Abstract
Interrupted dimeric coiled coil segments are found in a broad range of proteins and generally confer selective functional properties such as binding to specific ligands. However, there is only one documented case of a basic-helix–loop–helix leucine zipper transcription factor—microphthalmia-associated transcription factor (MITF)—in which an insertion of a three-residue stammer serves as a determinant of conditional partner selectivity. To unravel the molecular principles of this selectivity, we have analyzed the high-resolution structures of stammer-containing MITF and an engineered stammer-less MITF variant, which comprises an uninterrupted symmetric coiled coil. Despite this fundamental difference, both MITF structures reveal identical flanking in-phase coiled coil arrangements, gained by helical over-winding and local asymmetry in wild-type MITF across the stammer region. These conserved structural properties allow the maintenance of a proper functional readout in terms of nuclear localization and binding to specific DNA-response motifs regardless of the presence of the stammer. By contrast, MITF heterodimer formation with other bHLH-Zip transcription factors is only permissive when both factors contain either the same type of inserted stammer or no insert. Our data illustrate a unique principle of conditional partner selectivity within the wide arsenal of transcription factors with specific partner-dependent functional readouts.
INTRODUCTION
Protein sequences that induce extended α-helical coiled coil arrangements represent one of the most frequently occurring structural motifs in the protein fold universe, covering ∼10% of the proteomes from various organisms (1). Perhaps the most prominent functional role of helical coiled coils is to act as molecular spacers and rulers with atom-level precision (2,3). They critically contribute to defining and controlling precise dimensions in biological processes such as vesicle tethering, chromosome segregation, the architecture of the centriole and DNA recognition and cleavage.
At the molecular level, helical coiled coils assemble in a range of two up to six helices that can be either parallel or antiparallel (2). Coiled coils can originate from identical or different sequences, leading to either protein/protein homo- or hetero-oligomerization, respectively. There are three widespread classes of coiled coil assembled transcription factors: the simplest category is represented by basic leucine zipper (bZip) transcription factors, in which one long chopstick-type of dimeric helix arrangement serves as both dimerization and DNA-binding module (4). In the two other categories, leucine zippers (Zip) are combined either with basic helix-loop-helix (bHLH) domains, which form a second distinct dimerization site, or—in plants only—with homeodomains (5–7). Transcription factors with bHLH domains are also found in combination with PAS domains or with the less-characterized Orange domains (5,8,9). In previous work on the bZip transcription factors MafB and c-Fos, we have shown how preferences for homo- and hetero-dimerization can be determined by single-residue mutations within a given set of coiled coil heptad interactions (10). This and various other studies have also demonstrated how coiled coil arrangements can be modularly combined with other unrelated structural motifs that define additional permissible protein/protein interactions and protein/DNA-binding interactions, further increasing combinatorial complexity for various distinct functional readouts in transcriptional profiles of multi-protein component complexes (4,11–15).
Within the family of bHLH-Zip transcription factors, which contains about a dozen distinct members identified in higher vertebrates (7), there is a small sub-group known as the microphthalmia-associated transcription factor (MITF)/TFE family with four closely related members—MITF, TFEB, TFE3 and TFEC. By contrast, in lower organisms such as Drosophila melanogaster and Caenorhabditis elegans only one single MITF/TFE orthologue is found, suggesting a common evolutionary origin (16,17). Members of this family are global regulators of cell survival and energy metabolism by promoting expression of autophagy and lysosomal genes, with targets that are involved in oxidative metabolism and oxidative stress response (18–20). MITF/TFE-type transcription factors share the ability with other bHLH-Zip transcription factors to bind DNA-recognition elements such as the E-, M- and CLEAR-boxes, with the respective consensus sequences GCACGTGC, TCATGTGC and TCACGTGA. The palindromic E- and CLEAR-boxes differ at the base pair flanking the CACGTG core motif whereas the related M-box presents an asymmetric sequence pattern. Nevertheless, according to available data, their dimerization ability is restricted to members of the MITF/TFE family (19–21). Previous work from our group revealed that the coiled coil leucine zipper in MITF is structurally interrupted by a three-residue insert (22). Systematic studies of known coiled coil protein structures led to the classification of such inserts as ‘stammers’ generating a −51° helical phase change that is generally compensated by neighboring residues to allow continuation of a regular coiled coil (2,23). Stammer-containing coiled coil proteins are one of the rarest categories of proteins with perturbed coiled coils (23), suggesting that conservation of a stammer at a well-defined coiled coil position within all members of the MITF/TFE family is due to functional reasons. To the best of our knowledge, the stammer insert of the coiled coil segments in members of the MITF/TFE family represents the only established example where this insert determines permissive assembly with other bHLH-Zip transcription factors.
To unravel the underlying principles of conditional partnering, we determined the high-resolution structure of a stammer-less MITF variant, in addition to the already known structure of the MITF(wt) dimer (22). The coiled coil segment of the stammer-less MITF structure is continuous and without any asymmetry. Comparison of the two structures reveals how the stammer in MITF(wt) is accommodated by over-winding and local asymmetry to allow an identical arrangement of the flanking regions while maintaining key functional properties such as the ability for nuclear localization and binding of specific DNA-response motifs. We show several lines of evidence, supported by our structural findings, that only in-phase heterodimerization with other bHLH-Zip transcription factors, both of which either contain a stammer or no insert, is permissive. Our data add a new mechanism to the combinatorial arsenal of transcription factor partnering.
MATERIALS AND METHODS
Protein expression, purification and biophysical characterization
Two fragments from the Mus musculus MITF cDNA (residues 217–296 and 180–296) were mutated to remove residues 259–261 (Figure 1A, colored in red) with the Quickchange protocol (Agilent) and purified as described (22). Purified protein was kept in storage buffer containing 150mM NaCl, 10mM Tris-HCl (pH 7.5). The proteins were concentrated up to 10 mg.ml−1, using a VIVASPIN concentrator membrane with a 10 kDa molecular weight cutoff (Sartorius Stedim Biotech). A Superdex 200 10/300 GL (GE Healthcare) was first equilibrated in Storage Buffer and calibrated with Gel Filtration Standard (BioRad). The chromatography was run with a flow of 0.5 ml.mn−1. Absorbance was recorded at 280 nm. For each measurement, 100 μl of purified protein at about 2 mg.ml−1 was injected.
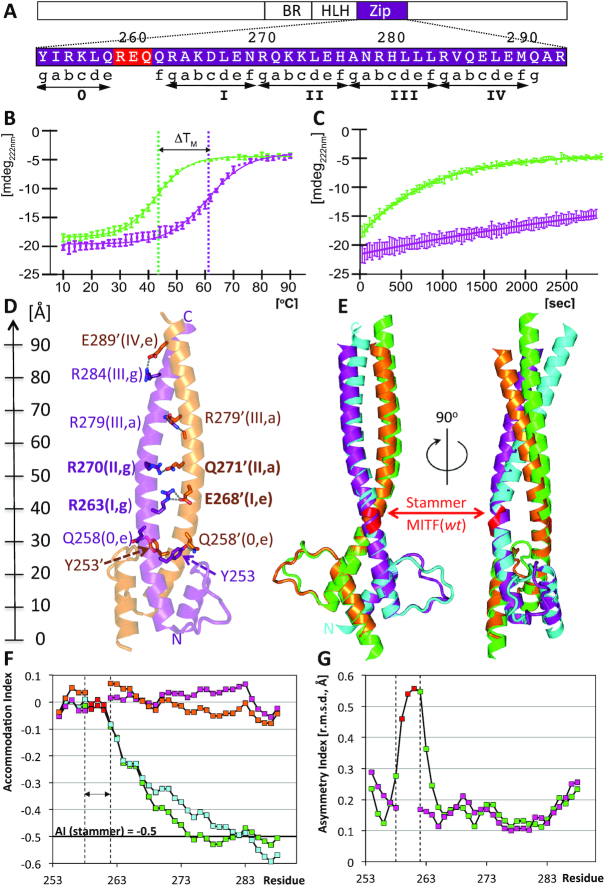
Figure 1.
Structural and biophysical characterization of stammer-less MITF (Δ). (A) Zoom into the MITF sequence of the structurally characterized coiled coil region, indicating the relationship between residue positions, heptad numbers and positions. The stammer residues of MITF(wt) removed in MITF(Δ) are in red. Heptad repeats are indicated by ‘gabcdef’ positions and are numbered. The location of this region within the overall MITF domain structure is schematically shown above. (B) Thermal stability of MITF(wt, green) and MITF(Δ, purple) measured by circular dichroism. Thermal unfolding midpoints are indicated with vertical lines. The melting temperature of MITF(Δ) is increased by 18°C when compared to MITF(wt). (C) Differences in MITF(wt, green) and MITF(Δ, purple) susceptibility to proteolysis by proteinase K. (D) Ribbon representation showing specific coiled coil interactions. Residue numbers, heptad numbers and positions (cf. Figure 3) are indicated for all specific interactions observed. A ruler to the left indicates the dimensions of the MITF coiled coil. (E) Superposition of MITF(wt) (PDB code 4ATH) with stammer-less MITF(Δ) in two different orientations, rotated by 90 degrees around a vertical axis. Color codes: polypeptide chains A and B of MITF(Δ): purple, orange; polypeptide chains A and B of MITF(wt): green, cyan. Residues 259–261, which have been removed in the stammer-less MITF variant, are in red. The color codes are used also in subsequent figures. (F) Change of Accommodation Index (23) of the two polypeptide chains of each MITF(wt) and MITF(Δ) along the coiled coil segment. The three-residue segment removed in MITF(Δ) is indicated by vertical lines. The theoretical change in Accommodation Index of a stammer-containing coiled coil is −0,5, indicated by a horizontal line. (G) Change of Asymmetry Index measured by the change of root mean squares deviations using a residue window of 7 is plotted for MITF(wt) and MITF(Δ), indicating a peak asymmetry across the stammer segment.
Prior to each measurement, samples of the protein fragments 217–296 were dialyzed against 10mM potassium phosphate (pH 7.5) and were diluted to 0.1mg.mL−1. Spectra were recorded at 10°C on a Chirascan CD Spectrometer (Applied Photophysics), between 185 and 260 nm in a 1 mm cuvette. Machine settings were as follows: 0.5 nm bandwidth, 0.5 s response and 0.5 nm data pitch. Spectra were background subtracted and the CD signal was converted to mean residue ellipticity. Each curve represents the mean of three separate measurements. Secondary structure content was estimated by CDNN (24).
Temperature scanning CD measurements were taken with simultaneous monitoring of the temperature and molar ellipticity at 222 nm at a heating rate of 1 K/min. Data were analyzed using Prism (GraphPad software, Inc.). We assumed a two-state folding process and therefore the point of transition was determined by a sigmoidal four-parameter fit: F(x) = Ymin + (Ymax − Ymin)/[1 + 10((Tm− X)*Hill slope)).
To measure susceptibility to proteolysis, 400 μl of 15 μM protein in Assay Buffer (200 mM NaCl, Tris–HCl pH 7.5 10 mM) were pre-incubated at a target temperature of 37°C. The protease solution was applied at a final concentration of 6 nM proteinase K from Engyodontium album (Sigma-Aldrich, Germany). The CD signal at 222 nm was recorded as described above in intervals of 30 s for 48 min. Full spectra from 280 to 200 nm were recorded before and after the kinetics.
X-ray structure determination and refinement
Hanging drop crystallization trials were carried out at 20°C, by mixing equal volumes of reservoir and MITF(Δ) protein fragment 217–296 solutions. Crystals grew in conditions of 2.0–2.4 M ammonium sulfate and 0.10–0.15 M cadmium sulfate. Crystals were soaked in cryo-solutions containing the crystallization mother liquor supplemented with 25% [v/v] glycerol, mounted onto a cryoloop (Hampton Research), and flash-cooled in liquid nitrogen. X-ray data were collected on the synchrotron radiation beamline ID23-2 at ESRF, Grenoble, France. Diffraction data were processed using iMOSFLM (25,26) and scaled with SCALA from the CCP4 suite (27). The structures were solved by molecular replacement with the program PHASER (28) using the HLH domain (residue range 219–258) of the MITF(wt) structure (4ath) as a search model. The final model was built with COOT (29) and refined with the PHENIX suite (30). The atomic coordinates and structure factors of MITF(Δ) have been deposited in the PDB with accession code 6FX5.
The dimerization interfaces were analyzed using the program PISA (31). The geometry parameters of the coiled coil structures were calculated with the program TWISTER (32) and AI (23). The program RAPIDO (33) was used to superimpose the structures and their different segments.
Homology model building and searches for related structures
For obtaining structural models of MITF(wt)/TFEB and MITF(Δ)/MAX, the structures of the MITF(wt) and MITF(Δ) dimers were used as templates, respectively. The sequence of one polypeptide chain in each of the two complexes was exchanged to the respective TFEB (UNIPROT: Q9R210) and MAX (UNIPROT: P28574) sequences. The resulting structural models of the MITF(wt)/TFEB and MITF(Δ)/MAX heterodimers were energy minimized performing 10 cycles of the GROMOS96 module in SwissPDBviewer 4.0 (34). The quality of the models was assessed with MOLPROBITY (35).
Structures from the PDB related to MITF were found using TopSearch, https://topsearch.services.came.sbg.ac.at (36). TopSearch is a web server that allows fast comparison of single- and multi-subunit protein structures with all structures in the PDB. It first compares the query structure with representative PDB structures (pre-selected through clustering of the PDB by structural similarity), and then, with all structures from the most similar clusters. For each pairwise structure comparison, an algorithm based on the singular value decomposition is used to minimize the root-mean-square error of two coordinate sets (ref: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3320710/), allowing permutations of the protein chains and fragments. To remove redundant structures, the TopSearch PDB database was filtered at 90% sequence identity and at 90% structure similarity, as defined by TopSearch. The hits containing a bHLH domain N-terminal to the coiled coil Zip segment were selected based on aligned residue ranges for further analysis.
Cell biology and imaging
HEK293T (obtained from ATCC, Manassas, USA) and 501Mel (a kind gift from Ruth Halaban, Yale University, USA) cells were cultured in DMEM with 10% FBS at 37°C with 5% CO2. Inducible 501Mel cells were generated by transfecting the cells with three piggybac vectors, containing a reverse-tetracyclin transcription activator, a transposase and GFP-tagged human MITF (M isoform), TFEB or TFE3. The piggybac vectors were a kind gift from Dr Kazuhiro Murakami (Hokkaido University). The selection was performed with 0.5 mg.mL−1 G418 (GIBCO) for 8 days obtaining stable cell lines. Q5 site-directed mutagenesis was used to generate MITF(Δ) in an expression vector (pcDNA3.1) containing the wild-type mouse gene. All constructs were verified by Sanger sequencing.
HEK293T cells (1.5 × 104) were seeded in 8-well glass chamber slides and transfected after 24 h with 0.23 μg DNA using FuGENE (Promega) transfection reagent. Cells were fixed 24 h after transfection using 4% formaldehyde, washed and blocked in PBS (containing 5% goat serum, 0.3% Triton X-100) for 1 h. Cells were washed with PBS and stained overnight using mouse monoclonal MITF C5 antibody (Abcam). Following washing, cells were incubated for 1 h with anti-mouse Alexa Fluor 546 (Thermo Scientific) and DAPI. Slides were washed with PBS and mounted using Fluoroshield (Sigma Aldrich) before imaging on an FV1200 Olympus inverted confocal microscope. Cells transfected with an empty pcDNA3.1 vector were used as controls.
HEK293T cells (2 × 104) were seeded in black 96-well plates and cultured for 24 h prior to transfection (FuGENE, Promega) with 35 ng luciferase-coupled promoter, 35 ng MITF construct and 35 ng pRL Renilla luciferase control reporter vectors. Cells were harvested 24 h after transfection using the Luciferase DualGlo kit (Promega) reagents according to the manufacturer's instructions. Promoter activity was measured in a microplate multimode reader (Modulus II, Turner BioSystems) with a 1-s reading per well. Luciferase activity was normalized to Renilla signals for each sample and results presented as fold increase over an empty vector control. Experiments were conducted in five technical replicates for each construct and repeated for at least three biological replicates. Error bars indicate standard error of the mean and a student's t-test was used to assess statistical differences.
Co-immunoprecipitation assay
The 501mel human melanoma cells expressing doxycycline-inducible human GFP-tagged MITF or TFEB were treated with 0.2 μl.ml−1 doxycycline and transfected with mouse FLAG-tagged MITF(wt), MITF(Δ), or human FLAG-tagged MAX 24 h after seeding. The 501 human melanoma cell line was co-transfected with mouse GFP-tagged MITF(Δ) and mouse FLAG-tagged MITF(wt) or human FLAG-tagged MAX. Cells were washed 48 h after transfection with ice-cold PBS and lysed on a shaker for 15 min with coIP lysis buffer (50 mM Tris–HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton) supplemented with PMSF and a protease inhibitor cocktail. The lysates were scraped and centrifuged at 14 000×g for 10 min. A fraction of each supernatant was collected and stored (input). The remaining lysates were incubated at 4°C on a rotating platform with 2.5 μg GFP polyclonal antibody (Abcam) for 3 h. Subsequently, 20 μl of A/G plus-agarose beads (Santa-Cruz Biotechnology) were added and samples incubated for 1 h at 4°C on a rotating platform. Samples were centrifuged (2500 rpm for 5 min) to collect the beads and then washed three times with TBS buffer. Samples were eluted with 25 μl of 2× Laemmli buffer, boiled for 5 min and run on a gel for immunoblotting with GFP, FLAG and actin antibodies.
Electrophoretic mobility shift assay (EMSA)
DNA fragments for the EMSA studies were generated by annealing the M-box-Fw (5′-AAAGTCAGTCATGTGCTTTTCAGA-3′) and M-box-Rv (5′-GTCTGAAAAGCACATGACTGACTTT-3′) oligos. The DNA fragment was labeled with [α-32P]-dCTP, (#BLU013H100UC, PerkinElmer) and the labeled oligo purified on Sephadex G-25 Quick Spin columns (#11273922001, Roche).
The EMSA was performed according to Pogenberg et al. (2012). Briefly, the proteins used were expressed from the respective DNA clones in the TNT-T7 Quick Coupled Transcription/Translation System (#L1170, Promega). Proteins were cotranslated when testing for heterodimerization. 2 μl of TNT cotranslated proteins were pre-incubated in a buffer containing 20 ng of poly(dI–dC), 10% fetal calf serum, 2 mM MgCl2 and 2 mM spermidine for 15 min on ice. For supershift assays, 0.5 μl of mouse monoclonal antiMITF (C5) antibody (#ab12039, Abcam) were added to the samples, followed by incubation on ice for 30 min. Then 50 000 counts per minute (cpm) of 32P-labeled probe in a binding buffer containing 10 mM Tris (pH 7.5), 100 mM NaCl, 2 mM dithiothreitol, 1 mM EDTA, 4% glycerol and 80 ng/ml salmon sperm DNA were added to the preincubated protein mix in a total reaction volume of 20 μl and incubated for 10 min at room temperature. The resulting DNA–protein complexes were resolved on 4.2% non-denaturing polyacrylamide gels, placed on a storage phosphorscreen, and scanned on a Typhoon PhosphorImager 8610 (Molecular Dynamics). The EMSA assay was performed in triplicate.
Isothermal titration calorimetry
ITC experiments were carried out with a VP-ITC system (MicroCal). Experiments were performed at 25°C in Assay Buffer. Purified MITF(wt) and MITF(Δ) protein fragments 180–296 were placed in the reaction cell at concentrations of 5–20 μM with each DNA oligonucleotide duplex (Metabion) at a concentration of 25–100 μM in the injection syringe. Injections of 10 μl of DNA solution were performed at 4-minute intervals. Data were processed with the Origin 7 software (MicroCal). The data were corrected by the heat of injection calculated from the basal heat remaining after saturation. A one-site binding mode was used to fit the data, using a non-linear least-squares algorithm. Experiments were repeated at least three times. The values reported are the average KD and ± values correspond to standard deviations.
Förster resonance energy transfer (FRET)
HEK293 cells (DSMZ, Braunschweig, Germany) were transfected with Transfectin (Biorad) with 4 μg human MITF(wt), MITF(Δ) or TFEB constructs, labeled N-terminally either with CFP as donor or YFP as acceptor. GFP-TFEB (#38119) and GFP-MITF-D (#38133) were purchased from Addgene and sub-cloned into peCFP/YFP C1 vectors (Clonetech). The MITF(Δ) mutant was designed using the QuickChange site-directed mutagenesis kit (Stratagene). The correctness of all constructs was confirmed by sequence analysis (Eurofins Genomics).
The transfected HEK293 cells were grown on coverslips for 24 h and afterwards transferred to an extracellular solution consisting of 140 mM NaCl, 5 mM KCl, 1 mM MgCl2, 2 mM CaCl2, 10 mM glucose and 10 mM HEPES buffer (pH 7.4, NaOH). A QLC100 Real-Time Confocal System (VisiTech Int.) connected to two Photometrics CoolSNAPHQ monochrome cameras (Roper Scientific) and a dual port adapter (dichroic: 505lp; cyan emission filter: 485/30; yellow emission filter: 535/50; Chroma Technology Corp.) was used for recording fluorescence images. This system was attached to an Axiovert 200M microscope (Zeiss, Germany) in conjunction with two diode lasers (445, 515 nm) (Visitron Systems). Image acquisition and control of the confocal system was performed with a Visiview 2.1.1 software (Visitron Systems). All images were recorded ∼600 ms illumination. Images were corrected for cross talk and cross-excitation. Specific cross-talk calibration factors were determined for each FRET experiment. After threshold determination and background subtraction after transfecting with YFP/CFP only, the corrected FRET (Eapp) was calculated on a pixel-to-pixel basis with a custom-made software (37,38) integrated in MatLab 7.0.4 with a microscope specific constant G value of 2.0. All experiments were performed at 22–24°C. All data are presented as mean ± standard error of mean (s.e.m.) for the indicated number of experiments. Statistical significance was determined by unpaired two-sided Student's t-test for comparisons of two groups using Origin Pro 2017. Statistical significance was set at P < 0.05.
RESULTS
The structure of a stammer-less MITF variant reveals an uninterrupted canonical coiled coil arrangement
To comparatively analyze the functional implications of the stammer insert in MITF, we first determined the overall properties of a stammer-less variant, referred to as MITF(Δ) (Figure 1), in addition to the wild-type version of MITF, referred to as MITF(wt) (22). In the MITF(Δ) variant, three residues 259–261 between the first heptad (heptad 0) and the remaining heptads (I-IV) of the coiled coil leucine zipper were removed (Figure 1A). As shown by analytical size exclusion chromatography, MITF(Δ) elutes as a single species at the same volume as the wt protein, indicating the same dimeric association state (Supplementary Figure S1A). The MITF(Δ) circular dichroism (CD) spectrum is also virtually identical with that of the wt protein, demonstrating that both MITF variants are equally folded at the secondary structure level (Supplementary Figure S1B). However, when we used CD for measuring the MITF melting temperature, we found an increase of 18°C for MITF(Δ) when compared to the wt protein (Figure 1B, Table 1), indicating a substantial stabilization of the overall structural arrangement of MITF(Δ). This is further supported by a substantial decrease of susceptibility to proteolysis of MITF(Δ) in comparison to the wt protein when treated with proteinase K (39) (Figure 1C).
Table 1.
Comparison of structural and biophysical parameters in MITF(wt) and MITF(Δ)
| MITF(wt) | MITF(Δ) | |
|---|---|---|
| PDB | 4ATH | 6FX5 |
| Resolution [Å] | 1.95 | 2.05 |
| Sequence segments | A: 216–295, B: 214–295 |
A: 220–295, B: 217–293 |
| Coiled coil interface [Å2] | 1,125 | 1,094 |
| Accommodation index | −0.5 | 0.0 |
| Maximum asymmetry index (residue) [Å] | 0.56 (261) | 0.21 (269) |
| Thermal unfolding midpoint [°C] | 42.7 | 60.9 |
| Permissive dimerization partners | MITF(wt), TFEB | MITF(Δ), MAX |
Next, we determined the crystal structure of the stammer-less MITF(Δ) variant at 2.05 Å resolution (Figure 1D, Supplementary Table S1). The final MITF(Δ) model comprises residues 220–294 and 217–293 of the two protomers referred to as chains A and B, which include the Basic Region (BR), Helix-Loop-Helix (HLH) segment and a substantial part of the coiled coil Zip segment. Unlike the asymmetric and segmented coiled coil arrangement found in MITF(wt), in MITF(Δ) the Zip region is composed of two symmetrically arranged 50-residues long α-helices that form a canonical coiled coil arrangement of five uninterrupted heptad repeats 0 and I–IV (residues 253–290), using the previously established numbering scheme (22).
Structure-based detection of distantly related bHLH-Zip transcription factors
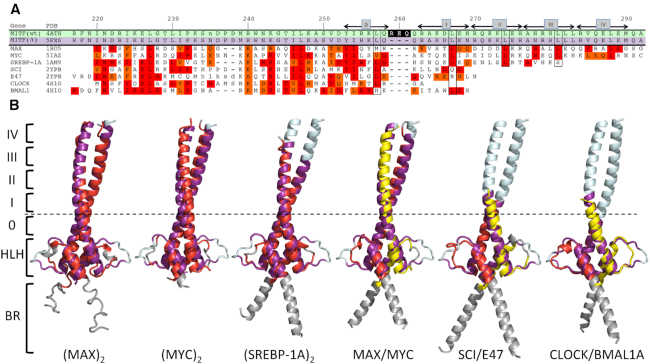
The availability of the structures of the same MITF sequence with and without the stammer between heptad 0 and the remaining heptads I–IV has allowed us to search for proteins with related structures, by exploiting conformational differences in the coiled coil segment caused by the stammer. For this analysis, we used the structures of both apo-MITF(wt) and MITF(Δ), as well as one representative MITF(wt) structure with the CLEAR box DNA recognition element. Similar to other MITF–DNA complexes, MITF in the presence of the CLEAR motif comprises a truncated coiled coil segment but includes an extended basic region involved in DNA binding (22,40). From the search results against the Protein Data Bank (PDB) we selected those structures that contain a bHLH domain N-terminal to the coiled coil Zip segment, reminiscent of that of MITF.
First, we analyzed whether different MITF structural variants detect each other in structural database searches. Indeed, MITF(wt) both in the absence and presence of DNA were among the 20 top hits for MITF(Δ) (Table 2, Supplementary Table S2). However, only the sequence segment N-terminal to the stammer in MITF(wt) including the bHLH domain aligns correctly, whereas the remaining part of the coiled coil still superimposes structurally but with a three-residue out-of-register sequence alignment (Figure 1E). This lowers the sequence identity in the respective alignment to 55% (Table 2). The level of structural similarity is comparable in all combinations of superimposed structures, reflected by r.m.s.d. values between 1.5 and 2.0 Å.
Table 2.
MITF-related HLH-bZip structures
| 4ATH - MITF(wt, apo) | 6G1L - MITF(wt, CLEAR) | 6FX5 (Δ, apo) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Query cover [%] | Sequence identity [%] | RMS [Å] | Query cover [%] | Sequence identity [%] | RMS [Å] | Query cover [%] | Sequence identity [%] | RMS [Å] | |
| Self-recognition | |||||||||
| 4ATH | 77.6 | 96.9 | 1.5 | 92.4 | 55.2 | 2.0 | |||
| 6G1L | 75.7 | 96.9 | 1.5 | 78.0 | 67.0 | 1.6 | |||
| 6FX5 | 83.4 | 55.2 | 2.0 | 72.0 | 67.0 | 1.6 | |||
| Related homodimer clusters | |||||||||
| MAX (1RO5) | 75.6 | 15.6 | 2.7 | 70.7 | 21.0 | 2.3 | 84.5 | 22.6 | 2.4 |
| MYC (5IAZ) | 69.5 | 24.2 | 2.2 | 66.2 | 25.9 | 2.3 | 81.8 | 22.6 | 2.7 |
| SREBP-1A (1AM9) | 67.7 | 34.7 | 2.0 | 85.6 | 36.4 | 1.7 | 75.1 | 33.6 | 1.8 |
| Related heterodimer clusters | |||||||||
| MYC/MAX (1NKP) | 71.4 | 20.9 | 2.8 | 77.3 | 26.8 | 1.9 | 82.9 | 21.4 | 2.0 |
| SCI/E47 (2YPB) | 56.8 | 25.3 | 2.0 | 75.7 | 26.2 | 1.7 | 58.6 | 28.6 | 1.8 |
| CLOCK/BMAL1 (4H1O) | 51.6 | 31.0 | 1.5 | 69.5 | 32.2 | 1.6 | 55.9 | 33.0 | 2.0 |
For further details, see Supplementary Table 2.
Based on these findings, it is not surprising that for all MITF structures used here, regardless of the presence of the stammer, related bHLH-Zip transcription factor structures are detected with similar similarity scores. We found three non-redundant clusters (<90% sequence identity) of homodimeric and three clusters of heterodimeric bHLH-Zip transcription factor pairs (Figure 2, Table 2). In contrast to our searches for self-identification of different MITF variants, the level of sequence identity between MITF and the other bHLH-Zip transcription factors is too low for confirming correctness of the coiled coil segment alignment. Therefore, validation of the alignments has been limited to the identification of potential coiled coil sequence abnormalities such as the stammer in MITF(wt) and established heptad sequence patterns that are, however, only partly conserved (Figure 2A). Interestingly, all detected homo- and heterodimers of bHLH-Zip transcription factors other than MITF are formed by structures with uninterrupted stammer-less coiled coil segments. Hence, the heptad periodicity is maintained in the respective structure-based sequence alignments only when using MITF(Δ) as the structural template whereas the use of MITF(wt) structure leads to out-of-register sequence alignments, reminiscent of the MITF(Δ)/MITF(wt) alignment. We therefore used the MITF(Δ) structure as the template for all illustrated superimpositions (Figure 2B). The level of sequence identity between these targets and MITF is between 15 and 37%, whereas the structural diversity is only slightly higher than superimposed MITF structural pairs with identical sequences (Table 2). The data thus show that the conformational plasticity that is inherent in coiled coil structures competes with or even overrules the structural diversity due to sequence diversity when using MITF as the template. We also noted that apart from both homo- and heterodimeric assemblies of MAX and MYC, the extent of superimposed heptad repeats with the coiled coil region is substantially reduced. For the heterodimeric bHLH-Zip transcription factor pairs SCI/E47 and CLOCK/BMAL1A, the superposition is limited to the first two heptad repeats, which are separated by the stammer in MITF(wt) (Figure 2B). We conclude that, as we were able to detect the same distantly related bHLH-Zip transcription factor pairs with three different MITF coordinate sets (Table 2, Supplementary Table S2), the search presented here is likely to cover all detectable bHLH-Zip transcription factor pairs whose molecular structures have been determined to date.
Figure 2.
Search for related bHLH-Zip transcription factors using MITF(Δ) as the template. (A) Structure-based sequence alignment. Gene names, as used in the text, and PDB codes of the target structures are indicated. Sequence numbers of the MITF sequence and the positions of heptad repeats 0, I–IV, as defined previously (22), are indicated above the alignment. Heptad positions d that are most conserved in known coiled coil structures are boxed to facilitate orientation. The three stammer residues that have been removed in MITF(Δ) are shown with a black background in the MITF(wt) sequence. Identical and conserved sequence positions are highlighted in red and orange, respectively. Note that the alignments for some bHLH-Zip transcription factors cover only a reduced number of heptad repeats. (B) Structural superimpositions. Color codes: MITF(Δ), purple (as in Figure 1) for superimposed parts, pale cyan for remaining parts; homodimeric bHLH-Zip transcription factor targets, red for superimposed parts, grey for remaining parts; heterodimeric bHLH-Zip transcription factor targets, red and yellow for superimposed parts, to emphasize the differences in the two chains. The approximate position of the stammer in MITF(wt) is indicated by a dashed line.
Quantitative comparison of stammer-containing and stammer-less MITF variant structures
The two atomic structures of MITF(wt) and the stammer-less MITF(Δ) variant allow a direct comparison to unravel a mechanistic understanding of the particular protein partner selectivity for coiled coil-based heterodimerization in MITF (19,21). To overcome the problem of out-of-register structure-based alignments when comparing MITF(wt) and MITF(Δ) (Table 2), we divided the respective MITF structures into two separate segments based on the HLH and Zip domains. Each of the two can be well aligned with a low r.m.s.d. equal to 0.57 Å for the HLH region (72 matching residues) and 0.67 Å for the coiled regions comprising heptad repeats I-IV (62 matching residues) (Supplementary Figure S2). The resulting values are considerably lower than those found in structural superposition using the entire MITF structure as one rigid template (Table 2), demonstrating the validity of dividing it into separate modular rigid bodies.
In comparison to MITF(Δ), there is a longitudinal shift of the residues distal to the stammer in MITF(wt) (residues 259–261) by 5–6 Å, which is in the range of the pitch of one α-helical turn of 5.4 Å, equivalent to one half heptad repeat (Figure 1E, Supplementary Figure S2). Hence, the radial orientation of the residues that form the C-terminal coiled coil segment (residue 261 to C-terminus) distal to the stammer insert in MITF(wt), remain practically the same in both structures (Figure 3A,B). As a result, the stammer insert also extends the overall length of the coiled coil segment in MITF(wt) (Figure 1E).
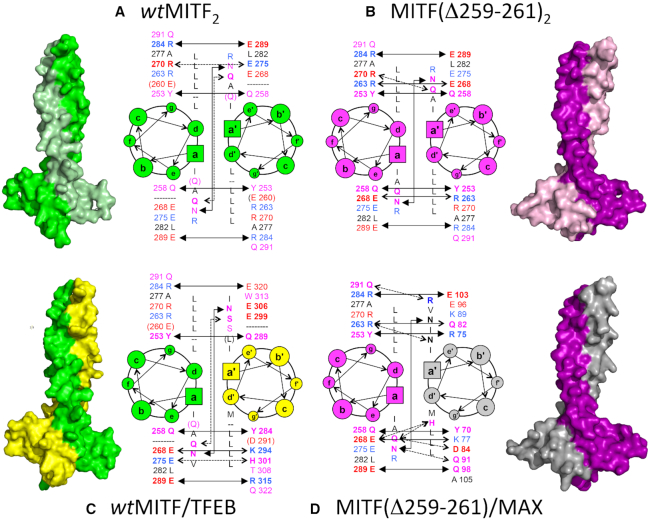
Figure 3.
Structural models and heptad wheel presentation of permissible complexes of MITF variant homodimers and heterodimers with bHLH-Zip transcription factors: (A) MITF2(wt), (B) MITF(Δ)2, (C) MITF(wt)/TFEB, (D) MITF(Δ)/MAX. Residue types in positions a/a’, d/d’, e/e’ and g/g’ of all visible heptad repeats are listed; for positions e/e’ and g/g’, in addition the residue numbers are given. To facilitate recognition of specific attractive interactions, acidic residues are in red, basic residues are in blue, and other hydrophilic residues are in pink. Those hydrogen bonds that are conserved in all four models are shown by solid lines with solid arrows; hydrogen bonds that are partly conserved are shown by solid lines with open arrows; non-conserved hydrogen bonds specific for one of the complexes illustrated are shown by dashed lines with open arrows. Additional color codes: TFEB, yellow; MAX, gray. The icon size for the different heptad positions differ to provide an intuitive impression of their location orthogonal to the document plane.
The area of the protein/protein interface of the coiled coil segment (residues 242 to C-terminus) is in the same range: 1125 Å2 in MITF(wt) and 1094 Å2 in MITF(Δ), which is slightly >200 Å2 per heptad repeat on average (Table 1). In both structures, it is mainly driven by knobs-into-holes packing for heptad positions a/a’ and d/d’ of all five coiled coil heptads 0 and I–IV (Figure 3A, B). In contrast to the MITF(wt) segmented coiled coil where we observed a hole in the dimeric interface next to the stammer insert (22), the arrangement of the coiled coil interface in MITF(Δ) is tight and without any major cavities or holes.
In both MITF structures, the Zip helices are connected by specific interactions over the entire coiled coil segment. As expected, the most significant conformational changes are observed in heptad 1, which is symmetric and tightly packed in the MITF(Δ) dimer and thus allows the formation of a new two-fold repeated salt bridge between Arg263 and Glu268 from both MITF(Δ) polypeptide chains (Figure 1D, Figure 3B). These interactions are not possible in the MITF(wt) structure, as the two residues are too far apart from each other to allow a specific interaction due to the asymmetry induced by the stammer (22). Changes in the overall arrangement caused by the stammer are extended into heptad 2, where different residue pairs are involved in specific interactions, whereas in heptads 3–5 only minor changes are observed. The initial coiled coil segment (heptad 0) between the HLH motif and the stammer segment is virtually identical in both structures, indicating that the main effect by the presence/absence of the stammer is propagated towards the C-terminal leucine zipper arrangement.
Comparison of the geometrical parameters used for the characterization of coiled coil structures demonstrates that removal of the stammer insert in the MITF(Δ) structure relieves the requirement of compensating for the expected −51° coiled coil phase shift, which is equivalent to an Insertion Index change IA of −0.5 according to a recent systematic analysis (23). In the structure of MITF(wt), a phase shift compensation is achieved by a change in Accommodation Index of −0.5 over a residue range of about 20 residues (Figure 1F). The structural irregularity induced by the stammer in MITF(wt) is also reflected by a peak in local structural asymmetry exceeding 0.5 Å r.m.s.d. within the coiled coil next to the stammer, whereas there is no significant asymmetry in the MITF(Δ) structure (Figure 1G, Table 1). Other geometric parameters such as coiled coil periodicity, radius, and axis curvature, also reveal substantial deviations for the stammer insert region in MITF(wt), which are not observed in the stammer-less MITF(Δ) structure (Supplementary Figure S3).
MITF has a modular functional domain arrangement
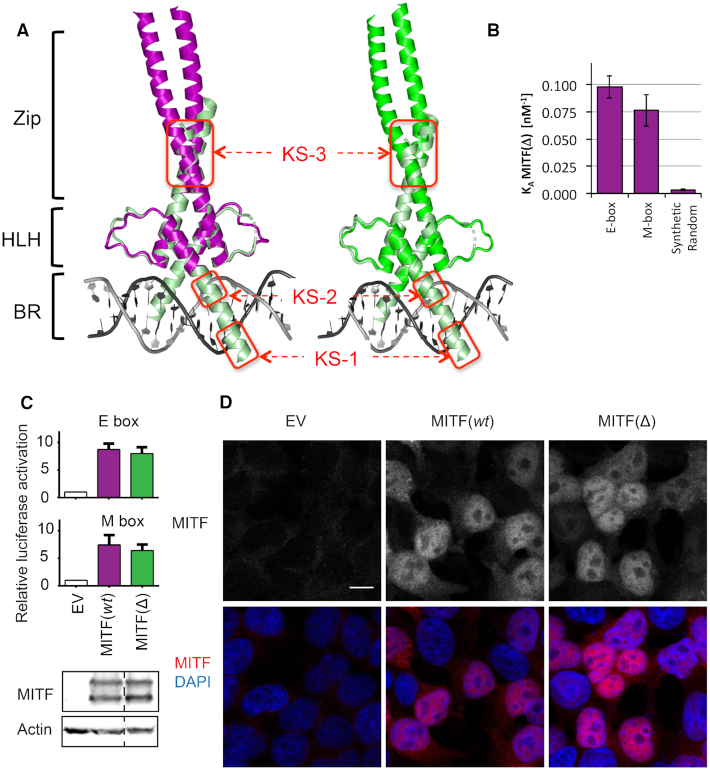
We next investigated whether MITF(Δ) retains the ability for nuclear localization and DNA binding. Composite structural models combining the structure of MITF(Δ) and previous structures of MITF(wt)–DNA complexes suggest a modular organization of the MITF region including the Basic Region (BR) and Helix-Loop-Helix (HLH) domain, which provide the DNA-binding properties of the protein, and the C-terminal coiled coil segment, which contains the region involved in homo- or heterodimerization with other bHLH-Zip transcription factors (Figure 4A). As measured by isothermal titration calorimetry (ITC), MITF(Δ) binds to DNA containing the specific E-box and M-box response motifs with dissociation constants of about 10 nM, which corresponds to a slightly lower affinity than those measured for MITF(wt) (22) (Figure 4B, Supplementary Figure S5, Table 3). The binding stoichiometry remains 2:1 in all measurements, representing the interactions of one MITF dimer with one DNA duplex. The selectivity for binding to the M- and E-box sequences over synthetic random DNA is ∼30-fold and thus is considerably lower than the values found for MITF(wt) (22). As the structures of the BR and HLH regions of both MITF variants are virtually identical (Figure 4A,B, Supplementary Figure S2), the most plausible explanation for this difference points to the increased thermal stability of the MITF(Δ) structure (Figure 1B). Thermal stabilization is generally caused by a rigidified structural arrangement (2,41) that may reduce the amount of structural flexibility required to generate several specific interactions to allow optimum DNA binding (42–44).
Figure 4.
Specific DNA-binding properties of MITF(wt) and MITF(Δ). (A) Composite structural models of the complete MITF(wt) and MITF(Δ) coiled coil segment superimposed with a MITF(wt)/M-box complex (PDB code: 4ATI) with a truncated coiled coil segment, demonstrating structural modularity of the BR, HLH and Zip (coiled coil) regions in MITF. Established karyophilic segments (45) are boxed in red and labeled KS-2 and KS-3. (B) Binding properties of MITF(Δ) to E-box, M-box and synthetic random DNA, measured by ITC (for further details, see Supplementary Figure 5). For binding data of MITF(wt) under identical experimental conditions, see (22). (C) Upper panel, transactivation assays in HEK293T cells in which the same MITF mutants were co-transfected together with TYR promoters containing either M- or E-boxes in comparison with an empty vector (EV) control. Lower panel, western blots show equivalent levels of expression of MITF(wt) and MITF(Δ). (D) Confocal imaging indicating proper localization of all MITF variants to the nucleus. Scale bar represents 10 μm and applies to all panels.
Table 3.
Binding of MITF to DNA recognition elements
| MITF (Δ) | MITF (wt) | ||
|---|---|---|---|
| M-box | K D [nM] | 1.3 ± 0.3 × 101 | 2.5 ± 0.5 |
| Binding stoichiometry (n) | 0.499 ± 0.008 | 0.505 ± 0.010 | |
| E-box | K D [nM] | 1.0 ± 0.1 × 101 | 1.7 ± 0.2 |
| Binding stoichiometry (n) | 0.503 ± 0.006 | 0.492 ± 0.018 | |
| Synthetic random | K D [nM] | 3.0 ± 0.6 × 102 | 4.9 ±0.4 × 102 |
| Binding stoichiometry (n) | 0.464 ± 0.016 | 0.505 ± 0.04 | |
| Selectivity | M-box / Synthetic random | 23 | 196 |
| E-box / Synthetic random | 30 | 288 |
We next performed a transactivation (TA) assay in HEK293T cells using a modified tyrosinase promoter containing M-box or E-box sequences and luciferase as a reporter (Figure 4C). These data indicate that MITF(Δ) is as efficient as the wt protein in activating the expression of MITF target promoters. This implies that the changes in terms of specific DNA binding observed with purified MITF(Δ) are less pronounced when performing complementary experiments with full-length protein in a cellular context.
One of our recent studies revealed that MITF has three sequence stretches with karyophilic properties in the bHLH-Zip domain, spanning residues 197–206, 214–217 and 255–265, that contribute to nuclear localization (45). Since the stammer insert in MITF(wt) is located within the third segment, we looked for specific structural differences within this segment in the respective MITF(wt) and MITF(Δ) structures. Whereas most of the positively charged residues within this segment (Arg255, Lys256, Arg263, Lys265) are exposed in both structures (Figure 1D), Arg259 is part of the triple residue deletion in MITF(Δ) and therefore absent in the respective structure. Confocal imaging revealed, however, that nuclear localization is not altered in the MITF(Δ) variant in HEK293T cells (Figure 4D), suggesting that a possible contribution of the stammer residue Arg259 to nuclear localization is dispensable.
The MITF stammer determines coiled coil protein partner specificity
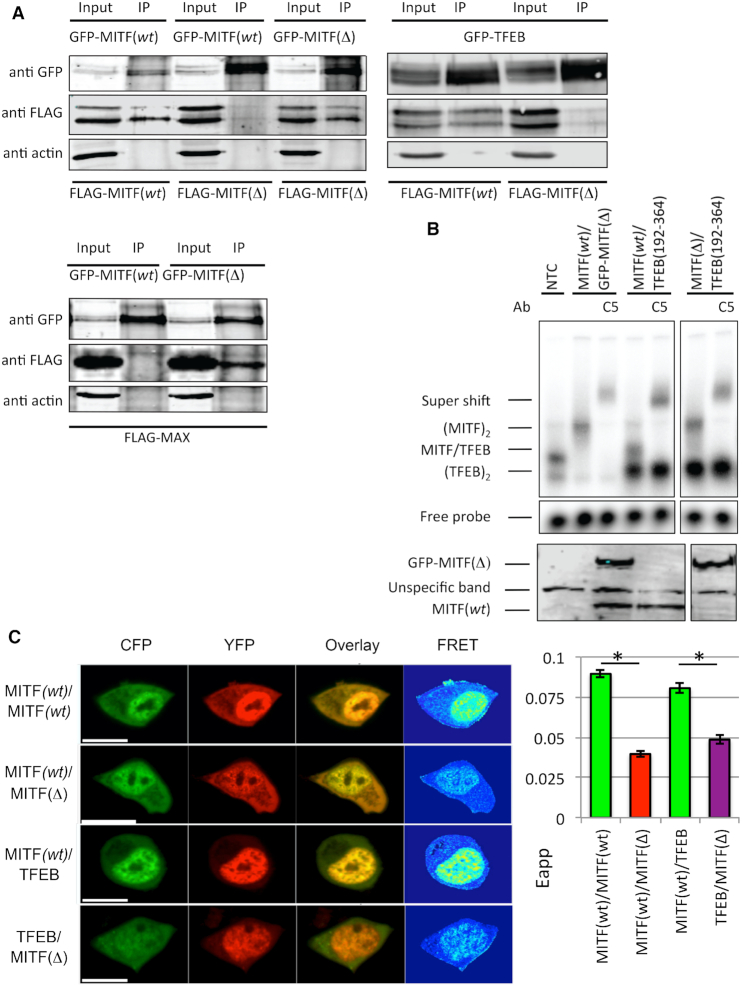
To determine whether staggering of heptad repeats induced by the MITF stammer insert provides a basis of exclusive selectivity of coiled coil protein partners with a strict requirement of in-phase heptad repeats, we used several bHLH-Zip transcription factors that are either in phase with stammer-inserted MITF(wt) or with the stammer-removed MITF(Δ) variant. To test in-phase partnering with MITF(wt) we used TFEB, another member of the MITF/TFE family that contains the same type of stammer insert as MITF (22). For the same type of experiment with the MITF(Δ) variant, we used MAX as a model (cf. Figure 2) since MAX has been established as a canonical bHLH-ZIP transcription factor with an uninterrupted coiled coil segment and an ability to promiscuously bind to several other bHLH-Zip transcription factors (5,46). We first assayed the ability of MITF(Δ) to homodimerize or heterodimerize with MITF(wt) and other members of the MITF/TFE subfamily by co-immunoprecipitation (coIP) in 501Mel human melanoma cells (Figure 5A). These data show that MITF(Δ), although capable of homodimerizing with itself (Figure 5A), did not dimerize with either MITF(wt) or TFEB (Figure 5A). Vice versa, only MITF(Δ) but not MITF(wt) dimerized with MAX.
Figure 5.
The effects of stammer removal on the ability of MITF to heterodimerize with other bHLH-ZIP transcription factors. (A) FLAG-tagged MITF(wt), MITF(Δ) and MAX were expressed together with either GFP-tagged MITF(wt), MITF(Δ) and TFEB in 501 Mel cells. Protein interactions were analyzed by co-immunoprecipitation (co-IP) by Western blot detection of either anti-FLAG or anti-GFP antibodies. Negative control (transfected EV-GFP) is shown in Supplementary Figure S4. (B) Upper panel, EMSA showing the binding of MITF(wt), MITF(Δ) and TFEB to the M-box recognition element. Supershifts with a MITF-specific antibody are indicated, confirming the specificity of the gel shifts. In the presence of M-box MITF(wt)/TFEB heterodimers were observed but no MITF(Δ)/TFEB heterodimers. Lower panel, Western blot confirming in vitro translation products of the GFP-tagged MITF(Δ) (top panel) and MITF(wt) (bottom panel) constructs. (C) FRET dimerization experiments in HEK 293 cells. Left panel, representative images of FRET experiments of MITF(wt)2, MITF(wt)/MITF(Δ), MITF(wt)/TFEB and MITF(Δ)/TFEB labeled with CFP and YFP, respectively. The lengths of scale bars correspond to 10 μm. Right panel, quantification of the data. Error bars are defined as standard error of the mean (SEM). Asterisks (*) indicate a significant difference (P < 0.05) of FRET values. Statistics are derived from student's t‐test distribution.
To further test whether MITF(Δ) was able to form heterodimers with TFEB in the presence of the M-box recognized by both transcription factors (21) we designed an EMSA assay using different sized MITF and TFEB constructs. To determine selectivity by differential EMSA mobility, we used GFP-tagged MITF as the larger partner and a truncated TFEB version (residues 192–364), comprising intact bHLH-Zip segments necessary for DNA binding and dimerization ability, as the smaller partner. The correctness of the constructs and their functional ability to bind DNA were verified in a series of control experiments (Supplementary Figure S4). The data show that only MITF(wt) heterodimerizes with TFEB in the presence of the M-box element but no heterodimeric assembly was detected when the mutant MITF(Δ) was co-translated with TFEB (Figure 5B). In conclusion, these data demonstrate that the stammer is critical for restricting the heterodimerization ability of the MITF-TFE proteins.
Next, we tested the ability for dimerization using an in vivo fluorescence resonance energy transfer (FRET) approach with HEK293 cells (Figure 5C). For these experiments, we used the MITF-D isoform, which remains cytosolic at resting conditions and translocates to the nucleus upon activation, such as starvation, which is reflected by high FRET levels in the nucleus (Figure 5C). As expected, we observed a strong FRET signal for in-phase CFP- and YFP-tagged MITF(wt) homodimers and MITF(wt)/TFEB heterodimers but not for MITF(Δ) in the presence of MITF(wt) or TFEB. These data demonstrate that the rules we derived from our structural data apply under physiological conditions as well.
Based on the experimental structures of the two MITF(wt) and MITF(Δ) homodimers, we built structural homology models of the two heterodimeric complexes that we functionally identified (Figure 3C, D, Table 1). For MITF(wt)/TFEB we created two closely related alternative models using the structure of the asymmetric MITF(wt) homodimer, either by replacing MITF chain A or MITF chain B by TFEB (only one is shown in Figure 3C). The MITF/MAX heterodimer was created using the structure of the MITF(Δ) homodimer. Since this template is highly symmetric, replacement of MITF(Δ) chains A or B by MAX led to virtually identical models.
When comparing the structural models, we observed three structurally conserved coiled coil hydrogen bond interactions in heptads 0, 3 and 4 in all four structures (Figure 3A–D, solid lines with filled arrow heads). The MITF heptad-0 g-e’/g’-e Tyr253–Glu258 interaction is also conserved in sequence in the MITF(wt)TFEB interaction. In MAX the residue equivalent to Tyr253 in MITF is conserved (Tyr70) whereas Glu258 of MITF is an arginine (Arg75). The MITF heptad-4 g-e’/g’-e Arg284–Glu289 interaction is also conserved across all four structural models. Both equivalent residues are identical in the TFEB sequence. In MAX, only the residue equivalent to MITF Glu289 is identical (Glu103), whereas the equivalent of MITF Arg284 is a glutamine (Gln98). Finally, there is an invariant heptad-3 a-a’/a’-a interaction in all four structures that is formed by Asn278–Asn278′ pair from both chains in MITF. The equivalent sequence positions in TFEB (Asn309) and MAX (Asn92) are conserved.
By contrast, there is one specific heptad-2 a-a’/a’-a interaction that is found only in the stammer-containing homodimeric MITF (wt) and heterodimeric MITF(wt)/TFEB complexes, formed by a pair of two identical glutamines (Gln271) from both coiled coil chains (Figure 3A, C; solid lines with open arrow heads). In TFEB the equivalent residue position is occupied by Ser302. Vice versa, there is a specific heptad-1 g-e’/g’-e interaction only found in the two stammer-less structures of homodimeric MITF(Δ) and heterodimeric MITF(Δ)/MAX (Figure 3B, D; solid lines with open arrow heads). In the MITF(Δ) homodimer, this interaction is formed by residues Arg263 and Glu268. In MAX, the two equivalent positions are the similar amino acids, Lys77 and Gln82.
In summary, our structural and functional findings provide novel insight into the underlying molecular parameters and rules that allow a mutually exclusive partner selection for the bHLH-Zip transcription factors.
DISCUSSION
The structural principles of interrupted helical coiled coil arrangements in proteins were unraveled more than two decades ago, mainly by the use of synthetic peptides and theoretical calculations (2,47,48). Within this framework, it was well established that a three-residue insert, also termed a ‘stammer’, leads to a decrease of superhelical periodicity compensated by helical overwinding. However, only a small number of protein structures containing an asymmetric stammer inserted coiled coil have been described to date. Known and well-characterized examples are the stammer inserts in PAN3 pseudokinase (49) and in the cyclin T binding domain of hexim (50), which are both essential for binding to cognate protein ligands. A stammer-less hexim variant interacts with 7SK snRNA in the same way as wt hexim (51).
The TFE family, including MITF and TFEB, provide the only reported evidence for stammer-dependent heterodimeric partner selection mediated by coiled coil interactions (21). Although the stammer motif itself is partly conserved in the different members of the TFE family (‘KEQ’ in TFEC and TFE3, ‘KDL’ in TFEB), previous data using a MITF variant in which three consecutive stammer residues were mutated into alanine showed no detectable effect altering the ability for selective homo/hetero-dimerization (22). The findings led to the conclusion that the observed sequence conservation is probably due to close evolutionary relations of different members of the TFE family rather than functional constraints.
Here, we show that both MITF(wt) and stammer-less MITF(Δ) retained their ability to bind DNA, suggesting that the sequence regions flanking the coiled coil segments are functionally autonomous, regardless of the presence of a stammer (Figure 4A, B). By contrast, heterodimeric out-of-phase interactions by a stammer-less and a stammer-containing partner are not permissive in the context of additional folding regions such as the BR and HLH segments in MITF that require an in-phase dimeric arrangement to be functional (22) (Figure 5). This model permits interactions of stammer-containing non-canonical and stammer-less, canonical bHLH-Zip transcription factors, as long as other heptad positions—in particular positions a, e and g—allow favorable interactions. By contrast, formation of pairs from stammer-containing and stammer-less bHLH-Zip transcription factors is, according to our findings, not permissive. Interestingly, this rule can be overcome in the absence of those regions, as shown for a truncated version of MITF containing only the coiled coil segment (39). In that study, using a selection screen focused on the MITF coiled coil as target, a synthetic helix sequence (iM10) without a stammer insert was enriched with a stammer-containing MITF coiled coil segment in a symmetric canonical arrangement (39). Similar to the substantial thermal stabilization by 18°C we found for the MITF(Δ) variant (Figure 1B), the melting temperature of the MITF/iM10 coiled coil was remarkably increased by 25°C when compared to the homodimeric MITF(wt) coiled coil. These findings confirm previous data demonstrating that a regular coiled coil pattern found in canonical, non-interrupted coiled coils substantially contributes to overall thermal stability and is therefore thermodynamically preferred in the absence of additional flanking dimerization modules such as the bHLH domain in MITF (47,48,52).
Ultimately, the precise readout of gene transcription relies on highly combinatorial interactions of multiple transcription factors and regulators prior to and during DNA binding, which also depends on DNA accessibility (53,54). Despite the inspiring early studies on the mechanism of heterodimerization of the two coiled coil transcription factors c-Jun and c-Fos (55,56), further structural studies to determine molecular rules by which homodimeric and heterodimeric coiled coil assemblies are discriminated have remained sparse. Of these, we have previously directly compared the structures of homodimeric MafB2 and heterodimeric MafB/c-Fos complexes to demonstrate how DNA recognition motif binding preferences change (10).
For the specific family of bHLH-Zip transcription factors, at present structures are only available for a few heterodimeric assemblies, all of which form uninterrupted coiled coils (Table 2) (5,46). These structures provide insights into how multiple partnerships with different functional readouts are possible, provided the respective participating sequences permit attractive interactions specifically in heptad positions a-a’/a’-a and g-e’/g’-e. Here we show that the presence of a stammer insert, as found within the coiled coils of the members of the MITF/TFE family, does not permit partnering with canonical bHLH-Zip transcription factors such as MAX, MAD and MYC, despite the presence of favorable coiled coil interactions C-terminal to the stammer. If we suppose that a shift of register would be possible, it would inevitably lead to dysfunctional neighboring modules including the DNA-binding domain, thus requiring proper dimerization architecture (Figure 4A). Whether this rule is less strict for non-permissive partnering with bHLH-Zip transcription factors with shorter coiled coil segments (Figure 2B) remains an open question at this point. Interestingly, heterodimeric partnering with a bZip transcription factor lacking the HLH motif has not been reported for any bHLH-Zip transcription factor, presumably due to a strict requirement for a dimeric arrangement of the HLH module (5,57).
Although many different mutations have been found in the MITF gene in mice, humans and other species, none affect the stammer sequence (22,58–60). We would expect such a mutation to exhibit severe effects on the phenotype, leading to microphthalmia and absence of melanocytes, since the protein would not be able to form homodimers or heterodimers with the TFE family.
Ultimately, our findings generate crucial questions for future research in the field. Searches for coiled coil structures with alternative modes for changing heptad register such as skips and stutters, equivalent to 1-residue and 4-residue insertions, in addition to stammers (three-residue insertion) (2,23,48) could provide new insight into how widespread restricted coiled coil heterodimerization may occur. It would also be revealing to investigate at which stage heterodimerization takes place under physiological conditions, whether at the level of ribosomal biosynthesis or by as yet uncharacterized disassembly/reassembly events post ribosomal biosynthesis. Finally, our data open up attractive possibilities in the rewiring of combinatorial interactions and functional readouts of different bHLH-Zip transcription factors by inserting or removing coiled coil phase interrupting sequence elements to design new circuits at a transcriptional level.
DATA AVAILABILITY
The atomic coordinates and structure factors of MITF(Δ) have been deposited in the PDB with accession code 6FX5.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the staff of the European Synchrotron Radiation Facility for assistance and support in using beamline ID23-2. Ariadna Terricabras, Jasmin Köhn and Violeta Iborra (EMBL, Hamburg, Germany), and Victoria Lunz (Johannes Kepler University Linz, Austria) are acknowledged for their technical assistance. We thank Nathan W. Schmidt (formally University of California, San Francisco) for using software developed by him to analyze coiled coil protein structures.
Author contributions: Structural, biophysics and bioinformatics experiments were performed by V.P., M.M. and A.O.-K. Cell biology, imaging, CoIPs and EMSA studies were performed by J.B.A., I.S. and M.H.O. FRET was performed by Ro.S. Study design and data interpretation by E.S., M.H.O., Ra.S., V.P. and M.W. V.P. and M.W. wrote the manuscript.
Notes
Present address: Vivian Pogenberg, University Hamburg Clinical Centre Hamburg-Eppendorf, Center for Experimental Medicine, Institute of Biochemistry and Signal Transduction, Martinistraße 52, 20246 Hamburg, Germany.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Icelandic Research Fund [163413 to E.S. and M.H.Ö.]; Austrian Science Foundation [FWF P28701 to Ra.S.]. Funding for open access charge: Icelandic Research Fund [163413 to E.S. and M.H.Ö.]; Austrian Science Foundation [FWF P28701 to Ra.S.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Rose A., Schraegle S.J., Stahlberg E.A., Meier I.. Coiled-coil protein composition of 22 proteomes–differences and common themes in subcellular infrastructure and traffic control. BMC Evol. Biol. 2005; 5:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lupas A.N., Gruber M.. The structure of alpha-helical coiled coils. Adv. Protein Chem. 2005; 70:37–78. [DOI] [PubMed] [Google Scholar]
- 3. Truebestein L., Leonard T.A.. Coiled-coils: the long and short of it. Bioessays. 2016; 38:903–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Miller M. The importance of being flexible: the case of basic region leucine zipper transcriptional regulators. Curr. Protein Pept. Sci. 2009; 10:244–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jones S. An overview of the basic helix-loop-helix proteins. Genome Biol. 2004; 5:226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ruberti I., Sessa G., Lucchetti S., Morelli G.. A novel class of plant proteins containing a homeodomain with a closely linked leucine zipper motif. EMBO J. 1991; 10:1787–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Skinner M.K., Rawls A., Wilson-Rawls J., Roalson E.H.. Basic helix-loop-helix transcription factor gene family phylogenetics and nomenclature. Differentiation. 2010; 80:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Steidl C., Leimeister C., Klamt B., Maier M., Nanda I., Dixon M., Clarke R., Schmid M., Gessler M.. Characterization of the human and mouse HEY1, HEY2, and HEYL genes: cloning, mapping, and mutation screening of a new bHLH gene family. Genomics. 2000; 66:195–203. [DOI] [PubMed] [Google Scholar]
- 9. Wu D., Rastinejad F.. Structural characterization of mammalian bHLH-PAS transcription factors. Curr. Opin. Struct. Biol. 2017; 43:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pogenberg V., Consani Textor L., Vanhille L., Holton S.J., Sieweke M.H., Wilmanns M.. Design of a bZip transcription factor with homo/heterodimer-induced DNA-binding preference. Structure. 2014; 22:466–477. [DOI] [PubMed] [Google Scholar]
- 11. Vinson C., Acharya A., Taparowsky E.J.. Deciphering B-ZIP transcription factor interactions in vitro and in vivo. Biochim. Biophys. Acta. 2006; 1759:4–12. [DOI] [PubMed] [Google Scholar]
- 12. Bhimsaria D., Rodriguez-Martinez J.A., Pan J., Roston D., Korkmaz E.N., Cui Q., Ramanathan P., Ansari A.Z.. Specificity landscapes unmask submaximal binding site preferences of transcription factors. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:E10586–E10595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hortschansky P., Ando E., Tuppatsch K., Arikawa H., Kobayashi T., Kato M., Haas H., Brakhage A.A.. Deciphering the combinatorial DNA-binding code of the CCAAT-binding complex and the iron-regulatory basic region leucine zipper (bZIP) transcription factor HapX. J. Biol. Chem. 2015; 290:6058–6070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rodriguez-Martinez J.A., Reinke A.W., Bhimsaria D., Keating A.E., Ansari A.Z.. Combinatorial bZIP dimers display complex DNA-binding specificity landscapes. Elife. 2017; 6:e19272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Inamoto I., Chen G., Shin J.A.. The DNA target determines the dimerization partner selected by bHLHZ-like hybrid proteins AhRJun and ArntFos. Mol. Biosyst. 2017; 13:476–488. [DOI] [PubMed] [Google Scholar]
- 16. Hallsson J.H., Haflidadottir B.S., Stivers C., Odenwald W., Arnheiter H., Pignoni F., Steingrimsson E.. The basic helix-loop-helix leucine zipper transcription factor Mitf is conserved in Drosophila and functions in eye development. Genetics. 2004; 167:233–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rehli M., Den Elzen N., Cassady A.I., Ostrowski M.C., Hume D.A.. Cloning and characterization of the murine genes for bHLH-ZIP transcription factors TFEC and TFEB reveal a common gene organization for all MiT subfamily members. Genomics. 1999; 56:111–120. [DOI] [PubMed] [Google Scholar]
- 18. Yang M., Liu E., Tang L., Lei Y., Sun X., Hu J., Dong H., Yang S.M., Gao M., Tang B.. Emerging roles and regulation of MiT/TFE transcriptional factors. Cell Commun. Signal. 2018; 16:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Napolitano G., Ballabio A.. TFEB at a glance. J. Cell Sci. 2016; 129:2475–2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Slade L., Pulinilkunnil T.. The MiTF/TFE family of transcription factors: master regulators of organelle signaling, metabolism, and stress adaptation. Mol. Cancer Res. 2017; 15:1637–1643. [DOI] [PubMed] [Google Scholar]
- 21. Hemesath T.J., Steingrimsson E., McGill G., Hansen M.J., Vaught J., Hodgkinson C.A., Arnheiter H., Copeland N.G., Jenkins N.A., Fisher D.E.. microphthalmia, a critical factor in melanocyte development, defines a discrete transcription factor family. Genes Dev. 1994; 8:2770–2780. [DOI] [PubMed] [Google Scholar]
- 22. Pogenberg V., Ogmundsdottir M.H., Bergsteinsdottir K., Schepsky A., Phung B., Deineko V., Milewski M., Steingrimsson E., Wilmanns M.. Restricted leucine zipper dimerization and specificity of DNA recognition of the melanocyte master regulator MITF. Genes Dev. 2012; 26:2647–2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schmidt N.W., Grigoryan G., DeGrado W.F.. The accommodation index measures the perturbation associated with insertions and deletions in coiled-coils: Application to understand signaling in histidine kinases. Protein Sci. 2017; 26:414–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bohm G., Muhr R., Jaenicke R.. Quantitative analysis of protein far UV circular dichroism spectra by neural networks. Protein. Eng. 1992; 5:191–195. [DOI] [PubMed] [Google Scholar]
- 25. Leslie A.G.W. Jnt CCP4/ESF-EACBM. Newsl. Protein Crystallogr. 1992; 26:https://www.ccp4.ac.uk/newsletters/No26.pdf. [Google Scholar]
- 26. Battye T.G., Kontogiannis L., Johnson O., Powell H.R., Leslie A.G.. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D. Biol. Crystallogr. 2011; 67:271–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Evans P. Scaling and assessment of data quality. Acta Crystallogr. D. Biol. Crystallogr. 2006; 62:72–82. [DOI] [PubMed] [Google Scholar]
- 28. Storoni L.C., McCoy A.J., Read R.J.. Likelihood-enhanced fast rotation functions. Acta Crystallogr. D. Biol. Crystallogr. 2004; 60:432–438. [DOI] [PubMed] [Google Scholar]
- 29. Emsley P., Lohkamp B., Scott W.G., Cowtan K.. Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W et al.. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Krissinel E., Henrick K.. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007; 372:774–797. [DOI] [PubMed] [Google Scholar]
- 32. Strelkov S.V., Burkhard P.. Analysis of alpha-helical coiled coils with the program TWISTER reveals a structural mechanism for stutter compensation. J. Struct. Biol. 2002; 137:54–64. [DOI] [PubMed] [Google Scholar]
- 33. Mosca R., Schneider T.R.. RAPIDO: a web server for the alignment of protein structures in the presence of conformational changes. Nucleic Acids Res. 2008; 36:W42–W46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schwede T., Kopp J., Guex N., Peitsch M.C.. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 2003; 31:3381–3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Williams C.J., Headd J.J., Moriarty N.W., Prisant M.G., Videau L.L., Deis L.N., Verma V., Keedy D.A., Hintze B.J., Chen V.B. et al.. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 2018; 27:293–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wiederstein M., Gruber M., Frank K., Melo F., Sippl M.J.. Structure-based characterization of multiprotein complexes. Structure. 2014; 22:1063–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zal T., Gascoigne N.R.. Photobleaching-corrected FRET efficiency imaging of live cells. Biophys. J. 2004; 86:3923–3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Derler I., Hofbauer M., Kahr H., Fritsch R., Muik M., Kepplinger K., Hack M.E., Moritz S., Schindl R., Groschner K. et al.. Dynamic but not constitutive association of calmodulin with rat TRPV6 channels enables fine tuning of Ca2+-dependent inactivation. J. Physiol. 2006; 577:31–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kukenshoner T., Wohlwend D., Niemoller C., Dondapati P., Speck J., Adeniran A.V., Nieth A., Gerhardt S., Einsle O., Muller K.M. et al.. Improving coiled coil stability while maintaining specificity by a bacterial hitchhiker selection system. J. Struct. Biol. 2014; 186:335–348. [DOI] [PubMed] [Google Scholar]
- 40. Moller K., Sigurbjornsdottir S., Arnthorsson A.O., Pogenberg V., Dilshat R., Fock V., Brynjolfsdottir S.H., Bindesboll C., Bessadottir M., Ogmundsdottir H.M. et al.. MITF has a central role in regulating starvation-induced autophagy in melanoma. Sci. Rep. 2019; 9:1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Karshikoff A., Nilsson L., Ladenstein R.. Rigidity versus flexibility: the dilemma of understanding protein thermal stability. FEBS J. 2015; 282:3899–3917. [DOI] [PubMed] [Google Scholar]
- 42. Bondos S.E., Swint-Kruse L., Matthews K.S.. Flexibility and disorder in gene regulation: LacI/GalR and Hox proteins. J. Biol. Chem. 2015; 290:24669–24677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Garton M., MacKinnon S.S., Malevanets A., Wodak S.J.. Interplay of self-association and conformational flexibility in regulating protein function. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2018; 373:20170190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Allemann R.K., Egli M.. DNA recognition and bending. Chem. Biol. 1997; 4:643–650. [DOI] [PubMed] [Google Scholar]
- 45. Fock V., Gudmundsson S.R., Gunnlaugsson H.O., Stefansson J.A., Ionasz V., Schepsky A., Viarigi J., Reynisson I.E., Pogenberg V., Wilmanns M. et al.. Subcellular localization and stability of MITF are modulated by the bHLH-Zip domain. Pigment Cell Melanoma Res. 2019; 32:41–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nair S.K., Burley S.K.. X-ray structures of Myc-Max and Mad-Max recognizing DNA. Molecular bases of regulation by proto-oncogenic transcription factors. Cell. 2003; 112:193–205. [DOI] [PubMed] [Google Scholar]
- 47. Brown J.H., Cohen C., Parry D.A.. Heptad breaks in alpha-helical coiled coils: stutters and stammers. Proteins. 1996; 26:134–145. [DOI] [PubMed] [Google Scholar]
- 48. Mason J.M., Arndt K.M.. Coiled coil domains: stability, specificity, and biological implications. ChemBioChem. 2004; 5:170–176. [DOI] [PubMed] [Google Scholar]
- 49. Christie M., Boland A., Huntzinger E., Weichenrieder O., Izaurralde E.. Structure of the PAN3 pseudokinase reveals the basis for interactions with the PAN2 deadenylase and the GW182 proteins. Mol. Cell. 2013; 51:360–373. [DOI] [PubMed] [Google Scholar]
- 50. Dames S.A., Schonichen A., Schulte A., Barboric M., Peterlin B.M., Grzesiek S., Geyer M.. Structure of the cyclin t binding domain of hexim1 and molecular basis for its recognition of P-TEFb. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:14312–14317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bigalke J.M., Dames S.A., Blankenfeldt W., Grzesiek S., Geyer M.. Structure and dynamics of a stabilized coiled-coil domain in the P-TEFb regulator Hexim1. J. Mol. Biol. 2011; 414:639–653. [DOI] [PubMed] [Google Scholar]
- 52. Schonichen A., Bigalke J.M., Urbanke C., Grzesiek S., Dames S.A., Geyer M.. A flexible bipartite coiled coil structure is required for the interaction of Hexim1 with the P-TEFB subunit cyclin T1. Biochemistry. 2010; 49:3083–3091. [DOI] [PubMed] [Google Scholar]
- 53. Reiter F., Wienerroither S., Stark A.. Combinatorial function of transcription factors and cofactors. Curr. Opin. Genet. Dev. 2017; 43:73–81. [DOI] [PubMed] [Google Scholar]
- 54. Sieweke M.H., Graf T.. A transcription factor party during blood cell differentiation. Curr. Opin. Genet. Dev. 1998; 8:545–551. [DOI] [PubMed] [Google Scholar]
- 55. Glover J.N., Harrison S.C.. Crystal structure of the heterodimeric bZIP transcription factor c-Fos-c-Jun bound to DNA. Nature. 1995; 373:257–261. [DOI] [PubMed] [Google Scholar]
- 56. Kerppola T.K., Curran T.. Fos-Jun heterodimers and Jun homodimers bend DNA in opposite orientations: implications for transcription factor cooperativity. Cell. 1991; 66:317–326. [DOI] [PubMed] [Google Scholar]
- 57. Murre C., Bain G., van Dijk M.A., Engel I., Furnari B.A., Massari M.E., Matthews J.R., Quong M.W., Rivera R.R., Stuiver M.H.. Structure and function of helix-loop-helix proteins. Biochim. Biophys. Acta. 1994; 1218:129–135. [DOI] [PubMed] [Google Scholar]
- 58. Steingrimsson E., Copeland N.G., Jenkins N.A.. Melanocytes and the microphthalmia transcription factor network. Annu. Rev. Genet. 2004; 38:365–411. [DOI] [PubMed] [Google Scholar]
- 59. Grill C., Bergsteinsdottir K., Ogmundsdottir M.H., Pogenberg V., Schepsky A., Wilmanns M., Pingault V., Steingrimsson E.. MITF mutations associated with pigment deficiency syndromes and melanoma have different effects on protein function. Hum. Mol. Genet. 2013; 22:4357–4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Goding C.R., Arnheiter H.. MITF-the first 25 years. Genes Dev. 2019; 33:983–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates and structure factors of MITF(Δ) have been deposited in the PDB with accession code 6FX5.