Summary
Generation of antigen‐specific humoral responses following vaccination or infection requires the maturation and function of highly specialized immune cells in secondary lymphoid organs (SLO), such as lymph nodes or tonsils. Factors that orchestrate the dynamics of these cells are still poorly understood. Currently, experimental approaches that enable a detailed description of the function of the immune system in SLO have been mainly developed and optimized in animal models. Conversely, methodological approaches in humans are mainly based on the use of blood‐associated material because of the challenging access to tissues. Indeed, only few studies in humans were able to provide a discrete description of the complex network of cytokines, chemokines and lymphocytes acting in tissues after antigenic challenge. Furthermore, even fewer data are currently available on the interaction occurring within the complex micro‐architecture of the SLO. This information is crucial in order to design particular vaccination strategies, especially for patients affected by chronic and immune compromising medical conditions who are under‐vaccinated or who respond poorly to immunizations. Analysis of immune cells in different human tissues by high‐throughput technologies, able to obtain data ranging from gene signature to protein expression and cell phenotypes, is needed to dissect the peculiarity of each immune cell in a definite human tissue. The main aim of this review is to provide an in‐depth description of the current available methodologies, proven evidence and future perspectives in the analysis of immune mechanisms following immunization or infections in SLO.
Keywords: computational immunology, cytometry, immune system, lymphocytes, microscopy, OMICs sciences
In this review we aim to dissect novel technologies to analyze the immune system plasticity at tissue level. Such tools are needed in order to ‘study immune compartments as one inter‐dependent functional unit'. Our purpose is to highlight the advantages and the limitations of the most widely used approaches focusing on the importance of tissues analysis and the need of a computational approach.

Introduction
The immune system is a complex biological system made of several factors, whose interplay in different body compartments such as blood and secondary lymphoid organs (SLO) is fundamental for the proper response against foreign antigens. This mechanism relies upon the interactions occurring among an intricate network of cytokines, chemokines and lymphocytes acting in tissues. These inter‐relationships lead to different types of immunological response, including generation of both antigen‐specific antibodies and long‐lived memory B cells able to respond to rechallenge 1.
Different samples, such as blood suspension and tissues from animal models, are essential to understand the immune system. These data can rarely be translated into human biology, for several reasons. For instance, inbred animals usually grow up in germ‐free conditions and are infected by a specific pathogen to mimic natural exposure. Conversely, the genetic background and environmental exposures from the first hours of life make every human being unique and not readily reproducible by animal models 2. Other major concerns include differences in the balance of leukocyte subsets, Toll‐like receptors and cytokines 3. Because access to SLO in humans is difficult, many studies have been focused upon the investigation of circulating lymphocyte subsets 4, leaving unresolved questions about their micro‐anatomical site of differentiation within tissues.
The introduction of multi‐parametric flow cytometry in the early 2000s 5 allowed the simultaneous detection of several different cell subtypes. Nevertheless, cell–cell interactions can only be partially explained by cell suspension analysis. The SLO and, in particular, lymph nodes (LNs) are key points in adaptive immunity and are essential for both the generation of protective immune response upon vaccination and the elimination of invading pathogens. Given the limited access to LNs, several investigators have focused their studies on understanding immune dynamics in tonsils. Despite the unique location of tonsils and the dominant presence of B cells compared to LNs, tonsillar lymphoid follicles share common features with LN follicles, such as the presence of a germinal center (GC) with distinct ‘dark' and ‘light' zones 6, 7, 8, 9.
A better understanding of the biology of human immune system within tissues, where the actual immune responses take place, is mandatory to fill the gap of knowledge between animal and human models and to gain more insight into the basic immunology. Moreover, during recent years the ‘omics' sciences are moving the field towards a personalized medicine strategy through the analysis of any biological event regulating genes, proteins and metabolite physiology. The huge amount of information potentially generated poses the need for advanced bioinformatics able to harmonize the produced data tools in order to gain an informative and comprehensive overview of the human immune system. Here we will review most of the available laboratory technologies able to dissect the immune function at different complexity levels.
Dissecting technologies
In general, the currently available immunoassays/platforms can be divided into (i) label‐free (i.e. deep seq, metabolomics) and (ii) label‐bound assays, where labeled probes [i.e. antibodies, messenger RNA (mRNA) probes] recognize specific protein/epitopes or amino acid sequences and can be revealed by different technologies, including light/fluorescence‐based techniques (i.e. confocal microscopy, enzyme‐linked immunoassays), mass spectrometry (i.e. CytoF, imaging mass cytometry) and radioactive tracers. Besides the high throughput potential, label‐free assays have the advantage to provide non‐hypothesis‐driven immunological insights leading to novel discoveries.
Phenotypical and functional characterization
Dissecting the complexity of immune reactions requires comprehensive analysis of the cellular populations mediating these interactions. To this end, the characterization of surface and intracellular molecules expressed by individual cells is of special interest. Development in this field has led to the design of multi‐parameter flow cytometry, which represents a milestone in the immunology field 10. Currently, the cells bound to antibodies can be detected by (i) immunofluorescence flow cytometry and (ii) mass spectrometry. Both platforms allow for the simultaneous quantitative analysis of several labels. Fluorescence‐activated cell‐sorting (FACS) permits the separation of cells with specific optical properties from the flow of the analyzed cells 11, 12 and their use for downstream analysis (proteomics, genomics, ex‐vivo culture) 13, 14, 15. Besides labeled antibodies, flow cytometry assays employing fluorescent mRNA or DNA probes have been recently developed for the detection of cell populations expressing specific nucleotide sequences 16. In addition to immunophenotyping, ex‐vivo flow cytometry assays have been used extensively for the functional assessment of relevant immune cell populations by detecting the de‐novo generation of biological factors such as cytokines, chemokines and killing mediators (GrzB, FasL) 17, 18. Analysis of the phosphorylation levels of intracellular proteins sheds further light on the signaling pathways operating in specific immune cells during the development of immune responses with respect to disease status 19. As well as the relative frequency, the expression level of molecules per cell (judged by mean fluorescence intensity) can be also analyzed. In addition to bulk populations, in‐depth immunophenotyping of T/B cells requires analysis of the development of antigen‐specific cells following infection or vaccination. The complexity of lymphocyte subsets can be also monitored in terms of specificity. In the mid‐1990s the introduction of human leukocyte antigen (HLA) tetrameric complexes revolutionized the field, allowing the detection of virus specific T cells 20. Additionally, probes specific for some viruses [i.e. cytomegalovirus (CMV)] or epitopes contained in vaccines (such as H1N1 in influenza vaccine) can be engineered to contain a fluorophore that can be monitored by flow cytometry in order to follow the development of antigen‐specific B/T cells in blood and tissues 21, 22, 23. Flow cytometry‐based assays also allow the analysis of hematopoietic neoplasms cells and epithelial neoplasms tracking circulating tumor single cells and clusters 24, 25, as well as different subsets of immune cells in the context of cancer 26. The overlapping wavelength spectra between fluorochromes limits the number of different labeled markers that could be used in an immunofluorescence flow cytometry assay. Recent development of novel fluorochromes enables the use of extra lasers and the detection of up to 30 parameters 27, 28.
In recent decades new approaches have been developed for the visualization and quantification of highly specialized cell subsets. Time‐of‐flight mass cytometry (CyTOF) is a relatively new technology that can potentially analyze more than 100 different markers coupled with elemental metal isotopes, which can be detected and quantified by time‐of‐flight mass spectrometry with a minimal overlap between channels 29, 30, 31, 32, 33. It should be noted that compensation still remains an issue that researchers deal with when using such approaches 34. Given the high number of markers currently available, CyTOF also permits a deep analysis of cell subsets in those settings where sample volume is limited, such as in pediatric populations 35. Moreover, CyTOF represents a useful tool to monitor immune responses in tissues after immunotherapy in the cancer field 36. Similarly to flow cytometry, metal isotopes can be bound to modified peptide‐MHC tetramers specific for different viruses to monitor lymphocyte differentiation upon vaccination or infection 37.
Both flow cytometry and CyTOF have been used for the analysis of cell suspension from peripheral blood [peripheral blood mononuclear cells (PBMC)] and tissues (i.e. gut, spleen or other SLO, lymphoid tissue mononuclear cells, LMCs) 9, 36, 38, 39, 40. Within the tissue microenvironment, immune cells are exposed to unique local signals mediated by their interaction with resident or recruited immune cells, stromal cells, extracellular matrix and, presumably, a unique cytokine/chemokine milieu. Furthermore, the magnitude and duration of stimulatory/inhibitory signals that the immune cells receive is highly dependent upon the local inflammation and tissue cellularity, and is presumably different between tissues from different anatomical sites. Therefore, it is reasonable to hypothesize that the phenotypical and molecular signature of tissue immune cells differs from the signature of their blood counterparts. Furthermore, ex‐vivo studies using fluorescence activated cell sorting (FACS) can inform the potential efficiency of specific immune functions 41. However, comparison between these two signatures could provide valuable information regarding the differentiation process and plasticity of relevant immune cells and lead to novel circulating biomarkers for the monitoring of the disease progression, efficacy of vaccination and other immunotherapies.
Microscopy tools
In‐depth molecular and functional/phenotypical characterization of cell populations cannot provide information concerning the actual topographic organization of the players involved in the complex orchestration of the immune response (not only myeloid and lymphoid cells but also the soluble factors, cytokines and chemokines) and the influence of the intricate micro‐architecture of SLO on this response. To this end, the use of microscopy‐based methods is of great importance. The development of equipment such as lasers and cameras, as well as the generation of novel labels, has led to the introduction of highly sophisticated microscopy tools allowing for the simultaneous detection of several molecules with high resolution. Wide‐field microscopy represents a basic but still valuable technique where the light source hits simultaneously the whole specimen with obvious resolution limits. Laser scanning confocal microscopy (LSCM) utilizes fluorochrome‐labeled antibodies for specific markers (42. Defined regions of interest are exposed to a laser light source, while only the light coming from the focal plane is collected. The use of several laser lines and ‘unmixing' algorithms, for the correction/compensation of obtained signals, allows for the simultaneous volumetric (use of z‐stacks) detection of multiple markers. Relevant platforms in combination with established antibodies or label‐stripping protocols, that remove the specific staining from a tissue and make possible several subsequent stainings on the same slide, can be used for the detection of a large number of molecules in the same tissue section, with obvious advantages in terms of phenotype characterization and retention of precious tissues 43. Although technically challenging, antigen‐specific probes have been successfully used for the detection of antigen‐specific CD8 T cells at tissue level 44, providing critical information on antigen specificity of tissue immune dynamics. Expanding the high volume imaging capacity, clearing‐enhanced three‐dimensional microscopy (Ce3D) represents, among others, a simple tissue‐clearing technique which enables further analysis of the relationship between immune cells and tissue microenvironments 45. The recent development of fluorescent mRNA/DNA probes has revolutionized the in‐situ detection of such molecules within tissues 46, 47. With regard to immunophenotyping, the CyTOF concept can be applied for the immunohistochemical characterization of frozen and formalin‐fixed/paraffin‐embedded tissues using antibodies bound to metal isotopes 48, 49. Current technology does not allow for a volumetric analysis. To overcome this limitation, investigators have used an alternative strategy where sequential slides are imaged and the 3D image can be reconstructed by computational tools.
Although providing critical knowledge for the dynamics of the immune system, analysis of tissue sections using conventional microscopy techniques does not provide real‐time assessment of the tissue immune dynamics. Intravital two‐photon laser scanning microscopy (TP‐LSM) permits direct visualization of immune cells in a living animal, exposing the organ of interest (i.e. LN, gut, eye, skin, etc.). This approach has obvious advantages in detecting the dynamics and plasticity of the cells as they occur, highlighting the relationship between cells and the environment in which they reside and specialize 50, 51, 52. The downside of intravital techniques is that they cannot be performed on human subjects, for obvious reasons. In addition, in‐vivo LSCM is a novel imaging technique that permits non‐invasive, morphological characterization of skin structures in humans 53. A more comprehensive analysis of microscopy technologies is reviewed elsewhere 54. Selecting an imaging platform depends upon the nature of the scientific question under investigation (i.e. requirement for resolution level and 3D representation). Furthermore, scanning time, automation of the process and the compatibility with other high‐throughput assays are important factors to consider.
Single‐cell technologies
As discussed above, the identification of cell–cell interaction in vivo, especially in humans, is still challenging, in particular in pediatric settings where blood samples are generally scarce in volume and tissues are not easily accessible to allow collection. Most of the above‐mentioned methodologies have been used for analysis of bulk populations, and they are not fully optimized to detect rare subsets of cells such as antigen‐specific cells. The uniqueness among immune cells in terms of phenotype, functionality and plasticity has advanced the field towards the development of single‐cell technologies or lab‐on‐a‐chip (LOC) able to dissect spatiotemporal dynamics of immune cells. Microfluidics‐based approaches have been applied in a variety of research lines, including genomics 55 and proteomics 56, and have already provided important insights into the study of T cell signaling and migration 57, 58, 59 or NK cells 60. Using a relevant platform, we recently reported the production of interleukin (IL)‐21 by peripheral T follicular helper cells (TfH) after in‐vitro stimulation in HIV‐infected patients responding to influenza vaccination 61. Considering the variety of data that could be collected from each single cell (DNA sequence, mRNA transcripts, proteins expression, etc.), it would be ideal to collect them all at once in a cost‐effective manner moving the field towards an ‘integrated single‐cell analysis' 62. Several methodologies have been recently implemented in order to perform single‐cell RNA‐Seq (scRNA‐seq). The most currently used are massively parallel RNA single‐cell sequencing (MARS‐seq), Fluidigm C1 single‐cell full‐length mRNA sequencing (Fig. 1) and ×10 genomic chromium single‐cell DNA sequencing. This last approach is able to perform a rapid droplet‐based encapsulation of single cells using a gel bead in emulsion (GEM) approach. Each gel bead is labeled with a unique barcode for molecular identification of data acquired through RNA Seq 63, 64, 65. Although this technique is extremely promising, it still needs to be validated in the context of rare antigen‐specific cells.
Figure 1.
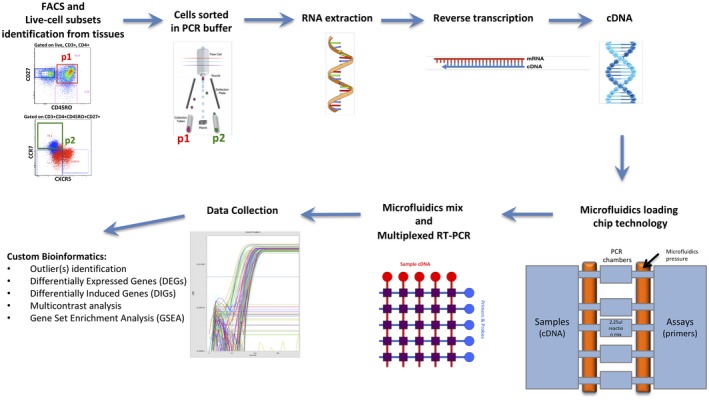
Tissue‐specific cell subsets at low numbers (as small as 1 cell), stained for surface molecules and analyzed by flow cytometry can be sorted from multiple subsets according to biological interest into tubes previously coated with specific polymerase chain reaction (PCR) buffer. After RNA extraction and cDNA reverse transcription, novel microfluidics‐based chip can be loaded with multiple assays and samples then mixed and analyzed for real time–PCR (RT–PCR). After data collection with specific r packages, the bioinformatics pipeline can be used to obtain multiple gene expression end‐points (e.g. outliers, differentially expressed genes (DEGs), differentially induced genes (DIGs) and pathway analysis by gene set enrichment analysis (GSEA).
Quantitative antibodies assay
As previously stated, soluble markers such as chemokines and cytokines interact with different cell types in the context of the complex tissue microanatomy for the proper functionality of the immune system. One of the most dated assays, although still valuable, is represented by the immunoblot and enzyme‐linked immunosorbent assay (ELISA). A possible application in SLO of such an assay could be the measurement of chemokines and cytokines with a critical role for GC function after vaccination. We recently demonstrated that, among the circulating cytokines/chemokines, the increase of CXCL13 after flu immunization is significantly correlated with the frequency of tonsillar TfH cells 9. Modern ELISA assays have been implemented to perform at high‐throughput levels, analyzing up to 100 cytokines per single assay 66 (Luminex technology). Despite the large use of these quantitative assays, concerns have been raised about measuring antibody production as the sole method for evaluating response to vaccines in particular populations such as immunocompromised patients 67, 68, 69, 70. Indeed, in these populations the achievement of a protective titer does not always correlate with a true protection of infection 71). Evaluation of antibody‐secreting cells (ASCs) at tissue level provides accurate information concerning the presence of memory response. Indeed, several factors may affect the levels of detected serum antibodies, such as the history of yearly vaccination (i.e. influenza), as well as the skewed immunity (in terms of immune activation/senescence) described in some groups of immunocompromised patients 72. From this perspective, several studies have been conducted to search for additional and more informative correlates of vaccination 73, 74. The enzyme‐linked immunospot (ELISPOT), and more recently the FluoroSpot assays, are some of the methods to investigate specific cell function (Fig. 2). Vaccine‐specific ASC has been detected by ELISPOT in SLO 1 week after vaccination in humans 9. In order to detect multiple cytokine production from the same cell, fluorescent ELISPOT assays have been introduced in the field of vaccine research, as simultaneous production of multiple cytokines have been correlated with vaccine efficacy.
Figure 2.
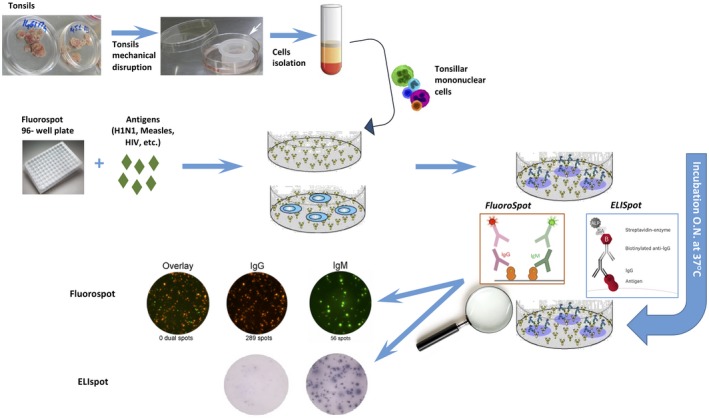
Enzyme‐linked immunospot (ELISPOT) and FluoroSpot assays can be used for the quantification of antibody‐secreting cells (ASCs) present in the tissue‐derived cells. Antigen‐specific responses are examined by wells coated with specific antigens at a definite dilution. ELISPOT allows the quantification of ASCs at the single‐cell level through a chromogenic detection, while FluoroSpot employs fluorescence detection allowing the quantification of immunoglobulin (Ig)G and IgM or IgA simultaneously. These assays can also be applied to the identification of specific cytokine‐secreting T cells.
All these assays, although informative, reflect a partial view of the complex cell interaction and need to be confirmed in vivo, where the microenvironment is much more intricate.
Computational data analysis
The performance of high‐throughput assays leads to generation of high‐volume data that need sophisticated computational, mathematical tools for their robust analysis.
This is an absolute requirement when deep sequencing data are under investigation. Although the volume of produced data is smaller, the analysis of generated by multi‐parametric flow cytometry or CytoF or multiplexed imaging platforms requires the development and application of advanced computational tools/algorithms. A recent example is the development of a method allowing for the conversion of imaging data to flow cytometry type of data (histocytometry) 75 and their further analysis using powerful programs such as FlowJo (Fig. 3). This method has found great applicability in numerous research fields 45, 46. Using a multi‐parametric approach combining flow cytometry, confocal microscopy and histocytometry, we recently showed for the first time immune mechanisms in the SLO underlying seasonal influenza vaccine response in tonsils isolated from vaccinated children. A significantly higher frequency of follicular helper CD4 T cells was found compared with the unvaccinated control group 9. More recently, a similar approach was used to highlight the dynamics of follicular cells in HIV‐infected individuals following influenza vaccination 76. Such tools allow for the quantitative analysis of multiplexed imaging data and their further process with modeling/statistical algorithms. Further development of this type of analysis (i.e. improving the time of data acquisition, data analysis, automation of the process) 77 is a critical factor for their wide application. During the past few decades, ‘systems biology' has been applied to vaccine research in an attempt to define molecular mechanisms orchestrating the immune response upon immunization or infections 78, 79. These high‐throughput approaches are now able to dissect every step involved in the process that lead genes (genomic), through gene expression (transcriptomic), to the production of proteins (proteomic) and their metabolism (metabolomic). The generation of high‐size, high‐complexity data using several platforms raises the need for computational tools allowing for a holistic approach, where ‘merging' data from different technologies can lead to deconvolution of immune responses to pathogens and immunogens with high resolution.
Figure 3.
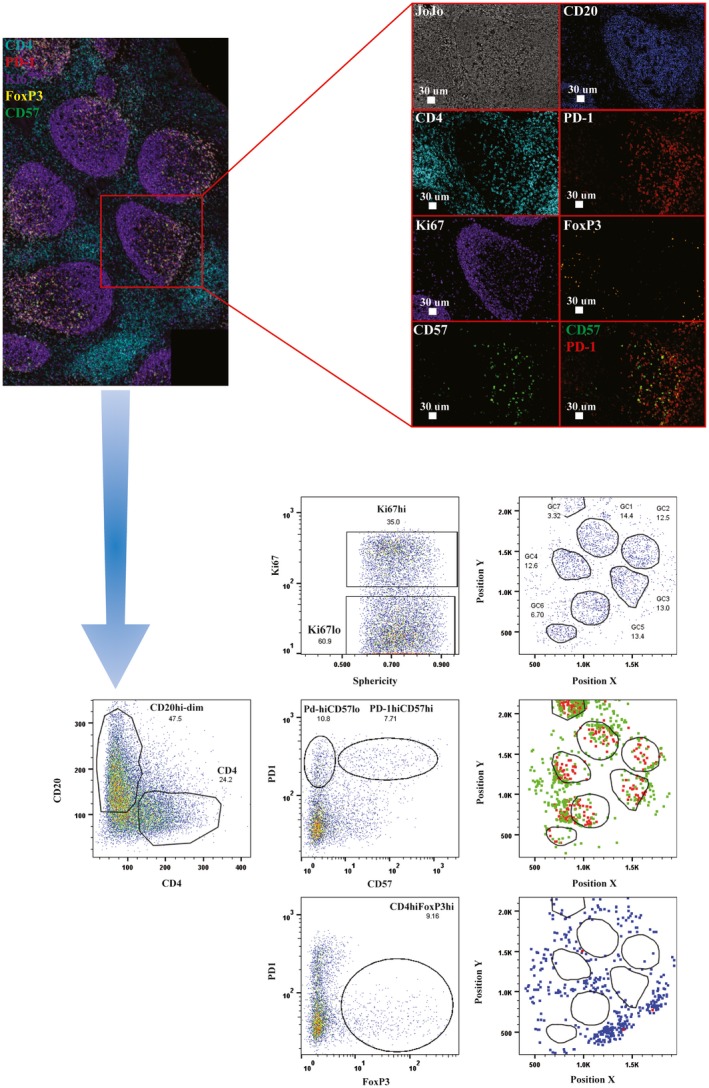
Specifically for lymph node (LN) immunology, multiplexed imaging allows for the evaluation of the heterogeneity within specific cell compartments [i.e. T follicular helper cells (Tfh) based on the expression of other markers‐CD57], the presence of potential suppressor CD4 T cells [forkhead box protein 3 (FoxP3+)] as well as the specific positioning of particular subset with respect to GC areas – dark zone versus light zone). The lower panels show the gating strategy that can be applied according to the markers used for the confocal image.
Understanding follicular immune dynamics
In the field of vaccinology and in infectious diseases in general, systems biology approaches have been employed to study the peripheral blood because of its easy accessibility and known correlates of vaccine response and protection. Although these peripheral correlates provide an interesting and highly reproducible opportunity to study immune responses following infections, they can only provide a description of the peripheral surrogates of the immune response development, which truly takes place in SLO. Indeed, following the antigen uptake by either antigen‐presenting cells or B cells in the periphery, the maturation of B cells takes place within the GCs through somatic hypermutation and class‐switch recombination; these pathways have only been partially explored by OMICs approaches. TfH cells are essential for the GC response. In recent years standard methods have dissected phenotype characteristics, transcriptional regulation and localization or migratory properties. In 2009, several studies reported how BCL6 intrinsic expression is crucial for TfH cell development and GC formation 80. In this context, the high‐dimensional OMIC approaches have led to important discoveries. A combined micro‐RNA study and gene expression analysis recently defined that BCL6 is silenced in its helper function for T cell maturation within the SLO by miR‐31 81. Global transcriptome analysis further revealed specific targets of the silencing action of miR‐31, thus showing crucial genes in the TfH activation (i.e. CXCR5, SAP, BTLA and CD28). A similar approach in the B cell counterpart was explored by McHeyzer‐Williams et al., who defined molecular programming driving the maturation of single B GC cells after vaccination booster 82. A two‐gene signature (CD83 and PolH) was able to discriminate the maturation phases which antigen‐specific B cells undergo within light and dark zones of the GC. This process can be triggered by an effective vaccination or booster and is able to induce high‐affinity memory B cells and durable antibody responses through a progressive rediversification of B cell receptors (BCRs) and natural selection of the BCRs. Transcriptional characteristics in the peripheral memory B cells 83, 84, as well as BCR sequences of memory B GC cells analyzed by large‐scale OMIC approaches, may serve as crucial prediction tools of vaccination efficacy and safety. However, this mechanism is not T cell‐independent, as TfH play an integral role in B cell differentiation and affinity maturation 85. An innovative OMICs approach, combining high‐dimensional mass cytometry with T cell receptor (TCR) repertoire sequencing, was recently used to interrogate the composition of gag‐specific TfH cells in primary human SLO in HIV‐infected patients (86. These data, confirming evidence produced in the peripheral blood, revealed that during HIV infection TfH cells remain capable of responding to HIV antigen, but are oligoclonally restricted despite persistent antigen stimulation, leading in turn to suboptimal B cell distribution and function. Another crucial point to stress is that most of these studies have been conducted on antigen‐specific cells rather than total subsets. This was mainly performed to overcome the heterogeneity problem found in cells of LMCs or PBMCs in order to gain high‐dimensional information from antigen‐specific cells involved in the maturation process within the SLO. Taken together, OMICs approaches, dissecting high‐dimensional molecular data within tissues and PBMCs, currently also represent essential tools for the proper characterization of cancer subtypes and pathways involved 87, 88.
Discussion
The complex microenvironment regulating the immune system is able to drive the maturation and differentiation of the immune cells through close cell–cell interactions. Thus, we need a comprehensive approach able to elucidate the intrinsic mechanisms leading to the proper generation of the immune response after antigen exposure.
Most studies focus their attention upon animal models where different types of samples are easily accessible; also, when performed in humans, the majority of these studies utilize blood samples for practical and ethical reasons. Both approaches present serious limitations. Animal experiments are carried out on subjects living in captivity and usually in germ‐free conditions, and thus with limited exposure to the influence of the environment (i.e. infections and vaccination); moreover, animal models do not reproduce the genetic inter‐ and intra‐individual variability among human populations. Of note, human studies using PBMCs analyze the immune response in the periphery, and this response is simply a blurred image of what has already happened, or is happening, in different tissues. Cell suspension analysis obtained from tissues can help, but without spatial and topographic data we risk missing some important pieces of the puzzle. Despite the huge contribution given to the field by the immunophenotyping technologies, these data alone are not sufficient to fully understand the great complexity of the immune system. Conversely, the microscopy technologies available and applicable to human samples are limited by the visualization of a steady state, which is far from the reality of an immune system in constant permutation.
Considering that the number of tools at our disposal is still increasing, the challenge resides in how to analyze, not in how to collect, data. Given the extraordinary quantity of data gathered from each experiment and the different ways in which we could respond to a biological question, we are facing the necessity of generating new approaches to decipher the complex immune system function and development. It is therefore mandatory to harmonize the tools currently available to answer the unresolved questions in the field of immunology. To do so, now more than ever, scientists from different fields – mathematics, bioinformatics, statistics, biology – must collaborate to build computational methods for more comprehensive and integrative data analysis. This is the purpose of systems biology 89, 90 (Fig. 4). Systems approaches, merging analytical, informatic and computational skills, can be used to characterize the status of the immune system and to predict its plasticity in response to different stimuli (infection, vaccination, cancer) in a uniform and reliable manner 90, 91, 92. Some groups are currently focusing their efforts on the generation of computational infrastructures and tools for immunology analyses. For example, the National Institute of Allergy and Infectious Diseases (NIAID) of the US National Institutes of Health (NIH) created the Human Immunology Project Consortium to build large data sets on human subjects under different conditions (HIPC; http://www.humanprofiling.org).
Figure 4.
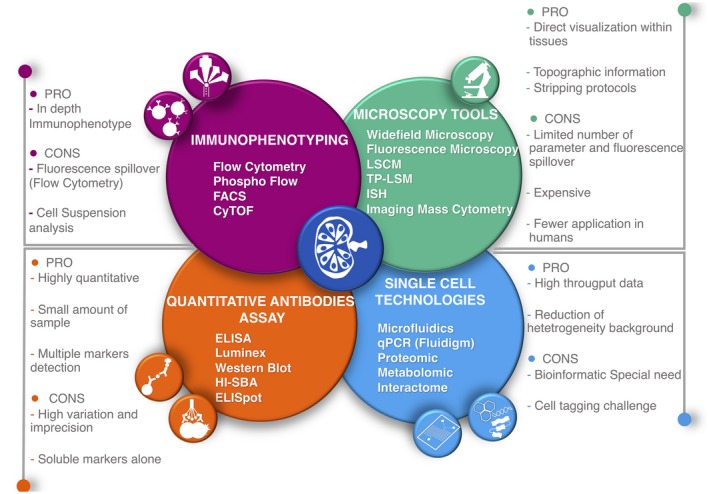
The illustration summarizes the most available technique at our disposal to study the immune system, highlighting the main advantages and limitations. These tools should be managed under the umbrella of the systems immunology. The lymph node at the center is to strengthen the necessity of performing studies on tissues. The lymph node image is taken and adapted from Servier Medical Art at: http://smart.servier.com/.
In addition, the complexity of the immune response and the different ways we can dissect it within modern immunology give rise to the necessity for scientists worldwide to share their knowledge using a more standardized approach. This is the only way to truly advance our understanding of immunity. Only with an in‐depth understanding of the immune system we can fuel the proper development of personalized medicine predicting the usefulness of a therapeutic intervention in a given patient.
Conclusion
In conclusion, most human studies are performed on blood for practical reasons, and most data derive from ex‐vivo experiments. Of note, it must be kept in mind that the trustworthiness of data coming from such ex‐vivo approaches needs to be confirmed in vivo, where the microenvironment is much more intricate. Human tissues samples are difficult to obtain, especially during wellbeing and in particular populations, such as pediatric populations. As pediatricians, our group is trying to validate tonsils as valuable SLO in children that can be used to investigate the intricate dynamics leading to the generation of an immune response. Considering that tonsils are routinely surgically removed, the use of these tissues could allow the performance of high‐standard immunological studies in those populations who have particular biological characteristics, but this is still poorly evaluated. Moreover, tissue samples are removed daily for various reasons, such as colon sections from newborns affected by necrotizing enterocolitis, gut biopsies during inflammatory bowel diseases evaluation, thymus following major cardiac surgery, spleen (post‐traumatic or in certain hematological conditions) and lymph nodes for diagnostic purposes, etc. In these selected situations, the problem does not reside in the availability of such tissues but on the laboratories' skills and the presence of personnel dedicated to such analysis. However, in other conditions (i.e. the study of HIV reservoirs in humans or the T/B cell interaction following immunization) the possibility of gaining access to tissues still remains challenging. Finally, high‐throughput data sets are generating more data than we are able to analyze. Systems immunology probably represents the key to elucidate how the actors of the immune response interact between them and in the microenvironment. The biology community in general, and the immunology community in particular, should apply itself to using standardized protocols and to share their knowledge. The time is ripe for a major leap forward.
Disclosures
The authors have no conflicts of interest to declare.
Author contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Acknowledgements
This work was supported by Associazione volontari Bambino Gesù and Ricerca Corrente Ospedale Pediatrico Bambino Gesù 2017/2018, the Intramural Research Program of the Vaccine Research Center, NIAID, National Institutes of Health and a CAVD grant (no. OP1032325) from the Bill and Melinda Gates Foundation.
References
- 1. Zhang Y, Garcia‐Ibanez L, Toellner KM. Regulation of germinal center B‐cell differentiation. Immunol Rev 2016; 270:8–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Davis MM. A prescription for human immunology. Immunity 2008; 29:835–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mestas J, Hughes CC. Of mice and not men: differences between mouse and human immunology. J Immunol 2004; 172:2731–8. [DOI] [PubMed] [Google Scholar]
- 4. Morita R, Schmitt N, Bentebibel SE et al Human blood CXCR5(+)CD4(+) T cells are counterparts of T follicular cells and contain specific subsets that differentially support antibody secretion. Immunity 2011; 34:108–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Roederer M. Multiparameter FACS analysis. Curr Protoc Immunol 2002; 49:5–8. [DOI] [PubMed] [Google Scholar]
- 6. Toellner KM, Scheel‐Toellner D, Sprenger R et al The human germinal centre cells, follicular dendritic cells and germinal centre T cells produce B cell‐stimulating cytokines. Cytokine 1995; 7:344–54. [DOI] [PubMed] [Google Scholar]
- 7. Steiniger B, Trabandt M, Barth PJ. The follicular dendritic cell network in secondary follicles of human palatine tonsils and spleens. Histochem Cell Biol 2011; 135:327–36. [DOI] [PubMed] [Google Scholar]
- 8. Johansson‐Lindbom B, Ingvarsson S, Borrebaeck CA. Germinal centers regulate human Th2 development. J Immunol 2003; 171:1657–66. [DOI] [PubMed] [Google Scholar]
- 9. Amodio D, Cotugno N, Macchiarulo G et al Quantitative multiplexed imaging analysis reveals a strong association between immunogen‐specific B cell responses and tonsillar germinal center immune dynamics in children after influenza vaccination. J Immunol 2018; 200:538–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fulwyler MJ. Electronic separation of biological cells by volume. Science 1965; 150:910–1. [DOI] [PubMed] [Google Scholar]
- 11. Melamed MR, Mullaney PF, Mendelsohn ML. An historical review of the development of flow cytometers and sorters In: Melamed MR, Mullaney PF, Mendelsohn ML, eds. Flow cytometry and sorting New York. Chichester: Wiley; 1979:3. [Google Scholar]
- 12. Herzenberg LA, Sweet RG, Herzenberg LA. Fluorescence‐activated cell sorting. Sci Am 1976; 234:108–17. [DOI] [PubMed] [Google Scholar]
- 13. Edwards AD, Manickasingham SP, Sporri R et al Microbial recognition via Toll‐like receptor‐dependent and ‐independent pathways determines the cytokine response of murine dendritic cell subsets to CD40 triggering. J Immunol 2002; 169:3652–60. [DOI] [PubMed] [Google Scholar]
- 14. Szaniszlo P, Wang N, Sinha M et al Getting the right cells to the array: Gene expression microarray analysis of cell mixtures and sorted cells. Cytometry A 2004; 59:191–202. [DOI] [PubMed] [Google Scholar]
- 15. Battye FL, Light A, Tarlinton DM. Single cell sorting and cloning. J Immunol Methods 2000; 243:25–32. [DOI] [PubMed] [Google Scholar]
- 16. El‐Naggar AK. Concurrent flow cytometric analysis of DNA and RNA. Methods Mol Biol 2004; 263:371–84. [DOI] [PubMed] [Google Scholar]
- 17. Brenchley JM, Karandikar NJ, Betts MR et al Expression of CD57 defines replicative senescence and antigen‐induced apoptotic death of CD8+ T cells. Blood 2003; 101:2711–20. [DOI] [PubMed] [Google Scholar]
- 18. Chattopadhyay PK, Melenhorst JJ, Ladell K et al Techniques to improve the direct ex vivo detection of low frequency antigen‐specific CD8+ T cells with peptide–major histocompatibility complex class I tetramers. Cytometry A 2008; 73:1001–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Krutzik PO, Nolan GP. Intracellular phospho‐protein staining techniques for flow cytometry: monitoring single cell signaling events. Cytometry A 2003; 55:61–70. [DOI] [PubMed] [Google Scholar]
- 20. Altman JD, Moss PA, Goulder PJ et al Phenotypic analysis of antigen‐specific T lymphocytes. Science 1996; 274:94–6. [PubMed] [Google Scholar]
- 21. Ogonek J, Verma K, Schultze‐Florey C et al Characterization of high‐avidity cytomegalovirus‐specific T cells with differential tetramer binding coappearing after allogeneic stem cell transplantation. J Immunol 2017; 199:792–805. [DOI] [PubMed] [Google Scholar]
- 22. Ferdman J, Palladino G, Liao HX et al Intra‐seasonal antibody repertoire analysis of a subject immunized with an MF59(R)‐adjuvanted pandemic 2009 H1N1 vaccine. Vaccine 2018; 36:5325–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Buggert M, Nguyen S, Salgado‐Montes de Oca G et al Identification and characterization of HIV‐specific resident memory CD8(+) T cells in human lymphoid tissue. Sci Immunol 2018; 3:eaar4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bhagwat N, Dulmage K, Pletcher CH Jr et al An integrated flow cytometry‐based platform for isolation and molecular characterization of circulating tumor single cells and clusters. Sci Rep 2018; 8. 5035–018‐23217‐5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pillai V, Dorfman DM. Flow cytometry of nonhematopoietic neoplasms. Acta Cytol 2016; 60:336–43. [DOI] [PubMed] [Google Scholar]
- 26. Saito T, Nishikawa H, Wada H et al Two FOXP3(+)CD4(+) T cell subpopulations distinctly control the prognosis of colorectal cancers. Nat Med 2016; 22:679–84. [DOI] [PubMed] [Google Scholar]
- 27. Liechti T, Roederer M. OMIP‐051‐28‐color flow cytometry panel to characterize B cells and myeloid cells. Cytometry A 2019; 95:150–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Telford W, Georges T, Miller C, Voluer P. Deep ultraviolet lasers for flow cytometry. Cytometry A 2019; 95:227–33. [DOI] [PubMed] [Google Scholar]
- 29. Bendall SC, Simonds EF, Qiu P et al Single‐cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science 2011; 332:687–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bandura DR, Baranov VI, Ornatsky OI et al Mass cytometry: technique for real time single cell multitarget immunoassay based on inductively coupled plasma time‐of‐flight mass spectrometry. Anal Chem 2009; 81:6813–22. [DOI] [PubMed] [Google Scholar]
- 31. Kay AW, Strauss‐Albee DM, Blish CA. Application of mass cytometry (CyTOF) for functional and phenotypic analysis of natural killer cells. Methods Mol Biol 2016; 1441:13–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Horowitz A, Strauss‐Albee DM, Leipold M et al Genetic and environmental determinants of human NK cell diversity revealed by mass cytometry. Sci Transl Med 2013; 5:208ra145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ornatsky O, Bandura D, Baranov V, Nitz M, Winnik MA, Tanner S. Highly multiparametric analysis by mass cytometry. J Immunol Methods 2010; 361:1–20. [DOI] [PubMed] [Google Scholar]
- 34. Chevrier S, Crowell HL, Zanotelli VRT, Engler S, Robinson MD, Bodenmiller B. Compensation of signal spillover in suspension and imaging mass cytometry. Cell Syst 2018; 6:612–620.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yao Y, Liu R, Shin MS et al CyTOF supports efficient detection of immune cell subsets from small samples. J Immunol Methods 2014; 15:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Spitzer MH, Carmi Y, Reticker‐Flynn NE et al Systemic immunity is required for effective cancer immunotherapy. Cell 2017; 168:487–502.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Newell EW, Sigal N, Bendall SC, Nolan GP, Davis MM. Cytometry by time‐of‐flight shows combinatorial cytokine expression and virus‐specific cell niches within a continuum of CD8+ T cell phenotypes. Immunity 2012; 36:142–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Papa I, Saliba D, Ponzoni M et al TFH‐derived dopamine accelerates productive synapses in germinal centres. Nature 2017; 547:318–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Leelatian N, Doxie DB, Greenplate AR et al Single cell analysis of human tissues and solid tumors with mass cytometry. Cytometry B Clin Cytom 2017; 92:68–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Godefroy E, Alameddine J, Montassier E et al Expression of CCR6 and CXCR6 by gut‐derived CD4(+)/CD8alpha(+) T‐regulatory cells, which are decreased in blood samples from patients with inflammatory bowel diseases. Gastroenterology 2018; 155:1205–17. [DOI] [PubMed] [Google Scholar]
- 41. Reuter MA, Del Rio Estrada PM, Buggert M et al HIV‐specific CD8(+) T cells exhibit reduced and differentially regulated cytolytic activity in lymphoid tissue. Cell Rep 2017; 21:3458–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gerner MY, Torabi‐Parizi P, Germain RN. Strategically localized dendritic cells promote rapid T cell responses to lymph‐borne particulate antigens. Immunity 2015; 42:172–85. [DOI] [PubMed] [Google Scholar]
- 43. Bolognesi MM, Manzoni M, Scalia CR et al Multiplex staining by sequential immunostaining and antibody removal on routine tissue sections. J Histochem Cytochem 2017; 65:431–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Skinner PJ, Daniels MA, Schmidt CS, Jameson SC, Haase AT. Cutting edge: in situ tetramer staining of antigen‐specific T cells in tissues. J Immunol 2000; 165:613–7. [DOI] [PubMed] [Google Scholar]
- 45. Li W, Germain RN, Gerner MY. Multiplex, quantitative cellular analysis in large tissue volumes with clearing‐enhanced 3D microscopy (Ce3D). Proc Natl Acad Sci USA 2017; 114:E7321–E7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ferrando‐Martinez S, Moysi E, Pegu A et al Accumulation of follicular CD8+ T cells in pathogenic SIV infection. J Clin Invest 2018; 128:2089–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fox CH, Cottler‐Fox M. In situ hybridization in HIV research. Microsc Res Tech 1993; 25:78–84. [DOI] [PubMed] [Google Scholar]
- 48. Giesen C, Wang HA, Schapiro D et al Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat Methods 2014; 11:417–22. [DOI] [PubMed] [Google Scholar]
- 49. Chang Q, Ornatsky O, Hedley D. Staining of frozen and formalin‐fixed, paraffin‐embedded tissues with metal‐labeled antibodies for imaging mass cytometry analysis. Curr Protoc Cytom 2017; 82:12–47. [DOI] [PubMed] [Google Scholar]
- 50. Park C, Hwang IY, Kehrl JH. The use of intravital two‐photon and thick section confocal imaging to analyze B lymphocyte trafficking in lymph nodes and spleen. Methods Mol Biol 2018; 1707:193–205. [DOI] [PubMed] [Google Scholar]
- 51. Bajenoff M, Germain RN. Seeing is believing: a focus on the contribution of microscopic imaging to our understanding of immune system function. Eur J Immunol 2007; 37(Suppl 1):S18–33. [DOI] [PubMed] [Google Scholar]
- 52. Miller MJ, Wei SH, Parker I, Cahalan MD. Two‐photon imaging of lymphocyte motility and antigen response in intact lymph node. Science 2002; 296:1869–73. [DOI] [PubMed] [Google Scholar]
- 53. Ilie MA, Caruntu C, Lixandru D et al In vivo confocal laser scanning microscopy imaging of skin inflammation: clinical applications and research directions. Exp Ther Med 2019; 17:1004–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sarris M, Betz AG. Shine a light: imaging the immune system. Eur J Immunol 2009; 39:1188–202. [DOI] [PubMed] [Google Scholar]
- 55. Tian J, Gong H, Sheng N et al Accurate multiplex gene synthesis from programmable DNA microchips. Nature 2004; 432:1050–4. [DOI] [PubMed] [Google Scholar]
- 56. Huang B, Wu H, Bhaya D et al Counting low‐copy number proteins in a single cell. Science 2007; 315:81–4. [DOI] [PubMed] [Google Scholar]
- 57. Faley S, Seale K, Hughey J et al Microfluidic platform for real‐time signaling analysis of multiple single T cells in parallel. Lab Chip 2008; 8:1700–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Long A, Mitchell S, Kashanin D et al A multidisciplinary approach to the study of T cell migration. Ann NY Acad Sci 2004; 1028:313–9. [DOI] [PubMed] [Google Scholar]
- 59. Varadarajan N, Julg B, Yamanaka YJ et al A high‐throughput single‐cell analysis of human CD8(+) T cell functions reveals discordance for cytokine secretion and cytolysis. J Clin Invest 2011; 121:4322–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Vanherberghen B, Olofsson PE, Forslund E et al Classification of human natural killer cells based on migration behavior and cytotoxic response. Blood 2013; 121:1326–34. [DOI] [PubMed] [Google Scholar]
- 61. de Armas LR, Cotugno N, Pallikkuth S et al Induction of IL21 in peripheral t follicular helper cells is an indicator of influenza vaccine response in a previously vaccinated HIV‐infected pediatric cohort. J Immunol 2017; 198:1995–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yalcin A, Yamanaka YJ, Love JC. Analytical technologies for integrated single‐cell analysis of human immune responses. Methods Mol Biol 2012; 853:211–35. [DOI] [PubMed] [Google Scholar]
- 63. Zheng GX, Terry JM, Belgrader P et al Massively parallel digital transcriptional profiling of single cells. Nat Commun 2017; 16:14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. See P, Lum J, Chen J, Ginhoux F. A single‐cell sequencing guide for immunologists. Front Immunol 2018; 23:2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Cotugno N, Ruggiero A, Santilli V et al OMICs technologies and vaccine development: from the identification of vulnerable individuals to the formulation of invulnerable vaccines. J Immunol Res 2019; 2019:8732191. doi: 10.1155/2019/8732191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Carson RT, Vignali DA. Simultaneous quantitation of 15 cytokines using a multiplexed flow cytometric assay. J Immunol Methods 1999; 227:41–52. [DOI] [PubMed] [Google Scholar]
- 67. Amanna IJ, Messaoudi I, Slifka MK. Protective immunity following vaccination: how is it defined? Hum Vaccin 2008; 4:316–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Trombetta CM, Perini D, Mather S, Temperton N, Montomoli E. Overview of serological techniques for influenza vaccine evaluation: past: present and future. Vaccines (Basel) 2014; 2:707–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Cagigi A, Cotugno N, Giaquinto C et al Immune reconstitution and vaccination outcome in HIV‐1 infected children: present knowledge and future directions. Hum Vaccin Immunother 2012; 8:1784–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Rinaldi S, Cagigi A, Santilli V et al B‐sides serologic markers of immunogenicity in kidney transplanted patients: report from 2012–2013 flu vaccination experience. Transplantation 2014; 98:259–66. [DOI] [PubMed] [Google Scholar]
- 71. Rinaldi S, Zangari P, Cotugno N et al Antibody but not memory B‐cell responses are tuned‐down in vertically HIV‐1 infected children and young individuals being vaccinated yearly against influenza. Vaccine 2014; 32:657–63. [DOI] [PubMed] [Google Scholar]
- 72. Cagigi A, Rinaldi S, Santilli V et al Premature ageing of the immune system relates to increased anti‐lymphocyte antibodies (ALA) after an immunization in HIV‐1‐infected and kidney‐transplanted patients. Clin Exp Immunol 2013; 174:274–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Cotugno N, De Armas L, Pallikkuth S, Rossi P, Palma P, Pahwa S. Paediatric HIV infection in the 'omics era: defining transcriptional signatures of viral control and vaccine responses. J Virus Erad 2015; 1:153–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Cagigi A, Cotugno N, Rinaldi S, Santilli V, Rossi P, Palma P. Downfall of the current antibody correlates of influenza vaccine response in yearly vaccinated subjects: toward qualitative rather than quantitative assays. Pediatr Allergy Immunol 2016; 27:22–7. [DOI] [PubMed] [Google Scholar]
- 75. Gerner MY, Kastenmuller W, Ifrim I, Kabat J, Germain RN. Histo‐cytometry: a method for highly multiplex quantitative tissue imaging analysis applied to dendritic cell subset microanatomy in lymph nodes. Immunity 2012; 37:364–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Moysi E, Pallikkuth S, De Armas LR et al Altered immune cell follicular dynamics in HIV infection following influenza vaccination. J Clin Invest 2018; 128:3171–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Kotov DI, Pengo T, Mitchell JS, Gastinger MJ, Jenkins MK. Chrysalis: a new method for high‐throughput histo‐cytometry analysis of images and movies. J Immunol 2019; 202:300–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Li S, Rouphael N, Duraisingham S et al Molecular signatures of antibody responses derived from a systems biology study of five human vaccines. Nat Immunol 2014; 15:195–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Tsang JS, Schwartzberg PL, Kotliarov Y et al Global analyses of human immune variation reveal baseline predictors of postvaccination responses. Cell 2014; 157:499–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Nurieva RI, Chung Y, Martinez GJ et al Bcl6 mediates the development of T follicular helper cells. Science 2009; 325:1001–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Ripamonti A, Provasi E, Lorenzo M et al Repression of miR‐31 by BCL6 stabilizes the helper function of human follicular helper T cells. Proc Natl Acad Sci USA 2017; 114:12797–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. McHeyzer‐Williams LJ, Milpied PJ, Okitsu SL, McHeyzer‐Williams MG. Class‐switched memory B cells remodel BCRs within secondary germinal centers. Nat Immunol 2015; 16:296–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Cotugno N, De Armas L, Pallikkuth S et al Perturbation of B cell gene expression persists in HIV‐infected children despite effective antiretroviral therapy and predicts H1N1 response. Front Immunol 2017; 11:1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. de Armas LR, Pallikkuth S, Pan L et al Cell profiling reveals PTEN overexpression in influenza‐ specific B cells in aging HIV‐infected individuals on antiretroviral therapy. Sci Rep 2019; 9:2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Qi H. T follicular helper cells in space–time. Nat Rev Immunol 2016; 16:612–25. [DOI] [PubMed] [Google Scholar]
- 86. Wendel BS, Del Alcazar D, He C et al The receptor repertoire and functional profile of follicular T cells in HIV‐infected lymph nodes. Sci Immunol 2018; 3:eaan8884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Ramazzotti D, Lal A, Wang B, Batzoglou S, Sidow A. Multi‐omic tumor data reveal diversity of molecular mechanisms that correlate with survival. Nat Commun 2018; 9:4453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Ressom HW, Di Poto C, Ferrarini A et al Multi‐omic approaches for characterization of hepatocellular carcinoma In: 2016 38th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC). Orlando, FL, 2016:3437–40. doi: 10.1109/EMBC.2016.7591467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Ideker T, Galitski T, Hood L. A new approach to decoding life: systems biology. Annu Rev Genomics Hum Genet 2001; 2:343–72. [DOI] [PubMed] [Google Scholar]
- 90. Chuang HY, Hofree M, Ideker T. A decade of systems biology. Annu Rev Cell Dev Biol 2010; 26:721–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Brusic V, Gottardo R, Kleinstein SH, Davis MM, HIPC Steering Committee. Computational resources for high‐dimensional immune analysis from the Human Immunology Project Consortium. Nat Biotechnol 2014; 32:146–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Germain RN, Meier‐Schellersheim M, Nita‐Lazar A, Fraser ID. Systems biology in immunology: a computational modeling perspective. Annu Rev Immunol 2011; 29:527–85. [DOI] [PMC free article] [PubMed] [Google Scholar]


