Abstract
In all organisms, a selected type of proteins accomplishes critical roles in cellular processes that govern gene expression. The multifunctional protein Gemin5 cooperates in translation control and ribosome binding, besides acting as the RNA-binding protein of the survival of motor neuron (SMN) complex. While these functions reside on distinct domains located at each end of the protein, the structure and function of the middle region remained unknown. Here, we solved the crystal structure of an extended tetratricopeptide (TPR)-like domain in human Gemin5 that self-assembles into a previously unknown canoe-shaped dimer. We further show that the dimerization module is functional in living cells driving the interaction between the viral-induced cleavage fragment p85 and the full-length Gemin5, which anchors splicing and translation members. Disruption of the dimerization surface by a point mutation in the TPR-like domain prevents this interaction and also abrogates translation enhancement induced by p85. The characterization of this unanticipated dimerization domain provides the structural basis for a role of the middle region of Gemin5 as a central hub for protein-protein interactions.
INTRODUCTION
RNA-binding proteins (RBPs) play a pivotal role in gene expression control and cell homeostasis (1). Generally, RBPs comprise RNA-binding domains (RBD) and protein-protein interaction modules (2–5), such that the combination of distinct domains provides multiple features to these factors. Gemin5 is a predominantly cytoplasmic RBP that forms part of the survival of motor neuron (SMN) complex in metazoan organisms (6,7). This multi-protein complex plays a critical role in the biogenesis of small nuclear ribonucleoproteins (snRNPs) (8), the components of the splicing machinery. However, Gemin5 is mainly found outside of the SMN complex (9), suggesting that it may have additional functions. In agreement with this view, Gemin5 acts as a scaffold protein, serving as a hub for distinct ribonucleoprotein (RNP) networks. Indeed, Gemin5 has been identified as a down-regulator of translation (10–12), and as a ribosome-interacting factor (13,14).
RBPs perform critical functions in all organisms, including viruses. However, viruses are obliged pathogens with reduced coding capacity and thus, have developed various strategies to subvert essential host factors into their own benefit, which include the proteolysis of specific RBPs and initiation factors (eIFs) (15). In particular, RNA viruses exemplified by picornaviruses, modify host factors to promote translation of the viral RNA using cap-independent mechanisms governed by Internal Ribosome Entry Site (IRES) elements (16), evading the inhibition of cap-dependent translation occurring in infected cells. Consistent with its role in key cellular processes, Gemin5 is proteolytically cleaved in picornavirus-infected cells, producing a polypeptide of 85 kDa (thereafter p85) (Figure 1A) (17). Importantly, contrary to the negative effect of Gemin5 in translation (10), expression of p85 in human cells stimulates IRES-driven translation (18).
Figure 1.
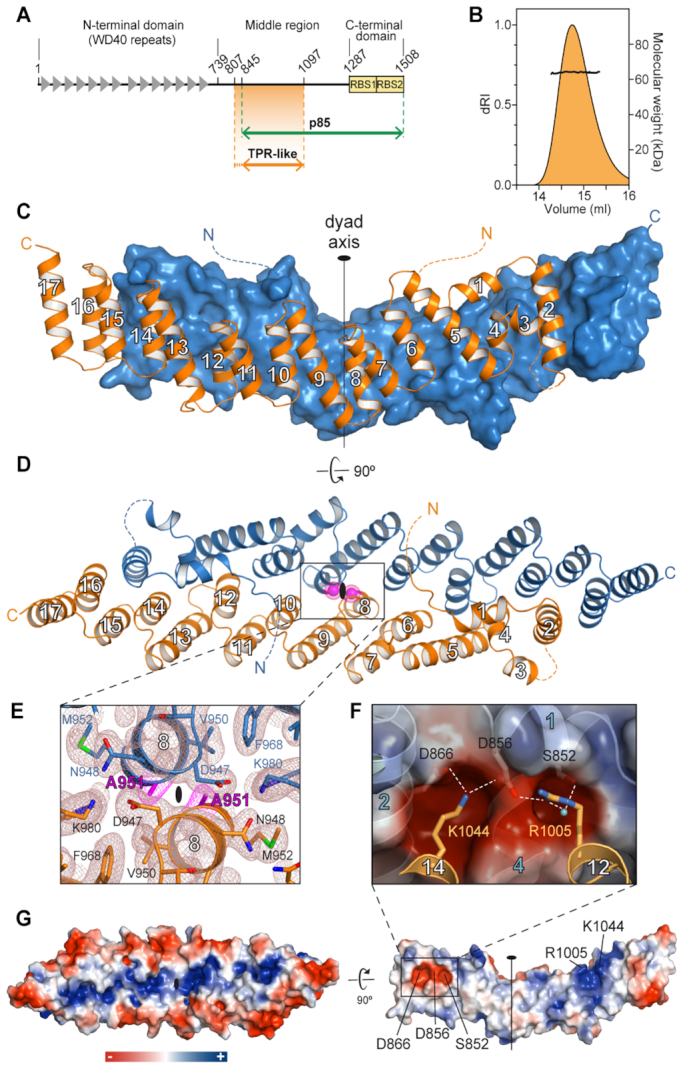
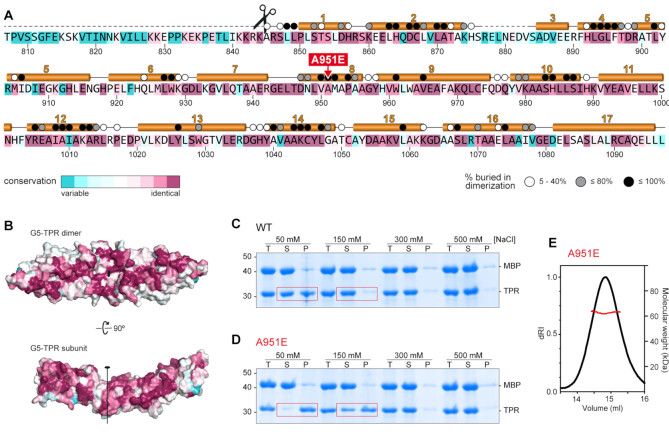
Crystal structure of Gemin5 TPR-like dimerization module. (A) Scheme of Gemin5 protein. WD40 repeats and RBS domains are indicated with gray triangles and yellow boxes, respectively. The p85 fragment and the TPR-like domain are indicated with green and orange arrows, respectively. (B) SEC-MALS analysis proves that G5-TPR is a dimer in solution, with a molecular weight of 64 kDa (±0.01%). (C) Structure of G5-TPR dimer with the subunit at front depicted in orange cartoon and the subunit at the back shown in blue surface representation. (D) Perpendicular view of (C) with both subunits represented in cartoon. The position of residues A951 in both subunits across the dyad axis are represented with magenta spheres. Dashed lines indicate regions not seen in the electron density maps. (E) Detail of the intersubunit interactions across the dyad axis and localization of residue A951. The 2Fobs− Fcalc electron density map at 1σ is shown in brown mesh. Residue A951 is highlighted in magenta. (F) Electrostatic interactions between residues D866 and D856 in one subunit, and R1005 and K1044 in the other subunit. A water molecule bridging the side chains of R1005 and S852 is shown as a cyan sphere. (G) Surface representation of the protein dimer (left) and of the dimerization interface of one of the subunits (right), colored according to the electrostatic potential. Blue and red contours begin at +2.5 and −2.5 kT, respectively.
Gemin5 contains two distinct functional regions at the protein ends (Figure 1A). The N-terminal part (amino acids 1–739) is composed of two juxtaposed seven-bladed WD40 domains (19) that recognize the Sm-site of small nuclear RNAs (snRNAs) and the m7G cap via base-specific interactions (20,21). The C-terminal part (amino acids 1287–1508) on the other hand, harbors a non-canonical RNA-binding site (RBS), with two moieties designated as RBS1 and RBS2 (18), which differ in RNA-binding capacity as well as in the ability to modulate selective translation. NMR structural analysis of the RBS1 polypeptide revealed a mixture of conformations in solution (18), frequently found in unstructured protein domains (22). However, the biological relevance of the ∼550 amino acids bridging the two functional ends of Gemin5 remains elusive.
Here, to better understand the potential role of the middle region of Gemin5, we isolated a fragment of the protein spanning amino acids 807–1097 and demonstrated that it forms a stable homodimer in solution. The crystal structure revealed a compact elongated canoe-shaped dimer with each subunit folding into a tetratricopeptide repeat (TPR)-like domain. Based on the structure, we designed a mutation A951E that disrupts the formation of the dimer. We further proved by pull-down and mass spectrometry analysis that the cleavage product p85 wild-type (wt), but not the p85 bearing mutation A951E, interacts with Gemin5 in living cells. These data also revealed that dimerization driven by p85-wt recruits cellular proteins involved in RNA splicing and translation processes, in contrast to p85-A951E. Furthermore, while p85-wt stimulated IRES- and cap-dependent translation, p85-A951E failed to enhance translation in human cells. Together, our results uncover a crosstalk between the ability of the dimerization module to associate with the full-length protein and the capacity of p85 to modulate translation.
MATERIALS AND METHODS
Protein engineering for structural and functional studies
The bioinformatic servers HHPred (23), Phyre2 (24) and I-TASSER (25) were used to predict the folding of the entire Gemin5 protein and to determine new possible construct targets. The gene fragment covering the predicted middle domain (residues 807–1097) was amplified by PCR using Phusion High-Fidelity DNA Polymerase (New England Biolabs), using pcDNA3-Xpress-G5 as template (13). Specific reverse and forward oligonucleotides (Sigma) are detailed in Supplementary Table S1. Using the In-Fusion technology (Clontech), the gene fragment was inserted into the pOPINM vector (Oxford Protein Production Facility) that contains a MAH6SSG_MBP tag followed by a region cleavable by the PreScission protease.
The sequence encoding the p85 polypeptide present in pcDNA3-Xpress-G5 (18) was amplified by PCR with specific oligonucleotides (Supplementary Table S1) and transferred to pcDNA3-CTAP (26) via NotI–PacI to generate the construct pcDNA3-CTAP-p85. Construct pcDNA3-Xpress-p85ΔRBS1/2 was generated by PCR amplifying the sequence present in pcDNA3-Xpress-G5 with specific primers (Supplementary Table S1) and transferred to pcDNA3-Xpress via BamHI-NotI. Constructs pcDNA3-CTAP-p85A951E, pcDNA3-Xpress-p85A951E, pcDNA3-Xpress-p85ΔRBS1/2A951E and pcDNA3-Xpress-G5A951E were generated by QuickChange mutagenesis (Agilent Technologies) using specific primers (Supplementary Table S1). Construct pOPINM_TPR-A951E was performed using PCR on pcDNA3-Xpress-p85A951E and In-Fusion cloning. All plasmids were confirmed by DNA sequencing (Macrogen). The construct expressing RBS1 protein was previously described (12).
Expression and purification of G5-TPR
BL21 Rosetta(DE3)pLysS cells transformed with the pOPINM_G5-TPR plasmid or the mutated variant pOPINM_G5-TPR-A951E were grown at 37°C in LB petri dishes supplemented with 2% glucose, 35 μg/ml ampicillin and 15 μg/ml chloramphenicol. For protein expression, the cell culture growing to mid-exponential phase (OD600 = 0.6–0.8) was induced adding 0.5 mM isopropyl-d-thiogalactopyranoside (IPTG), overnight at 20°C. The pellet from 1 l of culture was resuspended in 40 ml of buffer A (20 mM Tris–HCl pH 8.0, 0.5 M NaCl, 10 mM imidazole, 5% glycerol and 2 mM β-mercaptoethanol) supplemented with 1 pill of cOmplete™ EDTA-free Protease Inhibitor cocktail (Sigma-Aldrich). Cells were sonicated and the lysate was clarified by centrifugation in a Beckman JA-25.50 rotor at 50 000 × g for 1 h at 4°C. The supernatant was filtered (0.45 μm pore) and loaded onto a 5 ml HisTrap FF column (GE Healthcare, USA). After extensively washing with buffer A supplemented with 35 mM imidazole, the protein was eluted increasing the imidazole concentration to 250 mM. G5-TPR wild-type was cleaved overnight with GST-tagged PreScission protease (in a relation of 1/20th of the protein weight) and dialyzed against buffer B50 (20 mM Tris–HCl pH 6.8, 50 mM NaCl, 1 mM DTT and 5% glycerol). Cleaved G5-TPR was loaded onto a 5 ml HiTrap S HP column (GE Healthcare, USA). Whereas the MAH6SSG_MBP tag is recovered in the flow-through, G5-TPR wt is retained in the column and eluted by increasing the salt concentration at 150–200 mM NaCl. After concentration through an Amicon Ultra system (10 kDa cut-off), the sample was further purified by size-exclusion chromatography on a Superdex 200 Increase 10/300 column (GE Healthcare) pre-equilibrated in buffer B50. The G5-TPR A951E mutant eluted from the HisTrap FF column was digested with PreScission, dialyzed against buffer B500 (20 mM Tris–HCl pH 6.8, 0.5 M NaCl, 1 mM DTT and 5% glycerol), and loaded onto a second 5 ml HisTrap FF column. The protein collected in the flow-through was concentrated as before and further purified through a Superdex 200 increase 10/300 column equilibrated in buffer B500. The purified proteins were concentrated and used for further studies or supplemented with 20% glycerol, flash-frozen in liquid nitrogen and stored at −80°C. All purification steps were carried out at 4°C. Sample purity was evaluated by SDS-PAGE with Coomassie staining. Protein concentration was determined by Bradford assay.
Gel-filtration coupled to multi-angle light-scattering (SEC-MALS) measurements
300 μl of purified G5-TPR at 3 mg ml−1 was fractionated by gel filtration on a Superdex 200 10/300 column equilibrated in buffer B50, using ÄKTA purifier at a flow rate of 0.5 ml min−1. The eluted sample was characterized by in-line measurement of the refractive index and multi-angle light scattering using Optilab T-rEX and DAWN 8+ instruments, respectively (Wyatt). Data were analyzed using the ASTRA 6 software (Wyatt) to obtain the molar mass. Similar experiments were performed with the G5-TPR A951E mutant at concentrations of 5 and 0.65 mg ml−1 using the buffer B500 with increased salt concentration.
Crystallization
Initial crystallization screenings were performed at room temperature with drops of 0.7 μl protein solution at 4.8 mg ml−1 plus 0.7 μl reservoir solution equilibrated against 60 μl of reservoir solution from JCSG+, PACT, MPD suite (Qiagen) and Crystal Screen (Hampton Research) commercial screens. Initial hits were further optimized in MRC 48-well sitting-drop plates (Molecular Dimensions). Best-diffracting plate-shaped crystals appeared after 3–5 days in 200 mM Na/K Tartrate, 25% PEG 3350 and 100 mM Bis–Tris–methane pH 8.5. Other plate crystals grew in 200 mM NaI, 25% PEG 3350 and 100 mM Bis–Tris–methane pH 8.5. In both cases, cryo-protection was reached by directly soaking the crystals in a solution containing the mother liquor supplemented with 20% glycerol. Crystals were then fished with cryo-loops and flash-cooled in liquid nitrogen.
Data collection and structure determination
X-ray diffraction data were remotely collected at BL13-XALOC (ALBA synchrotron, Barcelona) using a Pilatus 6M detector. For each set, a total wedge of 180° of data was collected with 0.15° oscillation and 0.1 s exposure per frame. Data processing and scaling were performed with XDS (27). Taking advantage of the anomalous edge scattering of the iodine present in one of the crystallization conditions, crystallographic phases were determined by SAD using a wavelength of λ = 2.03 Å, where f″ for iodide is 10.5 e− (6.1 keV; http://skuld.bmsc.washington.edu/scatter/). SAD phases were calculated with AutoSol in Phenix (28), which identified 13 iodide sites. Initial model building using a poly-alanine sequence was performed with Autobuild and refined by iterative cycles with Phenix and Coot (29). Suitable quality of the final models was validated according to MolProbity (30). Analysis of the macromolecular interfaces was performed with PDBePISA (31). Sequence alignment and surface conservation representation were performed with Treefam (family TF328886), Ensembl (Genetree ENSGT00620000088064) and Consurf (32). Electrostatic surface potential was calculated with APBS plug-in of PyMOL and figures were prepared with PyMOL.
RNA–DNA electrophoretic mobility shift assay
RNA probes were uniformly labeled using α32P-CTP (500 Ci/mmol), T7 RNA polymerase (10 U), and linearized DNA (1 μg), as described (33). Constructs expressing RNAs corresponding to domain 5 of the FMDV IRES or its single stranded region, the long structured RNA and the short hairpin, have been described (12,34). RNA was purified through MicroSpin G-25 Columns (GE Healthcare), ethanol precipitated and resuspended in TE (Tris 10 mM, pH 8, EDTA 1 mM) to a final concentration of 0.04 pmol/μl. ssDNA and dsDNA probes (Supplementary Table S1) were 5′-end labeled using T4 polynucleotide kinase and γ32P-ATP (500 Ci/mmol). RNA integrity and DNA probes were analyzed in 6% acrylamide 7 M urea denaturing gel electrophoresis.
RNA-binding and DNA-binding protein reactions were carried out as described (12) with small modifications. The reactions were carried out in 10 μl of RNA-binding buffer [40 mM Tris–HCl pH 7.5, 250 mM NaCl, 0.1% (w/v) β-mercaptoethanol] for 15 min at room temperature, using serial increased concentration of protein. Electrophoresis was performed in non-denaturing 6.0% (29:1) polyacrylamide gels at 4°C, run in TBE buffer.
Protein complexes isolation by tandem affinity purification (TAP)
HEK293 cells (4× P100), grown in Dulbecco's Modified Eagle Medium (DMEM), transfected with the plasmids expressing p85-wt-TAP or p85-A951E-TAP proteins, were harvested 24 h post-transfection. The complexes associated to the TAP-tagged constructs were purified as described (33). Briefly, the supernatant of the first IgG Sepharose purification was subsequently subjected to a second Calmodulin (Agilent Technologies) purification step. Purified proteins were precipitated with 10% trichloroacetic acid at 4°C overnight, pelleted at 14 000 g for 15 min at 4°C, washed three times with 1 ml of acetone and dissolved in SDS-loading buffer. An aliquot (25%) was analyzed on silver stained SDS-PAGE gels to visualize the purification of proteins associated to Gemin5 p85-wt-TAP or p85-A951E-TAP polypeptides. Immunodetection of Gemin5 and p85 was performed using anti-Gemin5 (Novus) antibody.
In-gel digestion and mass spectrometry analysis
Two independent biological replicates of TAP samples obtained for p85-wt-TAP and p85-A951E-TAP, were applied onto a 10% SDS-PAGE gel. The protein bands concentrated in the stacking/resolving gel interface were visualized by Coomassie staining. The gel pieces were destained in acetonitrile:water (ACN:H2O, 1:1), were reduced and alkylated, and then digested in situ with sequencing grade trypsin (Promega, Madison, WI, USA), as described (35). The gel pieces were dried and re-swollen in 50 mM ammonium bicarbonate pH 8.8 with 60 ng/μl trypsin at 5:1 protein:trypsin (w/w) ratio. The tubes were kept in ice for 2 h and incubated at 37°C for 12 h. Digestion was stopped by the addition of 1% TFA. The desalted protein digest was dried, resuspended in 10 μl of 0.1% formic acid and analyzed by RP-LC–MS/MS in an Easy-nLC II system coupled to an ion trap LTQ-Orbitrap-Velos-Pro hybrid mass spectrometer (Thermo Scientific). The peptides were concentrated (on-line) by reverse phase chromatography using a 0.1 mm × 20 mm C18 RP precolumn (Thermo Scientific), and then separated using a 0.075 mm × 250 mm C18 RP column (Thermo Scientific) operating at 0.3 μl/min. Peptides were eluted using a 120 min dual gradient from 5 to 25% solvent B in 90 min followed by gradient from 25 to 40% solvent B over 120 min (Solvent A: 0.1% formic acid in water, solvent B: 0.1% formic acid, 80% acetonitrile in water). ESI ionization was done using a Nano-bore emitters Stainless Steel ID 30 μm (Proxeon) interface. The Orbitrap resolution was set at 30.000. Peptides were detected in survey scans from 400 to 1600 amu (1 μscan), followed by 20 data dependent MS/MS scans (Top 20), using an isolation width of 2 u (in mass-to-charge ratio units), normalized collision energy of 35%, and dynamic exclusion applied during 30 s periods.
Peptide identification from raw data was carried out using PEAKS Studio X (36) search engine (Bioinformatics Solutions Inc). Database search was performed against UniProt-Homo sapiens FASTA (decoy-fusion database). The following constraints were used for the searches: tryptic cleavage after Arg and Lys, up to two missed cleavage sites, and tolerances of 20 ppm for precursor ions and 0.6 Da for MS/MS fragment ions and the searches were performed allowing optional Met oxidation and Cys carbamidomethylation. False discovery rates (FDR) for peptide spectrum matches (PSM) were limited to 0.01. Only those proteins with at least two distinct peptides being discovered from LC/MS/MS analyses were considered reliably identified.
GO analysis
Gene Ontology analyses were performed by the DAVID database (https://david.ncifcrf.gov) on the overlapping proteins identified. The significantly enriched biological processes (BP) were identified using as a cutoff criteria P value <10−5 and a gene count ≥3.
Gemin5 polypeptides expression and luciferase activity assays
HEK293 cell monolayers (2 × 105) were cotransfected with a plasmid expressing luciferase in cap-dependent or IRES-dependent manner (pCAP-luc, pIRES-luc) (37), and a plasmid expressing Xpress-p85, Xpress-p85-A951E, Xpress-p85ΔRBS1/2, Xpress-p85ΔRBS1/2-A951E, or the corresponding empty vector side by side using Lipofectamine LTX (Thermo Scientific). Cell lysates were prepared 24 h post-transfection in 100 μl lysis buffer (50 mM Tris-HCl pH 7.8, 100 mM NaCl, 0.5% NP40). The protein concentration in the lysate was determined by Bradford assay. Equal amounts of protein were loaded in SDS-PAGE and processed for western blotting to determine the expression of the polypeptides using anti-Xpress (Invitrogen) antibodies. Immunodetection of tubulin (Sigma) was used as loading control. Secondary antibodies (Thermo Scientific) were used according to the manufacturer's instructions. The signal detected was done in the linear range of the antibodies.
Luciferase activity (RLU)/μg of total protein was internally normalized to the value obtained with the empty vector performed side by side. Each experiment was repeated independently three times. Values represent the mean ± SEM. The steady-state levels of luciferase RNA in control cells and in cells expressing Xpress-p85-wt or Xpress-p85-A951E were verified by RTqPCR as described (12). We computed P values for a difference in distribution between two samples with the unpaired two-tailed Student's t-test. Differences were considered significant when P < 0.05. The resulting P values were graphically illustrated in figures with asterisks as described in figure legends.
RESULTS
The middle region of Gemin5 bears a novel dimerization domain
A comprehensive bioinformatic analysis of the middle region of Gemin5 predicted the existence of an α-helix rich domain comprising residues 807–1097 with highest similitude to the tetratricopeptide repeat (TPR)-like domain of the elongator complex protein 1 (Elp1) (38). Guided by the predictions, we produced in bacteria a construct covering the putative TPR-like region of human Gemin5 (residues 807–1097) (Figure 1A) fused at the N-terminus to a cleavable polyhistidine-tagged maltose binding protein (MBP). The protein without tag and purified to homogeneity was highly soluble and the analysis by size-exclusion chromatography coupled to multi-angle light scattering (SEC-MALS) indicated a molecular mass of 64 kDa (Figure 1B). This is twice the expected 32 kDa mass for a single copy of the protein, indicating that Gemin5 residues 807–1097 form a stable homodimer in solution.
Diffraction quality crystals grown in presence of sodium iodide allowed solving the structure of the protein at 2.7 Å resolution using single-wavelength anomalous dispersion (SAD) (Table 1). The protein model was used for phasing by molecular replacement a second data set collected up to 2 Å resolution from an isomorphous crystal grown with tartrate instead of iodide (Table 1). The crystal structure revealed a dimer formed by two molecules of the protein related by a twofold non-crystallographic symmetry (Figure 1C–E). The two subunits in the dimer are similar (Supplementary Figure S1), and consist of 17 α-helices forming a flat and elongated right-handed α-solenoid with a 40° angle kink between helices α1–8 on one side and helices α9–17 on the other (Figure 1C and D). This helical palisade is nucleated by six antiparallel double-helix repeats (helices α5–6, α7–8, α9–10, α11–12, α13–14 and α15–16) with structural and sequence similarity to the classical PP5 TPR motif (39) (Supplementary Figure S2). A C-terminal ‘capping’ helix (α17) (Figure 1C and D), present in most TPR domains (40), covers the hydrophobic surface of the last helical repeat at one end of the subunit. At the other end, the structure is completed by two additional N-terminal helical pairs: helix α1 lays perpendicular on ‘top’ of the palisade and makes a 90° angle with helix α2, which connects through a disordered region (residues 877–883) to a pair of short α-helices, α3 and α4, also related by a 90° angle (Figure 1C and D). There was no electron density attributable to the N-terminal residues 807–844, and thus, these 38 amino acids were not included in the final models. Indeed, we noticed that the protein reproducibly experimented a proteolytic degradation, yielding a ∼28 kDa product (Supplementary Figure S3) that agrees in size with the crystal model, suggesting that the N-terminal sequence is cleaved off during crystallization. Interestingly, the cleavage site coincides with the 843RKAR846 motif used by the L protease to release the p85 polypeptide during foot-and-mouth disease virus (FMDV) infection (Figure 1A) (17).
Table 1.
Data collection and refinement statistics
| G5-TPR (SAD) | G5-TPR (native) | |
|---|---|---|
| PDB ID | 6RNS | 6RNQ |
| Data Collection | ||
| Wavelength (Å) | 2.0330 | 0.9792 |
| Space group | P21 | P21 |
| Cell dimensions | ||
| a, b, c (Å) | 67.8, 54.6, 103.0 | 64.2, 55.0, 103.6 |
| α, β, γ (°) | 90, 108.8, 90 | 90, 104.3, 90 |
| Resolution (Å) | 97.52–2.89 (3.07–2.89) | 100.43–2.13 (2.19–2.13) |
| Total reflections | 51 512 (8372) | 131 796 (11 040) |
| Unique reflections | 16 049 (2604) | 39 306 (3208) |
| R meas | 0.126 (0.719) | 0.066 (0.542) |
| Mean I/σI | 7.5 (1.8) | 6.6 (2.0) |
| Completeness (%) | 98.7 (99.7) | 99.5 (99.8) |
| Redundancy | 3.2 (3.2) | 3.4 (3.4) |
| CC1/2 | 0.99 (0.91) | 1.00 (0.93) |
| Refinement | ||
| Resolution (Å) | 48.76–2.89 | 100.4–2.13 |
| No. of reflections used | 15 962 | 39 227 |
| R work/Rfree | 0.23/0.26 | 0.20/0.22 |
| Rmsd | ||
| Bond lengths (Å) | 0.004 | 0.011 |
| Bond angles (°) | 0.84 | 1.13 |
| No. of atoms | ||
| Protein | 3805 | 3894 |
| Ligand | 13 | 1 |
| Water | 26 | 74 |
| MolProbity analysis | ||
| Ramachandran favored | 96.24% | 97.72% |
| Ramachandran outliers | 0.00% | 0.00% |
| Rotamer outliers | 0.78% | 0.26% |
| Clashscore | 2.27 | 1.44 |
| C-beta outliers | 0 | 0 |
| overall score | 1.26 | 0.94 |
Values in parenthesis correspond to the highest resolution shell.
The two protein subunits are confronted in antiparallel orientation, forming an elongated canoe-shaped homodimer approximately 120 Å long × 35 Å wide, with helices α16 and α17 from one subunit overtaking the other subunit at the stern and bow ends (Figure 1C and D). The dimer is formed by the intertwined α-helices in the second position of the helical repeats, with contributions of side chains from the helices in the first position of the repeats, and also by the N-terminal helices α1, α2 and α4 (Figure 1D). The closest intersubunit distance occurs between the two A951 residues at helices α8 flanking the molecular dyad axis (Figure 1D and E). Overall, the dimerization interface buries ∼3400 Å2 per subunit as computed by PISA (31), which accounts for 24% of the total protein surface and suggests the formation of a stable dimer. This tight association occurs mostly through hydrophobic interactions but also includes thirteen hydrogen bonds and two pairs of salt bridges between patches of complementary charge formed by residues R1005 and K1044 (both in α12) in one subunit and D856 (α1) and D866 (α2) in the other (Figure 1F and Supplementary Tables S2 and S3). The solvent exposed area is hydrophilic, with a positively charged narrow groove on ‘top’ of the dimer surface delimited by a rim of acidic residues (Figure 1G).
A search with the DALI server (41) confirmed the structural similarity between the TPR-like domain of Gemin5 (hereafter named as G5-TPR) and other TPR-containing proteins, particularly with Elp1 (38), showing highest Z-score of 11.1 and RMSD of 5.2 Å for 200 Cα positions (Supplementary Figure S4A and B).
Mutation of a conserved residue at the dyad axis abrogates protein dimerization
The alignment of 145 Gemin5 sequences from vertebrates shows an identity in the TPR-like region of 55%, which increases to 86% within mammals (Figure 2A and Supplementary Figure S5). The most conserved residues are involved in interactions within the subunit and across the dimer interface (Figure 2B and C). These results strongly suggest that the dimerization module is an evolutionary preserved feature of Gemin5.
Figure 2.
The conserved dimerization module in Gemin5 is destabilized by mutation A951E. (A) Gemin5 TPR-like sequence colored according to the conservation, from magenta (identity) to cyan (variable), based on the alignment of 145 Gemin5 sequences. The α-helices are depicted above the sequence, and missing regions in the model are indicated with dashed lines. The scissors mark the L protease cleavage motif. Residues buried up to 40%, 80% or 100% in the dimerization interface are denoted respectively with a white, grey or black dot on top. The substitution of residue A951 by glutamate is indicated in red. (B) Sequence conservation plotted on the surface of the protein dimer and on the dimerization interface. The molecules are represented in the same orientation as in Figure 1G. (C, D) Solubility of the MBP-cleaved G5-TPR and A951E variant monitored by SDS-PAGE in conditions with different concentrations of NaCl, showing the total (T), soluble (S) and precipitated (P) fractions. Red boxes highlight the decreased solubility of mutant A951E at lower salt concentrations. (E) SEC-MALS analysis proves that under conditions with 0.5 M NaCl, the A951E mutant forms dimers with a molecular weight of 63 kDa (±0.05%).
To further study the functionality of this association, we attempted to disrupt the dimerization of the G5-TPR replacing by a glutamate the alanine residue A951 at the central and closest intersubunit distance between both subunits (Figures 1D, E and 2A). Despite the extensive dimerization surface, we expected that the bulkier and negatively charged side chains of two confronted glutamates would repel the two subunits due to steric clashes and charge repulsion. The G5-TPR A951E variant was successfully expressed in bacteria, but removal of the N-terminal MBP tag caused the protein to precipitate heavily under conditions with 50 or 150 mM NaCl in which G5-TPR wt is soluble up to 15 mg ml−1 (Figure 2C, D and Supplementary Figure S6). However, in conditions with higher concentration of salt (300–500 mM NaCl) the mutated protein was soluble (Figure 2C, D) and behaved as a dimer in solution, as shown by SEC-MALS analysis (Figure 2E).
These results indicate that mutation A951E weakens the dimerization of G5-TPR and that the isolated subunit has decreased solubility. Moreover, the insertion of the glutamate does not seemingly distort the folding of the mutated protein, which behaves as a stable dimer in conditions of higher ionic strength that must enhance the hydrophobic interactions between subunits.
The Gemin5 dimerization domain lacks RNA-binding ability
Since the functions reported for Gemin5 are directly connected to RNA-dependent pathways (13,42), we sought to investigate whether or not the dimerization module interacts with RNA. The RNA binding capacity of the purified G5-TPR was assessed in parallel to the RBS1 domain of Gemin5, previously shown to interact with domain 5 (d5) of the FMDV IRES (18), as well as to recognize multiple RNA targets in human cells (12). As shown in Figure 3A, G5-TPR was unable to form a retarded complex with d5 RNA, even at high protein concentration (1 μM), while RBS1 exhibited a robust RNA-binding affinity. To reinforce this result, we analyzed the capacity of the G5-TPR to interact with RNAs differing in sequence and secondary structure. All these RNAs (Figure 3B–D) [a hairpin of 22 nt, a 170 nt RNA adopting a complex stem-loop structure (12) and a single stranded RNA of 32 nt (34)] failed to form a retarded complex with the purified protein at high concentrations (1 μM). Furthermore, gel-shift assays carried out with ssDNA (29 nt) or dsDNA (29 bp) indicated that G5-TPR (up to 4.3 μM) does not recognize DNA (Figure 3D and E). Therefore, we conclude that the Gemin5 dimerization domain does not promote interaction with nucleic acids in vitro.
Figure 3.
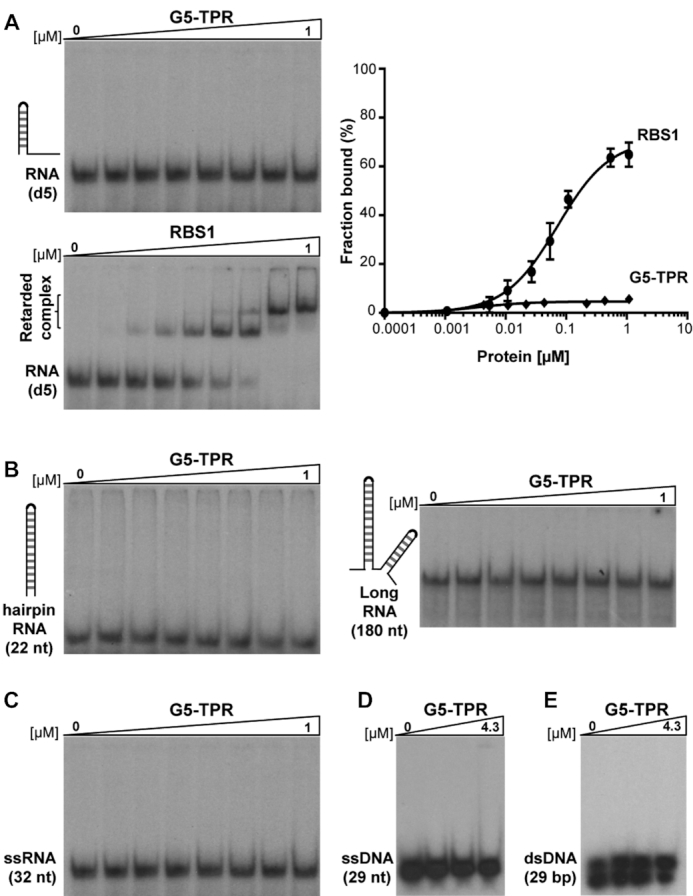
G5-TPR lacks nucleic acids binding capacity. (A) Representative example of a gel-shift assay conducted with increasing amounts of purified G5-TPR (0 to 1 μM) and labeled domain 5 (d5) RNA. Purified RBS1 (0 to 1 μM) was used as a positive control of the RNA-binding assay. The percentage of retarded complexes obtained in three independent assays is plotted on the right panel. Values represent the mean ± SEM. (B, C) RNA-binding assays performed with G5-TPR and various probes: a 22 nt hairpin RNA, a 170 nt RNA predicted to fold in several stem-loops and a single stranded 32 nt RNA. (D, E) Lack of retarded complex formation of G5-TPR with ssDNA (29 nt) or ds DNA (29 bp).
The Gemin5 cleavage product p85, but not p85-A951E, recruits the full-length protein in living cells
The finding of a novel dimerization module in the middle region of Gemin5 prompted us to study the functional relevance of this protein-protein interaction. Previous data showed the immunodetection of the endogenous Gemin5 in TAP-pull-down protein complexes isolated with a Gemin5 fragment spanning residues 1–1287 (13), suggesting that Gemin5 could oligomerize in the cell cytoplasm. However, the protein region responsible for this association was unknown. On the other hand, we have shown that Gemin5 is proteolytically cleaved in infected cells generating the polypeptide p85 (17), which includes the dimerization module (Figure 1A). Hence, we posit that the physiological relevance of the dimerization module could be linked to p85 function.
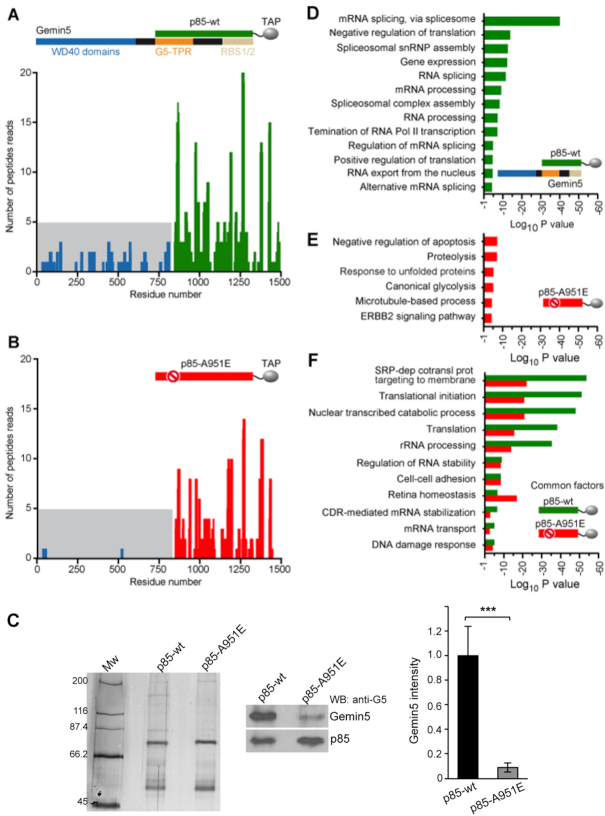
To determine whether p85 associates with full-length Gemin5 through the dimerization module in a cellular environment, we performed mass spectrometry analysis of proteins copurifying with p85-wt-TAP expressed in HEK293 cells (Dataset 1). Gemin5 was unequivocally identified in two replicates [score 375.86 and 344.37, coverage 55% and 42%, respectively]. Besides numerous peptides from the p85 region, analysis of Gemin5 amino acid sequences revealed the presence of trypsin fragments corresponding to residues 1–844, yielding a total of 33 peptides (Figure 4A and Supplementary Figure S7). Hence, these results show that the full-length Gemin5 copurifies with p85-wt-TAP, reflecting the formation of a stable complex with the endogenous protein.
Figure 4.
The p85 region of Gemin5 retrieves the endogenous Gemin5 in living cells. HEK293 cells were transfected with plasmids expressing p85-wt-TAP (A) or the mutant p85-A951E-TAP (B). Twenty-four hours later, cells were lysed, and the proteins copurifying with p85-wt or p85-A951E were identified by mass spectrometry. The number of peptide reads corresponding to Gemin5 determined in TAP samples of p85-wt (A; green bars) and p85-A951E (B; red bars) are represented according to their position. Peptide reads corresponding to Gemin5 residues 1–844 are colored in blue and highlighted in grey background. (C) Coimmunoprecipitation of Gemin5 with p85-wt. TAP samples obtained from HEK293 soluble cell extracts expressing p85-wt-TAP or p85-A951E-TAP were analyzed by silver staining of SDS-PAGE (left panel) to visualize the levels of purified TAP proteins. Coimmunoprecipitation of Gemin5 with p85 was then detected by western blot using anti-Gemin5 antibody (middle panel), and the results obtained in three independent assays (mean ± SEM) was represented (right panel). (D, E) Gene ontology (GO) classification obtained for the overlap of the pulldown replicates obtained with p85-wt only (D; green), or p85-A951E only (E; red). The top GO terms are represented according to P value; cut-off was set to 10−5. The legend depicts p85-Gemin5 association; p85-A951E fails to capture Gemin5. (F) GO terms shared by proteins p85-wt and p85-A951E.
To further investigate the biological relevance of the Gemin5 dimerization module we carried out mass spectrometry analysis of cellular proteins associated with p85-A951E-TAP mutant (Dataset 1). Analysis of the peptides copurifying with p85-A951E-TAP (score 330.38 and 292.0 for the biological replicate samples, similar to p85-wt-TAP) revealed only four peptides within the region 1–844 of Gemin5 (Figure 4B and Supplementary Figure S8), in contrast to the 33 peptides obtained for p85-wt-TAP. Hence, we conclude that the G5-TPR domain is necessary for the dimerization of Gemin5, and that mutation A951E disrupts the capacity of p85 to recruit the full-length protein in living cells.
Independent experimental data conducted to measure the intensity of co-immunoprecipitation of full-length Gemin5 protein in cells expressing p85-wt-TAP or the mutant p85-A951E-TAP, further confirmed that the recruitment of Gemin5 by p85-wt was 10-fold higher than that observed by p85-A951E (Figure 4C), reinforcing the data obtained by mass spectrometry.
The functional groups retrieved by p85 uncover the physiological relevance of Gemin5
Next, we sought to investigate the implication of protein-protein interactions on the functional groups copurifying with either p85-wt, which recruits the endogenous Gemin5 (Figure 4A), or p85-A951E, unable to do so (Figure 4B). The Gene Ontology (GO) annotation of the biological processes obtained with DAVID database (43) for the overlap of the biological replicates was classified according to P value for three separate groups: (i) proteins copurifying with p85-wt, which retrieves the full-length Gemin5 (Figure 4D), (ii) proteins exclusively associated with p85-A951E (Figure 4E) and (iii) proteins shared by p85-wt and p85-A951E, presumably interacting with residues 845–1508 (Figure 4F). The top GO functional groups copurifying exclusively with p85-wt are involved in mRNA splicing (10−40), negative regulation of translation (10−14), and snRNP assembly (10−13) (Figure 4D). These results are fully consistent with the functional properties reported for Gemin5 (44). This is followed by 10 additional GO groups related to RNA splicing, processing, and translation (gene expression, regulation of RNA splicing, mRNA processing, positive regulation of translation, and RNA export; P values ranging between 10−13 and 10−5).
In marked difference with p85-wt, none of the GO terms related to splicing and translation members were detected with the mutated p85-A951E protein (Figure 4E). In addition, the number of GO groups exclusively associated with the A951E mutant were fewer, and had lower statistical significance, although the protein was identified with similar score. Remarkably, p85-A951E copurified with members of the protein degradation pathway since the top GO terms are involved in negative regulation of apoptotic processes (10−7), proteolysis (10−7) and response to unfolded proteins (10−6), suggesting protein instability. None of these terms were observed with the p85-wt protein.
Besides the GO groups specific for the wt or the mutant version of p85, a large number of factors copurified with both proteins (Figure 4F). Thereby, these factors mostly interact with residues 845–1508 of Gemin5. GO terms shared by both p85-wt and p85-A951E do not involve RNA splicing, revealing distinct functional properties of the Gemin5 domain expressed in cells. According to P value, the top GO term was signal recognition particle (SRP)-dependent cotranslational protein targeting to membrane (P values for p85-wt and A951E 10−54 and 10−22, respectively), suggesting that Gemin5 interacts through the 845–1508 region with SRP. This result is in full agreement with a report showing that the Gemin5 alone was capable of interacting with SRP particle (45). The next interacting groups are involved in translation initiation (P value 10−52 wt, 10−21 A951E), nuclear transcribed metabolic processes, translation, and rRNA processing (P values ranging between 10−48 to 10−36 and 10−21 to 10−14 for wt and A951E, respectively), consistent with recently reported features of the protein (12,13,46). To our surprise, GO terms with significant P values are involved in regulation of RNA stability, cell-cell adhesion, retina homeostasis, coding region instability determinant (CRD)-mediated mRNA stabilization, mRNA transport and DNA damage response, suggesting the involvement of Gemin5 in still unknown functions.
In summary, there is a crosstalk between the ability of p85 to recruit Gemin5, driven by the dimerization module, and its capacity to associate with partners directly involved in RNA splicing, RNA processing, and translation control.
Disruption of the dimerization ability correlates with loss of translation stimulation by p85
We and others have shown that Gemin5 is involved in translation control (10–12). However, while the full-length protein down-regulates translation, the p85 product observed in infected cells enhances IRES-dependent translation (18). To determine the effect of p85-driven dimerization on translation control we carried out functional assays in HEK293 cells co-expressing Xpress-tagged p85 and luciferase downstream of the FMDV IRES (Figure 5A). These data revealed that expression of Xpress-p85-wt concurred with IRES-dependent stimulation of luciferase activity (Figure 5B) relative to control cells. In contrast, similar levels of expression of the Xpress-p85-A951E construct failed to do so (Figure 5B). Likewise, the stimulation of cap-dependent translation observed in p85-wt was not detected in the mutant p85-A951E (Figure 5B). In both cases, no significant differences in the steady-state levels of luciferase RNA were observed by RTqPCR at the time of cell harvesting (Supplementary Figure S9). Moreover, we used a p85 construct deleting the C-terminal region up to amino acid 1287 (Figure 5A) to determine the influence of the dimerization domain in translation control. Expression of Xpress-p85ΔRBS1/2 resulted in stimulatory effect on IRES- and cap-dependent translation regulation (Figure 5C), while the mutant Xpress-p85ΔRBS1/2A951E had no effect relative to control cells. Thus, we conclude that the A951E mutation disrupting the dimerization domain affects translation stimulation.
Figure 5.
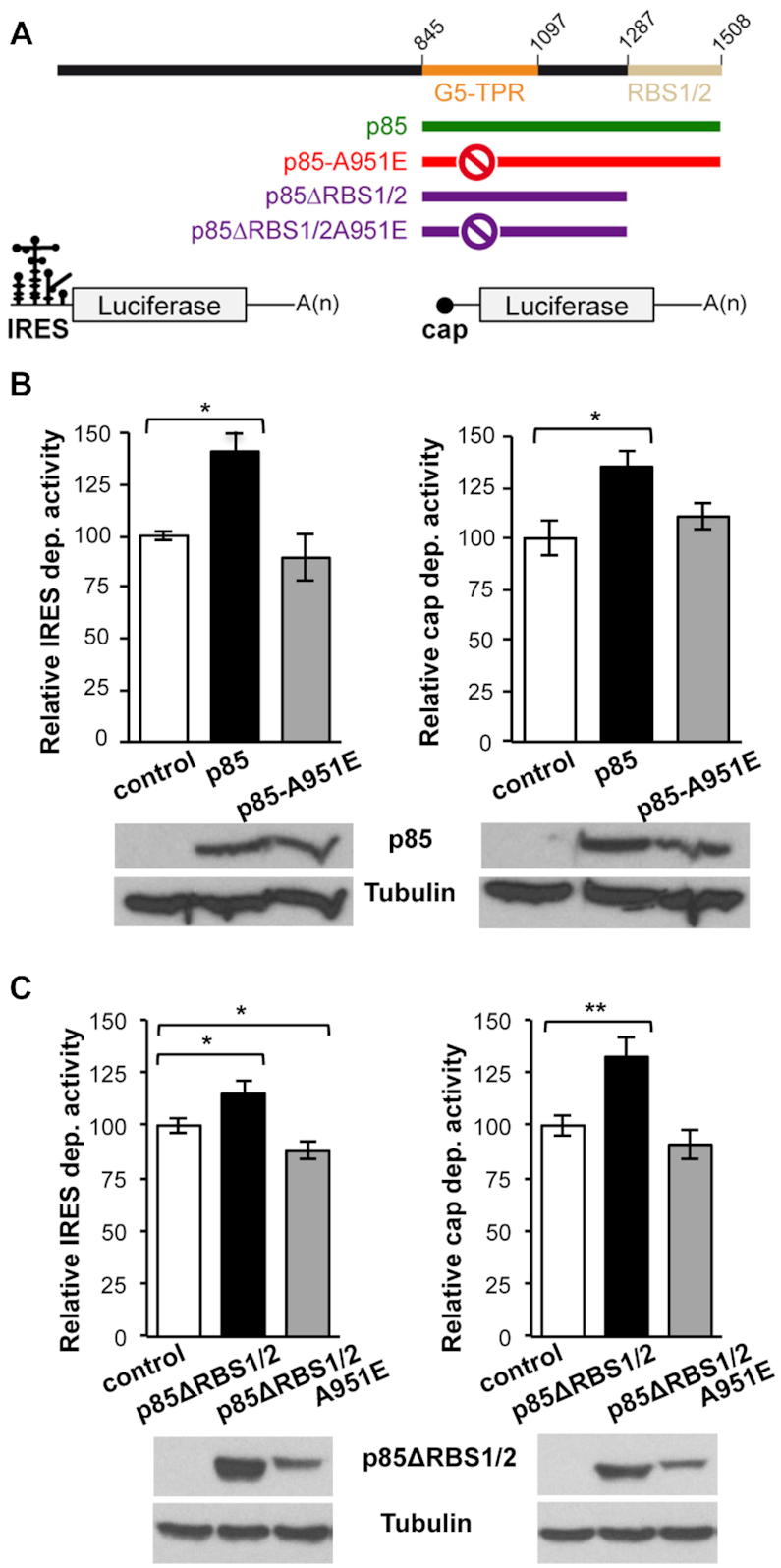
Translation stimulation by p85 depends on the TPR dimerization domain. (A) Diagram of the p85, p85-A951E, p85ΔRBS1/2, and p85ΔRBS1/2-A951E proteins expressed with the Xpress-tag (top), and of the luciferase reporter mRNAs carrying IRES or cap (bottom). (B) Luciferase activity measured in HEK293 cell lysates expressing IRES-mRNA or cap-mRNA co-transfected with Xpress-p85 and Xpress-p85-A951E. Expression of Xpress-p85 and Xpress-p85-A951E was monitored by western blot using anti-Xpress. Tubulin was used as loading control. (C) Luciferase activity assay using Xpress-p85ΔRBS1/2 and Xpress-p85ΔRBS1/2A951E constructs. Protein expression was monitored by western blot using anti-Xpress. In all cases, luciferase values are normalized to cells expressing the empty vector conducted side by side. Values represent the mean ± SEM obtained in three independent assays. Asterisks denote P-values (*P < 0.05, **P < 0.01).
DISCUSSION
In this study, we combined structural, proteomic and functional analysis to get insights into the multitasking protein Gemin5. We demonstrate that the middle region of Gemin5 contains a previously uncharacterized dimerization module, which also remains at the N-terminus of the viral-induced cleavage fragment p85 (Figure 6A,B). The TPR-like motifs of Gemin5 do not conform to the archetypical length of 34 amino acids, but rather alternate helical pairs of 32–33 and 27–29 amino acids that exhibit the loosely conserved TPR pattern of small and large hydrophobic residues (40,47) (Supplementary Figure S2). However, whereas most tandem TPR arrays assemble into a right-handed super helical structure with concave and convex surfaces for protein-protein and protein-RNA interactions (48), the helical repeats in Gemin5 are stretched in an α-solenoid rod that provides an extensive flat surface for self-dimerization. Importantly, and despite the tight association between G5-TPR subunits, a single point mutation in the dyad axis, A951E, was sufficient to destabilize the dimer (Figures 1D, E, 2D and Supplementary Figure S5).
Figure 6.
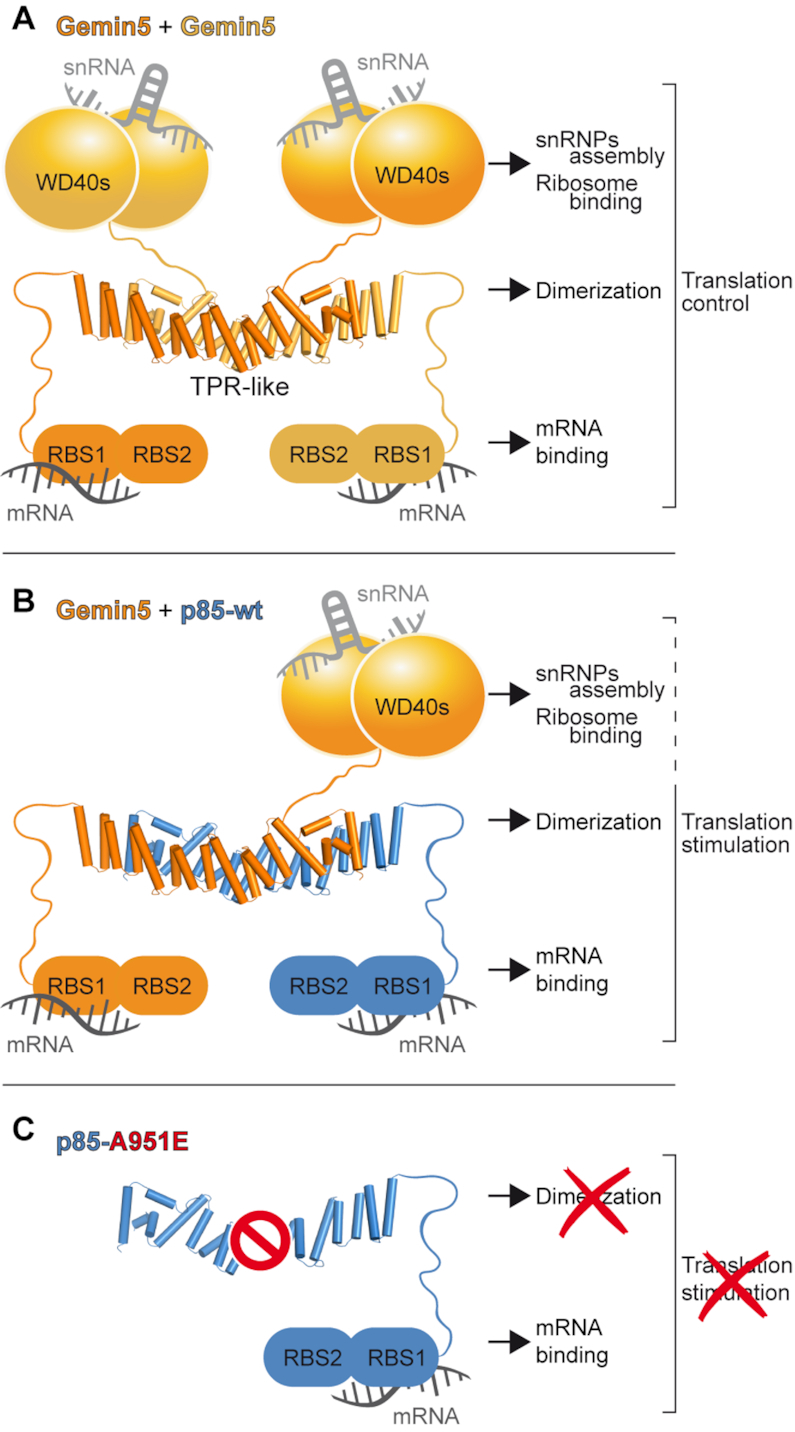
Model of Gemin5-p85 heterodimer driven by the TPR-like domain. (A) Schematic of Gemin5 dimer. The relative orientation between the WD40s, TPRs and RBS1/2 domains of Gemin5 is unknown, although they are represented at three different levels to highlight their distinct roles in snRNA and ribosome binding (WD40s), dimerization (TPR-like) and translation mRNA binding (RBS1/2). (B) Schematic representation of the association between Gemin5 (orange) and p85 (blueish). (C) Schematic representation of p85-A951E. The prohibition sign indicates that mutation A951E abrogates the interaction with Gemin5.
The canoe-shaped conformation of the G5-TPR homodimer is singular, and an exhaustive manual and computational search failed to retrieve other TPR motifs resulting in a similar association. Although a search with DALI highlighted the structural similarity between G5-TPR and the C-terminal region of Elp1, most interactions in the Elp1 homodimer occur between the TPR-like motifs in one subunit and the ‘capping’ helical bundle in the other (38) (Supplementary Figure S4B). Instead, 14 out of the 17 α-helices contribute directly to G5-TPR dimerization (Figures 1D, 2A and Supplementary Figure S4A). The dimerization mode of Gemin5 is more reminiscent of the association between the yeast vesicle coating proteins β’-COP and α-COP of the COPI complex (49), and Sec31 of the COPII complex (50). These proteins present α-solenoid regions that in α-COP and β’-COP interact in an antiparallel orientation with a 40° angle, forming a stable heterodimer (Supplementary Figure S4C) while in Sec31, the central α-solenoid regions of two subunits interact about a 2-fold axis, creating an interlocked homodimer (Supplementary Figure S4D). Curiously, alike in Gemin5, the α-solenoids of the mentioned proteins are preceded by N-terminal WD40 domains (Figures 1A, 6A), highlighting how a similar arrangement of these domains evolved into proteins with different functions.
We also show here that the G5-TPR alone does not interact with RNA, irrespectively of whether it consisted of single or double stranded molecules, short hairpins, long stem-loops, or DNA (Figure 3). This result establishes a main difference with proteins carrying TPR domains involved in RNA-dependent pathways or in direct RNA-binding (3,4,51–54). The high sequence conservation of the TPR-like domain strongly suggests that the dimerization module of Gemin5 plays a fundamental role for the architecture and activity of the protein. Interestingly, the first residue detectable on the crystal structure occurs immediately after residue K844, matching the L protease cleavage site that releases the p85 fragment during viral infection (17). Consequently, the dimerization module might play a central structural role also in the p85 product. Indeed, we have found that in living cells, full-length Gemin5 is captured with a p85 construct with a TAP-tag at the C-terminus (Figures 4A,C, 6B, Supplementary Figure S7). However, a p85-TAP variant carrying the point mutation A951E that prevents dimerization failed to recruit the endogenous Gemin5 protein (Figures 4B, C, 6B, Supplementary Figure S8). These data demonstrate that the TPR-like region is necessary for the interaction of p85 with the available Gemin5 protein in the cell environment, beyond its own dimerization. This physical interaction presumably contributes to the differential behavior of this viral-induced cleavage fragment in translation control. Among other possibilities, recruitment of Gemin5 by the p85 polypeptide might reduce the available pool of full-length protein to perform its different roles on RNA metabolism.
Here, we provide direct evidence for physical p85-Gemin5 interactions (Figure 4A, C), which in turn, unveiled fundamental properties of Gemin5. The top GO terms retrieved by p85-wt, which establishes protein-protein bridges with the endogenous Gemin5, are splicing-related components, translation control members, and snRNP assembly factors (Figure 4D). All these groups are consistent with the functions reported for this protein (44), and suggest that these components probably interact with the N-terminal region of Gemin5. In contrast, GO functional groups shared by p85-wt and p85-A951E constructs (thereby, factors shared with the C-terminal moiety 845–1508) are related to SRP-dependent cotranslational protein targeting to membrane, translation initiation and nuclear transcribed catabolic processes (Figure 4E). These families of factors confirm the known roles of Gemin5 and strongly suggest new potential functions for this versatile protein. Finally, GO terms associated uniquely to p85-A951E, which fails to retrieve the endogenous Gemin5 protein, are related to apoptotic processes, proteolysis, and response to unfolded proteins. We attribute this result to a protein instability problem caused by the destabilizing mutation on the dimerization domain. Together, these results corroborate that different domains of Gemin5 are responsible for its multiple functions, and highlight the biological relevance of the dimerization module of this protein.
The properties unveiled in our study of the dimerization module of Gemin5 shed new light on this little characterized protein. Gemin5 overexpression leads to protein synthesis decrease (Supplementary Figure S10), consistent with its previously reported role as a down-regulator of translation (44). Nonetheless, assays carried out with the full-length protein indicated no statistically significant differences in translation between the wt and the mutant A951E (Supplementary Figure S10). Given the modular organization of the protein and the different functions associated to each domain it is not surprising that expression of separate parts of the protein result in different effects. We show here that disruption of the complex formation by mutation A951E abolishes the translation enhancing effect of both p85 and p85ΔRBS1/2 proteins (Figure 5B, C), although the stimulatory effect exerted by p85ΔRBS1/2 is weaker than p85 protein. These results indicate that p85 comprises additional determinants for RNA-dependent pathways. Importantly, p85 harbors the bipartite non-canonical RBS1/2 domain at its C-terminus (18). The RBS1 domain not only displays robust RNA-binding capacity in vitro (Figure 3A), but also binds selective RNA targets in living cells (12). Therefore, the translation stimulatory capacity of p85 reflects the combined properties of the dimerization module and the RBS domains (Figure 6B).
Overall, these results led us to propose that mutations in the dimerization module of Gemin5 will result in loss of function. The protein is broadly expressed in all human tissues (55,56), and the loss of Gemin5/rigor mortis in Drosophila is lethal (57). On the other hand, altered levels of SMN proteins causing defects in SMN complex assembly lead to spinal muscular atrophy (SMA), a severe child disease (58). According to our data, mutation A951E disrupts the p85-Gemin5 interaction, hampers the association with proteins regulating splicing and translation processes, and abolishes translation control. Thus, it is plausible that defects in Gemin5 dimerization will course with severe disease. In support of this possibility, mutations affecting the TPR domains of other proteins have been shown to be functionally critical. Thus, splicing variants producing a protein lacking the TPR-like dimerization domain of Elp1 are involved in > 95% of all familial dysautonomia patients (38), whereas nonsense mutations in the TPR region of aryl-hydrocarbon-interacting protein-like 1 (AIPL1) cause inherited retinopathies (59), and missense mutations in the TPR domain of O-linked N-acetyl-glucosamine transferase cause intellectual disability (60).
In summary, we have disclosed an unprecedented dimerization module in Gemin5 and provided direct evidence for physical p85-Gemin5 interactions in the cell and its relationship to translation control. Further work will be needed to explore the implications of Gemin5 dimerization in other activities of this multifaceted protein.
DATA AVAILABILITY
The coordinates and structure factors for G5-TPR (SAD) and G5-TPR (native) were deposited in the Protein Data Bank under accession codes 6RNS and 6RNQ, respectively.
Supplementary Material
ACKNOWLEDGEMENTS
X-ray diffraction experiments were performed at XALOC beamline at ALBA Synchrotron with the collaboration of ALBA staff, with special thanks to Fernando Gil. We thank the CBMSO Proteomics Unit for help with the MS/MS analysis, the CNIO Spectroscopy Unit for SEC-MALS analysis, Jorge Ramajo for technical assistance, and Crisanto Gutierrez for valuable comments on the manuscript.
Notes
Present address: María Moreno-Morcillo. Spanish National Cancer Research Centre (CNIO), Melchor Fernández Almagro 3, 28029 Madrid, Spain.
Present address: Javier Fernández-Chamorro. Garvan Institute of Medical Research, 384 Victoria St, Darlinghurst NSW 2010, Australia.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
MINECO [BFU2016-80570-R, BFU2017-84492-R, RTI2018-098084-B-I00, AEI/FEDER, UE]; Comunidad de Madrid [B2017/BMD-3770]; Institutional grant from Fundación Ramón Areces. Funding for open access charge: MINECO [BFU2017-84492-R].
Conflict of interest statement. None declared.
REFERENCES
- 1. Gehring N.H., Wahle E., Fischer U.. Deciphering the mRNP Code: RNA-bound determinants of post-transcriptional gene regulation. Trends Biochem. Sci. 2017; 42:369–382. [DOI] [PubMed] [Google Scholar]
- 2. Lunde B.M., Moore C., Varani G.. RNA-binding proteins: modular design for efficient function. Nat. Rev. Mol. Cell Biol. 2007; 8:479–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Katibah G.E., Lee H.J., Huizar J.P., Vogan J.M., Alber T., Collins K.. tRNA binding, structure, and localization of the human interferon-induced protein IFIT5. Mol. Cell. 2013; 49:743–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yang Z., Liang H., Zhou Q., Li Y., Chen H., Ye W., Chen D., Fleming J., Shu H., Liu Y.. Crystal structure of ISG54 reveals a novel RNA binding structure and potential functional mechanisms. Cell Res. 2012; 22:1328–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ellisdon A.M., Dimitrova L., Hurt E., Stewart M.. Structural basis for the assembly and nucleic acid binding of the TREX-2 transcription-export complex. Nat. Struct. Mol. Biol. 2012; 19:328–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Battle D.J., Lau C.K., Wan L., Deng H., Lotti F., Dreyfuss G.. The Gemin5 protein of the SMN complex identifies snRNAs. Mol. Cell. 2006; 23:273–279. [DOI] [PubMed] [Google Scholar]
- 7. Matera A.G., Raimer A.C., Schmidt C.A., Kelly J.A., Droby G.N., Baillat D., Ten Have S., Lamond A.I., Wagner E.J., Gray K.M.. Composition of the survival motor neuron (SMN) complex in drosophila melanogaster. G3 (Bethesda). 2019; 9:491–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lau C.K., Bachorik J.L., Dreyfuss G.. Gemin5-snRNA interaction reveals an RNA binding function for WD repeat domains. Nat. Struct. Mol. Biol. 2009; 16:486–491. [DOI] [PubMed] [Google Scholar]
- 9. Battle D.J., Kasim M., Wang J., Dreyfuss G.. SMN-independent subunits of the SMN complex. Identification of a small nuclear ribonucleoprotein assembly intermediate. J. Biol. Chem. 2007; 282:27953–27959. [DOI] [PubMed] [Google Scholar]
- 10. Pacheco A., Lopez de Quinto S., Ramajo J., Fernandez N., Martinez-Salas E.. A novel role for Gemin5 in mRNA translation. Nucleic Acids Res. 2009; 37:582–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Workman E., Kalda C., Patel A., Battle D.J.. Gemin5 Binds to the survival motor neuron mRNA to Regulate SMN Expression. J. Biol. Chem. 2015; 290:15662–15669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Francisco-Velilla R., Fernandez-Chamorro J., Dotu I., Martinez-Salas E.. The landscape of the non-canonical RNA-binding site of Gemin5 unveils a feedback loop counteracting the negative effect on translation. Nucleic Acids Res. 2018; 46:7339–7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Francisco-Velilla R., Fernandez-Chamorro J., Ramajo J., Martinez-Salas E.. The RNA-binding protein Gemin5 binds directly to the ribosome and regulates global translation. Nucleic Acids Res. 2016; 44:8335–8351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Simsek D., Tiu G.C., Flynn R.A., Byeon G.W., Leppek K., Xu A.F., Chang H.Y., Barna M.. The mammalian Ribo-interactome reveals ribosome functional diversity and heterogeneity. Cell. 2017; 169:1051–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Walsh D., Mohr I.. Viral subversion of the host protein synthesis machinery. Nat. Rev. Microbiol. 2011; 9:860–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lozano G., Martinez-Salas E.. Structural insights into viral IRES-dependent translation mechanisms. Curr. Opin. Virol. 2015; 12:113–120. [DOI] [PubMed] [Google Scholar]
- 17. Pineiro D., Ramajo J., Bradrick S.S., Martinez-Salas E.. Gemin5 proteolysis reveals a novel motif to identify L protease targets. Nucleic Acids Res. 2012; 40:4942–4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fernandez-Chamorro J., Pineiro D., Gordon J.M., Ramajo J., Francisco-Velilla R., Macias M.J., Martinez-Salas E.. Identification of novel non-canonical RNA-binding sites in Gemin5 involved in internal initiation of translation. Nucleic Acids Res. 2014; 42:5742–5754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jin W., Wang Y., Liu C.P., Yang N., Jin M., Cong Y., Wang M., Xu R.M.. Structural basis for snRNA recognition by the double-WD40 repeat domain of Gemin5. Genes Dev. 2016; 30:2391–2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Xu C., Ishikawa H., Izumikawa K., Li L., He H., Nobe Y., Yamauchi Y., Shahjee H.M., Wu X.H., Yu Y.T. et al.. Structural insights into Gemin5-guided selection of pre-snRNAs for snRNP assembly. Genes Dev. 2016; 30:2376–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tang X., Bharath S.R., Piao S., Tan V.Q., Bowler M.W., Song H.. Structural basis for specific recognition of pre-snRNA by Gemin5. Cell Res. 2016; 26:1353–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jonas S., Izaurralde E.. The role of disordered protein regions in the assembly of decapping complexes and RNP granules. Genes Dev . 2013; 27:2628–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zimmermann L., Stephens A., Nam S.Z., Rau D., Kubler J., Lozajic M., Gabler F., Soding J., Lupas A.N., Alva V.. A completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. J. Mol. Biol. 2018; 430:2237–2243. [DOI] [PubMed] [Google Scholar]
- 24. Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J.. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015; 10:845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yang J., Yan R., Roy A., Xu D., Poisson J., Zhang Y.. The I-TASSER suite: protein structure and function prediction. Nat. Methods. 2015; 12:7–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen G.I., Gingras A.C.. Affinity-purification mass spectrometry (AP-MS) of serine/threonine phosphatases. Methods. 2007; 42:298–305. [DOI] [PubMed] [Google Scholar]
- 27. Kabsch W. Xds. Acta Crystallogr. D. Biol. Crystallogr. 2010; 66:125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W. et al.. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Emsley P., Lohkamp B., Scott W.G., Cowtan K.. Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen V.B., Arendall W.B. 3rd, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C.. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D. Biol. Crystallogr. 2010; 66:12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Krissinel E., Henrick K.. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007; 372:774–797. [DOI] [PubMed] [Google Scholar]
- 32. Ashkenazy H., Abadi S., Martz E., Chay O., Mayrose I., Pupko T., Ben-Tal N.. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016; 44:W344–W350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Francisco-Velilla R., Fernandez-Chamorro J., Lozano G., Diaz-Toledano R., Martinez-Salas E.. RNA-protein interaction methods to study viral IRES elements. Methods. 2015; 91:3–12. [DOI] [PubMed] [Google Scholar]
- 34. Pineiro D., Fernandez N., Ramajo J., Martinez-Salas E.. Gemin5 promotes IRES interaction and translation control through its C-terminal region. Nucleic Acids Res. 2013; 41:1017–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shevchenko A., Wilm M., Vorm O., Mann M.. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 1996; 68:850–858. [DOI] [PubMed] [Google Scholar]
- 36. Zhang J., Xin L., Shan B., Chen W., Xie M., Yuen D., Zhang W., Zhang Z., Lajoie G.A., Ma B.. PEAKS DB: de novo sequencing assisted database search for sensitive and accurate peptide identification. Mol Cell Prot: MCP. 2012; 11:M111 010587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lozano G., Francisco-Velilla R., Martinez-Salas E.. Ribosome-dependent conformational flexibility changes and RNA dynamics of IRES domains revealed by differential SHAPE. Sci. Rep. 2018; 8:5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Xu H., Lin Z., Li F., Diao W., Dong C., Zhou H., Xie X., Wang Z., Shen Y., Long J.. Dimerization of elongator protein 1 is essential for Elongator complex assembly. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:10697–10702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Das A.K., Cohen P.W., Barford D.. The structure of the tetratricopeptide repeats of protein phosphatase 5: implications for TPR-mediated protein-protein interactions. EMBO J. 1998; 17:1192–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. D’Andrea L.D., Regan L.. TPR proteins: the versatile helix. Trends Biochem. Sci. 2003; 28:655–662. [DOI] [PubMed] [Google Scholar]
- 41. Holm L., Sander C.. Dali: a network tool for protein structure comparison. Trends Biochem. Sci. 1995; 20:478–480. [DOI] [PubMed] [Google Scholar]
- 42. Yong J., Kasim M., Bachorik J.L., Wan L., Dreyfuss G.. Gemin5 delivers snRNA precursors to the SMN complex for snRNP biogenesis. Mol. Cell. 2010; 38:551–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Huang da W., Sherman B.T., Lempicki R.A.. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009; 4:44–57. [DOI] [PubMed] [Google Scholar]
- 44. Francisco-Velilla R., Azman E.B., Martinez-Salas E.. Impact of RNA-protein interaction modes on translation control: the versatile multidomain protein Gemin5. Bioessays. 2019; 41:e1800241. [DOI] [PubMed] [Google Scholar]
- 45. Piazzon N., Schlotter F., Lefebvre S., Dodre M., Mereau A., Soret J., Besse A., Barkats M., Bordonne R., Branlant C. et al.. Implication of the SMN complex in the biogenesis and steady state level of the signal recognition particle. Nucleic. Acids. Res. 2013; 41:1255–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Garcia-Moreno M., Noerenberg M., Ni S., Jarvelin A.I., Gonzalez-Almela E., Lenz C.E., Bach-Pages M., Cox V., Avolio R., Davis T. et al.. System-wide profiling of RNA-binding proteins uncovers key regulators of virus infection. Mol. Cell. 2019; 74:196–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhu H., Sepulveda E., Hartmann M.D., Kogenaru M., Ursinus A., Sulz E., Albrecht R., Coles M., Martin J., Lupas A.N.. Origin of a folded repeat protein from an intrinsically disordered ancestor. Elife. 2016; 5:e16761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kajander T., Cortajarena A.L., Mochrie S., Regan L.. Structure and stability of designed TPR protein superhelices: unusual crystal packing and implications for natural TPR proteins. Acta Crystallogr. D. Biol. Crystallogr. 2007; 63:800–811. [DOI] [PubMed] [Google Scholar]
- 49. Lee C., Goldberg J.. Structure of coatomer cage proteins and the relationship among COPI, COPII, and clathrin vesicle coats. Cell. 2010; 142:123–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fath S., Mancias J.D., Bi X., Goldberg J.. Structure and organization of coat proteins in the COPII cage. Cell. 2007; 129:1325–1336. [DOI] [PubMed] [Google Scholar]
- 51. Johnson B., VanBlargan L.A., Xu W., White J.P., Shan C., Shi P.Y., Zhang R., Adhikari J., Gross M.L., Leung D.W. et al.. Human IFIT3 modulates IFIT1 RNA binding specificity and protein stability. Immunity. 2018; 48:487–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Abbas Y.M., Pichlmair A., Gorna M.W., Superti-Furga G., Nagar B.. Structural basis for viral 5′-PPP-RNA recognition by human IFIT proteins. Nature. 2013; 494:60–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Grotwinkel J.T., Wild K., Segnitz B., Sinning I.. SRP RNA remodeling by SRP68 explains its role in protein translocation. Science. 2014; 344:101–104. [DOI] [PubMed] [Google Scholar]
- 54. Zhou X., Liao H., Chern M., Yin J., Chen Y., Wang J., Zhu X., Chen Z., Yuan C., Zhao W. et al.. Loss of function of a rice TPR-domain RNA-binding protein confers broad-spectrum disease resistance. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:3174–3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kim M.S., Pinto S.M., Getnet D., Nirujogi R.S., Manda S.S., Chaerkady R., Madugundu A.K., Kelkar D.S., Isserlin R., Jain S. et al.. A draft map of the human proteome. Nature. 2014; 509:575–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Uhlen M., Fagerberg L., Hallstrom B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson A., Kampf C., Sjostedt E., Asplund A. et al.. Proteomics. Tissue-based map of the human proteome. Science. 2015; 347:1260419. [DOI] [PubMed] [Google Scholar]
- 57. Gates J., Lam G., Ortiz J.A., Losson R., Thummel C.S.. rigor mortis encodes a novel nuclear receptor interacting protein required for ecdysone signaling during Drosophila larval development. Development. 2004; 131:25–36. [DOI] [PubMed] [Google Scholar]
- 58. Burghes A.H., Beattie C.E.. Spinal muscular atrophy: why do low levels of survival motor neuron protein make motor neurons sick. Nat. Rev. Neurosci. 2009; 10:597–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sacristan-Reviriego A., Bellingham J., Prodromou C., Boehm A.N., Aichem A., Kumaran N., Bainbridge J., Michaelides M., van der Spuy J.. The integrity and organization of the human AIPL1 functional domains is critical for its role as a HSP90-dependent co-chaperone for rod PDE6. Hum. Mol. Genet. 2017; 26:4465–4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Gundogdu M., Llabres S., Gorelik A., Ferenbach A.T., Zachariae U., van Aalten D.M.F.. The O-GlcNAc transferase intellectual disability mutation L254F distorts the TPR Helix. Cell Chem. Biol. 2018; 25:513–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The coordinates and structure factors for G5-TPR (SAD) and G5-TPR (native) were deposited in the Protein Data Bank under accession codes 6RNS and 6RNQ, respectively.