Abstract
Despite decades of research, efforts to directly target KRAS have been challenging. MRTX849 was identified as a potent, selective, and covalent KRASG12C inhibitor that exhibits favorable drug-like properties, selectively modifies mutant cysteine 12 in GDP-bound KRASG12C and inhibits KRAS-dependent signaling. MRTX849 demonstrated pronounced tumor regression in 17 of 26 (65%) of KRASG12C-positive cell line- and patient-derived xenograft models from multiple tumor types and objective responses have been observed in KRASG12C-positive lung and colon adenocarcinoma patients. Comprehensive pharmacodynamic and pharmacogenomic profiling in sensitive and partially resistant non-clinical models identified mechanisms implicated in limiting anti-tumor activity including KRAS nucleotide cycling and pathways that induce feedback reactivation and/or bypass KRAS dependence. These factors included activation of RTKs, bypass of KRAS dependence, and genetic dysregulation of cell cycle. Combinations of MRTX849 with agents that target RTKs, mTOR, or cell cycle demonstrated enhanced response and marked tumor regression in several tumor models, including MRTX849-refractory models.
Keywords: KRASG12C, MRTX849, Covalent, Inhibitor
Introduction
KRAS is one of the most frequently mutated oncogenes in cancer; however, efforts to directly target KRAS have been largely unsuccessful due to its high affinity for GTP/GDP and the lack of a clear binding pocket [1–4]. More recently, compounds were identified that covalently bind to KRASG12C at the cysteine 12 residue, lock the protein in its inactive GDP-bound conformation, inhibit KRAS-dependent signaling and elicit anti-tumor responses in tumor models [5–7]. Advances on early findings demonstrated that the binding pocket under the switch II region was exploitable for drug discovery culminating in the identification of more potent KRASG12C inhibitors with improved physiochemical properties which are now entering clinical trials. The identification of KRASG12C inhibitors provides a renewed opportunity to develop a more comprehensive understanding of the role of KRAS as a driver oncogene and to explore the clinical utility of direct KRAS inhibition.
KRASG12C mutations are present in lung and colon adenocarcinoma as well as smaller fractions of other cancers. The genetic context of KRASG12C alteration can vary significantly among tumors and is predicted to affect response to KRAS inhibition. KRAS mutations are often enriched in tumors due to amplification of mutant or loss of wild-type allele [8, 9]. In addition, KRAS mutations often co-occur with other key genetic alterations including TP53 and CDKN2A in multiple cancers, KEAP1 and/or STK11 in lung adenocarcinoma or APC and PIK3CA in colon cancer [3, 8–12]. Whether differences in KRAS mutant allele fraction or co-occurrence with other mutations influence response to KRAS blockade is yet not well understood. In addition, due to the critical importance of the RAS pathway in normal cellular function, there is extensive pathway isoform redundancy and a comprehensive regulatory network in normal cells to ensure tight control of temporal pathway signaling. RAS pathway negative feedback signaling is mediated by ERK1/2 and receptor tyrosine kinases (RTKs) as well as by ERK pathway target genes including dual-specificity phosphatases (DUSPs) and Sprouty (SPRY) proteins [13–17]. One important clinically relevant example is provided by the reactivation of ERK signaling observed following treatment of BRAFV600E-mutant cancers with selective BRAF inhibitors [18–20]. The observed intertumoral heterogeneity and extensive feedback signaling network in KRAS-mutant cancers may necessitate strategies to more comprehensively block oncogenic signal transduction and deepen the anti-tumor response in concert with KRAS blockade [15, 21, 22].
Potential strategies to augment the response to KRASG12C inhibitor treatment are evident at multiple nodes of the signaling pathway regulatory machinery. RAS proteins are small GTPases that normally cycle between an active, GTP-bound state and an inactive, GDP-bound state. RAS proteins are loaded with GTP through guanine nucleotide exchange factors (GEFs; e.g., SOS1) which are activated by upstream RTKs, triggering subsequent interaction with effector proteins that activate RAS-dependent signaling. RAS proteins hydrolyze GTP to GDP through their intrinsic GTPase activity which is dramatically enhanced by GTPase-activating proteins (GAPs). Mutations at codons 12 and 13 in RAS proteins impair GAP-stimulated GTP hydrolysis leaving RAS predominantly in the GTP-bound, active state.
Potent covalent KRASG12C inhibitors described to date bind only GDP-bound KRAS [5–7]. While codon 12 and 13 mutations decrease the fraction of GDP-bound KRAS, recent biochemical analyses revealed that KRASG12C exhibits the highest intrinsic GTP hydrolysis rate and highest nucleotide exchange rate among KRAS mutants [23]. Furthermore, the nucleotide-bound state of KRASG12C can be shifted toward the GDP-bound state by pharmacologically modulating upstream signaling with RTK inhibitors that increase the activity of KRASG12C inhibitors [7, 22, 24]. Likewise, SHP2 is a phosphatase that positively transduces RTK signaling to KRAS. Accordingly, SHP2 inhibitors are active in cancers driven by KRAS mutations that are dependent on nucleotide cycling, including KRASG12C [25–27].
MRTX849 is among the first KRASG12C inhibitors to advance to clinical trials. The comprehensive and durable inhibition of KRASG12C by MRTX849 provides a unique opportunity to understand the extent to which KRAS functions as an oncogenic driver. In addition, the observation that the response to blockade of KRAS is markedly different in vitro and in vivo indicates that evaluation of the consequences of KRAS blockade in in vivo model systems is critical to understand the role of KRAS-driven tumor progression. The demonstration of partial responses in lung and colon adenocarcinoma patients treated with MRTX849 in clinical trials indicates that results observed in tumor models extends to KRASG12C-positive human cancers. Our comprehensive molecular characterization of multiple tumor models at baseline and during response to KRAS inhibition has provided further insight toward the contextual role of KRAS mutation in the setting of genetic and tumoral heterogeneity. Finally, further interrogation of these genetic alterations and signaling pathways utilizing functional genomics strategies including CRISPR and combination approaches uncovered regulatory nodes that sensitize tumors to KRAS inhibition when co-targeted.
Results
MRTX849 is a Potent and Selective Inhibitor of KRASG12C, KRAS-Dependent Signal Transduction and Cell Viability In Vitro
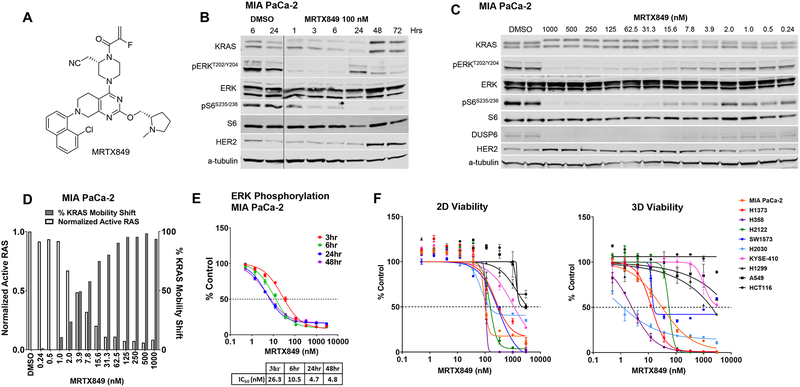
A structure-based drug design approach, including optimization for favorable drug-like properties, lead to the discovery of MRTX849 as a potent, covalent KRASG12C inhibitor (Figure 1A and Table S1). An LCMS-based KRASG12C protein modification assay revealed that MRTX849 demonstrated much greater modification of KRASG12C when preloaded with GDP compared with GTP (Table S2) supporting that MRTX849 binds to and stabilizes the inactive GDP-bound form of KRASG12C. Indeed, introducing a co-mutation that impairs the GTPase activity of KRASG12C [24] attenuated the inhibitory activity of MRTX1257, a close analogue of MRTX849 (Figure S1A). Secondary mutations that modulate the nucleotide exchange function of KRASG12C also affected inhibition by MRTX1257 supporting that the MRTX compound series is dependent on KRASG12C nucleotide cycling.
Figure 1.
MRTX849 is a potent, covalent KRASG12C inhibitor in vitro.
(A) Structure of MRTX849.
(B) Immunoblot protein western analyses of KRAS pathway targets in MIA PaCa-2 cells treated from 1 hours to 72 hours with MRTX849 at 100 nM.
(C) Immunoblot protein western analyses of KRAS pathway targets in MIA PaCa-2 cells treated for 24 hours with MRTX849 over a 13-point dose response.
|(D) Left y-axis shows Active RAS ELISA assay to determine the reduction in RAS-GTP abundance following MRTX849 treatment in MIA PaCa-2 cells for 24 hours. The vehicle value was normalized to 1 by dividing all average values by the vehicle value. Right y-axis shows quantitation of KRAS band shift by MRTX849 treatment in MIA PaCa-2 cells for 24 hours as assessed by western blot and densitometry.
(E) In-Cell Western blot assay to evaluate modulation of pERK in MIA PaCa-2 cells grown in standard tissue culture conditions treated with MRTX849 over a time course.
(F) CellTiter-Glo assay to evaluate cell viability performed on seven KRASG12C-mutant cell lines and three non-KRASG12C-mutant cell lines cells grown in 2D tissue culture conditions in a 3-day assay (left panel) or 3D conditions using 96-well, ULA plates in a 12-day assay (right panel).
We next determined the cellular activity of MRTX849 utilizing the KRASG12C-mutant H358 lung and MIA PaCa-2 pancreatic cancer cell lines. In both models, MRTX849 demonstrated an upward electrophoretic mobility shift of KRASG12C protein band migration by immunoblot, indicative of covalent modification of KRASG12C. A maximal mobility shift was observed by 1 hour, was maintained through 72 hours (Fig 1B and Fig S1B), and was evident at concentrations as low as 2 nM with near maximal modification observed at 15.6 nM (Figure 1C and S1C). Comparable inhibition of active RAS was observed as determined by a Raf RAS-binding domain (RBD) capture ELISA assay (Figure 1D and S1D). MRTX849 also inhibited KRAS-dependent signaling targets including ERK1/2 phosphorylation (Thr202/Tyr204 ERK1; pERK), S6 phosphorylation (RSK-dependent Ser235/236; pS6) and expression of the ERK-regulated DUSP6, each with IC50 values in the single-digit nanomolar range in both cell lines (Figure 1B, 1C, S1B and S1C). The evaluation of pERK over a time course of 48 hours indicated maximal inhibition was observed at 24 hours (Figure 1E and S1E). Treatment with the des-acrylamide version of MRTX849, which is unable to covalently bind to KRASG12C, did not demonstrate significant inhibition of ERK phosphorylation (Figure S1F). The H358 cell line was selected for determination of MRTX849 cysteine selectivity utilizing a LCMS-based proteomics approach able to detect approximately 6000 cysteine-containing peptides. After treatment for 3 hours, decreased KRASG12C Cys12 free peptide was detected with treated-to-control ratios of 0.029 and 0.008 determined at 1 μM and 10 μM, respectively, indicating near complete engagement of the intended target (Table S3). In contrast, the only other peptides identified were from lysine-tRNA ligase (KARS) at Cys209 near the detection limit indicating a high degree of selectivity toward KRASCYS12.
To evaluate the breadth of MRTX849 activity, its effect on cell viability was determined across a panel of 17 KRASG12C-mutant and three non-KRASG12C-mutant cancer cell lines using 2D (3-day, adherent cells) and 3D (12-day, spheroids) cell growth conditions. MRTX849 potently inhibited cell growth in the vast majority of KRASG12C-mutant cell lines with IC50 values ranging between 10 and 973 nM in the 2D format and between 0.2 and 1042 nM in the 3D format (Table S4 and Figure 1F). In agreement with prior KRASG12C inhibitor studies [5], MRTX849 demonstrated improved potency in the 3D assay format, as all but one KRASG12C-mutant cell line exhibited an IC50 value below 100 nM. Although MRTX849 was broadly effective in inhibiting viability of KRASG12C-mutant cell lines, IC50 values varied across the cell panel by 100-fold suggesting a differential degree of sensitivity to treatment. All three non-KRASG12C-mutant cell lines tested demonstrated IC50 values greater than 1 μM in 2D conditions and greater than 3 μM in 3D conditions suggesting the effect of MRTX849 on cell viability was dependent on the presence of KRASG12C.
To determine if the difference in sensitivity across the cell panel correlated with the ability of MRTX849 to bind to KRAS or inhibit KRAS-dependent signal transduction, seven KRASG12C-mutant cancer cell lines were selected from the panel for further evaluation. In each cell line, MRTX849 demonstrated a very similar concentration-dependent electrophoretic mobility shift (IC50) for KRASG12C protein migration suggesting that the ability to bind to and modify KRASG12C does not readily account for differences in response in viability studies (Figure 1B, 1C, S1B, S1C, S2A, and S2B). The effect of MRTX849 on selected phospho-proteins implicated in mediating KRAS-dependent signaling was also evaluated across the cell panel by immunoblot and/or reverse phase protein array (RPPA) following treatment for 6 or 24 hours. Notably, the concentration-response relationship and maximal effect of MRTX849 on inhibition of ERK and S6S235/236 phosphorylation varied across the cell panel (Figure S2A, S2C and Table S7). MRTX849 demonstrated only partial inhibition of phosphorylated ERK in KYSE-410 and SW1573 and a minimal effect on pS6S235/236 in SW1573, H2030, and KYSE-410 cells (Figure S2A and S2C). Each of these cell lines were among those that exhibited a submaximal response to MRTX849 in both 2D and 3D viability assays (Figure 1F). Although KRAS is implicated in mediating signal transduction through the PI3 Kinase and mTOR pathways, there was minimal evidence of a significant and/or durable effect of MRTX849 on Akt (S473, T308) or 4E-BP-1 (T37/T46, S65, T70) phosphorylation at any time point in any cell lines evaluated (Figure S2D). However, MRTX849 demonstrated concentration-dependent partial inhibition of the mTOR-dependent signaling targets, p70 S6 kinase (T412) and/or pS6 (S240/44), in the H358, MIA PaCa-2, H2122, and H1373 cell lines; each of which exhibited a maximal response to treatment. Together, these data suggest that maximizing inhibition of KRAS-dependent ERK and S6 signaling may be required to elicit a robust response in tumor cell viability assays.
MRTX849 Treatment In Vivo Leads to Dose-Dependent KRASG12C Modification, KRAS Pathway Inhibition and Anti-tumor Efficacy
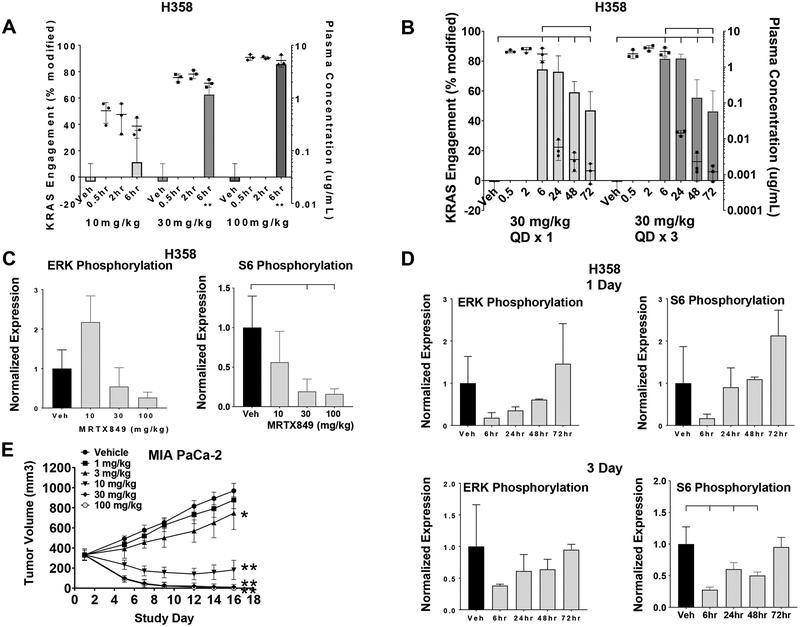
Studies were conducted to evaluate MRTX849 anti-tumor activity along with its pharmacokinetic and pharmacodynamic properties in vivo both to understand the clinical utility of this agent and to provide insight toward response to treatment. MRTX849 demonstrated moderate plasma clearance and prolonged half-life following oral administration (Table S1 and Figure S3). To evaluate the pharmacodynamic response to MRTX849 and to correlate drug exposure with target inhibition, MRTX849 was administered via oral gavage over a range of dose levels to H358 xenograft-bearing mice, and plasma and tumors were collected at defined time points. The fraction of covalently-modified KRASG12C protein was proportional to the plasma concentration of MRTX849 (Figure 2A). When evaluated over time after a single oral dose at 30 mg/kg the modified fraction of KRASG12C was 74% at 6 hours post-dose and gradually decreased to 47% by 72 hours (Figure 2B). This extended pharmacodynamic effect, despite declining levels of MRTX849 in plasma, was consistent with the irreversible inhibition of KRASG12C by MRTX849 and the relatively long half-life for the KRASG12C protein (~24 – 48 hours) (Table S5). The modification of KRASG12C was maximized after repeated daily dosing for 3 days at 30 mg/kg (Figure 2B) and higher dose levels did not demonstrate additional KRASG12C modification in multiple tumor models (data not shown). The maximum level of modification of ~80%, despite increasing dose and plasma levels of MRTX849 suggests that accurate measurement of complete inhibition of KRASG12C utilizing LCMS may not be attainable potentially due to a pool of active, non-cycling, or unfolded KRASG12C protein in tumors. Together, these studies demonstrated a dose-dependent increase in covalent modification of KRASG12C by MRTX849 and that the majority of targetable KRAS was covalently modified by MRTX849 over a repeated administration schedule at dose levels at or exceeding 30 mg/kg.
Figure 2.
MRTX849 modifies KRASG12C and inhibits KRAS signaling and tumor growth in vivo.
(A) MRTX849 was administered orally as a single dose to mice bearing established H358 xenografts (average tumor volume ~350 mm3) at 10, 30 and 100 mg/kg. KRAS modification and MRTX849 plasma concentration data from n=3 mice are shown as mean +/− standard deviation (SD). KRASG12C modification was statistically significant vs vehicle control using the two-tailed Student’s t-test. “**” indicates p-value < 0.01.
(B) MRTX849 was administered orally as a single dose or daily for three days to mice bearing established H358 xenografts (average tumor volume ~350 mm3) at 30 mg/kg. Plasma was collected 0.5, 2, 6, 24, 48 and 72 hours post administration of the last dose and tumors were collected 6, 24, 48 and 72 hours post dose. KRASG12C modification and MRTX849 plasma concentration data are shown from n=3 mice as mean +/− SD. Induction of modified KRASG12C protein at all time points was determined to be statistically significant vs. vehicle control using two-way ANOVA. In addition, induction of modified KRASG12C protein at 72 hours in Day 1 samples and 48 and 72 hours in Day 3 samples was statistically significant vs the 6-hour time point. Brackets indicate p-value < 0.05 as compared from left-most sample.
(C) MRTX849 was administered as in (A). Tumors were collected six hours post dose and total and phosphorylated ERK1/2 and total and phosphorylated S6 were analyzed by immunoblot and quantified by densitometric analysis. Relative fluorescent intensity of pERK1/2 and pS6 were normalized by dividing pERK1/2 and pS6 by total ERK1/2 and total S6, respectively. Vehicle tumors were normalized to 1 by dividing all average values by the vehicle value. Average pERK1/2 and pS6 values were divided by the average value in vehicle-treated tumors. Data shown represent the average of 2–3 tumors per treatment group plus SD. Reduction of pS6 relative fluorescent intensity was determined to be statistically significant vs vehicle control using the two-tailed Student’s t-test. Brackets indicate p-value <0.05 compared to left-most sample.
(D) MRTX849 was administered as in (B). Tumors were collected 6, 24, 48 or 72 hours post administration of the last dose and total and phosphorylated ERK1/2 and total and phosphorylated S6 were analyzed as in (C). Data shown represent the average of 3–4 tumors per treatment group plus SD. Reduction of pS6 relative fluorescent intensity on Day 3 was determined to be statistically significant vs vehicle control using two-way ANOVA. Brackets indicate p-value < 0.05 compared to left-most sample.
(E) MRTX849 was administered via daily oral gavage at the doses indicated to mice bearing established MIA PaCa-2 xenografts. Dosing was initiated when tumors were ~350 – 400 mm3. MRTX849 was administered to mice daily until Day 16. Data are shown as mean tumor volume +/− standard error of the mean (SEM). Tumor volumes at Day 16 were determined to be statistically significant vs vehicle control two-tailed Student’s t-test. “**” indicates p-value < 0.01. “*” indicates p-value < 0.05.
To evaluate the effect of MRTX849 on KRAS-dependent signal transduction in vivo, a single dose of MRTX849 at 10, 30 or 100 mg/kg was administered to H358 tumor-bearing mice. Dose-dependent inhibition of ERK1/2 and pS6S235/36 phosphorylation was observed at 6 hours post-dose based on immunoblot and densitometric analysis (Figure 2C). MRTX849 also demonstrated marked inhibition of ERK1/2 and S6S235/36 phosphorylation after one or three daily doses at 6 or 24 hours and levels gradually recovered by 72 hours after the final dose (Figure 2D). pERK1/2 and pS6S235/36 were further evaluated in formalin-fixed, paraffin-embedded sections from vehicle and MRTX849-treated xenografts in four tumor models utilizing immunohistochemical (IHC) methods coupled with image analysis algorithms. These studies demonstrated increased pERK1/2 and pS6 in non-tumor/stromal cells following MRTX849 administration indicating that immunoblotting studies with bulk tumor lysate likely underrepresent the degree of pathway inhibition in tumor cells, whereas IHC-based evaluation may more accurately reflect both the degree and spatial impact of pathway inhibition. Maximal inhibition was observed for both ERK and S6S235/36 phosphorylation after a single dose at the six-hour time point with a rebound in signaling evident 24 hours post-single dose in each model (Figure S4). Marked inhibition of ERK phosphorylation was observed at 6 hours post administration with 89%, 94%, and 94% inhibition observed compared with vehicle controls in MIA PaCa-2, H1373, and H2122 tumors, respectively (H358 pERK not quantifiable). This indicates that dose levels at or exceeding 30 mg/kg dose maximized inhibition of ERK phosphorylation in multiple models (Figure S4A and S4B). Inhibition of S6 phosphorylation at 6 hours was more variable with percent inhibition values of 76%, 50%, 86%, and 56% observed in MIA PaCa-2, H1373, H358, and H2122 tumors, respectively (Figure S4B). Together, these data indicate consistent acute (6 hr) inhibition of KRAS-dependent ERK phosphorylation was maximized in all evaluated models, whereas, inhibition of S6S235/36 was more variable, presumably due to varying degrees of KRAS-independent activation of this pathway in different tumor models.
MIA PaCa-2 and H358 were selected as MRTX849-responsive tumor models, thereby enabling a high-resolution understanding of dose-response relationships. Significant, dose-dependent, anti-tumor activity was observed at the 3, 10, 30 and 100 mg/kg dose levels in the MIA PaCa-2 model (Figure 2E). Evidence of rapid tumor regression was observed at the earliest post-treatment tumor measurement and animals in the 30 and 100 mg/kg cohorts exhibited evidence of a complete response at study Day 15. Dosing was stopped at study Day 16 and all 4 mice in the 100 mg/kg cohort and 2 out of 7 mice in the 30 mg/kg cohort remained tumor-free through study Day 70 (Figure S5A). In a second MIA PaCa-2 study, dose-dependent anti-tumor efficacy was observed at the 5, 10 and 20 mg/kg dose levels and 2/5 mice at the 20 mg/kg dose level exhibited complete tumor regression (Figure S5B). Significant dose-dependent anti-tumor efficacy was also observed in the H358 model, including 61% and 79% tumor regression at the 30 and 100 mg/kg dose levels, respectively, at Day 22 (Figure S5C). MRTX849 was well-tolerated and no effect on body weight was observed at all dose levels evaluated (Figure S5D). These studies indicated that MRTX849 demonstrated dose-dependent anti-tumor efficacy over a well-tolerated dose range and that the maximally efficacious dose of MRTX849 is between 30–100 mg/kg/day.
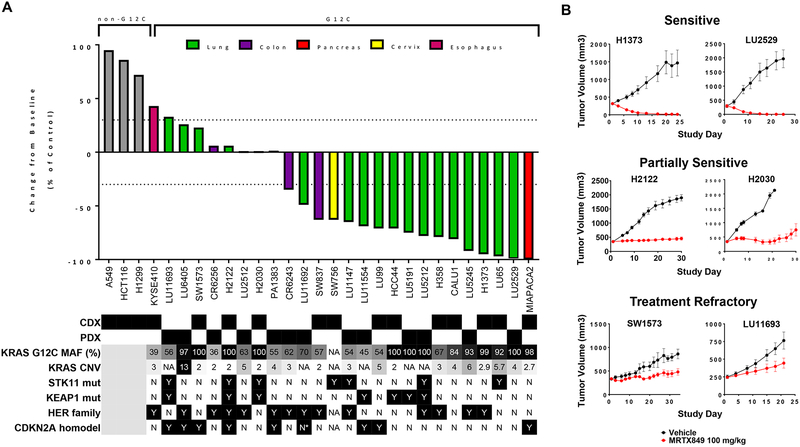
MRTX849 Demonstrates Broad Spectrum Tumor Regression in KRASG12C Cell Line and Patient-Derived Xenograft Models
To evaluate the breadth of anti-tumor activity across genetically and histologically heterogenous KRASG12C-mutant cancer models, MRTX849 was evaluated at a fixed dose of 100 mg/kg/day (a dose projected to demonstrate near maximal target inhibition in most models) in a panel of human KRASG12C-mutant cell line- (CDX) and patient-derived xenograft (PDX) models. MRTX849 demonstrated tumor regression exceeding 30% volume reduction from baseline in 17 out of 26 models (65%) at approximately three weeks of treatment (Figure 3A and Table S6). By comparison, MRTX849 did not exhibit significant anti-tumor activity at 100 mg/kg in three non-KRASG12C-mutant models (Figure 3A and Table S6). Together, these results indicate that KRASG12C-mutant tumors are broadly dependent upon mutant KRAS for tumor cell growth and survival and that MRTX849 elicits anti-tumor activity through a KRASG12C-dependent mechanism.
Figure 3.
Anti-tumor activity of MRTX849 in KRASG12C-mutant and non KRASG12C-mutant human tumor xenografts models.
(A) MRTX849 was administered via oral gavage at 100 mg/kg QD to mice bearing the cell line xenograft or PDX model indicated. Dosing was initiated when tumors were, on average, ~250 – 400 mm3. MRTX849 was formulated as a free base and resuspended as a solution in 10% Captisol, 50 mM citrate buffer, pH 5.0. The % change from baseline control was calculated at Day 19–22 for most models. Statistical significance was determined for each model and is shown in Table S6. Status of mutations and alterations in key genes are shown below each model. MAF (%) - Percent KRASG12C-mutant allele fraction by RNAseq; CNV – Copy number variation; * denotes very high CDK4 expression by RNAseq and possible amplification. HER family status was determined by averaging EGFR, ERBB2 and ERBB3 RNAseq expression for CDX (CCLE) or PDX (Crown huBase) models. Positive HER family calls denote greater than the median expression of the models tested. CDX and PDX model HER family calls were determined independently.
(B) Tumor growth inhibition plots from representative xenograft models that were categorized as sensitive, partially sensitive and treatment refractory.
While MRTX849 exhibited marked anti-tumor responses in the majority of models tested, a response pattern ranging from delayed tumor growth to complete regression was observed across the xenograft panel. The response to treatment was categorized as sensitive, partially sensitive and treatment refractory (Figure 3B). Rank order and Pearson statistical analyses were performed to evaluate the correlation between in vitro potency (IC50 in 2D or 3D viability assays) and anti-tumor response in vivo (% regression or progression on Day 22) and a significant correlation between response in cell lines compared with tumor models was not observed (Figure S6A and S6B). Thus, we focused on a comprehensive analysis of correlates with MRTX849 tumor response in vivo including tumor histology, co-occurring genetic alterations, as well as baseline or drug-induced changes in expression of KRAS-related genes (RNAseq) and/or protein signaling networks (RPPA [28] in 18 models) (Figure S7). No individual genetic alteration, including, but not limited to, KRAS mutant allele frequency, TP53, STK11, or CDKN2A predicted the anti-tumor activity of MRTX849. Interestingly, baseline gene and/or protein expression of selected members of the HER family of RTKs and of regulators of early cell cycle transition did exhibit a trend with the degree of anti-tumor response suggesting these pathways may influence the response to KRAS inhibitors (Figure S7A). Together, these data indicate that there are no individual binary biomarkers that clearly predict therapeutic response and that the molecular complexity and heterogeneity present in distinct KRAS mutated tumors may contribute to the response to target blockade.
MRTX849 Antitumor Activity Translates to RECIST Responses in Cancer Patients
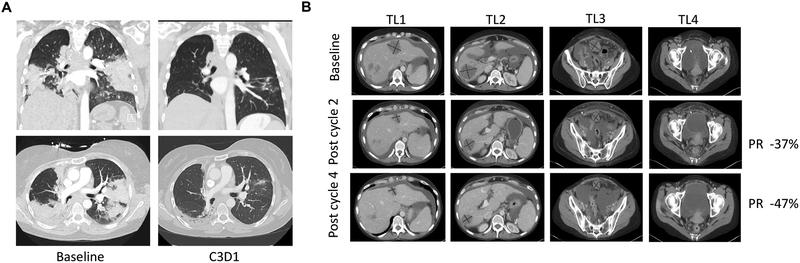
A 45-year old female former-smoker diagnosed with stage IV lung adenocarcinoma and refractory to multiple lines of therapy including carboplatin/pemetrexed/pembrolizumab, docetaxel, and investigational treatment with binimetinib and palbociclib was enrolled onto the MRTX849–001 Phase1b clinical trial with 2 bilateral lung lesions and mediastinal lymph node as target lesions. Targeted next-generation sequencing (NGS) demonstrated a KRAS G12C mutation (c.34G>T). In addition, loss-of-function KEAP1 (K97M) and STK11 (E223*) mutations were detected and are predicted to be deleterious to their respective proteins. The patient was administered MRTX849 (600 mg BID) and had marked clinical improvement within 2 weeks, including complete resolution of baseline cough and oxygen dependency. A RECIST-defined partial response of 33% reduction of target lesions was observed at Cycle 3 Day 1 (45 Days) and the patient continues on study (Figure 4A).
Figure 4.
Activity of MRTX849 in Lung and Colon Cancer Patients
(A) Pretreatment and 6-week scans of a heavily pretreated patient with a KRASG12C mutation-positive lung adenocarcinoma indicating 33% reduction of target lesions. Patient continues on study. The top panels show a coronal view and bottom panels an axial view of computed tomography (CT) chest images prior to MRTX849 treatment (left) and after two cycles of MRTX849 treatment (right).
(B) Baseline, 6-week (Cycle 2) and 12-week (Cycle 4) scans of a patient with a KRASG12C mutation-positive colon adenocarcinoma. Partial response confirmed at Cycle 4 and patient continues on study. Four lesions (TL1–4) are shown with axial views of CT images prior to MRTX849 treatment (top), after two cycles of MRTX849 treatment (center), and after four cycles of MRTX849 treatment (bottom).
A 47-year old female never-smoker with metastatic adenocarcinoma of the left colon who exhibited progressive disease after receiving multiple lines of systemic therapy including FOLFOX plus bevacizumab, single agent capecitabine, plus bevacizumab, and an investigational antibody drug conjugate was enrolled into the MRTX849–001 Phase1b clinical trial. This patient had extensive metastases involving the liver, peritoneum, ovaries and lymph nodes. Targeted NGS identified a KRAS G12C mutation. The patient was administered MRTX849 (600 mg BID) and demonstrated marked clinical improvement within 3 weeks and a visible decrease in size of her umbilical Sister Mary Joseph’s nodule. Her CEA levels decreased from 77 ng/ml at baseline to 11 ng/ml at Cycle 2 Day 1 and 3 ng/ml by Cycle 3 day 1 (normal range 0–5 ng/ml). A RECIST-defined partial response with 37% reduction of target lesions and complete response of a non-target lesion was observed at Cycle 3 Day 1 (Day 42). Confirmatory CT scans were conducted at Cycle 5, Day 1 (Day 84) and indicated a confirmed RECIST partial response with further reduction of target lesions at −47% from baseline (Figure 4B). The patient remains on treatment through Cycle 6.
Temporal Effects of MRTX849 on KRAS-Dependent Signaling and Feedback Pathways and Relationship to Anti-tumor Activity Following Repeat Dosing in Xenograft Models
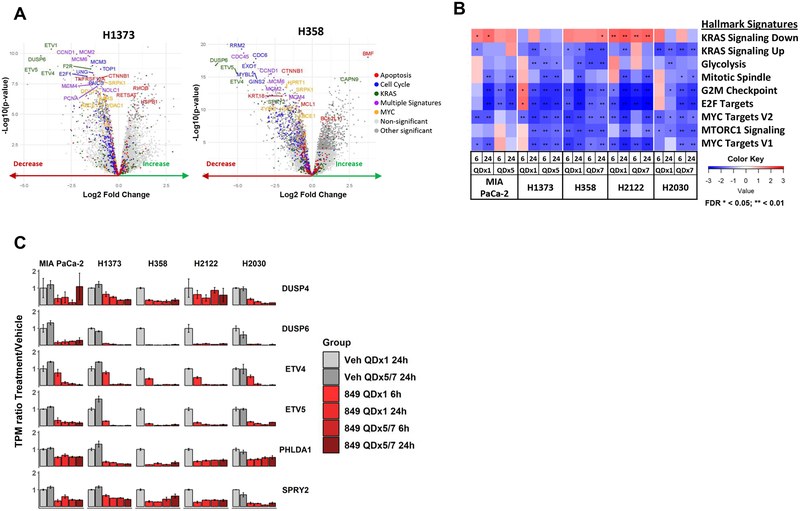
A comprehensive analysis was conducted to evaluate MRTX849-induced temporal molecular changes to further interrogate mechanisms of drug response across sensitive and partially sensitive models. To evaluate temporal changes in global gene expression, xenograft-bearing mice were administered vehicle or 100 mg/kg MRTX849 and RNAseq was performed on tumors at 6 and 24 hours post-treatment. Gene expression was evaluated at Day 1 and Day 5 for the sensitive models MIA PaCa-2 and H1373 to ensure sufficient tissue availability from regressing tumors, or at Day 7 in the partially sensitive models H358, H2122 and H2030 to coordinate with tumor stasis plateau. The top differentially expressed Gene Set Enrichment Analysis (GSEA) hallmark gene sets, regardless of tumor response, in all five models were several KRAS-annotated gene sets confirming MRTX849 selectively inhibits multiple genes directly related to KRAS signaling. MYC, mTOR, cell cycle, and apoptosis/BCL-2 pathway gene sets were also strongly differentially expressed confirming MRTX849 broadly impacted multiple well-established, KRAS-regulated pathways, several of which have proven difficult to directly inhibit with previous targeted therapies (Figure 5A, 5B, S8A–D). The marked impact of MRTX849 on a large number of genes that regulate cell cycle and apoptosis provides further insight into molecular mechanisms which mediate its antitumor activity.
Figure 5.
MRTX849 treatment in vivo regulates KRAS-dependent oncogenic signaling and feedback inhibitory pathways.
(A) Volcano plots displaying differentially expressed genes in xenograft tumors 24 hours after oral administration of vehicle or 100 mg/kg MRTX849 in a representative MRTX849-sensitive (H1373) and MRTX849-partially sensitive (H358) model. Significance denoted in the legend (adjusted p-value < 0.01).
(B) GSEA heatmaps depicting Hallmark Signature pathways differentially regulated in at least one model 24 hours following oral administration of a single 100 mg/kg MRTX849 dose compared to vehicle. Normalized Enrichment Score shown in all models 6 or 24 hours after a single dose (QDx1) or five (QDx5) or seven (QDx7) days dosing.
(C) Genes that feedback inhibit MAP kinase signaling are down regulated following MRTX849 treatment in all five cell line xenografts assessed by RNA-sequencing.
Targeted RNAseq analysis was performed on genes implicated in the temporal regulation of external signaling inputs and feedback pathways which collectively temper signaling flux through the RAS/RAF/MEK/ERK MAP kinase (MAPK) pathway including DUSP, SPRY and Pleckstrin Homology Like Domain Family A Member (PHLDA) family genes [13, 18]. These MAPK pathway negative regulators were each ranked among the most strongly decreased genes following MRTX849 treatment providing evidence that ERK-dependent transcriptional output is blocked and that pathways involved in reactivation of RTK- and ERK -dependent signaling were activated (Figure 5C and S4A).
Based on the observation of dynamic changes in transcriptional programs linked to KRAS pathway reactivation, IHC plus quantitative imaging of tumor cell-specific phospho-ERK and -S6 was evaluated over a range of time points. In the sensitive MIA PaCa-2 and H1373 tumor models, treatment with MRTX849 (100 mg/kg) demonstrated ≥90% inhibition of ERK phosphorylation at 6 and 24 hours on both Day 1 and 5 (Figure S4). In contrast, in the partially sensitive H358 and H2122 models, robust inhibition of ERK phosphorylation was observed at 6 hours after a single dose; however, marked recovery of ERK phosphorylation was observed at 24 hours post-single dose and at both 6 and 24 hours following 7 days of repeat-dose administration. Because DUSP, SPRY, and ETV family transcripts remain downregulated through 5–7 days in all models, it is evident that other independent factors contribute to temporal reactivation of ERK (Figure 5C). Similar to what was observed with single dose administration, the effect of MRTX849 on phosphorylated S6 was variable over time and did not track with the anti-tumor activity of MRTX849. Together, these results suggest that the extent and duration of inhibition of phosphorylated ERK may track with the magnitude of anti-tumor efficacy of KRASG12C inhibitors and that further evaluation of the role of the S6 is required to understand if it plays a role in drug sensitivity.
The effect of MRTX849 on cell proliferation and apoptosis was characterized by IHC analysis of Ki67 or cleaved caspase-3 after a single dose or repeat administration. The fraction of Ki67-positive cells was significantly reduced in tumors after repeat administration in all four models tested further supporting a broadly operative anti-proliferative mechanism, independent of the magnitude of MRTX849 anti-tumor response (Figure S4). Induction of apoptosis as determined by cleaved caspase-3 immunostaining was also evident on Day 1 of treatment (6 and/or 24 hr post-treatment) in the sensitive H358, MIA PaCa-2, and H1373 models (79–100% maximal regression) but not in the partially sensitive H2122 model (Figure S4). An expanded RPPA-based pathway analysis of several models also indicated a correlation between anti-tumor activity of MRTX849 and decreased survivin (statistically significant at Day 5/7 in 7 models evaluated) (Figure S7B) and a trend toward increased cleaved caspase-3 induction (Day 1, p = 0.08, 16 models) supporting the induction of apoptosis as a key mediator of a cytoreductive anti-tumor response (Figure S7C). Interestingly, the magnitude of reduction of MYC and cyclin B1 protein levels at Day 5/7 also closely correlated with MRTX849 anti-tumor activity, consistent with their roles as a critical regulators of KRAS-mediated cell growth and survival pathways (Figure S7B). Collectively, these data support that durable inhibition of ERK activity and that maximal inhibition of ERK-regulated outputs including MYC and E2F-mediated transcription are associated with induction of apoptosis and maximal response to MRTX849 treatment.
CRISPR/Cas9 Screen Identifies Vulnerabilities and Modifiers of Response to MRTX849 in KRASG12C-Mutant Cancer Cell Lines In Vitro and In Vivo
The correlative analysis of genomic or proteomic markers with response to MRTX849 in the defined panel of models provided only limited insight toward mechanism of therapeutic response or resistance. Therefore, we directly interrogated the role of selected genes in mediating therapeutic response utilizing a focused CRISPR/Cas9 knockout screen targeting approximately 400 genes including many genes involved in KRAS signaling. This was conducted in H358 and H2122 cells in vitro and in H2122 xenografts in vivo in presence and absence of MRTX849 treatment (Figure S9A–F). In MRTX849-anchored screens in vitro, sgRNAs that target RAS signaling pathways genes including MYC, SHP2 (H2122), mTOR pathway (MTOR and RPS6), and cell cycle genes (CDK1, CDK2, CDK4/6 and RB1) were identified to impact cell fitness. sgRNAs that target KEAP1 and Cbl were enriched in the H2122 model demonstrating cell-specific genetic routes towards improved fitness through loss of classical tumor suppressor genes, including in the context of MRTX849 treatment. KRAS sgRNA dropout was less pronounced in the MRTX849-treated cells compared with DMSO control-treated cells as would be expected with redundant depletion of the drug target (Figure S9C and S9D). To evaluate whether a distinct KRAS dependence or modulation of MRTX849 therapeutic response was observed in vitro vs in vivo, xenograft-bearing mice bearing H2122 cells (~250 mm3) transduced with the sgRNA library were orally administered vehicle or MRTX849 for two weeks (Figure S9A, S9E and S9F). In MRTX849-treated xenografts, sgRNAs targeting cell cycle, SHP2, MYC and mTOR pathway genes remained among the top depleted sgRNAs demonstrating inhibition of these targets in vivo, in the context of KRAS inhibition, leads to further tumor growth inhibition over and above the effects of KRAS inhibition alone (Figure S9E and S9F). sgRNAs targeting the tumor suppressor KEAP1 were enriched in MRTX849-treated xenografts suggesting loss of KEAP1 may represent a mechanism of intrinsic or acquired resistance. Interestingly, NRAS was one of the top enriched genes in the vehicle-treated xenografts suggesting NRAS functions as a tumor suppressor in this context, however enrichment was not as pronounced in the MRTX849-treated xenografts suggesting NRAS may compensate for KRAS in the context of KRAS inhibition (Figure S9F). Collectively, these data demonstrate the importance of selected proteins that regulate RTK and RAS-dependent signaling and cell cycle transition in mediating the oncogenic effects of mutant KRAS, and also provide a catalogue of potentially druggable vulnerabilities that complement KRAS blockade.
Cancer Therapeutic Combination Screen to Identify Rational and Clinically Tractable Strategies to Address Feedback and Resistance Pathways
To further interrogate pathways that mediate the anti-tumor response to MRTX849 and to identify combinations capable of enhancing response to MRTX849, a combination screen was conducted in vitro using a focused library of small molecule inhibitors across a panel of cell lines (Figure S10A, S10B, and Table S8). Approximately 70 compounds targeting relevant pathways (RTKs, MAPK/ERK, PI3K, mTOR, cell cycle) were tested in a 3 or 7-day viability assay and synergistic combinations were identified and ranked. Multiple hits from this screen were then identified for additional evaluation in combination studies with MRTX849, including the HER family inhibitor afatinib, the CDK4/6 inhibitor palbociclib, the SHP2 inhibitor RMC-4550, and mTOR pathway inhibitors.
Combination Strategies That Target Upstream Signaling Pathways Implicated in Extrinsic Regulation of KRAS Nucleotide Cycling and Feedback/Bypass Pathways
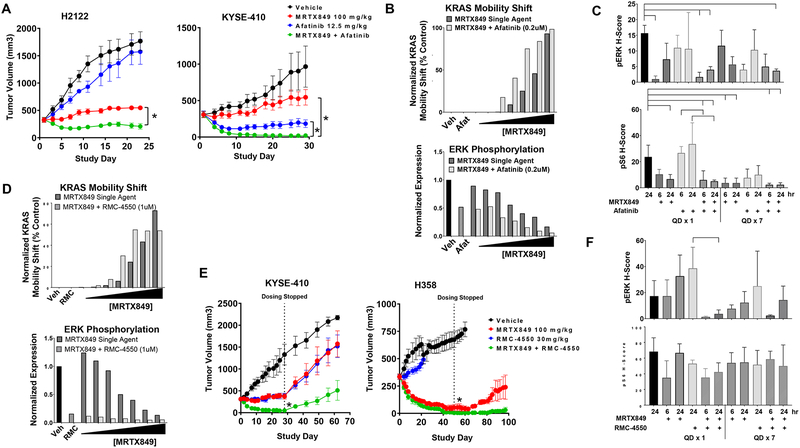
MRTX849 in combination with HER family inhibitors synergistically inhibited tumor cell viability in the majority of cell lines evaluated and were the top hit in the combination screen in vitro (Figure S10). Cell lines with highest (top 50th percentile) average composite baseline RNA expression values of selected HER family members exhibited the highest synergy scores to these combinations (Figure S11A). Afatinib was selected as a prototype HER family inhibitor based on its broad in vitro combination activity. Combination studies were conducted with MRTX849 and afatinib in five tumor models that were partially sensitive or treatment refractory to single agent MRTX849. The MRTX849 and afatinib combination demonstrated significantly greater anti-tumor efficacy compared with either single agent in all five models evaluated including multiple models exhibiting complete or near complete responses to the combination (Figure 6A and S11B
Figure 6.
HER family and SHP2 inhibitor combinations further inhibit KRAS signaling and exhibit increased anti-tumor responses.
(A) MRTX849 at 100 mg/kg, afatinib at 12.5 mg/kg or the combination was administered daily via oral gavage to mice bearing the H2122 or KYSE-410 cell line xenografts (n=5). Combination treatment led to a statistically significant decrease in tumor growth compared to either single agent treatment. “*” denotes adjusted p-value < 0.01.
(B Quantification of KRAS mobility shift and pERK in H2122 cells treated for 24 hours with MRTX849 (0.1 – 73 nM), afatinib (200 nM) or the combination assessed by western blot densitometry.
(C) MRTX849 at 100 mg/kg, afatinib at 12.5mg/kg, or the combination was administered once or daily for 7 days via oral gavage to mice bearing H2122 cell line xenografts (n=3/group). Tumors were harvested at 6 and 24 hours following the final dose. Tumor sections were stained with pERK and pS6 via immunohistochemistry methods. Quantitation of images shown by H-score in tumor tissue. Reduction of pERK or pS6 staining intensity was determined to be statistically significant relative to vehicle or either single agent using one-way ANOVA. Brackets indicate p-value <0.05 compared to left-most sample.
(D) Quantitation of KRAS band shift and pERK after 24 hour treatment with MRTX849 (0.1 – 73 nM), RMC-4550 (1 uM) or the combination in H358 cells assessed by western blot densitometry.
(E) MRTX849 at 100 mg/kg, RMC-4550 at 30 mg/kg or the combination was administered daily via oral gavage to mice bearing the KYSE-410 or H358 cell line xenografts (n=5/group). Combination treatment led to a statistically significant reduction in tumor growth compared to either single agent on the last day of dosing. “*” denotes adjusted p-value < 0.05.
(F) MRTX849 at 100 mg/kg, RMC-4550 at 30mg/kg, or the combination was administered via oral gavage to mice bearing KYSE-410 cell line xenografts (n=3/group) and tumors were harvested at 6 and 24 hours post dose. Tumor sections were stained with pERK, pS6 via immunohistochemistry methods. Quantitation of images shown by H-score in tumor tissue. Reduction of pERK staining intensity was determined to be statistically significant relative to RMC-4550 alone using one-way ANOVA. Brackets indicate p-value <0.05 compared to left-most sample.
To evaluate whether afatinib impacted covalent modification of KRASG12C by MRTX849, partially sensitive H2122 cells were treated with increasing concentrations of MRTX849 alone or in the presence of afatinib (200 nM, IC90) and the mobility shift in KRAS protein was densitometrically determined from immunoblots. A clear shift in the concentration-response to MRTX849 was apparent in the presence of afatinib indicating that the combination increased the fraction of modified KRASG12C consistent with the putative role of HER family receptors in extrinsic regulation of KRASG12C GTP loading (Figure 6B). The concentration-response relationship for inhibition of ERK phosphorylation was also clearly shifted in the presence of afatinib. To further evaluate the effect of the combination on KRAS-dependent signaling, four cell lines (H2030, H2122, H358, and KYSE-410) were treated over a range of MRTX849 concentrations in presence or absence of afatinib for 6 or 24 hours and key signaling molecules were evaluated by RPPA. Afatinib demonstrated clear inhibition of EGFR (pY1068) and HER2 (pY1248) activity and partial inhibition of ERK, Akt (S473), and p70S6K phosphorylation at both time points (Figure S11C). The effect of afatinib on S6 (S235/236, S240/244) and p90 RSK (S380) phosphorylation was variable and exhibited only minimal inhibition in most of the cell lines evaluated. The combination of afatinib and MRTX849 demonstrated markedly enhanced concentration-dependent inhibition and/or a greater magnitude of effect on ERK, RSK, p70 S6K, and S6 (S235/236) phosphorylation compared with MRTX849 alone at both 6 and 24 hours. Of note, neither afatinib, nor MRTX849 alone inhibited S6 phosphorylation at the S240/244 site regulated by mTOR/S6K whereas the combination demonstrated marked inhibition at 24 hours.
In vivo, the combination also exhibited a trend towards increased pERK and pS6 (S235/236) inhibition in the partially sensitive H2122 model in combination groups as determined by quantitation of immunostaining after one- or seven-days administration (Figure 6C). Similar results were observed in the MRTX849-refractory KYSE-410 model and the combination also increased the number of apoptotic cells in this model (Figure S12A–C). Collectively, these data indicate that upstream baseline HER family activation may limit the ability of MRTX849 to achieve robust inhibition of the ERK and mTOR/S6 signaling pathways. Accordingly, the combination of afatinib and MRTX849 can limit feedback reactivation of ERK and demonstrate complementary inhibition of Akt/mTOR/S6 signaling resulting in significantly improved anti-tumor activity.
SHP2 inhibition has been shown to inhibit the growth of cells that harbor KRASG12C mutations and this effect is likely mediated, in part, by decreasing KRAS GTP loading [25–27]. To evaluate whether SHP2 inhibition enhanced covalent modification of KRASG12C by MRTX849, H358 and H2122 cells were incubated with increasing MRTX849 concentrations with or without the SHP2 inhibitor, RMC-4550. In both cell lines, co-treatment with RMC-4550 (1 μM, IC90) demonstrated a MRTX849 concentration-dependent increase in KRASG12C protein modification and concomitant decrease in ERK phosphorylation compared with MRTX849 alone (Figure 6D and S13A). RPPA analysis of KRAS-dependent signaling was conducted at 6 or 24 hours post treatment in three cell lines (H358, H2030, H2122) over a range of MRTX849 concentrations in the presence or absence of RMC-4550. RMC-4550 demonstrated robust inhibition of ERK phosphorylation and partial inhibition of p90 RSK (S380) and p70 S6K (T412) at both time points (Figure S13B). The combination of RMC-4550 and MRTX849 demonstrated incrementally increased concentration-dependent inhibition of ERK and RSK phosphorylation in all cell lines at both 6 and 24 hours and markedly improved inhibition of S6 (S235/236) phosphorylation compared with MRTX849 alone in H2122 and H358 cells at 24 hours. In addition, the combination demonstrated near complete inactivation of KRAS in MRTX849 refractory KYSE-410 xenografts as determined using an active RAS ELISA assay and this was significant compared with single agents (Figure S13C). Based on these findings, combination studies were conducted with MRTX849 and RMC-4550 in six KRASG12C-mutated tumor models in vivo and the combination demonstrated significantly greater anti-tumor efficacy compared with either single agent in 4/6 models evaluated (Figure 6E and S13D). Consistent with the in vitro data, the combination also demonstrated a significant decrease in ERK phosphorylation compared with either single agent in the KYSE-410 model as determined by quantitation of tumor cell immunostaining on Day 1 at 6 and 24 hours and Day 7 at 6 hours post-dose (Figure 6F). Together, these data indicate that EGFR family and SHP2 blockade can augment the anti-tumor activity of KRASG12C inhibitors through enhancing covalent target modification and establishing a more comprehensive blockade of KRAS-dependent signaling.
Combinations That Inhibit Bypass Pathways Downstream of KRAS and Exhibit Increased Anti-tumor Activity in Xenograft Models
KRAS is implicated in regulation of the oncogenic S6 protein translation pathway through both ERK-dependent activation of RSK, which phosphorylates S6 at Ser235/236, and through crosstalk with the PI3 kinase and mTOR pathway that additionally phosphorylates S6 at Ser240/244 [29]. However, the S6 pathway can also be activated independently of mutated KRAS in tumor cells through hyperactivated RTK signaling, PI3 kinase activation, or STK11 mutations each of which converge on mTOR-mediated activation of S6. In the in vitro combination screen, mTOR inhibitors demonstrated synergy in a subset of evaluated cell lines (Figure S14A). To further evaluate the effect of the combination on KRAS and mTOR pathway-dependent signaling, four cell lines were treated with MRTX849 in presence or absence of the selective ATP-competitive mTOR inhibitor, vistusertib (1 μM) for 6 or 24 hours and several signaling molecules were evaluated by RPPA. Vistusertib demonstrated clear and robust inhibition of several components of the PI3 kinase/mTOR signaling pathway including Akt (S473), p70 S6K (T412), S6 (pS235/236, S240/244), and 4E-BP1 (S65, T70) phosphorylation in each cell line at both time points consistent with its mechanism of action (Figure S14B). MRTX849 alone did not affect 4E-BP1 or S6 (S240/244) activity and it exhibited a variable and cell line-dependent effect on p70 S6K and S6 (pS235/236) phosphorylation in these cell lines. Vistusertib also demonstrated marked induction of ERK phosphorylation, often several fold over vehicle control, at both time points in all four cell lines consistent with prior reports [30]. The combination of vistusertib and MRTX849 demonstrated a comparable level of inhibition of ERK phosphorylation compared to single agent MRTX849 indicating that the activation of ERK signaling by vistusertib was impeded by the combination of the two agents. In addition, MRTX849 combined with vistusertib demonstrated further inhibited p70 S6K and Akt S473 phosphorylation compared with either single agent. Near complete inhibition of S6 (S235/236, S240/244) phosphorylation at limit of detection was observed for the combination in each cell line at evaluated time points.
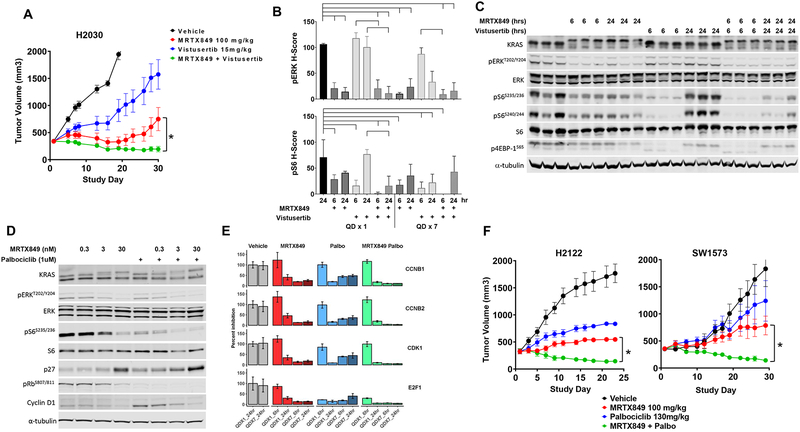
Consequently, a cohort of tumor models were identified and the combination of MRTX849 with the selective mTOR inhibitor vistusertib demonstrated marked tumor regression and significantly improved anti-tumor activity compared with either single agent in all six models evaluated (Figure 7A and S14C). MRTX849 in combination with a second, differentiated mTOR inhibitor everolimus, which inhibits TORC1 but not TORC2, in the H2030 xenograft model also demonstrated a striking combination effect (Figure S14D). In the KRASG12C, STK11-mutant H2030 model, MRTX849 demonstrated marked inhibition of ERK phosphorylation through 24 hours, but only exhibited partial inhibition of pS6235/36 at 6 hours post-dose, on Day 1 and 7 (Figure 7B and 7C). Vistusertib demonstrated marked inhibition of pS6235/36 at 6 hours post-treatment with evidence of recovery by 24 hours. The combination of vistusertib and MRTX849 did not have a further effect on ERK phosphorylation but demonstrated a significant reduction in pS6235/36 at Day 1 at 24 hours compared with vistusertib alone and a trend towards reduced pS6235/36 at both Day 1 and Day 7 at 6 hours compared with either single agent (Figure 7B and S14E). Together, these data indicate that MRTX849 and mTOR inhibitor combination demonstrates complementary inhibition of the ERK and mTOR/S6 signaling pathways resulting in broad anti-tumor activity in KRASG12C-mutant tumor models.
Figure 7.
CDK4/6 and mTOR combinations suppress independently hyperactivated downstream pathways and exhibit increased anti-tumor responses.
(A) MRTX849 at 100 mg/kg, vistusertib at 15 mg/kg or the combination was administered daily via oral gavage to mice bearing the H2122 or H2030 cell line xenografts (n=5/group). Combination treatment led to a statistically significant decrease in tumor growth compared to either single agent treatment. “*” denotes adjusted p-value < 0.05.
(B) MRTX849 at 100 mg/kg, vistusertib at 15 mg/kg, or the combination was administered once or daily for 7 days via oral gavage to mice bearing H2030 cell line xenografts (n=3/group). Tumors were harvested at 6 and 24 hours following the final dose. Tumor sections were stained with pERK and pS6 via immunohistochemistry methods. Quantitation of images shown by H-score in tumor tissue. Reduction of pERK or pS6 staining intensity was determined to be statistically significant relative to vehicle or either single agent using one-way ANOVA. Brackets indicate p-value <0.05 compared to left-most sample.
(C) Protein western blot analysis of KRAS pathway targets in H2030 xenografts treated with MRTX849 (100 mg/kg), vistusertib (15 mg/kg) or the combination, six or 24 hours after a single dose.
(D) Protein western blot analysis of KRAS pathway and cell cycle targets in H2122 cells treated for 24 hours with MRTX849, palbociclib or the combination.
(E) Normalized RNAseq gene expression data on E2F targets in H2122 xenografts treated with MRTX849, palbociclib or the combination, six and 24 hours after a single or seven daily doses.
(F) MRTX849 at 100 mg/kg, palbociclib at 130 mg/kg or the combination was administered daily via oral gavage to mice bearing the H2122 or SW1573 cell line xenografts (n=5). Combination treatment led to a statistically significant decrease in tumor growth compared to either single agent treatment. “*” denotes adjusted p-value < 0.05
Signaling through KRAS is known to mediate cell proliferation, at least in part, through the regulation of the cyclin D family and triggering Rb/E2F-dependent entry of cells into cell cycle. Loss of function mutations and homozygous deletions in the cell cycle tumor suppressor CDKN2A (p16) are co-incident in a subset of KRAS-mutant NSCLC and hyperactivate CDK4/6-dependent Rb phosphorylation and cell cycle transition. In the CDKN2A-null H2122 and SW1573 cell lines in vitro, MRTX849 demonstrated concentration-dependent partial inhibition of Rb phosphorylation (pRB pS807/811) and concurrent increase in p27 in H2122, but not SW1573 cells, at 24 hours (Figure 7D and S15A). MRTX849 in combination with the CDK4/6 inhibitor palbociclib (1 μM), demonstrated near complete inhibition of pRb in both H2122 and SW1573 cells and further induced p27 in H2122 cells. Interestingly, pS6 (S235/236) was also much more effectively suppressed by the combination in both H2122 and SW1573 cells, which is consistent with a recent report [31]. RNA expression of target genes and RPPA analysis of target protein signaling events was also used as a readout of cell cycle inhibition in the H2122 tumor model in vivo, and the combination of MRTX849 and palbociclib significantly inhibited E2F1 and selected E2F family target genes and induced p27 protein expression to a greater degree compared to either single agent and further reduced the number of Ki67 positive cells after 7 days of administration (Figure 7E, S15B and S15C). In addition, the combination demonstrated a significant decrease in pRB (S780) compared with either single agent after 7 days of administration in SW1573 tumors in vivo (Figure S15D). This combination also induced tumor regression in five tumor xenograft models which was significant compared with either single agent control (Figure 7F and S15E). While not significant, a trend was noted in which models with CDKN2A homozygous deletion exhibited an increased anti-tumor response to the combination of MRTX849 and CDK4/6 inhibition compared with models lacking evidence of genetic dysregulation of key cell cycle genes (Figure S15F and S15G).
Discussion
The identification of MRTX849 as a highly selective KRASG12C inhibitor capable of near complete inhibition of KRAS in vivo provides a renewed opportunity to better understand the role of this mutation as an oncogenic driver in various cancers and to guide rational clinical trial design. The lack of a significant correlation between sensitivity to MRTX849 anti-tumor activity in in vitro versus in vivo model systems made it necessary to further study KRAS oncogene dependence in tumor models in vivo, a more clinically relevant setting. The demonstration that MRTX849 exhibited significant anti-tumor efficacy in all evaluated KRASG12C-mutated cancer models and demonstrated marked regression in the majority (65%), confirms that this mutation is a broadly operative oncogenic driver and that MRTX849 represents a compelling therapeutic opportunity. This evidence of activity extended to patients as demonstrated by RECIST partial responses in two patients enrolled in a Phase I clinical trial of MRTX849. Collectively; however, these data also illustrate that the degree of dependence of cancer cells on presence of a KRASG12C mutation for growth and survival can vary across tumors and that co-occurring genetic alterations observed in KRAS-mutated cancers may influence response to direct targeted therapy. The further observation that KRAS mutations occur across different cancers and that no single co-occurring genetic alteration predicted response to treatment illustrates the genetic heterogeneity of KRAS-driven cancers. Findings in the present studies are consistent with other functional genomics or therapeutic strategies to block KRAS function across panels of cell lines or models which demonstrated a highly significant response of KRAS mutant cells to target knockdown, a heterogenous magnitude of response, and no clear co-occurring aberrations that predict resistance to target blockade [5, 32, 33]. Interestingly, despite the implication that certain mutations that co-occur with KRAS including TP53, STK11, and KEAP1 may limit therapeutic response in KRASG12C-positive lung cancers, none of these mutations correlated with response or resistance in the cell line panel. In addition, the partial response we reported in the lung adenocarcinoma patient was observed in a patient harboring deleterious co-mutations in both STK11 and KEAP1. Together, these data further illustrate the heterogeneity and complexity of KRAS-mutated cancers and suggest that no binary co-occurring genetic event may be predictive of therapeutic response.
Temporal and dose-response analysis indicated maximal modification of KRASG12C and durable inhibition of KRAS-dependent signaling was important in maximizing therapeutic response. The recovery of ERK signaling and the inability to inhibit mTOR/S6 signaling despite continued treatment were each associated with transient or submaximal response to MRTX849. ERK1/2 is implicated in direct phosphorylation and negative feedback regulation of EGFR (T669), FGFR1 (S777), and SOS1 and each of these targets may facilitate KRASG12C-independent resetting of ERK signaling flux [34–36]. The rapid and remarkable suppression of ERK pathway-regulated transcripts such as DUSP and SPRY/SPRED family members by MRTX849 in all models evaluated is consistent with that observed for RAF inhibitors and is implicated in reactivation of ERK and RTK signaling [18, 19]. The dual-specificity phosphatases, DUSP4 and 6, were strongly suppressed by MRTX849 and are implicated in dephosphorylating and inactivating ERK1/2 [14, 18, 37] whereas Spry family members are implicated in the negative regulation of RTKs, adaptor proteins (e.g., Grb2), and may participate in modifying RAS family nucleotide exchange and effector binding (e.g., Raf1) [38]. Although suppression of DUSP and SPRY/SPRED were broadly observed in all models, the magnitude of signaling reactivation and response to MRTX849 varied across models. This suggests some tumor models harbor additional factors that bypass KRAS dependence or impact RAS pathway signaling flux, such as expression or activation of selected RTKs (e.g., ERBB2 amplification in the KYSE-410 model) or STK11 loss-of-function mutations and may be primed for feedback reactivation of RAS-dependent signaling and/or limit the degree of signaling inhibition by MRTX849. This phenomenon was observed for BRAF V600E mutant colon cancer (but not melanoma) which exhibits high baseline EGFR expression, is primed for rapid feedback activation of this RTK, and is resistant to single agent inhibition but highly responsive to co-targeting BRAF (and/or MEK) and EGFR [20]. In addition, blockade of BRAF or MEK1/2 resulted in feedback-mediated activation of the PI3 kinase/mTOR signaling pathway in concert with the co-activation of upstream RTKs (e.g., EGFR) resulting in bypass of ERK pathway dependence and therapeutic resistance [17, 20, 39]. The observation that baseline expression of HER family RTKs trended with MRTX849 anti-tumor activity and that CRISPR-based drug-anchored screens implicated EGFR, SHP2, and mTOR/S6 pathways as co-targetable vulnerabilities both support the hypothesis that these targets act as conditional response modifiers.
Activation of RTK signaling in the context of KRASG12C-mutant cancer was predicted to limit MRTX849 therapeutic response both by enhancing extrinsic regulation of GTPase activity and initiating KRAS-independent ERK and mTOR/S6 pathway activation. Therefore, HER family and SHP2 inhibition were employed as strategies to either block the critical RTK family in KRAS-mutant cells or to block collective RTK signaling downstream, respectively. As MRTX849 only binds GDP-KRASG12C, both HER family and SHP2 inhibition each enhanced KRASG12C modification by MRTX849 and significantly improved anti-tumor activity. This observation is consistent with the putative role of activated RTKs in the engagement of SHP2 to mediate SOS1-dependent RAS GTP loading and to diminish RAS GAP activity each of which converge on enhanced RAS activation state [40]. The afatinib combination demonstrated a clear and marked inhibition of both the ERK/RSK and Akt/mTOR/S6 signaling pathways whereas the SHP2 inhibitor combination demonstrated a clear impact on ERK/RSK signaling and relatively less prominent impact on mTOR/S6 signaling. Although afatinib may more effectively address mTOR/S6 bypass signaling, SHP2 inhibition should be an effective combinatorial strategy to combat other RTKs outside of the HER family, such as FGFRs or MET, that could impact KRAS dependence. To further address bypass signaling mediated by RTK activation or STK11 mutations, each of which activate the mTOR/S6 signaling pathway independently of KRAS, mTOR inhibition in combination with MRTX849 was also evaluated. MRTX849 in combination with vistusertib, in fact, demonstrated significantly improved anti-tumor activity in vivo compared with either single agent in all six tumor models evaluated, regardless of STK11 mutational status. Consistent with the mechanism of action of vistusertib, comprehensive inhibition of Akt/mTOR/S6 signaling was observed for vistusertib alone and near complete inhibition of pS6S235/36 & 240/44 was observed in combination. In addition, the marked feedback reactivation of ERK by vistusertib was relieved by the combination. The induction of ERK activity has been observed in tumor cells following mTORC1 inhibition by rapalogs or ATP-competitive inhibitors and has been implicated in limiting antitumor activity of this class of agents [30, 41, 42] supporting the suppression of ERK signaling by MRTX849 as a key mechanism of response to the combination. Notably, all three combination strategies converge on more comprehensive inhibition of KRAS-dependent signaling converging on ERK and S6 activity. In addition, although the inhibition of the Akt/mTOR/S6 did not correlate with model response to MRTX849 (potentially due to tumor heterogeneity), the observation that both MTOR and RPS6 drop out in drug anchored CRISPR screens and that effective combination strategies more comprehensively block this pathway illustrate its likely importance in maximizing therapeutic response in KRAS mutated cancers.
Cell cycle dysregulation due to genetic alterations in cell cycle regulators identified additional factors that could modify the therapeutic response to MRTX849. In addition, CDKN2A, RB1, CDK4 and CDK6 were all identified as gene targets that impacted cell fitness in CRISPR screens. Genetic alterations including homozygous deletion of CDKN2A or amplification of CDK4 or CCND1 comprise up to 20% of KRAS-mutated NSCLC [43]. Combination studies with MRTX849 and palbociclib in vivo demonstrated more comprehensive inhibition of Rb and E2F family target genes and increased anti-tumor activity compared with either single agent in NSCLC models. In addition, these studies indicated that the combination resulted in more effective inhibition of S6 (S235/236) phosphorylation establishing a previously unappreciated connection between cell cycle blockade and protein translation pathways. Notably, this combination was especially effective in CDKN2A-deleted models suggesting that this combination strategy may be primarily beneficial in a molecularly-defined subset of patients characterized by decoupling of cell cycle regulation from KRAS.
Collectively, models exhibiting a cytoreductive response to single agent MRTX849 demonstrated a more comprehensive and durable inhibition of KRAS-dependent signaling and induction of an apoptotic response. These data suggest maintaining durable inhibition of KRAS-dependent signaling below a defined threshold is required to elicit tumor regression. The elucidation of mechanisms that limit the therapeutic response to single agent KRAS inhibition has provided insight toward strategies to enhance therapeutic activity in KRAS-mutant tumors. Of the 35% of models (9/26) that did not exhibit durable regression with single agent MRTX849 treatment, 5 models (KYSE410, SW1573, H2122, H2030, LU6405) were selected for rational combination studies and at least one combination demonstrated significant improvement in anti-tumor efficacy and elicited a >50% tumor regression in all five models evaluated. These results suggest essentially all KRASG12C-mutated cancers can derive clinical benefit from direct KRAS inhibitor-directed therapy either alone or in combination. Furthermore, rational pathway-centric combination regimens directed at hallmark signaling nodes may be directed to genetically-defined patient subsets. For example, KRAS-mutated NSCLC exhibits mutually exclusive, co-occurring genetic alterations in STK11 and CDKN2A [43]. The present data suggest that KRASG12C/STK11-mutated NSCLC could be readily addressed by combining a KRASG12C inhibitor with an RTK or mTOR inhibitor whereas KRASG12C/CDKN2A-mutated NSCLC could be more effectively addressed by combining with a CDK4/6 inhibitor. Collectively, the present studies support the broad utility of covalent KRASG12C inhibitors in treating KRASG12C mutated cancers and provide defining strategies to identify patients likely to benefit from single agent or rationally directed combinations.
Material and Methods
Reagents and Cell Lines
MRTX849 was synthesized at Array Biopharma, Inc., (Boulder, CO) or WuXi AppTec (Wuhan, China). MRTX849 in powder form was stored at room temperature and protected from light. MRTX849 was formulated in 100% DMSO and aliquoted for long term storage at −20°C. Roswell Park Memorial Institute 1640 medium (RPMI; #11875–093), Dulbecco’s Modified Eagle’s Me1dium (DMEM; #10566–016), penicillin and streptomycin (#15070–063), HEPES ((4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid); #15630–080), Dulbecco’s Phosphate-buffered Saline (DPBS; #14190–136) and sodium pyruvate (#11360–070) were obtained from Gibco/Thermo Fisher Scientific (Waltham, MA). Fetal Bovine Serum (FBS) was obtained from Corning (#35–011-CV, Corning, NY) and Nucleus Biologics (1824–001, San Diego, CA). MIA-PaCa-2, NCI-H358, SW837, NCI-H2122, SW756, Calu-1, SW1573, NCI-H1373, NCI-H2030, NCI-H1792, NCI-H23, UM-UC-3, A549, H1299 and HCT 116 cell lines were obtained from American Type Culture Collection (ATCC; Manassas, VA) between April 2014 and August 2015. Cell lines are abbreviated without the NCI prefix for brevity. The HCC-44 cell line was obtained in August 2015 from Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ; Braunschwieg, Germany). The LU99, LU65, and IALM cell lines were obtained in September 2017 from RIKEN (Tokyo, Japan). The KYSE-410 cell line was obtained in August 2015 from Sigma/Millipore (St. Louis, MO). Human cancer cell lines were maintained at 37°C in a humidified incubator at 5% CO2 and were periodically checked for mycoplasma. Cell lines used for in vivo studies were confirmed pathogen and mycoplasma-free by IMPACT 1 assessment (IDEXX BioAnalytics) prior to implant. Cell lines were carried for no more than 15 cell passages in this work.
In Vivo Studies
All mouse studies were conducted in compliance with all applicable regulations and guidelines of the Institutional Animal Care and Use Committee (IACUC) from the National Institutes of Health (NIH). Mice were maintained under pathogen-free conditions, and food and water was provided ad libitum. 6 – 8-week-old, female, athymic nude-Foxn1nu mice (Envigo, San Diego) were injected subcutaneously with tumor cells in 100 μl of PBS and Matrigel matrix in the right hind flank with 5.0e6 cells (Corning #356237; Discovery Labware, MA) 50:50 cells:Matrigel. Mouse health was monitored daily, and caliper measurements began when tumors were palpable. Tumor volume measurements were determined utilizing the formula 0.5 × L × W2 in which L refers to length and W refers to width of each tumor. When tumors reached an average tumor volume of ~350 – 400 mm3, mice were randomized into treatment groups. Mice were treated by oral gavage with either vehicle consisting of 10% research grade Captisol (CyDex Pharmaceuticals, KS) in 50 mM citrate buffer pH 5.0 or MRTX849 in vehicle at indicated doses. For efficacy studies, animals were orally administered MRTX849 or vehicle and monitored daily, tumors were measured 3 times per week and body weights were measured 2 times per week. Study Day on efficacy plots indicates the day after which MRTX849 treatment was initiated.
For studies conducted at Crown Biosciences China, 4–5-week-old female BALB/c nude mice were implanted with tumor fragments 2–3 mm in diameter into the right flank via trocar implant.
For studies conducted at Crown Biosciences San Diego, tumor cells were thawed, washed in phosphate-buffered saline (PBS), counted and resuspended in cold PBS at between ~50,000 – 100,000 cells per 100 μL. Cell suspensions were mixed with an equal volume of Cultrex extracellular matrix (ECM) (Trevigen; Gaithersburg, MD; #3432-005-01) and kept on ice. 6 – 8-week-old, female, non-obese diabetic severe combined immunodeficiency (NOD scid) mice were shaved prior to injection and 100 μL of the ECM-cell mixture was injected in the rear flank using a chilled 26 7/8-gauge syringe.
Mice were randomized, and dosing was initiated when the mean tumor volume was between ~250 – 300 mm3. Five mice were dosed with either vehicle alone (10% Captisol in 10 mM citrate buffer, pH 5.0; Teknova, Hollister, CA; #Q2443) or 100 mg/kg MRTX849 daily by oral gavage for 21 days (n=5 per group).
Statistical analysis of differences in mean tumor volume between vehicle and MRTX849-treated cohorts was run using a two-tailed Student’s t-test with equal variance in Excel (Microsoft; Redmond, WA). P-value < 0.05 was considered statistically significant.
Supplementary Material
Significance.
The discovery of MRTX849 provides a long-awaited opportunity to selectively target KRASG12C in patients. The in-depth characterization of MRTX849 activity, elucidation of response and resistance mechanisms, and identification of effective combinations provides new insight toward KRAS dependence and the rational development of this class of agents.
Acknowledgments
The authors thank Channing J. Der and Adrienne D. Cox for critical review of the manuscript. The authors thanks Molecular Diagnostic Services and Crown Biosciences for animal study support and Flagship Biosciences for immunohistochemistry and image analysis. Support for collaborative research to evaluate the impact of KRAS co-mutations on KRAS inhibitor activity with the Lito laboratory was provided, in part, by NIH/NCI (1R01CA23074501; 1R01CA23026701A1), The Pew Charitable Trusts, and the Damon Runyon Cancer Research Foundation. J. X. is supported by a National Research Service Award from the NIH/NCI (1F30CA232549-01).
Footnotes
Disclosure of conflict of interest: J.H., L.D.E., L.H., A.C., R.A., D.M.B., N.S., V.B., K.V., R.C., J.B., M.A.M., P.O, and J.G.C. are employees and shareholders of Mirati Therapeutics, Inc. A.P. is a shareholder of Mirati Therapeutics, Inc. B.R.B. M.R.B., J.B.F., J.P.F., and G.P.V. are employees and shareholders of Array BioPharma. J.B. is a former employee and shareholders of Array BioPharma. MSKCC has received research funds from Mirati Therapeutics which have been applied, in part, to research directed by P.L. P.A.J. is a compensated Scientific Advisor for Mirati Therapeutics.
Clinical Trials
MRTX849 clinical trials () were conducted in accordance with recognized US ethical guidelines (i.e., U.S. Common Rule) and per local institutional review board guidelines. All patients included in the clinical trial were subjected informed written consent and consented prior to study enrolment. MRTX849 was administered in 21-Day cycles to patients included in present studies per protocol.
References
- 1.John J, Sohmen R, Feuerstein J, Linke R, Wittinghofer A, and Goody RS, Kinetics of interaction of nucleotides with nucleotide-free H-ras p21. Biochemistry, 1990. 29(25): p. 6058–65. [DOI] [PubMed] [Google Scholar]
- 2.Matikas A, Mistriotis D, Georgoulias V, and Kotsakis A, Targeting KRAS mutated non-small cell lung cancer: A history of failures and a future of hope for a diverse entity. Crit Rev Oncol Hematol, 2017. 110: p. 1–12. [DOI] [PubMed] [Google Scholar]
- 3.Sanchez-Vega F, Mina M, Armenia J, Chatila WK, Luna A, La KC, et al. , Oncogenic Signaling Pathways in The Cancer Genome Atlas. Cell, 2018. 173(2): p. 321–337 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simanshu DK, Nissley DV, and McCormick F, RAS Proteins and Their Regulators in Human Disease. Cell, 2017. 170(1): p. 17–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Janes MR, Zhang J, Li LS, Hansen R, Peters U, Guo X, et al. , Targeting KRAS Mutant Cancers with a Covalent G12C-Specific Inhibitor. Cell, 2018. 172(3): p. 578–589 e17. [DOI] [PubMed] [Google Scholar]
- 6.Ostrem JM, Peters U, Sos ML, Wells JA, and Shokat KM, K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature, 2013. 503(7477): p. 548–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patricelli MP, Janes MR, Li LS, Hansen R, Peters U, Kessler LV, et al. , Selective Inhibition of Oncogenic KRAS Output with Small Molecules Targeting the Inactive State. Cancer Discov, 2016. 6(3): p. 316–29. [DOI] [PubMed] [Google Scholar]
- 8.Ambrogio C, Kohler J, Zhou ZW, Wang H, Paranal R, Li J, et al. , KRAS Dimerization Impacts MEK Inhibitor Sensitivity and Oncogenic Activity of Mutant KRAS. Cell, 2018. 172(4): p. 857–868 e15. [DOI] [PubMed] [Google Scholar]
- 9.Burgess MR, Hwang E, Mroue R, Bielski CM, Wandler AM, Huang BJ, et al. , KRAS Allelic Imbalance Enhances Fitness and Modulates MAP Kinase Dependence in Cancer. Cell, 2017. 168(5): p. 817–829 e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cancer Genome Atlas, N., Comprehensive molecular characterization of human colon and rectal cancer. Nature, 2012. 487(7407): p. 330–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schabath MB, Welsh EA, Fulp WJ, Chen L, Teer JK, Thompson ZJ, et al. , Differential association of STK11 and TP53 with KRAS mutation-associated gene expression, proliferation and immune surveillance in lung adenocarcinoma. Oncogene, 2016. 35(24): p. 3209–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Skoulidis F, Goldberg ME, Greenawalt DM, Hellmann MD, Awad MM, Gainor JF, et al. , STK11/LKB1 Mutations and PD-1 Inhibitor Resistance in KRAS-Mutant Lung Adenocarcinoma. Cancer Discov, 2018. 8(7): p. 822–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brant R, Sharpe A, Liptrot T, Dry JR, Harrington EA, Barrett JC, et al. , Clinically Viable Gene Expression Assays with Potential for Predicting Benefit from MEK Inhibitors. Clin Cancer Res, 2017. 23(6): p. 1471–1480. [DOI] [PubMed] [Google Scholar]
- 14.Caunt CJ and Keyse SM, Dual-specificity MAP kinase phosphatases (MKPs): shaping the outcome of MAP kinase signalling. FEBS J, 2013. 280(2): p. 489–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Merchant M, Moffat J, Schaefer G, Chan J, Wang X, Orr C, et al. , Combined MEK and ERK inhibition overcomes therapy-mediated pathway reactivation in RAS mutant tumors. PLoS One, 2017. 12(10): p. e0185862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sturm OE, Orton R, Grindlay J, Birtwistle M, Vyshemirsky V, Gilbert D, et al. , The mammalian MAPK/ERK pathway exhibits properties of a negative feedback amplifier. Sci Signal, 2010. 3(153): p. ra90. [DOI] [PubMed] [Google Scholar]
- 17.Turke AB, Song Y, Costa C, Cook R, Arteaga CL, Asara JM, et al. , MEK inhibition leads to PI3K/AKT activation by relieving a negative feedback on ERBB receptors. Cancer Res, 2012. 72(13): p. 3228–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lito P, Pratilas CA, Joseph EW, Tadi M, Halilovic E, Zubrowski M, et al. , Relief of profound feedback inhibition of mitogenic signaling by RAF inhibitors attenuates their activity in BRAFV600E melanomas. Cancer Cell, 2012. 22(5): p. 668–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lito P, Rosen N, and Solit DB, Tumor adaptation and resistance to RAF inhibitors. Nat Med, 2013. 19(11): p. 1401–9. [DOI] [PubMed] [Google Scholar]
- 20.Prahallad A, Sun C, Huang S, Di Nicolantonio F, Salazar R, Zecchin D, et al. , Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature, 2012. 483(7387): p. 100–3. [DOI] [PubMed] [Google Scholar]
- 21.Chandarlapaty S, Sawai A, Scaltriti M, Rodrik-Outmezguine V, Grbovic-Huezo O, Serra V, et al. , AKT inhibition relieves feedback suppression of receptor tyrosine kinase expression and activity. Cancer Cell, 2011. 19(1): p. 58–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Misale S, Fatherree JP, Cortez E, Li C, Bilton SJ, Timonina D, et al. , KRAS G12C NSCLC models are sensitive to direct targeting of KRAS in combination with PI3K inhibition. Clin Cancer Res, 2019. 25(2): 769–807. [DOI] [PubMed] [Google Scholar]
- 23.Hunter JC, Manandhar A, Carrasco MA, Gurbani D, Gondi S, and Westover KD, Biochemical and Structural Analysis of Common Cancer-Associated KRAS Mutations. Mol Cancer Res, 2015. 13(9): p. 1325–35. [DOI] [PubMed] [Google Scholar]
- 24.Lito P, Solomon M, Li LS, Hansen R, and Rosen N, Allele-specific inhibitors inactivate mutant KRAS G12C by a trapping mechanism. Science, 2016. 351(6273): p. 604–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mainardi S, Mulero-Sanchez A, Prahallad A, Germano G, Bosma A, Krimpenfort P, et al. , SHP2 is required for growth of KRAS-mutant non-small-cell lung cancer in vivo. Nat Med, 2018. 24(7): p. 961–967. [DOI] [PubMed] [Google Scholar]
- 26.Nichols RJ, Haderk F, Stahlhut C, Schulze CJ, Hemmati G, Wildes D, et al. , RAS nucleotide cycling underlies the SHP2 phosphatase dependence of mutant BRAF-, NF1- and RAS-driven cancers. Nat Cell Biol, 2018. 20(9): p. 1064–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruess DA, Heynen GJ, Ciecielski KJ, Ai J, Berninger A, Kabacaoglu D, et al. , Mutant KRAS-driven cancers depend on PTPN11/SHP2 phosphatase. Nat Med, 2018. 24(7): p. 954–960. [DOI] [PubMed] [Google Scholar]
- 28.Baldelli E, Bellezza G, Haura EB, Crino L, Cress WD, Deng J, et al. , Functional signaling pathway analysis of lung adenocarcinomas identifies novel therapeutic targets for KRAS mutant tumors. Oncotarget, 2015. 6(32): p. 32368–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roux PP, Shahbazian D, Vu H, Holz MK, Cohen MS, Taunton J, et al. , RAS/ERK signaling promotes site-specific ribosomal protein S6 phosphorylation via RSK and stimulates cap-dependent translation. J Biol Chem, 2007. 282(19): p. 14056–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rastogi R, Jiang Z, Ahmad N, Rosati R, Liu Y, Beuret L, et al. , Rapamycin induces mitogen-activated protein (MAP) kinase phosphatase-1 (MKP-1) expression through activation of protein kinase B and mitogen-activated protein kinase kinase pathways. J Biol Chem, 2013. 288(47): p. 33966–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lou K, Steri V, Ge AY, Hwang YC, Yogodzinski CH, Shkedi AR, et al. , KRAS(G12C) inhibition produces a driver-limited state revealing collateral dependencies. Sci Signal, 2019. 12(583). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McDonald ER 3rd, de Weck A, Schlabach MR, Billy E, Mavrakis KJ, Hoffman GR, et al. , Project DRIVE: A Compendium of Cancer Dependencies and Synthetic Lethal Relationships Uncovered by Large-Scale, Deep RNAi Screening. Cell, 2017. 170(3): p. 577–592 e10. [DOI] [PubMed] [Google Scholar]
- 33.Tsherniak A, Vazquez F, Montgomery PG, Weir BA, Kryukov G, Cowley GS, et al. , Defining a Cancer Dependency Map. Cell, 2017. 170(3): p. 564–576 e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li X, Huang Y, Jiang J, and Frank SJ, ERK-dependent threonine phosphorylation of EGF receptor modulates receptor downregulation and signaling. Cell Signal, 2008. 20(11): p. 2145–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Porfiri E and McCormick F, Regulation of epidermal growth factor receptor signaling by phosphorylation of the ras exchange factor hSOS1. J Biol Chem, 1996. 271(10): p. 5871–7. [DOI] [PubMed] [Google Scholar]
- 36.Zakrzewska M, Haugsten EM, Nadratowska-Wesolowska B, Oppelt A, Hausott B, Jin Y, et al. , ERK-mediated phosphorylation of fibroblast growth factor receptor 1 on Ser777 inhibits signaling. Sci Signal, 2013. 6(262): p. ra11. [DOI] [PubMed] [Google Scholar]
- 37.Huang CY and Tan TH, DUSPs, to MAP kinases and beyond. Cell Biosci, 2012. 2(1): p. 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hanafusa H, Torii S, Yasunaga T, and Nishida E, Sprouty1 and Sprouty2 provide a control mechanism for the Ras/MAPK signalling pathway. Nat Cell Biol, 2002. 4(11): p. 850–8. [DOI] [PubMed] [Google Scholar]
- 39.Villanueva J, Vultur A, Lee JT, Somasundaram R, Fukunaga-Kalabis M, Cipolla AK, et al. , Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell, 2010. 18(6): p. 683–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang J, Zhang F, and Niu R, Functions of Shp2 in cancer. J Cell Mol Med, 2015. 19(9): p. 2075–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Svejda B, Kidd M, Kazberouk A, Lawrence B, Pfragner R, and Modlin IM, Limitations in small intestinal neuroendocrine tumor therapy by mTor kinase inhibition reflect growth factor-mediated PI3K feedback loop activation via ERK1/2 and AKT. Cancer, 2011. 117(18): p. 4141–54. [DOI] [PubMed] [Google Scholar]
- 42.Zeng Y, Tian X, Wang Q, He W, Fan J, and Gou X, Attenuation of everolimus-induced cytotoxicity by a protective autophagic pathway involving ERK activation in renal cell carcinoma cells. Drug Des Devel Ther, 2018. 12: p. 911–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, et al. , Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med, 2017. 23(6): p. 703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.