Abstract
Background
The germline genetic events underpinning medulloblastoma (MB) initiation, and therefore the ability to determine who is at risk, are still unknown for the majority of cases. Microsatellites are short repeated sequences that make up ~3% of the genome. Repeat lengths vary among individuals and are often nonrandomly associated with disease, including several cancers such as breast, glioma, lung, and ovarian. Due to their effects on gene function, they have been called the “tuning knobs of the genome.”
Methods
We have developed a novel approach for identifying a microsatellite-based signature to differentiate MB patients from controls using germline DNA.
Results
Analyzing germline whole exome sequencing data from a training set of 120 MB subjects and 425 controls, we identified 139 individual microsatellite loci whose genotypes differ significantly between the groups. Using a genetic algorithm, we identified a subset of 43 microsatellites that distinguish MB subjects from controls with a sensitivity and specificity of 92% and 88%, respectively. This microsatellite signature was validated in an independent dataset consisting of 102 subjects and 428 controls, with comparable sensitivity and specificity of 95% and 90%, respectively. Analysis of the allele genotypes of those 139 informative loci demonstrates that their association with MB is a consequence of individual microsatellites' genotypes rather than their hypermutability. Finally, an analysis of the genes harboring these microsatellite loci reveals cellular functions important for tumorigenesis.
Conclusion
This study demonstrates that MB-specific germline microsatellite variations mark those at risk for MB development and suggests mechanisms of predisposition.
Keywords: germline predisposition, medulloblastoma, microsatellites
Key Points.
1. Microsatellites in germline DNA differentiate children with medulloblastoma.
2. Children with medulloblastoma do not have constitutional microsatellite instability.
3. Genes harboring the tumor associated microsatellites are cancer associated.
Importance of the Study.
This is the first study establishing a link between germline microsatellite genotypes and the presence of a pediatric cancer. Given the dearth of environmental influences and the relative paucity of germline mutations associated with pediatric cancers, microsatellites constitute an important novel and understudied source of genetic predisposition. Future screening and prevention strategies applied to rare diseases will require the means to identify populations at risk, a purpose that microsatellite signatures could fulfill. In addition to this potential, the establishment of the biological influence of specific microsatellites upon their companion genes could delineate avenues for preventative intervention.
Medulloblastoma (MB) is the most common malignant childhood brain tumor. Extensive genomic characterization has divided MB tumors into 4 molecular subgroups: wingless (WNT), sonic hedgehog (SHH), Group 3, and Group 4, with distinct transcriptional profiles, copy number alterations, somatic mutations, and clinical outcomes.1 Overall, 5–6% of MB patients harbor germline mutations that are considered to be pathogenic, including in adenomatous polyposis coli (APC), breast cancer 2 (BRCA2), partner and localizer of BRCA (PALB2), Patched 1 (PTCH1), suppressor of fused homolog (SUFU), and tumor protein 53 (TP53).2 TP53 germline mutations are present in 1% of MB cases, although this incidence rises to 8% of SHH cases and even further to 20% if one limits the analysis to SHH cases aged between 5 and 16 years.2 These predisposing mutations are individually insufficient to efficiently cause MB in animal models and require a potentiating background.3 Numerous genome-wide association studies in MB have focused on single nucleotide variants, ignoring noncoding regions and repetitive DNA. However, many studies have shown linkage between insertion/deletions (indels) of germline microsatellites (MS) and a number of neurological disorders such as Huntington disease and Friedreich ataxia—caused by an MS variant in the former in the coding sequence and in the latter in a noncoding intronic sequence.4 Furthermore, recent studies have shown that MS variations can contribute to the genetic background of several adult cancers.5–9 Many cancer-associated genes contain MS loci10 (eg, phosphatase and tensin homolog [PTEN], neurofibromatosis type 1 [NF1]), and in some cases somatic MS indels have been causally implicated in cancer.11 Based on these findings, we hypothesize that a permissive constitutional genetic environment may be created by the cooperation of germline DNA MS repeat elements affecting the transcriptional and translational landscape of individuals, making them susceptible to tumor formation through modulation of foundational cellular processes.
MS consist of a 1–6 base pair unit repeated in tandem to form an array.12 Over 600 000 unique MS exist in the human genome, often embedded in introns, exons, and regulatory regions.13 The length of MS loci frequently change due to strand slip replication and heterozygote instability,13 varying between alleles and between individuals. These changes can influence gene expression by inducing Z-DNA and H-DNA folding,14 altering nucleosome positioning14,15 and changing the spacing of DNA binding sites.13,16,17 Noncoding variations can alter DNA secondary structure and protein/RNA binding of proximate genes, resulting in changes in transcriptional and translational activity as well as alternative splicing.18 For these reasons, MS have been called the “tuning knobs” of the genome.14,19,20 Exonic MS loci containing repeated elements of 3 or 6 base pairs often cause amino acid gain or loss; other non-modulo-3 lengths typically cause frameshift mutations.21 Genes harboring MS contribute disproportionately to nervous system disorder, suggesting an importance in neurodevelopment.22 In fact, the role of repetitive elements is well established for some neurological diseases; polyglutamate repeats in particular play a role in Huntington disease, spinocerebellar ataxia, and spinobulbar muscular atrophy. Bioinformatic studies indicate that many genes hosting tandem repeats have a neural function.23 A thorough review of MS and their impact on disease has recently been published.24
Recent developments in MS genotyping algorithms and advances in genome sequencing have allowed the identification of germline MS genotypes that can distinguish healthy from affected individuals with different types of cancers (breast, colon, glioma, etc7,9,25–27). Here, for the first time in a pediatric cancer, we present a set of MS genotypes able to differentiate children with MB from healthy individuals based upon their germline DNA.
Methods
Patient Samples
The majority of sequencing data used in this work has been previously published and can be found in Supplementary Table 1 and the Supplementary Methods. Additionally, whole-exome sequencing (WES) from 6 MB patients' blood DNA was newly generated using the TruSeq exome target enrichment kit and Illumina Sequencer HiSeq 2500 (data available upon request). All patient material was collected after informed consent under a CNMC institutional review board approved protocol. Power calculations were performed to determine the sample size of training and validation cohorts (see Supplementary Methods).
Microsatellite Genotyping
A list of 625 195 unique MS in version GRCh38/hg38 of the human reference genome was generated with a custom Perl script ‘searchTandemRepeats.pl’ using default parameters28 (Supplementary Methods).
Reads from WES and whole-genome sequencing (WGS) were mapped to the human GRCh38/hg38 reference genome using Bowtie2,29 and reads mapping to MS locations were locally realigned using the Genome Analysis Toolkit IndelRealigner tool30 after duplicated reads were marked. We used the program Repeatseq31 to determine the genotype of MS in WES or WGS (Supplementary Methods).
For each microsatellite, we calculated whether the distribution of genotypes differs in the germline DNA from 2 groups of samples in the training dataset. For each microsatellite, a contingency table is populated with genotype counts for the 2 groups: MB and normal (Supplementary Fig. 1). Then, statistical differences were quantified using a generalized Fisher's exact test. The Benjamini–Hochberg multiple testing correction (n = 43 457 tested MS) was applied to control the false discovery rate. The numbers used to describe each genotype denote the length of that microsatellite in each allele.
Microsatellite Filtering to Control for Age, Ethnicity, and Sequencing Protocol
This study was designed to identify germline variations of MS specific to MB. The publicly available sequence data used contain limited metadata (sex, age, and ethnicity), variation in the sequencing protocol used, and relatively few children. Thus it was not possible to confine our analysis to age and ethnically matched controls, and therefore our analysis ran the risk of identifying MS with age, sequencing, and ethnic bias rather than disease status alone. In an attempt to eliminate this risk, we identified MS with potential bias—for age, sequencing protocol, or ethnicity—and excluded them from subsequent analysis. Details about how this filtering was done can be found in the Supplementary Methods and Supplementary Figures 2–4.
Metric to Score Samples and ROC Analysis
Metric to score samples
We designed a metric to score samples based on their unique distribution of MS genotypes. Essentially, the metric is a weighted sum of the genotypes belonging to each sample: weights stem from the difference in frequency for each genotype in the MB and healthy groups. A visual summary of the metric is provided in Supplementary Fig. 5. Scores calculated for every sample in our cohorts can be found in Supplementary Table 1.
ROC analysis
We used receiver operating characteristic (ROC) analysis to design a classification scheme capable of differentiating samples with MB from healthy controls. The area under the ROC curve (AUC) was used as a measure of how well scores differentiate the 2 groups. Then, a cutoff was selected for all future classification (see the “Validation” section below). Here, the cutoff is a single score that minimizes sensitivity and simultaneously maximizes specificity; it was identified using the Youden index (ie, sensitivity + specificity – 1)32 (Supplementary Methods).
Subset of Microsatellites (Genetic Algorithm)
Genetic algorithms are a class of biologically inspired algorithms and have been described extensively elsewhere.33 In this work we use a genetic algorithm to identify the most predictive subset of markers—from the set of 139 found to be correlated with MB—using a 2-step process. In initialization, an initial population is generated with random subsets of the 139 MS markers. In optimization, subsets are iteratively ranked, recombined, and replaced (see Supplementary Fig. 6). The algorithm has 4 adjustable hyperparameters: (i) the maximum population size, (ii) the size of each subset, (iii) the metric used to rank each subset, and (iv) the number of subsets replaced in each iteration. Details of each step and hyperparameters are provided in the Supplementary Methods.
Validation
Samples used for validation can be found in Supplementary Table 1 and the Supplementary Methods. Each validation sample was scored with the same metric used for the training samples (see above). The cutoff (identified in training) was used to predict which of the 530 validation samples had MB and which were healthy controls. MB was predicted for validation samples above the cutoff and compared with the known identity of the validation samples.
Additionally, an extended validation cohort using a recently published MB germline WGS dataset contained 290 samples that did not overlap with the samples we previously used. These samples were scored with the procedure described above.
Microsatellite Mutability
In order to test whether individuals with MB are more prone to MS variation, we used the total number of alleles genotyped for each microsatellite as a measure of its mutability and compared this metric across disease and control cohorts. Details regarding the identification of minor alleles can be found in the Supplementary Methods.
Downstream Analysis
For functional analysis, we used genes associated with the 139 loci of MS whose genotypes are significantly different between MB subjects and controls. Pathway analysis was performed using Ingenuity Pathway Analysis (Qiagen, https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis). Mutations and co-occurrence were analyzed using PedcBioPortal (www.cbioportal.org). In the mutation analysis, the numbers indicate the percent of tumors in each dataset with mutations in at least 1 of the 124 genes associated with the identified MS. Construction of a protein-protein interaction (PPI) network was conducted with STRING (Search Tool for the Retrieval of Interacting Genes/Proteins). Parameters and details can be found in the Supplementary Methods.
RNA Sequencing
In order to investigate the effect of the MS genotypes on their associated genes, we downloaded 170 files from RNA sequencing (RNA-seq) (European Genome-phenome Archive dataset EGAD00001003279; Supplementary Table 1) partially overlapping with our validation and extended validation cohorts. Details of the expression analysis can be found in the Supplementary Methods.
Stability Prediction of mRNA
To analyze the effect of the untranslated region (UTR)/exonic MS on the stability of the RNA, we predicted the minimum free energy (MFE) secondary structures for each of the MS variants using the RNAfold tool from the ViennaRNA package.34 Because the MFE depends on the length of the RNA, we normalized the MFE by the size of the RNA and then calculated the difference in MFE between the different RNA variants (ΔMFE).
Results
Identification of Medulloblastoma Microsatellite Informative Loci
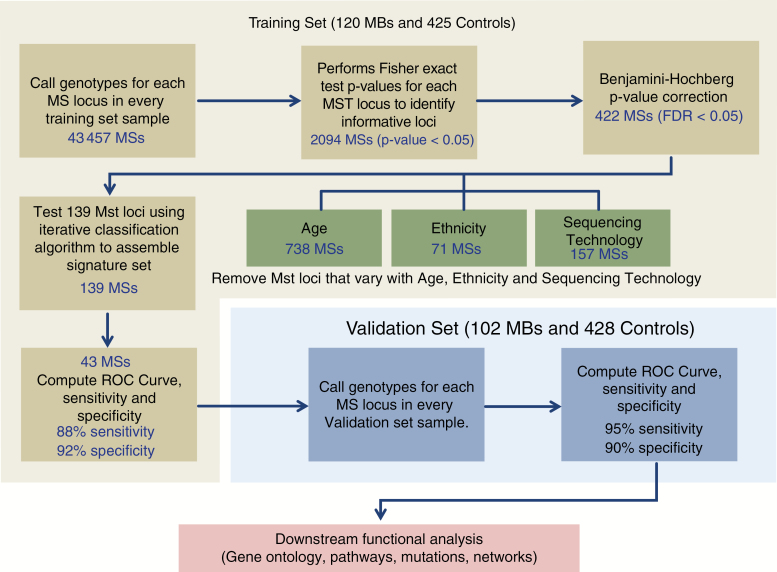
To explore the impact of variations of MS in MB predisposition, we have developed a new computational workflow to identify germline MS whose genotypes differ between children with MB and control subjects (Fig. 1). We have applied this approach to germline DNA sequencing data divided into training and validation groups. The training set contained 120 MB and 425 control individuals, and the validation set contained 102 MB and 428 control individuals (Supplementary Table 1). Using the training set, we first genotyped 43 457 different MS present in both the 120 MB samples and 425 healthy controls. For each of these microsatellites, a generalized Fisher's exact test was used to assess the statistical difference in genotype distribution between the 2 groups, revealing 2094 MS with P < 0.05. After Benjamini–Hochberg multiple testing correction (α = 0.05), 422 passed false discovery. We then performed 3 additional steps to remove MS that nonrandomly vary with age, ethnicity, and DNA sequencing protocol (Fig. 1, Supplementary Figures 2–4 and Supplementary Methods). In total, 283 MS were removed from the list of 422, resulting in a reduced list of 139 markers (Supplementary Table 2). In summary, our approach identified 139 MS from germline DNA whose genotypes were significantly different between MB subjects and healthy controls.
Fig. 1.
Schematic representation of the approach used for the identification and validation of medulloblastoma associated microsatellites. The approach used in this work is divided into 3 stages: computational identification of informative microsatellite loci using the training set, validation of the microsatellite markers in an independent validation cohort, and finally downstream analysis of the genes associated with those microsatellites. The first stage includes a filter to eliminate microsatellites that vary with age, ethnicity, and sequencing technology.
Medulloblastoma Microsatellite Classifier Set
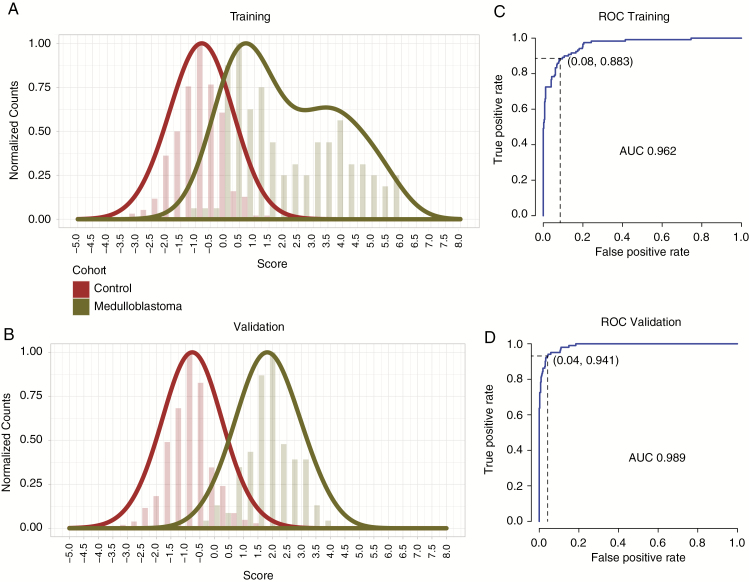
In order to identify a subset of MS with the best performance in distinguishing MB samples and healthy controls, we used the set of 139 MS to train an MB classifier. First, we designed a metric to score each sample based on the genotypes of the 139 MS (Supplementary Methods, Supplementary Fig. 5). We used a genetic algorithm33 to identify a subset of 43 MS that best distinguish MB samples from healthy controls (Supplementary Methods, Supplementary Fig. 6). Briefly, subsets of MS (drawn from the set of 139) are ranked by their precision and recall. Subsets with the lowest ROC results are discarded; those that remain are continuously mixed and re-ranked. The algorithm converged in 87 cycles to reveal a subset of 43 MS with an F-measure of 0.90 and an AUC of 0.962 (Fig. 2, Table 1, Supplementary Table 3, Supplementary Fig. 6). Using Youden's index,32 we determined that the optimal cutoff score for differentiating MB samples from healthy controls is 0.155 (Supplementary Fig. 7). Thus, we have identified a set of 43 MS whose genotype distributions are able to distinguish MB patients from healthy controls with 88% sensitivity and 92% specificity (Fig. 2A, C and Supplementary Fig. 8).
Fig. 2.
Distribution of metric scores in the training (A) and validation (B) cohorts. ROC analysis performed on training (C) and validation (D) cohorts. The y-axis indicates the normalized proportion of individuals in each cohort per score value bin on the x-axis.
Table 1.
List of 43 MS loci in the MB germline DNA classifier
| Position | Fisher's P-value | Benjamini Adj. P-value | Repeat Unit | Gene | Region |
|---|---|---|---|---|---|
| chr20:30215031-30215040 | 0 | 0 | A | – | Intergenic |
| chr4:71755149-71755177 | 0 | 3.60E-07 | TATT | GC | Intron |
| chr2:216610248-216610286 | 0 | 3.05E-06 | TTTTC | – | Intergenic |
| chr4:102949102-102949116 | 1.00E-08 | 5.17E-06 | AC | SLC9B1 | Intron |
| chr13:44943352-44943377 | 2.00E-08 | 1.21E-05 | AC | NUFIP1 | Intron |
| chr22:50215636-50215648 | 1.60E-07 | 7.59E-05 | G | SELENOO | Intron |
| chr17:17793780-17793820 | 3.54E-06 | 1.07E-03 | CAG | RAI1 | Exon |
| chr1:153770135-153770152 | 3.76E-06 | 1.11E-03 | GT | INTS3 | Intron |
| chr6:106518976-106519008 | 4.94E-06 | 1.36E-03 | CA | CRYBG1 | Intron |
| chr2:68217012-68217040 | 6.94E-06 | 1.83E-03 | AC | PPP3R1 | Intron |
| chr17:64504983-64504996 | 8.09E-06 | 2.03E-03 | TC | DDX5 | Intron |
| chr18:21540014-21540032 | 1.10E-05 | 2.58E-03 | AT | ESCO1 | Intron |
| chr5:1278442-1278456 | 1.10E-05 | 2.59E-03 | CA | TERT | Intron |
| chr18:42923771-42923785 | 1.16E-05 | 2.69E-03 | A | RIT2 | Intron |
| chr17:7024701-7024730 | 1.58E-05 | 3.51E-03 | CAG | BCL6B | Exon |
| chr16:4407278-4407290 | 1.81E-05 | 3.98E-03 | A | CORO7 | Intron |
| chr1:8946030-8946056 | 1.83E-05 | 3.99E-03 | CTT | CA6 | Intron |
| chr4:112653323-112653334 | 2.12E-05 | 4.39E-03 | T | LARP7 | Intron |
| chr4:77773174-77773189 | 2.42E-05 | 4.88E-03 | A | CNOT6L | Intron |
| chr5:64507682-64507694 | 2.69E-05 | 5.31E-03 | A | RGS7BP | Intron |
| chr1:3836469-3836492 | 3.11E-05 | 5.99E-03 | T | CEP104 | Intron |
| chr20:33015600-33015613 | 3.24E-05 | 6.11E-03 | A | BPIFB2 | Intron |
| chr10:26223687-26223728 | 6.49E-05 | 1.06E-02 | GT | GAD2 | Intron |
| chr15:91749825-91749843 | 6.80E-05 | 1.10E-02 | GGTGGA | – | Intergenic |
| chr15:64395361-64395385 | 7.25E-05 | 1.16E-02 | TG | TRIP4 | Intron |
| chr10:36522760-36522794 | 8.32E-05 | 1.29E-02 | GT | – | Intergenic |
| chr3:189907237-189907252 | 9.32E-05 | 1.40E-02 | T | – | Intergenic |
| chr5:91149985-91150025 | 1.04E-04 | 1.54E-02 | TTCTTT | ADGRV1 | Intron |
| chr20:33623955-33623983 | 1.07E-04 | 1.58E-02 | A | CBFA2T2 | Intron |
| chr16:75234097-75234113 | 1.47E-04 | 2.07E-02 | AC | BCAR1 | Intron |
| chr4:151662756-151662771 | 2.06E-04 | 2.71E-02 | T | FAM160A1 | 3′ UTR |
| chr7:37033493-37033506 | 2.43E-04 | 3.03E-02 | A | ELMO1 | Intron |
| chr10:27145290-27145311 | 2.51E-04 | 3.11E-02 | A | YME1L1 | Intron |
| chr5:79088359-79088398 | 2.81E-04 | 3.37E-02 | TG | BHMT2 | Intron |
| chr4:52062187-52062204 | 2.99E-04 | 3.51E-02 | T | SPATA18 | Intron |
| chr1:153645035-153645049 | 3.45E-04 | 3.92E-02 | T | CHTOP | Intron |
| chr15:85123338-85123386 | 3.44E-04 | 3.92E-02 | AC | PDE8A | Intron |
| chr17:80822939-80822950 | 3.56E-04 | 3.99E-02 | GT | RPTOR | Intron |
| chr20:5106079-5106120 | 3.76E-04 | 4.17E-02 | AC | TMEM230 | Intron |
| chr10:104037288-104037301 | 3.86E-04 | 4.25E-02 | T | COL17A1 | Intron |
| chr4:6084968-6084994 | 4.24E-04 | 4.55E-02 | A | JAKMIP1 | Intron |
| chr14:80905756-80905768 | 4.63E-04 | 4.84E-02 | A | CEP128 | Intron |
| chr6:49470141-49470153 | 4.84E-04 | 4.99E-02 | T | CENPQ | Intron |
In order to validate this result, we used an independent cohort of germline DNA including 102 MB and 428 control subjects. This number of sample was selected to ensure that the study would be robustly powered (Supplementary Methods). We applied the optimal cutoff (0.155), using Youden's index, to the independent validation sample set and found that the classifier could distinguish cases from controls with a sensitivity of 95% and specificity of 90% (Fig. 2B, D). We further tested our signature in a larger, recently published independent dataset of 290 germline MB WGS with deeper coverage35 (Supplementary Table 1) and found it to perform similarly well (Supplementary Fig. 9). In summary, we have identified and validated a set of 43 MS whose genotype distributions are able to distinguish MB patients from healthy controls using germline DNA with high sensitivity and specificity.
To determine whether the performance of the signature set varies across the 4 consensus molecular subgroups of MB, we tested the 43 MS signature across MB subgroups combining the training and the validation cohorts to achieve adequate sample sizes. It performed equally well for all MB subgroups (Kolmogorov–Smirnov test) (Supplementary Fig. 10).
Medulloblastoma Informative Microsatellite Loci Mutability
In the germline, indels of MS are significantly more common than elsewhere in the genome, 10−4 to 10−3 compared with 10−8 per locus per generation respectively.36 However, mutation rates also vary for different MS based on the length of the repeat, their repetitive motif, and influence on DNA folding.37 We hypothesized that the differences found for the 139 MB-associated MS (Supplementary Table 2) could be the result of increased MS genotype variation inherent in the individual with MB. In order to test whether individuals with MB are more prone to MS variation, we used the total number of alleles genotyped for each microsatellite as a measure of its mutability and compared this metric across disease and control cohorts. We didn't find significant differences in the number of genotyped alleles between healthy and MB individuals, supporting the conclusion that there is not a general instability of MS in MB patients. This is consistent with a previous report that found only 1 case out of 36 with increased MS instability.38 We then investigated whether the predictive capability could be related to a characteristic of the informative MS themselves by ranking all identified MS by allelic load. We found that while the 139 were among the more mutable MS (higher number of alleles), they did not include the most mutable sites. Additionally, we compared the number of homozygote and heterozygote genotypes and the MS array lengths as potential sources of variability and found no statistically significant differences between MB and control germline DNA. From these data, we conclude that the association of the 139 MS with MB is a consequence of those individual microsatellite genotypes rather than simply being a result of constitutional hypermutability.
Specificity of Microsatellite Informative Loci for Medulloblastoma
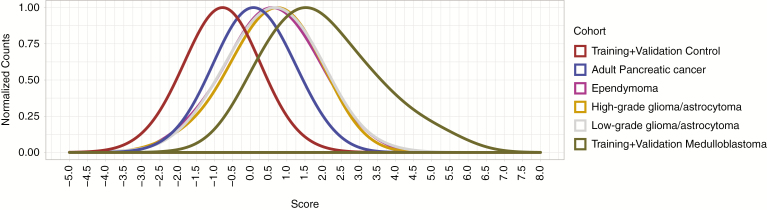
Interestingly, the group of 139 MS significantly associated with MB contained some intronic MS previously implicated in other cancer types, nuclear fragile X mental retardation protein interacting protein 1 (NUFIP1) and kinesin family member 1B (KIF1B),6 raising the possibility that the effects of germline MS genotypes are not histologically restricted. To analyze the specificity of the 43 signature MS markers for MB, we tested their performance on a cohort of 153 adult pancreatic ductal adenocarcinoma germline samples from The Cancer Genome Atlas. The signature does differentiate pancreatic cancer samples from the MB validation cohort; however, the pancreatic cancer sample scores are clearly elevated compared with healthy controls (Fig. 3). From this, one may expect other pediatric brain tumors to also have elevated prediction scores. To test this, we performed MS genotyping on 72 ependymomas, 65 high-grade gliomas, and 230 low-grade gliomas from the Children's Brain Tumor Tissue Consortium and observed elevated MS genotype scores compared with normal controls and pancreatic cancer samples (Fig. 3). Finally, we cross-referenced our set of 139 markers with 55 previously published informative MS markers for breast cancer7 and 105 for lung cancer.9 Two markers are in common with breast cancer, NUFIP1 and KIF1B, and 3 markers are shared with lung cancer: NUFIP1, KIF1B, and UBXN7. Interestingly, NUFIP1 and KIF1B had been identified as pan-cancer markers before.6 In summary, these findings support the overarching hypothesis that variations of MS play a role in cancer susceptibility at an organismal level and, like predisposing gene mutations, do not respect histologic boundaries.
Fig. 3.
Distribution of metric scores for the signature set of 43 MS in the normal, adult pancreatic ductal adenocarcinoma, ependymoma, high-grade glioma, low-grade glioma, and MB cohorts. The y-axis indicates the normalized proportion of individuals in each cohort per score value bin on the x-axis.
Role of the Informative MS Associated Genes
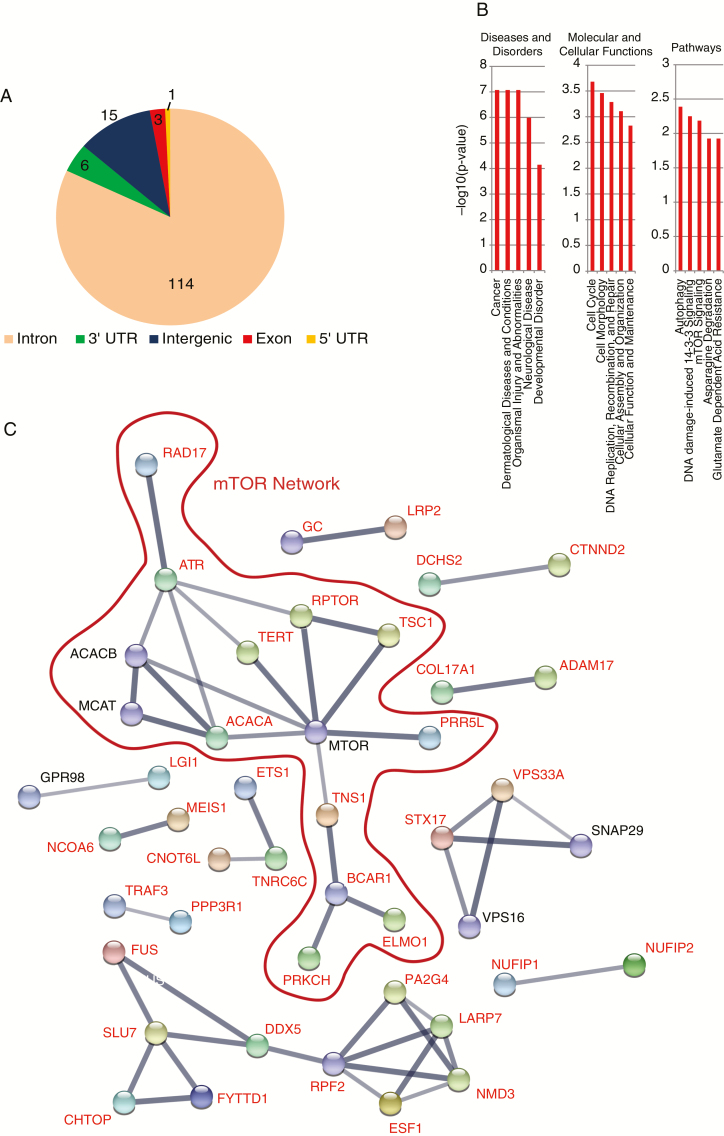
Of the 139 MS whose genotypes differed between MB and control samples, 114 were located in intronic regions, 15 in intergenic regions, 6 in 3′ UTRs, 3 in exonic regions, and 1 in a 5′ UTR (Fig. 4A). To understand the potential mechanistic roles of these genes, we conducted an Ingenuity Pathway Analysis of the 124 genes associated with non-intergenic informative MS loci. The analysis revealed statistically significant associations with cancer and molecular cellular functions such as cell cycle, DNA replication, recombination and repair, and cellular growth and proliferation (Fig. 4B, Supplementary Table 4). The occurrence of mutations in these 124 genes was examined in 4 MB cohorts available in cBioPotal. In spite of the known low mutation rate in MB tumors,2 we found that on average 17% of the MB cancer samples contained mutations in at least one of these 124 genes (Supplementary Table 5) compared with 4.5% of neuroblastoma tumors. An analysis of mutational co-occurrence, using the Sick Kids 2016 dataset within cBioPortal, indicates that 135 out of all possible (9591 = 139*(139-1)/2) MS pairs were found to significantly co-occur (P < 0.05). Interestingly, we found 2 patients with co-occurrence of mutations in 20 and 10 MB informative MS loci, respectively (Supplementary Table 6).
Fig. 4.
(A) Pie chart displaying the genomic locations of the 139 informative MS loci. (B) Gene Ontology analysis of the informative MB MS. Y-axis indicates the −log10(p-value) (C) PPI network of the 124 genes associated with the informative MS loci.
A PPI network comprising the 124 genes associated with the informative MS loci (Fig. 4C) resulted in a network with a PPI enrichment P-value of 0.0007. Despite the low number of proteins used as input, we found a significant hub related to mammalian target of rapamycin (mTOR), a prominent pathway (phosphatidylinositol-3 kinase/AKT/mTOR) in MB tumors that has been proposed as a promising therapeutic target.39,40 While it is interesting to speculate regarding a role for these MS impacting such a central cancer pathway, these are germline findings and therefore it will be necessary to first demonstrate a functional effect of the MS genotypes in the context of tumor cell biology.
It has been described that MS proximate to splice sites can alter mRNA splicing by altering immature RNA secondary structure.18 Out of 139 informative MS, 114 are located in intronic regions. As shown in Supplementary Fig. 11, all the intronic MS are within 1 to 260 bp of an exon and thus a splice site. To detect a possible effect on splicing, we determined the genotypes of the 114 intronic MS in RNA-seq data from 170 MB tumors and then correlated them with the abundance of each splice isoform (Supplementary Table 1); 49 of the 837 isoforms were significantly correlated (ANOVA P < 0.05; Supplementary Table 7). Exonic or UTR MS can affect the stability of the mature mRNA. We found 6 in 3′ UTRs, 3 in exonic regions, and 1 in a 5′ UTR (Fig. 4A). To analyze the effect of the UTR/exonic MS on the stability of the RNA, we predicted the MFE secondary structures for each of the MS variants using the RNAfold tool from the ViennaRNA package (see Supplementary Methods). We calculated the difference in MFE between each RNA variant and the shortest (Supplementary Fig. 12a). We found that differing MS lengths affect the predicted stability of the RNA. For example, we found that the retinoic acid induced 1 (RAI1) mRNA with a microsatellite of 38 nucleotides is the most stable, as opposed to that with a 41 nucleotide microsatellite, which is the least stable. The insertion of an extra CAG triplet increases the positional entropy of the loop formed by the CAG MS in RAI1 mRNA, resulting in the lower predicted stability (Supplementary Fig. 12b). We found that the genotypes associated with MB have the least stable mRNAs for the exonic MS in the RAI1 and B-cell chronic lymphocytic leukemia/lymphoma 6 member B (BCL6B) genes, as well as for the 3′ UTR MS in the ZBTB3 and MIDN genes. On the other hand, the genotype associated with MB on the 3′ UTR of the FAM160A1 gene has the most stable mRNA. To extend this analysis to mRNA abundance, we then analyzed RNA-seq data from 170 MB tumors (Supplementary Table 1) and correlated them with the MS genotypes in tumor DNA but did not find any significant correlation with gene expression (ANOVA; Supplementary Fig. 12c).
In this study, we have identified a set of 139 MS whose genotypes differ between MB patients and healthy controls. We have shown that a subset of 43 of these MS is able to differentiate MB individuals from controls based upon their germline DNA with a sensitivity and specificity of 95% and 90%, respectively. Although the prospective validation of a diagnostic test for MB predisposition will need to wait for a time when germline sequencing early in life is more ubiquitous, these findings represent the first evidence of germline predisposition to a pediatric cancer that is not attributable to a germline coding mutation.
Discussion
This study identifies a subset of MS that has genotypes that statistically differ between MB samples and healthy controls. Beyond marking individuals with MB, the role these MS may play in medulloblastoma etiology is not yet defined. MS array length variations are known to affect nucleosome positioning, the spacing of DNA binding sites, DNA secondary structure alteration, mRNA and protein stability, gene expression, alternative splicing, and protein sequence. MSs located in intronic regions can alter mRNA splicing by altering immature RNA secondary structure.18 An analysis of the location of these intronic MS showed that they are located in close proximity to splicing sites (1–260 bp) (Supplementary Fig. 11). Although this is expected as the training cohort is based on WES, we find significant correlations between isoform expression and MS genotypes supporting the hypothesis that MS close to splice sites can alter mRNA splicing. Although this result is appealing, we cannot demonstrate that MS genotypes are causal. Exonic and UTR MS can also alter mRNA stability, while exonic MS can have a direct impact on protein translation, function, or stability. Values of MFE of the predicted secondary structures for each of the exonic/UTR MS variants showed that MS genotype variants can change the stability of mRNA, although we haven't found any significant correlations between gene expression and MS genotypes (Supplementary Fig. 12).
Three informative MS loci are located in protein coding sequences (Fig. 4A); all of them are CAG trinucleotide repeats (RAI1, BCL6B, TNS1). The BCL6B gene has been implicated in colon, gastric, and hepatic cancer.41–43 RAI1 encodes for a nuclear protein with unknown function whose haploinsufficiency causes Smith–Magenis syndrome.44 Of note, there is no predisposition to medulloblastoma or any other cancer inherent in Smith–Magenis syndrome. However, both RAI1 and BCL6B are transcription factors located in the short arm of chromosome 17, the deletion of which is a recurrent alteration in the most common subgroups of MB tumors.45 The frame of the CAG repeat in RAI1 MS encodes a polyglutamine run. Apart from inducing changes in protein structure, short polyglutamine expansions are also thought to modulate transcription factor activity.46 Interestingly, the RAI1 protein is highly expressed in cerebellum, the region where MB tumors arise.47 Still, it's unclear how a polyglutamine variant in RAI1 may contribute to MB. Most polyglutamine diseases are characterized by insoluble protein aggregates, something not seen in most cancers. On the other hand, polyglutamine expansions have been shown to confer both gain and loss of function depending upon the affected protein.48 It's conceivable that this phenomenon plays a role in MB formation and/or maintenance.
The 43 germline MS identified may assist in differentiating individuals with MB from healthy controls; our classification scheme reached a sensitivity and specificity of 95% and 90%, respectively. Despite this, high sensitivity and specificity are not always indicative of high predictive value when applied to rare diseases such as MB, where false positives may outnumber true positives. Thus, more prospective development is necessary to determine the true predictive power of MS in MB formation.
The treatment for medulloblastoma leaves survivors with lifelong burdens, including hearing loss, cognitive deficits, endocrinopathies, and a heightened risk of stroke and secondary malignancies. Screening strategies for rare diseases present significant challenges, which could be mitigated by the ability to quickly identify a population at increased risk. If health care continues to evolve toward a standard of routine germline DNA sequencing at an early age, our predictive signature could be used to demarcate such a population. Early detection strategies, perhaps based upon advanced imaging or emerging liquid biopsy technologies, could allow for less invasive, more localized means of tumor control. However, the single best way to improve the lives of these children would be to prevent their tumors from forming in the first place, a dream that until recently had no conceivable means of achievement. Advances in immunotherapies including cancer vaccines create the potential to immunize an individual against tumor-specific antigens. Such a strategy would require the selection of individuals appropriate for such an intervention. Our work using MS based risk prediction could begin to provide this piece of the tumor prevention puzzle.
Funding
This work was supported by Hyundai Hope on Wheels and the Prevent Cancer Foundation, as well as by a Bradley Foundation grant to the Edward Via College of Osteopathic Medicine.
Supplementary Material
Acknowledgments
We wish to thank Dr Stefan M. Pfister (Hopp Children’s Cancer Center at the NCT Heidelberg, Heidelberg, Germany) and Paul A. Northcott (Department of Oncology, St Jude Children’s Research Hospital, Memphis, Tennessee) for kindly providing us with the germline DNA WES from healthy childhood controls. This study was made possible in part due to The Children’s Brain Tumor Tissue Consortium (CBTTC). The Gabriella Miller Kids First Data Resource Center and its properties (kidsfirstdrc.org) are supported by the NIH Common Fund under Award Number U2CHL138346, which is administered by the National Heart, Lung, And Blood Institute of the National Institutes of Health. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest statement. The authors declare no potential conflicts of interest.
Authorship statement. Experimental design: SR-H, NK, HRG, BRR. Experimental implementation: NK, SR-H. Data analysis: SR-H, NK, BRR, HRG. Manuscript preparation: NK, SR-H, HRG, BRR.
References
- 1. Taylor MD, Northcott PA, Korshunov A, et al. Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol. 2012;123(4):465–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Waszak SM, Northcott PA, Buchhalter I, et al. Spectrum and prevalence of genetic predisposition in medulloblastoma: a retrospective genetic study and prospective validation in a clinical trial cohort. Lancet Oncol. 2018;19(6):785–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Neumann JE, Swartling FJ, Schüller U. Medulloblastoma: experimental models and reality. Acta Neuropathol. 2017;134(5):679–689. [DOI] [PubMed] [Google Scholar]
- 4. Pearson CE, Nichol Edamura K, Cleary JD. Repeat instability: mechanisms of dynamic mutations. Nat Rev Genet. 2005;6(10):729–742. [DOI] [PubMed] [Google Scholar]
- 5. Maruvka YE, Mouw KW, Karlic R, et al. Analysis of somatic microsatellite indels identifies driver events in human tumors. Nat Biotechnol. 2017;35(10):951–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Karunasena E, Mciver LJ, Bavarva JH, Wu X, Zhu H, Garner HR. ‘Cut from the same cloth': shared microsatellite variants among cancers link to ectodermal tissues-neural tube and crest cells. Oncotarget. 2015;6(26):22038–22047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. McIver LJ, Fonville NC, Karunasena E, Garner HR. Microsatellite genotyping reveals a signature in breast cancer exomes. Breast Cancer Res Treat. 2014;145(3):791–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kinney N, Varghese RT, Anandakrishnan R, Garner HRS. ZDHHC3 as a risk and mortality marker for breast cancer in African American women. Cancer Inform. 2017;16:1176935117746644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Velmurugan KR, Varghese RT, Fonville NC, Garner HR. High-depth, high-accuracy microsatellite genotyping enables precision lung cancer risk classification. Oncogene. 2017;36(46):6383–6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nik-Zainal S, Davies H, Staaf J, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016;534(7605):47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Giannakis M, Hodis E, Jasmine Mu X, et al. RNF43 is frequently mutated in colorectal and endometrial cancers. Nat Genet. 2014;46(12):1264–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li YC, Korol AB, Fahima T, Beiles A, Nevo E. Microsatellites: genomic distribution, putative functions and mutational mechanisms: a review. Mol Ecol. 2002;11(12):2453–2465. [DOI] [PubMed] [Google Scholar]
- 13. Ellegren H. Microsatellites: simple sequences with complex evolution. Nat Rev Genet. 2004;5(6):435–445. [DOI] [PubMed] [Google Scholar]
- 14. Sawaya SM, Bagshaw AT, Buschiazzo E, Gemmell NJ. Promoter microsatellites as modulators of human gene expression. In: Hannan AJ, ed. Tandem Repeat Polymorphisms: Genetic Plasticity, Neural Diversity and Disease. New York, NY: Springer New York; 2012:41–54. [DOI] [PubMed] [Google Scholar]
- 15. Vinces MD, Legendre M, Caldara M, Hagihara M, Verstrepen KJ. Unstable tandem repeats in promoters confer transcriptional evolvability. Science. 2009;324(5931):1213–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gymrek M, Willems T, Guilmatre A, et al. Abundant contribution of short tandem repeats to gene expression variation in humans. Nat Genet. 2016;48(1):22–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bacolla A, Wells RD. Non-B DNA conformations as determinants of mutagenesis and human disease. Mol Carcinog. 2009;48(4):273–285. [DOI] [PubMed] [Google Scholar]
- 18. Lian Y, Garner HR. Evidence for the regulation of alternative splicing via complementary DNA sequence repeats. Bioinformatics. 2005;21(8):1358–1364. [DOI] [PubMed] [Google Scholar]
- 19. Li YC, Korol AB, Fahima T, Nevo E. Microsatellites within genes: structure, function, and evolution. Mol Biol Evol. 2004;21(6):991–1007. [DOI] [PubMed] [Google Scholar]
- 20. Kashi Y, King DG. Simple sequence repeats as advantageous mutators in evolution. Trends Genet. 2006;22(5):253–259. [DOI] [PubMed] [Google Scholar]
- 21. Borstnik B, Pumpernik D. Tandem repeats in protein coding regions of primate genes. Genome Res. 2002;12(6):909–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hannan AJ. Tandem repeats mediating genetic plasticity in health and disease. Nat Rev Genet. 2018;19(5):286–298. [DOI] [PubMed] [Google Scholar]
- 23. Nithianantharajah J, Hannan AJ. Dynamic mutations as digital genetic modulators of brain development, function and dysfunction. Bioessays. 2007;29(6):525–535. [DOI] [PubMed] [Google Scholar]
- 24. Hannan AJ. Tandem repeats mediating genetic plasticity in health and disease. Nat Rev Genet. 2018;19(5):286–298. [DOI] [PubMed] [Google Scholar]
- 25. Galindo CL, McIver LJ, Tae H, et al. Sporadic breast cancer patients' germline DNA exhibit an AT-rich microsatellite signature. Genes Chromosomes Cancer. 2011;50(4):275–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Karunasena E, McIver LJ, Rood BR, et al. Somatic intronic microsatellite loci differentiate glioblastoma from lower-grade gliomas. Oncotarget. 2014;5(15):6003–6014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vaksman Z, Garner HR. Somatic microsatellite variability as a predictive marker for colorectal cancer and liver cancer progression. Oncotarget. 2015;6(8):5760–5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tae H, Kim DY, McCormick J, Settlage RE, Garner HR. Discretized Gaussian mixture for genotyping of microsatellite loci containing homopolymer runs. Bioinformatics. 2014;30(5):652–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Van der Auwera GA, Carneiro MO, Hartl C, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43:11.10.1–11.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Highnam G, Franck C, Martin A, Stephens C, Puthige A, Mittelman D. Accurate human microsatellite genotypes from high-throughput resequencing data using informed error profiles. Nucleic Acids Res. 2013;41(1):e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Youden WJ. Index for rating diagnostic tests. Cancer. 1950;3(1):32–35. [DOI] [PubMed] [Google Scholar]
- 33. Anderson-Cook CM. Practical genetic algorithms. J Am Stat Assoc. 2005;100(471):1099–1099. [Google Scholar]
- 34. Lorenz R, Bernhart SH, Höner Zu Siederdissen C, et al. ViennaRNA Package 2.0. Algorithms Mol Biol. 2011;6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Northcott PA, Buchhalter I, Morrissy AS, et al. The whole-genome landscape of medulloblastoma subtypes. Nature. 2017;547(7663):311–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sun JX, Helgason A, Masson G, et al. A direct characterization of human mutation based on microsatellites. Nat Genet. 2012;44(10):1161–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Brinkmann B, Klintschar M, Neuhuber F, Hühne J, Rolf B. Mutation rate in human microsatellites: influence of the structure and length of the tandem repeat. Am J Hum Genet. 1998;62(6):1408–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Viana-Pereira M, Almeida I, Sousa S, et al. Analysis of microsatellite instability in medulloblastoma. Neuro Oncol. 2009;11(5):458–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Guerreiro AS, Fattet S, Fischer B, et al. Targeting the PI3K p110alpha isoform inhibits medulloblastoma proliferation, chemoresistance, and migration. Clin Cancer Res. 2008;14(21):6761–6769. [DOI] [PubMed] [Google Scholar]
- 40. Aldaregia J, Odriozola A, Matheu A, Garcia I. Targeting mTOR as a therapeutic approach in medulloblastoma. Int J Mol Sci. 2018;19(7):1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hu S, Cao B, Zhang M, et al. Epigenetic silencing BCL6B induced colorectal cancer proliferation and metastasis by inhibiting P53 signaling. Am J Cancer Res. 2015;5(2):651–662. [PMC free article] [PubMed] [Google Scholar]
- 42. Wang W, Huang P, Wu P, et al. BCL6B expression in hepatocellular carcinoma and its efficacy in the inhibition of liver damage and fibrogenesis. Oncotarget. 2015;6(24):20252–20265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Xu L, Li X, Chu ES, et al. Epigenetic inactivation of BCL6B, a novel functional tumour suppressor for gastric cancer, is associated with poor survival. Gut. 2012;61(7):977–985. [DOI] [PubMed] [Google Scholar]
- 44. Slager RE, Newton TL, Vlangos CN, Finucane B, Elsea SH. Mutations in RAI1 associated with Smith-Magenis syndrome. Nat Genet. 2003;33(4):466–468. [DOI] [PubMed] [Google Scholar]
- 45. Thompson MC, Fuller C, Hogg TL, et al. Genomics identifies medulloblastoma subgroups that are enriched for specific genetic alterations. J Clin Oncol. 2006;24(12):1924–1931. [DOI] [PubMed] [Google Scholar]
- 46. Gerber HP, Seipel K, Georgiev O, et al. Transcriptional activation modulated by homopolymeric glutamine and proline stretches. Science. 1994;263(5148):808–811. [DOI] [PubMed] [Google Scholar]
- 47. Fragoso YD, Stoney PN, Shearer KD, et al. Expression in the human brain of retinoic acid induced 1, a protein associated with neurobehavioural disorders. Brain Struct Funct. 2015;220(2):1195–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. La Spada AR, Taylor JP. Repeat expansion disease: progress and puzzles in disease pathogenesis. Nat Rev Genet. 2010;11(4):247–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.