Abstract
Meningitis due to Cryptococcus neoformans is responsible for upwards of 180,000 deaths worldwide annually, mostly in immunocompromised individuals. Currently there are no licensed fungal vaccines, and even with anti-fungal drug treatment, cryptococcal meningitis is often fatal. Our lab previously demonstrated vaccination with recombinant cryptococcal proteins delivered in glucan particles (GPs) protects mice against an otherwise lethal infection. The aim of the present study was to discover additional cryptococcal antigens affording vaccine-mediated protection. Sixteen proteins, each with evidence of extracellularity, were selected for in vivo testing based on their abundance in protective alkaline extracts of an acapsular C. neoformans strain, their known immunogenicity, and/or their high transcript level during human infection. Candidate antigens were recombinantly expressed in E. coli, purified and loaded into GPs. BALB/c and C57BL/6 mice received three subcutaneous injections of GP-based vaccine, and survival was assessed for 84 days following a lethal orotracheal challenge with strain KN99. As with our six published GP-vaccines, we saw differences in overall protection between mouse strains such that BALB/c mice typically demonstrated better survival than C57BL/6 mice. From these studies, we identified seven new proteins which, when administered as GP-vaccines, protect BALB/c and/or C57BL/6 mice against cryptococcal infection. With these results, we expand the pool of novel protective antigens to eleven proteins and demonstrate the potential for selection of highly transcribed extracellular proteins as vaccine targets. These screens highlight the efficacy of GP-subunit vaccines and identify promising antigens for further testing in anti-cryptococcal, multi-epitope vaccine formulations.
Keywords: Cryptococcus, fungal vaccine, T cell vaccine, C57BL/6 mouse, BALB/c mouse, acquired immune deficiency syndrome
1. Introduction
Cryptococcus neoformans and its closely related sister species, Cryptococcus gattii are the main etiologic agents of cryptococcosis, a fungal infection occurring predominately in immunocompromised populations. Inhalation of the yeast and its subsequent colonization of the lungs can present as fungal pneumonia. Dissemination to the central nervous system results in cryptococcal meningitis (CM) which is estimated to cause nearly 200,000 deaths annually or approximately 15% of AIDS-related mortalities [1]. The case fatality rate of CM in persons with AIDS is estimated to be upwards of 70% and survivors often are left with permanent neurological disabilities [1]. Moreover, the anticryptococcal drug armamentarium is limited and the most effective medications are often not available in the resource-poor countries experiencing the heaviest burden of cryptococcal disease. Vaccines to protect at risk individuals from developing cryptococcal disease are clearly needed but are not available.
Glucan particles (GPs) are porous cell wall shells derived from baker’s yeast (Saccharomyces cerevisiae) and composed primarily of β−1,3-glucan with small amounts of chitin [2]. They are recognized by the β−1,3-glucan receptor, Dectin-1, on phagocytes and are also potent activators of the complement system [3, 4]. GPs can be loaded with protein antigens within the hollow GPs, enabling delivery to phagocytes, including dendritic cells [2]. In initial studies, we found that immunization of mice with the model antigen ovalbumin encased in GPs elicited robust and long-lasting antibody and Th1- and Th17-biased CD4 T cell responses [2, 5]. Subsequent studies demonstrated vaccination of mice with a crude antigen extract encased in glucan particles (GP) protected mice against an otherwise lethal challenge with the highly virulent C. neoformans strain KN99 [6].
While these studies provided a proof of principle that GP-based vaccines could protect mice against experimental cryptococcosis, crude antigen preparations may be difficult to get approved for use in human vaccines due to concerns about potential toxicity including autoimmunity and reactogenicity. Therefore, an active focus of our laboratory is the identification of protein antigens for use in subunit vaccines to protect at risk individuals from developing cryptococcal disease. Our approach has been to recombinantly express candidate cryptococcal antigens in E. coli, load the antigens into GPs, vaccinate mice and then assess protection following a lethal fungal challenge. Using biased and unbiased approaches for antigen identification, we previously tested six such antigens and found four of them afforded significant protection when used in a GP-based vaccine in at least one mouse model of infection [7]. Here, we expanded our search for protective protein antigens using new approaches for candidate antigen identification. We predicted extracellular protein antigens would demonstrate an increased potential for detection by host immune surveillance over those internal to the pathogen and serve as better vaccine candidates. We therefore focused on proteins predicted to be extracellular, whether secreted or localized to the cell surface or wall. This paradigm has been taken for other subunit vaccines currently licensed for human use, targeting secreted toxins (tetanus), capsid proteins (HPV), and surface proteins (pertussis) [8, 9]. In our studies, we assessed the ability of 16 additional E. coli-derived recombinant cryptococcal proteins delivered as GP-vaccines to protect BALB/c and C57BL/6 mice against challenge with C. neoformans strain KN99. Seven of the new vaccines protected at least one mouse strain. With the four previously described, we now have a pool totaling 11 recombinant antigens which can be further analyzed for the development of a multi-epitope cryptococcal vaccine.
2. Materials and methods
2.1. Chemicals and culture media.
Reagents were from Thermo Fisher Scientific (Pittsburgh, PA), unless stated otherwise. Media was YPD (Difco Yeast Extract, Bacto Peptone, Dextrose with and without 2% agar); and Sabouraud Dextrose Agar (Remel) for culture of Cryptococcus. The BL21 strain of E. coli was used for protein expression by culturing in Overnight Express Instant TB medium (EMD Millipore, Billerica, MA) for 18h at 30°C with shaking. Antibiotic selection was with ampicillin (100 μg/ml) or kanamycin (100 μg/ml).
2.2. Strains of Cryptococcus.
C. neoformans var grubii strain KN99 [10] was maintained as a glycerol stock at −80°C and cultured initially on YPD agar. For in vivo challenge, KN99 was cultured in liquid YPD at 30°C with shaking for 18–20 h. Yeast cells were harvested by centrifugation and washed twice with phosphate-buffered saline (PBS). After counting with a hemocytometer, cells were suspended in PBS at a concentration of 4 × 105 cells/mL to challenge BALB/c mice and 2 × 105 cells/mL for C57BL/6 mice. CFUs were verified by culturing on Sabouraud dextrose agar.
2.3. Expression of cryptococcal proteins in E. coli and purification of protein.
The cDNA for each new protein, excluding sequence encoding an N-terminal signal peptide, C-terminal serine/threonine-rich region, and/or a GPI anchor was synthesized and cloned in pET19b by GenScript (Piscattaway, NJ), Cloning was done so as to fuse the vector-encoded HIS tag to the N-terminus of the cDNA. Methods for purification of recombinant protein on His Bind resin (EMD Millipore, Burlington, MA) were done as previously described [7] ; 6M Urea was included in the purification buffers to retain protein solubility. Following dialysis against 6M urea/ 20 mM Tris-HCl to remove imidazole, protein concentration was determined by the BCA assay and concentrated to 10 mg/ml using Amicon Ultra-15 centrifugal filters (10 kDA cutoff, Merck Millipore, Cork, Ireland). Protein purity was further assessed by SDS-PAGE followed by staining with Coomassie InstantBlue (Expedeon, Ltd. Cambridgeshire, UK).
2.4. Glucan particle (GP) vaccines.
Each recombinant protein was encapsulated in GPs as described [6, 7]. In addition to recombinant protein, each vaccine included mouse serum albumin (MSA; Equitech-Bio, Kerrville, TX) and yeast RNA (yRNA; Sigma-Aldrich, St. Louis, MO). A vaccine dose of 0.1 ml contained 10 μg of recombinant protein, 25 μg of MSA complexed with yRNA in 200 μg GPs (approximately 108 particles) suspended in sterile 0.9% saline. A control vaccine without recombinant protein included MSA and yRNA was designated as GP-MSA.
2.5. Mice.
BALB/c and C57BL/6 mice were from Jackson Laboratories and Charles River Laboratories. Mice were housed in a pathogen-free environment at the University of Massachusetts Medical School. All experimental procedures were approved by the University of Massachusetts Medical School Institutional Use and Care of Animals Committee.
2.6. Vaccination and challenge studies.
Vaccination was done by subcutaneous injection in the abdomen. Each mouse received 0.1 mL (1 dose) of GP vaccine with each vaccination. Three vaccinations were administered at two-week intervals. Two weeks after the third vaccination, the mice were challenged by orotracheal infusion into the lungs with 2×104 CFU (BALB/c mice) or 1×104 CFU (C57BL/6 mice) of C. neoformans strain KN99 in 50 μL PBS [7]. Mice were anesthetized with 2% isoflurane (Piramal Health Care, Andrah Pradesh, India) for inoculation. At 84 days post infection (DPI), surviving mice were euthanized. Dissected lungs were homogenized in 4 mL PBS with100 U/mL penicillin and 100 μg/mL streptomycin and plated on Sabouraud dextrose agar. CFUs were counted following incubation at 30°C for 2–3 days. Detection limits were 20 CFU/lung.
2.7. Bioinformatics.
cDNA and protein sequences were retrieved using respective CNAG numbers from the NCBI database for C. neoformans var. grubii strain H99 (taxid:235443). Homology determinations were based on Basic Local Alignment Search Tool protein (BLASTP) results of NCBI protein databases [11]. SignalP v3.0 was used to query each protein for a cleavable signal peptide [12].
2.8. Statistics.
Statistical analyses were performed and graphs generated using GraphPad Prism V 7.04. Kaplan-Meier survival curves were compared using the Mantel-Cox, log-rank test; p-values were assigned based on pair-wise comparisons to survival data for the control vaccine without recombinant protein, GP-MSA. As multiple comparisons were made, before statistical significance was denoted, the Bonferroni correction was applied.
3. Results and Discussion
3.1. Antigen Selection
We previously demonstrated the potential of subunit vaccines delivered by GPs to protect mice against an otherwise lethal infection with the highly virulent C. neoformans strain KN99 [6]. Those studies examined the first six recombinant protein antigens listed in Table 1. Of the six proteins, Cda2, MP88, and Fpd1 were selected for their known immunogenicity [6, 7, 13, 14]. The immunoreactivity of Cda2 coupled with the well-documented importance of chitosan for cryptococcal virulence prompted the testing of Cda1 and Cda3, the other two members of the cryptococcal chitin deacetylase family. The sixth protein, Sod1, is a superoxide dismutase and was selected for its abundance in alkaline extracts [6]. Four of the six antigens were protective when administered as GP-based vaccines [7].
Table 1.
Description of Selected Proteins
| CNAG | Name1 | Description | Recombinant Protein (kDa) |
Mass Spec Abundance2 |
RNA Seq Rank3 |
C. gattii
Ortholog4 |
FPKM Rank5 |
Human Homology6 |
|---|---|---|---|---|---|---|---|---|
| 05799 | Cda1 | chitin deacetylase | 42.4 | not found | 37 | CNBG_1745 | 344 | no |
| 01230 | Cda2 | chitin deacetylase | 43.8 | 24 | 1600 | CNBG_9064 | 226 | no |
| 01239 | Cda3 | chitin deacetylase | 37.9 | not found | 5344 | CNBG_0806 | 331 | no |
| 06291 | Fpd1 | deacetylase | 29.3 | <1 | 4362 | CNBG_5149 | 6355 | no |
| 01019 | Sod1 | superoxide dismutase | 20.1 | 97 | 84 | CNBG_0599 | 820 | yes |
| 00776 | MP88 | mannoprotein | 32.9 | <1 | 346 | CNBG_1155 | 555 | no |
| 00919 | Cpd1 | carboxypeptidase | 62.8 | 19 | 5209 | CNBG_6045 | 5616 | yes |
| 05097 | YjeF | ribokinase | 41.5 | 13 | 1024 | CNBG_4593 | 1607 | yes |
| 07442 | PTP | phosphotidylinositol transfer protein | 21.8 | 100 | 325 | CNBG_0642 | 709 | no |
| 07422 | DHA1 | delayed-type hypersensitivity protein | 36.9 | not found | 5502 | CNBG_0934 | 6285 | no |
| 00264 | Nuc | S1 nuclease/phospholipase C | 46.8 | 11 | 121 | CNBG_0307 | 1334 | no |
| 02030 | Glo1 | glyoxal oxidase | 66.2 | not found | 261 | CNBG_5182 | 768 | no |
| 03223 | 3223 | unknown | 24.4 | 6 | 128 | CNBG_2366 | 140 | no |
| 06432 | ACK | acetate kinase | 48.1 | 50 | 476 | CNBG_5028 | 1041 | yes |
| 01562 | Blp4 | unknown | 34.2 | not found | 101 | CNBG_3874 | 246 | no |
| 06267 | Rds1 | ferritin-like | 42.7 | not found | 6 | CNBG_9636 | 149 | no |
| 00581 | Sacch | saccharopepsin, aspartic protease | 48.6 | 27 | 59 | CNBG_1355 | 102 | yes |
| 04625 | Cerev | cerevisin, peptidase | 54.8 | 23 | 47 | CNBG_1027 | 86 | yes |
| 04735 | Mep1 | metallo proteinase | 92.7 | not found | 3 | CNBG_6001 | 13 | no |
| 03492 | 3492 | unknown | 45.2 | <1 | 66 | CNBG_2122 | 388 | no |
| 05872 | May1 | aspartic protease | 47.8 | not found | 152 | CNBG_1672 | 677 | yes |
| 03143 | 3143 | heat shock protein 9/12 | 11.5 | not found | 1 | CNBG_2441 | 3 | no |
| 04874 | 4874 | glucanase | 41.5 | not found | 60 | CNBG_5802 | 104 | no |
The first 6 proteins listed were previously tested and published [7]. 3223 was selected as a vaccine candidate but not tested due to insufficient protein yields using the methods described.
Mass spec data is normalized such that the top hit, PTP, has an abundance of 100 (bold) [6].
C. neoformans RNA seq reads from CSF of two human infections [18] were averaged to generate rank; 1 indicates the most abundant transcript (bold).
C. neoformans genes were transformed by orthology to C. gattii R265 using OrthoMCL Version 2.0.
C. gattii RNA seq. data from the bronchoalveolar lavage fluid of mice 24 hours post infection [29] were sorted so that the most fragments per kilobase of transcript per million mapped reads (FPKM) is ranked 1.
Human homology was determined using NCBI BLASTP; any homologs detected are designated “yes” regardless of score.
Given human leukocyte antigen diversity and the lack of known cryptococcal antigens that are immunodominant in humans, we reasoned that a protective T cell vaccine would need to contain epitopes from multiple proteins. Therefore, in the present study, we sought to identify additional protective protein antigens. Due to the larger size of the proteome and the probable homology to human proteins, vaccine protein antigen discovery for eukaryotic pathogens is considerably more complex compared with viral and bacterial pathogens. The C. neoformans genome is predicted to contain 6,967 protein-coding genes, of which >99% contain introns [10]. Thus, we needed to devise strategies to prioritize proteins for testing as candidate vaccine antigens.
A top priority for selection was evidence of extracellularity based on the presence of a predicted signal peptide in the protein sequence. C. neoformans H99 encodes 978 proteins with a predicted signal peptide, as determined by SignalP analysis through FungiDB [15]. In fungi, proteins with a signal peptide can be targeted to the cell membrane, to the cell wall, or for secretion as an extracellular protein [16]. Fungal proteins lacking signal peptides may also be secreted in vesicles, such as Sod1 [17]. We chose 17 extracellular cryptococcal protein antigens for testing in GP-based vaccines (Table 1, Table S1). Their selection was based on criteria including: 1) the presence of the protein in protective alkaline extracts of an acapsular C. neoformans strain [6]; 2) relative abundance of transcript in RNA isolated from C. neoformans collected from the cerebrospinal fluid (CSF) of patients with cryptococcosis [18]; and 3) previously published immunogenicity. Some of the proteins tested matched more than one criterion for their selection. To reduce the risk of vaccine-induced autoimmunity, we prioritized antigens that lacked significant homology with human proteins, as defined by the absence of hits on a BLASTP search [11].
We used the RNAseq data from human CSF in the selection of Glo1, 3223, Blp4, Rds1, Sacch, Mep1, 3492, May1, 3143, and 4874 [18]. We prioritized transcripts which were highly represented in the patient samples and that were predicted to encode for extracellular proteins with minimal homology to human proteins. The 6,778 transcripts from the CSF samples (GEO data GSE51573, [18]) were averaged and ranked by quantity of RNA seq reads such that the most abundant transcript was ranked “1” and the least abundant “6,778”. We determined the presence of a signal peptide for the 300 most abundant transcripts by analyses through the FungiDB website [15]. Additional favorable, but not requisite, criteria for promising candidates were defined. Prioritization was given to proteins that were members of protein families (Table S2), reasoning that cross-reactive vaccine responses to homologous cryptococcal proteins might occur and augment protection. Each protein tested as a vaccine had an ortholog in C. gattii for the potential development of a cross-species cryptococcal vaccine.
In addition to Sod1, the proteins, PTP, ACK, Cerev, Cpd1, YjeF, and Nuc were found to be present in alkaline extracts by mass spectrometry analysis [6]. Although initially selected for their transcript abundance and/or their immunogenicity, Cda2, Sacch, 3223, Fpd1, MP88, and 3492 were also identified in these extracts. Analyses of the transcriptome also identified previously selected proteins as highly transcribed during human infection, including Cda1, Sod1, and Cerev. Combined, these methods and analyses culminated in testing 16 of the 17 selected proteins as new GP-vaccines in mice; recombinant protein yield for 3223 was insufficient for testing as a vaccine. We did not use previous characterization as a virulence factor as a selection criterion for the new proteins.
3.2. Assessment of vaccine efficacy in BALB/c and C57BL/6 mice
Each of the candidate proteins, absent a signal peptide and/or serine/threonine-rich region, was recombinantly expressed with a His-tag in E. coli (Table 1). The recombinant proteins were then purified over nickel columns, concentrated and loaded into GPs. As serine/threonine-rich regions of fungal proteins are often O-glycosylated, and proteins generated in E. coli likely lack glycosylation, we did not include such portions of sequence. We have not yet defined the impact of glycosylation on vaccine efficacy for protein antigens delivered by GPs. While mannose receptor recognition of glycosylated regions of cryptococcal proteins does enhance immune responses compared to that seen with corresponding chemically deglycosylated proteins, we speculate the use of GP technology may abate this potential impact as GP phagocytosis is mediated by Dectin-1 and complement receptors [4, 19]. Cda2 (MP98) is an example of a protein which is heavily glycosylated in its native form, yet its production in E. coli still generates a protective vaccine [20]. Nevertheless, glycosylation and other posttranslational modifications can affect antigen presentation and processing, as well as inform antibody responses [21, 22]. Therefore, it is possible that had we used native glycosylated proteins in our vaccines, protection would have differed from that seen with the E. coli-derived recombinant proteins.
Mice received 3 biweekly subcutaneous injections of GP-vaccine containing 10 μg of recombinant protein. Two weeks after the third vaccination, the mice were challenged orotracheally with a lethal dose of C. neoformans strain KN99. Survival was monitored for 84 DPI, at which point the fungal burden in the lungs of each surviving mouse was assessed. Survivors had on average fewer CFUs than what was administered as a lethal challenge (1–2×104 CFUs); many had <20 CFU’s, which indicated the infection likely was cleared from the lungs (Figure S2). For perspective, a typical unprotected mouse dies with a fungal load of about 109 CFUs [7]. We observed distinct groups of mice that cleared infection and mice that succumbed to infection, but there was a third group of survivors at 84 DPI with a high fungal burden in the lungs (Figure S2). For these mice with high CFUs, progression to fatal cryptococcosis may have occurred had the experiment been extended.
Vaccines were tested in BALB/c and C57BL/6 mice, and, consistent with our published data on Cda1, Cda2, Cda3, Fpd1, Sod1 and MP88, the protection conferred by each GP-based vaccine varied as a function of mouse strain and candidate antigen (Figures 1 and Figure 2) [6]. Control groups included mice left unimmunized and mice immunized with GPs loaded with mouse serum albumin (GP-MSA). These control groups succumbed to infection by 30 DPI for both mouse strains.
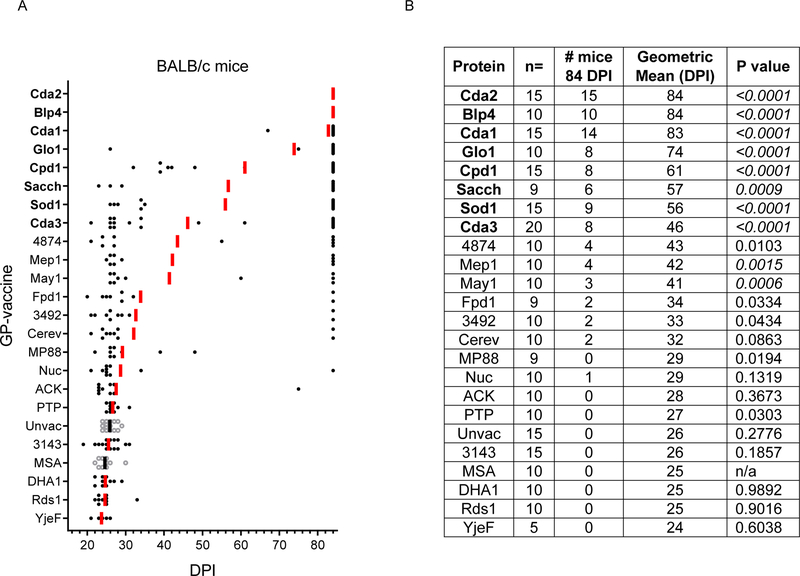
Figure 1: Survival following vaccination with GP-recombinant proteins in BALB/c mice.
Mice received 3 vaccinations with the indicated recombinant protein followed by an orotracheal challenge with 2×104 CFU of C. neoformans strain KN99. Mice were then assessed for survival over the next 84 days. Protective proteins are listed in bold. (A) Each dot represents one mouse, and the bars denote the geometric mean survival. Experimental groups are depicted as solid black circles with red bars. Unvaccinated and GP-MSA vaccinated controls are shown as hollow circles with black bars. (B) Descriptive statistics of candidate GP-vaccines. n = number of mice in the group. # mice 84 DPI indicates the number of survivors at d84 when the study ended. Geometric mean (DPI) refers to geometric mean survival in DPI. P values were calculated by Kaplan-Meier survival with Mantel-Cox significance compared to GP-MSA vaccinated mice. After applying the Bonferroni correction, significant comparisons (P<0.0023) are denoted in italics.
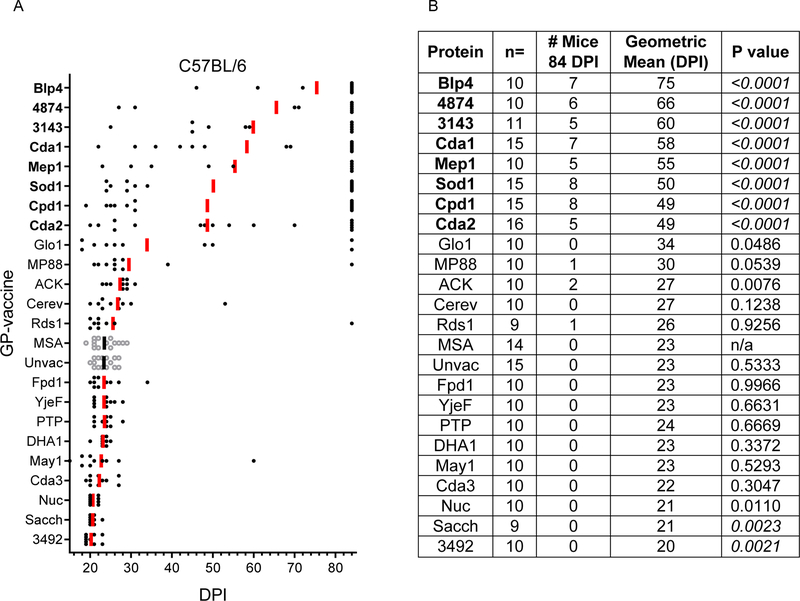
Figure 2: Survival following vaccination with GP-recombinant proteins in C57BL/6 mice.
As in Figure 1, except C57BL/6 mice were studied, and the challenge inoculum was 1×104 CFU.
Remarkably, BALB/c mice vaccinated with either GP-Cda2 or GP-Blp4 demonstrated 100% survival at the end of study (Figure 1). Six other GP-vaccines (GP-Cda1, GP-Glo1, GP-Cpd1, GP-Sacch, GP-Sod1, and GP-Cda3) afforded protection against fungal challenge, as judged by 84-day survival. For all the vaccinated animals, there were few deaths at intermediate time points; if mice survived 30 DPI, they were highly likely to still be alive at the end of the experiment (84 DPI). Intermediate geometric means reflect biphasic survival data wherein there are distinct groups of early deaths compared to survivors.
Eight of the GP-based vaccines conferred protection in C57BL/6 mice at 84 DPI (Figure 2). GP-Blp4 conferred the best protection in this mouse line, with 70% overall survival, followed by GP-4874, GP-3143, GP-Cda1, GP-Mep1, GP-Sod, GP-Cpd1, and GP-Cda2. None of the vaccines tested elicited 100% survival in C57BL/6 mice. Interestingly, vaccination with GP-Sacch or GP-3492 resulted in significantly less protection compared with GP-MSA. However, the difference in geometric mean survival was only two or three days and the biological relevance of this finding is unclear.
For most GP-vaccines in which protection was observed, efficacy was greater in BALB/c mice compared with C57BL/6 mice. (Figure 1 and Figure 2, Figure S1). That BALB/c mice are easier to protect is perhaps not surprising given that this mouse strain is generally less susceptible to cryptococcosis compared with C57/BL6 mice [23, 24]. However, in our studies using the highly virulent C. neoformans KN99 strain, the geometric mean survival of unvaccinated BALB/c and C57BL/6 mice did not greatly differ (26 and 23 DPI, respectively). Of interest, for three of the vaccines, GP-3143, GP-4874, and GP-Mep1, protection was greater in C57BL/6 mice. We speculate this finding could be due to these antigens having protective epitopes that stimulate more robust CD4+ T cells responses in C57BL/6 mice compared with BALB/c mice. As BALB/c and C57BL/6 mice have different MHC Class II and Class I haplotypes, it is predicted that some antigens will stimulate disparate strain-dependent T cell responses [7].
3.3. Evaluation of antigen selection criteria
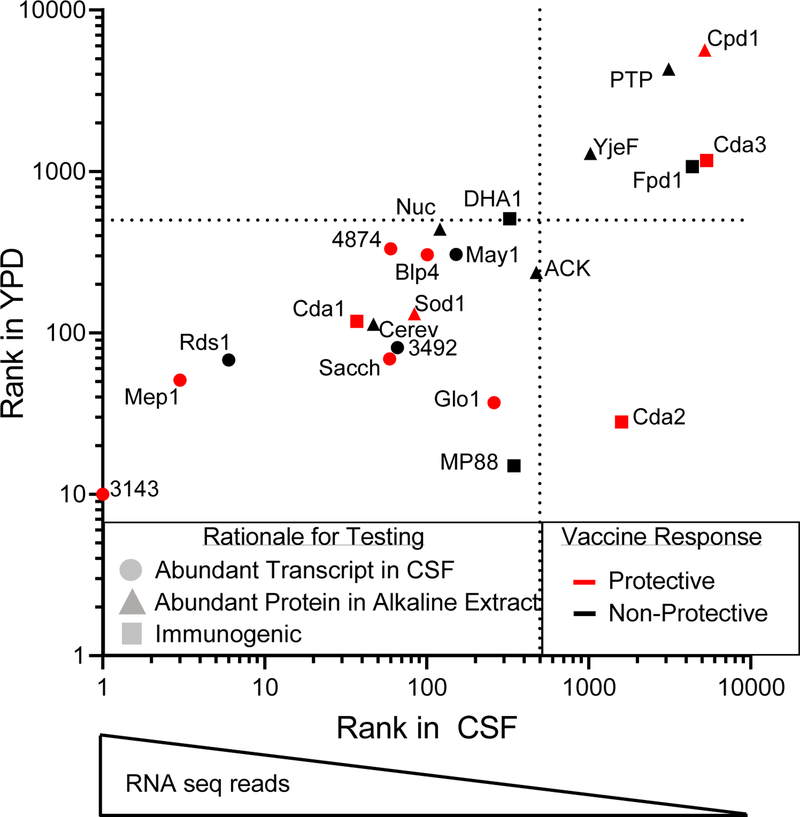
These studies identify seven novel GP-vaccines, which protect BALB/c and/or C57BL/6 mice against challenge with C. neoformans strain KN99. These vaccines (GP-Cpd1, GP-Sacch, GP-Mep1, GP-Glo1, GP-Blp4, GP-3143, and GP-4874), combined with the four previously described vaccines (GP-Cda1, GP-Cda2, GP-Cda3, and GP-Sod1), comprise a pool of eleven protective protein antigens. A variety of overlapping methods was used to select antigens for testing (Figure 3). For example, Cda2, selected for its known immunogenicity, was also identified as abundant by mass spectrometry analysis of an alkaline extract from a cultured acapsular strain [6, 13]. Despite being one of the most protective antigens, however, Cda2 transcripts are not abundantly expressed by Cryptococcus from the CSF of human patients (Figure 3) or from infected mice [25]; Cda1, which yields comparable protection, is the more abundant transcript in this family. Rds1, contrastingly, is one of the most abundantly transcribed cryptococcal proteins in the CSF during human infection, yet it does not generate a protective vaccine response. Each of the methods for selection yielded promising experimental vaccines as well as non-protective antigens, which we believe reinforces the importance of utilizing a multi-faceted approach in the identification of candidate antigens. While our selection process utilized transcriptomic data obtained from human infections [18], comparing rank of transcript abundance for these samples to those from YPD-cultured C. neoformans demonstrates a highly similar trend between conditions (Figure 3, Figure S1). This may aid in future screens as it suggests that transcript or proteomic data obtained from cell culture may suffice for the selection of candidate antigens when human or mouse derived samples are not available for such analyses. Eight of the eleven protective cryptococcal antigens are members of protein families (Table S2), perhaps highlighting the importance of protein cross-reactivity in amplifying a productive vaccine response. A larger sample size would be needed to determine the true value of each criterion for selection of antigens. Other factors, such as glycosylation and MHCII-restricted antigen presentation undoubtedly play a role.
Figure 3: Criteria for selection of extracellular protein antigens.
Initial selection of candidate antigens was based on transcript abundance (●), presence in alkaline extracts (▲), and/or known immunogenicity (■). Each method of selection yielded both protective (red) and non-protective (black) vaccines. Dotted lines indicate the cutoff for the top 500 most abundant transcripts, wherein a rank of “1” represents the highest number of transcripts. Transcriptome data from YPD-cultured Cryptococcus are graphed against those obtained from human CSF [18].
Recently, four cryptococcal proteins were identified that elicited predominantly IgG2a responses in the sera of infected mice [26]. As IgG2a responses are a surrogate for protective Th1 responses, IgG2a-associated antigens make promising candidates for testing in vaccine models. Of these four, three exhibit significant homology to human proteins. The fourth, however, is both abundantly transcribed in YPD and a member of a protein family; it may therefore be the most attractive prospect. Interestingly, 21 of the proteins we tested as vaccines are absent from several publications, which focused on identifying immunodominant cryptococcal proteins [26–28]. YjEf is the only protein of the 22 antigens deemed immunodominant based on serum antibodies in these studies [27], but GP-YjEf is not a protective vaccine in either mouse strain tested.
Conclusions
We have identified 7 novel cryptococcal protein antigens which, when formulated in GP-based vaccines, confer protection in BALB/c and/or C57BL/6 mice against an otherwise lethal challenge with C. neoformans strain KN99. These antigens (Cpd1, Glo1, Blp4, Sacch, Mep1, 3143, and 4874), combined with those previously published (Cda1, Cda2, Cda3, and Sod1), comprise a pool of 11 candidate antigens which warrant further study as components of subunit cryptococcal vaccines. Ongoing and future studies will determine if protection is enhanced by co-loading GPs with multiple, protective antigens, whether other adjuvants can substitute for GPs, and the immunological mechanism of protection.
Supplementary Material
Figure S1: Comparison of transcript abundance from C. neoformans var. grubii strains directly isolated from human CSF obtained from persons with cryptococcal meningitis compared to these same isolates following inoculation into YPD [18]. Dotted lines indicate the top 500 most abundant transcripts. Grey dots represent all the transcripts generated, and red dots represent the selected candidate antigens in Table 1. R=0.8272 as determined by Spearman Rank Order Correlation.
Figure S2: Fungal burden in the lungs of mice that survived cryptococcal challenge. Survivors at 84 DPI were euthanized, and their lungs were homogenized and plated to determine CFU (BALB/c, top; C57BL/6, bottom). Dotted horizontal line denotes the CFU administered for KN99 challenge. Each dot represents the CFU in the lungs of one mouse, the red bars denote the geometric mean.
Highlights.
-
-
Cryptococcal protein antigens were recombinantly expressed in E. coli.
-
-
Two mouse strains were vaccinated with recombinant protein-filled glucan particles.
-
-
After a lethal C. neoformans challenge, vaccine efficacy was assessed by survival.
-
-
Protection achieved in at least one mouse strain with 11/22 experimental vaccines.
Acknowledgements
We acknowledge NIH grants R01 AI025780, R01 AI102618, R01AI125045, R01 HL112671, and R01 AI072195. MMH was partially supported by NIH Training Grant T32 AI095213. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Abbreviations
- CM
cryptococcal meningitis
- DPI
days post infection
- GPs
glucan particles
- NCBI
National Center for Biotechnology Information
Footnotes
Declaration of Competing Interest
The authors report no competing interests.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Rajasingham R, Smith RM, Park BJ, Jarvis JN, Govender NP, Chiller TM, et al. Global burden of disease of HIV-associated cryptococcal meningitis: an updated analysis. Lancet Infect Dis. 2017;17:873–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Huang H, Ostroff GR, Lee CK, Specht CA, Levitz SM. Characterization and optimization of the glucan particle-based vaccine platform. Clinical and Vaccine Immunology. 2013;20:1585–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Agarwal S, Specht CA, Huang H, Ostroff GR, Ram S, Rice PA, et al. Linkage specificity and role of properdin in activation of the alternative complement pathway by fungal glycans. mBio. 2011;2:e00178–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Huang H, Ostroff GR, Lee CK, Agarwal S, Ram S, Rice PA, et al. Relative contributions of Dectin-1 and complement to immune responses to particulate β-glucans. The Journal of Immunology. 2012;189:312–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Huang H, Ostroff GR, Lee CK, Specht CA, Levitz SM. Robust stimulation of humoral and cellular immune responses following vaccination with antigen-loaded beta-glucan particles. mBio. 2010;1:e00164–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Specht CA, Lee CK, Huang H, Tipper DJ, Shen ZT, Lodge JK, et al. Protection against experimental cryptococcosis following vaccination with glucan particles containing Cryptococcus alkaline extracts. mBio. 2015;6:e01905–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Specht CA, Lee CK, Huang H, Hester MM, Liu J, Luckie BA, et al. Vaccination with recombinant Cryptococcus proteins in glucan particles protects mice against cryptococcosis in a manner dependent upon mouse strain and cryptococcal species. mBio. 2017;8:e01872–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Nascimento IP, Leite LC. Recombinant vaccines and the development of new vaccine strategies. Brazilian journal of medical and biological research = Revista brasileira de pesquisas medicas e biologicas. 2012;45:1102–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Patel SS, Wagstaff AJ. Acellular Pertussis Vaccine (Infanrix™-DTPa; SB-3). Drugs. 1996;52:254–75. [DOI] [PubMed] [Google Scholar]
- [10].Janbon G, Ormerod KL, Paulet D, Byrnes EJ, III, Yadav V, Chatterjee G, et al. Analysis of the genome and transcriptome of Cryptococcus neoformans var. grubii reveals complex RNA expression and microevolution leading to virulence attenuation. PLOS Genetics. 2014;10:e1004261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins.
- [12].Dyrløv Bendtsen J, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. Journal of Molecular Biology. 2004;340:783–95. [DOI] [PubMed] [Google Scholar]
- [13].Huang C, Nong S-h, Mansour MK, Specht CA, Levitz SM. Purification and characterization of a second immunoreactive mannoprotein from Cryptococcus neoformans that stimulates T-cell responses. Infection and Immunity. 2002;70:5485–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Levitz SM, Nong S-h, Mansour MK, Huang C, Specht CA. Molecular characterization of a mannoprotein with homology to chitin deacetylases that stimulates T cell responses to Cryptococcus neoformans. Proceedings of the National Academy of Sciences. 2001;98:10422–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Basenko EY, Pulman JA, Shanmugasundram A, Harb OS, Crouch K, Starns D, et al. FungiDB: An integrated bioinformatic resource for fungi and oomycetes. J Fungi (Basel). 2018;4:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Delic M, Valli M, Graf AB, Pfeffer M, Mattanovich D, Gasser B. The secretory pathway: exploring yeast diversity. FEMS Microbiology Reviews. 2013;37:872–914. [DOI] [PubMed] [Google Scholar]
- [17].Rodrigues ML, Nakayasu ES, Oliveira DL, Nimrichter L, Nosanchuk JD, Almeida IC, et al. Extracellular vesicles produced by Cryptococcus neoformans contain protein components associated with virulence. Eukaryotic Cell. 2008;7:58–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Chen Y, Toffaletti DL, Tenor JL, Litvintseva AP, Fang C, Mitchell TG, et al. The Cryptococcus neoformans transcriptome at the site of human meningitis. mBio. 2014;5:e01087–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Mansour MK, Schlesinger LS, Levitz SM. Optimal T cell responses to Cryptococcus neoformans mannoprotein are dependent on recognition of conjugated carbohydrates by mannose receptors. The Journal of Immunology. 2002;168:2872–9. [DOI] [PubMed] [Google Scholar]
- [20].Specht CA, Nong S, Dan JM, Lee CK, Levitz SM. Contribution of glycosylation to T cell responses stimulated by recombinant Cryptococcus neoformans mannoprotein. The Journal of Infectious Diseases. 2007;196:796–800. [DOI] [PubMed] [Google Scholar]
- [21].Zhou JY, Oswald DM, Oliva KD, Kreisman LSC, Cobb BA. The glycoscience of immunity. Trends in Immunology. 2018;39:523–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wolfert MA, Boons G-J. Adaptive immune activation: glycosylation does matter. Nature Chemical Biology. 2013;9:776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Chen G-h McNamara DA, Hernandez Y Huffnagle GB, Toews GB Olszewski MA. Inheritance of immune polarization patterns is linked to resistance versus susceptibility to Cryptococcus neoformans in a mouse model. Infection and Immunity. 2008;76:2379–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Huffnagle GB, Boyd MB, Street NE, Lipscomb MF. IL-5 is required for eosinophil recruitment, crystal deposition, and mononuclear cell recruitment during a pulmonary Cryptococcus neoformans infection in genetically susceptible Mice (C57BL/6). The Journal of Immunology. 1998;160:2393–400. [PubMed] [Google Scholar]
- [25].Upadhya R, Baker LG, Lam WC, Specht CA, Donlin MJ, Lodge JK. Cryptococcus neoformans Cda1 and its chitin deacetylase activity are required for fungal pathogenesis. mBio. 2018;9:e02087–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Firacative C, Gressler AE, Schubert K, Schulze B, Müller U, Brombacher F, et al. Identification of T helper (Th)1- and Th2-associated antigens of Cryptococcus neoformans in a murine model of pulmonary infection. Scientific Reports. 2018;8:2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Young M, Macias S, Thomas D, Wormley FL Jr.. A proteomic-based approach for the identification of immunodominant Cryptococcus neoformans proteins. PROTEOMICS. 2009;9:2578–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Chaturvedi AK, Weintraub ST, Lopez-Ribot JL, Wormley FL Jr.. Identification and characterization of Cryptococcus neoformans protein fractions that induce protective immune responses. PROTEOMICS. 2013;13:3429–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Ferrareze PAG, Streit RSA, Santos PRd, Santos FMd, Almeida RMCd, Schrank A, et al. Transcriptional analysis allows genome reannotation and reveals that Cryptococcus gattii VGII undergoes nutrient restriction during infection. Microorganisms. 2017;5:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Eigenheer RA, Lee YJ, Blumwald E, Phinney BS, Gelli A. Extracellular glycosylphosphatidylinositol-anchored mannoproteins and proteases of Cryptococcus neoformans. FEMS Yeast Research. 2007;7:499–510. [DOI] [PubMed] [Google Scholar]
- [31].Reuwsaat JCV, Motta H, Garcia AWA, Vasconcelos CB, Marques BM, Oliveira NK, et al. A predicted mannoprotein participates in Cryptococcus gattii capsular structure. mSphere. 2018;3:e00023–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Geddes JMH, Croll D, Caza M, Stoynov N, Foster LJ, Kronstad JW. Secretome profiling of Cryptococcus neoformans reveals regulation of a subset of virulence-associated proteins and potential biomarkers by protein kinase A. BMC Microbiology. 2015;15:206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Campbell LT, Simonin AR, Chen C, Ferdous J, Padula MP, Harry E, et al. Cryptococcus strains with different pathogenic potentials have diverse protein secretomes. Eukaryot Cell. 2015;14:554–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Clarke SC, Dumesic PA, Homer CM, O’Donoghue AJ, La Greca F, Pallova L, et al. Integrated activity and genetic profiling of secreted peptidases in Cryptococcus neoformans reveals an aspartyl peptidase required for low pH survival and virulence. PLOS Pathogens. 2016;12:e1006051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Park Y-D, Shin S, Panepinto J, Ramos J, Qiu J, Frases S, et al. A role for LHC1 in higher order structure and complement binding of the Cryptococcus neoformans capsule. PLOS Pathogens. 2014;10:e1004037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Mandel MA, Grace GG, Orsborn KI, Schafer F, Murphy JW, Orbach MJ, et al. The Cryptococcus neoformans gene DHA1 encodes an antigen that elicits a delayed-type hypersensitivity reaction in immune mice. Infect Immun. 2000;68:6196–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Comparison of transcript abundance from C. neoformans var. grubii strains directly isolated from human CSF obtained from persons with cryptococcal meningitis compared to these same isolates following inoculation into YPD [18]. Dotted lines indicate the top 500 most abundant transcripts. Grey dots represent all the transcripts generated, and red dots represent the selected candidate antigens in Table 1. R=0.8272 as determined by Spearman Rank Order Correlation.
Figure S2: Fungal burden in the lungs of mice that survived cryptococcal challenge. Survivors at 84 DPI were euthanized, and their lungs were homogenized and plated to determine CFU (BALB/c, top; C57BL/6, bottom). Dotted horizontal line denotes the CFU administered for KN99 challenge. Each dot represents the CFU in the lungs of one mouse, the red bars denote the geometric mean.