Summary
Differentiation of proinflammatory CD4+ conventional T cells (Tconv) are critical for productive antitumor responses yet their elicitation remains poorly understood. We exhaustively characterized myeloid cells in tumor draining lymph nodes (tdLN) of mice and identified two subsets of conventional type-2 dendritic cells (cDC2) that traffic from tumor to tdLN and present tumor-derived antigens to CD4+ Tconv, but then fail to support antitumor CD4+ Tconv differentiation. Regulatory T cell (Treg) depletion enhanced their capacity to elicit strong CD4+ Tconv responses and ensuing antitumor protection. Analogous cDC2 populations were identified in patients, and as in mice their abundance relative to Treg predicts protective ICOS+ PD-1lo CD4+ Tconv phenotypes and survival. Further, in melanoma patients with low Treg abundance, intratumoral cDC2 density alone correlates with abundant CD4+ Tconv and with responsiveness to anti-PD-1 therapy. Together, this highlights a pathway which restrains cDC2, and whose reversal enhances CD4+ Tconv abundance and controls tumor growth.
Introduction
Adaptive T cell responses are critical for controlling tumor growth through production of inflammatory cytokines and direct cytolytic targeting. Recent therapeutic advances that block inhibitory T cell checkpoint molecules like CTLA-4 or PD-1/PD-L1 have demonstrated clinical success, but in only a subset of cancer patients (Hodi et al., 2008; Huang et al., 2017; Larkin et al., 2015; Topalian et al., 2014). Recent evidence suggests that tumors often promote the generation of dysfunctional and exhausted T cells with deficient effector capacity reminiscent of exhaustion observed following chronic viral infection (Schietinger et al., 2016). T cell exhaustion is enforced at the chromatin level such that many T cells in the tumor microenvironment (TME) are likely not able to be rescued by immune checkpoint blockades (ICB) (Pauken et al., 2016; Philip et al., 2017). Thus, in those patients with poor T cell infiltration or irreversibly exhausted T cells, additional steps, such as improving de novo priming of effector T cells, may be necessary to engage effective antitumor immunity (Philip et al., 2017; Tumeh et al., 2014).
While CD8+ T cells are considered a primary immunotherapeutic target due to their classic role in tumor cell cytolysis, CD4+ Tconv are emerging as an important contributor to antitumor responses. In immunogenic settings, effector CD4+ Tconv augment immunity through licensing of dendritic cells (DC) (Behrens et al., 2004) and stimulating pro-inflammatory myeloid cell programs (Corthay et al., 2005). CD4+ Tconv have also been documented to improve the quality of effector CD8+ T cell responses to apoptotic cell antigens (a common source of tumor antigen) and contribute to T cell memory programming and maintenance (Laidlaw et al., 2016). Intriguingly, CD4+ Tconv have been described as having direct antitumor cytolytic function(Curran et al., 2013; Quezada et al., 2010) and HLA-DR expression on human tumor cells (MHC-II in mouse) has been identified as a biomarker for anti-PD-1/PD-L1 responsiveness (Johnson et al., 2016). Notably, effective anti-CTLA-4 therapy results in a systemically circulating population of ICOS+ PD-1lo CD4+ T helper 1-like (Th1-like) effector CD4+ Tconv critical for an antitumor response (Fan et al., 2014). Conversely, presence of a PD-1hi CD4+ Tconv phenotype, correlated with extensive tumor burden and likely T cell exhaustion, has been shown to be a negative prognostic indicator for checkpoint blockade (Zappasodi et al., 2018). As such, the processes that contributes to antitumor CD4+ Tconv activation and differentiation merit further investigation.
Generation of newly activated antitumor T cell clones typically requires their activation in secondary lymphoid organs such as the tumor-draining lymph node (tdLN), followed by subsequent infiltration into the tumor mass (Chen and Mellman, 2013). Initiation of an adaptive T cell response is driven by one or more types of innate myeloid antigen-presenting cells (APC) such as conventional dendritic cells (cDC) that present tumor antigen, co-stimulatory molecules, and cytokines to cognate antigen-specific T cells. Given the shortcomings in endogenously-generated antitumor T cell responses, there has long been therapeutic interest to improve cDC numbers and functionality as a means to boost T cell effector potential. Approaches such as cellular vaccines or administration of cDC growth factors, however, remain susceptible to endogenous immunosuppressive cells such as Treg (Josefowicz et al., 2012) which can potently suppress cDC (Bauer et al., 2014), although given the complexity of cDC populations, it is currently unclear if specific populations of cDC are selectively impacted.
Diverse in nature, cDC can be broadly divided into cDC1 and cDC2 populations that arise through distinct pre-DC lineages (Schlitzer et al., 2015) and can be either resident to the LN, or migrate in from peripheral tissues bearing antigen (Merad et al., 2013). Importantly, cDC1 and cDC2 often take on specialized roles in CD8+ T cell and CD4+ Tconv priming processes through their differential use of antigen processing and presentation pathways (Gutierrez-Martinez et al., 2015), production of effector cytokines (Merad et al., 2013), and spatial localization within the LN (Gerner et al., 2017). cDC1 have been identified as critical for directing CD8+ T cell immunity to various pathogens (Bedoui et al., 2009; Belz et al., 2005) and in mediating spontaneous antitumor CD8+ T cell responses (Broz et al., 2014; Roberts et al., 2016; Ruffell et al., 2014; Salmon et al., 2016; Spranger et al., 2017). In contrast, cDC2 contain substantial heterogeneity and they preferentially initiate CD4+ Tconv responses in a variety of immunological models (Gao et al., 2013; Krishnaswamy et al., 2017). While the division of labor between cDC1 and cDC2 in engaging CD8+ T cells and CD4+ Tconv, respectively, is an established phenomenon, this may depend on the tissue type and each may have multiple capabilities to tolerize or activate respective cells types, depending on the nature of the immune challenge. On the whole, the specific cDC roles in eliciting antitumor CD4+ Tconv immunity remains unresolved.
We therefore applied single-cell RNA-sequencing (scRNA-seq) to myeloid populations from tdLNs in mouse and human to understand the true diversity and function of cell types present, how they differ with cancer, and how the variance might affect the nature of the CD4+ Tconv that are available for tumor efficacy. Key in this was to understand how therapeutic intervention might alter the outcome of CD4+ Tconv priming. Additionally, we sought to understand whether human cancer biology paralled the mouse and assembled cohorts of patient biopies to determine how CD4+ Tconv phenotype and cDC composition were connected.
Results
Myeloid heterogeneity at single cell resolution
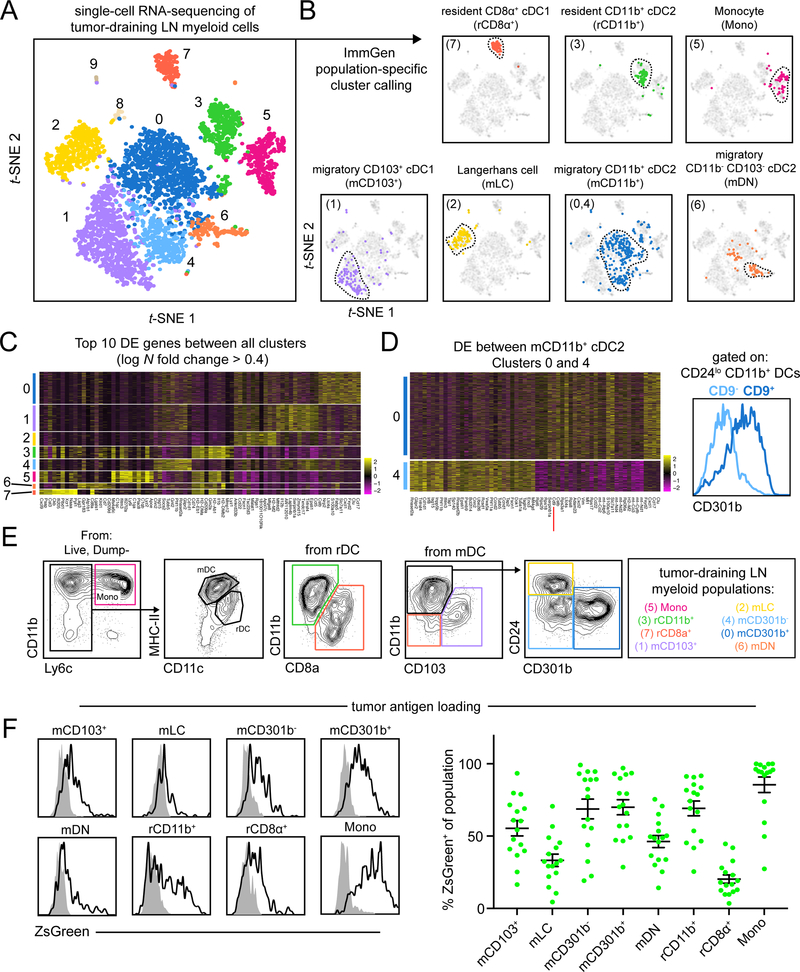
To comprehensively study the myeloid populations capable of priming anti-tumor CD4+ Tconv, we queried myeloid heterogeneity in the tdLN by sorting non-lymphocyte (CD90.2– B220− NK1.1−) myeloid cells (CD11c+ or CD11b+) from the tdLN of B16-F10 tumor-bearing mice. We performed scRNA-seq using the 10X Genomics Chromium platform paired with deep sequencing. Analysis of 4133 tdLN myeloid cells yielded 10 high quality and unique population clusters (Figure 1A, S1A).
Figure 1.
Unbiased scRNA-seq of myeloid cells in the tdLN reveals extensive heterogeneity.
(A) t-SNE display and graph-based clustering of CD90.2− B220− NK1.1− CD11b+ and/or CD11c+ myeloid cells sorted from B16F10 tdLN and processed for scRNA-seq. Each dot represents a single cell. (B) Expression of ImmGen population-specific gene signatures distributed across t-SNE plot of (A). (C) Heatmap displaying top 10 DE genes for each cluster when comparing clusters 0 through 7 (ranked by fold change) (D) (left) A heatmap displaying the top 30 DE genes between clusters 0 and 4, with Cd9 highlighted by a red line. (right) A flow cytometry histogram displaying the differential surface expression of CD301b between CD9− and CD9+ CD11b+ CD24− DCs (E) Representative gating strategy used to identify myeloid populations in the tdLN (F) Representative flow cytometry histograms displaying levels of ZsGreen tumor antigen within myeloid populations in the tdLN (left). Frequency of ZsGreen+ cells within t dLN myeloid populations (right). Data pooled from two independent experiments. Figure 2
To rigorously identify the myeloid populations and determine how they related to those previously described in other settings, we generated gene signatures of cell populations expected to be present in the LN from samples available from the Immunological Genome Project (ImmGen) database (Heng and Painter, 2008) and plotted expression of these signatures on the tdLN t-SNE plot. This allowed us to assign cellular identities to each cluster (Figure 1B and Table 1), apart from clusters 8 and 9, as they appeared to be lymphocyte contaminates and were excluded from further analysis (data not shown).
We then utilized gene overlays of individual canonical myeloid markers to further explore the cluster identities. Ccr7 and Itgax demarcarcated migratory (clusters 0, 1, 2, 4 and 6) and resident (clusters 3, 5 and 7) DCs, consistent with our assignments and the known biology (Figure S1B). DC clusters (0–4, 6–7) were further confirmed using canonical genes Zbtb46 and Flt3, whereas monocytes and T cell zone macrophages (TZ Macrophages), which are unable to prime CD4+ Tconv (Baratin et al., 2017), both occupied cluster 5, but localized to opposite sides of the cluster (Figure S1C).
We performed differential expression (DE) analysis for each myeloid cluster versus all other clusters and generated heatmaps for the top 10 most differentially expressed genes (Figure 1C and Table 2). In addition to highlighting key genes that contributed to the unbiased segregation of these populations, a number of markers also validated previous reports of specialized cellular functions such as production of Il12b in mCD103+ cDC1(Miller et al., 2012) or Ccl17 in CD11b+ cDC2 and mLC (Alferink et al., 2003). Moreover, there was a general pattern of shared transcriptional identity within resident and migratory populations, which was further elucidated by performing DE analysis between migratory (clusters 0, 1, 2, 4 and 6) and resident (clusters 3 and 7) DC populations (Figure S1D and Table 3). Expression was largely uniform within migratory and resident DC, with enrichment of genes previously associated with migratory populations such as Socs2 or Fscn1 (Miller et al., 2012).
Populations identified through unbiased clustering largely mirrored those identified using ImmGen-based criteria. However, of specific note, and in contrast to migratory cDC1, the canonical signature for migratory cDC2 applied to multiple clusters in our unbiased analysis. This indicated substantial and unresolved heterogeneity within this cDC class. DE analysis between migratory CD11b+ cDC2 clusters 0 and 4 identified the gene Cd9, a surface molecule, to be expressed specifically on cluster 0 (Figure 1D and Table 4). We determined that surface expression of CD9 parsed the two CD11b+ cDC2 populations and with further investigation we found that expression was concordant with a previously identified molecule that distinguished cDC2 subsets, CD301b (Kumamoto et al., 2016). Within the migratory CD11b+ cDC2 gate, CD9− cells were CD301b− (mCD301b−), whereas CD9+ cells were found to be CD301b+ (mCD301b+) (Figure 1D and S1E). Due to the robustness of staining and parity with existing literature, CD301b was thus used for subsequent parsing of CD11b+ cDC2 populations.
CD301b expression is often attributed to cells of monocyte/macrophage lineage and so we assessed expression of other monocyte/macrophage-related molecules on mCD301b− and mCD301b+. While both mCD301b− and mCD301b+ cells expressed CD135/FLT3 and SIRPα, consistent with cDC2 assignment (Miller et al., 2012), mCD301b+ expressed higher surface levels of markers generally associated with cells of a monocyte/macrophage lineage (Gautier et al., 2012), including CD14 (which we find later useful for parsing human cDC2 populations), CD16/32 (FcgRIII/II), CD200R and CD206 (Figure S1F). Furthermore, mCD301b+ expressed higher levels of inhibitory receptors PD-L2 and LILRB4 (Figure S1G). Despite some of these markers being associated with cells of macrophage lineage, CD11b+ cDC2 are phenotypically DC, based on expression of Zbtb46 (Meredith et al., 2012; Satpathy et al., 2012) (Figure S1H).
With the assistance of unbiased scRNA-seq on bulk myeloid cells from the tdLN, we were able to derive a flow cytometry panel that encompasses this heterogeneity (Figure 1E) and determined the frequency of these populations (Figure S1I). With this comprehensive delineation of major myeloid populations within the tdLN, we next sought to identify the exact APC(s) responsible for anti-tumor CD4+ Tconv priming by using the markers to track, isolate or genetically deplete distinct populations.
Requisite Migration of tdLN Populations
Previous work has highlighted the importance of CD103+ cDC1 migration to the tdLN for productive antitumor CD8+ T cell responses (Roberts et al., 2016). Less is known, however, about CD11b+ cDC2 migration from the tumor and we sought to identify whether these two cDC2 populations were tumor-originating and tumor-antigen bearing. Consistent with our scRNA-seq analysis, populations identified as migratory were found to express surface CCR7 within the tdLN (Figure S1J), consistent with previous migration from a peripheral tissue.
We then assessed the levels of tumor antigen within myeloid cells of tdLN from B16-ZsGreen (B16ZsGr) tumor-bearing animals (Figure 1F) and found that mCD103+, mCD301b− and mCD301b+ were the most dominant ZsGreen+ migratory populations, while resident populations generally had lower and heterogeneous levels of uptake, consistent with previous findings (Roberts et al., 2016). Notably, CD301b− and CD301b+ cDC2 are also present within the TME with fractions of both populations expressing CCR7 (Figure S1K), indicating their migratory capacity and providing confirmation of the abundant ZsGreen tumor antigen detected in these populations within the tdLN. We then generated B16-mCherry-OVA (B16ChOVA) tumor-bearing Ccr7−/− mice, and confirmed that these lacked normal frequencies of all migratory DC in the tdLN (Figure S2A). Furthermore, in this context, adoptively transferred CD4+ OT-II T cells were nearly completely unable to initiate proliferation (Figure S2B). This confirmed that migratory DC populations were critical but did not identify which cell population(s) could directly present antigens to drive proliferation nor how these populations would induce CD4+ Tconv differentiation.
De novo priming of CD4+ Tconv by cDC2 in the tdLN
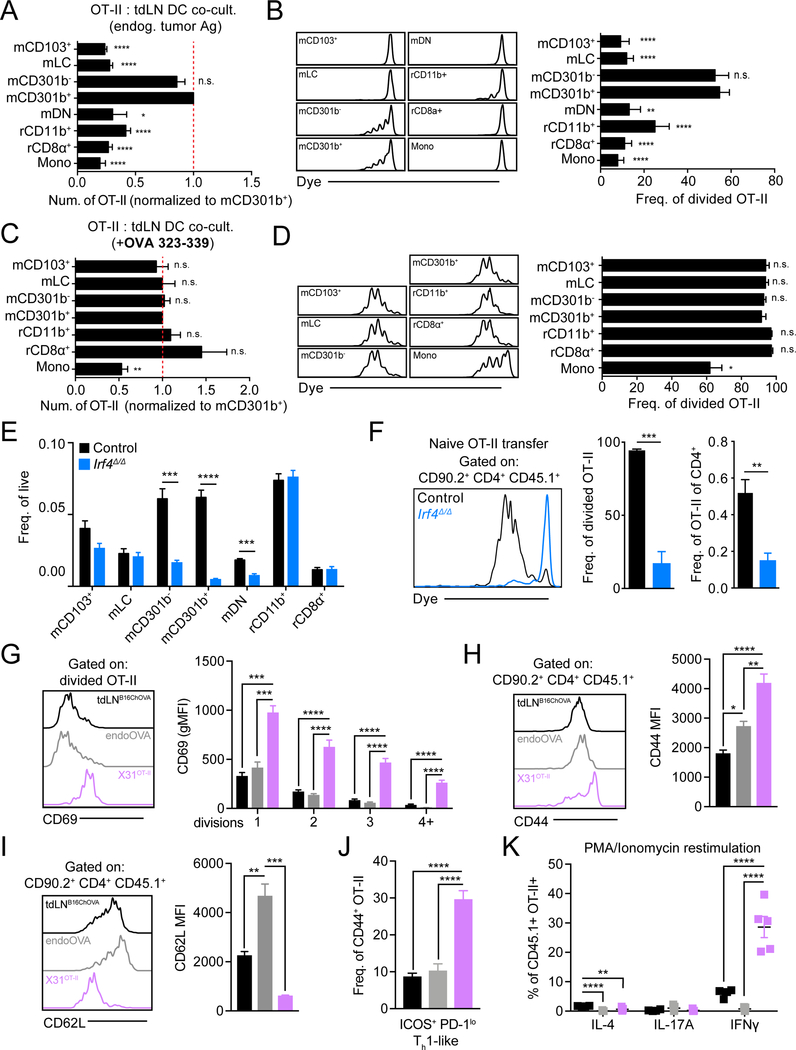
Using mice bearing B16ChOVA tumors, we sorted each of the identified tdLN myeloid populations, each of which contained in vivo acquired and processed tumor antigen, and co-cultured them ex vivo with naïve CD4+ OT-II T cells. This demonstrated that migratory CD11b+ cDC2, whether they be mCD301b− or mCD301b+, supported CD4+ OT-II T cell expansion based on absolute cell number (Figure 2A) and frequency of cells undergoing cell division (Figure 2B). Importantly, despite similar antigen loading (Figure 1F), rCD11b+ induced little proliferation of OT-II cells. Addition of exogenous OT-II OVA peptide (OVA323–339) resulted in comparable activation and proliferation across myeloid populations, indicating that other populations are viable and otherwise capable of engaging CD4+ Tconv, but likely simply do not process and present tumor antigen on MHC-II, restricting their ability to prime CD4+ Tconv (Figure 2C, 2D).
Figure 2.
mCD301b−/+ cDC2 are uniquely able to induce anti-tumor CD4+ Tconv proliferation but fail to initiate CD4+ Tconv differentiation.
(A-D) Purified CD4+ OT-II T cells were co-cultured ex vivo with sorted APC populations from tdLN and analyzed at 3 days. (A) Absolute number of live OT-II T cells recovered from co-culture, normalized and statistically compared to mCD301b+ condition (t-test). (B) Histograms of OT-II T cell dye dilution (left). Frequency of recovered OT-II T cells that had undergone division with statistical comparison to mCD301b+ condition (t-test) (right). (C) Absolute number of live OT-II T cells recovered from co-culture containing exogenous OVA peptide (323–339), normalized and statistically compared to mCD301b+ condition (t-test). (D) Histograms of OT-II T cell dye dilution (left). Frequency of recovered OT-II T cells that had undergone division with statistical comparison to mCD301b+ condition (t-test) (right). (E) Frequency of tdLN DC populations in control or Irf4−/− tumor-bearing mice. (F) Purified CD45.1+ OT-II T cells were adoptively transferred to control or Irf4−/− B16ChOVA tumor-bearing mice with tdLN harvested 3 days later to assess OT-II T cell dye dilution (left) and quantify the frequency of cells that had divided (middle) and their frequency of endogenous CD4+ T cells (right). (G-K) CD45.1+ CD4+ OT-II T cells were transferred to wild-type mice that were inoculated with B16chOVA (tdLNB16ChOVA), endoOVA, or X31pOVA and draining LNs were harvested for analysis. (G) Cell surface Cd69 levels on divided CD45.1+ CD4+ OT-II T cells (left) and quantification of MFI with each cell division as determined by dye dilution (right) 3 days following transfer. Surface CD44 (H) and CD62L (I) levels on transferred CD45.1+ CD4+ OT-II T cells (left) and quantification of MFI (right) 3 days following transfer. (J) Frequency of transferred CD45.1+ CD4+ CD44+ OT-II T cells that are ICOS+PD-1lo Th1-like. (K) Frequency of transferred CD45.1+ CD4+ CD44+ OT-II T cells that produce IL-4, IL-17A and IFNγ following PMA/Ionomycin restimulation with detection by intracellular antibody staining 7 days after transfer. Data are represented as average ± SEM unless explicitly specified. *P <0.05, **P<0.01, ***P<0.001, ****P<0.0001.
To extend this study in vivo, we next tested whether CD11b+ cDC2 were required for initiating CD4+ Tconv priming within the tdLN. Mice lacking Irf4 in DC have been shown to lack LN cDC2 (Krishnaswamy et al., 2017), however use of Irf4flox/floxItgaxCre resulted in consistent spontaneous germline deficiency (data not shown), complicating our efforts to delete Irf4 specifically in the myeloid compartment. We instead used globally deficient Irf4Δ/Δ (Irf4flox/floxActBCre) B16ChOVA-tumor-bearing animals, with adoptively transferred wild-type OT-II T cells, wherein we observed a reduction of all migratory cDC2 populations in the tdLN (Figure 2E). Transferred OT-II cells in Irf4Δ/Δ mice failed to proliferate as assessed by dye dilution and similarly failed to accumulate in the tdLN (Figure 2F). In contrast, in Xcr1DTR mice, depletion of mCD103+ and rCD8α+ did not impact OT-II proliferation (Figure S2C, S2D). By exploiting differential expression of Cx3cr1 in rCD11b+ (Supplemental Table 2), we generated Cx3cr1lsl-DTRCD11cCre animals that allowed for specific depletion of rCD11b+ following DT administration (Figure S2E). Consistent with our in vitro findings, depletion of rCD11b+ did not reduce OT-II proliferation (Figure S2F). Both in vitro and in vivo, migratory CD11b+ cDC2, but not other cDC populations, were found to be the primary inducers of antitumoral CD4+ Tconv priming.
Tolerogenic CD4+ Tconv priming in the tdLN
In our tumor-bearing mice, effective anti-tumor immunity is not occurring despite evident initiation of CD4+ Tconv priming and we hypothesized that CD4+ Tconv differentiation by the identified cDC2 might not be generating effector differentiation. To examine this, we directly compared in vivo activation and differentiation of adoptively transferred OT-II T cells in the context of anti-tumor priming (tdLNB16ChOVA) with tolerance-inducing priming via injection of adjuvant-free antigen (endoOVA) and with robust effector CD4+ Tconv priming via infection by an influenza virus (X31pOVA). We found that CD69 expression on OT-II at day 3, representing a marker of the strength of T cell activation (Allison et al., 2016) and/or exposure to inflammatory cytokines (Shiow et al., 2006; Sun et al., 1998), was similar in tdLNB16ChOVA and tolerizing endoOVA and much lower as compared to inflammatory priming with X31pOVA (Figure 2G). Correspondingly, other markers of activation observed in robust X31pOVA activation, namely CD44 upregulation and CD62L downregulation, were not observed to the same extent on the tdLNB16ChOVA OT-II T cells, which were largely similar to those primed by endoOVA (Figure 2H, 2I).
Finally, we found minimal downstream differentiation toward a protective ICOS+ PD-1lo Th1 surface phenotype in both tdLNB16ChOVA and endoOVA conditions when examined day 7 post-transfer, compared to X31pOVA (Figure 2J). This also coincided with little to no cytokine production, notably IFNγ, following restimulation (Figure 2K). While we found that cDC2 initiate CD4+ Tconv priming in the tdLN, such defective effector Tconv differentiation predicts that therapeutic improvement of CD4+ Tconv priming might either function through alterations in cDC2 phenotype, or via the licensing of other cell types to become APCs for CD4+ Tconv.
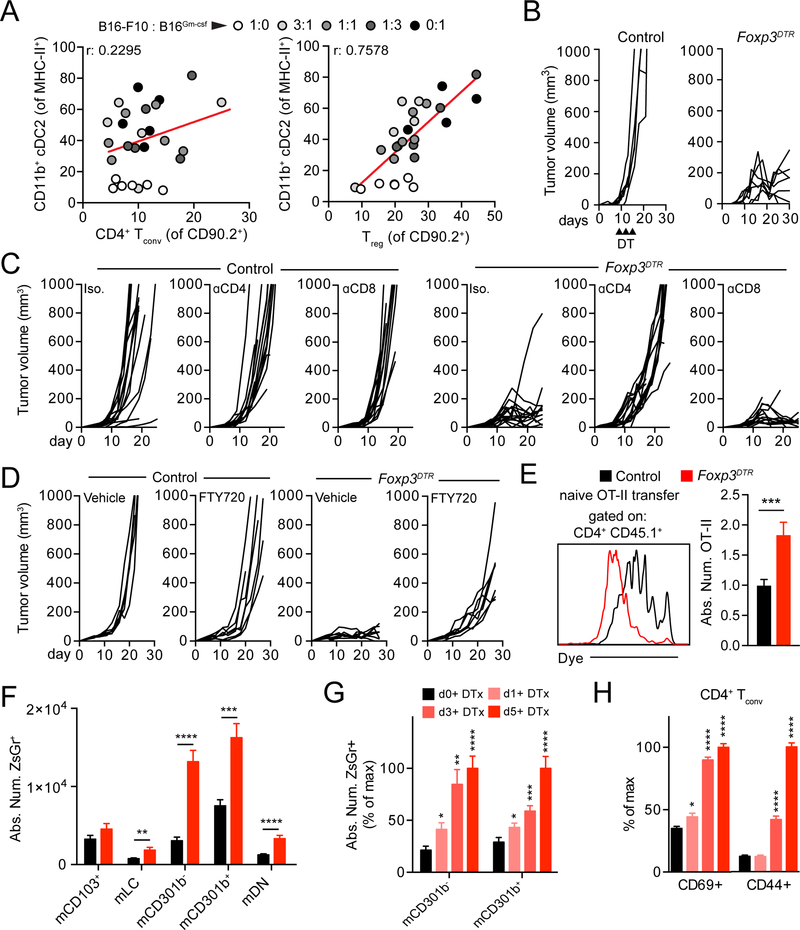
Concommittant expansion of Treg and CD11b+ cDC2 in the TME
While examining the expansion of cDC2 in the TME of tumors with variable proportions of tumor cells secreting GM-CSF (B16Gm-csf) (Broz et al., 2014), we found the surprising result that CD4+ Tconv numbers did not rise appreciably with the induced increase in CD11b+ cDC2 (considering both CD301b− and CD301b+ cDC2 subsets) (Figure 3A, S3A). Given that these populations clearly express epitopes on MHC-II to CD4+ T cells (Figure 2), we hypothesized that Treg may prefentially expand in response to CD11b+ cDC2 and may act as a feedback mechanism to suppress CD11b+ cDC2 function and thus effective antitumor CD4+ Tconv priming. Analyzing the same mice for Treg proportion we found, indeed, a positive correlation between Treg frequencies in the TME and cDC2 number. Given previous data suggesting that general DC may be altered or deleted by Treg (Bauer et al., 2014), we sought to test whether Treg might be restricting the trafficking and/or phenotype of CD11b+ cDC2, thereby generating poorly differentiated CD4+ Tconv.
Figure 3.
Regulatory T cell depletion enhances cDC2 migration to the tdLN and unleashes an anti-tumor CD4+ Tconv response. (A) Dot plot correlation of intratumoral CD11b+ CD301b−/+ cDC2 frequency within MHC-II+ cells against CD4+ Tconv within CD90.2+ (left) or Treg (right) within CD90.2+. Dots colored according to ratio of B16-F10:B16Gm-csf cells in the tumor. Two pooled experiments displayed. (B) Tumor growth from control and Foxp3DTR mice. Upward facing black arrowheads indicate DT treatment. Results depict tumor growth curves of individual mice. (C) Tumor growth from control or Foxp3DTR mice injected with isotype/anti-CD4/anti-CD8 depleting antibodies. Results depict tumor growth curves of individual mice. Two pooled experiments displayed. (D) Tumor growth from control or Foxp3DTR mice injected with with vehicle or FTY720. Results depict tumor growth curves of individual mice. Two pooled experiment displayed. (E) CD45.1+ CD4+ OT-II T cells were adoptively transferred into DT-treated control or Foxp3DTR B16ChOVA-tumor-bearing mice and recovered 3 days later for analysis of dye dilution (left) and quantification of absolute number of OT-II present within the tdLN (right). Three pooled experiments displayed with normalization to control. (F) Control and Foxp3DTR B16ZsGreen tumor-bearing mice were treated with DT and absolute number of ZsGreen+ migratory DC in the tdLN were analyzed at day 5 post-DT. (G) Control and Foxp3DTR B16ZsGreen tumor-bearing mice were treated with DT and absolute number of ZsGreen+ CD11b+ cDC2 in the tdLN were analyzed at day 0, 1, 3 and 5 post-DT. Data displayed as percent of maximum absolute number. Samples statistically compared to day 0 DT condition. (H) Control and Foxp3DTR B16ZsGreen tumor-bearing mice were treated with DT and analyzed for the frequency of CD4+ Tconv expressing CD69 and CD44 at day 0, 1, 3 and 5 post-DT. Data displayed as frequency of maximum expression. Samples statistically compared to day 0 DT condition. Data are represented as average ± SEM. *P <0.05, **P<0.01, ***P<0.001, ****P<0.0001.
Therapeutic benefits of Treg depletion rely on de novo CD4+ Tconv priming
Diptheria Toxin treatment of Foxp3DTR mice (Kim et al., 2007) led to robust acute Treg depletion and potent tumor rejection (Figure 3B and S3B, S3C) which required CD4+ Tconv as previously described (Bos et al., 2013) (Figure 3C). To examine the role of tdLN CD4+ Tconv priming specifically, we tested whether rejection depended on reactivation of CD4+ Tconv already present in the TME or expansion and infiltration of recently activated CD4+ Tconv. For this, we employed the use of the S1PR antagonist, FTY720, to block CD4+ Tconv egress from the tdLN (Chiba et al., 1998). While FTY720 treatment had little effect on tumor growth in progressing tumors, FTY720/DT-treated Foxp3DTR mice were unable to reject tumors in contrast to their DT-treated Foxp3DTR controls (Figure 3D), demonstrating CD4+ Tconv tdLN priming and egress is required for tumor rejection following Treg depletion.
Treg depletion induces enhancement of both CD11b+ cDC2 and CD4+ Tconv
Transfer of CD4+ OT-II into B16ChOVA tumor-bearing Treg-depleted animals led to greatly enhanced proliferation and expansion of OT-II in the tdLN at day 3 post-transfer compared to control (Figure 3E). Enhanced CD4+ Tconv proliferation led us to hypothesize that Treg depletion relieved suppression of the CD11b+ cDC2/CD4+ Tconv axis in the TME and tdLN. To test this, we first examined DC antigen trafficking to the tdLN following Treg depletion in B16ZsGr tumor-bearing control and Foxp3DTR mice. Following Treg depletion, ZsGreen+ mCD301b− and mCD301b+ were greatly increased in absolute number in the tdLN, while other migratory populations were only weakly increased or unchanged (Figure 3F). We found that Treg-mediated suppression of CD11b± cDC2 trafficking was not mediated through Il-10, Itgb8 (Tgfb1-related) nor Area as knockout of these genes had no impact on the number of ZsGreen± CD11b± cDC2 in the tdLN (Figure S4A–S4C). Trafficking of these cells, however, depended upon chemokine-mediated tumor to tdLN trafficking as treating mice with pertussis toxin (PTX), which blocks Gai signaling, blocked the rise in cDC2 in the Treg deplete condition to a similar degree as the non-depleted controls (Figure S3D). Furthermore, we observed neither an increase in proliferation of CD11b± cDC2 in the tdLN (Figure S3E) nor profound changes in DC precursors in the bone marrow of Foxp3DTR mice (Figure S3F,S3G), suggesting the expansion of CD11b± cDC2 was due to enhanced migration and not in situ proliferation in the tdLN or increased generation of DC. By analyzing changes in the abundance of CD11b+ cDC2 in the tdLN over time following Treg depletion (Figure 3G), we were also able to determine that the wave of enhanced CD11b+ cDC2 migration temporally coincided with increases in the poorly-upregulated activation markers identified in Figure 2 (CD69 and CD44) (Figure 3H, S3H) consistent with a model in which a new wave of CD11b+ cDC2 carried the capacity to improve the quality of priming.
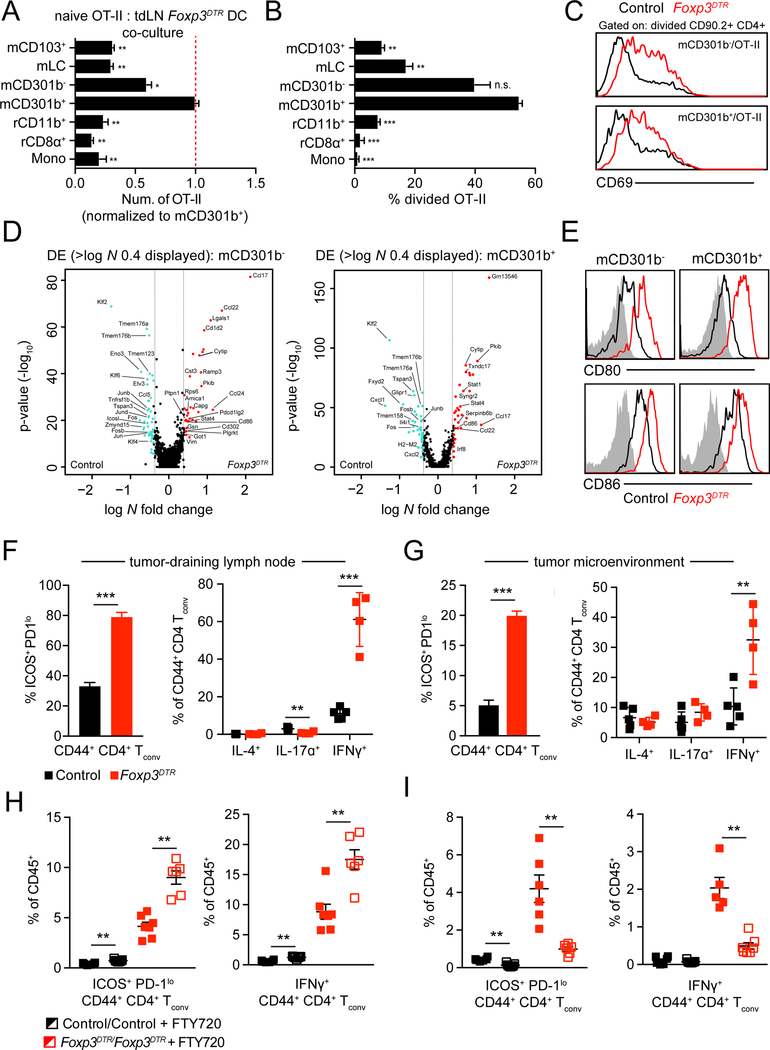
In order to test whether APC populations that trafficked to the tdLN in the absence of Treg were capable of priming CD4+ Tconv, we co-cultured each APC isolated from Foxp3DTR tdLN with CD4+ OT-II in vitro and measured their proliferation. This demonstrated that mCD301b− and mCD301b+ remained the only cells capable of supporting T cell division and accumulation (Figure 4A, 4B), while other cells were still only able to prime CD4 T cells if provided exogenous antigen (Figure S5A). Furthermore, by crossing Xcr1DTR with Foxp3DTR, we were able to genetically exclude that neither mCD103+ nor rCD8α+ were now required for improved CD4+ Tconv priming and tumor rejection following Treg depletion (Fig S5B–S5D).
Figure 4.
Regulatory T cell depletion enhances cDC2 function and CD4+ Tconv differentiation. (A-C) Purified CD4+ OT-II T cells were co-cultured with tdLN APC populations sorted from tdLN of control or Foxp3DTR B78chOVA-bearing animals and harvested 3 days after plating for analysis. (A) Absolute number of live OT-II T cells recovered, normalized and statistically compared to mCD301b+ condition. (B) Frequency of recovered OT-II T cells that had undergone division, statistically compared to mCD301b+ condition (t-test). (C) Cell surface CD69 levels on divided OT-II. (D) Volcano plots displaying DE expressed genes comparing control and Foxp3DTR tdLN mCD301b− (left) and mCD301b+ (right). Log N fold cutoff of 0.4 used. Genes of interest labelled. (E) Cell surface levels of CD80 and CD86 on mCD301b− and mCD301b+ in control and Foxp3DTR tdLN. (F) Frequency of CD44+ CD4+ Tconv that are ICOS+ PD-1lo in control and Foxp3DTR tdLN (left). Frequency of tdLN CD44+ CD4+ Tconv producing IL-4, IL-17A or IFNγ from control or Foxp3DTR tumor-bearing mice following ex vivo restimulation (right). (G) Frequency of CD44+ CD4+ Tconv that are ICOS+ PD-1lo in control and Foxp3DTR TME (left). Frequency of CD44+ CD4+ Tconv producing IL-4, IL-17A or IFNγ in control or Foxp3DTR TME following ex vivo restimulation (right). (H, I) Control and Foxp3DTR tumor-bearing mice were treated with FTY720 or vehicle and tdLN (H) or tumor (I) were harvested to quantify frequency of CD45+ cells that are CD44+ CD4+ ICOS+ PD-1lo Tconv (left) and are IFNγ-producing CD44+ CD4+ Tconv following ex vivo restimulation (right). Representative experiment displayed. Data are represented as average ± SEM. *P <0.05, **P<0.01, ***P<0.001, ****P<0.0001.
To then determine whether CD11b+ cDC2 generated in the absence of Treg, enhanced the quality of the priming reaction, we measured augmentation of CD69 expression on divided OT-II co-cultured with CD11b+ cDC2 in vitro (Figure 4C), and on divided OT-II transferred in vivo (Figure S5E). In both settings we found that, similar to CD4+ Tconv response in influenza (Figure 2), primed OT-II exhibited increased expression of CD69 compared to control tdLN conditions.
To directly measure the change in phenotype of CD11b+ cDC2 in the absence of Treg, we examined transcriptional changes in bulk myeloid cells from Foxp3DTR tdLN with scRNA-seq. This confirmed normal representation of myeloid populations within the Foxp3DTR tdLN (Figure S5F) and comparable UMI within each cluster (Figure S5G). We first compared gene expression differences between mCD301b- and mCD301b± from tdLN of Foxp3DTR mice and found that mCD301b- expressed higher levels of costimulatory molecules (e.g. Cd40, Cd70, Cd86) in addition to key CD4± Tconv chemoattractant chemokines (e.g. Ccl17, Ccl22), while mCD301b± expressed higher levels of AP-1 family members (e.g. Jun, Fos, Junb) (Figure S5H and Supplemental Table 5A). We then aggregated scRNA-seq from tdLN and Foxp3DTRtdLN data and performed DE analysis on mCD301b− and mCD301b+ from the two conditions and found pronounced increases in costimulatory genes (Cd80, Cd86), genes involved in T cell chemoattraction (Ccl17, Ccl22) and genes expressed in response to pro-inflammatory cytokines (Stati, Stat4) on CD11b+ cDC2 in Foxp3DTR tdLN (Figure 4D and Supplementary Table 5A). The increase in expression of both Cd80 and Cd86 was also verified by flow cytometry (Figure 4E), although we found costimulatory levels to also be increased on other DC populations (Figure S5I).
We next assessed whether enhanced CD11b+ cDC2 functionality coincided with improved CD4+ T cell differentiation in vivo. We observed profound increases in CD44+ ICOS+ PD-1lo Th1-like cells in the tdLN and tumor following Treg depletion, similar to the cells found following X31pOVA (Figure 2, Figure 4F,4G). Importantly, we observed that these cells expressed T-bet, which is required for Th1 differentiation (Figure S4J). However, loss of expression of the canonical cytokines in Treg, namely II-10, Itgb8 (involved in processing of TGF-β1) or Area, had minimal impact on the proportion of ICOS± PD-1lo Th1-like cells in the tdLN (Figure S4A–S4C), suggesting Treg utilize another mechanism to suppress the CD11b± cDC2/CD4± Tconv axis.
To confim that the increase in Th1-like cells in the tumor was due to enhanced de novo priming and differentiation, we again treated tumor-bearing control and Foxp3DTR mice with DT and FTY720 and analyzed immune composition in both tdLN and TME. In the tdLN, FTY720 treatment had minimal impact in control mice but led to significant increases of Th1-like cells in Foxp3DTR tdLN (Figure 4H). In contrast, the proportion of Th1-like CD4+ Tconv dropped precipitously in the TME of Foxp3DTR mice following FTY720 treatment (Figure 4I), indicating that the increased Th1-like CD4+ Tconv observed in the TME was due to enhanced de novo priming and subsequent tumor infiltration.
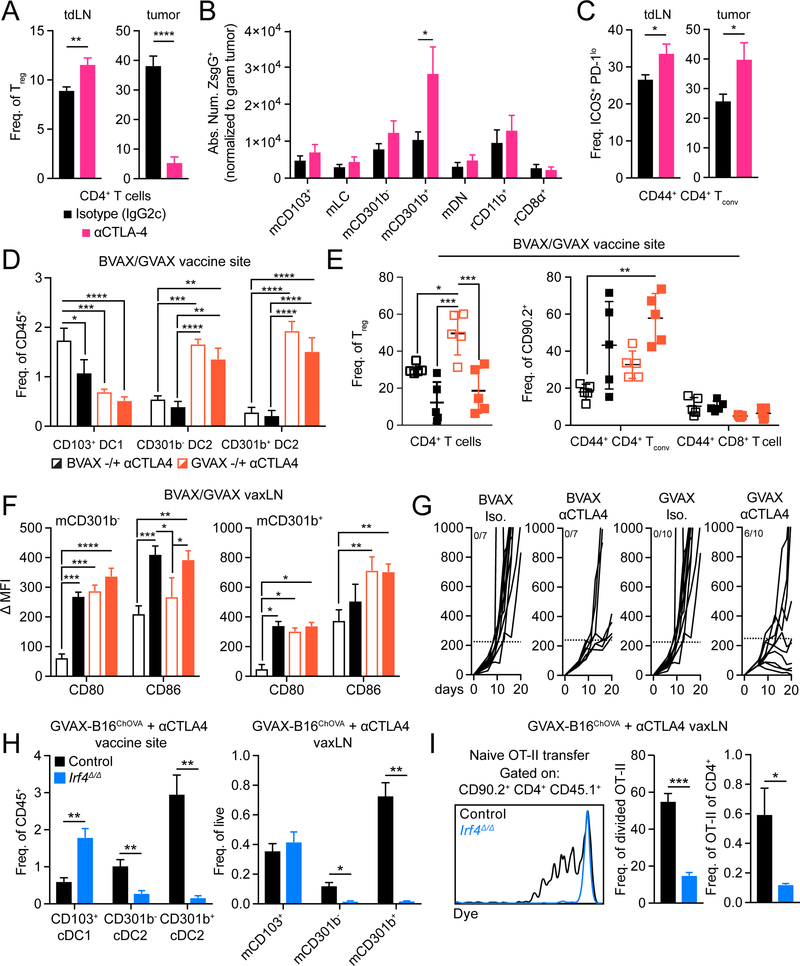
Anti-CTLA-4 based therapies function to induce expansion and functional enhancement of CD11b+ cDC2
Based our data in Figures 3 and 4, we hypothesized that Treg depletion within the TME, more than systemic effects associated with systemic depletion, was critical for optimal enhancement CD11b+ cDC2 migration and enhanced CD4+ Tconv differentiation. To address this we tested various effector forms of anti-CTLA-4, either those that only blocked CTLA-4 or a form that also resulted in intratumoal Treg depletion (cite Selby MS). Treatment of B16ZsGreen tumor-bearing animals with a depleting anti-CTLA-4 clone (mouse IgG2c, clone: 9D9) led to a reduction of Treg within the TME but not the tdLN (Figure 5A), and corresponded to increased migration of mCD301b+ to the tdLN, as compared to isotype (Figure 5B). This treatment elicited a significant increase in the proportion of ICOS+ PD-1lo CD4+ Tconv in the tdLN and tumor (Figure 5C), similar to that seen in the Foxp3DTR model (Figure 4). To assess whether these effects were due to Treg depletion and not merely blockade of CTLA-4, we compared treatment with a non-depleting anti-CTLA-4 clone (mouse IgG1, clone: 9D9) and found, here, that Treg were unchanged in the TME and tdLN, migration of either CD11 b+ cDC2 subset was unchanged and there was not an induction of ICOS+ PD-1lo CD4+ Tconv in the tdLN and tumor (Supplemental Figure 6A–C). Furthermore, levels of costimulatory molecules CD80 and CD86 were only increased on mCD301b− and mCD301b+ upon depletion of Treg within the TME (Supplemental Figure 6D,E). Together these data strongly support that Treg depletion in the TME is a primary driver of cDC2 migration and functional enhancement.
Figure 5.
Anti-CTLA-4 induces expansion and functional enhancement of CD11b+ cDC2. (A) Frequency of Treg within CD4+ T cells in tdLN (left) or tumor (right) from B16ZsGreen tumor-bearing mice treated with mouse IgG2c isotype or anti-CTLA-4 with a mouse IgG2c Fc. (B) Absolute number of ZsGreen+ DC in tdLN (normalized to weight of associated tumor). (C) Frequency of CD44+ CD4+ Tconv with ICOS+ PD-1lo surface phenotype in tdLN (left) or tumor (right). (D) DC frequency of CD45+ cells within the vaccine site of mice treated with BvAX+/− αCTLA-4 or GVAX +/− αCTLA-4. (E) Frequency of Treg amongst CD4+ T cells (left) and frequency of CD44+ CD4+ Tconv or CD44+ cD8+ T cells amongst CD90.2+ T cells within the vaccine site of mice treated with BVAX +/− αCTLA-4 or GVAX +/− αCTLA-4 (right). (F) Quantification of CD80 and CD86 DMFI on mCD301b− (left) or mCD301b+ (right) within the vaxLN of mice treated with BVAX +/− αCTLA-4 or GVAX +/− αCTLA-4. (G) Tumor growth from mice treated with BVAX +/− αCTLA-4 or GVAX +/− αCTLA-4. Ratio represents number of mice with tumors that displayed profound response (< 250mm3). Dotted line signifies 250 mm3. Representative experiment displayed. (H) DC frequency of either CD45+ or live cells within the vaccine site (left) or vaxLN (right) of control or Irf4Δ/Δ mice treated with GVAX-B16ChOVA and anti-CTLA-4. (I) Purified CD45.1+ OT-II T cells were adoptively transferred to control or Irf4Δ/Δ mice treated with GVAX-B16ChOVA and anti-CTLA-4 and vavLN were harvested 3 days later to assess OT-II T cell dye dilution (left) and quantify the frequency of cells that had divided (middle) and their frequency of endogenous CD4+ T cells (right). Data are represented as ± average SEM. *P <0.05, **P<0.01, ***P<0.001, ****P<0.0001.
As we found GM-CSF can induce CD11b+ cDC2 expansion, we hypothesized that combination GVAX (irradiated B16Gm-csf) and anti-CTLA-4 therapy potentiates CD4+ Tconv immunity through concurrent expansion of CD11b+ cDC2 and release of their suppression through Treg depletion at the vaccine site. To assess this, we compared the immune composition of the vaccine site between BVAX (irradiated B16-F10) +/− anti-CTLA-4 and GVAX +/− anti-CTLA-4 (clone 9H10). In either GVAX condition, we observed significant increases of both CD11b+ cDC2 subsets (Figure 5D), but anti-CTLA-4 treatment led to a reduction in Treg and expansion of CD4+ Tconv. (Figure 5E). We found that GVAX/anti-CTLA-4 functionally enhances CD11b+ cDC2 as expression of both CD80 and CD86 on mCD301b− within the vaccine-draining LN (vaxLN) were most improved following combination GVAX/anti-CTLA-4, whereas mCD301b+ benefitted primarily from GVAX alone (Figure 5F), indicating that perhaps Treg more specifically suppress CD301b− cDC2. When we compared tumor growth between BVAX +/− anti-CTLA-4 and GVAX +/− anti-CTLA-4 we observed that BVAX alone or in combination with anti-CTLA-4 was ineffective at inducing robust antitumor immunity. In contrast, GVAX combined with anti-CTLA-4 lead to a reduction in tumor growth rates during the course of the experiment (Figure 5G). To assess CD11b+ cDC2 depedency for CD4+ Tconv priming against tumor vaccine antigen, we analyzed cDC composition in the vaccine site and vaxLN of control and Irf4Δ/Δ mice treated with anti-CTLA-4 and a combination of GVAX and irradiated B16ChOVA. Similar to tumor-bearing animals, loss of Irf4 greatly reduced the presence of CD11b+ cDC2 in either site (Figure 5H), which corresponded to a near complete loss of CD4+ OT-II proliferation in the vaxLN (Figure 5I). Taken together, these data along with our prior findings indicate that CD11b+ cDC2 are active targets of Treg-mediated suppression and are central to the initiation of CD4+ Tconv antitumor immunity following therapeutic intervention.
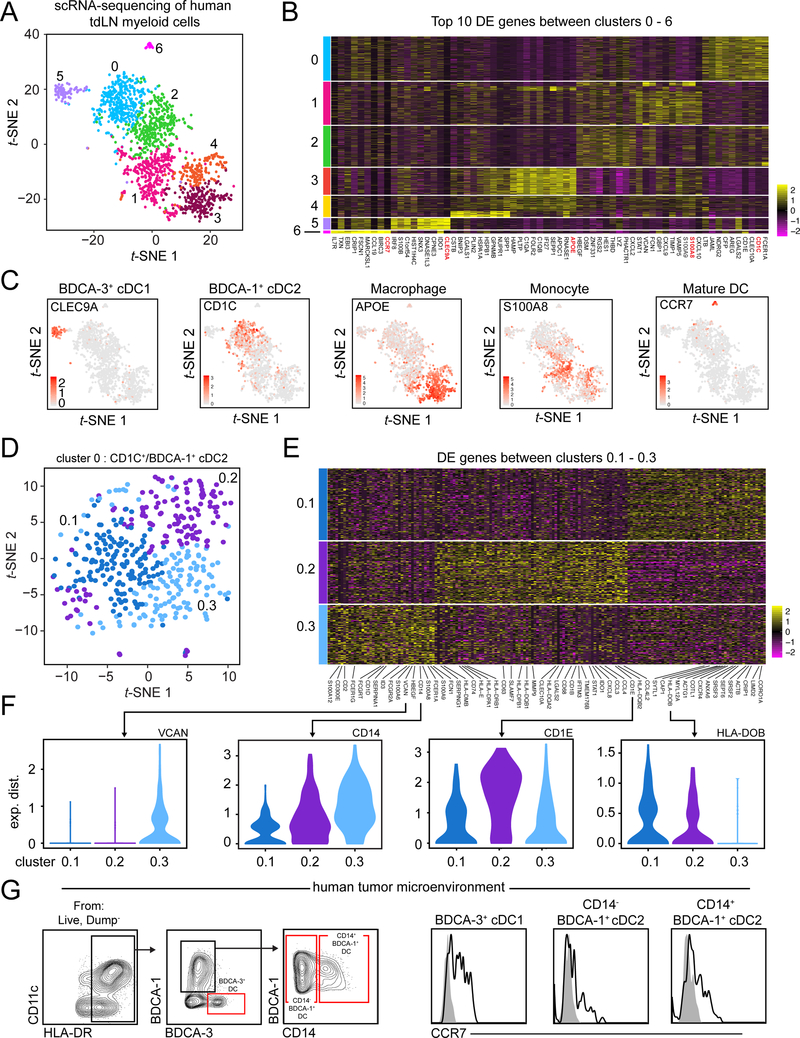
scRNA-seq of the human tdLN reveals similar heterogeneity within cDC2 subset between mouse and human.
Recent scRNA-seq on normal human blood has highlighted heterogeneity within human cDC2 (here defined by CD1c+/BDCA-1+) (Villani et al., 2017), although the existence of these populations within the human tdLN has not been assessed in an unbiased manner. To determine whether human tdLN had similar populations and heterogeneity to that of mouse and human blood, we performed scRNA-seq on myeloid populations isolated from a patient’s melanoma-draining LN. Following removal of non-APC cellular contaminants, we observed 7 unique clusters from 1,710 input cells (Figure 6A). Using DE and gene overlays, we were able to establish expressing cDC1 (hereby referred to as BDCA-3+ cDC1) occupied cluster 5 and CD1C expressing cDC2 (hereby referred to as BDCA-1+ cDC2) occupied cluster 0. The panel of genes expressed on cluster 0 and 5 were very similar to those identified previously for BDCA-1+ cDC2 and BDCA-3+ cDC1 (Villani et al., 2017), respectively, serving as confirmation of our initial identification (Figure 6B, 6C, Supplementary Table 6). To assess additional hetereogeneity within BDCA-1+ cDC2, we reclustered BDCA-1+ cDC2/cluster 0 and identified 3 populations (Figure 6D) that were transcriptionally distinct based on DE analysis (Figure 6E, 6F and Supplementary Table 7). Cluster 0.2 expressed high levels of CD1E, SLAMF7 and HLA-DQB2, genes that had been identified on a subset of blood cDC2 previously (Villani et al., 2017). Cluster 0.3, similar to mCD301b+ cDC2 in mouse, expressed genes often associated with cells of a monocyte/macrophage lineage, including CD14, VCAN and S100A8 and like cluster 0.2, resembled a previously identified cDC2 population (Villani et al., 2017). Cluster 0.1 was unique in that it was enriched for genes associated with cell motility (CORO1A, CRIP1, SEPT6, ANXA6) and may represent cellular status opposed to a bona fide distinct cellular population. We found that in addition to their presence within the human tdLN, CD14− BDCA-1+ cDC2 and CD14+ BDCA-1+ cDC2 were present within the TME of a human head and neck squamous cell carcinoma (HNSC) tumor and both expressed CCR7, indicating their migratory potential (Figure 6G). While previously identified in blood, our data suggest that cDC2 subsets in human tdLN or TME have similar characteristics to mouse cDC2 subsets, though it remains a possibility that further heterogeneity still exists within this compartment in humans, particularly across individuals.
Figure 6.
scRNA-seq of the human tdLN reveals heterogeneity within BDCA-1+ cDC2. (A) t-SNE display of CD45+ CD3− CD19/20− CD56− myeloid cells sorted from a human melanoma tdLN and processed for scRNA-seq with pDC, neutrophil, NK cell, T cell and B cell contaminants removed from graph-based clustering analysis. (B) Heatmap displaying top 10 DE genes for each cluster when comparing clusters 0 through 6 (ranked by fold change). (C) Gene overlays of markers associated with various myeloid cell types on human tdLN t-SNE. (D) t-SNE display and graph-based clustering of BDCA-1+ cDC2 (cluster 0) from (A). (E) Heatmap displaying DE genes between clusters 0.1–0.3 with genes of interested labelled. (F) Violin plots displaying expression probability differences for denoted genes within clusters 0.1–0.3. (G) Gating strategy (left) in human TME to identify BDCA-3+ cDC1, CD14− BDCA-1+ cDC2, CD14+ BDCA-1+ cDC2. Cell surface expression of CCR7 on BDCA-3+ cDC1, CD14− BDCA-1+ cDC2, CD14+ BDCA-1+ cDC2 (right).
Parsing the predictive nature of BDCA-1+ cDC2 in the human TME
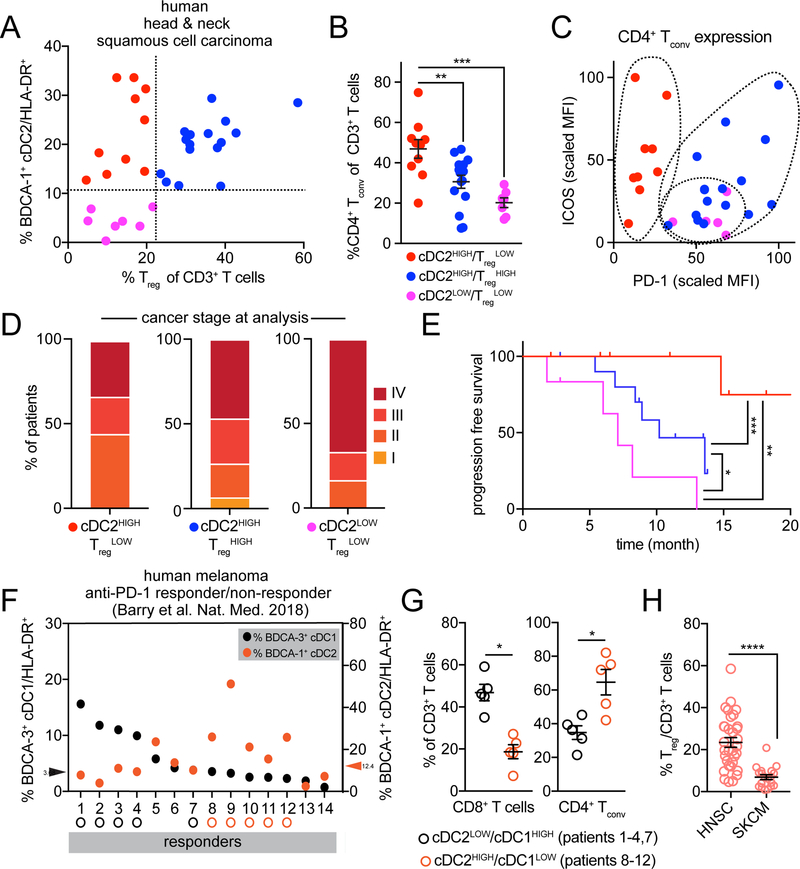
The results of our mouse models have specific predictions about the functional outcomes based on intratumoral cellular abundance. Data from Figure 3 and Figure 5 predict that CD4+ Tconv quantity and quality will vary with intatumoral cDC2 and Treg density. To assess this, we obtained 32 primary tumors from the head and neck region, a tumor type known to be rich in Treg (Mandal et al., 2016), and analyzed their immune composition with two independent flow cytometry panels (Figure 6G, S7A). In plots of BDCA-1+ cDC2 and Treg frequencies, we found three distinct patient TME with varied abundance of BDCA-1+ cDC2 or Treg (Figure 7A). As predicted from the mouse models, patient TME with low representation of BDCA-1+ cDC2 demonstrated the lowest level of CD4+ Tconv infiltration (Figure 7B). Consistent with Treg suppressing CD4+ Tconv immunity through BDCA-1+ cDC2, patients that were BDCA-1+ cDC2HIGH/TregLOW had greater CD4+ Tconv infiltration than patients with BDCA-1+ cDC2HIGH/TregHIGH. To ensure that differences in CD4+ Tconv were not merely due to a proportional shift, we analyzed CD8+ T cell frequencies which we found to vary independently to either Treg or CD4+ Tconv frequency (Figure S7B).
Figure 7.
BDCA-1+ cDC2 proportion in the human TME impacts CD4+ Tconv proportion and quality. (A) Dot plot of BDCA-1+ cDC2 frequency of HLA-DR+ cells and Treg frequency of CD3+ T cells as quantified by flow cytometry in 32 human HNSC tumor samples. Dotted lines represent demarcation of samples divided based on proportion of BDCA-1+ cDC2 (CD14−/+) and Treg. (B) The frequency of CD4+ T cells (of CD3+ T cells) within each type of TME identified in (A). (C) Surface expression of ICOS and PD-1 on CD4+ Tconv, as a normalized geometric MFI, within each type of human TME identified in (A). (D) Percent of patients with a given stage of cancer at the time of flow cytometric analysis. (E) Progression-free survival since disease diagnosis. Mantel-Cox test performed between groups. (F) 19 human melanoma tumor samples (14 anti-PD-1 responder, 5 anti-PD-1 non-responders – see S6E) were parsed based on abundance of BDCA-3+ cDC1 and plotted for proportions of both BdCA-3+ cDC1 (black) and BDCA-1+ cDC2 (orange). Responders were parsed based on those high for either BDCA-3+ cDC1 (above median split of 3.63) or BDCA-1+ cDC2 (above median split of 12.4). (G) Frequency of CD3+ T cells that are CD8+ T cells (left) and CD4+ Tconv (right) from the two groups identified in (F). (H) Proportions of Treg amongst CD3+ T cells in samples from HNSC (A-E) and skin cutaneous melanoma (SKCM) (includes anti-PD-1 responders and non-responders) (F, G). Data are represented as ± average SEM. *P <0.05, **P<0.01, ***P<0.001, ****P<0.0001.
Beyond total numbers, our model predicts improvements in ICOS+ PD-1loCD4+ Tconv phenotype would align with specific densities of BDCA-1+ cDC2 and Treg. Patients whose biopsies (TME) were low for both BDCA-1+ cDC2 and Treg had CD4+ Tconv that lacked ICOS but expressed high levels of PD-1 while CD4+ Tconv from tumors with high BDCA-1+ cDC2 and high Treg had high PD-1 as well, though expressed intermediate amounts of ICOS, perhaps reflective of cells previously reported (Zappasodi et al., 2018) (Figure 7C). In contrast, CD4+ Tconv from TME with abundant BDCA-1+ DC and low Treg frequencies had significantly higher surface expression of ICOS, paired with decreased PD-1 (Figure 7C, S7C, S7D). While cancer staging at the time of analysis was fairly similar across classes of TME (Figure 7D), progression-free survival was significantly better in patients whose TME had abundant BDCA-1+ cDC2 and low Treg than either of the other two TME classes (Figure 7E). Together these data suggest that the content of immune infiltrate informs not only the quality of an immune response (ICOS+ PD-1lo CD4+ Tconv) but also the capacity of antitumor immunity (progression-free survival).
The presence of high BDCA-3+ cDC1 and NK cells within the melanoma (SKCM) TME has been described as a general prognostic indicator of anti-PD-1 responsiveness (Barry et al., 2018), presumably due to their profoundly better ability to prime CD8+ T cells. However, in that study, we also identified patients with higher densities of CD4+ Tconv amongst responders and these did not have higher densities of cDC1. We reasoned that perhaps those responders were primed for CD4+ Tconv immunity and that this might instead rely upon cDC2. To thus assess whether BDCA-1+ cDC2 could also contribute to anti-PD-1 responsiveness, we re-gated flow cytometry data from patient biopsies to reflect the recent heterogeneity identified within BDCA-1+ cDC2. The frequency of BDCA-3+ cDC1 and BDCA-1+ cDC2 (both CD14−/+) of HLA-DR were plotted (Figure 7F). Non-responder TME were generally lower for DC of both subtypes (Figure S7E). Responder TME were then divided based on the abundance of either BDCA-3+ cDC1 or BDCA-1+ cDC2. We found that responders high for BDCA-1+ cDC2, compared to responders high for BDCA-3+ cDC1 had a significantly lower proportion of CD8+ T cells but significantly higher CD4+ Tconv within their TME (Figure 7G), promoting the hypothesis that these patients have improved CD4+ Tconv activity even with lower overall CD8+ T cell abundance. We reasoned that BDCA-1+ cDC2 abundance alone may predict CD4+ Tconv quantity in melanoma (SKCM), as proportions of Treg were significantly lower than in HNSC. (Figure 7H). These data suggest that classes of human TME can be divided based on the abundance of BDCA-1+ cDC2 and that this can be an indicator for both CD4+ Tconv quality and represent patients likely to respond to ICB.
Discussion
Here, we define the cell type(s) necessary for de novo priming of new antitumor CD4+ Tconv. A fundamental conclusion is that MHC-II presentation of peptides to prime naive CD4+ Tconv is heavily biased to CD11b+ cDC2, with the distinction between tumor control and tumor tolerance being determined by the phenotype of these cells. CD4+ Tconv priming in tdLN most resembles that of non-inflamed lymph nodes, where CD4+ Tconv are generated with a depressed activation state, with little or no evidence of the Th1 -like ICOS+ PD-1lo phenotype (Figure 2). While the TME may futher drive exhaustion, this conclusion suggests that efficacy of immunotherapies for CD4+ Tconv will rely on modulation of this defective step of priming. Indeed, given the apparent irreversibility of certain forms of exhaustion (Philip et al., 2017), it is possible that the efficacy of ICB is linked to ongoing de novo lymph node priming rather than only blockade of checkpoint ligands in the tumor.
Our scRNA-seq and functional data demonstrate that two distinct populations of IRF4-dependent CD11b+ cDC2 – for which we found homologs in human tdLN and TME – are required in vivo for initiating activation of antitumor CD4+ Tconv. The complement of myeloid cells identified in mouse tdLN are consistent with previous reports (Ochiai et al., 2014; Salmon et al., 2016; Tussiwand et al., 2015), however, our approach assayed all populations in parallel allowing for unambiguous confirmation of CD4+ Tconv stimulatory function. While our data suggest mCD301b− and mCD301b+ play largely redundant roles in antitumor immunity, as each population was capable of supporting comparable OT-II proliferation ex vivo and collectively in vivo (Figure 2), we cannot exclude the possibility that these cell populations have disparate functions in other tumor models or following different treatments. Following Treg depletion, mCD301b- displayed enhanced expression of both chemokines linked to CD4± Tconv chemoattraction and costimulatory molecules as compared to mCD301b+ (Figure 4). Paired with our findings suggesting mCD301b± in tdLN express higher surface levels of inhibitory PD-L2 and LILRB4 (Figure 1), we postulate that perhaps mCD301b- are more supportive of productive antitumor CD4± Tconv differentiation. cDC2 have been previously associated with CD4+ Tconv priming, with CD301b+ cDC2 being shown to specifically induce Th2 responses to adjuvant (Kumamoto et al., 2013) and a second report suggesting that pan-cDC2 population in tumor was demonstrated to induce Th17 differentiation primarily in vitro (Laoui et al., 2016). However, our findings specifically document suboptimal triggering of early activation (e.g. CD69, CD44 levels) as well as poor induction of differentiation in OT-II from tdLN and non-inflammed LN compared to inflammatory conditions such as influenza or Treg-depletion (Figure 2). Previous work has identified an IFNγ-dependent homeostatic 200 gene program associated with poor DC:T cell priming, some of which are co-opted in tumors (Nirschl et al., 2017). We also found expression of some of these genes in tdLN mDC (e.g. Socs2, Fscn1) (Figure 1) and were able to further define a functional readout for phenotypic defects in CD11b+ cDC2. Whether expression of these genes is associated with Treg-mediated suppression or defects in CD4+ Tconv differentiation is unclear, although these data support a homeostatic phenotypic dampening of cDC function, in particular cDC2, that can be reverted during specific inflammatory settings.
Both systemic and tumor-specific therapeutic depletion of Treg enhanced cDC2 migration and reverted phenotypic dysfunction, which in turn allowed productive antitumor CD4+ Tconv priming to occur in the tdLN. Previous studies have demonstrated reactivated CD4+ Tconv immunity following Treg-depletion (Bos et al., 2013; Wallin et al., 2016), although the mechanism and the prominence of cDC2 in modulating differentiation, through which reactivation occured was unclear. While systemic inflammation in response to global Treg depletion may provide added modulation of myeloid development, our data supports a central role of Treg, largely at the tumor site, as critical modulators of cDC2. Although we cannot preclude the possibility that Treg suppress CD4+ Tconv directly (Pandiyan et al., 2007) or impact DC more generally (Bauer et al., 2014; Qureshi et al., 2011), our data demonstrates a potently immunosuppressive relationship between Treg and CD11b+ cDC2, although the specific means through which Treg exert this suppression on cDC2 is still unknown (Supplemental Figure 5). Expansion of CD11b+ cDC2, in either the TME (Figure 3) or GVAX site (Figure 5), induces concurrent increases in Treg which likely represents that cDC2 produced under these conditions undergo continued suppression, opposing their ability to drive productive priming of effector CD4+ Tconv. This is consistent with data from others showing Treg require MHC-II on DC to expand in the periphery (Zou et al., 2010). Enhanced cDC2 migration to the tDLN was observed following Treg depletion induced by either genetic or therapeutic means (Figure 3, Figure 4). While DC can upregulate CCR7 in response to inflammatory cues (Krappmann et al., 2004), DC can also migrate continuously under homeostatic conditions (Baratin et al., 2015). Given the rapid migration of of cDC2 from tumor to tdLN following Treg-depletion, we suspect that Treg may regulate homeostatic cDC2 trafficking to the tdLN.
Phenotypically enhanced CD11b+ cDC2 were better able to support CD4+ Tconv priming and support improved differentiation to a ICOS+ PD-1lo Th1-like phenotype (Figure 3, Figure 4). Although CD103+ cDC1 have been shown to induce Th1 immunity in specific inflammatory settings (Liang et al., 2016), they were dispensible for tumor rejection following Treg depletion, supporting our data demonstrating that CD11b+ cDC2 are uniquely able to initiate productive antitumor CD4+ Tconv priming in the absence of Treg (Figure 4). Efficacious anti-CTLA-4 treatment in both mouse and human is associated with the generation of ICOS+ PD-1lo Th1-like systemically (Fan et al., 2014; Wei et al., 2017), although the site of this population’s initial emergence was previously undefined. We found that antitumor ICOS+ PD-1lo CD4+ Tconv arise in tdLN during de novo priming and additionally that infiltration of these tdLN-derived antitumor CD4+ Tconv, as opposed to via local reactivation in the TME, was the dominant mechanism through which tumor rejection occurred (Figure 3, Figure 4). While we did not directly assess this, we hypothesize that CD4± Tconv downregulate CD62L in a Foxp3DTR tdLN, allowing for improved circulation and TME entry. Interestingly, CD4± Tconv as opposed to CD8± T cells were required for tumor rejection (Figure 3), although it is currently unclear whether CD4± Tconv are cytoxic themselves or induce other immune populations to participate in tumor cell elimination. Together, this highlights the importance of enhancing cDC2 phenotype in patients in order to improve distal priming for more effective immunotherapy.
In human HNSC TME, we found a remarkable concordance with our Treg depletion data whereby heterogeneity in Treg and cDC2 abundance parsed subsets of patients with distinct phenotypes and, in particular, the relationship between BDCA-1+ cDC2 and Treg informed both the quantity and character of CD4+ Tconv. This parallel with our data in mouse strongly suggests that a similar mechanism of Treg-mediated suppression exists in human. The use of Treg alone as a prognostic indicator has varying levels of predictive power (Shang et al., 2015) and this may in part be due to the fact tha low Treg abundance fails to differentiate cohorts that have or do not have requisite cDC2 populations for CD4+ Tconv priming. Our pairing of Treg abundance with the additional parameter (BDCA-1+ cDC2) unmasks hetereogeneity of TME, allowing for significant predictions of immune response quality and disease-free survival (Figure 7). Looking forward, this suggests that BDCA-1+ DC abundance is a biomarker for a primed microenvironment for response to ICB or to novel therapies targeting Treg suppression of cDC2. Indeed, a human TME dataset of anti-PD-1 responder/non-responders demonstrated that while BDCA-3+ cDC1 cellularity is largely associated with anti-PD-1 responsiveness (Barry et al., 2018), some patients were surprisingly BDCA-3+ cDC1 low, but contained higher proportions of BDCA-1+ cDC2 and CD4+ Tconv, suggesting that at least in some TME, such as those with tumor cells that express MHC-II (Johnson et al., 2016), CD4+ Tconv may be capable of playing a preeminent role in successful antitumor responses.
Taken together, our work highlights CD11b+ cDC2/BDCA-1+ cDC2 as a target of Treg suppression and as a necessary population for directing antitumor CD4+ Tconv immunity. Furthermore, cDC2 abundance in the human TME may act as a biomarker for not only CD4+ Tconv quality but also as a contributing indicator for responsiveness to ICB. Classifying TME based on immune infiltrate has predictive power (Binnewies et al., 2018) and thus recent (Azizi et al., 2018; Lavin et al., 2017; Puram et al., 2017) and future efforts to characterize disparate TME with unbiased high-dimensional techniques will undoubtable prove invaluable for identifying unique classes of patient TME that are profoundly immunosuppressed or poised for therapeutic response.
Materials and Methods
Human Tumor Samples
The human head and neck tumor set consisted of a total of 32 tumors removed from the head and neck region, agnostic to location. The anti-PD-1 responder/non-responder melanoma tumor set was published previously (Barry et al., 2018). All patients consented for tissue collection under a UCSF IRB approved protocol (UCSF IRB# 13– 12246 and 14–15342). Samples were obtained after surgical excision with biopsies taken by the Pathology Department to confirm the presence of tumor cells. Patients were selected without regard to prior treatment. Freshly resected samples were placed in ice-cold PBS or Leibovitz’s L-15 medium in a 50 mL conical tube and immediately transported to the laboratory for evaluation. Patient samples were coded and flow analysis was scored by separate individuals prior to data agglomeration. All samples were processed and analyzed by flow cytometry, but only those with at least 1,000 live CD45+ cell events were included in the analysis.
Mice
All mice were treated in accordance with the regulatory standards of the National Institutes of Health and American Association of Laboratory Animal Care and were approved by the UCSF Institution of Animal Care and Use Committee. The following mice were purchased for acute use or maintained under specific pathogen-free conditions at the University of California, San Francisco Animal Barrier Facility. We attempted to use Irf4flox/flox;CD11c-Cre but discovered independent breeding cages were producing germline Irf4 globally deficient pups, complicating our findings (data not shown).
Method Details
Tumor cell lines, tumor cell injections and tumor growth experiments
B16-F10 (ATCC, CRL-6475) was purchased. B16-ChOVA (B16ChOVA), a derivative of B16-F10, was created through transduction of B16-F10 with an mCherry-OVA (ChOVA) fusion construct identical to that used in previous studies in our lab (Engelhardt et al., 2012; Roberts et al., 2016). B78ChOVA, derived from the parental B78 subline of B16, was generated in our laboratory and described previously (Broz et al., 2014). B16-ZsGreen (B16ZsGr) was previously generated in our laboratory as described (Headley et al., 2016). B16GM-CSF (GVAX) (Dranoff et al., 1993) were acquired from the laboratory of Dr. Lawrence Fong at UC San Francisco. Adherent cell lines were cultured at 37°C in 5% CO2 in DMEM (Invitrogen), 10% FCS (Benchmark), Pen/Strep/Glut (Invitrogen).
For tumor cell injection, adherent tumor cells were lifted using 0.05% Trypsin-EDTA (Thermo Fisher Scientific) and washed 3X with DPBS (Thermo Fisher Scientific). 1.0×105 – 2.5×105 tumor cells were resuspended in DPBS and mixed 1:1 with Matrigel GFR (Corning) for a final injection volume of 50 μL. Mice anesthetized with isoflurane (Henry Schein) were shaved on their flank and injected subcutaneously either unilaterally or bilaterally depending on the experimental setup.
For tumor measurements, tumors were typically measured 3 times per week using electronic calipers. Tumor volume was calculated through the formula V = 0.5(w2 × l). Mice were removed from the study and euthanized when tumors exceeded a volume of 1000 mm3.
Single Cell RNA Sequencing (scRNA-Seq)
For mouse scRNA-seq, live CD90.2− B220− Ly6G− NK1.1−CD11b+ and/or CD11c+ cells were sorted from inguinal and axillary LN with a BD FACSAria Fusion. For human scRNA-seq, live CD3−CD19/20−CD56− cells were sorted from a melanoma-draining LN on a BD FACSAria Fusion. After sorting, cells were pelleted and resuspended at 1×103 cells/μl in 0.04%BSA/PBA and loaded onto the Chromium Controller (10X Genomics). Samples were processed for single-cell encapsulation and cDNA library generation using the Chromium Single Cell 3’ v2 Reagent Kits (10X Genomics). The library was subsequently sequenced on an Illumina HiSeq 4000 (Illumina).
Single Cell Data Processing
Sequencing data was processed using 10X Genomics Cell Ranger V1.2 pipeline. The Cell Ranger subroutine mkfastq converted raw, Illumina bcl files to fastqs which were then passed to Cell Ranger’s count, which aligned all reads using the aligner STAR (Dobin et al., 2013) against UCSC mm10 or GRCh38 genomes for mouse and human cells, respectively. After filtering reads with redundant unique molecular identifiers (UMI), count generated a final gene-cellular barcode matrix. Both mkfastq and count were run with default parameters.
Cellular Identification and Clustering
For each sample, the gene - barcode matrix was passed to the R (v. 3.4.3) software package Seurat (Satija et al., 2015) (http://satijalab.org/seurat) (v2.3.0) for all downstream analyses. We then filtered on cells that expressed a minimum of 200 genes and required that all genes be expressed in at least 3 cells. We also removed cells that contained > 5% reads associated with cell cycle genes (Kowalczyk et al., 2015; Macosko et al., 2015). Count data was then log2 transformed and scaled using each cell’s proportion of cell cycle genes as a nuisance factor (implemented in Seurat’s ScaleData function) to correct for any remaining cell cycle effect in downstream clustering and differential expression analyses. For each sample, principal component (PC) analysis was performed on a set of highly variable genes defined by Seurat’s FindVariableGenes function. Genes associated with the resulting PCs (chosen by visual inspection of scree plots) were then used for graph-based cluster identification and subsequent dimensionality reduction using t-distributed stochastic neighbor embedding (tSNE). Cluster-based marker identification and differential expression were performed using Seurat’s FindAllMarkers for all between-cluster comparisons.
ImmGen Signature Generation
To generate a priori signatures for the myeloid cell types that we expected to find in the mouse tdLN sample, we downloaded microarray based transcriptional profiles from the Immunological Genome Project data Phase 1(Heng and Painter, 2008) (GSE15907). See Supplementary Table 1 for the specific samples used.
For each ImmGen population, we performed DE analysis comparing samples from the population of interest to the aggregate of the remaining 6 groups using the R package limma (Ritchie et al., 2015). We ordered the top 20 genes with the smallest FDR values (Benjamini and Hochberg, 1995) by fold change (excluding any genes that were downregulated in the group of interest) and then cross referenced the resulting list with the single cell expression matrix from each sample. This left genes that were both highly differentially expressed in the IMMGEN profiles and expressed in our single cell data sets of interest. The top 10 genes (or fewer if less than 10 genes remained) by fold change were then median normalized and aggregated to create a single “signature gene” for each cell type. These signature genes were 0–1 scaled and plotted in the context of the t-SNE dimensionality reduction to show cellular location.
Sequencing Sample Aggregation
In order to generate pairwise aggregations between samples and control for potential batch effects, we used Seurat’s Canonical Correlation Analysis (CCA) functionality. All post-filtered cells from each of the single sample analyses were used in the aggregate. CCA was performed on the union of the 2000 genes with highest dispersions from each dataset. The number of canonical correlation vectors (CCVs) used in downstream clustering and t-SNE analyses was chosen by visual inspection of heatmaps of genes associated with those top CCVs. Results were robust to moderate changes in this final number of CCVs.
Mouse Tissue Digestion and Flow Staining
Tumor and LN tissues were harvested and enzymatically digested with 0.2 mg/ml DNAse I (Sigma-Aldrich), 100 U/ml Collagenase I (Worthington Biochemical), and 500 U/ml Collagenase Type IV (Worthington Biochemical) for 30–45 minutes at 37 °C. TdLN included inguinal and axillary LN. Tumor samples were subjected to consistent agitation during this time and LN samples were rapidly pipetted at the half-point time. Samples were filtered to generate a single-cell suspension and washed with stain media (PBS, 2% FCS). For bone marrow cells, mouse femurs and tibias were flushed with stain media and subsequently underwent red blood cell lysis.
Cells harvested from these tissues or in vitro culture were washed with PBS and stained with Zombie NIR fixable viability dye (BioLegend) for 30 minutes at 4°C to distinguish live and dead cells. Cells were then washed with stain media and non-specific binding was blocked with anti-CD16/32 (BioXCell), and 2% rat serum (Invitrogen) and 2% Armenian hamster serum (Innovative Research). Cell surface proteins were then stained on ice for 30 minutes. Cells were washed again and resuspended with stain media prior to collection and analysis on a BD Fortesssa or LSR-II flow cytometer. When applicable, black latex beads were added to the sample for quantification of absolute cell number. For intracellular stains, cells were fixed and permeabilized with the FoxP3/Transcription Factor Staining Buffer Set (Thermo Fisher Scientific) after surface marker staining. Intracellular antibodies were stained in permeabilization buffer with 2% rat serum for at least 30 minutes at room temperature.
Human Tissue Digestion and Flow Staining
Tumor or LN tissue was thoroughly chopped with surgical scissors and transferred to GentleMACs C Tubes (Miltenyi Biotec) containing 20 uL/mL Liberase TL (5 mg/ml, Roche) and 50 U/ml DNAse I (Roche) in RPMI 1640 per 0.3 g tissue. GentleMACs C Tubes were then installed onto the GentleMACs Octo Dissociator (Miltenyi Biotec) and incubated according to the manufacturer’s instructions. Samples were then quenched with 10 mL of sort buffer (PBS/2% FCS/2mM EDTA), filtered through 100 um filters and spun down. Red blood cell lysis was performed with 175 mM ammonium chloride.
Cells were then incubated with Human FcX (Biolegend) to prevent non-specific antibody binding. Cells were then washed in DPBS and incubated with Zombie Aqua Fixable Viability Dye (Biolegend). Following viability dye, cells were washed with sort buffer and incubated with cell surface antibodies for 30 minutes on ice and subsequently fixed in either Fixation Buffer (BD Biosciences) or in Foxp3/Transcription Factor Staining Buffer Set (ThermoFisher Scientific) if intracellular staining was required.
APC-T cell In Vitro Co-Culture Assays
APC populations were double-sorted (yield followed by purity) from tdLN using a BD FACSAria Fusion and co-cultured with 2×104 isolated eFluor670-labeled OT-II T cells at a 1:5 ratio in complete RPMI (Pen/Strep, NEAA, NaPyr, 2-ME, 10% FCS) in 96-well V-bottom plates. Cells were harvested for analysis 3 days later. OVA peptide 323–339 (GenScript) was added to wells at 1 μg/ml as a positive control.
Mouse T cell Isolation and In Vivo Adoptive T Cell Transfer
Inguinal, axillary, brachial, superficial cervical and mesenteric LN were isolated from CD45.1+ OT-II mice. LN were smashed through 100 um filters and subsequently spun down and counted. CD4+ T cells were then isolated using EasySep CD4 negativeselection kits (STEMCELL Technologies).
1×105 isolated CD45.1+ CD4+ OT-II T cells were either transferred immediately in cases of PMA/Ionomycin restimulation experiments or labeled with Cell Proliferation Dye eFluor670 (Thermo Fisher Scientific) and 1.0–5.0×105 cells were adoptively transferred to CD45.2+ mice. LN were harvested for proliferation analysis at day 3 posttransfer and for PMA/ionomycin re-stimulation at day 7 post-transfer. XCR1DTR and Cx3cr1iDTR mice were treated with 500 ng of DT every other day beginning the day prior to OT-II transfer through the experimental end point. Foxp3DTR mice were injected with DT for two days prior to OT-II transfer and then the day following OT-II transfer.
T Cell Cytokine Analysis
For cytokine analysis of endogenous or adoptively transferred T cells, cells from either LN or tumors were used for re-stimulation. Single cell suspensions were incubated with 50 ng/ml PMA (Sigma-Aldrich), 500 ng/ml ionomycin (Thermo Fisher Scientific), 3 μg/ml brefeldin A (Cayman Chemical Company), and 2μM monensin (Thermo Fisher Scientific) for 5–6 hours in complete RPMI and stained for surface and intracellular proteins using the Foxp3/Transcription Factor Staining Buffer Set (ThermoFisher Scientific).
In Vivo Treatments
For diphtheria toxin (DT), while treatment schedules varied depending upon mouse genetic strain or type of experiment, mice received 500 ng boluses of un-nicked DT (List Biologics, 150) intraperitoneally. Foxp3DTR, XCR1DTR and Cx3cr1iDTR mice were typically injected on days 9, 10 and 12 followed by flow cytometric analysis at day 14.
For comparisons of CD4+ Tconv priming between steady-state, tumor-bearing and influenza-infected conditions, mice were injected subcutaneously with either 20 μg of endotoxin-free ovalbumin (Invivogen) in 50 μl of PBS or 2.0×105 B16ChOVA. Mice receiving influenza were infected intranasally with 1×105 PFU of X31-OT-II (Thomas et al., 2006), prepared as previously described (GeurtsvanKessel et al., 2008).CD45.1+ OT-II+ CD4+ T cells were transferred intravenously 2 days after ovalbumin and X31-OT-II treatment and 14 days after B16ChOVA injection.
To assess CD4/CD8 T cell dependency for tumor rejection following Treg depletion or GVAX/anti-CTLA-4 treatment, mice were injected with 250 μg of isotype (Clone: LTF-2, BioXCell), anti-CD4 (Clone: GK1.5, BioXCell) or anti-CD8a (Clone: 2.43, BioXCell) was injected at days 10, 13 and 16 post-tumor injection for Foxp3DTR and days 4, 7 and 10 for GVAX/anti-CTLA-4 treatment.
To assess the requirement of T cell LN egress, control or Foxp3DTR mice were treated with 500 ng of DT on days 9, 10 and 12 post-tumor injection and with 200 μg FTY720 (Cayman Chemicals) every day beginning on day 8 post-tumor injection through the end of the experiment.
For Fc-modified anti-CTLA-4 experiments, mice were injected with 2×105 B16ZsGreen cells. On days 7, 9, 10, 11 and 13 post-tumor injection, mice received 250 μg of mouse IgG2c isotype, anti-CTLA-4 IgG2c (modified clone 9D9, Bristol-Myers-Squibb), mouse IgG1 isotype or anti-CTLA-4 IgG1 (modified clone 9D9, Bristol-Myers-Squibb).
For GVAX/anti-CTLA-4 experiments, mice were injected with either 1×105 (tumor growth) or 2×105 B16-F10 (cellular analysis). On days 3, 6 and 9 post-tumor injection, mice were injected subcutaneously on their contralateral flank with either PBS or 1×106 50 Gy-irradiated GVAX cells and received either 250 μg anti-CTLA-4 (9H10, BioXCell) or Syrian hamster IgG isotype (BioXcell) on day 3, and 100 μg of antibody on days 6 and 9.
Statistical analysis and experimental design
Unless specifically noted, data displayed is from a representative experiment of ≥ 2 independent experiments. Experimental group assignment was determined by genotype or, if all wild-type mice, by random designation. Error bars represent mean ± S.E.M. calculated using Prism unless otherwise noted. Statistical analyses were performed using GraphPad Prism software. For pairwise comparisons, unpaired T tests were used unless otherwise noted. For statistical measures between more than two groups, one-way ANOVA would be performed unless otherwise noted. Comparisons found to be nonsignificant are not shown. Investigators were not blinded to group assignment during experimental procedures or analysis.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-mouse CD11c BV650 (clone N418) | Biolegend | 117339 |
| anti-mouse/human CD11b BV605 (clone M1/70) | Biolegend | 101257 |
| anti-mouse CD103 BV421 (clone 2E7) | Biolegend | 121421 |
| anti-mouse Ly-6C BV711 (clone HK1.4) | Biolegend | 128037 |
| anti-mouse CD90.2 BV785 (clone 30-H12) | Biolegend | 105331 |
| anti-mouse/human CD45R/B220 BV785 (clone RA3-6B2) | Biolegend | 103246 |
| anti-mouse Ly-6G BV785 (clone IA8) | Biolegend | 127645 |
| anti-mouse Siglec F BV786 (clone E50-2440) | BD Biosciences | 740956 |
| anti-mouse NK1.1 BV785 (clone PK136) | Biolegend | 108749 |
| anti-mouse CD24 PE/Cy7 (clone M1/69) | Biolegend | 101822 |
| anti-mouse MHC-II AF700 (clone M5/114.15.2) | Biolegend | 107622 |
| anti-mouse CD301b PE or APC (clone URA-1) | Biolegend | 146814, 146803 |
| anti-mouse CD8a PerCP/Cy5.5 or PE/Cy7 (clone 53-6.7) | Biolegend | 100734, 100722 |
| anti-mouse F4/80 FITC (clone BM8) | Biolegend | 123107 |
| anti-mouse CD45 PerCP/Cy5.5 (clone A20) | Biolegend | 110727 |
| anti-mouse CD197/CCR7 PE | Biolegend | 120105 |
| anti-mouse CD9 AF647 (clone MZ3) | Biolegend | 124809 |
| anti-mouse CD135/FLT3 PE (clone A2F10) | Biolegend | 135305 |
| anti-mouse CD172a/SIRPA PE or AF488 (clone P84) | Biolegend | 144011,144023 |
| anti-mouse CD14 PE (clone Sa14-2) | Biolegend | 123309 |
| anti-mouse CD16/32 PE (clone 93) | Biolegend | 101307 |
| anti-mouse CD200R PE (clone OX110) | Biolegend | 123907 |
| anti-mouse CD206 PE (clone C068C2) | Biolegend | 141705 |
| anti-mouse B7-H1 PE (PD-L1/CD274) (clone 10F.9G2) | Biolegend | 124307 |
| anti-mouse B7-H2 PE (ICOS-L/CD275) (clone HK5.3) | Biolegend | 107405 |
| anti-mouse B7-H3 PE (CD276) (clone RTAA15) | Biolegend | 123507 |
| anti-mouse B7-DC (PD-L2/CD273) PE (clone TY25) | Biolegend | 107205 |
| anti-mouse CD85K (LILRB4) PE | Biolegend | 144903 |
| anti-mouse CD4 BUV395 (clone GK1.5) | BD Biosciences | 563790 |
| anti-mouse/rat/human CD278/ICOS APC (clone C398.4A) | Biolegend | 313510 |
| anti-mouse CD279/PD-1 PE (clone RMP1-14) | Biolegend | 114118 |
| anti-mouse/human CD44 BV711 (clone IM7) | Biolegend | 103057 |
| anti-mouse CD69 BV650 (clone H1.2F3) | Biolegend | 104541 |
| anti-mouse Foxp3 eF450 (clone FJK-16s) | Thermo Fisher | 48-5773-82 |
| anti-mouse/rat/human Foxp3 AF647 (clone 150D) | Biolegend | 320014 |
| anti-mouse IL-4 PE (clone 11B11) | Biolegend | 504104 |
| anti-mouse IL-17a BV421 (clone TC11-18H10.1) | Biolegend | 506926 |
| anti-mouse IFNg PE/Cy7 (clone XMG1.2) | Biolegend | 505825 |
| anti-mouse/human T-bet BV605 (clone 4B10) | Biolegend | 644817 |
| anti-mouse CD117 APC (clone 2B8) | Biolegend | 105811 |
| anti-mouse CD115 PerCP/Cy5.5 (clone AFS98) | Biolegend | 135525 |
| anti-mouse Ly6-G (clone IA8) | Biolegend | 127603 |
| anti-mouse CD3e (clone 145-2C11) | Biolegend | 100303 |
| anti-mouse CD127 (clone A7R34) | Biolegend | 135005 |
| anti-mouse NK1.1 (clone PK136) | Biolegend | 108703 |
| anti-mouse/human CD45R (RA3-6B2) | Biolegend | 103203 |
| anti-mouse TER-119 (clone TER-119) | Biolegend | 116203 |
| anti-mouse TCR γ/δ (clone GL3) | Biolegend | 118103 |
| Streptavidin BV421 | Biolegend | 405226 |
| anti-human CD45 APC/e780 (clone HI30) | Thermo Fisher | 47-0459-42 |
| anti-human CD3e PerCP/e710 (clone OKT3) | Thermo Fisher | 46-0037-42 |
| anti-human HLA-DR BUV395 (clone G46-6) | BD Biosciences | 564040 |
| anti-human CD56 BUV737 (clone NCAM16.2) | BD Biosciences | 564448 |
| anti-human CD4 PE/Dazzle 594 (clone S3.5) | Biolegend | 100455 |
| anti-human CD8a BV605 (clone RPA-T8) | Biolegend | 301039 |
| anti-human CD127 BV650 (clone HIL-7R-M21) | BD Biosciences | 563225 |
| anti-human CD38 AF700 (clone HIT2) | Biolegend | 303523 |
| anti-human CD25 APC (clone 2A3) | BD Biosciences | 340939 |
| anti-human CD45RO PE (clone UCHL1) | BD Biosciences | 561889 |
| anti-human PD-1 BV786 (clone EH12) | BD Biosciences | 563789 |
| anti-human ICOS BV711 (clone DX29) | BD Biosciences | 563833 |
| anti-human FoxP3 PE/Cy7 (clone 236A/E7) | Thermo Fisher | 25-4777-41 |
| anti-human CTLA-4 BV421 (clone BNI3) | BD Biosciences | 565931 |
| anti-human/mouse/rat Ki67 AF488 (clone SolA15) | Thermo Fisher | 11-5698-82 |
| anti-human CD19 PerCP/e710 (clone H1B19) | Thermo Fisher | 45-0199-42 |
| anti-human CD20 PerCP/e710 (clone 2H7) | Thermo Fisher | 45-0209-42 |
| anti-human CD56 PerCP/e710 (clone CMSSB) | Thermo Fisher | 46-0567-42 |
| anti-human CD64 BUV737 (clone 10.1) | BD Biosciences | 564425 |
| anti-human CD11c AF700 (clone 3.9) | Thermo Fisher | 56-0116-42 |
| anti-human CD16 BV605 (clone 3G8) | Biolegend | 302039 |
| anti-human CD273/PDL2 BV650 (clone MIH18) | BD Biosciences | 563844 |
| anti-human/mouse TREM2 APC (clone 237920) | R&D Systems | FAB17291A |
| anti-human CD304 PE (clone 12C2 ) | Biolegend | 354503 |
| anti-human CD1C/BDCA-1 PE/Cy7 (clone L161) | Biolegend | 331515 |
| anti-human CD197 BV421 (clone G043H7) | Biolegend | 353207 |
| anti-human BDCA-3 FITC (clone AD5-14H12) | Miltenyi | 130-098-843 |
| anti-human PDL1 BV786 (clone MIH1) | BD Biosciences | 563739 |
| anti-human CD14 BV711 (clone M5E2) | Biolegend | 301837 |
| PE Rat IgG2a, k Isotype Ctrl Antibody (clone RTK2758) | Biolegend | 400508 |
| PE Rat IgG1, k Isotype Ctrl Antibody (clone RTK2071) | Biolegend | 400408 |
| APC Armenian Hamster IgG Isotype Ctrl Antibody (clone HTK888) | Biolegend | 400912 |
| BV605 Mouse IgG1, k Isotype Ctrl Antibody (clone MOPC-21) | Biolegend | 400162 |
| BV421 Mouse IgG2a, k Isotype Ctrl Antibody (clone MOPC-173) | Biolegend | 400259 |
| anti-mouse CD4 InVivoMab (clone GK1.5) | BioXCell | BE0003-1 |
| anti-mouse CD8 InVivoMab (clone 2.43) | BioXCell | BE0061 |
| anti-mouse CTLA-4 (CD152) InVivoMab (clone 9H10) | BioXCell | BE0131 |
| Rat IgG2b, k InVivoMab | BioXCell | BE0090 |
| Polyclonal Syrian hamster IgG InVivoMab | BioXCell | BE0087 |
| anti-mouse CD16/32 InVivoMab | BioXCell | BE0307 |
| anti-mouse CTLA-4 mouse IgG2c (modified clone 9D9) | Bristol-Myers-Squibb | |
| anti-mouse CTLA-4 mouse IgG1 (modified clone 9D9) | Bristol-Myers-Squibb | |
| mouse IgG2c isotype | Bristol-Myers-Squibb | |
| mouse IgG1 isotype | Bristol-Myers-Squibb | |
| Normal Rat Serum | Thermo Fisher | 10710C |
| Armenian Hamster Serum | Innovative Research | IGHMA-SER |
| Biological Samples | ||
| Human tumor samples | UC San Francisco | IRB # 13-12246 and 14-15342 |
| Mouse tissue samples (LN, tumor) | UC San Francisco | IACUC: AN170208 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Matrigel GFR, Phenol-red free | Corning | 356231 |
| Collagenase, Type I | Worthington Biochemical | LS004197 |
| Collagenase, Type IV | Worthington Biochemical | LS004189 |
| DNAse I | Roche | 10104159001 |
| Liberase TL | Roche | 5401020001 |
| Human TruStain FcX | Biolegend | 422302 |
| Zombie Aqua Fixable Viability Kit | Biolegend | 423102 |
| Zombie NIR Fixable Viability Kit | Biolegend | 423106 |
| Brilliant Stain Buffer Plus | BD Biosciences | 566385 |
| Brefeldin A (BFA) | Sigma-Aldrich | B7651 |
| Phorbol 12-myristate 13-acetate (PMA) | Sigma-Aldrich | P8139 |
| Ionomycin | Invitrogen | I24222 |
| Monensin Solution (1000X) | Thermo Fisher | 00-4505-51 |
| Diphtheria Toxin (unnicked) | List Biological Laboratories | 150 |
| FTY720 | Cayman Chemical Company | 10006292 |
| eBioscience™ Cell Proliferation Dye eFluor™ 670 | 65-0840-85 | |
| Ovalbumin Endofit | Invivogen | Vac-pova |
| OVA peptide (323-339) | Genscript | RP10610-1 |
| Critical Commercial Assays | ||
| Chromium Single Cell 3’ Library & Gel Bead Kit V2 | 10X Genomics | 120237 |
| BD Cytofix | BD Biosciences | 554655 |
| Foxp3/Transcription factor staining buffer set | Thermo Fisher | 00-5523-00 |
| EasySep Mouse CD4+ T Cell Isolation kit | STEMCELL Technologies | 19852 |
| Deposited Data | ||
| GEO XXXXXX | ||
| Experimental Models: Cell Lines | ||
| B16-F10 | ATCC | CRL-6475 |
| B16-ChOVA | UC San Francisco | N/A |
| B78-ChOVA | UC San Francisco | N/A |
| B16-GM-CSF | UC San Francisco | N/A |
| B16-ZsGreen | UC San Francisco | N/A |
| Experimental Models: Organisms/Strains | ||
| Mouse: C57BL/6J | The Jackson Laboratory | 000664 |
| Mouse: B6 CD45.1 (B6.SJL-Ptprca Pepcb/BoyJ) | The Jackson Laboratory | 002014 |
| Mouse: OT-II (B6.Cg-Tg(TcraTcrb)425Cbn/J | The Jackson Laboratory | 004194 |
| Mouse: Irf4 fl/fl (B6.129S1-Irf4tm1Rdf/J) | The Jackson Laboratory | 009380 |
| Mouse: ActB-Cre (FVB/N-Tmem163Tg(ACTB-cre)2Mrt/J) received backcrossed to C57/Bl6 | The Jackson Laboratory | 003376 |
| Mouse: CD11c-Cre (B6.Cg-Tg(Itgax-cre)1-1Reiz/J) | The Jackson Laboratory | 008068 |
| Mouse: Cx3cr1iDTR (B6N.129P2-Cx3cr1tm3(DTR)Litt/J) | The Jackson Laboratory | 025629 |
| Mouse: Ccr7−/− (B6.129P2(C)-Ccr7tm1Rfor/J) | The Jackson Laboratory | 006621 |
| Mouse: Zbtb46GFP (B6.129S6(C)-Zbtb46tm1.1Kmm/J) | The Jackson Laboratory | 027618 |
| Mouse: Foxp3DTR (B6.129(Cg)-Foxp3tm3(DTR/GFP)Ayr/J) | The Jackson Laboratory | 016958 |
| Mouse: Xcr1DTR (Xcr1tm2(HBEGF/Venus)Ksho) | Tsuneyasu Kaisho, Osaka University | MGI: 5544058 |
| Mouse: Foxp3Cre/YFP (B6.129(Cg)-Foxp3tm4(YFP/icre)Ayr/J) | The Jackson Laboratory | 016959 |
| Mouse: Itgb8fl/fl (Itgb8tm2Lfr) | Michael Rosenblum, UC San Francisco | MGI: 3608910 |
| Mouse: Il-10fl/fl (Il-10 flox) | Susan Kaech, Salk Institute. Generated by Werner Müller. | PMID: 15534372 |
| Mouse: Areg−/− (Aregtm1Dle) | Marco Conti, UC San Francisco. Generated by David C. Lee. | MGI: 2176531 |
| Software and Algorithms | ||
| CellRanger 2.0 | 10X Genomics | 10xgenomics.com |
| STAR | Dobin et.al. 2013 | code.google.com/p/rna-star/. |
| Seurat | Satija et al. 2015 | satijalab.org/seurat/ |
| R: The Project for Statistical Computing | r-project.org | |
Acknowledgements
We thank E. Wan in the Institute for Human Genetics at UCSF for helping prepare samples for single-cell RNA-sequencing. We would also like to thank the UCSF Parnassus Flow Cytometry Core for maintenance of flow cytometers and sorters. J.J. Engelhardt and Bristol-Myers-Squibb for Fc-modified anti-CTLA-4 antibodies. S.M. Kaech, M. Conti, and M.D. Rosenblum for various mouse strains. L. Rodda for advice related to single-cell RNA sequencing and M.L. Broz and M.B. Headley for guidance, advice and critical reading of the manuscript. Acquisition and analysis of certain human samples described in this study was partially funded by contributions from AbbVie, Amgen, Bristol-Myers Squibb, and Pfizer. This work was supported in part by the NSF Graduate Research Fellowship Program awarded to M.B and NIH grants U54 CA163123, R21CA191428 and R01 CA197363.
Footnotes
Data and Software Availability
Data Resources
The accession number for the expression matrix for the single-cell RNA-sequencing reported in this paper is GEO: XXXXXXXXX
References
- Alferink J, Lieberam I, Reindl W, Behrens A, Weiss S, Huser N, Gerauer K, Ross R, Reske-Kunz AB, Ahmad-Nejad P, et al. (2003). Compartmentalized production of CCL17 in vivo: strong inducibility in peripheral dendritic cells contrasts selective absence from the spleen. The Journal of experimental medicine 197, 585–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison KA, Sajti E, Collier JG, Gosselin D, Troutman TD, Stone EL, Hedrick SM, and Glass CK (2016). Affinity and dose of TCR engagement yield proportional enhancer and gene activity in CD4+ T cells. eLife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azizi E, Carr AJ, Plitas G, Cornish AE, Konopacki C, Prabhakaran S, Nainys J, Wu K, Kiseliovas V, Setty M, et al. (2018). Single-Cell Map of Diverse Immune Phenotypes in the Breast Tumor Microenvironment. Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baratin M, Foray C, Demaria O, Habbeddine M, Pollet E, Maurizio J, Verthuy C, Davanture S, Azukizawa H, Flores-Langarica A, et al. (2015). Homeostatic NF-kappaB Signaling in Steady-State Migratory Dendritic Cells Regulates Immune Homeostasis and Tolerance. Immunity 42, 627–639. [DOI] [PubMed] [Google Scholar]
- Baratin M, Simon L, Jorquera A, Ghigo C, Dembele D, Nowak J, Gentek R, Wienert S, Klauschen F, Malissen B, et al. (2017). T Cell Zone Resident Macrophages Silently Dispose of Apoptotic Cells in the Lymph Node. Immunity 47, 349–362.e345. [DOI] [PubMed] [Google Scholar]
- Barry KC, Hsu J, Broz ML, Cueto FJ, Binnewies M, Combes AJ, Nelson AE, Loo K, Kumar R, Rosenblum MD, et al. (2018). A natural killer-dendritic cell axis defines checkpoint therapy-responsive tumor microenvironments. Nature medicine. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer CA, Kim EY, Marangoni F, Carrizosa E, Claudio NM, and Mempel TR (2014). Dynamic Treg interactions with intratumoral APCs promote local CTL dysfunction. The Journal of clinical investigation 124, 2425–2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedoui S, Whitney PG, Waithman J, Eidsmo L, Wakim L, Caminschi I, Allan RS, Wojtasiak M, Shortman K, Carbone FR, et al. (2009). Cross-presentation of viral and self antigens by skin-derived CD103+ dendritic cells. Nat Immunol 10, 488–495. [DOI] [PubMed] [Google Scholar]
- Behrens G, Li M, Smith CM, Belz GT, Mintern J, Carbone FR, and Heath WR (2004). Helper T cells, dendritic cells and CTL Immunity. Immunology and cell biology 82, 84–90. [DOI] [PubMed] [Google Scholar]
- Belz GT, Shortman K, Bevan MJ, and Heath WR (2005). CD8alpha+ dendritic cells selectively present MHC class I-restricted noncytolytic viral and intracellular bacterial antigens in vivo. Journal of immunology (Baltimore, Md : 1950) 175, 196–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binnewies M, Roberts EW, Kersten K, Chan V, Fearon DF, Merad M, Coussens LM, Gabrilovich DI, Ostrand-Rosenberg S, Hedrick CC, et al. (2018). Understanding the tumor immune microenvironment (TIME) for effective therapy. Nature medicine 24, 541–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos PD, Plitas G, Rudra D, Lee SY, and Rudensky AY (2013). Transient regulatory T cell ablation deters oncogene-driven breast cancer and enhances radiotherapy. The Journal of experimental medicine 210, 2435–2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broz ML, Binnewies M, Boldajipour B, Nelson AE, Pollack JL, Erle DJ, Barczak A, Rosenblum MD, Daud A, Barber DL, et al. (2014). Dissecting the tumor myeloid compartment reveals rare activating antigen-presenting cells critical for T cell immunity. Cancer Cell 26, 638–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen DS, and Mellman I. (2013). Oncology meets immunology: the cancer-immunity cycle. Immunity 39, 1–10. [DOI] [PubMed] [Google Scholar]
- Chiba K, Yanagawa Y, Masubuchi Y, Kataoka H, Kawaguchi T, Ohtsuki M, and Hoshino Y. (1998). FTY720, a novel immunosuppressant, induces sequestration of circulating mature lymphocytes by acceleration of lymphocyte homing in rats. I. FTY720 selectively decreases the number of circulating mature lymphocytes by acceleration of lymphocyte homing. Journal of immunology (Baltimore, Md : 1950) 160, 5037–5044. [PubMed] [Google Scholar]
- Corthay A, Skovseth DK, Lundin KU, Rosjo E, Omholt H, Hofgaard PO, Haraldsen G, and Bogen B. (2005). Primary antitumor immune response mediated by CD4+ T cells. Immunity 22, 371–383. [DOI] [PubMed] [Google Scholar]
- Curran MA, Geiger TL, Montalvo W, Kim M, Reiner SL, Al-Shamkhani A, Sun JC, and Allison JP (2013). Systemic 4–1BB activation induces a novel T cell phenotype driven by high expression of Eomesodermin. The Journal of experimental medicine 210, 743–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X, Quezada SA, Sepulveda MA, Sharma P, and Allison JP (2014). Engagement of the ICOS pathway markedly enhances efficacy of CTLA-4 blockade in cancer immunotherapy. The Journal of experimental medicine 211, 715–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Nish SA, Jiang R, Hou L, Licona-Limon P, Weinstein JS, Zhao H, and Medzhitov R. (2013). Control of T helper 2 responses by transcription factor IRF4-dependent dendritic cells. Immunity 39, 722–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier EL, Shay T, Miller J, Greter M, Jakubzick C, Ivanov S, Helft J, Chow A, Elpek KG, Gordonov S, et al. (2012). Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat Immunol 13, 1118–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerner MY, Casey KA, Kastenmuller W, and Germain RN (2017). Dendritic cell and antigen dispersal landscapes regulate T cell immunity. The Journal of experimental medicine 214, 3105–3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Martinez E, Planes R, Anselmi G, Reynolds M, Menezes S, Adiko AC, Saveanu L, and Guermonprez P. (2015). Cross-Presentation of Cell-Associated Antigens by MHC Class I in Dendritic Cell Subsets. Frontiers in immunology 6, 363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng TS, and Painter MW (2008). The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol 9, 1091–1094. [DOI] [PubMed] [Google Scholar]
- Hodi FS, Butler M, Oble DA, Seiden MV, Haluska FG, Kruse A, Macrae S, Nelson M, Canning C, Lowy I, et al. (2008). Immunologic and clinical effects of antibody blockade of cytotoxic T lymphocyte-associated antigen 4 in previously vaccinated cancer patients. Proceedings of the National Academy of Sciences of the United States of America 105, 3005–3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang AC, Postow MA, Orlowski RJ, Mick R, Bengsch B, Manne S, Xu W, Harmon S, Giles JR, Wenz B, et al. (2017). T-cell invigoration to tumour burden ratio associated with anti-PD-1 response. Nature 545, 60–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DB, Estrada MV, Salgado R, Sanchez V, Doxie DB, Opalenik SR, Vilgelm AE, Feld E, Johnson AS, Greenplate AR, et al. (2016). Melanoma-specific MHC-II expression represents a tumour-autonomous phenotype and predicts response to anti-PD-1/PD-L1 therapy. Nature communications 7, 10582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josefowicz SZ, Lu LF, and Rudensky AY (2012). Regulatory T cells: mechanisms of differentiation and function. Annual review of immunology 30, 531–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krappmann D, Wegener E, Sunami Y, Esen M, Thiel A, Mordmuller B, and Scheidereit C. (2004). The IkappaB kinase complex and NF-kappaB act as master regulators of lipopolysaccharide-induced gene expression and control subordinate activation of AP-1. Molecular and cellular biology 24, 6488–6500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnaswamy JK, Gowthaman U, Zhang B, Mattsson J, Szeponik L, Liu D, Wu R, White T, Calabro S, Xu L, et al. (2017). Migratory CD11b(+) conventional dendritic cells induce T follicular helper cell-dependent antibody responses. Science immunology 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumamoto Y, Hirai T, Wong PW, Kaplan DH, and Iwasaki A. (2016). CD301b(+) dendritic cells suppress T follicular helper cells and antibody responses to protein antigens. eLife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumamoto Y, Linehan M, Weinstein JS, Laidlaw BJ, Craft JE, and Iwasaki A. (2013). CD301b(+) dermal dendritic cells drive T helper 2 cell-mediated immunity. Immunity 39, 733–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laidlaw BJ, Craft JE, and Kaech SM (2016). The multifaceted role of CD4(+) T cells in CD8(+) T cell memory. Nat Rev Immunol 16, 102–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laoui D, Keirsse J, Morias Y, Van Overmeire E, Geeraerts X, Elkrim Y, Kiss M, Bolli E, Lahmar Q, Sichien D, et al. (2016). The tumour microenvironment harbours ontogenically distinct dendritic cell populations with opposing effects on tumour immunity. Nature communications 7, 13720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin J, Chiarion-Sileni V, Gonzalez R, Grob JJ, Cowey CL, Lao CD, Schadendorf D, Dummer R, Smylie M, Rutkowski P, et al. (2015). Combined Nivolumab and Ipilimumab or Monotherapy in Untreated Melanoma. The New England journal of medicine 373, 23–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin Y, Kobayashi S, Leader A, Amir ED, Elefant N, Bigenwald C, Remark R, Sweeney R, Becker CD, Levine JH, et al. (2017). Innate Immune Landscape in Early Lung Adenocarcinoma by Paired Single-Cell Analyses. Cell 169, 750–765.e717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Huang HI, Benzatti FP, Karlsson AB, Zhang JJ, Youssef N, Ma A, Hale LP, and Hammer GE (2016). Inflammatory Th1 and Th17 in the Intestine Are Each Driven by Functionally Specialized Dendritic Cells with Distinct Requirements for MyD88. Cell reports 17, 1330–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal R, Senbabaoglu Y, Desrichard A, Havel JJ, Dalin MG, Riaz N, Lee KW, Ganly I, Hakimi AA, Chan TA, et al. (2016). The head and neck cancer immune landscape and its immunotherapeutic implications. JCI insight 1, e89829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merad M, Sathe P, Helft J, Miller J, and Mortha A. (2013). The dendritic cell lineage: ontogeny and function of dendritic cells and their subsets in the steady state and the inflamed setting. Annual review of immunology 31, 563–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meredith MM, Liu K, Darrasse-Jeze G, Kamphorst AO, Schreiber HA, Guermonprez P, Idoyaga J, Cheong C, Yao KH, Niec RE, et al. (2012). Expression of the zinc finger transcription factor zDC (Zbtb46, Btbd4) defines the classical dendritic cell lineage. The Journal of experimental medicine 209, 1153–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JC, Brown BD, Shay T, Gautier EL, Jojic V, Cohain A, Pandey G, Leboeuf M, Elpek KG, Helft J, et al. (2012). Deciphering the transcriptional network of the dendritic cell lineage. Nat Immunol 13, 888–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nirschl CJ, Suarez-Farinas M, Izar B, Prakadan S, Dannenfelser R, Tirosh I, Liu Y, Zhu Q, Devi KSP, Carroll SL, et al. (2017). IFNγamma-Dependent Tissue-Immune Homeostasis Is Co-opted in the Tumor Microenvironment. Cell 170, 127–141.e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochiai S, Roediger B, Abtin A, Shklovskaya E, Fazekas de St Groth B, Yamane H, Weninger W, Le Gros G, and Ronchese F. (2014). CD326(lo)CD103(lo)CD11b(lo) dermal dendritic cells are activated by thymic stromal lymphopoietin during contact sensitization in mice. Journal of immunology (Baltimore, Md : 1950) 193, 2504–2511. [DOI] [PubMed] [Google Scholar]
- Pandiyan P, Zheng L, Ishihara S, Reed J, and Lenardo MJ (2007). CD4+CD25+Foxp3+ regulatory T cells induce cytokine deprivation-mediated apoptosis of effector CD4+ T cells. Nat Immunol 8, 1353–1362. [DOI] [PubMed] [Google Scholar]
- Pauken KE, Sammons MA, Odorizzi PM, Manne S, Godec J, Khan O, Drake AM, Chen Z, Sen DR, Kurachi M, et al. (2016). Epigenetic stability of exhausted T cells limits durability of reinvigoration by PD-1 blockade. Science (New York, NY) 354, 1160–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philip M, Fairchild L, Sun L, Horste EL, Camara S, Shakiba M, Scott AC, Viale A, Lauer P, Merghoub T, et al. (2017). Chromatin states define tumour-specific T cell dysfunction and reprogramming. Nature 545, 452–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puram SV, Tirosh I, Parikh AS, Patel AP, Yizhak K, Gillespie S, Rodman C, Luo CL, Mroz EA, Emerick KS, et al. (2017). Single-Cell Transcriptomic Analysis of Primary and Metastatic Tumor Ecosystems in Head and Neck Cancer. Cell 171, 1611–1624.e1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quezada SA, Simpson TR, Peggs KS, Merghoub T, Vider J, Fan X, Blasberg R, Yagita H, Muranski P, Antony PA, et al. (2010). Tumor-reactive CD4(+) T cells develop cytotoxic activity and eradicate large established melanoma after transfer into lymphopenic hosts. The Journal of experimental medicine 207, 637–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi OS, Zheng Y, Nakamura K, Attridge K, Manzotti C, Schmidt EM, Baker J, Jeffery LE, Kaur S, Briggs Z, et al. (2011). Trans-endocytosis of CD80 and CD86: a molecular basis for the cell-extrinsic function of CTLA-4. Science (New York, NY) 332, 600–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts Edward W., Broz Miranda L., Binnewies M, Headley Mark B., Nelson Amanda E., Wolf Denise M., Kaisho T, Bogunovic D, Bhardwaj N, and Krummel Matthew F. (2016). Critical Role for CD103+/CD141+ Dendritic Cells Bearing CCR7 for Tumor Antigen Trafficking and Priming of T Cell Immunity in Melanoma. Cancer Cell 30, 324–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffell B, Chang-Strachan D, Chan V, Rosenbusch A, Ho CMT, Pryer N, Daniel D, Hwang ES, Rugo HS, and Coussens LM (2014). Macrophage IL-10 blocks CD8+ T cell-dependent responses to chemotherapy by suppressing IL-12 expression in intratumoral dendritic cells. Cancer Cell 26, 623–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon H, Idoyaga J, Rahman A, Leboeuf M, Remark R, Jordan S, Casanova-Acebes M, Khudoynazarova M, Agudo J, Tung N, et al. (2016). Expansion and Activation of CD103+ Dendritic Cell Progenitors at the Tumor Site Enhances Tumor Responses to Therapeutic PD-L1 and BRAF Inhibition. Immunity 44, 924–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satpathy AT, Kc W, Albring JC, Edelson BT, Kretzer NM, Bhattacharya D, Murphy TL, and Murphy KM (2012). Zbtb46 expression distinguishes classical dendritic cells and their committed progenitors from other immune lineages. The Journal of experimental medicine 209, 1135–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schietinger A, Philip M, Krisnawan VE, Chiu EY, Delrow JJ, Basom RS, Lauer P, Brockstedt DG, Knoblaugh SE, Hämmerling GJ, et al. (2016). Tumor-Specific T Cell Dysfunction Is a Dynamic Antigen-Driven Differentiation Program Initiated Early during Tumorigenesis. Immunity 45, 389–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlitzer A, Sivakamasundari V, Chen J, Sumatoh HR, Schreuder J, Lum J, Malleret B, Zhang S, Larbi A, Zolezzi F, et al. (2015). Identification of cDC1- and cDC2-committed DC progenitors reveals early lineage priming at the common DC progenitor stage in the bone marrow. Nat Immunol 16, 718–728. [DOI] [PubMed] [Google Scholar]
- Shang B, Liu Y, Jiang SJ, and Liu Y. (2015). Prognostic value of tumor-infiltrating FoxP3+ regulatory T cells in cancers: a systematic review and meta-analysis. Scientific reports 5, 15179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiow LR, Rosen DB, Brdickova N, Xu Y, An J, Lanier LL, Cyster JG, and Matloubian M. (2006). CD69 acts downstream of interferon-alpha/beta to inhibit S1P1 and lymphocyte egress from lymphoid organs. Nature 440, 540–544. [DOI] [PubMed] [Google Scholar]
- Spranger S, Dai D, Horton B, and Gajewski TF (2017). Tumor-Residing Batf3 Dendritic Cells Are Required for Effector T Cell Trafficking and Adoptive T Cell Therapy. Cancer Cell 31, 711–723 e714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Zhang X, Tough DF, and Sprent J. (1998). Type I interferon-mediated stimulation of T cells by CpG DNA. The Journal of experimental medicine 188, 2335–2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topalian SL, Sznol M, McDermott DF, Kluger HM, Carvajal RD, Sharfman WH, Brahmer JR, Lawrence DP, Atkins MB, Powderly JD, et al. (2014). Survival, durable tumor remission, and long-term safety in patients with advanced melanoma receiving nivolumab. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 32, 1020–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJ, Robert L, Chmielowski B, Spasic M, Henry G, Ciobanu V, et al. (2014). PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 515, 568–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tussiwand R, Everts B, Grajales-Reyes GE, Kretzer NM, Iwata A, Bagaitkar J, Wu X, Wong R, Anderson DA, Murphy TL, et al. (2015). Klf4 expression in conventional dendritic cells is required for T helper 2 cell responses. Immunity 42, 916–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villani AC, Satija R, Reynolds G, Sarkizova S, Shekhar K, Fletcher J, Griesbeck M, Butler A, Zheng S, Lazo S, et al. (2017). Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science (New York, NY) 356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallin JJ, Bendell JC, Funke R, Sznol M, Korski K, Jones S, Hernandez G, Mier J, He X, Hodi FS, et al. (2016). Atezolizumab in combination with bevacizumab enhances antigen-specific T-cell migration in metastatic renal cell carcinoma. Nature communications 7, 12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei SC, Levine JH, Cogdill AP, Zhao Y, Anang NAS, Andrews MC, Sharma P, Wang J, Wargo JA, Pe’er D, et al. (2017). Distinct Cellular Mechanisms Underlie Anti-CTLA-4 and Anti-PD-1 Checkpoint Blockade. Cell 170, 1120–1133.e1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappasodi R, Budhu S, Hellmann MD, Postow MA, Senbabaoglu Y, Manne S, Gasmi B, Liu C, Zhong H, Li Y, et al. (2018). Non-conventional Inhibitory CD4(+)Foxp3(−)PD-1(hi) T Cells as a Biomarker of Immune Checkpoint Blockade Activity. Cancer Cell 33, 1017–1032.e1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou T, Caton AJ, Koretzky GA, and Kambayashi T. (2010). Dendritic cells induce regulatory T cell proliferation through antigen-dependent and -independent interactions. Journal of immunology (Baltimore, Md : 1950) 185, 2790–2799. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.