Abstract
Transcriptional networks supervising class IIa HDAC expression are poorly defined. Here we demonstrate that MEF2D is the key factor controlling HDAC9 transcription. This control, which is part of a negative feed-back loop during muscle differentiation, is hijacked in cancer. In leiomyosarcomas the MEF2D/HDAC9 vicious circuit sustains proliferation and cell survival, through the repression of the death receptor FAS. Comprehensive genome-wide studies demonstrate that HDAC4 and HDAC9 control different genetic programs and show both specific and common genomic binding sites. Although the number of MEF2-target genes commonly regulated is similar, only HDAC4 represses many additional genes that are not MEF2D targets. As expected, HDAC4−/− and HDAC9−/− cells increase H3K27ac levels around the TSS of the respective repressed genes. However, these genes rarely show binding of the HDACs at their promoters. Frequently HDAC4 and HDAC9 bind intergenic regions. We demonstrate that these regions, recognized by MEF2D/HDAC4/HDAC9 repressive complexes, show the features of active enhancers. In these regions HDAC4 and HDAC9 can differentially influence H3K27 acetylation. Our studies describe new layers of class IIa HDACs regulation, including a dominant positional effect, and can contribute to explain the pleiotropic actions of MEF2 TFs.
INTRODUCTION
Class IIa HDACs are important regulators of different adaptive and differentiative responses. During embryonic development, these deacetylases influence specific differentiation pathways and tissue morphogenesis (1–3). In vertebrates HDAC4, HDAC5, HDAC7 and HDAC9 constitute the class IIa subfamily. Because of the Tyr/His substitution in the catalytic site, they exhibit a negligible lysine-deacetylase activity (2,3). However, the deacetylase domain, through the recruitment of the NCOR1/NCOR2/HDAC3 complex, can influence histones modifications, including acetylation (4–6). The repressive influence of class IIa HDACs can also be exploited independently from HDAC3 recruitment. In fact MITR, a HDAC9 splicing variant, can still repress transcription in the absence of the deacetylase domain (7). The amino-terminus of class IIa HDACs is dedicated to the binding of different transcription factors (TFs), among which MEF2 family members are the foremost characterized (3). Overall, class IIa HDACs genomic activities require their assembly into multiprotein complexes where they operate as platforms coordinating the activity of TFs, as well as of other epigenetic regulators (1–3,8).
These deacetylases are subjected to multiple levels of regulation. The phosphorylation-dependent control of the nuclear/cytoplasmic shuttling has been the most commonly investigated (3,9). Curiously, although the lineage-dependent expression is a main feature of class IIa, signalling pathways and mechanisms controlling their transcription are largely unknown (3). An exception is the muscle tissue. Here HDAC9 transcription is under the direct control of MEF2D. In this manner, the MEF2D-HDAC9 axis sustains a negative-feedback loop in the transcriptional circuit of muscle differentiation to buffer MEF2D activities (10). Importantly, in specific cancer types, this circuit seems to be misused. In pre-B acute lymphoblastic leukaemia MEF2D oncogenic fusions dramatically upregulate HDAC9 expression (11,12). Abrogation of the MEF2D-HDAC9 negative circuit was also observed in highly aggressive malignant rhabdoid tumor, non-small cell lung cancer, oral squamous cell carcinoma and leiomyosarcoma (13). Since the pro-oncogenic roles of class IIa HDAC have been proved by different studies, understanding the reasons and the importance of such abrogation is of primary interest in cancer research (14–18).
In this manuscript, we have investigated the MEF2-HDAC axis in cellular models of leiomyosarcoma (LMS). LMS are rare highly malignant tumors of mesenchymal origin, with cells presenting features of the smooth muscle lineage (19). We have demonstrated that the MEF2D-HDAC9 axis plays a key role in the maintenance of the transformed phenotype and deciphered the genomic, epigenomic, and transcriptomic landscapes under the control of class IIa HDACs.
MATERIALS AND METHODS
Cell cultures and cytofluorimetric analysis
Leiomyosarcomas cells (LMS), SK-UT-1, SK-LMS-1, MES-SA and DMR were grown as previously described (15). HEK-293T and AMPHO cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS. For PI staining, cells were collected and resuspended in 0.1 ml of 10 μg/ml propidium iodide (PI) (Sigma-Aldrich), in PBS and incubated for 10 min at RT. After washes, cells were fixed with 1% formaldehyde (Sigma-Aldrich) and treated with 10 μg/ml RNase A. Fluorescence was determined with a FACScan™ (Beckman Dickinson).
CRISPR/Cas9 technology
The generation of HDAC4 and HDAC9 null SK-UT-1 cells was previously described (6). SK-UT-1 cells mutated in the MEF2-binding sites within the HDAC9 promoter were obtained after co-transfection of the pSpCas9-2A Puro plasmid expressing the two sgRNA (GGTCGGCCTGAGCCAAAAAT, CTGGACAGCTGGGTTTGCTG) and the ssODN repair templates (20) (AAAGATAGAGGCTGGACAGCTGGGTTTGCTCGCGTAGGATCCAATGCATTAATGCAGGCT, AATCACTCGGCCATGCTTGACCTAGGATCCGCTCAGGCCGACCATTGTTCTATTTCTGTG) (ratio 10:1). After selections, clones were screened by PCR and immunoblot. Sanger sequencing was applied for the final validation.
Immunofluorescence, random cell motility and immunoblotting
Cells were fixed with 3% paraformaldehyde and permeabilized with 0.1% Triton X-100. The secondary antibodies were Alexa Fluor 488-, 546- or 633-conjugated anti-mouse and anti-rabbit secondary antibodies (Molecular Probes). Actin was labelled with phalloidin-AF546 (Molecular Probes). Cells were imaged with a Leica confocal scanner microscopy SP2. Nuclei were stained with Hoechst 33258 (Sigma-Aldrich). For S phase analysis, cells were grown for 3 h with 50 μM bromodeoxyuridine (BrdU). After fixation, coverslips were treated with HCl and processed for immunofluorescence. For random cell motility measurements, cells were seeded in six-well plates coated with fibronectin and subjected to time-lapse analysis. Images were recorded every 15 min for 6 h with a Leica AF6000 station. Time-lapse experiments were analyzed with the Metamorph software (Molecular Devices).
Cell lysates after SDS-PAGE and immunoblotting on nitrocellulose (Whatman) were incubated with primary antibodies. HPR-conjugated secondary antibodies were obtained from Sigma-Aldrich and blots were developed with Super Signal West Dura (Pierce), as previously described (20).
Antibodies and chemicals
The primary antibodies used were anti: MEF2D (BD Bioscience); MEF2A (C-21), Caspase-3 (E-8) and FAS (M-20) (Santa Cruz Biotechnology); RAN and Caspase 8 (D35G2) (Cell Signaling Technology); Actin, BrdU, and FLAG/M2 (Sigma-Aldrich); GFP, HDAC4 and Caspase-9 (15); HDAC5 (21); H3K27ac (ab4729) and H3K27me3 (ab6002) (Abcam); H3K4me3 (GTX128954, GeneTex). The anti-HDAC9 antibody was produced in rabbit by injecting a His-tagged fragment of HDAC9 (aa 275–600) expressed in Escherichia coli. The antiserum was affinity purified against the same fragment GST-tagged of HDAC9.
The following chemicals were used: Doxorubicin and Metformin (Alexis); Lapatinib and Imatinib (LC Laboratories); MKK2206 and SAHA (Cayman Chemicals); BrdU (Sigma-Aldrich); FasL (Peprotech).
Plasmid construction, transfection, retroviral and lentiviral infection, silencing
pLENTI-CRISPR/V2 (Plasmid #52961) and pSpCas9(BB)-2A-GFP(PX458) (Plasmid #48138) were from Addgene. pWZL-HYGRO FLIPs FLAG was obtained by a restriction-based approach from pcDNA3-FLIPs. pWZL-HYGRO FLAG plasmid was used as acceptor plasmid and as control for infection. The knock-down of MEF2D and MEF2A was achieved by using pLKO.1 shRNAs (TRCN0000015897, TRCN00000274054, TRCN00000432718, TRCN000005133), as already described (6). For the Luc assay on HDAC9 promoter the 15897 and 432718 shRNAs were selected and used in consideration of their higher efficiency (6). HDAC9 promoter (6) (bp –1160/+23) activity was measured in transfected 293-T or SK-UT-1 cells according to the manufacturer (Dual-Glo Luciferase Assay System, Promega) and expressed as a ratio to the luciferase activity of pRenilla. Transfections, viral infections and siRNA delivery were done as previously described (20,21). The following Invitrogen Stealth RNAi siRNAs (148 pmol) were used:
HDAC4 (CCACCGGAAUCUGAACCACUGCAUU),
HDAC9 1 (GAACAAACUGCUUUCGAAAUCUAUU),
HDAC9 2 (UGGGCCAACUGGAAGUGUUACUGAA).
Caspase and Resazurin reduction assays
The caspase activity was evaluated using the Apo-ONE caspase-3/7 homogeneous assay (Promega). Cells grown in 96-well plates were treated with the different insults and tested for caspase activity as recommended by the vendor. Resazurin assay was done as already described (22). Briefly, cells were incubated for 150 min. at 37°C with resazurin solution (0.15 mg/ml) (Sigma-Aldrich). The product of reduction was quantified by using the PerkinElmer EnSpire 2300 Multilabel Reader.
ChIP, library construction, ChIP-seq and NGS data analysis
ChIP was performed as previously described (18). Chromatin was immunoprecipitated with 2 μg of anti-H3K27ac, 2.5 μg of anti-MEF2D, 4 μg of anti-HDAC4 and anti-HDAC9 antibodies or control IgG. Three independent experiments were pulled and 5 ng of total DNA were used to prepare ChIP-seq libraries, according to TruSeq ChIP Sample Preparation guide (Illumina). Libraries were sequenced on the Illumina HiSeq 2000 sequencer. The quality of sequencing reads was evaluated using the ShortRead R/Biocoductor package (23). Sequencing reads from ChIP-seq experiments were aligned to the NCBI GRCh38 human reference with Bowtie 2 (24). Peak calling was performed against input sequences using MACS2 (25). Gene annotation, Venn diagrams and bar plots representing the peak localization in genomic elements/distance from TSS were obtained using the ChIPseeker R/Bioconductor package (26). Peak heatmaps and genomic loci visualization were generated using the gplots, biomaRt and Gviz R/Bioconductor packages (27–30). H3K27ac signals were normalized using the MAnorm method for quantitative comparison of ChIP-seq data (31).
RNA extraction and quantitative qRT-PCR
Cells were lysed using Tri-Reagent (Molecular Research Center). 1.0 μg of total RNA was retro-transcribed by using 100 units of M-MLV Reverse transcriptase (Life Technologies) in the presence of 1.6 μM oligo(dT) and 4 μM Random hexamers. qRT-PCRs were performed using SYBR green technology (KAPA Biosystems). Data were analyzed by comparative threshold cycle (delta delta Ct ΔΔCt) using HPRT and GAPDH as normalizer. A list of the primers used for qRT-PCR and ChIP-qPCR is provided in Supplementary Table S1.
RNA expression array and data analysis
Aliquots of RNAs, purified using RNeasy columns (Qiagen), were amplified according to the specifications of the Illumina TotalPrep RNA Amplification Kit (Ambion). Hybridization on Illumina whole-genome HumanHT-12 v 4.0 chip (Illumina), scanning and background subtraction were done according to the manufacturer's specification. Fold-change and P-values for each probe set were calculated as previously described (18). P-values data were then corrected for multiple hypotheses testing using the Benjamini–Hochberg methods. Differentially expressed genes (DEGs) were selected based on fold changes and adjusted P-values <0.05. Gene set enrichment analysis (GSEA) and the MSigDB database http://software.broadinstitute.org/gsea/index.jsp (32,33) were used to investigate statistically relevant biological associations.
Gene lists were analyzed separately using the GeneOntology (BiologicalProcess and ImmuneSystemProcess; Min GO Level = 3 and Max GO Level = 8), KEGG, WikiPathways, CORUM-FunCat-MIPS, REACTOME_Pathways and REACTOME_Reactions databases as source of information. A right-sided hypergeometric test (corrected using the Benjamini–Hochberg) was applied to find enriched terms.
Statistics
For experimental data Student t-test was employed. Mann–Whitney test was applied when normality could not be assumed. P < 0.05 was chosen as statistical limit of significance. For comparisons between samples >2, the Anova test was applied coupled to Kruskal–Wallis and Dunn's Multiple Comparison Test. We marked with *P < 0.05, **P < 0.01, ***P < 0.001. Unless otherwise indicated, all the data in the figures were represented as arithmetic means ± the standard deviations from at least three independent experiments.
RESULTS
HDAC9 is a MEF2-target gene highly transcribed in leiomyosarcomas cells
Class IIa HDACs and particularly HDAC5 and HDAC9, are overexpressed in approximately 30% of leiomyosarcomas (LMS) (20,34). The mechanisms responsible for this up-regulation are not defined. The TCGA data analysis (Figure 1A) shows that in LMS patients the co-overexpression of one class IIa HDAC and one MEF2 TF is frequent. In fact, a significant co-occurrence of MEF2D and HDAC9 overexpression (P-value 0.035, log odds ratio 1427) and a trend of co-occurrence for MEF2A and HDAC5 mark these patients. ENCODE data evidence the presence of MEF2 binding sites, which are conserved through evolution, in the proximal promoter of HDAC9 (Figure 1B). We proved that MEF2D up-regulates the transcription from the HDAC9 promoter in different cell lines including LMS cells (Figure 1C). SK-UT-1 cells faithfully reflect LMS in terms of MEF2D and HDAC9 de-regulation. Both proteins are highly up-regulated in these aggressive cells (6). When MEF2D was silenced, HDAC9 expression was down-regulated at the mRNA and protein levels (Figure 1D, F). By contrast, MEF2A silencing does not influence HDAC9 levels (Supplementary Figure S1A, B). Finally, in SK-UT-1 cells transcription from the HDAC9 promoter was strongly dependent on MEF2D (Figure 1E).
Figure 1.
The MEF2D-HDAC9 circuit in leiomyosarcomas. (A) Oncoprint of mRNA expression variations for the indicated MEF2 and class IIa HDACs family members. Data were obtained from the TCGA database and include RNAseq data of 100 patients with LMS. The heatmap shows the expression levels (z-score normalized log2 (FPKM) values) and was generated through cBioPortal (http://www.cbioportal.org). (B) The evolutionary conserved binding site, validated by ENCODE, for MEF2A (blue) and for MEF2C (red) in the proximal promoter of HDAC9 are shown. (C) Luciferase assay for HDAC9 promoter activity in HEK-293 cells transfected with MEF2D-GFP or GFP and the promoter regions (bp –1160/+23) amplified from IMR90, SK-LMS-1 and SK-UT-1 cells. 3xMEF construct, presenting three binding sites for MEF2, was used as positive control. Data were normalized by co-transfecting pRenilla and expressed as mean ± S.D., n = 3. (D) mRNA expression levels of the indicated genes, as measured by qRT-PCR, in MEF2D knock-down cells with respect to control. Two independent shRNA were used. Data are expressed as mean ± S.D., n = 3. (E) Luciferase assay for HDAC9 or control promoter activities in SK-UT-1 cells silenced for MEF2D expression. Data were normalized by co-transfecting pRenilla and expressed as mean ± S.D., n = 3. (F) Immunoblot analysis in SK-UT-1 MEF2D knock-down cells, using the indicated antibodies. Actin was used as loading control. (G) Chromatin was immunoprecipitated from SK-UT-1 cells WT or silenced for MEF2D, using the anti-MEF2D antibodies. Normal rabbit IgGs were used as control. The HDAC9 promoter region (-758:-528) containing the MEF2 binding sites was amplified. Data are presented as mean ± S.D., n = 4. (H) Chromatin was immunoprecipitated using the anti-H3K4me3, anti-H3K27ac and anti-H3K27me3 antibodies from SK-UT-1 cells and SK-UT-1 silenced for MEF2D. Data are presented as mean ± S.D, n = 3.
ChIP experiments demonstrated that MEF2D binds the promoter of HDAC9 (Figure 1G). MEF2D is required to sustain an open chromatin status, characterized by high levels of H3K4me3 and H3K27ac, on the HDAC9 promoter but not on the control TK promoter (Figure 1H and Supplementary Figure S1C). MEF2D down-regulation favors the appearance of H3K27me3 (Figure 1H and Supplementary Figure S1C). In summary, MEF2D is a key TF that boosts HDAC9 transcription in SK-UT-1 cells.
HDAC4 and HDAC9 show different subcellular localizations in LMS cells
To study the role of class IIa HDACs in LMS, we generated SK-UT-1 cells knocked-out for HDAC4 and HDAC9 using the CRISPR/Cas9 technology (6). Two different clones, generated using two different guides, were selected for each KO. Immunoblot analysis shows that LMS cells express HDAC9 and high levels of its splicing variant MITR (Figure 2A). As predicted, in HDAC9 KO cells both isoforms are absent. Curiously, in HDAC4−/− cells a different pattern of HDAC9 can be appreciated. Levels of the full-length HDAC9 are reduced and a shorter isoform is increased. Similarly, also the levels of MITR are augmented. In HDAC9−/− cells, a fast migrating HDAC4 isoform is detectable, which shows a size similar to the caspase-cleaved fragment of this deacetylase (35) (see below). The levels of HDAC5 are augmented in the two KO cells, more strongly in HDAC9−/− cells, possibly as part of a compensatory mechanism. HDAC7 is expressed at extremely low/undetectable levels in SK-UT-1 cells. Similarly, MEF2A and MEF2D levels are augmented in the KO cells, with MEF2A showing a higher increase in HDAC9−/− cells. These increases correlate with elevated levels of the corresponding mRNAs (Supplementary Figure S3A).
Figure 2.

Characterization of SK-UT-1 cells knocked-out for HDAC4 and HDAC9. (A) Immunoblot analysis of HDAC9, MITR, HDAC4, HDAC5, MEF2A and MEF2D in SK-UT-1 cells WT and in two KO clones for HDAC4 (125 and 26) and HDAC9 (167 and D43). Asterisk points to a non-specific band. Actin was used as loading control. (B) Immunofluorescence analysis in SK-UT-1 WT, HDAC4−/−, HDAC9−/− cells stained with the indicated antibody. Where indicated, cells were treated for 2 h with Leptomycin B (Lept. 50 ng/ml) to inhibit the nuclear export. The anti-RAN antibody was used to stain nuclei. Bar: 50 μm.
Regulation of class IIa HDACs nuclear-cytoplasmic shuttling is a key aspect for the control of their repressive activities. Immunofluorescence analysis proved that HDAC9 is prevalently nuclear in these LMS cells (Figure 2B). As a consequence, its localization is not influenced by leptomycin B treatment (Figure 2B). By contrast, HDAC4 shows a pan/diffused localization, which can be converted into nuclear after the inhibition of the CRM1-dependent nuclear export (Figure 2B). Ran localization was used as counterstaining. The KOs cells have proved the antibodies specificity. In summary, while HDAC9 is largely nuclear resident, HDAC4 is constantly subjected to nuclear/cytoplasmic shuttling.
Transcriptomes under HDAC4 and HDAC9 regulation in LMS cells
To investigate the genetic repertoire under the control of HDAC4 and HDAC9 in LMS, the transcriptome of two HDAC4−/− clones (26 and 125) and two HDAC9−/− clones (167 and D43) generated with independent guide pairs were compared.
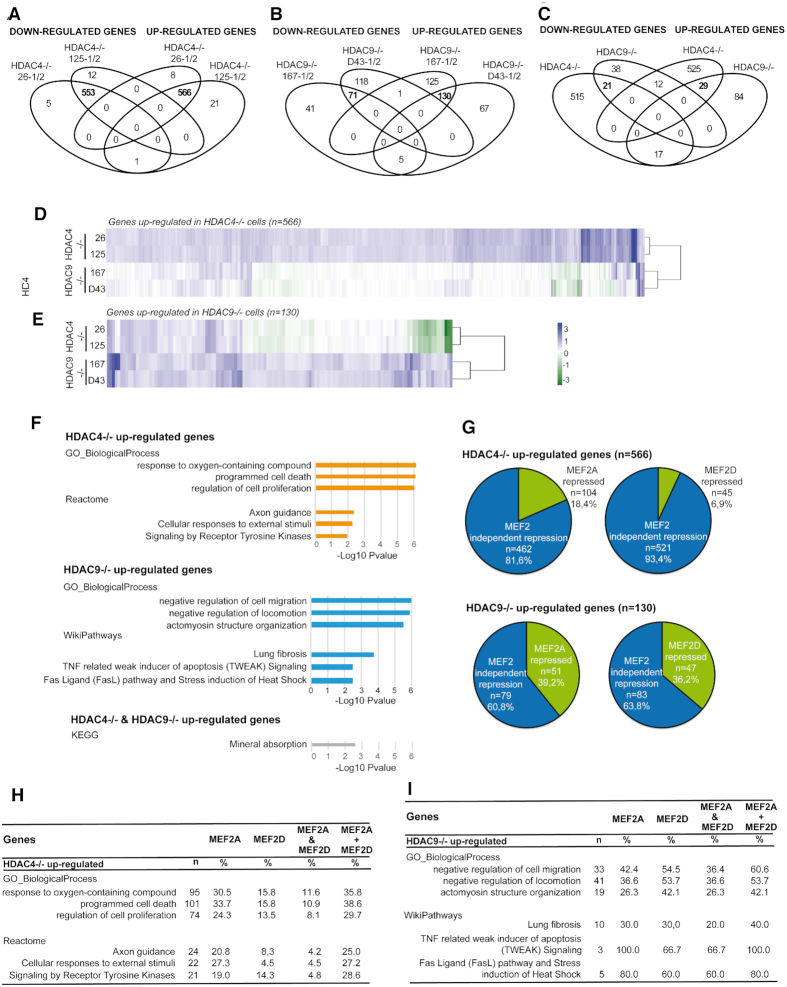
The vast majority of the up- and down-regulated genes (n = 566 and n = 533, respectively), were shared between the two HDAC4−/− clones (Figure 3A). By contrast, HDAC9−/− clones have much less commonly regulated genes (n = 130 up- and n = 71 down-regulated) and a consistent number (n = 192 up- and n = 159 down-regulated) were clone-specific (Figure 3B, Supplementary Tables S2, S3 and Figure S2).
Figure 3.
HDAC4 and HDAC9 regulate different patterns of genes, only partially overlapping. (A) Venn diagrams showing the number of transcripts commonly and differentially up-regulated or downregulated between the two clones of HDAC4−/− cells. (B) Venn diagrams showing the number of transcripts commonly and differentially up-regulated or downregulated between the two clones of HDAC9−/− cells. (C) Venn diagrams showing the number of transcripts commonly and differentially up-regulated or downregulated among the different clones of HDAC4−/− and HDAC9−/− cells. (D) Heatmap of the 566 significantly up-regulated genes in HDAC4−/− cells and their expression levels in HDAC9−/− cells. In the heatmap genes up-regulated are shown in green and down-regulated in blue, as fold changes. (E) Heatmap of the 130 significantly up-regulated genes in HDAC9−/− cells and their behavior in HDAC4−/− cells. In the heat map genes up-regulated are shown in green and down-regulated in blue, as fold changes. (F) Bar plots of the Cytoscape-ClueGO most significantly enriched functional terms according to the GO: Biological Process, Reactome or WikiPathways databases. Analysis were performed for the indicated groups of up-regulated genes, retaining the top 3 terms defined by the two most informative functional databases. (G) Pie charts illustrating the dependency on MEF2 of the genes up-regulated in HDAC4−/− and HDAC9−/− cells. The dependency on MEF2 was scored by looking at the genes affected by MEF2A/D knock-down (6). (H) Effect of MEF2A and MEF2D silencing on the gene lists associated with the most significantly enriched functional terms of the Cytoscape-ClueGO analysis as shown in (A) in SK-UT-1 HDAC4−/− cells. The dependency on MEF2 was expressed as percentage. (I) Effect of MEF2A and MEF2D silencing on the gene lists associated with the most significantly enriched functional terms of the Cytoscape-ClueGO analysis as shown in (A) in SK-UT-1 HDAC9−/− cells. The dependency on MEF2 was expressed as percentage.
In order to define a common gene signature, we compared the lists of up- and down-regulated genes between HDAC4−/− and HDAC9−/− cells. Twenty nine induced and 21 repressed genes represent the common signature of the two class IIa HDACs (Figure 3C). This result suggests that the two HDACs play distinct roles in LMS cells.
Since class IIa HDACs are well-known repressors of transcription, we focused the attention on transcripts whose levels increased after the knock-outs. To further confirm the specific activities of the two HDACs, we compared the levels of genes up-regulated in HDAC4−/− cells with those in HDAC9−/− cells and vice versa. Heatmaps show that although a group of genes was similarly up-regulated by the knock-out of the two HDACs, many genes were specific for each HDAC, as they were unperturbed in the other KO or, in some instances, repressed (Figure 3D and E). qRT/PCR analysis on a panel of these differentially regulated genes further validated these results (Supplementary Figure S3A).
The genetic programs regulated by HDAC4 and HDAC9
We used the Cytoscape plugin ClueGO (36,37) to understand the functions of genes up-regulated in HDAC4−/− and HDAC9−/− cells, as well as of genes induced in both conditions. As expected, the most significantly enriched functional terms differ from HDAC4 and HDAC9 (Figure 3F). HDAC4-repressed genes are involved in the oxidative stress response, proliferation and programmed cell death. By contrast, HDAC9 repressed genes include negative regulators of cell migration/locomotion and regulators of actin cytoskeleton. Interestingly, death receptor signaling emerged as a pathway under HDAC9 influence. A common genetic program regards the mineral absorption.
In order to identify genes under the influence of the MEF2-HDAC axis, we compared the lists of genes up-regulated in HDAC4−/− and HDAC9−/− cells with genes that are repressed by MEF2A and MEF2D in the same cells (6). Figure 3G highlights such overlaps, indicating a similar and strong contribution of these TFs to HDAC9-mediated repression. Although the majority of the HDAC4-repressed genes are not under the influence of MEF2A or MEF2D, the absolute number of MEF2A/D target genes is similar to HDAC9. Furthermore, within the HDAC4-repressed genes, MEF2A influence is more pronounced compared to MEF2D. This observation was confirmed by GSEA (Genes Set Enrichment Analysis). Here significant enrichments were obtained only when the comparisons were performed between HDAC9 and MEF2A or MEF2D regulated genes (Supplementary Figure S3B).
Subsequently, we wondered to what extent MEF2-HDAC co-targets could recapitulate the previously identified biological functions associated with genes repressed by HDAC4 or HDAC9. Figures 3H/I summarizes the number (n) of genes associated with each enriched term, the percentage of genes that are also MEF2A or MEF2D targets, the percentage of those in common between MEF2A and MEF2D (MEF2A & MEF2D) and the percentage of those that are targets of at least one MEF2 (MEF2A + MEF2D).
Finally, we used a right-sided Fisher's exact test to determine the probability that each biological function was significantly enriched in one of the HDAC/MEF2 co-target subsets with respect to the corresponding list of upregulated genes in HDAC4−/− or HDAC9−/− cells. Firstly, this allowed the identification of ‘response to oxygen-containing compound’ and ‘programmed cell death’ as terms enriched in both MEF2A (P-value = 7e–03 and P-value = 7e–04, respectively) and MEF2D targets common to HDAC4 (P-value = 7.5e–03 and P-value = 5.3e–03, respectively). We also defined ‘negative regulation of locomotion’ as specifically enriched (P-value = 4.6e–02) in the HDAC9/MEF2A targets subset, representing the only HDAC9-related biological process showing specificity for one of the two MEF2 regulators.
In summary our data demonstrate that different MEF2-HDAC complexes can regulate distinct gene-networks.
Mapping the genomic regions bound by HDAC4, HDAC9 and MEF2D
ChIP-seq experiments were performed to investigate, at a genomic level, HDAC4 and HDAC9 binding in relation to MEF2D. As expected from the transcriptomic analysis, HDAC4 shows a higher genomic binding (n = 7732) compared to HDAC9 (n = 1257), while MEF2D peaks are 2214, in agreement with previous studies (38). 68% of MEF2D peaks contains at least a MEF2 binding sites (Supplementary Table S4). By comparing the binding locations, 45% of MEF2D peaks co-localize with HDAC4, HDAC9 or both (Figure 4A). In particular, 56% of these events are in common with HDAC4 and HDAC9, 43% are MEF2D/HDAC4 specific and only 1.2% are MEF2D/HDAC9 specific. HDAC4 displays a substantial MEF2D-independent activity as 82% of its peaks are bound neither by MEF2D nor by HDAC9. On the contrary, HDAC9 specific peaks are less frequent (20%). DNA motif analysis on the HDAC4-specific peaks showed an enrichment for SMAD3 and CENPB binding motifs. When the same analysis was performed on HDAC4-specific peaks localized in H3K27ac enriched regions, BACH2 and ZNF384 showed the highest enrichments (Supplementary Figure S4). The ChIP-seq data for MEF2D, HDAC4, HDAC9 and the variations in H3K27ac were confirmed by ChIP-qPCR for 11 distinct genomic regions (Supplementary Figure S5). Overall, the analysis of the genomic binding sites confirms the different activities of the two HDACs, as emerged from the transcriptomic analysis.
Figure 4.
Defining the genomic binding sites and influences of MEF2D, HDAC4 and HDAC9 on H3K27ac. (A) Venn diagrams showing the overlaps between the MACS2-defined MEF2D, HDAC4 and HDAC9 ChIP-seq enriched peaks in SK-UT-1 cells. (B) Genomic distribution of the MACS2-defined MEF2D, HDAC4 and HDAC9 ChIP-seq enriched peaks in SK-UT-1 cells. (C) Proximity, expressed as percentage, of H3K27ac marks to the MEF2D, HDAC4 and HDAC9 ChIP-seq enriched peaks in SK-UT-1 cells. The minimum distance is 0bp (overlapping) and 1kbp, respectively, for the left and right panel. (D) Heatmaps showing the differences in H3K27ac distribution between HDAC4−/− and HDAC4+/+ or HDAC9−/− and HDAC9+/+ SK-UT-1 cells. The displayed regions are located ±3kb around the TSS of a subset of 475 microarray-defined HDAC4 repressed genes (left panel) and of 118 microarray-defined HDAC9 repressed genes (right panel). Binding peaks for MEF2D, HDAC4 and HDAC9 in the same regions are also provided. The differences in mRNAs levels are indicated by the heatmaps at the bottom.
The majority of HDAC4 and HDAC9 peaks localize in intergenic regions, (68% and 76%, respectively), as previously observed for other class IIa HDACs (18,39,40). By contrast only 36% of MEF2D peaks map in intergenic regions while 27% are in promoter regions (Figure 4B). Interestingly, MEF2D peaks localization undergoes significant changes when associated with common binding of HDAC4/HDAC9. In these cases 64% occur in intergenic, 14% in introns and only 18% in promoter regions. This re-localization seems to only partially depend on the presence of HDAC4 alone, since MEF2D/HDAC4 common mapping is 47% intergenic, 29% intronic and 17% in promoter regions. To further evaluate the chromatin status around these peaks, we mapped the H3K27 acetylation either as a perfect overlap (min. 1 bp in common) or in a 2 kb interval from the peak summit (Figure 4C). Overall, the presence of HDAC4 frequently correlates with an open chromatin status, while HDAC9 peaks are commonly confined in regions marked by poor H3K27 acetylation. These correlations are MEF2-independent. In general, the co-presence of MEF2D increases H3K27 acetylation incidence (Figure 4C).
It is plausible that the genomic binding sites of the two class IIa HDACs and possibly of MEF2D, within the regulative elements of the genes up-regulated after the KO of HDAC4 and HDAC9, may influence their epigenetic status and subsequently the transcriptional output. Hence, we evaluated variations of the H3K27ac status around the TSS (-/+3kb) of these genes. In the same regions, we also investigated the presence of MEF2D, HDAC4 and HDAC9 peaks. We excluded from the analysis transcripts with undefined functional annotations, thus resulting in 475 genes for HDAC4−/− and 118 genes for or HDAC9−/− cells.
The levels of H3K27ac were augmented around the TSS of several genes up-regulated in HDAC4−/− and HDAC9−/− cells. This increase was evident also in regions more distal with respect to the TSS (Figure 4D). Frequently, acetylation spreading emerged as a consequence of HDAC4 and HDAC9 deletions.
Fourteen genes are characterized by HDAC4 binding within 3 kb from the TSS and six of them also show co-binding with MEF2D. Among these six genes, ARMC4 and MPP7 evidence multiple binding events for MEF2D and HDAC4 (Figure 4D). These two genes are marked by intense spreading of H3K27ac in the absence of HDAC4. Surprisingly, peaks for HDAC9 were not found around the TSS of genes up-regulated in knocked-out cells, even though for 4 genes (CXCL1, ENC1, PLK2 and SORT1) MEF2D binding was observed.
Next, we expanded the analysis up to ±30 kb from the TSS to find evidence of distal regulative elements. The increase of H3K27ac and the spreading effects elicited by the absence of the two HDACs was confirmed (Figure 5A). ARMC4, MKX, MPP7, NFIB, ROR1, ZNRF3 are among the most evident examples of this behavior in the absence of HDAC4. CXCL1, CXCL8/IL8 and SMAD3 in the case of HDAC9 absence. Importantly, CXCL1 and IL8 are among the highest up-regulated genes in HDAC9−/− cells (Supplementary Table S2).
Figure 5.
Impact of HDAC4 and HDAC9 on H3K27 acetylation at genomic regions distal from the TSS. (A) Heatmaps showing the differences in H3K27ac distribution between HDAC4−/− and HDAC4+/+ or HDAC9−/− and HDAC9+/+ SK-UT-1 cells. The displayed regions are located ±30kb around the TSS of a subset of 475 microarray-defined HDAC4 repressed genes (top panel) and of 118 microarray-defined HDAC9 repressed genes (mid panel). Binding peaks for MEF2D, HDAC4 and HDAC9 in the same regions are also provided. The differences in mRNAs levels are indicated by the heatmaps at the right side. (B) Acetylation status of MEF2D/HDAC4/HDAC9 co-localizing peaks. Distances from the TSS of the closest coding genes are shown. The increase of the relative H3K27ac signal after the knock-out of HDAC4 (orange) or HDAC9 (light blue) is indicated. H3K27ac peaks that are exclusively enriched only after the knock-out of either HDAC4 or of HDAC9 are shown at the edges of the charts as HDAC4 or HDAC9 specifics.
Twenty five genes show HDAC4 binding within 30kb from the TSS and frequently with multiple binding events. Peaks for HDAC9 were again rare, with only RAB31 with a positive hit. MEF2D genomic binding was found in 28 genes up-regulated in HDAC4−/− cells, of which seven shared with HDAC4 and in eight genes up-regulated in HDAC9−/− cells (Figure 5A).
HDAC4 and HDAC9 regulate H3K27ac levels in regulative regions distal from the TSS
The previous analysis has revealed that, although changes in H3K27ac are prominent in regions around the TSS of genes up-regulated after the KO of the two HDACs, only a fraction of these genes displays the binding of HDAC4 (5,3%) and rarely of HDAC9 (0.8%). Certainly, some of these genes could be indirect target of the deacetylases. However, since several HDAC4 and HDAC9 peaks were found in the intergenic regions, we hypothesized that HDAC4 and HDAC9 in particular, could preferentially influence gene expression from distal regulative elements such as enhancers.
To explore this possibility, we investigated the variations of H3K27ac status after the knock-out of the two HDACs, at the distal regions marked by the co-presence of MEF2D, HDAC4 and HDAC9 peaks (Figure 4A; n = 510). Through this strategy we should identify the functional/active distal regulative regions of MEF2D target genes.
Approximately 42% of these common peaks lie in H3K27 acetylated regions. 57% of these acetylated regions are found away from a TSS (>30 kb from the TSS).
Next, we investigated whether some of these distal regions showed HDAC4 or HDAC9 dependent regulation of H3K27ac levels. Three different groups of peaks can be identified (Figure 5B). A first group comprises common peaks that do not show strong variations in relative H3K27ac fold increases, after the knock-out of both deacetylases (gray dots). A second group of common peaks shows increases in relative H3K27ac, which are more marked in the absence of one of the two deacetylases, more frequently in HDAC4−/− cells. The third group comprises H3K27ac peaks that appeared exclusively enriched after the knock-out of either HDAC4 or of HDAC9.
Within these distal regions differentially modulated by the two class IIa HDACs, we could expect to find regulative elements that orchestrate the expression of genes differentially regulated by HDAC9 and responsible for the different impact on the proliferation of SK-UT-1 cells.
Chromosome conformation capture (3C) assay, combined with Next Generation Sequencing (Hi-C) can provide a global view of all chromosomal interactions across the genome and maps Topologically Associated Domains (TADs) (41,42). Within these domains distal regulative regions can be identified. A distal intergenic region, where H3K27ac was modulated by HDAC9, is located almost 100kb downstream from ARHGEF28 locus. To understand whether it could act as a distal enhancer, we compared our ChIP-seq data to available Hi-C data obtained from the IMR90 cell line (43).
Figure 6 shows that a region, marked by multiple binding sites for MEF2D, HDAC4 and HDAC9 dramatically increases H3K27ac acetylation levels only after the knock-out of HDAC9. Hi-C data indicate that this region lies within a defined TAD and that, through chromatin looping, it could act distally to influence a region close to the TSS of ENC1. As a matter of fact, the ablation of HDAC9 augments H3K27ac levels in the proximity of ENC1.
Figure 6.
Example of a distal regulative region under HDAC9-specific regulation. Detailed view of the MEF2D, HDAC4, HDAC9 and H3K27ac tracks at the ARHGEF28 locus. H3K27ac normalized tracks are shown for WT, HDAC4−/− and HDAC9−/− SK-UT-1 cells. Gene structure and chromosomal location are shown, with the red boxes highlighting the presence of enhancer RNAs. The boxes (light blue) evidence the chromatin looping between the enhancer, under specific regulation of HDAC9 and the promoter of ENC1 whose expression is specifically up-regulated in HDAC9−/−cells. Hi-C data (43) were used to represent the TADs within the ARHGEF28/ENC1 locus (http://promoter.bx.psu.edu/hi-c/view.php).
Overall this analysis suggests that an intergenic region, where H3K27ac acetylation is modulated by HDAC9, could act as distal regulative region (enhancer) for the ENC1 gene. The detection of eRNAs in the same region regulated by HDAC9, as defined by SlideBase – FANTOM5 Human Enhancers Selector, further supports this possibility (44). Importantly, the ENC1 transcript is specifically up-regulated in HDAC9−/− cells (Supplementary Figures S3B and S5).
Another example of HDAC9-controlled, distal regulative region is represented by the intragenic locus of INSYN2B, which perfectly superimposes to a sub-TAD characterized by the presence of eRNAs (Supplementary Figure S6A). Finally, an example of a distal regulative region under the specific influence of HDAC4 is represented in Supplementary Figure S6B. This regulative intergenic region is located ∼35 kb upstream from the SLC8A1 locus. Within the TAD, binding of MEF2D, HDAC4 and HDAC9 can be found. In this case, however, it is the KO of HDAC4 that causes a diffuse spreading of H3K27ac throughout the sub-TAD (Supplementary Figure S6B).
HDAC9 influences actin cytoskeleton organization, cell spreading and motility
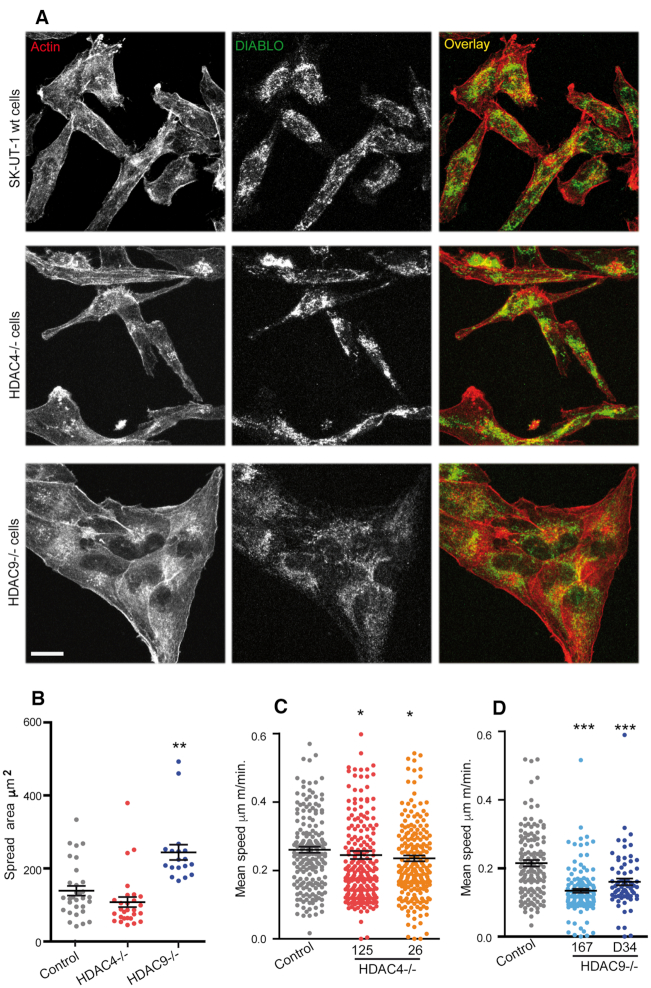
The gene expression studies discovered a role of HDAC9 as a coordinator of actin-cytoskeleton organization, cell adhesion and migration. We proved the relevance in vivo of this result by comparing the actin cytoskeleton among the different engineered LMS cell lines. SK-UT-1 wt and HDAC4−/− cells show a similar phenotype. Cells are bipolar and scattered. Accumulation of F-actin is well evident in specific localizations at the cell periphery, indicative of membrane ruffles, filopodia and lamellipodia (Figure 7A). In HDAC9−/− cells, these structures are much less evident. Cells establish stable contacts resembling epithelial colonies. We also compared the mitochondrial network using an antibody against DIABLO. Although mitochondria were highly fragmented in SK-UT-1 cells, in wt and HDAC4−/− cells they cluster in the perinuclear regions whereas in HDAC9−/− cells they are distributed throughout the cytoplasm (Figure 7A). Quantitative morphometric analysis was performed to compare the spread area in the different cells. As suggested by the immunofluorescence studies, the spreading was markedly increased in HDAC9−/− cells (Figure 7B). We also performed time-lapse studies to compare the random cell motility of the different cell lines. HDAC9−/− but not HDAC4−/− cells are characterized by a dramatically reduced random cell motility (Figure 7C, D). Overall these studies demonstrate that HDAC9, by repressing the expression of cytoskeletal components, controls cell adhesion, morphology and motility.
Figure 7.
HDAC9 controls spreading and motility in LMS cells. (A) Confocal images of the indicated LMS cell lines stained for actin (red), using phalloidin and DIABLO (green), by immunofluorescence to visualize mitochondria. Bar 50 μm. (B) Dot plot representing the spread area of the indicated SK-UT-1 cells. The median and the first and third quartiles are indicated; n > 25. (C) Dot plot representing the mean speed of the indicated SK-UT-1 cells. Time-lapse experiments were performed over a period of 6 h. The median and the first and third quartiles are indicated n > 134. (D) Dot plot representing the mean speed of the indicated SK-UT-1 cells. Time-lapse experiments were performed over a period of 6 hours. The median and the first and third quartiles are indicated; n > 89.
HDAC9 sustains cell survival by repressing FAS expression
The transcriptomic studies indicate that HDAC9 could repress apoptosis and particularly the activation of the extrinsic apoptotic pathway (Figure 3I). FAS emerged as a gene specifically up-regulated in HDAC9−/−cells. Analysis of Hi-C data shows that the FAS locus is embedded within a subTAD adjacent to the subTAD containing the ACTA2 locus, which is transcribed in the opposite orientation. Interestingly, a specific ACTA2 isoform share with FAS the promoter region (Figure 8A). The ChIP-seq did not identify peaks for MEF2D and HDAC9 within the FAS locus. Despite this, H3K27 acetylation is clearly augmented around the TSS and throughout the first intron of FAS, in the absence of HDAC9. Similarly a downstream intergenic region (approx. 40kb from the FAS TSS) is highly acetylated in HDAC9−/− cells. Interestingly, Hi-C data indicate that this intergenic region can make contact with the FAS promoter. H3K27 acetylation in HDAC9−/− cells was augmented also at the ACTA2 locus (Figure 8A). We validated the contribution of HDAC9 in the control of FAS and ACTA2 mRNA levels by qRT-PCR. As expected, FAS and ACTA2 expression was up-regulated in HDAC9−/− cells (Figure 8B). Up-regulation of the FAS protein was also verified by immunoblot (Figure 8C).
Figure 8.
HDAC9 promotes cell survival by repressing FAS transcription. (A) Genomic view of the ACTA2/FAS locus on chromosome 1 (GRCh38). Detailed view of H3K27ac normalized tracks (green) for WT, HDAC4−/− and HDAC9−/− SK-UT-1 cells. The boxes (light blue) evidence the chromatin looping between a distal regulative element and the FAS promoter. Hi-C data were used to define the TADs within the FAS locus. (B) mRNA expression levels of ACTA2, FAS and GAPDH, as measured by qRT-PCR. Data are presented as mean ± S.D relative to wild-type SK-UT-1 cells, n = 3. (C) Immunoblot analysis of FAS levels in the indicated SK-UT-1 clones. Actin was used as loading control. (D) Histogram representing the percentage of PI positivity in the indicated SK-UT-1 cells, treated with 25 and 50ng/ml of FASL Data are presented as mean ± S.D. n = 3. (E) Scatter dot plot representing the z-scores of FAS mRNA levels in individual TCGA tumors (n = 100) divided in two classes accordingly to HDAC9 levels. (F) Histogram representing the percentage of PI positivity in the indicated SK-UT-1 cells, expressing or not FLIPs and treated for 24 hours with 25ng/ml FASL. Data are expressed as mean ± S.D. n = 3. (G) Caspases activation (DEVDase activity) in the indicated SK-UT-1 cells, expressing or not FLIPs and treated for 24 hours with 25ng/ml of FASL. Data are expressed as mean ± S.D. n = 3. (H) Histogram representing the percentage of PI positivity in the indicated SK-UT-1 cells, expressing or not FLIPs. Data are expressed as mean ± S.D. n = 3. (I) Caspases activation (DEVDase activity) in the indicated SK-UT-1 cells, expressing or not FLIPs and treated for 24 hours with the indicated drugs. Data are expressed as mean ± S.D. n = 3.
Overall these data suggest that HDAC9 can sustain SK-UT-1 transformation by repressing the extrinsic apoptotic pathway. To prove this hypothesis, we analyzed the proliferative features of the different engineered SK-UT-1 cells. HDAC9 absence marginally reduced the percentage of cells in S phase. HDAC4−/− cells did not show overt defects (Supplementary Figure S7A). Time-course experiments showed that HDAC4−/− cells have a partial proliferative deficit, whereas proliferation of HDAC9−/− cells was dramatically impaired (Supplementary Figure S7B) and marked by an increased level of cell death. This low constitutive activation of the apoptotic machinery was confirmed by the Caspase-9 and Caspase-3 processing observed in the absence of apoptotic insults (Supplementary Figure S7C). When the different LMS cells were grown without serum, cell death was dramatically induced in the absence of HDAC9 (Supplementary Figure S7D). Finally, we compared the proliferation of the different SK-UT-1 cells when incubated with different pro-apoptotic drugs including: tyrosine-kinase, HDACs, isopeptidases (22) and Akt inhibitors or metmorfin. With the exclusion of the two TK inhibitors, only in HDAC9−/− cells all drugs showed a significant stronger anti-proliferative outcome (Supplementary Figure S7E). As expected, apoptosis elicited by FAS ligand (FASL) was clearly augmented in HDAC9−/− SK-UT-1 cells (Figure 8D). In vivo, analysis of TCGA data on leiomyosarcomas showed low levels of FAS mRNA and a significant anti-correlation between the FAS and HDAC9 mRNA levels (Figure 8E).
The extrinsic apoptotic pathway is activated in the absence of HDAC9
To prove the involvement of the extrinsic pathway we expressed the inhibitor of DISC activation FLIPs, the short isoform of CFLAR/FLIP (45). FAS-induced caspase activation was strongly attenuated in the presence of FLIPs (Figure 8F). Importantly, also the increase of caspase activity observed in the KO cells in the absence of added apoptotic insults, was blunted by FLIPs (Figure 8F). Next, we evaluated the percentage of cell death in untreated cells. The increase of cell death observed in the HDAC9−/− cells was completely abrogated by the presence of FLIPs (Figure 8G).
To exclude that the HDAC9-dependent regulation of FAS levels was a peculiar aspect of SK-UT-1 cells, we analyzed a panel of LMS cells for HDAC9 expression. HDAC9 levels were abundant also in DMR cells but not in SK-LMS-1 and MES-SA uterine sarcoma cells (Supplementary Figure S8A). When HDAC9 was downregulated by two different siRNAs in DMR cells, FAS, ACTA2 and IL8 levels were all augmented (Supplementary Figure S8B). The up-regulation of these genes was not observed after HDAC4 silencing. In agreement with SK-UT-1 cells, apoptosis was increased after HDAC9 silencing also in DMR cells (Supplementary Figure S8C). We also investigated whether an anti-correlation between class IIa HDACs and FAS levels could be observed in leiomyosarcomas in vivo. Supplementary Figure S8D shows that a significant anti-correlation with FAS expression in LMS involves also HDAC5, in addition to HDAC9.
We also investigated whether HDAC9 absence increased the apoptotic susceptibility of SK-UT-1 cells to different apoptotic insults such as: DNA damage (doxorubicin), Akt inhibition (MKK2206) and the proteotoxic stressor G5 (Figure 8H). Only apoptosis triggered by doxorubicin and G5 was potentiated by HDAC9 deletion. This increase was largely suppressed by the presence of FLIPs, thus suggesting an involvement of the extrinsic pathway. Analysis of caspase activation confirmed that the up-regulation of the extrinsic pathway characterizes the increased apoptotic susceptibility of HDAC9−/− cells in response to doxorubicin (Figure 8I).
Deletion of the MEF2-binding sites in the HDAC9 promoter recapitulates the knock-out of HDAC9
Our manuscript opened with the demonstration that MEF2D is the critical TFs involved in the up-regulation of HDAC9. Subsequently, we proved that HDAC9 plays a critical role in FAS expression and in the regulation of SK-UT-1 survival. To conclude our study we needed to demonstrate that the abrogation of the vicious loop between MEF2D and HDAC9 suppresses cell proliferation, up-regulates FAS expression and triggers apoptosis.
To this purpose we used the CRISPR/Cas9 technology to mutagenize the MEF2 binding sites in the HDAC9 promoter (Figure 9A). ChIP experiment demonstrated the absence of MEF2D binding in the HDAC9 promoter of engineered SK-UT-1 cells (ΔMEF2) (Figure 9B). As a consequence, epigenetic markers of open and active chromatin are reduced (H3K27ac and H3K4me3) while marker of closed/repressed chromatin (H3K27me3) appeared once MEF2 binding site were removed from the HDAC9 promoter. HDAC9 mRNA levels were dramatically decreased (Figure 9C). Immunoblot analysis confirmed the strong downregulation of HDAC9 expression, the concomitant up-regulation of FAS and the activation of Caspase-8 (Figure 9D). Similarly to FAS, other HDAC9 target genes (IL8 and ACTA2) were up-regulated at the mRNA level when the MEF2 binding sites in the HDAC9 promoter were mutagenized (Figure 9E). Finally, cell spreading, growth arrest and cell death were all up-regulated in SK-UT-1 cells with mutated HDAC9 promoter.
Figure 9.
MEF2D regulative elements in the HDAC9 promoter are required for HDAC9 overexpression, FAS silencing and cell survival (A) Scheme of the CRISPR-mediated mutagenesis of the two MEF2-binding sites in the HDAC9 promoter (B) Chromatin was immunoprecipitated from SK-UT-1 cells WT or mutagenized in the 2 MEF2D binding sites, using anti-MEF2D antibody. Normal rabbit IgGs were used as control. The HDAC9 promoter region containing the MEF2 binding sites was amplified. Data are presented as mean ± S.D., n = 3. (C) Chromatin was immunoprecipitated using the anti-H3K4me3, anti-H3K27ac and anti-H3K27me3 antibodies from SK-UT-1 cells WT or mutagenized in the 2 MEF2D binding sites Data are presented as mean and standard error. n = 3. (D) Relative mRNA expression levels of GAPDH and HDAC9, as measured by qRT-PCR in WT and SK-UT-1 cells with mutated MEF2 binding sites in the HDAC9 promoter (ΔMEF2). Data are presented as mean ± S.D. n = 3. (E) Immunoblot analysis of HDAC9, FAS, Caspase-8 and Actin in SK-UT-1 cells WT and with mutated MEF2 binding sites in the HDAC9 promoter (ΔMEF2). Actin was used as loading control. Asterisks point to non-specific bands, arrowhead to the caspase-8 cleaved form. (F) Relative mRNA expression levels of GAPDH, IL8, FAS and ACTA2, as measured by qRT-PCR in WT and SK-UT-1 cells with mutated MEF2 binding sites in the HDAC9 promoter (ΔMEF2). Data are presented as mean ± S.D. n = 3. (G) Dot plot representing the spread area of the indicated SK-UT-1 cells. The median and the first and third quartiles are indicated. n>70. (H) Histogram representing the doubling time of the indicated SK-UT-1 cells over a period of three days in culture. Data are presented as mean ± S.D. n = 3. (I) Histogram representing the percentage of PI positivity in the indicated SK-UT-1 cells, growing in the absence of any added apoptotic insult. Data are presented as mean ± S.D. n = 3.
In summary, abrogation of MEF2 binding at the HDAC9 promoter mirrors the effect of HDAC9 deletion on SK-UT-1 cells survival.
DISCUSSION
Dysregulations of class IIa HDACs expression have been reported in different tumors (1,3,6,13,46). How these alterations influence the epigenetic plasticity of cancer cells is still unknown. In this manuscript we have investigated the altered expression and dissected the functions of class IIa HDACs in LMS. These tumors are considered genetically complex soft tissue sarcomas, with a high mutational burden and a complex karyotype with several losses, gains and amplifications (19,47). Alterations in the PI3K/AKT/PTEN pathway, deletions of the tumor suppressors TP53 and RB1 and mutations in ATRX and MED12 (34,48,49) are frequent in LMS. Up-regulation of miR-143 and miR-145 and low expression of inflammatory response genes are also common in LMS (34). Approximately 30% of LMS express high levels of a class IIa HDACs, and of HDAC9 in particular (6).
We have demonstrated that the increased expression of HDAC9 stems from an alteration of the MEF2-HDAC feed-back loop. Silencing of MEF2D causes a parallel downregulation of HDAC9 levels and it is coupled to the appearance of a repressive epigenetic state at its promoter. Furthermore, the CRISPR-mediated deletion of the MEF2D binding sites in the HDAC9 promoter switches off HDAC9 transcription and perfectly recapitulates the proliferative defects of SK-UT-1 cells knocked out for HDAC9.
Our in vitro studies find substantiation in vivo. In LMS patients the high levels of HDAC9 are significantly correlated with high levels of MEF2D expression. A similar alteration in the MEF2-class IIa HDACs loop could be responsible for the overexpression of HDAC5. In this case MEF2A seems to be involved. Dysfunctions of the MEF2D-HDAC9 circuit might be common to other tumor types (50). For example in pre-B acute lymphoblastic leukemia, characterized by MEF2D translocations, HDAC9 is frequently up-regulated (11–13).
Three important questions still deserve specific answers: i) which is the mechanism that up-regulates MEF2D expression in LMS? ii) why does not the MEF2D-HDAC9 complex repress HDAC9 transcription, by binding the HDAC9 promoter? iii) why is the relationship between MEF2D and HDAC9 so exclusive? Does HDAC9 play a different epigenetic role with respect to the other class IIa HDACs?
We do not have an answer to the first question. However, by defining the genomic binding sites for MEF2D, HDAC4 and HDAC9 we provide some cues to the others points.
Class IIa HDACs have been described as tissue specific deacetylases (1–3,46). Certain phenotypes, observed after members specific knock-outs in mice or ablation in human cells, have been explained as the consequence of their lineage-dependent expression (6,18,21,51–54). Our study demonstrates that further levels of complexity do exist. The first level concerns the genomic bindings. HDAC4 and HDAC9 share several genomic binding sites but HDAC4, in particular, binds many additional regions, which possibly escape a HDAC9-dependent regulation.
The second level of complexity concerns MEF2D. This important class IIa HDACs partner can assemble onto specific genomic regions both the HDAC4 and the HDAC9 repressive complexes, while in other regions only a specific repressive complex is recruited, most frequently containing HDAC4 rather than HDAC9. Through the selective recruitment of a specific HDAC or a combination of more HDACs, MEF2 can monitor different patterns of gene expression, as verified by the transcriptomic analysis and proved by the different genetic programs under the control of the two HDACs examined. Within this level of complexity also the MITR isoform can be included. In fact, although MITR has been described as a transcriptional repressor, it contains16 aa of unknown function at the carboxy-terminus that differ from the HDAC9 canonical isoform (55). Perhaps this complex, when assembled on the HDAC9 promoter, could impair the manifestation of a full repressive state.
A third level of complexity is obtained by a locus-dependent influence of a specific class IIa HDAC. In fact, even though a region is characterized by the co-presence of MEF2D/HDAC4/HDAC9 complexes, frequently only a family member plays an active role in the epigenetic regulation. We define this phenomenon as the dominant positional effect of a specific class IIa HDAC. An example is the intergenic region distal from the AHRGEF28 locus. This region shows features of an enhancer and through chromatin looping could regulate the expression of ENC1. Although both HDAC4 and HDAC9 bind this region, only the knock-out of HDAC9 increases H3K27ac levels at the enhancer and promoter sites thus augmenting ENC1 expression.
In general, the paucity of promoter regions bound by HDAC9 suggests that this HDAC is recruited by MEF2D or by alternative TFs to repress gene expression principally from distal regulative elements showing features of enhancers.
Another important difference marks HDAC9 with respect to HDAC4. HDAC9 very rarely binds the genome in open chromatin regions, as defined by H3K27ac. This data indicates that HDAC9 typifies stably repressed chromatin domains while HDAC4 influences more dynamic genomic regions, or could suggest that HDAC4 acts as the priming pioneering repressive factor for HDAC9.
Finally, a further layer of regulation is represented by MEF2D complexes that are not recognized by both HDAC4 and HDAC9. Although a contribution of HDAC5 should be taken into account, the demonstration that MEF2A and MEF2D can still sustain transcription on specific loci discourages the HDAC5 hypothesis (6). On these loci, MEF2D acts positively by promoting the transcription of the associated genes. This evidence demonstrates that MEF2D has the capability to selectively escape the surveillance of class IIa HDACs on some well-defined genomic regions.
Despite the less numerous genomic binding sites and reduced number of genes under its influence, HDAC9 shows a strong impact on LMS cells proliferation. Cells deprived of HDAC9 show a higher apoptotic index that limits their prolonged maintenance in tissue culture. In this respect, we speculate that a certain degree of transcriptional heterogeneity, observed among the different HDAC9−/− clones, could reflect adaptive mechanisms that cells engage to survive the strong apoptotic pressure.
FAS up-regulation and the activation of the extrinsic pathway are responsible for the increased apoptotic pressure. Cytokines such as TNF-α, IL12 and IFNγ can sustain FAS transcription through the engagement of NF-kB, SP1, STAT1 and IRF8 (56–60). Also TP53 family members are involved in FAS transcription (61,62).
We could not find HDAC9 and MEF2D binding sites at the promoter region of FAS. However H3K27ac was increased after the deletion of HDAC9, both at the promoter and at a distal site within the subTAD. This distal region can undergo looping with the FAS promoter.
How HDAC9 could influence H3K27 acetylation at the FAS/ACTA2 locus is mysterious. It could operate through a distal enhancers or indirectly by controlling the expression of specific TFs. In fact, approx. 10% of the genes repressed by HDAC9 encode for TFs or epigenetic regulators and among them SMAD3 has been demonstrated to up-regulate FAS transcription (63). Although FAS is a key element of the increased apoptotic response in HDAC9 deficient SK-UT-1 cells, MEF2 are pleiotropic TFs and in other contexts their pro-survival activities can be mediated by the regulation of different pro-survival or pro-death genes (64–66).
The impact of HDAC9 on LMS cells proliferation/survival is not limited to SK-UT-1 cells. DMR cells similarly overexpress HDAC9 and HDAC9 downregulation up-regulates FAS levels and triggers apoptosis in DMR cells too. We are confident that the repressive influence of HDAC9 on FAS transcription is a critical event for LMS survival/aggressiveness, as proved by the anti-correlation observed in tumors.
CONCLUSIONS
LMS are aggressive cancers, local recurrence and metastasis develop in approximately 40% of cases, which drastically reduce survival (19,34,47–49). Under these conditions available therapies are largely ineffective and the identification of new therapeutic targets is mandatory. Our studies point to class IIa HDACs and HDAC9 in particular as interesting targets to revitalize the extrinsic apoptotic pathway. Furthermore, the simultaneous up-regulation of chemokines, cytokines and secreted factors, exemplified by CXCL1, IL8, THBS1 and CYR61, after HDAC9 depletion, can sustain FAS-induced apoptosis and the action of the immune system for a better elimination of the neoplastic cells (67–69). This represent a strong incitement to evaluate HDAC9 inhibitors as possible onco-immunological drugs. Finally, we are confident that our deep investigation and description of the genomic preferences of MEF2D-HDAC4-HDAC9 complexes can fit to other models and help researchers in finding a genetic and epigenetic explanation to the pleiotropic actions of MEF2 TFs.
DATA AVAILABILITY
The transcriptomic raw data are available as GEO accession GSE132569: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc = GSE132569.
Raw data corresponding to ChIP-seq experiments are uploaded with GEO accession GSE132622: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc = GSE132622.
The link to a UCSC genome browser session displaying the uploaded sequence tracks is http://genome.ucsc.edu/s/DameBioinfo/DiGiorgio_et_al.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Michele Gentile for the help in some experiments and Dr Ed Seto for providing HDAC9 cDNA. We also thank ‘Studio Associato Molaro Pezzetta Romanelli Del Fabbro e associati’ for the donation of computing devices
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
AIRC Associazione Italiana per la Ricerca sul Cancro [IG 15640]; POR-FESR regione Friuli-Venezia Giulia ATeNA and Sarcoma Foundation of America (to C.B.); EDG received an AIRC fellowship (Volontari Jesolo). Funding for open access charge: Sarcoma Foundation of America (SFA) Ricerca Libera Claudio Brancolini.
Conflict of interest statement. None declared.
REFERENCES
- 1. Parra M. Class IIa HDACs - new insights into their functions in physiology and pathology. FEBS J. 2015; 282:736–744. [DOI] [PubMed] [Google Scholar]
- 2. Fitzsimons H.L. The Class IIa histone deacetylase HDAC4 and neuronal function: Nuclear nuisance and cytoplasmic stalwart. Neurobiol. Learn. Mem. 2015; 123:149–145. [DOI] [PubMed] [Google Scholar]
- 3. Di Giorgio E., Brancolini C.. Regulation of class IIa HDAC activities: it is not only matter of subcellular localization. Epigenomics. 2016; 8:251–269. [DOI] [PubMed] [Google Scholar]
- 4. Lahm A., Paolini C., Pallaoro M., Nardi M.C., Jones P., Neddermann P., Sambucini S., Bottomley M.J., Lo Surdo P., Carfi A. et al.. Unraveling the hidden catalytic activity of vertebrate class IIa histone deacetylases. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:17335–17340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Park S.-Y., Kim G.S., Hwang H.-J., Nam T.-H., Park H.-S., Song J., Jang T.-H., Lee Y.C., Kim J.-S.. Structural basis of the specific interaction of SMRT corepressor with histone deacetylase 4. Nucleic Acids Res. 2018; 926:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Di Giorgio E., Franforte E., Cefalù S., Rossi S., Dei Tos A.P., Brenca M., Polano M., Maestro R., Paluvai H., Picco R. et al.. The co-existence of transcriptional activator and transcriptional repressor MEF2 complexes influences tumor aggressiveness. PLoS Genet. 2017; 13:1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sparrow D.B., Miska E.A., Langley E., Reynaud-Deonauth S., Kotecha S., Towers N., Spohr G., Kouzarides T., Mohun T.J.. MEF-2 function is modified by a novel co-repressor, MITR. EMBO J. 1999; 18:5085–5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Harris L.G., Wang S.H., Mani S.K., Kasiganesan H., Chou C.J., Menick D.R.. Evidence for a non-canonical role of HDAC5 in regulation of the cardiac Ncx1 and Bnp genes. Nucleic Acids Res. 2016; 44:3610–3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mathias R.A., Guise A.J., Cristea I.M.. Post-translational modifications regulate class IIa histone deacetylase (HDAC) function in health and disease. Mol. Cell Proteomics. 2015; 14:456–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Haberland M., Arnold M.A., McAnally J., Phan D., Kim Y., Olson E.N.. Regulation of HDAC9 gene expression by MEF2 establishes a negative-feedback loop in the transcriptional circuitry of muscle differentiation. Mol. Cell Biol. 2007; 27:518–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gu Z., Churchman M., Roberts K., Li Y., Liu Y., Harvey R.C., McCastlain K., Reshmi S.C., Payne-Turner D., Iacobucci I. et al.. Genomic analyses identify recurrent MEF2D fusions in acute lymphoblastic leukaemia. Nat. Commun. 2016; 7:13331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Suzuki K., Okuno Y., Kawashima N., Muramatsu H., Okuno T., Wang X., Kataoka S., Sekiya Y., Hamada M., Murakami N. et al.. MEF2D-BCL9 fusion gene is associated with high-risk acute B-cell precursor lymphoblastic leukemia in adolescents. J. Clin. Oncol. 2016; 34:3451–3459. [DOI] [PubMed] [Google Scholar]
- 13. Di Giorgio E., Hancock W.W., Brancolini C.. BBA - reviews on cancer MEF2 and the tumorigenic process, hic sunt leones. BBA - Rev. Cancer. 2018; 1870:261–273. [DOI] [PubMed] [Google Scholar]
- 14. Rad R., Rad L., Wang W., Cadinanos J., Vassiliou G., Rice S., Campos L.S., Yusa K., Banerjee R., Li M.A. et al.. PiggyBac transposon mutagenesis: A tool for cancer gene discovery in mice. Science. 2010; 330:1104–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Di Giorgio E., Clocchiatti A., Piccinin S., Sgorbissa A., Viviani G., Peruzzo P., Romeo S., Rossi S., Dei Tos A.P., Maestro R. et al.. MEF2 is a converging hub for HDAC4 and PI3K/Akt-induced transformation. Mol. Cell Biol. 2013; 22:4473–4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lei Y., Liu L., Zhang S., Guo S., Li X., Wang J., Su B., Fang Y., Chen X., Ke H. et al.. Hdac7 promotes lung tumorigenesis by inhibiting Stat3 activation. Mol. Cancer. 2017; 16:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Paluvai H., Giorgio E.Di., Brancolini C.. Unscheduled HDAC4 repressive activity in human fibroblasts triggers TP53-dependent senescence and favors cell transformation. Mol. Oncol. 2018; 10:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cutano V., Di Giorgio E., Minisini M., Picco R., Dalla E., Brancolini C.. HDAC7-mediated control of tumor microenvironment maintains proliferative and stemness competence of human mammary epithelial cells. Mol. Oncol. 2019; 13:1651–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Duffaud F., Ray-Coquard I., Salas S., Pautier P.. Recent advances in understanding and managing leiomyosarcomas. F1000Prime Rep. 2015; 7:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F.. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013; 8:2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Clocchiatti A., Di Giorgio E., Ingrao S., Meyer-Almes F.J., Tripodo C., Brancolini C.. Class IIa HDACs repressive activities on MEF2-depedent transcription are associated with poor prognosis of ER+ breast tumors. FASEB J. 2013; 27:942–954. [DOI] [PubMed] [Google Scholar]
- 22. Cersosimo U., Sgorbissa A., Foti C., Drioli S., Angelica R., Tomasella A., Picco R., Semrau M.S., Storici P., Benedetti F. et al.. Synthesis, characterization, and optimization for in vivo delivery of a nonselective isopeptidase inhibitor as new antineoplastic agent. J. Med. Chem. 2015; 58:1691–1704. [DOI] [PubMed] [Google Scholar]
- 23. Morgan M., Anders S., Lawrence M., Aboyoun P., Pagès H., Gentleman R.. ShortRead: a bioconductor package for input, quality assessment and exploration of high-throughput sequence data. Bioinformatics. 2009; 25:2607–2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W. et al.. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yu G., Wang L., He Q.. Genome analysis ChIPseeker: an R / Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 2015; 31:2382–2383. [DOI] [PubMed] [Google Scholar]
- 27. Durinck S., Moreau Y., Kasprzyk A., Davis S., De Moor B., Brazma A., Huber W.. BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics. 2005; 21:3439–3440. [DOI] [PubMed] [Google Scholar]
- 28. Durinck S., Spellman P.T., Birney E., Huber W.. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 2009; 4:1184–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hahne F., Ivanek R.. Visualizing genomic data using Gviz and bioconductor. Methods Mol. Biol. 2016; 1418:335–351. [DOI] [PubMed] [Google Scholar]
- 30. Warnes G.R., Bolker B., Bonebakker L., Gentleman R., Huber W., Liaw A., Lumley T., Maechler M., Magnusson A., Moeller S. et al.. 2019; gplots: Various R Programming Tools for Plotting Data. R package version 3.0.1.1.
- 31. Shao Z., Zhang Y., Yuan G.C., Orkin S.H., Waxman D.J.. MAnorm: a robust model for quantitative comparison of ChIP-Seq data sets. Genome Biol. 2012; 13:R16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S. et al.. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liberzon A., Birger C., Thorvaldsdóttir H., Ghandi M., Mesirov J.P., Tamayo P.. The molecular signatures database hallmark gene set collection. Cell Syst. 2015; 6:417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cancer Genome Atlas Research Network Comprehensive and integrated genomic characterization of adult soft tissue sarcomas. Cell. 2017; 171:950–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Paroni G., Mizzau M., Henderson C., Del Sal G., Schneider C., Brancolini C.. Caspase-dependent regulation of histone deacetylase 4 nuclear-cytoplasmic shuttling promotes apoptosis. Mol. Biol. Cell. 2004; 15:2804–2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T.. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003; 13:2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bindea G., Mlecnik B., Hackl H., Charoentong P., Tosolini M., Kirilovsky A., Fridman W.H., Pagès F., Trajanoski Z., Galon J.. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009; 25:1091–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wales S., Hashemi S., Blais A., McDermott J.C.. Global MEF2 target gene analysis in cardiac and skeletal muscle reveals novel regulation of DUSP6 by p38MAPK-MEF2 signaling. Nucleic Acids Res. 2014; 42:11349–11362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Taniguchi M., Carreira M.B., Cooper Y.A., Bobadilla A.C., Heinsbroek J.A., Koike N., Larson E.B., Balmuth E.A., Hughes B.W., Penrod R.D. et al.. HDAC5 and its target gene, Npas4, function in the nucleus accumbens to regulate cocaine-conditioned behaviors. Neuron. 2017; 96:130–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Azagra A., Román-González L., Collazo O., Rodríguez-Ubreva J., de Yébenes V.G., Barneda-Zahonero B., Rodríguez J., Castro de Moura M., Grego-Bessa J., Fernández-Duran I. et al.. In vivo conditional deletion of HDAC7 reveals its requirement to establish proper B lymphocyte identity and development. J. Exp. Med. 2016; 213:2591–2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rao S.S.P., Huntley M.H., Durand N.C., Stamenova E.K., Bochkov I.D., Robinson J.T., Sanborn A.L., Machol I., Omer A.D., Lander E.S. et al.. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014; 159:1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Roy S.S., Mukherjee A.K., Chowdhury S.. Insights about genome function from spatial organization of the genome. Hum. Genomics. 2018; 12:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang Y., Song F., Zhang B., Zhang L., Xu J., Kuang D., Li D., Choudhary M.N.K., Li Y., Hu M. et al.. The 3D Genome Browser: a web-based browser for visualizing 3D genome organization and long-range chromatin interactions. Genome Biol. 2018; 19:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ienasescu H., Li K., Andersson R., Vitezic M., Rennie S., Chen Y., Vitting-Seerup K., Lagoni E., Boyd M., Bornholdt J., de Hoon M.J. et al.. On-the-fly selection of cell-specific enhancers, genes, miRNAs and proteins across the human body using SlideBase. Database. 2016; 26:2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Thome M., Schneider P., Hofmann K., Fickenscher H., Meinl E., Neipel F., Mattmann C., Burns K., Bodmer J.L., Schröter M. et al.. Viral FLICE-inhibitory proteins (FLIPs) prevent apoptosis induced by death receptors. Nature. 1997; 386:517–521. [DOI] [PubMed] [Google Scholar]
- 46. Clocchiatti A., Florean C., Brancolini C.. Class IIa HDACs: From important roles in differentiation to possible implications in tumourigenesis. J. Cell Mol. Med. 2011; 15:1833–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Miettinen M. Smooth muscle tumors of soft tissue and non-uterine viscera: biology and prognosis. Mod Pathol. 2014; 27(Suppl. 1):S17–S29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hoang N.T., Acevedo L.A., Mann M.J., Tolani B.. A review of soft-tissue sarcomas: translation of biological advances into treatment measures. Cancer Manag. Res. 2018; 10:1089–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mäkinen N., Aavikko M., Heikkinen T., Taipale M., Taipale J., Koivisto-Korander R., Bützow R., Vahteristo P.. Exome sequencing of uterine leiomyosarcomas identifies frequent mutations in TP53, ATRX, and MED12. PLoS Genet. 2016; 12:e1005850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gil V.S., Bhagat G., Howell L., Zhang J., Kim C.H., Stengel S., Vega F., Zelent A., Petrie K.. Deregulated expression of HDAC9 in B cells promotes development of lymphoproliferative disease and lymphoma in mice. Dis. Model Mech. 2016; 9:1483–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chang S., Young B.D., Li S., Qi X., Richardson J.A., Olson E.N.. Histone deacetylase 7 maintains vascular integrity by repressing matrix metalloproteinase 10. Cell. 2006; 126:321–334. [DOI] [PubMed] [Google Scholar]
- 52. Pigna E., Simonazzi E., Sanna K., Bernadzki K.M., Proszynski T., Heil C., Palacios D., Adamo S., Moresi V.. Histone deacetylase 4 protects from denervation and skeletal muscle atrophy in a murine model of amyotrophic lateral sclerosis. EBioMedicine. 2019; 40:717–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Vega R.B., Matsuda K., Oh J., Barbosa A.C., Yang X., Meadows E., McAnally J., Pomajzl C., Shelton J.M., Richardson J.A. et al.. Histone deacetylase 4 controls chondrocyte hypertrophy during skeletogenesis. Cell. 2004; 119:555–566. [DOI] [PubMed] [Google Scholar]
- 54. Chang S., McKinsey T.A., Zhang C.L., Richardson J.A., Hill J.A., Olson E.N.. Histone deacetylases 5 and 9 govern responsiveness of the heart to a subset of stress signals and play redundant roles in heart development. Mol. Cell Biol. 2004; 24:8467–8476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Petrie K., Guidez F., Howell L., Healy L., Waxman S., Greaves M., Zelent A.. The histone deacetylase 9 gene encodes multiple protein isoforms. J. Biol. Chem. 2003; 278:16059–16072. [DOI] [PubMed] [Google Scholar]
- 56. Chan H., Bartos D.P., Owen-Schaub L.B.. Activation-dependent transcriptional regulation of the human Fas promoter requires NF-kappaB p50-p65 recruitment. Mol. Cell Biol. 1999; 19:2098–2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kühnel F., Zender L., Paul Y., Tietze M.K., Trautwein C., Manns M., Kubicka S.. NFkappaB mediates apoptosis through transcriptional activation of Fas (CD95) in adenoviral hepatitis. J. Biol. Chem. 2000; 275:6421–6427. [DOI] [PubMed] [Google Scholar]
- 58. Li-Weber M., Laur O., Dern K., Krammer P.H.. T cell activation-induced and HIV tat enhanced CD95(APO-1/Fas) ligand transcription involves NF-kappaB. Eur. J. Immunol. 2000; 30:661–670. [DOI] [PubMed] [Google Scholar]
- 59. Zhou Z., Lafleur E.A., Koshkina N.V., Worth L.L., Lester M.S., Kleinerman E.S.. Interleukin-12 up-regulates Fas expression in human osteosarcoma and Ewing's sarcoma cells by enhancing its promoter activity. Mol. Cancer Res. 2005; 3:685–691. [DOI] [PubMed] [Google Scholar]
- 60. Yang D., Thangaraju M., Browning D.D., Dong Z., Korchin B., Lev D.C., Ganapathy V., Liu K.. IFN regulatory factor 8 mediates apoptosis in nonhemopoietic tumor cells via regulation of Fas expression. J. Immunol. 2007; 179:4775–4782. [DOI] [PubMed] [Google Scholar]
- 61. Gressner O., Schilling T., Lorenz K., Schulze Schleithoff E., Koch A., Schulze-Bergkamen H., Lena A.M., Candi E., Terrinoni A., Catani M.V. et al.. TAp63alpha induces apoptosis by activating signaling via death receptors and mitochondria. EMBO J. 2005; 24:2458–2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Schilling T., Schleithoff E.S., Kairat A., Melino G., Stremmel W., Oren M., Krammer P.H., Müller M.. Active transcription of the human FAS/CD95/TNFRSF6 gene involves the p53 family. Biochem. Biophys. Res. Commun. 2009; 387:399–404. [DOI] [PubMed] [Google Scholar]
- 63. Kim S.G., Jong H.S., Kim T.Y., Lee J.W., Kim N.K., Hong S.H., Bang Y.J.. Transforming growth factor-beta 1 induces apoptosis through Fas ligand-independent activation of the Fas death pathway in human gastric SNU-620 carcinoma cells. Mol. Biol. Cell. 2004; 15:420–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Estrella N.L., Clark A.L., Desjardins C.A., Nocco S.E., Naya F.J.. MEF2D deficiency in neonatal cardiomyocytes triggers cell cycle re-entry and programmed cell death in vitro. J. Biol. Chem. 2015; 290:24367–24380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Tobin S.W., Hashemi S., Dadson K., Turdi S., Ebrahimian K., Zhao J., Sweeney G., Grigull J., McDermott J.C.. Heart Failure and MEF2 Transcriptome Dynamics in Response to β-Blockers. Sci. Rep. 2017; 7:4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Brown F.C., Still E., Koche R.P., Yim C.Y., Takao S., Cifani P., Reed C., Gunasekera S., Ficarro S.B., Romanienko P. et al.. MEF2C Phosphorylation Is Required for Chemotherapy Resistance in Acute Myeloid Leukemia. Cancer Discov. 2018; 8:478–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Volpert O.V., Zaichuk T., Zhou W., Reiher F., Ferguson T.A., Stuart P.M., Amin M., Bouck N.P.. Inducer-stimulated Fas targets activated endothelium for destruction by anti-angiogenic thrombospondin-1 and pigment epithelium-derived factor. Nat. Med. 2002; 8:349–357. [DOI] [PubMed] [Google Scholar]
- 68. Juric V., Chen C.C., Lau L.F.. Fas-mediated apoptosis is regulated by the extracellular matrix protein CCN1 (CYR61) in vitro and in vivo. Mol. Cell Biol. 2009; 29:3266–3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Cullen S.P., Henry C.M., Kearney C.J., Logue S.E., Feoktistova M., Tynan G.A., Lavelle E.C., Leverkus M., Martin S.J.. Fas/CD95-induced chemokines can serve as “find-me” signals for apoptotic cells. Mol. Cell. 2013; 49:1034–1048. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The transcriptomic raw data are available as GEO accession GSE132569: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc = GSE132569.
Raw data corresponding to ChIP-seq experiments are uploaded with GEO accession GSE132622: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc = GSE132622.
The link to a UCSC genome browser session displaying the uploaded sequence tracks is http://genome.ucsc.edu/s/DameBioinfo/DiGiorgio_et_al.