Abstract
Discovering new long non-coding RNAs (lncRNAs) has been a fundamental step in lncRNA-related research. Nowadays, many machine learning-based tools have been developed for lncRNA identification. However, many methods predict lncRNAs using sequence-derived features alone, which tend to display unstable performances on different species. Moreover, the majority of tools cannot be re-trained or tailored by users and neither can the features be customized or integrated to meet researchers’ requirements. In this study, features extracted from sequence-intrinsic composition, secondary structure and physicochemical property are comprehensively reviewed and evaluated. An integrated platform named LncFinder is also developed to enhance the performance and promote the research of lncRNA identification. LncFinder includes a novel lncRNA predictor using the heterologous features we designed. Experimental results show that our method outperforms several state-of-the-art tools on multiple species with more robust and satisfactory results. Researchers can additionally employ LncFinder to extract various classic features, build classifier with numerous machine learning algorithms and evaluate classifier performance effectively and efficiently. LncFinder can reveal the properties of lncRNA and mRNA from various perspectives and further inspire lncRNA–protein interaction prediction and lncRNA evolution analysis. It is anticipated that LncFinder can significantly facilitate lncRNA-related research, especially for the poorly explored species. LncFinder is released as R package (https://CRAN.R-project.org/package=LncFinder). A web server (http://bmbl.sdstate.edu/lncfinder/) is also developed to maximize its availability.
Keywords: sequence intrinsic composition, multi-scale secondary structure, EIIP physicochemical property, machine learning, predictive modeling
Introduction
Long non-coding RNAs (lncRNAs), one kind of transcripts that are longer than 200 nucleotides and unable to encode proteins in the intracellular space, have been at the forefront in recent years [1–3]. Several studies indicate that more than 80% of the human genome has biochemical functions, whereas only less than 2% of the genome can be translated into proteins [4, 5]. Furthermore, up to 70% of the non-coding sequences are transcribed into lncRNAs [6]. All these figures suggest that lncRNAs embrace lots of valuable information awaiting our exploration. Only a small fraction of lncRNAs have been studied, but scientists have discovered a wide range of biological processes that lncRNAs involved, such as epigenetic regulation, metabolic processes, chromosome dynamics and cell differentiation [7–12]. Lots of evidence have indicated that lncRNAs are highly relevant to various complex human diseases [13] such as lung cancer [14] Alzheimer diseases [15] and cardiovascular diseases [16]. Database LncRNADisease [17] and Lnc2Cancer [18] have collected thousands of experimentally verified relations between lncRNAs and diseases, which have also confirmed the intimate connections between lncRNAs and diseases.
Nowadays, next-generation sequencing technologies have furnished us thousands and thousands of unclassified transcripts, demanding prompt studies. From identification to annotation, many platforms and databases have been developed to facilitate research on lncRNAs [19, 20]. LncRNA identification is the fundamental step of lncRNA research. Many methods and tools have been developed using machine learning techniques. CPC (Coding Potential Calculator) [21] aligns sequences against reference protein database, which is highly representative of the alignment-based tools. As an extremely powerful tool for coding potential assessment, CPC is not tailored for lncRNA identification, but it can predict lncRNAs according to the open reading frame (ORF) information and alignment results of BLASTX [22]: transcripts with great coding potential tend to possess HITs and ORFs with relatively high quality and thus being classified as protein-coding RNAs. Nonetheless, several major limitations can hardly be avoided because of CPC’s great reliance upon reference protein databases. First, lncRNAs are less conserved than mRNAs. A high proportion of lncRNAs display many features similar to protein-coding sequences [23], which may mislead CPC and make it incorrectly categorize lncRNAs as mRNAs. Second, CPC requires a high-quality and rather comprehensive database, but many species have only insufficient annotations. Moreover, CPC relies heavily on the outputs of BLASTX, but multiple-sequence alignment tools cannot guarantee optimal alignments [24]. Finally, the extremely time-consuming process of alignment makes the use of CPC on massive-scale data difficult. Apart from BLASTX, other types of alignment are also employed to identify lncRNAs. PhyloCSF [25] is based on multiple alignments and phylogenetic model comparison; COME [26] uses features from BLASTX and phastCons score [27]; lncRNA-ID [28] and lncRScan-SVM [29] employ profile-hidden Markov model-based alignment [30] and PhastCons score, respectively.
Owing to the restrictions of CPC, several alignment-free approaches are developed afterward. CPAT (Coding Potential Assessment Tool) [24], CNCI (Coding-Non-Coding Index) [31] and PLEK (predictor of long noncoding RNAs and messenger RNAs based on an improved k-mer scheme) [32] are the typical examples of this category. The main advantage of alignment-free tools is high efficacy without loss of accuracy. CPAT calculates Fickett TESTCODE score [33, 34] and hexamer score on ORF region to measure the differences of nucleotide position and codon usage between non-coding transcripts and protein-coding transcripts. The features of CNCI are based on adjoining nucleotide triplets (ANTs) matrix and unequal distribution of codons (codon bias). PLEK employs improved k-mer scheme to classify the sequences. COME selects Infernal result [48], expression data [49, 50] and histone modification [4] as features. To overcome the drawbacks of CPC, a new lncRNA identification tool named CPC2 [35] was developed recently. Unlike CPC, CPC2 is an alignment-free tool and is based on only sequence-intrinsic features. Compared with CPC, CPC2 displays substantial improvement in accuracy and efficiency. In addition to several classic features such as ORF information and Fickett TESTCODE score, CPC2 also utilizes isoelectric point (pI) [36, 37] to calculate coding potential and thus predicts lncRNAs. Alignment-free lncRNA identification tools also include DeepLNC [38], lncScore [39] and FEElnc [40]. All these popular lncRNA identification methods are summarized in Table 1. From Table 1, it can be observed that features of many methods are based upon adjoining nucleotide frequencies, directly or indirectly. The essence of these kinds of features is to evaluate the differences in intrinsic composition between lncRNAs and mRNAs. However, the problem is the sequence compositions varies from species to species, and thus these methods provide very unstable performances on different species [41]. One possible way to cushion this negative effect is re-training the machine learning model for different species, although only CPAT and PLEK can be re-trained by users. However, the limitation of this remedy includes insufficient sequences of many species that makes it impossible to tailor the model specifically for every species. Thus, the pre-built model should be well qualified for various species.
Table 1.
Overview of machine learning-based lncRNA identification tools
| Methods | CPC [21] | CPAT a [24] | CNCI [31] | PLEK [32] | LncRNA-ID [28] | lncRScan-SVM [29] | DeepLNC [ 38 ] | COME [ 26 ] | CPC2 [ 35 ] |
|---|---|---|---|---|---|---|---|---|---|
| Year | 2007 | 2013 | 2013 | 2014 | 2015 | 2015 | 2016 | 2016 | 2017 |
| Category | Protein-coding potential calculator | Protein-coding potential calculator | lncRNA predictor | lncRNA predictor | lncRNA prediction method | lncRNA predictor | lncRNA predictor | Protein-coding potential calculator | Protein-coding potential calculator |
| Input Format | FASTA | FASTA, BED | FASTA, GTF | FASTA | GTF | FASTA | GTF | FASTA, GTF | |
| Species | Multi-species | Human, mouse, | Vertebrate, plant | Vertebrate, plant | Human, mouse | human | human, mouse, | Multi-species | |
| fly, zebrafish | fly, worm, plant | ||||||||
| Requirements | Linux, BLAST, | Linux, Python2.7, | Linux, Python2.7 | Linux, Python2.7 | Linux, Python2.7, | Linux, R | Linux, Python, | ||
| Protein database | R | Biopython | Biopython | ||||||
| Model | SVM | Logistic regression | SVM | SVM | Balanced random forest | SVM | deep neural network | balanced random forest | support vector machine |
| Features | ORF information, BLASTX [22] | ORF length, transcript length, Fickett TESTCODE score [33, 34], Hexamer score | ANT information, codon bias | Improved k-mer frequencies | ORF length and coverage, ribosome interaction [42–44], profile HMM-based alignment [30] | Count of stop codon, exon information, txCdsPredict score [45], PhastCons score [27] | k-mer frequencies | GC content, BLASTX [22], PhastCons score [27], ribosome profiling [3], INFERNAL result [48], expression data [49, 50], histone modification [4] | ORF information, Fickett TESTCODE score [33, 34], isoelectric point [36, 37] |
| Re-training | |||||||||
| PubMed | https://www.ncbi.nlm.nih.gov/pubmed/17631615 | https://www.ncbi.nlm.nih.gov/pubmed/23335781 | https://www.ncbi.nlm.nih.gov/pubmed/23892401 | https://www.ncbi.nlm.nih.gov/pubmed/25239089 | https://www.ncbi.nlm.nih.gov/pubmed/26315901 | https://www.ncbi.nlm.nih.gov/pubmed/26437338 | https://www.ncbi.nlm.nih.gov/pubmed/27608726 | https://www.ncbi.nlm.nih.gov/pubmed/28521017 | |
| Software | http://cpc.cbi.pku.edu.cn/download | https://sourceforge.net/projects/rna-cpat | https://github.com/www-bioinfo-org/CNCI | https://sourceforge.net/projects/plek | https://sourceforge.net/projects/lncrscansvm | https://bioserver.iiita.ac.in/deeplnc | https://github.com/lulab/COME | http://cpc2.cbi.pku.edu.cn/download.php | |
| Web Server | http://cpc.cbi.pku.edu.cn | http://lilab.research.bcm.edu/cpat | http://cpc2.cbi.pku.edu.cn |
A brief summary of several lncRNA identification tools. Among the methods summarized above, the majority of tools identify lncRNAs with sequence-derived features alone, and only CPAT and PLEK can be re-trained by users. DeepLNC only provides web server, while CNCI, PLEK, lncRScan-SVM and COME can be downloaded for local use. CPC, CPAT and CPC2 are released as stand-alone application as well as web server. All the stand-alone tools listed in the table require Linux operating system.
CPAT has updated its features in the latest version. Original feature” coverage of ORF” has been replaced with feature ‘length of the transcript’. Features ‘Fickett TESTCODE score’ and ‘hexamer score’ are calculated on ORF region.
Different tools also select different machine learning algorithms to construct a classifier: CPC, CNCI, PLEK, lncRScan-SVM and CPC2 use support vector machine (SVM); CPAT and lncScore employ logistic regression, whereas lncRNA-ID, COME and FEElnc are based on random forest or balanced random forest [46, 47]. It is also worth mentioning that DeepLNC is constructed using deep learning algorithm, deep neural network (DNN).
All these approaches can conduct lncRNA identification, but it is difficult for users to customize the tools for non-model organism transcriptomes or analyze sequences with specific features. In addition, different tools employ different machine learning algorithms. But to what extent will different machine learning algorithms alter the classifiers’ performances? In this study, we establish an integrated lncRNA identification package LncFinder, which could help users extract features from different feature categories, construct classifiers with various machine learning algorithms and evaluate the performances of different feature combinations or machine learning algorithms. Besides, methods based on k-mer frequencies often have a large number of features and demonstrate unstable results on different species. LncFinder includes two schemes to refine the features and enhance the stability. Experimental results show that the schemes we proposed achieve satisfactory accuracy and stability. Apart from sequence composition features used by existing tools, we are also wondering whether there are any other kinds of critical information embodied in a sequence that can be explored to predict lncRNA. Thus, features from secondary structure and physicochemical property are explored and introduced to conduct lncRNA identification. In light of the comprehensive feature exploration and selection, a novel lncRNA identification method is also developed. Benchmarked against several state-of-the-art tools, our method presents the most satisfactory overall result on multiple species.
Hexamer score [24], as the most discriminating feature of CPAT, is an ingenious method to streamline the features of the k-mer scheme. In this article, we defined two measurements Euclidean-distance and Logarithm-distance to capture the differences of sequence-intrinsic composition between lncRNA and protein-coding RNA. Having been evaluated on the human data set, the Euclidean-distance scheme achieved an accuracy of 0.8484 and Logarithm-distance achieved 0.8521, while the hexamer score of CPAT obtained an accuracy of 0.6416. We next investigated other kinds of features from the perspective of secondary structure and physicochemical property. In the secondary structure-derived feature group, multi-scale structural information was introduced. Furthermore, electron–ion interaction pseudo-potential (EIIP) [51] was employed to calculate physicochemical features. These two feature groups were evaluated comprehensively using recursive feature elimination (RFE) and the feature selection algorithm we designed. Tested on the same data set as sequence-derived features, feature combinations from multi-scale secondary structure category obtained an accuracy of 0.8525, while EIIP-based physicochemical features obtained an accuracy of 0.8853. Together with sequence-intrinsic composition features, three feature groups were incorporated into five different machine learning classifiers constructed with logistic regression, SVM, random forest, extreme learning machine (ELM) and deep learning algorithms to assess to what degree can different machine learning algorithms affect the performance of these features. The novel lncRNA identification method we proposed was developed using the optimal feature combination and machine learning algorithm. We finally compared this new method with several widely used tools on the data sets of human (Homo sapiens), mouse (Mus musculus), wheat (Triticum aestivum), zebrafish (Danio rerio), and chicken (Gallus gallus). On the one hand, we expect our method can show improvements in accuracy and efficiency, and on the other hand, we intend to evaluate each tool’s stability on different species.
LncFinder is an integrated package that can be used to predict lncRNA and analyze the properties of lncRNA. LncFinder aims to offer new perspectives to capture the differences between lncRNAs and mRNA. Compared with existing lncRNA identification tools, LncFinder has the following merits:
LncFinder includes an innovative algorithm predicting lncRNAs using heterologous features from three different categories: intrinsic composition of sequence, multi-scale structural information and physicochemical property based on EIIP and fast Fourier transform (FFT). This novel method outperforms several widely used tools on multiple species with more robust and reliable performances.
The machine learning model used by LncFinder is determined with comprehensive comparisons. Five classifiers, logistic regression, SVM, random forest, ELM and deep learning are evaluated with parameter tuning. The classifier is constructed with the algorithm that obtains the highest accuracy.
LncFinder is highly flexible and remarkably user-friendly. Almost all classic alignment-free features proposed by other methods can be extracted with LncFinder. As a one-stop platform, LncFinder can complete feature extraction, feature selection, classifier construction and performance evaluation easily and efficiently. LncFinder’s customization of features and machine learning algorithm will effectively facilitate research on poorly explored species and lncRNA properties analysis. The support of parallel computing will also greatly accelerate the process of feature selection and classifier construction.
LncFinder is readily accessible and convenient to use. Virtually all lncRNA identification tools require UNIX/Linux operating system (OS) and several hundred megabytes (MB), even several gigabytes (GB), of storage space to compute the sequences locally, whereas LncFinder is released as R package and is compatible with almost all widely used OS platforms, such as Windows, UNIX/Linux and Mac OS X. Accepted by Comprehensive R Archive Network (CRAN), LncFinder can be installed conveniently in R with only one command, and the size of LncFinder is only 2.7 MB. In addition, a web server is also developed to provide a practical and effective alternative for lncRNA identification. The web server can classify lncRNAs of multiple species and calculate sequence coding potential. Informative lncRNA-related tools, databases and research progress are also summarized on our web server for users’ reference. The summaries have revised some outdated details and are updated regularly.
Feature exploration
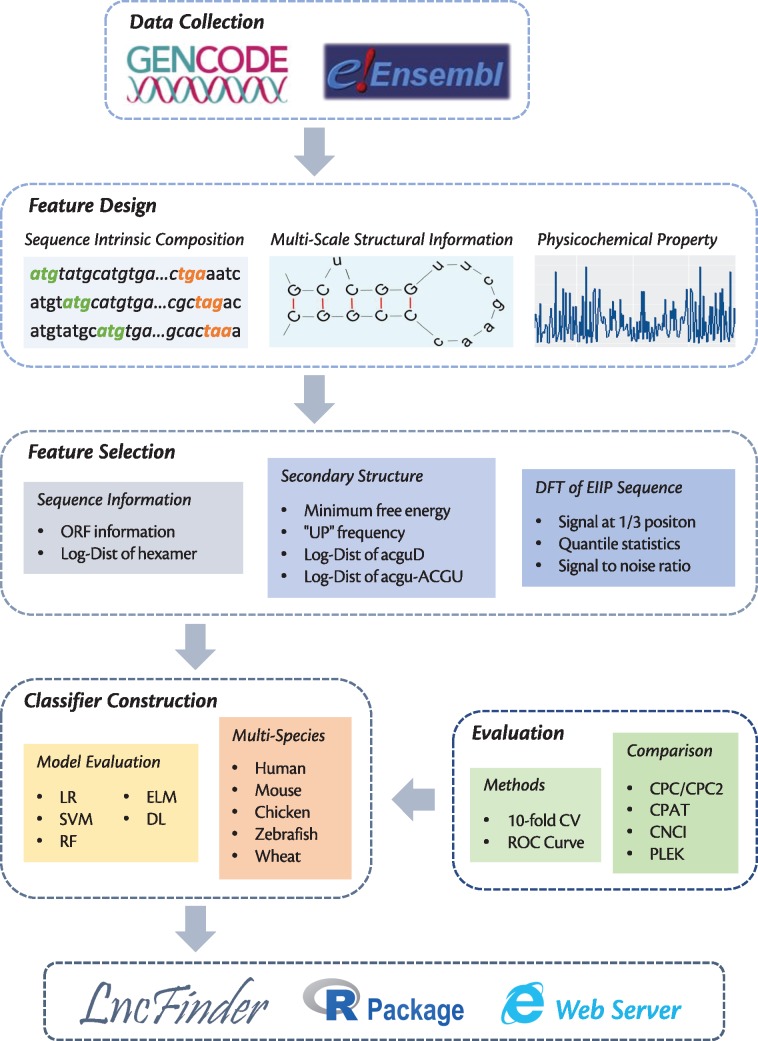
In this article, we discuss the discriminating power of three kinds of feature categories, especially secondary structural and physicochemical features. Classical features employed by existing tools are reviewed, and new features are also designed to offer a new perspective on lncRNA identification. The framework of this research is displayed in Figure 1.
Figure 1.
Framework of this study. Data sets used in our experiments are collected from GENCODE and Ensembl. Only one transcript from each gene is used. In addition to sequence-intrinsic composition, features are also extracted from multi-scale secondary structure and EIIP-based physicochemical property using two feature selection schemes. Evaluated with 10-fold CV and ROC curve, the optimal feature combination and machine learning algorithm are obtained to develop a new method for lncRNA identification. This method is benchmarked against five popular tools on five species, and it is finally included in LncFinder, which is a highly flexible package for lncRNA identification and analysis. LncFinder is published as R package as well as web server.
Features are evaluated using the data sets of multiple species. Data sets of human and mouse used in our experiments are the same as those of Achawanantakun et al., 2015 [28], which are collected from GENCODE [2, 52] and experimentally verified data [3]. Data sets of wheat, chicken and zebrafish are collected from Ensembl [53]. In these data sets, only one transcript from each gene is used. Detailed information has been summarized in Table S1 (Supplementary File 1 - Methods). All data sets can be downloaded from our web server.
Features of sequence intrinsic composition
Several studies have demonstrated that the distribution of adjoining bases is different in non-coding RNAs (ncRNAs) and protein-coding transcripts [24, 31]. The most general method to capture the distribution differences is k-mer scheme, which is employed by CNCI (k = 3), PLEK (k = 1–5), DeepLNC (k = 2, 3, 5) and some other tools. Nevertheless, the feature number rises dramatically with the increase of k. Considering that protein-coding transcripts are finally translated into amino acid sequences, the combination of two adjoining amino acids should have some patterns, hence a biased usage of these nucleotides (A, C, G and T). We can therefore distinguish lncRNAs from protein-coding transcripts by measuring hexamer usage. However, there will be 46 hexamer features if we extract features utilizing k-mer scheme. Too many features will lead to extremely cumbersome processes of classification and classifier construction.
Hexamer score [24] of CPAT is a useful way to measure hexamer frequencies without a large number of features, but this method only computes the average hexamer frequencies of the training data sets. For any unknown transcript, the hexamer patterns are scanned, but their frequencies are abandoned. Here, we propose the following two new measurements to quantify the usage bias of hexamer: Euclidean-distance and Logarithm-distance. Each scheme has three features: distance to lncRNA (Dist.LNC), distance to protein-coding transcript (Dist.PCT) and distance ratio (Dist.Ratio). We define these features as follows:
where freq.seq are the k-adjoining base(s) frequencies of one unevaluated sequence; freq.lnc denotes the average frequencies of lncRNAs’ k-adjoining base(s); i denotes the different types of k-adjoining base(s), and n is the total number of the k-adjoining base(s) in one sequence. Based on first two equations, EucDist.LNC and LogDist.LNC can be computed. EucDist.PCT and LogDist.PCT can be obtained similarly. The main idea underlying the proposed measurements is to estimate the unevaluated sequence is ‘close to’ lncRNA or protein-coding sequence. The two measurements and hexamer score will be evaluated in our experiments with 10-fold cross validation (10-fold CV). Because both the hexamer frequencies in training set and unevaluated sequences are considered by our measurements, we expect they can display more stable performances than hexamer score on multiple species.
For protein-coding transcripts, the longest ORF closely resembles coding sequence (CDS), which is the region that can be translated to amino acids. Although lncRNAs are non-coding transcripts, the longest ORF can also be regarded as the most potential region for encoding amino acids. Because the hexamers in CDS encode amino acids for some purposes, we expect that calculating hexamer frequencies on the longest ORF region is more sensible than on full sequence. Thus, we will evaluate the performances of three schemes (Euclidean-distance, Logarithm-distance and hexamer score) on the full sequence as well as on the longest ORF region. The sliding window will slide 1 nt each step on full sequence but slide 3 nt each step on the longest ORF region to simulate the translation process.
Features of secondary structural information
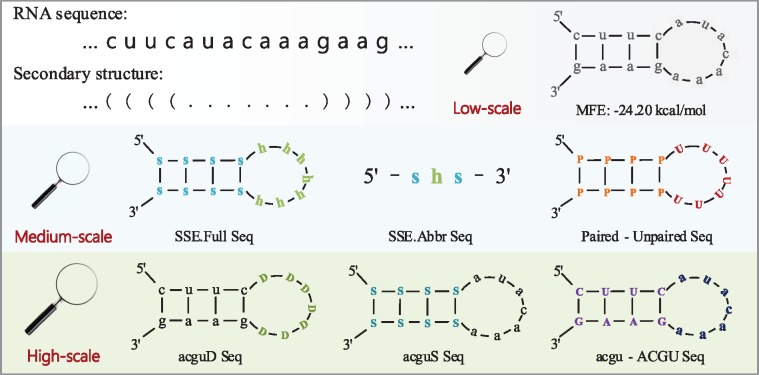
The secondary structure plays important roles in multiple biological functions and is considered more conserved than primary sequence [54, 55]. But seldom has structural information been employed to predict lncRNA. To explore the discriminating power of this category, we here introduce multi-scale secondary structural features that portray the structural information of one RNA sequence from the following three levels: stability, secondary structure elements (SSEs) combined with pairing condition and structure-nucleotide sequences. RNA secondary structures can be obtained from program RNAfold of ViennaRNA Package [56], which calculates secondary structures using the minimum free energy (MFE)-based algorithm.
MFE is a basic structural outline displaying the stability of the RNA structure and is thus being selected as the low-scale feature. Although only few lncRNAs are unstable, lncRNAs are, on average, less stable than mRNAs [57]. The box plots of the MFE of lncRNAs and mRNAs are displayed in Figure S1-1 (a), which show that mRNAs generally tend to possess lower MFE.
To extract features from higher levels, we first design six multi-scale secondary structure-derived sequences. Let seq[n] be an RNA sequence of length N, and the nucleotides are denoted in lowercase . Let SS[n] be the secondary structure sequence of seq[n], and SS[n] is defined using dot-bracket notation . SSEs can depict one RNA’s basic components, and here we employ the following four SSEs: stem (s), bulge (b), loop (l) and hairpin (h). Figure 2 illustrates how six multi-scale secondary structural sequences are obtained from seq[n] and SS[n]. Replacing nucleotides of seq[n] with corresponding SSEs, we can obtain the first secondary structure-derived sequence—SSE full sequence (SSE.Full Seq). Regarding the continuous identical SSE as one SSE, another sequence—SSE abbreviated sequence (SSE.Abbr Seq)—is obtained. In the dot-bracket notation, a dot ‘.’ means unpaired base and brackets ‘(‘or’)’ represent paired base (also the SSE stem). Thus, SS[n] can be converted to Paired-Unpaired Seq using the following formula:
Figure 2.
Illustration of multi-scale secondary structure-derived sequences. As a low-scale feature, MFE is a basic structural outline presenting one RNA sequence’s stability. Medium-scale sequences briefly sketch RNA information and can be obtained from dot-bracket notation sequence alone without using sequence nucleotide composition. High-scale sequences can be viewed as a high-resolution panorama displaying the integration of sequence and structural information.
These three sequences can sketch basic RNA information and be viewed as medium-scale structural information that is converted from SS[n] alone without using the nucleotide composition of seq[n].
Just like observing an object using different magnifiers, we can perceive more details with a high-power magnifier. On a high-scale level, three secondary structure-derived sequences, namely, acgu-Dot Sequence (acguD Seq), acgu-Stem Sequence (acguS Seq) and acgu-ACGU Seq, can be obtained by combining secondary structure sequence SS[n] and primary sequence seq[n]:
In acguD Seq, unpaired nucleotides are replaced with character ‘D’, thus acguD Seq can be regarded as a portrait describing the percentage of the unpaired base and the intrinsic composition of the SSE stem. Similarly, acguS Seq can be viewed as a portrait serving the complementary roles. The third sequence acgu-ACGU Seq is obtained by converting nucleotides of seq[n] into uppercase if they are paired bases. The combination of these three sequences can be considered a high-resolution panorama presenting the integration of sequence and structural information.
In our study, two strategies, improved k-mer scheme [58] and Logarithm-distance of k-adjoining bases, are employed to extract features from six multi-scale secondary structural sequences. The optimal k will be determined by 10-fold CV.
Features of EIIP-derived physicochemical features
CPC2 uses pI values to reveal the physicochemical differences between lncRNAs and protein-coding transcripts. CPC2 attempts to theoretically translate RNA sequence into protein sequence and applies pI values to the new obtained protein sequence. In this article, we explore the physicochemical property from another viewpoint, namely, EIIP values. EIIP was initially used to locate exons. Each nucleotide (a, c, g and t) has one EIIP value, and these values indicate the energy of delocalized elections in nucleotides [51]. For any DNA sequence, nucleotides can be replaced with the following EIIP values: . Compared with pI values, EIIP values are directly applied to RNA sequences, which can avoid the potential bias caused by the speculated translation process.
Let Xe[n] be the EIIP indicator sequence of Seq[n]. Using FFT on Xe[n], we can get the corresponding power spectrum :
For protein-coding transcripts, an obvious peak usually appears at the N∕3 position, but no such peak can be found in non-coding transcripts [59] (see Figure S1-2 for example). Moreover, the power of the protein-coding transcript is generally higher than that of lncRNA. Thus, we can capture these differences with the following features: the signal at 1/3 position (), average power () and signal-to-noise ratio (SNR). and SNR are defined as follows:
From the box plots in Figure S1-1 (b, c, d), it can be noted that most of lncRNAs possess lower and SNR values.
We additionally sort the power spectrum in descending order and calculate the quantiles statistics (Q1, Q2, Q3, minimum and maximum values) of power values on different ranges. The ranges are designed with two different ways. The ranges in the first group varies from the top 10 to top 100 of the sorted power spectrum, and the ranges are also from the top 10% to 100% of the sorted power spectrum. As the signals of mRNAs are generally stronger than those of lncRNAs, protein-coding transcripts should tend to have higher values of quantiles statistics than lncRNAs. EIIP-based features embody the physicochemical as well as 3-base periodicity properties of protein-coding sequences [60, 61], and we anticipate that features from this category can present robust results on non-model data sets.
Feature selection and model validation
Feature selection is conducted with 10-fold CV to determine the optimal feature extraction scheme as well as to evaluate the performance of different feature groups. The performances are evaluated with the following five standard metrics: sensitivity, specificity, accuracy, F-measure and Cohen's kappa coefficient [62].
In Cohen’s kappa coefficient, Pr(o) denotes the proportion of units in which the judges agreed, and Pr(e) is the proportion of units for which agreement is expected by chance. In our evaluation, lncRNAs are labeled as positive class, and protein-coding transcripts are labeled as negative class. Based on the results of RFE and our feature selection algorithm (see Algorithm S3), we can finally obtain the optimal feature combination, which comprises 19 features (see Table 2).
Table 2.
Features selected from three feature groups
| Sequence-intrinsic composition | Multi-scale structural information | EIIP-based physicochemical property |
|---|---|---|
| Logarithm-distancea of hexamer on ORF | Minimum free energy (MFE) | Signal at 1/3 position (Se[N∕3]) |
| Length of the longest ORF | UP frequency of paired–unpaired sequence | SNR |
| Coverage of the longest ORF | Logarithm-distancea of acguD sequence | Quantile statistics (Q1, Q2, min and max) |
| Logarithm-distancea of acgu-ACGU sequence |
Logarithm-distance consists of three features: LogDist.LNC, LogDist.PCT and LogDist.Ratio.
We integrated these features into the models built with several widely used machine learning algorithms to assess the performance of the features as well as the machine learning algorithms. Logistic regression [63], SVM [64, 65], random forest [66], ELM [67, 68] and deep learning [69] were evaluated with 10-fold CV in our experiments. According to the results, SVM was superior to other models, but the differences in the performances were quite subtle (see Result Section).
Result analysis
Feature selection and model validation are conducted using human data set A with 10-fold CV. The selected features and classic alignment-free features are evaluated on human data set B. After determining the optimal feature combination and machine learning algorithm, we actually obtain a novel lncRNA identification method. Our method is benchmarked against five popular machine learning-based lncRNA identification tools, namely, CPC, CPAT, CNCI, PLEK and CPC2, on human data set B, as well as data sets mouse, wheat, zebrafish and chicken to assess each tool’s performances and stabilities.
Feature selection
Features discussed in this article can be divided into the following three groups: sequence-intrinsic composition, secondary structural information and EIIP-based physicochemical property. For the sequence-derived features, features of Logarithm-distance on ORF region achieved an accuracy of 0.9598, while features of Euclidean-distance on ORF region obtained 0.9596. On the whole sequence, Logarithm-distance features obtained an accuracy of 0.8521 and Euclidean-distance features obtained 0.8484. The difference between the two measurements’ performances was minor, though Logarithm-distance had higher accuracy and F-measure (see Figure S2-1, Supplementary File 1 for detailed information). As the most discriminating feature of CPAT, the hexamer score had an accuracy of 0.8458 and 0.6416 on ORF region and the whole sequence, respectively. Measurements of Logarithm-distance and Euclidean-distance greatly outperform CPAT’s hexamer score. We further combined Logarithm-distance features with the following two classic ORF-related features: the length and coverage of the longest ORF. The RFE results are displayed in Table S2-1 (Supplementary File 1). None of the five features is redundant. High importance scores of Logarithm-distance features (see Table S2-2) indicate that these three features are of critical importance in the lncRNA identification. Only five features, Logarithm-distance of hexamer on ORF region (consists of three features, namely, LogDist.LNC, LogDist.PCT and LogDist.Ratio), the length and coverage of the longest ORF, can highly represent the sequence-intrinsic information of one RNA sequence. Five sequence-derived features presented an accuracy of 0.9630 and an F-measure of 0.9628 on human data set A (Table 3).
Table 3.
Performances of each feature group on human data set A
| Feature group | Sensitivity | Specificity | Accuracy | F-measure | Kappa |
|---|---|---|---|---|---|
| Sequence | 0.9555 | 0.9705 | 0.9630 | 0.9628 | 0.9261 |
| SSa | 0.8129 | 0.8921 | 0.8525 | 0.8464 | 0.7050 |
| EIIP | 0.9021 | 0.8686 | 0.8853 | 0.8872 | 0.7706 |
| All features | 0.9642 | 0.9726 | 0.9684 | 0.9682 | 0.9368 |
Multi-scale structural features. The results are obtained from 10-fold CV. Bold numbers indicate the highest value.
Figures S2-2 to S2-4 show the performances of k-mer features extracted from multi-scale secondary structure-derived sequences. It seems that features based on the k-mer scheme displayed a passable result. Nonetheless, the accuracy dropped when secondary structure-based features were combined with sequence feature group (see Table S2-3). Figures S2-5 and S2-6 display the performances of multi-scale secondary structural features extracting with Logarithm-distance measurement. Except for subgroup SSE.Abbr Seq, the performances showed no major difference with those of k-mer features, but the features are refined, and the feature number of each subgroup is reduced to 3. Moreover, Logarithm-distance features of subgroups acguD Seq, acguS Seq and acgu-ACGU Seq boosted the accuracy of sequence-derived features (Table S2-4), which confirmed the discriminating power of secondary structure and the feasibility of Logarithm-distance measurement. Hence, we selected scheme Logarithm-distance to extract features of these three subgroups. Although subgroups SSE.Full Seq, SSE.Abbr Seq and Paired-Unpaired Seq, regardless of calculating k-mer frequencies or Logarithm-distance, cannot improve the performance further, some useful information can still be extracted. According to the results of Figure S2-3, we calculated the importance scores of 4-mer frequencies of Paired-Unpaired Seq and 2-mer frequencies of SSE.Abbr Seq by conducting RFE algorithm, and the feature with the highest score of each subgroup were included in this feature group (see Table S2-5).
Now three feature subgroups (Logarithm-distance of acguD Seq, acguS Seq and acgu-ACGU Seq) and three features (MFE, UP frequency of Paired-Unpaired Seq and bulge frequency of SSE.Abbr Seq) derived from the multi-scale secondary structure may enhance the performance of sequence-derived feature. However, it is still necessary to perform feature selection to determine the optimal feature combination. Because the information of secondary structure-derived sequence has been embodied in Logarithm-distance features, we avoid selecting key features by performing RFE algorithm, which may detach three Logarithm-distance features. We re-evaluated the feature group, which consists of three subgroups and three features with a new algorithm displayed in Algorithm S2. This algorithm ranks different feature groups according to their average performances of 10-fold CV. For each iteration, the feature group that shows the best improvement in accuracy will be added to the selected feature set. One feature group alone may not improve the performance, but it may boost the result by combining with other feature groups. To avoid missing potential feature groups, all the feature groups with the highest score of each iteration will be evaluated. The final results of feature selection were summarized in Table S2-6. Eight multi-scale secondary structural features were determined, namely, MFE, UP frequency of Paired-Unpaired Seq, Logarithm-distance of acguD Seq and acgu-ACGU Seq (See Table 2). Eight secondary structural features obtained an accuracy of 0.8525 and an F-measure of 0.8464 on human data set A.
As to the features based on EIIP values, subgroup quantile statistics on the position of top 10% presented the best performance in our experiments (see Figure S2-7). Depending on the results of RFE, the signal at 1/3 position (), SNR and quantile statistics (Q1, Q2, min and max values) were selected as EIIP-based features (see Tables S2-7 for RFE result). Six EIIP-based physicochemical features achieved an accuracy of 0.8853 and an F-measure of 0.8872 on human data set A.
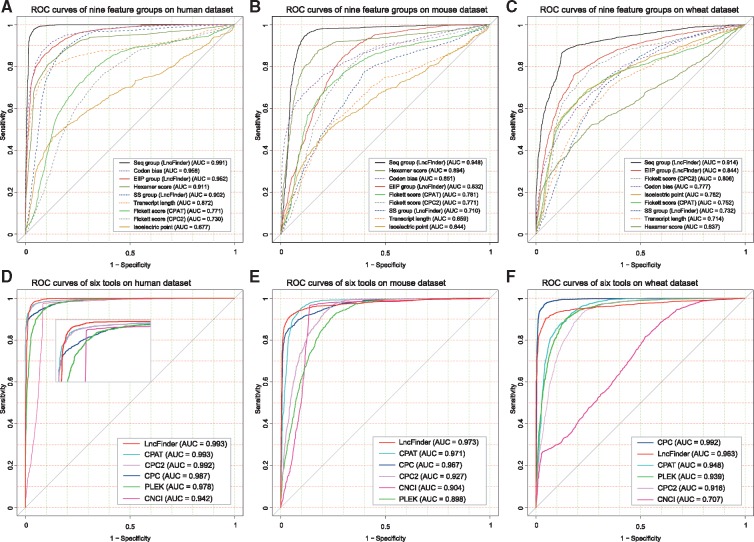
To assess the performances and cross-species’ stabilities of different feature groups, three feature groups we designed and six other classic alignment-free feature groups (codon bias, hexamer score, Fickett TESTCODE score of CPAT and CPC2, pI and transcript length) were evaluated on the following three species: human, mouse and wheat. All feature groups were used to build SVM classifiers separately with the training set of human data set B. Then the classifiers built with human data set were used to predict the test set of human data set B and the test sets of mouse and wheat. Feature groups’ receiver operating characteristic (ROC) curve [70, 71] on three test sets were shown in Figure 3 (A), (B), and (C). Figure 3 (A) displays each feature group’s performance on human species, while Figure 3 (B) and (C) presents their cross-species stabilities. From Figure 3 (A), it can be observed that the top five feature groups are Logarithm-distance features (Seq group), codon bias, EIIP-based features, hexamer score of CPAT and multi-scale secondary structural features (SS group). Three feature groups among the top five were extracted from sequence-intrinsic composition. The sequence-derived features we designed outperformed other sequence-intrinsic features such as codon bias and hexamer score with the AUC (area under curve) of 0.991. Six EIIP-based features had performance comparable to that of 64 codon bias features. Secondary structural features surpassed features’ transcript length, Fickett TESTCODE score and pI value with the AUC of 0.902, which demonstrates the discriminating power of structural features. The Fickett TESTCODE score methods employed by CPAT and CPC have some minor differences. CPAT calculates Fickett TESTCODE score on ORF region and obtains AUC 0.781. Figure 3 (B) and (C) shows the results of each feature groups on data sets of mouse and wheat. All feature groups showed some fluctuations in their performances, but sequence-derived features still achieved the highest AUC. Sequence-derived features and EIIP-based features displayed better performances than other feature groups. Multi-scale secondary structural features only had average cross-species results, but this feature category was among the top five feature groups on human data set B. Based on a comprehensive evaluation of different feature groups, 19 critical features are selected from sequence-intrinsic composition, multi-scale secondary structural information and EIIP-based physicochemical property (see Table 2). All three feature groups achieved an accuracy of 0.9684 and an F-measure of 0.9682 on human data set A (see Table 3). Using LncFinder, users can extract various features and construct their own classifiers for different purposes.
Figure 3.
ROC curves of different feature groups and different tools on three species. (A) Sequence-derived (Seq Group), EIIP-derived (EIIP Group), secondary structure-derived (SS Group) and other six classic feature groups were evaluated on human data set B. All three feature categories we proposed were among the top five feature groups. Logarithm-distance features outperformed other sequence-intrinsic features such as codon bias and hexamer score with the highest AUC. Six EIIP-based features even had performance comparable to that of 64 codon bias features. (B) Nine feature groups were extracted from the training set of human data set B and were used to build classifiers. Figure (B) shows the nine classifiers’ performances on mouse test set. All feature groups showed some fluctuations in performances, but sequence-derive features still achieved the best AUC. (C) Classifiers built on human data set B were evaluated with a test set of wheat data set. Compared with Figure 3 (A), AUC of codon bias features decreased about 18%, while AUC of hexamer score decreased about 30%. EIIP-based feature group surpassed codon bias features and demonstrated its satisfactory cross-species performance. Sequence-derived features still obtained the best AUC. (D) LncFinder and other five tools, namely, CPC (offline version), CPAT (re-trained model), CNCI, PLEK (re-trained model) and CPC2, were tested on human data set B. LncFinder and CPAT had the best AUC, but the accuracy of CPAT was lower than that of LncFinder. (E) LncFinder and other five tools were tested on mouse data set. LncFinder achieved the best result. (F) LncFinder and other five tools were tested on wheat data set. LncFinder achieved the best AUC on human and mouse data sets. Although the accuracy of CPC on human and mouse data sets was inferior to that of other tools, CPC surpassed all alignment-free tools on wheat data set. LncFinder had the best performance among alignment-free tools. We cannot know which tool is best for one specific species in advance. A tool that can present robust and stable results on multiple species is of crucial importance. LncFinder had the most stable and reliable performances among these tools.
Model validation
The results of five machine learning models are displayed in Figure S2-8. The parameters of different machine learning models were tuned with 10-fold CV. The performances of each model under different parameters are displayed in Table S3-16 and Table S3-17.
The classifier based on SVM achieved the highest accuracy, 0.9687, while deep learning had the lowest, 0.9523. In fact, most of the models’ accuracies ranged from 0.965 to 0.968. The difference of performances between the SVM model and the random forest model was even negligible: the accuracy of the random forest model was 0.9681. The stable results of different classifiers reflect that the critical features we designed are of a high standard and classifier-neutral. SVM had the best accuracy, and random forest achieved the best F-measure. In this experiment, we selected SVM to build the classifier. But researchers can also use LncFinder to construct models with other machine learning algorithms. The detailed procedures and results of feature selection and model validation are included in the Result Section in Supplementary File 1. After evaluating the features and obtaining the SVM classifier, we obtain a novel lncRNA predictor. In the next section, we will benchmark our predictor against several widely used tools to further evaluate the discriminating power of different methods.
Evaluations by comparison with popular tools
In this section, our lncRNA identification method was benchmarked against CPC, CPAT, CNCI, PLEK and CPC2 on five species, namely, human (Homo sapiens), mouse (Mus musculus), wheat (Triticum aestivum), zebrafish (Danio rerio) and chicken (Gallus gallus). The novel lncRNA identification method is one of the main functions of LncFinder package, and here we use LncFinder to denote the method we developed. In our experiments, we used UniRef90 [72] as the protein reference database of CPC. Because CPAT and PLEK can be trained with users’ sequences, the re-trained models were built with the data sets that are identical to the training sets of LncFinder. As suggested in their documentations, the parameters of PLEK were tuned with grid search, and the cut-off of CPAT was determined using 10-fold CV. Both the new trained and pre-built models were evaluated to have a comprehensive and fair comparison. CPC, CPAT and CPC2 provide a web server and a standalone version; both versions were tested in our experiments. The web server of CPC2 presented the results that are identical to the standalone version. For CPC and CPAT, however, the results of the web server and standalone version showed some minor differences, which may result from different genome assemblies of the training set. Additionally, considering that the secondary structure calculated by RNAfold may not present the actual structure, LncFinder can be configured to predict lncRNAs using the structural information provided by users or simply without using the multi-scale secondary structural features. In our evaluation, the structural features’ excluded version of LncFinder was also benchmarked against other tools.
Performance evaluation of human data set B
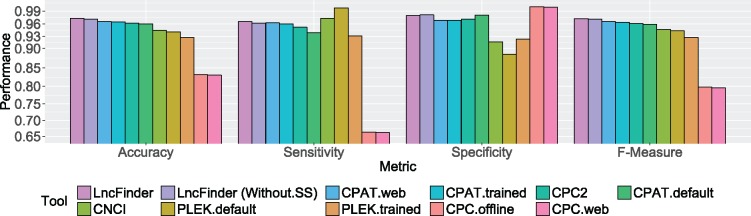
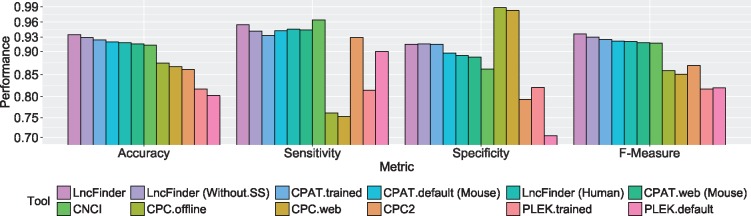
Figure 4 displays the performances of different tools on human data set B. It can be noted that CPC had the best specificity (1.00 of standalone version and 0.9988 of the web server). However, the accuracy (0.8318 of standalone version and 0.8304 of the web server) was not that excellent owing to the low sensitivity (0.6636 of standalone version and 0.6620 of the web server). As an alignment-based method, CPC is mainly designed to assess the coding potential, and it is very useful to evaluate highly conserved protein-coding transcripts. Nevertheless, many lncRNAs overlap protein-coding genes, which could make CPC incorrectly classify long non-coding transcripts as protein-coding sequences. As the upgraded version of CPC, CPC2 showed considerable improvement (accuracy, 0.9614; F-measure, 0.9610) on its predecessor. Compared with CPC, CPC2 achieved much better sensitivity and thus much better accuracy. CPAT had relatively high accuracy and F-measure on the web server, 0.9654 and 0.9653, respectively. CNCI surpassed PLEK (accuracy of pre-build model, 0.9410) with an accuracy of 0.9450. Because the default models were trained with a large scale of sequences, which may have some overlaps with our test sets, CPAT and PLEK were evaluated with the re-trained models as well. The accuracy of CPAT (re-trained model) was 0.9642 and the accuracy of PLEK was 0.9274. LncFinder achieved the highest accuracy and F-measure, 0.9728 and 0.9726, respectively. The high accuracy and F-measure imply LncFinder is provided with a better balance between precision and recall.
Figure 4.
Performances of different tools on human data set B. LncFinder had the best accuracy of 0.9728. CPC had a strong tendency to classify lncRNAs as protein-coding transcripts and thus having low accuracy of 0.8304 (web server). As an upgraded version, CPC2 presented accuracy of 0.9614. CPC2 was a big improvement on its predecessor and also outperformed CNCI and PLEK that obtained accuracies of 0.9450 and 0.9274, respectively. CPAT (re-trained model) was inferior to only LncFinder and obtained an accuracy of 0.9642. Even when secondary structure-derived features were excluded, LncFinder (Without.SS) can still surpass other tools with an accuracy of 0.9716.
Even when secondary structure-derived features were excluded, LncFinder still outperformed other tools. Figure 3 (D) displays the ROC curves of CPC (offline version), CPAT (re-trained model), CNCI, PLEK (re-trained model), CPC2 and LncFinder on human data set B. Both LncFinder and CPAT had the best AUC, but the accuracy of CPAT was lower than that of LncFinder. For detailed data of the evaluation on human data set B, please refer to Table S3-18.
Performance evaluation of mouse data set
We additionally evaluated the performance of different tools on the mouse data set because it is one of the most studied species. CPC predicts sequences largely depending on the reference data set; thus, CPC can be applied to various species with one default model. According to the manuals, the default models of CNCI and PLEK are competent to predict sequences of other vertebrate species; CPC2 is a species-neutral classification tool that can be used for non-model organism transcriptomes. We, therefore, compared CNCI, PLEK (default model), CPC2 with LncFinder (default model for human) to have a fair evaluation. CPAT is the only alignment-free tool that has the pre-built model for mouse, and both the default model for mouse and the re-trained model were included in our tests.
Figure 5 displays the performances of different tools on the mouse data set. CPC still obtained the highest specificity (0.9883 of standalone version), but the accuracy (0.8750 of standalone version) was affected by the low sensitivity (0.7617 of standalone version). CPC2, however, had a high sensitivity of 0.9289; because of its poor specificity of 0.7933, it could obtain an accuracy of only 0.8611. CPAT with a re-trained model obtained an accuracy of 0.9242, while PLEK with re-trained model had an accuracy of 0.8178. LncFinder achieved the best result with an accuracy 0.9347 and an F-measure of 0.9360, which indicates its satisfactory overall performance. Furthermore, LncFinder (without secondary structure-derived features) surpassed other tools with an accuracy of 0.9286. When the model for human was used, LncFinder achieved an accuracy 0.9186 and an F-measure 0.9207, which still surpassed other tools’ default models (accuracy of PLEK, 0.8025; accuracy of CPC2, 0.8611; accuracy of CNCI, 0.9133; accuracy of CPAT’s web server, 0.9161). Although using the model for the human data set, LncFinder was only inferior to the re-trained model of CPAT. Figure 3 (E) displays the ROC curves of CPC (offline version), CPAT (re-trained model), CNCI, PLEK (re-trained model), CPC2 and LncFinder on the mouse data set. LncFinder had the best AUC and presented a satisfactory trade-off between sensitivity and false-positive rate (FPR, 1-specificity). CNCI and PLEK had much higher FPR and lower AUC. The original data of this evaluation are listed in Table S3-19.
Figure 5.
Performances of different tools on mouse data set. LncFinder achieved the best accuracy of 0.9347, while PLEK (re-trained model) had an accuracy 0.8178. The accuracy of CPAT (re-trained model) was 0.9242 and better than CNCI’s 0.9133, CPC’s 0.8678 (web server) and CPC2’s 0.8611. LncFinder (Without.SS) outperformed other tools with an accuracy of 0.9286 even without secondary structure-derived features. When using the model for human, LncFinder outperforms CPC/CPC2, CPAT (web server), CNCI and PLEK (re-trained model) with a satisfactory accuracy of 0.9186. Under this circumstance, LncFinder can even rival CPAT (re-trained model for mouse), which demonstrates LncFinder’s robustness and high cross-species stability.
Performance evaluation of wheat data set
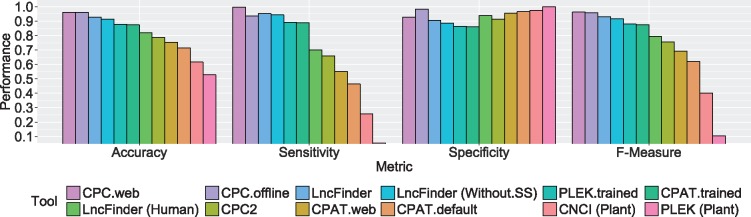
We further compared different tools on plant data set. The data set was constructed with the sequences of wheat because of its sufficient lncRNA sequences. According to the manuals of CNCI and PLEK, both tools provide models for plant sequences prediction. Thus, their pre-built models for plants were included in our tests. Because CPAT has no model for plant species, we additionally compared CPAT with LncFinder by employing their default models that are built with human data sets.
Figure 6 shows the performances of different tools on the wheat data set. CPC outperformed all the alignment-free tools with the highest accuracy and F-measure, 0.9595 and 0.9585, respectively. Its successor, CPC2, nonetheless, had an accuracy of 0.7870 and an F-measure of 0.7560. The accuracy of CNCI (default model for plant) was 0.6158, and the accuracy of PLEK (default model for plant) was 0.5275. Although CNCI and PLEK provide models for plant, their results were not that favorable. Using the re-trained model, the accuracy of PLEK increased from 0.5275 to 0.8773. The performance of CPAT (re-trained model) was slightly inferior to that of PLEK (re-trained model) with an accuracy of 0.8743. LncFinder obtained the best performance among alignment-free tools with an accuracy of 0.9283. LncFinder also presented satisfactory sensitivity and specificity. From Figure 6, it can be seen that all the tools had high specificity, but different tools had various sensitivity. When each tool’s default model was used for this test set, LncFinder (default model for human) had the best sensitivity of 0.7000, while PLEK (default model for plant) only got 0.0550. Both LncFinder and CPAT were tested using their default models for the human sequences. The performance of LncFinder (accuracy, 0.8190; F-measure, 0.7946) was much better than that of CPAT (accuracy, 0.7145; F-measure, 0.6188). CPAT employs logistic regression, and the best cutoffs of different species vary considerably. CPAT’s suggested cutoff for human is 0.364, while the best cutoff for mouse is 0.440. In our experiments, the optimal cutoff for wheat even reached 0.537. An inappropriate cutoff can lead to an inferior performance. Figure 3 (F) displays ROC curves of CPC (offline version), CPAT (re-trained model), CNCI, PLEK (re-trained model), CPC2 and LncFinder on the wheat data set. CPC surpassed all alignment-free tools on wheat, although it presented unsatisfactory results on human and mouse. Among alignment-free tools, LncFinder achieved the best AUC of 0.983. It is reasonable to assume that the poor performance of CNCI can be ameliorated if CNCI can be re-trained with new data sets. The original data of the evaluation of wheat are listed in Table S3-20.
Figure 6.
Performances of different tools on wheat data set. Although CPC had inferior performances on human and mouse, it achieved the best accuracy on wheat. CPC obtained an accuracy of 0.9595, but his alignment-free successor CPC2 only had an accuracy of 0.7870. The accuracies of CPAT (re-trained model) and PLEK (re-trained model) were 0.8743 and 0.8773, respectively, while LncFinder obtained an accuracy of 0.9283. When default models were used, CPAT (model for human), CNCI (default model for plants) and PLEK (default model for plants) had accuracies of 0.7145, 0.6158 and 0.5275, respectively. LncFinder (model for human) had an accuracy of 0.8190. Although CNCI and PLEK provide default models for plants, the performances were substandard. LncFinder has the best performance among alignment-free tools. Even using the model for human, LncFinder still outperformed CPC2, CNCI and the default models of CPAT and PLEK.
Performance evaluation of zebrafish and chicken data sets
We finally evaluated the stabilities and performances of CPC, CPAT, CNCI, PLEK, CPC2 and LncFinder on zebrafish and chicken data sets. The results are displayed in Table 4.
Table 4.
Performances of different tools on zebrafish and chicken data sets
| Methods | Zebrafish (Danio rerio) |
Chicken (Gallus gallus) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sensitivity | Specificity | Accuracy | F-measure | Kappa | Sensitivity | Specificity | Accuracy | F-measure | Kappa | |
| CPC | 0.6728 | NA | NA | NA | NA | 0.5784 | 0.9888 | 0.7836 | 0.7277 | 0.5671 |
| CPAT | 0.8668 | 0.8660 | 0.8664 | 0.8663 | 0.7328 | 0.9189 | 0.9178 | 0.9183 | 0.9183 | 0.8366 |
| CNCI | 0.8535 | 0.8728 | 0.8631 | 0.8618 | 0.7263 | 0.9128 | 0.9051 | 0.9089 | 0.9093 | 0.8179 |
| PLEK | 0.8715 | 0.8255 | 0.8485 | 0.8519 | 0.6970 | 0.9346 | 0.9124 | 0.9235 | 0.9244 | 0.8740 |
| CPC2 | 0.8948 | 0.7835 | 0.8391 | 0.8476 | 0.6783 | 0.7650 | 0.9235 | 0.8443 | 0.8308 | 0.6885 |
| LncFinder | 0.8815 | 0.8838 | 0.8826 | 0.8825 | 0.7653 | 0.9491 | 0.9321 | 0.9406 | 0.9411 | 0.8813 |
Bold numbers indicate the highest value. LncFinder has the best performance. In our test, CPC could not process the protein-coding transcripts of zebrafish; thus, only the result of lncRNAs is obtained.
Because CPC needs to align the sequences against the reference database and CNCI has to calculate the most-like CDS (MLCDS), these two tools have strict requirements for sequence quality. For the sequences containing some non-nucleotide characters (such as ‘X’), which are very common for some poorly explored species, CPC may throw an error and stop the computation, and CNCI may omit these sequences automatically. In this test, tools CPAT, PLEK and LncFinder functioned normally, but CPC could not identify the protein-coding transcripts of zebrafish. Thus, only the result of lncRNAs was obtained. We also noticed that CNCI omitted 7 lncRNAs and 6 protein-coding transcripts of chicken and 13 protein-coding transcripts of zebrafish automatically.
From Table 4, it can be observed that LncFinder outperformed other tools with the highest accuracy and F-measure. The tool CPC had the best performance on wheat, but the sensitivity of CPC was much lower on human, mouse, zebrafish and chicken than the sensitivity of other tools; therefore, CPC had low accuracy. CPC2 had much better overall performance than CPC. For the zebrafish data set, CPAT achieved an accuracy of 0.8664 and was better than CNCI, PLEK and CPC2. But LncFinder surpassed CPAT with an accuracy of 0.8826. PLEK performed better than CPC, CPAT, CNCI and CPC2 on the chicken data set and had an accuracy of 0.9235. But LncFinder obtained about 1.7% higher accuracy than PLEK. According to the results of the five species, LncFinder displayed the most stable and satisfactory performance. The robustness and fault-tolerance capability make LncFinder a valuable and practical lncRNA identification tool for multiple species, especially for those poorly explored species.
Evaluation of computational speed
The running times of six tools were evaluated on the same platform. We here avoid using large servers for computational speed evaluation. An average hardware environment can assess each tool’s efficiency and usability much clearly. The platform configurations are Intel[textregistered] CoreTM i7-2600 processor @ 3.40 GHz, 8 GB memory and 64 bits Linux OS. Human data set B, which contains 2500 long non-coding transcripts and 2500 protein-coding transcripts, was used to evaluate six tools. CPC2 used 8.87 seconds to complete the prediction, while CPC needed 4675.45 min to complete the process of alignment and identification. CPAT was only slightly inferior to CPC2 and used 9.05 s to identify 5000 sequences. With the help of parallel computing, it took CNCI, PLEK and LncFinder 1333.19 s, 83.67 s and 56.01 s, respectively to complete the identification. If predicting sequences without using secondary structure features, it took LncFinder 35.76 s to finish the process. LncFinder is more efficient than CPC, CNCI and PLEK. Although slower than CPC2 and CPAT, LncFinder can still predict several thousand sequences within 1 min and present more reliable results. For detailed data, please refer to Table S2-8.
Discussion
In this study, we reviewed several widely used lncRNA identification tools and their features. Numerous alignment-free feature groups, such as codon bias, Fickett TESTCODE score and pI were evaluated on three data sets to assess their performances and cross-species stabilities. Additionally, we also comprehensively explored the following three feature categories: sequence-intrinsic composition, multi-scale secondary structural information and physicochemical features obtained from EIIP and FFT. Based on the feature selection process, 19 heterologous features were extracted. We incorporated the 19 features into the following five popular machine learning algorithms: logistic regression, SVM, random forest, ELM and deep learning to validate the heterologous features we designed as well as assess the effect of different machine learning algorithms on lncRNA prediction. The stable performances of different classifiers indicated that the features are critical and reliable. According to the experiments’ results, we proposed a novel lncRNA identification method. Benchmarked against several state-of-the-art tools, our method displayed more accurate and stable performances on multiple species with acceptable time costs. An integrated package LncFinder is finally established to facilitate the research on lncRNA. Various classic features as well as features we designed can be extracted with LncFinder. Users can use LncFinder to build the predictor with other feature groups or machine learning algorithms. As a one-stop package for lncRNA identification and analysis, LncFinder can effectively and efficiently complete the main steps of predictor construction including feature extraction, feature selection, model construct and performance evaluation. LncFinder was released as R package. To maximize its availability, a web server was also developed for lncRNA prediction.
Euclidean-/Logarithm-distance, two new measurements, were designed to capture the sequence-intrinsic composition. Compared with other sequence-derived features, Logarithm-distance can achieve high accuracy as well as simplify the features markedly. Our designed multi-scale structural features capture structural information at different resolution levels by integrating sequence composition with MFE and structural sequences. EIIP-derived features based on FFT can provide another view from the prospect of physicochemical property. The sequence-derived features are based upon linguistic meaning, whereas the features extracted from the secondary structure and EIIP can be further interpreted as semantic annotations, which implies higher-level information of biological functions.
According to our experiments, features of Logarithm-distance of hexamer on ORF region performed satisfactorily with an accuracy of 0.9598, and the accuracy of all features from three categories combined was 0.9687 with parameter tuning. It seems that the improvement of secondary structural and EIIP-derived features was trivial. However, six EIIP-derived features achieved an accuracy of 0.8853, and eight secondary structure-related features obtained an accuracy of 0.8525. In contrast, the accuracy of the hexamer score on ORF region, the most discriminating feature of CPAT, was only 0.8458. From Figure 3, secondary structural and EIIP-derived features outperformed features of Fickett TESTCODE score, transcript length and pI value. The performance of EIIP-derived features was even better than that of tool CNCI [see Figure 3 (A) and (D)]. The performances of the secondary structure and EIIP-based features are not far inferior to those of sequence-derived features, but sequence-derived features have achieved fairly high accuracy, thus leaving limited room for other features to enhance. Nineteen features from these three categories were used to build our method. The secondary structure calculated by RNAfold may not completely reflect the actual structural information of one sequence. Therefore, LncFinder can predict lncRNA with sequence-derived features and EIIP features only.
Five widely used machine learning algorithms, namely, logistic regression, SVM, random forest, ELM and deep learning were compared to determine how much will different machine learning algorithms affect the performance of lncRNA identification using the features we designed. Deep learning in our test had the lowest accuracy of 0.9523, while SVM obtained 0.9687. Because there were only 19 features used to build classifiers, it may be unnecessary to employ deep learning for such a small scale of features. It is also worth mentioning that many of the species have only limited lncRNA sequences. The insufficient training data may lead to overfitting of the deep learning model. Also, deep learning requires tuning of many parameters, which requires a much longer time than other models to perform parameter tuning and obtain the optimal model. Only minor distinctions existed among logistic regression, SVM, random forest and ELM. The difference in accuracy between the SVM model and the random forest model was merely 0.0006, which suggests that these 19 features are very robust, and the fundamental features are of crucial importance in lncRNA identification. In our experiments, SVM displayed the highest accuracy and F-measure; random forest presented the best AUC; logistic regression is fast and easy to build. Our lncRNA identification method is developed using SVM not only because SVM achieved the highest accuracy but also for its small size and convenient application. If we apply random forest algorithm, the size of the final package will be about 25 times as large as that of the current version. As to logistic regression, the best cutoffs of different species may vary widely, which may produce an adverse effect on the tool’s generalization ability. Using LncFinder, users can construct different classifiers with various machine learning algorithms.
We further compared our method (denoted by LncFinder) with five popular lncRNA identification tools, namely CPC, CPAT, CNCI, PLEK and CPC2. These five tools are selected because they are typical and considered state of the art. CPC is a classic alignment-based tool, whereas the other four tools are alignment-free. CPAT and PLEK can be re-trained by new data sets, which can also present a comprehensive comparison. Because results of BLASTX largely depend on the protein reference database and play an important role in CPC prediction, CPC does not have to train several models for different species as long as the reference database is large and comprehensive enough. Nonetheless, CPC requires about 90 GB of free space for storing the reference database of NCBI or more than 20 GB for the database of UniRef90. Additionally, CPC needs a lot of time to complete the process of alignment, which makes CPC less efficient than other alignment-free tools. For human and mouse data sets, CPC had the highest specificity but the lowest sensitivity. This imbalanced performance has led to unsatisfactory accuracy. CPC2 predicted lncRNAs with sequence-intrinsic features alone and had the result much better than CPC on the human data set. However, the performance of CPC2 was slightly lower than that of CPC on the mouse data set. For other alignment-free identification tools, CNCI and PLEK (pre-built model) had comparable results. The accuracy of CPAT was higher than that of CPC, CNCI and PLEK, but lower than that of LncFinder. LncFinder achieved the best performances on human and mouse data sets, even when the secondary structure-derived features were excluded.
As to plant species, we observed some intriguing phenomena from each tool’s performance on wheat data set. CPC obtained the best result on wheat data set, despite its lower sensitivity and accuracy on human and mouse data sets. CNCI and PLEK, though provide models that can be used to predict lncRNA of plant, their performances on wheat were hardly acceptable. One possible explanation is that there are fewer similarities between protein-coding transcripts and lncRNAs in wheat than in human. For instance, 38.44% (961/2500) of lncRNAs from human test set B has BLASTX HITS but only 1.95% (39/2000) of lncRNAs from wheat test set has BLASTX HITS. Consequently, CPC finds fewer HITS in lncRNAs in wheat, and thus avoids classifying lncRNAs as mRNA and has low sensitivity. Moreover, the nucleotide usages of different plants may be less conserved than those of vertebrates. Thus, the tools greatly depending on nucleotide composition features, such as CNCI and PLEK, displayed poor results when the species of test set largely differs from the species of their pre-built models’ training set. Gene structure is closely related to evolutionary changes and protein functionality. The differences in gene structure may also affect the classifier’s performance. Compared with the popular alignment-free tools, LncFinder displayed the best accuracy and F-measure. LncFinder and CPAT avoid using every nucleotide composition frequency to construct the model, which helps cushion the effect of various intrinsic compositions of different species. Unlike hexamer score that only uses the hexamer frequencies of reference data set, LncFinder also considers the frequencies of unevaluated sequences, thus showing more stable performances than CPAT. According to our evaluation, we can find that it is essential to build new models for different species, but only CPAT, PLEK and LncFinder support model re-training. For some poorly explored species, limited lncRNA may be not sufficient to train a new model, and we need to employ models trained on other species. In that case, CPAT may present inadequate results owing to its wide range of cutoffs for different species. But LncFinder’s default model (trained with human data set) showed more reliable cross-species performances than the default models of CPC, CPAT, CNCI, PLEK and CPC2.
The computational time of each tool was also evaluated. LncFinder is more efficient than CPC, CNCI and PLEK. CPC is the slowest owing to the process of alignment. CNCI is less efficient than other alignment-free tools mainly because it takes more time to find the MLCDS region. PLEK employs 1364 features that slow the prediction and make the process of model re-training extremely time-consuming. CPAT and CPC2 are faster than LncFinder mainly because (1) the source codes of CPAT and CPC2 were implemented in C and Python, which are faster than R, and (2) CPAT used logistic regression to build machine learning model, which is faster than SVM. Nonetheless, LncFinder is qualified to predict lncRNA at a large-scale level with an acceptable time cost.
In this study, we comprehensively reviewed and evaluated different lncRNA identification features and tools. And we also developed a valuable and user-friendly package LncFinder. However, there remain some tough challenges. The sequence compositions of different species showed varying degrees of differences, which entails intrinsic composition-based tools supporting model re-training. Nonetheless, the further question is that we cannot build models for all species. Hence, more critical features that can be applied to multiple species needed to be explored, especially for plants. Furthermore, the performances of each tool vary from species to species, and it is practically impossible to know in advance which tool can achieve the highest accuracy on a specific species. Thus, a tool with stable and satisfactory results on multiple species is highly essential for lncRNA research.
In this study, 19 critical features are obtained from feature selection and 10-fold CV, which could reveal some valuable distinctions between lncRNAs and mRNAs from the perspective of sequence-intrinsic composition, secondary structure and EIIP-based physicochemical property. These features are expected to play positive roles in other lncRNA-related research, such as interaction, annotation and evolution. As an integrated lncRNA identification platform, LncFinder can facilitate relevant research and provide scientists with useful information.
Application of LncFinder
Functions of LncFinder are not limited to lncRNA identification. The stand-alone version of LncFinder is a one-stop package for feature extraction, feature selection, model validation, classifier construction and performance evaluation. LncFinder’s lncRNA identification algorithm is developed based on the optimal feature combination and the most appropriate classifiers. A web server is provided for lncRNA identification to make LncFinder a highly flexible and remarkably user-friendly tool.
R package of LncFinder
R package of LncFinder has been included in CRAN. Users can simply install LncFinder by entering the command ‘install.packages(“LncFinder”)’ in R, and an appropriate version will be installed automatically. Package and reference manual can also be downloaded from CRAN (stable version): https://CRAN.R-project.org/package=LncFinder or GitHub (Dev version): https://github.com/HAN-Siyu/LncFinder.
The stand-alone version of LncFinder provides a batch of practical functions to facilitate lncRNA identification and analysis. (1) LncFinder provides a novel lncRNA identification method. Models for multiple species are provided. Two modes can be selected to identify lncRNA with or without using secondary structure-derived features. Secondary structure sequences can be loaded from external files, in case users should have structural data obtained from experiments or other reliable sources. (2) LncFinder can be used to build new machine learning classifiers. Features and classifiers can all be customized, which helps users construct models with various feature groups or machine learning algorithms. LncFinder can extract various alignment-free features such as GC content, k-mer frequencies, hexamer score, Fickett TESTCODE score, length and coverage of ORF, Euclidean-/Logarithm-distance of k-mer frequencies, EIIP-derived features, multi-scale secondary structural features and pI value. Machine learning algorithms such as logistic regression, SVM, random forest can be employed to construct models with parameter tuning. (3) Machine learning-related functions such as feature selection, k-fold CV and parameter tuning are also included in LncFinder to help users select the optimal feature combination and machine learning algorithm. The functions, descriptions and options of LncFinder R package have been briefly summarized in Table 5. Please refer to Supplementary File 2—R Package and Supplementary File 4—R Package Manual for examples and detailed information. The Documentation of LncFinder is generated with R package” roxygen2” [73].
Table 5.
Functions and descriptions of LncFinder R package
| Function | Description | Option |
|---|---|---|
| Functions for classic features extraction | ||
| compute_EIIP() | Compute EIIP-derived features | 1. spectrum.percent: set the percentage of the sorted power spectrum; 2. quantile.probs: set the quantile interval |
| compute_EucDist() | Compute Euclidean-distance features | 1. k: set the sliding window size;2. step: set the sliding window step; 3. on.ORF: calculate features on ORF region |
| compute_FickettScore() | Compute Fickett TESTCODE Score | on.ORF: calculate Fickett TESTCODE Score on ORF region |
| compute_GC() | Compute GC content | on.ORF: calculate GC content on ORF region |
| compute_hexamerScore() | Compute hexamer score | see compute_EucDist() |
| compute_kmer() | Compure k-mer features | improved.mode: use the improved method proposed by PLEK; other options see compute_EucDist() |
| compute_LogDist() | Compure Logarithm-distance | see compute_EucDist() |
| compute_pI() | Compure isoelectric point | 1. on.ORF: calculate isoelectric point on ORF region;2. ambiguous.base: take ambiguous bases into account |
| find_orfs() | Find ORFs | reverse.strand: find ORFs on the reverse strand |
| Functions for LncRNA identification and new classifier construction | ||
| lnc_finder() | Identify lncRNAs using LncFinder | 1. svm.model: select species, such as human, mouse and wheat; 2. SS.features: use multi-scale secondary structure features |
| build_model() | Build new model using LncFinder | see lnc_finder() |
| extract_features() | Extract features proposed by LncFinder | SS.features: extract multi-scale secondary structure features |
| read_SS() | Load external secondary structure information | |
| run_RNAfold() | Run RNAfold and capture the results | |
| svm_cv() | Perform cross validation for SVM model | 1. folds.num: set the number of folds for cross-validation;2. seed: set the seed for random number generation; other parameters for SVM model training |
| svm_tune() | Tune SVM model | see svm_cv() |
This table briefly summaries the main functions of LncFinder R package. All functions and descriptions are based on LncFinder R Package (version 1.1.2). The package will be updated regarlarly. Refer to Supplementary File 4 - R package Manual for detailed descriptions and examples.
Web server of LncFinder
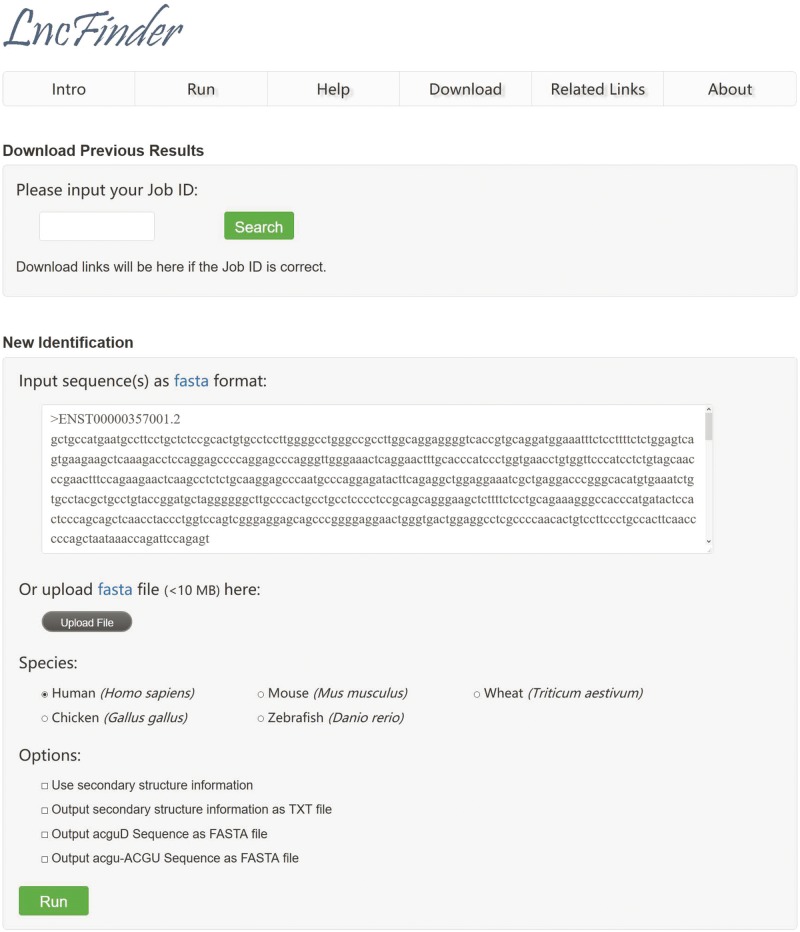
The web interface of LncFinder is developed to suit the convenience of the users. This web server is available at http://bmbl.sdstate.edu/lncfinder/. A backup server is also established, which can be accessed via http://csbl.bmb.uga.edu/mirrors/JLU/lncfinder/. Figure 7 is a screenshot of LncFinder’s web server. The web server provides the following three functional modules: (1) lncRNA identification for multiple species; (2) downloads of multi-species models, data sets and secondary structural sequences; and (3) an instructive summary of lncRNA-related tools, databases and news.
Figure 7.
Screenshot of LncFinder’s web server.
The web server of LncFinder supports sequences in FASTA format as input. Users can input sequences in the text area or just upload a FASTA file. Users can also select to identify lncRNAs with or without multi-scale secondary structural features. Now five species, namely, human, mouse, chicken, zebrafish and wheat, are available on our web server. The results will be displayed after the identification is complete. The original results and structure-related sequences can be exported and downloaded. Each prediction task will be assigned a Job ID, and users can use the Job ID to download previous results and secondary structure-derived sequences. Moreover, additional models for other species can be downloaded for local use. The web server also provides an informative summary for users’ convenience, which includes the updated information on lncRNA prediction tools, various kinds of databases and lncRNA research progress. The summaries will be updated regularly. See Supplementary File 3 - Web Server for detailed information.
Supplementary Data
Supplementary data are available online at https://academic.oup.com/bib.
Supplementary Material
Acknowledgements
The authors are grateful to Li YL. (Hokkaido University), Sun Y. (Beijing Normal University) and Wang RY. (Nihon University) for assisting with data collection; to Cheng M., Fan LR., Guo Y. and Tao MX. from Jilin University for the tests of R package and web server. The authors also thank the editor and anonymous reviewers for handling this manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (61472158, 61402194, and 71774154), Natural Science Foundation of Jilin Province (20180101331JC, 20170520063JH and 20180101050JC), Zhuhai Premier-Discipline Enhancement Scheme, Guangdong Premier Key-Discipline Enhancement Scheme and Graduate Innovation Fund of Jilin University (2017124).
Key Points
Many classic features were reviewed and discussed. Features from the following three categories: Euclidean-/Logarithm-distance of hexamer, multi-scale secondary structural information and EIIP-based physicochemical property were also explored to enhance the accuracy of lncRNA prediction.
Based on these three feature categories, a novel lncRNA identification algorithm was developed with the comprehensive processes of feature selection and model validation. Benchmarked against several state-of-the-art methods, our algorithm presented the most robust and satisfactory performance on multiple species.
An integrated platform LncFinder was developed to facilitate lncRNA identification and analysis. LncFinder can effectively perform feature extraction, feature selection, classifier construction and performance evaluation. Our lncRNA identification algorithm was included in LncFinder as well.
Released as R package and web server, LncFinder can be run on multiple OS platforms. LncFinder is a flexible and useful tool for coding/non-coding sequence prediction, coding potential assessment, lncRNA property analysis, machine learning model construction and performance evaluation.
It is necessary for tools to support model retraining, which could significantly improve their performance. More critical features need to be designed to develop robust and species-neutral tools.
Siyu Han is a graduate student at the College of Computer Science and Technology, Jilin University, Changchun, China. His research interests include computational biology and machine learning methods.
Yanchun Liang is a professor at the College of Computer Science and Technology, Key Laboratory of Symbol Computation and Knowledge Engineering of Ministry of Education, Jilin University, Changchun, China. He is also a professor at Zhuhai Laboratory of Key Laboratory of Symbol Computation and Knowledge Engineering of Ministry of Education, Zhuhai College of Jilin University, Zhuhai, China. His research interests include computational intelligence, machine learning methods, text mining, and bioinformatics.
Qin Ma is the director of the Bioinformatics and Mathematical Biosciences Lab and an assistant professor in the Department of Agronomy, Horticulture and Plant Science at South Dakota State University. He is also an adjunct faculty member in the Department of Mathematics and Statistics at South Dakota State University and BioSNTR, SD, USA.
Yangyi Xu is a graduate student at the College of Computer Science and Technology, Jilin University, Changchun, China.
Yu Zhang is an associate professor at the College of Computer Science and Technology, Jilin University, Changchun, China.
Wei Du is an associate professor at the College of Computer Science and Technology, Jilin University, Changchun, China.
Cankun Wang is a graduate student in the Department of Mathematics and Statistics, South Dakota State University, SD, USA.
Ying Li is an associate professor at the College of Computer Science and Technology, Key Laboratory of Symbol Computation and Knowledge Engineering of Ministry of Education, Jilin University, Changchun, China. Her research topics include machine learning, bioinformatics and computational biology.
References
- 1. Harrow J, Frankish A, Gonzalez JM, et al. GENCODE: the reference human genome annotation for the ENCODE project. Genome Res 2012;22(9):1760–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Derrien T, Johnson R, Bussotti G, et al. The GENCODE v7 catalogue of human long non-coding RNAs: analysis of their structure, evolution and expression. Genome Res 2012;22(9):1775–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Guttman M, Russell P, Ingolia NT, et al. Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell 2013;154(1):240–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Djebali S, Davis CA, Merkel A, et al. Landscape of transcription in human cells. Nature 2012;489(7414):101–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pennisi E. Genomics. Encode project writes eulogy for junk DNA. Science 2012;337(6099):1159, 1161. [DOI] [PubMed] [Google Scholar]
- 6. Yang Q, Zhang S, Liu H, et al. Oncogenic role of long noncoding RNA AF118081 in anti-benzo[a]pyrene-trans-7, 8-dihydrodiol-9, 10-epoxide-transformed 16HBE cells. Toxicol Lett 2014;229(3):430–9. [DOI] [PubMed] [Google Scholar]
- 7. Bhartiya D, Kapoor S, Jalali S, et al. Conceptual approaches for lncRNA drug discovery and future strategies. Expert Opin Drug Discov 2012;7(6):503–13. [DOI] [PubMed] [Google Scholar]
- 8. Rinn JL, Chang HY.. Genome regulation by long noncoding RNAs. Ann Rev Biochem 2012;81:145–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lu Q, Ren S, Lu M, et al. Computational prediction of associations between long non-coding RNAs and proteins. BMC Genomics 2013;14:651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. da Rocha ST, Boeva V, Escamilla-Del-Arenal M, et al. Jarid2 is implicated in the initial xist-induced targeting of PRC2 to the inactive X chromosome. Mol Cell 2014;53(2):301–16. [DOI] [PubMed] [Google Scholar]
- 11. O’Leary VB, Ovsepian SV, Carrascosa LG, et al. PARTICLE, a triplex-forming long ncRNA, regulates locus-specific methylation in response to low-dose irradiation. Cell Rep 2015;11(3):474–85. [DOI] [PubMed] [Google Scholar]
- 12. Zhang Y, Tao Y, Liao Q.. Long noncoding RNA: a crosslink in biological regulatory network. Brief Bioinform 2017, in press. doi: 10.1093/bib/bbx042. [DOI] [PubMed] [Google Scholar]
- 13. Chen X, Yan CC, Zhang X, You ZH.. Long non-coding RNAs and complex diseases: from experimental results to computational models. Brief Bioinform 2017;18(4):558–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shi X, Sun M, Liu H, et al. A critical role for the long non-coding RNA GAS5 in proliferation and apoptosis in non-small-cell lung cancer. Mol Carcinog 2015;54(Suppl 1):E1–12. [DOI] [PubMed] [Google Scholar]
- 15. Ng SY, Lin L, Soh BS, et al. Long noncoding RNAs in development and disease of the central nervous system. Trends Genet 2013;29(8):461–8. [DOI] [PubMed] [Google Scholar]
- 16. Congrains A, Kamide K, Oguro R, et al. Genetic variants at the 9p21 locus contribute to atherosclerosis through modulation of ANRIL and CDKN2A/B. Atherosclerosis 2012;220(2):449–55. [DOI] [PubMed] [Google Scholar]
- 17. Chen G, Wang Z, Wang D, et al. LncRNADisease: a database for long-non-coding RNA-associated diseases. Nucleic Acids Res 2013;41(Database issue):D983–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ning S, Zhang J, Wang P, et al. Lnc2Cancer: a manually curated database of experimentally supported lncRNAs associated with various human cancers. Nucleic Acids Res 2016;44(D1):D980–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xu J, Bai J, Zhang X, et al. A comprehensive overview of lncRNA annotation resources. Brief Bioinform 2016;18(2):236–49. [DOI] [PubMed] [Google Scholar]
- 20. Yotsukura S, duVerle D, Hancock T, et al. Computational recognition for long non-coding RNA (lncRNA): software and databases. Brief Bioinform 2017;18(1):9–27. [DOI] [PubMed] [Google Scholar]
- 21. Kong L, Zhang Y, Ye ZQ, et al. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res 2007;35(Suppl 2):W345–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Altschul SF, Madden TL, Schäffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 1997;25(17):3389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ulitsky I, Bartel DP.. lincRNAs: genomics, evolution, and mechanisms. Cell 2013;154(1):26–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang L, Park HJ, Dasari S, et al. CPAT: coding-potential assessment tool using an alignment-free logistic regression model. Nucleic Acids Res 2013;41(6):e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lin MF, Jungreis I, Kellis M, et al. PhyloCSF: a comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics 2011;27(13):i275–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hu L, Xu Z, Hu B, Lu ZJ.. COME: a robust coding potential calculation tool for lncRNA identification and characterization based on multiple features. Nucleic Acids Res 2017;45(1):e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Siepel A, Bejerano G, Pedersen JS, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res 2005;15(8):1034–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Achawanantakun R, Chen J, Sun Y, et al. LncRNA-ID: long non-coding RNA IDentification using balanced random forests. Bioinformatics 2015;31(24):3897–905. [DOI] [PubMed] [Google Scholar]
- 29. Sun L, Liu H, Zhang L, et al. lncRScan-SVM: a tool for predicting long non-coding RNAs using support vector machine. PLoS One 2015;10(10):e0139654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Finn RD, Clements J, Eddy SR, et al. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res 2011;39:W29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sun L, Luo H, Bu D, et al. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res 2013;41(17):e166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li A, Zhang J, Zhou Z, et al. PLEK: a tool for predicting long non-coding RNAs and messenger RNAs based on an improved k-mer scheme. BMC Bioinformatics 2014;15:311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fickett JW. Recognition of protein coding regions in DNA sequences. Nucleic Acids Res 1982;10(17):5303–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fickett JW, Tung CSS.. Assessment of protein coding measures. Nucleic Acids Res 1992;20(24):6441–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kang YJ, Yang DC, Kong L, et al. CPC2: a fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res 2017;45(W1):W12–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bjellqvist B, Hughes GJ, Pasquali C, et al. The focusing positions of polypeptides in immobilized pH gradients can be predicted from their amino acid sequences. Electrophoresis 1993;14(10):1023–31. [DOI] [PubMed] [Google Scholar]
- 37. Bjellqvist B, Basse B, Olsen E, et al. Reference points for comparisons of two-dimensional maps of proteins from different human cell types defined in a pH scale where isoelectric points correlate with polypeptide compositions. Electrophoresis 1994;15(1):529–39. [DOI] [PubMed] [Google Scholar]
- 38. Tripathi R, Patel S, Kumari V, et al. DeepLNC, a long non-coding RNA prediction tool using deep neural network. Netw Model Anal Health Inform Bioinforma 2016;5(1):21. [Google Scholar]
- 39. Zhao J, Song X, Wang K, et al. lncScore: alignment-free identification of long noncoding RNA from assembled novel transcripts. Sci Rep 2016;6(1):34838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wucher V, Legeai F, Hédan B, et al. FEELnc: a tool for long non-coding RNAs annotation and its application to the dog transcriptome. Nucleic Acids Res 2017;45(8):e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Han S, Liang Y, Li Y, et al. Long noncoding RNA identification: comparing machine learning based tools for long noncoding transcripts discrimination. Biomed Res Int 2016;2016:8496165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kozak M. Recognition of AUG and alternative initiator codons is augmented by G in position +4 but is not generally affected by the nucleotides in positions +5 and +6. EMBO J 1997;16(9):2482–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kozak M. Initiation of translation in prokaryotes and eukaryotes. Gene 1999;234(2):187–208. [DOI] [PubMed] [Google Scholar]
- 44. Ingolia NT, Lareau LF, Weissman JS, et al. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 2011;147(4):789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kent WJ, Sugnet CW, Furey TS, et al. The human genome browser at UCSC. Genome Res 2002;12(6):996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hu L, Di C, Kai M, et al. A common set of distinct features that characterize noncoding RNAs across multiple species. Nucleic Acids Res 2015;43(1):104–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chen C, Liaw A, Breiman L. Using Random Forest to Learn Imbalanced Data. Technical Report 1999, Department of Statistics, UC Berkeley Andy, 2004.
- 48. Nawrocki EP, Eddy SR.. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 2013;29(22):2933–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang L, Feng Z, Wang X, et al. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2010;26(1):136–8. [DOI] [PubMed] [Google Scholar]
- 50. Necsulea A, Soumillon M, Warnefors M, et al. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature 2014;505(7485):635–40. [DOI] [PubMed] [Google Scholar]
- 51. Nair AS, Sreenadhan SP.. A coding measure scheme employing electron-ion interaction pseudopotential (EIIP). Bioinformation 2006;1(6):197–202. [PMC free article] [PubMed] [Google Scholar]
- 52. Harrow J, Denoeud F, Frankish A, et al. GENCODE: producing a reference annotation for ENCODE. Genome Biol 2006;7(Suppl 1):S4.1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yates A, Akanni W, Amode MR, et al. Ensembl 2016. Nucleic Acids Res 2016;44:D710–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Burge SW, Daub J, Eberhardt R, et al. Rfam 11.0: 10 years of RNA families. Nucleic Acids Res 2013;41(D1):D226–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mattei E, Ausiello G, Ferrè F, et al. A novel approach to represent and compare RNA secondary structures. Nucleic Acids Res 2014;42(10):6146–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lorenz R, Bernhart SH, Höner zu Siederdissen C, et al. ViennaRNA Package 2.0. Algorithms Mol Biol 2011;6(1):26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Clark MB, Johnston RL, Inostroza-Ponta M, et al. Genome-wide analysis of long noncoding RNA stability. Genome Res 2012;22(5):885–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Charif D, Lobry JR.. SeqinR 1.0-2: a contributed package to the R Project for statistical computing devoted to biological sequences retrieval and analysis In: Bastolla U, Porto M, Roman HE, Vendruscolo M (eds), Structural Approaches to Sequence Evolution: Molecules, Networks, Populations Biological and Medical Physics, Biomedical Engineering. New York: Springer Verlag, 2007, 207–232. [Google Scholar]
- 59. Silverman BD, Linsker R.. A measure of DNA periodicity. J Theor Biol 1986;118(3):295–300. [DOI] [PubMed] [Google Scholar]
- 60. Tsonis AA, Elsner JB, Tsonis PA, et al. Periodicity in DNA coding sequences: implications in gene evolution. J Theor Biol 1991;151(3):323–31. [DOI] [PubMed] [Google Scholar]
- 61. Tiwari S, Ramachandran S, Bhattacharya A, et al. Prediction of probable genes by fourier analysis of genomic sequences. Comput Appl Biosci 1997;13(3):263–70. [DOI] [PubMed] [Google Scholar]
- 62. Cohen J. A coefficient of agreement for nominal scales. Educ Psychol Meas 1960;20(1):37–46. [Google Scholar]
- 63. Kuhn M. Building predictive models in R using the caret package. J Stat Soft 2008;28(5):1–26. [Google Scholar]
- 64. Chang CC, Lin CJ.. LIBSVM. A library for support vector machines. ACM Trans Intell Syst Technol 2011;2(3):27. [Google Scholar]
- 65. Meyer D, Dimitriadou E, Hornik K. et al. e1071: misc Functions of the Department of Statistics, Probability Theory Group (Formerly: e 1071), TU Wien R package version 1.6–8. 2017. https://CRAN.R-project.org/package=e1071.
- 66. Liaw A, Wiener M.. Classification and Regression by randomForest. R News 2002;2(3):18–22. [Google Scholar]
- 67. Huang GB, Wang DH, Lan Y, et al. Extreme learning machines: a survey. Int J Mach Learn Cybern 2011;2(2):107–22. [Google Scholar]
- 68. Gosso A. elmNN: Implementation of ELM (Extreme Learning Machine) algorithm for SLFN (Single Hidden Layer Feedforward Neural Networks). R package version 1.0. 2012. https://CRAN.R-project.org/package=elmNN.
- 69.H2O.ai. R Interface for H2O, R package version 3.10.0.8 2016. https://github.com/h2oai/h2o-3.
- 70. Robin X, Turck N, Hainard A, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 2011;12:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wickham H. ggplot2: Elegant Graphics for Data Analysis. New York: Springer-Verlag, 2009. [Google Scholar]
- 72. Suzek BE, Huang H, McGarvey P, et al. UniRef: comprehensive and non-redundant UniProt reference clusters. Bioinformatics 2007;23(10):1282–8. [DOI] [PubMed] [Google Scholar]
- 73. Wickham H, Danenberg P, Eugster M; RStudio. roxygen2: In-Line Documentation for R R package version 6.0.1. 2017. https://CRAN.R-project.org/package=roxygen2.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.