Abstract
Many enzymes assemble into defined oligomers, providing a mechanism for cooperatively regulating activity. Recent studies have described a mode of regulation in which enzyme activity is modulated by polymerization into large-scale filaments. Here we describe an ultrasensitive form of polymerization-based regulation employed by human CTP synthase 2 (CTPS2). Cryo-EM structures reveal that CTPS2 filaments dynamically switch between active and inactive forms in response to changes in substrate and product levels. Linking the conformational state of many CTPS2 subunits in a filament results in highly cooperative regulation, greatly exceeding the limits of cooperativity for the CTPS2 tetramer alone. The structures reveal a link between conformation and control of ammonia channeling between the enzyme’s active sites, and explain differences in regulation of human CTPS isoforms. This filament-based mechanism of enhanced cooperativity demonstrates how the widespread phenomenon of enzyme polymerization can be adapted to achieve different regulatory outcomes.
CTP synthase (CTPS) is the key regulatory enzyme in pyrimidine biosynthesis, with critical roles in regulation of nucleotide balance1, maintenance of genome integrity2,3, and synthesis of membrane phospholipids4. CTPS catalyzes the conversion of UTP to CTP in an ATP-dependent process, the rate-limiting step in CTP synthesis. CTPS is regulated through feedback inhibition by CTP binding, and is allosterically regulated by GTP, making it sensitive to levels of the four essential ribonucleotides, reflecting its role as a critical regulatory node in nucleotide metabolism5–8. CTPS is a homotetramer, with each monomer composed of a glutaminase and an amidoligase domain connected by a helical linker9. Ammonia is generated from glutamine then transfered to the amidoligase domain, where it is ligated to UTP to form CTP; while both of these catalytic mechanisms are well understood, the mechanism of ammonia transfer between the two separated active sites has not yet been described. Previously, we showed that CTPS undergoes a conserved conformation cycle controlled by substrate and product binding, involving two major structural changes: upon substrate binding, the glutaminase domain rotates towards the amidoligase domain, bringing the two active sites closer, and the tetramer interface rearranges to accommodate UTP binding10.
Humans have two CTPS isoforms encoded on separate genes, CTPS1 and CTPS2, that share 75% identity. Their relative roles remain unclear. CTPS1 plays a specific and central role in lymphocyte proliferation, and its loss in humans causes severe immune deficiency11,12. CTPS is frequently misregulated in cancer2,13, with CTPS2 misregulation specifically implicated in osteosarcoma14. Given these roles in health and disease, how the two human enzymes are differentially regulated remains an open question of clinical significance.
Polymerization into filaments provides an additional layer of CTPS regulation. For many filament-forming enzymes, polymerization is driven by binding to substrates, products, or allosteric regulators, and tunes activity by locking the enzyme in high or low activity states10,15–18. For CTPS, the mechanisms by which filaments modulate enzyme activity vary among species. In E. coli, CTPS filaments stabilize a product-bound, inactive conformation of the enzyme, leading to enhanced inhibition in the filament10,15. By contrast, human CTPS1 forms hyper-active filaments composed of enzyme in an active, substrate-bound conformation that disassemble on CTP binding10. CTPS filaments appear in response to cellular stress, during particular developmental stages, and in tumor tissue, suggesting a role in adaptation to changing metabolic needs19–23.
Given the importance of understanding the regulatory differences between the two human isoforms and the observed variation in filament-based regulation among species, here we aimed to determine whether there are differences in filament structure and function between CTPS1 and CTPS2.
RESULTS
CTPS2 forms distinct substrate- and product-bound filaments
We imaged CTPS2 by negative stain EM in the presence of substrates UTP and ATP or products CTP and ADP (Fig. 1a). Surprisingly, unlike CTPS1, which only assembles in the substrate-bound conformation and disassembles upon CTP addition, either substrates or products promoted CTPS2 filament assembly, suggesting a novel mode of regulation.
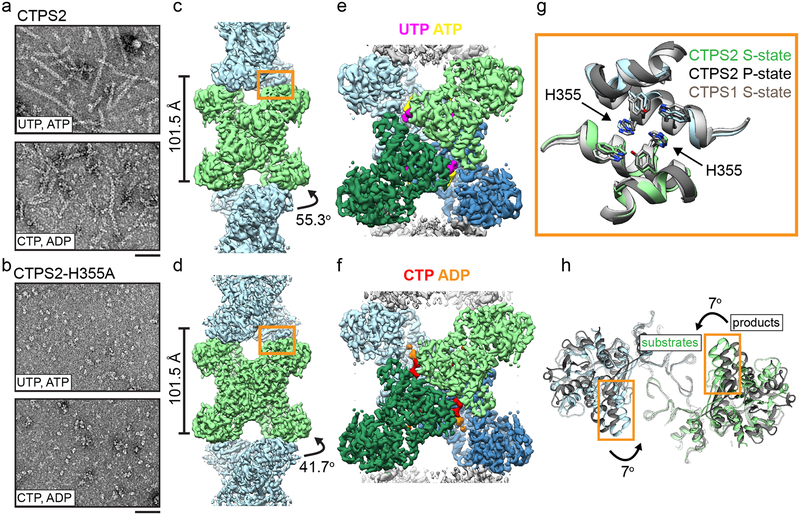
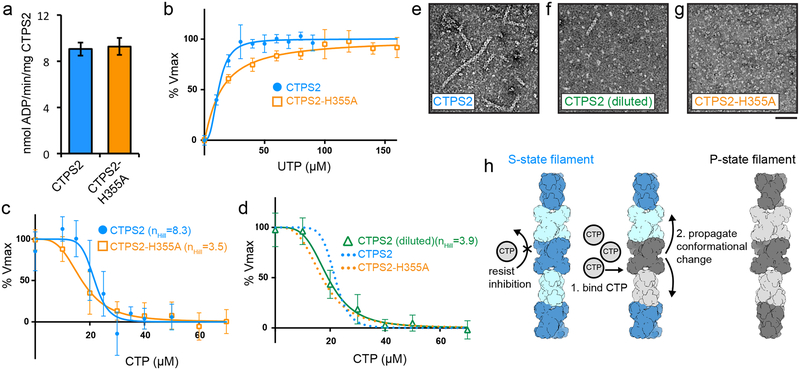
Fig. 1: CTPS2 forms distinct S and P-state filaments using the same filament interface.
(a,b) Negative stain EM images of CTPS2 wild-type (a) or CTPS2-H355A non-polymerizing mutant (b) in the presence of substrates or products. Scale bars are 50 nm. (c,d) Initial cryo-EM reconstructions of S-state (c, 4Å resolution) and P-state (d, 3.6 Å resolution) CTPS2 filaments showing differing helical symmetry, colored by tetramer. Helical rise and rotation are indicated. (e,f) High-resolution reconstructions of S-state (e, 3.5Å resolution) and P-state (f, 3.1 Å resolution) CTPS2 filaments focused on a single tetramer, colored by protomer. Nucleotides are colored as indicated. (g) The filament interface (orange box in c,d) is conserved in the CTPS2 S-state (color), CTPS2 P-state (dark grey), and CTPS1 S-state (light grey) structures. Position of conserved residue H355 is indicated. (h) View down the helical axis comparing the positions of the filament contacts (orange box) in the S-state (color) and P-state (grey) filaments.
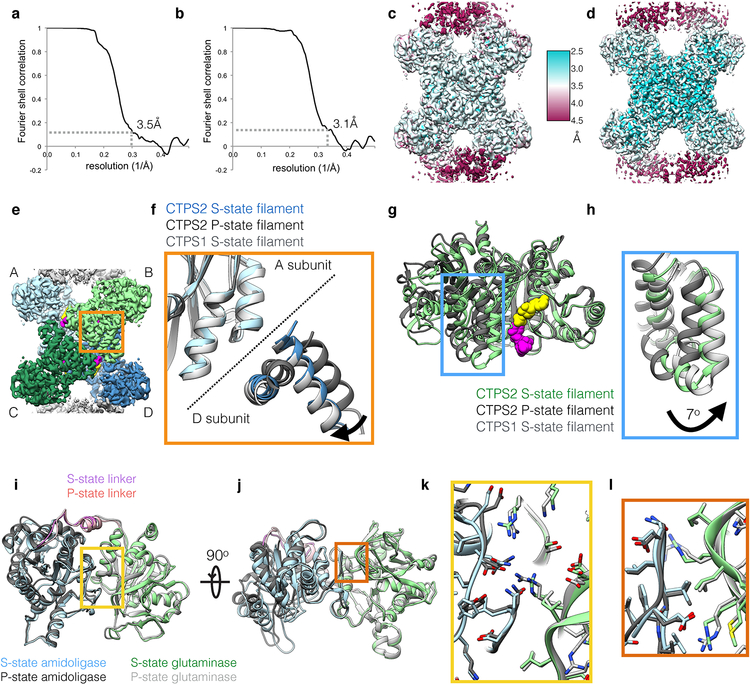
We solved cryo-EM structures of substrate-bound (S-state) and product-bound (P-state) CTPS2 filaments (Fig. 1). Initial reconstructions encompassing multiple tetramers revealed the helical architecture of the filaments (Fig. 1c, d), while masked refinements focused on single tetramers produced higher resolution structures at 3.5Å and 3.1Å of the S-state and P-state filaments, respectively (Fig. 1e, f, Extended Data Fig. 1, Extended Data Fig. 2, Extended Data Fig. 3a–d, Table 1). Both CTPS2 filaments are composed of stacked tetramers, with a eukaryote-specific helical insert in the glutaminase domain forming the interfaces between tetramers. The filament assembly interactions are identical in both CTPS2 filament states (Cα RMSD 0.8 Å), and are the same as the CTPS1 interface10 (Cα RMSD 1.3 Å) (Fig. 1g). We previously showed that mutation of conserved H355 at the filament interface completely abolishes CTPS1 polymerization10. This mutation has the same effect on both S- and P-state CTPS2 polymerization (Fig. 1b).
Table 1:
Cryo-EM data collection, refinement and validation statistics
| S state CTPS2 filament (EMD-20354, PDB 6PK4) | P state CTPS2 filament (EMD-20355, PDB 6PK7) | |
|---|---|---|
| Data collection | ||
| Magnification | 130000 | 130000 |
| Voltage (kV) | 300 | 300 |
| Electron fluence (e–/Å2) | 90 | 90 |
| Defocus range (μm) | 1.0–2.5 | 1.0–2.5 |
| Pixel size (Å) | 0.525 | 0.525 |
| Reconstruction | ||
| Pixel size (Å) | 1.05 | 1.05 |
| Symmetry imposed | D2 | D2 |
| Helical symmetry | ||
| Rise (Å) | 101.5 | 101.5 |
| Rotation (°) | 55.3 | 41.7 |
| Particle images (no.) | 53964 | 22705 |
| Map resolution (0.143 fsc) (Å) | 3.5 | 3.1 |
| Refinement | ||
| Initial model used (PDB code) | 2V4U, 2VO1 | 2V4U, 2VO1 |
| Map sharpening B factor (Å2) | −50 | −50 |
| Model composition | ||
| Protein residues | 559 | 557 |
| Ligands | UTP, ATP | CTP, ADP |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.41 | 0.52 |
| Bond angles (°) | 0.69 | 0.66 |
| Validation | ||
| MolProbity score | 2.15 | 2.06 |
| Clashscore | 12 | 11 |
| Poor rotamers (%) | 1 | 0.4 |
| Ramachandran plot | ||
| Favored (%) | 92 | 91 |
| Allowed (%) | 8 | 9 |
| Outliers (%) | 1 | 0 |
While the filament assembly interfaces are identical in both CTPS2 filament states, the conformation of the enzyme and the helical symmetry are strikingly different. The S-state CTPS2 and CTPS1 filaments are very similar at the level of monomer, tetramer, and filament10 (Fig. 1c,e, Extended Data Fig. 3e–h). By contrast, tetramers in the CTPS2 P-state filament are in an inactive, CTP-inhibited conformation, similar to that observed in bacterial CTPS homologs5,9,10 (Fig. 1d,f, Extended Data Fig. 3e–h). These differences in conformation result in different helical architectures. The two domains of each protomer rotate relative to each other by 7° between the S- and P-states; the interactions at the interdomain interface remain fixed (Cα r.m.s.d. 0.8 Å), with the rotation arising from flexing of residues 40–87 relative to the core of the amidoligase domain (Extended Data Fig. 3i–l). The interdomain rotation alters the positions of filament contacts around the helical axis, leading to a 14° difference in the helical rotation per tetramer between the S-state and P-state filaments (Fig. 1c,d,h). Rare CTPS2 filaments observed in the absence of nucleotides had S-state architecture in negative stain reconstructions, suggesting this may be a somewhat more stable conformation of the enzyme (Extended Data Fig. 4). CTPS2 therefore assembles into active and inactive filaments with unique architectures, depending on the ligand-binding and conformational state of constituent tetramers, while maintaining a fixed filament interface.
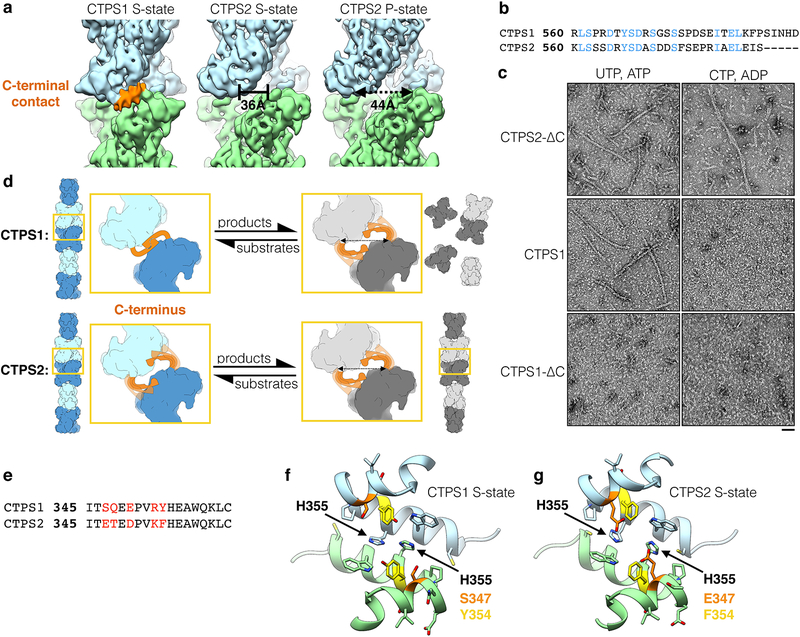
CTPS1 filaments are stabilized by a C-terminal interface
Given that the primary filament interface is conserved between CTPS1 and CTPS2, we aimed to address why we observe S-state but not P-state CTPS1 filaments (Extended Data Fig. 5, Extended Data Fig. 6). We considered that large-scale conformational changes or disruption of tetramerization could be the reason product-bound CTPS1 does not form filaments. However, negative stain 2D averages and a 3D reconstruction revealed that product-bound CTPS1 formed tetramers with normal overall morphology (Extended Data Fig. 5). Further, product-bound CTPS1 tetramers very occasionally associated into pairs with the same assembly contacts observed in filaments (Extended Data Fig. 5a–c). Coupled with our previous observation that addition of CTP causes S-state CTPS1 filaments to disassemble10, this suggests that while CTPS1 can form filament contacts in the product-bound state, these contacts are much less stable than in S-state CTPS1 filaments. S-state CTPS1 filaments have additional interactions between poorly ordered C-terminal tails of adjacent tetramers10, which we did not observe in either CTPS2 filament (Extended Data Fig. 6a); compared to the full-length sequences, the C-termini are poorly conserved between CTPS1 and CTPS2 (75% overall sequence identity versus 41% C-terminal sequence identity) (Extended Data Fig. 6b). We therefore suspected that differences in the C-terminal tails could account for differences in the assembly of CTPS1 and CTPS2 filaments.
To test this hypothesis, we analyzed CTPS1-ΔC and CTPS2-ΔC C-terminal truncation mutants (both truncated to residue 558) by negative stain EM. CTPS2-ΔC behaved like full-length CTPS2, forming filaments with either substrates or products, consistent with the lack of density for a C-terminal contact in either CTPS2 filament structure (Extended Data Fig. 6a,c). By contrast, unlike full-length CTPS1, CTPS1-ΔC no longer formed S-state filaments (Extended Data Fig. 6c). Overall, this indicates that the additional C-terminal contact is required to stabilize CTPS1 filaments, but is likely sterically incompatible with the P-state conformation; based on the CTPS2 structures, rotation of the glutaminase domains upon transition from the S-state to the P-state increases the distance between adjacent C-termini in the filament by 8Å (Extended Data Fig. 6a,d).
Novel product binding site in the P-state CTPS2 filament
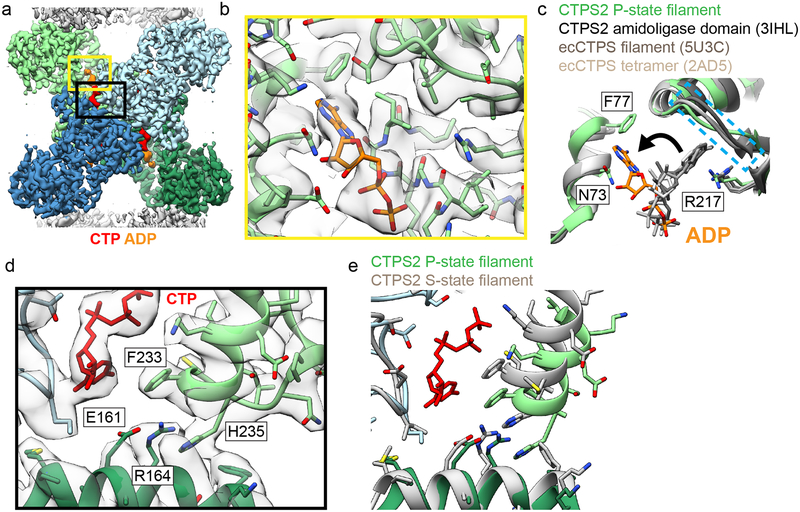
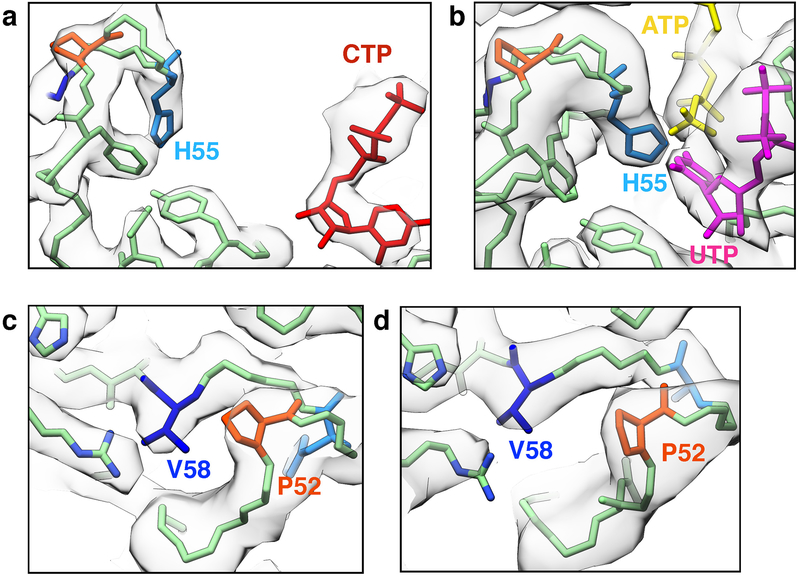
The P-state CTPS2 filament structure revealed a novel ADP binding site. In all existing crystal and cryo-EM structures of CTPS homologs with adenine nucleotides bound, the adenine ring binds to a pocket formed by R211 and the “lid” residues 238–2445,10. In S-state CTPS2 filaments ATP is bound in the same position as in previous structures. By contrast, in the P-state CTPS2 filament, while the ADP phosphates are bound in the conventional position, the adenine base is reoriented by approximately 90° towards the glutaminase domain, and packs in a new site between residues N73 and F77 (Fig. 2a–c). This suggests that the adenine base can bind both sites in CTPS2, and switches to the second site upon transition to the P-state. Furthermore, the overlap between the ATP and ADP binding sites could allow ADP to act as an allosteric regulator, similar to the allosteric regulation observed with CTP at the partially overlapping UTP/CTP binding site.
Fig. 2: Product binding and changes in the tetramer interface in CTPS2 structures.
(a) CTPS2 P-state tetramer, colored by monomer. (b) Zoomed-in view of the yellow box in (a), showing ADP (orange) bound in a novel conformation. Cryo-EM density is shown in transparent grey. (c) Comparison of ADP conformations in the CTPS2 P-state filament (color) with existing ADP-bound CTPS structures (grey). ADP in the P-state filament is packed between residues F77 and N73. In other CTPS structures, ADP is bound to a pocket formed by R217 and lid residues 244–250 (dashed blue box). (d) Zoomed-in view of the black box in (a), showing the CTP binding site. (e) Comparison of the tetramer interface around the CTP binding site in the P-state (blue and green) and S-state (grey) structures.
The CTP-binding mode in the P-state filament structure is the same as that in existing CTP-bound E. coli CTPS structures5,10: helix 224–234 is pulled towards CTP, with F233 packing against the CTP base, producing a hydrogen-bonding network amongst residues E161, R164, and H235 at the tetramerization interface (Fig. 2d, e). Consistent with this binding mode, mutation of these residues has been shown to eliminate feedback inhibition of CTPS15, including of CTPS1 in CHO cells that results in resistance to chemotherapeutic drugs2.
Substrate binding opens a tunnel in the CTPS monomer
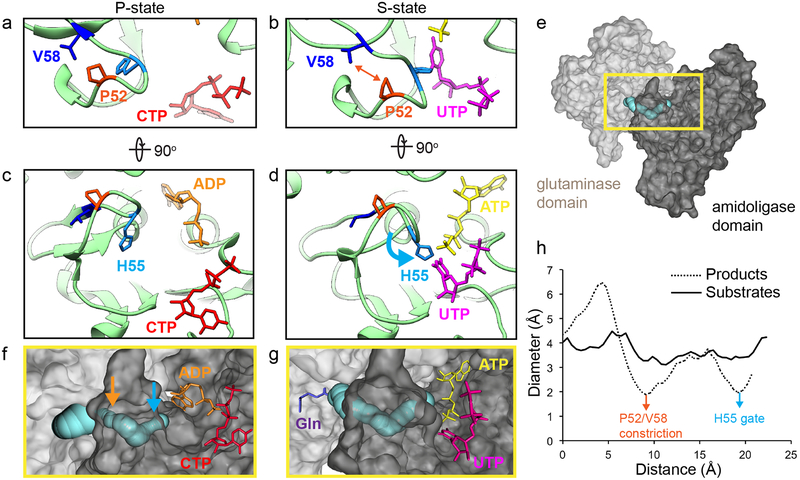
The S-state CTPS2 filament structure is the first near-atomic resolution structure of any CTPS in the substrate-bound conformation, providing insight into the mechanism of ammonia transfer between the two active sites. Previous studies have identified a putative ~25 Å tunnel required to facilitate ammonia transfer between the glutaminase and amidoligase active sites9,24 (Fig. 3, Extended Data Fig. 7). However, in the P-state CTPS2 structure as well as existing P-state bacterial structures, this tunnel is blocked by a constriction formed by conserved residues V58, P52, and H55 (V60, P54, and H57 in E. coli)9 (Fig. 3a, c, Extended Data Fig. 7a, c). Based on a crystal structure of E. coli CTPS, Endrizzi et al.9 predicted that H57 may act as a “gate” at the exit of the ammonia tunnel, with UTP binding altering the orientation of H57, causing the gate to open. Indeed, in the S-state CTPS2 filament, H55 reorients to interact with the UTP base, pulling loop P52-V58 towards the amidoligase active site (Fig. 3b, d, Extended Data Fig. 7b, d) (Supplementary Video 1). This conformational change opens the H55 gate and relieves the P52-V58 constriction, providing a tunnel with a nearly uniform ~4 Å diameter for ammonia transfer between the two active sites (Fig. 3e–h). This structural coupling of substrate binding with opening of the ammonia tunnel likely provides the mechanistic basis for the observed coupling of the two enzymatic activities of CTPS, which ensures ammonia is only released into the active site when a UTP substrate is present to accept it25.
Fig. 3: UTP binding opens a tunnel connecting the amidoligase and glutaminase active sites in the S-state CTPS2 filament.
(a,b) Conformation of residues at the P52-V58 constriction in the P-state (a) and S-state (b) CTPS2 filaments. (c,d) Position of the H55 gate in the P-state (c) and S-state (d) CTPS2 filaments. (e) CTPS2 monomer showing the position of the tunnel (blue) linking the glutaminase (light grey) and amidoligase (dark grey) domains. (f,g) Zoomed-in view of the yellow box in (e), showing tunnels identified by CAVER software in the P-state (f) and S-state (g) CTPS2 structures. Positions of the H55 gate (blue arrow) and P52-V58 constriction (orange arrow) in the P-state structure are indicated. The expected position of glutamine is shown in purple (based on PDB 1VCO). (h) Plot showing the diameter of the tunnel along its length for P- and S-state structures, with positions of the H55 gate (blue arrow) and P52-V58 constriction (orange arrow) indicated.
Regulation of CTPS2 filaments is highly cooperative
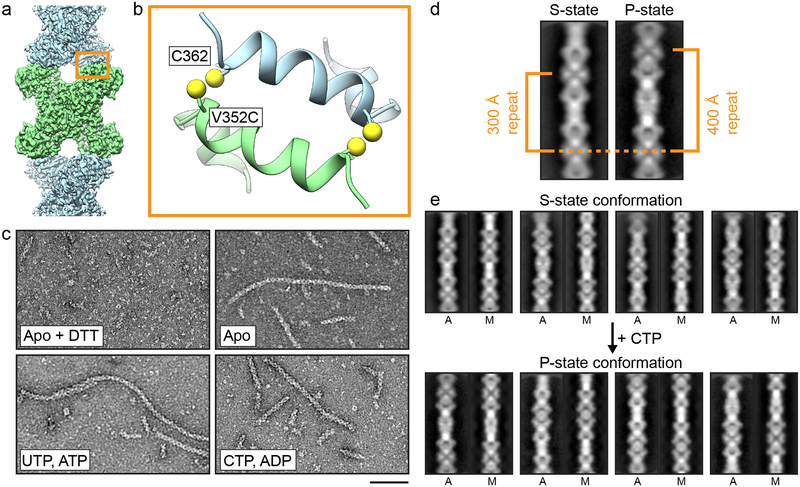
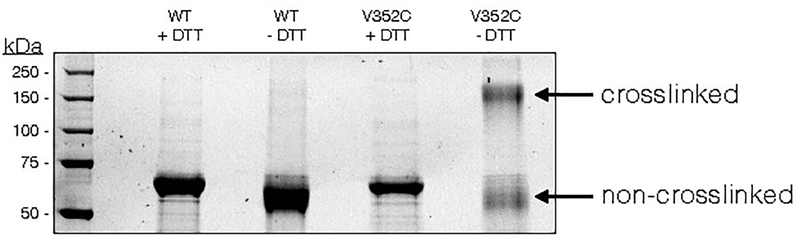
Given that the filament interface is identical in the S-state and P-state structures, we hypothesized that CTPS2 filaments could directly switch between the S-state and P-states while remaining polymerized, perhaps allowing for coordinated conformational changes along entire filaments. To test this hypothesis, we trapped CTPS2 in filaments by engineering cysteine disulfide crosslinks at the filament interface, yielding the CTPS2CC mutant (Fig. 4a, b, Extended Data Fig. 8). In the absence of ligands CTPS2CC spontaneously and robustly polymerized into filaments under non-reducing conditions (Fig. 4c), with approximately 50% of CTPS2CC forming crosslinks, suggesting that even substoichiometric formation of disulfide bonds is sufficient to dramatically stabilize filament assembly (Extended Data Fig. 8). We generated 2D averages of crosslinked CTPS2CC in different ligand states to probe for conformational changes within the filaments. Because their different helical symmetries give rise to a characteristic ~180° repeated view every 300 Å (S-state) or 400 Å (P-state), the different architectures are readily distinguishable in 2D averages (Fig. 4d). Classification and alignment of 2D averages to low-pass filtered projections of the CTPS2 structures revealed that apo CTPS2CC filaments had S-state architecture and transitioned to the P-state upon addition of CTP, confirming that conformational switching within intact filaments is possible (Fig. 4e) (Supplementary Video 2).
Fig. 4: Disulfide-crosslinked CTPS2 filaments can switch conformations.
(a) CTPS2 filament showing the position of the CTPS2CC mutant at the filament interface. (b) Zoomed-in view of the orange box in (a), showing the design of the CTPS2CC mutant. The V352C mutation is across from native C362. (c) Negative stain EM images of hCTPSCC under reducing conditions (+DTT), and under non-reducing conditions in the presence or absence of nucleotides. Scale bars are 50 nm. (d) 2D averages of S- and P-state CTPS2CC filaments, with characteristic 180° repeat distances indicated. (e) 2D class averages of cross-linked CTPS2CC filaments before and after addition of CTP. 2D averages (A) are aligned to low-pass filtered projections of CTPS2 cryo-EM models (M) in the S-state (top) and P-state (bottom) conformations.
We suspected that linking the conformational state of many CTPS2 subunits within a filament could lead to enhanced cooperativity in CTPS2 regulation. We therefore compared the UTP and CTP kinetic parameters of CTPS2 filaments with those of the CTPS2-H355A non-polymerizing mutant (Fig. 5, Extended Data Fig. 9). Unlike CTPS110, polymerization did not increase the Vmax of CTPS2 (Fig. 5a). Further, CTPS2 filaments and CTPS2-H355A homotetramers exhibited nearly identical S0.5 and IC50 values for UTP and CTP (Fig. 5b–d, Supplementary Table 1). CTPS2-H355A inhibition is highly cooperative, with nHill of 3.5 that approaches the theoretical limit for a tetramer (Fig. 5c). CTPS kinetic parameters reported for various species vary significantly, but hill coefficients close to 4 have been reported for both activation and inhibition6,8,26–28. Remarkably, CTPS2 filaments exhibited an even higher nHill of 8.3, providing a switchlike response to changes in CTP concentrations (Fig. 5c). Dilution of wild-type CTPS2 below its critical concentration for assembly caused disassembly into tetramers and occasional short filaments, resulting in a nHill of 3.9, similar to that observed for the non-polymerizing mutant (Fig. 5d, f). Polymerization therefore greatly increases the cooperativity of CTPS2 regulation, likely due to concerted conformational changes within filaments (Fig. 5h).
Fig. 5: Regulation of CTPS2 filaments is highly cooperative.
(a-d) ADP-Glo enzyme assays comparing kinetics of CTPS2 filaments and tetramers. (a) Vmax is the same for CTPS2 filaments and CTPS2-H355A tetramers. (b) UTP kinetic curves for CTPS2 and CTPS2-H355A. Assays were performed with 1500 nM enzyme. (c) CTP inhibition curves for CTPS2 and CTPS2-H355A at 1500 nM protein. nHill values are indicated. (d) CTP inhibition curve for CTPS2 at 300 nM protein. Fits for CTPS2 and CTPS2-H355A inhibition from panel (c) are reproduced for comparison. nHill value is indicated. (e-g) Negative stain EM images of CTPS2 and CTPS2-H355A under ADP-Glo assay conditions. CTPS2 forms filaments at 1500 nM concentration (e), but disassembles into tetramers and rare short filaments at 300 nM concentration (f). CTPS2-H355A does not form filaments at 1500 nM concentration (g). Scale bars are 50 nm. (h) Model for cooperative regulation in CTPS2 filaments. CTP binding induces conformational changes which propagate along filaments. Data shown in graphs are mean and s.d. of n = 3 technical replicates for panels a,b and d, and n = 6 technical replicates for CTPS2 assays in panel (c).
DISCUSSION
It is intriguing that CTPS1, unlike CTPS2, requires an additional C-terminal contact in order to robustly polymerize into filaments. This suggests that the affinity of the primarty contact (containing H355) is reduced in CTPS1 compared to CTPS2. Although the residues that make direct contact across the filament assembly interface are conserved between the two isoforms, there are a number of amino acid substitutions nearby10 that may account for reduced affinity in CTPS1 (Extended Data Fig. 6e–g). In particular, there are differences in two positions that flank the critical assembly residue H355, 354 (CTPS1:Tyr, CTPS2:Phe) and 347 (CTPS1:Ser, CTPS2:Glu), which may create subtle structural differences that shift the interaction affinity at this interface (Extended Data Fig. 6f,g).
Both CTPS1 and CTPS2 contain a number of identified and putative phosphorylation sites in their C-terminal tails29,30. Phosphoregulation via the CTPS1 C-terminus may therefore provide an additional mechanism for regulating CTPS1 polymerization. Differences in the regulatory role of filaments between CTPS1 and CTPS2 may reflect their different physiological roles: consistent with its role in cellular proliferation during the immune response CTPS1 may be induced into filaments that reduce sensitivity to feedback inhibition and allow expansion of CTP pools to meet increased demand, whereas under more homeostatic conditions the extreme sensitivity of CTPS2 fiilaments may help maintain a strictly defined UTP/CTP balance. The possibility also exists that CTPS1 and CTPS2 co-assemble, either at the level of mixed tetramers or mixed filaments; how co-assembly might affect filament structure and biochemical activity will be interesting avenues of future investigation.
Cooperativity in biological systems often arises from the association of protein subunits into oligomeric complexes, allowing for coordinated regulation through coupling of conformational states. Typically, cooperativity is associated with assemblies of relatively few protein subunits, with hemoglobin tetramers providing a canonical example. Large protein arrays and polymers hold the potential for massive cooperativity, with conformational information integrated across hundreds or thousands of protein subunits31–33. Several examples of this phenomenon occur in membrane-embedded systems where two-dimensional arrays can exhibit switchlike transitions, like the chemostatic network controlling bacterial flagellar motion where conformational states propagate across clusters of membrane-bound receptors to amplify external signals34–36, or the coupled gating of ryanodine receptor arrays in muscle cells37. In the case of CTPS2, highly cooperative enzyme regulation results from ligand-induced propagation of conformational changes along linear polymers. This allows for increased sensitivity to substrate and product balance. Sensitivity in the regulation of nucleotide biosynthesis is important to the maintenance of genomic integrity as imbalances in nucleotide pools are linked to increased mutagenesis, sensitization to DNA damaging agents, and multidrug resistance38,39.
Many enzymes in a broad range of metabolic pathways form filamentous polymers19,21,40–43. In the few examples that have been biochemically and structurally characterized to date, enzyme regulation arises from assembling filaments that stabilize particlular conformational states10,15–18. The enhanced cooperativity of CTPS2 is a novel filament-based mechanism of enzyme regulation, which likely serves to stabilize nucleotide levels over a narrow concentration range. This new function for metabolic filaments highlights the diversity of ways in which self-assembly can be adapted to allosterically fine-tune enzyme regulation and improve the efficiency of metabolic control.
METHODS
Purification of CTPS
CTPS1 and CTPS2 were expressed in Saccharomyces cerevisiae strains GHY55 and GHY56, as described by Han et al44. In GHY55 and GHY56, both endogenous Saccharomyces cerevisiae CTP synthase genes URA7 and URA8 are deleted. This lethal deletion is rescued by plasmids pDO105-CTPS1 or pDO105-CTPS2, which direct expression of C-terminally His6-tagged CTPS1 or CTPS2 from the ADH1 promoter. Wild-type and mutant CTPS1 and CTPS2 were all purified by the same method: Cells carrying the pDO105-CTPS constructs were grown overnight in 4X YPD at 30°C, harvested by centrifugation at 4°C at 4000–g, frozen as droplets in liquid nitrogen, and then ground into powder while frozen. Cell powder (~20g) was resuspended in lysis buffer (50 mM Tris-HCl, 200 mM NaCl, 0.3M sucrose, 20 mM imidazole, pH 8.0), and centrifuged at 33,764–g for 40 minutes at 4°C in a Thermo Scientific Fiberlite F14–14 × 50cy rotor. Clarified lysate was loaded onto a 5 mL HisTrap FF Crude column (GE) on an ÄKTA Start chromatography system (GE) pre-equilibrated in column buffer (20 mM Tris-HCl, 0.5M NaCl, 45 mM imidazole, 10% glycerol, pH 7.9). The column was washed with 25 column volumes (CV) of column buffer, and CTPS was eluted with 5CV of elution buffer (20 mM Tris-HCl, 0.5M NaCl, 250 mM imidazole, 10% glycerol, pH 7.9) as 1 mL fractions. Fractions containing CTPS were pooled and dialyzed into storage buffer (20 mM Tris-HCl, 0.5M NaCl, 10% glycerol, 7 mM β-mercaptoethanol, pH 7.9) using Snakeskin 3500 MWCO dialysis tubing (Thermo Scientific). Dialyzed CTPS was then concentration approximately 5-fold using a 3 kDa cut-off centrifugal filter unit (Millipore), flash-frozen in liquid nitrogen, and stored at −80°C.
Cloning of mutants
CTPS2-H355A and CTPS2CC mutants were generated using the Gibson assembly method45. PCR was used to amplify two separate fragments of the pDO105-CTPS2 plasmid backbone flanking the mutation site (forward primer: G CTA TGC AAA GCT GAT GGT ATT C; reverse primer: GTC CTC GGT TTC AGT GAT CTT CTC), which were then ligated together with ~60 base pair DNA fragments containing the desired H355A or V352C mutations (for H355A: C ACT GAA ACC GAG GAC CCT GTG AAA TTT GCT GAA GCT TGG CAG AAG CTA TGC AAA GCT G; for V352C: CTG AAA CCG AGG ACC CTT GTA AAT TTC ATG AAG CTT GGC AGA AGC TAT GCA AAG CTG). To generate the CTPS1-ΔC and CTPS2-ΔC C-terminal truncation mutants, PCR was used to amplify CTPS1(1–558) and CTPS2(1–558) sequences from pDO105-CTPS1 and pDO105-CTPS2 with flanking XhoI and XbaI sites, which were then cloned back into the XhoI and XbaI sites of pDO105 (CTPS1 and CTPS2 forward primer: GCT AAG CTC TAG AAA TAA TTT TGT; CTPS1 reverse primer: G GGG CTC GAG GCC TTT CTG GAG GTA ATG TGA GAG; CTPS2 reverse primer: G GGG CTC GAG ACC CTG TTG CAA GTA GGC). Mutations were confirmed by Sanger sequencing.
Negative-stain electron microscopy
Prior to imaging, CTPS in storage buffer was exchanged into imaging buffer (20 mM Tris-HCl, 100 mM NaCl, 7 mM β-mercaptoethanol, pH 7.9) using a 7K MWCO Zeba Spin Desalting Column (Thermo Scientific). For imaging CTPS2CC disulfide crosslinked filaments, protein was exchanged into non-reducing buffer (20 mM Tris-HCl, 100 mM NaCl, pH 7.9). 100 mM DTT was added to depolymerize CTPS2CC filaments. CTPS was applied to glow-discharged carbon-coated grids, stained with 0.7% uranyl formate, and imaged on a Tecnai G2 Spirit (FEI co.) operating at 120 kV. Images were acquired at 67,000X magnification on a US4000 4k × 4k CCD camera (Gatan, Inc.).
Negative-stain electron microscopy image processing and reconstructions
3D reconstruction of Apo CTPS2 filaments was performed using iterative helical real space reconstruction (IHRSR)46,47 in SPIDER, with hsearch_lorentz48 used to refine helical symmetry parameters, and with D2 point-group symmetry enforced. Cryo-EM structures of S-state or P-state CTPS2 filaments low-pass filtered to 40Å were used as starting models. 2D class averages of CTPS2CC were generated by manually picking particles and performing 2D classification in Relion49. CTPS2CC class averages were aligned to 30Å low-pass filtered projections of the S-state and P-state cryo-EM structures using e2classvsproj.py in EMAN250. For product-bound CTPS1, particles were picked using cisTEM51, then exported to Relion49 for 2D classification and 3D auto-refinment.
Cryo-electron microscopy
Cryo-EM samples were prepared by applying CTPS2 to glow-discharged CFLAT 1.2/1.3 holey-carbon grids (Protochips Inc.), blotting with a Vitrobot (FEI co.), and plunging into liquid ethane. CTPS2 was exchanged into imaging buffer and incubated with nucleotides for 5 minutes at 37°C before preparing cryo-EM samples. Conditions for the S-state filament structure were 7 μM CTPS2, 2 mM UTP, 2 mM ATP, 0.2 mM GTP, and 10 mM MgCl2. Conditions for the P-state filament structure were 8 μM CTPS2, 2 mM CTP, 2 mM ADP, and 10 mM MgCl2. Data for preliminary 3D reconstructions was collected on a Tecnai G2 F20 (FEI co.) operating at 200 kV. Movies were acquired on a K-2 Summit Direct Detect camera in counting mode with a pixel size of 1.26 Å/pixel, collecting 36 frames with a total dose of 45 electrons/Å2, with a defocus range of −1.0 to −2.5 μm. Data for the high-resolution S-State and P-State CTPS2 structures was collected on a Titan Krios (FEI co.) equiped with a Quantum GIF energy filter (Gatan Inc.) operating in zero-loss mode with a 20 eV slit width. Movies were acquired on a K-2 Summit Direct Detect camera in super-resolution mode with a pixel size of 0.525 Å/pixel, collecting 50 frames with a total dose of 90 electrons/Å2. Movies were collected within a defocus range of −1.0 to −2.5 μm. EPU (FEI co.) and Leginon52 software were used for automated data collection.
Cryo-electron microscopy image processing and reconstructions
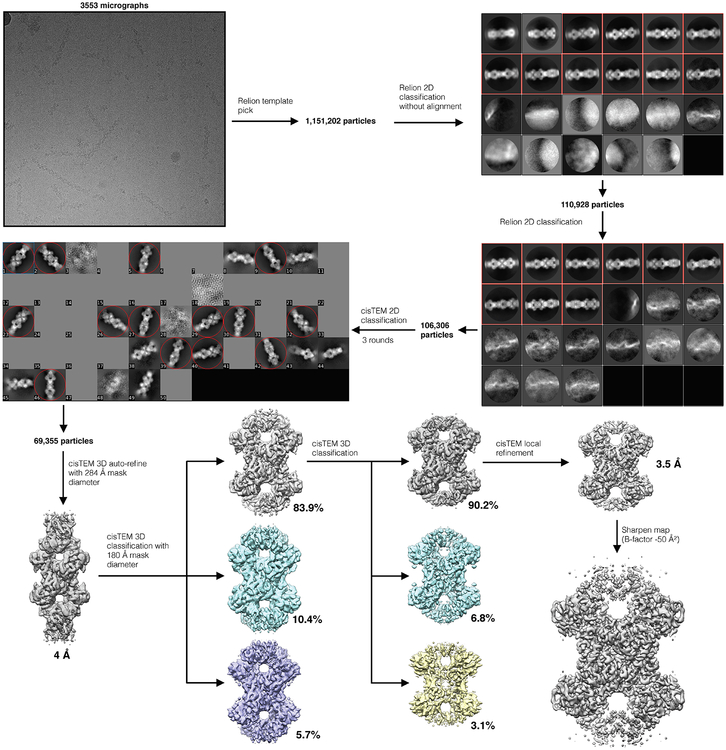
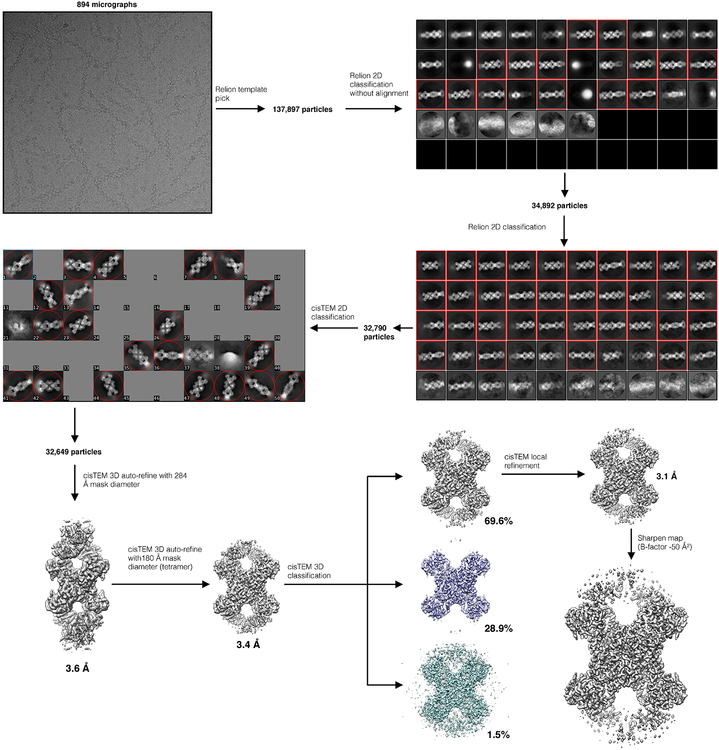
Movie frame alignment and dose-weighted summation of all 50 frames for each movie were performed using MotionCor253, and CTF parameters were estimated using GCTF54. For S-State and P-State CTPS2 data collected on the Tecnai G2 F20, particles were picked manually using Appion55. 3D reconstructions at ~8Å resolution were generated using IHRSR46,47 in SPIDER, using cylinders as starting models and imposing D2 symmetry, with helical symmetry refined using hsearch_lorentz48, providing starting models for subsequent 3D refinement. For Titan Krios data, particles were initially picked manually from a subset of images, and used to generate 2D averages in Relion49. These initial 2D averages were used as templates for Relion automated picking from all images. 2D classification in Relion was used to remove poorliy aligning particle picks, and well-defined particles were then exported to cisTEM51. Further 2D and 3D classification was then performed in cisTEM to remove any remaining poorly aligning particles. Final reconstructions of S-state and P-state CTPS2 were generated in cisTEM, using automatic refinement, followed by manual refinement with CTF refinement implemented. D2 symmetry was imposed during all 3D refinement. Maps were sharpened in cisTEM using a b-factor of −50 Å2. Resolutions were estimated using the FSC0.143 cutoff. Flowcharts of cryo-EM data processing are provided in Supplementary Figures 1 and 2, and additional details of cryo-EM data collection and structure determination are provided in Table 1.
Model building
MODELLER56 was used to generate an initial homology model of the full-length CTPS2 monomer, using partial crystal structures of the human CTPS glutaminase (PDB 2V4U) and amidoligase (PDB 2VO1) domains aligned to a crystal structure of the full-length E. Coli CTPS monomer (PDB 2AD5). The CTPS2 glutaminase, amidoligase, and linker domains were fit individually as rigid bodies into EM maps using Chimera. Structures were then refined using multiple cycles of real-space refinement in Phenix57 and Coot58.
Tunnel modelling
CAVER Analyst59 software was used to model tunnels through the CTPS2 atomic models. The same starting coordinates were used for the S-state and P-state filaments, at a site adjacent to the glutaminase domain catalytic cysteine 399. Probe radii of 0.5Å and 1.5Å were used for the P and S-state structures, respectively, and other tunnel computation parameters were set to default values. The use of a less restrictive, smaller probe radius for the inhibited state allowed us to define a continuous tunnel through the constriction points. Plots of tunnel diameter versus distance were also produced in CAVER Analyst.
CTPS2 kinetics
Kinetic parameters for CTPS2 and CTPS2-H355A were determined using the ADP-Glo assay (Promega), using similar conditions to those described by Sakamoto et al60. Assays were performed in CTPS assay buffer: 50 mM K-HEPES, 5 mM KCl, 0.01% tween 20, 0.01% BSA, 20 mM MgCl2, pH 8.0. All steps of the assay were performed at room temperature in black, low volume 384 well plates (Corning). UTP kinetic assays were performed with 1500 nM CTPS2, 0–150 μM UTP, 500 μM ATP, 5 μM GTP, and 500 μM Glutamine. CTP inhibition assays were performed with 300 nM or 1500 nM CTPS2, 0–70 μM CTP, 100 μM UTP, 100 μM ATP, 5 μM GTP, and 100 μM Glutamine. The total volume of the CTPS2 assays was 6 μl, and reactions were run for 60 minutes (300 nM CTPS2) or 12 minutes (1500 nM CTPS2). CTPS2 reactions were terminated by addition of 6 μl ADP-Glo reagent and incubated for 1 hour, after which 12 μl of kinase detection reagent was added. After one hour, luminescence was recorded using a Varioskan Lux (Thermo Scientific) microplate reader. Assays were performed in triplicate, and three luminescence readings were taken for each assay and averaged. Vmax values were calibrated by comparison to a standard curve with various ADP:ATP ratios in CTPS assay buffer. Kinetics data were fit by 4 parameter logistic regression, solving for maximum rate, minimum rate, hill number, and IC50 or S0.5 (for UTP kinetics: V0 = Vmax+(Vmin - Vmax)/[1+([UTP])/S0.5)^nHill]; for CTP inhibition V0 = Vmin + (Vmax - Vmin)/[1 + ([CTP]/IC50)^nHill)]). Data were plotted as percent maximum rate, according to the formula: 100*[(V0 - Vmin)/(Vmax - Vmin)].
Reporting summary
Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Data availability
Cryo-EM structures and atomic models have been deposited in the EMDB and wwPDB, respectively, with the accession codes EMD-20354, PDB 6PK4 (S-state CTPS2 filament), and EMD-20355, PDB 6PK7 (P-state CTPS2 filament).
Extended Data
Extended Data Fig. 1. CryoEM data processing flowchart for the S-state CTPS2 filament.
Additional details are provided in the Methods section.
Extended Data Fig. 2. CryoEM data processing flowchart for the P-state CTPS2 filament.
Additional details are provided in the Methods section.
Extended Data Fig. 3. Details of CTPS2 cryoEM structures.
(a,b) FSC curves for the S-state (a) and P-state (b) CTPS2 filament structures, showing resolutions of 3.5Å and 3.1Å, respectively, by the FSC0.143 criteria. (c,d) ResMap local resolution maps of the S-state (c) and P-state (d) CTPS2 filament structures. (e) CTPS2 tetramer, with monomers A-D shown in different colors. (f) Zoomed-in view of the orange box in (e), showing S-state CTPS2 (blue), P-state CTPS2 (dark grey), and S-state CTPS1 (light grey) filament structures aligned on the amidoligase domain of subunit A. S-state CTPS1 and CTPS2 are extended across the tetramer interface, compared to P-state CTPS2. (g) Monomers from the S-state (green) and P-state (dark grey) CTPS2 filament structures aligned on the amidoligase domain. (h) Zoomed-in view of the blue box in (g), with S-State CTPS1 also shown in light grey. The glutaminase domain is rotated by 7° towards the amidoligase domain in the S-state structure. (i,j) Two views of the S-state (color) and P-state (grey) CTPS2 monomers aligned on the glutaminase-amidoligase interface. (k) Zoomed-in view of the yellow box in (i). (l) Zoomed-in view of the orange box in (j). The glutaminase-amidoligase interface is essentially identical (Cα RMSD 0.8 Å) in the S-state and P-state filaments.
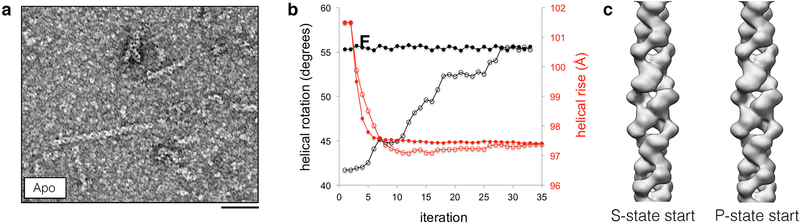
Extended Data Fig. 4. Rare apo CTPS2 filaments have S-state filament architecture.
(a) Negative stain EM images of apo CTPS2. Occasional filaments are observed. (b) Helical rise (red) and rotation (black) values plotted over multiple rounds of iterative helical real space reconstruction of apo CTPS2 filaments in stain. Starting helical symmetry values and models from cryoEM structures of S-state (closed circles) or P-state (open circles) CTPS2 filaments were used. Both reconstructions converge on the S-state filament helical symmetry values. (c) The apo CTPS2 filament structures from the reconstructions described in (b) are the same and have the S-state helical rotation, regardless of which starting symmetries and models are used. Scale bars are 50 nm.
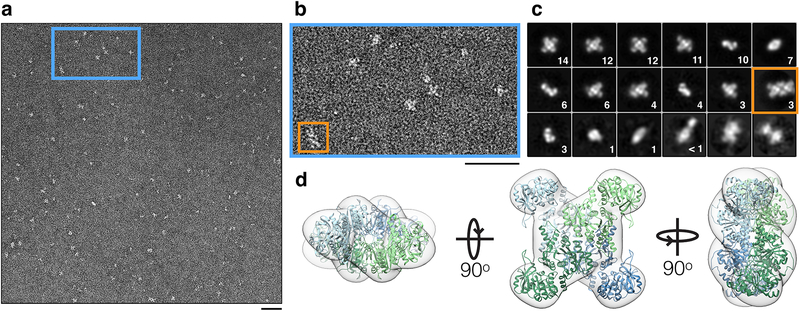
Extended Data Fig. 5. Product-bound CTPS1 tetramers occasionally associate.
(a) Negative stain EM images of CTPS1 in the presence of CTP and ADP. (b) Zoomed-in view of the blue box in (a). The majority of CTPS1 is tetrameric, but rare pairs of tetramers are also observed (orange box). (c) Reference-free 2D averages of CTPS1 in the presence of CTP and ADP. The percentage of all particles in each class is indicated. A single class containing pairs of tetramers (orange box) accounts for 3% of all particles. (d) Negative stain EM reconstruction of the product-bound CTPS1 tetramer at 18Å resolution. The overall architecture resembles structures of existing CTPS tetramers. A CTPS1 tetramer from PBD 5U03 is fit for comparison. Scale bars are 50 nm.
Extended Data Fig. 6. The CTPS1 C-terminus is required for filament formation.
(a) CryoEM maps of CTPS1 and CTPS2 filaments, low-pass filtered to 8Å for comparison. CTPS1 filaments have an additional C-terminal filament contact (orange) that is not observed in S- or P-state CTPS2 filaments. In the CTPS2 filament structures, the distance between adjacent C-termini increases by 8Å upon transition from the S-state to the P-state conformation. (b) Sequence alignment of the CTPS1 and CTPS2 C-termini. Conserved residues are shown in blue. The C-termini have 41% sequence identity, compared with 75% overall sequence identity. (c) Negative stain EM images of CTPS2-ΔC, wild-type CTPS1, and CTPS1-ΔC in the presence of substrates or products. CTPS1-ΔC does not form filaments. (d) Model for the role of the C-terminus in CTPS1 and CTPS2 polymerization. The CTPS1 C-terminus forms a contact that stabilizes the S-state filament, but is incompatible with the P-state conformation. The CTPS2 C-terminus is not required for polymerization. The lack of density for the CTPS2 C-terminus in the S- and P-state structures suggests it is disordered. (e) Sequence alignment of CTPS1 and CTPS2 primary filament contacts, with differing residues highlighted in red. (f,g) CTPS1 (f) and CTPS2 (g) primary filament contact. Residues 347 and 354 adjacent to conserved H355, which differ between the two structures, are shown in orange and yellow. Scale bars are 50 nm.
Extended Data Fig. 7. CryoEM density at the P52-V58 and H55 constriction points.
(a,b) CryoEM density (grey) and atomic models (color) at the H55 gate in the P-state (a) and S-state (b) CTPS2 filaments. (c,d) CryoEM density (grey) and atomic models (color) at the P52-V58 constriction in the P-state (c) and S-state (d) CTPS2 filaments.
Extended Data Fig. 8. CTPS2cc forms disulfide crosslinks.
SDS-PAGE gel of wild-type CTPS2 and CTPS2CC under reducing (100 mM DTT) and non-reducing conditions. Bands for crosslinked CTPS2CC are visible under non-reducing conditions.
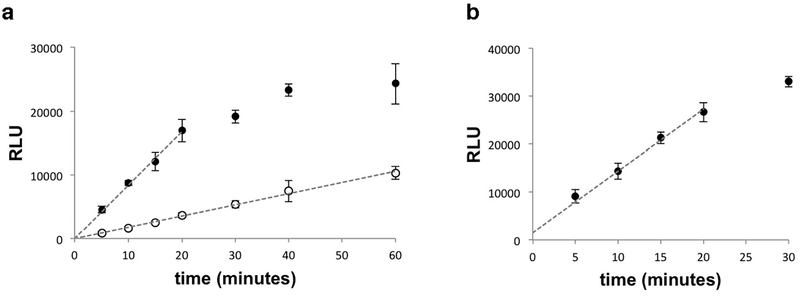
Extended Data Fig. 9. ADP-Glo assays show a linear response in relative light units (RLU) over the time used for CTPS2 kinetics assays.
(a) Time-course of ADP-Glo assay under the substrate conditions used for CTP inhibition assays at 300 nM CTPS2 (open circles) and 1500 nM CTPS2 (closed circles). Rates were calculated from 12 minute timepoints for 1500 nM CTPS2 and 60 minutes for 300 nM CTPS2. (b) Time-course of ADP-Glo assay at 1500 nM CTPS2 with the lowest UTP concentration used in UTP kinetics assays (10 μM UTP). Dashed lines indicate the linear portion of the assay used to measure reaction velocity. Data shown are mean and s.d. of n = 3 technical replicates for all panels.
Supplementary Material
Animation of the CTPS2 active site morphing between the S-state and P-state conformations. The glutaminase domain (green) rotates towards the amidoligase domain (blue), while loop P52-V58 (orange) is pulled towards the UTP base (magenta) in the S-state. ATP (yellow) is also shown in the active site.
Animation of CTPS2 morphing between the S-state and P-state filament conformations. Colored by tetramer. Filament contact sites (orange) remain the same despite conformational changes between the two filament states.
Acknowledgments
We thank G. Carman (Rutgers University) for the CTPS-expressing S. cerevisiae strains. We thank Enoch Baldwin (UC Davis), who provided valuable feedback on our experimental results and early drafts of this manuscript. We are grateful to the Arnold and Mabel Beckman Cryo-EM Center at the University of Washington for use of electron microscopes. This work was supported by the US National Institutes of Health (R01 GM118396 to J.M.K.).
Footnotes
Competing financial interests
The authors declare no competing interests.
REFERENCES
- 1.Trudel M, Van Genechten T & Meuth M Biochemical characterization of the hamster thy mutator gene and its revertants. The Journal of biological chemistry 259, 2355–2359 (1984). [PubMed] [Google Scholar]
- 2.Whelan J, Phear G, Yamauchi M & Meuth M Clustered base substitutions in CTP synthetase conferring drug resistance in Chinese hamster ovary cells. Nat Genet 3, 317–322, doi: 10.1038/ng0493-317 (1993). [DOI] [PubMed] [Google Scholar]
- 3.Aronow B, Watts T, Lassetter J, Washtien W & Ullman B Biochemical phenotype of 5-fluorouracil-resistant murine T-lymphoblasts with genetically altered CTP synthetase activity. The Journal of biological chemistry 259, 9035–9043 (1984). [PubMed] [Google Scholar]
- 4.Carman GM & Zeimetz GM Regulation of phospholipid biosynthesis in the yeast Saccharomyces cerevisiae. The Journal of biological chemistry 271, 13293–13296, doi: 10.1074/jbc.271.23.13293 (1996). [DOI] [PubMed] [Google Scholar]
- 5.Endrizzi JA, Kim H, Anderson PM & Baldwin EP Mechanisms of product feedback regulation and drug resistance in cytidine triphosphate synthetases from the structure of a CTP-inhibited complex. Biochemistry 44, 13491–13499, doi: 10.1021/bi051282o (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Habrian C et al. Inhibition of Escherichia coli CTP Synthetase by NADH and Other Nicotinamides and Their Mutual Interactions with CTP and GTP. Biochemistry 55, 5554–5565, doi: 10.1021/acs.biochem.6b00383 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goto M, Omi R, Nakagawa N, Miyahara I & Hirotsu K Crystal structures of CTP synthetase reveal ATP, UTP, and glutamine binding sites. Structure 12, 1413–1423, doi: 10.1016/j.str.2004.05.013 (2004). [DOI] [PubMed] [Google Scholar]
- 8.MacDonnell JE, Lunn FA & Bearne SL Inhibition of E. coli CTP synthase by the “positive” allosteric effector GTP. Biochim Biophys Acta 1699, 213–220, doi: 10.1016/j.bbapap.2004.03.002 (2004). [DOI] [PubMed] [Google Scholar]
- 9.Endrizzi JA, Kim H, Anderson PM & Baldwin EP Crystal structure of Escherichia coli cytidine triphosphate synthetase, a nucleotide-regulated glutamine amidotransferase/ATP-dependent amidoligase fusion protein and homologue of anticancer and antiparasitic drug targets. Biochemistry 43, 6447–6463, doi: 10.1021/bi0496945 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lynch EM et al. Human CTP synthase filament structure reveals the active enzyme conformation. Nature structural & molecular biology 24, 507–514, doi: 10.1038/nsmb.3407 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martin E et al. CTP synthase 1 deficiency in humans reveals its central role in lymphocyte proliferation. Nature 510, 288–292, doi: 10.1038/nature13386 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kucuk ZY, Zhang K, Filipovich L & Bleesing JJ CTP Synthase 1 Deficiency in Successfully Transplanted Siblings with Combined Immune Deficiency and Chronic Active EBV Infection. Journal of clinical immunology 36, 750–753, doi: 10.1007/s10875-016-0332-z (2016). [DOI] [PubMed] [Google Scholar]
- 13.Williams JC, Kizaki H, Weber G & Morris HP Increased CTP synthetase activity in cancer cells. Nature 271, 71–73 (1978). [DOI] [PubMed] [Google Scholar]
- 14.Fan H, Lu S, Wang S & Zhang S Identification of critical genes associated with human osteosarcoma metastasis based on integrated gene expression profiling. Mol Med Rep, doi: 10.3892/mmr.2019.10323 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barry RM et al. Large-scale filament formation inhibits the activity of CTP synthetase. eLife 3, e03638, doi: 10.7554/eLife.03638 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anthony SA et al. Reconstituted IMPDH polymers accommodate both catalytically active and inactive conformations. Molecular biology of the cell, doi: 10.1091/mbc.E17-04-0263 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Webb BA, Dosey AM, Wittmann T, Kollman JM & Barber DL The glycolytic enzyme phosphofructokinase-1 assembles into filaments. The Journal of cell biology 216, 2305–2313, doi: 10.1083/jcb.201701084 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hunkeler M et al. Structural basis for regulation of human acetyl-CoA carboxylase. Nature 558, 470–474, doi: 10.1038/s41586-018-0201-4 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Carcamo WC et al. Induction of cytoplasmic rods and rings structures by inhibition of the CTP and GTP synthetic pathway in mammalian cells. PloS one 6, e29690, doi: 10.1371/journal.pone.0029690 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Strochlic TI et al. Ack kinase regulates CTP synthase filaments during Drosophila oogenesis. EMBO reports 15, 1184–1191, doi: 10.15252/embr.201438688 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petrovska I et al. Filament formation by metabolic enzymes is a specific adaptation to an advanced state of cellular starvation. eLife, doi: 10.7554/eLife.02409 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aughey GN et al. Nucleotide synthesis is regulated by cytoophidium formation during neurodevelopment and adaptive metabolism. Biology open 3, 1045–1056, doi: 10.1242/bio.201410165 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Calise SJ et al. Glutamine deprivation initiates reversible assembly of mammalian rods and rings. Cellular and molecular life sciences : CMLS 71, 2963–2973, doi: 10.1007/s00018-014-1567-6 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCluskey GD & Bearne SL “Pinching” the ammonia tunnel of CTP synthase unveils coordinated catalytic and allosteric-dependent control of ammonia passage. Biochim Biophys Acta Gen Subj 1862, 2714–2727, doi: 10.1016/j.bbagen.2018.08.008 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Willemoes M & Sigurskjold BW Steady-state kinetics of the glutaminase reaction of CTP synthase from Lactococcus lactis. The role of the allosteric activator GTP incoupling between glutamine hydrolysis and CTP synthesis. Eur J Biochem 269, 4772–4779 (2002). [DOI] [PubMed] [Google Scholar]
- 26.Kang GJ et al. Cyclopentenylcytosine triphosphate. Formation and inhibition of CTP synthetase. The Journal of biological chemistry 264, 713–718 (1989). [PubMed] [Google Scholar]
- 27.Long CW & Pardee AB Cytidine triphosphate synthetase of Escherichia coli B. I. Purification and kinetics. The Journal of biological chemistry 242, 4715–4721 (1967). [PubMed] [Google Scholar]
- 28.McCluskey GD, Mohamady S, Taylor SD & Bearne SL Exploring the Potent Inhibition of CTP Synthase by Gemcitabine-5’-Triphosphate. Chembiochem 17, 2240–2249, doi: 10.1002/cbic.201600405 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Kassel KM, Au da R, Higgins MJ, Hines M & Graves LM Regulation of human cytidine triphosphate synthetase 2 by phosphorylation. The Journal of biological chemistry 285, 33727–33736, doi: 10.1074/jbc.M110.178566 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choi MG & Carman GM Phosphorylation of human CTP synthetase 1 by protein kinase A: identification of Thr455 as a major site of phosphorylation. The Journal of biological chemistry 282, 5367–5377, doi: 10.1074/jbc.M610993200 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bray D & Duke T Conformational spread: the propagation of allosteric states in large multiprotein complexes. Annu Rev Biophys Biomol Struct 33, 53–73, doi: 10.1146/annurev.biophys.33.110502.132703 (2004). [DOI] [PubMed] [Google Scholar]
- 32.Wyman J Possible allosteric effects in extended biological systems. J Mol Biol 39, 523–538 (1969). [DOI] [PubMed] [Google Scholar]
- 33.Changeux JP, Thiery J, Tung Y & Kittel C On the cooperativity of biological membranes. Proceedings of the National Academy of Sciences of the United States of America 57, 335–341, doi: 10.1073/pnas.57.2.335 (1967). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sourjik V & Berg HC Functional interactions between receptors in bacterial chemotaxis. Nature 428, 437–441, doi: 10.1038/nature02406 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Bai F et al. Conformational spread as a mechanism for cooperativity in the bacterial flagellar switch. Science 327, 685–689, doi: 10.1126/science.1182105 (2010). [DOI] [PubMed] [Google Scholar]
- 36.Cluzel P, Surette M & Leibler S An ultrasensitive bacterial motor revealed by monitoring signaling proteins in single cells. Science 287, 1652–1655 (2000). [DOI] [PubMed] [Google Scholar]
- 37.Marx SO, Ondrias K & Marks AR Coupled gating between individual skeletal muscle Ca2+ release channels (ryanodine receptors). Science 281, 818–821 (1998). [DOI] [PubMed] [Google Scholar]
- 38.Meuth M The molecular basis of mutations induced by deoxyribonucleoside triphosphate pool imbalances in mammalian cells. Exp Cell Res 181, 305–316 (1989). [DOI] [PubMed] [Google Scholar]
- 39.Mathews CK Deoxyribonucleotide metabolism, mutagenesis and cancer. Nat Rev Cancer 15, 528–539, doi: 10.1038/nrc3981 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Noree C, Sato BK, Broyer RM & Wilhelm JE Identification of novel filament-forming proteins in Saccharomyces cerevisiae and Drosophila melanogaster. The Journal of cell biology 190, 541–551, doi: 10.1083/jcb.201003001 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu JL Intracellular compartmentation of CTP synthase in Drosophila. Journal of genetics and genomics 37, 281–296, doi: 10.1016/S1673-8527(09)60046-1 (2010). [DOI] [PubMed] [Google Scholar]
- 42.Ingerson-Mahar M, Briegel A, Werner JN, Jensen GJ & Gitai Z The metabolic enzyme CTP synthase forms cytoskeletal filaments. Nature cell biology 12, 739–746, doi: 10.1038/ncb2087 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shen QJ et al. Filamentation of Metabolic Enzymes in Saccharomyces cerevisiae. Journal of genetics and genomics 43, 393–404, doi: 10.1016/j.jgg.2016.03.008 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Han GS et al. Expression of Human CTP synthetase in Saccharomyces cerevisiae reveals phosphorylation by protein kinase A. The Journal of biological chemistry 280, 38328–38336, doi: 10.1074/jbc.M509622200 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gibson DG et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nature methods 6, 343–345, doi: 10.1038/nmeth.1318 (2009). [DOI] [PubMed] [Google Scholar]
- 46.Egelman EH The iterative helical real space reconstruction method: surmounting the problems posed by real polymers. Journal of structural biology 157, 83–94, doi: 10.1016/j.jsb.2006.05.015 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Sachse C et al. High-resolution electron microscopy of helical specimens: a fresh look at tobacco mosaic virus. J Mol Biol 371, 812–835, doi: 10.1016/j.jmb.2007.05.088 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Egelman EH A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy 85, 225–234 (2000). [DOI] [PubMed] [Google Scholar]
- 49.Scheres SH RELION: implementation of a Bayesian approach to cryo-EM structure determination. Journal of structural biology 180, 519–530, doi: 10.1016/j.jsb.2012.09.006 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tang G et al. EMAN2: an extensible image processing suite for electron microscopy. Journal of structural biology 157, 38–46, doi: 10.1016/j.jsb.2006.05.009 (2007). [DOI] [PubMed] [Google Scholar]
- 51.Grant T, Rohou A & Grigorieff N cisTEM, user-friendly software for single-particle image processing. eLife 7, doi: 10.7554/eLife.35383 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Suloway C et al. Automated molecular microscopy: the new Leginon system. Journal of structural biology 151, 41–60, doi: 10.1016/j.jsb.2005.03.010 (2005). [DOI] [PubMed] [Google Scholar]
- 53.Zheng SQ et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nature methods 14, 331–332, doi: 10.1038/nmeth.4193 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang K Gctf: Real-time CTF determination and correction. Journal of structural biology 193, 1–12, doi: 10.1016/j.jsb.2015.11.003 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lander GC et al. Appion: an integrated, database-driven pipeline to facilitate EM image processing. Journal of structural biology 166, 95–102 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Webb B & Sali A Comparative Protein Structure Modeling Using MODELLER. Curr Protoc Protein Sci 86, 2 9 1–2 9 37, doi: 10.1002/cpps.20 (2016). [DOI] [PubMed] [Google Scholar]
- 57.Adams PD et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66, 213–221, doi: 10.1107/S0907444909052925 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Emsley P, Lohkamp B, Scott WG & Cowtan K Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501, doi: 10.1107/S0907444910007493 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jurcik A et al. CAVER Analyst 2.0: analysis and visualization of channels and tunnels in protein structures and molecular dynamics trajectories. Bioinformatics 34, 3586–3588, doi: 10.1093/bioinformatics/bty386 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sakamoto K et al. Identification of cytidine-5-triphosphate synthase1-selective inhibitory peptide from random peptide library displayed on T7 phage. Peptides 94, 56–63, doi: 10.1016/j.peptides.2017.06.007 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Animation of the CTPS2 active site morphing between the S-state and P-state conformations. The glutaminase domain (green) rotates towards the amidoligase domain (blue), while loop P52-V58 (orange) is pulled towards the UTP base (magenta) in the S-state. ATP (yellow) is also shown in the active site.
Animation of CTPS2 morphing between the S-state and P-state filament conformations. Colored by tetramer. Filament contact sites (orange) remain the same despite conformational changes between the two filament states.
Data Availability Statement
Cryo-EM structures and atomic models have been deposited in the EMDB and wwPDB, respectively, with the accession codes EMD-20354, PDB 6PK4 (S-state CTPS2 filament), and EMD-20355, PDB 6PK7 (P-state CTPS2 filament).