Abstract
Background
Chromosomal inversion polymorphisms play a role in adaptation to heterogeneous environments. Inversion polymorphisms are implicated in the very high ecological flexibility of the three main malaria vector species of the Afrotropical Anopheles gambiae complex, facilitating the exploitation of anthropogenic environmental modifications and promoting a strong association with humans. In addition to extending the species’ spatial and temporal distribution, inversions are associated with epidemiologically relevant mosquito behavior and physiology, underscoring their medical importance. We here present novel PCR-RFLP based assays strongly predictive of genotype for the cosmopolitan 2Rb inversion in An. coluzzii and An. gambiae, a development which overcomes the numerous constraints inherent to traditional cytological karyotyping.
Methods
We designed PCR-RFLP genotyping assays based on tag SNPs previously computationally identified as strongly predictive (> 95%) of 2Rb genotype. We targeted those tags whose alternative allelic states destroyed or created the recognition site of a commercially available restriction enzyme, and designed assays with distinctive cleavage profiles for each inversion genotype. The assays were validated on 251 An. coluzzii and 451 An. gambiae cytologically karyotyped specimens from nine countries across Africa and one An. coluzzii laboratory colony.
Results
For three tag SNPs, PCR-RFLP assays (denoted DraIII, MspAI, and TatI) reliably produced robust amplicons and clearly distinguishable electrophoretic profiles for all three inversion genotypes. Results obtained with the DraIII assay are ≥ 95% concordant with cytogenetic assignments in both species, while MspAI and TatI assays produce patterns highly concordant with cytogenetic assignments only in An. coluzzii or An. gambiae, respectively. Joint application of species-appropriate pairs of assays increased the concordance levels to > 99% in An. coluzzii and 98% in An. gambiae. Potential sources of discordance (e.g. imperfect association between tag and inversion, allelic dropout, additional polymorphisms in the restriction target site, incomplete or failed restriction digestion) are discussed.
Conclusions
The availability of highly specific, cost effective and accessible molecular assays for genotyping 2Rb in An. gambiae and An. coluzzii allows karyotyping of both sexes and all developmental stages. These novel tools will accelerate deeper investigations into the role of this ecologically and epidemiologically important chromosomal inversion in vector biology.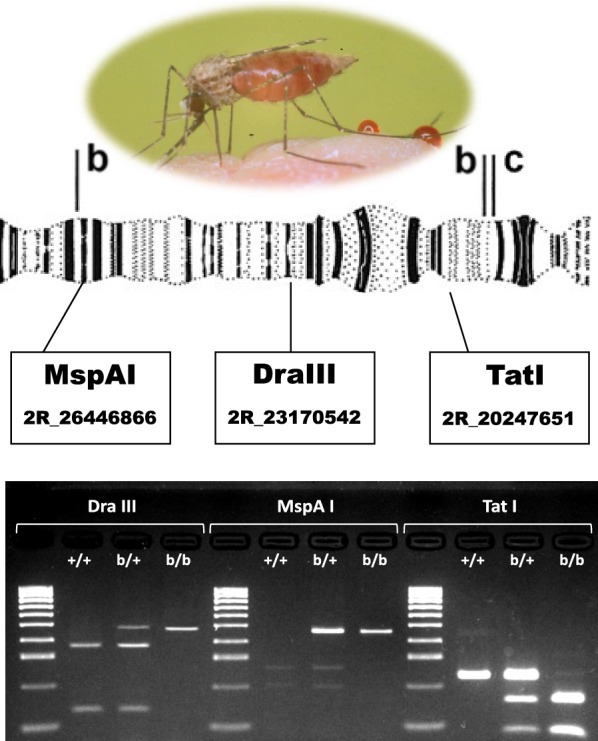
Keywords: Anopheles gambiae complex, Chromosomal inversion, Inversion genotyping, Malaria vector, Molecular karyotyping, PCR-RFLP, Tag SNP
Background
The three main malaria vector species belonging to the Afrotropical Anopheles gambiae complex, An. coluzzii, An. gambiae and An. arabiensis, are characterized by extensive paracentric inversion polymorphisms mostly involving the right arm of chromosome 2 [1, 2]. By suppressing recombination between alternative arrangements in heterokaryotypes and maintaining blocks of locally adapted genes within the breakpoints [3, 4], these paracentric inversions have enabled an extraordinary ecological flexibility, allowing colonization of different habitats across sub-Saharan Africa and facilitating ecological differentiation [5–8]. Inversion polymorphisms also are implicated in the efficient exploitation of anthropogenic environmental modifications and ecological disturbances such as irrigation and deforestation [1, 2, 9–13], helping to promote a strong association between these mosquitoes and humans. This has not only extended their spatial and temporal distribution but also helped transform these species into the most efficient malaria vectors worldwide.
The adaptive value of inversion polymorphisms is particularly evident in the case of the 2La arrangement in An. coluzzii and An. gambiae, whose temporal and spatial distribution is strongly correlated with degree of aridity [2, 14–16]. This strong correlation was first shown 40 years ago based on the demanding cytological karyotyping of thousands of polytene chromosome slides obtained from ovarian nurse cells of females at the half-gravid stage after blood meal - the only adult stage with sufficient chromosome polytenization to allow examination of the banding pattern [17]. Only subsequent to the relatively recent molecular characterization of the 2La breakpoint regions [18] did it become possible to develop a reliable PCR-based diagnostic assay [19] that made karyotyping accessible to non-cytogeneticist and allowed the scoring of large numbers of specimens irrespective of gender, life stage, physiological status, or method of specimen storage. Since then, application of this assay has facilitated the study of ecologically relevant phenotypes associated with the 2La inversion in both laboratory and field settings, such as enhanced desiccation resistance and response to thermal stress [20–26]. Initial cytogenetic observations made decades ago also associated inversion 2La with phenotypes of epidemiological importance, such as susceptibility to Plasmodium infection [27], indoor resting behaviour and response to vector control interventions [2]. Development of molecular diagnostics for inversions like 2La overcomes an important obstacle to follow-up association studies [28] that have been unfeasible before now. Future applications of this assay and others under development for additional inversions will foster a deeper understanding of already recognized or suspected phenotypic associations, and boost the discovery and dissection of unsuspected physiological and behavioural traits of epidemiological and ecological relevance determined by inversions.
Non-random spatial and temporal distribution with respect to degree of aridity also characterizes inversion frequencies on the right arm of chromosome 2, where up to five common inversion polymorphisms segregate in An. coluzzii and An. gambiae. Here, we focus on 2Rb because, aside from 2La, it is the only other inversion in these species with a cosmopolitan distribution across sub-Saharan Africa [1]. Despite molecular characterization of its breakpoints, complex repetitive flanking sequence precluded the development of a robust PCR-based karyotyping assay for this inversion via the same breakpoint-spanning strategy used for the 2La inversion [29]. The breakpoint-proximal 2Rb PCR diagnostic that was developed is not applicable for natural populations due to poor accuracy [29]. Without modern molecular tools that are widely accessible, current understanding of the phenotypic and epidemiological significance of the 2Rb inversion is largely limited to a few historical cytogenetic studies, mostly focused on the association of this polymorphism with dry environments or seasons [2, 13, 27, 30]. The same 2Rb inversion is polymorphic in An. arabiensis, where it has been associated with host-choice [27, 31], underscoring its broader epidemiological relevance in the An. gambiae complex and the importance of studying it more closely.
Recently, multiple tag single nucleotide polymorphisms (SNPs) significantly associated with inversions across geography were computationally identified [32] in a database of genomic variation (Ag1000G) based on deep genome re-sequencing of thousands of specimens from natural An. coluzzii and An. gambiae populations spanning Africa [33]. These tag SNPs are suitable for in silico karyotyping of individual fully sequenced An. gambiae and An. coluzzii mosquitoes (not An. arabiensis, as it was underrepresented in Ag1000G at the time of tag ascertainment). They are also under development as tools for high throughput molecular karyotyping of unsequenced mosquitoes, using targeted approaches such as amplicon sequencing [32]. However, the need remains for inexpensive and widely accessible approaches for genotyping of individual inversions. Amplicon sequencing is ideally suited to large-scale studies, which may not serve more focused needs or smaller budgets. Equally important, those planning to embark on major GWAS studies using amplicon sequencing for inversion genotyping will need to make sure ahead of their sequencing investment that the inversions of interest are sufficiently polymorphic in their populations to give them adequate power to find significant associations if they exist, a goal well-suited to inexpensive PCR assays.
Here we present novel PCR-RFLP based assays that exploit three of the SNPs previously identified [32] as strongly predictive of 2Rb inversion status in An. coluzzii and An. gambiae. We validated these assays on hundreds of cytologically karyotyped An. coluzzii and An. gambiae samples collected across Africa. These assays fill an important gap in available resources required to further our understanding of behavioural, physiological, and epidemiological traits conferred by this widespread inversion, potentially revealing heterogeneities relevant to the success of vector control interventions.
Methods
Cytological karyotyping
Anopheles coluzzii and An. gambiae field-collected specimens were molecularly identified and cytologically karyotyped either specifically for this study or in the framework of previously published studies (Additional file 1: Table S1). In addition, An. coluzzii specimens from the Banfora M colony were karyotyped. This colony was established in 2014 from collections made in the Banfora District of Burkina Faso by the Liverpool School of Tropical Medicine and Hygiene with support from the Centre National de Recherche et de Formation sur le Paludisme. Polytene chromosome preparations followed della Torre [17], extending the hydration of ovarian follicles up to 4 h where necessary, to compensate for the several years of preservation in Carnoy’s solution for historical samples. Paracentric inversion karyotypes were scored according to established nomenclature [2, 13]. All chromosomal slides specifically prepared as part of this study were karyotyped by two independent experts and polytene complements were documented with photomicrographs. Micrographs were retained to allow reassessment of the cytogenetic karyotype in the event of incongruent cytogenetic and molecular results.
Design of PCR-RFLP genotyping assays for 2Rb
Tag single nucleotide polymorphisms (SNPs) predictive of 2Rb genotype were computationally identified previously [32]. Briefly, fully sequenced specimens of An. coluzzii and An. gambiae from the Ag1000G database of natural variation [33] were assigned a presumptive 2Rb inversion genotype using local principal components analysis (PCA) of biallelic SNPs in a window of the genome corresponding to 2Rb. Tag SNPs in that window were those whose genotypes were highly concordant with PCA-based inversion genotypes, such that for most specimens (> 80%), the number of alternate alleles at that site (0, 1 or 2) matched the number of chromosomes inverted for 2Rb carried by the corresponding specimen (0, 1 or 2). For the purposes of designing robust PCR-RFLP genotyping assays from a small subset of the resulting 349 tag SNPs identified by Love et al. [32], we began with the ten tags that showed the highest degree of concordance (> 95%) between SNP- and inversion genotype. Among these ten tag SNPs, we screened for those in which alternative allelic states destroyed or created a restriction enzyme recognition site cleavable by a commercially available enzyme (n = 5), using RestrictionMapper v3 software [34]. Using the An. gambiae and An. coluzzii reference genomes (AgamP4 and AcolM1, respectively) accessed through VectorBase [35] and Primer3Plus v.2.4.2 software [36], we designed primer pairs expected to anneal in both species, that flanked each tag SNP and produced amplicons 200–500 bp in length. We avoided primer binding sites containing either high frequency variants (> 5%, as judged from Ag1000G variation data) or repetitive sequence (as judged from softmasking of AgamP4). We prioritized those assays with robust amplification and enzyme cleavage, and whose electrophoretic profiles provided optimal contrast between inversion genotypes.
PCR-RFLP genotyping
Genomic DNA extraction was conducted from individual cytogenetically karyotyped specimens following a variety of standard protocols, including DNAzol Reagent (Thermo Fisher Scientific, Waltham, MA, USA), DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany), CTAB, and other approaches. Concentration and quality of a subset of genomic DNA samples was assessed using Quant-iT PicoGreen dsDNA Reagent (Thermo Fisher Scientific) or the Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific). The mean concentration was 26 ng/µl based on PicoGreen quantification.
PCR was carried out in 25 µl reactions containing 20 mM Tris-HCl (pH 8.3), 50 mM KCl, 200 µM of each dNTP, 2 mM MgCl2, 5–10 pmol of each primer, 1 U of Taq polymerase and 1 µl of template genomic DNA. PCR conditions included an initial incubation at 94 °C for 2 min, 35 cycles of 94 °C for 30 s, 58 °C for 30 s and 72 °C for 45 s, followed by 72 °C for 2 min and a 4 °C hold.
Restriction digests were performed in 20 µl reactions with 0.5 µl of the appropriate restriction enzyme, following manufacturer recommendations (DraIII and MspAI in 1× CutSmart Buffer at 37 °C for 1 h, (New England Biolabs, Ipswich, MA, USA); TatI in 1× Tango Buffer at 65 °C for 1 h (ThermoFisher Scientific). The amount of PCR product added to each reaction varied from 5 µl for DraIII and MspA1 digests, to 8–10 µl for TatI digests. Optionally, DraIII and MspAI digests were inactivated at 65 °C for 20 min. Results were analyzed by electrophoresis through agarose gels stained with SYBR Safe, using TBE buffer (2% agarose and 0.5× TBE at the University of Notre Dame; 3% agarose and 1× TBE at the University of Rome). Optionally, SDS loading dye was prepared (10 µl of 10%SDS per 1 ml of 6× loading dye) and added to samples prior to electrophoresis to eliminate protein-DNA interactions and prevent gel shifts, as recommended by Thermo Fisher Scientific.
Amplicon sequencing
Enzymatic cleanup of the amplified PCR product was achieved in reactions containing 2 U of Exonuclease 1 (USB Corporation, Cleveland, OH), 1 U of Shrimp Alkaline Phosphate (USB Corporation), 1.8 µl of ddH2O, and 8 µl of the PCR product. After incubation at 37 °C for 15 min, the enzymes were inactivated at 80 °C for 15 min. Sanger sequencing was performed directly on the resulting samples, using one PCR primer and the ABI 3730X1 DNA Analyzer Platform (PE Applied Biosystems, Warrington, England).
Results and discussion
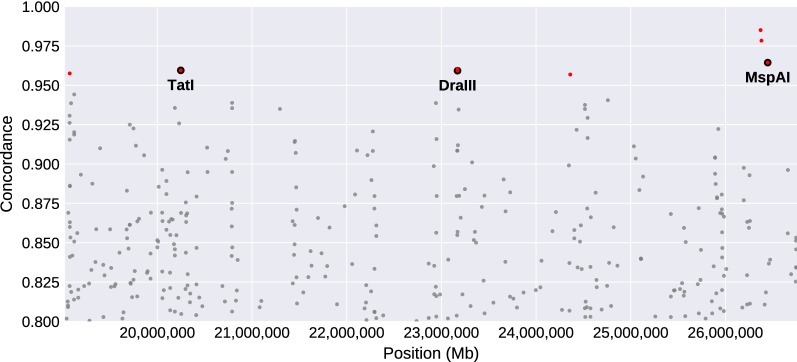
Of the 349 tag SNPs computationally identified as predictive of 2Rb genotype by Love et al. [32], we focused on those whose concordance with PCA-based inversion genotype in Ag1000G was > 95% and whose alternative alleles created or destroyed the recognition sequence of a commercially available restriction enzyme. For three of five such tags, it was possible to design PCR-RFLP assays that reliably produced robust amplicons and distinctive electrophoretic profiles for all three inversion genotypes (Table 1, Fig. 1). For simplicity and brevity, we refer to these three assays by the names of the restriction enzymes each assay employs: DraIII, MspAI and TatI. The chromosomal location of the three tag SNPs targeted by each assay, shown in relation to the 2Rb inversion breakpoints and other 346 tags, is shown in Fig. 2. Overall, the set of 349 tags is not noticeably skewed toward inversion breakpoints, and one of the assay tags (DraIII) is centrally located inside the inversion. Each of the three assays was tested on cytologically karyotyped specimens sampled independently of Ag1000G, from nine countries across Africa (251 An. coluzzii and 451 An. gambiae), and one chromosomally polymorphic An. coluzzii laboratory colony recently established from Burkina Faso (Table 2, Additional file 1: Table S1).
Table 1.
PCR-RFLP genotyping assays for inversion 2Rb in An. gambiae and An. coluzzii
| Tag position | Concord | Ref/Alt | Restriction enzyme | Chromosome cut | Primer pairs (5’-3’) | Amplicon size (bp) | Cleavage products (bp) |
|---|---|---|---|---|---|---|---|
| 23170542 | 96.7% | C/T | DraIII | 2R+b | F: GCCGTTCTCCAGGCTCAG | 481 | 348, 133 |
| R: AAACTCCATTGTACTGGCTGAA | |||||||
| 26446866 | 96.7% | G/A | MspA1 | 2R+b | F: TTCACAACGAAATGGCAAGA | 463 | 264, 199 |
| R: TAGGGCAGTGTTGAGGGAAC | |||||||
| 20247651 | 96.3% | G/A | TatI | 2Rb | F: AATGGCCAGTCTCGAAAGAA | 227 | 153, 74 |
| R: GAAGGGAACGTATGATAATGCAG |
Abbreviations: Tag position, chromosome coordinate; Concord, minimum percent concordance with inversion genotype based on Love et al. [32]; Ref/Alt, reference and alternate allele at tag SNP
Fig. 1.
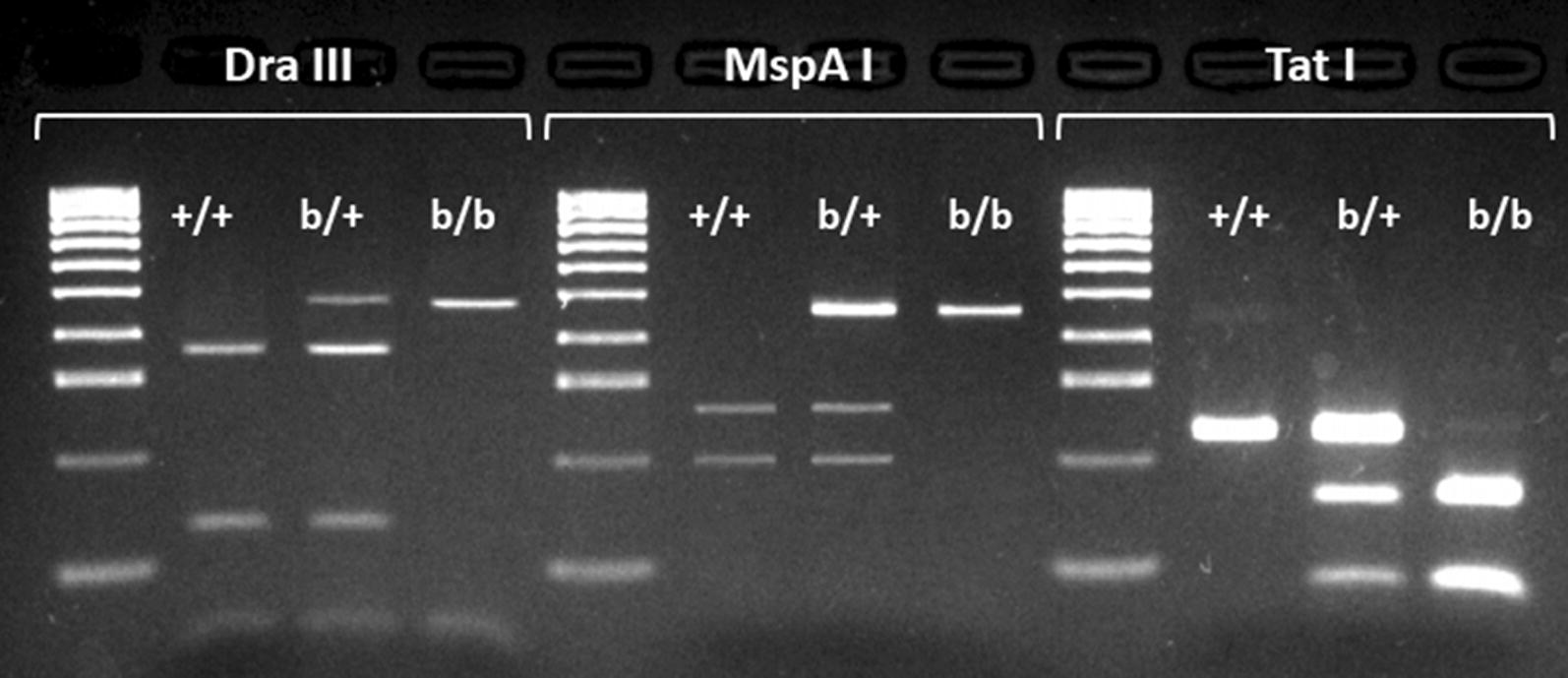
Representative electrophoretic profiles of the DraIII, MspAI and TatI assays for inversion genotyping of 2Rb. Standard (un-inverted) homozygotes for 2Rb, +/+; heterozygotes, b/+; inverted homozyogtes, b/b. Molecular weight marker (Lanes 1, 5, 9), HyperLadder 100 bp (Bioline, Memphis, TN, USA): 100–1000 bp in increments of 100 bp
Fig. 2.
Position of tag SNPs within 2Rb. Scatterplot of genomic location and SNP genotype-inversion genotype concordance for tag SNPs identified for 2Rb. SNPs with concordance > 95% are in red. Those targeted by PCR-RFLP assays are circled and labelled by assay name
Table 2.
Degree of concordance between cytological karyotype and individual PCR-RFLP genotyping assay
| Country | DraIII concordance (%) | MspAI concordance (%) | TatI concordance (%) | |||
|---|---|---|---|---|---|---|
| An. coluzzii | An. gambiae | An. coluzzii | An. gambiae | An. coluzzii | An. gambiae | |
| Benin | 19/19 (100) | na | 19/19 (100) | na | 15/19 (78.9) | na |
| Burkina Faso | na | 19/20 (95.0) | na | 14/20 (70) | na | 17/20 (85) |
| Burkina Faso (lab) | 25/25 (100) | na | 26/27 (96.3) | na | 16/24 (66.6) | na |
| Cameroon | 10/10 (100) | 263/276 (95.2) | 10/10 (100) | 208/274 (75.9) | 10/10 (100) | 253/275 (92) |
| Gambia + Senegala | 6/6 (100) | 56/58 (96.6) | 5/6 (83.3) | 52/56 (92.9) | 4/6 (66.7) | 47/47 (100) |
| Guinea Bissau | 3/3 (100) | 18/18 (100) | 3/3 (100) | 16/16 (100) | 3/3 (100) | 16/16 (100) |
| Mali | 201/211 (95.3) | 55/57 (96.5) | 128/131 (97.7) | 22/26 (84.6) | 104/132 (78.8) | 21/28 (75) |
| Tanzania + DRCa | na | 11/13 (84.6) | na | 13/13 (100) | na | 13/13 (100) |
| Total | 264/274 (96.4) | 422/443 (95.3) | 191/196 (97.4) | 325/405 (80.2) | 152/194 (78.3) | 367/399 (92.0) |
aSpecimens were combined into one sample due to geographical proximity and small sample size
Abbreviations: DRC, Democratic Republic of Congo; na, not available
Sources of discordance and their mitigation
Before detailing the results of each assay, we first consider the factors that could produce disagreement between cytogenetic and PCR-RFLP evidence, and the approaches we took to limit this where feasible. Although we predicted strong agreement between cytological and PCR-RFLP genotype assignments on the basis of > 95% concordance between the component tag SNPs and 2Rb inversion status in Ag1000G (Table 1), the association between tag and inversion is inherently imperfect. Given this unavoidable limitation, multiple PCR-RFLP assays can be combined on the same specimen to increase confidence in the genotypic assignment (see below). In addition, evidence from Table 3 of Love et al. [32] suggests that the rate of human error in 2Rb cytogenetic karyotyping and/or metadata recording is ~4%. We were able to address this issue for the cytogenetic karyotyping performed specifically for this study, by preserving slides that were used to make assignments as well as by preparing an extensive photomicrographic record, allowing us to re-examine (confirm) the cytological assignments in the event of disagreements. This was possible for 227 specimens, but not for the remaining specimens that were processed during preceding studies that did not take the same precautionary measures. Finally, the PCR-RFLP process may also produce artifactual results for technical reasons or due to genetic polymorphisms. The possibility of incomplete or failed restriction digestion is a technical issue that we mitigated by repeating PCR-RFLP assays in the presence of controls, whenever genotypic mismatches were encountered. Substitutions elsewhere in the restriction enzyme recognition site, even if the allelic state of the tag matches the enzyme recognition sequence, can prevent enzymatic cleavage. To determine whether conflicting cytogenetic and PCR-RFLP assignments could be attributed to non-focal (i.e. non-tag) nucleotide polymorphisms in the enzyme recognition sites, we sequenced a subset of PCR amplicons (n = 80). When designing PCR primers flanking the tag SNP, we avoided known polymorphic sites with frequencies 5% in Ag1000G, but in highly polymorphic species like An. gambiae and An. coluzzii [33], the occurrence of polymorphisms in the primer binding sites that could prevent or hinder primer annealing and extension in a fraction of specimens is plausible, and could lead to underrepresentation or elimination of the affected allele (‘allelic dropout’). Allelic dropout, commonly observed in microsatellite data from a broad variety of organisms including An. gambiae [37, 38], is manifested by the underrepresentation of heterozygotes in a population sample. Genetic evidence of a heterozygote deficit typically comes from tests of Hardy-Weinberg equilibrium (HWE), but in the present study, application of this test is complicated by small sample sizes, sourced from many distinct localities even within the same country, challenging the assumption of HWE. Direct evidence (and mitigation) of allelic dropout by designing and applying alternative primers, is a viable but labor-intensive option not adopted here.
DraIII
The overall rate of concordance between the DraIII assay and cytological karyotypes was comparably high in both species, 96.4% for An. coluzzii and 95.5% for An. gambiae (Table 2, Additional file 1: Table S1). This performance is not substantially different from the degree of concordance between the tag SNP and inversion status in the Ag1000G database (96.7%; Table 1). The small total number of discordant cytogenetic and DraIII assignments can be explained at least in part by the fact that the association between tag and inversion is imperfect.
We considered other sources of discordance between DraIII and cytogenetics among the 10 An. coluzzii and 20 An. gambiae specimens with conflicting assignments (Additional file 1: Table S1). Allelic dropout is the most plausible explanation for the five An. coluzzii and 12 An. gambiae in which a cytogenetically heterozygous karyotype (‘1’) disagreed with a DraIII homozygous profile (‘0’ or ‘2’). Moreover, in the 12 instances in which cytogenetic heterozygotes in either species were classified by DraIII as homozygous inverted (‘2’), another (not mutually exclusive) explanation could be failure of enzymatic digestion of true 2R+b amplicons, either for technical reasons or due to the presence of additional SNPs in the recognition sequence apart from the tag itself. Sequencing of five An. gambiae amplicons from specimens typed as ‘1’ cytogenetically and as ‘2’ by their DraIII profiles revealed no evidence of sequence heterozygosity at the tag SNP position, as would have been expected for a true heterozygote. While we cannot rule out that we may have failed to detect true heterozygotes due to strong allelic imbalance in the sequencing reaction, all amplicon sequences appeared to be homozygous at the tag SNP for the uncleavable ‘2’ allele. This suggests that the discrepancies are not due to technical problems with restriction digestion, but more likely are due to allelic dropout and/or incomplete association of the tag with the inversion. Interestingly, in one of these five specimens we did detect a different polymorphism in the DraIII recognition site apart from the tag position, but because the genotype at the tag already rendered it uncleavable by DraIII, this substitution did not impact the expected DraIII profile.
We also sequenced four amplicons from An. coluzzii and An. gambiae derived from specimens whose cytogenetic assignment was homozygous ‘0’, but whose DraIII profile was heterozygous. In one case, sequencing confirmed the cytogenetic assignment, revealing another SNP in the DraIII recognition sequence of one allele that explained the DraIII restriction profile of ‘1’. For two other specimens, sequencing validated the DraIII profile, a result consistent with incomplete association of the tag with the inversion or with partial digestion. The fourth specimen with an unconfirmed cytological assignment of ‘2’ had a DraIII profile of ‘1’, but sequencing revealed that the tag SNP genotype was ‘0’, with no indication of additional SNPs in the recognition sequence. The underlying conflict between cytology (‘2’) and sequence (‘0’) is unresolved, but the DraIII profile of ‘1’ is consistent with partial digestion.
MspAI
In our previous work, the 349 tag SNPs developed for 2Rb proved highly concordant with inversion status in both species and worked well for in silico karyotyping regardless of taxon [32]. It therefore surprised us initially that in the present study, the performance of the MspAI assay depended strongly on taxonomic status (Table 2, Additional file 1: Table S1). Whereas the agreement between cytological and MspAI assignments was 97.4% for An. coluzzii, with only five specimens showing mismatches, much lower agreement (80.2%) was measured for An. gambiae. Close scrutiny suggests that the An. gambiae discrepancies were most likely caused by allelic dropout rather than a failure of the tag SNP itself to predict inversion status. In fact, 66 of 80 An. gambiae specimens with discordant genotypic assignments (among 405 scored) had a cytogenetic karyotype of ‘1’ and a MspAI profile of ‘0’ or ‘2’. Moreover, 56 of those 66 had MspAI profiles of ‘2’, further suggesting that the standard (uninverted) chromosome was the more likely to be affected by allelic dropout. Sequence analysis of the amplicons from 34 An. gambiae specimens with discrepant MspAI profiles of ‘0’ (n =8) and ‘2’ (n = 26) revealed tag genotypes consistent with the MspAI assay.
We also sequenced representative amplicons of five An. gambiae specimens manifesting other discrepancies, in which a homozygous cytogenetic karyotype (‘0’ or ‘2’) disagreed with a heterozygous MspAI profile (two cytologically confirmed examples were sequenced), or an MspAI profile of the opposite homozygote (three examples were sequenced; cytological confirmation was lacking). Sequencing revealed no polymorphisms in the MspAI restriction site apart from the tag itself, and the tag status was fully concordant with the MspAI digestion profile.
For the five An. coluzzii (of 196 scored) with mismatches between cytogenetic and molecular profiles, the cytogenetic assignment was double-checked and confirmed in all cases. Three of these had a cytogenetic karyotype of ‘1’ accompanied by a homozygous MspAI profile of either ‘0’ or ‘2’. In all three, sequencing confirmed the homozyogous MspAI profile, with no additional SNPs in the recognition sites. The other two mismatches involved a cytologically homozygous karyotype (‘0’ or ‘2’) with a heterozygous MspAI profile that was confirmed by sequencing.
If it is assumed that the cytogenetic karyotype was the correct one in each of the above instances of conflict, allelic dropout is one possible explanation when a cytogenetic heterozygote assignment disagrees with a molecular homozygote assignment, but this possibility is smaller if the tag genotype is ‘1’. Whatever the conflict, incomplete association of the tag with the inversion is another, non-exclusive, explanation.
TatI
Overall concordance between cytogenetic karyotype and the TatI assay was lower than for the other two assays, but as was the case for MspAI, there was also a pronounced difference between species. Agreement between cytogenetic and TatI assignments was 92% for An. gambiae, but only 78.4% for An. coluzzii. Unlike the MspIA assay, discordances consistent with allelic dropout (i.e. cytogenetic assignment of ‘1’ and TatI assignment of ‘0’ or ‘2’) were not disproportionate to other types of discordance in either species. Instead, An. coluzzii simply had a higher rate of conflicts of all types (Table 2, Additional file 1: Table S1).
Sequencing of amplicons from 15 specimens (12 An. gambiae and 3 An. coluzzii) with heterozygous cytogenetic assignments and homozygous TatI profiles invariably confirmed that the tag SNP genotype matched the TatI profile, and no other SNPs were identified in the restriction site, consistent with allelic dropout and/or incomplete association with the tag and the inversion.
Sequencing of amplicons from eight An. gambiae with homozygous cytogenetic assignments discordant with heterozygous TatI profiles, revealed four cases in which the tag genotype agreed with the TatI digestion profile. Of these, one could be explained by another SNP in the restriction site, and the remaining three implicated an incomplete TatI digest leading to an inaccurate TatI profile. In three An. coluzzii specimens with homozygous cytogenetic assignment and heterozygous TatI profile, sequencing confirmed the TatI profile, suggesting that if the cytological assignment is assumed correct, these represent the incomplete association of the tag with the inversion.
Finally, sequencing of the amplicon from one An. gambiae specimen with opposite homozygote assignments (cytogenetic ‘2’ vs TatI profile ‘0’), confirmed the TatI assignment, suggesting incomplete association between tag and inversion.
Combinatorial approaches
The DraIII assay is ≥ 95% concordant with cytogenetic assignments in both species, a level that should be adequate for most applications. However, if additional confidence is desired, two assays could be applied jointly on the same specimen. This might be advisable for molecular karyotyping of mosquito populations from regions underrepresented in the Ag1000G database (at the time we accessed it for our work), or underrepresented in the present study, in which An. gambiae samples from Cameroon and An. coluzzii samples from Mali predominate.
Our data suggest that the combination of DraIII and MspAI for An. coluzzii (individually concordant with cytogenetics at 96.4% and 97.4%, respectively) and of DraIII and TatI for An. gambiae (individually concordant at 95.5% and 92%) would be most effective. Joint application of these pairs increased the concordance between cytogenetic and molecular assignments to > 99% (185/186) in An. coluzzii and 98% (354/361) in An. gambiae. In practice, specimens with conflicting molecular assignments (6 of 192 for An. coluzzii and 29 of 390 for An. gambiae) would be considered ambiguous and should be excluded.
Conclusions
Here we have developed three cost effective and accessible molecular assays that can be used individually or in combination for genotyping 2Rb in An. gambiae and An. coluzzii with high specificity. Their performance metrics are based on the conservative assumption that the cytogenetic karyotype is the correct one in the case of conflict between cytogenetic and molecular assignments. Indeed, our results suggest that a variety of phenomena (e.g. imperfect association between tag and inversion, allelic dropout, polymorphisms in the enzyme recognition and/or primer binding sites) contribute to incorrect molecular assignments. However, cytogenetic karyotyping is not infallible, and our experimental design allowed for the validation of only a fraction of the cytogenetic assignments used in this study. From the 1970s to the 1990s, a series of double-blind checks by cytogeneticists at the University of Rome La Sapienza (including the present authors) produced error estimates ranging from 0% to 5%, depending on slide quality. Errors were mainly due to mismatch between the actual reading and karyotype encoding, either on the preparation slide or on the recording sheets, rather than actual banding pattern misinterpretations (V. Petrarca, personal communication). Other groups with less extensive cytogenetic skill and experience might encounter higher error rates. Accordingly, the true accuracy of the PCR-RFLP assays may exceed what we report here. The MspAI assay performed relatively poorly in An. gambiae largely due to allelic dropout. Compared to the other two assays, MspAI targets a SNP very near one of the 2Rb breakpoints (Fig. 2), where the recombination rate is expected to be relatively low. Low recombination should enhance population structure, both between opposite orientations of 2Rb and between the two taxa. Future directions include designing PCR-RFLP assays to genotype 2Rb in An. arabiensis, once this species is adequately represented in Ag1000G. In addition, 2Rc is an inversion locally common in West Africa that, like 2Rb, is implicated in environmental adaptation and ecotypic differentiation. Based on the tag SNPs previously identified in An. gambiae and An. coluzzii [32], efforts are underway to develop PCR-RFLP assays for 2Rc genotyping. Together, these assays will accelerate deeper investigations into the role of these ecologically and epidemiologically important chromosomal inversions in vector biology.
Supplementary information
Additional file 1: Table S1. Detailed genotypic concordance between cytological karyotype and individual PCR-RFLP genotyping assay.
Acknowledgements
The authors thank M. Kern for assistance with PCR-RFLP assays at the University of Notre Dame, and H. Ranson (The London School of Hygiene and Tropical Medicine) for sharing the Banfora colony of An. coluzzii.
Authors’ contributions
NJB and AdT conceived the experiments and obtained the funding. RRL contributed the tag SNPs during their development. RMG and V Pichler designed the assays. RMG, V Pichler, MC, AV and LS performed the molecular assays. RMG and V Pichler performed the data analyses. MC performed cytological karyotyping. V Petrarca, MP and BC provided the samples and confirmed of cytological karyotyping. V Pichler, AdT, NJB, MC and RMG wrote the first draft of the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by the US National Institutes of Health (R01 AI125360). During this work, NJB was supported by Target Malaria, which receives core funding from the Bill & Melinda Gates Foundation and from the Open Philanthropy Project Fund, an advised fund of Silicon Valley Community Foundation. AdT was also supported by Progetti di Ricerca Università SAPIENZA 2018. AV received summer support from the Glynn Family Honors Program of the University of Notre Dame. RMG was supported in part by a Kinesis-Fernández Richards Family Fellowship from the University of Notre Dame.
Availability of data and materials
Data supporting the conclusions of this article are included in the article and its supplementary files. In addition, amplicon sequences determined from this study are available in GenBank under accession numbers MN599476-MN599555.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Raquel Montanez-Gonzalez, Verena Pichler and Maria Calzetta contributed equally to this work
Contributor Information
Raquel Montanez-Gonzalez, Email: Raquel.Montanez-Gonzalez.1@nd.edu.
Verena Pichler, Email: verena.pichler@uniroma1.it.
Maria Calzetta, Email: maria.calzetta@uniroma1.it.
Rachel R. Love, Email: Rachel.R.Love.33@nd.edu
Alexandra Vallera, Email: avaller1@nd.edu.
Lydia Schaecher, Email: lschaech@nd.edu.
Beniamino Caputo, Email: beniamino.caputo@uniroma1.it.
Marco Pombi, Email: marco.pombi@uniroma1.it.
Vincenzo Petrarca, Email: vincenzo.petrarca@uniroma1.it.
Alessandra della Torre, Email: alessandra.dellatorre@uniroma1.it.
Nora J. Besansky, Email: nbesansk@nd.edu
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13071-019-3877-x.
References
- 1.Coluzzi M, Sabatini A, della Torre A, Di Deco MA, Petrarca V. A polytene chromosome analysis of the Anopheles gambiae species complex. Science. 2002;298:1415–1418. doi: 10.1126/science.1077769. [DOI] [PubMed] [Google Scholar]
- 2.Coluzzi M, Sabatini A, Petrarca V, Di Deco MA. Chromosomal differentiation and adaptation to human environments in the Anopheles gambiae complex. Trans R Soc Trop Med Hyg. 1979;73:483–497. doi: 10.1016/0035-9203(79)90036-1. [DOI] [PubMed] [Google Scholar]
- 3.Wellenreuther M, Bernatchez L. Eco-evolutionary genomics of chromosomal inversions. Trends Ecol Evol. 2018;33:427–440. doi: 10.1016/j.tree.2018.04.002. [DOI] [PubMed] [Google Scholar]
- 4.Kirkpatrick M. How and why chromosome inversions evolve. PLoS Biol. 2010;8:e1000501. doi: 10.1371/journal.pbio.1000501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Costantini C, Ayala D, Guelbeogo WM, Pombi M, Some CY, Bassole IHN, et al. Living at the edge: biogeographic patterns of habitat segregation conform to speciation by niche expansion in Anopheles gambiae. BMC Ecol. 2009;9:16. doi: 10.1186/1472-6785-9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simard F, Ayala D, Kamdem GC, Etouna J, Ose K, Fotsing JM, et al. Ecological niche partitioning between the M and S molecular forms of Anopheles gambiae in Cameroon: the ecological side of speciation. BMC Ecol. 2009;9:17. doi: 10.1186/1472-6785-9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ayala D, Acevedo P, Pombi M, Dia I, Boccolini D, Costantini C, et al. Chromosome inversions and ecological plasticity in the main African malaria mosquitoes. Evolution. 2017;71:686–701. doi: 10.1111/evo.13176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coluzzi M. Spatial distribution of chromosomal inversions and speciation in anopheline mosquitoes. In: Barigozzi C, editor. Mechanisms of Speciation. New York: Alan R. Liss, Inc.; 1982. pp. 143–153. [PubMed] [Google Scholar]
- 9.Coluzzi M. Heterogeneities of the malaria vectorial system in tropical Africa and their significance in malaria epidemiology and control. Bull WHO. 1984;62(Suppl.):107–113. [PMC free article] [PubMed] [Google Scholar]
- 10.Coluzzi M. Malaria and the Afrotropical ecosystems: impact of man-made environmental changes. Parassitologia. 1994;36:223–227. [PubMed] [Google Scholar]
- 11.Coluzzi M, Sabatini A, Petrarca V, Di Deco MA. Behavioural divergences between mosquitoes with different inversion karyotypes in polymorphic populations of the Anopheles gambiae complex. Nature. 1977;266:832–833. doi: 10.1038/266832a0. [DOI] [PubMed] [Google Scholar]
- 12.Coluzzi M, Petrarca V, DiDeco MA. Chromosomal inversion intergradation and incipient speciation in Anopheles gambiae. Boll Zool. 1985;52:45–63. doi: 10.1080/11250008509440343. [DOI] [Google Scholar]
- 13.Toure YT, Petrarca V, Traore SF, Coulibaly A, Maiga HM, Sankare O, et al. The distribution and inversion polymorphism of chromosomally recognized taxa of the Anopheles gambiae complex in Mali. West Africa. Parassitologia. 1998;40:477–511. [PubMed] [Google Scholar]
- 14.Coluzzi M. Malaria vector analysis and control. Parasitol Today. 1992;8:113–118. doi: 10.1016/0169-4758(92)90277-9. [DOI] [PubMed] [Google Scholar]
- 15.Cheng C, White BJ, Kamdem C, Mockaitis K, Costantini C, Hahn MW, et al. Ecological genomics of Anopheles gambiae along a latitudinal cline: a population-resequencing approach. Genetics. 2012;190:1417–1432. doi: 10.1534/genetics.111.137794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petrarca V, Sabatinelli G, Di Deco MA, Papakay M. The Anopheles gambiae complex in the Federal Islamic Republic of Comoros (Indian Ocean): some cytogenetic and biometric data. Parassitologia. 1990;32:371–380. [PubMed] [Google Scholar]
- 17.della Torre A. Polytene chromosome preparation from anopheline mosquitoes. In: Crampton JM, Beard CB, Louis C, editors. Molecular biology of disease vectors: a methods manual. London: Chapman & Hall; 1997. pp. 329–336. [Google Scholar]
- 18.Sharakhov IV, White BJ, Sharakhova MV, Kayondo J, Lobo NF, Santolamazza F, et al. Breakpoint structure reveals the unique origin of an interspecific chromosomal inversion (2La) in the Anopheles gambiae complex. Proc Natl Acad Sci USA. 2006;103:6258–6262. doi: 10.1073/pnas.0509683103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.White BJ, Santolamazza F, Kamau L, Pombi M, Grushko O, Mouline K, et al. Molecular karyotyping of the 2La inversion in Anopheles gambiae. Am J Trop Med Hyg. 2007;76:334–339. doi: 10.4269/ajtmh.2007.76.334. [DOI] [PubMed] [Google Scholar]
- 20.Gray EM, Rocca KA, Costantini C, Besansky NJ. Inversion 2La is associated with enhanced desiccation resistance in Anopheles gambiae. Malar J. 2009;8:215. doi: 10.1186/1475-2875-8-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fouet C, Gray E, Besansky NJ, Costantini C. Adaptation to aridity in the malaria mosquito Anopheles gambiae: chromosomal inversion polymorphism and body size influence resistance to desiccation. PLoS One. 2012;7:e34841. doi: 10.1371/journal.pone.0034841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rocca KA, Gray EM, Costantini C, Besansky NJ. 2La chromosomal inversion enhances thermal tolerance of Anopheles gambiae larvae. Malar J. 2009;8:147. doi: 10.1186/1475-2875-8-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cassone BJ, Molloy MJ, Cheng C, Tan JC, Hahn MW, Besansky NJ. Divergent transcriptional response to thermal stress by Anopheles gambiae larvae carrying alternative arrangements of inversion 2La. Mol Ecol. 2011;20:2567–2580. doi: 10.1111/j.1365-294X.2011.05114.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng C, Tan JC, Hahn MW, Besansky NJ. A systems genetic analysis of inversion polymorphisms in the malaria mosquito Anopheles gambiae. Proc Natl Acad Sci USA. 2018;115:E7005–E7014. doi: 10.1073/pnas.1806760115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reidenbach KR, Cheng C, Liu F, Liu C, Besansky NJ, Syed Z. Cuticular differences associated with aridity acclimation in African malaria vectors carrying alternative arrangements of inversion 2La. Parasit Vectors. 2014;7:176. doi: 10.1186/1756-3305-7-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ayala D, Zhang S, Chateau M, Fouet C, Morlais I, Costantini C, et al. Association mapping desiccation resistance within chromosomal inversions in the African malaria vector Anopheles gambiae. Mol Ecol. 2018;28:1333–1342. doi: 10.1111/mec.14880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petrarca V, Beier JC. Intraspecific chromosomal polymorphism in the Anopheles gambiae complex as a factor affecting malaria transmission in the Kisumu area of Kenya. Am J Trop Med Hyg. 1992;46:229–237. doi: 10.4269/ajtmh.1992.46.229. [DOI] [PubMed] [Google Scholar]
- 28.Riehle MM, Bukhari T, Gneme A, Guelbeogo WM, Coulibaly B, Fofana A, et al. The Anopheles gambiae 2La chromosome inversion is associated with susceptibility to Plasmodium falciparum in Africa. Elife. 2017;6:e25813. doi: 10.7554/eLife.25813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lobo NF, Sangare DM, Regier AA, Reidenbach KR, Bretz DA, Sharakhova MV, et al. Breakpoint structure of the Anopheles gambiae 2Rb chromosomal inversion. Malar J. 2010;9:293. doi: 10.1186/1475-2875-9-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rishikesh N, Di Deco MA, Petrarca V, Coluzzi M. Seasonal variations in indoor resting Anopheles gambiae and Anopheles arabiensis in Kaduna, Nigeria. Acta Trop. 1985;42:165–170. [PubMed] [Google Scholar]
- 31.Main BJ, Lee Y, Ferguson HM, Kreppel KS, Kihonda A, Govella NJ, et al. The genetic basis of host preference and resting behavior in the major African malaria vector, Anopheles arabiensis. PLoS Genet. 2016;12:e1006303. doi: 10.1371/journal.pgen.1006303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Love RR, Redmond SN, Pombi M, Caputo B, Petrarca V, della Torre A, et al. In silico karyotyping of chromosomally polymorphic malaria mosquitoes in the Anopheles gambiae complex. G3 (Bethesda) 2019;9:3249–3262. doi: 10.1534/g3.119.400445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miles A, Harding NJ, Bottà G, Clarkson CS, Antão T, Kozak K, et al. Genetic diversity of the African malaria vector Anopheles gambiae. Nature. 2017;552:96–100. doi: 10.1038/nature24995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.RestrictionMapper. http://www.restrictionmapper.org/. Accessed 7 Aug 2019.
- 35.Giraldo-Calderon GI, Emrich SJ, MacCallum RM, Maslen G, Dialynas E, Topalis P, et al. VectorBase: an updated bioinformatics resource for invertebrate vectors and other organisms related with human diseases. Nucleic Acids Res. 2015;43:D707–D713. doi: 10.1093/nar/gku1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, et al. Primer3 - new capabilities and interfaces. Nucleic Acids Res. 2012;40:e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lehmann T, Licht M, Elissa N, Maega BT, Chimumbwa JM, Watsenga FT, et al. Population structure of Anopheles gambiae in Africa. J Hered. 2003;94:133–147. doi: 10.1093/jhered/esg024. [DOI] [PubMed] [Google Scholar]
- 38.Pinto J, Egyir-Yawson A, Vicente J, Gomes B, Santolamazza F, Moreno M, et al. Geographic population structure of the African malaria vector Anopheles gambiae suggests a role for the forest-savannah biome transition as a barrier to gene flow. Evol Appl. 2013;6:910–924. doi: 10.1111/eva.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Detailed genotypic concordance between cytological karyotype and individual PCR-RFLP genotyping assay.
Data Availability Statement
Data supporting the conclusions of this article are included in the article and its supplementary files. In addition, amplicon sequences determined from this study are available in GenBank under accession numbers MN599476-MN599555.