Abstract
Massive generation of health-related data has been key in enabling the big data science initiative to gain new insights in healthcare. Nursing can benefit from this era of big data science, as there is a growing need for new discoveries from large quantities of nursing data to provide evidence-based care. However, there are few nursing studies using big data analytics. The purpose of this article is to explain a knowledge discovery and data mining approach that was employed to discover knowledge about hospital-acquired catheter-associated urinary tract infections from multiple data sources, including electronic health records and nurse staffing data. Three different machine learning techniques are described: decision trees, logistic regression, and support vector machines. The decision tree model created rules to interpret relationships among associated factors of hospital-acquired catheter-associated urinary tract infections. The logistic regression model showed what factors were related to a higher risk of hospital-acquired catheter-associated urinary tract infections. The support vector machines model was included to compare performance with the other two interpretable models. This article introduces the examples of cutting-edge machine learning approaches that will advance secondary use of electronic health records and integration of multiple data sources as well as provide evidence necessary to guide nursing professionals in practice.
Keywords: Catheter-associated urinary tract infections, Data mining, Machine learning
Big data science is a key component to the attainment of new insights about healthcare enabled by large amounts of health-related data generated from electronic health records (EHRs), genomics, sensors, ubiquitous devices, and social media. Nursing can benefit from this era of big data science, as there is a growing need for new insights from nursing data to provide evidence-based care. It is imperative that the nursing profession lead in big data science to ensure that the knowledge is not only shaped by the priority of person-centric care but also that knowledge is useful to nursing.1 However, there are few nursing studies using big data analytics.2 There are many complex clinical problems associated with numerous factors that would benefit from big data analytics. This article demonstrates the application of data science methods to a complex clinical problem, hospital-acquired catheter-associated urinary tract infection (HA-CAUTI), which is one of the major nosocomial infections significantly affecting patient outcomes.3,4 Although many past studies identified risk factors and strategies to lower HA-CAUTI incidents, it is still a prevalent nosocomial infection.5 There is a need to explore additional factors associated with HA-CAUTI, such as nurse staffing, and develop a predictive model to detect patients at risk in the hospital. This study uses data from EHRs, which contain patient clinical data, ICU data for CAUTI occurrence, and nurse staffing data to discover new insights for HA-CAUTIs.
The variables related to HA-CAUTIs are quite diverse, making the import of such variables difficult to discern and results inconsistent among studies. Traditional statistical methods have limitations for investigating potentially numerous and complex interactions among many variables and providing in-depth knowledge from copious data.6,7 Big data research using EHRs and nurse staffing data has merit for such new knowledge discovery from large-scale data.
The purpose of this article is to explain the knowledge discovery and data mining (KDDM) approach using machine learning that was employed to discover knowledge about HA-CAUTI from multiple data sources and predict the patients at risk for HA-CAUTI. Clinical results of our study have been reported elsewhere.8 However, not many nursing studies have used large data analytics such as the machine learning approach in their research.2 The focus of this article is to explain the methods employed that can serve as a guide for others interested in using these methods for analyzing complex health conditions and to increase understanding among nurses about this approach. The KDDM process can be described as a search to find information that was previously unnoticed in a database. It is an iterative process of identifying valid and useful patterns from large quantities of data and provides a framework for understanding new knowledge. Three different machine learning methods—decision trees (DTs), logistic regression (LR), and support vector machines (SVMs)—were used. The strengths and weaknesses of the three approaches for knowledge discovery associated with HA-CAUTI are discussed.
A methodological challenge faced in our study and addressed in this article consisted of creating a dataset from multiple sources including EHRs and a national-level nursing quality indicator database. Multiple data sources allow investigation of complex multidimensional interactions that can provide in-depth insights.9 Another challenge was evaluating a rare event, which HA-CAUTI proved to be.
KNOWLEDGE DISCOVERY AND DATA MINING APPROACH
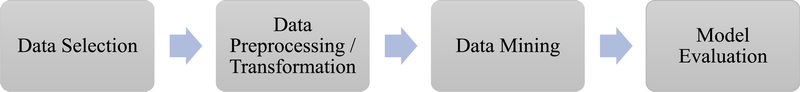
The KDDM approach encompasses a variety of statistical analyses, pattern recognition, and machine learning techniques.10 The KDDM approach is often used for prediction, in which some variables in a database predict the values of other variables of interest. The first step of KDDM is selecting a target dataset. It is essential to identify a subset of data from a source, such as EHRs, on which data mining is to be performed. The next step of KDDM is preparation of the data, which involves data cleaning, such as removing noisy data, selecting the necessary information for model construction, and compensating for missing data fields. It is important to have a compact, clean dataset to build a model and decrease the effect of noisy data, including meaningless data, corrupt information, and missing data, which hinders a model’s performance. The third step is the transformation of data, including reducing data, discretizing numeric variables, putting data in a unified format, selecting variables, and transforming multiple classes in a variable into binary ones. Generally, preprocessing accounts for roughly 80% of the KDDM process.11 The fourth step of KDDM is data mining using algorithms to identify patterns of interest. This process consists of applying data analysis and discovery algorithms that produce particular patterns or models from the data.12 Interpreting the mined patterns and evaluating the model are the fifth step; at this stage, one or more of the previous steps may be repeated to improve the performance. The sixth step is using the knowledge discovered, which involves incorporation into another system for further action.
With the wide implementation of EHRs, machine learning is emerging as a useful tool for large-scale health data analysis. Because EHRs contain a large quantity of patient information and provide massive amounts of structured and unstructured data, the use of EHRs increases the potential for efficient access to comprehensive and standardized data. Large sample sizes and the integration of multiple data sources using a KDDM approach have advantages in discovering hidden information from data and adding new predictors such as social, behavioral, and environmental factors to the analysis. It allows nursing researchers to generate new insights and knowledge and to create evidence-based guidelines for the clinical field. Each KDDM step and methods applied in this study are presented in Figure 1 and described in the following section.
FIGURE 1.
Steps in the KDDM approach.
STEPS OF THE KNOWLEDGE DISCOVERY AND DATA MINING METHOD
Selecting a Target Data Set
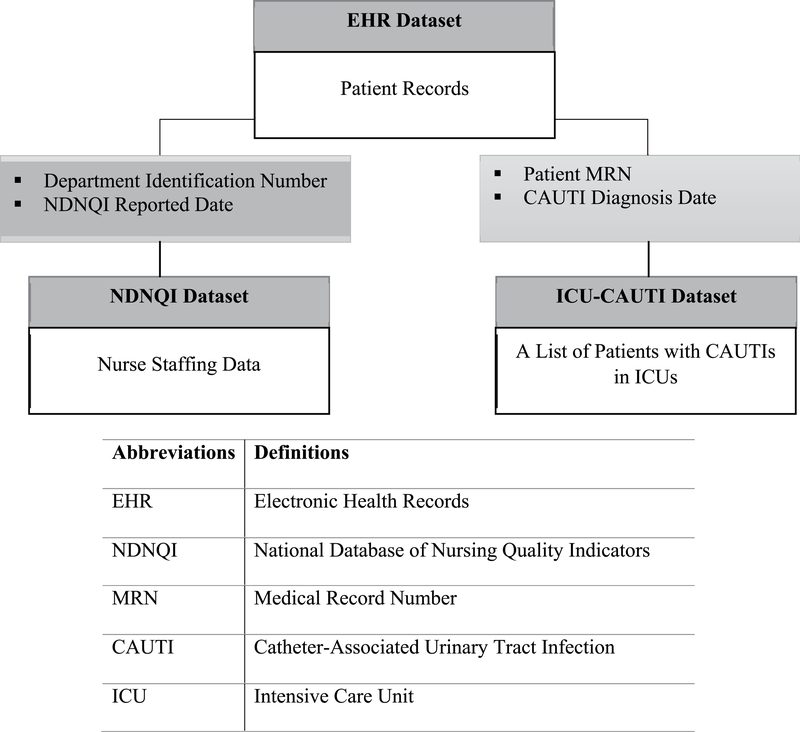
Three different datasets from EHRs, ICUs, and nurse staffing data were combined to develop new knowledge from clinical practice. The relationships of the three datasets are displayed in Figure 2. The three datasets provide unique information such as clinical predictors from the EHR, nurse staffing data including quality care measures, and ICU data for a list of patients who acquired CAUTIs. Dataset 1 from the University of Minnesota (UMN) Academic Health Center-Information Exchange (AHC-IE) Clinical Data Repository (CDR; hereafter referred to as the EHR dataset) includes EHR data of ICU patients. The AHC-IE CDR includes the data of more than 2.4 million patients from seven hospitals and 40 clinics, with the following data categories: allergies, claims, demographics, diagnoses and problems, encounters and visits, flowsheets, histories, immunizations and vaccinations, institutions and locations, laboratory results, medications, procedures, providers, and vitals. Dataset 2 includes unit-level nurse staffing data from the University of Minnesota Medical Center (UMMC), submitted to the National Database of Nursing Quality Indicators (NDNQI) on a quarterly basis (the NDNQI dataset). The NDNQI data are collected to measure nursing care quality and the nursing work environment and are aggregated and compared at a national level.13 Dataset 3 (the ICU-CAUTI dataset) includes a list of patients who acquired CAUTIs in the ICUs at the UMMC. The ICU setting was chosen because CAUTIs were the most prevalent type of HAI in UMMC ICUs. The list of patients with CAUTIs reflects the continuous changes in definitions of CAUTIs that occurred over time and is not easily determined in an EHR. Three datasets were extracted and delivered to the UMN secure data shelter, accessible only via a virtual private network. The EHR dataset was then combined with the NDNQI data and the list of CAUTI patients in ICUs to create an integrated dataset.
FIGURE 2.
Relationship of the three datasets of the study.
The selected subset of the EHR data for this study contained data on adult patients (aged 18 years or older) admitted between January 1, 2012, and June 30, 2015, to any of three ICUs (medical, surgical, and cardiovascular) at the UMMC. Patients who did not have indwelling urinary catheters were excluded. The total number of patients in the three ICUs was 8496. The total number of unique ICU admissions for final analysis was 11 226, because a patient could have multiple ICU admissions during the relevant time period.
The NDNQI dataset consists of nurse staffing data. The nurse staffing data submitted to NDNQI between January 1, 2012, and June 30, 2015, in three ICUs were extracted and then included in the UMN data shelter for mapping with other datasets. The ICU-CAUTI dataset includes a list of patients who acquired CAUTIs during their hospitalization in any of the three UMMC ICUs. The list was maintained separately in the UMMC ICU database to track patients with CAUTIs in the ICUs, and includes the patient medical record number (MRN), department name, and date of CAUTI diagnosis for each patient. The list was used to determine which patients acquired CAUTIs during hospitalization in the three ICUs. The list is the gold standard for the outcome measure in this article, because it was created after manual chart reviews using the Centers for Disease Control and Prevention definition for CAUTI.
The outcome variable in the analysis referred to patients who acquired a CAUTI during a hospitalization between January 1, 2012, and June 30, 2015, at one of three UMMC ICUs whose patient MRN appeared on the list of patients who had CAUTIs in the ICUs and who did not have a CAUTI or UTI when admitted.
Integration of Multiple Data Sources
To create an integrated dataset from the three different data sources, data were extracted and then mapped using shared key components. The EHR and NDNQI datasets were linked using department ID and NDNQI reported date. The ICU-CAUTI dataset was matched to the EHR and NDNQI datasets using patient MRN and date of CAUTI diagnosis.
The variables in the EHR dataset were linked with each other using the patient MRN and master service IDs. Because data in the EHR dataset were delivered in multiple tables, integrating the variables in the EHR dataset was the first step before mapping this dataset to other datasets. A master service ID represents a unique type of encounter such as hospitalization or clinic. In this dataset, a master service ID was used as a unique hospitalization with admission and discharge dates. Once the data from the EHR dataset were prepared, the dataset was mapped to the ICU-CAUTI dataset, which contained patient MRNs, department IDs, and CAUTI diagnosis dates. These variables were used as key components to link the two datasets. Because a patient could have multiple hospitalizations—that is, one patient MRN could be linked to multiple hospitalizations—the diagnosis date in the ICU-CAUTI dataset was used to determine when the CAUTI occurred among multiple hospitalizations. The quarterly reported NDNQI dataset was linked to the previous two datasets—the EHR and ICU-CAUTI datasets—using report dates and department IDs. Mapping the two datasets with the NDNQI datasets resulted in a larger sample (n = 11 226); this reflected that patients had stayed in different ICUs during one hospitalization. Therefore, a unique ICU admission was the unit of analysis in the final integrated dataset.
Data in the EHR and NDNQI datasets were reviewed, and variables potentially useful for this study based on the researcher’s literature review and domain expertise selected or added. Nine variables from the EHR and four from the NDNQI were identified.8 Individual patient-level and unit-level variables for analysis are included in Table 1.
Table 1.
Variables for Analysis
| Individual Patient-Level Variables | |
| Age (≥18 y) | Charlson Comorbidity Index score |
| Gender | Hospitalization within previous 6 mo |
| Immunosuppression | Rationale for continued use of catheter |
| Pre-existing Catheter | Lab result—glucose |
| Length of hospital stay | Presence of HA-CAUTI |
| Unit-level variables | |
| Total nursing hours per patient day | |
| Percent of direct care RNs with associate’s degree in nursing | |
| Percent of direct care RNs with BSN, MSN, or PhD | |
| Percent of direct care RNs with specialty nursing certification | |
Data Preparation and Transformation
Before conducting the analysis, data were preprocessed to enhance the quality of the final dataset. Missing values were imputed using k-nearest neighbors algorithm, which is widely used to impute new cases based on a similarity of available cases. All categorical data were transformed into a binary format because LR in Weka outperforms when the data format is binary. Examples include age, gender, laboratory results, immunosuppression, preexisting catheter, and Charlson Comorbidity Index score.
Analyzing Rare Events
Skewed distribution of rare events, known as unbalanced classes, may lower a model’s predictive power. This was an important step in the methods because there was a critical gap between the number of ICU admissions without HA-CAUTIs (n = 10 365) and ICU admissions with HA-CAUTIs (n = 55). When the target events (HA-CAUTI) are rare and there are unbalanced classes, machine learning requires steps to correctly classify these rare events. In this study, a cost-sensitive classification method was used to counter the class imbalance. Cost-sensitive classification refers to a classification method that uses misclassification costs as a penalty to correct class imbalance; the goal is to minimize the total cost. Most classification algorithms are programmed to minimize the error rate regardless of misclassification errors. That is, whether the error is false positive or false negative, misclassification errors are treated equally. However, in healthcare, different types of misclassification errors have different consequences. For example, false negative is more important than false positive in HA-CAUTI event; if a patient with an HA-CAUTI is the positive class and a patient without an HA-CAUTI is negative, then misclassifying an HA-CAUTI (eg, the patient is positive but classified as negative—a false negative) is far more serious, and therefore expensive, than a false-positive error. The delay in proper diagnosis and treatment can even endanger the patient. Therefore, cost-sensitive classification in this study gave much higher cost (penalty) to false negatives than to false positives. In cost-sensitive classification, any given instance should be classified into the class that has the minimum expected cost. This principle prevents an instance from being classified as a false negative. To find the optimal cost for the model, different values were tested (100, 200, and 300), and the number of false negatives and accuracy were compared. Since the ratio between the number of ICU admissions without HA-CAUTI (n = 10 365) and the number of ICU admissions with HA-CAUTI (n = 55) was approximately 190:1, the values around 200 were tested.
Data Mining Modeling
For data mining, the integrated dataset was divided into training and testing datasets. The training set was used to construct the prediction model, and test set was used to evaluate the prediction performance of the model. The standard way to estimate a model’s performance is to use a 10-fold cross-validation.14 Therefore, the data were divided randomly into 10 approximately equal partitions (folds); nine of the 10 partitions were used for training (creating) the model, while the remaining one was used for testing (validating) the model. The procedure was repeated 10 times so that each partition was used once for testing.
Three data mining models were developed using the following machine learning techniques: DTs, LR, and SVM. The machine learning techniques are explained below.
Decision Trees
Decision trees are tree-like structures that start from root nodes and end with leaf nodes. The model has several branches consisting of different variables, and the leaf node on each branch represents a class or a kind of class distribution. Decision trees describe the relationship among variables and the relative importance of variables. This method uses recursive data separation to construct a tree, by repeatedly splitting the branches into subgroups until splitting no longer adds any information to the predictions. Mathematical algorithms are used to identify a variable and corresponding threshold that splits the input observation into two or more subgroups. The Gini index is a widely used split criterion in DTs, a statistical measure of distribution to evaluate how mixed the classes are split into two groups. A maximized value of the Gini index can be reached when the observations are equally distributed, whereas a minimized value is 0 when all values belong to one class. The threshold maximizes the homogeneity of the resulting subgroups. This step is repeated at each leaf node until the complete tree is constructed. Decision tree models are easy to interpret, not sensitive to outliers. However, the reliability of prediction models declines when there is overfitting. Overfitting occurs when the model is overly complex with too many parameters. To avoid this, appropriate pruning is necessary. Pruning is a technique that reduces the DT size to identify a smaller tree with the lowest training error rate.
Logistic Regression
Logistic regression is an extension of traditional regression wherein a set of independent variables is usually used to model a binary outcome. Logistic regression is an appropriate method for this study to model the dichotomous variable of patients with and without HA-CAUTIs. Logistic regression builds the model to predict the odds of an event’s occurrence (HA-CAUTI) using weights to maximize the likelihood of reproducing the data. An odds ratio (OR) is a measure of association between a variable and an outcome. When the OR is greater than 1, a variable is associated with higher odds of an outcome; when the OR is less than 1, a variable is associated with lower odds of an outcome; and when the OR equals 1, a variable does not influence the odds of the outcome. Although LR is vulnerable when there are too many parameters, it has proven to be robust in a number of domains and is an effective method of estimating probabilities from dichotomous variables.15
Support Vector Machines
Support vector machines are among the most powerful classification algorithms for predictive accuracy.16 Support vector machines are becoming popular in the clinical field because of their robust performance and strong mathematical and statistical foundations. Support vector machines work well with high-dimensional data and complex modeling and are durable for the overfitting problem. The core of the SVM method is a process that finds a hyperplane, which separates the examples into different outcomes. When separated for two-class problems (HA-CAUTI vs no HA-CAUTI), SVMs find an optimal hyperplane with a maximum distance to the closest point of each of the two classes. When separated, similarity of the points to each other is important. The similarity is computed by a kernel function. The linear kernel function that leads to a linear decision hyperplane was used in this study. However, SVMs require extensive computing memory and a considerable amount of computational time to deal with the large amount of data. Additionally, the decisions during the SVM modeling process are not easy to understand, whereas the modeling process of DTs and LRs is considered transparent. Despite its low interpretability, the SVM model was included in this study to compare the predictive performance with the other two data mining models.
Model Evaluation
The evaluation and comparison criteria for three data mining models in this study were (1) number of false negatives, (2) accuracy, (3) specificity, (4) sensitivity, (5) the area under the receiver operating characteristic (ROC) curve, and (6) clinical interpretability. The number of false negatives refers to the number of patients who were classified as not having an HA-CAUTI but who did have a CAUTI; false negatives are especially important in clinical settings because they prevent the administration of proper care to patients who need acute treatment. Therefore, having fewer false-negative values was preferred. The accuracy reflects a model’s overall performance, that is, the percentage of cases that are correctly predicted in the test dataset. A minimum accuracy threshold for a model was 75%. Both sensitivity and specificity are used to measure true-positive rates from models. The area under the ROC curve indicates a model’s performance; it is acceptable when it is over 0.7.17 Clinical interpretability for the models is also important because part of the purpose of this study was to discover and actually translate new knowledge on HA-CAUTIs for clinical care.
Analytic Software
Three software packages were used. Data preparation was carried out using two software suites, structured query language (SQL; currently maintained by International Standards Organization/International Electrotechnical Commission, Geneva, Switzerland) and Python (Python Software Foundation, Wilmington, DE); SQL was used for data extracting and combining data, and Python was used for data preprocessing and transformation. The data were then collapsed into an integrated dataset, because many data mining techniques require the data to be in this flat file format. The analysis was carried out using Weka, a commonly used data mining software developed by a machine learning group at the University of Waikato in New Zealand (https://www.cs.waikato.ac.nz/ml/weka/).
MODELS RESULTING FROM EACH MACHINE LEARNING TECHNIQUE
The three machine learning techniques (DT, LR, and SVM) produced three data mining models, which are discussed and compared in the following sections. An in-depth report of the factors associated with HA-CAUTIs derived from these models has been published elsewhere.8 Because there is a critical gap between the number of patients with CAUTIs and patients without CAUTIs in the population, unbalanced classes were corrected using a cost-sensitive classification method. Different cost values for the classifier were examined to find the optimal cost for the predictive models. The cost was set to 200 for the modeling because 200 had the fewest possible false negatives.
Decision Trees
Weka supports a number of DT models such as J48, RepTree, and BFTree. BFTree showed the best performance in terms of accuracy and the number of false negatives. BFTree creates DTs using a best-first expansion of nodes. The best-first method explores all variables and sorts them in order of performance and finds the most promising node. A heuristic search was used for binary split for nominal variables. A heuristic function was used to search available information at each branching step in the DT to decide which node to follow. The Gini index was used for the splitting criterion. The model used a postpruning strategy for finding the best number of branches. The postpruning operates after recursive splitting of DTs and selects branches that have the best validation accuracy with the fewest number of leaves.
Logistic Regression
The LR model reported each variable’s OR. The ORs show the magnitude of each variable’s contribution in modeling. Weka does not report confidence intervals (CIs) or P value for each variable; instead, the program provided an overall average CI, which was 95% to compare the predictive models.
Support Vector Machines
The SVM model uses a “black-box” approach, which means the decisions during the process are unknown and not easily explainable. However, the SVM model shows robust performance in classification. The output of the SVM model provided weights for each variable. Although there was a limitation in terms of interpretation, the variable weight indicated the variable’s relevance for modeling. That is, a larger absolute value for the weight meant that the variable was relatively important in discrimination of the two classes. The variables with a positive weight have association with the classification of HA-CAUTI as “yes,” whereas negative weighted variables have association with the classification of HA-CAUTI as “no.”
Model Evaluation
Model evaluation and comparison used the number of false negatives, accuracy, sensitivity, specificity, precision, and the area under the ROC curve. The evaluation results of each model using the 200 cost are shown in Table 2.
Table 2.
Evaluation Results From the Predictive Models
| Evaluation Criteria | DT | LR | SVMs |
|---|---|---|---|
| No. of false negatives | 13 | 17 | 13 |
| Accuracy (%) | 75.87 | 75.83 | 71.50 |
| Sensitivity | 0.81 | 0.75 | 0.80 |
| Specificity | 0.76 | 0.76 | 0.71 |
| Precision | 0.02 | 0.02 | 0.02 |
| Area under the ROC curve | 0.78 | 0.85 | 0.76 |
| Clinical interpretability | Yes | Yes | No |
The results showed that the DT model outperformed the LR model for the reduction of false negatives, accuracy, and sensitivity, but it performed less well for the area under the ROC curve. In terms of clinical interpretability, the results of the DT model showed associated factors of HA-CAUTI and the relationships among those variables. The results of the LR model showed the ORs of each variable to represent risk factors of HA-CAUTI. The DT model outperformed the SVM model for accuracy, sensitivity, specificity, and the area under the ROC curve. Additionally, the DT model was clinically interpretable, whereas the SVM model was not.
DISCUSSION
In this article, a KDDM approach and three cutting-edge machine learning methods for predicting the patients at risk for HA-CAUTI and analyzing factors associated with HA-CAUTI using multiple data sources were presented. The article provides an example of using combined datasets and machine learning techniques, as demonstrating each step for integrating different datasets, employing a data mining approach, and evaluating resulting models. By analyzing large amounts of data generated from EHRs, nurse researchers may discover new insights for developing interventions supporting evidence-based nursing practice. Additionally, the use of EHR and big data analytics can enhance the visibility of nursing research and the care that nurses provide.
Evaluation of the three models generated for this study shows that the DT model created rules that combine multiple variables, making it easy to interpret relationships and interactions among the associated factors of HA-CAUTI.8 The model did automate feature selection, visualizing the most discriminatory factors and the relative importance of the associated factors. The DT model outperformed the other two data mining models in most of the evaluation criteria. However, this technique requires appropriate pruning to avoid trees that are too complex and improve model accuracy.
The LR model showed the effect of individual variables associated with a higher risk of HA-CAUTI rather than the combined relationship of these variables. The ORs showed the magnitude of each variable’s contribution in modeling. A limitation of the analytic software, Weka, is that it did not provide CIs and P values for each variable in the LR model. There is a need for future studies using different analytic software to look at detailed results of variables for the LR model.
The SVM model worked well with large amounts of data and showed robust modeling performance. The model had the fewest false negatives, but its accuracy was the lowest among the three models. Further, the “black box” method makes interpretation of results difficult.
Combination of data from multiple sources enabled interoperable data use on a large scale. Big data analysis using machine learning techniques enables the investigation of unknown patterns and relationships among numerous factors. Because there are many factors associated with HA-CAUTIs. it is important to look at each factor’s association in addition to the interactions of those factors. There were limitations, however, in combining multiple data sources. Although Dataset 1 (EHR dataset) contained data from 2.4 million patient records, Dataset 2 (NDNQI dataset) was collected at only one hospital, and Dataset 3 (ICU-CAUTI dataset) was limited to ICUs in hospitals because that is where the majority of HA-CAUTIs occur. Combining the datasets limited the amount of data for analysis. The study population began with a large number, but the useable cases were culled when matching patients and determining the best source for reliable data. Although unbalanced data were corrected using the cost-sensitive approach, the validity of the outcome would have been improved if there had been more patients with CAUTI.
Another limitation of the methods is that they did not include unstructured data, such as text or provider’s notes. The EHR in the UMN AHC-IE contains a substantial amount of data that were semistructured or unstructured. Since HA-CAUTIs are nursing-sensitive outcomes, there may be some factors relevant to CAUTI occurrence that are embedded in the nursing notes. Therefore, using all the data documented in the EHRs would have provided more in-depth knowledge of HA-CAUTIs.
CONCLUSION
Despite the fact that there are a number of clinical guidelines and studies about CAUTIs, it is still one of the major nosocomial infections. The factors associated with CAUTIs are diverse, requiring investigation of associations among multiple variables and its interactions. This article shows that the KDDM approach using three machine learning techniques and multiple data sources offered the ability to investigate unknown patterns and relationships among the factors associated with HA-CAUTIs. Nursing research using the KDDM approach may lead to more in-depth discoveries and insight, higher-quality nursing care, and more tailored intervention and management strategies for patients with complex health problems.
Acknowledgments
The authors have disclosed that they have no significant relationships with, or financial interest in, any commercial companies pertaining to this article.
Contributor Information
Jung In Park, School of Nursing, University of California, Irvine.
Donna Z. Bliss, School of Nursing, University of Minnesota, Minneapolis.
Chih-Lin Chi, School of Nursing, University of Minnesota, Minneapolis.
Connie W. Delaney, School of Nursing, University of Minnesota, Minneapolis.
Bonnie L. Westra, School of Nursing, University of Minnesota, Minneapolis.
References
- 1.Brennan PF, Bakken S. Nursing needs big data and big data needs nursing. Journal of Nursing Scholarship. 2015;47(5): 477–484. [DOI] [PubMed] [Google Scholar]
- 2.Westra BL, Sylvia M, Weinfurter EF, et al. Big data science: a literature review of nursing research exemplars. Nursing Outlook. 2017;65(5): 549–561. [DOI] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. National and State Healthcare Associated Infections Progress Report. Atlanta, GA: CDC; 2016. [Google Scholar]
- 4.Burton DC, Edwards JR, Srinivasan A, Fridkin SK, Gould CV. Trends in catheter-associated urinary tract infections in adult intensive care units—United states, 1990–2007. Infection Control & Hospital Epidemiology. 2011;32(8): 748–756. [DOI] [PubMed] [Google Scholar]
- 5.Tambyah PA. Prevention and control of catheter associated urinary tract infection. Journal of Microbiology, Immunology and Infection. 2015; 48(2): S16. [Google Scholar]
- 6.Murdoch TB, Detsky AS. The inevitable application of big data to health care. Journal of the American Medical Association. 2013;309(13): 1351–1352. [DOI] [PubMed] [Google Scholar]
- 7.Zhao C, Luan J. Data mining: going beyond traditional statistics. New Directions for Institutional Research. 2006;2006(131): 7–16. [Google Scholar]
- 8.Park JI, Bliss DZ, Chi C, Delaney CW, Westra BL. Factors associated with healthcare-acquired catheter-associated urinary tract infections: analysis using multiple data sources and data mining techniques. Journal of Wound Ostomy & Continence Nursing. 2018;45(2): 168–173. [DOI] [PubMed] [Google Scholar]
- 9.Ng K, Kakkanatt C, Benigno M, et al. Curating and integrating data from multiple sources to support healthcare analytics. Studies in Health Technology and Informatics. 2015;216: 1056. [PubMed] [Google Scholar]
- 10.Freitas AA. Data Mining and Knowledge Discovery With Evolutionary Algorithms. Berlin, Germany: Springer Science & Business Media; 2013. [Google Scholar]
- 11.Zhang S, Zhang C, Yang Q. Data preparation for data mining. Applied Artificial Intelligence. 2003;17(5–6): 375–381. [Google Scholar]
- 12.Fayyad U, Piatetsky-Shapiro G, Smyth P. From data mining to knowledge discovery in databases. AI Magazine. 1996;17(3): 37. [Google Scholar]
- 13.Montalvo I. The National Database of Nursing Quality Indicators™ (NDNQI®). OJIN: The Online Journal of Issues in Nursing. 2007;12(3): 112–214. [Google Scholar]
- 14.Witten IH, Frank E, Hall MA, Pal CJ. Data Mining: Practical Machine Learning Tools and Techniques. Burlington, MA: Morgan Kaufmann; 2016. [Google Scholar]
- 15.Long WJ, Griffith JL, Selker HP, D’agostino RB. A comparison of logistic regression to decision-tree induction in a medical domain. Computers and Biomedical Research. 1993;26(1): 74–97. [DOI] [PubMed] [Google Scholar]
- 16.Cristianini N, Shawe-Taylor J. An Introduction to Support Vector Machines and Other Kernel-Based Learning Methods. Cambridge, England: Cambridge University Press; 2000. [Google Scholar]
- 17.Redon J, Coca A, Lazaro P, et al. Factors associated with therapeutic inertia in hypertension: validation of a predictive model. Journal of Hypertension. 2010;28(8): 1770–1777. [DOI] [PubMed] [Google Scholar]