Summary
The B cell response to Ehrlichia muris is dominated by plasmablasts (PB), with few - if any - germinal centers (GC), yet it generates protective IgM memory B cells (MBC) that express the transcription factor T-bet and harbor V region mutations. As Ehrlichia prominently infects the liver, we investigated the nature of liver B cell response and that of the spleen. B cells within infected livers proliferated and underwent somatic hypermutation (SHM). Vh region sequencing revealed trafficking of clones between the spleen and liver that was often followed by local clonal expansion and intraparenchymal localization of T-bet+ MBC. T-bet+ MBC expressed MBC subset markers CD80 and PD-L2. Many T-bet+ MBC lacked CD11b or CD11c expression but had marginal zone (MZ) B cell phenotypes and colonized the splenic MZ, revealing T-bet+ MBC plasticity. Hence, liver and spleen are generative sites of B cell responses that include V region mutation and result in liver MBC localization.
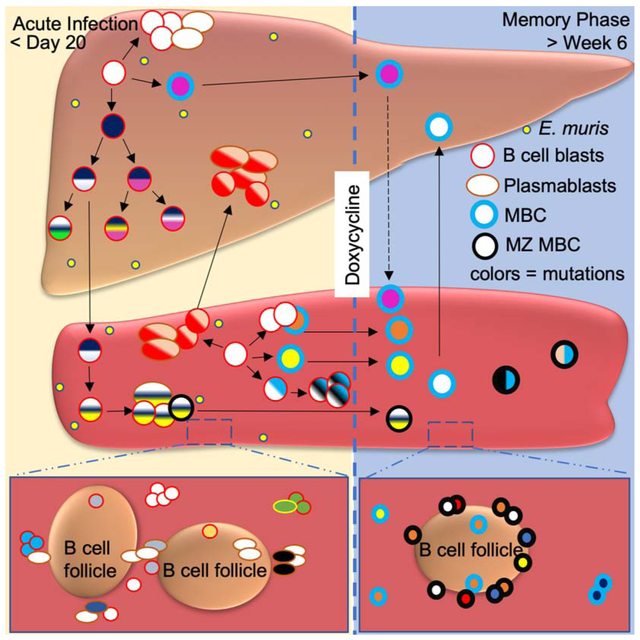
Graphical Abstract
eTOC Blurb
Infection by the intracellular bacterium Ehrlichia induces few - if any - germinal centers, yet it generates protective IgM memory B cells (MBC). Trivedi et al. show that the liver and spleen are generative sites of B cell responses to E. muris, including V region mutation and long-term MBC localization.
INTRODUCTION
The conventional B cell response to pathogens such as the influenza virus and the malarial parasite is dependent on a GC pathway that results in the production of antibody forming cells (AFC) and MBC (Coro et al., 2006, Stephens et al., 2009). However, certain pathogens such as Borrelia burgdorferi, Salmonella typhimurium and Ehrlichia muris suppress or delay the onset of a GC response; B cell responses instead follow a non-canonical pathway (Hastey et al., 2012, Cunningham et al., 2007, Racine et al., 2010, Di Niro et al., 2015). Ehrlichia is a gram-negative, obligate intracellular bacterium that causes a tick-borne infection (Anderson et al., 1991, Dawson et al., 1991). In humans, infection by Ehrlichia chaffeensis causes human monocytotropic ehrlichiosis, which is characterized by flu-like symptoms such as fever, headache, myalgia, and hematological abnormalities (Ismail and McBride, 2017). In both humans and mice, liver is a prominent site of Ehrlichia infection ((Sehdev and Dumler, 2003, Ismail et al., 2004, Ismail et al., 2010)). Ehrlichia induces a B cell response in humans, with antibodies detected in the serum of infected patients (Standaert et al., 2000). In mice, Ehrlichia infection induces large numbers of IgM AFC and considerable yet comparatively lower numbers of IgG AFC (Racine et al., 2008, Racine et al., 2010, Winslow et al., 2000).
Ehrlichia infection induces the expression of the transcription factor T-bet in AFC and a subset of splenic memory B cells (MBC) (Winslow et al., 2017). While T-bet expression in B cells was originally documented as a regulator of isotype switch induced in response to TLR9 signals (Peng et al., 2002, Jegerlehner et al., 2007), its expression has been closely associated with so-called age-associated B cells (ABC) (Rubtsov et al., 2011, Hao et al., 2011) . ABC are found especially in older female mice and in autoimmune-prone mice (Hao et al., 2011, Rubtsov et al., 2011). These T-Bet+ ABC are typically CD11b+ and CD11c+, but lack expression of CD21 and CD23 (Hao et al., 2011). A similar population has been identified in humans and is associated with lupus. T-bet+ B cells can also be induced by various infections and T-bet can also be expressed in PB. (Rubtsova et al., 2013, Barnett et al., 2016, Moir et al., 2008, Rubtsov et al., 2011, Rubtsova et al., 2017, Rubtsov et al., 2013). A subset of MBC formed during certain conditions, including Ehrlichia infection, can express T-bet as well. The role of T-bet in B cells and its relationship to ABC, MBC and PB development and function is an active area of research, and the relationships among these cells and processes is not fully clear.
Despite the fact that liver is a primary site for infection in humans and mice (Ismail et al., 2010, Ismail et al., 2004, Sehdev and Dumler, 2003), there is limited information on hepatic B cell responses to Ehrlichia (Miura and Rikihisa, 2009, Habib et al., 2016). Here we examined the extent to which the B cell response to Ehrlichia occurs in the liver and the consequences of this local response. We found that the liver was a major locus for B cell proliferation and SHM during the acute phase of the immune response. High throughput sequencing (HTS) analyses revealed bi-directional trafficking of mutated B cell blasts and PB between the spleen and liver. After pathogen clearance, we observed T-bet expressing MBC that persisted in the spleen and that were localized in the liver, including some that were histologically intraparenchymal and resisted intravascular labeling with i.v. anti-CD19. In the spleen, Ehrlichia infection remodeled the MZ compartment, which initially dissolved and was later reconstituted by a majority of T-bet expressing MBC induced by infection. Although T-bet expressing MBC populations have generally been phenotyped as CD11b+CD11c+CD21−CD23−, many formed post-Ehrlichia infection have a CD21hiCD23low MZ phenotype. Further, only a fraction of these are CD11b+CD11c+. Hence, Ehrlichia infection elicits unusual populations of T-bet+ MBC and reveals additional plasticity among these cells.
RESULTS
E. muris infection induces a robust B cell response marked by T-bet expression
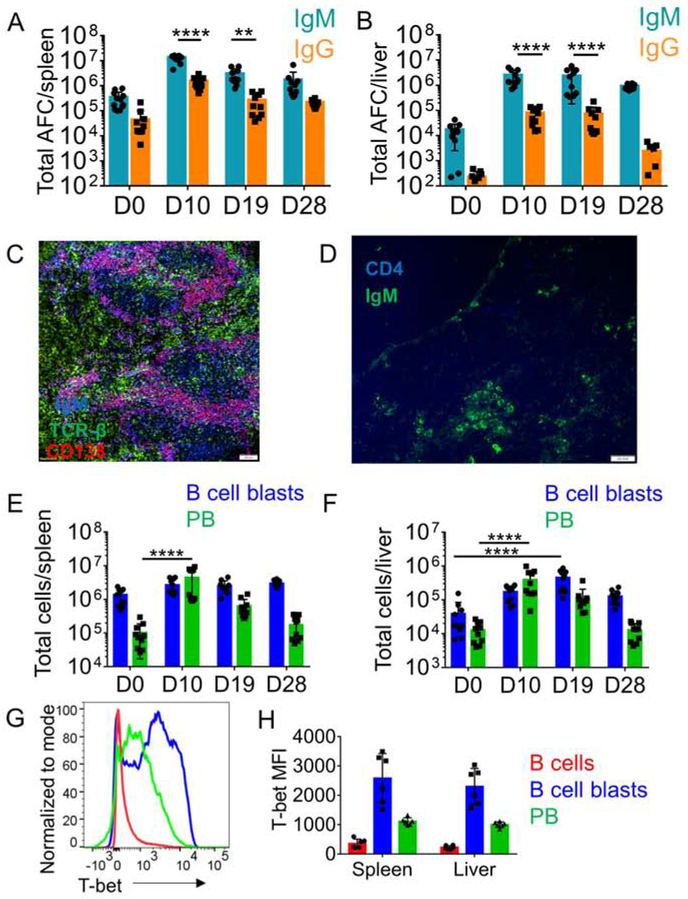
Inoculation of mice with E. muris leads to systemic infection of spleen, and liver (Ismail et al., 2004, Olano et al., 2004). In response to Ehrlichia, splenic B cells do not form GCs and instead respond by rapid extrafollicular expansion (Racine et al., 2010). In agreement with previous reports (Ismail et al., 2004, Olano et al., 2004), we found that E. muris infection was marked by considerable bacterial burden in spleen and liver along with enlargement of these organs (figure S1A-B). In both spleen and liver, infection elicited large numbers of AFC by day 10, which remained elevated until at least day 28 post-infection (figure 1A-B). There was a substantial IgG response, but the number of IgG AFC was lower than IgM AFC (figure 1A-B). Responding B cells were located within patches at extrafollicular sites in spleen and in the liver parenchyma (figure 1C-D, figure S1D-E). Flow cytometric analysis of spleen and liver from infected mice revealed expanded populations of B cell blasts (CD44+) and PB (CD44+CD138+PB) (figure S1F and figure 1E-F). Consistent with previous reports (Racine et al., 2010), we did not find induction of PNA-positive GC B cells by flow cytometry (FC) (figure S1C). Confirming previous studies (Winslow et al., 2017), we found that PB in E. muris-infected mice expressed T-bet. B cell blasts also expressed high amounts of T-bet. (figure 1G-H).
Figure 1: E. muris infection induces a robust B cell response marked by T-bet expression.
(A,B) Total IgM and IgG AFC measured by ELISpot assay during E. muris infection in spleen (A) and liver (B), (C,D) Immunofluorescence staining of cryo-sections from spleens (C) of infected mice (D10) for PB and cryosections of liver (D) for B cells and T cells. In C scale bars represent 100μm and D scale bars represent 100 pixels. (E,F) Total B cell blasts (blue) and PB (green) measured by FC over the course of E. muris infection in spleen (E) and liver (F), (G, H) Histogram (G) and Mean Fluorescence Intensity (H) of T-bet expression in B cells during acute E. muris infection. Data are representative of at least two independent experiments and in (A, B, E, F, H) data are represented as mean with SD of groups of at least two mice. *p <0.05, **p<0.01, ***p<0.001, ****p<0.0001 Statistics for A, B, E, F were done by two-way ANOVA. See figure S1-S2.
To decipher the role of T-bet in the acute B cell response, we used Tamoxifen inducible Cre mediated deletion of T-bet during an ongoing E. muris infection (figure S2A). Assessment of T-bet expression on D11 post-infection revealed that treatment with tamoxifen reduced T-bet expression on splenic B cell blasts and PB and showed a similar trend in hepatic B cell populations (figure S2B). Since liver is a metabolically active site, we suspect that tamoxifen got metabolized rapidly by liver cells and was not available for B cells. We assessed the total number of responding B cells and found that splenic PB were higher in T-bet deficient mice compared to the T-bet sufficient group (figure S2C). There was no difference in the total numbers of splenic B cell blasts, hepatic B cell blasts, and hepatic PB upon T-bet deletion (figure S2C). Enzyme-linked Immunosorbent Spot (ELISpot) assay showed a trend of increased IgM AFC in the spleen and liver but there was no difference in total and antigen-specific splenic and hepatic AFC upon T-bet deletion (figure S2 D-I). Unlike influenza infection and mouse models of lupus disease (Stone et al., 2019, Rubtsova et al., 2017), we did not observe a dampened AFC response in the absence of T-bet. It is possible that even though we did observe lower T-bet expression in responding populations, that deletion during B cell development, as performed in the influenza and lupus models, differed from inducible deletion we performed here.
A Diverse B cell receptor (BCR) Repertoire is required for the AFC response to E. muris
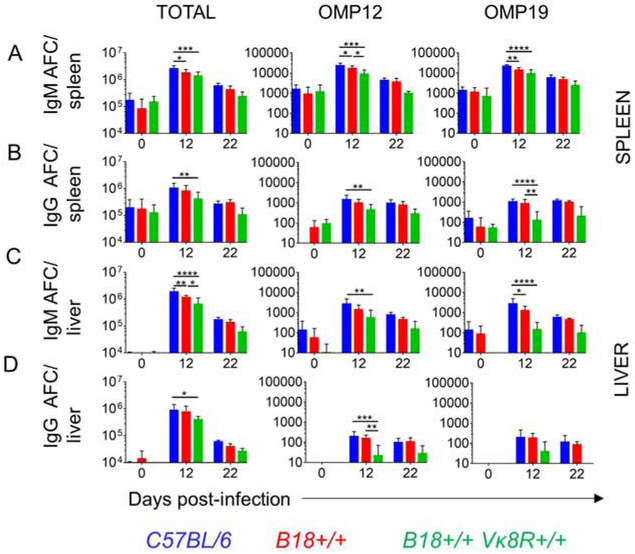
We examined the specificity of the responding PB using E. muris outer membrane proteins Omp 12 and Omp19 as coating antigens (Ags) in ELISpot assays (Crocquet-Valdes et al., 2011). Although these Ags are reported to be immunodominant E. muris Ags (Racine et al., 2008, Yates et al., 2013), we found that less than 1% of IgM and IgG AFC were specific to these Ags in both spleen and liver (figure 2 A-D). This result is consistent with non-specific expansion of B cells mediated by signals through different pathogen recognition receptors without relying on BCR recognition or stimulation. However, if the B cell response were indeed specific, then restricting the BCR repertoire would negatively impact the AFC response. To examine this, we used heavy chain-restricted B18+/− mice that utilize B18 heavy chain gene paired with diverse light chains as well as heavy and light chain restricted B18+/− Vκ8R +/− mice that utilize B18 heavy chain and Vκ8R light chain genes. In general, we observed a trend of reduced total and antigen specific AFC response in the spleens and livers of the BCR restricted mice when compared to wild-type (WT) counterparts (figure 2). In spleen, we observed that heavy and light chain restriction led to reduced total and Omp12 and Omp19 AFC at day 12 post infection (figure 2A-B). The splenic total and antigen specific IgM AFC were also reduced merely by heavy chain restriction at day 12 post infection (figure 2A). The hepatic total IgM and IgG AFC, Omp12 specific IgM and IgG AFC and Omp19 specific IgM AFC were reduced in B18+/− Vκ8R+/− mice compared to WT mice at day 12 post infection (figure 2C-D). In all scenarios, BCR restricted mice mounted an increased response in comparison to baseline, however, failed to mount an AFC response that matched the magnitude of the WT repertoire (figure 2). No differences were observed in the AFC response between WT and BCR restricted mice at D22 post-infection as the acute response had subsided (figure 2). These data demonstrate that antigen sensing through the BCR is required for the massive AFC response that is seen upon E. muris infection and suggest that while it may be of low affinity, the majority of it depends on specific BCR recognition.
Figure 2: BCR restriction reduces the magnitude of B cell response to E. muris.
(A-D panel 1) Total IgM and IgG response to E. muris in spleen (A-B) and liver (C-D) measured by ELISpot assay. (A-D panels 2 and 3) Antigen-specific IgM and IgG response to E. muris in spleen (A-B) and liver (C-D) measured by ELISpot assay. Data are representative of at least two independent experiments and in (A-D) data are represented as mean with SD of groups of at least two mice. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. Statistics for panel A-D were done by two-way ANOVA.
Localized proliferation and SHM of B cells in the liver
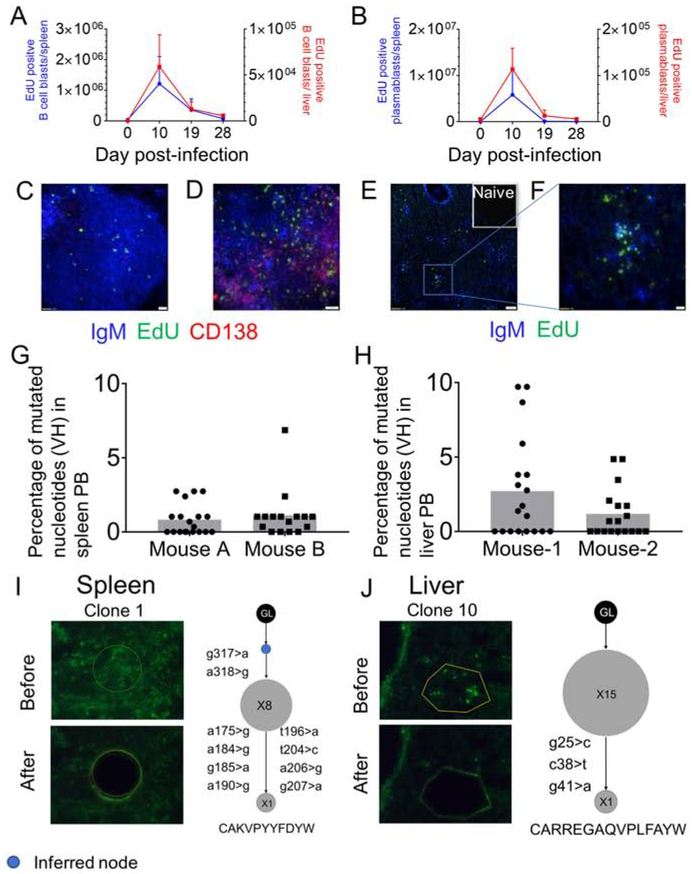
While increased numbers of B cell blasts and PB were found in the livers of E. muris-infected mice, it was not clear whether the hepatic B cell response is a product of infiltrating B cells derived from lymphoid organs or from local proliferation and differentiation of B cells in the liver. To investigate this, we injected the mice with 5-Ethynyl-2’-deoxyuridine (EdU) 30 minutes prior to the harvest of organs to label cells that were actively undergoing DNA synthesis during that period. As expected, there was a substantial increase in EdU-positive splenic B cell blasts and PB compared to the naïve control at 10 days post-infection (figure 3A-B). Hepatic B cell blasts and PB were proliferating to an extent comparable to the splenic responders (figure 3A-B). Histologic analysis showed that proliferation of IgM and CD138 positive B cells in the spleen occurred within and outside of B cell follicles (figure 3C-D, figure S3A). Proliferation of IgM positive B cells in the liver was seen in the liver parenchyma and around the portal triads (figure 3E-F, figure S3B). These data indicate that liver is a generative site for the B cell response to E. muris.
Figure 3. Localized proliferation and SHM of B cells in the spleen and liver.
EdU positive B cell blasts (A) and PB (B) of spleen (blue) and liver (red) over the course of infection measured by FC. (C-D) 40X image of proliferating B cells in a naïve spleen (C) and E. muris infected spleen at D10 post infection (D). (E, F) 10X (E) and 40X (F) images of proliferating B cells in the liver parenchyma at D10 post infection. In C and F, scale bars represent 20μm, in D scale bars represent 200 pixels and in E, scale bars represent 100μm. (G-H) Percentage of mutated nucleotides in PB from spleen (G) and liver (H). (I-J) Laser microdissections of IgM-positive PB patches (green) and corresponding clonal trees from Ig region sequencing from the spleen (I) and the liver (J). Inferred nodes are blue. Node size is proportional to the number of sequences. The CDR3 amino acid sequence of each clone shown at the bottom. Data are representative of at least two independent experiments. See figure S3 and table S1.
To investigate the presence of SHM in PB responses that lacked GCs, we assessed the expression of the enzyme Activation Induced Cytidine Deaminase (AID). We observed that both splenic and hepatic B cell blasts and PB expressed higher amounts of AID than naïve B cells and T cells (figure S3 C-E). To further asses SHM, we amplified and sequenced V region genes from DNA of splenic and hepatic FACS sorted PB. The mutation rate was 1% for PB from spleen and 1.5% from liver (figure 3G-H). However, approximately 50% of V genes sequences were unmutated at both sites (figure 3G-H); indicating that the cells harboring these did not induce the SHM program. Mutated PB could either be generated during local proliferation or undergo mutation at a separate site and then migrate. Since liver is a generative site, we hypothesized that mutation could occur within locally responding foci. To address where mutations were actually occurring, we used laser microdissection of IgM patches from the liver and spleen parenchyma. These microdissections typically captured ~20 cells, of which the full nucleus would be present in about ½ of them. The finding of intraclonal diversity among small groups of isolated cells demonstrates ongoing mutation at that site (Jacob et al., 1991, William et al., 2002, Di Niro et al., 2015). Of 11 microdissections from spleen, 6 had clones with sequences that were mutated from the closest germline (GL) Vh gene (Table S1). Of 10 microdissections from the liver, 7 had sequences with mutations when compared to the closest GL Vh gene (Table S1). These clonally related sequences could be assembled into 4 clonal lineage trees from the spleen (figure 3I, and figure S3F) and 4 clonal lineage trees from the liver (figure 3J, figure S3G). The finding in multiple cases at each site that there were cells that shared mutations (i.e. the clones had trunks) and that also differed by other mutations (i.e. the clones had branches) provides evidence that SHM occurred locally in the spleen and the liver parenchyma.
Repertoire characteristics of the acute B cell response in spleen and liver
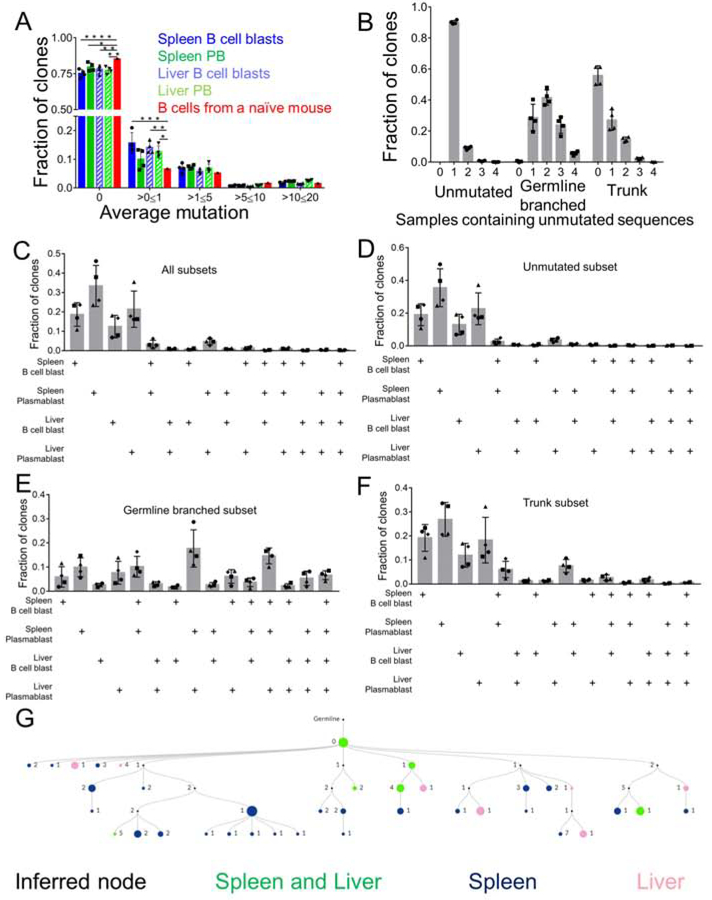
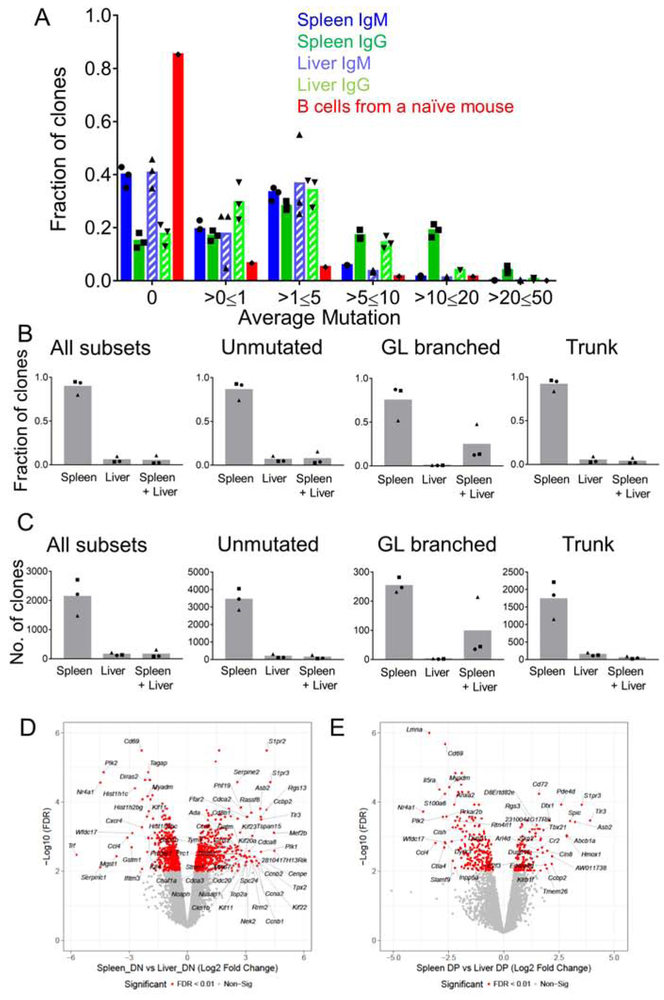
Next, to more deeply investigate the extent of SHM and the magnitude of overlap of the responding B cell repertoire in different B cell populations at both sites, we performed high throughput sequencing of the heavy chain VDJ mRNA, as described previously (Di Niro et al., 2015). We FACS sorted splenic and hepatic B cell blasts and PB at D13 post-infection and created libraries with V region sequences of these 4 populations. HTS analysis was done as previously described (Rosenfeld et al., 2018, Sheneman et al., 2006). Clones were assembled using ImmuneDB (see Methods), but to avoid spurious results were included in this analysis only if they had between 1 and 4 non-templated Complementarity-Determining Region 3 (CDR3) residues (as measured from the first non-GL-encoded nucleotide) or had at least 4 common V-gene mutations across all sequences. We found that about 75% of the clones within the 4 populations were unmutated (figure 4A), suggesting that Ehrlichia infection induced SHM in only a subset of B cells, which is consistent with our microdissection results (figure 3G-J). Overall, 10-15% of the clones had an average mutation of >0 and <=1 and 5-10% of the clones had an average mutation between 1-5 (figure 4A). To gain insight into how clones grew and migrated, we categorized the clones according to their clonal lineage characteristics into 3 groups that might reflect different origins or patterns of clonal expansion: Unmutated, GL branched (mutated clones that do not have a common shared mutation) and Trunk (clones that have a common shared mutation and bifurcate further). Although the total number of Unmutated clones was the greatest (at ~75%, Fig. 4A), GL branched clones were substantially larger in terms of both clone size and “instances” (defined as total number of unique sequences), while Trunk clones were intermediate (figure S4 A-B). Hence, while there were a greater number of unmutated clones, those that did mutate had expanded considerably in terms of numbers of sequences and thus presumably numbers of cells.
Figure 4. SHM in spleen and liver B cell blasts and PB.
(A) Mutation distribution in splenic and hepatic B cell blasts and PB populations. (B) Fraction of unmutated, GL-branched and trunk clones found in any of the 4 populations of hepatic and splenic B cell blasts and PB. (C-F) Fraction of clones shared within splenic and hepatic B cell blasts and PB populations of all clones (C), Unmutated clones (D), GL branched clones (E), and Trunk clones (F). (G) An example of a multi-tiered clonal lineage found in both spleen and liver. Nodes found in both organs are green, nodes found in spleen only are blue, nodes found in liver only are pink, inferred nodes are black. Node size is proportional to the number of sequences. *p <0.05, **p<0.01, ***p<0.001, ****p<0.0001 Statistics for A were done by two-way ANOVA. See figure S4.
Within each category we then assessed the extent of sharing of clones between the 4 populations that were sequenced. Among Unmutated clones, the vast majority (~90%) were found in only one population and only 10% were found in as many as 2 populations (figure 4B). However, the GL branched clones demonstrated extensive spread among the different populations, with 40% of clones found in 2 populations, ~25% clones found in 3 populations and ~5% shared among all 4 populations (figure 4B). In the case of the Trunk clones, we found that about 20% of the clones were shared between 2 populations (figure 4B).
To determine patterns of migration and differentiation we assessed the fraction of clones that overlapped among all the populations, a minimal estimate due to limited sampling. Overall, splenic and hepatic PB demonstrated the most overlap (~5%) (figure 4C). Upon breaking down the clones based on their unique clonal lineage categories defined above, we found that splenic and hepatic PB populations exhibited the most overlap that amounted to ~20% of the GL branched subset and 5% of the Trunk subset (figure 4D-F). The GL branched category demonstrated the most overlap, wherein, 15% of the clones were shared amongst splenic B cell blast and PB along with hepatic PB and 5% of the clones were shared by all 4 populations (figure 4E). Analysis of the selection pressure on the B cell clones revealed that the responding GL branched clones exhibited a greater degree of selection pressure in comparison to Trunk clones (figure S4 C-F). Overall, these data demonstrated that Ehrlichia infection induced SHM in a subset of responding B cells and that within the mutated clones, there was both differentiation and spreading.
These conclusions were further supported by genealogies of representative larger expanded clones that were found in both spleen and liver (figures 4G and S4 G-I). In figures 4G, S4G, and S4I the clones depicted had a GL sequence that was found in both organs; however, the clone further diversified, accumulating additional mutations that demarcated nodes and branches that were found only in spleen or liver. In figure S4H, the GL sequence was found only in the liver, but other nodes of the clone overlapped between the spleen and liver and a branch was found to present only in the spleen. These data are most consistent with bidirectional spreading of B cell clones along with continued local mutation and expansion after dissemination.
Memory like T-bet positive B cells persist in the spleen and liver after infection subsides
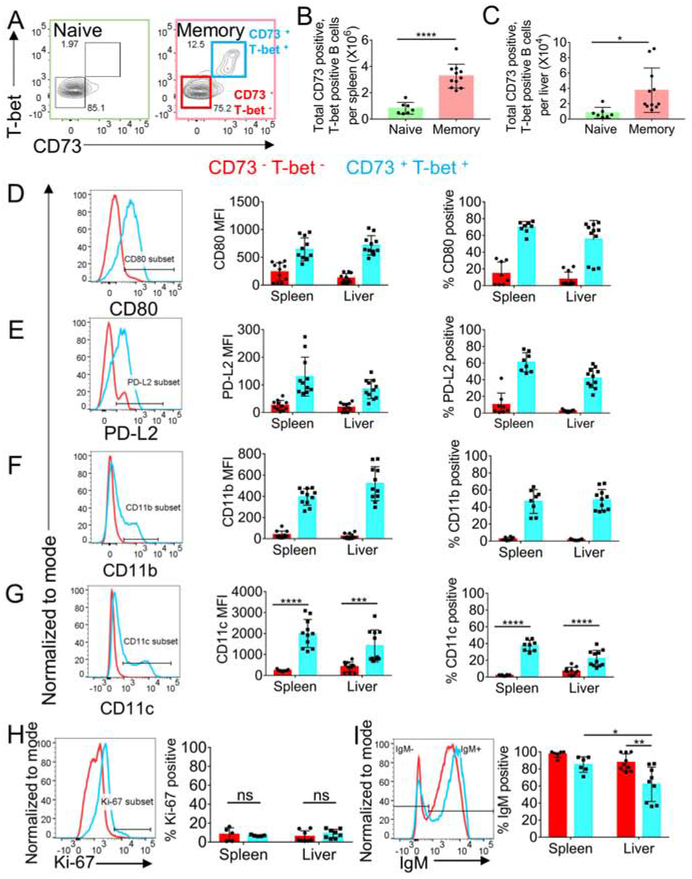
Previously, CD19+CD80+CD11c+T-bet+ B cells have been shown to persist in spleen for substantial time periods after initial Ehrlichia infection (Papillion et al., 2017, Racine et al., 2008, Racine et al., 2010, Winslow et al., 2017, Kenderes et al., 2018). These were interpreted to depend on inflammation or chronic low-level antigen stimulation, since Ehrlichia muris causes persistent infection. We sought to determine whether generation and maintenance of both splenic and hepatic MBC populations depends on antigen persistence. To eliminate persistent Ehrlichia infection, we treated mice with doxycycline at 4 weeks post-infection, and then examined the MBC responses in the spleen and livers between weeks 6 and 12 post-infection. We refer to these mice as “memory mice”. We gated on MBC by using a commonly used marker that includes classical MBC markers, CD73 (Anderson et al., 2007), along with T-bet expression (figure 5A). We found MBC in both spleen and liver of the memory mice compared to their naïve counterparts (figure 5A-C). In both spleen and liver, these cells expressed the classical MBC markers CD80 and PD-L2 in a unimodal fashion (figure 5 D-E). Whereas, a distinct subpopulation of the CD73+T-bet+ cells expressed the ABC markers CD11b and CD11c (figure 5F-G). Moreover, these MBC were quiescent, with few expressing Ki67 (figure 5H). IgM was expressed by most of the MBC, with a fraction lacking IgM being more predominant in the liver (figure 5I). These findings revealed that CD11b and CD11c, which up to now have been used to identify ABC-type MBC (Winslow et al., 2017), only identify a fraction—in fact less than half—of all T-bet+ elicited MBC, raising the question of whether there is additional heterogeneity among T-bet expressing MBC.
Figure 5. Memory-like T-bet positive B cells persist in the spleen and liver after infection clearance.
(A) Gating strategy for CD73+ and T-bet+ B cells spleen and liver of naïve and memory mice. (B-C) CD73+ and T-bet+ B cells in spleen (B) and liver (C) harvested after perfusion with PBS of naïve and memory mice. (D-G) Histogram, Mean fluorescence intensity and percentage of cells positive for CD80 (D), PD-L2 (E), CD11b (F), CD11c (G) in CD73− and T-bet− and CD73+ and T-bet+ B cells in the spleen and liver of memory mice. (H-I) Histogram and percentage of Ki-67+ cells (H) and Histogram and percentage of IgM+ cells in CD73− and T-bet− and CD73+ and T-bet+ B cells in the spleen and liver of memory mice. Data are representative of at least two independent experiments and in (B-H) data are represented as mean with SD of groups of at least two mice. *p <0.05, **p<0.01, ***p<0.001, ****p<0.0001 Statistics for 5B, C, I were done by t-test and 5G were done by two-way ANOVA. See figure S5.
To rule out any contamination from circulating MBC in the liver that were not removed by perfusion, we employed an approach that involved labeling all circulating B cells via an intravenous injection of CD19-PE. We harvested spleen, blood and liver 3 minutes after the injection. Though all the B cells in the blood became labeled, given the short window of time and because of the large size and molecular weight of the fluorophore PE, the antibody (Ab) should not be able to reach the liver parenchyma to a substantial extent. So, any liver-localized MBC should be CD19-PE low or negative. In practice, during tissue processing, tissue-localized cells encounter CD19-PE released from the substantial vascular compartment of the liver (which is un-perfused) and thereby become stained, albeit to a substantially lower level. Post-processing, we further stained the cells with anti-CD19 in a different color (BUV395). In the liver there was a clear population of cells that are much more dimly stained with CD19-PE (~5x dimmer) but more brightly stained for CD19-BUV395 (figure S5A-B). No such population existed in the blood (figure S5A-B). As expected, spleen had mostly CD19-PE-low cells as follicular B cells are not directly accessible to blood and were not quickly stained by an Ab (figure S5A-B). There were about 10% of liver MBC in un-perfused liver that weakly stained for CD19-PE and brightly stained for CD19-BUV395. Moreover, the number of these cells (i.e. frequency of them times total cells recovered) was comparable to the number of MBC that we observed in the liver after perfusion (figure S5C and figure 5C). These data provided support for the interpretation that there is a tissue-localized MBC population that was observed as persistent after perfusion of livers. The other ~90% of cells in un-perfused liver that stain brightly with anti-CD19-PE within 3 minutes were presumably in intravascular spaces, such as sinusoids, and these would very largely be washed away by perfusion. Moreover, by histology, we found CD19- and CD11c-expressing MBC in the liver parenchyma of immune mice (figure S5D). Upon comparison to livers from naïve mice, we observed more CD19- and CD11c-double positive cells per field in the immune mouse livers (figure S5E-F). Taken together, these data suggested that there was a liver-localized MBC population induced by E. muris infection.
Repertoire and Phenotypic Characteristics of the MBC response in spleen and liver
The validation of CD73 as a marker for T-bet+ MBC in this setting allowed us to FACS sort CD73 positive hepatic and splenic MBC from memory mice and perform V region HTS to assess both SHM and clonal sharing among sites and subsets. As seen from the fraction of clones that are unmutated (figure 6A), 60% IgM MBC and 80% of IgG MBC clones were mutated in both organs. Overall, IgG clones harbored more mutations than IgM clones and 40% of splenic IgG clones and 20% of hepatic IgG clones had an average mutation between 5 and 20 (figure 6A). A substantial fraction of clones had 1-5 mutations per sequence, which is comparable to levels seen in MBC after primary responses to model antigens that occurred in GCs (Kaji et al., 2012, Anderson et al., 2007). In addition, these data established that the MBC compartment localized in the liver was similarly mutated to that in the spleen. Approximately 90% of the clones were found only in spleen with about 5% found in only in liver and 5% shared between spleen and liver (figures 6B-C). The GL-branched category had the most clones shared in spleen and liver, indicative of the fact that this category includes clones with more mutation and expansion, though a smaller category overall in terms of numbers of clones (figure 6B-C). A major reason for the relative lack of overlap of clones in spleen and liver could be the much larger size of the spleen MBC pool and differences in sampling depth. Alternatively, it is possible that only few splenic MBC engrafted the liver and/or the majority of liver-derived MBC engrafted the splenic MBC compartment. Analysis of clonal trees from large clones did reveal trees that were consistent with both migration after differentiation as well as extensive local production of diversified MBC clonal populations (figure S6).
Figure 6. Repertoire and phenotypic characteristics of the MBC population of spleen and liver.
(A) Mutation distribution of MBC population in spleen and liver from memory mice and B cells from a naïve mouse. (B-C) Overlap within different subsets of MBC clones analyzed by fraction of clones (B), and number of clones (C). (D-E) Volcano plots demonstrating highly expressed genes in spleen and liver DN (D) and DP (E) MBC subsets. See figure S6-7 and table S2.
To phenotypically characterize the splenic and hepatic MBC, we performed RNA-seq analysis of CD11b− and CD11c− double negative (DN) and double positive (DP) MBC on FACS sorted populations from memory mice (figure S7A). We found 730 genes that were differentially and significantly expressed (FDR<0.01, 2-Fold change) in the splenic and hepatic MBC populations (figure S7B, Table S2). Gene clusters marked with navy blue, red and dark red were significantly and differentially expressed in the DN subsets compared to the DP subsets regardless of the site of origin (figure S7B). Gene clusters marked with yellow, purple, pink, turquoise blue and light pink were significantly and differentially expressed in the liver MBC compared to the splenic counterparts (figure S7B). We found several genes that were differentially and significantly expressed in spleen DN and DP subsets in comparison to their liver counterparts (figure 6 D-E). Liver MBC subsets both expressed CD69 when compared to splenic MBC subsets (figure 6D-E), consistent with expression patterns of bona fide T resident memory cells (Sathaliyawala et al., 2013). We verified the CD69 expression on the liver MBC subsets during acute response (D10 post infection) by FC (figure S7 C-D). Together, these data demonstrate that while there were shared genes that were expressed in MBC subsets regardless of their tissue localization, there were several genes unique to the subset and site of origin. This suggests that the local tissue microenvironment may be shaping the MBC differently in the spleen and liver, much as it does for T cells and macrophages (Kumar et al., 2017, Lavin et al., 2014).
We performed gene set enrichment analysis (GSEA) in comparison to a published dataset from CD11c-expressing splenic B cells isolated 30 days after Ehrlichia infection that were presumptive MBC (Winslow et al., 2017). GSEA revealed that genes found in that database were enriched in transcriptomes of splenic and hepatic DP MBC compared to DN MBC (figure S7E-F). This suggests that our data are consistent with the previously published dataset with reference to Ehrlichia infection. We further performed differential analysis of splenic and liver memory subsets with respect to naive B cells from a previously published study (Barnett et al., 2016) (see Methods of inter-study normalization detail). Using a set of genes that have increased expression in nitrophenyl hapten-induced MBC (data not shown), we found that splenic DN, splenic DP, and hepatic DP MBC subsets had an enrichment of memory genes, while hepatic DN MBC had a similar trend (figure S7 G-J). These data suggest that Ehrlichia-induced splenic DP and DN along with hepatic DP MBC subsets express genes that are characteristically expressed by classical, GC-induced MBC.
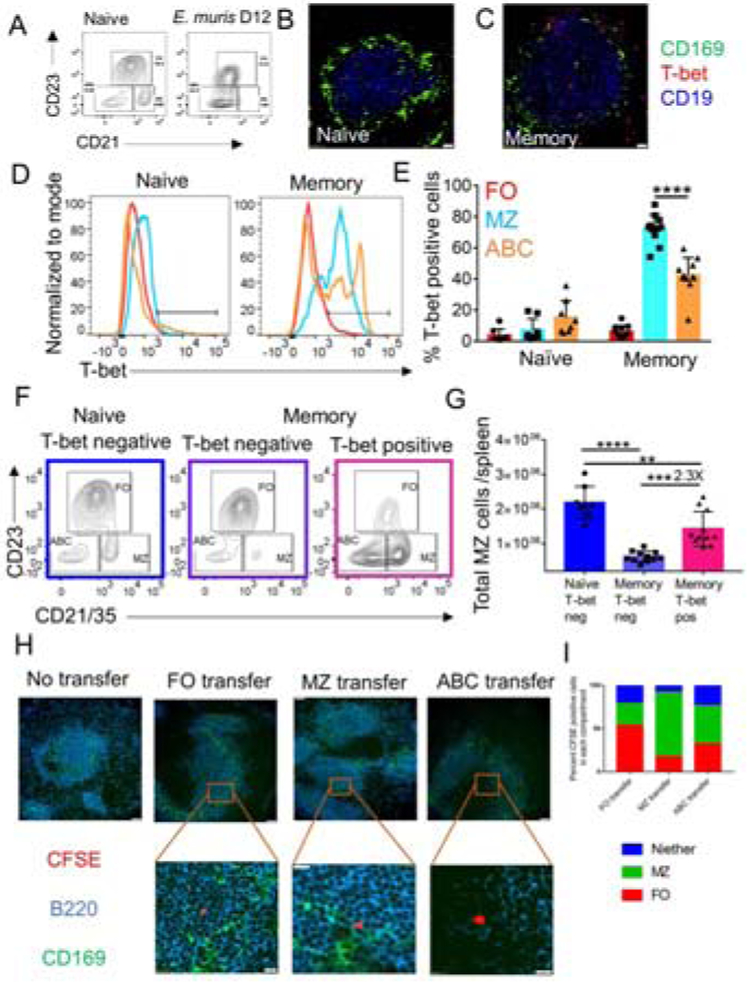
T-bet positive B cells dominantly repopulate the splenic marginal zone (MZ) post-infection
During the acute phase of Ehrlichia infection, the histologic MZ architecture is disrupted, with loss of CD169 positive metallophilic macrophages that demarcate the MZ (data not shown). By FC, CD23lo CD21+ MZ phenotype B cells were also absent (figure 7A). At a memory time point the histologic MZ was largely regenerated with a border of CD169+ cells, albeit somewhat less organized than in a naïve animal (figure 7B-C). Nonetheless, at this memory time point, very few of the CD23lo CD21+ MZ phenotype cells that had repopulated this compartment were T-bet negative, unlike in the naïve mouse in which essentially all MZ phenotype B cells lacked T-bet expression (figure 7D-E). On average there were 2.3-fold more T-bet+ than T-bet-MZ B cells in the memory mice (figure 7F-G). Histologically, in memory mice T-bet+ cells were found abundantly in the MZ region and were also scattered in the follicular (FO) region (figure 7C). A prior report had identified CD11c+ B cells in or near the MZ at day 63 post-infection (Yates et al., 2013), although as noted earlier, CD11c would only pick up <40% of total T-bet+ MBC. Generally, ABC are thought to lack the expression of B cell subset markers CD21 (high on MZ B cells) and CD23 (high on FO B cells) (Rubtsova et al., 2015, Rubtsova et al., 2017, Hao et al., 2011). However, a large fraction of the T-bet+ MBC formed in this setting post-Ehrlichia infection were of a MZ phenotype, with strong expression of CD21; in addition, some had an ABC (CD21negCD23neg) or FO phenotype.
Figure 7: After infection resolves, T-bet positive MBC dominate the MZ of the spleen.
(A) B cell subset gating on CD19 positive B cells in naïve or D12 post E. muris infection. (B-C) CD169 (green), T-bet (red) and CD19 (blue) staining in Naïve (B) and memory spleens at 40X magnification (C). In B-C scale bars represent 20μm. (D-E) Histogram (D) and percentage (E) of T-bet+ B cells in Naïve and memory mice (F) B cell subset gating on naïve and memory T-bet+ B cells. (G) Quantification of T-bet+ MZ B cells in naïve and memory mice. (H) 20X images of CFSE-labeled CD19+ CD73+ CD45.2/2 splenic MBC populations sorted as CD23+ CD21− (FO), CD21+ CD23− (MZ) and CD21− CD23− (ABC) and transferred into CD45.1/2 mice, shown at 42 hours post-transfer. Scale bars in the top and bottom panels represent 50μm and 10μm, respectively. (I) From the recipient mice as in (H), 67 CFSE labeled cells from the FO transfer group were counted over 42 images, 108 CFSE labeled cells from the MZ transfer group were counted over 21 images, and 105 CFSE labeled cells from the ABC transfer group were counted over 44 images. Percentages of the CFSE positive transferred MBC subsets found in FO or MZ or neither of those locations are presented in the bar graphs. Data are representative of at least 2 independent experiments with groups of at least two mice. In (E) and (G) data are represented as mean with SD and statistics were done by two-way ANOVA. *p <0.05, **p<0.01, ***p<0.001, ****p<0.0001.
To assess whether the FO, MZ and ABC MBC subsets retain their phenotype and preferentially lodge to those sites upon transfer, we used FACS to purify FO, MZ and ABC CD45.2 MBC using the expression of CD21 and CD23. We carboxyfluorescein succinimidyl ester (CFSE)-labeled the MBC subsets and transferred into a B18+/− Vκ8R+/− CD45.1/2 naïve mouse. We harvested the spleens of the host 42 hours after the transfer to assess homing of the MBC subsets. Based on histologic analysis, >50% FO MBC preferentially populated the follicle, >70% MZ MBC populated the MZ area and the ABC populated all the zones (figure 7H-I). These data suggest that the MBC subsets could rehome to the compartment that they originated in and that their localization is a relatively stable characteristic.
DISCUSSION
Here we found that liver is a generative site of B cell responses during local infection of the liver. Local B cell responses in non-lymphoid organs have been reported in several other contexts. Influenza infection yields lung responses that involve generation of tertiary lymphoid tissue (Moyron-Quiroz et al., 2004). Local production of IgA AFC in the lamina propria (Fagarasan et al., 2001) is likely driven by commensal flora (Kunisawa et al., 2013, Fagarasan et al., 2002). However, unlike lung and gut, liver has no direct mucosal interface with the environment.
Hepatitis infection in humans also can generate B cell infiltration, again involving tertiary lymphoid tissue and even local GC formation (Farci et al., 2010, Murakami et al., 1999, Racanelli et al., 2001, Sansonno et al., 2004). In contrast, hepatic E. muris infection did not induce tertiary lymphoid tissue or GCs, yet SHM and class switch were induced in a fraction of responding cells in the liver. Intrahepatic responses to E. muris involved both portal triads and foci that were within the parenchyma itself. Thus, under circumstances of infection, robust and mature local B cell responses can ensue directly within parenchymal tissue.
The advantages of mounting local B cell response are several. Some pathogens may not infect or spread well to lymphoid tissue. Pathogens such as E. muris and Plasmodium, may clear systemically but persist in the liver as low-level infections (Dumler et al., 1993, Mueller et al., 2009, Crocquet-Valdes et al., 2011, Thirumalapura et al., 2009, Thirumalapura et al., 2008). Therefore, if local B cell responses were not possible, there might be little if any Ab produced against such pathogens. Intraparenchymal T cell responses and infiltration are more commonly seen; local B cell responses might optimize these, as they would allow for enhanced Ag presentation to T cells. In addition, B cells produce substantial amounts of inflammatory cytokines, such as IL-6 and TNF-α, which could stimulate other arms of the immune system and be directly protective in a local manner.
HTS of V regions revealed extensive cross-site clonal mixing. Although limited sampling depth led to a general underestimation of the extent of clonal size, diversification and cross-seeding of tissues., it is reasonable to assume that not all clones disseminated. Overall, mutation was only observed in about 25% of clones and it was these clones, which we termed “GL Branched”, that tended to be found in multiple sites and in both PB and B cell blast compartments. These clones were evidently much larger than those in either the Unmutated or Trunk subsets. It is not clear what controls whether clones initiate SHM and expand to a greater extent; it could be affinity-driven and/or dependent on proximity to sites of proliferating bacteria and thereby the degree of persistent antigen-stimulation as well as T cell help.
Patterns of distribution of clone members were most consistent with bidirectional interchange of both PB and B cell blasts in clones that were expanding and undergoing SHM. Regardless of how such clones evolved, their disseminating nature illuminates a feature of the immune system that would anticipate pathogen dissemination by enabling actively responding B cell clones to populate sites distant from where they received their initial stimulus, thus creating a more comprehensive and adaptive immune response. Compared to responding B cells at early time points post infection, MBC clones had more V region mutation. This suggests that either most MBC were formed at later time points in the response, after more mutations accumulated, or that the more expanded clones that had more mutation were more likely to spawn longer-lived MBC.
At steady state, expanded B cell clones are also disseminated in normal humans (Meng et al., 2017). Cerutti and colleagues identified an IgM MBC subset disseminated throughout human gut, which does not share clonal origin with most peripheral MBC but is the precursor of local IgM and IgA production (Magri et al., 2017). These MBC are likely driven by gut commensals, but their precise site of origin remains unclear. In analogous elegant studies, Lindner et al. found dispersed clones of MBC in multiple Peyer’s patches, though they did not examine lamina propria; these MBC migrate to spleen and they are clonally related to mammary gland IgA plasma cells (Lindner et al., 2012). In these settings, the inductive phase was considered to occur in secondary lymphoid tissue, with subsequent spread of both MBC and plasma cells.
We conclude that many of the B cells in the liver detected at memory time points are MBC; it is unlikely that they were passenger B cells from blood that remained despite our perfusion of the liver prior to harvest. Their memory characteristics, including isotype switch in a fraction, V region mutation, and particularly expression of T-bet, CD73 and CD80, are all unlike B cells found in blood. Intravenous injection of anti-CD19-PE also demonstrated the presence of a liver-localized MBC population that was resistant to rapid labeling. Because we have not directly studied their migration, we prefer to call these cells “tissue-localized” MBC to reflect that they were not in lymphoid structures or intravascular, rather than tissue-resident. The length of residence of the MBC we identified remains to be determined. While tissue MBC are relatively understudied compared to their T cell counterparts, prior work has identified such cells in gut at steady state in both mice and humans and they can be induced by influenza infection or oral immunization in mice (Onodera et al., 2012, Bemark et al., 2016).
As discussed for the primary response, the presence of MBC localized in the liver after the clearance of the infection is distinctive, in that it is not a mucosal barrier site, unlike lung and gut. However, MBC populations are clearly identified during chronic liver disease in humans, including chronic hepatitis and autoimmunity. In these cases, continuous immune stimulation is likely responsible for the development and maintenance of such populations. By analogy, MBC in liver post E. muris infection may have been formed from prior local acute responses and/or may have migrated from splenic populations, as exemplified by clones that were both liver-specific and found in both spleen and liver. The functional implications of liver-localized MBC for subsequent infection and protection are unclear, but it is tempting to speculate that they can provide protection for certain types of local reinfection, as is proposed for resident memory T cells (Kumar et al., 2017) and certain types of tissue localized MBC (Onodera et al., 2012, Bemark et al., 2016).
Our findings also have implications for understanding T-bet+ cells in terms of their origin, identity, location and function. In agreement with others (Kenderes et al., 2018, Winslow et al., 2017), we confirmed that these cells were indeed MBC, and largely non-proliferative. While previous reports in the context of E. muris have relied on CD11c expression as a surrogate marker, we found fully 50% of T-bet+ MBC did not express CD11c or CD11b. Conversely, there were also many that do express CD21 and CD23, although the canonical ABC are described as not expressing those markers (Rubtsova et al., 2015, Rubtsova et al., 2017). This basic immunophenotyping revealed considerable heterogeneity among the E. muris-induced T-bet+ MBC. Moreover, RNA-seq analysis demonstrated that certain shared genes were expressed in MBC subsets regardless of their tissue location but also a substantial number of genes unique to the particular subset and site of origin. This suggests that the local tissue microenvironment may be shaping the MBC differently in the spleen and liver.
Finally, in the course of tracking the B cell response to E. muris, we unexpectedly found that infection caused a wholesale remodeling of the splenic MZ. MZ disruption, as part of more pervasive splenic architecture disruption, has been reported in the context of multiple infections, including lymphocytic choriomeningitis virus, Salmonella, and malaria (Rosche et al., 2015, Muller et al., 2002, Urban et al., 2005). However, after E. muris infection, during the eventual reorganization the MZ was repopulated by MBC induced by infection, rather than by more typical primary MZ B cells. Hence, the repertoire and presumably the functional capacity of the MZ B cell compartment was markedly reprogrammed, with ~70% of MZ phenotype B cells expressing T-bet post-infection, compared to a negligible proportion prior to infection. This major alteration lasted for at least 8 weeks post-infection, and we assume for much longer. It is unclear why T-bet+ B cells, rather than naïve MZ B cells, preferentially repopulate the MZ. This type of mechanism might explain the presence of mutated IgM MBC in the MZ observed in human spleens (Weller et al., 2004). We further propose that, given the importance of MZ B cells in responding acutely to various infections (Martin et al., 2001, Bankoti et al., 2012), the remodeling of the MZ by E. muris, and potentially other similar primary infections, could be a mechanism by which a significant initial infection could alter or impair the response to subsequent infection.
STAR METHODS
Lead contact and materials availability
Requests for resources and reagents should be directed to and will be fulfilled by the lead contact for this study, Mark Shlomchik, (mshlomch@pitt.edu).
Experimental model and subject details
The mice used in this study were bred under specific pathogen free conditions in the animal facility at the University of Pittsburgh. All mouse work was done according to the protocols approved by the University of Pittsburgh Institutional Animal Care and Use Committee. The following mouse strains were used: C57BL/6 (Jackson Laboratories), B18+/+ and B18+/+ Vk8R+/+ (Sonoda et al., 1997), huCD20 TamCre (Khalil et al., 2012), T-bet fl/fl (Intlekofer et al., 2008), huCD20 TamCre T-bet fl/fl, and B18+/− Vk8R+/− CD45.1/2.
E. muris was used for infection of the above-mentioned mouse strains (Thirumalapura et al., 2009).
Method Details
Infection and Treatment Procedures:
E. muris infections were done according to procedures described previously (Stevenson et al., 2006, Thirumalapura et al., 2009). Briefly, E. muris inoculum was prepared by passage through wild-type C57BL/6 carrier mice. Single cell suspensions from spleens harvested from carrier mice were used for infection of experimental mice. Mice were infected intraperitoneally with 105 E. muris/mouse (Thirumalapura et al., 2009). The bacterial burden was assessed by quantitative RT-PCR as described previously (Stevenson et al., 2006). Tamoxifen treatments were done at day 3, 5 and 7 post infection at a dose of 1 or 2 mg orally in corn oil. For labeling B cells in the circulation, 1μg CD19 PE was injected intravenously. After 3 minutes, blood, spleen and liver were harvested and analyzed. For MBC homing experiments, CD45.2 MBC subsets were FACS sorted, CFSE labeled and transferred intravenously into B18+/− Vk8R+/− CD45.1/2 mice.
ELISpot Assays:
Single cell suspensions from spleen were obtained by mechanical disruption of the tissue, followed by treatment with ACK buffer for lysis of red blood cells. Single cell suspension of the liver was prepared by mechanical disruption using the MACS dissociator along with use of 50KU/mL DNase and collagenase 100U/mL, followed by treatment with ACK buffer for lysis of red blood cells. The single cell suspension from the liver was re-suspended in 20% Percoll and underlaid with 80% Percoll. The Percoll gradient was spun at 450 rcf for 20 minutes at room temperature. The lymphocytes were collected from the interface of the Percoll gradients and washed with media before further use in various assays described below. For ELISpot assay, Immulon 4-HBX plates were coated with the following antigens: anti-kappa at 5 mg/ml, Omp12 at 4mg/mL and Omp19 at 4mg/mL. E. coli strains containing recombinant plasmids for E. muris antigens Omp 12 and Omp 19 were provided by David Walker at University of Texas Medical Branch (Crocquet-Valdes et al., 2011). The recombinant His tagged proteins were produced and purified using Ni-NTA spin kit from Qiagen as per standard protocols (Crocquet-Valdes et al., 2011). For ELISpot assay, non-specific binding was blocked with 1% bovine serum albumin (BSA) in PBS. Splenocytes were incubated overnight at 37°C. AFC were detected by using alkaline phosphatase-conjugated secondary Ab (to IgG or IgM, Southern Biotech) and 5-bromo-4-chloro-3-indolyl-phosphate in agarose.
Flow Cytometry
For FC staining, non-specific binding was blocked using anti-FcR clone 2.4G2 and dead cells were excluded using cell viability dye (Tonbo Biosciences). The Abs were either purified in our lab or purchased and are as follows. Anti-B220 (Biolegend, RA3-6B2), anti-CD19 (BD ID3), anti-IgM (homemade, B7-6), anti-CD45 (home-made 30-F11), anti-CD21 (homemade, 7G6), anti-CD23 (ebioscience, B3B4) for B cells, anti-CD4 (Biolegend, GK1.5), anti-TCR-β (Biolegend H57-597) for T cells, anti-CD138 (Biolegend, 281-2) and anti-CD44 (Biolegend, IM7) for B cell blasts and PB, anti-CD73 (Biolegend, TY-11.8), anti-CD80 (ebioscience, 16-1oA1), anti-PD-L2 (ebioscience, TY25) for MBC, PNA (vector labs), anti-CD95 (BD, Jo2) for GCBC, anti-CD169 (Biolegend, 3D6.112) for metalophillic macrophages, anti-CD11b (Biolegend M1-70), anti-CD11c (ebioscience, N418), anti-CD69 (ebioscience, H1.2F3), anti-AID (ebioscience, mAID-2) and anti-T-bet (Biolegend, 4B10). The click IT Plus Edu kit was purchased from Invitrogen and the staining was done according to the recommended protocol. The cells were analyzed on LSR II or Fortessa instruments (BD) and the data were analyzed on FlowJo software.
Immunofluorescence imaging
7 mm spleen sections were prepared from OCT-frozen tissues, fixed in acetone for 10 min, and stored at −80°C. The slides were thawed and re-hydrated using PBS and blocked using PBS+1% BSA and 2.4G2 for 10 minutes. The slides were then stained with relevant Abs as described in the figure legends in a dark humid chamber for 30 minutes. The Abs were either purified in our lab or purchased and are as follows. Anti-B220 (Biolegend RA3-6B2), anti-CD19 (BD ID3), anti-IgM (home-made B7-6), for B cells, anti-CD4 (Biolegend GK1.5), anti-TCR-β (Biolegend H57-597) for T cells, anti-CD138 (Biolegend 281-2) for PB, anti-CD169 (Biolegend 3D6.112) for metalophillic macrophages, anti-CD11c (ebioscience N418) and anti-T-bet (Biolegend 4B10). The slides were washed thrice using PBS and the same steps were followed for secondary Ab. For intracellular staining, the sections were permeabilized using 0.3% Triton X-100 reagent before staining. Sections were mounted using ProLong Anti-fade Gold (Life Technologies) and imaged using Olympus Fluorescence Microscope IX3-BSW and acquired using Cell Sens Dimension software.
V region sequencing
V region sequencing was done on FACS sorted PB or micro-dissected B cell patches. 7 mm spleen sections were prepared from OCT-frozen tissues on the membrane-coated PEN slides (Leica). PB patches were detected using anti-IgM Alexa488 staining. Microdissections were performed using Zeiss PALM MicroBeam Laser Capture Microdissection System. Dissected patches were collected in the cap of PCR microtubes in 12uL of digestion buffer (50 mM Tris-HCl, 50mM KCl, 0.63 mM EDTA, 0.22% Igepal, 0.22% Tween20, 0.8 mg/ml proteinase K). The patches or FACS sorted PB were digested at 37°C for 4 hours and at then at 90°C for 5 minutes. Primers, V region amplification and data analysis has been described previously (Di Niro et al., 2015). Briefly, a primary PCR was performed using the primers MsVHE-short: 5-GGGAATTCGAGGTGCAGCTGCAG-3 and a mix of 4 JH region anti-sense primers 3 SalI P-mJH01: 5-TGCGAAGTCGACGCTGAGGAGACGGTGACCGTGG-3 3SalIP-mJH02: 5-TGCGAAGTCGACGCTGAGGAGACTGTGAGAGTGG-3 3SalIP-mJH03: 5-TGCGAAGTCGACGCTGCAGAGACAGTGACCAGAG-3 3SalIP-mJH04: 5-TGCGAAGTCGACGCTGAGGAGACGGTGACTGAGG-3. A second nested PCR was performed using 1uL of the product from the 1st PCR using the primers MsVHE: 5-GGGAATCGAGGTGCAGCTGCAGGAGTCTGG-3 and a mix of JH antisense primers 5’- TGGTCCCTGTGCCCCAGACATCG -3’, 5’- GTGGTGCCTTGGCCCCAGTAGTC -3’, 5’- AGAGTCCCTTGGCCCCAGTAAGC -3’ and 5’- GAGGTTCCTTGACCCCAGTAGTC -3. High fidelity polymerase PfU Turbo (Agilent) was used for the PCR amplification to minimize the possibility of PCR error while generating the V region sequence. The resulting PCR products were cloned and sequenced using Zero Blunt PCR cloning kit (Thermofisher). Sequence analysis was done by using http://www.imgt.org/IMGT_vquest/analysis.
Cell Sorting and RNA preparation:
PB were sorted as TCR-β negative, CD138 positive, CD44 positive, CD19 intermediate, B cell blasts were sorted as TCR-β negative, CD44 positive, CD138 negative, CD19 positive, naïve B cells were sorted as CD19 positive, CD138 negative and CD44 negative, CD73 negative. For HTS, B cells were sorted from naïve mice as CD45 positive, CD19 positive, CD73 negative, and MBC were sorted from memory mice as CD45 positive, CD19 positive and CD73 positive. For mRNA sequencing analysis, B220 positive, CD73 positive MBC were sorted as CD11b, CD11c double negative and CD11b, CD11c double positive from memory mice. The staining was done as described earlier for FC. After sorting, cells were spun down and washed with PB and re-suspended in RLT+1% beta-mercaptoethanol. RNA was prepared with RNAeasy microplus kits from Qiagen, according to recommended protocol.
HTS library preparation and analysis
For HTS, B cell blasts, PB, and MBC were FACS sorted and RNA was prepared as described above. The method for high-throughput sequencing of the B cell repertoire was performed as previously described (Di Niro et al., 2015, Tsioris et al., 2015). Briefly, RNA was reverse-transcribed into cDNA using a biotinylated oligo dT primer. An adaptor sequence was added to the 3' end of all cDNA, which contains the Illumina P7 universal priming site and a 17-nucleotide unique molecular identifier (UMI). Products were purified using streptavidin-coated magnetic beads followed by a primary PCR reaction using a pool of primers targeting the IGHA, IGHD, IGHE, IGHG, IGHM, IGKC and IGLC regions, as well as a sample-indexed Illumina P7C7 primer. The immunoglobulin-specific primers contained tails corresponding to the Illumina P5 sequence. PCR products were then purified using AMPure XP beads. A secondary PCR was then performed to add the Illumina C5 clustering sequence to the end of the molecule containing the constant region. The number of secondary PCR cycles was tailored to each sample to avoid entering plateau phase, as judged by a prior quantitative PCR analysis. Final products were purified, quantified with Agilent Tapestation and pooled in equimolar proportions, followed by high-throughput paired-end sequencing on the Illumina MiSeq platform. For sequencing, the Illumina 600 cycle kit was used with the modifications that 325 cycles were used for read 1, 6 cycles for the index reads, 300 cycles for read 2 and a 20% PhiX spike-in to increase sequence diversity.
FASTA files provided by Juno were analyzed with ImmuneDB v0.24.0 using default parameters for all stages (Rosenfeld et al., 2018).The GL reference sequences were acquired from IMGT's GENE-DB https://www.imgt.org/genedb/. After clonal assignment, lineages were generated with clearcut (Sheneman et al., 2006) using neighbor-joining, excluding mutations that occurred in only one sequence. Data were then exported from ImmuneDB for downstream analysis. Clones were included in this analysis only if they had between 1 and 4 non-templated CDR3 residues (as measured from the first non-GL-encoded nucleotide) or at least 4 common V-gene mutations across all sequences. ETE3 v3.1.1 (Huerta-Cepas et al., 2016) was used to calculate tree metrics and numpy v1.15.0 was used for all statistical testing.
RNA seq analysis
MBC were FACS sorted and RNA was prepared as described above. cDNA libraries were constructed using SMARTer low input kit for mRNA seq (Clontech). Samples were sequenced using Illumina NextSeq 500 with 75 bp paired-end reads and aligned to the mm10 genome using the STAR aligner (Dobin et al., 2013). The number of uniquely aligned reads ranged from 22 to 32 million. Gene-level counts were determined using featureCounts (Liao et al., 2014), and raw counts were quantile normalized to each other for differential expression using the voom method (Law et al., 2014) in the limma R package (Ritchie et al., 2015) The presence of certain liver-specific transcripts indicated unavoidable liver cell contamination in the liver sample; to prevent this from confounding our analysis, differential expression was performed only on genes, which were having at least 30 counts in the all liver and spleen samples. All RNA-seq data were deposited in the NCBI’s Gene Expression Omnibus database (GEO) with accession ID GSE137154. All gene-set enrichments were preformed using the rankSumTestWithCorrelation function in limma, which explicitly corrects for correlation among genes in the gene set being interrogated. For differential analysis of splenic and liver memory subsets in figure S7, the naive B cell transcriptional profile was extracted from a previously published microarray study (Barnett et al., 2016). For normalization of the datasets, the Quantile method was used.
Quantification and Statistical Analysis
Statistical analysis was performed with Prism (GraphPad Software). P values were determined using Student’s t tests (two-tailed). For multiple comparisons, Two-Way ANOVA or Mann Whitney tests or Chi square analysis were applied. Differences between groups were considered significant for P values < 0.05 (* p < 0.05; ** p < 0.01; *** p < 0.001; **** p < 0.0001).
Data and Code Availability
The RNA seq data generated during this study is available in the NCBI’s Gene Expression Omnibus database (GEO) with accession ID GSE137154.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rat anti B220 (clone: RA3 6B2) PerCPCy5.5 | BD Biosciences | Catalog # 561101 RRID AB_10565970 |
| Rat anti CD19 (clone 1D3) FITC | BD Biosciences | Catalog # 553785 RRID AB_395049 |
| Rat anti IgM (clone B7-6) A488 | Prepared in the lab | N/A |
| Rat anti CD21 (clone 7G6) A488 | Prepared in the lab | N/A |
| Rat anti CD23 (clone B3B4) PE | eBioscience | Catalog # 12-0232-82 RRID AB_465595 |
| Rat anti CD4 (clone GK1.5) BV421 | Biolegend | Catalog # 100401 RRID AB_10900241 |
| Hamster anti TCR-β (clone H57-597) PECy7 | Biolegend | Catalog # 109222 |
| Rat anti CD138 (clone 281-2) APC | Biolegend | Catalog # 142506 RRID AB_10960141 |
| Rat anti CD44 (IM7) APCCy7 | Biolegend | Catalog # 103028 RRID AB_830785 |
| Rat anti CD73 (clone TY-11.8) bi | Biolegend | Catalog # 127204 RRID AB_1089062 |
| Hamster anti CD80 (clone 16-10A1) APC | eBioscience | Catalog # 16-0801-82 RRID AB_467348 |
| Rat anti PD-L2 (clone TY25) PE | eBioscience | Catalog # 12-5986-82 RRID AB_466845 |
| Hamster anti CD95 (clone Jo2) PECy7 | BD Biosciences | Catalog # 557653 RRID AB_2739999 |
| Rat anti CD169 (clone 3D6.112) A647 | Biolegend | Catalog # 142402 RRID AB_2563620 |
| Anti CD11b (clone M1-70) APCCy7 | Biolegend | Catalog # 101214 RRID AB_830641 |
| Anti CD11c (clone N418) A647 | eBioscience | Catalog # 14-0114-82 RRID AB_469346 |
| Anti CD69 (H1.2F3) PE | eBioscience | Catalog # 12-0691-82 RRID AB_467326 |
| Rat anti AID (clone mAID2) bi | eBioscience | Catalog # 14-5959-82 RRID AB_468662 |
| Anti T-bet (clone 4B10) PECy7 | Biolegend | Catalog # 644824 RRID AB_2561760 |
| Bacterial and Virus Strains | ||
| Ehrlichia muris | This paper and Thirumalapura et al., 2009 | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| E. muris antigens Omp 12 and Omp 19 | This paper and Crocquet-Valdes et al., 2011 | N/A |
| PNA | Vector laboratories and conjugated in the lab | Catalog # L1070 RRID AB_2315097 |
| Oligonucleotides | ||
| 5-GGGAATTCGAGGTGCAGCTGCAG-3 | This paper and Di Niro et al., 2015 | N/A |
| 5-TGCGAAGTCGACGCTGAGGAGACGGTGACCGTGG-3 | ||
| 5-TGCGAAGTCGACGCTGAGGAGACTGTGAGAGTGG-3 | ||
| 5-TGCGAAGTCGACGCTGCAGAGACAGTGACCAGAG-3 | ||
| 5-TGCGAAGTCGACGCTGAGGAGACGGTGACTGAGG-3 | ||
| 5-GGGAATCGAGGTGCAGCTGCAGGAGTCTGG-3 | ||
| 5- TGGTCCCTGTGCCCCAGACATCG -3 | ||
| 5- GTGGTGCCTTGGCCCCAGTAGTC -3 | ||
| 5- AGAGTCCCTTGGCCCCAGTAAGC -3 | ||
| 5- GAGGTTCCTTGACCCCAGTAGTC -3 | ||
| Critical Commercial Assays | ||
| Click IT Plus Edu kit | Thermofisher | Catalog # C10634 |
| Ni NTA Spin kit | Qiagen | Catalog # 31314 |
| RNeasy plus micro kit | Qiagen | Catalog # 74034 |
| Zero Blunt PCR Cloning kit | Thermofisher | Catalog # 450031 |
| Deposited Data | ||
| mRNA-seq data | Gene Expression Omnibus database | GSE137154 |
| Experimental Models: Organisms/Strains | ||
| C57BL/6 mice | Jackson Laboratories | Catalog # 000664 |
| B18+/+ C57BL/6 mice | This paper and Sonoda et al., 1997 | N/A |
| B18+/+ Vk8R+/+ C57BL/6 mice | This paper and Sonoda et al., 1997 | N/A |
| huCD20 TamCre C57BL/6 mice | This paper and Khalil et al., 2012 | N/A |
| T-bet fl/fl C57BL/6 mice | This paper and Intlekofer et al., 2008 | N/A |
| huCD20 TamCre T-bet fl/fl C57BL/6 mice | This paper | N/A |
| B18+/− Vk8R+/− CD45.1/2 C57BL/6 mice | This paper | N/A |
| Software and Algorithms | ||
| Flow Jo version 10 | Flow Jo |
https://www.flowjo.com/ RRID SCR_008520 |
| GraphPad Prism 7 | GraphPad |
https://www.graphpad.com/scientific-software/prism/ RRID SCR_002798 |
| IMGT V quest | IMGT |
http://www.imgt.org/ RRID SCR_012780 |
| MUSCLE | European Bioinformatics |
https://www.ebi.ac.uk/Tools/msa/muscle/ RRID SCR_011812 |
| FastQC v0.11.7 | Babraham Institute |
https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ RRID SCR_014583 |
| Trimgalore v0.3.8 | Babraham Institute |
https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ RRID SCR_016946 |
| cutadapt v1.18 | Babraham Institute |
https://github.com/marcelm/cutadapt/ RRID:SCR_011841 |
| STAR v2.6.1a | Dobin et al., 2013 | https://github.com/alexdobin/STAR |
| Subread v1.6.2; featureCounts | Liao et al., 2014 |
http://subread.sourceforge.net RRID SCR_009803 |
| Limma v3.20.9-voom | Law et al., 2014; Ritchie et al., 2015 |
https://ucdavis-bioinformatics-training.github.io/2018-June-RNA-Seq-Workshop/thursday/DE.html RRID SCR_010943 |
| Limma v3.20.9-rankSumTestWithCorrelation | Barry et al. 2008 |
https://www.rdocumentation.org/packages/limma/versions/3.28.14/topics/rankSumTestWithCorrelation RRID SCR_010943 |
| ImmuneDB v0.24.0 | Rosenfeld, et al., 2018 |
https://immunedb.readthedocs.io/en/latest/ RRID:SCR_017125 |
Highlights.
E. muris induces localized B cell proliferation, differentiation and SHM in liver
Primary response and MBC clones interchange between liver and spleen
E. muris induces T-bet+ MBCs that adopt follicular, ABC and MZ phenotypes
E muris dissolves the MZ, which is subsequently reconstituted by T-bet+ MBCs
Acknowledgements:
We thank Dr. Amanda Poholek for providing the Tbx21 fl/fl mice. We thank Dr. David Walker at the University of Texas Medical Branch for providing E. muris antigens Omp 12 and Omp19. We thank the Histology Core at Magee Research Institute for laser micro-dissections. We are thankful to Laura Conter and Rachel Green for helping with the experiments.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests: The authors declare no competing interests.
REFERENCES
- Anderson BE, Dawson JE, Jones DC & Wilson KH 1991. Ehrlichia chaffeensis, a new species associated with human ehrlichiosis. J Clin Microbiol, 29, 2838–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson SM, Tomayko MM, Ahuja A, Haberman AM & Shlomchik MJ 2007. New markers for murine memory B cells that define mutated and unmutated subsets. J Exp Med, 204, 2103–14. 10.1084/jem.20062571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankoti R, Gupta K, Levchenko A & Stager S 2012. Marginal zone B cells regulate antigen-specific T cell responses during infection. J Immunol, 188, 3961–71. 10.4049/jimmunol.1102880 [DOI] [PubMed] [Google Scholar]
- Barnett BE, Staupe RP, Odorizzi PM, Palko O, Tomov VT, Mahan AE, Gunn B, Chen D, Paley MA, Alter G, Reiner SL, Lauer GM, Teijaro JR & Wherry EJ 2016. Cutting Edge: B Cell-Intrinsic T-bet Expression Is Required To Control Chronic Viral Infection. J Immunol, 197, 1017–22. 10.4049/jimmunol.1500368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker AM, Dao KH, Han BK, Kornu R, Lakhanpal S, Mobley AB, Li QZ, Lian Y, Wu T, Reimold AM, Olsen NJ, Karp DR, Chowdhury FZ, Farrar JD, Satterthwaite AB, Mohan C, Lipsky PE, Wakeland EK & Davis LS 2013. SLE peripheral blood B cell, T cell and myeloid cell transcriptomes display unique profiles and each subset contributes to the interferon signature. PLoS One, 8, e67003. 10.1371/journal.pone.0067003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bemark M, Hazanov H, Stromberg A, Komban R, Holmqvist J, Koster S, Mattsson J, Sikora P, Mehr R & Lycke NY 2016. Limited clonal relatedness between gut IgA plasma cells and memory B cells after oral immunization. Nat Commun, 7, 12698. 10.1038/ncomms12698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coro ES, Chang WL & Baumgarth N 2006. Type I IFN receptor signals directly stimulate local B cells early following influenza virus infection. J Immunol, 176, 4343–51. 10.4049/jimmunol.176.7.4343 [DOI] [PubMed] [Google Scholar]
- Crocquet-Valdes PA, Thirumalapura NR, Ismail N, Yu X, Saito TB, Stevenson HL, Pietzsch CA, Thomas S & Walker DH 2011. Immunization with Ehrlichia P28 outer membrane proteins confers protection in a mouse model of ehrlichiosis. Clin Vaccine Immunol, 18, 2018–25. 10.1128/CVI.05292-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham AF, Gaspal F, Serre K, Mohr E, Henderson IR, Scott-Tucker A, Kenny SM, Khan M, Toellner KM, Lane PJ & Maclennan IC 2007. Salmonella induces a switched antibody response without germinal centers that impedes the extracellular spread of infection. J Immunol, 178, 6200–7. 10.4049/jimmunol.178.10.6200 [DOI] [PubMed] [Google Scholar]
- Dawson JE, Anderson BE, Fishbein DB, Sanchez JL, Goldsmith CS, Wilson KH & Duntley CW 1991. Isolation and characterization of an Ehrlichia sp. from a patient diagnosed with human ehrlichiosis. J Clin Microbiol, 29, 2741–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Niro R, Lee SJ, Vander Heiden JA, Elsner RA, Trivedi N, Bannock JM, Gupta NT, Kleinstein SH, Vigneault F, Gilbert TJ, Meffre E, Mcsorley SJ & Shlomchik MJ 2015. Salmonella Infection Drives Promiscuous B Cell Activation Followed by Extrafollicular Affinity Maturation. Immunity, 43, 120–31. 10.1016/j.immuni.2015.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M & Gingeras TR 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics, 29, 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumler JS, Sutker WL & Walker DH 1993. Persistent infection with Ehrlichia chaffeensis. Clin Infect Dis, 17, 903–5. 10.1093/clinids/17.5.903 [DOI] [PubMed] [Google Scholar]
- Fagarasan S, Kinoshita K, Muramatsu M, Ikuta K & Honjo T 2001. In situ class switching and differentiation to IgA-producing cells in the gut lamina propria. Nature, 413, 639–43. 10.1038/35098100 [DOI] [PubMed] [Google Scholar]
- Fagarasan S, Muramatsu M, Suzuki K, Nagaoka H, Hiai H & Honjo T 2002. Critical roles of activation-induced cytidine deaminase in the homeostasis of gut flora. Science, 298, 1424–7. 10.1126/science.1077336 [DOI] [PubMed] [Google Scholar]
- Farci P, Diaz G, Chen Z, Govindarajan S, Tice A, Agulto L, Pittaluga S, Boon D, Yu C, Engle RE, Haas M, Simon R, Purcell RH & Zamboni F 2010. B cell gene signature with massive intrahepatic production of antibodies to hepatitis B core antigen in hepatitis B virus-associated acute liver failure. Proc Natl Acad Sci U S A, 107, 8766–71. 10.1073/pnas.1003854107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habib S, El Andaloussi A, Hisham A & Ismail N 2016. NK Cell-Mediated Regulation of Protective Memory Responses against Intracellular Ehrlichial Pathogens. PLoS One, 11, e0153223. 10.1371/journal.pone.0153223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y, O'neill P, Naradikian MS, Scholz JL & Cancro MP 2011. A B-cell subset uniquely responsive to innate stimuli accumulates in aged mice. Blood, 118, 1294–304. 10.1182/blood-2011-01-330530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastey CJ, Elsner RA, Barthold SW & Baumgarth N 2012. Delays and diversions mark the development of B cell responses to Borrelia burgdorferi infection. J Immunol, 188, 5612–22. 10.4049/jimmunol.1103735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J, Serra F & Bork P 2016. ETE 3: Reconstruction, Analysis, and Visualization of Phylogenomic Data. Mol Biol Evol, 33, 1635–8. 10.1093/molbev/msw046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Intlekofer AM, Banerjee A, Takemoto N, Gordon SM, Dejong CS, Shin H, Hunter CA, Wherry EJ, Lindsten T & Reiner SL 2008. Anomalous type 17 response to viral infection by CD8+ T cells lacking T-bet and eomesodermin. Science, 321, 408–11. 10.1126/science.1159806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail N, Bloch KC & Mcbride JW 2010. Human ehrlichiosis and anaplasmosis. Clin Lab Med, 30, 261–92. 10.1016/j.cll.2009.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail N & Mcbride JW 2017. Tick-Borne Emerging Infections: Ehrlichiosis and Anaplasmosis. Clin Lab Med, 37, 317–340. 10.1016/j.cll.2017.01.006 [DOI] [PubMed] [Google Scholar]
- Ismail N, Soong L, Mcbride JW, Valbuena G, Olano JP, Feng HM & Walker DH 2004. Overproduction of TNF-alpha by CD8+ type 1 cells and down-regulation of IFN-gamma production by CD4+ Th1 cells contribute to toxic shock-like syndrome in an animal model of fatal monocytotropic ehrlichiosis. J Immunol, 172, 1786–800. 10.4049/jimmunol.172.3.1786 [DOI] [PubMed] [Google Scholar]
- Jacob J, Kelsoe G, Rajewsky K & Weiss U 1991. Intraclonal generation of antibody mutants in germinal centres. Nature, 354, 389–92. 10.1038/354389a0 [DOI] [PubMed] [Google Scholar]
- Jegerlehner A, Maurer P, Bessa J, Hinton HJ, Kopf M & Bachmann MF 2007. TLR9 signaling in B cells determines class switch recombination to IgG2a. J Immunol, 178, 2415–20. 10.4049/jimmunol.178.4.2415 [DOI] [PubMed] [Google Scholar]
- Kaji T, Ishige A, Hikida M, Taka J, Hijikata A, Kubo M, Nagashima T, Takahashi Y, Kurosaki T, Okada M, Ohara O, Rajewsky K & Takemori T 2012. Distinct cellular pathways select germline-encoded and somatically mutated antibodies into immunological memory. J Exp Med, 209, 2079–97. 10.1084/jem.20120127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenderes KJ, Levack RC, Papillion AM, Cabrera-Martinez B, Dishaw LM & Winslow GM 2018. T-Bet(+) IgM Memory Cells Generate Multi-lineage Effector B Cells. Cell Rep, 24, 824–837 e3. 10.1016/j.celrep.2018.06.074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AM, Cambier JC & Shlomchik MJ 2012. B cell receptor signal transduction in the GC is short-circuited by high phosphatase activity. Science, 336, 1178–81. 10.1126/science.1213368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar BV, Ma W, Miron M, Granot T, Guyer RS, Carpenter DJ, Senda T, Sun X, Ho SH, Lerner H, Friedman AL, Shen Y & Farber DL 2017. Human Tissue-Resident Memory T Cells Are Defined by Core Transcriptional and Functional Signatures in Lymphoid and Mucosal Sites. Cell Rep, 20, 2921–2934. 10.1016/j.celrep.2017.08.078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunisawa J, Gohda M, Hashimoto E, Ishikawa I, Higuchi M, Suzuki Y, Goto Y, Panea C, Ivanov II, Sumiya R, Aayam L, Wake T, Tajiri S, Kurashima Y, Shikata S, Akira S, Takeda K & Kiyono H 2013. Microbe-dependent CD11b+ IgA+ plasma cells mediate robust early-phase intestinal IgA responses in mice. Nature Communications, 4, 1772. 10.1038/ncomms2718 https://www.nature.com/articles/ncomms2718#supplementary-information [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson JD, Thurman JM, Rubtsov AV, Claypool D, Marrack P, Van Dyk LF, Torres RM & Pelanda R 2012. Murine gammaherpesvirus 68 infection protects lupus-prone mice from the development of autoimmunity. Proc Natl Acad Sci U S A, 109, E1092–100. 10.1073/pnas.1203019109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin Y, Winter D, Blecher-Gonen R, David E, Keren-Shaul H, Merad M, Jung S & Amit I 2014. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell, 159, 1312–26. 10.1016/j.cell.2014.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law CW, Chen Y, Shi W & Smyth GK 2014. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol, 15, R29. 10.1186/gb-2014-15-2-r29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK & Shi W 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics, 30, 923–30. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Lindner C, Wahl B, Fohse L, Suerbaum S, Macpherson AJ, Prinz I & Pabst O 2012. Age, microbiota, and T cells shape diverse individual IgA repertoires in the intestine. J Exp Med, 209, 365–77. 10.1084/jem.20111980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magri G, Comerma L, Pybus M, Sintes J, Llige D, Segura-Garzon D, Bascones S, Yeste A, Grasset EK, Gutzeit C, Uzzan M, Ramanujam M, Van Zelm MC, Albero-Gonzalez R, Vazquez I, Iglesias M, Serrano S, Marquez L, Mercade E, Mehandru S & Cerutti A 2017. Human Secretory IgM Emerges from Plasma Cells Clonally Related to Gut Memory B Cells and Targets Highly Diverse Commensals. Immunity, 47, 118–134 e8. 10.1016/j.immuni.2017.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin F, Oliver AM & Kearney JF 2001. Marginal zone and B1 B cells unite in the early response against T-independent blood-borne particulate antigens. Immunity, 14, 617–29. 10.1016/s1074-7613(01)00129-7 [DOI] [PubMed] [Google Scholar]
- Meng W, Zhang B, Schwartz GW, Rosenfeld AM, Ren D, Thome JJC, Carpenter DJ, Matsuoka N, Lerner H, Friedman AL, Granot T, Farber DL, Shlomchik MJ, Hershberg U & Luning Prak ET 2017. An atlas of B-cell clonal distribution in the human body. Nat Biotechnol, 35, 879–884. 10.1038/nbt.3942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K & Rikihisa Y 2009. Liver transcriptome profiles associated with strain-specific Ehrlichia chaffeensis-induced hepatitis in SCID mice. Infect Immun, 77, 245–54. 10.1128/IAI.00979-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moir S, Ho J, Malaspina A, Wang W, Dipoto AC, O'shea MA, Roby G, Kottilil S, Arthos J, Proschan MA, Chun TW & Fauci AS 2008. Evidence for HIV-associated B cell exhaustion in a dysfunctional memory B cell compartment in HIV-infected viremic individuals. J Exp Med, 205, 1797–805. 10.1084/jem.20072683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyron-Quiroz JE, Rangel-Moreno J, Kusser K, Hartson L, Sprague F, Goodrich S, Woodland DL, Lund FE & Randall TD 2004. Role of inducible bronchus associated lymphoid tissue (iBALT) in respiratory immunity. Nat Med, 10, 927–34. 10.1038/nm1091 [DOI] [PubMed] [Google Scholar]
- Mueller I, Galinski MR, Baird JK, Carlton JM, Kochar DK, Alonso PL & Del Portillo HA 2009. Key gaps in the knowledge of Plasmodium vivax, a neglected human malaria parasite. Lancet Infect Dis, 9, 555–66. 10.1016/S1473-3099(09)70177-X [DOI] [PubMed] [Google Scholar]
- Muller S, Hunziker L, Enzler S, Buhler-Jungo M, Di Santo JP, Zinkernagel RM & Mueller C 2002. Role of an intact splenic microarchitecture in early lymphocytic choriomeningitis virus production. J Virol, 76, 2375–83. 10.1128/jvi.76.5.2375-2383.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami J, Shimizu Y, Kashii Y, Kato T, Minemura M, Okada K, Nambu S, Takahara T, Higuchi K, Maeda Y, Kumada T & Watanabe A 1999. Functional B-cell response in intrahepatic lymphoid follicles in chronic hepatitis C. Hepatology, 30, 143–50. 10.1002/hep.510300107 [DOI] [PubMed] [Google Scholar]
- Naradikian MS, Hao Y & Cancro MP 2016. Age-associated B cells: key mediators of both protective and autoreactive humoral responses. Immunol Rev, 269, 118–29. 10.1111/imr.12380 [DOI] [PubMed] [Google Scholar]
- Olano JP, Wen G, Feng HM, Mcbride JW & Walker DH 2004. Histologic, serologic, and molecular analysis of persistent ehrlichiosis in a murine model. Am J Pathol, 165, 997–1006. 10.1016/S0002-9440(10)63361-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onodera T, Takahashi Y, Yokoi Y, Ato M, Kodama Y, Hachimura S, Kurosaki T & Kobayashi K 2012. Memory B cells in the lung participate in protective humoral immune responses to pulmonary influenza virus reinfection. Proc Natl Acad Sci U S A, 109,2485–90. 10.1073/pnas.1115369109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papillion AM, Kenderes KJ, Yates JL & Winslow GM 2017. Early derivation of IgM memory cells and bone marrow plasmablasts. PLoS One, 12, e0178853. 10.1371/journal.pone.0178853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng SL, Szabo SJ & Glimcher LH 2002. T-bet regulates IgG class switching and pathogenic autoantibody production. Proc Natl Acad Sci U S A, 99, 5545–50. 10.1073/pnas.082114899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racanelli V, Sansonno D, Piccoli C, D'amore FP, Tucci FA & Dammacco F 2001. Molecular characterization of B cell clonal expansions in the liver of chronically hepatitis C virus-infected patients. J Immunol, 167, 21–9. 10.4049/jimmunol.167.1.21 [DOI] [PubMed] [Google Scholar]
- Racine R, Chatterjee M & Winslow GM 2008. CD11c expression identifies a population of extrafollicular antigen-specific splenic plasmablasts responsible for CD4 T-independent antibody responses during intracellular bacterial infection. J Immunol, 181, 1375–85. 10.4049/jimmunol.181.2.1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racine R, Jones DD, Chatterjee M, Mclaughlin M, Macnamara KC & Winslow GM 2010. Impaired germinal center responses and suppression of local IgG production during intracellular bacterial infection. J Immunol, 184, 5085–93. 10.4049/jimmunol.0902710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W & Smyth GK 2015. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res, 43, e47. 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosche KL, Aljasham AT, Kipfer JN, Piatkowski BT & Konjufca V 2015. Infection with Salmonella enterica Serovar Typhimurium Leads to Increased Proportions of F4/80+ Red Pulp Macrophages and Decreased Proportions of B and T Lymphocytes in the Spleen. PLoS One, 10, e0130092. 10.1371/journal.pone.0130092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld AM, Meng W, Luning Prak ET & Hershberg U 2018. ImmuneDB, a Novel Tool for the Analysis, Storage, and Dissemination of Immune Repertoire Sequencing Data. Front Immunol, 9, 2107. 10.3389/fimmu.2018.02107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubtsov AV, Rubtsova K, Fischer A, Meehan RT, Gillis JZ, Kappler JW & Marrack P 2011. Toll-like receptor 7 (TLR7)-driven accumulation of a novel CD11c(+) B-cell population is important for the development of autoimmunity. Blood, 118, 1305–15. 10.1182/blood-2011-01-331462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubtsov AV, Rubtsova K, Kappler JW & Marrack P 2013. TLR7 drives accumulation of ABCs and autoantibody production in autoimmune-prone mice. Immunol Res, 55, 210–6. 10.1007/s12026-012-8365-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubtsova K, Rubtsov AV, Cancro MP & Marrack P 2015. Age-Associated B Cells: A T-bet-Dependent Effector with Roles in Protective and Pathogenic Immunity. J Immunol, 195, 1933–7. 10.4049/jimmunol.1501209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubtsova K, Rubtsov AV, Thurman JM, Mennona JM, Kappler JW & Marrack P 2017. B cells expressing the transcription factor T-bet drive lupus-like autoimmunity. J Clin Invest, 127, 1392–1404. 10.1172/JCI91250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubtsova K, Rubtsov AV, Van Dyk LF, Kappler JW & Marrack P 2013. T-box transcription factor T-bet, a key player in a unique type of B-cell activation essential for effective viral clearance. Proc Natl Acad Sci U S A, 110, E3216–24. 10.1073/pnas.1312348110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansonno D, Lauletta G, De Re V, Tucci FA, Gatti P, Racanelli V, Boiocchi M & Dammacco F 2004. Intrahepatic B cell clonal expansions and extrahepatic manifestations of chronic HCV infection. Eur J Immunol, 34, 126–36. 10.1002/eji.200324328 [DOI] [PubMed] [Google Scholar]
- Sathaliyawala T, Kubota M, Yudanin N, Turner D, Camp P, Thome JJ, Bickham KL, Lerner H, Goldstein M, Sykes M, Kato T & Farber DL 2013. Distribution and compartmentalization of human circulating and tissue-resident memory T cell subsets. Immunity, 38, 187–97. 10.1016/j.immuni.2012.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehdev AE & Dumler JS 2003. Hepatic pathology in human monocytic ehrlichiosis. Ehrlichia chaffeensis infection. Am J Clin Pathol, 119, 859–65. 10.1309/F7EA-B5P7-3217-16LJ [DOI] [PubMed] [Google Scholar]
- Sheneman L, Evans J & Foster JA 2006. Clearcut: a fast implementation of relaxed neighbor joining. Bioinformatics, 22, 2823–4. 10.1093/bioinformatics/btl478 [DOI] [PubMed] [Google Scholar]
- Sonoda E, Pewzner-Jung Y, Schwers S, Taki S, Jung S, Eilat D & Rajewsky K 1997. B cell development under the condition of allelic inclusion. Immunity, 6, 225–33. 10.1016/s1074-7613(00)80325-8 [DOI] [PubMed] [Google Scholar]
- Standaert SM, Yu T, Scott MA, Childs JE, Paddock CD, Nicholson WL, Singleton J Jr. & Blaser MJ 2000. Primary isolation of Ehrlichia chaffeensis from patients with febrile illnesses: clinical and molecular characteristics. J Infect Dis, 181, 1082–8. 10.1086/315346 [DOI] [PubMed] [Google Scholar]
- Stephens R, Ndungu FM & Langhorne J 2009. Germinal centre and marginal zone B cells expand quickly in a second Plasmodium chabaudi malaria infection producing mature plasma cells. Parasite Immunol, 31, 20–31. 10.1111/j.1365-3024.2008.01066.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson HL, Jordan JM, Peerwani Z, Wang HQ, Walker DH & Ismail N 2006. An intradermal environment promotes a protective type-1 response against lethal systemic monocytotropic ehrlichial infection. Infect Immun, 74, 4856–64. 10.1128/IAI.00246-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SL, Peel JN, Scharer CD, Risley CA, Chisolm DA, Schultz MD, Yu B, Ballesteros-Tato A, Wojciechowski W, Mousseau B, Misra RS, Hanidu A, Jiang H, Qi Z, Boss JM, Randall TD, Brodeur SR, Goldrath AW, Weinmann AS, Rosenberg AF & Lund FE 2019. T-bet Transcription Factor Promotes Antibody-Secreting Cell Differentiation by Limiting the Inflammatory Effects of IFN-gamma on B Cells. Immunity, 50, 1172–1187 e7. 10.1016/j.immuni.2019.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thirumalapura NR, Crossley EC, Walker DH & Ismail N 2009. Persistent infection contributes to heterologous protective immunity against fatal ehrlichiosis. Infect Immun, 77, 5682–9. 10.1128/IAI.00720-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thirumalapura NR, Stevenson HL, Walker DH & Ismail N 2008. Protective heterologous immunity against fatal ehrlichiosis and lack of protection following homologous challenge. Infect Immun, 76, 1920–30. 10.1128/IAI.01293-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsioris K, Gupta NT, Ogunniyi AO, Zimnisky RM, Qian F, Yao Y, Wang X, Stern JN, Chari R, Briggs AW, Clouser CR, Vigneault F, Church GM, Garcia MN, Murray KO, Montgomery RR, Kleinstein SH & Love JC 2015. Neutralizing antibodies against West Nile virus identified directly from human B cells by single-cell analysis and next generation sequencing. Integr Biol (Camb), 7, 1587–97. 10.1039/c5ib00169b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban BC, Hien TT, Day NP, Phu NH, Roberts R, Pongponratn E, Jones M, Mai NT, Bethell D, Turner GD, Ferguson D, White NJ & Roberts DJ 2005. Fatal Plasmodium falciparum malaria causes specific patterns of splenic architectural disorganization. Infect Immun, 73, 1986–94. 10.1128/IAI.73.4.1986-1994.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller S, Braun MC, Tan BK, Rosenwald A, Cordier C, Conley ME, Plebani A, Kumararatne DS, Bonnet D, Tournilhac O, Tchernia G, Steiniger B, Staudt LM, Casanova JL, Reynaud CA & Weill JC 2004. Human blood IgM "memory" B cells are circulating splenic marginal zone B cells harboring a prediversified immunoglobulin repertoire. Blood, 104, 3647–54. 10.1182/blood-2004-01-0346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- William J, Euler C, Christensen S & Shlomchik MJ 2002. Evolution of autoantibody responses via somatic hypermutation outside of germinal centers. Science, 297, 2066–70. 10.1126/science.1073924 [DOI] [PubMed] [Google Scholar]
- Winslow GM, Papillion AM, Kenderes KJ & Levack RC 2017. CD11c+ T-bet+ memory B cells: Immune maintenance during chronic infection and inflammation? Cell Immunol, 321, 8–17. 10.1016/j.cellimm.2017.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winslow GM, Yager E, Shilo K, Volk E, Reilly A & Chu FK 2000. Antibody-mediated elimination of the obligate intracellular bacterial pathogen Ehrlichia chaffeensis during active infection. Infect Immun, 68, 2187–95. 10.1128/iai.68.4.2187-2195.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates JL, Racine R, Mcbride KM & Winslow GM 2013. T cell-dependent IgM memory B cells generated during bacterial infection are required for IgG responses to antigen challenge. J Immunol, 191, 1240–9. 10.4049/jimmunol.1300062 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA seq data generated during this study is available in the NCBI’s Gene Expression Omnibus database (GEO) with accession ID GSE137154.