Abstract
Motivation
The complexity of protein–protein interactions (PPIs) is further compounded by the fact that an average protein consists of two or more domains, structurally and evolutionary independent subunits. Experimental studies have demonstrated that an interaction between a pair of proteins is not carried out by all domains constituting each protein, but rather by a select subset. However, determining which domains from each protein mediate the corresponding PPI is a challenging task.
Results
Here, we present domain interaction statistical potential (DISPOT), a simple knowledge-based statistical potential that estimates the propensity of an interaction between a pair of protein domains, given their structural classification of protein (SCOP) family annotations. The statistical potential is derived based on the analysis of >352 000 structurally resolved PPIs obtained from DOMMINO, a comprehensive database of structurally resolved macromolecular interactions.
Availability and implementation
DISPOT is implemented in Python 2.7 and packaged as an open-source tool. DISPOT is implemented in two modes, basic and auto-extraction. The source code for both modes is available on GitHub: https://github.com/korkinlab/dispot and standalone docker images on DockerHub: https://hub.docker.com/r/korkinlab/dispot. The web server is freely available at http://dispot.korkinlab.org/.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Large-scale characterization of protein–protein interactions (PPIs) using high-throughput interactomics approaches, such as yeast-two-hybrid and tandem-affinity purification/mass spectrometry methods (Gavin et al., 2002; Rolland et al., 2014), have provided the scientists with the new insights of the cell functioning at the systems level and allowed to better understand the molecular machinery underlying complex genetic disorders (Barabasi and Oltvai, 2004; Cui et al., 2015; Mitra et al., 2013). Structural studies of PPIs have revealed that a PPI is often carried out by smaller structural protein subunits, the protein domains (Ekman et al., 2005; Jin et al., 2009; Vogel et al., 2004). Roughly two-thirds of eukaryotic and more than one-third of prokaryotic proteins are estimated to be multi-domain proteins (Ekman et al., 2005), and thus it is not surprising that ≈ 46% of structurally resolved interactions are domain–domain interactions (Kuang et al., 2016). A high-throughput breakdown of the interactome at this, domain-level, resolution is a much more experimentally challenging task, currently unfeasible at the whole-system level and requiring computational methods to step in (Deng et al., 2002; Finn et al., 2005; Ohue et al., 2014; Segura et al., 2015).
Here, we present a simple knowledge-based domain interaction statistical potential (DISPOT), a tool that leverages the statistical information on interactions shared between the homologous domains from structurally defined domain families. The knowledge-based potentials are extracted from our comprehensive database of structurally resolved macromolecular interactions, DOMMINO (Kuang et al., 2016). Our statistical potential can be integrated into PPI prediction methods that deal with multi-domain proteins by ranking all possible pairwise combinations of domain interactions between two or more proteins. We want to stress that although DISPOT potentials provide some insight into PPI, it is not a classification method, and data provided by it should be used in conjunction with additional information, e.g. a specific pathway (Fig. 1E).
Fig. 1.
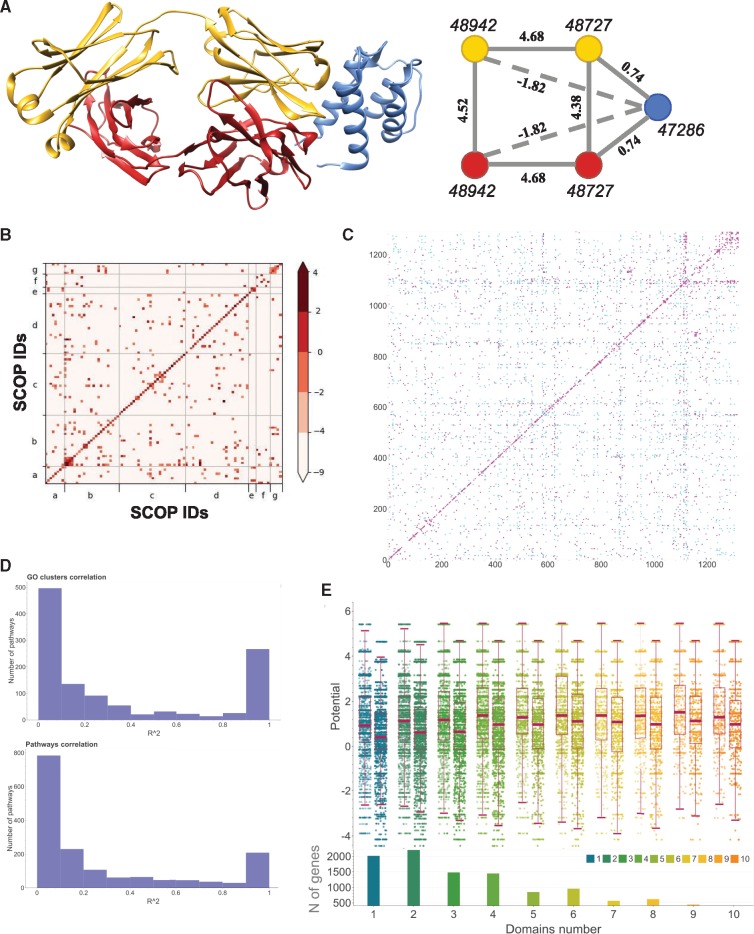
DISPOT statistical potential and its application. (A) A crystal structure (left) of the protein complex between CNTO607 Fab human monoclonal antibody (yellow and red colors denote two different chains) and interleukin-13 (IL-13, shown in blue), and the corresponding domain–domain interaction network (right). Shown in italics are SCOP family IDs, and in bold are DISPOT values for the corresponding interactions. Nodes colored with the same color belong to the same chain. Solid lines connecting nodes correspond to the physical interactions, while dashed lines connect nodes corresponding to the protein domains that do not physically interact. (B) A heatmap showing DISPOT values calculated for each pair of SCOP families, where only potentials for pairs of SCOP families with five and more non-redundant interactions are plotted. The families are grouped based on the SCOP class (a–g) and are ordered within each fold based on their IDs. (C) A contact map showing the correlation between experimentally obtained human interactome HI-I-05 and DISPOT-based PPI prediction. A prediction that calls a PPI correctly is shown in magenta, while PPIs that were missed are shown in cyan. (D) Correlation calculated using R2 correlation coefficient between the hu.MAP interaction probability score and DISPOT statistical potential for KEGG pathways (bottom) and GO clusters (top). (E) Distribution of the protein-level DISPOT statistical potentials grouped by the number of SCOP domains in a protein defined using SUPERFAMILY
2 Methodology
The development of DISPOT is driven by several observations. First, an average interaction between a pair of proteins is not carried out by all domains constituting each protein, but only by a select subset. Indeed, each domain has its unique structure and biological function and may not be designed to interact with a particular domain from another protein (Banappagari et al., 2010; Shimizu et al., 2016). Second, the domain–domain interactions often share homology: when two homologous domains interact with their partners, these partners frequently also share the homology with each other (Kuang et al., 2016). Thus, one can introduce the domain–domain interaction propensity in terms of the frequency of domain–domain interactions between the two domain families. Lastly, the propensity of domains to interact is expected to vary across different families, thus allowing to provide the finer resolution of the PPI network.
The quantification of the odds for a domain from one domain family to interact with a domain from another family is defined in this work as a knowledge-based statistical potential. Statistical potentials are widely used in biophysical applications, often for characterizing the residue contacts between the protein chains (Huang and Zou, 2008; Krüger et al., 2014; Lu et al., 2003). One of the main applications of the residue-level statistical potentials is in protein docking (Kozakov et al., 2006). Our domain–domain statistical potential complements the residue-level potentials by considering structural units from the higher-level of protein structure hierarchy and requiring no structural information about the protein domains. Specifically, the input for DISPOT includes the protein sequences of the two proteins interacting with each other.
First, the domain architecture of each protein is obtained. To do so, a region of the protein sequence is annotated to a family of homologous domains. For the definition of domain families, we leverage the structural classification of proteins (SCOP) family-level classification (Andreeva et al., 2004). SCOP represents a structure-based hierarchical classification of relationships between protein domains or single-domain proteins with ‘family’ being the first level of SCOP classification and ‘superfamily’ being the second level. Protein domains from the same SCOP family are evolutionary closely related and often share the same function. Since a protein with no structural information cannot be directly annotated by SCOP, we use SUPERFAMILY (Gough and Chothia, 2002), a Hidden Markov Model (HMM)-based approach that maps regions of a protein sequence to one or several SCOP families or superfamilies. SUPERFAMILY allows us to cover a substantial subset of known proteins: the HMM coverage at the protein sequence and overall amino acid levels for the UniProt database were reported at 64.73% and 58.78%, respectively, in 2014 (Oates et al., 2015).
Second, for each pair of SCOP families we count a number of non-redundant PPIs between the members of these families that have been experimentally determined. Our source of data is DOMMINO (Kuang et al., 2012, 2016) a comprehensive database of structurally resolved macromolecular interactions. It contains information about interactions between the protein domains, interdomain linkers, terminal sequences, and protein peptides. In this work, we use exclusively domain–domain interactions because the data about this type of interactions is the most abundant. To remove redundancy in the data, we use ASTRAL compendium (Brenner et al., 2000), which is integrated into the SCOPe database (Fox et al., 2014). From ASTRAL, we obtain a set of domains, where each domain shares <95% sequence identity to any other domain in the set. This set is then used to determine pairs of redundant domain–domain interactions in the original DOMMINO dataset. Two domain–domain interactions are determined as redundant if both corresponding pairs of domains share 95% or more sequence identity. For each pair of redundant domain–domain interactions, one interaction is randomly removed. The process continues until no pair of redundant interactions can be detected.
Third, for each domain family from each protein, a statistical potential is calculated (Fig. 1A). There are two types of statistical potentials introduced in this work: (i) calculated for a domain from a specific domain family and (ii) calculated for a pair of domains, one domain from each of the two interacting proteins. The statistical potential Pi for a single domain Di is calculated based on the total number of interactions extracted from the non-redundant DOMMINO dataset for the specific SCOP family this domain belongs to. The statistical potential Pij for a pair of domains, Di and Dj, is calculated based on the total number of occurrences Nij of the interactions between all domains from the same two SCOP families as Di and Dj. Those numbers are then transformed into probabilities as follows:
where is an average number of interactions for a domain family and is an average number of interactions for a pair of domain families, both calculated from the non-redundant DOMMINO set.
DISPOT potentials are derived following a standard strategy for calculating a statistical potential. The statistical potentials for the atomic contact pairs are traditionally derived based on Boltzmann relation (Huang and Zou, 2008):
where k is the Boltzmann constant, T is the system’s temperature, pij is an experimentally observed density of atom pairs from different partners in a complex at distance and is corresponding density in the reference state. Since we do not work with the atomic-level physical interactions, we replace the Boltzmann constant from DISPOT equations and substitute temperature with the inverse of normalization constant Z. In addition, pij and are substituted with the number of interactions between domains in DOMMINO database.
DISPOT can also provide integrated protein-level statistics. There are multiple ways to combine the domain-level statistics into a protein-level statistics. Two simple approaches to integrate domain–domain interactions for a given PPI in terms of a standalone (single protein) and interaction (protein pair) potentials are:
respectively, where i and j correspond to the domains from protein u and v. The rationale behind these definitions lies in the assumption that a single strongest domain–domain interaction is the one of the most important defining factor for the PPI. These definitions of cumulative potentials were tested in terms of their ability to predict a PPI using several experimental sources. First, we obtained the coverage landscape by the cumulative potentials on the experimental protein–protein interactomes one obtained using high-throughput yeast-two-hybrid screening (HI-I-05) (Rual et al., 2005) and another one obtained using curated literature-based search (LitBM-17, http://interactome.baderlab.org/data/LitBM-17.psi). As expected, while this naïve method was able to recover 2944 PPIs in HI-I-05, it missed 1188 PPIs even using a lenient threshold of −20 (Fig. 1C). Similarly, the cumulative potential was able to recover only 1718 PPIs while 1453 PPIs were not recovered (Supplementary Fig. S1). We then apply the same pairwise cumulative potential to the large-scale mass spectrometry study (Drew et al., 2017). Specifically we study the correlation between the hu.MAP probability score and cumulative pairwise score among KEGG pathways (Kanehisa and Goto, 2000) and GO clusters produced by GeneSCF on 13 855 genes with SUPERFAMILY annotation (Subhash and Kanduri, 2016) (Fig. 1D). While the number of highly correlated pairs was substantial, the number of pairs with very little correlation still prevailed. Finally, the analysis of the cumulative single potential for a protein showed that it can obtain a diverse range of values and this property seems to be independent of how many domains this protein has (Fig. 1E). Similar behavior was observed when looking at the other basic cumulative measures (Supplementary Fig. S3).
Overall, we have analyzed and summarized interactions from 3619 SCOP family pairs that were extracted from 352 199 PPIs. In total, domains from 1384 SCOP families were characterized that form domain–domain interactions in 1384 ‘homo-SCOP’ interaction pairs (i.e., both domains are annotated with the same SCOP family) and 2235 ‘hetero-SCOP’ pairs (Fig. 1B and Supplementary Fig. S1). The analysis of the calculated statistical potentials showed a wide diversity across different families.
Finally, we would like to make a cautionary note of using the developed tool. DISPOT was designed not as a PPI prediction tool, but rather a tool that provides additional information on the likelihood of specific domain–domain interactions in a given physical PPI. The main reason is the fact that structural coverage of the PPI space is still far from being full, which leads to the presence of a high number of false negatives if one was to use DISPOT as a standalone predictor. This intuition has been supported by our evaluation of DISPOT against the two interactomics golden standards. Thus, if a researcher wants to employ DISPOT in a PPI prediction method, we recommend adding the DISPOT potentials as features to the overall feature vector, that would include other parameters, such as secondary structure, evolutionary conservation of the sequence, predicted residue hydrophobicity, etc.
3 Implementation and usage
The basic mode is implemented in Python with the dependency on packages pandas and numpy. It takes SCOP identifiers (IDs) for either ‘family’ (fa) or ‘superfamily’ (sf) hierarchy levels as an input and produces statistical potential for corresponding pair of domains. Switching between the SCOP levels is implemented in command line option sf. One of the possible input options is a command line option domains, which provides a list of space-separated SCOP identifiers. Based on this list, the program produces all possible unique pairwise combinations of identifiers and the corresponding statistical potentials. Option max produces the highest value of statistical potential for a selected domain and an SCOP ID for the corresponding interaction domain partner. Option output specifies the output file. If no file path is specified, then program opens a console output prompting a user to input the data. A detailed description of all acceptable input formats and options is available in README file and help menu of the main script dispot.py.
The auto-extraction version relies on the SUPERFAMILY models and scripts and HMMER program for extracting the corresponding SCOP IDs for either family or superfamily levels of hierarchy. The Perl programming language interpreter is an additional dependency. HMMER is compatible with the major linux distributions (it has been tested on Ubuntu 16.04 and Alpine 3.7 with additional installation of alpine-glibc). Windows users are advised to use the docker image. The main script is dispot.py, and it includes several options: fasta_folder—to specify a path to the folder with FASTA files; output_folder—to specify a path to the results and max—to substitute the regular output of all pairwise statistical potentials with the highest statistical potential for a given domain family and an SCOP ID of the interaction partner on which this value is achieved. Additional script batch_process.py provides almost the same functionality, except it uses the default locations: ./data/for the input and ./data/results/for the output. For each FASTA sequence, we extract a SUPERFAMILY-derived SCOP ID and the location(s) of the corresponding domain on the protein sequence. It is stored in the ./tmp/folder and is available until the next run of any of the scripts mentioned in this section. The data are stored in the Python dictionary objects serialized by package pickle.
DISPOT has also been implemented as a web server that carries the full functionality of the developed methods and comes with a tutorial. The web server is freely available at http://dispot.korkinlab.org/.
Funding
This work was supported by the National Science Foundation (1458267) and National Institute of Health (LM012772-01A1) to D.K.
Conflict of Interest: none declared.
Supplementary Material
References
- Andreeva A. et al. (2004) SCOP database in 2004: refinements integrate structure and sequence family data. Nucleic Acids Res., 32, D226–D229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banappagari S. et al. (2010) A conformationally constrained peptidomimetic binds to the extracellular region of HER2 protein. J. Biomol. Struct. Dyn., 28, 289–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabasi A.-L., Oltvai Z.N. (2004) Network biology: understanding the cell’s functional organization. Nat. Rev. Genet., 5, 101.. [DOI] [PubMed] [Google Scholar]
- Brenner S.E. et al. (2000) The ASTRAL compendium for protein structure and sequence analysis. Nucleic Acids Res., 28, 254–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H. et al. (2015) The variation game: cracking complex genetic disorders with NGS and omics data. Methods, 79, 18–31. [DOI] [PubMed] [Google Scholar]
- Deng M. et al. (2002) Inferring domain-domain interactions from protein-protein interactions In: Proceedings of the Sixth Annual International Conference on Computational Biology, ACM, pp. 117–126. [Google Scholar]
- Drew K. et al. (2017) Integration of over 9,000 mass spectrometry experiments builds a global map of human protein complexes. Mol. Syst. Biol., 13, 932.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekman D. et al. (2005) Multi-domain proteins in the three kingdoms of life: orphan domains and other unassigned regions. J. Mol. Biol., 348, 231–243. [DOI] [PubMed] [Google Scholar]
- Finn R.D. et al. (2005) iPfam: visualization of protein–protein interactions in PDB at domain and amino acid resolutions. Bioinformatics, 21, 410–412. [DOI] [PubMed] [Google Scholar]
- Fox N.K. et al. (2014) SCOPe: structural classification of proteins-extended, integrating SCOP and ASTRAL data and classification of new structures. Nucleic Acids Res., 42, D304–D309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin A.-C. et al. (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature, 415, 141.. [DOI] [PubMed] [Google Scholar]
- Gough J., Chothia C. (2002) SUPERFAMILY: hMMs representing all proteins of known structure. SCOP sequence searches, alignments and genome assignments. Nucleic Acids Res., 30, 268–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S.-Y., Zou X. (2008) An iterative knowledge-based scoring function for protein–protein recognition. Proteins, 72, 557–579. [DOI] [PubMed] [Google Scholar]
- Jin J. et al. (2009) Eukaryotic protein domains as functional units of cellular evolution. Sci. Signal., 2, ra76.. [DOI] [PubMed] [Google Scholar]
- Kanehisa M., Goto S. (2000) KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res., 28, 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozakov D. et al. (2006) PIPER: an FFT-based protein docking program with pairwise potentials. Proteins, 65, 392–406. [DOI] [PubMed] [Google Scholar]
- Krüger D.M. et al. (2014) DrugScorePPI knowledge-based potentials used as scoring and objective function in protein-protein docking. PLoS One, 9, e89466.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang X. et al. (2012) DOMMINO: a database of macromolecular interactions. Nucleic Acids Res., 40, D501–D506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang X. et al. (2016) DOMMINO 2.0: integrating structurally resolved protein-, RNA-, and DNA-mediated macromolecular interactions. Database, 2016, bav114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H. et al. (2003) Development of unified statistical potentials describing protein-protein interactions. Biophys. J., 84, 1895–1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra K. et al. (2013) Integrative approaches for finding modular structure in biological networks. Nat. Rev. Genet., 14, 719.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oates M.E. et al. (2015) The SUPERFAMILY 1.75 database in 2014: a doubling of data. Nucleic Acids Res., 43, D227–D233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohue M. et al. (2014) MEGADOCK: an all-to-all protein-protein interaction prediction system using tertiary structure data. Protein Pept. Lett., 21, 766–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland T. et al. (2014) A proteome-scale map of the human interactome network. Cell, 159, 1212–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rual J.-F. et al. (2005) Towards a proteome-scale map of the human protein–protein interaction network. Nature, 437, 1173.. [DOI] [PubMed] [Google Scholar]
- Segura J. et al. (2015) Using neighborhood cohesiveness to infer interactions between protein domains. Bioinformatics, 31, 2545–2552. [DOI] [PubMed] [Google Scholar]
- Shimizu M. et al. (2016) Near-atomic structural model for bacterial DNA replication initiation complex and its functional insights. Proc. Natl. Acad. Sci. USA, 113, E8021–E8030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subhash S., Kanduri C. (2016) GeneSCF: a real-time based functional enrichment tool with support for multiple organisms. BMC Bioinform., 17, 365.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel C. et al. (2004) Structure, function and evolution of multidomain proteins. Curr. Opin. Struct. Biol., 14, 208–216. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.