Abstract
The objective of the study was to identify the mechanism of action for a radiation mitigator of the gastrointestinal (GI) acute radiation syndrome (ARS), identified in an unbiased high‐throughput screen. We used mice irradiated with a lethal dose of radiation and treated with daily injections of the radiation mitigator 1‐[(4‐nitrophenyl)sulfonyl]‐4‐phenylpiperazine to study its effects on key pathways involved in intestinal stem cell (ISC) maintenance. RNASeq, quantitative reverse transcriptase‐polymerase chain reaction, and immunohistochemistry were performed to identify pathways engaged after drug treatment. Target validation was performed with competition assays, reporter cells, and in silico docking. 1‐[(4‐Nitrophenyl)sulfonyl]‐4‐phenylpiperazine activates Hedgehog signaling by binding to the transmembrane domain of Smoothened, thereby expanding the ISC pool, increasing the number of regenerating crypts and preventing the GI‐ARS. We conclude that Smoothened is a target for radiation mitigation in the small intestine that could be explored for use in radiation accidents as well as to mitigate normal tissue toxicity during and after radiotherapy of the abdomen.
Keywords: acute radiation syndrome, developmental signaling, intestinal stem cells, radiation
The intestinal stem cell niche responds to radiation with apoptosis of intestinal stem cells and subsequent loss of tissue integrity. Treatment with compound #5 activates Hedgehog signaling, expands the number of intestinal stem cells and regenerating crypts, and allows for survival of the animals after a lethal dose of radiation.
1.
Significance statement.
Exposure to lethal doses of ionizing radiation manifests in a symptom complex that is summarized as acute radiation syndrome (ARS) and the extent of radiation damage to the hematopoietic and gastrointestinal (GI) systems codetermine the acute survival of the exposed individual. So far, no person who experienced a full‐scale GI‐ARS has ever survived. This creates a—so far—mostly unmet need to develop countermeasures that mitigate the effects of lethal radiation doses when given after radiation exposure. This study presents that a piperazine compound prevents GI‐ARS in mice when given 24 hours after a lethal dose of radiation and uncovers the underlying mechanism of action, thus providing a basis for a future rational development of effective radiation mitigators.
2. INTRODUCTION
Exposure to even moderate doses of radiation expected during nuclear accidents, after deployment of “dirty bombs” or attacks with nuclear weapons has severe effects on normal tissue compartments that will lead to the development of a set of symptoms summarized as acute radiation syndrome (ARS).1 In particular, damage to the hematopoietic system and the gut after total body radiation (TBI) exposure is often lethal and difficult to control even under optimal supportive care.2 The characteristic feature of the gastrointestinal ARS (GI‐ARS) is the rapid loss of the integrity of the epithelial layer lining the intestines. This results in poor resorption of nutrients and fluids in the gut and loss of the intestine's barrier function. Seven to 10 days after radiation exposure, exposed individuals experience dehydration, sepsis, and bleeding, cumulatively leading to death. Previous studies established that in mouse models of GI‐ARS the number of regenerating crypts per circumference at 5 days after irradiation is inversely correlated with the radiation dose and determines if the animals recover.3, 4
Generally, radiation‐induced death of differentiated cells and loss of tissue specific stem cells are the underlying mechanisms for radiation‐induced loss of tissue function. In the small intestines, finger‐like structures called villi form a large epithelial surface responsible for the absorption of nutrients. The epithelial cells of these villi originate from intestinal stem cells (ISCs) found at the base of crypt‐like structures surrounding the villi.5 Rapidly proliferating transiently amplifying cells produce differentiated progeny that migrate toward the tip of the villi in a conveyer belt‐like fashion where they shed into the intestinal lumen and die. In response to irradiation the villi are lost but depending on the radiation dose, the tissue is restored over time by the surviving ISCs able to initiate regenerating crypts.4
In the event of an exposure to radiation during nuclear accidents, or by nuclear explosive devices and “dirty bombs,” treatments are needed to mitigate radiation damage to the hematopoietic system and the gut, in order to prevent ARS.6, 7 Mass‐casualty scenarios come with multiple logistic challenges that make successful distribution of drugs to a large number of victims within the first 24 hours after an incident unlikely.8, 9, 10 Currently, there are very few drugs known to mitigate the effects of radiation when applied 24 hours after radiation exposure or later.7 A recent high‐throughput screen performed by the UCLA Center for Medical Countermeasures against Radiation identified compounds with radiation mitigator properties to the hematopoietic system and the gut.11, 12 Among these compounds, 1‐[(4‐nitrophenyl)sulfonyl]‐4‐phenylpiperazine (compound #5; Figure 1A) most efficiently mitigated hematopoietic and GI‐ARS. However, the underlying mechanisms of action were not investigated12 and are addressed in our present study.
Figure 1.
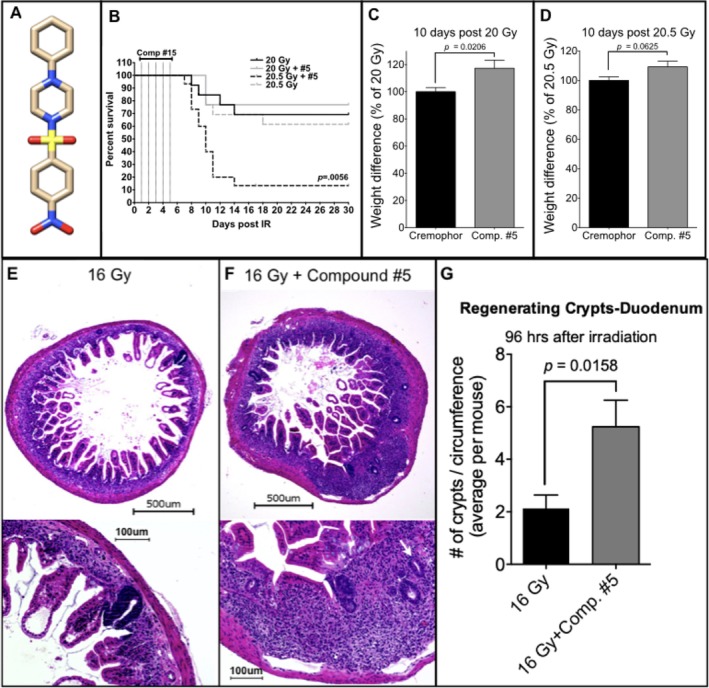
Compound #5 significantly improves survival and mitigates weight loss after a lethal dose of radiation. A, Structure of 1‐[(4‐nitrophenyl)sulfonyl]‐4‐phenylpiperazine/compound #5. B, Female 12‐week‐old C3H mice were abdominally irradiated with 20 Gy (n = 13 per group or n = 26 total) or 20.5 Gy (n = 15 per group or n = 30 total). Twenty‐four hours later, the animals were treated with compound #5 (5 mg/kg) or Cremophor/DMSO subcutaneously, followed by another four daily treatments. Animals treated with 20 Gy did not reach median survival. By day 11, after exposure to irradiation 80% of the mice succumb to a single dose of 20.5 Gy. However, only five daily treatments with compound #5, beginning 24 hours after irradiation exposure, significantly improved survival with only 31% of the mice succumbing by day 11 postirradiation (P = .0056, log‐rank [Mantel‐Cox]) test. C and D, Weights on day 10 after exposure to either 20 Gy (C) or 20.5 Gy (D) total abdominal irradiation were normalized to the animals starting weights and shown as a percent difference from the Cremophor/DMSO‐treated animals. Compound #5 significantly improved weight loss in animals exposed to 20 Gy (P = .0206, Student's t test) and trended similarly, but not significantly, in animals treated with 20.5 Gy (P = .0625, Student's t test). E‐G, Female, 12‐week‐old C3H mice received 16 Gy of total body irradiation, and beginning 24 hours after irradiation, were administered subcutaneous injections of compound #5 (5 mg/kg) or Cremophor/DMSO daily. Ninety‐six hours after irradiation, the duodenums were removed, fixed, embedded in paraffin, and thin cross‐sections were stained with H&E. E and F, The number of regenerating crypts (E, F; white arrows, bottom panels) present in at least five cross‐sections per mouse were counted at high magnification (×20; E, F; bottom panels). Treatment with compound #5 resulted in a significant increase in the average number of regenerating crypts per circumference after exposure to a lethal dose (16 Gy) of total body irradiation (G, n = 8 mice per group, P = .0158, unpaired, two‐tailed, Student's t test). Scale bars in (E/F) upper images 500 μm, lower images 100 μM
Here, we hypothesized that drugs that successfully mitigate radiation damage in the small intestine impact ISC survival and/or expansion. We report that compound #5 modulates Hedgehog signaling in the crypts of the small intestines by binding to Smoothened and thereby leading to ISC compartment expansion, improved intestinal recovery, and survival of lethally irradiated animals.
3. MATERIALS AND METHODS
3.1. Animals
C3Hf/Sed/Kam were originally obtained from the MD Anderson Cancer Center. All mice were rederived, bred, and maintained in a pathogen‐free environment in the American Association of Laboratory Animal Care‐accredited Animal Facilities of Department of Radiation Oncology, University of California (Los Angeles, California) in accordance to all local and national guidelines for the care of animals.
3.2. Irradiation
Twelve‐week‐old C3Hf/Sed/Kam mice were irradiated in groups of five animals using an experimental x‐ray irradiator (Gulmay Medical Inc, Atlanta, Georgia) at a dose rate of 2.8159 Gy/minute for the time required to apply a prescribed dose. Radiation was administered between 11:30 a.m. and 1:30 p.m. to minimize variation of circadian effects on radiation responses.13 Control animals were sham‐irradiated. The x‐ray beam was operated at 300 kV and hardened using a 4 mm Be, a 3 mm Al, and a 1.5 mm Cu filter. For total abdominal irradiation (TAI) the rest of the body was shielded with 10 mm lead blocks.
3.3. In vivo drug administration
Twenty‐four hours after irradiation mice received a single subcutaneous injection of 5 mg/kg of 1‐[(4‐nitrophenyl)sulfonyl]‐4‐phenylpiperazine (compound #5, Vitascreen, Champaign, Illinois) dissolved in 15 μL DMSO (Dimethylsulfoxide) and then suspended in 1 mL of 1% Cremophor EL. Control animals received DMSO/Cremophor EL only. For survival studies mice received five daily doses of the drug spaced by 24‐hour intervals. To assess toxicity, the mice were weighed daily.
3.4. Microcolony stem cell assay
Mice were irradiated with 16 Gy TBI. Twenty‐four hours after irradiation, the animals were treated with compound #5, as described above, and were treated daily until tissue harvesting. The technique of Withers and Elkind4 was used to assay the survival of intestinal crypts 96 hour following TBI. Briefly, the entire small intestine was excised and divided into duodenum, jejunum, or ileum using a ratio of total small intestine length of 1:3:2, respectively. The central 2.5 cm of duodenum and jejunum were fixed in neutral buffered formalin, and 4‐6 segments (~0.5 cm in length) were taken from the fixed regions of each mouse and embedded in paraffin. Tissue sections were cut at a thickness of 4 μm and stained with hematoxylin and eosin. The numbers of regenerating crypts containing at least 10 viable cells were counted microscopically using ×20 magnification on a BZ‐9000 microscope (Keyence, Itasca, Illinois).
3.5. Quantitative reverse transcription PCR
Quantitative reverse transcriptase‐polymerase chain reaction (qRT‐PCR) was performed in the QuantStudio 3 Real‐Time PCR System (Applied Biosystems, Carlsbad, California) using the PowerUp SYBR Green Master Mix (Applied Biosystems). C t for each gene was determined and ΔΔC t was calculated relative to the designated reference sample. Gene expression values were then set equal to 2−ΔΔCt as described by the manufacturer of the kit (Applied Biosystems). PCR primers were synthesized by Invitrogen and the sequences are shown in Table S1. Hypoxanthine Phosphoribosyltransferase, ribosomal Protein Lateral Stalk Subunit P0, and peptidyl‐Prolyl‐Cis‐Trans Isomerase A were used as housekeeping genes.
3.6. RNASeq
Four hours after drug treatment, RNA was extracted from the proximal portion of the duodenum using Trizol. RNASeq analysis was performed by Novogene (Chula Vista, California). Quality and integrity of total RNA was controlled on a Bioanalyzer (Agilent Technologies, Waldbronn, Germany). The RNA sequencing library was generated using NEBNext Ultra RNA Library Prep Kit (New England Biolabs, Ipswich, Massachusetts). The library concentration was quantified using a Qubit 3.0 fluorometer (Life Technologies, Carlsbad, California), and then diluted to 1 ng/μL. Insert size was checked (2100 Bioanalyzer, Agilent Technologies; Waldbronn, Germany) and quantified by qPCR (library molarity >2 nM). The library was sequenced (Illumina NovaSeq6000) with an average of 50 M reads per sample.
Downstream analysis was performed using STAR, HTseq, and Cufflink. Alignments were parsed using Tophat and differential expressions were determined through DESeq2. Reference genome and gene model annotation files were downloaded from genome website browser (University of California, Santa Cruz) directly. Indexes of the reference genome were built using spliced transcripts alignment to a reference (STAR) and paired‐end clean reads were aligned to the reference genome, using STAR (v2.5). HTSeq v0.6.1 was used to count the read numbers mapped of each gene. The Fragments per Kilobase of Transcript per Million Mapped Reads of each gene was calculated based on the length of the gene and reads count mapped to this gene.
Differential expression analysis between was performed using the DESeq2 R package (2_1.6.3). The resulting P‐values were adjusted using the Benjamini and Hochberg's approach for controlling the false discovery rate. Genes with an adjusted P‐value of <.05 were assigned as differentially expressed. Gene set enrichment analysis of differentially expressed genes was performed using the Broad Institute's Molecular Signature Database (MSigDB).14, 15 Gene sets with corrected P‐values less than .05 were considered significantly enriched by differential expressed genes.
3.7. Immunohistochemistry
Cross‐sections of the small intestines were fixed in formalin, embedded in paraffin, and 4 μm sections were stained for Ki67, β‐catenin, and the intracellular domain of Notch (Notch‐ICD; Translational Pathology Core Laboratory at UCLA). Briefly, slides were placed in xylenes to remove paraffin, then a series of ethanol, washed in tap water, and incubated in 3% hydrogen peroxide/methanol for 10 minutes. After a wash in distilled water, the slides were incubated for 25 minutes in citrate buffer, pH 6 at 95°C. The slides rinsed in phosphate buffered saline containing 0.05% tween‐20 (PBST), and then incubated with a rabbit anti‐mouse Ki67 antibody (Cell Signaling, Danvers, MA, #12202; 1:200) or rabbit anti‐β‐catenin antibody (Cell Signaling, #9562; 1:50) at RT for 1 hour and with a rabbit anti‐Notch1‐ICD antibody (Abcam, #ab8925, 1:50, Cambridge, UK) overnight at 4°C. The slides were rinsed with PBST, and were incubated with secondary polymer at RT for 30 minutes; the polymers used were Dako EnVision + System‐HRP Labeled Polymer anti‐rabbit (Dako, Carpinteria, CA, #K4003) for Ki67 or MACH3 rabbit HRP polymer (Biocare Medical, Concord, California) for β‐catenin. Slides were rinsed with PBST and incubated with 3,3′‐diaminobenzidine, washed in tap water, counterstained with Harris' hematoxylin, dehydrated in ethanol, and mounted with media. Slides were visualized using a BZ‐9000 fluorescence microscope (Keyence, Osaka, Japan) at ×60 magnification. The numbers of Ki67+ nuclei were counted per region of interest for a total of 8‐10 images per animal. Slides were stained for YAP (Yes‐associated Protein)/TAZ using a rabbit anti‐human antibody (Cell Signaling, #8418; 1:200) and secondary anti‐rabbit FITC‐conjugated secondary antibody, the nuclei counterstained with DAPI and analyzed using a confocal microscope.
3.8. Enteroid isolation
Intestinal enteroids were prepared as previously described.16 Briefly, mice were sacrificed and the small intestine was harvested, flushed with phosphate buffered saline (PBS) and opened longitudinally. The villi were then scraped off using an ice‐cold glass slide, washed with PBS (×3), chopped into small segments, dropped in a 50 mL tube containing 20 mL of ice‐cold PBS with Pen/Strep and washed by pipetting up and down. After segments were allowed to settle by gravity, the supernatant was discarded and the procedure was repeated approximately 15‐20 more times until the solution was clear. The intestinal segments were resuspended in PBS with 5 mM EDTA and rocked at 20 rpm for 15 minutes at room temperature. The segments were then allowed to settle by gravity and the pellets resuspended in 10 mL of ice‐cold PBS with 0.1% BSA, pipetted three times and allowed to settle. The supernatant was collected, passed through a 70 μm filter into a 50 mL tube and saved as fraction 1 (F1). The procedure was repeated to obtain fractions 2‐4 (F2‐F4). The fractions were centrifuged at 290g for 5 minutes at 4°C. The supernatant was discarded and the pellet was resuspended in 10 mL of PBS + 0.1% BSA. The suspension was centrifuged at 200g at 4°C. The pellet was resuspended in 5 mL of Dulbecco's modified Eagle's medium (DMEM)/F12 medium (Gibco). The fractions containing the most crypts were pelleted and resuspended in 50% complete IntestiCult Organoid Growth Medium (STEMCELL Technologies, Vancouver, British Columbia, Canada) and 50% Matrigel (Corning, Corning, New York). All experiments were performed on cultured crypts on passage #2‐7. For passaging, growth media was removed and cold PBS with 5 mM EDTA was added to each well. The Matrigel was mechanically disrupted using a P1000 pipette tip and the solution was rocked at 20 rpm at RT for 15 minutes. The disrupted crypts were pelleted by centrifugation at 290g for 5 minutes at 4°C.
3.9. Quantitative budding crypt assay
Budding crypt assays were conducted by plating 60 μL of crypt mixture, containing 150 crypts (for wells treated with radiation) or 50 crypts (for nonirradiated controls), into each well of a prewarmed 96‐well plate. The Matrigel was allowed to solidify and then complete IntestiCult Organoid Growth Medium was slowly added to cover the embedded crypts. The plate was incubated at 37°C overnight and irradiated the following day with 0 Gy or 3 Gy. Compound #5 was added 3 hours or 24 hours later and the media was changed daily for 6‐8 days. The number of enteroids with budding crypts was determined once the crypts reached maturity.
3.10. R‐Spondin1 replacement assay
Intestinal enteroids from 6 to 8‐week‐old female C3H mice were prepared as described above. Enteroids were dissociated into crypts and resuspended in 1:1 Matrigel:enteroid growth medium (advanced DMEM/F12 supplemented with 1% penicillin/streptomycin, 10 mM HEPES, 1% GlutaMAX [all from Gibco], 1× N‐2MAX [R&D Systems Minneapolis, MN], 1× SM1 [STEMCELL Technologies], 1 μM N‐acetylcysteine [Sigma], 50 ng/mL EGF, 100 ng/mL Noggin, 10 μM Y‐27632 [all from STEMCELL Technologies]) in the presence or absence of 1 μg/mL R‐Spondin1 (PeproTech, Westlake village, California). Crypts were plated in a 96 well plate in triplicates at the density of 50 crypts per well for nonirradiated wells and 150 crypts irradiated wells. After the Matrigel solidified, fresh enteroid growth media supplemented with 10 μM compound #5 or DMSO was added and the crypts were incubated at 37°C overnight. The following day, the plates were irradiated with 0 Gy or 3 Gy. The media was replaced daily for 6 days after which the number of budding crypts was determined.
3.11. Smoothened binding assay
HEK293 cells were cultured in DMEM supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin under standard conditions. Cells were transfected with a plasmid coding for mCherry‐tagged murine Smoothened protein (mCherry‐Smoothened1‐N‐18 was a gift from Michael Davidson, Addgene plasmid #55134). For binding studies cells were plated and treated with BODIPY‐cyclopamine (Biovision, Milpitas, California) at 5 nM for 2 hours in the presence or absence of compound #5. After 2 hours, cells were washed in PBS, fixed with paraformaldehyde and imaged using a fluorescence microscope (BZ‐9000) or analyzed by flow cytometry (LSR II, Becton Dickinson, Franklin Lakes, NJ and FlowJo v10).
3.12. Gli reporter luciferase assay
A Gli reporter luciferase assay was performed following the manufacturer's protocol. Briefly, Gli Reporter—NIH3T3 cells (catalog no. 60409, BPS Bioscience, San Diego, California) were harvested and seeded in a white clear bottom 96‐well plate at a density of 25,000 cells per well in 100 μL of the growth medium. The cells were incubated at 37°C overnight in a CO2 incubator. The following day, fresh media with compound #5 (0.1, 1, 10, and 100 μM) or Vismodegib (5 and 10 μM) were added. Cells were incubated for 2 hours after which recombinant murine sonic hedgehog (mShh) protein (1 μg/mL) was added to the positive control wells and the Vismodegib wells. Cells were incubated for an additional 30 hours. The luciferase assay was performed using the ONE‐Step Luciferase Assay System (catalog no. 60690, BPS Bioscience). Hundred microliters of the substrate was added to each well and rocked at RT for 15 minutes. The resulting luminescence was assessed using a plate reader (SpectraMax M5, Molecular Devices, Sunnyvale, California) at room temperature.
3.13. In silico docking experiments
Docking was performed using Autodock 4.2.17 The crystal structure of human Smoothened in complex with TC114 (5V57) was downloaded from the protein database (http://www.rcsb.org/pdb). The ligands were removed in Chimera.18 The receptor data was imported into AutoDockTools (vers 1.5.6, Scripps Institute), water was removed, hydrogens were added and polar hydrogens merged. The ligand 3D structure for 1‐[(4‐nitrophenyl)sulfonyl]‐4‐phenylpiperazine was downloaded from Chemspider (ID672004), converted using OpenBabel19 and entered into AutoDockTools. A grid box of 126 Å × 126 Å × 126 Å with a gridcenter at −103.522, −112.608, 51.04, and grid point spacing of 0.2 Å was generated to cover the known binding site for Smoothened antagonists and agonists within the transmembrane domain of Smoothened. Affinity maps of the present atoms and an electrostatic map were calculated using AutoGrid. The protein structure was considered rigid and the ligand was considered to be flexible. Every docking result was derived from 25 genetic algorithm runs, which terminated after a maximum of 2.5 × 106 energy evaluations and a maximum of 2.7 × 104 generations with a population size of 150 and a rate of gene mutation of 0.02 and a crossover rate of 0.8. The results were ranked based on their predicted binding energy.
3.14. Statistics
All analyses were performed in the GraphPad Prism software package. Two‐sided Student's t tests and a two‐way analysis of variance for multiple comparisons were used. A log‐rank test was used to determine a P‐value for the Kaplan‐Meier survival curves in Figure 1. A P‐value equal to or smaller than .05 was considered statistically significant.
4. RESULTS
4.1. Compound #5 mitigates the effects of TAI
C3H mice were subjected to TAI of 20.5 Gy at the published LD70/10 for this mouse strain.20 A second set of animals were irradiated with a dose of 20 Gy, an LD70/10. Twenty‐four hours after irradiation, mice were treated with daily subcutaneous injections of compound #5 at 5 mg/kg, solubilized in DMSO/Cremophor, for five consecutive days. Control animals were treated with radiation and DMSO/Cremophor only. Median survival in animals treated with 20 Gy and Cremophor/DMSO (n = 13) or 20 Gy and compound #5 (n = 13) was not reached. Seventy percent (9/13) of the animals survived in the 20 Gy/Cremophor/DMSO group and 77% (10/13) survived in the 20 Gy/compound #5 group. The median survival of control mice treated with 20.5 Gy (n = 15) was 10 days, thus indicating death due to an GI‐ARS and confirming the very steep dose response curve for TAI.20 Only 2/15 animals were alive 30 days after irradiation with 20.5 Gy. Five daily injections of compound #5 starting 24 hours after irradiation resulted in 61% (8/13) of the animals surviving 30 days after being exposed to 20.5 Gy (Figure 1B, P = .0056, log‐rank test) and prevented weight loss in animals irradiated with 20 Gy (Figure 1C, P = .0206). The same trend was observed in animals treated with 20.5 Gy but did not meet the threshold for statistical significance (Figure 1D, P = .0625).
4.2. Compound #5 promotes crypt regeneration and expands the stem cell compartment
Accidental radiation exposure will rarely be restricted to the abdomen and Mason et al demonstrated that at high radiation doses ISC survival in mice is independent of the irradiation technique (TBI or TAI).20 Therefore, to test the effect of compound #5 on the survival of ISCs, animals were treated with 16 Gy TBI (LD50/10).20 Twenty‐four hours after irradiation administration of compound #5 (5 mg/kg) in DMSO/Cremophor, or DMSO/Cremophor only was started and administered daily. Animals were euthanized 96 hours after irradiation and cross‐sections of the small intestines were analyzed for the appearance of regenerating intestinal crypts (Figure 1E,F). In animals irradiated with 16 Gy, followed 24 hours later by four daily treatments with compound #5, the number of regenerating crypts4 at 96 hours after irradiation (72 hours after first application of compound #5) increased twofold (P = .0158; Figure 1G), suggesting an increased number of surviving or newly formed ISCs in compound #5‐treated animals. In the absence of irradiation, treatment with compound #5 resulted also in a change in distribution of the villi length, with a higher percentage of villi having a shorter length in mice treated with compound #5. In Cremophor‐treated animals, 46.3% of villi were between 100 to 300 μm in length, whereas 24 hours after a single dose of compound #5, 58.6% of the villi were in this size range (not significant, Figure 2A,B,E). Irradiation with 16 Gy decreased the number of villi longer than ~400 μm, and treatment with a single dose of compound #5 (24 hours postirradiation) had minimal effect on villi length at 48 and 96 hours postirradiation and 24 and 72 hours after compound #5 treatment (Figure 2C‐E). In addition, treatment with compound #5 resulted in no change in the thickness of the submucosa (Figure 2F).
Figure 2.
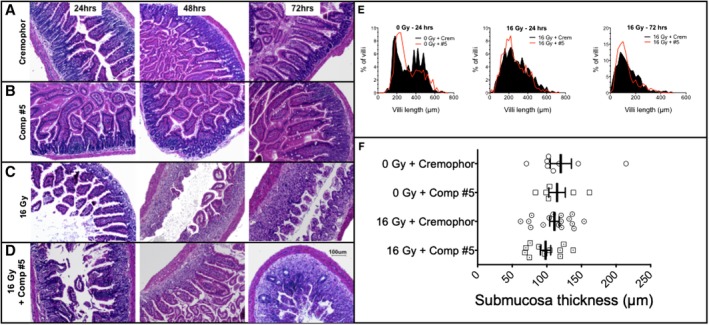
Compound #5 does not significantly alter villi length or submucosa thickness in the small intestines of irradiated mice. Female 12‐week‐old C3H mice received 16 Gy of total body irradiation or sham irradiation and 24 hours later were administered a single dose of compound #5 (5 mg/kg) or Cremophor/DMSO (crem) subcutaneously. A‐D, Mice were sacrificed 24 hours, 48 hours, and 72 hours after administering compound #5 and the small intestines were removed, fixed, and small cross‐sections were stained with H&E. A scale bar for (A)‐(D) representing 100 μM is depicted in (D). E, The length of the villi lining the entire circumference was measured in at least five cross‐sections per mouse. The percent of villi with a certain length was plotted against the villi length to create a distribution of villi length for each treatment group. At 24 and 72 hours after compound #5 treatment, there was a modest increase in the percentage of shorter villi (E, black filled plots are the distributions of villi length for Cremophor‐treated animals and the red lines represent compound #5 treated animals). F, The submucosa thickness was measured in the tissue sections from mice at 24 hours after compound #5 or Cremophor treatment. There was no significant effect on submucosa thickness after treatment with compound #5 (Student's t tests, P > .2 for all comparisons)
In agreement with the observed increase in the number of regenerating crypts in the intestines of mice treated with compound #5, we observed a significant increase in the number of proliferating (Ki67‐positive) cells within the crypt compartment of the small intestines of mice irradiated with 16 Gy followed by treatment with compound #5, 24 hours after irradiation (Figure 3C‐E, P < .0001). However, compound #5 did not affect the number of Ki67‐positive nuclei in unirradiated tissues (Figure 3A,B,E, P = .3612). In nonirradiated animals, Ki67 staining was most prominent in the region of transiently amplifying cells and—although weaker—also present in crypt base columnar cells (CBC cells; Figure 3B).
Figure 3.
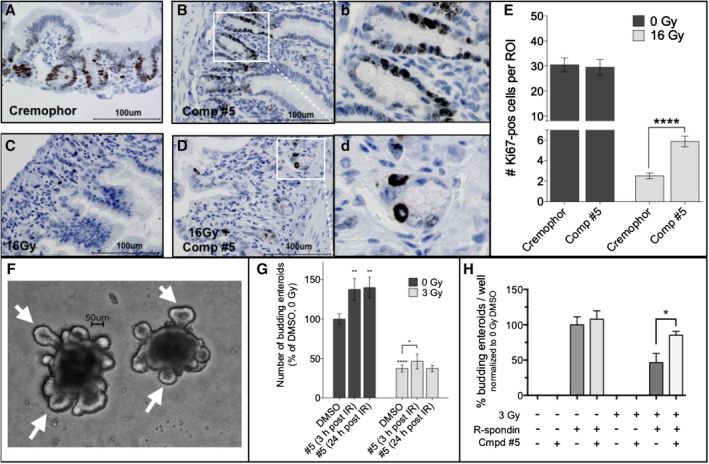
Compound #5 stimulates proliferation in crypt cells in the small intestines of irradiated mice. A‐D, C3H mice were sham‐irradiated (A, B) or received 16 Gy of total body irradiation (C, D) and 24 hours later were administered Cremophor/DMSO (A, C) or a single dose of compound #5 (5 mg/kg; B, D) subcutaneously. Twenty‐four hours after administering compound #5 (48 hours after irradiation exposure), the duodenums were removed, fixed, and thin cross‐sections were stained for the proliferation marker Ki67 (A‐D). The number of Ki67‐pos cells (brown stain; B, D) per region of interest present in at least five circumferences per mouse were counted. Scale bar: 100 μM. (b) and (d) are full size images of the regions indicated in (B) and (D). E, A single treatment with compound #5 resulted in a significant increase in the number of Ki67‐pos cells after exposure to a lethal dose (16 Gy) of total body irradiation (P < .0001, two‐tailed Student's t test) in the small intestines of irradiated mice (E, light bars) but no significant change in the intestines of nonirradiated animals (E, dark bars). F, In order to test if compound #5 affected intestinal stem cells, small intestines of C3H mice were removed and crypts were isolated and cultured for propagating intestinal enteroids in vitro. The three‐dimensional in vitro enteroid cultures form crypt‐like budding structures (white arrows) that recapitulate the in vivo crypt structures contain the intestinal stem cell compartment, thus providing a method for testing the effect of different treatments on the intestinal stem cells in vitro. Scale bar: 50 μM. G, Treatment of in vitro intestinal enteroids with a single dose of 3 Gy significantly decreases the number of budding enteroids, indicating a significant loss or damage of intestinal stem cells. This radiation‐induced intestinal stem cell loss was mitigated by daily treatment with compound #5 (10 μM). G, Graphs represent the average percent budding enteroids per well ± SEM. *, P < .05; **, P < .01; ****, P < .0001. H, In order to test if compound #5 can substitute for R‐Spondin1, intestinal enteroids isolated from C3H mice were propagated and placed in media made with or without the critical enteroid growth factor R‐Spondin1 and/or compound #5 (10 μM). The cells were sham‐irradiated or treated with a single dose of 3 Gy and the compound #5 was replaced daily, after 6 days the budding enteroids were counted. Compound #5 could not substitute for the lack of R‐Spondin1 but had an additive effect on enteroid budding in the presence of R‐Spondin1
To test the effect of compound #5 on the function of ISCs more directly, we generated intestinal enteroids from the small intestines of C3H mice and treated the enteroid cultures with a single dose of 3 Gy in vitro, followed by addition of compound #5 either 3 hours or 24 hours postirradiation. The number of enteroids with budding crypts was determined after 6 days. A significant increase in the number of functional enteroids (able to generate crypt structures, Figure 3F, white arrows) was observed when compound #5 was added 3 hours after irradiating the intestinal enteroids in vitro (P = .0479, Figure 3G).
Piperazines are known scaffolds used in the drug development of agonists and antagonists of G‐protein coupled receptors (GPCR) and since leucine rich repeat containing G protein‐coupled receptor 5 (Lgr5) belongs to the family of GPCRs, we next tested if compound #5 could substitute for R‐spondin1, the natural ligand of Lgr5. When established intestinal enteroids were dissociated and plated into R‐Spondin1‐free media and treated with R‐Spondin1 and/or compound #5, no secondary enteroid formation was observed in cultures treated with only compound #5 (Figure 3H). Cultures continuously treated with R‐Spondin1 formed budding enteroids. Similar results were obtained when the enteroids were irradiated with a single dose of 3 Gy but the combined treatment with R‐Spondin1 and compound #5 significantly increased the number of budding enteroids. This indicated that compound #5 has a different molecular target than R‐Spondin1, which sustains and amplifies Wnt signaling.
4.3. Compound #5 induces an acute immunological response in the small intestines
At this point in our study the molecular target for compound #5 was unknown, which hindered further optimization of the drug. Therefore, we decided to study acute gene expression changes in the murine small intestines after a single dose of compound #5. Animals were treated with s.c. injections of compound #5 or Cremophor/DMSO. The small intestines were harvested 4 hours later, flushed with PBS and total RNA was extracted from the proximal portion of the duodenum. Surprisingly, RNASeq resulted in the differential expression of only 15 genes in animals treated with compound #5 (Figure 4A,B). The upregulation of six of these genes was confirmed using qRT‐PCR (Figure 4C) Gene set enrichment analysis computed overlaps with immunological signatures (Figure 4D), which was in agreement with our previous report of increased cellularity in Peyer's patches, the immune system of the gut, after treatment with compound #5.21
Figure 4.
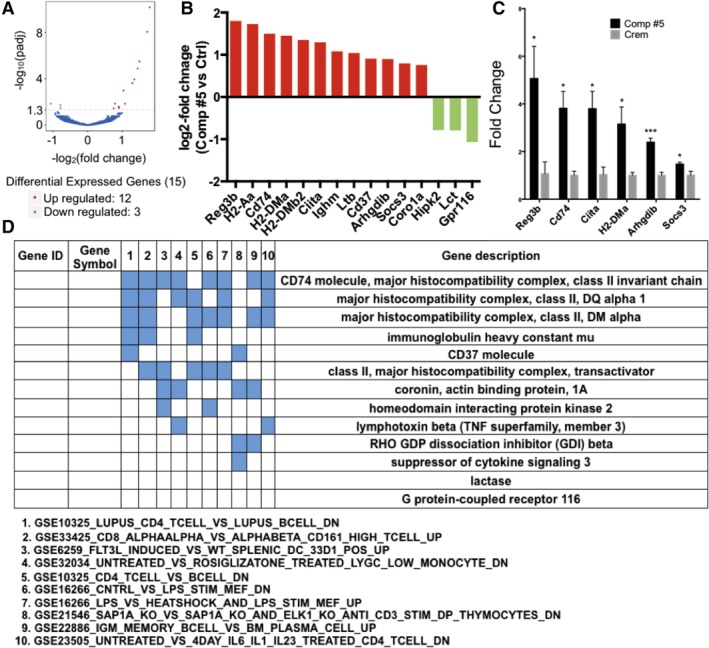
Compound #5 induces an immunological gene expression signature. Female, 12‐week‐old C3H mice were treated with Cremophor/DMSO or compound #5. Four hours after treatment the small intestines were harvested and subjected to RNASeq analysis to identify pathways engaged early after treatment with compound #5. Only 15 genes were differentially expressed. A, Volcano plot of genes up or downregulated. B, Log2‐fold change in expression of individual genes. C, qRT‐PCR validation of six genes differentially expressed (*, P < .05 and ***, P < .001, Student's t test; D) gene set enrichment analysis of overlapping genes
4.4. Effects of compound #5 on developmental signaling pathways in the intestinal crypt
Since the increased number of regenerating crypts after treatment with compound #5 pointed to increased numbers of ISCs we next studied if pathways known to be involved in ISC renewal were activated in response to compound #5 treatment. Because ISCs are particularly dependent upon Wnt pathway activation and the Wnt pathway drives many of the canonical stem cell markers of the small intestine, we assessed whether compound #5 activated Wnt signaling in vivo. Staining against β‐catenin at 48 hours after irradiation and 24 hours after drug treatment revealed weak stabilization and nuclear translocation of β‐catenin (Figure 5A‐D) thus, indicating that compound #5 does not significantly activate the Wnt signaling pathway directly or indirectly. Furthermore, this analysis was hindered by the variation of β‐catenin staining in each circumference and the known gradual 10‐fold decrease of Wnt signaling from the proximal to the distal small intestines.22
Figure 5.
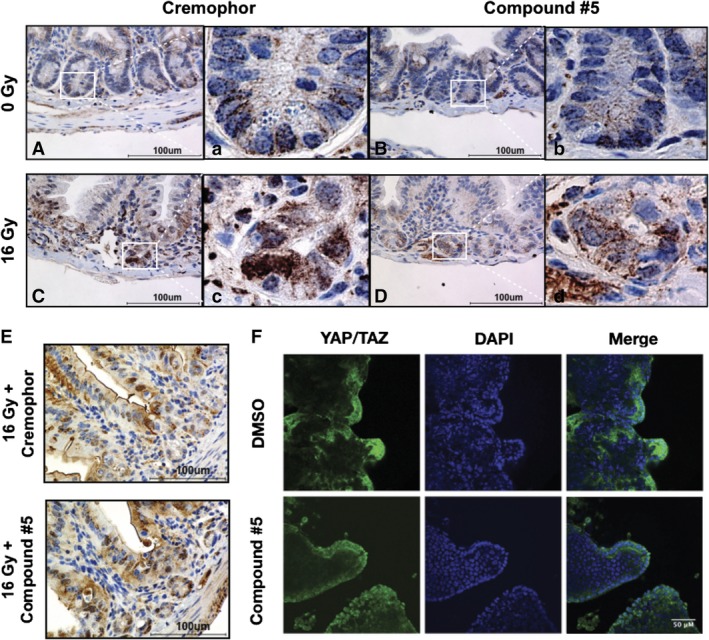
Compound #5 does not significantly induce Wnt, notch, or hippo signaling. A‐D, Female, 12‐week‐old C3H mice were sham‐irradiated (A, B) or subjected to 16 Gy total body irradiation (C, D). Twenty‐four hours later, the mice were treated with Cremophor/DMSO (A, C) or compound #5 (5 mg/kg; B, D) subcutaneously. Twenty‐four hours after compound #5 treatment (48 hours after irradiation), intestines were removed, fixed, and thin cross‐sections were stained for β‐catenin. Compound #5 modestly increased β‐catenin staining in vivo with minimal induction of nuclear localization. Scale bar: 100 μM. (a)‐(d) are full size images of the regions indicated in (A)‐(D). E, Female, 12‐week‐old C3H mice were subjected to 16 Gy total body irradiation. Twenty‐four hours later, the mice were treated with Cremophor/DMSO or compound #5 (5 mg/kg) subcutaneously. Twenty‐four hours after compound #5 treatment (48 hours after irradiation), intestines were removed, fixed, and thin cross‐sections were stained for Notch1‐ICD, the active form of Notch1. Compound #5 did not increase Notch1‐ICD staining in vivo, thus excluding Notch1 as a target for compound #5. Scale bar: 100 μM. F, Small intestines of C3H mice were removed and crypts were isolated and cultured for propagating intestinal enteroids in vitro. Enteroids were treated with DMSO or compound #5 (10 μM) for 24 hours. The enteroids were removed from the matrix and attached to slides using a cytospin (Shandon Elliot, Tacoma, Washington). The slides were fixed, permeabilized, and stained for YAP/TAZ and a nuclear counterstain. The slides were imaged using confocal microscopy. Compound #5 not affect the nuclear localization of YAP/TAZ, suggesting that the Hippo signaling pathway was not affected by compound #5. Scale bar: 50 μM
Next, we tested if compound #5 activated Notch signaling, a second pathway that is involved in the maintenance of the intestinal crypt.23 Staining compound #5‐treated section with an antibody against Notch1‐ICD, the intracellular domain of the cleaved, active form of Notch1 showed variation of Notch activation in the individual circumferences but no clear Notch activation pattern in response to compound #5 (Figure 5E).
The Hippo pathway is also critical during crypt regeneration after injury, including radiation injury.24 Hippo pathway components YAP/TAZ are activated and localized to the nucleus during recovery from radiation injury. We assayed for Hippo pathway activation using in vitro enteroids treated for 24 hours with compound #5 or DMSO. The enteroids were fixed, stained with antibodies against YAP/TAZ and a nuclear counterstain (DAPI) and analyzed by confocal microscopy. Compound #5 had no apparent effect on the nuclear translocation of YAP/TAZ proteins in crypt‐like structures (Figure 5F).
To further investigate potential mechanisms of action of compound #5, we created a qPCR array of ISC genes and components of major stem cell signaling pathways (Table S1). Gene expression analysis of the proximal portions of the duodenums of mice treated with compound #5 for 24 hours versus Cremophor/DMSO‐treated animals revealed a significant downregulation of Cysteine‐rich angiogenic inducer 61 (Cyr61), Axin related protein 2 (Axin2), connective tissue growth factor (Ctgf), SPARC realted modular calcium binding 2 (Smoc2), Octamer‐binding transcription factor 4 (Oct4), Yap1, Dachshund family transcription factor 1 (Dach1), Musashi RNA binding prorein 1 (Msi1), and Hairy/enhancer of split related with YRPW motif like protein (HeyL; Figure 6A), while only the expression level of the hedgehog‐driven gene Gli1 (GLI family zinc finger 1) was significantly upregulated (Figure 6B).
Figure 6.
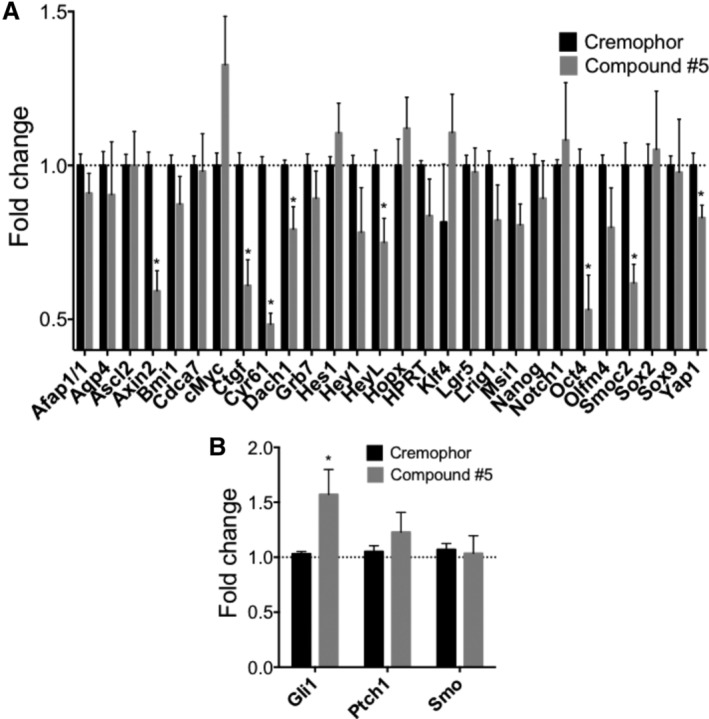
Compound #5 induces Gli1 expression. Female, 12‐week‐old C3H mice were treated with Cremophor or compound #5. Twenty‐four hours after drug treatment the small intestines and the proximal portion of the duodenum was harvested and subjected to qRT‐PCR analysis using a custom intestinal stem cell PCR primer array. Treatment with compound #5 significantly downregulated the expression of Cyr61, Axin2, Ctgf, Smoc2, Oct4, Yap1, Dach1, Msi1, and HeyL (A) but led to an upregulation of Gli1 (B; *, P < .05, Student's t test)
In order to gain further insight into the mechanism by which compound #5 targets the Hedgehog pathway, we overexpressed Smoothened‐mCherry (SMO‐mCherry) in HK293 cells. When treated with 5 nM boron‐dipyrromethene (BODIPY)‐cyclopamine (a green‐fluorescent form of the well‐known Smoothened inhibitor cyclopamine), SMO‐mCherry and cyclopamine colocalized (Figure 7A). Addition of compound #5 displaced BODIPY‐cyclopamine from SMO‐mCherry in a concentration‐dependent manner (Figure 7B). In order to quantify this effect, we repeated the experiment and analyzed the cells using flow cytometry, which determined an IC50 of 88 ± 3.7 μM for compound #5 (Figure 7C). This indicated that compound #5 could potentially occupy the same binding region as cyclopamine, which binds to the transmembrane region of Smoothened.25 When Smoothened is activated it translocates to the primary cilium where it prevents proteasomal degradation of the zinc‐finger transcription factor Gli.26 To further confirm engagement of the Hedgehog pathway we treated Gli1‐Luc reporter cells, in which luciferase expression is under control of a Gli binding element, with compound #5. This led to a dose‐dependent activation of the reporter construct, thus supporting activation of the Hedgehog pathway by compound #5. Treatment of the cells with recombinant murine sonic hedgehog (Shh), a known Hedgehog agonist, activated the Gl1‐responsive element to the same order of magnitude as compound #5 and this activation was inhibited by the Hedgehog antagonist Vismodegib (Figure 7D). In order to test functionally if compound #5 binds to SMO, murine small intestinal enteroids were treated with Vismodegib, another Hedgehog pathway inhibitor. Addition of compound #5 attenuated the effects of Vismodegib on budding crypt formation in this model (Figure 7E).
Figure 7.
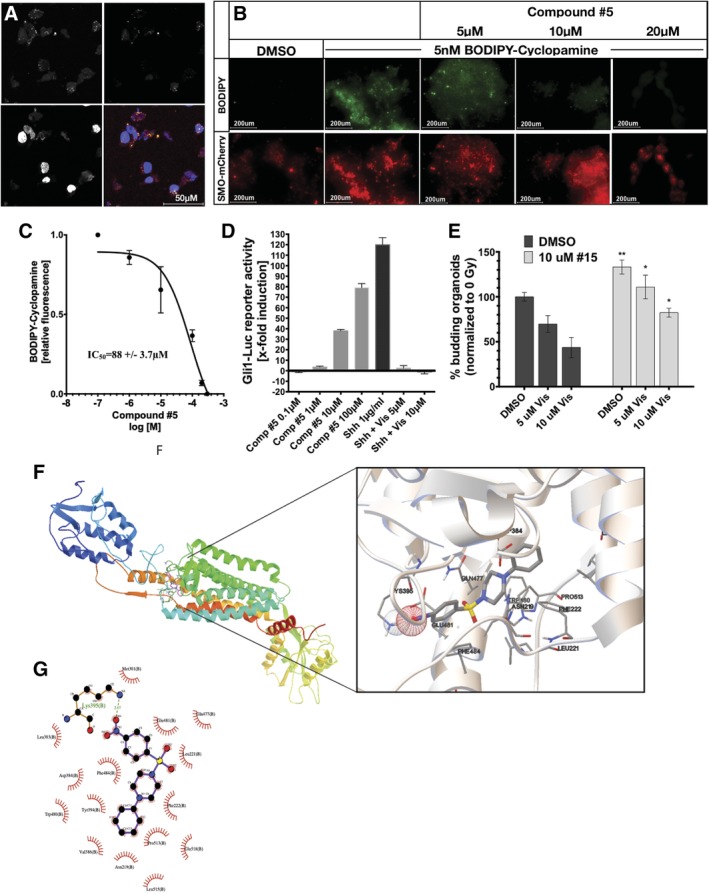
Compound #5 binds to Smoothened and activates Gli signaling. A, HEK293 cells were transiently transfected with an expression construct for mCherry‐tagged Smoothened protein. Smoothened‐mCherry overexpressing cells (red) were treated with BODIPY‐cyclopamine (green), fixed, counterstained with DAPI and imaged using confocal microscopy (×63 magnification, scale bar: 50 μM). The BODIPY signal colocalized with the Smoothened protein. B, HEK293 cells, overexpressing Smoothened‐mCherry were pretreated with BODIPY‐cyclopamine (green). Treatment with compound #5 led to a concentration‐dependent replacement of cyclopamine from its binding site. Images show clusters of HEK293 cells at ×4 magnification. Scale bar: 200 μM. C, Quantification of the replacement of BODIPY‐cyclopamine by compound #5 in mCherry‐tagged Smoothened protein overexpressing HEK293 using flow cytometry determined an IC50 of 88 μM ± 3.7 μM for compound #5 (three biologically independent repeats). D, Compound #5 caused a concentration‐dependent activation of a Gli signaling (*, P < .05 and **, P < .01, Student's t test). E, Compound #5 significantly attenuated the inhibitory effect of the Smoothened inhibitor Vismodegib in a budding enteroid assay. F, In silico docking results for compound #5 predicted binding to the transmembrane domain of Smoothened. G, LigPlot 2D presentation of the interactions of compound #5 with the transmembrane domain of Smoothened
Finally, we performed in silico docking studies, which predicted binding of compound #5 to the transmembrane domain of Smoothened with a hydrogen bond to Lys395 and a binding free energy of −10.01 kCal/mol (Figure 7F,G).
5. DISCUSSION
Nuclear accidents, terrorist attacks with dirty bombs, or deployed nuclear weapons will potentially expose large numbers of the general public and first responders to substantial doses of ionizing radiation.27 Even in developed countries, the resulting number of victims suffering from ARS will outnumber the capacity of facilities able to provide high‐end supportive care or bone marrow transplants.28 Providing care for an overwhelming number of people that would be exposed to radiation in such a scenario would be further complicated by the necessity to deliver treatment as early as possible after exposure. Yet, in a mass‐casualty scenario delivering a treatment within the first 24 hours after exposure would be impossible for the majority of the victims.8, 9, 10 Therefore, current approaches aiming at discovering effective radiation mitigators focus on drugs that show efficacy when given 24 hours (or later) after radiation exposure in order to be of use under such circumstances.6
Severe damage to the gut will be one of the life‐threatening radiation injuries that will require immediate attention.7 Within hours to days after radiation exposure, the intestinal epithelium responds with massive cell death, villi are lost and the barrier function of the intestines is compromised. Left untreated, victims will succumb to massive loss of fluids, bleeding, malabsorption, and sepsis.
Hypothetically, the intestine's response to radiation could be modified in different ways. A first approach could be through facilitating the repair of DNA damage caused by radiation exposure and prevention of apoptosis. However, in the small intestines, repair of potentially lethal DNA damage after exposure to radiation is a process that starts early after exposure and is mostly completed after 4 hours.3 Therefore, if a treatment aims at improving or promoting DNA repair any successful intervention would have to be administered in advance or at least rapidly after radiation exposure, which would face the above‐mentioned major logistic obstacles if a larger number of people are exposed at the same time. Likewise, apoptosis in the small intestines is a rapid process that peaks at 4 hours after irradiation is completely initiated at 24 hours,29 thus excluding at least the primary wave of apoptosis as a potential intervention point for radiation mitigators. However, compounds aiming at preventing apoptosis could still be useful in reducing secondary or tertiary waves of programmed cell death in the intestines.
An alternative approach could be the expansion of the ISC pool either from surviving ISCs by stimulation of symmetric divisions, recruitment and interconversion of reserve stem cell populations, or dedifferentiation of differentiating progeny into a stem cell state.30, 31, 32 Treatments to achieve these goals would allow for a wider time window and would most likely rely on activating pathways involved in ISC and TA cell pool expansion and/or ISC and TA cell interconversion, a process thought to be driven by Wnt, Notch, Hippo, and Hedgehog signaling.23
Here, we report that treatment with the radiation mitigator compound #5 24 hours after exposure to a lethal dose of radiation led to an increased proliferation of crypt cells 48 hours after irradiation (Figure 3A‐D), a significant increase in the number of regenerating crypts at 96 hours after irradiation (Figure 3E) and a significant increase in survival of the animals (Figure 1). The microcolony assay for regenerating crypts used in our study is the gold standard for assessing the number of surviving ISCs4 and thus, the increase in numbers of regenerating crypts indicated that compound #5 indeed affected the size of the ISC pool (Figure 1G). This was further supported by the finding that compound #5 significantly increased the number of budding enteroids in an in vitro intestinal enteroid assay. In these three‐dimensional intestinal enteroid (mini‐guts) assays the “budding” structures (Figure 3F,G) are believed to recapitulate the in vivo crypt structures, which contain the ISC compartment within a niche of other supporting cells.5
In search for the molecular mechanisms behind our in vitro and in vivo observations we next performed RNASeq. Our results showed that the expression levels of only a surprisingly small number of genes were altered 4 hours after compound #5 treatment (Figure 4A). It has to be noted that the whole proximal portion of the duodenum was used for this experiment, which included not only crypts but also the mucosa and submucosa portion of the intestines. This experimental route was chosen to test for global effects of compound #5 on the intestines and because the target cell population was not narrowed down at this point. By design, this experiment did not have the required signal‐to‐noise ratio that would allow for detection of gene expression changes in only the small population of crypt cells and we interpreted these results as an indicator for a small target cell population for compound #5.
Consequently, we next explored the developmental pathways that govern ISC renewal (Figure 5) as potential targets for compound #5 and demonstrated that compound #5 activated the Hedgehog pathway, as the mRNA of the downstream target Gli1 was significantly upregulated 24 hours after treatment of the animals with compound #5 and reporter cells showed activation of a Gli1‐responsive element, comparable to activation by recombinant mShh (Figures 6B and 7D). Competition experiments and in silico docking studies supported direct binding of compound #5 to the transmembrane domain of Smoothened (Figure 7). Smoothened is known to be expressed in only a small population of pericryptal stromal cells,33 bone marrow‐derived stromal cells34 and in follicular dendritic cells in the germinal centers.35 In mouse models of inflammatory bowel disease Hedgehog signaling in stromal cells at the base of the crypt leads to increased IL‐10 production as a major anti‐inflammatory mechanism.33 This is in agreement with our observation that compound #5 not only mitigated the GI‐ARS but also led to increased cellularity in the germinal centers of Peyer's patches,21 prevented hematopoietic ARS,12 increased the number of peripheral CD11b+Ly6C+Ly6G+ myeloid cells and stimulated IL‐10 production.12 At this point it is unclear if compound #5 acts on ICSs directly or indirectly. Compound #5 increased ICSs in vitro but early enteroid passages will most likely still contain stromal components and is quite likely that compound #5 creates an anti‐inflammatory environment permissive for ICS expansion or interconversion. Future studies are warranted to determine the target cell population for Piperazines in the small intestines.
6. CONCLUSION
We conclude that Piperazines are a novel class of Smoothened agonists and that Smoothened is an attractive and druggable target to mitigate the GI‐ARS.
CONFLICT OF INTEREST
The authors declare no potential conflict of interest.
AUTHOR CONTRIBUTIONS
S.D.M., K.B., E.V.: collection and assembly of data acquisition, data analysis and interpretation, manuscript writing, final approval of the manuscript; P.M., F.C., L.H., C.A., M.S., S.D.M., G.R., M.E.: collection and assembly of data acquisition, data analysis and interpretation, final approval of the manuscript; F.P.: manuscript writing, conception and design, financial support, final approval of the manuscript.
7.
Supporting information
Table S1. Primers used for qRT‐PCR
ACKNOWLEDGMENTS
F.P. was supported by a grant from the National Institute of Allergy and Infectious Diseases (AI067769) and by grants from the National Cancer Institute (CA211015, CA200234).
Duhachek‐Muggy S, Bhat K, Medina P, et al. Radiation mitigation of the intestinal acute radiation injury in mice by 1‐[(4‐nitrophenyl)sulfonyl]‐4‐phenylpiperazine. STEM CELLS Transl Med. 2020;9:106–119. 10.1002/sctm.19-0136
Sara Duhachek‐Muggy and Kruttika Bhat contributed equally to the study.
Funding information National Cancer Institute, Grant/Award Numbers: CA200234, CA211015; National Institute of Allergy and Infectious Diseases, Grant/Award Number: AI067769
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Warren S, Bowers JZ. The acute radiation syndrome in man. Ann Intern Med. 1950;32:207‐216. [DOI] [PubMed] [Google Scholar]
- 2. Weisdorf D, Chao N, Waselenko JK, et al. Acute radiation injury: contingency planning for triage, supportive care, and transplantation. Biol Blood Marrow Transplant. 2006;12:672‐682. [DOI] [PubMed] [Google Scholar]
- 3. Withers HR, Elkind MM. Radiosensitivity and fractionation response of crypt cells of mouse jejunum. Radiat Res. 1969;38:598‐613. [PubMed] [Google Scholar]
- 4. Withers HR, Elkind MM. Microcolony survival assay for cells of mouse intestinal mucosa exposed to radiation. Int J Radiat Biol Relat Stud Phys Chem Med. 1970;17:261‐267. [DOI] [PubMed] [Google Scholar]
- 5. Sato T, Vries RG, Snippert HJ, et al. Single Lgr5 stem cells build crypt‐villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262‐265. [DOI] [PubMed] [Google Scholar]
- 6. Brenner DJ, Chao NJ, Greenberger JS, et al. Are we ready for a radiological terrorist attack yet? Report from the centers for medical countermeasures against radiation network. Int J Radiat Oncol Biol Phys. 2015;92:504‐505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Williams JP, McBride WH. After the bomb drops: a new look at radiation‐induced multiple organ dysfunction syndrome (MODS). Int J Radiat Biol. 2011;87:851‐868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Department of Homeland Security USA , Homeland Security Council. National Planning Scenarios, version 20.2. 2009.
- 9. Davis LE, LaTourrette T, Mosher DE, Davis LM, Howell DR. Individual Preparedness and Response to Chemical, Radiological, Nuclear, and Biological Terrorist Attacks. Santa Monica, CA: RAND Corporation; 2003. [Google Scholar]
- 10. Carter AB, May MM, Perry WJJF. The day after: action following a nuclear blast in a U.S. city. Washington Q. 2007;30:19‐32. [Google Scholar]
- 11. Kim K, Damoiseaux R, Norris AJ, et al. High throughput screening of small molecule libraries for modifiers of radiation responses. Int J Radiat Biol. 2011;87:839‐845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Micewicz ED, Kim K, Iwamoto KS, et al. 4‐(Nitrophenylsulfonyl)piperazines mitigate radiation damage to multiple tissues. PLoS One. 2017;12:e0181577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Plett PA, Sampson CH, Chua HL, et al. Establishing a murine model of the hematopoietic syndrome of the acute radiation syndrome. Health Phys. 2012;103:343‐355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge‐based approach for interpreting genome‐wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545‐15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mootha VK, Lindgren CM, Eriksson KF, et al. PGC‐1alpha‐responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267‐273. [DOI] [PubMed] [Google Scholar]
- 16. Sato T, Clevers H. Primary mouse small intestinal epithelial cell cultures. Methods Mol Biol. 2013;945:319‐328. [DOI] [PubMed] [Google Scholar]
- 17. Morris GM, Huey R, Lindstrom W, et al. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem. 2009;30:2785‐2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pettersen EF, Goddard TD, Huang CC, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605‐1612. [DOI] [PubMed] [Google Scholar]
- 19. O'Boyle NM, Banck M, James CA, et al. Open babel: an open chemical toolbox. J Chem. 2011;3:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mason KA, Withers HR, McBride WH, et al. Comparison of the gastrointestinal syndrome after total‐body or total‐abdominal irradiation. Radiat Res. 1989;117:480‐488. [PubMed] [Google Scholar]
- 21. Bhat K, Duhachek‐Muggy S, Ramanathan R, et al. 1‐(4‐Nitrobenzenesulfonyl)‐4‐phenylpiperazine increases the number of Peyer's patch‐associated regenerating crypts in the small intestines after radiation injury. Radiother Oncol. 2019;132:8‐15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Davies PS, Dismuke AD, Powell AE, Carroll KH, Wong MH. Wnt‐reporter expression pattern in the mouse intestine during homeostasis. BMC Gastroenterol. 2008;8:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. de Sousa EMF, de Sauvage FJ. Cellular plasticity in intestinal homeostasis and disease. Cell Stem Cell. 2019;24:54‐64. [DOI] [PubMed] [Google Scholar]
- 24. Gregorieff A, Liu Y, Inanlou MR, Khomchuk Y, Wrana JL. Yap‐dependent reprogramming of Lgr5(+) stem cells drives intestinal regeneration and cancer. Nature. 2015;526:715‐718. [DOI] [PubMed] [Google Scholar]
- 25. Byrne EF, Luchetti G, Rohatgi R, et al. Multiple ligand binding sites regulate the Hedgehog signal transducer Smoothened in vertebrates. Curr Opin Cell Biol. 2018;51:81‐88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Briscoe J, Therond PP. The mechanisms of Hedgehog signalling and its roles in development and disease. Nat Rev Mol Cell Biol. 2013;14:416‐429. [DOI] [PubMed] [Google Scholar]
- 27. Jordan BR. The Hiroshima/Nagasaki survivor studies: discrepancies between results and general perception. Genetics. 2016;203:1505‐1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Majhail NS, Mau LW, Chitphakdithai P, et al. National survey of hematopoietic cell transplantation center personnel, infrastructure, and models of care delivery. Biol Blood Marrow Transplant. 2015;21:1308‐1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Potten CS, Grant HK. The relationship between ionizing radiation‐induced apoptosis and stem cells in the small and large intestine. Br J Cancer. 1998;78:993‐1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yousefi M, Li L, Lengner CJ. Hierarchy and plasticity in the intestinal stem cell compartment. Trends Cell Biol. 2017;27:753‐764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schmitt M, Schewe M, Sacchetti A, et al. Paneth cells respond to inflammation and contribute to tissue regeneration by acquiring stem‐like features through SCF/c‐kit signaling. Cell Rep. 2018;24:2312.e2317‐2328.e2317. [DOI] [PubMed] [Google Scholar]
- 32. Beumer J, Clevers H. Regulation and plasticity of intestinal stem cells during homeostasis and regeneration. Development. 2016;143:3639‐3649. [DOI] [PubMed] [Google Scholar]
- 33. Lee JJ, Rothenberg ME, Seeley ES, et al. Control of inflammation by stromal Hedgehog pathway activation restrains colitis. Proc Natl Acad Sci USA. 2016;113:E7545‐E7553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shi S, Sun J, Meng Q, et al. Sonic hedgehog promotes endothelial differentiation of bone marrow mesenchymal stem cells via VEGF‐D. J Thorac Dis. 2018;10:5476‐5488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sacedon R, Diez B, Nunez V, et al. Sonic hedgehog is produced by follicular dendritic cells and protects germinal center B cells from apoptosis. J Immunol. 2005;174:1456‐1461. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Primers used for qRT‐PCR
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


