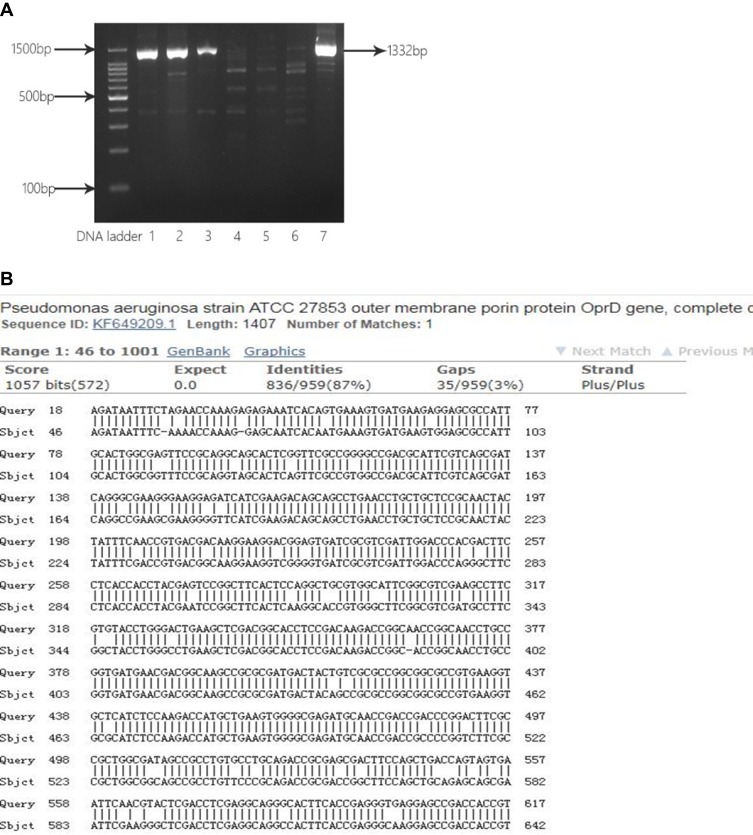
OprD2 gene from five CRPA strains was amplified (Figure 4A), and DNA sequencing showed partial deletions or point mutations at multiple locations in this gene (Figure 4B).
Figure 4.
Analysis of partial sequence of OprD2 gene from clinical isolates. (A) Electrophoresis of PCR products for OprD2. Lane 1, 2, 3 and 7: CRPA43, CRPA 52, CRPA 56 and. CRPA 60 isolates with OprD2 PCR products (1332 bp); Lane 4, 5, 6: CRPA01, CRPA02, and CRPA03 isolates without OprD2 PCR products (1332 bp). (B) BLAST of OprD2 PCR products against P. aeruginosa standard strain ATCC27853.
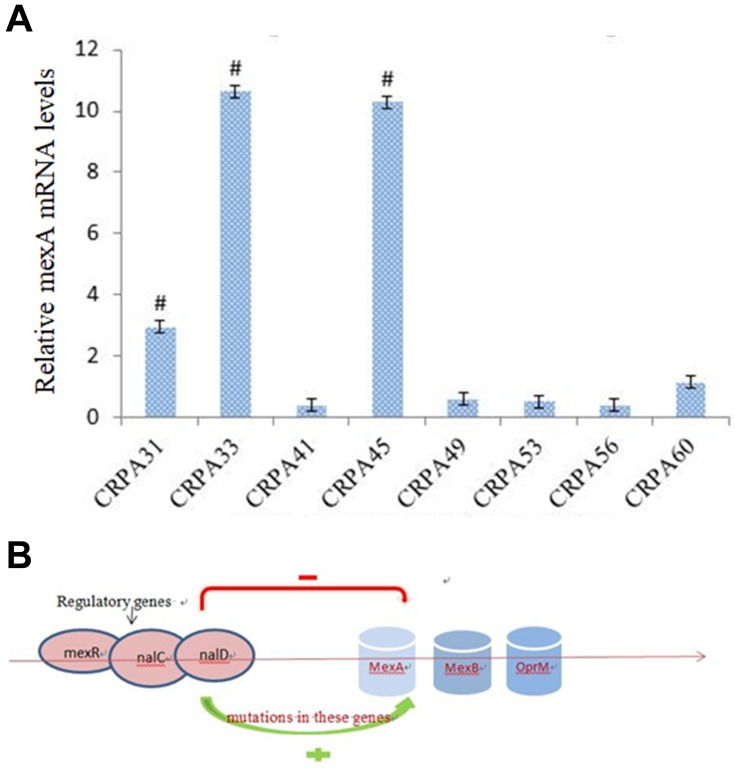
A sharp reduction in MIC was observed in eight isolates when MC207110 was added, indicating that these eight CRPA isolates had efflux pump activity against meropenem. qRT-PCR showed that three strains among them had significantly higher mexA expression compared to other five strains (Figure 5A), but we did not find mutations in mexR from both clinical isolates or the control standard strain.
Figure 5.
(A) Real-time PCR analysis of mexR mRNA levels from a clinical isolate. # p<0.05 compared to other five strains. (B) Mutations in regulatory genes such as mexR, nalD and nalC may contribute to the overexpression of mexAB-OprM and increased resistance to carbapenem.
Discussion
P. aeruginosa is a common pathogen during clinical infections. A survey showed that P. aeruginosa was the most common pathogen (17%) for nosocomial-acquired pneumonia (HAP) infection in hospitals.11 Data from South Africa from 2011 to 2015 demonstrated that the clinical isolation rate for PA was 17.4%, lower than that of Staphylococcus aureus (38%) and Klebsiella pneumoniae (22.2%).12 In our study, the detection rate for PA was 15% in 2015, second only to E. coli (26%). The emergence of CRPA makes it very difficult to treat infectious diseases and limits the choice of appropriate antibiotic therapy.
The production of metallo‑beta‑lactamase is the most common mechanism of antibiotic resistance. In this study, we used IPM-EDTA disk assay and identified 32 MBLs-positive strains. The positive rate was 51.6%, higher than the rate of 21.8% and 37.5% reported in other studies.13 In the present study, there were five IMP-positive strains (15.5%) and four VIM-positive strains (12.5%), similar to the results reported previously.14,15 However, 27 strains were SIM-positive (84.4%), which was the most prevalent MBL gene in our study. This result is inconsistent with previous studies, maybe due to the difference in antibiotic usage in different areas. Collectively, these results suggest that MBL is one of the mechanisms by which P. aeruginosa acquires resistance to carbapenem, and bla SIM is the most prevalent gene that encodes MBLs in the strains isolated in our area.
Outer membrane protein OprD2 is important for carbapenem such as imipenem and meropenem to transfer through the cell membrane.16 Loss of OprD2 expression can decrease the susceptibility of P. aeruginosa to carbapenem. Both deletion and insertional mutations of OprD2 gene have been reported to affect the expression of OprD2 and the resistance of P. aeruginosa to carbapenem. A study in Iran showed that 10.52% of CRPA strains had a 1179 bp insertional sequence ISPpu21 at position 728 of the OprD2 gene.5 Our study demonstrated that oprD expression was decreased in 5 of the 62 clinical isolates, and DNA sequencing showed that the 5 isolates had partial deletions of small fragments and point mutations at multiple locations. Therefore, OprD2 plays an important role in the emergence of carbapenem resistance in CRPA strains.
Efflux pump mediated resistance is a significant threat to the treatment of infections. The overexpression of mexAB-oprM efflux pumps has been associated with multiple drug resistance in P. aeruginosa.17 Several mutations (insertion, deletion and point mutations) in mexR gene have been shown to play an important role in the regulation of the expression of efflux pumps in P. aeruginosa. Our study showed three strains had a high expression of mexA. However, we found no mutations in mexR gene in these strains after sequencing. It is worthy to investigate whether mutations in other regulatory genes such as nalD and nalC genes contribute to the overexpression of mexAB-OprM (Figure 5B).18,19
In conclusion, the dose of antibiotics and the period of administration differ from country to country. Our study demonstrated that the resistance of P. aeruginosa to carbapenem antibiotics is synergistic action of several resistance mechanisms, including but not limited to metallo-β-lactamase production, the loss of OprD2 and the overexpression of efflux pump mexAB-oprM. Therefore, it is important to explore antimicrobial resistance in strains isolated locally to develop targeted and personalized therapy for infectious diseases.
Acknowledgments
This study was supported by the Natural Science Foundation of Inner Mongolia Autonomous Region of China (No. 2017MS(LH)0803).
Disclosure
All authors report no conflicts of interest in this work.
References
- 1.Pendleton JN, Gorman SP, Gilmore BF. Clinical relevance of the ESKAPE pathogens. Expert Rev Anti Infect Ther. 2013;11:297–308. doi: 10.1586/eri.13.12 [DOI] [PubMed] [Google Scholar]
- 2.Mensa J, Barberán J, Soriano A, et al. Antibiotic selection in the treatment of acute invasive infections by Pseudomonas aeruginosa: guidelines by the Spanish Society of chemotherapy. Rev Esp Quimioter. 2018;31:78–100. [PMC free article] [PubMed] [Google Scholar]
- 3.Hong DJ, Bae IK, Jang IH, Jeong SH, Kang HK, Lee K. Epidemiology and characteristics of Metallo-β-Lactamase-producing Pseudomonas aeruginosa. Infect Chemother. 2015;47:81–97. doi: 10.3947/ic.2015.47.2.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Viedma E, Juan C, Villa J, et al. VIM-2-producing multidrug-resistant Pseudomonas aeruginosa ST175 clone, Spain. Emerg Infect Dis. 2012;18:1235–1241. doi: 10.3201/eid1808.111234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shariati A, Azimi T, Ardebili A, et al. Insertional inactivation of oprD in carbapenem-resistant Pseudomonas aeruginosa strains isolated from burn patients in Tehran, Iran. New Microbes New Infect. 2018;21:75–80. doi: 10.1016/j.nmni.2017.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Piddock LJ. Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin Microbiol Rev. 2006;19:382–402. doi: 10.1128/CMR.19.2.382-402.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.George AM, Levy SB. Amplifiable resistance to tetracycline, chloramphenicol, and other antibiotics in Escherichia coli: involvement of a non-plasmid-determined efflux of tetracycline. J Bacteriol. 1983;155:531–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poole K, Krebes K, McNally C, Neshat S. Multiple antibiotic resistance in Pseudomonas aeruginosa: evidence for involvement of an efflux operon. J Bacteriol. 1993;175:7363–7372. doi: 10.1128/jb.175.22.7363-7372.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nikaido H. Structure and mechanism of RND-type multidrug efflux pumps. Adv Enzymol Relat Areas Mol Biol. 2011;77:1–60. doi: 10.1002/9780470920541.ch1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pourakbari B, Yaslianifard S, Yaslianifard S, Mahmoudi S, Keshavarz-Valian S, Mamishi S. Evaluation of efflux pumps gene expression in resistant Pseudomonas aeruginosa isolates in an Iranian referral hospital. Iran J Microbiol. 2016;8:249–256. [PMC free article] [PubMed] [Google Scholar]
- 11.Walter J, Haller S, Quinten C, et al. Healthcare-associated pneumonia in acute care hospitals in European Union/European Economic Area countries: an analysis of data from a point prevalence survey, 2011 to 2012. Euro Surveill. 2018;23:1700843. doi: 10.2807/1560-7917.ES.2018.23.32.1700843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramsamy Y, Essack SY, Sartorius B, Patel M, Mlisana KP. Antibiotic resistance trends of ESKAPE pathogens in Kwazulu-Natal, South Africa: a five-year retrospective analysis. Afr J Lab Med. 2018;7:887. doi: 10.4102/ajlm.v7i2.887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaur A, Singh S. Prevalence of Extended Spectrum Betalactamase (ESBL) and Metallobetalactamase (MBL) producing Pseudomonas aeruginosa and acinetobacter baumannii isolated from various clinical samples. J Pathog. 2018;2018:6845985. doi: 10.1155/2018/6845985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jabalameli F, Taki E, Emaneini M, Beigverdi R. Prevalence of metallo-β-lactamase-encoding genes among carbapenem-resistant Pseudomonas aeruginosa strains isolated from burn patients in Iran. Rev Soc Bras Med Trop. 2018;51:270–276. doi: 10.1590/0037-8682-0044-2018 [DOI] [PubMed] [Google Scholar]
- 15.Azimi A, Peymani A, Pour PK. Phenotypic and molecular detection of metallo-β-lactamase-producing Pseudomonas aeruginosa isolates from patients with burns in Tehran, Iran. Rev Soc Bras Med Trop. 2018;51:610–615. doi: 10.1590/0037-8682-0174-2017 [DOI] [PubMed] [Google Scholar]
- 16.Li J, Hu W, Li M, Deng S, Huang Q, Lu W. Evaluation of matrix-assisted laser desorption/ionization time-of-flight mass spectrometry for identifying VIM- and SPM-type metallo-β-lactamase-producing Pseudomonas aeruginosa clinical isolates. Infect Drug Resist. 2019;12:2781–2788. doi: 10.2147/IDR.S211984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li XZ, Nikaido H, Poole K. Role of mexA-mexB-oprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:1948–1953. doi: 10.1128/AAC.39.9.1948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cao L, Srikumar R, Poole K. MexAB-OprM hyperexpression in NalC-type multidrug-resistant Pseudomonas aeruginosa: identification and characterization of the nalC gene encoding a repressor of PA3720-PA3719. Mol Microbiol. 2004;53:1423–1436. doi: 10.1111/j.1365-2958.2004.04210.x [DOI] [PubMed] [Google Scholar]
- 19.Sobel ML, Hocquet D, Cao L, Plesiat P, Poole K. Mutations in PA3574 (nalD) lead to increased MexAB-OprM expression and multidrug resistance in laboratory and clinical isolates of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2005;49:1782–1786. doi: 10.1128/AAC.49.5.1782-1786.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]