Abstract
WhiB1 is a monomeric iron–sulfur cluster-containing transcription factor in the WhiB-like family that is widely distributed in actinobacteria including the notoriously persistent pathogen Mycobacterium tuberculosis (M. tuberculosis). WhiB1 plays multiple roles in regulating cell growth and responding to nitric oxide stress in M. tuberculosis, but its underlying mechanism is unclear. Here we report a 1.85 Å-resolution crystal structure of the [4Fe–4S] cluster-bound (holo-) WhiB1 in complex with the C-terminal domain of the σ70-family primary sigma factor σA of M. tuberculosis containing the conserved region 4 (σA4). Region 4 of the σ70-family primary sigma factors is commonly used by transcription factors for gene activation, and holo-WhiB1 has been proposed to activate gene expression via binding to σA4. The complex structure, however, unexpectedly reveals that the interaction between WhiB1 and σA4 is dominated by hydrophobic residues in the [4Fe–4S] cluster binding pocket, distinct from previously characterized canonical σ704-bound transcription activators. Furthermore, we show that holo-WhiB1 represses transcription by interaction with σA4in vitro and that WhiB1 must interact with σA4 to perform its essential role in supporting cell growth in vivo. Together, these results demonstrate that holo-WhiB1 regulates gene expression by a non-canonical mechanism relative to well-characterized σA4-dependent transcription activators.
INTRODUCTION
Mycobacterium tuberculosis is the main causative pathogen of tuberculosis and the second greatest killer after the Human Immunodeficiency Virus (HIV) among single infectious agents (1). M. tuberculosis causes disease by adapting to the host immune response using dedicated regulatory systems that include the White B-like (Wbl) family proteins. Wbl proteins contain a [4Fe–4S] cluster, were first identified in Streptomyces coelicolor, and are widely distributed in actinobacteria (2,3). Seven Wbl proteins are found in M. tuberculosis, and they play versatile roles in diverse biological processes including redox homeostasis, cell division, nutrition starvation and antibiotic resistance (4–6).
Structural and functional analyses indicate that M. tuberculosis Wbl proteins are monomeric transcription factors with three conserved motifs (4): (i) a [4Fe–4S] cluster binding motif consisting of four cysteines in the N-terminus, (ii) a DNA binding motif (also known as AT-hook) in the C-terminus and (iii) an intervening Gly-rich motif (β-turn) that links the two functional domains. M. tuberculosis Wbl proteins interact with the primary sigma factor σA of the RNA polymerase (RNAP) holoenzyme in their [4Fe–4S] cluster-bound (holo-) form, but not in their cluster-free (apo-) form (7,8). σA is a member of the σ70-family primary sigma factors possessing four conserved regions (region 1–4), of which region 4 (σ704) recognizes the −35 element and is a highly conserved hub for direct interaction with many transcription factors including the M. tuberculosis Wbl proteins (7,9,10). Two Wbl members (WhiB3 and WhiB7) have been shown to require interaction with σA4 to activate transcription (7,11,12).
Among the M. tuberculosis Wbl proteins, WhiB1 is of particular interest because it is essential for cell growth and it is suggested to have a role in the initiation of dormancy in response to nitric oxide (NO) (6). NO is a potent antimicrobial chemical produced by hosts to combat tuberculosis infection (13,14). Consequently, the transcription factors in the defense system of M. tuberculosis that swiftly sense and respond to NO are critical for the survival and pathogenesis of the bacterium. The [4Fe–4S] cluster in WhiB1 is highly reactive to NO over O2 (15,16), making it a specific NO sensor in aerobic bacteria. NO disrupts the WhiB1 [4Fe–4S] cluster by initially forming a [Fe-S] cluster–NO complex, and eventually resulting in cluster degradation and formation of apo-WhiB1 (15). Binding of WhiB1 to σA significantly decreases the O2 reactivity of the [4Fe–4S] cluster in the WhiB1:σA complex, which is reportedly stable under aerobic conditions for two weeks while retaining high reactivity to NO (8). Upon exposure to NO both in vitro and in Escherichia coli, the WhiB1:σA complex was observed to rapidly react with NO and disassociate (8). This observation indicates a potential role of WhiB1 as a NO sensor and regulator in mycobacteria considering that the WhiB1:σA complex is reactive to NO in the presence of the native NO-sensing and detoxification system in E. coli (17). The molecular basis for the O2 stability of the [4Fe–4S] cluster in the WhiB1:σA complex is unknown because the structure of the complex has not been determined.
Both NO-treated holo-WhiB1 and apo-WhiB1, but not holo-WhiB1, have been shown to bind to and repress at least two essential genes: groEL2, encoding a chaperonin; and whiB1 itself (6,18). The positively charged residues in the C-terminal DNA binding motif of apo-WhiB1, including Lys72, Arg73, Arg74, Lys79 and Arg81, are required for DNA binding (19). For this reason, WhiB1 was defined as a transcriptional repressor in the past (6), while the active form of WhiB1 that supports active mycobacterial growth remains elusive. Based on its interaction with σA4, holo-WhiB1 has been proposed to activate gene expression using a canonical mechanism utilized by Class II transcription activators such as the cyclic AMP receptor protein (CRP) and the fumarate and nitrate reduction regulatory protein (FNR) (10,20–24). However, holo-WhiB1 possesses characteristics that are different from previously characterized Class II transcription activators. First, WhiB1 is a monomeric transcription factor with a single DNA binding motif, while canonical Class II transcription activators are either homodimers or contain multiple DNA binding motifs that confer high specificity and affinity for target DNA (10,23). Second, holo-WhiB1 does not bind to its own promoter and no target gene of holo-WhiB1 has been identified thus far (6), while σ704-binding transcription activators are typically self-regulated, activating gene expression by binding to specific promoter sequences and recruiting RNAP to the target genes (10,25). An in-depth mechanistic understanding of transcriptional regulation by WhiB1 requires atomic-resolution structural information showing how holo-WhiB1 interacts with σA4. An NMR structure of free holo-WhiB1 has been reported (8), but there is no 3D structural information available for σA-bound WhiB1 or any other Wbl proteins.
In the course of defining the molecular mechanism of holo-WhiB1 in σA4-dependent transcriptional regulation, we solved a crystal structure of holo-WhiB1 in complex with the C-terminal domain of σA (σACTD) containing the intact σA4 region. Our structural analysis reveals that hydrophobic residues unexpectedly dominate the interaction between holo-WhiB1 and σA. This is distinctly different from previously characterized Class II transcription activators, which typically interact loosely with region 4 of σ70-family primary sigma factors through polar residues on the surface (11,12,26,27). In vitro, the observed transcription repression by holo-WhiB1 via interaction with σA4 further supports the hypothesis that WhiB1 regulates gene expression in a manner distinct from the canonical transcription activation by σA4-bound Class II activators. In addition, the local protein environment of the Fe−S cluster in the WhiB1:σACTD complex explains how the complex formation renders this typically O2-labile cluster into a remarkably O2-stable [4Fe–4S] cluster that can sense NO in aerobic bacteria.
MATERIALS AND METHODS
Bacterial strains and growth media
The bacterial strains and plasmids used in this study are listed in Supplementary Table S1. All the E. coli strains used for cloning and protein over-expression were grown in Luria–Bertani (LB) media supplemented with suitable antibiotics as needed at 37 °C with shaking at 200 rpm, unless otherwise specified.
Mycobacterium smegmatis MC2 155 (M. smegmatis) and the related whiB1 mutants generated in this study were grown in either Middlebrook 7H9 broth or on 7H10 agar (BD DifcoTM) supplemented with 10% (v/v) ADC (2% dextrose, 5% bovine serum albumin, 0.85% NaCl and 0.003% catalase), 0.2% (v/v) glycerol, 0.05% (v/v) Tween 80 (Sigma) and suitable antibiotics as needed.
Creation of a conditional whiB1 knockout mutant in M. smegmatis
The whiB1 (MSMEG_1919) knockout mutant in M. smegmatis was generated using homologous recombination-based in-frame unmarked deletion as previously described (28). Briefly, a 844-bp DNA fragment upstream of whiB1 and a 1135-bp DNA fragment downstream of whiB1 were amplified by PCR from the genomic DNA of M. smegmatis and cloned into pUC-Zeo flanking the zeocin resistance gene to construct the recombinant plasmid pUC-Zeo+MsmwhiB1LR. After confirming the construct by DNA sequencing, a linear recombinant DNA fragment was prepared by double digestion of pUC-Zeo+MsmwhiB1LR with KpnI and HindIII. The resulting linear DNA fragment was electroporated into an M. smegmatis strain containing the helper plasmid pJV53-GFP to allow the expression of recombination proteins gp60 and gp61 of mycobacteriophage Che9c (29). Transformants were selected on 7H10 agar plates supplemented with 25 μg/ml kanamycin and 50 μg/ml zeocin. As reported for M. tuberculosis (6), no viable whiB1 deletion mutants were obtained after multiple attempts, although single crossover was observed.
To confirm that whiB1 is essential in M. smegmatis, we created a conditional whiB1 knockout mutant as shown in Supplementary Figure S1. For this purpose, the DNA fragment containing the 3FLAG-tagged whiB1 gene with its flanking sequences (1159 bp upstream and 638 bp downstream of whiB1, respectively) was cloned into the integration plasmid pKW08-Lx-Int (30). The resulting plasmid pKW08 + 3FLAGwhiB1LR was transformed and integrated in the genome of M. smegmatis to obtain a complementary copy of whiB1 under its native promoter in the genome of M. smegmatis. This strain was used for subsequent deletion of whiB1 at its native location as described above. Several colonies from the screening plates were streaked on 7H10 agar plates without antibiotics to facilitate excision of the zeocin cassette. The conditional whiB1 knockout mutant from the resulting colonies was verified by PCR and further confirmed by DNA sequencing (Supplementary Figure S1).
Site-directed mutagenesis of whiB1 in M. smegmatis
To generate site-directed whiB1 mutants in M. smegmatis, a template plasmid pUC-Hyg+MsmwhiB1SDM was first constructed with a 612-bp DNA fragment containing the whiB1 gene and its upstream DNA flanking the left arm, and a 1135-bp DNA fragment downstream of whiB1 flanking the right arm of the hygromycin resistance gene, respectively. Desired single point mutations of whiB1 in pUC-Hyg+Msm-whiB1SDM were constructed using Q5® Site-Directed Mutagenesis Kit (New England Biolabs) according to the manufacturer's instructions and confirmed by DNA sequencing. Linear recombinant DNA fragments were then prepared from the resulting plasmids by double digestion with KpnI and HindIII, and electroporated into an M. smegmatis strain containing the helper plasmid pJV53-GFP. The homologous recombination procedures for generating site-directed whiB1 mutants were similar to those used in creating the whiB1 deletion mutant, as previously described (28). The resulting whiB1 mutants were verified by PCR and further confirmed by DNA sequencing.
Plasmid construction for protein over-expression in E. coli
The DNA sequences encoding the wildtype WhiB1 (Rv3219, 1–84 aa) and the C-terminal domain of σA (Rv2703) containing the C-terminal residues 359–528 of the protein (denoted σAC170) were amplified by PCR from M. tuberculosis H37Rv genomic DNA (a gift from Dr. Midori Kato-Maeda's group at the University of California, San Francisco). The whiB1 gene was cloned into pET19b and the σAC170 gene was cloned into pCDF-1b to express His10-WhiB1 and His6-σAC170, respectively. These plasmids were then further modified for expressing the truncated and mutant proteins used for crystallization and pull-down assays.
For preparing tagless WhiB1 used in in vitro pull-down assays and in vitro transcription, the whiB1 gene was cloned into pETDuet-His6-SUMO vector (modified from pETDuet). For co-expression and purification with His6-σAC170, the whiB1 gene was cloned into the pET28b vector to express the tagless WhiB1 wildtype and mutant proteins. The pET21b-MtbWhiB1–76 and pET28b-6HisMtbσAC112 plasmids were used for over-expressing and purifying the WhiB1:σAC112 complex used for crystallization.
All the plasmids were confirmed by DNA sequencing before being transformed into E. coli BL21-Gold (DE3) strain for protein expression.
General procedures for protein expression, purification and analysis
Over-expression of proteins for structural and biochemical studies was done in E. coli BL21-Gold (DE3). Protein expression was induced by adding 100 μM isopropyl β-d-thiogalactopyranoside (IPTG, Sigma-Aldrich) to the cell culture at 16 °C for 12–15 h grown in Terrific Broth. For over-expression of proteins containing a [4Fe–4S] cluster, 100 μM ferric ammonium sulfate was added to the cell culture before induction. Cells were collected by centrifugation and kept at −80 °C for storage.
Affinity purification of His-tagged proteins containing WhiB1 (wildtype and mutants) was carried out using a HisTrap HP column (GE Healthcare Life Sciences) at room temperature either in an anaerobic chamber (Coy Lab) or under a customized Schlenk Line, unless otherwise specified. All the buffers used for protein purification were thoroughly degassed under the Schlenk Line and supplemented with 1 mM 1,4-dithiothreitol (DTT) immediately before use. Purification of His6-σA or His6-σAC170 was performed at 4°C under aerobic conditions to minimize degradation. Phenylmethanesulfonyl fluoride (PMSF at 1 mM, Sigma-Aldrich) and the Roche Protease Inhibitor Cocktail (at 1 tablet/100 ml buffer) were added to the lysis buffer to reduce degradation of target proteins. Imidazole in the eluted fractions was removed by passing through either a desalting column or a Superdex 200 column (GE Healthcare Life Sciences). Samples after each step of purification were analyzed by SDS-PAGE and by UV-Visible (UV-Vis) spectroscopy. Unless otherwise specified, the final purified proteins in 50 mM Tris−HCl, pH 8.0, 100 mM NaCl, 1mM DTT were stored in liquid nitrogen until use.
The selenomethionine (SeMet)-substituted WhiB1:σAC112 protein used for single wavelength anomalous diffraction (SAD) phasing was expressed in E. coli BL21-Gold (DE3) by the metabolic inhibition method as previously described (31) and purified by Ni2+-affinity chromatography and gel filtration as described above.
For preparing tagless WhiB1 used in the pull-down assays and in vitro transcription, the SUMO tag was removed from the purified His6-SUMO-WhiB1 by the SUMO protease Ulp1 (Thermo Fisher Scientific). The resulting sample was passed through HisTrap HP column to remove undigested protein and Ulp1, and further purified by gel filtration.
Expression and purification of the M. tuberculosis RNA polymerase core enzyme from E. coli was carried out as previously described (32). The plasmid used for expression of the M. tuberculosis RNA polymerase core enzyme was received from Dr Robert Landick's group at the University of Wisconsin–Madison and used without any modification. M. tuberculosis RNA polymerase holoenzyme was prepared by mixing the purified M. tuberculosis RNA polymerase core enzyme with excess His6-σA, and passing the mixture through a gel filtration column (Superose 6 increase 10/300 GL, GE Healthcare Life Sciences) to remove unbound His6-σA.
UV-Vis spectra of the purified proteins were recorded using an HP 8452a diode array UV-Vis spectrophotometer (Agilent Technologies Inc.). Protein concentrations were estimated by the Pierce Bradford Assay Kit (Thermo Fisher Scientific) and by the absorption at 280 nm. The absorption at 410 nm, which is characteristic of proteins containing [4Fe–4S]2+ clusters (15,33), was used to estimate the occupancy of the [4Fe–4S]2+ cluster in the protein samples containing WhiB1.
Western blot analysis of 3FLAG-tagged proteins was detected by chemiluminescence using mouse anti-FLAG M2 antibody (Sigma) as the primary antibody and the horseradish peroxidase-coupled goat anti-mouse IgG antibody (Jackson ImmunoResearch Laboratories) as the secondary antibody.
Crystallization
Initial crystallization screens of the WhiB1:σAC112 complex were carried out at 18°C in a Coy anaerobic chamber using the sitting-drop vapor diffusion method, followed by optimization of the crystallization hits. High-quality crystals were obtained by mixing 1 μl of WhiB1:σAC112 at 70 mg/ml with an equal volume of the reservoir solution containing 100 mM Bis-Tris pH 5.5, 200 mM MgCl2 and 25% (w/v) PEG3350. Crystals of the SeMet-substituted WhiB1:σAC112 were produced under the same conditions. All the crystals were briefly soaked in the reservoir solution supplemented with 20% glycerol for cryoprotection before flash-cooling in liquid nitrogen.
The σAC112 protein in the complex was partially degraded during the crystallization process based on SDS-PAGE analysis and peptide identification by mass spectrometry. In the high-quality single crystals used for X-ray crystallography, partially degraded σAC112 showed a similar apparent size as WhiB1 on the SDS-PAGE gel (Supplementary Figure S2).
X-ray crystallographic data collection, structural determination and analysis
X-ray diffraction data were collected at the beamlines 9-2 and 12-2 of the Stanford Synchrotron Radiation Lightsource from single crystals maintained at 100 K using a 6M Pixel Array Detector. The SAD data were collected from a single SeMet-substituted WhiB1:σAC112 crystal at the Se K-edge absorption peak (12,679 eV). The diffraction data were indexed, integrated and scaled with either HKL2000 or XDS (34,35). Two different crystal forms, C2221 and P21, were found in the crystals produced from different batches of crystallization.
Experimental phases were determined by SAD with Phenix.Autosol (36), using the SeMet-substituted WhiB1:σAC112 diffraction data up to 2.1 Å resolution. Since no crystal of the native WhiB1:σAC112 diffracted better than 2.3 Å, the same dataset used for experiment phasing was used for structure refinement with a resolution cutoff extended to 1.85 Å considering the high multiplicity of the data and the CC1/2 of 0.882 in the highest resolution shell.
Model building and structure refinement were carried out using COOT and Phenix (37,38). Two copies of the WhiB1:σACTD complex molecules were found in each asymmetrical unit of the C2221 form and four copies in the P21 form. Residues 2–73 of WhiB1 and residues 454–521 of σACTD were included in the final model for the complexes of both crystal forms, while residues 2–75 of WhiB1 and 454–523 of σACTD were included in at least one subunit of the refined model in the C2221 form. Notably, the structures of the WhiB1:σACTD complex determined from the C2221 and P21 forms agree closely, except for the flexible loop region containing residues Val20-Ala26 in WhiB1. The data collection and refinement statistics are summarized in Table 1. Because of the better-defined electron density of residues at the N- and C-terminus, the refined structure in the C2221 form was used for generating the figures and analyzing the structure as described in the main text.
Table 1.
Data collection and refinement statistics
| Crystal 1 | Crystal 2 | Crystal 1 data for phasing | |
|---|---|---|---|
| Data collection a | |||
| Space group | C2221 | P21 | C2221 |
| Cell dimensions | |||
| a, b, c (Å) | 37.4, 114.1, 144.7 | 37.8, 144.4, 59.9 | 37.4, 114.1, 144.7 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 106.8, 90.0 | 90.0, 90.0, 90.0 |
| Wavelength (Å) | 0.9793 | 0.9793 | 0.9793 |
| Resolution (Å) | 50–1.85 (1.88–1.85) | 50–1.85 (1.87–1.85) | 50–2.10 (2.16–2.10) |
| R merge | 0.095 (0.894) | 0.061 (1.42) | 0.073 (0.629) |
| I / σI | 36.8 (0.84) | 22.3 (1.9) | 24.3 (4.7) |
| Completeness (%) | 91.5 (56.4) | 98.3 (98.3) | 99.3 (93.7) |
| Multiplicity* | 22.1 (13.2) | 14.0 (14.4) | 26.4 (26.8) |
| CC1/2(%) | 99.8 (82.2) | 99.9 (86.2) | 99.3 (98.3) |
| Refinement | |||
| Resolution (Å) | 50–1.85 (1.95–1.85) | 50–1.85 (1.92–1.85) | |
| No. reflections | 20218 | 51328 | |
| No. molecules per asymmetric unit | 2 | 4 | |
| R work/Rfree | 0.189/0.236 | 0.193/0.234 | |
| No. atoms | 2531 | 4821 | |
| Protein | 2266 | 4472 | |
| Ligand | 30 | 39 | |
| Water | 235 | 310 | |
| B-factors | 34.94 | 50.54 | |
| Protein | 34.47 | 50.64 | |
| Ligand | 34.82 | 42.34 | |
| Water | 39.54 | 50.12 | |
| R.m.s deviations | |||
| Bond lengths (Å) | 0.007 | 0.01 | |
| Bond angles (°) | 1.16 | 1.21 | |
| Ramachandran statistics | |||
| Favored regions (%) | 99.63 | 98.87 | |
| Allowed regions (%) | 0.37 | 1.13 | |
| Outliers (%) | 0 | 0 | |
| PDB ID | 6ONO | 6ONU |
aThe highest-resolution shell statistics are shown in parentheses.
*For the data used for phasing, the anomalous multiplicity was shown.
Structural figures were prepared with the PyMol Molecular Graphics System v 2.3 (http://www.pymol.org). Sequence and structural alignments were performed in Clustal Omega and ESpript online server (http://espript.ibcp.fr), respectively (39,40). Tunnel dimensions were calculated using the Caver 3.0 software with probe radii indicated in the figures (41).
In vitro and in vivo protein-protein interaction assays
The interactions between WhiB1 and His6-σAC170 (wildtype and mutants) were carried out using two different approaches—in vitro pull-down assay and co-expression and affinity purification—to verify the effect of single mutations in either WhiB1 or σAC170 on the WhiB1:σAC170 complex formation.
For the in vitro pull-down assays, an equal amount of the purified His6-σAC170 protein (wildtype or mutant) in 50 mM Tris–HCl, pH 8.0, 100 mM NaCl, 1 mM DTT was pre-loaded onto Ni2+-NTA Sepharose resin (GE Healthcare Life Sciences). An excess of tagless WhiB1 (either purified wildtype or the whole cell supernatant containing an over-expressed WhiB1 mutant) protein was then mixed with immobilized His6-σAC170 on the Ni2+ Sepharose resin. After incubating for 5 minutes with gentle mixing followed by extensive washing with the same buffer containing 50 mM imidazole, His6-σAC170 (alone or in complex with WhiB1) was eluted in the same buffer containing 250 mM imidazole and analyzed by SDS-PAGE. A negative control of the Ni2+ Sepharose resin without pre-incubated with His6-σAC170 was used to exclude the possibility that tagless WhiB1 binds nonspecifically to the Ni2+ resin.
For the co-expression and affinity purification method, two plasmids containing genes encoding His6-σAC170 and tagless WhiB1 (either wildtype or mutant), respectively, were co-transformed in E. coli BL21-Gold (DE3) strain. Protein co-expression and purification was performed similarly as described above, except that loose Ni2+ Sepharose HP resin was used for batch affinity purification of the His6-σAC170-bound tagless WhiB1 (either the wildtype or mutants) simultaneously. For each sample, the whole cell supernatant from an equal amount of the cell pellet was mixed with Ni2+ Sepharose HP resin that had been pre-equilibrated in buffer containing 50 mM Tris–HCl, pH 8.0, 100 mM NaCl, 1 mM DTT. The resin was then washed with the same buffer containing 50 mM imidazole, followed by eluting the target proteins with the same buffer containing 250 mM imidazole. The eluted protein samples were analyzed by UV-Vis absorption spectroscopy and SDS-PAGE.
Electrophoretic mobility shift assay (EMSA)
The whiB1 promoter (PwhiB1) was amplified from the M. tuberculosis H37Rv genomic DNA (from 3595416 to 3595712 bp) and cloned into the EcoRI/HindIII site of the pUC18 vector. The resulting plasmid, pUC18-PwhiB1, was used as the template for preparing a biotin-labeled DNA probe using the 5′-biotin labeled M13F and M13R primers. The EMSA experiments were performed using the LightShift Chemiluminescent EMSA Kit (Thermo Fisher Scientific) as previously described (6,19). The biotin-labeled probe (0.2 nM) was incubated with 0–12 μM of His6-σA or His6-σAC170, either alone or pre-mixed with equimolar amounts of WhiB1, in 100 mM NaCl, 40 mM Tris–HCl, pH 8.0, 10 mM MgCl2, 1 mM EDTA, 0.25 mg/ml BSA and 1 mM DTT in a total volume of 20 μl. 50 ng/μl Poly (dI-dC) was included in all reactions as a non-specific competitor. After incubation on ice for 20 min, the reaction mix was subjected to electrophoretic separation in a 6% native polyacrylamide gel at 100 V in 0.5 × TBE buffer. DNA and DNA-protein complexes were transferred from the gel to a Hybond-N+ nylon membrane (GE Healthcare Life Sciences) and UV cross-linked to the membrane. The biotin-labeled DNA was detected using the LightShift Chemiluminescent EMSA Kit according to the manufacturer's instructions.
In vitro transcription assays
The rrnAP3 DNA template used for in vitro transcription assays was amplified by PCR from the M. tuberculosis H37Rv genomic DNA (from 1471524 to 1471892 bp with an expected transcription product of 245 nt including 8 nt from the primer in the 3’ end) and confirmed by DNA sequencing. For each 20-μl reaction, 250 nM RNAP holoenzyme was mixed with the desired amount of tagless WhiB1 (wildtype or F17A mutant) in the KCl assay buffer (10 mM Tris–HCl, pH 8.0, 50 mM KCl, 10 mM MgCl2, 0.1 mM EDTA, 0.1 mM DTT, 50 μg/ml BSA). A negative control reaction was performed simultaneously using 250 nM RNAP core enzyme without σA and WhiB1. The reaction mixtures were incubated for 10 min at room temperature under anaerobic conditions. The rrnAP3 DNA template was then added to each reaction mixture at a final concentration of 50 nM and incubated at room temperature for an additional 10 min. Transcription was initiated by adding NTP mix (ATP, CTP and GTP at a final concentration of 100 μM) and 1.0 μCi of [α-32P]UTP (800 Ci mmol−1, PerkinElmer Life Sciences). After incubation at 37°C for 5 min, the reaction was stopped with a 2X Stop buffer (45 mM Tris–HCl, 45 mM boric acid, 8 M urea, 30 mM EDTA, 0.02% bromophenol blue, 0.02% xylene cyanol). Transcription products were denatured by heating at 95°C, separated on polyacrylamide gels, and visualized using a phosphorimager.
RESULTS
Crystallization and structural determination of the M. tuberculosis WhiB1:σACTD complex
To improve protein stability and enable crystallization of the WhiB1:σACTD complex, we made a truncated WhiB1 (aa 1–76) construct that lacks the unstructured C-terminal loop, and a σA construct that contains only the last 112 residues (aa 417–528). The constructs were designed based on the previously reported NMR structure of free WhiB1 and the cryo-EM structure of σA in the M. tuberculosis RNAP holoenzyme (8,32). The resulting WhiB1 (without a tag) and σA (with a His6 tag) constructs were then co-expressed and purified from E. coli, and crystallized under anaerobic conditions (see MATERIALS AND METHODS).
Experimental phases were determined by single-wavelength anomalous dispersion (SAD) from the SeMet-substituted WhiB1:σAC112 complex, and the structure was solved and refined at 1.85 Å resolution using Phenix and COOT (see MATERIALS AND METHODS, Supplementary Figure S3) (37,38). The data processing and refinement statistics are summarized in Table 1.
Overall structure of the WhiB1:σACTD complex
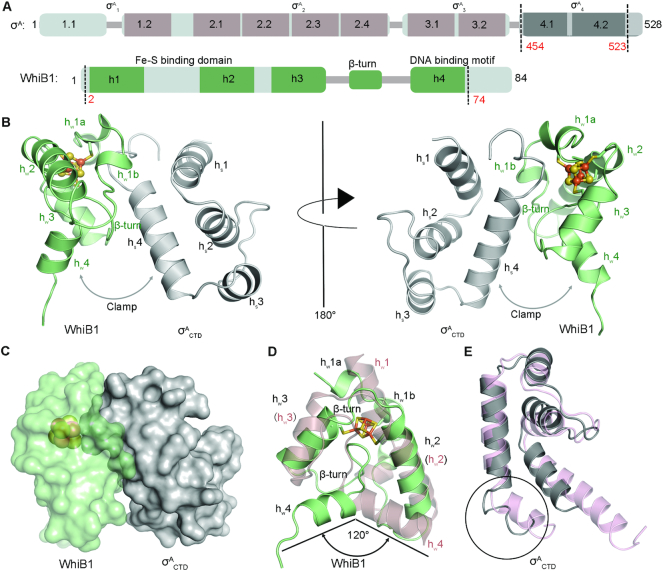
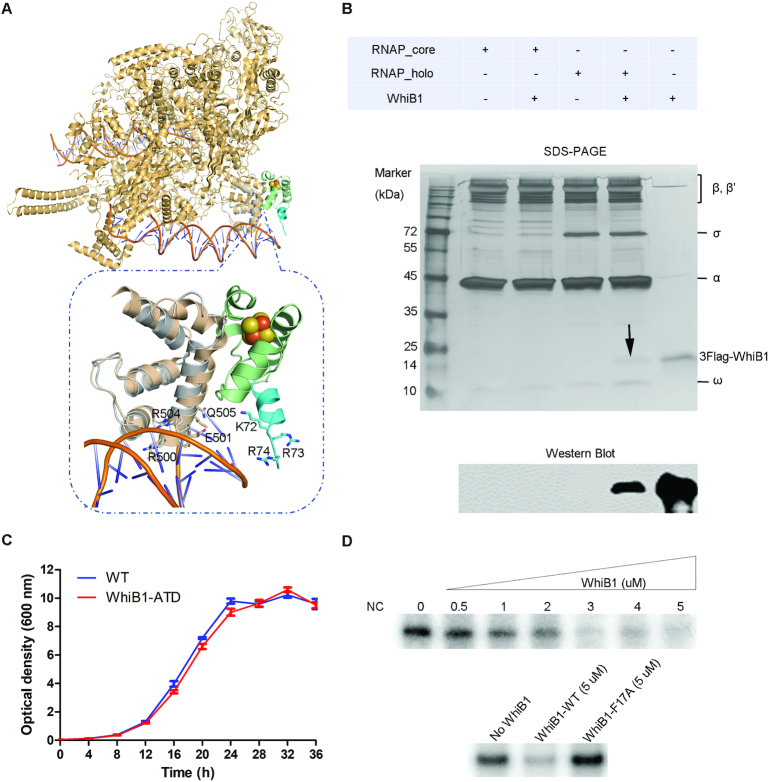
The structure of the WhiB1:σACTD complex shows that WhiB1 and σACTD interact with each other at the [4Fe–4S] cluster binding pocket of WhiB1 (Figure 1). An all-helical fold is adopted by both WhiB1 and σACTD, with helices hw1–4 of WhiB1 and hs1–4 of σACTD, respectively (Figure 1B and C). The complex is hinged at the [4Fe–4S] cluster and forms a clamp between the C-terminal helices of WhiB1 (hw4) and σACTD (hs4) (Figure 1B), both of which have been implicated in DNA binding (19,32,42,43).
Figure 1.
Overview of the WhiB1:σACTD complex from M. tuberculosis. (A) Domain organization of σA and WhiB1 in M. tuberculosis based on the primary and 3D structural analysis. The conserved domains are shown in rectangles, while the variable regions are represented by thin sticks. The regions included in the refined crystal structure are indicated by the dash lines. (B and C) Cartoon and surface representation, respectively, of the WhiB1:σACTD complex. The [4Fe–4S] cluster of WhiB1 is shown in ball-and-stick representation, with Fe atoms colored orange and yellow for S. The clamp shape formed between the C-terminal helices of WhiB1 (hw4) and σACTD (hs4) is highlighted by the arrow. (D) Crystal structure of WhiB1 in the complex is superimposed with the NMR structure of free WhiB1 (PDB ID: 5OAY, in pink). Only the first model of the NMR structure is shown for clarity. (E) Crystal structure of σACTD in the complex is superimposed to the cryo-EM structure of M. tuberculosis RNA polymerase – DNA complex (PDB ID:6C04, in pink). The region different between the two structures at the C-terminus of σACTD is highlighted in the circle.
Structural comparison demonstrates that both WhiB1 and σACTD undergo conformational changes upon forming the complex. An overlay of the WhiB1:σACTD complex with the NMR structure of free WhiB1 reveals a global structural rearrangement upon σACTD binding (8). This includes changes in the inter-helical and helix-loop interactions, and the breaking of helix hw1 into two shorter helical regions (hw1a and b) (Figure 1D). Helix hw4 and the following C-terminal DNA binding motif move concomitantly with the structural rearrangement. In the NMR structure of free WhiB1, helix hw4 is interlocked with hw2, which has been cited to explain why free holo-WhiB1 does not bind to DNA (8). In the WhiB1:σACTD complex helix hw4 of WhiB1 is about 120° apart from its corresponding position in free WhiB1 and is released from hw2 (Figure 1D). The conformation of hw4 in the complex appears stabilized by the interaction with hw3. The hw4 conformation may also be stabilized in part by crystal contacts between WhiB1 and σACTD′ in the second complex molecule of the asymmetric unit (Supplementary Figure S3), while it is expectedly more flexible in solution. The overall conformation of σACTD in the complex is similar to that in the Mtb RNAP holoenzyme, except for the C-terminus of helix hs4 (Figure 1E).
The molecular interface of WhiB1 and σACTD
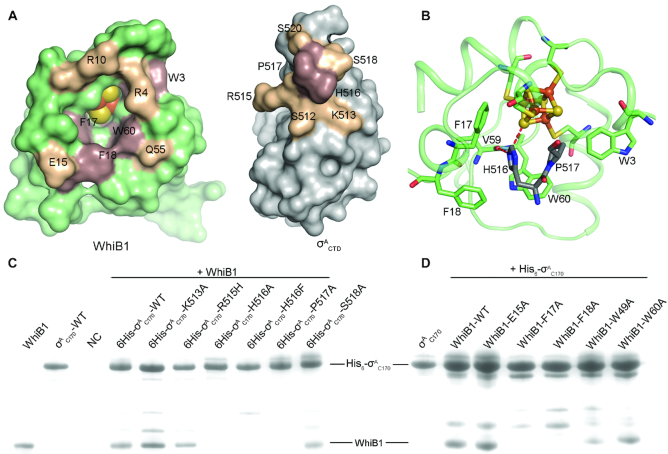
Our structural analysis indicates that the interaction between WhiB1 and σACTD involves considerable hydrophobic contacts around the [4Fe–4S] cluster binding pocket. As shown in Figure 2, His516 and Pro517 in the C-terminal loop of σACTD insert into the [4Fe–4S] cluster binding pocket of WhiB1. There is a cluster of hydrophobic residues around the Fe–S cluster binding pocket, including Trp3, Phe17, Trp60 and Phe18 (Figure 2, Supplementary Table S2). Each of these aromatic residues is highly conserved in WhiB1 across different classes of the phylum Actinobacteria (Supplementary Figure S4).
Figure 2.
Molecular interface of WhiB1 and σACTD. (A) Open-book view of Figure 1C. Residues in contact at the interface of WhiB1 and σACTD are labeled, with the hydrophobic residues colored brown and the charged residues at the surface colored wheat, respectively. (B) Close-up view of the insertion of His516 and Pro517 of σACTD into the [4Fe–4S] cluster binding pocket of WhiB1, surrounded by four aromatic residues (Trp3, Phe17, Phe18 and Trp60) shown in sticks. The polar interactions of the His516 imidazole ring with Val59 and the [4Fe–4S] cluster, respectively, in WhiB1 are highlighted by red dash lines. The [4Fe–4S] cluster of WhiB1 is shown in ball-and-stick representation, with the carbon atoms of WhiB1 and in σACTD colored palegreen and grey, respectively; Fe in orange; N in blue; O in red; and S in yellow. (C) and (D) are the SDS-PAGE analyses of the samples from in vitro pull-down assays. In Panel C, the purified His6-σAC170 proteins (either wildtype or mutants as indicated) were used as the bait, and the purified tagless wildtype WhiB1 protein was used as the prey. In Panel D, the purified wildtype His6-σAC170 protein was used as the bait, and the whole cell supernatant containing a tagless WhiB1 mutant was used as the prey. The purified tagless WhiB1 and His6-σAC170 were loaded in the first two lanes in Panel C as reference. In the negative control (NC), only tagless WhiB1 was used in the pull-down assay without the presence of the bait (His6-σAC170).
To test the importance of these hydrophobic residues for the interaction of WhiB1 with σACTD, His516 and Pro517 were substituted with Ala in σAC170 (containing the last 170 residues 359–528 of σA and the intact σA4). Protein-protein interaction assays with σAC170 H516A and P517A mutants indicate that these hydrophobic residues are critical for WhiB1 binding. Substitution of either His516 or Pro517 by Ala completely abolishes the interaction between WhiB1 and σAC170 as observed both in vitro pull-down assay and in co-expression and affinity purification, establishing the functional importance of these residues (Figure 2C, Supplementary Figure S5). Likewise, an F17A or F18A substitution in WhiB1 abolishes the interaction between WhiB1 and σAC170, while neither shows a significant effect on [4Fe–4S] cluster binding in WhiB1 (Figure 2D, Supplementary Figures. S5 and S6). Substitution of the Trp residues in WhiB1 shows varied effects on complex formation as well as the stability of the [4Fe–4S] cluster, which is discussed in detail in the following section.
Besides the aforementioned hydrophobic interactions, a His516-centered hydrophilic network is also required for the interaction between WhiB1 and σACTD. As highlighted in Figure 2B, Nϵ2 of His516 forms a 2.8-Å hydrogen bond with the carbonyl O of Val59. In addition, Nϵ2 of His516 is 3.5 Å away from a bridging sulfide (S4) in the [4Fe–4S] cluster and thus it may form a polar interaction with the cluster. To evaluate the importance of the His516-centered hydrophilic network for the complex formation, we substituted His516 with Phe in σAC170. The σAC170 H516F mutant completely abolishes binding to WhiB1 (Figure 2C, Supplementary Figure S5). When replacing His516 with Phe in the crystal structure, the shortest distance between the modeled Phe and any non-bonded residues is ∼2.4 Å, suggesting that the observed effect of the H516F mutation is more likely due to disruption of the His516-centered polar interactions rather than due to a steric effect considering the flexibility of the loops in this region.
To test whether disruption of the WhiB1:σA4 interaction affects WhiB1’s essential role in mycobacteria, residues Phe17 and Phe18 in whiB1, both of which are required for σA4 binding in vitro, were mutated to Ala and the impacts on the cell growth in the non-pathogenic mycobacterial model Mycobacterium smegmatis (M. smegmatis) were assessed. All M. tuberculosis Wbl proteins except WhiB5 are highly conserved in M. smegmatis. In particular, the WhiB1 proteins from M. Smegmatis and M. tuberculosis share 96% sequence identity, with variations in only three residues (Supplementary Figure S4). Moreover, as demonstrated in M. tuberculosis (6), deletion of whiB1 is lethal in M. smegmatis unless a complementary copy of whiB1 is present (see MATERIALS and METHODS, Supplementary Figure S1). These results establish that M. smegmatis is a suitable mycobacterial model for the functional studies of WhiB1. We found that substitution of either Phe17 or Phe18 with Ala in whiB1 is lethal in M. smegmatis, consistent with their essential role in the WhiB1:σAC170 interaction in vitro. These results indicate that the interaction between WhiB1 and σA4 is required for WhiB1’s essential role in mycobacterial growth.
Surface charge complementarity is a common mechanism used by σ704-bound transcription activators (7,11,12) but it does not seem to be a critical mechanism for stabilizing the WhiB1:σACTD complex. As shown in Figure 2A, there are two regions in the complex interface that contain oppositely charged residues (Arg4 and Arg10 in WhiB1 versus Ser518 and Ser520 in σACTD; Glu15 and Gln55 in WhiB1 versus Lys513 and Arg515 in σACTD). However, the residue contacts in these two regions vary significantly between two copies of the complex molecules in the asymmetric unit of the crystal structure (Supplementary Table S3), suggesting that they do not drive complex formation. As expected, a single mutation of the selected residues (Glu15 in WhiB1; Lys513, Arg515, Ser518 and Ser520 in σACTD) in these two regions did not show significant effects on the WhiB1:σAC170 complex formation (Figure 2C and D, Supplementary Figure S5). Arg515 lies close to the WhiB1:σACTD interface. While this residue is required for σA binding and transcription activation by several M. tuberculosis Wbl proteins, it is not required by WhiB1 (7,11,12). In the crystal structure of WhiB1:σACTD, the sidechain of Arg515 protrudes out towards the solvent and does not make close contacts with any residues in WhiB1, explaining why Arg515 in WhiB1 is not required for σA binding.
Taken together, our structural analyses and protein-protein interaction assays demonstrate that the formation of the WhiB1:σACTD complex is driven by a combination of hydrophobic contacts and His516-centered polar interactions inside the Fe–S cluster binding pocket, instead of surface charge complementarity. The site-directed mutagenesis studies on whiB1 in M. smegmatis support that the interaction with σA is required for the essential role of WhiB1, revealing a potential mechanism of action for holo-WhiB1 in supporting mycobacterial growth.
The local environment of the [4Fe–4S] cluster binding pocket in the WhiB1:σACTD complex
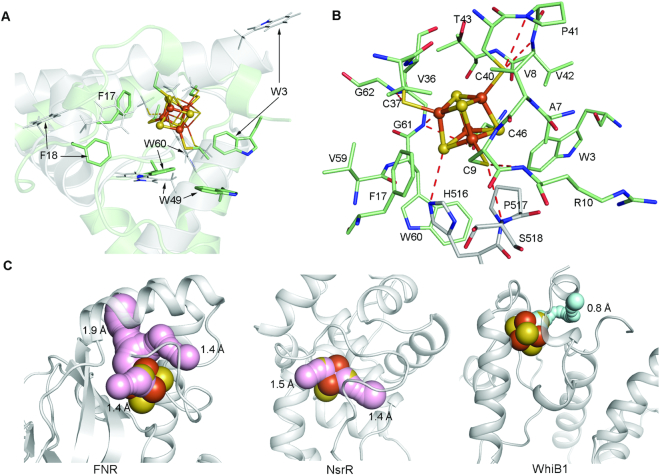
The WhiB1:σA complex has been reported to be O2–stable (8). This differs from other structurally-characterized gas molecule sensors containing a [4Fe–4S] cluster, for example the O2/NO sensor FNR and the NO sensor NsrR, both of which are highly sensitive to O2 (44–47). Examination of the local environment surrounding the [4Fe–4S] cluster in the WhiB1: σACTD complex reveals the structural basis for its exceptional O2 stability. As shown in Figure 3A, the protein environment surrounding the [4Fe–4S] cluster binding site is altered in the WhiB1:σACTD complex relative to free WhiB1. The most noticeable differences include four aromatic residues (Trp3, Phe17, Phe18 and Trp60) that are closer to the Fe–S cluster binding pocket, while the side chain of Trp49 faces away from the cluster in the WhiB1:σACTD complex relative to free WhiB1.
Figure 3.
Structural basis for the stability of the [4Fe–4S] cluster in the WhiB1:σACTD complex. (A) Comparison of the crystal structure of the σACTD–bound WhiB1 with the NMR structure of free WhiB1 (PDB ID: 5OAY) aligned at the Fe-S cluster binding site. Only the first model of the NMR structure is shown for clarity. The [4Fe–4S] cluster and the aromatic residues near the cluster for the σACTD–bound WhiB1 are shown in the ball-and-stick and stick representation, respectively; while their corresponding residues in free WhiB1 are shown in thin lines. The carbon atoms are colored pale green for the σACTD–bound WhiB1 and grey for free WhiB1. Other non-carbon atoms in this figure are colored by the element type: blue for N, red for O, orange for Fe and orange and yellow for S. (B) Stick representation of the local environment of the [4Fe–4S] cluster binding pocket. The [4Fe–4S] cluster is shown in ball-and-stick representation. The carbon atoms are colored pale green for WhiB1 and grey for σACTD. (C) Solvent-accessible paths from the surface towards the [4Fe–4S] cluster in FNR (PDB ID: 5E44) and NsrR (PDB ID:5N07) were calculated by CAVER with a shell radius of 1.4 Å, which is the assumed radius cutoff for solvent molecules. The paths are highlighted in pink and the minimum bottleneck of each path is indicated. No solvent-accessible path is found in the WhiB1:σACTD complex with a shell radius of 1.4 Å. Instead the largest tunnel found in the WhiB1:σACTD complex with the minimum bottleneck of 0.8 Å is shown for demonstration and is highlighted in cyan.
As a result of the conformational changes upon σA4 binding, the [4Fe–4S] cluster of the complex is enclosed in a hydrophobic core consisting of several short-chain hydrophobic residues besides the three aromatic residues of WhiB1 (Trp3, Phe17 and Trp60) and His516 and Pro517 of σACTD mentioned above (Figure 3B). The side chains of the polar residues (Arg4, Arg10 and Thr43 in WhiB1 and Ser518 in σACTD) around the [4Fe–4S] cluster all protrude from the cluster, with the sole exception of His516 in σACTD. Apart from the imidazole ring of His516, six backbone amides are found within 4 Å of the sulfur ligands of the [4Fe–4S] cluster in the complex. The dipoles of these peptide backbone amides may stabilize the cluster via a protein-derived electrostatic field (48,49). Cavity analysis indicates that the biggest channel extending from the WhiB1:σACTD surface to the [4Fe–4S] cluster has a minimum bottleneck of ∼0.8 Å. Assuming a 1.4-Å radius cutoff for water molecules, this indicates that the solvent accessibility to the [4Fe–4S] cluster in the complex is highly limited (Figure 3C). In contrast, multiple channels with a minimum bottleneck of ∼1.4 Å or larger to the [4Fe–4S] cluster in either FNR or NsrR homodimer structures have been identified by cavity analysis, suggesting that the Fe-S clusters of these two proteins remain fully accessible to solvent. Our cavity analysis indicates that solvent accessibility is correlated with the O2 stability of the [4Fe–4S] clusters in these proteins.
The characteristics of the local environment around the [4Fe–4S] cluster in the WhiB1:σACTD complex resemble that of the high potential iron-sulfur proteins (HiPIPs) involved in electron transport (50,51). As indicated by the name, HiPIPs have an unusually high redox potential compared to many other [4Fe–4S] ferredoxins, and undergo a 3+/2+ rather than 2+/1+ redox cycle under physiological conditions (48). HiPIPs are O2-stable owing to their high redox potentials, but remain reactive to NO (52,53). The presence of a hydrophobic barrier composed of aromatic residues, solvent inaccessibility of the [4Fe–4S] cluster, and electrostatic interactions with local backbone amides have been suggested to play a major role in improving the O2 stability of HiPIPs (48,49,51,54). We predict that similar features of the local environment surrounding the [4Fe–4S] cluster in the WhiB1:σACTD complex may increase its redox potential as in HiPIPs and thus improve the cluster stability in the presence of O2.
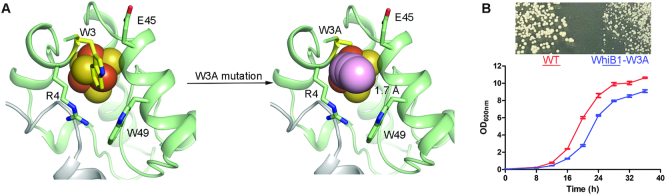
To identify the critical residues near the Fe–S cluster and to test whether aromatic residues and/or solvent accessibility contribute to the stability of the [4Fe–4S] cluster in the WhiB1:σACTD complex as demonstrated in HiPIPs, we carried out an alanine scanning mutagenesis of the three Trp residues near the cluster in WhiB1. The first Trp residue we examined is Trp3. As shown in Figure 3A, Trp3 is close to the [4Fe–4S] cluster of the complex and stands at the protein surface facing bulk solvent in this crystal structure. Cavity analysis predicts that a W3A mutation would open a pore in the cluster binding pocket large enough for solvent access to the cluster in the complex (Figure 4A). However, Trp3 is far from the Fe–S cluster (>7.5 Å in all models) in the NMR structure of free WhiB1, and no conformational change of Trp3 was detected upon σA binding by the NMR chemical shift perturbation analysis (8). To evaluate the effect of Trp3 on the Fe-S cluster stability in free WhiB1 and in the WhiB1:σACTD complex, we characterized the WhiB1 W3A mutant in vitro and in M. smegmatis. As shown in Supplementary Figure S5, we found that the W3A mutation in WhiB1 results in a severe loss of the [4Fe–4S] cluster and diminishes the stability of WhiB1 when expressed and purified alone. The W3A mutation also disrupts WhiB1’s interaction with σAC170, most likely due to the instability of the cluster (Supplementary Figure S5, Supplementary Figure S6B). In M. smegmatis, the whiB1-W3A mutant showed a significant defect in cell growth under optimal growth conditions (Figure 4B). These observations unambiguously indicate, that Trp3 is crucial for the stability of the Fe–S cluster in WhiB1 and the interaction between WhiB1 and σA, consistent with our crystal structural analysis but different from the reported NMR studies (8). These results also corroborate that binding to σA is required for WhiB1’s role in mycobacterial growth as described above in the mutagenesis studies on Phe17 and Phe18 in WhiB1.
Figure 4.
Effect of Trp3 on the stability of the [4Fe–4S] cluster in the WhiB1:σACTD complex. (A) Solvent-accessible path from the surface towards the [4Fe–4S] cluster in the mutated model of the WhiB1-W3A:σACTD complex was calculated by CAVER 3.0 with a shell radius of 1.4 Å. The minimum bottleneck of the path is 1.7 Å. (B) Cell growth of M. smegmatis wildtype (WT) and the whiB1-W3A mutant on either a 7H10 agar plate for 3 days (top) or in 7H9 broth under optimal growth conditions. Error bars in the growth curves represent the standard deviation of triplicate samples.
In contrast to Trp3, the WhiB1-W49A mutant protein has properties comparable to those of the wildtype WhiB1 in SDS-PAGE and UV-Visible spectral analyses when co-expressed and purified as a complex with His6-σAC170 from E. coli (Supplementary Figures S5C and S6B). However, when expressed and purified alone from E. coli, WhiB1-W49A exhibits a less stable [4Fe–4S] cluster based on the UV-Visible spectra and has weaker binding to σAC170 in the in vitro pull-down assay (Figure 2D, Supplementary Figures S5 and S6). These results indicate that Trp49 in WhiB1 undergoes a conformational change upon σACTD binding as suggested by the structural comparison between the NMR structure of free WhiB1 and our crystal structure of the WhiB1:σACTD complex (Figure 3A). The mutation of Trp60 (W60A), on the other hand, has little effect either on the stability of the [4Fe–4S] cluster or on its binding to σAC170 (Figure 2D, Supplementary Figures. S5 and S6). This is likely because Trp60 is buried inside the Fe–S cluster binding pocket in the complex and is not exposed to the bulk solvent.
Altogether, our structural and biochemical analysis indicate that the unusually hydrophobic, solvent-inaccessible local environment surrounding the [4Fe–4S] cluster in the WhiB1:σACTD complex makes the cluster remarkably stable to O2 and thus favorable for NO sensing in aerobic environments.
WhiB1 represses transcription by interacting with σA4 in the RNAP holoenzyme
When superimposing σACTD in the WhiB1:σACTD complex on the cryo-EM structure of the M. tuberculosis RNAP–DNA complex, we find no steric clash between WhiB1 and the core subunits in the RNAP holoenzyme (Figure 5A) (32). We thus predict that WhiB1 can bind to σA4 in the RNAP holoenzyme. To test this, we first incubated either the RNAP holoenzyme or core enzyme with and without σA, respectively, in the presence of an excess amount of the 3FLAG-tagged holo-WhiB1 under anaerobic conditions. The mixtures were then separated by size exclusion chromatography, and the peak fractions were collected for SDS-PAGE and western blot analysis. The results clearly demonstrate that WhiB1 forms a stable complex with the RNAP holoenzyme in a σA–dependent manner (Figure 5B).
Figure 5.
WhiB1 represses transcription from the rrnAP3 promoter in vitro by interaction with σA in the RNA polymerase holoenzyme. (A) Superimposition of the WhiB1-bound σACTD and σACTD onto the M. tuberculosis RNAP–DNA complex (PDB ID: 6C04). The residues implicated in DNA binding in WhiB1 and σA4 are highlighted. (B) Detection of complex formation between WhiB1 and σA in RNAP by size-exclusion chromatography. 3FLAG-WhiB1 was detected by SDS PAGE (as indicated by the arrow) and western blot analysis using anti-FLAG M2 antibody (Sigma) in the peak fraction of the sample containing the RNAP holoenzyme preincubated with 3FLAG-WhiB1, but not in the sample either containing the RNAP holoenzyme alone or containing the RNAP core enzyme (without σA) either or not preincubated with 3FLAG-WhiB1. (C) Cell growth curves of M. smegmatis wildtype (WT) and the whiB1 truncation mutant (WhiB1-ATD) without the C-terminal DNA binding motif, AT-hook, in 7H9 broth under optimal growth conditions. (D) In vitro transcription from the rrnAP3 promoter in the absence and presence of the wildtype WhiB1 or the WhiB1-F17A mutant. Each 20-μl reaction contains the RNAP holoenzyme (250 nM), the rrnAP3 DNA template (10 nM), and varying amounts of the wild-type WhiB1 or WhiB1-F17A mutant as indicated. The reaction buffer contains 10 mM Tris-HCl, pH 8.0, 50 mM KCl, 10 mM MgCl2, 0.1 mM EDTA, 0.1 mM DTT, and 50 μg/ml BSA. The RNAP holoenzyme was pre-incubated with the WhiB1 proteins under anaerobic conditions before adding the rrnAP3 DNA template. The RNAP core enzyme (250 nM) was used as a negative control (NC). The in vitro transcript products were separated on 8M-6% polyacrylamide gels, and the full gel images are shown in Supplementary Figure S8.
The superimposed structure also reveals that the clamp comprising the C-terminal helix hw4 in WhiB1 and helix hs4 in σACTD, is positioned across the DNA helix of the -35 element in the RNAP–DNA complex (Figure 5A). Several residues of σACTD implicated in DNA binding align along the inner interface of the clamp and are in close contact with DNA (the inset of Figure 5A) (32). Helix hw4 of WhiB1 and the DNA binding motif immediately following the C-terminus of this region straddle the DNA helix (the inset of Figure 5A). However, none of the residues in the C-terminal DNA binding motif of WhiB1 seem to be close enough for DNA binding in this superimposed model. Considering the flexibility of the loop structure on both ends of WhiB1 hw4, we could not rule out the possibility that σA–bound WhiB1 is potentially involved in DNA binding in solution. To address this possibility, we examined the effect of WhiB1 on σA binding to the whiB1 promoter (PwhiB1) by electrophoretic mobility shift assays (EMSAs). The results from EMSA assays indicate that addition of WhiB1 to either σAC170 or σA does not enhance its binding to PwhiB1 under the experimental conditions (Supplementary Figure S7).
To test whether the C-terminal DNA binding motif (AT-hook) of WhiB1 is required for its essential role in mycobacterial growth, we constructed a truncation mutant of whiB1 in M. smegmatis with a deletion of the AT-hook (whiB1-ATD). In this mutant, the last 13 residues (Lys72–Val84), including five Lys and Arg residues that are required for DNA binding by apo-WhiB1 (19), are removed from the whiB1 gene by homologous recombination-based in-frame unmarked deletion (see MATERIALS AND METHODS, Supplementary Figure S1D). As shown in Figure 5C, we find that the growth rate of the whiB1-ATD mutant was comparable to the wildtype strain under optimal growth conditions, indicating that the single DNA binding motif of WhiB1 is dispensable to M. smegmatis viability under non-stressed conditions.
The results from our structural analysis and the mutagenesis study on the single DNA binding motif in WhiB1 described above led us to search for an alternative mechanism of transcriptional regulation by holo-WhiB1 different from that of a canonical σA4-dependent transcription activator. A close examination of the overlaid structures between the WhiB1:σACTD complex and M. tuberculosis RNAP indicates that the WhiB1 binding site on σA4 overlaps with the C-terminal domain of the α subunit (αCTD) in RNAP. αCTD is separated from the N-terminal domain of the α subunit by a long flexible linker that is absent in most mycobacterial RNAP structures (55). In E. coli and Bacillus subtilis, αCTD binds to the A/T-rich regions upstream of the −35 element (UP element) for transcription factor–independent activation of many promoters, including the gene encoding 16S rRNA (56,57). A similar mechanism of transcriptional regulation by αCTD likely exists in M. tuberculosis based on the 3D structure of RNAP and sequence analysis of potential UP elements in promoters (55). If so, WhiB1 binding to σACTD may block the access of αCTD to the UP element and thus interfere with the αCTD-dependent transcription.
To test this hypothesis, we examined the effect of WhiB1 on the in vitro transcription of the M. tuberculosis rrs gene encoding 16S rRNA from the rrnAP3 promoter in M. tuberculosis (58). The results shown in Figure 5D indicate a clear concentration–dependent transcription repression from rrnAP3 by the wildtype WhiB1 but not the WhiB1-F17A mutant, which does not bind to σA4. Thus, we conclude that the transcription repression from rrnAP3 by holo-WhiB1 requires its interaction with σA4 under these experimental conditions. To evaluate the effect of WhiB1 on the promoter escape, we examined the abortive transcripts from the in vitro transcription reaction mixtures. As shown in Supplementary Figure S9, we find that addition of WhiB1 does not cause an increase in abortive transcripts, which would be expected if WhiB1 repressed gene expression by inhibiting promoter escape. This observation is consistent with our hypothesis that WhiB1 represses transcription from rrnAP3 by preventing αCTD from binding to the UP element.
DISCUSSION
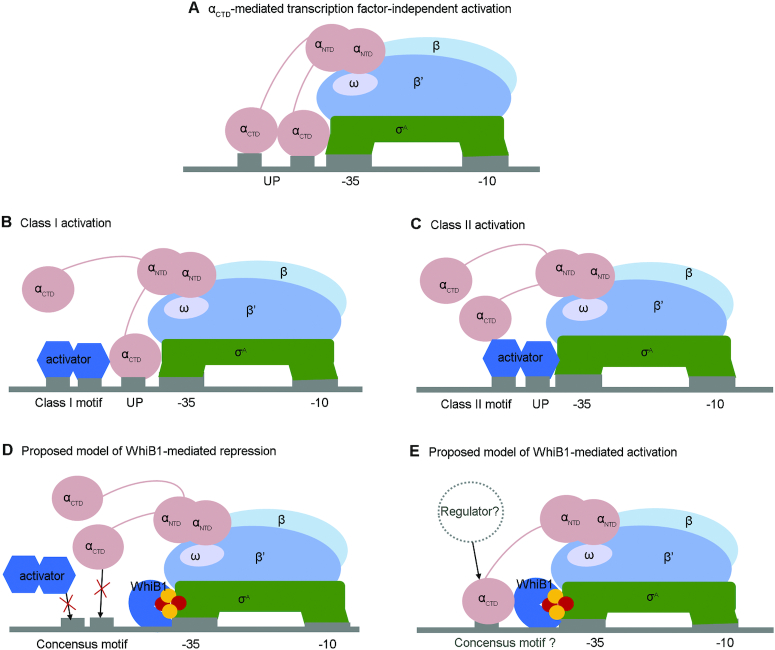
In the present report, we provide the first atomic view of the molecular interface between WhiB1 and σACTD and establish the structural basis for the O2 stability of the [4Fe–4S] cluster in the σACTD–bound WhiB1. We also demonstrate that WhiB1 can repress gene expression by interaction with σA in the RNAP holoenzyme in vitro, and that the interaction between WhiB1 and σA4 is essential for supporting mycobacterial growth in vivo. As discussed below, these data suggest that WhiB1 regulates gene expression by a non-canonical mechanism that is distinguished from to the promoter-centered, Class II transcription activation typically observed with σ704–bound transcription activators (Figure 6) (10,23).
Figure 6.
Proposed working model of holo-WhiB1 in transcriptional regulation via interaction with σA4. (A) Diagram that demonstrates the αCTD-mediated transcription factor–independent activation by recognizing the up element (UP) of the target promoters. (B and C) Diagrams of the Class I and Class II transcription activation, respectively. The 3D structures of the cyclic AMP receptor protein (CRP) bound RNAP–DNA complexes for the CRP-dependent Class I and Class II transcription activation (PDB IDs: 3IYD and 5I2D, respectively) are used as models. (D and E) Diagrams of the proposed working model for holo-WhiB1 in transcriptional regulation via interaction with σA4. Panel D illustrates that WhiB1 binding to σA4 can block the binding site for activators or αCTD, and thus repress transcription. Panel E shows that WhiB1 binding to σA4 in RNAP, either alone or in concert with a to-be-defined transcription regulator, may stabilize the interaction between RNAP and a specific subset of promoters, and thus activate transcription.
The crystal structure of the WhiB1:σACTD complex reported here not only provides long-sought atomic detail about the interaction between σA4 and a Wbl protein, but also reveals how a monomeric transcription regulator with a single DNA binding motif interacts with σA4 in bacteria. A genome-wide survey of oligomerization states of transcription factors from M. tuberculosis suggests that monomeric transcription factors are more abundant than the few that have been characterized (7). For example, two essential monomeric transcription factors, RbpA and CarD, have recently been identified as global transcription regulators. They stabilize the RNAP–DNA open complex in M. tuberculosis via interaction with RNAP (including regions 2 and 3 of σA) and the fork-junction promoter DNA near the -10 element with low sequence specificity (59–61). Considering the structure of WhiB1, we expect that holo-WhiB1 may have low DNA specificity like RbpA and CarD, if it binds to DNA at all. This speculation is supported by our observation that WhiB1 does not significantly enhance the binding affinity of σA to the whiB1 promoter in the EMSA assays, and that the single DNA binding motif of WhiB1 is not required for its essential role in supporting mycobacterial growth. Although we cannot rule out the possibility that σA4–bound WhiB1 may recognize target gene promoters other than whiB1 itself, such a case has not yet been reported for a canonical Class II transcription activator in bacteria. As a σA4-bound transcription regulator, WhiB1 is more likely to be involved in differentiation of gene expression by modulating the DNA binding preference of RNAP, rather than facilitating the transition and stabilizing the open state of the RNAP–DNA complex as RbpA and CarD do.
A key finding in this report is the unusual hydrophobic residue-driven interaction between WhiB1 and σA4, which induces conformational changes in both binding partners. This distinguishes WhiB1 from other well-characterized Class II transcription activators (i.e. CRP and FNR) that bind directly to σ704 (10,23). In the canonical Class II activation mechanism, transcription activators recognize and bind to the target DNA sequence adjacent to the -35 element, and then recruit RNAP to the site for the initiation of transcription via contact with σ704 (Figure 6). Although less common, there are examples of transcription activators, such as SoxS involved in redox stress response, that can bind to σ704 in RNAP without the presence of target DNA via the pre-recruitment mechanism (27). In either case, these transcription activators are in loose contact with σ704 through surface charge complementarity, and typically do not induce conformational changes in either protein (11,12,21). The loose interaction with σ704 is consistent with the promoter-centered activation mechanism, which requires rapid disassociation between the transcription activator and RNAP upon initiation of transcription, and release of RNAP for recruitment by other σ704–dependent transcription factors. The hydrophobic residue-driven interaction between WhiB1 and σA4, on the other hand, is incompatible with such a mechanism. Interestingly, a similar interaction with σ704 has been observed with the so-called “phage appropriator” AsiA, a viral transcription regulator from the bacteriophage T4 that remodels the DNA preference of host RNAP by a mechanism of σ appropriation (62,63). In this mechanism, AsiA binds tightly to σ704 in E. coli RNAP and induces conformational changes in σ704 so that it cannot recognize the –35 element of host genes; instead the phage activator MotA makes use of the remodeled σ704 in host RNAP to express genes in the middle stage of the phage T4 life cycle.
Besides the σA4–bound transcription activators, the interaction interface of WhiB1 and σA also overlaps with αCTD in RNAP. In bacteria, αCTD not only binds to the UP elements for transcription factor–independent activation of many promoters, but also plays a major role in transcriptional regulation through interaction with Class I and Class II transcription factors (Figure 6A-C) (64–66). The effect of WhiB1 on in vitro transcription that we observe aligns with previous studies on the 3D structure of RNAP and sequence analysis of potential UP elements in promoters in M. tuberculosis (55), suggesting that αCTD may play a similar role in transcriptional regulation in mycobacteria as reported in E. coli and other bacteria. How the interaction between WhiB1 and σA influences the functional role of αCTD and σA4–bound transcription activators warrants further investigation.
Considering all the evidence together, we propose that holo-WhiB1 regulates gene expression by modulating the promoter recognition preference of RNAP during transcription initiation (Figure 6D and E). This working model accounts for our observation of an unusual hydrophobic residue-driven interaction between holo-WhiB1 and σA4 and high levels of expression of WhiB1 relative to σA in M. tuberculosis, as reported previously (5). On one hand, owing to its tight interaction with σA4, it is plausible that WhiB1 could block Class II transcription activators and/or αCTD from binding to σA4, thus suppressing expression of genes regulated by these transcription activators and/or αCTD if at high cellular levels (Figure 6D). We have experimentally demonstrated that WhiB1 can interact with σA to repress transcription from rrnAP3 in vitro. Moreover, it has been reported that under-expression of whiB1 in M. tuberculosis caused more genes to be up-regulated than down-regulated (20 versus 3) in a comparative transcriptomic experiment (8). Our model is also consistent with the previous report that WhiB1 was post-translationally regulated by the Clp protease, and disruption of the WhiB1 degradation by the Clp protease inhibited cell growth in M. tuberculosis (67). On the other hand, our site-directed mutagenesis of whiB1 in M. smegmatis suggests that the interaction with σA4 is required for the essential role of WhiB1 in supporting mycobacterial growth. This may imply that the interaction between WhiB1 and σA4 is required for the activation of essential genes, either alone or in concert with other regulators or αCTD (Figure 6E). Candidate genes for this mode of regulation include the essential gene encoding salicylate synthase, mbtI, which was down-regulated with under-expression of WhiB1 in M. tuberculosis (8,68).
Although the proposed working model of holo-WhiB1 is unprecedented among the σA4–bound bacterial transcription factors, it closely resembles the mode of action of Spx, a global regulator that is responsive to heat and oxidative stress in Bacillus subtilis (69–71). Similar to holo-WhiB1, Spx is a monomeric transcription factor without known DNA binding capacity (57). It was first characterized as an anti-α factor because Spx was found to bind to αCTD of RNAP and interfere with αCTD-dependent transcription of genes (57,72,73). Subsequently, it was reported that Spx also activate redox stress response genes and its interaction with αCTD is required for transcription activation (73,74). Like WhiB1, Spx is also regulated by the Clp protease (75), presumably to balance its dual activities.
In summary, our results indicate that holo-WhiB1 is a non-canonical transcription factor that interacts with σA4 using a newly characterized interface dominated by hydrophobic contacts and represses transcription from rrnAP3 in vitro, distinguishing it from previously characterized σ704-dependent transcription activators (7,11,12). The interaction with σA4 also renders the Fe–S cluster in WhiB1 remarkably stable to O2, which is needed for NO sensing in aerobic bacteria. Future investigations of the genetic and functional aspects of WhiB1 and the structural characterization of WhiB1 interaction with the RNAP holoenzyme will further advance the mechanistic understanding of WhiB1 in NO sensing and transcriptional regulation in M. tuberculosis.
DATA AVAILABILITY
Atomic coordinates and structure factors have been deposited in the RCSB Protein Data Bank (PDB) under the accession codes 6ONO and 6ONU for the WhiB1:σACTD complex in the C2221 form and P21 form, respectively.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Prof. M.A. Wilson and Prof. J.B. Howard for insightful discussions, Prof. M. Kato-Maeda for the M. tuberculosis H37Rv genomic DNA, Prof. Y. Sun for the plasmids used for site-directed mutagenesis in M. smegmatis, Prof. R. Landick for the plasmid and protocols for purification of the RNA polymerase and in vitro transcription experiment, Prof. C. Woods and Prof. L. Zhang for sharing equipment. The authors thank staff at beamlines 9-2 and 12-2 of SSRL for the assistance during the X-ray diffraction data collection.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Foundation CAREER Award [CLP 1846908 to L-M.Z.]; National Institute of Health [P20 GM113126 to L-M.Z. through the Nebraska Center for Integrated Biomolecular Communication, and P30 GM103335 to L-M.Z. and D.F.B. through the Nebraska Redox Biology Center]; Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences [DE-AC02–76SF00515]. Funding for open access charge: National Science Foundation CAREER Award (to L-M.Z.).
Conflict of interest statement. None declared.
REFERENCES
- 1. World Health Organization Global Tuberculosis Report 2016. 2016;
- 2. Chandra G., Chater K.F.. Developmental biology of Streptomyces from the perspective of 100 actinobacterial genome sequences. FEMS Microbiol. Rev. 2014; 38:345–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Davis N.K., Chater K.F.. The Streptomyces coelicolorwhiB gene encodes a small transcription factor-like protein dispensable for growth but essential for sporulation. Mol. Gen. Genet. 1992; 232:351–358. [DOI] [PubMed] [Google Scholar]
- 4. Saini V., Farhana A., Steyn A.J.. Mycobacterium tuberculosis WhiB3: a novel iron-sulfur cluster protein that regulates redox homeostasis and virulence. Antioxid Redox Signal. 2012; 16:687–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Larsson C., Luna B., Ammerman N.C., Maiga M., Agarwal N., Bishai W.R.. Gene expression of Mycobacterium tuberculosis putative transcription factors whiB1-7 in redox environments. PLoS One. 2012; 7:e37516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Smith L.J., Stapleton M.R., Fullstone G.J., Crack J.C., Thomson A.J., Le Brun N.E., Hunt D.M., Harvey E., Adinolfi S., Buxton R.S. et al.. Mycobacterium tuberculosis WhiB1 is an essential DNA-binding protein with a nitric oxide-sensitive iron-sulfur cluster. Biochem. J. 2010; 432:417–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Feng L., Chen Z., Wang Z., Hu Y., Chen S.. Genome-wide characterization of monomeric transcriptional regulators in Mycobacterium tuberculosis. Microbiology. 2016; 162:889–897. [DOI] [PubMed] [Google Scholar]
- 8. Kudhair B.K., Hounslow A.M., Rolfe M.D., Crack J.C., Hunt D.M., Buxton R.S., Smith L.J., Le Brun N.E., Williamson M.P., Green J.. Structure of a Wbl protein and implications for NO sensing by M. tuberculosis. Nat. Commun. 2017; 8:2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gomez M., Doukhan L., Nair G., Smith I.. sigA is an essential gene in Mycobacterium smegmatis. Mol. Microbiol. 1998; 29:617–628. [DOI] [PubMed] [Google Scholar]
- 10. Lee D.J., Minchin S.D., Busby S.J.. Activating transcription in bacteria. Annu. Rev. Microbiol. 2012; 66:125–152. [DOI] [PubMed] [Google Scholar]
- 11. Burian J., Yim G., Hsing M., Axerio-Cilies P., Cherkasov A., Spiegelman G.B., Thompson C.J.. The mycobacterial antibiotic resistance determinant WhiB7 acts as a transcriptional activator by binding the primary sigma factor SigA (RpoV). Nucleic Acids Res. 2013; 41:10062–10076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Steyn A.J., Collins D.M., Hondalus M.K., Jacobs W.R. Jr., Kawakami R.P., Bloom B.R.. Mycobacterium tuberculosis WhiB3 interacts with RpoV to affect host survival but is dispensable for in vivo growth. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:3147–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gusarov I., Shatalin K., Starodubtseva M., Nudler E.. Endogenous nitric oxide protects bacteria against a wide spectrum of antibiotics. Science. 2009; 325:1380–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. MacMicking J., Xie Q.W., Nathan C.. Nitric oxide and macrophage function. Annu. Rev. Immunol. 1997; 15:323–350. [DOI] [PubMed] [Google Scholar]
- 15. Crack J.C., Smith L.J., Stapleton M.R., Peck J., Watmough N.J., Buttner M.J., Buxton R.S., Green J., Oganesyan V.S., Thomson A.J. et al.. Mechanistic insight into the nitrosylation of the [4Fe–4S] cluster of WhiB-like proteins. J. Am. Chem. Soc. 2011; 133:1112–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Crack J.C., Green J., Thomson A.J., Le Brun N.E.. Iron-sulfur clusters as biological sensors: the chemistry of reactions with molecular oxygen and nitric oxide. Acc. Chem. Res. 2014; 47:3196–3205. [DOI] [PubMed] [Google Scholar]
- 17. Spiro S. Nitric oxide-sensing mechanisms in Escherichia coli. Biochem. Soc. Trans. 2006; 34:200–202. [DOI] [PubMed] [Google Scholar]
- 18. Stapleton M.R., Smith L.J., Hunt D.M., Buxton R.S., Green J.. Mycobacterium tuberculosis WhiB1 represses transcription of the essential chaperonin GroEL2. Tuberculosis. 2012; 92:328–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Smith L.J., Stapleton M.R., Buxton R.S., Green J.. Structure-function relationships of the Mycobacterium tuberculosis transcription factor WhiB1. PLoS One. 2012; 7:e40407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Feng Y., Zhang Y., Ebright R.H.. Structural basis of transcription activation. Science. 2016; 352:1330–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lonetto M.A., Rhodius V., Lamberg K., Kiley P., Busby S., Gross C.. Identification of a contact site for different transcription activators in region 4 of the Escherichia coli RNA polymerase σ70 subunit. J. Mol. Biol. 1998; 284:1353–1365. [DOI] [PubMed] [Google Scholar]
- 22. Mettert E.L., Kiley P.J.. Reassessing the structure and function relationship of the O2 sensing transcription factor FNR. Antioxid Redox Signal. 2018; 29:1830–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Browning D.F., Busby S.J.. Local and global regulation of transcription initiation in bacteria. Nat. Rev. Microbiol. 2016; 14:638–650. [DOI] [PubMed] [Google Scholar]
- 24. Niu W., Kim Y., Tau G., Heyduk T., Ebright R.H.. Transcription activation at Class II CAP-dependent promoters: Two interactions between CAP and RNA polymerase. Cell. 1996; 87:1123–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Burian J., Ramon-Garcia S., Sweet G., Gomez-Velasco A., Av-Gay Y., Thompson C.J.. The mycobacterial transcriptional regulator whiB7 gene links redox homeostasis and intrinsic antibiotic resistance. J. Biol. Chem. 2012; 287:299–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Taliaferro L.P., Keen E.F. 3rd, Sanchez-Alberola N., Wolf R.E. Jr.. Transcription activation by Escherichia coli Rob at Class II promoters: protein-protein interactions between Rob's N-terminal domain and the σ70 subunit of RNA polymerase. J. Mol. Biol. 2012; 419:139–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zafar M.A., Shah I.M., Wolf R.E. Jr.. Protein-protein interactions between σ70 region 4 of RNA polymerase and Escherichia coli SoxS, a transcription activator that functions by the prerecruitment mechanism: evidence for “off-DNA” and “on-DNA” interactions. J. Mol. Biol. 2010; 401:13–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mao X.J., Yan M.Y., Zhu H., Guo X.P., Sun Y.C.. Efficient and simple generation of multiple unmarked gene deletions in Mycobacterium smegmatis. Sci. Rep. 2016; 6:22922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. van Kessel J.C., Hatfull G.F.. Recombineering in Mycobacterium tuberculosis. Nat. Methods. 2007; 4:147–152. [DOI] [PubMed] [Google Scholar]
- 30. Williams K.J., Joyce G., Robertson B.D.. Improved mycobacterial tetracycline inducible vectors. Plasmid. 2010; 64:69–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Doublie S. Preparation of selenomethionyl proteins for phase determination. Methods Enzymol. 1997; 276:523–530. [PubMed] [Google Scholar]
- 32. Boyaci H., Chen J., Lilic M., Palka M., Mooney R.A., Landick R., Darst S.A., Campbell E.A.. Fidaxomicin jams Mycobacterium tuberculosis RNA polymerase motions needed for initiation via RbpA contacts. Elife. 2018; 7:e34823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Johnson M.K. Iron-sulfur proteins: new roles for old clusters. Curr. Opin. Chem. Biol. 1998; 2:173–181. [DOI] [PubMed] [Google Scholar]
- 34. Otwinowski Z., Minor W.. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997; 276:307–326. [DOI] [PubMed] [Google Scholar]
- 35. Kabsch W. XDS. Acta Crystallogr. D. 2010; 66:125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W. et al.. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Emsley P., Lohkamp B., Scott W.G., Cowtan K.. Features and development of Coot. Acta Crystallogr D. 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W. et al.. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sievers F., Higgins D.G.. Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 2018; 27:135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Robert X., Gouet P.. Deciphering key features in protein structures with the new ENDscript server. Nucleic. Acids. Res. 2014; 42:W320–W324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chovancova E., Pavelka A., Benes P., Strnad O., Brezovsky J., Kozlikova B., Gora A., Sustr V., Klvana M., Medek P. et al.. CAVER 3.0: a tool for the analysis of transport pathways in dynamic protein structures. PLoS Comput. Biol. 2012; 8:e1002708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lonetto M., Gribskov M., Gross C.A.. The σ70 family: sequence conservation and evolutionary relationships. J. Bacteriol. 1992; 174:3843–3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Campbell E.A., Muzzin O., Chlenov M., Sun J.L., Olson C.A., Weinman O., Trester-Zedlitz M.L., Darst S.A.. Structure of the bacterial RNA polymerase promoter specificity sigma subunit. Mol. Cell. 2002; 9:527–539. [DOI] [PubMed] [Google Scholar]
- 44. Volbeda A., Dodd E.L., Darnault C., Crack J.C., Renoux O., Hutchings M.I., Le Brun N.E., Fontecilla-Camps J.C.. Crystal structures of the NO sensor NsrR reveal how its iron-sulfur cluster modulates DNA binding. Nat. Commun. 2017; 8:15052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kiley P.J., Beinert H.. Oxygen sensing by the global regulator, FNR: the role of the iron-sulfur cluster. FEMS Microbiol. Rev. 1998; 22:341–352. [DOI] [PubMed] [Google Scholar]
- 46. Volbeda A., Darnault C., Renoux O., Nicolet Y., Fontecilla-Camps J.C.. The crystal structure of the global anaerobic transcriptional regulator FNR explains its extremely fine-tuned monomer-dimer equilibrium. Sci Adv. 2015; 1:e1501086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Partridge J.D., Bodenmiller D.M., Humphrys M.S., Spiro S.. NsrR targets in the Escherichia coli genome: new insights into DNA sequence requirements for binding and a role for NsrR in the regulation of motility. Mol. Microbiol. 2009; 73:680–694. [DOI] [PubMed] [Google Scholar]
- 48. Jensen G.M., Warshel A., Stephens P.J.. Calculation of the redox potentials of iron-sulfur proteins: the 2-/3- couple of [Fe4S*4Cys4] clusters in Peptococcus aerogenes ferredoxin, Azotobacter vinelandii ferredoxin I, and Chromatium vinosum high-potential iron protein. Biochemistry. 1994; 33:10911–10924. [DOI] [PubMed] [Google Scholar]
- 49. Langen R., Jensen G.M., Jacob U., Stephens P.J., Warshel A.. Protein control of iron-sulfur cluster redox potentials. J. Biol. Chem. 1992; 267:25625–25627. [PubMed] [Google Scholar]
- 50. Hirano Y., Takeda K., Miki K.. Charge-density analysis of an iron-sulfur protein at an ultra-high resolution of 0.48 Å. Nature. 2016; 534:281–284. [DOI] [PubMed] [Google Scholar]
- 51. Dey A., Jenney F.E. Jr., Adams M.W., Babini E., Takahashi Y., Fukuyama K., Hodgson K.O., Hedman B., Solomon E.I.. Solvent tuning of electrochemical potentials in the active sites of HiPIP versus ferredoxin. Science. 2007; 318:1464–1468. [DOI] [PubMed] [Google Scholar]
- 52. Foster H.W., Cowan J.A.. Chemistry of nitric oxide with protein-bound iron sulfur centers. Insights on physiological reactivity. J. Am. Chem. Soc. 1999; 121:4093–4100. [Google Scholar]
- 53. Crack J.C., Stapleton M.R., Green J., Thomson A.J., Le Brun N.E.. Mechanism of [4Fe–4S](Cys)4 cluster nitrosylation is conserved among NO-responsive regulators. J. Biol. Chem. 2013; 288:11492–11502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Agarwal A., Li D.W., Cowan J.A.. Role of aromatic residues in stabilization of the [Fe4S4] cluster in high-potential iron proteins (HiPIPs): physical characterization and stability studies of Tyr-19 mutants of Chromatium vinosum HiPIP. Proc. Natl. Acad. Sci. U.S.A. 1995; 92:9440–9444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hubin E.A., Lilic M., Darst S.A., Campbell E.A.. Structural insights into the mycobacteria transcription initiation complex from analysis of X-ray crystal structures. Nat. Commun. 2017; 8:16072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Rao L., Ross W., Appleman J.A., Gaal T., Leirmo S., Schlax P.J., Record M.T., Gourse R.L.. Factor-independent activation of rrnB P1 - an “extended” promoter with an Upstream Element that dramatically increases promoter strength. J. Mol. Biol. 1994; 235:1421–1435. [DOI] [PubMed] [Google Scholar]
- 57. Schafer H., Heinz A., Sudzinova P., Voss M., Hantke I., Krasny L., Turgay K.. Spx, the central regulator of the heat and oxidative stress response in B. subtilis, can repress transcription of translation-related genes. Mol. Microbiol. 2019; 111:514–533. [DOI] [PubMed] [Google Scholar]
- 58. Gonzalez-y-Merchand J.A., Colston M.J., Cox R.A.. The rRNA operons of Mycobacterium smegmatis and Mycobacterium tuberculosis: comparison of promoter elements and of neighbouring upstream genes. Microbiology. 1996; 142:667–674. [DOI] [PubMed] [Google Scholar]
- 59. Hubin E.A., Tabib-Salazar A., Humphrey L.J., Flack J.E., Olinares P.D., Darst S.A., Campbell E.A., Paget M.S.. Structural, functional, and genetic analyses of the actinobacterial transcription factor RbpA. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:7171–7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hubin E.A., Fay A., Xu C., Bean J.M., Saecker R.M., Glickman M.S., Darst S.A., Campbell E.A.. Structure and function of the mycobacterial transcription initiation complex with the essential regulator RbpA. Elife. 2017; 6:e22520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Boyaci H., Chen J., Jansen R., Darst S.A., Campbell E.A.. Structures of an RNA polymerase promoter melting intermediate elucidate DNA unwinding. Nature. 2019; 565:382–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lambert L.J., Wei Y., Schirf V., Demeler B., Werner M.H.. T4 AsiA blocks DNA recognition by remodeling σ70 region 4. EMBO J. 2004; 23:2952–2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Gregory B.D., Nickels B.E., Garrity S.J., Severinova E., Minakhin L., Urbauer R.J.B., Urbauer J.L., Heyduk T., Severinov K., Hochschild A.. A regulator that inhibits transcription by targeting an intersubunit interaction of the RNA polymerase holoenzyme. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:4554–4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Gourse R.L., Ross W., Gaal T.. UPs and downs in bacterial transcription initiation: the role of the alpha subunit of RNA polymerase in promoter recognition. Mol. Microbiol. 2000; 37:687–695. [DOI] [PubMed] [Google Scholar]
- 65. Hudson B.P., Quispe J., Lara-Gonzalez S., Kim Y., Berman H.M., Arnold E., Ebright R.H., Lawson C.L.. Three-dimensional EM structure of an intact activator-dependent transcription initiation complex. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:19830–19835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ross W., Schneider D.A., Paul B.J., Mertens A., Gourse R.L.. An intersubunit contact stimulating transcription initiation by E coli RNA polymerase: interaction of the alpha C-terminal domain and σ region 4. Genes Dev. 2003; 17:1293–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Raju R.M., Jedrychowski M.P., Wei J.R., Pinkham J.T., Park A.S., O’Brien K., Rehren G., Schnappinger D., Gygi S.P., Rubin E.J.. Post-translational regulation via Clp protease is critical for survival of Mycobacterium tuberculosis. PLoS Pathog. 2014; 10:e1003994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Harrison A.J., Yu M.M., Gardenborg T., Middleditch M., Ramsay R.J., Baker E.N., Lott J.S.. The structure of MbtI from Mycobacterium tuberculosis, the first enzyme in the biosynthesis of the siderophore mycobactin, reveals it to be a salicylate synthase. J. Bacteriol. 2006; 188:6081–6091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zuber P. Spx-RNA polymerase interaction and global transcriptional control during oxidative stress. J. Bacteriol. 2004; 186:1911–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Rojas-Tapias D.F., Helmann J.D.. Identification of novel Spx regulatory pathways in Bacillus subtilis uncovers a close relationship between the CtsR and Spx regulons. J. Bacteriol. 2019; 201:e00151-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Schafer H., Turgay K.. Spx, a versatile regulator of the Bacillus subtilis stress response. Curr. Genet. 2019; 65:871–876. [DOI] [PubMed] [Google Scholar]
- 72. Nakano S., Nakano M.M., Zhang Y., Leelakriangsak M., Zuber P.. A regulatory protein that interferes with activator-stimulated transcription in bacteria. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:4233–4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Newberry K.J., Nakano S., Zuber P., Brennan R.G.. Crystal structure of the Bacillus subtilis anti-alpha, global transcriptional regulator, Spx, in complex with the alpha C-terminal domain of RNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:15839–15844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Nakano S., Kuster-Schock E., Grossman A.D., Zuber P.. Spx-dependent global transcriptional control is induced by thiol-specific oxidative stress in Bacillus subtilis. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:13603–13608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Nakano M.M., Hajarizadeh F., Zhu Y., Zuber P.. Loss-of-function mutations in yjbD result in ClpX- and ClpP-independent competence development of Bacillus subtilis. Mol. Microbiol. 2001; 42:383–394. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors have been deposited in the RCSB Protein Data Bank (PDB) under the accession codes 6ONO and 6ONU for the WhiB1:σACTD complex in the C2221 form and P21 form, respectively.