Abstract
Our understanding of the role of folate one-carbon metabolism in colon carcinogenesis remains incomplete. Previous studies indicate that a methyl donor deficient (MDD) diet lacking folic acid, choline, methionine, and vitamin B12 is associated with long-lasting changes to the intestinal epithelium and sustained tumor protection in Apc-mutant mice. However, the metabolic pathways by which the MDD diet effects these changes are unknown. Colon samples harvested from ApcΔ14/+ mice fed the MDD diet for 18 weeks were profiled using a GC-MS and LC-MS/MS metabolomics platform. Random forest and pathway analyses were used to identify altered metabolic pathways and associated gene expression changes were analyzed by RT-PCR. Approximately 100 metabolites impacted by the MDD diet were identified. As expected, metabolites within the methionine cycle, including methionine (−2.9-fold, P<0.001) and betaine (−3.3-fold, P<0.001), were reduced. Elevated homocysteine (110-fold, P<0.001) was associated with increased flux through the transsulfuration pathway. Unexpectedly, levels of deoxycholic acid (−4.5-fold, P<0.05) and several other secondary bile acids were reduced. There were also unexpected reductions in the levels of carnitine (−2.0-fold, P<0.01) and a panel of acylcarnitines involved in fatty acid β-oxidation. Finally, metabolites involved in redox balance, including ascorbate and hypotaurine, were found to be persistently elevated. These findings provide clues to the molecular changes underlying MDD-mediated tumor protection and identify regulatable metabolic pathways that may provide new targets for colon cancer prevention and treatment.
Keywords: Colorectal cancer, methyl donor, folate, metabolism
Introduction
Investigations into the effects of folate consumption on colorectal cancer (CRC) risk have yielded conflicting results. Early epidemiological studies suggested that folate deficiency increases the risk of CRC in the general population.(1–3) However, recent meta-analyses found no effect on CRC incidence.(4,5) Furthermore, clinical chemoprevention studies suggest that elevated folate intake in certain high-risk individuals may actually increase CRC risk.(6,7) Thus, it has been proposed that the dosage and timing of folate intervention must be carefully evaluated when developing strategies for chemoprevention.(1,8) Preclinical studies from our laboratory(9,10) have shown that a diet deficient in the methyl donor nutrients folate, choline, methionine and vitamin B12 prevents tumor development in Apc-mutant mice. These studies suggest that reduced methyl donor intake may benefit individuals at elevated risk of colon cancer. Our most recent study(9) demonstrated that even temporary dietary methyl donor deficiency (MDD) provides lasting tumor protection that is associated with persistent changes to the intestinal epithelium.
Potential mechanisms that may account for the tumor protection afforded by methyl donor restriction have not been adequately explored. Each of the methyl donor nutrients removed from the MDD diet play a role in folate one-carbon metabolism (FOCM). FOCM is responsible for fundamental biosynthetic processes, including the production of nucleotides and the generation of S-adenosylmethionine (SAM), the “universal methyl donor” used in DNA and histone methylation.(11) FOCM has also been shown to influence other key metabolic processes, including lipid and energy metabolism.(12) These wide-ranging roles underscore the importance of FOCM to cellular homeostasis; ultimately, disruption of FOCM can compromise DNA stability, alter gene expression, and inhibit cellular proliferation and survival.(13) Furthermore, methyl donor nutrients contribute to many metabolic functions independent of their role in FOCM, including the synthesis of phospholipids and sphingolipids, proteins, hormones and other small molecules.(12)
Our previous studies(9,10) suggested that the manipulation of FOCM is a promising approach for colon cancer prevention. Our earliest study demonstrated that administration of an MDD diet to Apc-mutant mice was associated with reductions in the number and size of tumors in the small intestine (SI) and colon and a concurrent reduction in serum folate.(10) Microarray analysis revealed that dietary MDD was associated with reduced expression of genes involved in the inflammatory response, cell-cycle progression, and mitosis.(10) Our follow-up study recapitulated the tumor-protective effects of dietary MDD in a different Apc-mutant mouse model and demonstrated that these effects persisted beyond the period of active nutrient deficiency.(9) Furthermore, histological analyses revealed that MDD was associated with reduced SI and colon crypt length, decreased rates of crypt fission, reduced expression of epithelial cell proliferation markers (ki-67 and phospho-histone H3), and increased markers of epithelial cell apoptosis (cleaved caspase 3), together suggesting that dietary MDD alters intestinal epithelial cell turnover.(9) Finally, this study found a reduction in SI and colon cells expressing the putative tumor stem cell marker doublecortin like kinase 1 (Dclk1), suggesting that dietary MDD may provide tumor protection by depleting a population of cancer-initiating cells within the colon.(9)
However, our previous study(9) also revealed that methyl donor restriction is associated with adverse effects including disruption of normal body weight gain and hepatic steatosis. While these effects were reversible, they may limit clinical utility of dietary methyl donor depletion. Investigation of the specific metabolic pathways that are disrupted under conditions of dietary MDD may identify novel targets for more effective cancer prevention and treatment strategies. The present study uses an untargeted metabolomic profiling platform to quantify ~400 biochemicals present within samples of grossly normal whole colon tissue obtained from ApcΔ14/+mice used in our previous study.(9) We show that dietary MDD is associated with many metabolic alterations, a subset of which persists following methyl donor repletion. These metabolic changes were enriched in several key pathways, including the methionine cycle, the transsulfuration pathway, secondary bile acid synthesis, and fatty acid β-oxidation. These observations provide clues as to the molecular changes underlying MDD-induced tumor protection and identify metabolomic features of the MDD diet that may provide new targets for CRC prevention.
Materials and Methods
Animal Treatment and Sample Collection
Dietary protocols using ApcΔ14/+ and ApcMin/+mice and the collection of colonic tissue samples have previously been described.(9,10) All animal experiments were conducted with approval from the Institutional Animal Care and Use Committee (IACUC), University of Connecticut Health. Dietary studies used the following amino-acid defined experimental diets: Methyl Donor Sufficient (MDS; TD.99366, Harlan Laboratories, Madison WI) diet and Methyl Donor Deficient (MDD; TD.00605) diet, which was identical to the MDS diet except for the depletion of folate, choline, methionine and vitamin B12, and the addition of homocysteine. Complete diet compositions are shown in Supplementary Table 1. ApcΔ14/+ mice were randomized at 4 weeks of age and placed into four groups; each group was placed on a specific dietary regimen for 18 weeks.(9) Group I (MDS ad libitum) mice were fed the MDS diet ad libitum for the entire 18-week period. Group II (MDD) mice were fed the MDD diet ad libitum for 18 weeks. Group III (MDS-PF) mice were pair-fed the MDS diet in an amount equivalent to levels consumed by group II mice. Finally, mice in group IV (MDD:MDS-repletion), mice were placed on the MDD diet for 11 weeks and then transferred to the MDS diet for an additional 7 weeks. All mice were sacrificed at 22 weeks of age. Upon sacrifice, colons were harvested, flushed with ice-cold PBS and opened longitudinally. Normal-appearing segments of whole tissue (approximately 30 mg each) were collected from the distal 2–3 cm of the colon and were immediately snap-frozen and stored at −80°C until analysis. The TBARS assay (Cayman Chemical, Ann Arbor, MI) was used to quantify levels of malondialdehyde according to the manufacturer’s protocol.
Metabolite Profiling
Colon tissue samples from a representative subset of mice (n=7/group) were selected for metabolite profiling such that the subset had the same average body weight, tumor count, and tumor size as the group at large (Supplementary Table 2). All tissue analyzed had a normal appearance and no visible tumors upon gross examination. Thirty-mg of snap-frozen, normal-appearing whole colon tissue was shipped to Metabolon, Inc. (Durham, NC) on dry ice analysis. The platform used for sample preparation and analysis has been previously described.(14–16) Briefly, tissue samples were processed using the automated MicroLab STAR system (Hamilton Company). Each sample was spiked with an internal standard prior to processing to monitor extraction efficiency. Samples were mechanically disrupted and homogenized and metabolites were extracted using cold methanol precipitation under vigorous shaking for 2 minutes (GenoGrinder2000, Glen Mills). Tissue extracts were divided into three aliquots for analysis by UPLC-MS/MS with positive ion electrospray ionization (ESI), UPLC-MS/MS with negative ion ESI and GC-MS. For LC-MS/MS, the samples were stored overnight under nitrogen before being dried and then reconstituted in LC-compatible acidic or basic solvents containing a cocktail of 12 injection standards at fixed concentrations. Each sample was analyzed by ultra-performance liquid chromatography (Waters ACQUITY) and mass spectrometry (Thermo-Finnigan LTG), using a mass spectrometer consisting of an ESI source and linear ion-trap mass analyzer operated at nominal mass resolution. Each sample was analyzed under acidic positive ion-optimized conditions and basic negative ion-optimized conditions in two independent runs using dedicated chromatography columns (Waters UPLC BEH C18–2.1×100 mm, 1.7 μm). Extracts analyzed under acidic conditions were gradient eluted in a mixture of water and ethanol containing 0.1% formic acid, while basic extracts were analyzed in water/methanol containing 6.5 mM ammonium bicarbonate. MS analysis used both MS and MS/MS scans using dynamic exclusion, with a scan range of 80–1000 m/z. For GC, each sample was dried under vacuum overnight before being derivatized under dried nitrogen using bistrimethyl-silyltrifluoroacetamide (BSTFA). Samples were then separated on a 5% diphenyl/95% dimethyl polysiloxane fused silica column (20 m × 0.18 mm ID; 0.18 um film thickness) under helium and temperature gradient from 60° to 340°C. Samples were analyzed using a fast-scanning single-quadrupole mass spectrometer (Thermo-Finnigan Trace DSQ) operated at unit mass resolving power; the scan range was 50–750 m/z. Metabolites were identified by comparing GC/LC-MS parameters, including column retention time, spectral peaks, and mass-to-charge ratio to a proprietary reference chemical library.
Quantitative Real-Time PCR
Total RNA was extracted from samples of whole colon using the RNA isolation kit (RNeasy Mini Kit, QIAGEN). 250ng of total RNA was used as a template for cDNA synthesis using the iScript cDNA synthesis kit (BIO-RAD) following the manufacturer’s instructions. cDNA was diluted in nuclease-free water before it was utilized for gene expression by quantitative RT-PCR (qRT-PCR). All qRT-PCR reactions involved the use of CFX real-time PCR (Bio-RAD), iTaq SYBR Green supermix and pre-designed, validated primers for Cpt1a (Fwd: 5’-CTCCGCCTGAGCCATGAAG-3’, Rev: 5’-CACCAGTGATGATGCCATTCT-3’), Cpt2 (Fwd: 5’-CAGCACAGCATCGTACCCA-3’, Rev: 5’-TCCCAATGCCGTTCTCAAAAT-3’), and Ppara (Fwd: 5’-TTTCGGCGAACTATTCGGCTG-3’, Rev: 5’-GGCATTTGTTCCGGTTCTTCTT-3’) (Integrated DNA technologies). Relative quantification of mRNA levels by the ΔΔCT method was done after normalization of total cDNA to endogenous 18s rRNA.
Bioinformatics and Biostatistics
MS peaks representing metabolites of interest were quantified by calculating the area under the curve. Missing values were assumed to be below the limit of detection and were imputed with the minimum observed value for each compound. Biochemical data was normalized to total protein to account for differences in metabolite levels due to differences in the amount of material present in each sample. For analyses performed over several days, a data normalization step was included to correct for potential variation from instrument inter-day variability. Each compound was corrected in run-day blocks by setting the medians equal to 1 and normalizing each data point proportionately (termed the “block correction”). Statistical analysis of mean metabolite levels was performed using Welch’s two-sample t-test followed by false discovery rate (FDR) correction using the Benjamini-Hochberg procedure. Differences with an FDR-corrected P-value < 0.05 were considered to be statistically significant, as this value indicates that for a given pair-wise comparison the probability under the null hypothesis that the variate would be observed as a value equal to or more extreme than the observed value is < 5% after adjusting for the total number of pair-wise comparisons made in the overall analysis. Pearson’s correlation analysis was performed in R. Random Forest Analysis (RFA) was used to generate metabolite signatures that allowed accurate classification of de-identified samples into experimental groups. Mean Decrease Accuracy (MDA) analysis was used to quantify the importance of individual metabolites to accurate classification by RFA. Hierarchical clustering was performed by calculating Euclidean distance between samples, and dimensionality was reduced using Principal Component Analysis (PCA). RFA, MDA, clustering and PCA were performed using R-programming language and software. Pathway analysis was accomplished by calculating relative-betweenness centrality using MetaboAnalyst 3.0.(17) For qRT-PCR and the TBARS assay, statistical comparisons between groups were made using the Kruskal-Wallis test with Dunn’s post-test. For IHC, comparisons between groups were made using one-way ANOVA with Bonferroni post-test.
Results
Dietary methyl donor deficiency alters the colonic metabolome
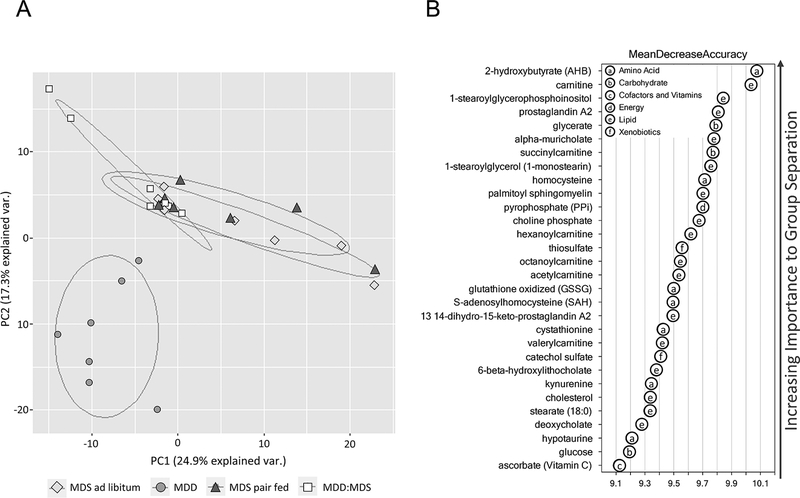
Using a well-characterized, integrated metabolomics platform(15), a total of 388 unique metabolites were quantified from a subset of mice (n=7 mice/group) from each of the 4 dietary groups. Resultant “metabolite profiles” were used for all subsequent analyses. As shown by the PCA plot in Figure 1A, MDD samples were separated from the other dietary groups, indicating the presence of a distinct metabolic profile. Furthermore, the metabolite profiles in the MDD:MDS-repletion group were partially overlapping with the MDS ad libitum and MDS-PF groups, suggesting that some metabolic changes may be sustained beyond active methyl donor restriction.
Figure 1. Principal component analysis (PCA) biplot analysis of experimental diets groups.
(A) Principal component analysis (PCA) biplot indicating a distinct metabolic profile associated with MDD. Partial overlap of MDD:MDS samples with both MDS ad libitum and MDS pair-fed controls indicates metabolic changes that persist at least 7 weeks beyond methyl donor repletion. (B) Top 30 metabolites ranked by their contribution to separation of MDD and MDS-PF samples by Random Forest Analysis (RFA). These metabolites were used as a starting point in the identification of metabolic pathways most significantly affected by dietary MDD.
The distinct metabolic signature associated with the MDD diet is further illustrated by RFA, which differentiated metabolites between the MDD and each of the other dietary groups with 100% accuracy (Fig. 1B). A comparison of metabolite profiles between the MDD and MDS-PF mice identified changes specifically associated with methyl donor depletion; these metabolites are shown in Table 1. Linear correlation analysis revealed that most of these metabolites were significantly correlated with colon tumor number (Pearson’s r, Table 1). These top metabolites were classified in subgroups according to function and used to identify metabolic pathway alterations associated with dietary MDD. Three altered metabolic pathways most closely associated with the MDD diet and consistently correlated with colon tumor multiplicity were identified; methionine and cysteine metabolism, bile acid metabolism and carnitine-dependent fatty acid oxidation (FAO).
Table 1.
Top 30 significantly altered metabolites associated with MDD diet and their correlations with colon tumor count.
| Metabolite | KEGG ID | Subgroup | Fold change (MDD vs MDS PF) | Q-value* | Pearson’s r ([Metabolite] vs Tumor Count) | Q-valuet† |
|---|---|---|---|---|---|---|
| Ascorbate | C00072 | Ascorbate and Aldarate Metabolism | 1.6 | 2.9E-03 | −0.49 | 6.0E-03 |
| catechol sulfate | C00090 | Benzoate metabolism | −12.5 | 1.0E-02 | 0.20 | 2.5E-01 |
| alpha-muricholate | C17647 | Bile Acid Metabolism | −10.0 | 2.5E-05 | 0.59 | 1.0E-03 |
| 6-beta-hydroxylithocholate | C15515 | Bile Acid Metabolism | −7.1 | 2.7E-05 | 0.46 | 8.0E-03 |
| deoxycholate | C04483 | Bile Acid Metabolism | −10.0 | 1.9E-03 | 0.43 | 1.0E-02 |
| carnitine | C00318 | Carnitine-Dependent Fatty Acid Oxidation | −2.4 | 5.5E-07 | 0.39 | 1.9E-02 |
| hexanoylcarnitine | -- | Carnitine-Dependent Fatty Acid Oxidation | −2.6 | 4.0E-04 | 0.38 | 2.2E-02 |
| octanoyl carnitine | C02838 | Carnitine-Dependent Fatty Acid Oxidation | −2.6 | 7.6E-05 | 0.32 | 6.3E-02 |
| acetylcarnitine | C02571 | Carnitine-Dependent Fatty Acid Oxidation | −1.9 | 1.0E-04 | 0.45 | 8.0E-03 |
| valerylcarnitine | -- | Carnitine-Dependent Fatty Acid Oxidation | −4.2 | 5.0E-04 | 0.41 | 1.3E-02 |
| thiosulfate | C05529 | Chemical | 1.7 | 1.0E-03 | −0.52 | 2.0E-03 |
| prostaglandin A2 | C05953 | Eicosanoid | 3.1 | 6.5E-05 | −0.44 | 8.0E-03 |
| 13,14-dihydro-15-keto-prostaglandin A2 | -- | Eicosanoid | 3.4 | 6.3E-05 | −0.28 | 1.0E-01 |
| glycerate | C00258 | Glycolysis, Gluconeogenesis, and Pyruvate Metabolism | −1.7 | 4.7E-03 | 0.39 | 1.8E-02 |
| glucose | C00031 | Glycolysis, Gluconeogenesis, and Pyruvate Metabolism | −12.5 | 8.3E-03 | 0.26 | 1.3E-01 |
| stearate | C01530 | Long Chain Fatty Acid | 1.5 | 9.9E-03 | −0.34 | 4.7E-02 |
| 1-stearoylglycerophosphoinositol | -- | Lysolipid | 2.8 | 9.0E-05 | −0.41 | 1.4E-02 |
| 2-hydroxybutyrate (AHB) | C05984 | Methionine, Cysteine and Taurine Metabolism | 6.1 | 5.0E-04 | −0.43 | 1.0E-02 |
| homocysteine | C00155 | Methionine, Cysteine and Taurine Metabolism | 111.2 | 6.3E-05 | −0.46 | 7.0E-03 |
| glutathione, oxidized (6SSG) | C00127 | Methionine, Cysteine and Taurine Metabolism | 1.4 | 1.2E-02 | −0.38 | 2.1E-02 |
| S-adenosylhomocysteine (SAH) | C00021 | Methionine, Cysteine and Taurine Metabolism | 14.5 | 2.7E-05 | −0.52 | 2.0E-03 |
| cystathionine | C02291 | Methionine, Cysteine and Taurine Metabolism | 12.7 | 1.9E-03 | −0.44 | 8.0E-03 |
| hypotaurine | C00519 | Methionine, Cysteine and Taurine Metabolism | 5.2 | 4.9E-05 | −0.45 | 8.0E-03 |
| 1-stearoylglycerol (1-monostearin) | D01947 | Monoacylglycerol | 1.9 | 1.9E-03 | −0.16 | 3.5E-01 |
| pyrophosphate (PPi) | C00013 | Oxidative Phosphorylation | 1.9 | 9.0E-04 | −0.52 | 3.0E-03 |
| choline phosphate | C00588 | Phospholipid Metabolism | −2.4 | 8.1E-05 | 0.39 | 1.8E-02 |
| palmitoyl sphingomyelin | -- | Sphigolipid Metabolism | 3.7 | 4.0E-04 | −0.36 | 2.9E-02 |
| cholesterol | C00187 | Sterol | 1.6 | 2.1E-03 | −0.41 | 1.4E-02 |
| succinylcarnitine | -- | TCA Cycle | −2.2 | 4.9E-05 | 0.47 | 7.0E-03 |
| kynurenine | C00328 | Tryptophan Metabolism | 3.1 | 7.9E-03 | −0.29 | 9.0E-02 |
KEGG = Kyoto Encyclopedia of Genes and Genomes
Statistical comparisons between MDD and MDS PF groups made using Welch’s two-sample t-test. False discovery rate (FDR)-adjusted Q-values were derived from P-values using the Benjamini-Hochberg procedure.
Significance of Pearson’s correlations was determined by comparison to the t-distribution assuming 30 degrees of freedom. False discovery rate (FDR)-adjusted Q-values were derived from P-values using the Benjamini-Hochberg procedure.
MDD reduces methionine cycle flux and increases the transsulfuration pathway
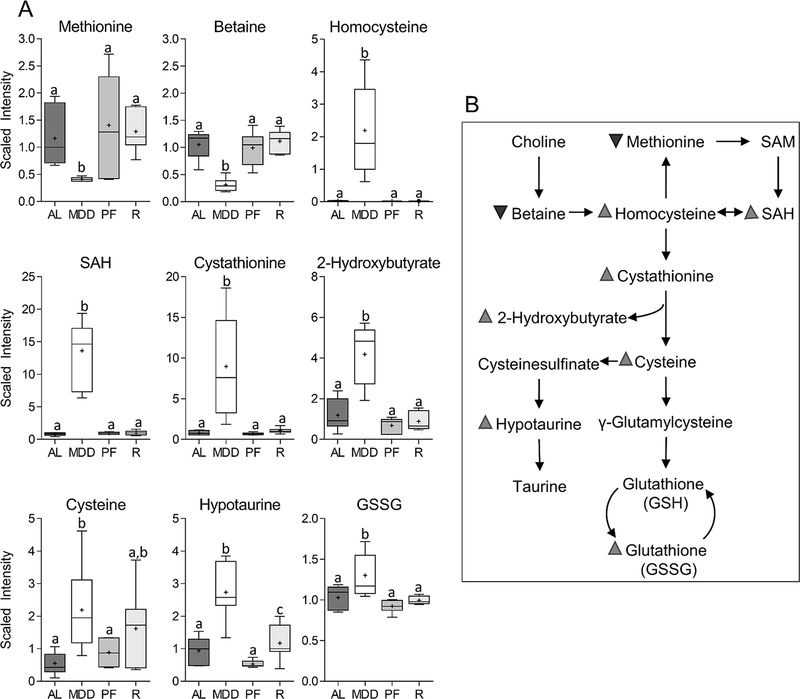
We anticipated that the MDD diet would reduce the levels of methyl donor nutrients within the colon. As shown in Figure 2A, MDD-fed mice showed significant reductions in the colonic levels of methionine (−2.9-fold, P<0.001) and betaine (−3.3-fold, P<0.001), the metabolite immediately downstream of choline, validating our prediction. Furthermore, there were marked increases in the levels of homocysteine (110-fold, P<0.001) and S-adenosylhomocysteine (SAH) (17-fold, P<0.001) (Figure 2A), a metabolite normally produced by demethylation of the “universal methyl donor”, S-adenosylmethionine (SAM).(11) MDD also caused increases in cystathionine (11.5-fold, P<0.001), 2-hydroxybutyrate (3.5-fold, P<0.01) and cysteine (4.0-fold, P<0.05), indicating increased flux through the cysteine-generating transsulfuration pathway.(18) MDD mice also exhibited increases in hypotaurine (2.9-fold, P<0.01) and glutathione disulfide (1.3-fold, P<0.05), changes associated with increased activity of the taurine and glutathione synthesis pathways. As shown in Supplementary Table 3, levels of transsulfuration pathway-associated metabolites, including homocysteine, SAH, cystathionine, and hypotaurine exhibited significant inverse correlations with positive immunohistochemical staining for the proliferation marker phospho-histone H3 (PHH3) and the stem cell marker doublecortin-like kinase 1 (DCLK1). These metabolites also exhibited significant positive correlations with the apoptotic marker cleaved caspase 3 (CC3). There were no significant differences in any metabolites in the methionine cycle or transsulfuration pathway between the MDS ad libitum and MDS-PF groups.
Figure 2. Changes in the levels of metabolites involved in the methionine cycle and the transsulfuration pathway.
Levels of methionine (−2.9-fold, P<0.001), betaine (−3.3-fold, P<0.001), homocysteine (109-fold, P<0.001), s-adenosylmethionine (17.1-fold, P<0.001), cystathionine (11.5-fold, P<0.001), 2-hydroxybutyrate (3.5-fold, P<0.01), cysteine (4.0-fold, P<0.05), hypotaurine (2.9-fold, P<0.01), and glutathione disulfide (1.3-fold, P<0.05) are all altered under conditions of dietary methyl donor restriction. Whiskers on Tukey’s box plots represent Interquartile Range (IQR). Statistically significant differences (P<0.05) between groups, measured by Welch’s two-sample t-test, are indicated within bar graphs by differences in the letter placed above each group. “AL” = MDS ad libitum, “MDD” = Methyl donor deficient, “PF” = MDS Pair-fed, “R” = MDD:MDS Repletion group.
In the MDD:MDS repletion group, most of the metabolites within the methionine cycle and transsulfuration pathways had levels equivalent to the MDS ad libitum controls, indicating that these MDD-induced changes were fully reversible upon methyl donor repletion (Figure 2A). The one exception was hypotaurine, which remained significantly elevated upon methyl donor repletion. A schematic depiction of the changes to methionine and cysteine metabolism associated with methyl donor restriction is shown in Figure 2B.
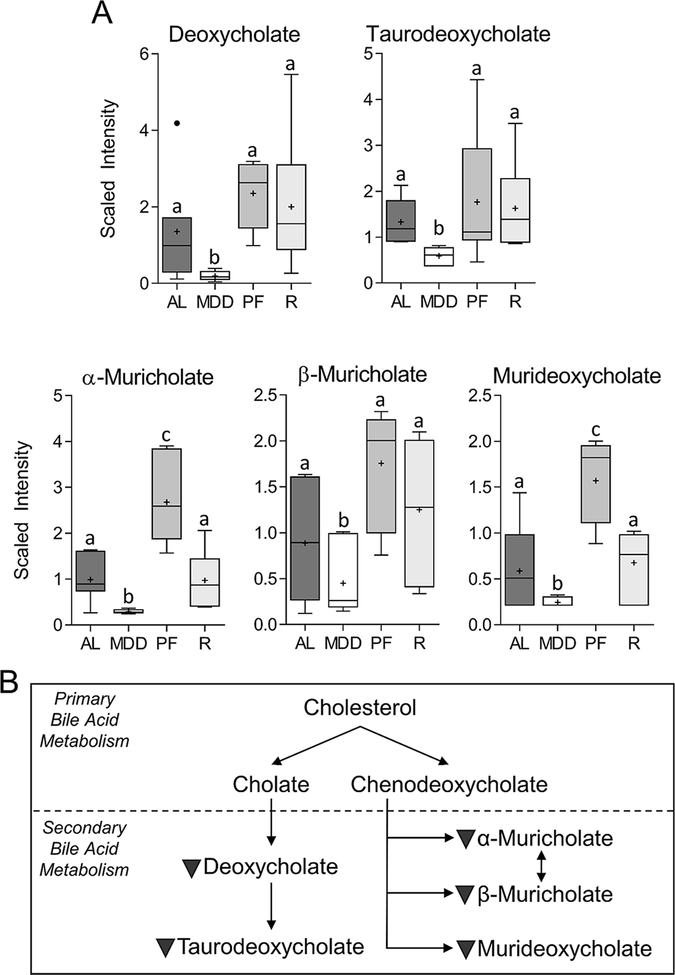
MDD inhibits formation of secondary bile acids (BAs)
As shown in Figure 3A, methyl donor restriction reduced the levels of the secondary bile acids (BAs), deoxycholate (−4.5-fold, P<0.05) and its taurine-conjugated bile salt, taurodeoxycholate (−2.2-fold, P<0.01), within the colon, relative to the MDS ad libitum controls. MDD also reduced the levels of α-muricholate (−3.8-fold, P<0.005), β-muricholate (−2.2-fold, P<0.05) and murideoxycholate (−2.6-fold, P<0.05), secondary bile acids that are formed in the mouse. Secondary bile acid levels returned to normal levels following methyl donor repletion. Colon bile acids levels exhibited a moderate, but significant, inverse correlation with the proportion of cells positive for ki67 (Supplementary Table 3). In contrast to the reductions associated with MDD, α-muricholate (2.7-fold, P<0.05) and murideoxycholate (2.7-fold, P<0.05) were found to be significantly elevated in the MDS-PF group relative to MDS ad libitum; non-significant increases were also observed for deoxycholate, taurodeoxychlate, and β-muricholate. These metabolite changes caused by MDD are summarized in Figure 3B.
Figure 3. Changes in the levels of secondary bile acids.
Levels of deoxycholate (−4.5-fold, P<0.05), taurodeoxycholate (−2.2-fold, P<0.01), alpha-muricholate (−3.8-fold, P<0.005), beta-muricholate (−2.2-fold, P<0.05), and murideoxycholate (−2.6-fold, P<0.05) are reduced under conditions of dietary methyl donor restriction, suggesting inhibition of secondary BA synthesis. Whiskers on Tukey’s box plots represent Interquartile Range (IQR). Statistically significant differences (P<0.05) between groups, measured by Welch’s two-sample t-test, are indicated within bar graphs by differences in the letter placed above each group. “AL” = MDS ad libitum, “MDD” = Methyl donor deficient, “PF” = MDS Pair-fed, “R” = MDD:MDS Repletion group.
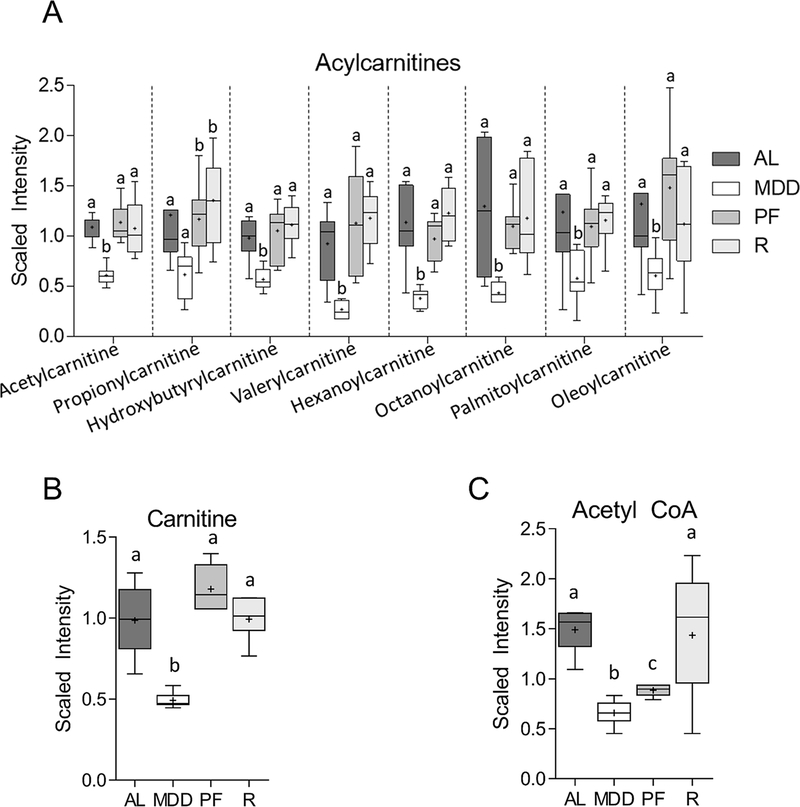
MDD inhibits carnitine-dependent FAO
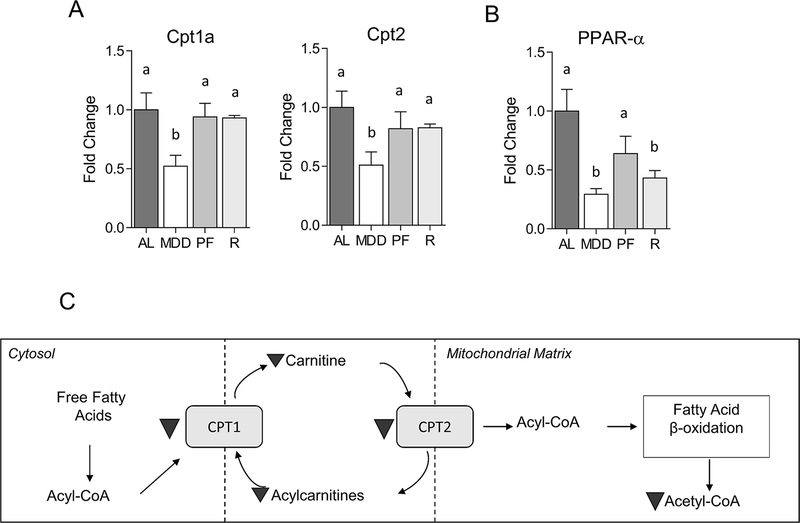
Metabolite subgroup analysis identified significant associations between MDD and a number of acylcarnitines, fatty acid-derived acyl groups conjugated to the shuttle molecule carnitine, as well as carnitine itself. These fatty acid metabolites are transported into the mitochondrial matrix and used for energy production via fatty acid β-oxidation.(19) As shown in Figure 4A, several acylcarnitines were reduced in the colons of MDD-fed mice, including acetylcarnitine (−1.8-fold, P<0.001), propionylcarnitine (−2.0-fold, P<0.05), hydroxybutyrylcarnitine (−1.7-fold, P<0.01), valerylcarnitine (−3.4-fold, P<0.001), hexanoylcarnitine (−3.0-fold, P<0.01), octanoylcarnitine (−3.0-fold, P<0.05), palmitoylcarnitine (−2.1-fold, P<0.05) and oleoylcarntine (−2.2-fold, P<0.05). Decreases in acylcarnitine levels are a likely consequence of the limited availability of carnitine, an indirect metabolic product of methionine.(20) In fact, carnitine levels were significantly reduced (−2.0-fold, P<0.01) by methyl donor restriction (Figure 4B). Mitochondrial oxidation of the acylcarnitines is an important source of acetyl CoA, the levels of which were reduced in MDD-fed mice (−2.5-fold, P<0.05) and MDS-PF mice (−1.8-fold, P<0.05; Figure 4C). As shown in Figure 5A&B, MDD was also associated with decreased expression of two key enzymatic regulators the acylcarnitine system, carnitine palmitoyl transferase 1a (Cpt1a; −1.9-fold, P<0.05) and Cpt2 (−2.0-fold, P<0.05), as well as peroxisome proliferator-activated receptor α (Ppara), a canonical regulator of Cpt1 transcription (−3.4-fold, P<0.05). Levels of carnitine and several acylcarnitines were associated with large and significant positive correlations with PHH3 and DCLK1 staining, as well as inverse correlations with CC3 staining. Trends towards reduced expression of Cpt2 and Ppara were also observed in the MDS-PF group, though these changes failed to achieve statistical significance.
Figure 4. Changes in the levels of metabolites involved in fatty acid β-oxidation (FAO).
Levels of a panel of acylcarnitines (A), as well as carnitine (B; −2.0-fold, P<0.01) and acetyl CoA (−2.5-fold, P<0.05), are reduced under conditions of dietary methyl donor restriction, suggesting inhibition of FAO. Whiskers on Tukey’s box plots represent Interquartile Range (IQR). Statistically significant differences (P<0.05) between groups, measured by Welch’s two-sample t-test, are indicated within bar graphs by differences in the letter placed above each group. “AL” = MDS ad libitum, “MDD” = Methyl donor deficient, “PF” = MDS Pair-fed, “R” = MDD:MDS Repletion group.
Figure 5. Changes in the expression of FAO-regulatory genes.
Dietary MDD was associated with reduced expression of carnitine palmitoyl transferase 1a (CPT1A, −1.9-fold, P<0.05), CPT1B (−2-fold, P<0.05), and peroxisome proliferator-activated receptor α (PPARα, −3.3-fold, P<0.01). Bar graphs indicate mean mean fold change normalized to control ± SEM. Statistically significant differences (P<0.05) between groups, measured by Kruskal-Wallis test with Dunn’s post-test, are indicated within bar graphs by differences in the letter placed above each group. “AL” = MDS ad libitum, “MDD” = Methyl donor deficient, “PF” = MDS Pair-fed, “R” = MDD:MDS Repletion group.
It has been reported that CPT1 directly interacts with the mitochondrial membrane protein BCL-2, and that this interaction promotes resistance to apoptosis.(21) As shown in Supplementary Figure 1, analysis of colon tissue from our previously-published study revealed that MDD is associated with reduced BCL-2 expression.(10) Furthermore, electron scanning microscopy of these same tissues revealed significant mitochondrial swelling associated with methyl donor restriction (Figure S1). Immunohistochemistry revealed that ki-67+ proliferative cells were drastically reduced in the colonic epithelium under conditions of MDD. A sustained reduction in the ki-67+ was still evident upon methyl donor repletion (Supplementary Figure 2). Together, these results suggest that methyl donor restriction inhibits mitochondrial FAO, leading to impaired acetyl CoA-dependent energy metabolism, reduced proliferation and increased apoptosis. Upon methyl donor repletion, however, the levels of FAO-related metabolites returned to normal. A schema depicting metabolic alterations to carnitine-dependent FAO are shown in Figure 5C.
MDD causes persistent changes to intestinal redox balance
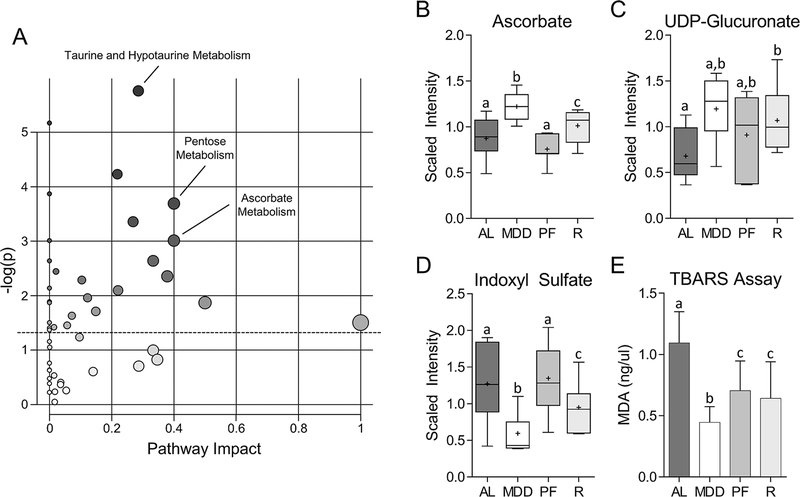
The metabolic changes described up to this point were identified by RFA. The following data describe the results of a second analysis designed specifically to identify sustained metabolic alterations that may persist beyond active methyl donor restriction. As shown in Figure 6A, pathway analysis of ‘retained’ changes in the MDD:MDS-repletion and MDD-fed mice, relative to the MDS ad libitum group, were identified using MetaboAnalyst 3.0. These alterations in MDD-fed mice were enriched in several key metabolic pathways involved in redox balance, including taurine and hypotaurine metabolism (Figure 2A), and ascorbate metabolism. As shown in Figure 6B and C, the levels of ascorbate, a potent antioxidant(22), were elevated in both the MDD (1.4-fold, P<0.05) and MDD:MDS-repletion mice (1.2-fold, P<0.05), as was UDP-glucuronate (MDD, 1.8-fold, P<0.05; MDD:MDS-R, 1.6-fold, P<0.05), an intermediate in ascorbate synthesis. As shown in Supplementary Figure 3, the levels of indoleproprionate (IPA), a potent, bacterially-derived oxygen radical scavenger(23), were increased in MDD-fed mice (8.3-fold, P<0.01), and in the MDD:MDS-repletion group (3.2-fold, P=NS), although the latter group did not achieve statistical significance. These findings suggest that dietary MDD affects bacterial metabolism, and that these changes may also persist beyond active methyl donor restriction. The levels of indoxyl sulfate, a pro-oxidant toxin produced by intestinal microbes(24), were reduced in both the MDD (−2.1-fold, P<0.05) and MDD:MDS-repletion mice (−1.3-fold, P<0.05) (Figure 6D). Finally, we used the TBARS assay to quantify the levels of malondialdehyde, a product of lipid peroxidation that is used as a marker for oxidative stress(25), within the colon. As shown in Figure 6E, malondialdehyde levels were significantly reduced in both the MDD (−2.5-fold, P<0.05) and MDD:MDS-repletion groups (−1.7-fold, P<0.05).
Figure 6. Identification of MDD-associated metabolic changes that persist during methyl donor repletion.
(A) Biplot of metabolite set enrichment analysis (MSEA) depicting metabolic pathways significantly enriched for persistent changes. Overall, three pathways involved in redox balance were significantly enriched for persistent changes: taurine and hypotaurine metabolism, pentose metabolism, and ascorbate metabolism. The size and color of each point represents the number of metabolites within the pathway and the enrichment score (ES), respectively. Pathway Impact is representative of the number of unique metabolic pathways that interact with a given pathway. (B) Levels of ascorbate, a potent antioxidant, are increased in both MDD (1.4-fold, P<0.05) and MDD:MDS-Repletion mice (1.2-fold, P<0.05). (C) Levels of UDP-glucoronate, and intermediate in the synthesis of ascorbate are also elevated in MDD (1.8-fold, P<0.05) and MDD:MDS-Repletion mice (1.2-fold, P<0.05), suggesting persistent upregulation of ascorbate synthesis following temporary MDD. (D) Levels of indoxyl sulfate, a pro-oxidant bacterial toxin, are reduced in both MDD (−2.1-fold, P<0.05) and MDD:MDS-Repletion mice (−1.3-fold, P<0.05). (E) Levels of malondialdehyde (MDA), a marker of oxidative stress measured by the TBARS assay are significantly reduced in both MDD (−2.5-fold, P<0.05) and MDD:MDS-Repletion mice (−1.7-fold, P<0.05). Together, these results suggest oxidative stress is persistently reduced within the colon following temporary MDD. Whiskers on Tukey’s box plots represent Interquartile Range (IQR). Error bars in (E) represent SEM. Statistically significant differences (P<0.05) between groups, measured by Welch’s two-sample t-test, are indicated within bar graphs by differences in the letter placed above each group. “AL” = MDS ad libitum, “MDD” = Methyl donor deficient, “PF” = MDS Pair-fed, “R” = MDD:MDS Repletion group.
Discussion
Our understanding of the role of one-carbon metabolism in cancer is incomplete, but accumulating evidence suggests that high folate intake in individuals harboring neoplastic foci may promote cancer development.(6,9,10) The controversy surrounding folate supplementation in individuals harboring neoplastic foci prompted our development of an experimental mouse diet in which we depleted folic acid, methionine, choline, and vitamin B12. This methyl donor restricted diet showed striking tumor protection in two Apc mouse models.(9,10) The present study expands upon this previous work by examining diet-induced alterations to the colonic metabolome that may be associated with tumor protection.(9,10) Untargeted metabolomic analyses revealed that dietary methyl donor restriction is associated with significant changes to multiple metabolic pathways implicated in colorectal carcinogenesis. While mostly reversible, a subset of these metabolic alterations persist within colon for at least seven weeks after mice are returned to a methyl donor replete diet. Ultimately, we believe that these sustained metabolic changes may provide new targets for cancer prevention and treatment strategies based upon manipulation of FOCM.
As expected, dietary methyl donor deficiency was associated with extensive alterations to FOCM within the colon, including reduced levels of methionine and choline, together with marked increases in homocysteine. While several earlier studies have examined the individual impact of methionine, choline and folate, our study is relatively unique in focusing on the effects of combined restriction of these nutrients on cancer development. Dietary choline and betaine were shown in several epidemiological studies to have no direct effect on CRC risk in both women and men(26,27), although a recent study of a Chinese population found that high intake of total choline was associated with a lower risk of colorectal cancer, an effect that was not modified by folate intake.(28) With respect to methionine, the increased demand of this micronutrient by cancer cells has been well-established, whereby cancer cells are more sensitive to methionine restriction than normal cells.(29) As shown by Komninou et al.(30), dietary methionine restriction reduced the multiplicity of precancerous aberrant crypt foci (ACF) in azoxymethane-treated F344 rats, suggesting a potential role for methionine restriction in cancer prevention. Although our results are not specific to methionine, they do raise the possibility that methionine restriction may contribute in part to cancer protection. Future studies will be needed to determine whether methionine restriction, either singly or in combination with other methyl donor nutrients, may also affect chemotherapeutic efficacy. Finally, while multiple metabolites in the methionine cycle and transsulfuration pathway were altered under conditions of MDD, no changes in these metabolites were observed in the MDS pair fed group, suggesting that they are specific to dietary methyl donor restriction. Thus, these metabolic changes may help to explain why dietary methyl donor restriction is associated with greater tumor protection than caloric restriction (CR) alone.(9,10)
Methyl donor restriction was associated with marked increases in the levels of SAH, which is produced directly from homocysteine via SAH hydrolase.(31) SAH has previously been shown to inhibit the activity of the DNA methyltransferases(32), and elevated levels of SAH are associated with widespread DNA hypomethylation.(33) As shown by Eads et al.(34) and Weis et al.(35), genetic ablation of Dnmt1 and 3a, respectively, prevent the formation of tumors in Apc-mutant mice, indicating a requirement for de novo DNA methylation in the formation of intestinal polyps. Our data raise the possibility that elevated levels of SAH may contribute to cancer suppression by inhibiting DNMTs, preventing the aberrant DNA methylation associated with polyp formation. Furthermore, epigenetic modifications to DNA can have long-term impacts on gene expression.(36) Thus, it is possible that these alterations to SAH may directly contribute to the sustained cancer protection observed in the MDD-MDS repletion group.
Methyl donor restriction was also associated with reduced levels of the secondary bile acids deoxycholate, taurodeoxcycholate and muricholate. Secondary bile acids have been implicated in the initiation and progression of colon cancer.(37) Exposure of intestinal epithelial cells to secondary bile acids causes oxidative stress, DNA damage and apoptosis, with prolonged exposures leading to genomic instability and acquired apoptotic resistance.(38–40) Thus, reduced levels of secondary bile acids caused by our dietary intervention protocol may contribute in part to the observed tumor protection. Since there were no changes to the levels of the primary bile acids caused by the MDD diet, other mechanisms must be considered, including possible changes to the luminal microbiota. Secondary bile acids are predominantly synthesized by anaerobic bacteria that reside within the colon and the composition of the intestinal microbiome has been shown to play an important role in regulating the secondary bile acid pool.(41) Future studies investigating the interplay between a methyl donor restricted diet, changes to microbial community structure and the control of microbiome-derived secondary bile acids may elucidate novel mechanisms for cancer protection. One interesting possibility focuses on a recent study by Yoshimoto et al.(42), showing that enterohepatic circulation of secondary BAs promotes tumor formation in an obesity-associated dimethylbenz(α)anthracene hepatocellular carcinoma mouse model. While the pro-tumorigenic effect of secondary bile acids was dependent upon bacterial colonization, these secondary BAs also induced a senescence-associated secretory phenotype (SASP), characterized by an increased production of pro-inflammatory and pro-tumorigenic cytokines. We recently demonstrated that human colonic aberrant crypt foci (ACF)(43), the earliest CRC precursor lesion, exhibit SASP-like transcriptional changes. Thus, it may be reasonable to consider the potential downstream effects of methyl donor restriction on pro-inflammatory cytokines and senescence-associated gene expression within the colon, particularly at early stages of intestinal neoplasia. Unexpectedly, significant increases in α-muricholate and murideoxycholate were observed in the MDS-PF group, indicating that CR in the absence of methyl donor restriction increases intestinal bile acids. While short-term CR has been shown to elevate intestinal BAs (44), these data show that this elevation persists with longer-term CR. It is unclear what effect elevated BAs in the context of CR may have on intestinal tumorigenesis given that BAs are pro-tumorigenic while CR is thought to be tumor protective.(37) However, this may help explain why CR is associated with a smaller reduction in tumor burden than dietary methyl donor restriction in our previous studies.(9,10)
Metabolite profiling further revealed that MDD may inhibit FAO by reducing the availability of carnitine, thereby restricting transport of fatty acids (FAs) into the mitochondrial matrix.(45) As reviewed by Carracedo et al(19)., several recent studies have shown that cancer cells are dependent upon FAO to meet their increased energy demand and there is a growing consensus that targeting FAO may offer a viable therapeutic strategy for cancer treatment.(19) Etomoxir, an irreversible FAO inhibitor, increases sensitivity of human leukemia cells to apoptosis via the intrinsic death pathway and inhibits tumor growth in a prostate cancer xenograft model.(46,47) Notably, MDD was associated with reduced expression of Ppara and its targets Cpt1a and Cpt2, which regulate the rate of fatty acid transport into the mitochondria.(48) These data indicate that, in addition to limiting the availability of FAO substrates, dietary MDD impacts the transcription of FAO-regulatory genes. CPT1a, which is overexpressed in many human tumors, has been shown to promote apoptotic resistance via interaction with BCL-2.(49,50) The association of MDD with decreased BCL-2 expression and mitochondrial swelling, consistent with activation of the intrinsic apoptosis pathway, suggests that MDD may sensitize cells to apoptosis by disrupting CPT1-BCL2 interactions. This hypothesis is consistent with the observation that levels of FAO-associated metabolites are correlated with immunohistochemical markers of proliferation and stemness, and inversely correlated with markers of apoptosis, as well as with our previous observation that MDD is associated with significant increases in apoptotic cells within intestinal tumors.(9) Finally, significant reductions in acetyl CoA, and trends towards reduced expression of Cpt2 and Ppara were observed in the MDS-PF group, suggesting that CR impacts FAO. This observation is consistent with previous data indicating that severe short-term CR reduces availability of acetyl-CoA.(51) However, in contrast to MDD, MDS-PF was not associated with reductions in upstream FAO metabolites. Thus, methyl donor restriction and CR may impact acetyl CoA levels via distinct mechanisms, for example suppression of FAO and acetyl CoA synthesis versus increased demand for acetyl CoA under conditions of starvation. This observation may also help explain why dietary MDD has a greater impact on tumor burden than CR in this model. Overall, we believe that our findings provide additional evidence to support the FAO pathway as a potential target for cancer therapy, most likely to be applied in combination with existing drug protocols.
Finally, our analysis of metabolic changes associated with the MDD:MDS-repletion group defined a set of alterations that persisted in the tissue beyond active methyl donor restriction. Unexpectedly, these retained metabolic changes occurred predominantly within pathways related to oxidative stress, including hypotaurine synthesis, glutathione synthesis and ascorbate metabolism. Increased oxidative stress can stimulate a host of metabolic changes that activate proliferative, pro-inflammatory and angiogenic signaling pathways.(52) Given these strong associations, many studies have attempted to use dietary interventions as a means of controlling redox balance within the tissue.(53) However, as reviewed by Papaioannou et al.(53), clinical trials that have tested antioxidants such as vitamins A, C and E, selenium and β-carotene, either as single agents or in combination with other antioxidants, have proven ineffective for reducing CRC risk within the general population. In the present study, we demonstrate that a sustained increase in the levels of multiple endogenous antioxidants, such as ascorbate, hypotaurine, and indole-3-proprionate, within the colon may provide long-lasting activation of innate antioxidant pathways. Furthermore, the observed increase in these endogenous antioxidants was accompanied by a persistent reduction in the oxidative stress marker malondialdehyde within the colon. Taken together, these findings suggest that dietary interventions designed to activate innate antioxidant mechanisms and increase tissue levels of endogenous antioxidants may be a more effective approach than the administration of exogenous antioxidant compounds. Future studies designed to unravel the mechanism underlying MDD-associated increases in endogenous antioxidants, and to elucidate the contribution of individual methyl donor nutrients, may lead to the development of safer, targeted dietary chemoprevention strategies.
In conclusion, the present study extends our previous findings in ApcMin/+ and ApcΔ14/+ mice and identifies several key metabolic pathways that are altered under conditions of dietary methyl donor restriction. It is entirely possible that these metabolic alterations underlie the diet-related tumor suppression observed earlier. Specific targeting of these selected metabolic pathways may provide novel mechanisms for therapeutic and/or chemopreventive interventions, offering the potential to disrupt neoplastic growth without the associated adverse effects caused by dietary methyl donor deficiency. In addition, we believe these data afford additional mechanistic insights into the dramatic tumor suppression associated with MDD. For example, diet-induced changes to SAH biosynthesis and suppression of FAO may have clear clinical benefit when considering new therapeutic strategies. Our observation that methyl donor restriction reduces the tissue levels of several important secondary bile acids, changes that are likely mediated by altered microbiota, warrants further investigation. Overall, additional studies are needed to evaluate the efficacy of targeting these metabolic pathways, either individually or collectively, and to better define their relative contributions to the tumor-suppressive properties of methyl donor restriction.
Supplementary Material
Implications:
Metabolomic profiling reveals molecular changes underlying MDD-induced tumor protection and may provide new targets for CRC prevention and treatment.
Acknowledgments
This work is supported by NIH grant CA159976 and 1 R21 CA231255-01 to DWR.
List of Abbreviations:
- ACF
aberrant crypt foci
- BA
bile acid
- CRC
colorectal cancer
- FA
fatty acid
- FAO
fatty acid oxidation
- FOCM
folate one-carbon metabolism
- IPA
indole-3-propionate
- MDA
mean decrease in accuracy
- MDD
methyl donor deficient
- MDS
methyl donor sufficient
- PCA
principal component analysis
- PF
pair-fed
- RFA
random forest analysis
- SAH
S-adenosylhomocysteine
- SAM
S-adenosylmethionine
- SASP
senescence-associated secretory phenotype
Footnotes
The authors declare no potential conflicts of interest.
References
- 1.Mason JB. Folate, cancer risk, and the Greek god, Proteus: a tale of two chameleons. Nutr Rev 2009;67:206–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giovannucci E. Epidemiologic studies of folate and colorectal neoplasia: a review. J Nutr. 2002;132:2350S–2355S. [DOI] [PubMed] [Google Scholar]
- 3.Harnack L, Jacobs DR, Nicodemus K, Lazovich D, Anderson K, Folsom AR. Relationship of folate, vitamin B-6, vitamin B-12, and methionine intake to incidence of colorectal cancers. Nutr Cancer 2002;43:152–8. [DOI] [PubMed] [Google Scholar]
- 4.Vollset SE, Clarke R, Lewington S, Ebbing M, Halsey J, Lonn E, et al. Effects of folic acid supplementation on overall and site-specific cancer incidence during the randomised trials: meta-analyses of data on 50,000 individuals. Lancet Lond Engl 2013;381:1029–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qin T, Du M, Du H, Shu Y, Wang M, Zhu L. Folic acid supplements and colorectal cancer risk: meta-analysis of randomized controlled trials. Sci Rep 2015;5:12044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cole BF, Baron JA, Sandler RS, Haile RW, Ahnen DJ, Bresalier RS, et al. Folic acid for the prevention of colorectal adenomas: a randomized clinical trial. JAMA 2007;297:2351–9. [DOI] [PubMed] [Google Scholar]
- 7.Figueiredo JC, Mott LA, Giovannucci E, Wu K, Cole B, Grainge MJ, et al. Folic acid and prevention of colorectal adenomas: a combined analysis of randomized clinical trials. Int J Cancer 2011;129:192–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim Y-I. Folic acid fortification and supplementation--good for some but not so good for others. Nutr Rev 2007;65:504–11. [DOI] [PubMed] [Google Scholar]
- 9.Hanley MP, Kadaveru K, Perret C, Giardina C, Rosenberg DW. Dietary Methyl Donor Depletion Suppresses Intestinal Adenoma Development. Cancer Prev Res Phila Pa 2016;9:812–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kadaveru K, Protiva P, Greenspan EJ, Kim Y-I, Rosenberg DW. Dietary methyl donor depletion protects against intestinal tumorigenesis in Apc(Min/+) mice. Cancer Prev Res Phila Pa 2012;5:911–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mentch SJ, Locasale JW. One-carbon metabolism and epigenetics: understanding the specificity. Ann N Y Acad Sci 2016;1363:91–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Obeid R. The metabolic burden of methyl donor deficiency with focus on the betaine homocysteine methyltransferase pathway. Nutrients 2013;5:3481–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gonen N, Assaraf YG. Antifolates in cancer therapy: structure, activity and mechanisms of drug resistance. Drug Resist Updat Rev Comment Antimicrob Anticancer Chemother 2012;15:183–210. [DOI] [PubMed] [Google Scholar]
- 14.Montrose DC, Zhou XK, Kopelovich L, Yantiss RK, Karoly ED, Subbaramaiah K, et al. Metabolic Profiling, a Non-invasive Approach for the Detection of Experimental Colorectal Neoplasia. Cancer Prev Res Phila Pa 2012;5:1358–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Evans AM, DeHaven CD, Barrett T, Mitchell M, Milgram E. Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Anal Chem 2009;81:6656–67. [DOI] [PubMed] [Google Scholar]
- 16.Ohta T, Masutomi N, Tsutsui N, Sakairi T, Mitchell M, Milburn MV, et al. Untargeted metabolomic profiling as an evaluative tool of fenofibrate-induced toxicology in Fischer 344 male rats. Toxicol Pathol 2009;37:521–35. [DOI] [PubMed] [Google Scholar]
- 17.Xia J, Sinelnikov IV, Han B, Wishart DS. MetaboAnalyst 3.0--making metabolomics more meaningful. Nucleic Acids Res 2015;43:W251–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McBean GJ. The transsulfuration pathway: a source of cysteine for glutathione in astrocytes. Amino Acids 2012;42:199–205. [DOI] [PubMed] [Google Scholar]
- 19.Carracedo A, Cantley LC, Pandolfi PP. Cancer metabolism: fatty acid oxidation in the limelight. Nat Rev Cancer 2013;13:227–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vaz FM, Wanders RJA. Carnitine biosynthesis in mammals. Biochem J 2002;361:417–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paumen MB, Ishida Y, Han H, Muramatsu M, Eguchi Y, Tsujimoto Y, et al. Direct interaction of the mitochondrial membrane protein carnitine palmitoyltransferase I with Bcl-2. Biochem Biophys Res Commun 1997;231:523–5. [DOI] [PubMed] [Google Scholar]
- 22.Beyer RE. The role of ascorbate in antioxidant protection of biomembranes: interaction with vitamin E and coenzyme Q. J Bioenerg Biomembr 1994;26:349–58. [DOI] [PubMed] [Google Scholar]
- 23.Poeggeler B, Pappolla MA, Hardeland R, Rassoulpour A, Hodgkins PS, Guidetti P, et al. Indole-3-propionate: a potent hydroxyl radical scavenger in rat brain. Brain Res 1999;815:382–8. [DOI] [PubMed] [Google Scholar]
- 24.Bolati D, Shimizu H, Yisireyili M, Nishijima F, Niwa T. Indoxyl sulfate, a uremic toxin, downregulates renal expression of Nrf2 through activation of NF-κB. BMC Nephrol 2013;14:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gaweł S, Wardas M, Niedworok E, Wardas P. [Malondialdehyde (MDA) as a lipid peroxidation marker]. Wiad Lek Wars Pol 1960 2004;57:453–5. [PubMed] [Google Scholar]
- 26.Cho E, Willett WC, Colditz GA, Fuchs CS, Wu K, Chan AT, et al. Dietary choline and betaine and the risk of distal colorectal adenoma in women. J Natl Cancer Inst 2007;99:1224–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee JE, Giovannucci E, Fuchs CS, Willett WC, Zeisel SH, Cho E. Choline and betaine intake and the risk of colorectal cancer in men. Cancer Epidemiol Biomark Prev Publ Am Assoc Cancer Res Cosponsored Am Soc Prev Oncol 2010;19:884–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu M-S, Fang Y-J, Pan Z-Z, Zhong X, Zheng M-C, Chen Y-M, et al. Choline and betaine intake and colorectal cancer risk in Chinese population: a case-control study. PloS One 2015;10:e0118661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cellarier E, Durando X, Vasson MP, Farges MC, Demiden A, Maurizis JC, et al. Methionine dependency and cancer treatment. Cancer Treat Rev 2003;29:489–99. [DOI] [PubMed] [Google Scholar]
- 30.Komninou D, Leutzinger Y, Reddy BS, Richie JP. Methionine restriction inhibits colon carcinogenesis. Nutr Cancer 2006;54:202–8. [DOI] [PubMed] [Google Scholar]
- 31.Kusakabe Y, Ishihara M, Umeda T, Kuroda D, Nakanishi M, Kitade Y, et al. Structural insights into the reaction mechanism of S-adenosyl-L-homocysteine hydrolase. Sci Rep 2015;5:16641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oliva A, Galletti P, Zappia V, Paik WK, Kim S. Studies on substrate specificity of S-adenosylmethionine: protein-carboxyl methyltransferase from calf brain. Eur J Biochem FEBS 1980;104:595–602. [DOI] [PubMed] [Google Scholar]
- 33.Caudill MA, Wang JC, Melnyk S, Pogribny IP, Jernigan S, Collins MD, et al. Intracellular S-adenosylhomocysteine concentrations predict global DNA hypomethylation in tissues of methyl-deficient cystathionine beta-synthase heterozygous mice. J Nutr 2001;131:2811–8. [DOI] [PubMed] [Google Scholar]
- 34.Eads CA, Nickel AE, Laird PW. Complete genetic suppression of polyp formation and reduction of CpG-island hypermethylation in Apc(Min/+) Dnmt1-hypomorphic Mice. Cancer Res 2002;62:1296–9. [PubMed] [Google Scholar]
- 35.Weis B, Schmidt J, Maamar H, Raj A, Lin H, Tóth C, et al. Inhibition of intestinal tumor formation by deletion of the DNA methyltransferase 3a. Oncogene 2015;34:1822–30. [DOI] [PubMed] [Google Scholar]
- 36.Environmental Feil R. and nutritional effects on the epigenetic regulation of genes. Mutat Res 2006;600:46–57. [DOI] [PubMed] [Google Scholar]
- 37.Ajouz H, Mukherji D, Shamseddine A. Secondary bile acids: an underrecognized cause of colon cancer. World J Surg Oncol 2014;12:164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Payne CM, Bernstein C, Dvorak K, Bernstein H. Hydrophobic bile acids, genomic instability, Darwinian selection, and colon carcinogenesis. Clin Exp Gastroenterol 2008;1:19–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martinez JD, Stratagoules ED, LaRue JM, Powell AA, Gause PR, Craven MT, et al. Different bile acids exhibit distinct biological effects: the tumor promoter deoxycholic acid induces apoptosis and the chemopreventive agent ursodeoxycholic acid inhibits cell proliferation. Nutr Cancer 1998;31:111–8. [DOI] [PubMed] [Google Scholar]
- 40.Yui S, Saeki T, Kanamoto R, Iwami K. Characteristics of apoptosis in HCT116 colon cancer cells induced by deoxycholic acid. J Biochem (Tokyo) 2005;138:151–7. [DOI] [PubMed] [Google Scholar]
- 41.Ridlon JM, Kang DJ, Hylemon PB, Bajaj JS. Bile acids and the gut microbiome. Curr Opin Gastroenterol 2014;30:332–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoshimoto S, Loo TM, Atarashi K, Kanda H, Sato S, Oyadomari S, et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature 2013;499:97–101. [DOI] [PubMed] [Google Scholar]
- 43.Mo A, Jackson S, Varma K, Carpino A, Giardina C, Devers TJ, et al. Distinct Transcriptional Changes and Epithelial-Stromal Interactions Are Altered in Early-Stage Colon Cancer Development. Mol Cancer Res MCR 2016;14:795–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fu ZD, Klaassen CD. Increased bile acids in enterohepatic circulation by short-term calorie restriction in male mice. Toxicol Appl Pharmacol 2013;273:680–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Flanagan JL, Simmons PA, Vehige J, Willcox MD, Garrett Q. Role of carnitine in disease. Nutr Metab 2010;7:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Samudio I, Harmancey R, Fiegl M, Kantarjian H, Konopleva M, Korchin B, et al. Pharmacologic inhibition of fatty acid oxidation sensitizes human leukemia cells to apoptosis induction. J Clin Invest 2010;120:142–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schlaepfer IR, Rider L, Rodrigues LU, Gijón MA, Pac CT, Romero L, et al. Lipid catabolism via CPT1 as a therapeutic target for prostate cancer. Mol Cancer Ther 2014;13:2361–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Song S, Attia RR, Connaughton S, Niesen MI, Ness GC, Elam MB, et al. Peroxisome proliferator activated receptor alpha (PPARalpha) and PPAR gamma coactivator (PGC-1alpha) induce carnitine palmitoyltransferase IA (CPT-1A) via independent gene elements. Mol Cell Endocrinol 2010;325:54–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Y-N, Zeng Z-L, Lu J, Wang Y, Liu Z-X, He M-M, et al. CPT1A-mediated fatty acid oxidation promotes colorectal cancer cell metastasis by inhibiting anoikis. Oncogene 2018; [DOI] [PubMed] [Google Scholar]
- 50.Yao C-H, Liu G-Y, Wang R, Moon SH, Gross RW, Patti GJ. Identifying off-target effects of etomoxir reveals that carnitine palmitoyltransferase I is essential for cancer cell proliferation independent of β-oxidation. PLoS Biol 2018;16:e2003782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mariño G, Pietrocola F, Eisenberg T, Kong Y, Malik SA, Andryushkova A, et al. Regulation of autophagy by cytosolic acetyl-coenzyme A. Mol Cell 2014;53:710–25. [DOI] [PubMed] [Google Scholar]
- 52.Bhattacharyya A, Chattopadhyay R, Mitra S, Crowe SE. Oxidative stress: an essential factor in the pathogenesis of gastrointestinal mucosal diseases. Physiol Rev 2014;94:329–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Papaioannou D, Cooper KL, Carroll C, Hind D, Squires H, Tappenden P, et al. Antioxidants in the chemoprevention of colorectal cancer and colorectal adenomas in the general population: a systematic review and meta-analysis. Colorectal Dis Off J Assoc Coloproctology G B Irel 2011;13:1085–99. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.