Summary
There have been many studies on the mechanisms of internalization of DNA–anti‐DNA immune complexes by cells, including the one used for rheumatoid factor‐expressing mouse B cells. In parallel, studies on the role of intracellular DNA sensors in the pathogenesis of systemic lupus erythematosus (SLE) have been conducted, including the one using a mouse model lacking one of the sensors. These and other data have established a framework for understanding the pathogenic role of anti‐DNA antibodies, but studies on normal cells are limited. Here, we used the monoclonal anti‐dsDNA antibody 2C10, 2‐kbp dsDNA and healthy human peripheral blood mononuclear cells (PBMCs) to test whether and how 2C10 and/or DNA cause pathology in normal cells. We found that on culture with PBMCs, 2C10 preferentially entered monocytes and that DNA enhanced this internalization. In contrast, DNA alone was not significantly internalized by monocytes, but 2C10 facilitated its internalization. This was suppressed by cytochalasin D, but not by methyl‐β‐cyclodextrin, chloroquine or an Fc blocker, suggesting the involvement of macropinocytosis in this process. Internalization of 2C10 and DNA together resulted in production of interferon (IFN)‐α, IFN‐γ, tumor necrosis factor (TNF)‐α, monocyte chemoattractant protein‐1 (MCP‐1), interleukin (IL)‐1β, IL‐6, IL‐10 and IL‐33 by PBMCs. Cytokine production was suppressed by chloroquine and shikonin, but not by RU.521, suggesting dependence on activation of the Toll‐like receptor (TLR)‐9 and absent in melanoma 2 (AIM‐2) pathways. These results established a simple model to demonstrate that anti‐DNA antibodies can cause dysregulation of cytokine network mimicking systemic lupus erythematosus in culture of normal PBMCs, and emphasize again the importance of maintaining anti‐DNA antibodies at low levels by treatment.
Keywords: anti‐DNA antibodies, endocytosis, macropinocytosis, systemic lupus erythematosus
Fluorescence‐labeled DNA (D) entered normal monocytes with the help of anti‐DNA antibody 2C10 (2).

Introduction
Systemic lupus erythematosus (SLE) is a prototypical systemic autoimmune disease that affects many different organs, including the kidney, heart, lung, central nervous system, skin, joints and blood cells 1. Although its etiology is not fully elucidated, genetic susceptibility and environmental factors are thought to lead to a breakdown of self‐tolerance mechanisms, and a variety of autoantibodies are produced 2. Of these, antibodies against nuclear antigens such as DNA and Sm are adopted in widely used classification criteria for SLE 3. Antibodies reactive to double‐stranded (ds) DNA show high disease specificity, and their changing titers reflect disease activity in many patients. However, details of the pathogenic activities of anti‐dsDNA antibodies remain to be clarified 4.
Many cytokines are implicated in loss of tolerance and tissue damage in SLE. Among them, interferon (IFN)‐α drives persistent self‐directed immune reactions by inhibiting regulatory T cells and up‐regulating B cell activation factor (BAFF), and is recognized as a central mediator of lupus pathophysiology 5. Additionally, levels of many cytokines and chemokines, including tumor necrosis factor (TNF)‐α, IL‐1β, IL‐6, IL‐8, IL‐10, IL‐17 and monocyte chemoattractant protein‐1 (MCP‐1) are elevated in SLE 2. The role, if any, of anti‐dsDNA antibodies in the dysregulation of this cytokine network is therefore a matter of prime importance. More than two decades ago it was found that some anti‐DNA antibodies induce expression of IFN‐α or inflammatory cytokines by normal mononuclear cells, but at that time little was known about intracellular nucleic acid sensors 6, 7. More recently, a clearer understanding of the manner by which innate immune cells detect intracellular DNA or RNA introduced by pathogens has emerged. Toll‐like receptor (TLR)‐9 was first reported to recognize bacterial DNA in 2000 8, followed by the identification of TLR‐3 as a sensor of dsRNA 9, and TLR‐7 and TLR‐8 as sensors of single‐stranded (ss) RNA 10. Cyclic GMP–AMP synthase‐stimulator of interferon genes (cGAS‐STING) 11 and absent in melanoma‐2 (AIM‐2) 12, 13 were also found to act as sensors of cytoplasmic DNA. However, how innate immune systems are activated in a pathophysiological setting of autoimmune diseases remains to be clarified.
In light of recent developments, we have used a well‐characterized monoclonal anti‐dsDNA antibody together with synthetic dsDNA to investigate whether and how they induce production of potentially pathogenic cytokines by peripheral blood mononuclear cells (PBMCs) from healthy subjects. The results show that DNA facilitated the internalization of the anti‐dsDNA antibody preferentially by the monocyte‐rich fraction and, reciprocally, the anti‐dsDNA antibody facilitated internalization of DNA. As a consequence, secretion of cytokines related to lupus pathogenesis was elevated in PBMCs. Internalization by the Fc receptor‐independent process of macropinocytosis and activation via TLR‐9 and AIM‐2 by the internalized DNA appeared to be involved in this phenomenon.
Materials and methods
Cells and monoclonal antibody
The study protocol was approved by the Ethics Committee of TMDU Faculty of Medicine (M2000‐1480). PBMCs from healthy volunteers were isolated by density gradient centrifugation over Ficoll‐Paque Plus (GE Healthcare, Chicago, IL, USA). PBMCs and the human monocytic leukemia cell line THP‐1 were cultured in RPMI‐1640 medium supplemented with 10% fetal bovine serum, 100 u/ml penicillin, 100 μg/ml streptomycin and 10 mM non‐essential amino acids. The anti‐dsDNA monoclonal antibody 2C10 [immunoglobulin (Ig)G2b,κ] was generated from an MRL/lpr mouse, and its fine specificity and amino acid sequence of the variable regions have been previously reported 14, 15. It was purified from the culture supernatant of the hybridoma cells grown in Dulbecco’s modified Eagle’s medium (DMEM) medium supplemented with 10% fetal bovine serum, 100 u/ml penicillin, 100 μg/ml streptomycin and 10 mM non‐essential amino acids, by salting‐out with half‐saturated ammonium sulfate followed by column chromatography with Protein G HP Spin Trap (GE Healthcare). Final concentrations of lipopolysaccharide (LPS) in the preparation were confirmed to be < 0·1 pg/ml by the Limulus Color KY Test (Fujifilm Wako Chemical, Osaka, Japan).
Reagents
Using a pcDNA3.1/Zeo(+) vector (Thermo Fisher Scientific, Waltham, MA, USA) as a template, a 2‐kilo base pairs (kbp) DNA fragment was amplified by polymerase chain reaction (PCR) using the following primers: sense: 5′‐TAATACGACTCACTATAGGG‐3′ and anti‐sense: 5′‐CTAGAGGTCGACGGTATACAG‐3′. In some experiments for detection of internalized DNA, the DNA fragment was fluorescently labeled using ChromaTide AlexaFluor 488‐5‐dUTP (Thermo Fisher Scientific). Other reagents were purchased as follows: cytochalasin D from Fujifilm Wako Chemical, methyl‐β‐cyclodextrin from Merck (Kenilworth, NJ, USA), Dynasore and shikonin from Adipogen Life Sciences (San Diego, CA, USA), chloroquine and RU.521 from Invivogen (San Diego, CA, USA) and human BD Fc block from BD Biosciences (San Jose, CA, USA).
Detection of internalized antibody
THP‐1 cells or PBMCs were seeded into 48‐well culture plates. Following a 10‐min incubation with the fluorescence‐labeled or ‐unlabeled 2‐kbp DNA described above, 2C10 (final concentration 5–10 μg/ml, unless otherwise indicated) or isotype‐matched control IgG (R&D Systems, Minneapolis, MN, USA) was added to the wells and incubated for 1 or 2 h at 37°C in a CO2 incubator. Unbound DNA and antibody were removed by washing with ice‐cold phosphate‐buffered saline (PBS), and the cells were fixed and permeabilized using a fixation/permeabilization kit (BD Biosciences). Cells were then stained with phycoerythrin (PE)‐labeled goat anti‐mouse IgG (Abcam, Cambridge, UK) for THP‐1 or Alexa Fluor 488‐labeled goat anti‐mouse IgG (Abcam) for PBMCs for 20 min at room temperature. In inhibition experiments, cells were treated with 10 μg/ml cytochalasin D, 5 mM methyl‐β‐cyclodextrin or 10 μM chloroquine for 30 min, or with 25 μg/ml human BD Fc block for 10 min. After the supernatants were replaced with fresh medium, DNA and 2C10 were added as described above. The results were analyzed using a flow cytometer (CytoFLEX; Beckman Coulter, Bream, CA, USA) and a fluorescence microscope (Keyence, Osaka, Japan).
Measurement of cytokines
For quantifying cytokine content in the supernatants of PBMCs, cells were seeded in a 96‐well plate (1 × 106 cells/well). Following a 10‐min incubation with 2‐kbp DNA, 5 μg/ml 2C10 or isotype‐matched control IgG was added to the wells. In inhibition experiments, cells were pretreated with 10 μg/ml cytochalasin D, 5 mM methyl‐β‐cyclodextrin, 80 μM Dynasore, 10 μM chloroquine, 2 μM shikonin or 2 μg/ml RU.521 for 30 min and the supernatants were replaced with fresh medium before DNA and 2C10 were added, as described above. After 4 or 48 h of culture, cytokine content in the supernatants was determined by multi‐analyte flow assays using the Legendplex Human Inflammation Panel 13‐plex (Biolegend, San Diego, CA, USA).
Statistical analysis
Data are expressed as the mean ± standard error of the mean (s.e.m.). P‐values were calculated using a two‐tailed Student’s t‐test (Fig. 5), a one‐tailed Mann–Whitney U‐test (Fig. 7) or a one‐way analysis of variance (anova) followed by Tukey’s multiple comparison test (Figs 2d, 3b, 4c, 6) using Prism (GraphPad Software, La Jolla, CA, USA). P < 0·05 was considered statistically significant.
Figure 5.
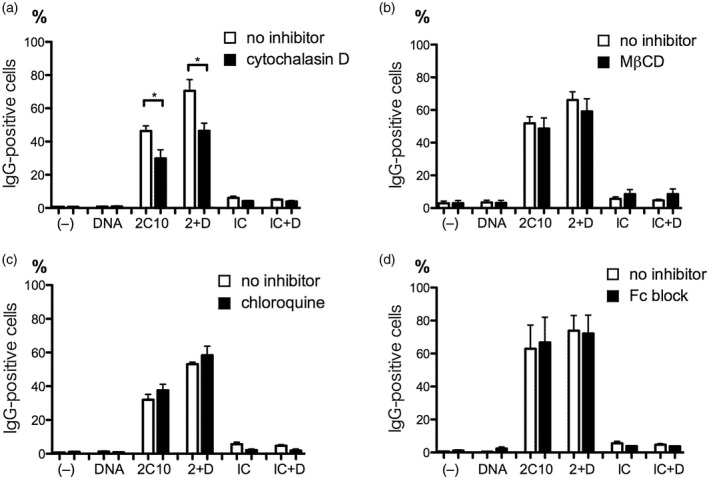
Internalization of 2C10 and DNA into monocytes is mediated by macropinocytosis. Peripheral blood mononuclear cells (PBMCs) from healthy volunteers were treated for 30 min with cytochalasin D (a), methyl‐β‐cyclodextrin (MβCD) (b) or chloroquine (c), or for 10 min with human BD Fc blockTM (d). After washing out the inhibitors, cells were incubated with or without 20 ng/ml unlabeled DNA for 10 min. Then, 5 μg/ml 2C10 or isotype‐matched control (IC) was added and incubated for 1 h. Internalized IgG was detected using Alexa Fluor 488‐labeled goat anti‐mouse IgG, and the percentage of IgG‐positive cells in the monocyte population was estimated. (–): no stimulation; 2 + D: 2C10 + DNA; IC + D: isotype control + DNA. *P < 0·05 versus the cells not treated with inhibitors; n = 3–6.
Figure 7.
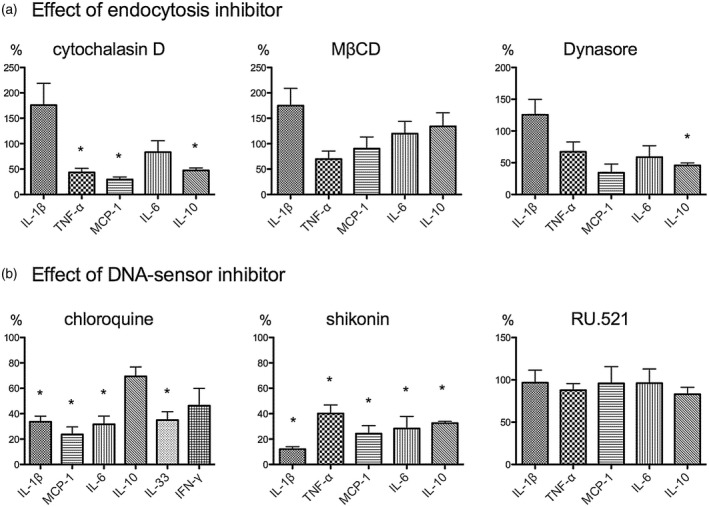
Cytokine expression induced by DNA and 2C10 results from internalization via macropinocytosis and activation of Toll‐like receptor (TLR)‐9 and absent in melanoma 2 (AIM‐2). Peripheral blood mononuclear cells (PBMCs) were treated with endocytosis inhibitors (a) or DNA sensor inhibitors (b) for 30 min. After washing out the inhibitors, cells were incubated with 200 ng/ml DNA for 10 min, then 5 μg/ml 2C10 was added and incubated for 4 h (with cytochalasin D, MβCD, Dynasore, shikonin or RU.521) or 48 h (with chloroquine). Cytokine levels in the supernatants are expressed as the percentage of each cytokine relative to cells incubated with DNA and 2C10 without pretreatment with inhibitors. Date represent the mean ± standard error of the mean (s.e.m.) of four experiments using PBMCs from a single donor. *P < 0·05.
Figure 2.
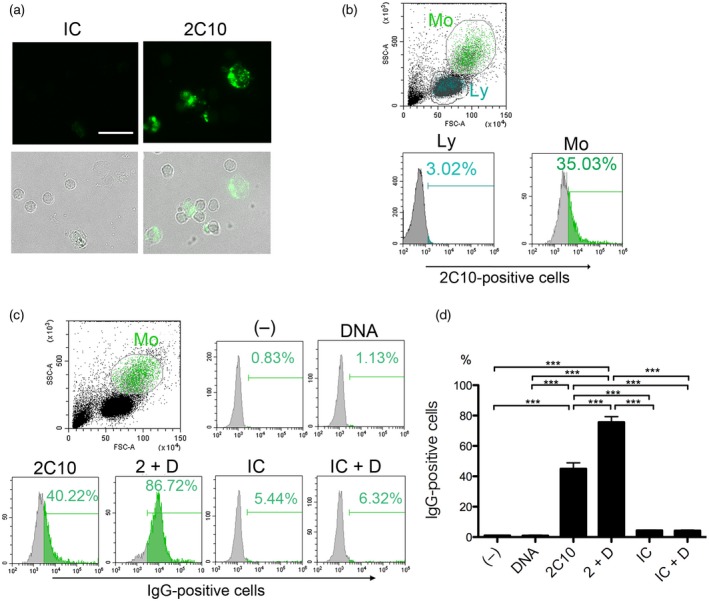
Anti‐DNA antibody 2C10 enters peripheral blood mononuclear cells (PBMCs), which is facilitated by DNA. PBMCs from healthy volunteers were incubated with 5 μg/ml 2C10 or isotype‐matched control (IC) for 1 h. Before detection of internalized antibodies using Alexa Fluor 488‐labeled goat anti‐mouse immunoglobulin (Ig)G secondary antibody, cells were fixed and permeabilized. (a) Microscopic image. Upper row: IgG; lower row: merged IgG and transmitted light images. Scale bar = 20 μm. (b) PBMCs were incubated with 2C10 and analyzed by flow cytometry. Monocytes (Mo) and lymphocytes (Ly) were demarcated by a forward‐ and side‐scatter plot. The histograms are representative results showing the percentages of 2C10‐positive cells. (c) PBMCs from healthy volunteers were incubated with or without 20 ng/ml unlabeled DNA for 10 min. Thereafter, 5 μg/ml 2C10 or IC was added and incubated for 1 h. Internalized IgG was detected using Alexa Fluor 488‐labeled goat anti‐mouse IgG. Cells were gated on the monocyte population. The histograms are representative results showing the percentage of IgG‐positive cells. (d) Mean ± standard error of the mean (s.e.m.) of the percentage of IgG‐positive cells calculated from three independent experiments. (–): no stimulation, 2 + D: 2C10 + DNA; IC + D: isotype control + DNA. ***P < 0·001, n = 7.
Figure 3.
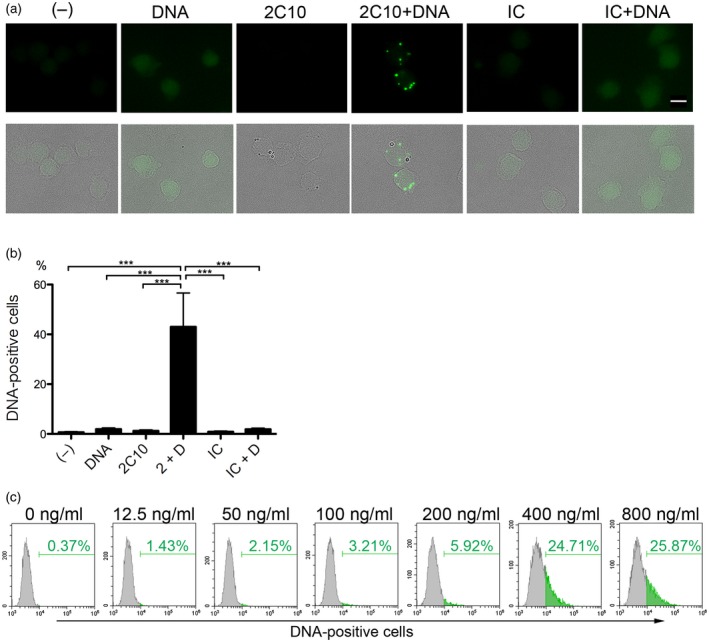
Anti‐DNA antibody 2C10 facilitates the internalization of DNA into THP‐1 cells. (a) THP‐1 cells were incubated with or without 400 ng/ml Alexa Fluor 488‐labeled DNA for 10 min, and then 10 μg/ml 2C10 or isotype‐matched control (IC) was added. After 2 h, internalized DNA was assessed. Upper row: DNA; lower row: merged DNA and transmitted light images. Scale bar = 10 μm. (b) Internalized DNA was quantified by flow cytometry and the mean ± standard error of the mean (s.e.m.) of the percentage of DNA‐positive cells was calculated from three independent experiments. ***P < 0·001, n = 5. (c) THP‐1 cells were incubated with the indicated concentrations of Alexa Fluor 488‐labeled DNA for 10 min, and then 10 μg/ml 2C10 was added. After incubation for 2 h, the percentage of DNA‐positive cells was assessed by flow cytometry. (–): no stimulation; 2 + D: 2C10 + DNA; IC + D: isotype control + DNA.
Figure 4.
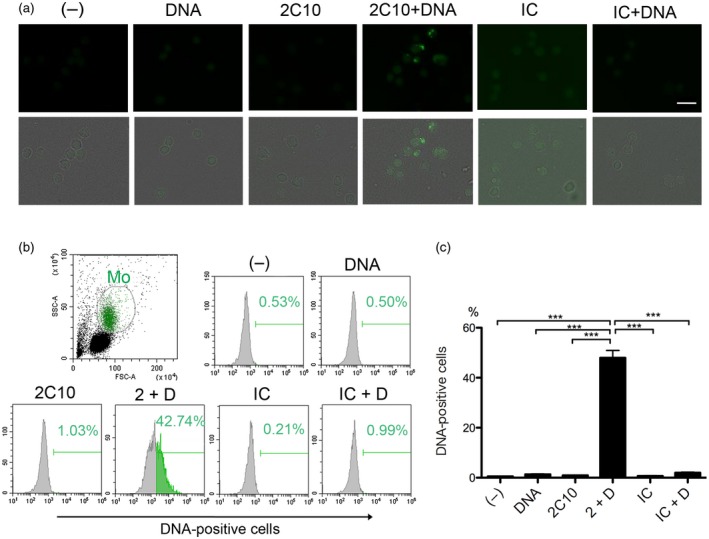
Anti‐DNA antibody 2C10 facilitates the internalization of DNA into normal peripheral blood mononuclear cells (PBMCs). (a) PBMCs were incubated with or without 400 ng/ml Alexa Fluor 488‐labeled DNA for 10 min, and then 10 μg/ml 2C10 or isotype‐matched control (IC) was added. After 1 h, internalized DNA was assessed. Upper row: DNA; lower row: merged DNA and transmitted light images. Scale bar = 20 μm. (b) The percentage of DNA‐positive cells in the monocyte‐rich population determined by flow cytometry. (c) Mean ± standard error of the mean (s.e.m.) of the percentage of DNA‐positive cells in the monocyte‐rich population calculated from three independent experiments. (–): no stimulation; 2 + D: 2C10 + DNA; IC + D: isotype control + DNA. ***P < 0·001, n = 5.
Figure 6.
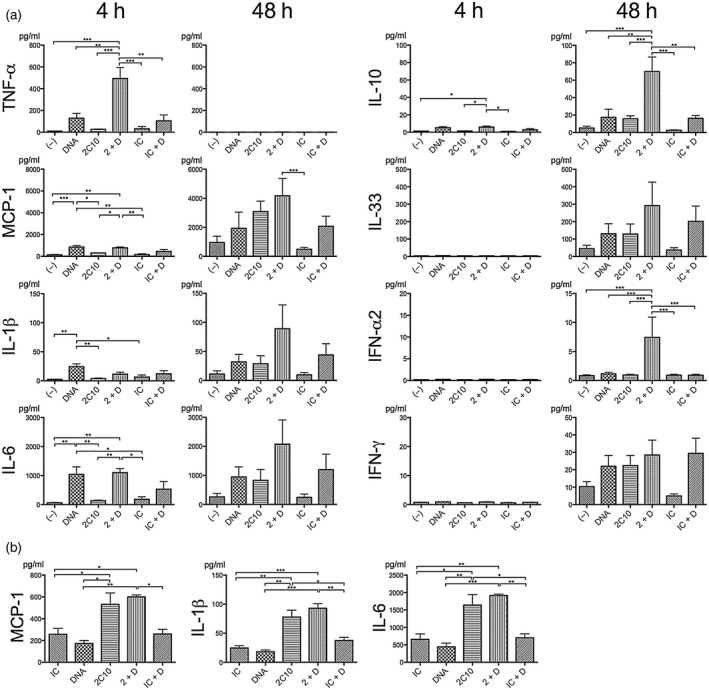
Internalization of 2C10 and DNA induces expression of interferon (IFN)‐α and proinflammatory cytokines by peripheral blood mononuclear cells (PBMCs). (a) PBMCs were incubated with 5 μg/ml 2C10 or isotype control (IC) for 4 or 48 h, with or without prior 10‐min incubation with 200 ng/ml unlabeled DNA, and cytokine levels in the supernatants were assessed. Data represent the mean ± standard error of the mean (s.e.m.) of four to seven experiments using PBMCs from two to three donors. (b) PBMCs were incubated with 5 μg/ml 2C10 or IC for 4 h, with or without prior 10‐min incubation with 10 ng/ml unlabeled DNA. Data represent the mean ± s.e.m. of three experiments using PBMCs from a single donor. (–): no stimulation; 2 + D: 2C10 + DNA; IC + D: isotype control + DNA. Every figure was analyzed by a one‐way analysis of variance (anova) followed by Tukey’s multiple comparison test, and only significantly different pairs are indicated by asterisks. *P< 0·05; **P < 0·01; ***P < 0·001.
Results
DNA enhances the internalization of monoclonal anti‐dsDNA antibody 2C10 by live cells
As we have observed previously, anti‐dsDNA antibody 2C10 enters live monocytes and the monocytic cell line THP‐1 16. To confirm if the current batch of 2C10 actually enters live cells, we tested the effect of fixation and permeabilization (F/P) of THP‐1 cells. Following incubation with 2C10, if we performed F/P before the reaction of fluorescence‐labeled anti‐mouse IgG secondary antibody, small specks and dim cytoplasmic staining were observed (Fig. 1a). In contrast, when F/P was performed after the secondary antibody reaction, the fluorescence was markedly diminished, indicating that the secondary antibody did not penetrate the intact cell membrane, and the major part of 2C10 was in the cells. When THP‐1 cells were incubated with different concentrations of 2C10, it was incorporated into the cells in a dose‐dependent manner (Fig. 1b). Because immune complexes are easily engulfed by phagocytic cells, and anti‐DNA antibodies are still associated with small amounts of DNA even after purification using protein A or protein G 17, 18, we hypothesized that DNA could be involved in the process of 2C10 internalization. To test this, we added 2‐kbp dsDNA to the cultures 10 min prior to the incubation of the THP‐1 cells with 2C10. This resulted in an increased internalization of 2C10, but not the isotype‐matched control IgG, in the presence of DNA (Fig. 1c).
Figure 1.
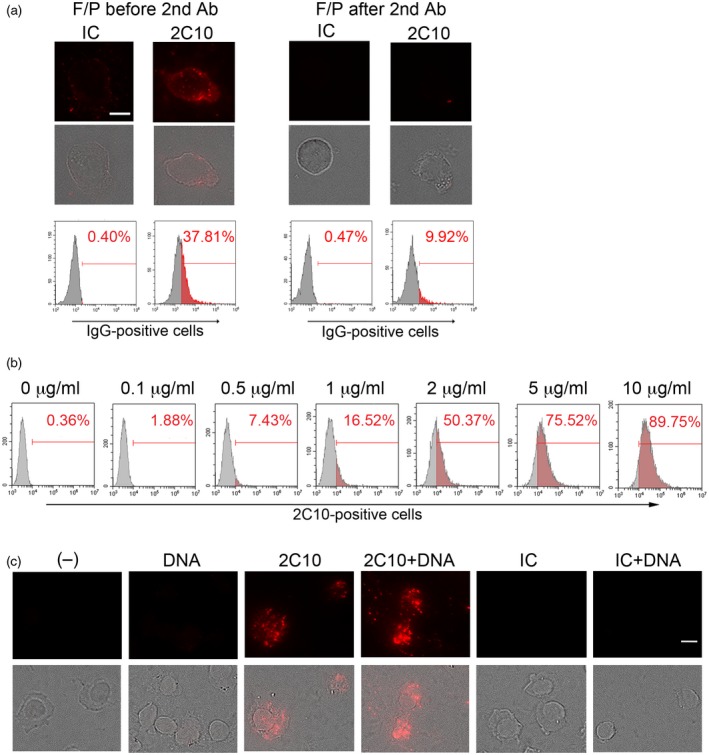
Anti‐DNA antibody 2C10 enters THP‐1 cells, which is facilitated by DNA. (a) THP‐1 cells were incubated with 2 μg/ml 2C10 or isotype‐matched control (IC) for 1 h. On the left, before detection of internalized antibodies using fluorescence‐labeled goat anti‐mouse immunoglobulin (Ig)G secondary antibody, cells were fixed and permeabilized (F/P). In the right, antibodies on the cell surface were detected by fluorescence‐labeled goat anti‐mouse IgG secondary antibody, then cells were F/P. Upper row: IgG; lower row: merged IgG and transmitted light images. Scale bar = 10 μm. Histograms are representative results showing the percentages of IgG‐positive cells. (b) THP‐1 cells were incubated with the indicated concentrations of 2C10 for 1 h. Internalized 2C10 was detected by phycoerythrin (PE)‐labeled goat anti‐mouse IgG and analyzed by flow cytometry. Histograms are representative results showing the percentage of 2C10‐positive cells. (c) THP‐1 cells were incubated with or without 400 ng/ml unlabeled DNA for 10 min. Thereafter, 10 μg/ml 2C10 or IC was added and incubated for 2 h. Internalized IgG was detected by PE‐labeled goat anti‐mouse IgG. Upper row: IgG; lower row: merged IgG and transmitted light images. Scale bar = 10 μm.
To confirm whether 2C10 also enters normal monocytes, we incubated PBMCs from healthy donors with 2C10 and observed that the antibody was incorporated mainly into slightly large cells (Fig. 2a). When the secondary antibody was added without preceding F/P, virtually no staining was observed, indicating that 2C10 entered the cells (data not shown). When monocytes and lymphocytes were demarcated by a forward‐ and side‐scatter plot of flow cytometry, it was clear that 2C10 preferentially entered the monocyte fraction (Fig. 2b). Although more than 84% of the cells in the monocyte fraction were CD14‐positive, some other cell types were thought to be included, and thus we describe the population as the ‘monocyte‐rich fraction’ hereafter. Incubating PBMCs with 5 μg/ml 2C10 alone resulted in approximately 40% of the monocyte‐rich population taking up the antibody, but when DNA was added to PBMCs prior to 2C10 the percentage increased to approximately 80%, again indicating that DNA‐enhanced internalization of 2C10 (Fig. 2c,d). After a 2‐h incubation with 2C10 there was no annexin V staining, suggesting that the cells were viable, not apoptotic, at this time‐point (data not shown).
dsDNA enters live cells in the presence of 2C10
We next tested whether 2C10 facilitates internalization of DNA. THP‐1 cells were incubated with 2C10 in the presence of Alexa 488‐labeled 2‐kbp DNA. Thereafter, distinct specks of labeled DNA could be seen in the cytoplasm (Fig. 3a). However, cells incubated with labeled DNA alone or DNA plus isotype‐matched control IgG showed only a faint fluorescence, probably reflecting non‐specific cell surface DNA binding. Quantitative analysis by flow cytometry clearly showed that the labeled DNA alone was not incorporated by THP‐1 cells to any significant extent, whereas 2C10, but not isotype‐matched IgG, strongly facilitated DNA internalization (Fig. 3b). In the presence of 10 μg/ml 2C10, different concentrations of labeled DNA entered THP‐1 cells in a dose‐dependent manner, reaching a plateau at 400 ng/ml (Fig. 3c).
To verify the facilitation effect of 2C10 on DNA internalization into normal human cells, PBMCs were incubated with 2C10 in the presence or absence of Alexa 488‐labeled DNA. Similar to THP‐1 cells, a fraction of cells in PBMCs incorporated DNA into the cytoplasm, but only when also incubated with 2C10 (Fig. 4a). Flow cytometric analysis revealed that labeled DNA mainly entered the monocyte‐rich fraction in the presence of 2C10, with the proportion of DNA‐containing cells in that fraction reaching 40–50% (Fig. 4b, c), whereas that in the lymphocyte‐rich fraction was 3–4%.
Macropinocytosis is involved in the internalization of 2C10 and DNA
To identify the pathways involved in the internalization of 2C10 and DNA, PBMCs were treated for 30 min with the macropinocytosis inhibitor cytochalasin D, the caveolae/lipid raft endocytosis inhibitor methyl‐β‐cyclodextrin (MβCD) or the clathrin‐dependent endocytosis inhibitor chloroquine 19 prior to the incubation with DNA and 2C10. Internalization of 2C10 into monocytes was found to be significantly suppressed by cytochalasin D, irrespective of the presence or absence of DNA (Fig. 5a). In contrast, neither MβCD nor chloroquine significantly inhibited internalization (Fig. 5b,c). Because murine IgG is known to bind to the human Fcγ receptor (FcγR) II 20, we tested whether a human FcγR blocker would inhibit the internalization of 2C10, but no significant suppression was observed (Fig. 5d). These results suggest that macropinocytosis plays a role in the internalization of 2C10 and DNA.
2C10 and DNA induce expression of cytokines implicated in lupus pathogenesis
To study whether or not PBMCs are activated as a consequence of incorporation of 2C10 together with DNA, cytokine levels in the culture supernatants were measured by multi‐analyte flow assays (Fig. 6a). After a 4‐h incubation, TNF‐α was present at significantly higher concentrations in supernatants of cells stimulated with 2C10 (5 μg/ml) and DNA (200 ng/ml), relative to 2C10 alone, or DNA alone. In contrast, after 4 h even in the presence of 200 ng/ml DNA alone, the amounts of MCP‐1, IL‐1β and IL‐6 were greatly increased. We therefore repeated the experiments using 10 ng/ml DNA. At this dose, 2C10 together with the DNA induced these cytokines more effectively than did DNA alone, but there were no differences between 2C10 alone and 2C10 + DNA (Fig. 6b). These results suggest that 2C10 itself may still be carrying a small amount of DNA. Extending the incubation time to 48 h resulted in secretion of MCP‐1, IL‐1β, IL‐6, IL‐10, IL‐33, IFN‐α (α2 subtype) and IFN‐γ into the PBMC supernatants. Although the manner by which each of these cytokines contributes to SLE pathogenesis has not been fully elucidated, serum levels of all these cytokines are known to be elevated in this disease. With the exception of IFN‐γ, cytokine concentrations tended to be higher in supernatants of cells cultured with both 2C10 and DNA (200 ng/ml). These results suggest that the DNA induced the expression of multiple different cytokines when incorporated into cells with the assistance of the antibody 2C10. The effect of 2C10 and DNA on IFN‐γ production was not clear, possibly because this cytokine is not produced by monocytes, but by T cells indirectly stimulated by monokines. Nevertheless, IFN‐γ plays an important role in lupus pathogenesis by conversely stimulating monocytes to express BAFF 21.
Expression of cytokines is suppressed by endocytosis inhibitors and DNA sensor inhibitors
To confirm whether endocytosis contributes to the expression of cytokines, PBMCs were treated with cytochalasin D, MβCD or the clathrin‐dependent endocytosis inhibitor Dynasore. Treating PBMCs with cytochalasin D before addition of DNA and 2C10 resulted in a significant reduction in the expressions of TNF‐α, MCP‐1 and IL‐10 relative to untreated cells (Fig. 7a). However, MβCD and Dynasore showed no significant suppressive effect on these cytokine expressions except for Dynasore on IL‐10. Dynasore tended to suppress MCP‐1, IL‐6 and TNF‐α, but P‐values for them were 0·057, 0·10 and 0·10, respectively. These results indicate that endocytosis, especially macropinocytosis, is necessary as an initial step for the cytokine expression.
Further, to study whether the internalized DNA is detected by intracellular DNA sensors, PBMCs were treated with the TLR‐9 inhibitor chloroquine, which was also used as a clathrin‐dependent endocytosis inhibitor in the above experiments with no effect (Fig. 5c), or the AIM‐2/nucleotide‐binding oligomerization domain, leucine rich repeat and pyrin domain‐containing protein 3 (NLRP‐3)‐inflammasome inhibitor shikonin 22 or the cGAS inhibitor RU.521 23. Treatment of PBMCs with chloroquine resulted in reduction of IL‐1β, MCP‐1, IL‐6 and IL‐33 levels in the supernatants (Fig. 7b). Shikonin suppressed secretion of IL‐1β, TNF‐α, MCP‐1, IL‐6 and IL‐10, while RU.521 had no significant suppressive effects. These results suggest that internalized DNA was detected by TLR‐9 in endosomes, and after leaking into the cytoplasm it was also detected by AIM‐2.
Discussion
In this study, we showed that the anti‐dsDNA monoclonal antibody 2C10 was preferentially taken up by the monocyte‐rich fraction of normal human PBMCs by the process of macropinocytosis and that this was facilitated by DNA. In contrast, and more important from the clinical point of view, internalization of DNA by the cells was significant only in the presence of 2C10. We found that following the internalization of both antibody and DNA, secretion by the PBMCs of multiple cytokines relating to SLE pathogenesis was increased, and that this was suppressed by the macropinocytosis inhibitor cytochalasin D, as well as the TLR‐9 inhibitor chloroquine and the AIM‐2/NLRP‐3 inflammasome inhibitor shikonin. These results suggest that in SLE, DNA in the plasma or on the cell surface can enter monocytes through endocytosis with the help of anti‐DNA antibodies. Thereafter, multiple intracellular DNA sensors can be activated, leading to production of cytokines relevant to disease pathogenesis. Thus, anti‐DNA antibodies, once produced, play a pivotal role in promoting a vicious circle in the lupus pathogenesis.
As long as four decades ago, Alarcón‐Segovia et al. first observed that anti‐nuclear antibodies enter living cells 24, 25. Subsequently, numerous studies referring to cell‐internalizable anti‐DNA antibodies have been conducted using different assay conditions. Many possible means by which antibodies can enter cells have been implicated, including via Fcγ receptors 24, 26, an unidentified nucleosome receptor 27, DNA‐histone complexes 28, glycosaminoglycans 29 and myosin 1 30. In SLE, apoptosis is enhanced 31 and clearance of the apoptotic debris is compromised 2, 32, resulting in higher concentrations of DNA or nucleosomes in the sera of patients relative to healthy controls. Estimated mean levels of 40–240 ng/ml of cell‐free DNA in serum samples from SLE patients have been reported 33, 34, 35. As a consequence, DNA or nucleosomes are attached to the cell surface both in‐vivo and in in‐vitro culture systems 27, 36, 37. Although the precise mechanism of the DNA binding remains obscure, certain positively charged cell surface molecules are supposed to mediate this interaction. 2C10 makes contact with dA‐dT base pairs in or over the minor groove of dsDNA 14, such that a single molecule of 2‐kbp dsDNA can be bound by dozens of 2C10 antibodies. Furthermore, multiple DNA molecules can be cross‐linked to form large immune complexes. Together with the results of suppression by cytochalasin D, binding of the immune complexes or cross‐linking of the cell surface DNA is likely to ruffle the cell membrane and result in triggering macropinocytosis. Fc receptors are unlikely to be involved, because an Fc‐blocker did not suppress the internalization, and consistent with this, 2C10 can enter Fc‐receptor‐negative HeLa cells (data not shown). Fc‐receptor‐independent internalization of anti‐DNA antibodies has been observed by other investigators, including Jang et al. 38, who used the single‐chain variable fragment of a monoclonal antibody. Apart from anti‐DNA antibodies, small amount of free DNA added to the culture was thought to enter the cells by pinocytosis because DNA alone could induce production of some cytokines, as shown in Fig. 6a. This DNA internalization was below detection sensitivity of our assay system, and could not be observed by microscopy or flow cytometry as shown in Fig. 4.
Since the beginning of this century, studies on the innate immune system have revealed that many different intracellular molecules recognize nucleic acids derived from viruses or bacteria. These sensors definitely act as an essential defense mechanism, but if self‐DNA or RNA were to be taken up into the cells, as shown in the following studies, they might be activated and contribute to the pathogenesis of autoimmune diseases 39. For example, the TLR‐9 pathway of mouse autoreactive B cells which express rheumatoid factor on the surface was activated by chromatin–IgG complexes 40. Activation of mouse dendritic cells was also caused by dual engagement of TLR‐9 and FcγRIII or another unidentified receptor 41. Similar studies using human plasmacytoid dendritic cells (pDCs) showed activation of TLR‐9 by DNA–anti‐DNA immune complexes which bound to FcγRIIa 26. Another study showed that DNA–anti‐DNA immune complexes entered pDCs and activated them via TLR‐9, but only when neutrophil‐derived anti‐microbial peptides were attached to the immune complexes 42. Thus, most previous studies have suggested a significant role in the pathogenesis of SLE of Fc receptors and TLR‐9 in pDCs. In this study, we adopted a simple assay system composed of a monoclonal antibody, homogeneous dsDNA and PBMCs from healthy subjects, and observed that the DNA entered monocytes only when it was associated with the anti‐dsDNA antibody 2C10. This internalization resulted in increased secretion of TNF‐α, MCP‐1, IL‐1β, IL‐6, IL‐10, IL‐33, IFN‐α and IFN‐γ by the PBMCs. Because chloroquine inhibited secretion of most of these cytokines, it was thought that activation by the internalized DNA proceeded via TLR‐9. Using TLR‐9 agonist oligodeoxynucleotides (ODNs), it has been suggested that large multimeric ODNs tend to be retained in early endosomes for a long period and activate the TLR‐9–IFN regulatory factor 7 (IRF‐7) pathway leading to IFN‐α production, whereas smaller ODNs move on to late endosomes and stimulate the TLR‐9–nuclear factor (NF)‐κB pathway to produce proinflammatory cytokines 43. In our model, using 2‐kbp DNA, both pathways seem to be activated in monocytes. Another possible explanation is that while proinflammatory cytokines were produced by monocytes, IFN‐α was released from a different cell type. This could well be the pDCs, because activation of TLR‐9 in conventional dendritic cells (DCs) and macrophages is reported to drive a strong proinflammatory response, whereas in pDCs the same receptor triggers a potent type I interferon response 39. Furthermore, we have noticed that 2C10 entered the lymphocyte‐rich fraction at small, but not negligible, percentages (Fig. 2b). The whole picture of interaction among different cell types harboring DNA/anti‐DNA remains to be clarified.
In addition to TLR‐9, we concluded that AIM‐2 was also involved in our model, because it can recognize cytoplasmic DNA to form inflammasomes and lead to the activation of caspase‐1, cleavage of pro‐IL‐1β and secretion of IL‐1β 13. Shikonin, an ingredient of a traditional Chinese herbal medicine with anti‐inflammatory effects, was recently reported to inhibit caspase‐1 directly 22. It inhibited secretion of IL‐1β in our system, suggesting a role of caspase‐1 in the AIM‐2 inflammasome formed on recognition of DNA. An involvement of the NLRP‐3 inflammasome cannot be excluded, but NLRP‐3 does not directly recognize DNA. Internalization of dsDNA has recently been suggested to activate NLRP‐3 via induction of reactive oxygen species and K+ efflux, but the precise mechanism of this process remains uncertain 44.
In conclusion, by establishing a simple assay system with normal mononuclear cells, we demonstrated that just a single anti‐dsDNA antibody can complete a model of lupus pathogenesis, from facilitation of DNA internalization through to secretion of multiple cytokines, including IFN‐α and proinflammatory cytokines. These results again emphasize the importance of maintaining anti‐dsDNA antibodies at low levels in the treatment of SLE.
Disclosure
All the authors have no conflicts of interest to disclose.
Author contributions
K. I. performed the experiments and wrote the paper; M. I. performed the experiments; T. K. designed the study and revised the paper.
Acknowledgement
This study was supported by JSPS KAKENHI Grant number JP16K089029.
References
- 1. Oku K, Atsumi T. Systemic lupus erythematosus: nothing stales her infinite variety. Mod Rheumatol 2018; 28:758–65. [DOI] [PubMed] [Google Scholar]
- 2. Tsokos GC, Lo MS, Reis PC, Sullivan KE. New insights into the immunopathogenesis of systemic lupus erythematosus. Nat Rev Rheumatol 2016; 12:716–30. [DOI] [PubMed] [Google Scholar]
- 3. Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum 1997; 40:1725. [DOI] [PubMed] [Google Scholar]
- 4. Pisetsky DS. Anti‐DNA antibodies – quintessential biomarkers of SLE. Nat Rev Rheumatol 2016; 12:102–10. [DOI] [PubMed] [Google Scholar]
- 5. Crow MK. Type I interferon in the pathogenesis of lupus. J Immunol 2014; 192:5459–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vallin H, Perers A, Alm GV, Ronnblom L. Anti‐double‐stranded DNA antibodies and immunostimulatory plasmid DNA in combination mimic the endogenous IFN‐alpha inducer in systemic lupus erythematosus. J Immunol 1999;163:6306–13. [PubMed] [Google Scholar]
- 7. Sun KH, Yu CL, Tang SJ, Sun GH. Monoclonal anti‐double‐stranded DNA autoantibody stimulates the expression and release of IL‐1 beta, IL‐6, IL‐8, IL‐10 and TNF‐alpha from normal human mononuclear cells involving in the lupus pathogenesis. Immunology 2000; 99:352–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hemmi H, Takeuchi O, Kawai T et al A Toll‐like receptor recognizes bacterial DNA. Nature 2000; 408:740–5. [DOI] [PubMed] [Google Scholar]
- 9. Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double‐stranded RNA and activation of NF‐kappaB by Toll‐like receptor 3. Nature 2001; 413:732–8. [DOI] [PubMed] [Google Scholar]
- 10. Heil F, Hemmi H, Hochrein H et al Species‐specific recognition of single‐stranded RNA via Toll‐like receptor 7 and 8. Science 2004; 303:1526–9. [DOI] [PubMed] [Google Scholar]
- 11. Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 2008; 455:674–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hornung V, Ablasser A, Charrel‐Dennis M et al AIM2 recognizes cytosolic dsDNA and forms a caspase‐1‐activating inflammasome with ASC. Nature 2009; 458:514–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Monteith AJ, Kang S, Scott E et al Defects in lysosomal maturation facilitate the activation of innate sensors in systemic lupus erythematosus. Proc Natl Acad Sci USA 2016; 113:E2142–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kubota T, Watanabe N, Kanai Y, Stollar BD. Enhancement of oxidative cleavage of DNA by the binding sites of two anti‐double‐stranded DNA antibodies. J Biol Chem 1996; 271:6555–61. [DOI] [PubMed] [Google Scholar]
- 15. Jang YJ, Lecerf JM, Stollar BD. Heavy chain dominance in the binding of DNA by a lupus mouse monoclonal autoantibody. Mol Immunol 1996; 33:197–210. [DOI] [PubMed] [Google Scholar]
- 16. Virachith S, Saito M, Watanabe Y et al Anti‐β2‐glycoprotein I antibody with DNA binding activity enters living monocytes via cell surface DNA and induces tissue factor expression. Clin Exp Immunol 2019; 195:167–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Satake F, Watanabe N, Miyasaka N et al Induction of anti‐DNA antibodies by immunization with anti‐DNA antibodies: mechanism and characterization. Lupus 2000; 9:489–97. [DOI] [PubMed] [Google Scholar]
- 18. Fenton KA, Tommeras B, Marion TN, Rekvig OP. Pure anti‐dsDNA mAbs need chromatin structures to promote glomerular mesangial deposits in BALB/c mice. Autoimmunity 2010; 43:179–88. [DOI] [PubMed] [Google Scholar]
- 19. Dutta D, Donaldson JG. Search for inhibitors of endocytosis: intended specificity and unintended consequences. Cell Logist 2012; 2:203–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Werwitzke S, Trick D, Sondermann P et al Treatment of lupus‐prone NZB/NZW F‐1 mice with recombinant soluble Fc gamma receptor II (CD32). Ann Rheum Dis 2008; 67:154–61. [DOI] [PubMed] [Google Scholar]
- 21. Harigai M, Kawamoto M, Hara M et al Excessive production of IFN‐gamma in patients with systemic lupus erythematosus and its contribution to induction of B lymphocyte stimulator/B cell‐activating factor/TNF ligand superfamily‐13B. J Immunol 2008; 181:2211–9. [DOI] [PubMed] [Google Scholar]
- 22. Zorman J, Susjan P, Hafner‐Bratkovic I. Shikonin suppresses NLRP3 and AIM2 inflammasomes by direct Inhibition of caspase‐1. PLOS ONE 2016; 11:e0159826. doi: 10.1371/journal.pone.0159826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vincent J, Adura C, Gao P et al Small molecule inhibition of cGAS reduces interferon expression in primary macrophages from autoimmune mice. Nat Commun 2017; 8:750. doi: 10.1038/s41467-017-01770-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Alarcon‐Segovia D, Ruizarguelles A, Fishbein E. Antibody to nuclear ribonucleoprotein penetrates live human mononuclear cells through Fc receptors. Nature 1978; 271:67–9. [DOI] [PubMed] [Google Scholar]
- 25. Alarcón‐segovia D, Llorente L, Fishbein E, Diazjouanen E. Abnormalities in the content of nucleic acids of peripheral blood mononuclear cells from patients with systemic lupus erythematosus relationship to DNA antibodies. Arthritis Rheum 1982; 25:304–17. [DOI] [PubMed] [Google Scholar]
- 26. Means TK, Latz E, Hayashi F et al Human lupus autoantibody‐DNA complexes activate DCs through cooperation of CD32 and TLR9. J Clin Invest 2005; 115:407–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Koutouzov S, Cabrespines A, Amoura Z et al Binding of nucleosomes to a cell surface receptor: redistribution and endocytosis in the presence of lupus antibodies. Eur J Immunol 1996; 26:472–86. [DOI] [PubMed] [Google Scholar]
- 28. Zannikou M, Bellou S, Eliades P et al DNA‐histone complexes as ligands amplify cell penetration and nuclear targeting of anti‐DNA antibodies via energy‐independent mechanisms. Immunology 2016; 147:73–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Avrameas A, Gasmi L, Buttin G. DNA and heparin alter the internalization process of anti‐DNA monoclonal antibodies according to patterns typical of both the charged molecule and the antibody. J Autoimmun 2001; 16:383–91. [DOI] [PubMed] [Google Scholar]
- 30. Yanase K, Smith RM, Puccetti A et al Receptor‐mediated cellular entry of nuclear localizing anti‐DNA antibodies via myosin 1. J Clin Invest 1997; 100:25–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Perniok A, Wedekind F, Herrmann M et al High levels of circulating early apoptotic peripheral blood mononuclear cells in systemic lupus erythematosus. Lupus 1998; 7:113–8. [DOI] [PubMed] [Google Scholar]
- 32. Sisirak V, Sally B, D'Agati V et al Digestion of chromatin in apoptotic cell microparticles prevents autoimmunity. Cell 2016; 166:88–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang SG, Lu X, Shu XM et al Elevated plasma cfDNA may be associated with active lupus nephritis and partially attributed to abnormal regulation of neutrophil extracellular traps (NETs) in patients with systemic lupus erythematosus. Intern Med 2014; 53:2763–71. [DOI] [PubMed] [Google Scholar]
- 34. Tug S, Helmig S, Menke J et al Correlation between cell free DNA levels and medical evaluation of disease progression in systemic lupus erythematosus patients. Cell Immunol 2014; 292:32–9. [DOI] [PubMed] [Google Scholar]
- 35. Kato Y, Park J, Takamatsu H et al Apoptosis‐derived membrane vesicles drive the cGAS‐STING pathway and enhance type I IFN production in systemic lupus erythematosus. Ann Rheum Dis 2018; 77:1507–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kubota T, Kanai Y, Miyasaka N. Interpretation of the cross‐reactivity of anti‐DNA antibodies with cell surface proteins: the role of cell surface histones. Immunol Lett 1990; 23:187–94. [DOI] [PubMed] [Google Scholar]
- 37. Laktionov PP, Tamkovich SN, Rykova EY et al Cell‐surface‐bound nucleic acids: free and cell‐surface‐bound nucleic acids in blood of healthy donors and breast cancer patients. Ann NY Acad Sci 2004; 1022:221–7. [DOI] [PubMed] [Google Scholar]
- 38. Jang JY, Jeong JG, Jun HR et al A nucleic acid‐hydrolyzing antibody penetrates into cells via caveolae‐mediated endocytosis, localizes in the cytosol and exhibits cytotoxicity. Cell Mol Life Sci 2009; 66:1985–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Crowl JT, Gray EE, Pestal K et al Intracellular nucleic acid detection in autoimmunity. Annu Rev Immunol 2017; 35:313–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Leadbetter EA, Rifkin IR, Hohlbaum AM et al Chromatin‐IgG complexes activate B cells by dual engagement of IgM and Toll‐like receptors. Nature 2002; 416:603–7. [DOI] [PubMed] [Google Scholar]
- 41. Boule MW, Broughton C, Mackay F et al Toll‐like receptor 9‐dependent and ‐independent dendritic cell activation by chromatin‐immunoglobulin G complexes. J Exp Med 2004; 199:1631–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lande R, Ganguly D, Facchinetti V et al Neutrophils activate plasmacytoid dendritic cells by releasing self‐DNA–peptide complexes in systemic lupus erythematosus. Sci Transl Med 2011;3:73ra19–73ra19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Okuya K, Tamura Y, Saito K et al Spatiotemporal regulation of heat shock protein 90‐chaperoned self‐DNA and CpG‐oligodeoxynucleotide for type I IFN induction via targeting to static early endosome. J Immunol 2010; 184:7092–9. [DOI] [PubMed] [Google Scholar]
- 44. Shin MS, Kang Y, Lee N et al Self double‐stranded (ds) DNA induces IL‐1 beta production from human monocytes by activating NLRP3 inflammasome in the presence of anti‐dsDNA antibodies. J Immunol 2013; 190:1407–15. [DOI] [PMC free article] [PubMed] [Google Scholar]


