Abstract
Objectives
Pseudomonas aeruginosa is known as a leading cause of nosocomial infections worldwide. Antimicrobial resistance and biofilm production, as two main virulence factors of P. aeruginosa, are responsible for the persistence of prolonged infections. In this study, antimicrobial susceptibility pattern and phenotypic and genotypic characteristics of biofilm of P. aeruginosa were investigated.
Results
A total of 80 clinical P. aeruginosa isolates were obtained. Isolates showed resistance to all antibiotics with a rate from 12.5% (n = 10) against amikacin and piperacillin/tazobactam to 23.75% (n = 19) to levofloxacin. Multidrug-resistant P. aeruginosa accounted for 20% (n = 16). 83.75% (n = 67) of isolates showed biofilm phenotype. All three biofilm-related genes were found simultaneously in 87.5% (n = 70) of P. aeruginosa and 13.5% (n = 10) of the isolates had none of the genes tested. From the results of the present study, combination therapy including an anti-pseudomonal beta-lactam (piperacillin/tazobactam or ceftazidime) and an aminoglycoside or carbapenems (imipenem, meropenem) with fluoroquinolones in conjunction with an aminoglycoside can be used against Pseudomonas infections. However, reasonable antimicrobial use and high standards of infection prevention and control are essential to prevent further development of antimicrobial resistance. Combination strategies based on the proper anti-pseudomonal antibiotics along with anti-biofilm agents can also be selected to eradicate biofilm-associated infections.
Keywords: Antimicrobial resistance, Biofilm formation, Biofilm genes, Pseudomonas aeruginosa
Introduction
Pseudomonas aeruginosa, as one of the most common hospital pathogens, is involved in a wide of severe opportunistic infections, particularly in immunocompromised patients [1]. As a global problem, the increasing rate of multidrug-resistance (MDR) strains has resulted in the medical therapy against P. aeruginosa be complicated [2–4]. In addition, the ability of P. aeruginosa to produce biofilm is thought to be a main factor involved in chronic infections. Biofilms are complex of microbial cells embedded in an extracellular matrix composed of proteins, extracellular DNA, and exopolysaccharides, providing a protective life-style for bacteria and are extremely challenging and costly to treat by antimicrobial compounds [5].
The biofilm components of P. aeruginosa are composed of at least three distinct exopolysaccharides, including alginate, Psl and Pel [6]. Alginate is mainly produced by P. aeruginosa clinical isolates originated from the lungs of cystic fibrosis (CF) patients [7]. It is a linear polymer consisting of β-d-mannuronic acid and α-l-guluronic acid and has an important role in structural stability and protection of biofilm [8]. Alginate synthesis in P. aeruginosa is controlled by the algACD operon. AlgD encoded by algD is a GDP-mannose dehydrogenase that catalyzes the production of GDP-mannuronic acid from GDP-mannose [9]. The algD gene mediates the control of alginate biosynthesis and transcription of the Alg proteins, and also is responsible for the final production of precursor GDP-mannuronic acid, the foundation molecule for polymerization and alginate synthesis [7].
P. aeruginosa isolates obtained from environments often produce two different exopolysaccharides. Psl (polysaccharide synthesis locus) is a neutral polysaccharide composed of a repeating pentasaccharide, consisting of d-mannose, d-glucose, and l-rhamnose. Psl has been shown that provides cell–cell and cell–surface interactions during biofilm formation, thereby playing an important in the initiation of biofilm formation and protection of biofilm structure [10]. The psl operon consisting of 15 co-transcribed genes (pslA to pslO) is required for Psl synthesis. PslD protein, encoded by pslD gene, is localized in the periplasm/outer membrane and is required for biofilm formation, probably by the export of a biofilm-relevant exopolysaccharide [11]. The formation of a layer of polymer/cells at the air–liquid interface of a P. aeruginosa standing culture is termed pellicle formation that is controlled by the pel (pellicle) operon [12]. Pel is a cellulose-sensitive exopolysaccharide composed of 1→4 linked partially acetylated galactosamine and glucosamine sugars [13]. The pel operon is composed of seven genes (pelA to pelG) [14]. PelF has been suggested to be a soluble glycosyltransferase using UDP-glucose as a donor substrate toward the Pel exopolysaccharide biosynthesis [12].
Given the potential of biofilm in increasing antimicrobial resistance and, as a result, the persistence of infections caused by P. aeruginosa, identification of the isolates possessing such factor will help us to better understand the pathogenesis of the organism. This study aimed to evaluate the antimicrobial susceptibility pattern as well as the phenotypic and genotypic characteristics of biofilm in P. aeruginosa isolates.
Main text
Methods
Bacterial isolates were collected during September 2017 to August 2018 from clinical specimens of patients in three university affiliated hospitals in Gorgran, Iran. Laboratory identification of P. aeruginosa isolates were performed by standard microbiological and biochemical methods, including pigment production in agar, oxidase and catalase tests, reactions in triple sugar iron (TSI) agar, SIM (sulfide, indole, motility), and oxidative-fermentative (OF) media (Merck, Darmstadt, Germany), and finally, growth at 42 °C [2].
Susceptibility of isolates to different antibiotics was determined by disk diffusion agar method on cation-adjusted Mueller–Hinton agar (Merck, Darmstadt, Germany) according to the Clinical and Laboratory Standards Institute (CLSI) recommendations [15]. Antibiotic disks (MAST Diagnostics, Merseyside, UK) tested were ceftazidime (CAZ, 30 μg), piperacillin/tazobactam (PTZ, 100 μg/10 μg), ciprofloxacin (CIP, 5 μg), levofloxacin (LEV, 5 μg), gentamicin (GM, 10 μg), amikacin (AK, 30 μg), tobramycin (TOB, 10 μg), imipenem (IMI, 10 μg), and meropenem (MEM, 10 μg). Escherichia coli ATCC 25922 was used as a control for susceptibility testing. Multidrug-resistant P. aeruginosa (MDR-PA) was defined as isolate resistant to more than one antimicrobial agent in three or more antimicrobial categories [16].
Quantitative assessment of biofilm formation was performed by the colorimetric microtiter plate assay as described previously by Stepanović et al. [17] with some modifications. An overnight culture of P. aeruginosa was adjusted to the turbidity of a 1 McFarland standard. Suspensions were diluted 1:100 in 200 μL tryptic soy broth (TSB) containing 1% glucose (Merck, Darmstadt, Germany), and were then transferred into the sterile flat-bottomed 96-well polystyrene microplates (JET Biofil, Guangzhou, China). Wells were gently washed three times with sterile phosphate buffered saline (PBS, pH 7.3) after 24 h of incubation at 37 °C. Adherent biofilms were fixed by 99% methanol for 15 min, the solutions were removed, and the plate was air-dried. Biofilms were stained by 200 μL of crystal violet 0.1% (Sigma Chemical Co., St Louis, MO, USA) for 5 min at room temperature, and then rinsed by water and allowed to dry. Biofilm in each well was destained by treatment with 200 μL of 95% ethanol for 30 min. The optical density (OD) was measured at 570 nm using a microtiter plate reader (BioTek, Bad Friedrichshall, Germany). All experiments were performed in triplicate and repeated three times. In addition, a cut-off value (ODc) was established. It is defined as three standard deviations (SD) above the mean OD of the negative control: Odc = average OD of negative control + (3 × SD of negative control). The isolates were classified into the four following categories based upon the OD: non-biofilm producer (OD < ODc); weak-biofilm producer (ODc < OD < 2 × ODc); moderate-biofilm producer (2 × ODc < OD < 4 × ODc); strong-biofilm producer (4 × ODc < OD).
All P. aeruginosa isolates were evaluated for three biofilm-encoding genes, algD, pslD, and pelF by polymerase chain reaction (PCR) method, using specific primers [8] synthesized by Metabion company (Metabion international AG, Germany). DNA extraction was performed from bacterial colonies by boiling method. All three genes were amplified under the following thermal conditions: initial denaturation at 95 °C for 5 min, followed by 30 cycles of denaturation at 94 °C for 30 s, annealing at 60 °C for 40 s, extension at 72 °C for 40 s, and a final elongation step at 72 °C for 5 min. PCR products were analyzed with UV light after running at 100 V for 1 h on a 1% agarose gel stained with DNA safe stain (SinaClon, Tehran, Iran).
Chi squared test was performed on the relationship between categorical variables, including biofilm characteristics and antimicrobial resistance using SPSS software, 18.0 (SPSS Inc., Chicago, IL, USA). A p value < 0.05 was considered as statistically significant.
Results
A total of 80 distinct P. aeruginosa isolates were obtained from patients, of which 44 (55%) were from males and 36 (45%) were from females. Analysis of P. aeruginosa distribution in clinical specimens indicated that the most isolates (n = 29, 36.25%) were originated from endotracheal secretions, followed by urine (n = 26, 32.5%), blood (n = 11, 13.75%), wound (n = 8, 10%), CSF (n = 4, 5%), and ear (n = 2, 2.5%).
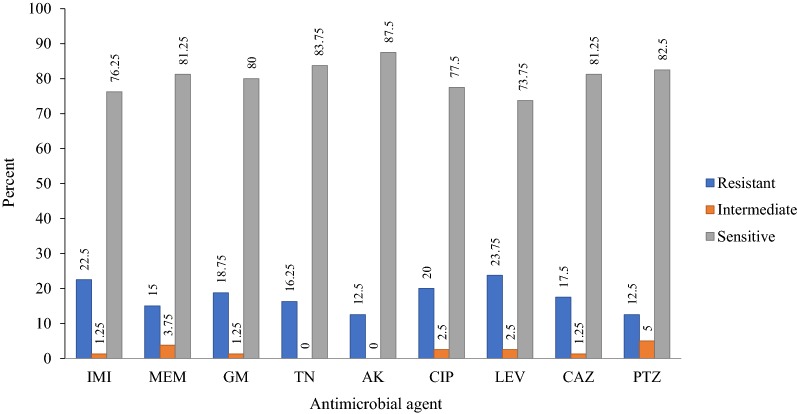
Based on the CLSI interpretive criteria [15], resistance rate among P. aeruginosa isolates to antibiotics tested was as follow (Fig. 1): IMI 22.5% (n = 18), MEM 15% (n = 12), GM 18.75% (n = 15), TN 16.25% (n = 13), AK 12.5% (n = 10), CIP 20% (n = 16), LEV 23.75 (n = 19), CAZ 17.5% (n = 14), and PTZ 12.5% (n = 10). The prevalence of MDR-PA and non-MDR-PA was 20% (n = 16) and 80% (n = 64), respectively.
Fig. 1.
Antibiotic susceptibility patterns of P. aeruginosa clinical isolates. IMI imipenem, MEM meropenem, GM gentamicin, TOB tobramycin, AK amikacin, CIP ciprofloxacin, LEV levofloxacin, CAZ ceftazidime, PTZ piperacillin/tazobactam
Biofilm phenotypes accounted for 83.75% (n = 67), being distributed in the following categories: 16.25% (n = 13) produced strong biofilm; 33.75% (n = 27) produced moderate biofilm; 33.75% (n = 27) produced weak biofilm, whilst 16.25% of isolates (n = 13) were identified as non-biofilm producer (Table 1). A high occurrence of biofilm-encoding genes was found (Fig. 2): 87.5% (n = 70) of the isolates presented all three algD, pslD, and pelF genes, simultaneously (identified as algD +/pslD +/pelF + genotypic pattern), while 12.5% (n = 10) had none and considered as algD −/pslD −/pelF − pattern. In addition, isolates were divided to four groups based on both phenotypic and genotypic characteristics of biofilm: biofilm positive/gene positive (n = 59, 73.75%); biofilm negative/gene positive (n = 11, 13.75%); biofilm positive/gene negative (n = 8, 10%); biofilm negative/gene negative (n = 2, 2.5%). MDR phenotype accounted for 17.91% (n = 12) of 67 biofilm producers and 20% (n = 14) of 70 genotypically positive isolates.
Table 1.
Relationship between biofilm characteristic and antibiotic susceptibility pattern among P. aeruginosa clinical isolates
| Phenotypic pattern of biofilm, no. (%) | Genotypic pattern of biofilm, no. (%) | ||
|---|---|---|---|
| AlgD +/pslD +/pelF + | AlgD −/pslD −/pelF − | p-value | |
| Strong, 13 (16.25) | 10 (76.92) | 3 (23.07) | 0.001 |
| Moderate, 27 (33.75) | 25 (92.59) | 2 (7.41) | 0.0001 |
| Weak 27 (33.75) | 24 (88.89) | 3 (11.11) | 0.0001 |
| Non-biofilm, 13 (16.25) | 11 (84.61) | 2 (15.38) | 0.0001 |
| Total, 80 (100) | 70 (87.5) | 10 (12.5) | 0.0001 |
Fig. 2.
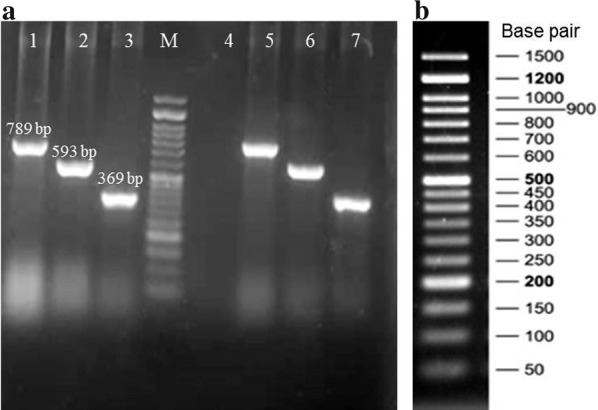
PCR amplification of biofilm-encoding genes in one selected clinical isolate of P. aeruginosa as representative. a Lane 1–3: PCR products of the pelF, algD, and pslD genes, respectively. M: 50 bp DNA ladder. Lane 4: PCR mixture without DNA template as control negative. Lane 5–7: PCR products of the corresponding genes in P. aeruginosa PAO1 reference strain as control positive. b A 50 bp DNA ladder containing seventeen discrete fragments ranging from 50 to 1500 bp with double intensity reference bands at 200 bp, 500 bp, and 1200 bp
Discussion
Development of resistance by P. aeruginosa to many antimicrobial agents is a great challenge in controlling its infections [1, 18, 19]. Comprehensive surveillance of antimicrobial resistance in European countries for 2017 demonstrated a range of combined resistance (resistance to three or more antimicrobial groups, including piperacillin ± tazobactam, ceftazidime, fluoroquinolones, aminoglycosides and carbapenems) from 0% (Iceland) to 59.1% (Romania) [20]. The prevalence of MDR P. aeruginosa in Iran has been estimated at 58%, with a variation in geographical areas: the highest and lowest rates were observed in Tehran (100%) and Zahedan (16%), respectively [21]. In a recent study, Bavasheh et al. [22] found that 27.8% of clinical P. aeruginosa isolates were MDR. Similarly, the rate of isolates with resistance to at least three antimicrobial groups in our study was 20% that was lower than that reported from other studies [8, 23, 24]. Although the rate of multi-resistance in the present study was relatively low, this may be an alarming situation that reflects a threat limiting treatment options in therapeutic centers studied.
Similar to the results of other studies [25, 26], a significant number of included isolates (83.75%) formed biofilm. The present study revealed a high prevalence of algD, pslD, and pelF genes, being presented simultaneously in a considerable proportion (87.5%) of P. aeruginosa isolates, a finding that are similar to those found by Banar et al. [8]. Other genes associated with biofilm formation, such as pslA and pelA were detected by Ghadaksaz et al. [27] with a frequency of 83.7% and 45.2%, respectively, and Pournajaf et al. [25] with a frequency of 89.5% and 57.3%, respectively among P. aeruginosa clinical isolates. However, little data is available about the prevalence rate of pslD and pelF genes in different regions of the world.
In agreement with other studies [8, 27], our results revealed a significant correlation between the biofilm forming capacity and the presence of relevant genes (p-value < 0.0001). About 88.06% of 67 biofilm producer isolates showed algD +/pslD +/pelF + genotypic pattern, while 11.94% were algD −/pslD −/pelF −. On the other hand, 84.61% of 13 non-biofilm producers carried biofilm genes. The capacity of biofilm production despite the absence of biofilm genes studied indicates other genetic determinants of biofilm participate in matrix formation in P. aeruginosa [14, 28, 29]. By contrast, the presence of genes without biofilm production may be due to chromosomal mutations in different regulatory systems, affecting the production of functional biofilm-associated proteins. Hou et al. [30] reported that no P. aeruginosa isolates were phenotypically positive for biofilm formation in Congo red agar and microtiter plate assays, whereas 31.03% of isolates contained the pslA gene. Conformational changes in quorum sensing proteins due to mutations in lasI/lasR and rhlI/rhlR systems were suggested in previous studies [31–33] as the reason why these isolates are unable to produce biofilm.
According to the results of this study, P. aeruginosa that produced biofilm and also those carried biofilm-associated genes were mainly considered as non-MDR. This may cause a misunderstanding in the first place that biofilm production is not related to antibiotic resistance. It is noteworthy that all isolates in our study were subjected to antimicrobial susceptibility testing as planktonic cells and not in biofilm form. Thus, multiple mechanisms of biofilm and its architectural features, including glycocalyx matrix, outer membrane structure, heterogeneity in metabolism and growth rate, persister cells formation, genetic adaptation, stress responses as well as quorum sensing conferring MDR phenotype were not involved [34]. Regardless, Lima et al. [31] in Brazil found 48.4% of biofilm producer P. aeruginosa isolates were MDR and 51.6% were non-MDR. In another study, Abidi et al. [35] reported that biofilm production was significantly higher in MDR isolates.
In conclusion, combination therapy including an anti-pseudomonal beta-lactam (piperacillin/tazobactam or ceftazidime) and an aminoglycoside or carbapenems (imipenem, meropenem) with fluoroquinolones in conjunction with an aminoglycoside can be used against Pseudomonas infections. Although the rate of resistance to multiple antibiotics among the P. aeruginosa isolates was relatively low in the present study, prudent antimicrobial use and high standards of infection prevention and control are essential to prevent further development of resistant strains. In addition, combination strategies based on the proper anti-pseudomonal antibiotics with anti-biofilm agents can be used to enhance the treatment of biofilm-associated infections.
Limitations
This study may be limited by the lack of clinical information of the patients (treatment, prescription drugs, mortality rate, length of a hospital stay) from whom bacteria were isolated. Furthermore, this study indicates that determination of expression levels of biofilm-associated genes by quantitative real-time PCR may help to evaluate the role of each corresponding gene in biofilm production.
Acknowledgements
The authors would like to appreciate the staffs of hospitals and all colleagues at the Department of Microbiology, Golestan University of Medical Sciences, Gorgan, Iran, for their cooperation in the present study.
Abbreviations
- CF
cystic fibrosis
- CLSI
Clinical and Laboratory Standards Institute
- MDR-PA
multidrug-resistant P. aeruginosa
- OD
optical density
- OF
oxidative-fermentative
- Pel
pellicle
- PCR
polymerase chain reaction
- Psl
polysaccharide synthesis locus
- SIM
sulfide, indole, motility
- TSB
tryptic soy broth
- TSI
triple sugar iron
Authors’ contributions
AA conceived, designed, and supervised the study and revised the manuscript; EK and FE performed the experiments and drafted the manuscript; AA, and AJ analyzed and interpreted the data; AM contributed to statistical analysis. All authors read and approved the final manuscript.
Funding
The present study was financially supported by Golestan University of Medical Sciences, Gorgan, Iran, with Grant Number 110212.
Availability of data and materials
The data associated with this study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
All stages of this study was approved by the Ethics Committee of Golestan University of Medical Sciences with the ethical code number IR.GUMS.REC.1397.142. All included patients were informed orally of the purpose and experimental procedures of the study. However, they did not directly participate in this project. All experiments were performed on bacteria isolated from clinical samples of admitted patients to hospitals.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Esmat Kamali, Email: esmatkm.2222@gmail.com.
Ailar Jamali, Email: jamali@goums.ac.ir.
Abdollah Ardebili, Email: ardebili_abdollah57@yahoo.com.
Freshteh Ezadi, Email: fereshtehezadi92@gmail.com.
Alireza Mohebbi, Email: alirezaa2s@gmail.com.
References
- 1.Pachori P, Gothalwal R, Gandhi P. Emergence of antibiotic resistance Pseudomonas aeruginosa in intensive care unit; a critical review. Genes Dis. 2019;6(2):109–119. doi: 10.1016/j.gendis.2019.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dogonchi AA, Ghaemi EA, Ardebili A, Yazdansetad S, Pournajaf A. Metallo-β-lactamase-mediated resistance among clinical carbapenem-resistant Pseudomonas aeruginosa isolates in northern Iran: a potential threat to clinical therapeutics. Tzu Chi Med J. 2018;30(2):90–96. doi: 10.4103/tcmj.tcmj_101_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bijari A, Azimi L, Fallah F, Ardebili A, Rastegar Lari E, Rastegar Lari A. Involvement of the multidrug efflux pumps in betalactams resistant Pseudomonas aerugionsa clinical isolates collected from burn patients in Iran. Infect Disord Drug Targets. 2016;16(3):172–177. doi: 10.2174/1871526516666160829154302. [DOI] [PubMed] [Google Scholar]
- 4.Moreira MR, Guimarães MP, Rodrigues AA, Gontijo Filho PP. Antimicrobial use, incidence, etiology and resistance patterns in bacteria causing ventilator-associated pneumonia in a clinical-surgical intensive care unit. Rev Soc Bras Med Trop. 2013;46(1):39–44. doi: 10.1590/0037-868216722013. [DOI] [PubMed] [Google Scholar]
- 5.Baker P, Hill PJ, Snarr BD, Alnabelseya N, Pestrak MJ, Lee MJ, et al. Exopolysaccharide biosynthetic glycoside hydrolases can be utilized to disrupt and prevent Pseudomonas aeruginosa biofilms. Sci Adv. 2016;2(5):e1501632. doi: 10.1126/sciadv.1501632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Franklin MJ, Nivens DE, Weadge JT, Howell PL. Biosynthesis of the Pseudomonas aeruginosa extracellular polysaccharides, alginate, Pel, and Psl. Front Microbiol. 2011;2:167. doi: 10.3389/fmicb.2011.00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Govan JR, Deretic V. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol Mol Biol Rev. 1996;60(3):539–574. doi: 10.1128/mr.60.3.539-574.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Banar M, Emaneini M, Satarzadeh M, Abdellahi N, Beigverdi R, van Leeuwen WB, et al. Evaluation of mannosidase and trypsin enzymes effects on biofilm production of Pseudomonas aeruginosa isolated from burn wound infections. PLoS ONE. 2016;11(10):e0164622. doi: 10.1371/journal.pone.0164622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laverty G, Gorman SP, Gilmore BF. Biomolecular mechanisms of Pseudomonas aeruginosa and Escherichia coli biofilm formation. Pathogens. 2014;3(3):596–632. doi: 10.3390/pathogens3030596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma L, Conover M, Lu H, Parsek MR, Bayles K, Wozniak DJ. Assembly and development of the Pseudomonas aeruginosa biofilm matrix. PLoS Pathog. 2009;5(3):e1000354. doi: 10.1371/journal.ppat.1000354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campisano A, Schroeder C, Schemionek M, Overhage J, Rehm BH. PslD is a secreted protein required for biofilm formation by Pseudomonas aeruginosa. Appl Environ Microbiol. 2006;72(4):3066–3068. doi: 10.1128/AEM.72.4.3066-3068.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghafoor A, Jordens Z, Rehm BH. Role of PelF in Pel polysaccharide biosynthesis in Pseudomonas aeruginosa. Appl Environ Microbiol. 2013;79(9):2968–2978. doi: 10.1128/AEM.03666-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jennings LK, Storek KM, Ledvina HE, Coulon C, Marmont LS, Sadovskaya I, et al. Pel is a cationic exopolysaccharide that cross-links extracellular DNA in the Pseudomonas aeruginosa biofilm matrix. Proc Natl Acad Sci U S A. 2015;112(36):11353–11358. doi: 10.1073/pnas.1503058112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friedman L, Kolter R. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol Microbiol. 2004;51(3):675–690. doi: 10.1046/j.1365-2958.2003.03877.x. [DOI] [PubMed] [Google Scholar]
- 15.Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. 28th ed. M100. Wayne: Clinical and Laboratory Standards Institute (CLSI); 2018. p. 38.
- 16.Magiorakos AP, Srinivasan A, Carey R, Carmeli Y, Falagas M, Giske C, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 17.Stepanović S, Vuković D, Hola V, Bonaventura GD, Djukić S, Ćirković I, et al. Quantification of biofilm in microtiter plates: overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. APMIS. 2007;115(8):891–899. doi: 10.1111/j.1600-0463.2007.apm_630.x. [DOI] [PubMed] [Google Scholar]
- 18.Gill J, Arora S, Khanna S, Kumar KH. Prevalence of multidrug-resistant, extensively drug-resistant, and pandrug-resistant Pseudomonas aeruginosa from a tertiary level intensive care unit. J Glob Infect Dis. 2016;8(4):155–159. doi: 10.4103/0974-777X.192962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tümmler B. Emerging therapies against infections with Pseudomonas aeruginosa. F1000Res. 2019 doi: 10.12688/f1000research.19509.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.European Centre for Disease Prevention and Control. Surveillance of Antimicrobial Resistance in Europe 2017. https://ecdc.europa.eu/en/publications-data/surveillance-antimicrobial-resistance-europe-2017. Accessed 15 Nov 2018.
- 21.Vaez H, Salehi-Abargouei A, Ghalehnoo ZR, Khademi F. Multidrug resistant Pseudomonas aeruginosa in Iran: a systematic review and metaanalysis. J Glob Infect Dis. 2018;10(4):212–217. doi: 10.4103/jgid.jgid_113_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bavasheh N, Karmostaji A. Antibiotic resistance pattern and evaluation of blaOXA-10, blaPER-1, blaVEB, blaSHV genes in clinical isolates of Pseudomonas aeruginosa isolated from hospital in south of Iran in 2014–2015. Infect Epidemiol Microbiol. 2017;3(1):1–5. [Google Scholar]
- 23.Nikokar I, Tishayar A, Flakiyan Z, Alijani K, Rehana-Banisaeed S, Hossinpour M, et al. Antibiotic resistance and frequency of class 1 integrons among Pseudomonas aeruginosa, isolated from burn patients in Guilan, Iran. Iran J Microbiol. 2013;5(1):36–41. [PMC free article] [PubMed] [Google Scholar]
- 24.Faghri J, Nouri S, Jalalifar S, Zalipoor M, Halaji M. Investigation of antimicrobial susceptibility, class I and II integrons among Pseudomonas aeruginosa isolates from hospitalized patients in Isfahan, Iran. BMC Res Notes. 2018;11(1):806. doi: 10.1186/s13104-018-3901-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pournajaf A, Razavi S, Irajian G, Ardebili A, Erfani Y, Solgi S, et al. Integron types, antimicrobial resistance genes, virulence gene profile, alginate production and biofilm formation in Iranian cystic fibrosis Pseudomonas aeruginosa isolates. Infez Med. 2018;26(3):226–236. [PubMed] [Google Scholar]
- 26.Vasiljević Z, Jovčić B, Ćirković I, Đukić S. An examination of potential differences in biofilm production among different genotypes of Pseudomonas aeruginosa. Arch Biol Sci. 2014;66(1):117–121. [Google Scholar]
- 27.Ghadaksaz A, Fooladi AAI, Hosseini HM, Amin M. The prevalence of some Pseudomonas virulence genes related to biofilm formation and alginate production among clinical isolates. J App Biomed. 2015;13(1):61–68. [Google Scholar]
- 28.Müsken M, Di Fiore S, Dötsch A, Fischer R, Häussler S. Genetic determinants of Pseudomonas aeruginosa biofilm establishment. Microbiol. 2010;156(2):431–441. doi: 10.1099/mic.0.033290-0. [DOI] [PubMed] [Google Scholar]
- 29.Moradali MF, Ghods S, Rehm BH. Pseudomonas aeruginosa lifestyle: a paradigm for adaptation, survival, and persistence. Front Cell Infect Microbiol. 2017;7:39. doi: 10.3389/fcimb.2017.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hou W, Sun X, Wang Z, Zhang Y. Biofilm-forming capacity of Staphylococcus epidermidis, Staphylococcus aureus, and Pseudomonas aeruginosa from ocular infections. Invest Ophthalmol Vis Sci. 2012;53(9):5624–5631. doi: 10.1167/iovs.11-9114. [DOI] [PubMed] [Google Scholar]
- 31.Lima JLdC, Alves LR, Jacomé PRLA, Neto B, Pacífico J, Maciel MAV, et al. Biofilm production by clinical isolates of Pseudomonas aeruginosa and structural changes in LasR protein of isolates non biofilm-producing. Braz J Infect Dis. 2018;22(2):129–136. doi: 10.1016/j.bjid.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bjarnsholt T, Jensen PØ, Jakobsen TH, Phipps R, Nielsen AK, Rybtke MT, et al. Quorum sensing and virulence of Pseudomonas aeruginosa during lung infection of cystic fibrosis patients. PLoS One. 2010;5(4):e10115. doi: 10.1371/journal.pone.0010115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Senturk S, Ulusoy S, Bosgelmez-Tinaz G, Yagci A. Quorum sensing and virulence of Pseudomonas aeruginosa during urinary tract infections. J Infect Dev Ctries. 2012;6(6):501–507. doi: 10.3855/jidc.2543. [DOI] [PubMed] [Google Scholar]
- 34.Singh S, Singh SK, Chowdhury I, Singh R. Understanding the mechanism of bacterial biofilms resistance to antimicrobial agents. Open Microbiol. J. 2017;11:53–62. doi: 10.2174/1874285801711010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abidi SH, Sherwani SK, Siddiqui TR, Bashir A, Kazmi SU. Drug resistance profile and biofilm forming potential of Pseudomonas aeruginosa isolated from contact lenses in Karachi-Pakistan. BMC Ophthalmol. 2013;13:57. doi: 10.1186/1471-2415-13-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data associated with this study are available from the corresponding author on reasonable request.