Abstract
Versatile and precise genome modifications are needed for a wider range of adoptive cellular therapies1–5. Here we report two improvements that increase the efficiency of CRISPR-Cas9-based genome editing in clinically relevant primary cell types. Truncated Cas9 target sequences (tCTS) added at the ends of the homology-directed repair (HDR) template interact with Cas9 ribonucleoproteins (RNPs) to shuttle the template to the nucleus, enhancing HDR efficiency ~2–4 fold. Furthermore, stabilizing Cas9 RNPs into nanoparticles with poly(glutamic acid) improves editing efficiency an additional ~2-fold, reduces toxicity, and enables lyophilized storage without loss of activity. Combining the two improvements increases gene targeting efficiency even at reduced HDR template doses yielding ~2–6 times as many viable edited cells across multiple genomic loci in diverse cell types, such as bulk T cells, CD8+ T cells, CD4+ T cells, regulatory T cells (Tregs), γ-T cells, B cells, NK cells, and primary and iPS-derived6 hematopoietic stem progenitor cells (HSPCs).
We recently reported an approach to reprogram human T cells with CRISPR-based genome targeting without the need for viral vectors5, where we found that varying the relative concentrations of both Cas9 RNP and HDR template had significant effects on targeting efficiency and toxicity. However, many research and clinical applications still depend upon improving efficiency, cell viability, and generalizability of non-viral genome targeting across cell types1–4. Here, we set out to optimize the interactions between the HDR template and nanoparticle-stabilized RNPs to further improvement genome editing efficiency independent of cell type.
We devised a novel approach to promote nuclear entry of the template. Unlike previous efforts utilizing complex covalent linkages7, we attempted to recruit Cas9 RNPs with nuclear location sequences (NLS) to the HDR template by enhancing Watson-Crick interactions. CRISPR-Cas9 interacts specifically with both genomic and non-genomic dsDNA8, and nuclease-inactive dCas9 has been used in many applications to localize protein and RNA effectors to specific DNA sequences without cleaving the target sequence9. We therefore tested if we could enhance HDR by targeting a dCas9-NLS ‘shuttle’ to the ends of an HDR template by coding 20 bp Cas9 Target Sequences (CTS) at the ends of the homology arms (Supplementary Fig. 1). Indeed, CTS-modified HDR templates mixed with dCas9-NLS RNP did show mild improvements in HDR efficiency in primary human T cells (Supplementary Fig. 1) but required two distinct RNPs (with dCas9 and catalytically-active Cas9). These data encouraged us to search for a simplified method utilizing the same RNP to both cut a specified genomic site and recruit Cas9-NLS to HDR templates.
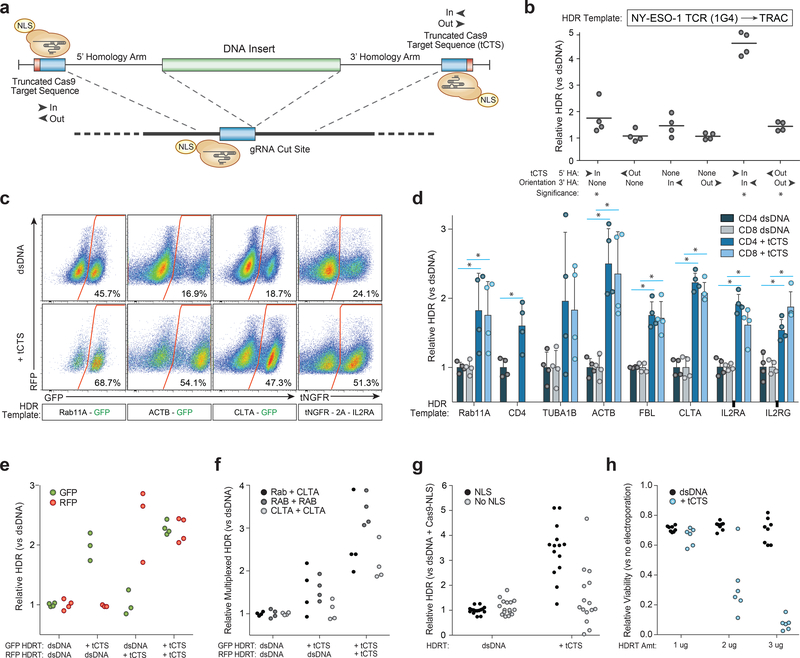
We hypothesized that a single catalytically-active Cas9-NLS RNP would suffice for both on-target genomic cutting and ‘shuttling’ if the HDR template was designed with truncated (16bp) Cas9 Target Sequences (tCTS) that enable Cas9 binding but do not enable cutting10 (Fig. 1a and Supplementary Fig. 2). With the proper sequence orientation, the addition of tCTS markedly improved knockin efficiency of a 1.5kb DNA sequence inserting a reprogrammed TCRα and TCRβ specificity at the endogenous TRAC (T-cell receptor α constant) locus (Fig. 1b and Supplementary Fig. 2). This tCTS shuttle system also improved genome targeting efficiencies across a variety of loci in different primary human T cell subsets (Fig. 1c,d and Supplementary Fig. 3). HDR templates with the tCTS achieved preferential targeting even in direct competition with unmodified dsDNA HDR templates simultaneously delivered to the same cells (Fig. 1e and Supplementary Fig. 4). Additionally, the tCTS shuttle improved efficiencies of bi-allelic and multiplexed targeting across different loci (Fig. 1f and Supplementary Fig. 4). The full HDR efficiency gains critically depended on the presence of an NLS in the Cas9 RNP, use of an on-target gRNA, and pre-incubation of the Cas9-NLS RNP with the tCTS-modified HDR template (Fig. 1g and Supplementary Fig. 5). Taken together these data demonstrate that coupling the HDR template with tCTS to a Cas9-NLS RNP can enhance genome targeting efficiency without requiring modification of the protein or gRNA.
Figure 1. Truncated Cas9 Targets Sequences (tCTS) in HDR templates increase large non-viral knockin efficiency.
(a) Enzymatically active Cas9-NLS RNPs can bind truncated Cas9 target sequences (tCTS) added to the ends of an HDR template.
(b) “In” facing orientation of the tCTSs (PAM facing in towards the center of the inserted sequence vs “out” away from the insert) on the edges of both the 5’ and 3’ homology arms improved knockin efficiency of a new TCRα-TCRβ specificity at the endogenous TRAC locus. *P<0.05 (Mann-Whitney test vs no tCTS control).
(c) Representative flow cytometry plots showed improved targeting efficiency across target genomic loci with the tCTS modifications compared to an unmodified dsDNA HDR template.
(d) The tCTS modifications improved targeting efficiencies of large knockins across eight genomic loci tested in both CD4+ and CD8+ T cells. Note CD4-GFP expression was not observed at relevant levels in CD8+ T cells, as expected. *P<0.05 (Two-way paired T test with Holm-Sidak multiple comparison correction).
(e) Multiplexed electroporation of GFP and RFP knockin templates to the RAB11A locus where neither, one, or both templates had a tCTS modification revealed direct competitive knockin advantage of ‘shuttle’ system compared to unmodified dsDNA template (technical replicates from n=2 donors).
(f) The tCTS modification improved multiplexed dual knockin at different genomic loci as well as bi-allelic knockin at a single target locus (technical replicates from n=2 donors).
(g) Full improvement of knockin efficiencies with the tCTS modifications (but not with unmodified dsDNA HDR templates) were dependent upon the presence of a NLS on Cas9 protein (multiple technical replicates from n=2 donors).
(h) Decreased viability was seen with the tCTS modifications at lower DNA concentrations compared to unmodified dsDNA HDR template.
The relative rates of HDR (b, d, e, g), multiplexed HDR (f), or viability (h) with the tCTS shuttle are displayed normalized to unmodified dsDNA HDR template (b, d-g) or to no electroporation controls (h) in n=4 biologically independent blood donors (For some templates different healthy donors were used) (b, c, d) or from n=2 biologically independent blood donors with multiple technical replicates shown to convey variance (e-h). Center lines indicate mean (b, d), error bars indicate standard deviation (d). HDR efficiency was measured 4 days post electroporation and viability (total number of live cells relative to no electroporation control) at 2 days post electroporation.
Exogenous DNA (including HDR templates) can be cytotoxic at high concentrations4,5,11. We therefore assayed the effects of the RNP-HDR template interactions on cell viability. Gene targeting using tCTS-modified HDR templates improved efficiency, but we observed decreased cell viability with tCTS-modified HDR template at doses lower than unmodified dsDNA HDR templates (Fig. 1h). Decreased viability was only observed with an on-target gRNA and pre-incubation of the RNP with tCTS-modified HDR template, but did not entirely depend on the presence of the NLS on Cas9 (Supplementary Fig. 5c) consistent with possible enhanced cytoplasmic delivery of DNA during electroporation due to the RNP-HDR template interaction.
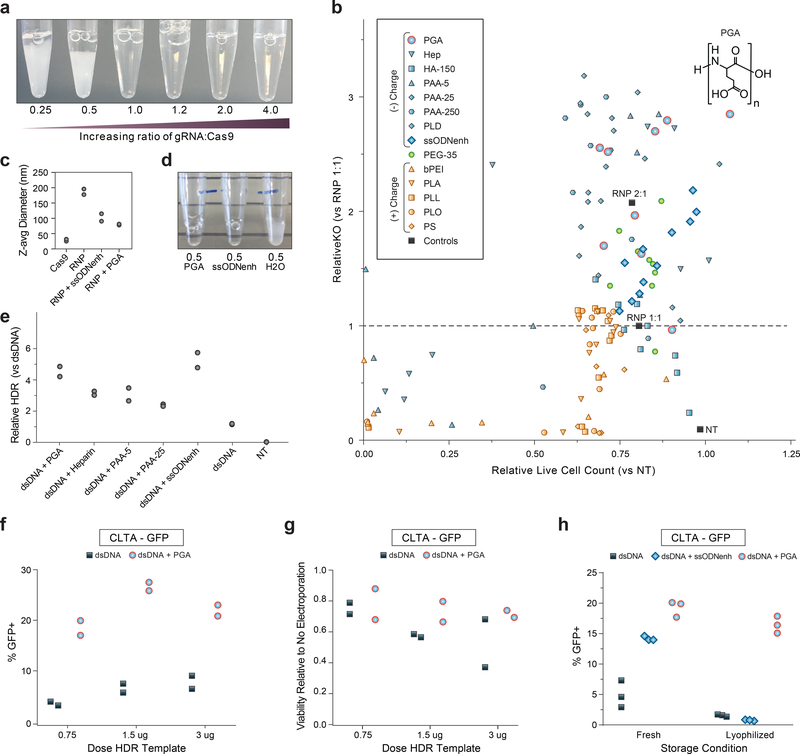
In previous experiments, we observed that Cas9 RNP co-delivery could mitigate exogenous DNA toxicity (from unmodified plasmids or dsDNA template) in human T cells5. We therefore wondered whether optimizing RNP delivery could improve effects on cell viability. We noted that the Cas9 protein itself appears to be only quasi-stable when complexed with guide RNA; a molar excess of protein (RNA to Cas9 protein molar ratio of <1.0) results in a milky opaque solution with rapid sedimentation (Fig. 2a) of poorly functional material (Supplementary Fig. 6a). Previous reports have suggested that RNP electroporation can be improved with addition of extra sgRNA or a non-homologous single strand DNA enhancer (ssODNenh)12. Excess guide RNA or addition of ssODNenh dispersed the Cas9 RNPs (Supplementary Fig. 6b) and boosted editing efficiency of electroporated RNPs (Supplementary Fig. 6c). However, combination of both guide RNA and ssODNenh in excess did not further improve editing (Supplementary Fig. 6c), suggesting a possible shared mechanism of action.
Figure 2: Stabilizing Cas9 RNP nanoparticles with anionic polymers improves editing outcomes.
(a) Photograph 15 minutes after mixing gRNA and Cas9 protein incubated at 37C to form RNPs. Cas9 RNPs prepared at low molar ratio of gRNA:protein appeared cloudy and rapidly settled out of solution. Representative photograph of 3 repeated independent experiments.
(b) Multiple polymers were screened for the ability to enhance CD4 gene knock-out editing when mixed with RNPs formulated at 1:1 gRNA:protein ratio then electroporated into primary human CD4+ T cells. Loss of surface CD4 expression at 3 days assessed by flow cytometry is normalized to unenhanced editing efficiency (RNP 1:1 without any additive) on the y-axis, and the live cell count is normalized to mock non-electroporated (NT) cells on the x-axis. Negatively charged polymers are shaded blue: poly(L-glutamic acid) (PGA), heparin sulfate (Hep), hyaluronic acid 150 kDa (HA-150), poly(acrylic acid) at 5 kDa (PAA-5) 25 kDa (PAA-25) or 250 kDa (PAA-250), poly(L-aspartic acid) (PLD), ssODNenh. Neutral polymers are shaded green: poly(ethylene glycol) 35 kDa (PEG-35), and positively charged polymers are shaded orange: polyethyleneimine 25 kDa (PEI), poly(L-arginine) 15–70 kDa (PLA), poly(L-lysine) 15–30 kDa (PLL), poly(L-ornithine) 30–70 kDa (PLO), and protamine sulfate (PS). PGA (blue circles outlined in red) with chemical structure shown inset above data point that corresponds to 100mg/mL concentration. Each polymer sample was tested at serial dilutions to avoid potential dose-dependent cytotoxicity falsely masking impact on editing efficiency, and each concentration is depicted as an individual point that is an average for two different blood donors.
(c) Cas9 RNPs at 0.5 molar ratio of gRNA:protein (prepared same as in (a)) could be further dispersed with addition of PGA or ssODNenh, whereas dilution with water alone had no visible benefit. Representative photograph of three repeated independent experiments.
(d) PGA and ssODNenh stabilized and reduced the size of RNP nanoparticles. Cas9 RNPs prepared at 2:1 molar ratio of gRNA:protein alone (RNP) or mixed with PGA or ssODNenh were assessed for hydrodynamic particle size by dynamic light scattering. Z-average particle size is shown for n=2 independent preparations (individual sample size distributions and peaks shown in Supplementary Fig. 8).
(e) Multiple anionic polymers boosted knockin editing efficiency. Polymers mixed with Cas9 RNPs prepared at a 2:1 gRNA:protein ratio were further mixed with 1ug unmodified dsDNA HDR templates (targeting insertion of an N-terminal fusion of GFP to Rab11a), electroporated into CD4+ T cells, and editing efficiency were assessed by flow cytometry at day 3. The relative rates of HDR is displayed compared to unmodified dsDNA HDR template without enhancer. Data shown for each of n=2 biologically independent blood donors.
(f) PGA-stabilized Cas9 RNPs prepared at a 2:1 ratio gRNA:protein markedly improved knockin editing in primary human Bulk (CD3+) T cells targeting a C-terminal fusion of GFP to clathrin and (g) improve viability of electroporated cells (compared to untreated cells). Data shown for each of n=2 biologically independent blood donors.
(h) Cas9 RNPs prepared at a 2:1 ratio gRNA:protein without or with PGA or ssODNenh were mixed with 1ug of unmodified dsDNA HDR template targeting an N-terminal fusion of GFP to RAB11A, lyophilized overnight, stored dry at −80C, then later reconstituted in water prior to electroporating into primary human CD3+ (Pan) T cells. PGA-stabilized Cas9 nanoparticles were protected through lyophilization and reconstitution and retained activity for robust knockin editing. Three technical replicates shown for n=1 blood donor, representative of two repeated independent experiments.
We hypothesized that the polymeric and anionic nature of nucleic acids shields excess positively-charged residues of the Cas9 protein from nearby exposed portions of Cas9-bound gRNA, thus preventing aggregation and improving RNP particle stability. We therefore screened various commercially-available water-soluble biological and synthetic polymeric materials for the ability to also enhance electroporation-mediated Cas9 knock-out editing. Multiple different anionic polymers such as poly(glutamic acid) (PGA), poly(aspartic acid), heparin, and poly(acrylic acid) all enhanced editing efficiency in a dose-dependent manner without addition of ssODNenh or excess gRNA (Fig. 2b). The charge-neutral material poly(ethyleneglycol) (PEG) had only minimal impact on RNP activity; positively charged polymers poly(ethylenimine), protamine sulfate, poly-L-lysine, poly-L-ornithine, and poly-L-arginine all reduced editing efficiency (and viability) (Fig. 2b), thus establishing anionic charge as a key factor for RNP enhancement. Further, enhancement of RNP-based editing depended upon polymer chain length (Supplementary Fig. 7), which is similar to the reported length-dependence of ssODNenh12 and consistent with colloid-stabilizing biomaterials13. Comparison of particle sizes by dynamic light scattering (DLS) revealed that Cas9 protein by itself is 10–15nm in diameter as expected for dispersed individual molecules, but aggregates of ~200+ nm size were formed when gRNA was added (Fig. 2c and Supplemental Fig. 8). However, the addition of either PGA or ssODNenh prevented aggregation into micron-sized particles (Fig. 2d) and improved the size distribution of RNP nanoparticles to less than 100nm on average, with peaks in the 20nm and 100–120nm ranges (Fig. 2c and Supplementary Fig. 8).
We next tested if anionic polymers could also enhance non-viral knockin genome targeting. PGA was effective at enhancing knockin editing in primary T cells (Fig. 2e) when mixed with Cas9 RNPs and a reduced concentration of an unmodified dsDNA template targeting integration of a Rab11a-GFP fusion. PGA-stabilized RNP nanoparticles promoted efficiency gains in primary human T cells (Fig. 2f) and appeared to reduce the toxicity of higher doses of HDR template (Fig. 2g). These efficiency gains were independent of the order of PGA addition, guide RNA source, or Cas9 nuclease manufacturer (Supplemental Figs. 9–11). Combining ssODNenh or higher concentrations of guide RNA with PGA did not further enhance knock-in efficiency, consistent with shared mechanisms due to the polymeric anionic charge of these molecules (Supplemental Fig. 12). This does not preclude contributions of additional material-specific properties, such as the stability of the PGA polymer during lyophilization. RNP stabilization with PGA (but not ssODNenh) permitted freeze-thaw cycles or lyophilization with retained knockout and knockin editing activity (Fig. 2h and Supplementary Fig. 13). PGA-stabilized RNP nanoparticles therefore enhance edited cell viability and could be stored dry enabling significant streamlining of gene-modified cell manufacturing for research or clinical translation.
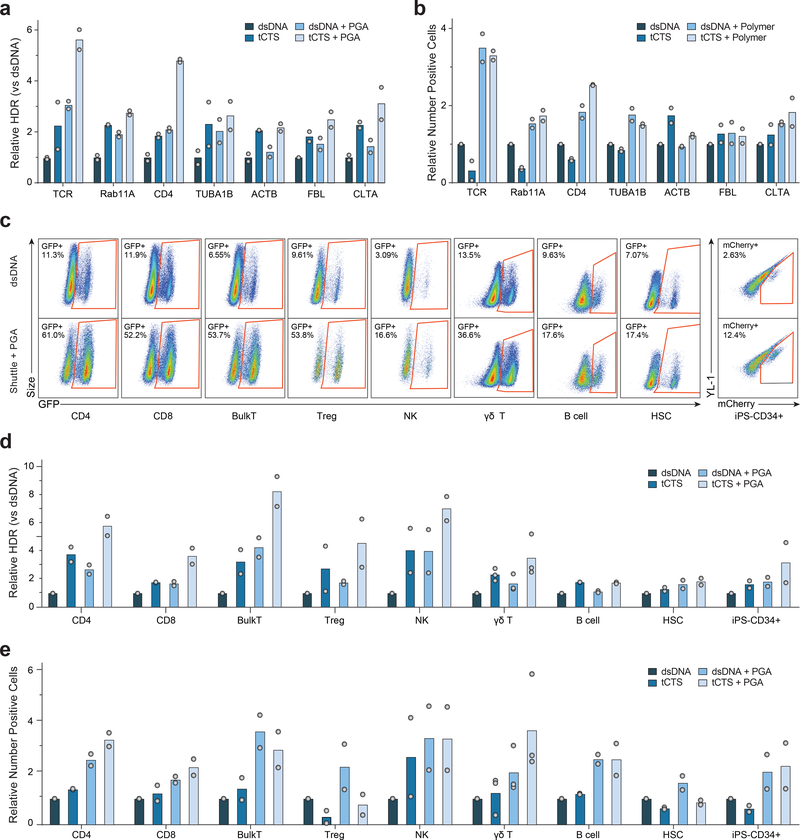
We further tested to ensure that combining the tCTS ‘shuttle’ and anionic polymers improved both the efficiency and resulting cell viability of non-viral genome targeting. Improved efficiency and cell recovery were consistent across human blood donors when replacing the endogenous TCR with a therapeutically-relevant TCR that recognizes the NY-ESO 1 tumor antigen (Supplementary 14). While increasing HDR template dose with or without the tCTS-modification could improve the fraction of knockin edited cells (Supplementary Fig. 14 a,c), the dose-dependent toxicity observed with the tCTS-modified HDR template (Fig. 1h) was mitigated by PGA-stabilized RNPs (Supplementary Fig. 14 b,d). The maximal knockin cell yield was achieved with a combination PGA-stabilized RNPs and a reduced dose of tCTS-modified HDR templates (Supplementary Fig. 14). Importantly, the tCTS-modified HDR templates with PGA-stabilized RNPs markedly enhanced both knockin efficiency and viability (Fig. 3a) and improved the recovery of viable T cells edited at a variety of endogenous genomic loci (Fig. 3b).
Figure 3: PGA-stabilized Cas9 RNP and tCTS-modified-HDR templates improved knockin gene editing outcomes across a variety of genetic loci and clinically-relevant immune cell types.
(a-b) Cas9 RNPs were prepared at a 2:1 ratio gRNA:protein with or without PGA polymer and mixed with high doses (2–4 ug) of regular dsDNA or tCTS-modified HDR template targeting knockin at multiple genomic loci: transgenic NY-ESO 1 tumor antigen TCR into the TRAC locus, or GFP fusion at the N- or C- of RAB11A, CD4, TUBA1B, ACTB, FBL, or CLTA genes. The combination of PGA-stabilized Cas9 RNP nanoparticles and ‘shuttle’ tCTS-modified-HDR template both improved relative frequency of HDR (a) and resulted in higher yield of successfully edited cells (b).
(c-e) Cas9 RNPs were prepared at 2:1 ratio gRNA:protein with or without PGA polymer and mixed with low doses (0.5 – 1 ug) of unmodified dsDNA or ‘shuttle’ tCTS-modified HDR templates targeting knockin of GFP or mCherry to the N-terminus of Clathrin. The PGA-stabilized Cas9 RNP nanoparticles and tCTS-modified HDR templates improved editing efficiency in a variety of primary human immune cell types as visualized in representative flow cytometry plots (after gating for live cells and respective cell type-specific surface markers) (c) or expressed as relative frequency of GFP or mCherry+ positive cells (d), and resulted in higher yield of number of successfully edited cells (e).
The relative rates of HDR (a,d) or edited cell recovery (b,e) are displayed normalized to unmodified dsDNA HDR template without enhancer for each given gene locus (a-b) or cell type (d-e). Data in shown for each of n=2 (for CD4, CD8, BulkT, Treg, γδ-T, B cell, HSC, iPS-CD34) or n=3 (for NK) biologically independent blood donors. Data in (c) representative of two repeated independent experiments.
We also assessed potential off-target genome editing events given concern that improved delivery of RNPs via stabilization into nanoparticles could increase both on- and off-target double-strand breaks. With PGA we observed only slightly increased off-target indel formation relative to RNP alone at previously identified off-target sites for the well-characterized guide RNA targeting EMX114,15 (Supplementary Fig 15a). We also investigated if increased HDR template delivery via tCTS modifications would increase off-target transgene integration. As we have previously described, linear dsDNA templates can integrate and express transgenes through non-homology directed mechanisms modeled by introducing a targeted cut at an off-target site5. Using this functional test for off-target integrations, we found that the addition of tCTS sequences for off-target guides or scrambled guides did not increase off-target expression of a GFP transgene compared to standard dsDNA HDR templates (Supplementary Fig. 15b–c). Although further work will be needed to assess the global off-target edits and integrations for specific RNPs and HDR templates, these results suggest that PGA and modified templates can boost knockin targeting markedly while minimizing effects on undesirable outcomes.
Encouraged by these results in T cells, we applied the combined system to enable non-viral genome targeting in a wider set of therapeutically relevant primary human hematopoietic cells. Using a template encoding a GFP fusion to the vesicle-coating protein Clathrin (encoded by the CLTA gene), which should be broadly expressed across cell types, we consistently observed improved editing efficiencies with the combined system across many cell types (Fig. 3c–e). Bulk CD3+ T cells, purified CD4+ T cells or CD8+ T cells, and purified CD127low CD25+ CD4+ regulatory T cells (Tregs) all achieved a similarly high knockin efficiency of up to > 50% of cells, with 3–8x increase in the percentage of knockin edited cells at reduced HDR template concentrations (Fig. 3c,d). While standard RNP and unmodified HDR template achieved only minimal knockin in isolated primary human NK cells or B cells, the combined system resulted in over 15% transgene-positive cells and a 2–5-fold increase in edited cell yield (Fig. 3c–e). In γδ-T cells the combined system exhibited improved editing efficiency from ~5% to ~28%, and 5–6 fold improved edited cell recovery compared to a standard RNP and unmodified HDR template. Finally, we were able to express large transgene insertions in over 15% of human CD34+ HSPCs without a viral vector, in both primary mobilized peripheral blood- and iPS-derived6 CD34+ HSPCs, with ~2–3x increased yield of knockin edited cells. The combined non-viral system was thus demonstrated to be effective in diverse human hematopoietic cell types.
Together, PGA as a RNP nanoparticle-stabilizing enhancer and the tCTS-modified HDR template enabled high percentage editing with improved edited cell yields in a variety of primary hematopoietic cell types opening the door to next-generation adoptive cell therapies beyond T cells. The combined nanoparticle-tCTS template system is a novel platform to explore gene function in clinically-relevant cell types that had been previously challenging to genome modify. The formation of PGA-stabilized RNP nanoparticles and the utilization of PCR primers to introduce tCTS modifications to HDR templates are both methods that can be rapidly adapted to any existing Cas9 RNP-based editing protocol. Notably, marked improvements in large gene targeting to endogenous loci were achieved without further optimizing cell cycle dynamics, small molecule modulation of DNA repair machinery, or specialized chemistries; these complementary strategies may eventually offer additional efficiency gains. Some variation in targeting success remains depending on locus, knockin sequence, electroporation parameters, and cell type. Further optimization of polymers or tCTS ‘shuttle’ configurations could offer even further improvements. Our data demonstrate a technically simple system that greatly enhances the capabilities of Cas9 RNP-mediated non-viral genome targeting in primary human hematopoietic cells and has direct translational potential for research, biotechnology, and clinical endeavors.
METHODS
Cell Culture
Primary adult cells were obtained from healthy human donors from leukoreduction chamber residuals after Trima Apheresis (Vitalant, formerly Blood Centers of the Pacific) or from freshly drawn whole blood under a protocol approved by the UCSF IRB (BU101283). Peripheral blood mononuclear cells (PBMCs) were isolated by Ficoll-Paque (GE Healthcare) centrifugation using SepMate tubes (STEMCELL, per manufacturer’s instructions). Specific lymphocytes were then further isolated by magnetic negative selection using an EasySep Human B Cell, CD4+ T Cell, Bulk (CD3+) T Cell, CD8+ T cell, CD4+ CD127low CD25+ Regulatory T Cell, Gamma/Delta T Cell, or NK Cell Isolation kit (STEMCELL, per manufacturer’s instructions).
Isolated CD4+, CD8+, CD3+ (Bulk T Cells), Regulatory (CD25hiCD127low), or γδ-T cells were activated and cultured for 2 days at 0.5 to 1.0 million cells/mL in XVivo15 medium (Lonza) with 5% Fetal Bovine Serum, 50 μM 2-mercaptoethanol, 10 mM N-Acetyl L-Cystine, anti-human CD3/CD28 magnetic Dynabeads (ThermoFisher) at a beads to cells ratio of 1:1, and a cytokine cocktail of IL-2 at 300 U/mL (UCSF Pharmacy), IL-7 at 5 ng/mL (R&D Systems), and IL-15 at 5 ng/mL (R&D Systems). Activated T cells were harvested from their culture vessels and Dynabeads were removed by placing cells on an EasySep cell separation magnet (STEMCELL) for 5 minutes. Isolated B cells were cultured in IMDM (ThermoFischer) with 5% Fetal Bovine Serum, 100 ng/mL MEGACD40L (Enzo), 1000 ng/mL CpG (InvivoGen), 500 U/mL IL-2 (UCSF Pharmacy), 50 ng/mL IL-10 (ThermoFischer), and 10 ng/mL IL-15 (R&D Systems). Freshly isolated NK cells were cultured in Xvivo15 medium (Lonza) with 5% Fetal Bovine Serum, 50 μM 2-mercaptoethanol, 10 mM N-Acetyl L-Cystine, together with IL-2 (at 1000 U/mL) and MACSiBead Particles pre-loaded with anti-human CD335 (NKp46) and anti-human CD2 antibodies (Miltenyi Biotech). Primary adult peripheral blood G-CSF-mobilized CD34+ hematopoietic stem cells were purchased from StemExpress Inc and cultured at 0.5e6 cells/mL in SFEM II media supplemented with CC110 cytokine cocktail (STEMCELL) for two days prior to electroporation. Induced pluripotent stem-cells were generated and differentiated into CD34+ HSPCs as previously described6 then cultured in SFEM media (STEMCELL) supplemented with IL-2 at 10ng/mL, IL-6 at 50ng/mL, SCF at 50ng/mL, FLT-3L at 50ng/mL, and TPO at 50ng/mL (Peprotech).
RNP Formulation with Polymers
Cas9 RNPs were formulated immediately prior to electroporation, except when frozen or lyophilized as indicated. Synthetic crRNA (with guide sequence listed in Supplementary Table 1) and tracrRNA were chemically synthesized (Edit-R, Dharmacon Horizon or Integrated DNA Technologies (IDT)), resuspended in 10 mM Tris-HCL (7.4 pH) with 150 mM KCl or IDT duplex buffer at a concentration of 160 μM, and stored in aliquots at −80C. To make gRNA, aliquots of crRNA and tracrRNA were thawed, mixed 1:1 by volume, and annealed by incubation at 37C for 30 min to form an 80 μM gRNA solution. For comparison, chemically modified gRNA was purchased from Synthego and resuspended according to manufacturer’s protocol. Cas9-NLS, dCas9-NLS, or Cas9 without NLS was purchased from the UC Berkeley QB3 MAcrolab; HiFiCas9 was purchased from IDT. To make RNPs, gRNA was then further diluted in buffer first, or directly mixed 1:1 by volume with 40 μM Cas9-NLS protein to achieve the desired molar ratio of gRNA:Cas9 (2:1 ratio unless otherwise stated). Unless otherwise stated, final dose of RNP per nucleofection was 50pmol on a Cas9 protein basis.
For initial screening, polymers were purchased dry (see Supplementary Table 2) and resuspended to 100mg/mL in water except as noted, passed through a 0.2 um syringe filter, and stored at −80C prior to use. The ssODNenh electroporation enhancer (with sequence listed in Supplementary Table 2) was synthesized (IDT) and resuspended to 100 uM in water. Serial dilutions of polymers or ssODNenh were made in water, then mixed 1:1 volume with preformed RNPs. For subsequent knockin experiments, 15–50 kDa poly(L-glutamic acid) (Sigma) was resuspended to 100mg/mL in water, sterile filtered, and mixed with freshly-prepared gRNA at 0.8:1 volume ratio prior to complexing with Cas9 protein for final volume ratio gRNA:PGA:Cas9 of 2:1.6:1. RNP particle size was measured by dynamic light scattering dispersed in PBS on a Zetasizer Nano ZS (Malvern Panalytical).
For RNP lyophilization, freshly prepared RNPs premixed with PGA or ssODNenh and HDR templates were diluted 1:1 v:v in 50mM trehalose, flash frozen in a liquid nitrogen bath then immediately dried on a Labconco Freeze Dry System-Freezone 4.5 lyophilizer for 24 hours, and stored at −20C until use. Dry RNP was resuspended in water to achieve the original concentration, incubated for 5 minutes at 37C, then mixed with cells for electroporation.
HDR Template Generation and Electroporation
Long double-strand HDR templates encoding various gene insertions (see Supplementary Table 1) and 300–350bp homology arms were synthesized as GeneBlocks (IDT) and cloned into a pUC19 plasmid, which then served as a template for generating a PCR amplicon. Specific PCR primers targeting the left and right homology arms and with additional described Cas9 Target Sequences (CTS) (see Supplementary Figure 2) were synthesized (IDT) without chemical modifications. Amplicons were generated as previously described5 with KapaHiFi polymerase (Kapa Biosystems), purified by SPRI bead cleanup, and resuspended in water to 0.5 to 2 ug/uL measured by light absorbance on a NanoDrop spectrophotometer (ThermoFisher).
HDR templates were mixed and incubated with RNPs for at least 5 minutes prior to mixing with and electroporating into cells. Immediately prior to electroporation in a Lonza 4D 96-well format Nucleofector (Lonza), cells were centrifuged for 10 minutes at 90g, media aspirated, and resuspended in the electroporation buffer P3 (Lonza) using 17–20 μL buffer per 0.5 to 1.0 e6 cells. T cells, NK cells, and B cells were electroporated with pulse code EH-115; primary HSPCs with pulse code ER-100, and iPS-derived CD34 HSPCs with pulse code EY-100. Immediately after electroporation, cells were rescued with addition of 80uL of growth media directly into the electroporation well, incubated for 10–20 minutes, then removed and diluted to 0.5–1e6 cells/mL in growth media. Additional fresh growth media and cytokines were added every 48 hours.
At 3–5 days post electroporation (except for NK cells collected at 5–7 days), cells were collected for staining and flow cytometry analysis. Briefly, cells were stained for cell type-specific surface markers and live-dead discrimination (see list of antibodies in Supplementary Table 3) then analyzed on an Attune NxT Flow Cytometer with automated 96-well sampler (ThermoFisher) sampling a defined volume (between 50–150uL per well) to obtain quantitative cell counts. Cytometry data was processed and analyzed using FlowJo software (BD Biosciences).
Amplicon Sequencing for Off-target Editing
CD3+ (Bulk) T cells were electroporated with various RNPs incorporating an EMX1 guide RNA previously shown to have high-levels of off-target cutting by CIRCLE-Seq15. At day 3 post-editing, genomic DNA was extracted and purified with a Zymo Quick-DNA MiniPrep kit. After normalizing input quantities of genomic DNA, a two-step PCR amplicon sequencing approach using NEB Q5 2X Master Mix Hot Start Polymerase with the manufacturer’s recommended thermocycler conditions. An initial 18 cycle PCR reaction using previously validated primers14 (see Supplementary Table 1) to an approximately 150–250 bp region centered on the predicted gRNA cut sites. After a 1.0X SPRI purification step, a second 14-cycle PCR was then performed to append P5 and P7 Illumina sequencing adaptors and sample-specific barcodes, followed again by a 1.0X SPRI purification. Concentrations were normalized across samples, pooled, and the amplicon library was sequenced on an Illumina Mini-Seq with paired 300 base reads run mode. Amplicon sequence reads were processed with the CRISPResso2 algorithm in batch mode16 using default parameters. We eliminated reads with potential sequencing errors detected as single base substitutions but no indels, and the remaining reads identified as indels were used to calculated the NHEJ percentage at each on- or off-target site. We excluded two off-target sites where amplicon sequencing was performed but control-treated cells from both donors contained sequence variants at >5% of reads that was attributable either to germline variants and/or PCR and sequencing errors at poly A/T sequences.
Statistical Analysis
Unless otherwise stated in the figure legend, all experiments were repeated at least twice with biologically independent samples, and data was aggregated for display and analysis. For Figure 1b, a two-way Mann-Whitney test was utilized to compare tCTS orientation and modifications to the control unmodified template, with n=4 different biologically independent blood donors. For Figure 1d, a two-way paired T test with Holm-Sidak multiple comparison correction was used to compare relative HDR at each locus. Exact p values are available in Supplemental Table 4.
Data Availability
Amplicon sequencing data has been deposited in the NIH NCBI SRA (Bioproject PRJNA564604) and flow cytometry raw data files are available upon request. Plasmids containing the HDR template sequences used in the study are available through AddGene (Supplementary Table 1), and annotated DNA sequences for all constructs are available upon request.
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of the Marson lab, Chris Jeans and the QB3 MacroLab, and Aaron Dolor for suggestions and technical assistance. This research was supported by NIH grants DP3DK111914–01, R01DK1199979 and P50GM082250 (A.M.), a grant from the Keck Foundation (A.M.), gifts from Jake Aronov, Barbara Bakar, and the American Endowment Foundation (A.M.), a gift from the UCSF Diabetes Center, a gift from the Jeffrey Modell Foundation, and a National Multiple Sclerosis Society grant (A.M.; CA 1074-A-21). D.N.N was supported by the UCSF Biology of Infectious Diseases Training Program (T32A1007641), an NIH Loan Repayment Program grant (L40 AI140341), and the CIS CSL Behring Fellowship Award. T.L.R. was supported by the UCSF Medical Scientist Training Program (T32GM007618), the UCSF Endocrinology Training Grant (T32 DK007418), and the NIDDK (F30DK120213). A.M. holds a Career Award for Medical Scientists from the Burroughs Wellcome Fund, is an investigator at the Chan Zuckerberg Biohub and has received funding from the Innovative Genomics Institute (IGI) and the Parker Institute for Cancer Immunotherapy (PICI). The UCSF Flow Cytometry Core was supported by the Diabetes Research Center grant NIH P30 DK063720.
COMPETING INTERESTS
The authors declare competing financial interests: A.M. is a co-founder of Spotlight Therapeutics. T.L.R. and A.M. are co-founders of Arsenal Therapeutics. J.A.B. is a founder of Sonoma Biotherapeutics. A.M. serves as on the scientific advisory board of PACT Pharma, is an advisor to Trizell, and was a former advisor to Juno Therapeutics. The Marson laboratory has received sponsored research support from Juno Therapeutics, Epinomics, Sanofi and gift from Gilead. Patents have been filed based on the findings described here.
REFERENCES
- 1.Yin H, Xue W & Anderson DG CRISPR-Cas: a tool for cancer research and therapeutics. Nat Rev Clin Oncol, doi: 10.1038/s41571-019-0166-8 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Dunbar CE et al. Gene therapy comes of age. Science 359, doi: 10.1126/science.aan4672 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Cornu TI, Mussolino C & Cathomen T Refining strategies to translate genome editing to the clinic. Nat Med 23, 415–423, doi: 10.1038/nm.4313 (2017). [DOI] [PubMed] [Google Scholar]
- 4.David RM & Doherty AT Viral Vectors: The Road to Reducing Genotoxicity. Toxicol Sci 155, 315–325, doi: 10.1093/toxsci/kfw220 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Roth TL et al. Reprogramming human T cell function and specificity with non-viral genome targeting. Nature 559, 405–409, doi: 10.1038/s41586-018-0326-5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vo LT et al. Regulation of embryonic haematopoietic multipotency by EZH1. Nature 553, 506–510, doi: 10.1038/nature25435 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pouton CW, Wagstaff KM, Roth DM, Moseley GW & Jans DA Targeted delivery to the nucleus. Adv Drug Deliv Rev 59, 698–717, doi: 10.1016/j.addr.2007.06.010 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Doudna JA & Charpentier E Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346, 1258096, doi: 10.1126/science.1258096 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Dominguez AA, Lim WA & Qi LS Beyond editing: repurposing CRISPR-Cas9 for precision genome regulation and interrogation. Nat Rev Mol Cell Biol 17, 5–15, doi: 10.1038/nrm.2015.2 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang F & Doudna JA CRISPR-Cas9 Structures and Mechanisms. Annu Rev Biophys 46, 505–529, doi: 10.1146/annurev-biophys-062215-010822 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Luecke S et al. cGAS is activated by DNA in a length-dependent manner. EMBO Rep 18, 1707–1715, doi: 10.15252/embr.201744017 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richardson CD, Ray GJ, Bray NL & Corn JE Non-homologous DNA increases gene disruption efficiency by altering DNA repair outcomes. Nat Commun 7, 12463, doi: 10.1038/ncomms12463 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bernkop-Schnurch A Strategies to overcome the polycation dilemma in drug delivery. Adv Drug Deliv Rev 136–137, 62–72, doi: 10.1016/j.addr.2018.07.017 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Vakulskas CA et al. A high-fidelity Cas9 mutant delivered as a ribonucleoprotein complex enables efficient gene editing in human hematopoietic stem and progenitor cells. Nat Med 24, 1216–1224, doi: 10.1038/s41591-018-0137-0 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsai SQ et al. CIRCLE-seq: a highly sensitive in vitro screen for genome-wide CRISPR-Cas9 nuclease off-targets. Nat Methods 14, 607–614, doi: 10.1038/nmeth.4278 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clement K et al. CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat Biotechnol 37, 224–226, doi: 10.1038/s41587-019-0032-3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ran FA et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154, 1380–1389, doi: 10.1016/j.cell.2013.08.021 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Amplicon sequencing data has been deposited in the NIH NCBI SRA (Bioproject PRJNA564604) and flow cytometry raw data files are available upon request. Plasmids containing the HDR template sequences used in the study are available through AddGene (Supplementary Table 1), and annotated DNA sequences for all constructs are available upon request.