Abstract
Histatin 5 (Hst‐5) is an antimicrobial peptide with strong antifungal activity against Candida albicans, an opportunistic pathogen that is a common cause of oral thrush. The peptide is natively secreted by human salivary glands and shows promise as an alternative therapeutic against infections caused by C. albicans. However, Hst‐5 can be cleaved and inactivated by a family of secreted aspartic proteases (Saps) produced by C. albicans. Single‐residue substitutions can significantly affect the proteolytic resistance of Hst‐5 to Saps and its antifungal activity; the K17R substitution increases resistance to proteolysis, while the K11R substitution enhances antifungal activity. In this work, we showed that the positive effects of these two single‐residue modifications can be combined in a single peptide, K11R–K17R, with improved proteolytic resistance and antifungal activity. We also investigated the effect of additional single‐residue substitutions, with a focus on the effect of addition or removal of negatively charged residues, and found Sap‐dependent effects on degradation. Both single‐ and double‐substitutions affected the kinetics of proteolytic degradation of the intact peptide and of the fragments formed during degradation. Our results demonstrate the importance of considering proteolytic stability and not just antimicrobial activity when designing peptides for potential therapeutic applications.
Keywords: antimicrobial peptides, Candida albicans, histatin 5, proteolysis kinetics
1. INTRODUCTION
Oral thrush is an infection commonly observed among HIV‐positive patients, and a common culprit of the infection is the commensal organism Candida albicans. Human saliva contains antimicrobial peptides, including histatin 51 (Hst‐5) (Figure 1), that can prevent the proliferation of the opportunistic pathogen.2 Hst‐5 is one of the 12 histatins secreted by the parotid and submandibular glands,3 and, of these histatins, the 24‐mer Hst‐5 has the strongest antifungal activity against C. albicans.3, 4 HIV‐positive patients have a reduced level of Hst‐5, and the low level of the peptide is hypothesized to play a role in the increased susceptibility of the HIV‐positive population to infection.5 The native antifungal activity of Hst‐5 in human saliva highlights its promise as a therapeutic peptide to target oral C. albicans infections.
Figure 1.
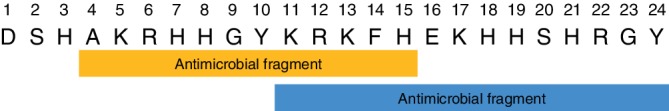
Hst‐5 sequence with reported antimicrobial fragments noted
Although the antifungal activity of Hst‐5 is well known, its exact mechanism of action in the cell is still debated. Once actively transported into the fungal cell, the peptide causes an ion imbalance and cell volume loss that leads to eventual cell death.6, 7 To understand how properties of Hst‐5 affect its antifungal activity, studies over the last three decades analyzed the peptide through a number of residue modifications,8, 9, 10, 11 peptide truncations,12, 13, 14 and residue substitutions in the truncated peptide.13, 14, 15 Peptide truncation studies proposed that Residues 4–1512 or 11–2413 are the minimal antifungal fragment of the peptide (Figure 1). Meanwhile, the residue substitution studies suggest that the tendency to form an α‐helical structure is not vital for the antifungal activity of the intact peptide, but the charge of a modified residue is important.13, 14, 15
While the antifungal activity and native presence of Hst‐5 make the peptide attractive as an antimicrobial agent, the peptide faces challenges due to its susceptibility to proteolysis by enzymes released by pathogens. In particular, Hst‐5 is cleaved by some of the 10 secreted aspartic proteases (Sap1‐10) produced by C. albicans.16, 17 Most of the Saps are fully secreted into the extracellular space (Sap1‐8), while two of them (Sap9‐10) are either bound to the cell membrane via a glycosylphosphatidylinositol (GPI) anchor or GPI proteins.18 Proteolytic target studies using antimicrobial peptides,16, 17, 19 peptide libraries,18, 20, 21, 22 and a protein library23 indicate that, generally, Saps have a preference for basic amino acids or large hydrophobic amino acids at the cleavage sites.20, 21, 22 A caveat of the synthetic libraries generally used in such studies, however, is that they are often composed of random sequences of peptides that may not exist as part of a protein or cell with which the Saps may interact. Furthermore, the studies do not relate the cleavage to the functionality of the peptide. Hst‐5‐specific studies revealed the presence of lysine residues at most of the proteolytic sites cleaved by Saps.16, 17 Bochenska et al. conducted a study of proteolysis of Hst‐5 over time and showed that, for a majority of the Saps, the peptide is first cleaved between residues K17 and H18.17 In our previous work, we showed that substitution of K17 to arginine significantly enhances the peptide's resistance to degradation at the site by Sap9, Sap2, and whole C. albicans cells without reducing the antifungal activity of the peptide.24 Our results demonstrated the potential of using residue modifications to improve the proteolytic stability of Hst‐5 and motivate evaluation of additional Hst‐5 modifications to inhibit proteolysis by fungal proteases and improve the design of antifungal peptides.
In this study, we further investigated the effect of residue modifications on the antifungal activity of Hst‐5 and its proteolysis by Saps, including the kinetics of proteolysis. We explored addition or removal of a negatively charged residue and showed the effect can be beneficial or detrimental to both proteolytic resistance and antifungal activity. Along with single residue mutations, we also combined two previously studied mutations (K11R and K17R) into one peptide variant to bring together the effect of modifications that improve antifungal activity (K11R) and increase proteolytic resistance (K17R).24 Our results show that the combined effects of individually beneficial modifications can further improve Hst‐5 and reveal the kinetics of proteolysis of Hst‐5 variants. These results will guide future peptide designs to improve the therapeutic potential of Hst‐5 and other antifungal peptides.
2. RESULTS
In our previous work, we discovered that a single residue substitution from a positively charged lysine to a positively charged arginine or uncharged leucine can significantly increase the resistance of Hst‐5 to proteolysis by Saps or enhance the peptide's antifungal activity.24 In this work, we explored the effect of additional modifications to the peptide on its proteolytic susceptibility and antifungal activity (Table 1).
Table 1.
Hst‐5 and variants with one or two amino acid substitutions
| Sequencea | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Peptide | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 |
| Hst‐5 | D | S | H | A | K | R | H | H | G | Y | K | R | K | F | H | E | K | H | H | S | H | R | G | Y |
| K13H | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | H | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ |
| K13E | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | E | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ |
| E16R | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | R | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ |
| E16L | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | L | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ |
| K11R–K17R | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | R | ‐ | ‐ | ‐ | ‐ | ‐ | R | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ |
Dash indicates residue was unchanged from the parent Hst‐5 peptide.
To determine whether the enhancements seen with single amino acid modifications can be combined to achieve further enhancement, both K11 and K17 were substituted to arginine (K11R–K17R). The K11R modification alone leads to enhanced antifungal activity, and the K17R modification alone leads to enhanced resistance to proteolysis.24 We hypothesized that a peptide containing both modifications would combine these two beneficial properties.
Since our previous Hst‐5 variants explored effects of substitutions at a positively charged residue, here we investigated the role of the negatively charged E16 residue on the peptide's properties. The E16 residue was chosen since the glutamic acid at the site is the only negatively charged residue that is part of one of the proposed minimal antifungal fragments of Hst‐5 (Residues 11–2413). To test the role of the negatively charged residue in the robustness and the activity of Hst‐5, we modified the residue to a positively charged arginine (E16R) or an uncharged leucine (E16L) (Table 1). The E16 residue is also next to the K17 site where, previously, the substitution to arginine or leucine significantly enhanced the peptide's resistance to Saps.24 Therefore, we expected that its modification would impact the peptide stability or activity.
Hst‐5 is a highly positively charged peptide, and the charge may play a role in its recognition by Saps and its mechanism of action within the cell. Thus, we investigated the effect of decreasing the charge of the peptide with a K13E substitution. The K13 residue is part of the proposed active fragments of the peptide, and we previously found its modification to a positively charged arginine or an uncharged leucine leads to a Sap‐dependent effect on proteolysis.24 While the K13L substitution increased resistance to degradation by Sap2, the same modification dramatically increased degradation by Sap9, and degradation by both Sap2 and Sap9 was increased for the K13R modification. Tsai et al. previously reported that the K13E substitution results in reduced antifungal activity,9 but the effect of the substitution on proteolysis was not explored. To further probe the effect of charged residues at the K13 site, we also included the K13H modification.
2.1. Modified peptides show a wide range of susceptibility to proteolysis
The effect of the amino acid modifications on the proteolytic susceptibility of Hst‐5 was evaluated by incubating Hst‐5 and its variants with purified, recombinant Sap9 and Sap2 as well as the supernatant of C. albicans culture and intact C. albicans cells (without supernatant). The culture supernatant and cells were used to study the proteolytic effect of native, fully secreted Saps and anchored Saps, respectively, on the peptides. The intact peptide and degradation products were separated via gel electrophoresis and the level of degradation was quantified through densitometric analysis of Coomassie‐stained gels (Figure 2, Figure S1).
Figure 2.
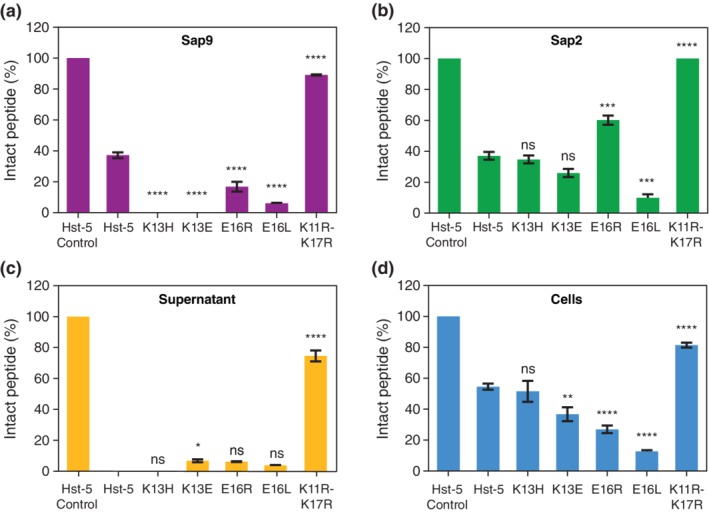
Extent of proteolysis of parent Hst‐5 and Hst‐5 variants by (a) recombinant Sap9, (b) recombinant Sap2, (c) C. albicans culture supernatant, and (d) intact C. albicans cells. After the peptides (50 μM) were incubated with the Saps (3 μg/ml Sap9 or 0.05 μg/ml Sap2), culture supernatant, or C. albicans cells (1 × 109 cells/ml), samples were run on a gel and stained with Coomassie. Images of representative Coomassie‐stained gels are provided in Figure S1. The amount of intact peptide remaining was quantified by comparing the intensity of the band for the intact peptide to the band(s) for the fragments through densitometric analysis. Error bars indicate standard error of the mean (n = 3 for (c) and n = 6 for remaining panels). The level of statistical significance relative to parent Hst‐5 is indicated by the number of asterisks: ns for no significance, * for p < .05, ** for p < .01, *** for p < .001, and **** for p < .0001
As we hypothesized during the peptide design, the double‐substituted peptide K11R–K17R showed significantly enhanced resistance in all the conditions tested. With recombinant Saps, only 37% of the parent Hst‐5 remained, while over 88% of intact K11R–K17R peptide remained after incubation with Sap9 (Figure 2a), and no detectable level of degradation was observed with Sap2 (Figure 2b). Incubation with culture supernatant and intact C. albicans cells showed a similar increase in resistance to proteolysis. Over 74% and 81% of intact K11R–K17R peptide remained after incubation with culture supernatant and intact cells, respectively (Figure 2c,d); in contrast, only 54% intact Hst‐5 remained after exposure to the cells and no detectable amount of intact peptide was measured in culture supernatant samples. The robustness of the K11R–K17R in all four conditions shows that the addition of the K11R substitution does not diminish the proteolytic resistance we originally discovered with the K17R substitution.24
Removal of a negatively charged residue and modifications at the E16 residue had mixed effects on the susceptibility of the peptides to proteolysis. While the E16R modification enhanced resistance to Sap2 (60% intact peptide, Figure 2b), significantly more degradation compared to unmodified Hst‐5 occurred after exposure to Sap9 (16% intact, Figure 2a) or whole cells (27% intact, Figure 2d). Removal of the charge at this site with the E16L substitution had a deleterious impact on proteolytic stability; the peptide was more degraded in all cases except with culture supernatant, which was inconclusive. The modifications at the K13 site led to a degradation pattern similar to the one for the E16R substitution. After exposure to Sap2, the K13H and K13E peptides retained an amount of intact peptide comparable to Hst‐5; however, these variants were completely degraded by Sap9. The K13E peptide was more susceptible to C. albicans cells, as it showed a significant decrease in the level of intact peptide compared to Hst‐5, while K13H showed no difference compared to the parent peptide. In general, both removal of an existing negative charge at the E16 site or addition of one at K13 site had a negative impact on the peptide proteolytic stability in the presence of Saps and C. albicans cells.
Overall, all the tested variants of Hst‐5 showed a deviation from the results for Hst‐5 in at least one of the conditions tested. The levels of intact peptide remaining after incubation with whole C. albicans cells more closely mirrored incubation with Sap2 rather than Sap9, despite Sap9 being the anchored Sap. It should be noted, however, that other Saps are present during incubation with culture supernatant and whole C. albicans cells. Furthermore, the concentration of Saps in the supernatant samples may be higher than those in the cell samples, since, to ensure sufficient presence of Saps, the cultures used to collect the supernatant were grown for a longer time than the cultures used to harvest cells. Consequently, some of the degradation is likely due to Saps other than Sap2 or Sap9. Removal or addition of negatively charged residues also appears to play a role in the susceptibility of the peptides to Saps, but the variability of the effect on degradation of the two Saps highlights the differences in cleavage of peptides by Saps.
2.2. Amino acid substitutions shift cleaved fragment abundance
The gel electrophoresis analysis presents the changes to the susceptibility of Hst‐5 to proteolysis due to the amino acid substitutions but does not indicate where cleavage occurs. To discern the impact on the cleavage sites and relative amount of the fragments produced, the peptides were analyzed with capillary electrophoresis‐mass spectrometry (CEMS) after incubation with the recombinant Saps. We tested the variants E16R, E16L, and K11R–K17R, in addition to Hst‐5, as they showed the most variable susceptibility to the purified Saps and C. albicans cells compared to parent Hst‐5 and to each other. The peptide MRFA was added to each sample as an internal standard prior to CEMS. The abundance of each intact peptide or its fragments was quantified by looking at the area of the peak for each peptide in the electrophoretogram relative to the peak area for the internal standard in the electrophoretogram (Figure S2). Fragments with a relative abundance of ≥0.01 compared to the MRFA signal are depicted in Figure 3. Other fragments detected at a lower level are listed in Figure S3.
Figure 3.
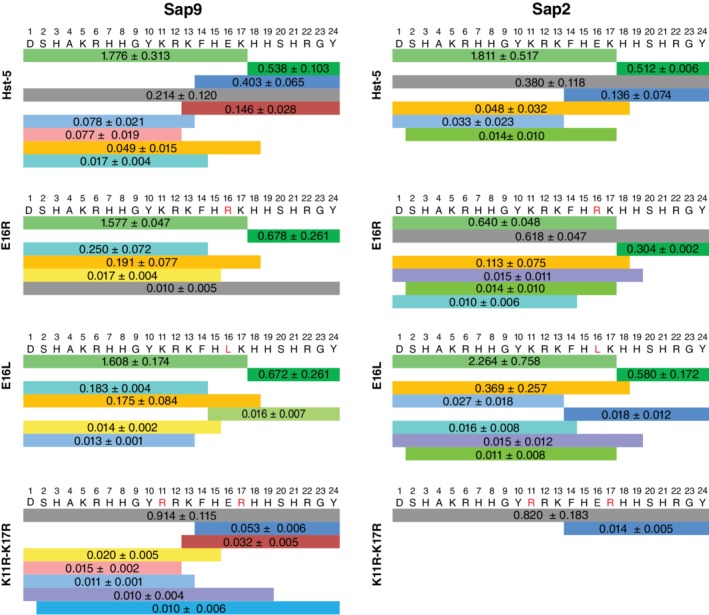
Peptide fragments resulting from degradation by Sap9 and Sap2. Each band color indicates a different fragment, and the number on the band shows the relative abundance of the fragment as the average relative peak area. Following incubation of 50 μM peptide with 3 μg/ml Sap9 and 0.05 μg/ml Sap2, peak areas for the intact peptide (gray) and degradation fragments were obtained from an electrophoretogram of the CEMS data. The data are shown as the peak area relative to the area for an internal standard. Errors are the standard error of the mean (n = 3). Fragments with relative peak area > .01 are displayed
For the relative abundance of intact peptide, the mass spectrometry results are consistent with the gel electrophoresis results for both Saps. The intact peptide was one of the most abundant peptides following incubation of the E16R peptide with Sap2 and the K11R–K17R peptide with Sap9 and Sap2 (Figure 3). The same samples showed enhanced resistance to proteolysis in the densitometry analysis of the electrophoresis gels (Figure 2a,b). The E16L peptide also followed the gel electrophoresis result in that, after incubation with either Sap, almost no intact E16L peptide was detected.
In addition to eliciting a shift in the signal of intact peptide, the E16R and E16L substitutions also led to a shift in the relative abundance of other fragments. While Hst‐5 and K11R–K17R produced fragments from cleavage at the K13 site (e.g., fragments with Residues 13–24 and 14–24), these fragments were less prevalent in the E16R and E16L peptides. In fact, we detected no significant cleavage around the K13 residue of the E16R peptide in either Sap sample. For the E16L peptide, only the fragment from cleavage on the C‐terminal side of residue K13 was detected at a high level.
The degradation pattern for the K11R–K17R variant was quite different from Hst‐5 and the other variants. Unlike the other variants, the most abundant peptide following cleavage of K11R–K17R by either Sap9 or Sap2 was the intact peptide, and substantially lower levels of cleavage fragments were detected. The lack of a significant signal from the fragment with Residues 1–17 after incubation with either Sap indicates that significantly less cleavage occurred at the K17 site. The outcome is consistent with our prediction during design of this peptide, since the individual K11R and K17R substitutions previously reduced degradation at the K17 site.24
2.3. Modifications modulate the rate of proteolysis by Sap9
The mass spectrometry analysis demonstrates that the residue substitutions affect the abundance of the intact peptides and fragments of the peptides after exposure to the Saps for 2 hr. End‐point comparison of fragments at only one incubation duration, however, does not fully capture the effect of residue substitution on the interaction of a peptide with a Sap enzyme. To gain more insight into what occurs during the 2 hr incubation period, a kinetic study was conducted with Sap9. We chose to use Sap9 in this study, because it had a more significant effect on the degradation of Hst‐5 variants than Sap2. The peptides Hst‐5, E16L, and K11R–K17R were incubated with Sap9 for 5, 30, and 120 min and analyzed with CEMS. For controls at 0 min, Sap9 that was first inactivated with heat was added to the peptide samples.
Each of the tested variants showed a unique pattern of degradation kinetics (Figure 4). Continuous depletion of Hst‐5 (Figure 4a) and E16L (Figure 4c) occurred throughout the experiment, while intact K11R–K17R (Figure 4e) maintained a steady presence. In the case of the E16L variant, the rate of depletion of the intact peptide in the first 5 min of incubation was significantly faster than the other two peptides. Over 90% of the signal from intact E16L, relative to the signal at 0 min, was lost within the first 30 min of incubation (Figure 4c), which is significantly more loss than intact Hst‐5 and K11R–K17R for the same incubation period. By the last incubation time point, almost no intact E16L peptide remained while nearly all intact K11R–K17R remained. The rapid degradation of E16L explains why almost no intact E16L was detected in the end‐point mass spectrometry analysis after a 120 min incubation with Saps (Figure 3).
Figure 4.
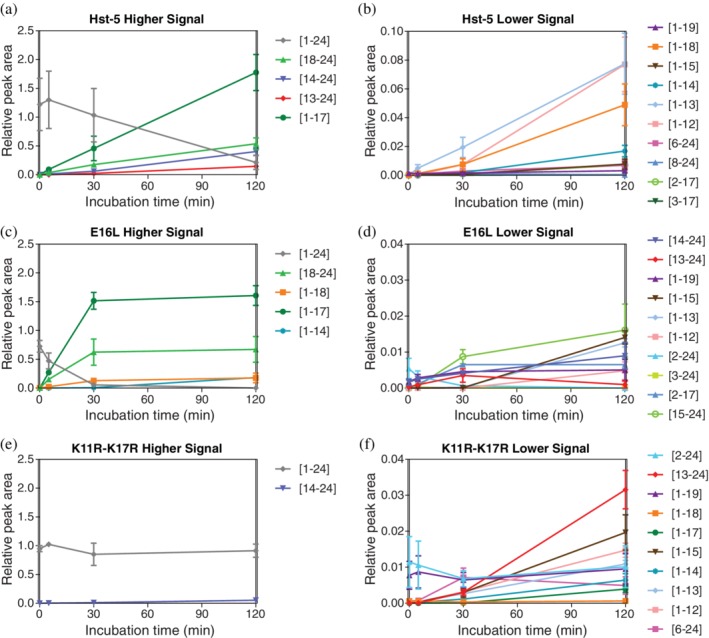
Kinetics of degradation of (a,b) Hst‐5, (c,d) E16L, and (e,f) K11R–K17R by Sap9. Degradation products with higher signals are shown in (a), (c), and (e), while those with lower signals are shown in (b), (d), and (f). Hst‐5 and the variants (50 μM) were incubated with 3 μg/ml Sap9 for 0, 5, 30, and 120 min. Data are shown as the average relative peak area, which was obtained from the electrophoretogram of the sample mass spectra relative to an internal standard. Fragments that showed a relative peak area > .001 for at least one of the time points are displayed. Errors are the standard error of the mean (n = 3)
The residue substitutions also affected the formation of peptide fragments and their subsequent degradation. The relative signal for the fragment containing Residues 1–17, along with its corresponding Residues 18–24 fragment, rapidly increased for the first 30 min in the Hst‐5 and E16L samples. After the 30‐min time point, however, the Hst‐5 and E16L peptides show a different trend. While the relative signal of the Residues 1–17 fragment from Hst‐5 continued to increase between 30 and 120 min, the amount of fragment formed from the E16L peptide reached a steady state, likely because the majority of intact E16L was already cleaved in the first 30 min of incubation.
We observed a similar pattern for the fragment containing Residues 13–24, a fragment that was observed after proteolysis by Sap9 but not Sap2 (Figure 3). While its abundance continuously increased in the Hst‐5 and K11R–K17R samples, this fragment increased in the first 30 min for the E16L sample and then decreased as the fragment was subsequently further degraded, explaining why only a low signal of the fragment was detected in the end‐point samples of E16L (Figures S3 and 4d). The results from the kinetic study show that the modifications to the peptides not only affect the rate of degradation of the intact peptide, but also the fragments formed from the degradation. Different rates of degradation lead to variation in the type and abundance of fragments present at each time point, which may play a role in antifungal activity in the presence of proteolytic enzymes.
2.4. Selected variants have enhanced antifungal activity
While the enhanced resistance to Saps exhibited by some of the Hst‐5 variants could indicate promise as a therapeutic, ensuring that the modified variants retain their antifungal activity is important. Previous work demonstrated that substitution of a single lysine residue to arginine or leucine at the K5, K11, K13, or K17 sites in Hst‐5 does not diminish the antifungal activity, even if the modifications lead to an increase in proteolysis by Saps.24 Therefore, all of the current Hst‐5 variants were tested for their antifungal activity, even if a modification led to an increase in the susceptibility to proteolysis. To determine the antifungal activity of Hst‐5 and the variants, the peptides were serially diluted and incubated with C. albicans cells for 30 min at 30°C in buffer. The mixtures were then diluted, inoculated into YPD media, and grown overnight to determine the reduction in cell viability due to exposure to the peptides.
As hypothesized during the peptide design, the K11R–K17R modification led to enhanced antifungal activity (Figure 5) with a minimum inhibitory concentration for 50% inhibition of growth (MIC50) of 25 μM, compared to 100 μM for Hst‐5. This improvement is comparable to the enhanced activity of the K11R modified peptide we previously observed.24 The enhanced antifungal activity of the K11R modification was preserved in the double‐substituted K11R–K17R peptide, while gaining proteolytic resistance from the K17R substitution.
Figure 5.
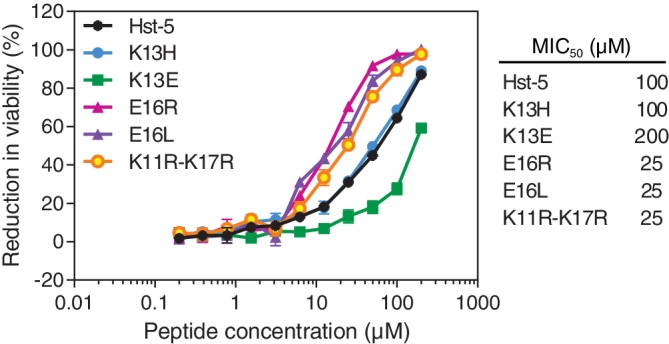
Antifungal activity of Hst‐5 and the modified variants. The peptides were serially diluted and incubated with 2.5 × 107 cells/ml C. albicans. The error bars indicate the standard error of the mean (n = 6), though many are too small to be visible. MIC50 (minimum inhibitory concentration for 50% inhibition of growth) values for the peptides are also provided
Surprisingly, both modifications at the E16 site led to enhanced antifungal activity despite increased susceptibility to degradation by C. albicans (Figure 2d). The proteolysis may not have affected the antifungal segment of the peptides and suggests that for the antifungal activity, a positively charged or noncharged residue is preferred over a negatively charged residue at the E16 site. While removal of the negative charge at E16 enhanced the antifungal activity, addition of a negative charge at the K13 site reduced the activity. The MIC50 of K13E increased to 200 μM, consistent with the decrease in activity observed by Tsai et al.9 The positive charge at the K13 site may be more important than the specific residue at this site, as the K13H modification showed similar antifungal activity to the parent Hst‐5. Our results highlight the complex relationship between peptide charge (and the specific residues present) and the antifungal activity of the peptides. To fully understand the relationship, additional studies that focus on the uptake and mechanism of activity of these peptides are needed.
2.5. K11R–K17R peptide maintain activity after exposure to Saps
Due to two possible regions of Hst‐5 being identified as the antifungal fragment of Hst‐5 (Residues 4–1512 and 11–2413), the sequence of the fragments formed from proteolytic cleavage is insufficient to determine if a mixture of cleavage fragments retains antifungal activity. To determine the antifungal activity of the proteolytic fragments, we conducted the antifungal activity assay on peptides that we first incubated with Saps. The peptides were incubated with Sap9 and Sap2 concentrations (6 μg/ml Sap9 or 0.2 μg/ml Sap2) that were two or four times higher, respectively, than the concentrations used for the gel electrophoresis assay to ensure the effect of proteolysis on antifungal activity could be observed. As expected from the antifungal activity assay with the intact peptides, the K11R–K17R, E16R, and E16L peptides retained the most antifungal activity after exposure to either Sap (Figure 6), while the K13H and K13E modified peptides completely lost their activity. Notably, the Sap‐treated K11R–K17R peptide retained antifungal activity similar to the untreated peptide, which is higher than the untreated parent Hst‐5. The gel electrophoresis analysis of the same samples showed that an ample amount of the intact K11R–K17R remained (76% with Sap9 and 82% with Sap2) even after incubation with the higher Sap concentrations (Figure S4). Based on these results, the antifungal activity of K11R–K17R is likely primarily from the intact peptide rather than degradation fragments. Furthermore, a greater amount of the activity is preserved in the Sap‐treated K11R–K17R peptide compared to the Sap‐treated K11R and K17R single substitutions reported in our previous work,24 demonstrating the potential significance of incorporating proteolytic resistance in therapeutic peptide design.
Figure 6.
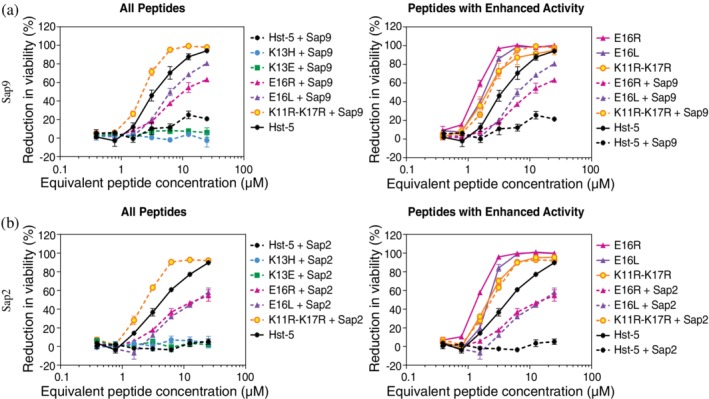
Antifungal activity of the peptides after incubation with or without (a) Sap9 or (b) Sap2. The peptides (50 μM) were incubated with Sap9 (6 μg/ml), Sap2 (0.2 μg/ml), or 1 mM NaPB buffer. The peptides were then serially diluted and incubated with 2.5 × 105 cells/ml C. albicans prior to analyzing viability. For each Sap, the left plot shows activity of Hst‐5 and all variants after exposure to Saps. The right plot compares the activity of Hst‐5 and peptides that showed enhanced activity (E16R, E16L, K11R–K17R) before and after incubation with Saps. The error bars indicate the standard error of the mean (n = 4)
Unlike the K11R–K17R peptide, the antifungal activity of the E16R and E16L variants following Sap incubation is likely from the proteolytic fragments. The E16‐substituted peptides were completely degraded at the higher concentrations of the Saps (Figure S4) and yet still retained a high level of antifungal activity. Interestingly, the fragments formed by incubation of the E16L variant with Sap9 have greater antifungal activity than the fragments of the E16R peptide (Figure 6), which could indicate that the E16L sample had more active fragments remaining after exposure to Sap9 or that the E16L substitution itself provides an advantage in the antifungal activity of the fragments formed. This assay demonstrates that residue substitutions can improve the antifungal activity of a peptide by enhancing the proteolytic resistance or by producing proteolytic fragments that maintain a higher level of activity.
2.6. Modified peptides do not exhibit increased cytotoxicity to mammalian cells
To investigate cytotoxicity of the variants toward human cells, the parent Hst‐5 and modified peptides were incubated with HEK293T cells. The percent of nonviable cells was quantified using propidium iodide (PI) staining and flow cytometry. PI can only enter cells when the cellular membrane is destabilized, indicating a damaged, non‐viable cell. The percent of PI‐positive cells for each Hst‐5 variant was compared to that of a buffer control, and no significant differences were observed (Table 2). Additionally, the percent of PI‐positive cells for each modified peptide was compared to that of Hst‐5 and the buffer control and no significant differences were observed (Table 2), suggesting that the modifications do not lead to toxicity of the Hst‐5 variants.
Table 2.
Percent nonviable (PI‐positive) HEK 293T cells after incubation with peptides
| Sample | PI‐positive cells (%)a |
|---|---|
| 1 mM NaPB | 5.11 ± 1.38 |
| Hst‐5 | 7.22 ± 1.03 |
| K13H | 4.69 ± 1.08 |
| K13E | 6.33 ± 1.30 |
| E16R | 10.74 ± 3.65 |
| E16L | 5.25 ± 0.76 |
| K11R–K17R | 4.63 ± 0.83 |
No statistically significant difference was found between any of the peptide variants and Hst‐5 or the buffer (1 mM NaPB) control.
3. DISCUSSION
This work explored the effects of amino acid substitutions on Hst‐5, with a focus on the additive effect of combining two individually beneficial modifications in the same variant and on the addition or removal of a negatively charged residue within the antifungal fragment. We evaluated the impact of the residue substitutions on the susceptibility and kinetics of Hst‐5 to proteolysis by Saps and C. albicans and the antifungal activity of the peptide. By enhancing resistance of the peptide to proteolysis, we aimed to create a peptide with prolonged antifungal activity against C. albicans.
The negatively charged E16 residue affects the peptide's resistance to proteolysis and its antifungal activity. The E16R substitution displayed a Sap‐dependent response, with the E16R modification leading to less proteolysis by Sap2 but increased proteolysis by Sap9 (Figure 2a,b). The increased degradation of E16R by Sap9 agrees with the previously identified preference of Sap9 for an arginine N‐terminal to lysine,22 since the E16R substitution produces an R‐K residue pair. The E16L substitution, meanwhile, led to an increase in proteolysis by Sap2, Sap9, and C. albicans cells. While the substitution of E16 had mixed effects on the proteolysis of Hst‐5, both modifications of the E16 residue had a profound impact on the antifungal activity. The substitutions not only enhanced the activity of the intact peptide (Figure 5) but also led to a higher level of activity retained after exposure to Saps (Figure 6). The retention of the activity likely comes from fragments containing the E16R or E16L residue, implicating the residue in the activity of the peptide. Work by Helmerhorst et al. bolsters this theory, as they enhanced the activity of modified Hst‐5 fragments dhvar1 and dhvar2 by substituting the glutamic acid within the fragments with lysine.15 Driscoll et al., on the other hand, found that an E16G substitution leads to reduced activity.8 The improved activity with arginine, leucine, and lysine substitutions but worsened activity with a glycine substitution suggests that identity of the residue at the E16 site and the presence of a side chain plays an important role for the antifungal activity of the peptide. Future work could be aimed at determining if that role is during the uptake of the peptide into the cell or in the mechanism of the antifungal activity.
Unlike the E16 residue, substitutions at the K13 residue did not lead to improvement in proteolytic resistance or antifungal activity. The K13E and K13H modifications did not significantly affect interactions with Sap2 (Figure 2b), but they made the peptides significantly more susceptible to Sap9 (Figure 2a). The increased degradation of K13E by Sap9 may be due to the presence of the negatively charged glutamic acid residue on the C‐terminal side of the R12 residue. Schild et al. previously reported enrichment of cleavage in peptides with an acidic amino acid C‐terminal to an arginine or lysine when exposed to Sap9.22 In terms of antifungal activity, the K13H substitution had no impact, but the K13E substitution led to a significant decrease in the activity (Figure 5), which was also observed by Tsai et al. for the same modification.9 The preservation of the antifungal activity by the K13H peptide at a comparable level to Hst‐5 supports the importance of the positive charge at this site, as suggested by Tsai et al.9 However, we previously observed reduced activity with K13R substitution,24 so the positive charge alone is not sufficient, and the identity of the residue at this site is also important.
In addition to single amino acid substitutions, we also investigated the additive effect of combining individual modification that we previously showed lead to more resistance to Saps (K17R) or enhanced antifungal activity (K11R).24 Previous studies have combined amino acid modifications to study the effect on antifungal activity of Hst‐5 or Hst‐5 fragments,9, 13 but no other work has attempted to combine enhancements in proteolytic resistance and antifungal activity. The double arginine‐modified K11R–K17R variant significantly enhanced resistance to proteolysis by both Saps and C. albicans (Figure 2) and showed almost no degradation around the R17 modified residue, as expected based on our previous study with the K17R modification.24 The antifungal activity of the intact peptide was similar to the previously tested K11R variant,24 which was higher than the parent Hst‐5 and the K17R variant. The improvements observed with the K11R–K17R peptide show that it is possible to combine enhancements from individual modifications into a single variant. Importantly, the results illustrate that considering multiple therapeutically relevant properties concurrently is beneficial in peptide design; probing residue modifications based on only the proteolytic resistance or only the activity would have missed the discovery of the more robust and active K11R–K17R peptide. The increased resistance to proteolysis and the improved activity of K11R–K17R, along with its minimal toxicity to HEK293T cells, show the promising potential for this peptide as a long‐lasting therapeutic peptide. Future work, however, is required to evaluate the resistance of the modified peptide to human proteases and, if needed, further design the peptide to enhance its resistance to the human proteases.
The kinetic study on the interaction of Sap9 with Hst‐5, E16L, and K11R–K17R revealed that the residue substitutions do indeed affect the kinetics of proteolysis (Figure 4). While a kinetic degradation study on Hst‐5 has been done with Saps17 and with whole saliva,25 no previous work explored the effect of amino acid substitutions on the kinetics. The initial degradation of Hst‐5 and the E16L variant led to rapid accumulation of the degradation fragment with Residues 1–17, suggesting that the first cleavage of the peptides occurs C‐terminal to the K17 residue. Bochenska et al. observed a similar initial rapid accumulation of the same Hst‐5 fragment during incubation with Sap9.17 However, they recorded a decrease of the fragment after 30 min,17 whereas the fragment accumulated throughout the incubation period in the current study. The difference in degradation is likely due to the enzyme:substrate ratio, buffer, and pH differences between the studies. For the E16L variant, the Residues 1–17 fragment reached a steady state level (Figure 4c) after 120 min of degradation, while the abundance of the same fragment continued to increase in Hst‐5. Furthermore, the Residues 13–24 fragment of E16L initially accumulated before decreasing in abundance, while the analogous fragment in Hst‐5 and K11R–K17R continued to accumulate. Based on these data, the residue substitutions incorporated in the variants of Hst‐5 modify not only the degradation kinetics of the intact peptide but also the degradation kinetics of the cleavage fragments. Evaluating the degradation kinetics of both intact peptide and the resulting degradation fragments is necessary to fully understand the effect of residue modification on proteolysis of Hst‐5 and other peptides.
The modifications to Hst‐5 that we studied show a clear advantage in antifungal activity against C. albicans compared to native Hst‐5. We explored whether our modifications are among the natural variations reported in the literature for the encoding gene in humans. Hst‐5 is generated as a proteolytic product of histatin 3 (Hst‐3), which is encoded by HST2.3, 26 Multiple alleles for HST2 have been identified in people of African descent,27, 28, 29 and one of the same alleles is also present in the Japanese population.30 However, the modifications we studied would not be encoded by these alleles; in fact, only one of the alleles (HST2, identified in people of African descent) would result in an amino acid mutation (R22Q) in the corresponding Hst‐5 peptide.27 Thus, our beneficial modifications to Hst‐5 have not been “discovered” by nature through evolution. A likely explanation for this is that our modifications could reduce other beneficial functions of Hst‐5 or its precursor Hst‐3. For example, Hst‐5 has activity against other fungal pathogens, including other Candida species and the fungus Aspergillus fumigatus,31 and a number of bacterial pathogens, including Streptococcus gordonii, Enterococcus faecium, Staphylococcus aureus, Acinetobacter baumanni, Pseudomonas aeruginosa, and Enterobacter species).32, 33, 34 Additionally, Hst‐3 (the precursor of Hst‐5) stimulates wound healing.35 The selective pressure related to antifungal activity of Hst‐5 against C. albicans may not have been strong enough to overcome the selective pressures related to these other functions. Our design focused specifically on protease resistance and improvement in activity against C. albicans, properties which will increase the potential for success in our application of the peptide as an antifungal agent targeting C. albicans. We showed with our K11R–K17R variant that this focused set of criteria can successfully be used to rationally design an improved peptide antifungal agent that nature's constraints did not evolve.
4. CONCLUSIONS
Our results demonstrate the importance of probing the effect of multiple properties of interest when designing peptides for improved therapeutic potential. We focused on proteolytic stability and antifungal activity and showed that modifications have varying effects on antifungal activity and overall proteolytic resistance. Furthermore, modifications that alter proteolysis affect not only the degradation of the intact peptide but its fragments as well. By considering both antifungal activity and proteolytic resistance, we successfully designed the K11R–K17R variant of Hst‐5 that offers improvements in both properties. The approaches and considerations used in this study are not limited to Hst‐5 and C. albicans and will also be beneficial in engineering other antimicrobial peptides to enhance their potential as alternative therapeutics.
5. MATERIALS AND METHODS
5.1. Peptides and enzymes
Hst‐5 and its variants were synthesized by Genscript (purity ≥95% with trifluoroacetic acid salt removal to hydrochloride). Recombinant Sap2 and Sap9 (without its GPI anchor) were expressed in Pichia pastoris and purified as described previously.18, 22, 36
5.2. Degradation of peptides by Saps and C. albicans
The extent of proteolysis of the peptides by purified Saps was evaluated through gel electrophoresis. Hst‐5 and its variants (20 μl) were mixed with Saps (20 μl) in 0.5 mL microcentrifuge tubes to final concentrations of 50 μM peptide and 3 μg/ml Sap9 or 0.05 μg/ml Sap2 in 1 mM sodium phosphate buffer (NaPB). As controls, the peptides were mixed with NaPB. Following incubation for 2 hr at 37°C, the samples were mixed with tricine sample buffer (200 mM Tris‐HCl, pH 6.8, 40% glycerol, 2% SDS, without Coomassie Blue G‐250) containing 2% β‐mercaptoethanol and heated at 100°C for 5 min to inactivate the Saps. To separate the intact peptide and its degradation products, the samples were run on 16.5% Tris‐tricine gels (Bio‐Rad) at 60 V. The gels were fixed for 30 min in a 10% acetic acid/40% methanol/50% water fixing solution, stained with Bio‐Safe Coomassie stain (Bio‐Rad) for 1 hr, and washed with fresh water four times. The stained gels were imaged on a ChemiDoc MP imager (Bio‐Rad). Image Lab software (Bio‐Rad) was used for densitometric analysis, where the upper band in each lane was analyzed as non‐degraded peptide and the lower band was analyzed as the degraded products. Degradation reactions were performed on three different days with two replicates within each day, for a total of n = 6.
For the study of proteolysis by C. albicans, the supernatant of the cell culture and the cells were tested separately. For the culture supernatant, a colony inoculation of ATCC 90028 strain (American Type Culture Collection) was subcultured to optical density (OD600) of 0.1 in YPD medium (10 g/L yeast extract, 20 g/L peptone, 20 g/L dextrose) and then grown at 30°C for 17.5 hr. The supernatant was collected, and the buffer was exchanged to 2 mM NaPB using a 10 kDa molecular weight cutoff column (GE). Equal volumes of supernatant and peptides (100 μM) were mixed, incubated for 2 hr at 37°C and boiled for 10 min at 100°C with tricine sample buffer. As controls, the peptides were mixed with NaPB only. For analysis of degradation by cells, a subculture of an overnight culture of C. albicans was grown to an OD600 of 1–1.2. Cells were harvested and washed three times in 100 mM NaPB, which has a high ionic strength to prevent internalization of the peptides into the cells.37 Washed cells were resuspended in the buffer and mixed with an equal volume of each peptide to final concentrations of 1 × 109 cells/mL C. albicans and 50 μM peptide. A control containing each peptide with 100 mM NaPB was included. After incubation for 2 hr at 37°C, cells were removed by centrifugation, and the supernatant was heated at 100°C for 10 min after addition of tricine sample buffer, as done with the culture supernatant samples. Following the protocol described above for degradation by Saps, samples were run on Tris‐tricine gels, stained, and analyzed. Three replicates of culture supernatant reactions were performed (n = 3), and the cell degradation reactions were performed on three different days with two replicates within each day (n = 6).
One‐way ANOVA tests with α < .05 and Dunnett's multiple comparison tests (Hst‐5 sample as the control) were performed for statistical analysis. The number of asterisks indicates the level of statistical significance in figures: * for p < .05, ** for p < .01, *** for p < .001, and **** for p < .0001.
5.3. Capillary electrophoresis‐mass spectrometry
CEMS was used to determine and quantify the fragments formed from cleavage by Saps. Once Saps (3 μg/ml Sap9 or 0.05 μg/ml Sap2) were heat inactivated after incubation with 50 μM peptides, 25 μl of each sample was desalted with a C‐18 TopTip micro‐spin column (Glygen Corp.) following the manufacturer's protocol. Solutions of 0.1% formic acid and 0.1% formic acid/60% acetonitrile (ACN) were used for binding and eluting, respectively. To each elution mixture, 1 μl of the peptide MRFA (0.1 mg/ml) was added as an internal standard. The samples were further diluted with 0.1% formic acid before CEMS analysis. CE separation of the degradation fragments was carried out at 30 kV with an ECE‐001 capillary electrophoresis system (CMP Scientific, Brooklyn, NY) using an etched capillary with PS1 neutral coating. A CMP Scientific EMASS‐II CE‐MS ion source was used to couple CE with the mass spectrometer inlet. A Thermo Scientific Orbitrap Fusion Lumos Tribrid mass spectrometer was used to acquire full mass spectra in the range of 200–1,450 m/z with resolution R = 120,000 (200 m/z). EThcD spectra at R = 120,000 (200 m/z) with a 3 s cycle time were recorded to identify fragments from degradation. The EThcD spectra of each sample were processed with Proteome Discoverer (V2.1) with the Prosight PD node to identify the degradation products. Full scan mass spectra were deconvoluted, and the intensities of the intact peptide, fragments, and internal standard MRFA were analyzed with the Thermo Xcalibur 3.0 software.
5.4. Kinetics of proteolysis
To gain an understanding of the kinetics of the interaction between the peptides and Saps, a time‐course study on the proteolysis of Hst‐5 and its variants by Sap9 was performed. As described for the proteolytic study with gel electrophoresis, a series of peptide and Sap9 mixtures with final concentrations of 50 μM peptide and 3 μg/ml Sap9 were prepared. Samples were incubated for 5, 30, or 120 min at 37°C and then heated at 100°C for 5 min to inactivate Sap9. For the 0 min time point, Sap9 was first heat inactivated and then added to the peptides. The samples were desalted with a C‐18 TopTip micro‐spin column and analyzed by CEMS as described above.
5.5. Antifungal activity assay with intact peptide and fragments
The antifungal activity of intact parent Hst‐5 and its variants was evaluated using an optical density‐based candidacidal assay. An overnight culture of C. albicans was subcultured and grown at 30°C in YPD media to an OD600 of 1–1.2. Cells were harvested, washed three times with 2 mM NaPB, and diluted to 5 × 107 cells/mL. Each of the peptides was serially diluted (0–400 μM) in water. Equal volumes (20 μl) of diluted cells and diluted peptide were then mixed in a round‐bottom 96‐well culture plate. As a control, an equal volume of water was mixed with the diluted cells. After 30 min of incubation at 30°C, 320 μl of 1 mM NaPB was added to stop additional interaction of peptides and cells.38 Samples were further diluted and approximately 250 cells were inoculated into an equal mixture of YPD (100 μl) and 1 mM NaPB (100 μl) in round‐bottom culture plates. The cells were then grown overnight at 30°C on a microplate shaker at 350 rpm. Wells with only YPD and NaPB provided the background signal. After overnight incubation, cells in each well were resuspended, and the OD600 was measured. The reduction in viability was calculated as
The activity assays were performed on three different days with two replicates within each day for a total of n = 6.
To determine the antifungal activity of the peptides after exposure to Saps, 50 μM of peptide was incubated with either 6 μg/ml Sap9 or 0.2 μg/ml Sap2 for 2 hr at 37°C. After the incubation, the Saps were inactivated by heating the samples at 100°C for 5 min. As controls, the peptides were mixed with NaPB. The antifungal activity of Sap‐incubated samples was assessed following the described protocol for intact peptide using a C. albicans concentration of 5 × 105 cells/ml. Degraded peptide samples were prepared on two different days for each of the Saps. The antifungal activity assays were performed on two different days for each Sap sample with two replicates within the same day for a total of n = 4.
5.6. Cytotoxicity of peptides to mammalian cells
Cytotoxicity of the peptides was assessed using PI staining with HEK293T cells (American Type Culture Collection). Cells were grown in 12‐well tissue culture plates to 90–100% confluency in Dulbecco's Modified Eagle Medium (DMEM) with high glucose (ThermoFisher) that was supplemented with 10% fetal bovine serum (FBS, ThermoFisher) and 1% penicillin–streptomycin (10,000 U/ml, ThermoFisher). The cell culture medium was replaced with serum‐free medium (DMEM without FBS supplementation) 16 hr prior to peptide incubation. Cells were rinsed with phosphate‐buffered saline (PBS) and incubated with 1 ml of peptide solution (200 μM in 1 mM NaPB) for 30 min at 37°C. Controls of 1 mM NaPB and ethanol were included. A solution of 250 μl of 0.25% trypsin‐ethylenediaminetetraacetic acid (Invitrogen) was used to dissociate cells from the tissue culture plate. Cells were collected by centrifugation and resuspended in 250 μl of PBS. Immediately before loading samples onto the flow cytometer (BD FACS Canto II Cell Analyzer, BD Biosciences), 1 μl of 1 mg/ml PI was added. Analysis was performed in FlowJo (FlowJo, LLC) by establishing PI‐positive and PI‐negative gating between the viable and non‐viable population peaks on the PI histogram for the ethanol control. The cytotoxicity assay was performed seven different times across four different days for n ≥ 7. One‐way ANOVA tests with α < .05 and Tukey multiple comparison tests were performed for statistical analysis.
Supporting information
Figure S1: Representative Coomassie‐stained gels showing proteolysis.
Figure S2: Representative electrophoretogram for CEMS of Hst‐5.
Figure S3: Low‐signal peptide fragments resulting from degradation.
Figure S4: Extent of proteolysis of with higher concentrations of Saps
ACKNOWLEDGMENTS
We thank Dr. James Xia and CMP Scientific Corp for access to their CE system and CE‐MS ion source. This work was supported by a National Institutes of Health training grant in Host‐Pathogen Interactions (T32AI089621B), a National Institutes of Health shared instrumentation grant (1S10OD019938), a Burroughs Wellcome Career Award at the Scientific Interface (to KMS), the National Science Foundation (CBET 1511718; to AJK), the Fischell Department of Bioengineering, and the University of Maryland.
Ikonomova SP, Moghaddam‐Taaheri P, Wang Y, et al. Effects of histatin 5 modifications on antifungal activity and kinetics of proteolysis. Protein Science. 2020;29:480–493. 10.1002/pro.3767
Funding information Burroughs Wellcome Fund, Grant/Award Number: Career Award at the Scientific Interface; Division of Chemical, Bioengineering, Environmental, and Transport Systems, Grant/Award Number: 1511718; National Institute of Allergy and Infectious Diseases, Grant/Award Number: T32AI089621B; NIH Office of the Director, Grant/Award Number: 1S10OD019938; University of Maryland
REFERENCES
- 1. Peters BM, Zhu J, Fidel PL Jr, et al. Protection of the oral mucosa by salivary histatin‐5 against Candida albicans in an ex vivo murine model of oral infection. FEMS Yeast Res. 2010;10:597–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hofs S, Mogavero S, Hube B. Interaction of Candida albicans with host cells: Virulence factors, host defense, escape strategies, and the microbiota. J Microbiol. 2016;54:149–169. [DOI] [PubMed] [Google Scholar]
- 3. Oppenheim FG, Xu T, McMillian FM, et al. Histatins, a novel family of histidine‐rich proteins in human parotid secretion. Isolation, characterization, primary structure, and fungistatic effects on Candida albicans . J Biol Chem. 1988;263:7472–7477. [PubMed] [Google Scholar]
- 4. Xu T, Levitz SM, Diamond RD, Oppenheim FG. Anticandidal activity of major human salivary histatins. Infect Immun. 1991;59:2549–2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Khan SA, Fidel PL, Thunayyan AA, Varlotta S, Meiller TF, Jabra‐Rizk MA. Impaired histatin‐5 levels and salivary antimicrobial activity against C. albicans in HIV infected individuals. J AIDS Clin Res. 2013;4:193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Puri S, Edgerton M. How does it kill?: Understanding the candidacidal mechanism of salivary histatin 5. Eukaryot Cell. 2014;13:958–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Baev D, Li XS, Dong J, Keng P, Edgerton M. Human salivary histatin 5 causes disordered volume regulation and cell cycle arrest in Candida albicans . Infect Immun. 2002;70:4777–4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Driscoll J, Duan C, Zuo Y, Xu T, Troxler R, Oppenheim FG. Candidacidal activity of human salivary histatin recombinant variants produced by site‐directed mutagenesis. Gene. 1996;177:29–34. [DOI] [PubMed] [Google Scholar]
- 9. Tsai H, Raj PA, Bobek LA. Candidacidal activity of recombinant human salivary histatin‐5 and variants. Infect Immun. 1996;64:5000–5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tsai H, Bobek LA. Studies of the mechanism of human salivary histatin‐5 candidacidal activity with histatin‐5 variants and azole‐sensitive and ‐resistant Candida species. Antimicrob Agents Chemother. 1997;41:2224–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Situ H, Balasubramanian SV, Bobek LA. Role of α‐helical conformation of histatin‐5 in candidacidal activity examined by proline variants. Biochim Biophys Acta Gen Subj. 2000;1475:377–382. [DOI] [PubMed] [Google Scholar]
- 12. Raj PA, Edgerton M, Levine MJ. Salivary histatin 5: Dependence of sequence, chain length, and helical conformation for candidacidal activity. J Biol Chem. 1990;265:3898–3905. [PubMed] [Google Scholar]
- 13. Rothstein DM, Spacciapoli P, Tran LT, et al. Anticandida activity is retained in P‐113, a 12‐amino‐acid fragment of histatin 5. Antimicrob Agents Chemother. 2001;45:1367–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Helmerhorst EJ, van't Hof W, Veerman EC, Simoons‐Smit I, Nieuw Amerongen AV. Synthetic histatin analogues with broad‐spectrum antimicrobial activity. Biochem J. 1997;326(1):39–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Helmerhorst EJ, Reijnders IM, van't Hof W, Veerman EC, Nieuw Amerongen AV. A critical comparison of the hemolytic and fungicidal activities of cationic antimicrobial peptides. FEBS Lett. 1999;449:105–110. [DOI] [PubMed] [Google Scholar]
- 16. Meiller TF, Hube B, Schild L, et al. A novel immune evasion strategy of Candida albicans: Proteolytic cleavage of a salivary antimicrobial peptide. PLoS One. 2009;4:e5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bochenska O, Rapala‐Kozik M, Wolak N, Aoki W, Ueda M, Kozik A. The action of ten secreted aspartic proteases of pathogenic yeast Candida albicans on major human salivary antimicrobial peptide, histatin 5. Acta Biochim Pol. 2016;63:403–410. [DOI] [PubMed] [Google Scholar]
- 18. Albrecht A, Felk A, Pichova I, et al. Glycosylphosphatidylinositol‐anchored proteases of Candida albicans target proteins necessary for both cellular processes and host‐pathogen interactions. J Biol Chem. 2006;281:688–694. [DOI] [PubMed] [Google Scholar]
- 19. Bochenska O, Rapala‐Kozik M, Wolak N, et al. Inactivation of human kininogen‐derived antimicrobial peptides by secreted aspartic proteases produced by the pathogenic yeast Candida albicans . Biol Chem. 2015;396:1369–1375. [DOI] [PubMed] [Google Scholar]
- 20. Koelsch G, Tang J, Loy JA, et al. Enzymic characteristics of secreted aspartic proteases of Candida albicans . Biochim Biophys Acta Prot Struct Mol Enzymol. 2000;1480:117–131. [DOI] [PubMed] [Google Scholar]
- 21. Aoki W, Kitahara N, Miura N, et al. Comprehensive characterization of secreted aspartic proteases encoded by a virulence gene family in Candida albicans . J Biochem. 2011;150:431–438. [DOI] [PubMed] [Google Scholar]
- 22. Schild L, Heyken A, de Groot PW, et al. Proteolytic cleavage of covalently linked cell wall proteins by Candida albicans Sap9 and Sap10. Eukaryot Cell. 2011;10:98–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Laskay ÜA, Srzentić K, Monod M, Tsybin YO. Extended bottom‐up proteomics with secreted aspartic protease Sap9. J Proteomics. 2014;110:20–31. [DOI] [PubMed] [Google Scholar]
- 24. Ikonomova SP, Moghaddam‐Taaheri P, Jabra‐Rizk MA, Wang Y, Karlsson AJ. Engineering improved variants of the antifungal peptide histatin 5 with reduced susceptibility to Candida albicans secreted aspartic proteases and enhanced antimicrobial potency. FEBS J. 2018;285:146–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sun X, Salih E, Oppenheim FG, Helmerhorst EJ. Kinetics of histatin proteolysis in whole saliva and the effect on bioactive domains with metal‐binding, antifungal, and wound‐healing properties. FASEB J. 2009;23:2691–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sabatini LM, Azen EA. Histatins, a family of salivary histidine‐rich proteins, are encoded by at least two loci (HIS1 and HIS2). Biochem Biophys Res Commun. 1989;160:495–502. [DOI] [PubMed] [Google Scholar]
- 27. Araki M, Anstey NM, Mwaikambo ED, Dua A, Amberger E, Azen EA. An expanded histatin gene polymorphism and test of a possible disease resistant phenotype. Hum Mutat. 1997;10:58–64. [DOI] [PubMed] [Google Scholar]
- 28. Azen EA. Genetic polymorphism of basic proteins from parotid saliva. Science. 1972;176:673–674. [DOI] [PubMed] [Google Scholar]
- 29. Sabatini LM, Azen EA. Two coding change mutations in the HIS2 2 allele characterize the salivary histatin 3‐2 protein variant. Hum Mutat. 1994;4:12–19. [DOI] [PubMed] [Google Scholar]
- 30. Fujigaki Y, Imamura Y, Oomori Y, Ouryouji K, Miyazawa H, Wang PL. Polymorphism of salivary histatin gene and periodontal disease in the Japanese population. J Int Acad Periodontol. 2009;11:220–225. [PubMed] [Google Scholar]
- 31. Helmerhorst EJ, Reijnders IM, van't Hof W, Simoons‐Smit I, Veerman EC, Amerongen AV. Amphotericin B‐ and fluconazole‐resistant Candida spp., Aspergillus fumigatus, and other newly emerging pathogenic fungi are susceptible to basic antifungal peptides. Antimicrob Agents Chemother. 1999;43:702–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Du H, Puri S, McCall A, Norris HL, Russo T, Edgerton M. Human salivary protein histatin 5 has potent bactericidal activity against ESKAPE pathogens. Front Cell Infect Microbiol. 2017;7:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Andrian E, Qi G, Wang J, Halperin SA, Lee SF. Role of surface proteins SspA and SspB of Streptococcus gordonii in innate immunity. Microbiology. 2012;158:2099–2106. [DOI] [PubMed] [Google Scholar]
- 34. Rydengard V, Andersson Nordahl E, Schmidtchen A. Zinc potentiates the antibacterial effects of histidine‐rich peptides against Enterococcus faecalis . FEBS J. 2006;273:2399–2406. [DOI] [PubMed] [Google Scholar]
- 35. Oudhoff MJ, Bolscher JG, Nazmi K, et al. Histatins are the major wound‐closure stimulating factors in human saliva as identified in a cell culture assay. FASEB J. 2008;22:3805–3812. [DOI] [PubMed] [Google Scholar]
- 36. Borg‐von Zepelin M, Beggah S, Boggian K, Sanglard D, Monod M. The expression of the secreted aspartyl proteinases Sap4 to Sap6 from Candida albicans in murine macrophages. Mol Microbiol. 1998;28:543–554. [DOI] [PubMed] [Google Scholar]
- 37. Jang WS, Li XS, Sun JN, Edgerton M. The P‐113 fragment of histatin 5 requires a specific peptide sequence for intracellular translocation in Candida albicans, which is independent of cell wall binding. Antimicrob Agents Chemother. 2008;52:497–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li XS, Reddy MS, Baev D, Edgerton M. Candida albicans Ssa1/2p is the cell envelope binding protein for human salivary histatin 5. J Biol Chem. 2003;278:28553–28561. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Representative Coomassie‐stained gels showing proteolysis.
Figure S2: Representative electrophoretogram for CEMS of Hst‐5.
Figure S3: Low‐signal peptide fragments resulting from degradation.
Figure S4: Extent of proteolysis of with higher concentrations of Saps


