Abstract
Introduction
Accumulating evidence suggests that cutaneous viral infections are risk factors for the development of keratinocyte carcinomas (KC). The Viruses in Skin Cancer (VIRUSCAN) Study, a prospective cohort study, was established in 2014 to investigate the risk of KC associated with cutaneous human papillomavirus and polyomavirus infection and the possible interaction with ultraviolet radiation exposure (UVR).
Methods/Results
VIRUSCAN incorporates repeated measures of viral infection using multiple markers of infection and quantitative measures of UVR using a spectrophotometer. Participants were recruited between July 14, 2014-August 31, 2017 at the University of South Florida Dermatology Clinic in Tampa, FL. After excluding 124 individuals with prevalent KC at baseline, 1,179 participants (53.2% women, 46.8% men, all ages 60 years and older) were followed for up to four years with routine skin exams occurring every 6–12 months. Here we present the VIRUSCAN Study design, methods and baseline characteristics including demographics, sun exposure behavior, quantitative UVR exposure measurements and cutaneous viral prevalence for the full study cohort.
Conclusions
The VIRUSCAN Study will provide critical temporal evidence needed to assess the causality of the role cutaneous viral infections play in the development of KC, as well as the potential interaction between cutaneous viral infections and UVR exposure.
Impact
Study findings will be valuable in future development of novel KC prevention strategies.
Keywords: Cutaneous human papillomavirus, epidemiology, non-melanoma skin cancer, polyomavirus, ultraviolet radiation exposure, VIRUSCAN
INTRODUCTION
Keratinocyte carcinoma (KC), comprised of basal cell carcinoma (BCC) and squamous cell carcinoma (SCC), is the most frequently occurring cancer in the United States (1). KC incidence increased by 35% between 2006 and 2012, and an estimated 5.4 million KC cases were diagnosed in 2012. Despite its low metastatic potential and mortality rates, KC results in over 2,000 deaths annually in the United States (2). Furthermore, individuals are frequently diagnosed with multiple KCs, requiring multiple surgeries throughout their lifetimes and resulting in significant patient morbidity and substantial economic burden at the national level (3, 4). Established host risk factors for KC include older age, male sex and sun-sensitive skin likely to freckle and burn (5, 6). Ultraviolet radiation (UVR) is an environmental risk factor for KC, with intermittent and childhood UVR associated with increased risk of BCC (7, 8) and chronic sun exposure associated with SCC (7). Immunologic deficiencies and chronic immunosuppression are also linked to the development of KC, particularly SCC (9, 10). The continued increase in KC incidence, despite general awareness of the harms of sun exposure (1), underscores the need to identify additional modifiable risk factors that may lead to the development of novel prevention strategies.
Several epidemiological studies have observed KC to be associated with markers of cutaneous viral infections, including cutaneous human papillomaviruses (cuHPV) (11–14) and Merkel cell polyomavirus (MCV), a cutaneous human papillomavirus (HPyV) (15). CuHPV infections are particularly common in immunodeficient (16–18) and immunosuppressed individuals (19–21), but are also observed in immunocompetent populations, with cuHPV infection occurring early in life: 55–70% cuHPV DNA prevalence has been reported for infants and children up to 4 years old with similar prevalence rates observed among their parents (22, 23). CuHPV seroprevalence increases with age, with rates of 32%, 58%, and 60% observed in children ages 1–14, young adults ages 15–34, and older adults ages >34, respectively (24). CuHPV DNA prevalence has also been shown to increase with age (25, 26). While there are no established risk factors for cuHPV infection, previous studies have reported associations between cuHPV prevalence and UVR exposure (27, 28), light skin phenotypes (29, 30), male gender (29, 31, 32) and smoking (29, 30). Similar to cuPHV, HPyV infections also occur in early childhood (33), with MCV seroprevalence rates increasing with age: 40%, 85%, 90% in ages 1–4, 15–19, 50–79, respectively (34). While seroprevalence rates suggest most adults have been exposed to cuHPV and MCV at some point in their lifetime, and presumably cleared, DNA-based markers of infection indicate that infections can be newly acquired in adulthood (27, 35).
Findings between cuHPV infection and KC have been most consistently observed for cuHPV types in genus β (“β-HPV”) and cutaneous SCC (36). Experimental studies have identified multiple signalling pathways disrupted by β-HPV oncoproteins, with both in vitro (37–40) and mouse models (41–43) demonstrating the cooperation between β-HPV infection and UVR in SCC development. In humans, cuHPV infections are more likely to be detected in tumors that occur in sun exposed areas of the body (28). A few epidemiologic studies have sought to investigate the interaction between UVR exposure and β-HPV infection by examining SCC risk in subgroups of β-HPV seroreactivity and skin phenotype (11, 32) or past self-reported UVR exposure (44). While positive interactions were observed, these studies were limited by subjective UVR measurements and reliance on single time point assessments of both UVR exposure and β-HPV seroreactivity. Furthermore, no studies have investigated the potential interaction between UVR and infection with cuHPV types in other genera.
The Viruses in Skin Cancer (VIRUSCAN) Study is a prospective cohort study being conducted at the Moffitt Cancer Center and the University of South Florida Dermatology Clinic, in Tampa, Florida, to investigate the risk of KC associated with cuHPV/HPyV infections. The primary study aims are to estimate baseline prevalence of type-specific cuHPV/HPyV infections using multiple biospecimens, determine the risk of incident KC associated with baseline infections, and evaluate UVR as a potential cofactor in virus-associated KC risk. Furthermore, longitudinally banked biospecimens will be used to estimate the risk of incident KC associated with incident and persistent cuHPV/HPyV infection and investigate the concordance between virus types present prior to KC diagnosis and viral DNA detected in subsequent incident tumors. The VIRUSCAN Study addresses several research needs, as it is one of the few prospective studies to incorporate repeated measures of cuHPV/HPyV infection, multiple biomarkers of cuHPV/HPyV infection and quantitative measurements of UVR exposure. Here we describe the VIRUSCAN Study design, methods and baseline characteristics.
MATERIALS AND METHODS
Participants and eligibility criteria
Participants were recruited from the University of South Florida Dermatology Clinic, a high volume dermatology clinic in which patients undergo routine total body skin exams for the early detection of skin cancers. University of South Florida Dermatology Clinic patients undergoing total body skin examination are often self-referred, including those who seek treatment of a dermatologic condition and are advised to undergo total body skin examination while they are in the clinic and those who attend the clinic specifically for skin cancer screening.
University of South Florida Dermatology Clinic patients aged 60 or older were eligible to enroll in the VIRUSCAN Study. The recruitment age was restricted to this age group to optimize the number of incident cancers detected during follow-up. Patients with a history of either BCC or SCC were eligible, but patients with a history of both BCC and SCC were excluded to ensure all study participants were naïve to at least one type of KC at baseline. To maximize the generalizability of study findings to broad populations of individuals at risk for KC, no additional exclusion criteria were applied. As such, organ transplant recipients were included but comprised only 1% of the VIRUSCAN Study population and will be excluded from a sensitivity analysis to assess whether associations observed differ from those observed in the full cohort. Eligible patients scheduled to undergo a total body skin examination were identified via daily medical chart reviews and approached for study participation during their clinic visit. The study was approved by the University of South Florida Institutional Review Board, and all participants provided written informed consent.
During the design phase of the VIRUSCAN Study, enrollment was projected to be 1,500 based on historical clinic records. Using this fixed sample size, previously published incidence rates of KC (45), and assuming an alpha error rate of 0.05, power of 80% and false discovery rate of < 10%, the calculated minimal detectable hazard ratios (MDHR) for the association between cuHPV/HPyV infection at baseline and SCC ranged from 1.89 to 2.82 for 40% to 10% of the cohort testing virus-positive at baseline, respectively, and 1.64 to 2.23 for KC overall. Over three years of enrollment, 2,929 patients were approached, of whom 1,303 (44.5%) consented. While the number of enrolled participants was less than projected, the observed SCC and KC incidence rates are trending higher than projected, therefore the final analyses will have more than adequate statistical power to detect the associations of interest.
While there were no differences in the age of participants (mean=69.1 years, SD=6.4) versus non-participants (mean=69.9 years, SD=7.5 P=0.06), females were more likely to enroll in the study than men (52.2% female vs. 47.8% men, P=0.03). Of the 1,303 participants enrolled, 124 (9.5%) underwent biopsies as a result of the baseline total body skin examination that were subsequently determined to be malignant after pathology review. These participants with prevalent KC at baseline (83 SCC, 78 BCC and 1 basosquamous carcinoma) were excluded from follow-up visits. The remaining 1,179 participants were followed for up to four years, depending on the year of enrollment in the study. Follow-up visits coincided with participants’ routine total body skin examination appointments at the University of South Florida Dermatology Clinic, the frequency of which varied, depending on the participant’s skin cancer risk profile, including skin type, family history of skin cancer and sun exposure history. Typically, participants returned to the clinic every 6–12 months for a total body skin examination. For example, among the 421 participants enrolled in the first year of the study, 87.9% returned to the clinic at least once during the subsequent three years of follow-up, with 69.4% returning at least twice and 53.9% returning three or more times.
Data collection
Table 1 summarizes data and biospecimen collection for VIRUSCAN Study participants at baseline and during follow-up. Variables were broadly classified as patient demographics, skin cancer risk factors, medical history, cutaneous viral infection status, tumor characteristics, and measures of UV exposure.
Table 1.
Variables collected in the VIRUSCAN Study, Tampa, FL, 2014–2018.
| Source | Variable details | Visit |
|---|---|---|
| Study questionnaire | B | |
| Demographics | Gender, age, race, ethnicity, education level, marital status, place of birth | |
| Sun protection | Use of sunscreen, long sleeves, pants, hats, sunglasses, and sun avoidance | |
| Sun exposures | Occupational sun exposure, history of sunburn, skin reaction to the sun, use of sunlamps or UV nail dryers | |
| Phenotypic factors | Eye color, hair color, untanned skin color, number of moles on arm/body | |
| Skin cancer screening | Number of skin exams per year | |
| Skin conditions | History of skin biopsies, eczema, psoriasis, acne, vitiligo, actinic keratosis, keratoacanthoma, warts, other skin conditions, treatment for skin conditions | |
| Cancer history | Melanoma, basal cell carcinoma, squamous cell carcinoma, other skin cancers or other cancers | |
| Other medical conditions | Organ transplant, Crohn's disease, Type I diabetes, Type II diabetes, heart disease, epidermodysplasia verruciformis, lupus, multiple sclerosis, Sezary syndrome, rheumatoid arthritis, osteoarthritis, xeroderma pigmentosum, asthma/bronchitis/emphysema, gout | |
| Smoking and alcohol | Smoking and drinking habits | |
| Steroid medication | Use of oral steroids or steroid creams | |
| Clinical findings of total body skin examinations | Identification of pre-malignant (actinic keratosis) and malignant lesions (tumor histology, anatomic site [head, neck, torso, arms, legs]) | B and FU |
| Biospecimens | ||
| Blood | HPV/HPyV serum antibodies | B |
| Eyebrow hair | HPV/HPyV viral DNA | B and FU |
| Skin swabs | HPV/HPyV viral DNA | B and FU |
| Tumor Tissue | HPV/HPyV viral DNA, solar elastosis, papillomatosis, hypergranulosis, and crateriform architecture (SCC only) | B and FU |
| Spectrophotometer | Inner upper arm, forearm and forehead spectrophotometer reading | B and FU |
| Sun exposure questionnaire | (subsample n=159) | FU |
| Sun exposure (past week) | Time outdoors, frequency of sunbathing and spending time in the sun sitting or lying | |
| Sun protection (past week) | Use of sunscreen, clothing, hat and sun avoidance |
Abbreviations: B, baseline visit; FU, follow-up visit.
Questionnaire
Participants completed an electronic questionnaire at baseline that included information on demographics, sun protective behaviors, skin cancer risk factors and medical history (Table 1). Participants unable to complete the questionnaire during the visit were emailed a link to complete the questionnaire online. Non-responders were sent up to five email reminders followed by a final phone call reminder. The overall questionnaire response rate was 97.5%, with no differences observed between the 1,150 participants who completed the questionnaire and the 29 who did not with respect to age (P=0.11) or gender (P=0.58).
Clinical findings of total body skin exams
The outcomes of the total body skin examination conducted at baseline and throughout follow-up were recorded in the study database, including negative findings, instances of actinic keratosis treated with liquid nitrogen (standard practice at the University of South Florida Dermatology Clinic) and biopsies of suspicious lesions. Medical records were reviewed throughout the follow-up period to document histopathological results corresponding to any skin biopsies conducted on participants. Characteristics of those lesions determined to be malignant were recorded in the study database, including tumor size, histology and anatomic location (head, neck, torso, arms, and legs).
UVR exposure assessment
To model the time-dependent dynamic of recent UVR exposure and cutaneous viral infection acquisition and persistence, as well as the interaction between recent UVR exposure and infection associated with subsequent KC development, an objective measure of UVR was needed - one that could be obtained over multiple time points in a manner least burdensome to study participants. As such, a spectrophotometer was used to quantify skin pigmentation at baseline and throughout study follow-up as a proxy for UVR exposure, similar to previous epidemiologic studies (46, 47). Skin pigmentation readings were obtained with the Konica Minolta CM-600D spectrophotometer using the specular component included mode with Spectra Magic NX Lite USB Ver. 2.5 software (48). This instrument measures color on three different axes: lightness on a scale of 0 (black) to 100 (white), axis a, indicating color within the red through green range, and axis b, measuring color within the yellow through blue range, with increasing values on a- and b-axes, indicating saturation of color (49). The instrument was calibrated using a white tile every morning per manufacturer guidelines. Each reading was obtained three times, and the average reading was recorded. The inter-user reliability of the spectrophotometer readings across two VIRUSCAN Study coordinators was determined to be 0.91.
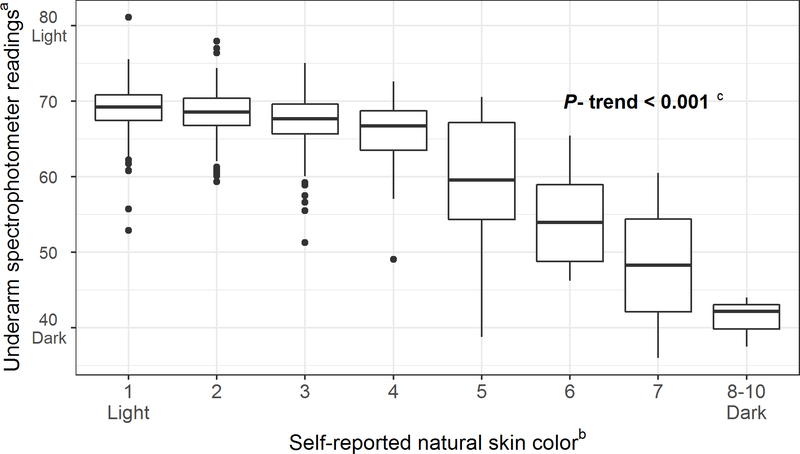
Natural skin tone was assessed using spectrophotometer readings of the sun-unexposed underside of the upper arm (i.e. the axilla), with higher readings indicating lighter natural skin tone. As shown in Figure 1, spectrophotometer readings significantly decreased with increasing category of self-reported skin tone (50). The degree of recent tanning in response to recent UVR exposure was measured by calculating the difference between the color readings (ΔE × ab) on an area of sun-exposed skin (top of the forearm or forehead) and the axilla. These anatomic sites were chosen to correspond with the areas where viral infection measurements were obtained and will be used to model the potential interaction between UVR exposure and cutaneous viral infections. The difference between the sun-exposed pigmentation readings (forearm and forehead) and the sun-unexposed pigmentation readings (underarm) was calculated and used as an objective measure of UVR exposure in response to more recent tanning (48). However, as the exact timing of UVR exposure relative to the spectrophotometer readings is unclear, we administered a questionnaire asking about sun exposures over the past week among a subsample of VIRUSCAN participants (n=159) to validate the spectrophotometer readings as a measure of more recent UVR exposure. The questionnaire included sun exposure and sun protection scales from a questionnaire previously validated against dosimeter-based measures of outdoor time in the past week (51).
Figure 1.
Association between underarm spectrophotometer reading and self-reported natural skin color in the VIRUSCAN Study, Tampa, FL, 2014–2017. A strong positive correlation was observed between the baseline underarm spectrophotometer readings and self-reported natural skin color.
aLower values of the underarm spectrophotometer readings represent darker skin pigmentation.
bSelf-reported natural skin color was assessed with the New Immigrant Survey Color Scale which included images of hands in a variety of skin shades categorized as ranging from 0 to 10, with 0 representing the lightest skin and 10 representing the darkest skin.
cThe Jonckheere-Terpstra test was used to determine if a trend existed between underarm spectrophotometer readings and self-reported natural skin color.
Biospecimens
Several biospecimens were obtained for the assessment of cutaneous viral infections. Peripheral blood samples were collected at baseline, processed and stored in aliquots of serum, plasma and mononuclear cells, with serum samples earmarked for the analysis of antibodies to cuHPV and HPyV. Plucked eyebrow hairs and normal forearm skin swabs (52) were collected at baseline and throughout follow-up for the measurement of cuHPV and HPyV DNA. For those 240 study participants who developed KC throughout the follow-up period, formalin-fixed paraffin-embedded tumor tissue sections and tumor tissue slides were obtained from the USF Pathology Department for the measurement of cuHPV/HPyV viral DNA and histopathologic grading, respectively. The study pathologist reviewed each hematoxylin and eosin-stained slide and graded the degree of solar elastosis present (0–3) in the adjacent normal tissues as a measure of cumulative UVR exposure (53, 54). For SCC, the pathologist described the presence of tumor architectural features possibly associated with cuHPV infection, including papillomatosis, hypergranulosis, and crateriform architecture.
Assessment of past and current cutaneous viral infections
Serum samples were analyzed for antibodies to the L1 capsid protein corresponding to 17 β-HPV types and 7 γ-HPV types, as well as antibodies to both the VP1 capsid protein and T-antigens corresponding to 4 cuHPyV types (Supplementary Table S1). Selected types were those shown to be associated with KC in the literature, in addition to the most prevalent types observed in tumor tissues obtained from our previous case-control study (14, 55). Multiplex serology assays were conducted at the German Cancer Research Center in Heidelberg, Germany, using methods previously described (56, 57).
Baseline and follow-up eyebrow hair, skin swab and tumor samples were shipped to the Infections and Cancer Biology Group at the International Agency for Research on Cancer for DNA extraction and genotyping. Multiplex/Luminex PCR-assays were used for the measurement of viral DNA corresponding to 46-β HPV types, 52 γ-HPV types and 5 polyomavirus types (Supplementary Table S1) using methods previously described (52). Analysis of the tumor samples as well as the eyebrow hair and skin swab samples collected throughout follow-up is ongoing. Repeated measures of cutaneous viral infections in the eyebrow hairs and skin swabs will be used to model incident and persistent infections and their association with subsequent KC risk.
Statistical analysis of baseline characteristics
Baseline questionnaire data were largely complete, as <5% of data were missing for each questionnaire item. Given the low rate of missing data, data for select variables were imputed among the 1,150 participants who completed the questionnaire to minimize bias introduced by list-wise deletion of incomplete data. The questionnaire data were complete for age, gender and ethnicity. However, other variables were imputed, including education level, marital status and place of birth, sun protective behaviors and skin cancer risk factors. No medical history-related variables were imputed. The imputation was performed using the multivariate imputation by chained equations algorithm within R, version 3.5.0 (R Foundation for Statistical Computing, Vienna, Austria), which imputes data on a variable basis (58). The algorithm specifies an imputation model for each target variable that takes into account all other variables with the assumption that data are missing at random (58). A different type of model was assigned to each variable based on the variables’ intrinsic distribution, e.g., continuous (predictive mean match), binary (logistic regression) and unordered categorical (polytomous logistic regression) or ordered categorical (proportional odds model). Five imputed data sets were generated and used to calculate pooled Chi-square p-values for all analyses here that used the imputed baseline questionnaire data.
To better understand how representative the VIRUSCAN Study population is of the general population, demographics, skin cancer risk factors and sun protective behaviors of study participants were compared to the general population, using publically available data from the 2015 National Health Interview Survey (NHIS) (59, 60). The NHIS is a health survey of the non-institutionalized U.S. population conducted annually by the Centers for Disease Control and Prevention (60). The NHIS dataset was restricted to adults ≥60 living in the Southern U.S. for the present analysis, since this age group and geographic region most closely relate to the VIRUSCAN Study population.
The correlation between the spectrophotometer readings obtained at the forearm and forehead were described using Spearman’s rank correlation. Associations between the spectrophotometer readings and tertiles of self-reported recent sun exposure and protection scales were examined using logistic regression, adjusted for age and gender. The baseline prevalence of cuHPV/HPyV infection was defined as the proportion of patients whose sample was positive for viral DNA (in the skin swabs and eyebrow hairs) or serum antibodies. Viral prevalence rates were calculated for all beta types tested and by species, type-specific prevalence rates are presented for the eyebrow hair and skin swab samples.
RESULTS
Descriptive profile and generalizability of the cohort
Baseline demographics, skin cancer risk factors and sun protective behaviors of the VIRUSCAN Study participants are presented in Table 2, compared to those of the general Southern United States population as measured by the NHIS population. VIRUSCAN Study participants were more likely to have sun-sensitive skin than the NHIS population. For example, 4.2% of VIRUSCAN participants reported no change to their skin after one hour sun exposure as compared to 17.9% of the NHIS population (Table 2). The VIRUSCAN Study participants were also more likely to report skin cancer risk factors such as use of tanning beds, a history of skin cancer and former smoking. Interestingly, 66.4% and 75.2% of VIRUSCAN Study participants reported use (always/often/sometimes) of sunscreen and hats as compared to 47.6% and 42.4% of the NHIS population, respectively.
Table 2.
Demographic Characteristics and Skin Cancer Risk Factors and Protective Behaviors in the VIRUSCAN Study, Tampa, FL, 2014–2017 and the Southern United States Population
| Demographic and skin cancer risk factors | VIRUSCAN (n=1,179) |
Southern NHIS 2015c (n=25,149,699) |
p-valued | |||
|---|---|---|---|---|---|---|
| N | % | n | % | |||
| Age in years | ||||||
| Mean SD | 69.0 | 6.3 | 70.2 | 7.6 | < 0.001 | |
| Gender | ||||||
| Female | 627 | 53.2 | 14,073,280 | 56.0 | 0.055 | |
| Male | 552 | 46.8 | 11,076,419 | 44.0 | ||
| Race | ||||||
| White | 1,132 | 96.1 | 20,421,079 | 81.2 | < 0.001 | |
| Others | 46 | 3.9 | 4,728,620 | 18.8 | ||
| Ethnicity | ||||||
| Non-Hispanic | 1,115 | 94.6 | 22,748,364 | 90.5 | < 0.001 | |
| Hispanic or Latino | 64 | 5.4 | 2,401,335 | 9.5 | ||
| Sunburn in the past 12 months | ||||||
| No | 1,061 | 92.5 | 20,241,154 | 86.1 | < 0.001 | |
| Yes | 86 | 7.5 | 3,273,041 | 13.9 | ||
| History of any cancer | ||||||
| No | 565 | 49.1 | 19,269,413 | 76.8 | < 0.001 | |
| Yes | 585 | 50.9 | 5,807,869 | 23.2 | ||
| History of skin cancer at study enrollment | ||||||
| No known skin cancer | 687 | 59.7 | 5,264,232 | 90.7 | < 0.001 | |
| Any type of skin cancer | 463 | 40.3 | 536,843 | 9.3 | ||
| Smoke status | ||||||
| Never smoked | 586 | 51.0 | 13,307,427 | 53.1 | < 0.001e | |
| Former smoker | 524 | 45.6 | 9,180,053 | 36.6 | ||
| Current smoker | 38 | 3.3 | 2,563,205 | 10.2 | ||
| Reaction to 1 hour sun exposure | ||||||
| No change | 48 | 4.2 | 3,430,354 | 17.9 | < 0.001e | |
| Tans without sunburn | 234 | 20.4 | 4,556,119 | 23.8 | ||
| Mild sunburn with tana | 488 | 42.6 | 4,898,328 | 25.6 | ||
| Sunburn without blisters | 315 | 27.5 | 4,198,736 | 21.9 | ||
| Blistering sunburn | 60 | 5.2 | 2,052,810 | 10.7 | ||
| Ever used a sun lamp or tanning bed | ||||||
| No | 905 | 78.8 | 1,943,216 | 97.2 | < 0.001e | |
| Yes | 243 | 21.2 | 55,992 | 2.8 | ||
| Sunscreen useb | ||||||
| Always | 109 | 10.5 | 4,295,888 | 21.2 | < 0.001f | |
| Often | 265 | 25.5 | 2,155,292 | 10.7 | ||
| Sometimes | 316 | 30.4 | 3,180,984 | 15.7 | ||
| Rarely | 221 | 21.3 | 2,022,689 | 10.0 | ||
| Never | 127 | 12.2 | 8,566,555 | 42.4 | ||
| Wore a hatb | ||||||
| Always | 245 | 23.6 | 3,617,098 | 17.9 | < 0.001f | |
| Often | 282 | 27.1 | 1,974,467 | 9.8 | ||
| Sometimes | 255 | 24.5 | 2,965,504 | 14.7 | ||
| Rarely | 141 | 13.6 | 1,949,292 | 9.7 | ||
| Never | 116 | 11.2 | 9,650,956 | 47.9 | ||
| Stayed in shadeb | ||||||
| Always | 109 | 10.5 | 4,303,200 | 21.3 | < 0.001f | |
| Often | 464 | 44.7 | 6,050,002 | 29.9 | ||
| Sometimes | 356 | 34.3 | 6,119,738 | 30.2 | ||
| Rarely | 81 | 7.8 | 2,011,929 | 9.9 | ||
| Never | 27 | 2.6 | 1,762,869 | 8.7 | ||
| Wore protective clothesb | Wore long sleeves | Wore long pants | ||||
| Always | 83 | 8.0 | 2,660,356 | 13.2 | 5,605,566 | 27.8 |
| Often | 248 | 23.8 | 1,537,657 | 7.6 | 2,545,902 | 12.6 |
| Sometimes | 339 | 32.6 | 3,404,590 | 16.9 | 3,666,794 | 18.2 |
| Rarely | 262 | 25.2 | 2,506,741 | 12.4 | 1,893,874 | 9.4 |
| Never | 109 | 10.5 | 10,027,229 | 49.8 | 6,475,233 | 32.1 |
Abbreviations: NHIS, National Health Interview Survey; SD, standard deviation; VIRUSCAN, Viruses in Skin Cancer.
The corresponding category for the NHIS data is: burn mildly with some or no tanning.
The sun exposure timeframe differed between the VIRUSCAN questionnaire (if outside in sun for 15 minutes or more) and the NHIS questionnaire (if outside in sun for 1 hour or more) for the sun protection variables.
The NHIS data set was restricted to adults ages 60+ and weighted using the “sampleweight” variable.
P-values were calculated using Chi-square tests without correction for continuity and confirmed by conditional exact tests.
The following variables were tested using a pooled Chi-square test from 5 imputed datasets including: smoke status, reaction to 1 hour sun exposure and ever use a sun lamp or tanning bed.
The following variables were tested using the Cochran-Armitage trend test: sunscreen use, wore a hat and stayed in shade.
Spectrophotometer readings of recent UVR exposure
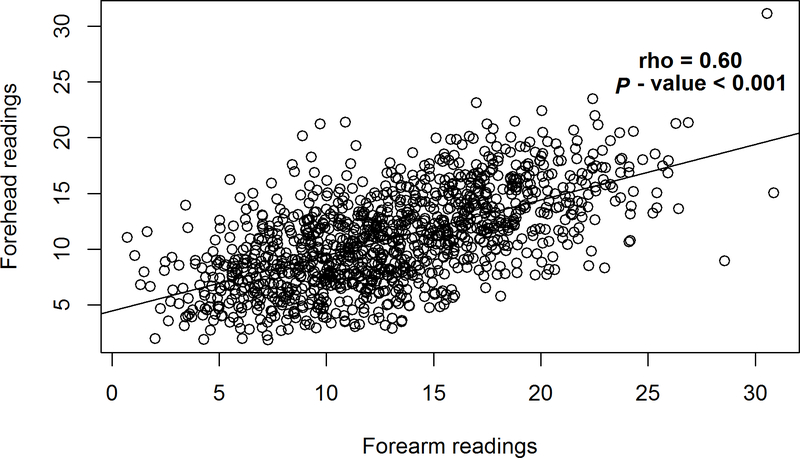
A strong, intra-individual correlation was observed between the spectrophotometer readings obtained at the forearm and forehead (Figure 2; rho=0.61; P < 0.001). Further analysis of baseline skin pigmentation readings revealed greater variation in the forearm spectrophotometer readings (median= 12.63, IQR=7.46) as compared to the forehead readings (median=10.76, IQR=6.03). Based on past-week exposure, individuals in the highest tertile of self-reported sun exposure were 2.5 times more likely to have a higher than median forearm spectrophotometer readings compared to those in the lowest tertile of sun exposure (Table 3; odds ratio (OR) 2.48, 95% CI: 1.01, 6.33, P-trend=0.06), after adjustment for age and sex. Similarly, when a ratio of the summary measures of sun exposure and sun protective behaviors was considered, a positive trend was observed between tertiles of the ratio and high versus low forearm spectrophotometer readings (Table 3; OR tertile 3 vs 1=2.89, 95% CI: 1.24, 6.95, P-trend=0.01). No associations were observed with the forehead spectrophotometer readings (Supplemental Table S2).
Figure 2.
Correlation between forearm and forehead spectrophotometer readings in the VIRUSCAN Study, Tampa, FL, 2014–2017. A strong positive correlation was observed between the baseline forearm and forehead spectrophotometer readings.
aRho and P value were calculated using Spearman correlation.
Table 3.
Correlation Between Forearm Spectrophotometer Readings and Past-Week Measures of Sun Exposure and Protection Among a Subsample of VIRUSCAN Study Participants (n=159), Tampa, FL, 2014–2017
| Forearm spectrophotometer readingsa |
||||||
|---|---|---|---|---|---|---|
| Questionnaire measurement of exposure/protection | low (<=11.55) |
high (>11.55) |
OR | 95% CIb | ||
| n | % | n | % | |||
| Exposure scale | ||||||
| T1 | 45 | 56.2 | 34 | 43.0 | 1.00 | Referent |
| T2 | 22 | 27.5 | 21 | 26.6 | 1.03 | 0.45, 2.32 |
| T3 | 13 | 16.2 | 24 | 30.4 | 2.48 | 1.01, 6.33 |
| P-trendc = 0.06 | ||||||
| Protection scale | ||||||
| T1 | 29 | 36.2 | 30 | 38.0 | 1.00 | Referent |
| T2 | 26 | 32.5 | 26 | 32.9 | 0.97 | 0.42, 2.24 |
| T3 | 25 | 31.2 | 23 | 29.1 | 0.76 | 0.32, 1.75 |
| P-trend = 0.54 | ||||||
| Ratio of exposure/protection scales | ||||||
| T1 | 37 | 46.8 | 23 | 29.1 | 1.00 | Referent |
| T2 | 26 | 32.9 | 24 | 30.4 | 1.14 | 0.49, 2.61 |
| T3 | 16 | 20.3 | 32 | 40.5 | 2.89 | 1.24, 6.95 |
| P-trend = 0.01 | ||||||
Forearm spectrophotometer readings were dichotomized based on the median value.
Odds ratio and 95% confidence interval were calculated using logistic regression, adjusted for age and gender.
The median score/ratio of each group was used to test the trend between score and spectrophotometer readings, using logistic regression adjusted for age and gender.
Baseline prevalence of cutaneous viral infections
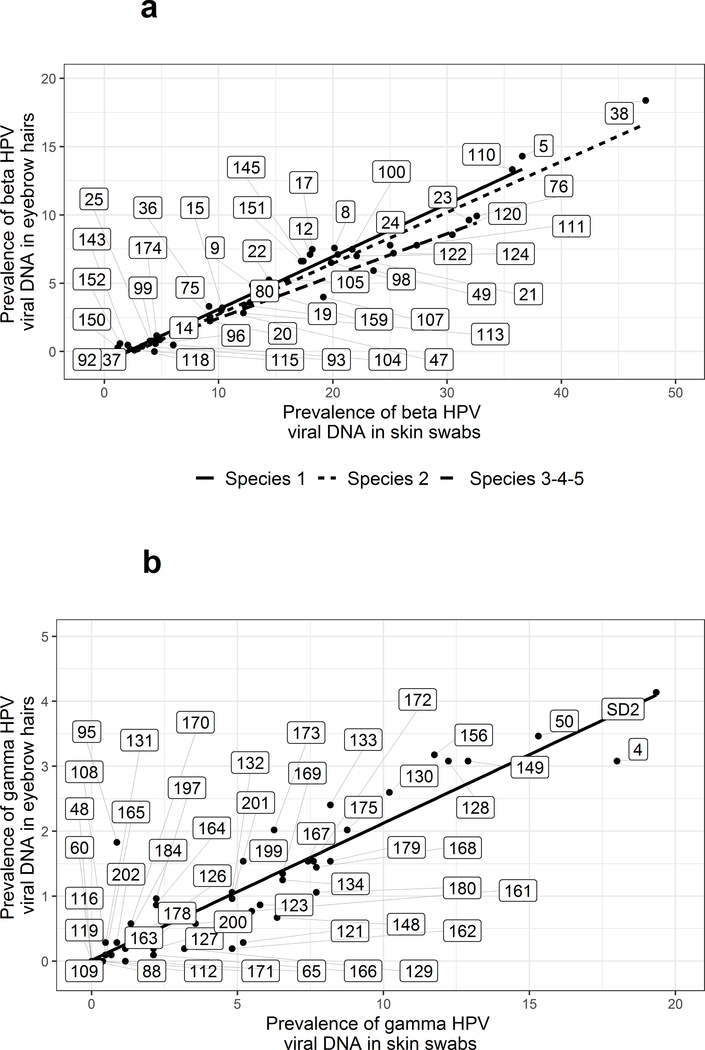
Based on the completed analysis of baseline samples obtained from the 370 VIRUSCAN Study participants enrolled in the first year of the study who were eligible for follow-up, the prevalence of cuHPV and HPyV was consistently higher in skin swabs than eyebrow hairs, with strong virus-type specific correlations across eyebrow hairs and skin swabs in terms of prevalence, degree of infection and number of types (52). When expanding the analysis to include baseline samples obtained from all VIRUSCAN Study participants eligible for follow-up, similar findings were observed, with higher cuHPV and HPyV prevalence in skin swabs compared to eyebrow hairs and strong type-specific viral prevalence across both sample types (Figure3). For example, DNA corresponding to at least one β-HPV type was detected in the skin swabs of 95.6% of participants and in the eyebrow hairs of 64.4% of participants, with the same trends observed for β-species 1 and 2 (80.5% and 41.2% vs. 90.1% and 53.0%, respectively). Similarly, the γ-HPV prevalence was also higher in the skin swabs (75.6%) than in the eyebrow hairs (29.7%). HPyV prevalence rates followed the same pattern with 82.1% of swabs showing infection and only 35.8% of eyebrow hairs. The type-specific cuHPV and HPyV prevalence rates for VIRUSCAN Study participants are presented in Supplementary Table S3. β-HPV types with the highest prevalence rates in both the skin swab and eyebrow hair were types 5 (36.6% and 14.3%), 38 (47.4% and 18.4%) and 110 (35.7% and 13.3%), with types 4 (18% and 3.1%), 50 (15.3% and 3.5%), and SD2 (19.3% and 4.1%) being the most prevalent γ-HPV types (Figure 3). Additionally, among the subset of cuHPV/HPyV types tested in serology, the seroprevalence was 73.6% for any β-HPV, 57.3% for any γ-HPV and 98.6% for any HPyV.
Figure 3.
Cutaneous Human papillomavirus (HPV) prevalence in skin swabs and eyebrow hairs among VIRUSCAN Study participants, Tampa, FL 2014–2017. The scatterplots display the correlation between type-specific HPV viral prevalence in skin swabs (x-axis) and eyebrow hairs (y-axis). The numbers within the rectangles indicate the HPV type tested for both (a) β-HPV types and (b) γ-HPV types.
aRho and P value were calculated using Spearman correlation.
DISCUSSION
The VIRUSCAN Study is the first prospective cohort study to assess the association between cutaneous viral infections and KC using multiple, repeated markers of viral infection and quantitative measures of ultraviolet radiation exposure. In this manuscript, we present detailed information on the design, methods and baseline characteristics of the VIRUSCAN Study. VIRUSCAN Study participants are at higher risk for skin cancer than the general population, but tend to be more conscious of skin health, engaging in sun protection and undergoing routine skin cancer screening exams. Therefore, this population is ideal for assessing the VIRUSCAN Study aims as we expect to observe a large number of incident KC cases throughout the follow up period.
The spectrophotometer-based skin pigmentation readings obtained at two anatomic sites (forehead and forearm) were highly correlated, although forearm readings varied more than forehead readings, perhaps reflecting differences in sun protective behaviors, as 50.7% of participants used hats always/often to protect their forehead but only 31.8% of participants used long sleeves always/often to protect their forearms (Table 2). Measuring exposure at the forearm maybe a better surrogate for overall sun exposure than forehead measurements, further evidenced by the positive associations observed between forearm spectrophotometer measurements and self-reported recent sun exposure that were not observed for forehead spectrophotometer measurements. Taken together, these results confirm that spectrophotometer based measurements of skin pigmentation are objective measures that may be useful for tracking an individual’s recent sun exposure over time in relation to other time-dependent biomarkers of disease risk and may be useful in future epidemiologic studies seeking to incorporate recent measures of UVR exposure.
A high prevalence of cutaneous viral infections was observed in both skin swabs and eyebrow hairs obtained from VIRUSCAN Study participants, consistent with previous epidemiologic studies of cutaneous viral infections (22, 27). Interestingly, regardless of the cuHPV species, viral prevalence was consistently higher in skin swabs than in eyebrow hairs, with HPyV types following this same pattern. We also observed a strong correlation between type-specific infections across skin swabs and eyebrow hairs. These full baseline viral prevalence results confirm the findings previously reported among a subset of VIRUSCAN Study participants enrolled in year one in which prevalence, as well as number of types and degree of infection, were correlated across skin swabs and eyebrow hairs (52).
The VIRUSCAN Study has some methodologic limitations. The study population is a select group, comprised of older adults residing in a geographic region with high ambient UVR exposure, engaging in sun protective behaviors more often than the general population. Therefore, the generalizability of study findings may be limited. However, if UVR is a necessary cofactor in virus-associated skin carcinogenesis, then examining virus-associated KC risk in populations with higher UVR exposure maximizes the likelihood of observing such associations. Furthermore, should cutaneous viral infections be associated with KC in the VIRUSCAN Study population, then future studies could examine similar associations in populations with lower UVR exposures. Importantly, the VIRUSCAN Study participants would be an ideal target population in which to test novel skin cancer prevention strategies, due to their high skin cancer risk profile and motivation to decrease their risk.
To minimize participant burden, blood samples were only collected at baseline. However, baseline seroreactivity will provide information about past viral infections, while the DNA-based markers of infection in eyebrow hairs and skin swabs will provide insight on current infection. The tumor samples collected were typically small in size since they are screen-detected cancers. However, β-HPV types have been shown to be more prevalent in earlier stages of skin carcinogenesis (61) and therefore, the viral DNA detection may be more feasible in smaller tumors compared to more advanced, larger tumors. We were unable to collect tissue samples from premalignant lesions such as actinic keratosis because the standard treatment protocol at the University of South Florida Dermatology Clinic is cryodestruction. However, incident actinic keratosis lesions are documented throughout follow-up, thus enabling the assessment of actinic keratosis risk associated with baseline viral infections. Finally, while VIRUSCAN Study participants were only followed for up to four years; longer term follow-up may be possible with additional grant funding.
A major strength of the VIRUSCAN Study is the incorporation of repeated measures of UV exposure and cutaneous viral infections that will be used to assess temporal patterns of exposure and their associations with incident KC, as well as acquisition of new viral infection. Additionally, the longitudinal biospecimens were tested for over 100 cutaneous virus types, facilitating the investigation of KC risk associated with persistence of viral infections, an aspect of HPV infection known to be associated with increased risk of other cancers (62). Furthermore, the banked samples will facilitate multi-disciplinary research involving measures of immune function and subsequent cancer development. Finally, our subsample validation study indicates that the spectrophotometer readings yield an objective and quantifiable measure of recent UVR exposure which will be used in the VIRUSCAN study to assess the possible interaction between UVR exposure and cuHPV/HPyV infection in relation to KC risk.
While the VIRUSCAN Study is ongoing, the longitudinal results will provide critical temporal evidence as to the role of cutaneous viral infections in the development of KC, as well as the potential interaction between viral infections and UVR. These findings will inform future KC prevention efforts, such as the development of a vaccine which could serve as a primary prevention strategy among individuals at high risk for KC.
Supplementary Material
ACKNOWLEDGEMENTS
Funding was provided by the National Cancer Institute at the National Institutes of Health (1R01-CA17758), awarded to D.E. Rollison, and supported all authors. This work was supported in part by the Survey Methods and Tissue Cores at the H. Lee Moffitt Cancer Center and Research Institute, a comprehensive cancer center designated by the National Cancer Institute (Grant P30-CA076292).
The authors would like to sincerely thank the USF Dermatology Clinic patients who contributed generously of their time to the VIRUSCAN Study.
Conflicts of interest: Dr. Anna R. Giuliano reports being a Merck & Co, Inc. grant recipient and advisory board member. The remaining authors state no relevant financial disclosures.
References
- 1.Rogers HW, Weinstock MA, Feldman SR, et al. Incidence Estimate of Nonmelanoma Skin Cancer (Keratinocyte Carcinomas) in the U.S. Population, 2012. JAMA Dermatol 2015;151(10):1081–6. [DOI] [PubMed] [Google Scholar]
- 2.Mudigonda T, Pearce DJ, Yentzer BA, et al. The economic impact of non-melanoma skin cancer: a review. J Natl Compr Canc Netw 2010;8(8):888–96. [DOI] [PubMed] [Google Scholar]
- 3.Rees JR, Zens MS, Gui J, et al. Non melanoma skin cancer and subsequent cancer risk. PLoS One 2014;9(6):e99674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen JG, Fleischer AB Jr., Smith ED, et al. Cost of nonmelanoma skin cancer treatment in the United States. Dermatol Surg 2001;27(12):1035–8. [DOI] [PubMed] [Google Scholar]
- 5.Acarturk TO, Edington H. Nonmelanoma skin cancer. Clin Plast Surg 2005;32(2):237–48. [DOI] [PubMed] [Google Scholar]
- 6.Fitzpatrick TB. The validity and practicality of sun-reactive skin types I through VI. Arch Dermatol 1988;124(6):869–71. [DOI] [PubMed] [Google Scholar]
- 7.Moan J, Grigalavicius M, Baturaite Z, et al. The relationship between UV exposure and incidence of skin cancer. Photodermatol Photoimmunol Photomed 2015;31(1):26–35. [DOI] [PubMed] [Google Scholar]
- 8.Rosso S, Zanetti R, Martinez C, et al. The multicentre south European study ‘Helios’. II: Different sun exposure patterns in the aetiology of basal cell and squamous cell carcinomas of the skin. Br J Cancer 1996;73(11):1447–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karagas MR, Cushing GL Jr., Greenberg ER, et al. Non-melanoma skin cancers and glucocorticoid therapy. Br J Cancer 2001;85(5):683–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Proby CM, Harwood CA, Neale RE, et al. A case-control study of betapapillomavirus infection and cutaneous squamous cell carcinoma in organ transplant recipients. Am J Transplant 2011;11(7):1498–508. [DOI] [PubMed] [Google Scholar]
- 11.Bouwes Bavinck JN, Neale RE, Abeni D, et al. Multicenter study of the association between betapapillomavirus infection and cutaneous squamous cell carcinoma. Cancer Res 2010;70(23):9777–86. [DOI] [PubMed] [Google Scholar]
- 12.Karagas MR, Waterboer T, Li Z, et al. Genus beta human papillomaviruses and incidence of basal cell and squamous cell carcinomas of skin: population based case-control study. BMJ (Clinical research ed) 2010;341:c2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Waterboer T, Abeni D, Sampogna F, et al. Serological association of beta and gamma human papillomaviruses with squamous cell carcinoma of the skin. Br J Dermatol 2008;159(2):457–9. [DOI] [PubMed] [Google Scholar]
- 14.Iannacone MR, Gheit T, Pfister H, et al. Case-control study of genus-beta human papillomaviruses in plucked eyebrow hairs and cutaneous squamous cell carcinoma. Int J Cancer 2014;17(10):28552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rollison DE, Giuliano AR, Messina JL, et al. Case-control study of Merkel cell polyomavirus infection and cutaneous squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev 2012;21(1):74–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jablonska S, Dabrowski J, Jakubowicz K. Epidermodysplasia verruciformis as a model in studies on the role of papovaviruses in oncogenesis. Cancer Res 1972;32(3):583–9. [PubMed] [Google Scholar]
- 17.Ramoz N, Rueda LA, Bouadjar B, et al. Mutations in two adjacent novel genes are associated with epidermodysplasia verruciformis. Nature genetics 2002;32(4):579–81. [DOI] [PubMed] [Google Scholar]
- 18.Majewski S, Jablonska S. Epidermodysplasia verruciformis as a model of human papillomavirus-induced genetic cancer of the skin. Arch Dermatol 1995;131(11):1312–8. [PubMed] [Google Scholar]
- 19.Boxman IL, Berkhout RJ, Mulder LH, et al. Detection of human papillomavirus DNA in plucked hairs from renal transplant recipients and healthy volunteers. J Invest Dermatol 1997;108(5):712–5. [DOI] [PubMed] [Google Scholar]
- 20.Harwood CA, Surentheran T, McGregor JM, et al. Human papillomavirus infection and non-melanoma skin cancer in immunosuppressed and immunocompetent individuals. Journal of medical virology 2000;61(3):289–97. [DOI] [PubMed] [Google Scholar]
- 21.Meyer T, Arndt R, Nindl I, et al. Association of human papillomavirus infections with cutaneous tumors in immunosuppressed patients. Transplant international : official journal of the European Society for Organ Transplantation 2003;16(3):146–53. [DOI] [PubMed] [Google Scholar]
- 22.Antonsson A, Karanfilovska S, Lindqvist PG, et al. General acquisition of human papillomavirus infections of skin occurs in early infancy. J Clin Microbiol 2003;41(6):2509–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weissenborn SJ, De Koning MN, Wieland U, et al. Intrafamilial transmission and family-specific spectra of cutaneous betapapillomaviruses. Journal of virology 2009;83(2):811–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Michael KM, Waterboer T, Sehr P, et al. Seroprevalence of 34 human papillomavirus types in the German general population. PLoS Pathog 2008;4(6):e1000091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Antonsson A, Forslund O, Ekberg H, et al. The ubiquity and impressive genomic diversity of human skin papillomaviruses suggest a commensalic nature of these viruses. Journal of virology 2000;74(24):11636–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boxman IL, Russell A, Mulder LH, et al. Association between epidermodysplasia verruciformis-associated human papillomavirus DNA in plucked eyebrow hair and solar keratoses. J Invest Dermatol 2001;117(5):1108–12. [DOI] [PubMed] [Google Scholar]
- 27.Hampras SS, Giuliano AR, Lin HY, et al. Natural history of cutaneous human papillomavirus (HPV) infection in men: the HIM study. PLoS One 2014;9(9):e104843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Forslund O, Iftner T, Andersson K, et al. Cutaneous human papillomaviruses found in sun-exposed skin: Beta-papillomavirus species 2 predominates in squamous cell carcinoma. J Infect Dis 2007;196(6):876–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Antonsson A, Green AC, Mallitt KA, et al. Prevalence and stability of antibodies to 37 human papillomavirus types--a population-based longitudinal study. Virology 2010;407(1):26–32. [DOI] [PubMed] [Google Scholar]
- 30.Iannacone MR, Michael KM, Giuliano AR, et al. Risk factors for cutaneous human papillomavirus seroreactivity among patients undergoing skin cancer screening in Florida. J Infect Dis 2010;201(5):760–9. [DOI] [PubMed] [Google Scholar]
- 31.Feltkamp MC, Broer R, di Summa FM, et al. Seroreactivity to epidermodysplasia verruciformis-related human papillomavirus types is associated with nonmelanoma skin cancer. Cancer Res 2003;63(10):2695–700. [PubMed] [Google Scholar]
- 32.Karagas MR, Nelson HH, Sehr P, et al. Human papillomavirus infection and incidence of squamous cell and basal cell carcinomas of the skin. J Natl Cancer Inst 2006;98(6):389–95. [DOI] [PubMed] [Google Scholar]
- 33.Kean JM, Rao S, Wang M, et al. Seroepidemiology of human polyomaviruses. PLoS Pathog 2009;5(3):e1000363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nicol JT, Robinot R, Carpentier A, et al. Age-specific seroprevalences of merkel cell polyomavirus, human polyomaviruses 6, 7, and 9, and trichodysplasia spinulosa-associated polyomavirus. Clinical and vaccine immunology : CVI 2013;20(3):363–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hampras SS, Giuliano AR, Lin HY, et al. Natural history of polyomaviruses in men: the HPV infection in men (HIM) study. J Infect Dis 2015;211(9):1437–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chahoud J, Semaan A, Chen Y, et al. Association between beta-genus human papillomavirus and cutaneous squamous cell carcinoma in immunocompetent individuals-A meta-analysis. JAMA Dermatol 2016;152(12):1354–64. [DOI] [PubMed] [Google Scholar]
- 37.Jackson S, Harwood C, Thomas M, et al. Role of Bak in UV-induced apoptosis in skin cancer and abrogation by HPV E6 proteins. Genes Dev 2000;14(23):3065–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hufbauer M, Cooke J, van der Horst GT, et al. Human papillomavirus mediated inhibition of DNA damage sensing and repair drives skin carcinogenesis. Mol Cancer 2015;14:183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Akgul B, Garcia-Escudero R, Ghali L, et al. The E7 protein of cutaneous human papillomavirus type 8 causes invasion of human keratinocytes into the dermis in organotypic cultures of skin. Cancer Res 2005;65(6):2216–23. [DOI] [PubMed] [Google Scholar]
- 40.Muench P, Probst S, Schuetz J, et al. Cutaneous papillomavirus E6 proteins must interact with p300 and block p53-mediated apoptosis for cellular immortalization and tumorigenesis. Cancer Res 2010;70(17):6913–24. [DOI] [PubMed] [Google Scholar]
- 41.Marcuzzi GP, Hufbauer M, Kasper HU, et al. Spontaneous tumour development in human papillomavirus type 8 E6 transgenic mice and rapid induction by UV-light exposure and wounding. J Gen Virol 2009;90(Pt 12):2855–64. [DOI] [PubMed] [Google Scholar]
- 42.Viarisio D, Muller-Decker K, Accardi R, et al. Beta HPV38 oncoproteins act with a hit-and-run mechanism in ultraviolet radiation-induced skin carcinogenesis in mice. PLoS Pathog 2018;14(1):e1006783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Viarisio D, Mueller-Decker K, Kloz U, et al. E6 and E7 from beta HPV38 cooperate with ultraviolet light in the development of actinic keratosis-like lesions and squamous cell carcinoma in mice. PLoS Pathog 2011;7(7):e1002125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hall L, Struijk L, Neale RE, et al. Re: Human papillomavirus infection and incidence of squamous cell and basal cell carcinomas of the skin. J Natl Cancer Inst 2006;98(19):1425–6. [DOI] [PubMed] [Google Scholar]
- 45.Clouser MC, Roe DJ, Foote JA, et al. Effect of non-steroidal anti-inflammatory drugs on non-melanoma skin cancer incidence in the SKICAP-AK trial. Pharmacoepidemiology and drug safety 2009;18(4):276–83. [DOI] [PubMed] [Google Scholar]
- 46.Jeon SY, Lee CY, Song KH, et al. Spectrophotometric measurement of minimal erythema dose sites after narrowband ultraviolet B phototesting: clinical implication of spetrophotometric values in phototherapy. Ann Dermatol 2014;26(1):17–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.John EM, Schwartz GG, Koo J, et al. Sun exposure, vitamin D receptor gene polymorphisms, and risk of advanced prostate cancer. Cancer Res 2005;65(12):5470–9. [DOI] [PubMed] [Google Scholar]
- 48.Hesterberg RS, Amorrortu RP, Zhao Y, et al. T regulatory cell subpopulations associated with recent ultraviolet radiation exposure in a skin cancer screening cohort. J Immunol 2018;201(11):3269–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Clarys P, Alewaeters K, Lambrecht R, et al. Skin color measurements: comparison between three instruments: the Chromameter(R), the DermaSpectrometer(R) and the Mexameter(R). Skin research and technology : official journal of International Society for Bioengineering and the Skin (ISBS) [and] International Society for Digital Imaging of Skin (ISDIS) [and] International Society for Skin Imaging (ISSI) 2000;6(4):230–8. [DOI] [PubMed] [Google Scholar]
- 50.Massey DS, Martin JA. The NIS Skin Color Scale. Princeton, NJ: Princeton University Press, 2003. [Google Scholar]
- 51.Koster B, Sondergaard J, Nielsen JB, et al. The validated sun exposure questionnaire: association of objective and subjective measures of sun exposure in a Danish population-based sample. Br J Dermatol 2017;176(2):446–56. [DOI] [PubMed] [Google Scholar]
- 52.Rollison DE, Schell MJ, Fenske NA, et al. Cutaneous viral infections across two anatomic sites among a cohort of skin cancer screening patients. J Infect Dis 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Karagas MR, Zens MS, Nelson HH, et al. Measures of cumulative exposure from a standardized sun exposure history questionnaire: a comparison with histologic assessment of solar skin damage. Am J Epidemiol 2007;165(6):719–26. [DOI] [PubMed] [Google Scholar]
- 54.Kuklinski LF, Zens MS, Perry AE, et al. Skin microtopography as a measure of photoaging and risk of squamous cell carcinoma of the skin in a US population. Photodermatol Photoimmunol Photomed 2017;33(1):41–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Iannacone MR, Gheit T, Waterboer T, et al. Case-control study of cutaneous human papillomaviruses in squamous cell carcinoma of the skin. Cancer Epidemiol Biomarkers Prev 2012;21(8):1303–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Waterboer T, Sehr P, Michael KM, et al. Multiplex human papillomavirus serology based on in situ-purified glutathione s-transferase fusion proteins. Clin Chem 2005;51(10):1845–53. [DOI] [PubMed] [Google Scholar]
- 57.Waterboer T, Sehr P, Pawlita M. Suppression of non-specific binding in serological Luminex assays. Journal of immunological methods 2006;309(1–2):200–4. [DOI] [PubMed] [Google Scholar]
- 58.van Buuren S, Groothuis-Oudshoorn K. mice: Multivariate Imputation by Chained Equations in R. J Stat Softw 2011;45(3):1–67. [Google Scholar]
- 59.Blewett LA, Drew JAR, Griffin R, et al. IPUMS Health Surveys: National Health Interview Survey, Version 6.3 [NHIS 2015 Sample Adult and Cancer Supplement]. Minneapolis, MN: IPUMS, 2018. [Google Scholar]
- 60.Miller D, Fernandez CA, Lee DJ. National Health Interview Survey In: Gellman MD, Turner JR, eds. Encyclopedia of Behavioral Medicine. New York, NY: Springer New York, 2013:1286–8. [Google Scholar]
- 61.Weissenborn SJ, Nindl I, Purdie K, et al. Human papillomavirus-DNA loads in actinic keratoses exceed those in non-melanoma skin cancers. J Invest Dermatol 2005;125(1):93–7. [DOI] [PubMed] [Google Scholar]
- 62.Koshiol J, Lindsay L, Pimenta JM, et al. Persistent human papillomavirus infection and cervical neoplasia: a systematic review and meta-analysis. Am J Epidemiol 2008;168(2):123–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.