Summary
Mucosal‐associated invariant T (MAIT) cells and Vδ2+ γδ T cells are anti‐bacterial innate‐like lymphocytes (ILLs) that are enriched in blood and mucosa. ILLs have been implicated in control of infection. However, the role of ILLs in community‐acquired pneumonia (CAP) is unknown. Using sputum samples from a well‐characterized CAP cohort, MAIT cell and Vδ2+ T cell abundance was determined by quantitative polymerase chain reaction (qPCR). Cytokine and chemokine concentrations in sputum were measured. The capacity of bacteria in sputum to produce activating ligands for MAIT cells and Vδ2+ T cells was inferred by 16S rRNA sequencing. MAIT cell abundance in sputum was higher in patients with less severe pneumonia; duration of hospital admission was inversely correlated with both MAIT and Vδ2+ T cell abundance. The abundance of both ILLs was higher in patients with a confirmed bacterial aetiology; however, there was no correlation with total bacterial load or the predicted capacity of bacteria to produce activating ligands. Sputum MAIT cell abundance was associated with interferon (IFN)‐α, IFN‐γ, and sputum neutrophil abundance, while Vδ2+ T cell abundance was associated with CXCL11 and IFN‐γ. Therefore, MAIT and Vδ2+ T cells can be detected in sputum in CAP, where they may contribute to improved clinical outcome.
Keywords: community‐acquired pneumonia, gamma delta T cells, MAIT cells, qPCR, severity, Vδ2+ T cells
Mucosal‐associated invariant T (MAIT) cells and Vδ2+ γδ T cells are innate‐like lymphocytes that are abundant in humans in blood and at mucosal surfaces, including the lungs. They provide rapid effector responses upon activation by specific ligands produced by a broad range of bacteria, including numerous pulmonary pathogens. Here we show that MAIT and Vδ2+ γδ T cells can be detected in sputum in patients with community acquired pneumonia, where they may contribute to improved clinical outcome.
Introduction
Community‐acquired pneumonia (CAP) is a leading cause of hospital admission 1. Innate immune defences are critical for the prevention and early control of pulmonary infection and for instructing the adaptive immune response 2. Innate and adaptive immunity are bridged by innate‐like lymphoid cells, such as mucosal‐associated invariant T (MAIT) cells and γδ T cells; however, their role in CAP is not yet fully understood.
MAIT cells occur at high frequencies in blood and at mucosal surfaces, including the lung 3. They have a semi‐invariant T cell receptor (TCR), Vα7.2‐Jα12/20/33, that is restricted by the highly conserved, major histocompatibility complex (MHC) class Ib‐like molecule, MR1, and recognizes derivatives of the riboflavin synthesis pathway 4, 5. This pathway is present in many bacteria, including numerous pulmonary pathogens, but not in humans 6. In addition, MAIT cells can be activated independently of their TCR by cytokines, in particular, by interleukin (IL)‐12 and ‐18, suggesting potential involvement in the immune response to non‐riboflavin‐synthesizing bacteria and viruses 7. Studies in mice suggest that MAIT cells play an important role in the protection against both bacterial and viral pulmonary pathogens and in the containment of chronic infection 8, 9, 10, 11. In humans, several observations suggest a potential role of MAIT cells in protection against pneumonia. First, MAIT cell frequency in blood is decreased in patients with pneumonia or active tuberculosis 12. Secondly, in critically ill patients, a persistently low frequency of MAIT cells increases the risk of subsequent nosocomial infections, including pneumonia 13. Thirdly, MAIT cells are depleted in HIV‐infected patients 14, who are 25 times more likely to develop bacterial pneumonia, an increased risk which is not completely explained by the loss of CD4+ T cells 15.
The Vγ9Vδ2 subset is the predominant γδ T cell population in humans, comprising 1–9% of circulating T cells. They are unique to humans and higher primates and are activated by (E)‐4‐hydroxy‐3‐methyl‐but‐2‐enyl pyrophosphate (HMB‐PP), an intermediate of the non‐mevalonate pathway of isoprenoid biosynthesis present in many bacteria, including pulmonary pathogens, but not in humans 16. Vγ9Vδ2 T cells may also play a role in the immune response to pulmonary infection. Expansion of the human Vγ9Vδ2 T cell population has been reported in several bacterial infections 16, 17, as well as their rapid trafficking to the lungs after activation 18. Adoptive transfer of Vγ9Vδ2 T cells in non‐human primates protected them from pulmonary Mycobacterium tuberculosis infection 19.
In this study, we used quantitative real‐time polymerase chain reaction (qPCR) to investigate the abundance of MAIT cells and Vγ9Vδ2 T cells in the sputum of patients with CAP. The abundance of MAIT cells and Vγ9Vδ2 T cells in sputum was correlated with clinical data.
Materials and methods
Sputum and blood samples
Frozen (–80°C) sputum samples (n = 88) from a previously reported cohort of patients with CAP were analysed 20. Ethical approval was granted by the Northern A Health and Disability Ethics Committee (12/NTA/30). Induced sputa were collected from healthy volunteers (n = 12) by nebulization of hypertonic saline 21. Ethical approval was granted by the University of Otago Human Research Ethics Committee (H19/053).
To validate the qPCR method, blood from healthy adult volunteers was used (n = 13). Peripheral blood mononuclear cells (PBMCs) were isolated using Lymphoprep (Alere Technologies AS, Oslo, Norway) and cryopreserved in liquid nitrogen until use. Collection of blood was approved by the University of Otago Ethics Committee (Health) (H14/046).
Quantitative real‐time polymerase chain reaction (qPCR)
DNA was extracted from sputa and PBMCs using the NucleoSpin Tissue DNA Kit (Macherey‐Nagel, Düren, Germany). Prior to DNA extraction, samples were defrosted and pretreated with dithiothreitol (0·1 mg/ml) at 37°C for 20 min or until fully digested; samples were considered fully digested when upon inspection they appeared to be a liquid and could be pipetted. The primers and probes used for quantification of MAIT cells (Vα7.2‐Jα12/20/33), Vδ2+ γδ T cells (Vδ2‐Jδ1/2/3/4), β2‐microglobulin (β2M), and bacteria (PAN23S rDNA) (all from Integrated DNA Technologies, Coralville, IA, USA) are shown in the Supporting information, Table S1. qPCR was performed using the KAPA Probe Fast qPCR kit (KAPA Biosystems, Wilmington, MA, USA) and an ABI Prism ViiA7 (Applied Biosystems, Foster City, CA, USA): 95°C for 3 min, then 40 cycles of 95°C for 3 s and 60°C for 20 s. Human DNA quantities were determined by amplifying the β2M gene 22. The amount of Vα7.2‐Jα12/20/33 and Vδ2‐Jδ1/2/3/4 relative to β2M was determined by the comparative threshold cycle (CT) method (2ΔΔCT). LinRegPCR was then used to calculate absolute β2M copy number 23, and this was multiplied by the relative amount of Vα7.2‐Jα12/20/33 or Vδ2‐Jδ1/2/3/4 to calculate the absolute abundance of MAIT or Vδ2+ γδ T cells (presented in arbitrary units). When no DNA was detected, a value less than the lowest amount detected was assigned (1 × 10–10 arbitrary units). To determine the bacterial load, 23S rDNA was detected using the KAPA SYBR FAST Universal kit (KAPA Biosystems): at 95°C for 3 min, then 40 cycles of 95°C for 3 s and 58°C for 30 s. A standard curve with Staphylococcus aureus DNA was used for quantification.
Flow cytometry and cell separation
PBMCs were stained with the following antibodies: CD3‐phycoerythrin (PE)/cyanin 7 (Cy7), Vδ2‐fluorescein isothiocyanate (FITC), Vα7.2‐PE (BioLegend, San Diego, CA, USA), CD8‐eFluor450 (eBioscience, San Diego, CA, USA) and CD161‐allophycocyanin (APC) (Miltenyi Biotech, Bergisch Gladbach, Germany). Live/Dead Fixable Near IR dye (Life Technologies, Carlsbad, CA, USA), and 123count eBeads (eBioscience) were included with each sample. Data was acquired on a FACSCanto II (BD Biosciences, San Jose, CA, USA) and analysed using FlowJo version 10 software (TreeStar, Inc., San Carlos, CA, USA). The gating strategy is shown in Supporting information, Fig. S1. For comparisons of qPCR and flow cytometry, DNA was extracted from the same cryovial of PBMCs as analysed by flow cytometry. In some experiments MAIT cells and Vδ2+ T cells were sorted for DNA extraction using the BD FACSAria I (BD Biosciences). Column‐based depletion of PBMCs labelled with Vα7.2‐PE or Vδ2‐PE (both BioLegend) was performed with anti‐PE microbeads and MS columns (both Miltenyi Biotech).
Analysis of cytokine and chemokine production
Cytokine and chemokine levels in sputum samples were measured using the LEGENDplex 13‐plex Human Inflammation Panel kit, measuring chemokine ligand (CCL)2 [Monocyte chemoattractant protein 1 (MCP‐1)], interferon‐α (IFN‐α), IFN‐γ, IL‐1β, IL‐6, chemokine (C‐X‐C motif) ligand 8 (CXCL8; IL‐8), IL‐10, IL‐12 (p70), IL‐17A, IL‐18, IL‐23, IL‐33, and tumour necrosis factor (TNF)‐α, or the LEGENDplex 13‐plex Human Proinflammatory Chemokine Panel Kit (BioLegend), measuring CCL2 (MCP‐1), CCL3 (macrophage inflammatory protein; MIP‐1α), CCL4 (MIP‐1β), CCL5 (regulated on activation, normal T cell expressed and secreted; RANTES), CCL11 (exotoxin), CCL17 (thymus and activation regulated chemokine; TARC), CCL20 (MIP‐3α), CXCL1 (growth‐regulated oncogene α; GROα), CXCL5 (epithelial neutrophil‐activating protein 78; ENA‐78), CXCL8 (IL‐8), CXCL9 (monokine induced by gamma; MIG), CXCL10 (IP‐10), and CXCL11 (IFN‐inducible T cell alpha chemoattractant; I‐TAC). Samples were acquired on a BD FACSCanto II, and analysed using LEGENDplex software (BioLegend).
Metagenomics
16S rRNA sequencing was performed on sputum‐extracted DNA at the Environmental Sample Preparation and Sequencing Laboratory at Argonne National Laboratories (Chicago, IL, USA), as described in the Supporting information. Briefly, libraries of the V4 hypervariable region were prepared using primers 515F and 806R 24 and samples were sequenced on an Illumina MiSeq using 2 × 250 base pairs (bp) read chemistry. The average number of reads per sample was 19 754 (range = 1485–63 876). Quality control, assembly and operational taxonomic units (OTU) assignment of sequences was performed using Mothur 25. Taxonomy was assigned using the GreenGenes database version 13.5.99 26. Picrust was used to infer metagenomic capacity of 16S rRNA data 27. Bacterial genes encoding for riboflavin and C5 isoprenoid biosynthesis were curated from the KEGG database (Supporting information, Table S2).
Statistics
Data were analysed in Prism version 7 (GraphPad Software, San Diego, CA, USA). Medians, interquartile range and all data points are shown. Comparisons between two groups were made with the Mann–Whitney U‐test. Comparisons between more than two groups were made with the Kruskal–Wallis test with Dunn’s multiple comparisons test. For continuous data, Spearman’s or Pearson’s correlations were calculated as indicated. Significance was defined as two‐sided P < 0·05.
Results
In this study, we sought to enumerate MAIT and Vδ2+ T cells at the site of infection in patients with CAP. Because flow cytometric analysis was not possible, as samples had been frozen without a cryoprotectant, we used qPCR on DNA extracted from sputum samples to quantify innate‐like lymphocyte (ILL)‐specific VDJ TCR recombinations 22. During VDJ recombination, variable and junctional segments are brought together in the genome to form a single exon. Here, we used a second generation qPCR assay capable of identifying the predominant MAIT cell TCRs (Vα7.2‐Jα33/12/20) and a new assay to detect Vδ2+ TCRs (Vδ2‐Jδ1/2/3/4) 28, 29, 30.
To confirm the specificity of the primers and probes, qPCR was performed on DNA isolated from FACS‐sorted PBMC populations. Significantly more Vα7.2‐Jα12/20/33 and Vδ2‐Jδ1/2/3/4 were detected in sorted MAIT cell (CD3+CD161++Vα7.2+ lymphocytes) and Vδ2+ T cell (CD3+Vδ2+ lymphocytes) populations, respectively, than in depleted populations (Fig. 1a,b). PCR was also performed on PBMCs with or without column‐based depletion of Vα7.2+ cells or Vδ2+ cells. With depletion, the magnitude of reduction of Vα7.2‐Jα12/20/33 (qPCR) and CD3+CD161++Vα7.2+ lymphocytes (flow cytometry), and Vδ2‐Jδ1/2/3/4 (qPCR) and CD3+Vδ2+ lymphocytes (flow cytometry) were similar (Supporting information, Fig. S2). To determine the quantitative accuracy of the assays, we compared the absolute cell abundance of MAIT cells and Vδ2+ T cells in PBMCs as determined by flow cytometry and qPCR. Strong linear correlations for the numbers of MAIT and Vδ2+ T cells determined by qPCR and flow cytometry were observed (Fig. 1c,d). The limit of detection of both assays was ≤ 10 cells. Taken together, these results validate the qPCR method for measuring absolute cell abundance.
Figure 1.
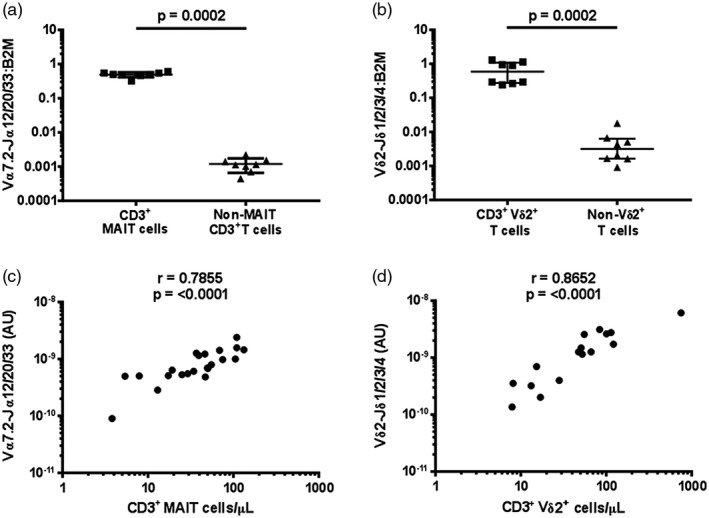
Validation of the quantitative polymerase chain reaction (qPCR) method to determine mucosal‐associated invariant T (MAIT) cell and Vδ2+ T cell abundance. The specificity of the qPCR assay was determined by measuring the abundance of (a) Vα7.2‐Jα12/20/33 gDNA in MAIT cells (live/CD3+/CD161++/Vα7.2+) and non‐MAIT CD3+ T cells (live/CD3+/CD161–/Vα7.2–) (n = 5) and (b) Vδ2‐Jδ1/2/3/4 gDNA in Vδ2+ cells (live/CD3+/Vδ2+) and Vδ2– CD3+ T cells (live/CD3+/Vδ2–) (n = 8), each sorted from peripheral blood mononuclear cells (PBMCs) and abundance expressed relative to β2 microglobulin (β2M) in arbitrary units. Correlation between the abundance of (c) MAIT (n = 22) and (d) Vδ2+ T cells (n = 16) in whole PBMC samples measured by qPCR and by flow cytometry; the absolute copy number of β2M was calculated with LinRegPCR 23 and the number of MAIT cells (Vα7.2‐Jα12/20/33 gDNA) and Vδ2+ T cells (Vδ2‐Jδ1/2/3/4 gDNA) determined relative to β2M and expressed in arbitrary units. In (a) and (b), individual data points representing the mean of technical triplicates of the qPCR assay, the median and interquartile ranges are shown; differences between groups were analysed by Mann–Whitney tests; (c) and (d) were analysed using Spearman correlations. **P < 0·01, ***P < 0·001 (Mann–Whitney test).
Sputum samples were available from 88 patients with CAP. Of these, 36 samples were excluded due to microscopic evidence of significant oropharyngeal contamination (≥ 10 squamous epithelial cells per ×100 field). Of the 52 samples included in the study, 43 (83%) were spontaneously produced and nine were (17%) induced sputa. The median patient age was 67·5 years (range = 36–101) and 27 (52%) were female. Eighteen (35%) patients had pre‐existing chronic obstructive pulmonary disease (COPD) or other structural lung disease and 11 (21%) had asthma. On chest X‐ray, 31 (60%) had lobar consolidation while 21 (40%) had multi‐lobar consolidation. The quality of the DNA obtained from the sputum samples was satisfactory: the median absorbance 260 : 280 ratio was 1·85 (IQR = 1·833–1·88) and the median Ct value for B2M was 23·6 (IQR = 22·8–25·5).
The abundance of MAIT cells, Vδ2+ T cells, and total ILLs, defined as the sum of MAIT and Vδ2+ T cell abundance in sputum, were significantly higher in patients with CAP than in healthy controls (Fig. 2). In patients with CAP, MAIT and Vδ2+ T cell abundance in sputum were significantly correlated (Supporting information, Fig. S3a). There was no evidence of a correlation between MAIT cell, Vδ2+ T cell, or ILL abundance in sputum and patient age (Supporting information, Fig. S3b,d). MAIT cell, Vδ2+ T cell, and ILL abundance in sputum did not significantly differ by specimen type, gender, pre‐existing lung disease, duration of illness prior to presentation, failure of out‐patient antibiotic therapy, or type of consolidation on chest X‐ray (Supporting information, Table S3).
Figure 2.
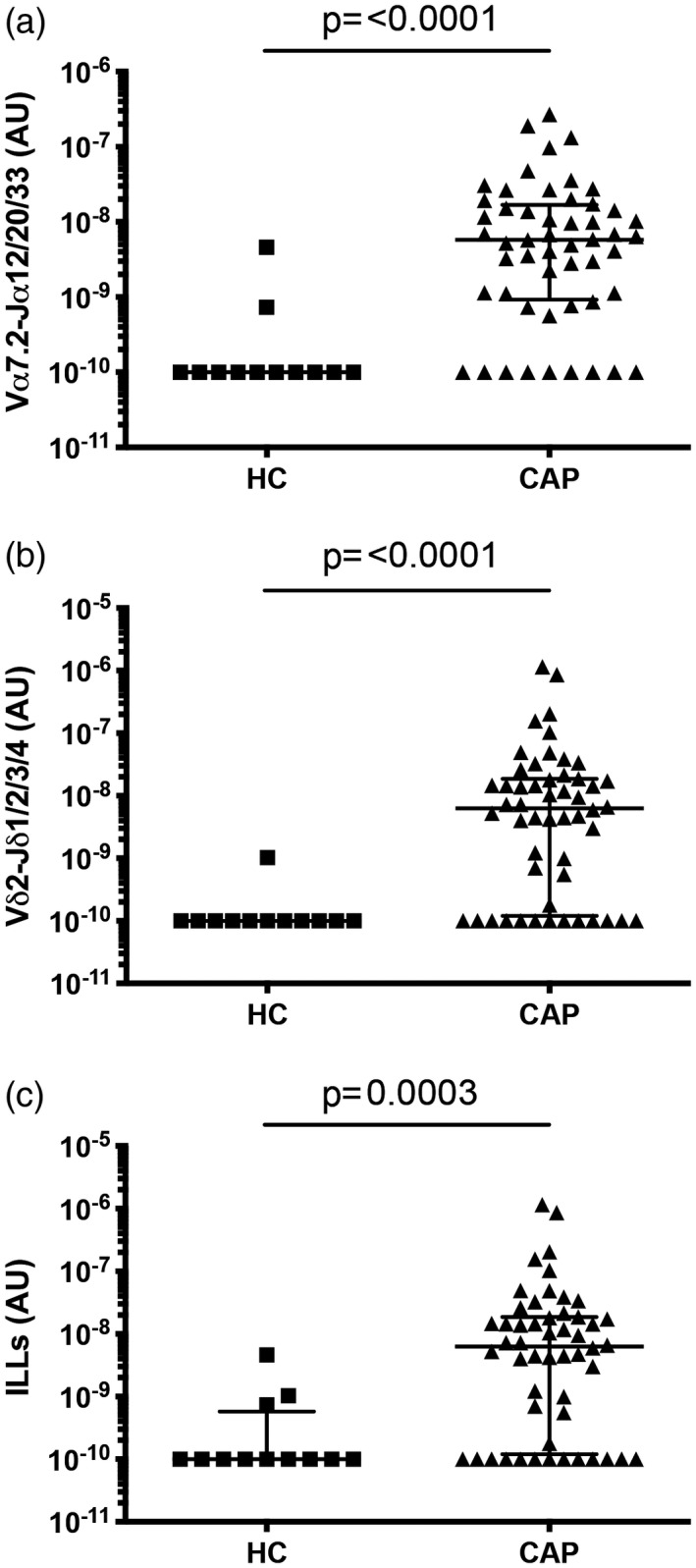
Mucosal‐associated invariant T (MAIT) cell and Vδ2+ T cell abundance in sputum is significantly higher in patients with community‐acquired pneumonia (CAP) than in healthy controls (HC). The abundance of (a) MAIT cells (Vα7.2‐Jα12/20/33 gDNA), (b) Vδ2+ T cells (Vδ2‐Jδ1/2/3/4 gDNA) and (c) innate‐like lymphocytes (ILLs) in sputum from healthy controls and patients with CAP. MAIT cell and Vδ2+ T cell abundance were measured by quantitative polymerase chain reaction (qPCR); the absolute copy number of β2 microglobulin (β2M) was calculated with LinRegPCR 23 and the number of MAIT cells (Vα7.2‐Jα12/20/33 gDNA) and Vδ2+ T cells (Vδ2‐Jδ1/2/3/4 gDNA) determined relative to β2M and expressed in arbitrary units. Individual data points (n = 12 for healthy controls, n = 52 for patients with CAP) representing the mean of technical triplicates of the qPCR assay, the median and interquartile ranges are shown. Groups were compared by the Mann–Whitney test.
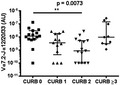
While there was no evidence of a difference in MAIT cell, Vδ2+ T cell, or total ILL abundance in patients requiring intensive care unit admission (n = 6) (Supporting information, Fig. S4), the abundance of MAIT cells and ILLs differed significantly, with pneumonia severity as measured by the CURB65 score (Fig. 3). CURB65 is a five‐point clinical scoring system that allows the stratification of patients into mortality risk groups 31. Patients with a CURB65 score of 0 had significantly more MAIT cells and total ILLs than those with a CURB65 score of 2; other comparisons were not significant (Fig. 3a, Supporting information, Fig. S5a). The Infectious Diseases Society of America/American Thoracic Society (IDSA/ATS) pneumonia severity criteria comprises an alternative clinical scoring system which is designed to identify patients who require management in an intensive care or high‐dependency unit 32. Patients with severe pneumonia by the IDSA/ATS criteria (n = 4) had a lower MAIT cell abundance (Supporting information, Fig. S3c), but total ILL abundance was not significantly different (Supporting information, Fig. S5b). There was no evidence of correlation between Vδ2+ T cell abundance and pneumonia severity, as calculated by the CURB65 or IDSA/ATS criteria (Fig. 3b,d). Duration of stay in hospital was negatively correlated with ILL, MAIT cell, and Vδ2+ T cell abundance (Fig. 4).
Figure 3.
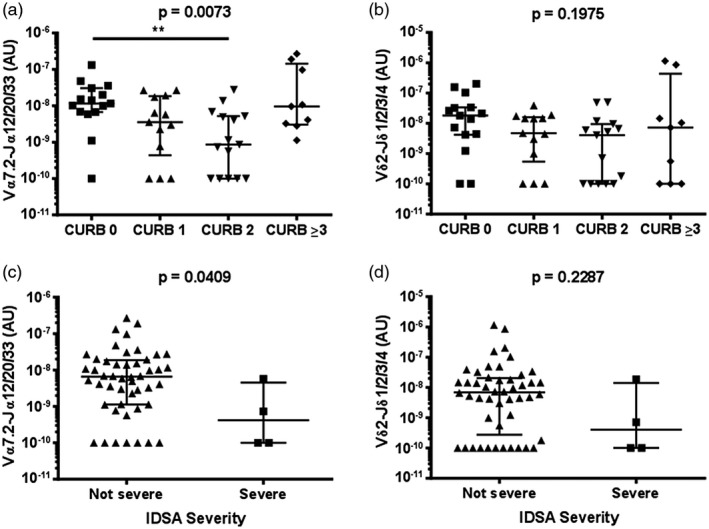
Reduced disease severity in patients with higher mucosal‐associated invariant T (MAIT) cell abundance in sputum. The abundance of (a) MAIT cells (Vα7.2‐Jα12/20/33 gDNA) and (b) Vδ2+ T cells (Vδ2‐Jδ1/2/3/4 gDNA) in sputum of patients by CURB‐65 score. Abundance of (c) MAIT cells and (d) Vδ2+ T cells in sputum of patients by the IDSA severity criteria. MAIT cell and Vδ2+ T cell abundance were measured by quantitative polymerase chain reaction (qPCR); the absolute copy number of β2 microglobulin (β2M) was calculated with LinRegPCR 23 and the number of MAIT cells (Vα7.2‐Jα12/20/33 gDNA) and Vδ2+ T cells (Vδ2‐Jδ1/2/3/4 gDNA) determined relative to β2M and expressed in arbitrary units. Individual data points (n = 52) representing the mean of technical triplicates of the qPCR assay, the median and interquartile ranges are shown. CURB‐65 data were analysed using Kruskal–Wallis one‐way analysis of variance (ANOVA) and Dunn’s multiple comparison test; IDSA severity data were analysed using the Mann–Whitney test. **P < 0·01.
Figure 4.
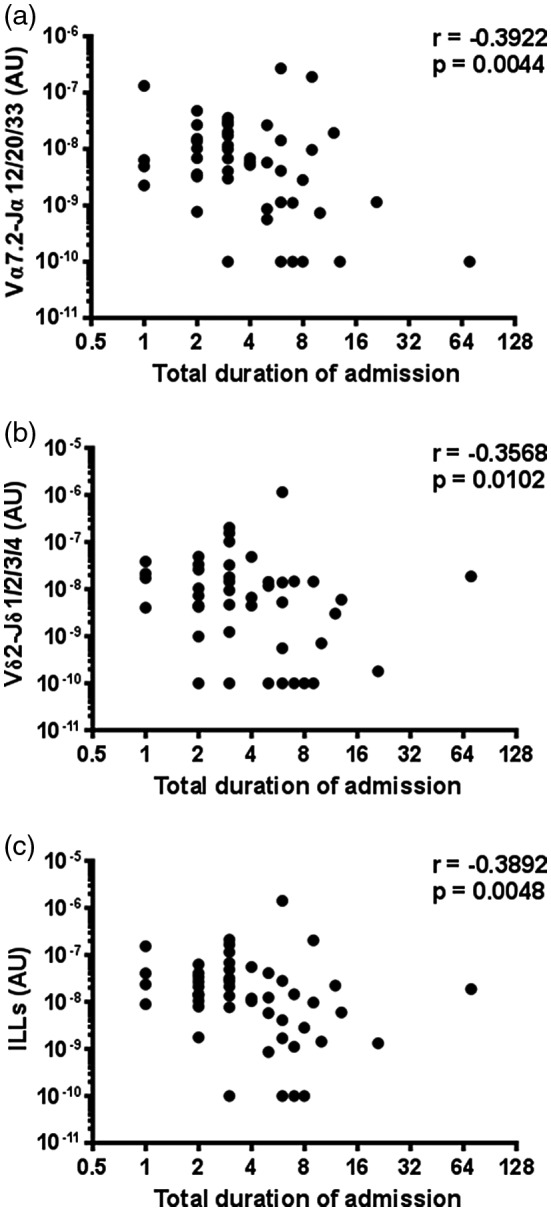
Duration of hospital admission is inversely correlated with the abundance of innate‐like lymphocytes (ILLs) in sputum. The abundance of (a) mucosal‐associated invariant T (MAIT) cells (Vα7.2‐Jα12/20/33 gDNA), (b) Vδ2+ T cells (Vδ2‐Jδ1/2/3/4 gDNA), and (c) ILLs were measured by quantitative polymerase chain reaction (qPCR) and the association with duration of admission to hospital assessed with Spearman correlations. The absolute copy number of β2 microglobulin (β2M) was calculated with LinRegPCR 23 and the number of MAIT cells (Vα7.2‐Jα12/20/33 gDNA) and Vδ2+ T cells (Vδ2‐Jδ1/2/3/4 gDNA) determined relative to β2M and expressed in arbitrary units. Individual data points (n = 52) representing the mean of technical triplicates of the qPCR assay are shown.
MAIT cells, but not Vδ2+ T cells, were more abundant in sputum with more neutrophils, as determined by microscopy performed by the diagnostic laboratory prior to freezing (Fig. 5). There was no evidence of a correlation between MAIT cell or Vδ2+ T cell abundance in sputum and the blood leucocyte count or C‐reactive protein (CRP) levels (Supporting information, Fig. S6).
Figure 5.
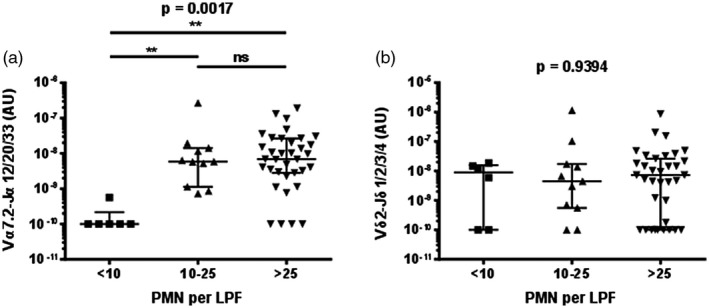
Mucosal‐associated invariant T (MAIT) cell abundance in sputum is associated with the number of neutrophils in sputum. The abundance of (a) MAIT cells (Vα7.2‐Jα12/20/33 gDNA) and (b) Vδ2+ T cells (Vδ2‐Jδ1/2/3/4 gDNA) by the number of neutrophils (PMN) per low‐powered field (LPF) in sputum. MAIT cell and Vδ2+ T cell abundance was determined by quantitative polymerase chain reaction (qPCR); the absolute copy number of β2 microglobulin (β2M) was calculated with LinRegPCR 23 and the number of MAIT cells (Vα7.2‐Jα12/20/33 gDNA) and Vδ2+ T cells (Vδ2‐Jδ1/2/3/4 gDNA) determined relative to β2M and expressed in arbitrary units. Microscopy was performed on sputum in the diagnostic laboratory prior to freezing. Individual data points (n = 52) representing the mean of technical triplicates of the qPCR assay, the median and interquartile ranges are shown. Data were analysed with the Kruskal–Wallis one‐way analysis of variance (ANOVA); n.s. = not significant.
A bacterial aetiology of infection was identified in 23 cases by routine diagnostic testing (Supporting information, Table S4). MAIT cell, Vδ2+ T cell, and overall ILL abundance in sputum were all higher in patients with an identified bacterial pathogen (Fig. 6a,b, Supporting information, Fig. S7a). Of note, no MAIT or Vδ2+ T cells were detected in several samples in which no bacterial pathogen was detected (Fig. 6a,b). Vδ2+ T cell, but not MAIT cell, abundance was higher in patients with pneumonia caused by either S. pneumoniae or Legionella spp. (Supporting information, Fig. S7b–e). In contrast, there was no indication of a correlation between either subset with total bacterial load (Fig. 6c,d).
Figure 6.
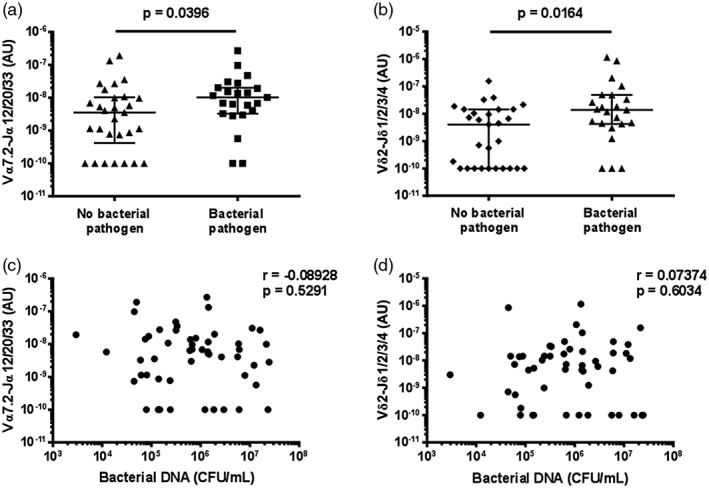
Mucosal‐associated invariant T (MAIT) cell and Vδ2+ T cell abundance in sputum is higher in patients with an identified bacterial pathogen. The abundance of (a) MAIT cells (Vα7.2‐Jα12/20/33 gDNA) and (b) Vδ2+ T cells (Vδ2‐Jδ1/2/3/4 gDNA) in sputum of patients with and without an identified bacterial pathogen. The association between (c) MAIT cell and (d) Vδ2+ T cell abundance in sputum and total bacterial load. MAIT cell and Vδ2+ T cell abundance and total bacterial load were measured by quantitative polymerase chain reaction (qPCR); the absolute copy number of β2 microglobulin (β2M) was calculated with LinRegPCR 23 and the number of MAIT cells (Vα7.2‐Jα12/20/33 gDNA) and Vδ2+ T cells (Vδ2‐Jδ1/2/3/4 gDNA) determined relative to β2M and expressed in arbitrary units. Bacterial pathogens were identified in the diagnostic laboratory by sputum culture, blood culture, PCR on sputum for Legionella spp., or by urinary antigen detection for Streptococcus pneumoniae or L. pneumophila serotype 1 (Supporting information, Table S4). Individual data points (n = 52) representing the mean of technical triplicates of the qPCR assay are shown; in (a) and (b) the median and interquartile ranges are shown. Differences between groups were analysed by Mann–Whitney tests (a,b). The association with total bacterial load was assessed with Spearman correlations (c,d). CFU/ml = colony‐forming units per millilitre.
As pneumonia can be a polymicrobial infection caused by components of the oral flora, we conducted 16S rRNA sequencing of samples to identify bacterial OTUs present in the sputum samples and predict the ability of those OTUs to make the metabolic ligands for ILLs. This revealed that there was no significant correlation in the inferred metagenomic data between the abundance of MAIT cells and the abundance of genes encoding for riboflavin synthesis or between the abundance of Vδ2+ T cells and the abundance of genes encoding for C5 isoprene synthesis (Supporting information, Table S5). Culture of pathogens from sputum samples correlated with the abundance of genera containing pathogens in the 16S rRNA data (Supporting information, Fig. S8).
Finally, where there was sufficient sputum sample available, we analysed cytokine (n = 40) and chemokine (n = 30) levels. The amount of IFN‐γ in sputum was significantly correlated with MAIT cell, Vδ2+ T cell, and overall ILL abundance (Table 1, Supporting information, Fig. S9a–c). The amount of IFN‐α in sputum was significantly correlated with MAIT cell abundance alone (Table 1, Supporting information, Fig. S9d–f). The amount of CXCL11 in sputum was significantly correlated with Vδ2+ T cell and overall ILL abundance, but not with MAIT cell abundance (Table 1, Supporting information, Fig. S9g–i). No other cytokines or chemokines analysed displayed evidence of a significant correlation with MAIT cell, Vδ2+ T cell or overall ILL abundance (Table 1).
Table 1.
Adssociation of ILL abundance in sputum with sputum concentrations of cytokines and chemokines
| Analyte | ILL | MAIT cells | Vδ2+ T cells | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Spearman’s r | 95% CI | P (two‐tailed) | Spearman’s r | 95% CI | P (two‐tailed) | Spearman’s r | 95% CI | P (two‐tailed) | |
| CCL2* | 0·026 | –0·347 to 0·392 | 0·891 | –0·031 | –0·397 to 0·343 | 0·869 | 0·098 | –0·283 to 0·451 | 0·608 |
| CCL2† | 0·167 | –0·162 to 0·463 | 0·303 | 0·171 | –0·158 to 0·465 | 0·293 | 0·201 | –0·128 to 0·489 | 0·215 |
| CCL3* | 0·030 | –0·344 to 0·396 | 0·875 | 0·106 | –0·275 to 0·458 | 0·577 | –0·029 | –0·395 to 0·345 | 0·879 |
| CCL4* | –0·038 | –0·402 to 0·337 | 0·842 | 0·071 | –0·307 to 0·430 | 0·710 | –0·101 | –0·454 to 0·280 | 0·596 |
| CCL5* | 0·220 | –0·164 to 0·545 | 0·243 | 0·251 | –0·131 to 0·568 | 0·181 | 0·088 | –0·292 to 0·444 | 0·644 |
| CCL11* | 0·027 | –0·346 to 0·393 | 0·888 | 0·120 | –0·261 to 0·469 | 0·527 | –0·116 | –0·466 to 0·265 | 0·540 |
| CCL17* | –0·192 | –0·525 to 0·191 | 0·309 | –0·243 | –0·562 to 0·140 | 0·196 | –0·248 | –0·566 to 0·134 | 0·186 |
| CCL20* | 0·153 | –0·230 to 0·495 | 0·419 | 0·168 | –0·216 to 0·506 | 0·376 | 0·127 | –0·255 to 0·474 | 0·505 |
| CXCL1* | 0·018 | –0·355 to 0·385 | 0·927 | 0·077 | –0·302 to 0·435 | 0·686 | 0·006 | –0·365 to 0·375 | 0·975 |
| CXCL5* | –0·169 | –0·507 to 0·214 | 0·372 | –0·197 | –0·528 to 0·187 | 0·297 | –0·118 | –0·468 to 0·263 | 0·534 |
| CXCL8* | –0·053 | –0·415 to 0·323 | 0·780 | 0·031 | –0·343 to 0·396 | 0·872 | –0·016 | –0·383 to 0·356 | 0·935 |
| CXCL8† | –0·040 | –0·356 to 0·284 | 0·806 | 0·073 | –0·253 to 0·384 | 0·653 | 0·006 | –0·314 to 0·326 | 0·969 |
| CXCL9* | 0·051 | –0·325 to 0·413 | 0·790 | –0·074 | –0·432 to 0·304 | 0·697 | 0·095 | –0·285 to 0·449 | 0·618 |
| CXCL10* | 0·154 | –0·229 to 0·496 | 0·417 | –0·112 | –0·463 to 0·269 | 0·554 | 0·240 | –0·143 to 0·560 | 0·202 |
| CXCL11* | 0·376 | 0·007 to 0·655 | 0·041 | 0·165 | –0·218 to 0·504 | 0·383 | 0·392 | 0·025 to 0·665 | 0·032 |
| IFNα† | 0·311 | –0·010 to 0·574 | 0·051 | 0·328 | 0·009 to 0·587 | 0·039 | 0·230 | –0·097 to 0·513 | 0·153 |
| IFNγ† | 0·488 | 0·200 to 0·699 | 0·001 | 0·421 | 0·117 to 0·653 | 0·007 | 0·487 | 0·198 to 0·698 | 0·001 |
| IL1β† | 0·200 | –0·129 to 0·489 | 0·217 | 0·296 | –0·026 to 0·563 | 0·063 | 0·083 | –0·243 to 0·393 | 0·609 |
| IL6† | 0·123 | –0·205 to 0·427 | 0·449 | 0·086 | –0·241 to 0·395 | 0·599 | 0·183 | –0·146 to 0·475 | 0·258 |
| IL10† | 0·067 | –0·259 to 0·379 | 0·684 | 0·176 | –0·153 to 0·469 | 0·278 | 0·079 | –0·248 to 0·389 | 0·630 |
| IL12p70† | 0·171 | –0·158 to 0·466 | 0·291 | 0·244 | –0·083 to 0·523 | 0·130 | 0·062 | –0·264 to 0·374 | 0·705 |
| IL17A† | 0·251 | –0·076 to 0·528 | 0·119 | 0·256 | –0·070 to 0·532 | 0·111 | 0·239 | –0·088 to 0·519 | 0·138 |
| IL18† | 0·214 | –0·114 to 0·500 | 0·186 | 0·197 | –0·132 to 0·486 | 0·224 | 0·241 | –0·086 to 0·521 | 0·134 |
| IL23† | –0·012 | –0·331 to 0·310 | 0·943 | 0·103 | –0·224 to 0·410 | 0·526 | –0·042 | –0·357 to 0·282 | 0·796 |
| IL33† | 0·200 | –0·128 to 0·490 | 0·216 | 0·164 | –0·164 to 0·460 | 0·311 | 0·234 | –0·093 to 0·515 | 0·146 |
| TNFα† | 0·282 | –0·042 to 0·552 | 0·078 | 0·287 | –0·036 to 0·556 | 0·072 | 0·249 | –0·077 to 0·527 | 0·121 |
IFN = interferon; IL = interleukin; TNF = tumour necrosis factor; MAIT = mucosal‐associated invariant T; ILL = innate‐like lymphocytes; CI = confidence interval. Significant P‐values are shown in bold type.
Measured with LEGENDplex 13‐plex Human Proinflammatory Chemokine Panel kit (BioLegend) (n = 30).
measured with LEGENDplex 13‐plex Human Inflammation Panel kit (BioLegend) (n = 40).
Discussion
In this study, we quantified MAIT cells and Vδ2+ T cells at the site of infection in patients with CAP and found their abundance to be inversely associated with clinical markers of severity. Higher numbers of MAIT cells and Vδ2+ T cells were found in sputum of CAP patients with a confirmed bacterial aetiology, but their abundance was not related to bacterial load or the predicted capacity of bacteria to produce the MAIT cell or Vδ2+ T cell activating ligands. Instead, Vδ2+ T cell abundance correlated with the concentration of CXCL11, MAIT cell abundance with the concentration of IFN‐α, and both cell populations with the amount of IFN‐γ in sputum. Overall, this suggests an important role for MAIT cells and Vδ2+ T cells in the immune response to CAP.
As the frozen sputum specimens were not suitable for flow cytometry, qPCR was used to quantify MAIT and Vδ2+ γδ T cell DNA. By using multiple junctional primers, this method can detect the most common recombined MAIT cell TCRs and all possible Vδ2+ γδ T cell TCRs 28, 29, 30. We elected to perform the analysis on DNA rather than RNA because DNA is more stable than RNA, an important consideration given that the samples had been frozen for 3 years, and assessment at the RNA level could be influenced by differences in TCR expression (down‐regulation of TCR expression upon activation could compromise the assessment of absolute cell numbers). Importantly, the abundance of MAIT and Vδ2+ cells determined by qPCR and flow cytometry were strongly correlated. This method could be used to complement flow cytometry and enable quantification of ILLs in a range of tissues and body fluids.
The abundance of MAIT and Vδ2+ T cells in sputum of patients with CAP was highly correlated. This suggests that common signals lead to the recruitment and/or proliferation of these ILLs. Indeed, both cell populations express chemokine receptors associated with trafficking to the lungs and sites of inflammation 3, 17, 33, and have the ability to proliferate 17, 34.
While the levels of ILLs were higher in those with an identified bacterial pathogen, there was no evidence of association with total bacterial load. An association between MAIT cell recruitment to the lungs and bacterial inoculum has recently been reported in a mouse model of L. longbeachae pneumonia 10. The lack of correlation between total bacterial load and MAIT cell, Vδ2+ T cell, or ILL abundance in our study may reflect heterogeneity in the duration of infection, prior antibiotic treatment, sample contamination with oropharyngeal microflora, and non‐bacterial causes of pneumonia. Indeed, the most commonly detected pathogens in pneumonia are viruses 35, 36, 37, 38. As this study was conducted outside the influenza season 39, patients were not tested for respiratory viruses; however, CAP resulting from other respiratory viruses, such as human rhinovirus, cannot be excluded 35. Identification of the causative agent in CAP is notoriously difficult; in a recent pneumonia aetiology study, extensive testing identified a pathogen in only 38% of patients 38.
Interestingly, not all bacterial pathogens identified are able to produce the ligands for MAIT and Vδ2+ T cells. Vδ2+ T cell abundance was higher in those with S. pneumoniae or Legionella infection, yet these species do not produce HMB‐PP 40. Further, neither MAIT cell nor Vδ2+ T cell abundance correlated with the quantity of bacteria in sputum with the capacity to produce riboflavin or HMB‐PP, respectively (as inferred from metagenomic analysis), although this may not reflect metabolic activity in vivo, which would be better assessed by RNA sequencing. Therefore, the abundance of TCR ligands is unlikely to be the sole determinant of MAIT and Vδ2+ T cell recruitment and/or proliferation, although their concentrations have not been measured. The abundance of Vδ2+ T cells in sputum correlated with the concentration of CXCL11 (I‐TAC), which is produced by monocytes, endothelial cells, and fibroblasts in response to IFN‐γ and IFN‐β and binds to CXCR3 41. Of note, we have recently shown that activated MAIT cells can also produce CXCL11 42. CXCR3 is expressed by Vδ2+ T cells and their chemotaxis in response to CXCR3 ligands has previously been reported 43. While the correlation of Vδ2+ T cell abundance with CXCL11 concentration may be confounded by the concentration of IFN‐γ, the lack of correlation of MAIT cell abundance with CXCL11 concentration argues against this. While MAIT cells express CXCR3, they also express high levels of CD26, which has been shown to inactivate CXCL11 and prevent T cell chemotaxis 44, 45. No chemokines were identified that correlated with MAIT cell abundance. However, IFN‐α concentration correlated with MAIT cell abundance but not Vδ2+ T cell abundance. IFN‐α2 has been shown to be a chemotactic factor for T cells 46. Alternatively, IFN‐α may result in activation and proliferation of MAIT cells in the lung 47, 48. In a mouse model of influenza infection, accumulation of MAIT cells in the lungs was dependent upon IL‐18 11. However, in our study, MAIT cell abundance was not associated with IL‐18 concentration.
MAIT cell abundance was positively correlated with neutrophil infiltration. Neutrophils are essential in the control of multiple bacterial pulmonary infections, and have a major role in the initiation of the adaptive immune response 49. MAIT cells produce IL‐17A, inducing production of CXCL‐8, resulting in neutrophil recruitment 50, 51. Although IL‐17A was detected in the sputum samples, it was detected at low concentration (median = 13·3 ng/μL, IQR = 7·2–30·6 ng/μl) and did not significantly correlate with MAIT cell abundance. Similarly, while γδ T cells have been found to produce IL‐17 in response to lung infection 52, we did not detect a significant correlation between the abundance of Vδ2+ T cells and IL‐17A production or neutrophil abundance. This suggests that other cell types may contribute to the production of IL‐17A and CXCL‐8, and hence to neutrophil recruitment. Of note, activated MAIT cells can produce CXCL‐8, and have been shown to induce neutrophil migration in a transwell assay, albeit in a IL‐8‐independent manner 42. The correlation between MAIT cell and neutrophil abundance may reflect co‐recruitment in response to inflammation and/or recruitment of neutrophils by activated MAIT cells.
We found that the levels of IFN‐γ in sputum were positively correlated with MAIT cell, Vδ2+ T cell, and overall ILL abundance in sputum, suggesting that these cells are a major source of IFN‐γ in CAP. In a previous study, IFN‐γ could be detected in the bronchoalveolar fluid at higher concentrations in patients with CAP than in healthy controls, although concentration did not correlate with severity 53. TNF‐α could only be detected in the bronchoalveolar fluid of some patients, while IL‐17A was not detected at all 53. IFN‐γ has an important role in the response to various pulmonary bacterial infections 54, 55. It has recently been reported that immune defence in pulmonary infection with L. longbeachae in Rag2−/−γC−/− mice is reliant upon IFN‐γ production by adoptively transferred MAIT cells; a significant reduction in survival was shown in mice where adoptively transferred MAIT cells were from IFN‐γ–/– mice 10. However, mortality was unchanged when MAIT cells from TNF‐/‐, IL‐17–/–, perforin–/– or granzyme A and B–/– mice were transferred 10. Production of IFN‐γ, TNF‐α, and IL‐17 by MAIT and γδ T cells has previously been reported in mouse lung infection models 56, 57.
The abundance of MAIT cells and Vδ2+ cells in sputum was inversely associated with the severity of infection. Higher numbers of MAIT cells were found in the sputum of patients with low‐severity pneumonia (CURB65 score and IDSA/ATS criteria). The median abundance of MAIT cells in sputum differed significantly by CURB65 score. Comparing the abundance of MAIT cells between patients with different CURB65 scores, MAIT cell abundance was significantly lower in patients with a CURB65 score of 2 than in patients with a score of 0, but other comparisons, including the difference between patients with a CURB65 score of ≥ 3 and 0 and ≥ 3 and 2 were not significant. This may be due to the low numbers of patients with severe pneumonia in this cohort, greater recruitment of MAIT cells to the lungs in patients with the most severe infections or confounding factors. Indeed, several parameters that were associated with greater MAIT cell abundance were also more common in patients with a CURB65 score of ≥ 3 (higher type I IFN levels, the presence of a bacterial pathogen, neutrophil abundance) and may be confounding. Therefore, it remains an open question which would require more and new data to address. While there was a significant difference in MAIT cell abundance between those with non‐severe and severe pneumonia, as classified by the IDSA/ATS criteria, we take this result as indicative due to the small sample size (n = 4) of one of the groups. No difference was seen in admission to the intensive care unit (n = 6), which may reflect low numbers of severe cases or other reasons for subsequent intensive care unit admission. Both MAIT cell and Vδ2+ T cell numbers in sputum were negatively correlated with the duration of hospital admission. Duration of admission may reflect severity of infection, but could be confounded by comorbidities, such as COPD and age 58. MAIT cells numbers in the blood decrease with age 59 and are also depleted from the blood and lungs of patients with COPD 60. However, neither MAIT cell nor Vδ2+ T cell abundance in sputum was correlated with age and there was no difference between those with or without COPD (data not shown).
Overall, our findings suggest that ILLs, particularly MAIT cells, may play a protective role against severe infection. This is consistent with recent data from a mouse model, where MAIT cells were shown to have a role in protecting against pulmonary infection with L. longbeachae; bacterial clearance was diminished in MAIT cell‐deficient mice and enhanced in mice with an expanded MAIT cell population 10. In humans, MAIT cell numbers were found to be reduced in the blood of severely unwell patients admitted to intensive care, especially those with a bacterial infection, and there was a non‐significant trend towards MAIT cell recovery protecting against subsequent nosocomial infection 13. In contrast, it has been suggested that MAIT and Vδ2+ T cells contribute to a poorer clinical outcome in patients with a first episode of continuous ambulatory peritoneal dialysis (CAPD) peritonitis 61. Of note, other bacterial factors, such as endotoxins, virulence factors, ability to form biofilms, and resistance to anti‐microbials may influence inflammatory cytokine production and the failure rate of peritoneal dialysis, and were not assessed in that study. Therefore, ILLs and the inflammatory response they induce may be protective in one clinical scenario (pneumonia) but deleterious in another (CAPD peritonitis).
Other studies have suggested a role for lymphocytes in the immune response to pneumonia. Lymphopaenia in blood has been associated with increased risk of death from CAP 62. Paats et al. found no difference in the relative frequencies of CD4+, CD8+ or γδ T cell populations (expressed as a proportion of the total CD3+ T cell population) in either blood or bronchoalveolar lavage fluid between healthy controls and patients with CAP 63. In patients with CAP, however, increased IL‐17A, IL‐22, and IL‐17A/IL‐22 production by CD3+CD4+ cells in bronchoalveolar lavage fluid and increased IL‐17A/IL‐22 production by CD3+CD4+ cells in PBMCs was seen with phorbol myristate acetate (PMA) stimulation; more IL‐17A/IL‐22‐producing CD3+CD4+ cells were seen in the blood of patients admitted to the intensive care unit, but no difference was seen between those with severe or non‐severe pneumonia. Of note, little IL‐17A and/or IL‐22 production was seen in γδ T cells in blood or bronchoalveolar lavage fluid. Increased IL‐17A, IL‐22, and IL‐17A/IL‐22 production by CD3+CD8+ cells was seen in bronchoalveolar lavage fluid but not PBMCs 63. Both MAIT cells and Vδ2+ T cells can produce IL‐17A and IL‐22, and will have contributed to the CD3+CD8+ and γδ T cell populations in the study by Paats et al. 51. In another study, Orlov et al. performed bronchoalveolar lavage on patients with suspected ventilator‐associated pneumonia 64. CD4+ cells were enriched from bronchoalveolar lavage fluid and then stimulated with PMA and ionomycin. Those with confirmed pneumonia had a reduced proportion of T helper (Th) type 17 cells (defined as CD45+CD4+CCR6+IL‐17A+ cells), while the proportion of Th1 cells (CD45+CD4+IFN‐γ+) was unchanged. Therefore IL‐17A‐producing T cells, in particular CD4+ T cells, contribute to the immune response to pneumonia. Unfortunately, because of the method of sample preservation, it was not possible to examine other lymphocyte subpopulations in our study.
Sputa, both spontaneously produced and induced are commonly used as proxies for more invasive respiratory samples, such as bronchoalveolar lavage, in pulmonary infection. A high rate of concordance for microbiological diagnosis between paired sputum and brochoalveolar lavage samples has been reported 65. There are no studies comparing the immune cell composition of sputa and bronchoalveolar lavage in pneumonia. Studies in healthy controls, patients with asthma or COPD (both stable and acute exacerbations), and patients with sarcoidosis or non‐granulomatous lung disease suggest that the immune cell composition of induced sputa may better represent the airways than the alveolar space, with a greater percentage of neutrophils and fewer alveolar macrophages and lymphocytes 66, 67, 68, 69, 70, although the profile of T cell subsets (as defined by CD4 and CD8 expression) is similar in induced sputa and bronchoalveolar lavage 69, 70. In contrast, lymphocytes are the predominant cell type in the bronchial submucosa 67. In a study of patients with COPD, there was no difference in the total or differential cell count between spontaneously produced and induced sputa 71. Therefore, the frequency of ILLs in sputa, whether spontaneously produced or induced, may not necessarily correlate with the frequency in other compartments.
Several limitations should be noted. First, the method of sample preservation excluded the possibility of phenotyping and in‐vitro functional assessment of MAIT cells and Vδ2+ T cells. This should be assessed in future prospective studies. Secondly, MAIT cells and Vδ2+ T cells were not detected in several samples, falling below the limit of detection of the assay; this prevented us from fitting multiple regression models. Thirdly, no paired blood samples were available to compare ILL abundance. Fourthly, as discussed above, this cohort lacked severe cases. Fifthly, we have not made an adjustment for multiple testing in our analysis, so replication in a prospective cohort will be important to assess the hypotheses generated by this study. Finally, there may be unmeasured confounders that account for our findings; therefore it is not possible to determine causation.
In conclusion, we have confirmed the presence of ILLs at the site of infection in CAP, and have identified an association of these cells with an improved clinical outcome. This should be investigated further in prospective studies. If these findings are confirmed, immunotherapies to enhance MAIT cell numbers and function could be considered for the prevention of severe CAP.
Disclosures
The authors have no conflicts of interest.
Author contributions
J. E. U., D. R. M., and X. C. M. conceived the study; J. E. U., R. F. H., D. R. M., X. C. M., and B. B. designed the study protocol; R. F. H. and J. C. conducted the experiments; R. F. H., X. W., X. C. M., M. S., and J. E. U. carried out the analysis and interpretation of data; M. R. S. provided statistical advice; R. F. H. and M. S. drafted the manuscript; R. F. H., M. S., and J. E. U. critically revised the manuscript for intellectual content. All authors read and approved the final manuscript.
Supporting information
Fig. S1. Gating strategy to determine MAIT and Vδ2+ T cell counts for comparison to qPCR data.
Fig. S2. Validation of the qPCR method to determine MAIT and Vδ2+ T cell abundance. The specificity of the qPCR assay was determined by measuring the abundance of (a) Vα7.2‐Jα12/20/33 gDNA in PBMCs and PBMCs depleted of Vα7.2+ cells and (c) Vδ2‐Jδ1/2/3/4 gDNA in PBMCs and PBMCs depleted of Vδ2+ cells; abundance is expressed relative to β2M in arbitrary units. The effectiveness of depletion of (b) MAIT cells (CD3+CD161++Vα7.2+ lymphocytes) and (d) Vδ2+ T cells (CD3+Vδ2+ lymphocytes) was assessed by flow cytometry. Each data point represents a different donor (n = 6), and in (a, c) represent the mean of technical triplicates of the qPCR assay. The median, and interquartile ranges are shown. Two independent experiments were performed. Differences between groups were analysed by Mann‐Whitney tests.
Fig. S3. MAIT cell and Vδ2+ T cell abundance in sputum are correlated, but no correlation with age is seen. MAIT cell (Vα7.2‐Jα12/20/33 gDNA) and Vδ2+ T cell (Vδ2‐Jδ1/2/3/4 gDNA) abundance in sputum of patients with CAP was measured by qPCR; the absolute copy number of β2M was calculated with LinRegPCR 23 and the number of MAIT cells (Vα7.2‐Jα12/20/33 gDNA) and Vδ2+ T cells (Vδ2‐Jδ1/2/3/4 gDNA) determined relative to β2M and expressed in arbitrary units. (a) The relationship between MAIT cell and Vδ2+ T cell abundance. (b‐d) The association between (b) MAIT cell, (c) Vδ2+ T cell, and (d) ILL abundance in sputum and patient age. Individual data points (n = 52) representing the mean of technical triplicates of the qPCR assay are shown. Relationships between variables were assessed with Spearman correlations.
Fig. S4. Abundance of ILLs does not differ with ICU admission. Abundance of (a) MAIT cells (Vα7.2‐Jα12/20/33 gDNA), (b) Vδ2+ T cells (Vδ2‐Jδ1/2/3/4 gDNA), and (c) ILLs in sputum of patients with CAP requiring admission to the intensive care unit (ICU) or not. Cell abundance was measured by qPCR; the absolute copy number of β2M was calculated with LinRegPCR 23 and the number of MAIT cells (Vα7.2‐Jα12/20/33 gDNA) and Vδ2+ T cells (Vδ2‐Jδ1/2/3/4 gDNA) determined relative to β2M and expressed in arbitrary units. Individual data points (n = 52) representing the mean of technical triplicates of the qPCR assay, the median, and interquartile ranges are shown. Data were analysed by Mann‐Whitney tests.
Fig. S5. Reduced disease severity in patients with higher ILL abundance in sputum. (a) The abundance of ILLs in sputum of patients by CURB‐65 score. (b) The abundance of ILLs in severe cases of pneumonia, as defined by the IDSA severity criteria. Cell abundance was measured by qPCR; the absolute copy number of β2M was calculated with LinRegPCR 23 and the number of MAIT cells (Vα7.2‐Jα12/20/33 gDNA) and Vδ2+ T cells (Vδ2‐Jδ1/2/3/4 gDNA) determined relative to β2M and expressed in arbitrary units. Individual data points (n = 52) representing the mean of technical triplicates of the qPCR assay, the median, and interquartile ranges are shown. CURB‐65 data was analysed using Kruskal‐Wallis 1‐way ANOVA and Dunn’s multiple comparison test; IDSA severity data was analysed by the Mann‐Whitney test. *P < 0.05.
Fig. S6. MAIT cell abundance in sputum is not associated with blood leukocyte count or with CRP. The association of (a) MAIT cell (Vα7.2‐Jα12/20/33 gDNA) and (b) Vδ2+ T cell (Vδ2‐Jδ1/2/3/4 gDNA) abundance in sputum with blood leukocyte count. The association of (c) MAIT cell and (d) Vδ2+ T cell abundance in sputum with blood CRP concentration (mg/L). MAIT cell and Vδ2+ T cell abundance was determined by qPCR; the absolute copy number of β2M was calculated with LinRegPCR 23 and the number of MAIT cells (Vα7.2‐Jα12/20/33 gDNA) and Vδ2+ T cells (Vδ2‐Jδ1/2/3/4 gDNA) determined relative to β2M and expressed in arbitrary units. Individual data points representing the mean of technical triplicates of the qPCR assay are shown. Blood leukocyte (n = 52) and CRP (n = 49) data were analysed using Spearman correlations. WCC = white cell (leukocyte) count; ns = not significant; CRP = C‐reactive protein.
Fig. S7. Vδ2+ T cell abundance in sputum is higher in patients with CAP caused by Streptococcus pneumoniae or Legionella spp. (a) The abundance of ILLs in sputum of patients with and without an identified bacterial pathogen. (b‐c) The abundance of MAIT cells (Vα7.2‐Jα12/20/33 gDNA) in sputum of patients with CAP caused by (b) S. pneumoniae or (c) Legionella spp. The abundance of Vδ2+ T cells (Vδ2‐Jδ1/2/3/4 gDNA) in sputum of patients with CAP caused by (d) S. pneumoniae or (e) Legionella spp. Cell abundance was measured by qPCR; the absolute copy number of β2M was calculated with LinRegPCR 23 and the number of MAIT cells (Vα7.2‐Jα12/20/33 gDNA) and Vδ2+ T cells (Vδ2‐Jδ1/2/3/4 gDNA) determined relative to β2M and expressed in arbitrary units. Individual data points (n = 52) representing the mean of technical triplicates of the qPCR assay, the median, and interquartile ranges are shown. Differences between groups were analysed by Mann‐Whitney tests.
Fig. S8. Relationship between cultured pathogens and 16S rRNA data. Genera of clinical interest are indicated on the X axis, while the Y axis shows the log relative abundance of each genera of interest within the 16S rRNA data. Facets correspond to groups of samples from which pathogen was cultured (HFLU = Haemophilus influenzae (n = 5); LGN = Legionella spp. (n = 14); MCAT = Moraxella catarrhalis (n = 5); PAER = Pseudomonas aeruginosa (n = 1); PNEU = Streptococcus pneumoniae (n = 4); SAUR = Staphylococcus aureus (n = 1); NSG = no significant growth (n = 41)). The mean abundance of genera is not significantly different between groups, as measured by the Kruskal‐Wallis and Wilcoxon signed‐rank tests, except for between samples +/‐ Legionella (P < 0.001).
Fig. S9. Association of ILL abundance in sputum with sputum concentrations of IFN‐γ, IFN‐α, and CXCL11. (a‐c) The association of sputum IFN‐γ concentrations with the abundance of (a) MAIT cells (Vα7.2‐Jα12/20/33 gDNA), (b) Vδ2+ T cells (Vδ2‐Jδ1/2/3/4 gDNA), and (c) ILLs in sputum. (d‐f) The association of sputum IFN‐α concentrations with the abundance of (d) MAIT cells, (e) Vδ2+ T cells, and (f) ILLs in sputum. (g‐i) The association of sputum CXCL11 concentrations with the abundance of (g) MAIT cells, (h) Vδ2+ T cells, and (i) ILLs in sputum. Cell abundance was measured by qPCR; the absolute copy number of β2M was calculated with LinRegPCR 23 and the number of MAIT cells (Vα7.2‐Jα12/20/33 gDNA) and Vδ2+ T cells (Vδ2‐Jδ1/2/3/4 gDNA) determined relative to β2M and expressed in arbitrary units. IFN‐γ, IFN‐α, and CXCL11 were measured by LEGENDplex bead array. Individual data points (n = 40 for IFN‐γ and IFN‐α; n = 30 for CXCL11) representing the mean of technical triplicates of the qPCR assay and the mean of technical duplicates of the LEGENDplex assay are shown. Associations between different cell populations and soluble mediators were assessed with Spearman correlations.
Acknowledgements
This work was supported by a University of Otago Research Grant (J. E. U., D.R.M.).
References
- 1. Pfuntner A, Wier LM, Stocks C. Most frequent conditions in U.S. hospitals, 2011. HCUP Statistical Brief #162. Rockville, MD: Agency for Healthcare Research and Quality; 2013:1–12. [PubMed] [Google Scholar]
- 2. Fearon D, Locksley R. The instructive role of innate immunity in the acquired immune response. Science 1996; 272:50–4. [DOI] [PubMed] [Google Scholar]
- 3. Ussher JE, Klenerman P, Willberg CB. Mucosal‐associated invariant T (MAIT) cells: new players in anti‐bacterial immunity. Front Immunol 2014; 5:450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Treiner E, Duban L, Bahram S et al Selection of evolutionarily conserved mucosal‐associated invariant T cells by MR1. Nature 2003; 422:164–9. [DOI] [PubMed] [Google Scholar]
- 5. Corbett AJ, Eckle SB, Birkinshaw RW et al T‐cell activation by transitory neo‐antigens derived from distinct microbial pathways. Nature 2014; 509:361–5. [DOI] [PubMed] [Google Scholar]
- 6. Kjer‐Nielsen L, Patel O, Corbett AJ et al MR1 presents microbial vitamin B metabolites to MAIT cells. Nature 2012; 491:717–23. [DOI] [PubMed] [Google Scholar]
- 7. Ussher JE, Bilton M, Attwod E et al CD161++CD8+T cells, including the MAIT cell subset, are specifically activated by IL‐12+IL‐18 in a TCR‐independent manner. Eur J Immunol 2014; 44:195–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chua W‐J, Truscott SM, Eickhoff CS, Blazevic A, Hoft DF, Hansen TH. Polyclonal mucosa‐associated invariant T cells have unique innate functions in bacterial infection. Infect Immun 2012; 80:3256–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Meierovics A, Yankelevich WJC, Cowley SC. MAIT cells are critical for optimal mucosal immune responses during in vivo pulmonary bacterial infection. Proc Natl Acad Sci USA 2013; 110:E3119–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang H, D'Souza C, Lim XY et al MAIT cells protect against pulmonary Legionella longbeachae infection. Nat Commun 2018; 9:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wilgenburg BV, Loh L, Chen Z et al MAIT cells contribute to protection against lethal influenza infection in vivo . Nat Commun 2018; 9:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Le Bourhis L, Martin E, Peguillet I et al Antimicrobial activity of mucosal‐associated invariant T cells. Nat Immunol 2010; 11:701–8. [DOI] [PubMed] [Google Scholar]
- 13. Grimaldi D, Bourhis L, Sauneuf B et al Specific MAIT cell behaviour among innate‐like T lymphocytes in critically ill patients with severe infections. Intens Care Med 2013; 40:1–10. [DOI] [PubMed] [Google Scholar]
- 14. Cosgrove C, Ussher JE, Rauch A et al Early and nonreversible decrease of CD161++/MAIT cells in HIV infection. Blood 2013; 121:951–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gohil S, Heo M, Schoenbaum E, Celentano D, Pirofski L‐A. CD8+ T cells and risk for bacterial pneumonia and all‐cause mortality among HIV‐infected women. J Acquir Immune Defic Syndr 2012; 60:191–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen ZW. Multifunctional immune responses of HMBPP‐specific Vg2Vd2 T cells in M. tuberculosis and other infections. Cell Mol Immunol 2013; 10:58–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Morita CT, Jin C, Sarikonda G, Wang H. Nonpeptide antigens, presentation mechanisms, and immunological memory of human Vγ2Vδ2 T cells: discriminating friend from foe through the recognition of prenyl pyrophosphate antigens. Immunol Rev 2007; 215:59–76. [DOI] [PubMed] [Google Scholar]
- 18. Ali Z, Shao L, Halliday L et al Prolonged (E)‐4‐hydroxy‐3‐methyl‐but‐2‐enyl pyrophosphate‐driven antimicrobial and cytotoxic responses of pulmonary and systemic Vγ2Vδ2 T cells in macaques. J Immunol 2007; 179:8287–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Qaqish A, Huang D, Chen CY et al Adoptive transfer of phosphoantigen‐specific gammadelta T cell subset attenuates Mycobacterium tuberculosis infection in nonhuman primates. J Immunol 2017; 198:4753–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Maze MJ, Slow S, Cumins AM et al Enhanced detection of Legionnaires’ disease by PCR testing of induced sputum and throat swabs. Euro Resp J 2014; 43:644–6. [DOI] [PubMed] [Google Scholar]
- 21. Chanez P, Holz O, Ind PW, Djukanović R, Maestrelli P, Sterk PJ. Sputum induction. Euro Resp J 2002; 20:3s–8s. [DOI] [PubMed] [Google Scholar]
- 22. Ussher JE, Phalora P, Cosgrove C et al Molecular analyses define Valpha7.2‐Jalpha33+ MAIT cell depletion in HIV infection: a case‐control study. Medicine 2015; 94:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ruijter JM, Ramakers C, Hoogaars WM et al Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res 2009; 37:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Caporaso JG, Lauber CL, Walters WA et al Ultra‐high‐throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 2012; 6:1621–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schloss PD, Westcott SL, Ryabin T et al Introducing mothur: open‐source, platform‐independent, community‐supported software for describing and comparing microbial communities. Appl Environ Microbiol 2009; 75:7537–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. DeSantis TZ, Hugenholtz P, Larsen N et al Greengenes, a chimera‐checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 2006; 72:5069–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Langille MG, Zaneveld J, Caporaso JG et al Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 2013; 31:814–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Reantragoon R, Corbett AJ, Sakala IG et al Antigen‐loaded MR1 tetramers define T cell receptor heterogeneity in mucosal‐associated invariant T cells. J Exp Med 2013; 210:2305–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Holtmeler W, Chowers Y, Lumeng A, Morzycka‐Wroblewska E, Kagnoff MF. The delta T cell receptor repertoire in human colon and peripheral blood is oligoclonal irrespective of V region usage. J. Clin Invest 1995; 96:1108–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kurioka A, Jahun AS, Hannaway RF et al Shared and distinct phenotypes and functions of human CD161++ Vα7.2+ T cell subsets. Front Immunol 2017; 8:1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lim WS, van der Eerden MM, Laing R et al Defining community acquired pneumonia severity on presentation to hospital: an international derivation and validation study. Thorax 2003; 58:377–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mandell LA, Wunderink RG, Anzueto A et al Infectious Diseases Society of America/American Thoracic Society consensus guidelines on the management of community‐acquired pneumonia in adults. Clin Infect Dis 2007; 44:S27–S72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Caccamo N, La Mendola C, Orlando V et al Differentiation, phenotype, and function of interleukin‐17‐producing human Vgamma9Vdelta2 T cells. Blood 2011; 118:129–38. [DOI] [PubMed] [Google Scholar]
- 34. Kurioka A, Ussher JE, Cosgrove C et al MAIT cells are licensed through granzyme exchange to kill bacterially sensitized targets. Mucosal Immunol 2015; 8:429–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Musher DM, Thorner AR. Community‐acquired pneumonia. N Engl J Med 2014; 371:1619–28. [DOI] [PubMed] [Google Scholar]
- 36. Zhang TG, Li AH, Lyu M, Chen M, Huang F, Wu J. Detection of respiratory viral and bacterial pathogens causing pediatric community‐acquired pneumonia in Beijing using real‐time PCR. Chronic Dis Transl Med 2015; 1:110–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wolff BJ, Bramley AM, Thurman KA et al Improved detection of respiratory pathogens by use of high‐quality sputum with TaqMan array card technology. J Clin Microbiol 2017; 55:110–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jain S, Self WH, Wunderink RG et al Community‐acquired pneumonia requiring hospitalization among U.S. adults. N Engl J Med 2015; 373:415–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lopez L, Huang QS. Influenza surveillance in New Zealand 2012. Wellington, New Zealand: Institute of Environmental Science and Research Ltd (ESR), 2013:1–74. [Google Scholar]
- 40. http://www.genome.jp/kegg-bin/show_pathway?map00900 (accessed 29 November 2017).
- 41. Tokunaga R, Zhang W, Naseem M et al CXCL9, CXCL10, CXCL11/CXCR3 axis for immune activation – a target for novel cancer therapy. Cancer Treat Rev 2018; 63:40–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lamichhane R, Schneider M, de la Harpe SM et al TCR‐ or cytokine‐activated CD8+ mucosal‐associated invariant T cells are rapid polyfunctional effectors that can coordinate immune responses. Cell Rep 2019; 28:3061–3076.e5. [DOI] [PubMed] [Google Scholar]
- 43. Poggi A, Carosio R, Fenoglio D et al Migration of Vδ1 and Vδ2 T cells in response to CXCR3 and CXCR4 ligands in healthy donors and HIV‐1‐infected patients: competition by HIV‐1 Tat. Blood 2004; 103:2205–13. [DOI] [PubMed] [Google Scholar]
- 44. Sharma PK, Wong EB, Napier RJ et al High expression of CD26 accurately identifies human bacteria‐reactive MR1‐restricted MAIT cells. Immunology 2015; 145:443–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ludwig A, Schiemann F, Mentlein R, Lindner B, Brandt E. Dipeptidyl peptidase IV (CD26) on T cells cleaves the CXC chemokine CXCL11 (I‐TAC) and abolishes the stimulating but not the desensitizing potential of the chemokine. J Leukoc Biol 2002; 72:183–91. [PubMed] [Google Scholar]
- 46. Foster GR, Masri SH, David R et al IFN‐α subtypes differentially affect human T cell motility. J Immunol 2004; 173:1663–70. [DOI] [PubMed] [Google Scholar]
- 47. van Wilgenburg B, Scherwitzl I, Hutchinson EC et al MAIT cells are activated during human viral infections. Nat Commun 2016; 7:11653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Crouse J, Kalinke U, Oxenius A. Regulation of antiviral T cell responses by type I interferons. Nat Rev Immunol 2015; 15:231–42. [DOI] [PubMed] [Google Scholar]
- 49. Jaillon S, Galdiero MR, Del Prete D, Cassatella MA, Garlanda C, Mantovani A. Neutrophils in innate and adaptive immunity. Semin Immunopathol 2013; 35:377–94. [DOI] [PubMed] [Google Scholar]
- 50. Laan M, Cui ZH, Hoshino H et al Neutrophil recruitment by human IL‐17 via C‐X‐C chemokine release in the airways. J Immunol 1999; 162:2347–52. [PubMed] [Google Scholar]
- 51. Billerbeck E, Kang Y‐H, Walker LJ et al Analysis of CD161 expression on human CD8+ T cells defines a distinct functional subset with tissue‐homing properties. Proc Natl Acad Sci USA 2010; 107:3006–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Cheng P, Liu T, Zhou WY et al Role of gamma‐delta T cells in host response against Staphylococcus aureus‐induced pneumonia. BMC Immunol 2012; 13:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Paats MS, Bergen IM, Hanselaar WEJJ et al Local and systemic cytokine profiles in nonsevere and severe community‐acquired pneumonia. Eur Respir J 2013; 41:1378–85. [DOI] [PubMed] [Google Scholar]
- 54. Deng JC, Tateda K, Zeng X, Standiford TJ. Transient transgenic expression of gamma interferon promotes Legionella pneumophila clearance in immunocompetent hosts. Infect Immun 2001; 69:6382–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Suarez‐Mendez R, Garcia‐Garcia I, Fernandez‐Olivera N et al Adjuvant interferon gamma in patients with drug‐resistant pulmonary tuberculosis: a pilot study. BMC Infect Dis 2004; 4:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lockhart E, Green AM, Flynn JL. IL‐17 production is dominated by T cells rather than CD4 T cells during Mycobacterium tuberculosis infection. J Immunol 2006; 177:4662–9. [DOI] [PubMed] [Google Scholar]
- 57. Meierovics A, Yankelevich WJ, Cowley SC. MAIT cells are critical for optimal mucosal immune responses during in vivo pulmonary bacterial infection. Proc Natl Acad Sci USA 2013; 110:E3119–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Suter‐Widmer I, Christ‐Crain M, Zimmerli W, Albrich W, Mueller B, Schuetz P. Predictors for length of hospital stay in patients with community‐acquired pneumonia: results from a Swiss multicenter study. BMC Pulm Med 2012; 12:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Novak J, Dobrovolny J, Novakova L, Kozak T. The decrease in number and change in phenotype of mucosal‐associated invariant T cells in the elderly and differences in men and women of reproductive age. Scand J Immunol 2014; 80:271–5. [DOI] [PubMed] [Google Scholar]
- 60. Hinks TSC, Wallington JC, Williams AP, Djukanović R, Staples KJ, Wilkinson TMA. Steroid‐induced deficiency of mucosal‐associated invariant T cells in the chronic obstructive pulmonary disease lung. Implications for nontypeable Haemophilus influenzae infection. Am J Respir Crit Care Med 2016; 194:1208–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Liuzzi AR, Kift‐Morgan A, Lopez‐Anton M et al Unconventional human T cells accumulate at the site of infection in response to microbial ligands and induce local tissue remodeling. J Immunol 2016; 197:2195–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bermejo‐Martin JF, Cilloniz C, Mendez R et al Lymphopenic community acquired pneumonia (L‐CAP), an immunological phenotype associated with higher risk of mortality. EBioMedicine 2017; 24:231–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Paats MS, Bergen IM, Hanselaar WEJJ et al T helper 17 cells are involved in the local and systemic inflammatory response in community‐acquired pneumonia. Thorax 2013; 68:468–74. [DOI] [PubMed] [Google Scholar]
- 64. Orlov M, Dmyterko V, Wurfel MM, Mikacenic C. Th17 cells are associated with protection from ventilator associated pneumonia. PLOS ONE 2017; 12:e0182966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Dubourg G, Abat C, Rolain J‐M, Raoult D. Correlation between sputum and bronchoalveolar lavage fluid cultures. J Clin Microbiol 2015; 53:994–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Keatings VM, Evans DJ, O’Connor BJ, Barnes PJ. Cellular profiles in asthmatic airways: a comparison of induced sputum, bronchial washings, and bronchoalveolar lavage fluid. Thorax 1997; 52:372–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Maestrelli P, Saetta M, Di Stefano A et al Comparison of leukocyte counts in sputum, bronchial biopsies, and bronchoalveolar lavage. Am J Respir Crit Care Med 1995; 152:1926–31. [DOI] [PubMed] [Google Scholar]
- 68. Lensmar C, Elmberger G, Sandgren P, Skold CM, Eklund A. Leukocyte counts and macrophage phenotypes in induced sputum and bronchoalveolar lavage fluid from normal subjects. Eur Respir J 1998; 12:595–600. [DOI] [PubMed] [Google Scholar]
- 69. Fireman E, Topilsky I, Greif J et al Induced sputum compared to bronchoalveolar lavage for evaluating patients with sarcoidosis and non‐granulomatous interstitial lung disease. Respir Med 1999; 93:827–34. [DOI] [PubMed] [Google Scholar]
- 70. Grootendorst DC, Sont JK, Willems LNA et al Comparison of inflammatory cell counts in asthma: induced sputum vs bronchoalveolar lavage and bronchial biopsies. Clin Exp Allergy 1997; 27:769–79. [PubMed] [Google Scholar]
- 71. Bhowmik A, Seemungal TAR, Sapsford RJ, Devalia JL, Wedzicha JA. Comparison of spontaneous and induced sputum for investigation of airway inflammation in chronic obstructive pulmonary disease. Thorax 1998; 53:953–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Gating strategy to determine MAIT and Vδ2+ T cell counts for comparison to qPCR data.
Fig. S2. Validation of the qPCR method to determine MAIT and Vδ2+ T cell abundance. The specificity of the qPCR assay was determined by measuring the abundance of (a) Vα7.2‐Jα12/20/33 gDNA in PBMCs and PBMCs depleted of Vα7.2+ cells and (c) Vδ2‐Jδ1/2/3/4 gDNA in PBMCs and PBMCs depleted of Vδ2+ cells; abundance is expressed relative to β2M in arbitrary units. The effectiveness of depletion of (b) MAIT cells (CD3+CD161++Vα7.2+ lymphocytes) and (d) Vδ2+ T cells (CD3+Vδ2+ lymphocytes) was assessed by flow cytometry. Each data point represents a different donor (n = 6), and in (a, c) represent the mean of technical triplicates of the qPCR assay. The median, and interquartile ranges are shown. Two independent experiments were performed. Differences between groups were analysed by Mann‐Whitney tests.
Fig. S3. MAIT cell and Vδ2+ T cell abundance in sputum are correlated, but no correlation with age is seen. MAIT cell (Vα7.2‐Jα12/20/33 gDNA) and Vδ2+ T cell (Vδ2‐Jδ1/2/3/4 gDNA) abundance in sputum of patients with CAP was measured by qPCR; the absolute copy number of β2M was calculated with LinRegPCR 23 and the number of MAIT cells (Vα7.2‐Jα12/20/33 gDNA) and Vδ2+ T cells (Vδ2‐Jδ1/2/3/4 gDNA) determined relative to β2M and expressed in arbitrary units. (a) The relationship between MAIT cell and Vδ2+ T cell abundance. (b‐d) The association between (b) MAIT cell, (c) Vδ2+ T cell, and (d) ILL abundance in sputum and patient age. Individual data points (n = 52) representing the mean of technical triplicates of the qPCR assay are shown. Relationships between variables were assessed with Spearman correlations.
Fig. S4. Abundance of ILLs does not differ with ICU admission. Abundance of (a) MAIT cells (Vα7.2‐Jα12/20/33 gDNA), (b) Vδ2+ T cells (Vδ2‐Jδ1/2/3/4 gDNA), and (c) ILLs in sputum of patients with CAP requiring admission to the intensive care unit (ICU) or not. Cell abundance was measured by qPCR; the absolute copy number of β2M was calculated with LinRegPCR 23 and the number of MAIT cells (Vα7.2‐Jα12/20/33 gDNA) and Vδ2+ T cells (Vδ2‐Jδ1/2/3/4 gDNA) determined relative to β2M and expressed in arbitrary units. Individual data points (n = 52) representing the mean of technical triplicates of the qPCR assay, the median, and interquartile ranges are shown. Data were analysed by Mann‐Whitney tests.
Fig. S5. Reduced disease severity in patients with higher ILL abundance in sputum. (a) The abundance of ILLs in sputum of patients by CURB‐65 score. (b) The abundance of ILLs in severe cases of pneumonia, as defined by the IDSA severity criteria. Cell abundance was measured by qPCR; the absolute copy number of β2M was calculated with LinRegPCR 23 and the number of MAIT cells (Vα7.2‐Jα12/20/33 gDNA) and Vδ2+ T cells (Vδ2‐Jδ1/2/3/4 gDNA) determined relative to β2M and expressed in arbitrary units. Individual data points (n = 52) representing the mean of technical triplicates of the qPCR assay, the median, and interquartile ranges are shown. CURB‐65 data was analysed using Kruskal‐Wallis 1‐way ANOVA and Dunn’s multiple comparison test; IDSA severity data was analysed by the Mann‐Whitney test. *P < 0.05.
Fig. S6. MAIT cell abundance in sputum is not associated with blood leukocyte count or with CRP. The association of (a) MAIT cell (Vα7.2‐Jα12/20/33 gDNA) and (b) Vδ2+ T cell (Vδ2‐Jδ1/2/3/4 gDNA) abundance in sputum with blood leukocyte count. The association of (c) MAIT cell and (d) Vδ2+ T cell abundance in sputum with blood CRP concentration (mg/L). MAIT cell and Vδ2+ T cell abundance was determined by qPCR; the absolute copy number of β2M was calculated with LinRegPCR 23 and the number of MAIT cells (Vα7.2‐Jα12/20/33 gDNA) and Vδ2+ T cells (Vδ2‐Jδ1/2/3/4 gDNA) determined relative to β2M and expressed in arbitrary units. Individual data points representing the mean of technical triplicates of the qPCR assay are shown. Blood leukocyte (n = 52) and CRP (n = 49) data were analysed using Spearman correlations. WCC = white cell (leukocyte) count; ns = not significant; CRP = C‐reactive protein.
Fig. S7. Vδ2+ T cell abundance in sputum is higher in patients with CAP caused by Streptococcus pneumoniae or Legionella spp. (a) The abundance of ILLs in sputum of patients with and without an identified bacterial pathogen. (b‐c) The abundance of MAIT cells (Vα7.2‐Jα12/20/33 gDNA) in sputum of patients with CAP caused by (b) S. pneumoniae or (c) Legionella spp. The abundance of Vδ2+ T cells (Vδ2‐Jδ1/2/3/4 gDNA) in sputum of patients with CAP caused by (d) S. pneumoniae or (e) Legionella spp. Cell abundance was measured by qPCR; the absolute copy number of β2M was calculated with LinRegPCR 23 and the number of MAIT cells (Vα7.2‐Jα12/20/33 gDNA) and Vδ2+ T cells (Vδ2‐Jδ1/2/3/4 gDNA) determined relative to β2M and expressed in arbitrary units. Individual data points (n = 52) representing the mean of technical triplicates of the qPCR assay, the median, and interquartile ranges are shown. Differences between groups were analysed by Mann‐Whitney tests.
Fig. S8. Relationship between cultured pathogens and 16S rRNA data. Genera of clinical interest are indicated on the X axis, while the Y axis shows the log relative abundance of each genera of interest within the 16S rRNA data. Facets correspond to groups of samples from which pathogen was cultured (HFLU = Haemophilus influenzae (n = 5); LGN = Legionella spp. (n = 14); MCAT = Moraxella catarrhalis (n = 5); PAER = Pseudomonas aeruginosa (n = 1); PNEU = Streptococcus pneumoniae (n = 4); SAUR = Staphylococcus aureus (n = 1); NSG = no significant growth (n = 41)). The mean abundance of genera is not significantly different between groups, as measured by the Kruskal‐Wallis and Wilcoxon signed‐rank tests, except for between samples +/‐ Legionella (P < 0.001).
Fig. S9. Association of ILL abundance in sputum with sputum concentrations of IFN‐γ, IFN‐α, and CXCL11. (a‐c) The association of sputum IFN‐γ concentrations with the abundance of (a) MAIT cells (Vα7.2‐Jα12/20/33 gDNA), (b) Vδ2+ T cells (Vδ2‐Jδ1/2/3/4 gDNA), and (c) ILLs in sputum. (d‐f) The association of sputum IFN‐α concentrations with the abundance of (d) MAIT cells, (e) Vδ2+ T cells, and (f) ILLs in sputum. (g‐i) The association of sputum CXCL11 concentrations with the abundance of (g) MAIT cells, (h) Vδ2+ T cells, and (i) ILLs in sputum. Cell abundance was measured by qPCR; the absolute copy number of β2M was calculated with LinRegPCR 23 and the number of MAIT cells (Vα7.2‐Jα12/20/33 gDNA) and Vδ2+ T cells (Vδ2‐Jδ1/2/3/4 gDNA) determined relative to β2M and expressed in arbitrary units. IFN‐γ, IFN‐α, and CXCL11 were measured by LEGENDplex bead array. Individual data points (n = 40 for IFN‐γ and IFN‐α; n = 30 for CXCL11) representing the mean of technical triplicates of the qPCR assay and the mean of technical duplicates of the LEGENDplex assay are shown. Associations between different cell populations and soluble mediators were assessed with Spearman correlations.


