Abstract
Molecular barcoding technologies that uniquely identify single cells are hampered by limitations in barcode measurement. Readout by sequencing does not preserve the spatial organization of cells in tissues, whereas imaging methods preserve spatial structure but are less sensitive to barcode sequence. Here we introduce a system for image-based readout of short (20bp) DNA barcodes. In this system, called Zombie, phage RNA polymerases transcribe engineered barcodes in fixed cells. The resulting RNA is subsequently detected by fluorescent in situ hybridization. Using competing match and mismatch probes, Zombie can accurately discriminate single-nucleotide differences in the barcodes. This method allows in situ readout of dense combinatorial barcode libraries and single-base mutations produced by CRISPR base editors without requiring barcode expression in live cells. Zombie functions across diverse contexts, including cell culture, chick embryos, and adult mouse brain tissue. The ability to sensitively read out compact and diverse DNA barcodes by imaging will facilitate a broad range of barcoding and genomic recording strategies.
Editorial summary:
The spatial location and sequence of DNA barcodes are detected with high sensitivity in fixed tissues.
Molecular recording systems could revolutionize the study of development and disease by allowing reconstruction of dynamic, single-cell developmental histories from end-point measurements1. In these systems, individual cells actively record information within their genome by continuous editing of uniquely identifiable engineered genomic target sites, or ‘barcodes’2-9. Multiple methods that use CRISPR/Cas9 or site specific recombinases to produce barcode diversity have now been developed2-4,10-13. One of the most promising approaches is the use of CRISPR base editors, in which catalytically impaired Cas9 is fused to deaminases and other enzymes to target mutations to specific nucleotides without generating double stranded breaks8,9.
In these approaches, readout of barcode edits is most often done by sequencing, which is sensitive to single nucleotide variations and can be performed at high throughput. However, sequencing-based approaches disrupt spatial organization of cells within tissues, and often recover information only from a minority of cells14. The ability to accurately and efficiently read out single cell barcode edits in situ would link dynamic developmental history with spatial multicellular organization that is essential for the function of many biological systems.
Recent work has produced an explosion of methods for in situ detection of nucleic acids. These include strategies for combinatorially encoding a large diversity of transcripts15-18, techniques for amplifying signal from single mRNA molecules19-23, and approaches for in situ sequencing24-28. These methods could be used to detect barcodes transcribed in living cells prior to fixation. However, ensuring detectable barcode expression across a diverse population of living cells can be challenging due to stochastic silencing, bursty expression, and unintended cell-type dependent promoter activity. Eliminating the need for expression in living cells could therefore simplify the design of barcode systems. In addition, some methods only detect large scale differences in target sequence and therefore cannot access single nucleotide variations. For example, a recent demonstration of recording was based on detection of large scale barcode deletions2. Thus, a simple and effective strategy for discriminating barcode edits in fixed tissues has been lacking.
Here, we introduce an in situ detection method that is sensitive to single nucleotide edits and can be applied in diverse organismal contexts. It uses well-characterized RNA polymerases from the bacteriophages T3, T7, and SP6 to transcribe genomically integrated barcodes in fixed cells, producing an amplified RNA product that can then be detected using single molecule FISH (smFISH)19 or Hybridization Chain Reaction (HCR)21,29. Phage polymerases are known to be efficient and specific for their target promoters30, but have not, to our knowledge, been previously applied in fixed cells. Because it is based on ‘waking up’ otherwise transcriptionally ‘dead’ (silent) barcodes in fixed cells, we term the technique Zombie, for ‘Zombie is Optical Measurement of Barcodes by In situ Expression’. We showed that Zombie efficiently detects short (20bp) barcodes, accurately discriminates single nucleotide variants (SNVs), and detects edits made by base editors, without requiring endogenous expression. These capabilities allow for compact virally delivered combinatorial barcode libraries, and set the stage for future recording applications. Furthermore, the simplicity and robustness of this system enables it to function not only in cell culture but also in chick embryos and adult mouse brain tissues.
Results
Phage RNA polymerases can transcribe synthetic DNA barcodes in fixed cells
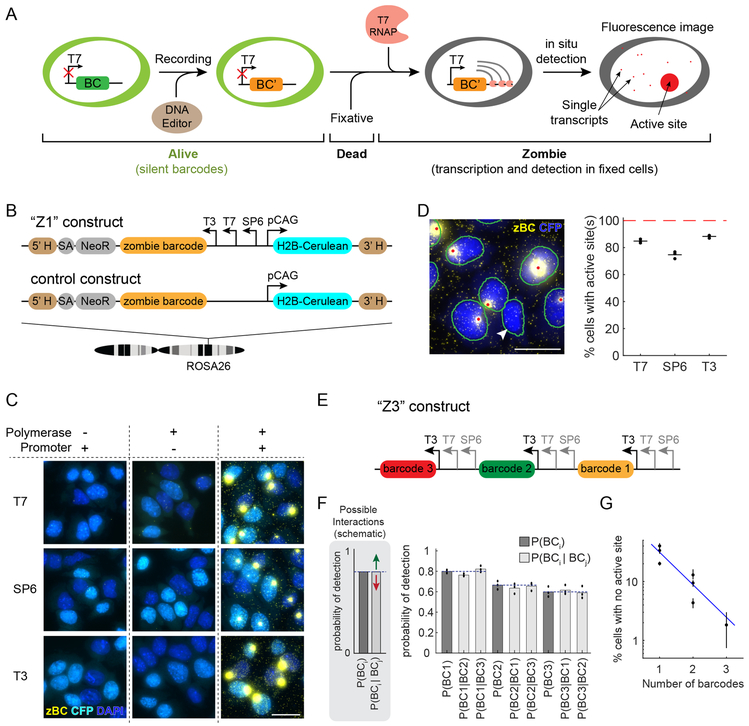
We first set out to develop a method for specifically amplifying and detecting barcodes integrated in the genome (Fig. 1A). We designed a construct, labeled Z1 (Fig. 1B), containing a previously described 900bp barcode sequence2 downstream of tandem SP6, T7, and T3 phage promoters, along with an H2B-Cerulean fluorescent protein under the control of the constitutive mammalian CAG promoter for imaging of cell nuclei. We integrated Z1 site-specifically at the ROSA26 locus in mouse embryonic stem (mES) cells. We also made a similar cell line with a control construct that lacks the phage promoters (Fig. 1B).
Figure 1. Phage RNA polymerases enable in situ readout of DNA barcodes without in vivo expression.
(A) Workflow for analysis of Zombie barcodes (left to right). First, barcode constructs containing a phage promoter, such as T7, that is inactive in live cells, are integrated in the genome. Second, and optionally, base editors or other DNA modifying enzymes (brown) can alter barcode sequence to increase barcode diversity. Third, cells are fixed and phage RNA polymerase (pink) is added. This enables transcription of the barcode to RNA (gray lines). RNA transcripts accumulate at the active site (large red dot), and also diffuse away from it (small red dots represent individual transcripts). (B) The Z1 construct was engineered to contain a barcode downstream of T3, T7, and SP6 phage promoters, and to express H2B-Cerulean fluorescent protein (CFP) in living cells from a divergently oriented mammalian promoter. Z1 was stably integrated in mouse ES cells at the ROSA26 locus (single integration per genome). This line was compared to a similar cell line containing the control construct lacking phage promoters. (C) Polyclonal control cells and Z1 cells (columns) were imaged with or without the indicated phage polymerases (rows). HCR was used to detect barcode RNA (zBC). Nuclei are visualized by native fluorescence of H2B-CFP (cyan) as well as DAPI staining (blue). Barcode transcripts appear only in Z1 cells with phage polymerase (yellow dots, right column). The experiment was independently repeated twice with similar results. Scale bar is 25 μm. (D) In monoclonal cultures, active sites can be detected in most cells (image). Nuclei (blue) and active sites (yellow) are segmented automatically (green outlines and red dots, respectively). One cell in this field of view does not show any active site (arrowhead). Scale bar is 25 μm. Percentages of cells with detectable active sites for each polymerase are shown on the right. Horizontal lines indicate the mean of replicates (n=3 biologically independent samples). Total of 3916 cells were analyzed, with at least 420 cells for each replicate. (E) The Z3 construct encodes three 900bp barcodes, each expressed from a distinct set of phage promoters. This construct was integrated at ROSA26, transcribed using T3 RNA polymerase, and imaged in all three color channels. T7 and SP6 promoters are shaded gray because they are not used in (F) and (G). Sizes of elements are not drawn to scale. (F) Schematic: Assuming independence, the conditional probability of detecting barcode i in a cell, given detection of another barcode (j), should equal the overall probability of barcode i detection, with deviations signifying either synergy (green arrow) or interference (red arrow) between barcodes. Bar plot: for Z3, the conditional probability analysis shows independent detection events for all three barcodes. Bars indicate mean of 3 replicates (points). (G) Fraction of Z3 cells with no detectable active sites declines with the number of barcodes analyzed, consistent with independent expression of different phage promoters in the same cell. Thus, detection efficiency can be increased with additional barcode copies. Dots represent the mean for different barcodes or barcode combinations and black vertical lines show the range over three replicates. Blue line indicates the exponential fit. Total of 564 cells were analyzed for plots in F and G.
To detect the barcode, we grew polyclonal populations of cells, fixed them, added each of the phage RNA polymerases, and performed HCR with a set of split initiator probes21 to detect RNA transcripts (see Methods for details). Fluorescence imaging revealed two types of dots: bright fluorescent dots within cell nuclei and more numerous, but considerably dimmer, diffraction-limited dots scattered throughout the nucleus and cytoplasm (Fig. 1C). Neither type of dot was observed when either the phage promoters or polymerase were omitted (Fig. 1C). Parental cells lacking a barcode exhibited no dots when cultured alone, but showed some overlapping dimmer dots when co-cultured with engineered cells (Fig. S1). These results suggest that the bright dots reflect phage polymerase-dependent transcription at the integration site, whereas the dimmer dots reflect individual transcripts that can diffuse away from the cell in which they were produced. Together, this barcode design and analysis protocol enable in situ expression and detection of genomically integrated barcodes at integration sites.
We next sought to quantify the efficiency of detection. We selected a monoclonal line with exactly one integration per diploid genome, termed mES-Z1. Within the clone, we consistently detected 1 or 2 bright dots in the majority of cells, likely due to cell cycle phase variation at the time of fixing, with a small fraction of cells missing any bright dots (Figs. 1D and S2). While we were able to detect the transcription active sites efficiently with all three phage RNA polymerases, the average detection efficiencies of T3 (88%) and T7 (85%) were higher than that of SP6 (75%) (Fig. 1D). Variations in efficiencies may reflect the relative positions of the promoters in the construct, relative amounts of active enzymes, as well as intrinsic differences between the polymerases.
A lack of barcode detection could result if certain cells are impermeable to polymerases or otherwise do not permit in situ transcription. Alternatively, it could reflect intrinsic stochasticity in the polymerization reaction. To distinguish these possibilities, we engineered a second line containing a single integration of a construct termed Z3, in which three barcodes are each controlled by a separate set of phage promoters and can be detected using distinct fluorescence channels (Fig. 1E). If non-detection is a property of the individual cells, we would expect to predominantly detect either all three barcodes or no barcodes (strong correlation). By contrast, in a stochastic transcription model, we would expect that detection of one barcode would not affect the probability of detecting another barcode (weak correlation).
Analysis of active site co-localization in 564 cells revealed no significant correlation or pairwise mutual information between any pair of barcodes (chi-squared test, p-values 0.7970, 0.1917, and 0.1256 for the three pairs; Fig. S3). The chance of detecting each barcode in a cell was independent of detection of the other barcodes (Fig. 1F). Consistent with this observation, the fraction of cells with no detected active sites declined exponentially with the number of barcodes analyzed in the same cell at the rate expected from the single barcode detection frequencies (Fig. 1G). Together, these data suggest that detection is a stochastic event that occurs independently at each barcode. Therefore, although a fraction of barcodes fail to produce detectable signal, the false negative rate per cell can be reduced by increasing the barcode copy number. This property may be valuable in the study of rare cell types, where capturing information from majority of cells is essential.
Zombie enables reliable in situ detection of 20bp DNA barcodes
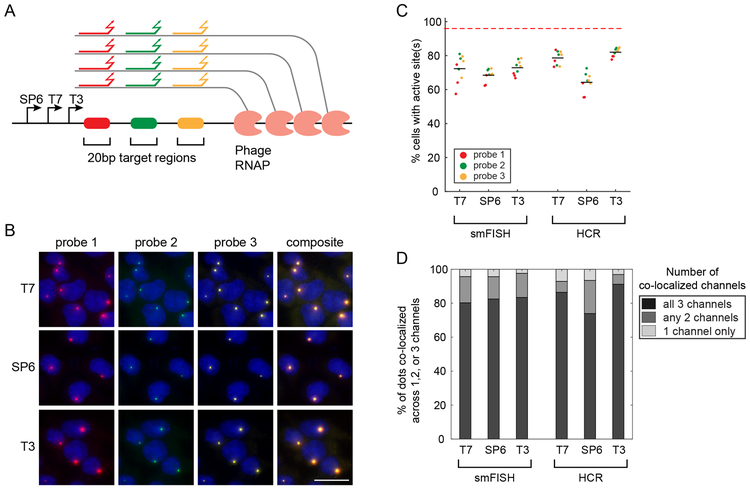
Barcode transcription produces multiple RNA molecules from the same template in close proximity, which effectively amplifies the barcode target and could facilitate robust detection of short barcodes. To test this, we hybridized fixed mES-Z1 cells after the in vitro transcription step with three orthogonal 20bp probes targeting regions downstream of the phage promoters (Fig. 2A). We then analyzed the binding of these probes, by both smFISH and HCR29,31. In both analyses, we observed easily detectable transcription active sites in all three channels (Figs. 2B and S4). For all three phage RNA polymerases, the active sites could be detected in a large fraction of cells (Fig. 2C), and most dots were redundantly detected in multiple channels (Fig. 2D). These results show that barcodes as short as 20bp can be efficiently and reliably detected in situ.
Figure 2. Reliable detection of short barcodes.
(A) Short probes (colored lines) target 20bp regions of the larger Z1 barcode sequence and can be detected in distinct fluorescence channels. Local accumulation of transcripts at the active site effectively amplifies signal and enables detection, even with a single probe per target site. (B) Z1 cells were treated with each polymerase (rows) and imaged in three channels (columns) after detection with individual fluorescently labeled probes (colors matching those in A). Final column shows composite images. The barcode in Z1 cells is integrated site-specifically at the ROSA26 locus. The experiment was independently repeated three times with similar results. Scale bar is 25 μm. (C) Signal from each individual probe can be detected in the majority of the cells by smFISH or HCR. Plot shows the percentage of Z1 cells with active sites detected using a single 20bp probe. Dots are color-coded based on probe identity. n=3 biologically independent samples. Lines show the average efficiency over three probes and three replicates. (D) Colocalization analysis shows that the majority of dots colocalize in multiple channels, indicating the reliability of single probe detection. For each condition, gray shades indicate fractions of dots that are detected in only 1 channel or co-detected in 2 or 3 channels. Data from three biologically independent samples are combined in each condition. For plots in C and D, total of 5097 cells were analyzed, with at least 669 cells for each condition.
Zombie enables in situ detection of single nucleotide mismatches
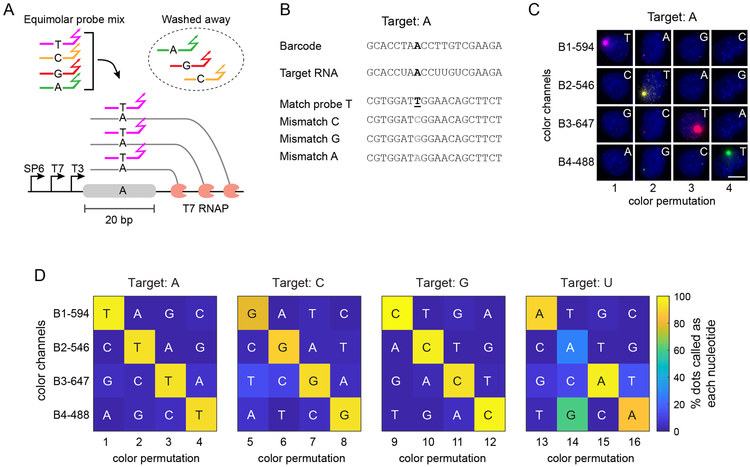
Discrimination of small sequence differences could facilitate imaging-based barcoding applications. While structured and toehold probes can be used to detect single nucleotide variations by leveraging base pairing within the probe32-36, traditional probes can bind to target sequences even when they contain a single nucleotide mismatch (Fig. S5)32. We hypothesized that simultaneously competing multiple probes, each containing a distinct nucleotide at a single site, for binding to the many transcripts present in an active site could lead to preferential binding of exact match probes over mismatch probes, and thereby enable nucleotide identification (Fig. 3A).
Figure 3. Probe competition accurately discriminates single nucleotide variants.
(A) Perfect match probes outcompete those with a single mismatch when an equimolar mixture of all 4 probe variants is used. This feature can be used to detect SNVs in situ. (B) Sequences of barcode, target RNA, and probes with SNV position indicated in bold underline (match) and brown (mismatch). (C) Representative images of Z1 cells showing detection of the correct target nucleotide in the barcode (see panel D for quantification of the results and Fig. S6 for representative images of other target nucleotides). All images were acquired under the same conditions and displayed with identical processing parameters for each channel (row). Each column represents one experiment in which four probes with a SNV and orthogonal HCR initiators (B1-4) were mixed and hybridized to the sample with the indicated color permutation. Letters indicate the probe variant in each image. HCR initiator and the fluorescence channel used for each probe are shown next to the rows. The barcode in Z1 cells is integrated site specifically in ROSA26 locus. Scale bar is 10 μm. (D) Probe competition can detect all four target nucleotides. Each matrix represents SNV analysis with four distinct color permutations, as in (C), with the indicated target nucleotide at distinct positions. For targeting U (right-most matrix), one permutation (14) is ambiguous due to wobble base pairing, but others (e.g. 15) provide accurate discrimination. Color scale represents the percentage of dots in which the indicated color channel has the highest rank of normalized brightness (see Methods). Total of 4009 cells were analyzed, with at least 135 cells for each color permutation.
To test this idea, we fixed mES-Z1 cells, performed in vitro transcription with T7 RNA polymerase, and targeted a 20bp region of the Z1 barcode with four probes, each containing a distinct nucleotide at a single position, and each detectable with orthogonal HCR initiators in different fluorescence channels (Fig. 3B). To control for systematic differences among fluorescent dyes, we performed each analysis with four different fluorescence channel permutations (Figs. 3C and S6, columns) and quantified the relative fluorescence intensities of each channel for each active site. We performed this analysis four times, once for each possible nucleotide at the variable position (Figs. 3C-D and S6).
When targeting A, C, or G, we observed a strong preference for the correct target nucleotide (Fig. 3D) across different color-HCR initiator permutations, ranging between 92 to 96% for A, 79 to 93% for C, and 93 to 99% for G (percentages indicate the fraction of fluorescent dots that are ‘called’ correctly by the algorithm). Some inaccurate calls may be explained by non-specific background HCR amplification in a region that overlaps with the cell nuclei but is not a true active site. However, when targeting U, in addition to the matched A probes, detectable signal was also observed from the mismatched G probes (Fig. S6), consistent with wobble base pairing between U and G37. Nevertheless, the base calling algorithm detected the correct match probe in three out of four permutations tested, with 90%, 97%, and 85% accuracy (Fig. 3D).
To investigate the dependence of SNV discrimination on the position of variant nucleotide within the probe, we performed a similar analysis with SNVs in positions 1 through 7 of the probes (Fig. S7). Positions 2 through 7 provided accurate SNV discrimination. Further, this analysis provided additional examples of accurate discrimination when U is the target (Fig. S7, position 6). These results indicate that probe competition can enable accurate in situ identification of SNVs.
Zombie reads out in vivo barcode base edits
CRISPR base editors have recently emerged as powerful tools for precise and predictable genome editing38-42. They can target and edit genomic DNA with single base pair resolution in a multiplexable manner. Heritable somatic mutations created by base editors could enable subsequent reconstruction of cell lineage and event histories1,8,9. The ability to read out base edits by imaging, rather than sequencing, would enable lineage and event history recording approaches that preserve spatial information, operate in individual cells, and could allow accurate recovery of sequence information from a high fraction of cells2. Since Zombie allows in situ detection of single nucleotide mismatches, we next asked whether it could be combined with base editors to read out single base pair changes in a synthetic memory unit.
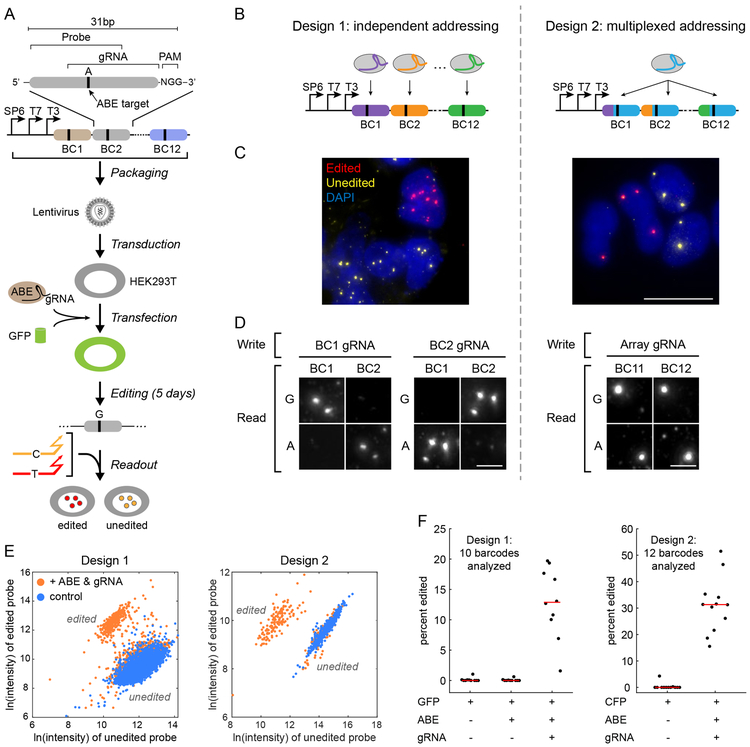
We engineered 31bp barcodes that could be edited by the Adenine Base Editor (ABE)40 and a corresponding gRNA (Fig. 4A). We concatenated these barcodes into ~500bp arrays, preceded by phage promoters. Using lentiviral vectors, we incorporated multiple array copies into the genome of HEK293T cells to create the Z-MEM cell lines (Fig. 4A). We then transiently co-transfected plasmids expressing the ABE (ABE7.10)40, the gRNA, and a fluorescent co-transfection marker (e.g. GFP) into Z-MEM cells, and cultured cells for five days. To analyze editing, we fixed cells, added T3 RNA polymerase, and detected transcribed barcodes using competing probes with distinct HCR initiators for edited and unedited states. This analysis was performed pair-wise, on adjacent barcodes. As a negative control, we also performed the analysis on cells that did not receive ABE or gRNA.
Figure 4. CRISPR base edits can be read out in situ.
(A) Arrays of 12 barcodes were designed so that, in each barcode, a single base pair (black vertical line) can be targeted by the adenine base editor (ABE) and a gRNA. The barcode arrays were packaged in lentivirus and transduced into HEK293T cells. ABE7.10, gRNA, and a fluorescent co-transfection marker (e.g. GFP), were transiently delivered as DNA into the cells, and editing was allowed to occur for 5 days. Finally, cells were fixed, treated with T3 RNA polymerase and read out by competing probes for original (orange) and edited (red) base variants. (B) Two designs of the memory array. Design 1 allows each barcode to be edited independently by a distinct gRNA, whereas all barcodes in design 2 are targeted by the same gRNA, providing more memory states for an individual gRNA. In both designs, the state of each individual barcode can be readout in situ, using Zombie. (C) Representative images, for design 1 (left) and design 2 (right), showing a mixture of edited (red) and unedited (yellow) active sites. Since barcodes are delivered by lentiviral transduction, cells can carry multiple copies of the barcode in their genome. The experiment was independently repeated twice with similar results. Scale bar is 10 μm. (D) Each barcode in design 1 (left) can be addressed independently using its corresponding gRNA. 2×2 matrices show results of targeting distinct barcodes. Edits are seen at the targeted barcode but not the adjacent non-targeted barcode. In contrast, design 2 gRNA (right) can edit all barcodes. The experiment was independently repeated twice with similar results. Scale bar is 3 μm. (E) Analysis of Barcode 1, Design 1 (left) and Barcode 10, Design 2 (right). Dots can be classified into distinct edited and unedited groups based on the signal intensity in edited and unedited channels. Scatter plots show the natural log of the intensity in edited versus unedited channels. Data from negative control samples (blue) are plotted on top of points from samples which received both ABE7.10 and gRNA plasmids. See Figures S9-10 for all barcodes in both designs. (F) Edits are detected when both ABE and gRNA are present. Each point represents one barcode, red lines show the median. Without ABE and barcode-specific gRNA, only a very small fraction of active sites are mis-identified as edited, indicating low false positive rates across barcodes. Note that editing rates differ among barcodes (vertical scatter). On average 1357 and 383 active sites were analyzed for each barcode at each condition, for design 1 and 2, respectively.
We designed two types of synthetic memory arrays (Fig. 4B). Design 1 enables independent addressing of different barcodes by distinct gRNAs, facilitating multi-channel recording. By contrast, design 2 uses one gRNA to edit all 12 barcodes, allowing a single gRNA to generate greater sequence diversity. In both cases, editing should result in single base pair changes in corresponding barcodes.
In both designs, individual barcodes showed an approximately binary response in imaging, appearing in either the edited or unedited channel, but not both (Fig. 4C). Moreover, pairwise analysis of the adjacent barcodes verified independent addressing in design 1 and multiplexed addressing in design 2 (Fig. 4D). We next quantified the signal intensity for each dot, in the edited and unedited channels, with or without co-transfection of ABE and gRNA (Figs. 4E and S8-10). Without ABE or gRNA most dots clustered in a single region (Fig. 4E, blue points). By contrast, when ABE and gRNA were both present a second cluster appeared, with a larger mean ratio of edited to unedited probe intensity (Fig. 4E, orange points), reflecting successful editing in a substantial fraction of cells (Fig. 4F). We observed similar behavior with the other analyzed barcodes (Figs. S9-10). We then used k-means clustering to classify the active sites as edited or unedited, with bootstrap resampling allowing determination of confidence for each assignment (Figs. S8-10). In both designs, except for a small subpopulation (yellow dots in Figs. S9-S10), active sites could be robustly classified based on their relative signal intensity.
A key parameter for recording is the edit rate, defined as the probability of an edit occurring at a given unedited target site per unit time. To estimate the relative edit rates of different barcodes, we tabulated the percentage of dots that were edited for each barcode in each design (Fig. 4F). These values varied widely across ten distinct design 1 barcodes, from 1.6% to 19.7% with a median of 12.9% (Probes for the two remaining units failed to generate signal and were not considered in the analysis). A broad range of edit rates, such as that observed here, has been shown to be advantageous in recording applications43. Similarly, design 2 units were edited at rates ranging from 15.5% to 51.5% with a median 31.3%. By contrast, memory units that were not targeted showed apparent edit rates close to 0 (Fig. 4F), consistent with both strong targeting specificity by ABE and accurate amplification and readout by Zombie. In a separate experiment, we showed that the edit rates measured by Zombie are similar to those measured by next generation sequencing for the same set of barcodes, further validating the accuracy of Zombie in situ readout (Fig. S11). Together, these results show that base editing can be targeted to distinct memory units and read out quantitatively in situ with high fidelity by Zombie.
Zombie identifies compact barcodes in embryonic and adult tissues
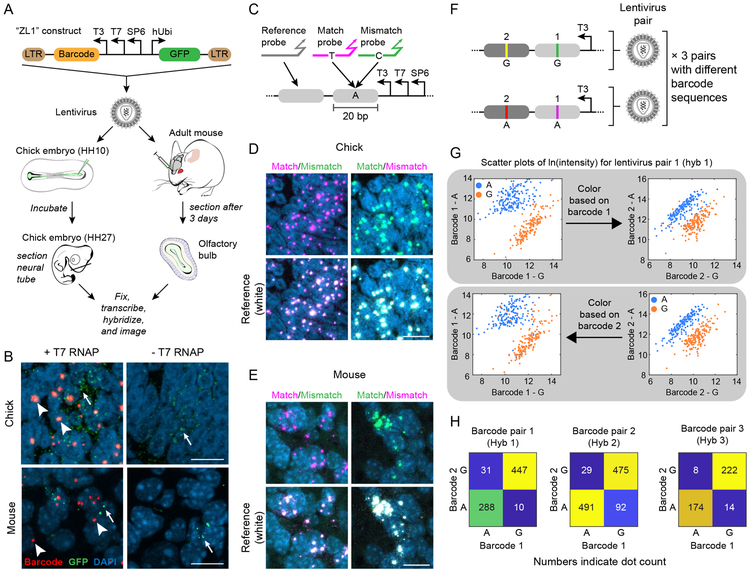
Reconstructing lineage information in embryos, brains, and tumors requires the ability to discriminate among a set of distinct barcodes or barcode edits in complex spatially organized contexts2-7,13. To test Zombie readout within tissues, we engineered a lentivirus, termed ZL1, containing probe target sequences downstream of phage promoters, along with a divergently oriented, constitutively expressed fluorescent protein reporter to enable identification of transduced cells (Fig. 5A). We first injected the lentivirus into the lumen of the developing chick neural tube at stage HH1044, and analyzed embryos 3 days later at stage HH27 (Fig. 5A, left). In a parallel study, we analyzed Zombie readout in adult mouse brain tissues, focusing on the olfactory bulb, which incorporates newly generated neurons in the adult stage45. We injected the ZL1 lentivirus into the granular cell layer of the olfactory bulb and sacrificed the mice for analysis 3 days later (Fig. 5A, right). In both cases, we observed robust, T7 polymerase-dependent in situ barcode transcription within the transduced regions (Fig. 5B). Together, these results show that Zombie can be used to detect viral barcodes in embryonic and adult tissue.
Figure 5. Zombie can detect barcodes and discriminate single nucleotide variants in chick embryo and adult mouse brain.
(A) The ZL1 construct includes a barcode downstream of phage promoters and a human Ubiquitin C promoter (hUbi) controlling GFP expression to allow identification of transduced cells. ZL1 was packaged in lentivirus and injected into the olfactory bulb of a 3 month old mouse or chick neural tube at embryonic stage HH10. Chick embryos were incubated for 3 days post-transduction, until stage HH27, and then frozen and sectioned for analysis of the neural tube. Mouse brains were frozen and sectioned 3 days post-transduction to analyze olfactory bulb. Both samples were then fixed, treated with T7 RNA polymerase, probed, and imaged. (B) In coronal sections through the diencephalon of chick embryos, we observed distinct active sites (arrowheads) with, but not without, transcription by T7 RNA polymerase. Similarly, Zombie active sites could also be detected, in a T7 dependent manner, in the granular cell layer of the olfactory bulb (arrowheads). Although the expression of GFP, detected by HCR, was sparse (arrows), the injection site could still be identified. All experiments were repeated on at least 3 sections with similar results. (C) To test for detection of single base pair mismatches in mouse and chicken tissue sections, samples were hybridized with match and mismatch probes (pink and green, respectively). A reference probe independently identified the active sites. (D, E) In both chicken and mouse samples, fluorescent signal at active sites was dominated by the match probe, regardless of channel assignments (columns). Match probes also co-localized with reference channels (bottom rows), indicating competition between match and mismatch probes does not reduce overall detection efficiency. All experiments were repeated on at least 3 sections with similar results. Since barcodes are delivered by lentiviral injection, cells can carry multiple copies of the barcode in their genome. Scale bars are 10 μm. (F) Pairs of barcoded lentiviral vectors were used to further assess the SNV detection capability in vivo. Each virus contains two distinct 20bp barcodes, denoted by 1 and 2. Within a pair, viruses have variants of these barcodes that differ with each other at only one base pair (A or G). A mix of three viral pairs, with different barcode sequences but the same SNV arrangement, was co-injected in the mouse olfactory bulb and read out in three rounds of hybridization and imaging, 12 days post-transduction. (G) Scatter plots showing natural log of signal intensity for two variants (A and G) of two barcodes (1 and 2) for lentivirus pair 1 (See Fig. S12 for the other pairs). Each point represents one active site. The points are color coded based on their barcode 1 state (top) or barcode 2 state (bottom) to show the concordance between the detected state of two barcodes. (H) In all pairs, the majority of active sites are classified as either A or G for both barcodes. Data are combined from two biological replicates.
We next tested the ability to discriminate single base pair mismatches in the same chick and mouse contexts. We analyzed tissues with an equimolar mixture of perfect match and single base mismatch probes, along with a third reference probe targeting a distinct downstream region, each in a distinct color channel (Fig. 5C). As a control, we also swapped color channels for the match and mismatch probes. Match probes strongly outcompeted mismatch probes, regardless of the color channel, in both organisms (Fig. 5D-E). Further, matching probes co-localized with reference probes, indicating that match-mismatch probe competition does not hinder detection efficiency (Fig. 5D-E). Taken together, these results demonstrate that Zombie can discriminate between single base pair mismatches in chick embryos and adult mouse brains.
Many in vivo barcoding and recording applications require simultaneous analysis of multiple barcode variants. To assess this capability, we designed three pairs of distinctly barcoded lentiviruses. Each virus contained two distinct 20bp barcodes, each containing an A or a G at a designated variable position. Critically, we designed these viruses such that the identity of the variable base in one barcode matched that of the other barcode in the same virus (Fig. 5F). With this design, two barcodes on the same virus should appear strongly correlated in the variable base, while barcodes on different viruses should vary independently. We selected A and G to mimic possible base editing outcomes (Fig. 4A).
We co-injected mouse olfactory bulbs with a mix of these three viral pairs. 12 days later we used Zombie with three consecutive rounds of hybridization and imaging to read out all pairs of viral barcodes. Single nucleotide differences between barcodes were readily identifiable based on the relative signal intensity of competing probes (Figs. 5G and S12). Further, as expected, we observed a strong correlation between the state of two barcodes appearing on the same virus, at each Zombie active site (Fig. 5G-H). Overall, 92% of sites were classified correctly as either A or G for both barcodes (Fig. 5H). Some of the remaining sites, classified as A for one barcode and G for another, might be explained by integration of both members of a lentivirus pair at sites too close to be spatially resolved (Fig. S13). Together, these results indicate that Zombie permits multiplexed barcode readout with single base discrimination in brain tissue.
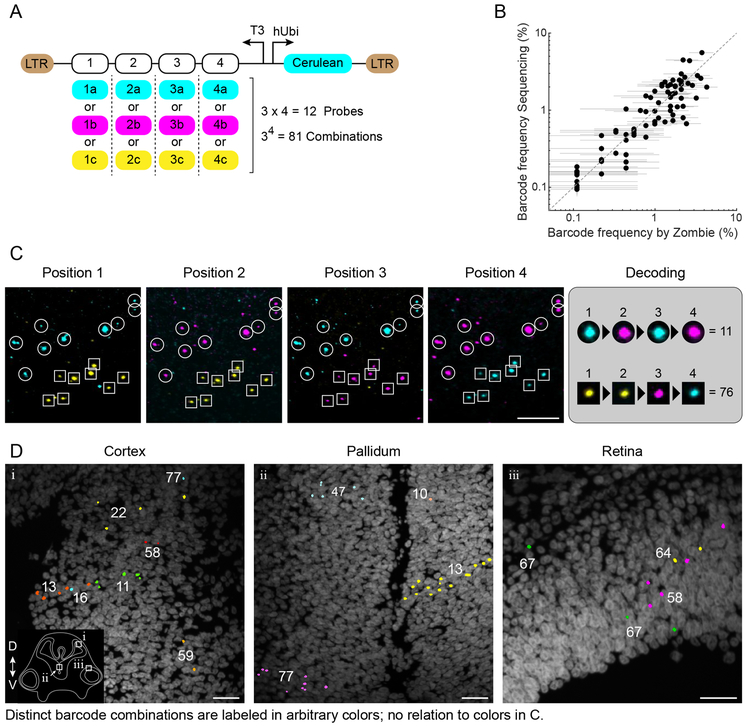
Combinatorial barcode libraries (Fig. 6A) could provide an exponentially increasing number of distinct barcodes with only a linear increase in the number of hybridization and imaging cycles needed to read them out46. The ability to detect short (20bp) DNA barcodes in situ should facilitate construction and delivery of such libraries. As a proof of principle, we constructed a lentiviral library containing 81 distinct combinations of 12 barcode sequences, each 20bp long (Fig. 6A). We transduced HEK293T cells with this library and read out the library in 3 rounds of hybridization and imaging, each one probing 4 out of 12 barcodes with orthogonal color channels (Fig. S14). In this analysis, barcode combinations were detected at frequencies consistent with those measured by next generation sequencing (Fig. 6B), corroborating the accuracy of in situ readout.
Figure 6. In situ readout of a combinatorial barcode library.
(A) A combinatorial lentiviral library in which each of 4 positions can take one of three distinct position-specific 20bp barcodes to generate 81 possible barcode combinations. The viruses also encode Cerulean downstream of hUbi promoter. (B) The frequency at which barcode combinations are detected in situ, in transduced HEK293T cells, is consistent with the frequency measured by next generation sequencing. Each point represents one barcode combination. 906 active sites were analyzed by Zombie. Error bars are 95% binomial confidence intervals, calculated using Clopper-Pearson method. Since the number of observations by imaging (906 active sites) is lower than the sequencing read count (102056 aligned reads), the horizontal error bars are wider than the vertical ones. (C) Detection of two clones of cells, labeled by two barcode combinations, in a coronal section of chick neural tube. Maximum intensity projected images corresponding to variants in each barcode position are merged in 3 color channels (cyan, magenta, and yellow, corresponding to A). Dots that do not appear consistently in all rounds are excluded from the analysis. (D) Examples of cells in developing chick cortex (i), pallidum (ii), and retina (iii) labeled with various barcode combinations (arbitrary colors). The inset shows the approximate location of the panels on a drawing of a coronal section through chick neural tube and indicates dorsal (D) and ventral (V) directions. For panels C and D, two embryos were analyzed. 39 out of 81 barcode combinations were identified in one embryo by analyzing 44 images acquired from 10 sections. In the other embryo, we identified 20 distinct barcode combinations in 11 images acquired from 6 consecutive sections. Scale bars are 25μm.
In a parallel, in vivo study, we injected the combinatorial library into the lumen of the developing neural tube at stage HH11 chick embryos. Three days later (stage HH27), we froze the embryos, performed the Zombie procedure, and analyzed in three rounds of hybridization, as with the HEK293T cells (Fig. 6C). We detected cells with distinct combinations of barcodes in both neural tube and retina of chick embryos (Fig. 6D). In many instances, cells labeled with the same barcode combination were observed close to each other and organized in a way that suggests clonal relationship (Fig. 6D, middle panel, clone 13). In other cases, despite relatively sparse labeling, cells with different barcode combinations were intermixed, indicating the necessity for high barcode diversity in establishing clonal relationships (Fig. 6D, left panel, clones 13, 16, and 11). These results demonstrate how Zombie can facilitate the use of combinatorial barcode libraries with imaging readout both in vitro and in vivo.
Finally, an ideal barcode readout system would be compatible with analysis of endogenous gene expression. To test this, we analyzed gene expression alongside barcode detection in the olfactory bulb of mice injected with the paired viruses (Fig. 5F). Using HCR, we confirmed that Tbx21 (expressed by projection neurons) and Tyrosine hydroxylase (Th; expressed by periglomerular cells) could be detected alongside barcodes, in the mitral and glomerular layers, respectively, as expected47,48(Fig. S15). This analysis demonstrates the suitability of Zombie for barcoding and recording applications that require readout of endogenous gene expression as well as barcodes in tissue samples.
Discussion
Here, we showed that phage RNA polymerases enable imaging-based barcode readout in individual fixed cells, producing easily detectable fluorescent dots localized to transcriptional sites (Fig. 1). Transcription enabled detection of 20bp barcodes (Fig. 2) with discrimination of single nucleotide variants using competing probes (Fig. 3). This capability further enabled recovery of edits made by a CRISPR base editor in live cells (Fig. 4). Finally, the system is versatile, operating not only in cell culture but also in chick embryos and adult mouse brain tissue (Fig. 5) and is therefore suitable for in vivo barcoding applications (Fig. 6). Taken together, these results indicate that this simple protocol allows high density barcoding and recording with in situ readout.
Concatenating multiple 20bp barcodes, as in Figure 6, can enable combinatorial libraries of distinct barcodes. We tested a modest library of 81 barcodes here. However, the same design could be scaled up to produce an exponential increase in coding capacity. For example, an array of 12 barcode positions, with 3 barcode variants per position, could achieve a potential barcode diversity of 531,441 variants, similar to that used in sequencing-based barcoding applications49-51, while requiring only 240bp of sequence and 9 rounds of imaging for read-out (An error correcting coding scheme would require additional hybridization rounds). Coding capacity could be further expanded by inserting multiple arrays at distinct, spatially resolvable genomic sites16.
Zombie should thus enable viral barcoding with imaging readout. In viral barcoding, cells are labeled at a single time-point or, more recently, at multiple time-points51, to enable subsequent identification of their descendants10. Viral barcoding methods have revolutionized the study of hematopoietic development49,52, neurobiology53,54, and cancer55. They have also enabled new high-throughput screening approaches56. However, so far, researchers have predominantly relied on sequencing for readout of virally delivered barcodes. Diverse combinatorial libraries of short Zombie-readable barcodes should enable simultaneous recovery of lineage, cell fate, and spatial organization in diverse settings, including development, regeneration, and cancer. Similarly, Zombie can facilitate multiplexed high-throughput screening, in which cellular phenotypes are assayed by imaging and connected to genetic or environmental perturbations that are identified by barcodes57.
An immediate application of Zombie will be to enable improved recording systems with image-based readout. In the previously described MEMOIR recording system, Cas9 stochastically and continuously edited ~1kb barcoded memory elements over multiple cell cycles2. These edits resulted in large scale sequence deletions, providing only a single binary memory state per kilobase of sequence. By contrast, in situ readout of base edits could provide a much higher memory density8,9. Additionally, by circumventing the need for barcode expression in living cells, this approach avoids issues with burstiness in expression and stochastic silencing. This approach should thus enable a more powerful imaging-based recording system, while maintaining compatibility with subsequent transcriptome readout, e.g. by seqFISH15,58, in the same cells.
There has appeared to be a general tradeoff between sequencing-based approaches that provide high throughput single nucleotide level readout but no spatial context and imaging approaches that preserve spatial information but lack the sensitivity of sequencing. Recent work has begun to bridge this gap in both directions46,59-61. By allowing imaging-based detection with sensitivity and scalability comparable to sequencing, we anticipate that Zombie will facilitate imaging-based barcoding, recording, and other applications currently dominated by sequencing.
Online Methods
Cell culture.
E14 mES cells (ATCC cat no. CRL-1821) were cultured in media containing GMEM (Sigma), 15% ES cell FBS qualified (Atlanta Biologicals), 1× MEM Non-Essential Amino Acids (ThermoFisher), 1mM Sodium Pyruvate (ThermoFisher), 100μM β-mercaptoethanol (ThermoFisher), 1x Penicillin - Streptomycin - L-Glutamine (ThermoFisher), and 1000 U/ml Leukaemia Inhibitory Factor (Millipore). Cells were maintained on polystyrene (Falcon) coated with 0.1% gelatin (Sigma) at 37°C and 5% CO2.
HEK293T cells were cultured in 1x DMEM (Corning), 10% FBS (Corning), 1x Penicillin - Streptomycin - L-Glutamine (Corning), 1mM Sodium Pyruvate (Corning), and 1x MEM Nonessential Amino Acids (Corning) on polystyrene (Falcon) plates at 37°C and 5% CO2. For transient transfections, HEK293T cells were plated in 48-well plates at the density of 125000 cells per well. The next day, cells were transfected with 1.5μl Lipofectamine 2000 (ThermoFisher) according to the manufacturer’s instruction. 350ng of ABE7.10 plasmid, 150ng of gRNA expression plasmid, and 100ng of GFP plasmid was used per well. In control wells, ABE7.10 and gRNA plasmids were replaced by pUC19 plasmid (NEB) to maintain the total amount of plasmids transfected at a constant level. Cells were then passaged to 24-well plates the day after transfection.
For in situ detection of barcodes, cells were plated on glass bottom 96-well plates (Cellvis) that were coated with 20 μg/ml laminin-511 (Biolamina) for at least 3 hours at 37°C.
Cell line engineering.
Sequences of all new constructs, barcodes, and probes used in this study are reported in Supplementary Table 1. To create stable polyclonal cell lines, mES cells were cultured in 24-well plates to approximately 70% confluency and co-transfected with 600ng of donor plasmid (Z1, control, or Z3) and 200ng of modified pX330 plasmid 62 (Addgene #42230) expressing Cas9 and a gRNA targeting ROSA26 locus (CAGGACAACGCCCACACACC). Transfection was performed using Lipofectamine LTX with Plus reagent (ThermoFisher) based on the manufacturer’s protocol. The cells were then passaged to a 6-well plate the next day and selected with 500ug/ml Geneticin starting at 2 days after transfection.
To establish Z1 and Z3 monoclonal cultures, approximately 1000 cells from the polyclonal population were cultured on a 10cm plate, from which individual colonies were picked and expanded. Clones were then genotyped by PCR to ensure that: the transgene is inserted properly in one of the ROSA26 loci, the other ROSA26 locus is intact, and there is no other integration of the transgene or Cas9 elsewhere in the genome.
Zombie procedure for cell culture samples.
Cells were washed with 1x PBS before fixation by 3:1 (v:v) mix of methanol and acetic acid (MAA) at room temperature for 20 minutes. Cross-linking fixation interferes with transcription by phage RNA polymerases, and therefore, should be avoided prior to the transcription step. Cells were then washed briefly first with 1x PBS and then with nuclease free water and subsequently were incubated with the transcription mix (MEGAscript Transcription Kit; Invitrogen) at 37°C for 3 hours. All three RNA polymerases used in this study (T3, T7, and SP6) work at comparable levels. The choice of one polymerase over another in different experiments was mostly arbitrary. After transcription, cells were fixed with 4% formaldehyde solution in PBS for 20 minutes at room temperature followed by two washes with 5X SSC, for 5 minutes each, to remove traces of formaldehyde.
The samples were then pre-incubated in hybridization buffer at 37°C for at least 10 minutes before overnight incubation, at 37°C, in hybridization buffer containing 4nM of each probe. When the experiment involved probe competition or split initiator probes with 25bp annealing region, 30% probe hybridization buffer (Molecular Technologies) was used for hybridization and, the next day, samples were washed four times, 15 minutes each, at 37°C with 30% probe wash buffer (Molecular Technologies) to remove excess probes, as previously described21. For probes with 20bp annealing region, in the absence of competition, 10% hybridization buffer (composed of 10% formamide, 10% Dextran Sulfate and 2X SSC in RNAse-Free water) was used for overnight hybridization as previously described29. These samples were then washed with a wash buffer, composed of 30% formamide, 2X SSC, and 0.1% Triton-X 100, at room temperature for 30 minutes, to remove excess probes, followed by a brief wash with 5X SSC.
HCR amplification was performed according to the manufacturer’s instruction. Briefly, samples were first washed with 5X SSCT (5X SSC + 0.1% Tween 20) for 5 minutes at room temperature and then incubated with amplification buffer (Molecular Technologies) for at least 10 minutes at room temperature. Meanwhile, each fluorescently labeled hairpin was prepared by snap cooling (heating at 95°C for 90 seconds and cooling to room temperature in a dark drawer for 30 minutes) in hairpin storage buffer. All the required hairpins were then added to the amplification buffer at the final concentration of 60μM each. Cells were then incubated, in the dark, with amplification buffer containing the hairpins for 45 minutes at room temperature. Subsequently, excess hairpins were removed by five washes with 5X SSCT over one hour. DAPI was added to the third wash to label nuclei. Nuclei could also be visualized using native fluorescent of H2B-CFP, when it was expressed in the cells (e.g. Fig. 1C-D). However, native fluorescence of cytoplasmically expressed fluorescent proteins could not be detected after the Zombie procedure. Samples were then kept in the dark at 4°C until imaging.
When additional rounds of hybridization and imaging was required, samples were incubated first with 1x DNase I buffer (Roche 4716728001) in nuclease free water at room temperature for 5 minutes and then with DNase I solution (2U/μl of the enzyme in 1x buffer) at 37°C for 3 hours, to digest probes and HCR hairpins from the previous round. Subsequently, samples were washed three times with pre-warmed 30% wash buffer at 37°C (first two washes for 5 min each and the third wash for 15 min). Another round of hybridization and HCR was then performed as described above.
The procedure described above is the main protocol we used in the cell culture experiments reported in this paper. See Supplementary Table 2 and Figures S16-18, for details regarding the variations to this main protocol.
Design of the synthetic memory arrays.
Each unit of the memory arrays includes a 20bp probe site that partially overlaps with a 20bp gRNA target site. gRNA target sites are followed by PAM sequence (NGG). To limit the possible outcome of base editing by ABE, gRNAs were designed so that from their position 2 to 10 there is only one “A” nucleotide, which occurs at position 5. We used Azimuth 2.0 software 63,64 to choose gRNA candidates with high on-target and low off-target scores. Each probe sequence is designed so that its GC content is 50% and its predicted Tm, calculated using nearest neighbor method65, is between 56 and 60°C. Sequences that form hairpins or dimers and homopolymeric tracts of 5bp or longer were avoided in the probes. We also avoided recognition sites of certain restriction enzymes (BsaI, BsmBI, BpiI, AarI, and XbaI) within the memory arrays to facilitate cloning. For design 1 array, probe sequences were chosen to differ from each other in at least 7 positions, to ensure specificity. For design 2, since all memory units are targeted with the same gRNA, 12 out of 20bp is shared among all probes. We chose the remaining 8bp so that all probes are different from each other in at least 2 positions of the first 4 nucleotides and at least another 2 positions among the second 4 nucleotides. Furthermore, to facilitate discrimination, we always mix probes targeting all 12 design 2 barcodes together, at equimolar ratio, with the ones not being analyzed in any given experiment at an orthogonal channel (e.g. B5 HCR initiator). See Supplementary Table 1 for full sequence of the arrays and their corresponding probes.
The combinatorial barcode library.
Synthetic gene fragments containing 81 barcode combinations were obtained from Twist Bioscience and cloned into a lentiviral transfer plasmid by golden gate cloning, using Esp3I and T7 DNA ligase (see Supplementary Table 1 for the sequence of plasmids and barcodes). After transformation into NEB 10-beta chemical competent E. coli (C3019I), more than 10,000 colonies were scraped off the plates and used to prepare DNA for lentiviral packaging.
Lentiviral delivery of barcodes.
Lentiviral vectors were produced and stored as previously described66 using the plasmids described above. The viral titer was determined by serial dilution. We only used viral preparations with at least 107 infectious units/μl. To establish stable cell lines, HEK293T cells were resuspended in the culture media, at a density of 500,000 cells per mL. 3μL of lentiviral prep was mixed in with 97μL of cell suspension. 10μL of this mix was then added to another 90μL of cell suspension in a separate tube. After mixing, the cells of the second tube were cultured in a 96-well plate for 3 days, without change of media. Subsequently, the cells were expanded in fresh media and used for the experiments.
To deliver barcodes to chicken embryos, fertilized eggs of white leghorn chickens were obtained from McIntyre Poultry & Fertile Eggs (Lakeside, California) and incubated in a humidified atmosphere at 38°C for 35 to 40 hours. The lentiviral prep was then injected in the neural tube of embryos ranging between stages HH10 and HH11 44. After injection, the eggs were closed with Parafilm and kept at 38°C. The embryos were analyzed 3 days after injection, at 5 days of incubation (stage HH27).
In mice, lentiviral injections were carried out stereotactically into the olfactory bulb of 3 month old male BL6 mice (JAX). Mice were anesthetized by single intraperitoneal injection with Ketamine/Xylazine solution. The stereotaxic coordinates were 5.5mm anterior from bregma, 1.2mm lateral from the midline, and 0.40mm ventral from the brain surface. We performed a single injection per olfactory bulb using 0.3 μl of the lentiviral prep. The mouse brains were analyzed either 3 or 12 days after injection, as described in the text.
Note that different viral integration sites or chromatin states could potentially vary in their accessibility to phage polymerases. All the experimental procedures performed on animal models was approved by the Institutional Animal Care and Use Committee of California Institute of Technology.
Next generation sequencing.
gDNA was extracted from cells using DNeasy Blood & Tissue kit (Qiagen) according to manufacturer instructions. Amplicon libraries containing the regions of interest (i.e. memory arrays or library barcodes) were then generated, from gDNA, with a two-step PCR protocol to add Illumina adapters and Nextera i5 and i7 combinatorial indices. Indexed amplicons were pooled and sequenced on the Illumina MiSeq platform with a 600-cycle, v3 reagent kit (Illumina, MS-102-3003). To analyze next generation sequencing data, raw FASTQ files were aligned to a FASTA-format reference file containing the expected amplicon sequences. Alignment was performed using the Burrows-Wheeler Alignment Tool (bwa-mem)67. For the combinatorial viral library (Fig. 6E), the number of reads aligning to each possible reference sequence was computed using a custom script in R, available here. For the base editing samples (Fig. S11), we extracted base calls from each read at the base editor target sites, as well as the quality scores at these sites. Paired-end reads were merged, accepting the base call with the highest quality score in overlapping regions. Reads with the quality score of more than 10, at the target site position, were included in the analysis.
Histology.
After harvesting, adult mouse brain and embryonic chicken tissues were washed with cold RNase free 0.1M phosphate-buffered saline solution (PBS, pH 7.4) at 4°C. Fresh tissues were then immersed into the Tissue-Tek O.C.T. Compound (#4583; Electron Microscopy Sciences) and werefrozen immediately for 3 minutes in isopentane cooled to −70°C in dry ice. Samples were then stored at −80°C until sectioning. 20μm thick sections were obtained using a Leica Cryostat, mounted on SuperFrost slides or coverslips coated with 2% v/v solution of (3-Aminopropyl)triethoxysilane in acetone. Sections were then stored at −80°C until use.
Zombie procedure for tissue sections.
The slides were first left to dry at room temperature for about 5 minutes and then fixed with MAA at room temperature in a glass staining jar for 3 hours. Subsequently, the slides were washed, by transfer to a new jar filled with PBS, three times for 5 minutes each. After a brief wash in nuclease free water, SecureSeal hybridization chambers (SKU:621501; Grace bio-labs) were put on the slides and transcription mix (MEGAscript T7 or T3 Transcription Kit; Invitrogen) was added on the sections and incubated for 3 hours at 37°C. After transcription, samples were fixed with 4% formaldehyde in PBS overnight at 4°C. Formaldehyde was then removed by three washes with 5X SSC at room temperature for 10 minutes each.
Hybridization was performed similar to what is described above for cell culture samples. Sections were pre-hybridized with probe hybridization buffer for at least 30 minutes at 37°C, before overnight incubation with probe hybridization buffer containing 4nM of each probe, at 37°C. When the experiment involved probe competition (e.g. Fig. 5C-H) or split initiator probes with 25bp annealing region (e.g. Figs. 5B and S15), 30% probe hybridization buffer (Molecular Technologies) was used for hybridization followed by 4 × 15min wash at 37°C with 30% probe wash buffer (Molecular Technologies). For probes with 20bp annealing region, in the absence of competition (e.g. Fig. 6), 10% hybridization buffer (composed of 10% formamide, 10% Dextran Sulfate and 2X SSC in RNAse-Free water) was used for overnight hybridization, followed by 2 × 30min wash in 30% formamide, 2X SSC, and 0.1% Triton-X 100, at room temperature. Then, after three brief washes with 5X SSCT at room temperature, sections were incubated with amplification buffer for 20 minutes, which was then replaced by amplification buffer containing snap cooled fluorescently labeled hairpins (Molecular Technologies), each at 60μM. After one hour incubation in the dark at room temperature, excess hairpins were removed by five washes with 5X SSCT over one hour. DAPI was added to the third wash to label nuclei.
For samples that required only one round of hybridization (e.g. Fig. 5B-E), hybridization chambers were removed at this point and sections were mounted in Aqua-mount (14-390-5; Thermo Scientific) and kept in the dark at 4°C until imaging. For multiple rounds of hybridization, 5X SSCT was replaced with anti-bleaching buffer16 (50 mM Tris-HCl pH 8.0, 300 mM NaCl, 2XSSC, 3 mM Trolox (Sigma 238813), 0.8% D-glucose (Sigma G7528), 100-fold diluted Catalase (Sigma C3155), 0.5 mg/mL Glucose oxidase (Sigma G2133) and 0.02 U/mL SUPERase In RNase Inhibitor (Invitrogen AM2694)) and samples were imaged as described below. After imaging, anti-bleaching buffer was washed first with 5X SSCT and then with 1x DNase I buffer (Roche 4716728001) in nuclease free water. Probes and HCR hairpins were then digested by 3 hours of incubation with DNase I solution (2U/μl of the enzyme in 1x buffer) at 37°C for 3 hours. Subsequently, the samples were washed three times with pre-warmed 30% wash buffer at 37°C (first two washes for 5 min each and the third wash for 15 min). Another round of hybridization and HCR was then performed as described above.
Imaging.
Cell culture samples were imaged on a Nikon Eclipse Ti inverted fluorescence microscope with a Zyla 4.2 sCMOS camera (Andor). We used a 60X oil objective (1.4 NA) and acquired 20 z-stacks with 0.5 micron spacing between them for each position. Positions were chosen solely based on DAPI channel to avoid bias. Imaging settings, including the exposure times, were kept the same for all the experiments involving cultured cells. Tissue sections were imaged either, using ZEN 2.3 (blue edition), on a Zeiss LSM800 confocal microscope with a 40X (Zeiss 1.2 NA), water immersion objective (Fig. 5B-E), or, using MetaMorph, on a Nikon Eclipse Ti inverted microscope, equipped with a Yokogawa CSU-W spinning disc unit (Andor) and an EMCCD camera (Andor iXon Ultra), using a 40X (Nikon 1.3 NA) oil objective (Figs. 6 and S15) or a 60X (Nikon 1.4 NA) oil objective (Fig. 5F-H). The same imaging setting was used for related samples to facilitate comparison between images.
Image analysis.
Images were processed and analyzed using MATLAB and Fiji68, mainly by custom scripts that are available here. For cell culture experiments, maximum intensity projection of the raw images were used in all analyses.
Segmentation.
Segmentation of nuclei and dots was done automatically in MATLAB by filtering and thresholding of the images. However, the results were manually inspected to ensure accuracy. Segmentation of nuclei was done based on either CFP (Figures 1, 2, 3, 5 and their related supplementary figures) or DAPI (Figs. 4, 6, and S8-11) channel. When relevant to the analysis (e.g. for efficiency calculations) incorrectly segmented nuclei were manually identified and removed from the analysis. Active site dots were considered to belong to a cell if their center overlapped with the nuclear segmentation of that cell.
Intensity measurement.
An estimate of dot intensity, used for Figures 4E, 5G-H, S2, S16-S18, and S8-12 was obtained by integration of pixel intensities over each dot’s segment. A more precise measure of dot intensity69 was used for Figures 3D and S7, which was based on fitting a 2D Gaussian to each dot’s filtered pixel intensity values and calculating the volume under the surface of the Gaussian.
Colocalization.
Colocalization of dots was identified based on close proximity (less than 4 pixels) of the center of segmented dots in two or more channels.
Classification.
For single nucleotide detection, where four probes compete for the same target site (Figs. 3D and S7), to assign a nucleotide to each dot, the natural log of intensity values for that dot in each channel were normalized linearly between 0 and 1, using the intensity values from all the dots detected in that channel across the experiment. The nucleotide associated with the channel that had the highest normalized intensity was then assigned to the dot. Calling the base edits (Fig. 4 and its related supplementary figures) as well as A and G classification in vivo (Figs. 5G-H and S12), was done by clustering natural log of intensity values in two groups using k-means clustering with cosine distance metric (kmeans function, MATLAB).
Registration.
Images of HEK293T cells transduced by the combinatorial viral library were registered initially based on CFP channel, using normalized cross-correlation method. A more refined registration was then achieved, using imregtform function in MATLAB, based on dots corresponding to different variant positions, regardless of their fluorescent channel, and using the CFP registration as the initial transformation.
Statistical analysis.
All experiments were performed in multiple distinct replicates, as indicated in the text and figure legends. Mutual information calculations in Figure S3 were performed as previously described70, by analyzing pairwise co-localization of barcodes in 564 cells across three replicates. Briefly, normalized mutual information (or uncertainty coefficient), U, between two barcodes, x and y, is defined as , where H is the entropy calculated by the formula . All statistics and tests are described fully in the text or figure legend.
Data availability.
Data that are not included in the paper are available at https://data.caltech.edu/records/1303 (https://doi.org/10.22002/D1.1303) or from the corresponding author.
Code availability.
Scripts for all analyses presented in this paper are available at https://data.caltech.edu/records/1303 (https://doi.org/10.22002/D1.1303) or from the corresponding author.
Material availability.
Plasmids and cell lines described in this paper are available, upon request, from the corresponding author.
Supplementary Material
Acknowledgements
We are grateful to M. Schwartzkopf, H. Choi, and N. Pierce for advice with HCR, K. Chow for help with cell culture, S. Shah for insightful discussions, and F. Ding for advice on image analysis. We also thank all the members of Elowitz, Cai, and Lois labs for helpful discussions and critical feedback. Some of the imaging for this paper was performed in the Biological Imaging Facility, with the support of the Caltech Beckman Institute and the Arnold and Mabel Beckman Foundation. The research was funded by NIH (R01 MH116508, M.B.E., C.L., & L.C.), The Paul G. Allen Frontiers Group and Prime Awarding Agency (UWSC10142, M.B.E., C.L., & L.C.), Jane Coffin Childs Memorial Fund for Medical Research (61-1650, A.A.), and NIH/NRSA training grant (T32 GM07616, D.M.C.). M.B.E. is a Howard Hughes Medical Institute Investigator.
Footnotes
Competing financial interests
Authors have submitted a provisional patent application based on the technology described in this manuscript.
References
- 1.Farzadfard F & Lu TK Emerging applications for DNA writers and molecular recorders. Science 361, 870–875 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frieda KL et al. Synthetic recording and in situ readout of lineage information in single cells. Nature 541, 107–111 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McKenna A et al. Whole-organism lineage tracing by combinatorial and cumulative genome editing. Science 353, aaf7907 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alemany A, Florescu M, Baron CS, Peterson-Maduro J & van Oudenaarden A Whole-organism clone tracing using single-cell sequencing. Nature 556, 108–112 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Kalhor R et al. Developmental barcoding of whole mouse via homing CRISPR. Science 361, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raj B et al. Simultaneous single-cell profiling of lineages and cell types in the vertebrate brain. Nat. Biotechnol 36, 442–450 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spanjaard B et al. Simultaneous lineage tracing and cell-type identification using CRISPR–Cas9-induced genetic scars. Nat. Biotechnol 36, 469–473 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farzadfard F et al. Single-Nucleotide-Resolution Computing and Memory in Living Cells. (2018). doi: 10.1101/263657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang W & Liu DR Rewritable multi-event analog recording in bacterial and mammalian cells. Science 360, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kebschull JM & Zador AM Cellular barcoding: lineage tracing, screening and beyond. Nat. Methods 15, 871–879 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Kalhor R, Mali P & Church GM Rapidly evolving homing CRISPR barcodes. Nat. Methods 14, 195–200 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Livet J et al. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature 450, 56–62 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Pei W et al. Polylox barcoding reveals haematopoietic stem cell fates realized in vivo. Nature 548, 456–460 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng GXY et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun 8, 14049 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shah S, Lubeck E, Zhou W & Cai L seqFISH Accurately Detects Transcripts in Single Cells and Reveals Robust Spatial Organization in the Hippocampus. Neuron 94, 752–758.e1 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Shah S et al. Dynamics and Spatial Genomics of the Nascent Transcriptome by Intron seqFISH. Cell 174, 363–376.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen KH, Boettiger AN, Moffitt JR, Wang S & Zhuang X RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348, aaa6090 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang X et al. Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science 361, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A & Tyagi S Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Methods 5, 877–879 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi HMT et al. Programmable in situ amplification for multiplexed imaging of mRNA expression. Nat. Biotechnol 28, 1208–1212 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choi HMT et al. Third-generation hybridization chain reaction: multiplexed, quantitative, sensitive, versatile, robust. Development 145, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rouhanifard SH et al. ClampFISH detects individual nucleic acid molecules using click chemistry-based amplification. Nat. Biotechnol (2018). doi: 10.1038/nbt.4286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marras SAE, Bushkin Y & Tyagi S High-fidelity amplified FISH for the detection and allelic discrimination of single mRNA molecules. Proc. Natl. Acad. Sci. U. S. A (2019). doi: 10.1073/pnas.1814463116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mitra RD, Shendure J, Olejnik J, Edyta-Krzymanska-Olejnik & Church, G. M. Fluorescent in situ sequencing on polymerase colonies. Anal. Biochem 320, 55–65 (2003). [DOI] [PubMed] [Google Scholar]
- 25.Ke R et al. In situ sequencing for RNA analysis in preserved tissue and cells. Nat. Methods 10, 857–860 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Lee JH et al. Highly multiplexed subcellular RNA sequencing in situ. Science 343, 1360–1363 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen X, Sun Y-C, Church GM, Lee JH & Zador AM Efficient in situ barcode sequencing using padlock probe-based BaristaSeq. Nucleic Acids Res. 46, e22 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feldman D et al. Pooled optical screens in human cells. (2018). doi: 10.1101/383943 [DOI] [Google Scholar]
- 29.Shah S et al. Single-molecule RNA detection at depth by hybridization chain reaction and tissue hydrogel embedding and clearing. Development 143, 2862–2867 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sousa R & Mukherjee S T7 RNA polymerase. Prog. Nucleic Acid Res. Mol. Biol 73, 1–41 (2003). [DOI] [PubMed] [Google Scholar]
- 31.Choi HMT et al. Mapping a multiplexed zoo of mRNA expression. Development 143, 3632–3637 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vieregg JR, Nelson HM, Stoltz BM & Pierce NA Selective nucleic acid capture with shielded covalent probes. J. Am. Chem. Soc 135, 9691–9699 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Levesque MJ, Ginart P, Wei Y & Raj A Visualizing SNVs to quantify allele-specific expression in single cells. Nat. Methods 10, 865–867 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sternberg JB & Pierce NA Exquisite sequence selectivity with small conditional RNAs. Nano Lett. 14, 4568–4572 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu LR et al. Continuously tunable nucleic acid hybridization probes. Nat. Methods 12, 1191–1196 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Symmons O et al. Allele-specific RNA imaging shows that allelic imbalances can arise in tissues through transcriptional bursting. (2018). doi: 10.1101/386359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mathews DH, Sabina J, Zuker M & Turner DH Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol 288, 911–940 (1999). [DOI] [PubMed] [Google Scholar]
- 38.Komor AC, Kim YB, Packer MS, Zuris JA & Liu DR Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Komor AC et al. Improved base excision repair inhibition and bacteriophage Mu Gam protein yields C:G-to-T:A base editors with higher efficiency and product purity. Sci Adv 3, eaao4774 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gaudelli NM et al. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 551, 464–471 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li X et al. Base editing with a Cpf1-cytidine deaminase fusion. Nat. Biotechnol 36, 324–327 (2018). [DOI] [PubMed] [Google Scholar]
- 42.Gehrke JM et al. An APOBEC3A-Cas9 base editor with minimized bystander and off-target activities. Nat. Biotechnol 36, 977–982 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chan M et al. Molecular recording of mammalian embryogenesis. (2018). doi: 10.1101/384925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hamburger V & Hamilton HL A series of normal stages in the development of the chick embryo. J. Morphol 88, 49–92 (1951). [PubMed] [Google Scholar]
- 45.Lois C & Alvarez-Buylla A Long-distance neuronal migration in the adult mammalian brain. Science 264, 1145–1148 (1994). [DOI] [PubMed] [Google Scholar]
- 46.Emanuel G, Moffitt JR & Zhuang X High-throughput, image-based screening of pooled genetic-variant libraries. Nat. Methods 14, 1159–1162 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Faedo A et al. Developmental expression of the T-box transcription factor T-bet/Tbx21 during mouse embryogenesis. Mech. Dev 116, 157–160 (2002). [DOI] [PubMed] [Google Scholar]
- 48.Baker H, Kawano T, Margolis FL & Joh TH Transneuronal regulation of tyrosine hydroxylase expression in olfactory bulb of mouse and rat. J. Neurosci 3, 69–78 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lu R, Neff NF, Quake SR & Weissman IL Tracking single hematopoietic stem cells in vivo using high-throughput sequencing in conjunction with viral genetic barcoding. Nat. Biotechnol 29, 928–933 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Naik SH et al. Diverse and heritable lineage imprinting of early haematopoietic progenitors. Nature 496, 229–232 (2013). [DOI] [PubMed] [Google Scholar]
- 51.Biddy BA et al. Single-cell mapping of lineage and identity in direct reprogramming. Nature (2018). doi: 10.1038/s41586-018-0744-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weinreb C, Rodriguez-Fraticelli AE, Camargo FD & Klein AM Lineage tracing on transcriptional landscapes links state to fate during differentiation. (2018). doi: 10.1101/467886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Walsh C & Cepko CL Widespread dispersion of neuronal clones across functional regions of the cerebral cortex. Science 255, 434–440 (1992). [DOI] [PubMed] [Google Scholar]
- 54.Kebschull JM et al. High-Throughput Mapping of Single-Neuron Projections by Sequencing of Barcoded RNA. Neuron 91, 975–987 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bhang H-EC et al. Studying clonal dynamics in response to cancer therapy using high-complexity barcoding. Nat. Med 21, 440–448 (2015). [DOI] [PubMed] [Google Scholar]
- 56.Dixit A et al. Perturb-Seq: Dissecting Molecular Circuits with Scalable Single-Cell RNA Profiling of Pooled Genetic Screens. Cell 167, 1853–1866.e17 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Boutros M, Heigwer F & Laufer C Microscopy-Based High-Content Screening. Cell 163, 1314–1325 (2015). [DOI] [PubMed] [Google Scholar]
- 58.Eng C-HL et al. Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH. Nature 568, 235–239 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Satija R, Farrell JA, Gennert D, Schier AF & Regev A Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol 33, 495–502 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen R et al. A Barcoding Strategy Enabling Higher-Throughput Library Screening by Microscopy. ACS Synth. Biol 4, 1205–1216 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weinstein JA, Regev A & Zhang F DNA microscopy: Optics-free spatio-genetic imaging by a stand-alone chemical reaction. (2018). doi: 10.1101/471219 [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only References
- 62.Cong L et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Doench JG et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol 34, 184–191 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Listgarten J et al. Prediction of off-target activities for the end-to-end design of CRISPR guide RNAs. Nat Biomed Eng 2, 38–47 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.SantaLucia J Jr. A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl. Acad. Sci. U. S. A 95, 1460–1465 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lois C, Hong EJ, Pease S, Brown EJ & Baltimore D Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science 295, 868–872 (2002). [DOI] [PubMed] [Google Scholar]
- 67.Li H Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv [q-bio.GN] (2013). [Google Scholar]
- 68.Schindelin J et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ding F & Elowitz MB Constitutive splicing and economies of scale in gene expression. Nat. Struct. Mol. Biol 26, 424–432 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Press William H. et al. Numerical Recipes in C: The Art of Scientific Computing. (Cambridge University Press, 1992). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data that are not included in the paper are available at https://data.caltech.edu/records/1303 (https://doi.org/10.22002/D1.1303) or from the corresponding author.