Abstract
Emerging viral outbreaks resulting from host switching is an area of continued scientific interest. Such events can result in disease epidemics or in some cases, clinically silent outcomes. These occurrences are likely relatively common and can serve as tools to better understand disease dynamics, and may result in changes in behavior, fecundity, and, ultimately survival of the host. Feline foamy virus (FFV) is a common retrovirus infecting domestic cats globally, which has also been documented in the North American puma (Puma concolor). The prevalent nature of FFV in domestic cats and its ability to infect wild felids, including puma, provides an ideal system to study cross-species transmission across trophic levels (positions in the food chain), and evolution of pathogens transmitted between individuals following direct contact. Here we present findings from an extensive molecular analysis of FFV in pumas, focused on two locations in Colorado, and in relation to FFV recovered from domestic cats in this and previous studies. Prevalence of FFV in puma was high across the two regions, ∼77 per cent (urban interface site) and ∼48 per cent (rural site). Comparison of FFV from pumas living across three states; Colorado, Florida, and California, indicates FFV is widely distributed across North America. FFV isolated from domestic cats and pumas was not distinguishable at the host level, with FFV sequences sharing >93 per cent nucleotide similarity. Phylogenetic, Bayesian, and recombination analyses of FFV across the two species supports frequent cross-species spillover from domestic cat to puma during the last century, as well as frequent puma-to-puma intraspecific transmission in Colorado, USA. Two FFV variants, distinguished by significant difference in the surface unit of the envelope protein, were commonly found in both hosts. This trait is also shared by simian foamy virus and may represent variation in cell tropism or a unique immune evasion mechanism. This study elucidates evolutionary and cross-species transmission dynamics of a highly prevalent multi-host adapted virus, a system which can further be applied to model spillover and transmission of pathogenic viruses resulting in widespread infection in the new host.
Keywords: feline foamy virus, retrovirus, cross-species transmission, puma, domestic cat, recombination
1. Introduction
Transmission of domestic animal pathogens at the wildlife-urban interface is highly relevant to disease emergence in new host populations (Wiethoelter et al. 2015; Chaudhary et al. 2018; Chiu et al. 2019), and is typically focused on virulent clinical outcomes in the new host. Enhanced ability to investigate the virome of an organism has improved our ability to evaluate transmission of viruses that do not result in acute death but result in chronic lifelong infection. Foamy viruses (FVs; family Retroviridae) are an ancient lineage of endemic animal-infecting retroviruses. Known to be largely host-specific, molecular evidence suggests FVs have co-evolved with their hosts, which include several non-human primate species, feline species, cattle, and horses (Switzer et al. 2005; Rethwilm and Bodem 2013; Khan et al. 2018). Although no overt diseases have been directly attributed to FV infection, coinfections and interactions with other viruses have been documented (Switzer et al. 2008; Choudhary et al. 2013; Alais et al. 2018; Powers et al. 2018). Numerous zoonotic transmissions of simian FV (SFV) from non-human primates to humans have been documented (Mouinga-Ondémé et al. 2012; Richard et al. 2015; Buseyne et al. 2018). Although not extensively investigated SFV transmissions have not been recorded from human to human. Endogenous relic FVs and FV-like elements have also been identified in numerous animals including the aye-aye (Daubentonia madagascariensis) genome (Han and Worobey 2012b), the ancient coelacanth fish (Han and Worobey 2012a), the Tuatara (Wei et al. 2019) and several other species (Xu et al. 2018).
FVs have linear ∼11–13 kb ssRNA genomes, containing two long terminal repeat regions and three major canonical retroviral genes, gag, pol, and env. At least two additional accessory proteins are encoded 3′ region to the env. Like all retroviruses, replication includes transcription from RNA to a DNA intermediate that integrates into the host genome. Although the viral entry receptor(s) for FFV has not yet been identified, studies have detected FVs in a broad range of tissue types and cells (Luther, Nuttall, and Gibbons 1978; Bouillant and Ruckerbauer 1984; Materniak, Bicka, and Kuźmak 2006; Murray and Linial 2006; Kehl, Tan, and Materniak 2013) with evidence suggests that the oral mucosa is the primary region for FV replication (Winkler, Löchelt, and Flower 1999; Calattini et al. 2006).
Of all the FVs, the primate FVs have been the most extensively researched to date, with several genetically distinct species groups having been identified in both Old World and New World primates. Phylogenetic analysis supports virus–host coevolution history, with additional evidence of occasional cross-species transmission mostly within the simian system (Switzer et al. 2004; Richard et al. 2015) and coinfection with multiple FV strains/genotypes (Liu et al. 2008). Based on the notion that FVs have co-evolved with their hosts, long-term molecular clock investigation indicates primate FVs have a slower nucleotide substitution rate than other RNA viruses and are the slowest evolving RNA viruses documented (Switzer et al. 2005). Recombination also plays an important role in FV evolution (Richard et al. 2015; Ensser et al. 2018). A presumed result of ancient recombination is the presence of two circulating serotypes which differ in a highly variable receptor binding encoding, also referred to as the surface unit (SU) region in the env gene observed in FV of both felines and primates (Winkler et al. 1998; Bleiholder et al. 2011; Galvin et al. 2013; Richard et al. 2015; Lambert et al. 2018).
Unlike primate FVs, very little is known about the genetic diversity, evolutionary dynamics, and ecology of feline FV (FFV). FFV infection of domestic cats is broadly distributed globally, with prevalence studies in Europe (Bleiholder et al. 2011), Australia (Winkler, Löchelt, and Flower 1999), Asia (Nakamura et al. 2000; Phung et al. 2001), and the USA (Powers et al. 2018), indicating FFV is endemic worldwide with these studies showing variable prevalence between 30 and 70 per cent. Although no consistent sex bias has been demonstrated, an increased prevalence is linked with aging of the animals (Winkler, Löchelt, and Flower 1999; Nakamura et al. 2000; Bleiholder et al. 2011). Oral mucosa is known as a key site of domestic cat FFV active replication (Cavalcante et al. 2018), with close social or conflict interactions such as grooming and/or biting likely primary transmission pathways (Winkler, Löchelt, and Flower 1999). This mode of transmission is also supported in simian populations, where biting and predation has been shown to be a major source of transmission within and between species (Calattini et al. 2004; Liu et al. 2008; Betsem et al. 2011).
Only four complete FFV genomes have been described from domestic cats that is, two from the USA (Helps and Harbour 1997; Winkler et al. 1998), and one each from Japan (Hatama et al. 2001), and Australia (Bodem et al. 1996). In wild felids, FFV infections have been documented using serology in European wildcats (Felis silvestris) from Scotland (Daniels et al. 1999) and leopard cats (Prionailurus bengalensis) in Vietnam (Nakamura et al. 2000). FFV has also been detected by partial genome sequencing in Iriomoteiriomote cats (subspecies P. bengalensis iriomotensis) in Japan and leopard cats in Argentina (Phung et al. 2001). A recent serologic survey of pumas in three USA states (California, Colorado, and Florida) is the first large-scale prevalence study in a wild felid species showing high levels of FFV seroconversion in the three areas, documenting ∼80 per cent positivity (Kechejian et al. 2019). Additionally, two full length FFV genomes have been recovered from the North American puma (Kehl et al. 2013), which genetically are highly similar to those found in domestic cats (Phung et al. 2001; Kehl et al. 2013).
In an effort to gain important insights into the origin, molecular ecology, and transmission dynamics of FFV in wild and domestic felids, we undertook the first extensive molecular survey and genetic analyses of FFV in wild felids, focusing on puma from two geographically distinct regions in Colorado, USA. As top predators, there is accumulating evidence that this species is at higher risk for pathogen spillover from prey (Franklin et al. 2007; Troyer et al. 2014; Lee et al. 2017a; Kellner et al. 2018), including spillover of feline leukemia virus from domestic cats to the endangered Florida panther with significant clinical impact (Cunningham et al. 2008; Chiu et al. 2019). We additionally undertook a comparative analysis to understand broader host–virus dynamics, analyzing FFV sequences from free-ranging feral domestic cats from Colorado and Australia, and free-roaming pumas from Florida and California. Our analysis supports frequent cross-species transmission of FFV during the last century along with puma-to-puma transmission, resulting in wide distribution and high prevalence of FFV in Puma across Colorado. As a result of these virus–host dynamics FFV sequences derived from puma living in Colorado, California, and endangered Florida panthers share high levels of similarity to FFV from domestic cats and do not form distinct species clades phylogenetically.
2. Materials and methods
2.1 Ethics statement
Puma samples were collected as part of ongoing studies at Colorado Parks and Wildlife (CPW) between 2005 and 2014, and provided to Colorado State University (CSU) for diagnostic evaluation. Domestic cat samples were collected by collaborating shelters and sent to CSU during the same period. Blood samples from these studies have been archived and reused for many unique analyses. CSU and CPW Institutional Animal Care and Use Committees reviewed and approved this work prior to initiation (CSU IACUC protocol 05-061A).
This work was performed in accordance with United States Department of Agriculture Animal Welfare Act and The Guide for the Care and Use of Laboratory Animals. CSU Public Health Assurance number is D16-00345. CSU is accredited by AAALAC International.
2.2 Sample collection
Whole blood samples from individual pumas and domestic cats were used for FFV analysis. Samples were collected opportunistically by collaborators and demographic information collected as previously described (Bevins et al. 2012; Carver et al. 2016), cold transported and stored at −80°C.
Colorado puma samples were collected by CPW as part of ongoing monitoring and study efforts in two regions of the Rocky Mountains in Colorado, USA (Fig. 1A). The Front Range (FR) is situated along the wildland–urban interface west of the city of Boulder, where animals were sampled as part of a study evaluating puma-human interactions. The Western Slope (WS) is a largely wildland/rural area on the Uncompahgre plateau near the towns of Montrose and Ridgway, where pumas were studied to evaluate impacts of hunting on population dynamics. Samples were collected 2007–13 in the FR and 2005–14 in the WS. Florida puma samples (whole blood) were collected by the Florida Fish and Wildlife Conservation Commission in an ongoing study to survey health and population management efforts.
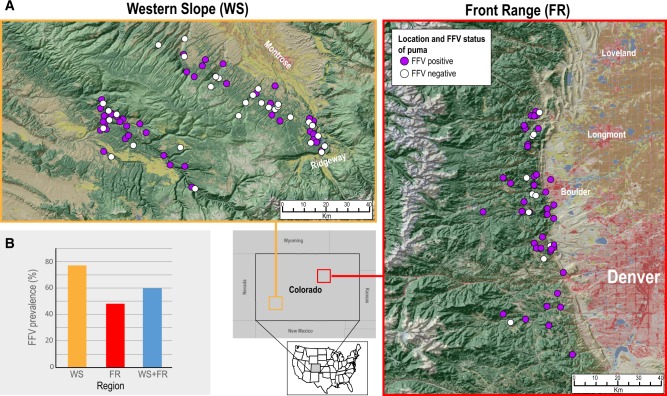
Figure 1.
FFV prevalence is high in pumas in the WS and FR. (A) Study areas and sampling sites for puma tested for FFV in Colorado, USA. (B) Bar graph showing prevalence of FFV across the two regions (WS and FR) and the average across both regions (WS+FR). Maps were sourced from the Multi-Resolution Land Characteristics consortium (Homer et al. 2015).
Colorado domestic cat samples were collected from free-ranging cats on entry to shelters close to the FR and WS regions in Colorado. Two shelters in Boulder (Boulder Humane Society and Community Cat Care Shelter) and one in Ridgway (Ridgway Second Chance Shelter) provided samples as previously described (Bevins et al. 2012; Carver et al. 2016). Australian domestic cat blood samples were opportunistically collected by the Beatty Lab at The University of Sydney, from pet domestic cats following routine clinical care at the university Veterinary Teaching Hospital.
2.3 DNA isolation
Whole blood samples were processed using a phenol-chloroform extraction as described by Di Pietro et al. (2011). Two hundred microliters of whole blood per sample were processed, and all subsequent reagent quantities were made proportional according to the recommended initial blood volume. Extracted DNA was quantified using a QuBit 2.0 fluorometer (Thermo Fisher Scientific, USA).
2.4 Screening puma samples for FFV using quantitative real-time PCR
A total of 169 Colorado puma samples (71 from the FR and 98 from the WS) were screened for FFV proviral DNA using methods outlined in Lee et al. (2017b) using a specific quantitative real-time (q)PCR screening protocol. The following protocol was used; 5 µl of iTaq™ Universal probe supermix (Bio-Rad, USA), 1 µl FFVgag F (10 µM) 5′-GGACGATCTCAACAAGGTCAACTAAA-3′, 1 µl FFVgag R (10 µM) 5′-TCCACGAGGAGGTTGCGA-3′, 0.2 µl FFVgag probe (10 µM), 5.8 µl sterile distilled water and 2 µl DNA (30–100 ng) template. DNA samples were run in triplicate alongside negative controls (DNA from known FFV negative domestic cats), positive controls (DNA from known experimentally infected FFV domestic cats), and plasmid dilution standards of 106–101 copies in duplicates. Reactions were run on a CFX Connect real-time PCR Detection System (Bio-Rad, USA) using the following cycling conditions: 95°C for 3 min followed by forty cycles of 95°C for 10 s and 60°C for 40 s (Lee et al. 2017b). Samples were considered positive only if two or more replicates were positive within the standard curve of detectable copies and within thirty-eight cycles.
2.5 Statistical analyses of the effects of sex and age on FFV status in puma
We evaluated the effects of sex (male/female) and age (young/adult) on FFV infection in pumas using Bayesian Generalized Linear Mixed Models (GLMMs), with location as a random effect, and sex, and age as fixed effects. Models tested whether a puma was qPCR negative or positive for FFV as a function of sex, age, and an interaction between sex and age. Using non-informative priors, models ran for 50,000 iterations, after a burn-in of 10,000 iterations and thinning of 10, yielding 4,000 values to derive parameter posterior means. Convergence was assessed visually by comparing the posterior distributions of multiple runs of each model. GLMM model weights and averaged coefficients were also calculated to examine the effect sizes of variables. All statistics were implemented in Program R (R Development Core Team, Vienna, Austria) version 3.3.3, using the ‘MCMCglmm’ package for the GLMMs (Supplementary Table S1).
2.6 Recovery of FFV genetic proviral sequences—full genome, pol, and env gene regions
Proviral sequences were recovered from a total of eighty-seven samples from Colorado pumas which were qPCR FFV positive. From these full genomes were isolated from twenty-eight individuals, and the pol and/or env gene regions from the remaining fifty-eight (Supplementary Table S2). Due to low DNA quantity, DNA degradation, or sample availability, these genetic regions (pol and env gene) could not be recovered from the remaining qPCR positive pumas. Additionally, pol and env gene sequences were amplified from three puma samples from Florida which were previously identified as FFV positive either by enzyme-linked immunosorbent assay (ELISA) or qPCR. Proviral sequences were recovered from nineteen domestic cat samples from Colorado and Australia. Six full genomes from Colorado domestic cats. FFV pol was recovered from an additional eleven Colorado cats, and env also was recovered from eight of these cats. FFV pol and env sequences from two domestic cats from Australia and three Florida panthers were also recovered and used in this analysis (Supplementary Table S2). Sequence data has been deposited in Genbank, accession numbers: MH633335-MH633442.
Nested PCR was used to recover proviral sequences for Sanger sequencing as follows: First Round 5 µl Kapa Hotstart Hifi polymerase (Kapa Biosystems, USA), 2 µl H2O, 1 µl of both the forward and reverse primer (Table 1) and 2 µl of DNA. Second Round—10 µl Kapa Hotstart Hifi polymerase (Kapa biosystems, USA), 6 µl H2O, 1 µl of both the forward and reverse primer (Table 1) and 2 µl of R1 PCR product. Cycling conditions were followed according to manufacturer’s specifications and an annealing temperature of 60°C. Reactions were run on a C1000 Touch Thermal Cycler (Bio-Rad, USA). Full genomes were amplified in two ∼6.3 kb fragments with ∼1 kb overlapping regions. PCR products were resolved on a 0.7 per cent agarose gel using electrophoresis for 30 min at 110 V. Bands of the correct size were excised from the gel and purified using a MEGAquick-spin™ Total Fragment DNA Purification Kit (iNtRON Biotechnology, Korea). Purified DNA was then cloned using pJET 1.2 blunt vector using the CloneJET PCR Cloning Kit (Thermo Fisher Scientific, USA) and XL1-Blue Escherichia coli competent cells (Agilent technologies, USA). Plasmids were isolated using DNA-spin Plasmid Purification Kit (iNtRON Biotechnology, Korea) and subsequently Sanger sequenced using primer walking at Quintarabio in San Francisco, CA. Genetic sequences were verified and assembled using Geneious 7.0.6 (Biomatters Ltd., New Zealand). Datasets of full genomes, pol and env (which both include pol and env regions from the full genome isolates) were compiled together with those downloaded from the public database GenBank. Nucleotide and amino acid datasets for both genes were used for downstream analyses.
Table 1.
Primer sequences used to recover the full genome, pol, and env proviral sequences.
| Primer name | Nested round | F/R primer | Product size of final amplicon | Primer sequence |
|---|---|---|---|---|
| Full FFV genome—5′ half | 1 | F | ∼6.3 kb | GAATTCTCACAGAGGAGAATACTCTCTGC |
| R | GCAACTCAGGATGAGTCAACTGAAGTTTCTG | |||
| 2 | F2 | GTCAACAAAAGCTCTTTTATCCCGGAG | ||
| R2 | TGGCCTAGATGGTCCACTATAATCACA | |||
| Full FFV genome—3′ half | 1 | F | ∼6.3 kb | GTTGGAGGGAAATTCCTCCTTCCCGAG |
| R | ACTGTCGTGGTCTATACCTGGGATAGGTTAG | |||
| 2 | F2 | AGTCATGCAAGACGAGAAGCCGT | ||
| R2 | CACTTTTCCCCAGGAATAGAGAAACAC | |||
| FFV env gene | 1 | F | ∼3.2 kb | CTAAGTGGAGGAAGCCTACAAAGG |
| R | CCACTTTCAGGAATTCCCTTATGACATTG | |||
| 2 | F2 | GATTATAGTGGACCATCTAGGCCAAC | ||
| R2 | CTGGTATGCATAGACAAAAGAGCTAAAG | |||
| FFV pol gene | 1 | F | ∼3.9 kb | GTAATCCTCAACAGCAGGGACC |
| R | GGAAGTATTTCCTCCCACGGTTAC | |||
| 2 | F2 | ATCAAGGACCTCGGCCAG | ||
| R2 | CAGGATGTGTTATTGCTTCTTTCCATTG |
2.7 Construction of phylogenetic trees
The full genomes and Pol and Env amino acid datasets were used to construct maximum likelihood phylogenetic trees. The best fit substitution models were inferred in MEGA 5.3 for the full genomes (GTR+I+G) using jModelTest (Posada 2009), and for the Pol and Env datasets (JTT+G) using ProtTest (Darriba et al. 2011). Trees were constructed using PhyML (Guindon et al. 2010) with approximate likelihood branch support. Branches presenting <80 per cent support were collapsed in TreeGraph2 (Stöver and Müller 2010). Trees were all midpoint rooted. Editing of trees was undertaken using FigTree V1.4.3 (Rambaut 2012) and demographic illustrative data were overlaid onto the Pol phylogenetic tree using ITOL v4.2.3 (Letunic and Bork 2016).
2.8 Recombination within pol and env gene regions
Recombination analyses for pol and env was undertaken using RDP4 software (Martin et al. 2015). Aligned datasets were uploaded and auto mask for optimal recombination detection applied. Events with three or more methods showing associated P-values <10−3 combined with strong phylogenetic support were considered as genuine events. Recombination-free datasets with recombination regions removed from all sequences were generated for downstream Bayesian evolutionary analysis by sampling trees (BEAST) analyses.
2.9 Bayesian analyses of FFV
2.9.1 FFV maximum likelihood phylogenetic tree
A maximum-likelihood phylogenetic tree was constructed from pol and env gene sequences using recombination-free datasets generated in RDP4 (Martin et al. 2015). This analysis was conducted using an Subtree pruning and regrafting (SPR) branch-swapping heuristic search in PhyML 3.0 (Guindon et al. 2010) and 1, 000 bootstrap replicates were used to estimate node robustness in the phylogeny. As part of this analysis, we specified the smart nucleotide selection model (Lefort, Longueville, and Gascuel 2017), which identified GTR + Γ4 model of nucleotide substitution model as most appropriate for this dataset. Using this tree, we performed a linear regression of root-to-tip divergences using the TempEst routine to check for temporal signal in the FFV sequences (Rambaut et al. 2016).
2.9.2 Time-calibrated phylogenetic trees and discrete trait analysis
To understand the temporal dynamics of FFV and to quantify host-switching over time, we constructed time-calibrated phylogenies in BEAST version 1.10 using the pol gene only; env was excluded from this analyses due to the high level of diversity resulting in a lack of temporal signal. Nonetheless, we do estimate the sub/site per year for both pol and env genes. Host and location were analyzed as discrete traits (Drummond and Rambaut 2007). Based on the two factors 1, domestic cat FFV likely represents a global endemic viral population due to domestic cat movement across the globe, and 2, we have a smaller relative representation of FFV from domestic cats compared with puma, we assigned sequences from domestic cats to one trait. To assess FFV movement between domestic cat and pumas across locations, FFV sequences from puma were assigned as traits based on their location (FR, WS, California, and Florida). We used a symmetric trait substitution model in which transition rates are assumed to occur at the same rate. Bayesian stochastic search variable selection procedure was employed to estimate host transition rates. SpreaD3 (Bielejec et al. 2016) was used to calculate Bayes Factor (BF) support for the most likely host transitions. Additionally, we used the same parameters but with a discrete trait asymmetrical model to test if cross-species transmission was more likely from domestic cat to puma or vice versa (i.e. we only used domestic cat and puma as traits, this data is shown as Supplementary Fig. S3), well-sampled puma populations were used for this, California and Florida puma were therefore excluded to simplify. Support for events with BF 3–10 = substantial, 10–30 = strong, 30–100 = very strong support, and >100 = decisive (Jarosz and Wiley 2014).
A relaxed (uncorrelated lognormal) molecular clock with continuous quantile parametrization was specified, and Bayesian skyline with time-aware Gaussian Markov random field coalescent prior was utilized (Minin, Bloomquist, and Suchard 2008). We used default priors and ran two independent MCMC (Markov chain Monte Carlo) analyses for 500 million steps and sampled every 500,000 states. Convergence of parameter estimates was assessed based effective sample size (ESS) (estimates with an ESS > 200 were considered converged).
3. Results
3.1 Prevalence, diversity, and demographics
qPCR screening of FFV in 169 pumas from two locations in Colorado revealed a higher prevalence of FFV in the FR of 55/71 (∼77%) compared with the WS which had a 48/98 (∼48%), and with a prevalence of 103/169 (∼60%) across both regions (Fig. 1B). Nucleotide similarity of FFV full genomes isolated from domestic cats and puma (including those from Florida, Australia, and USA, and international isolates available in the public database GenBank) range from 93 to 100 per cent pairwise nucleotide similarity (Supplementary Data S1). Amino acid pairwise comparison of Pol range from 94 to 100 per cent identity, whereas Env similarity ranges from 81 to 100 per cent (Supplementary Data S1). The SU in the Env protein is the most genetically diverse region on an amino acid level (Supplementary Data S1). Similar to findings for FFV from domestic cats (Winkler et al. 1998), two distinct Env genotypes were identified, with amino acid identity ranging from 93 to 100 per cent within genotypes, and 82 to 85 per cent identity between genotypes (Supplementary Data S1). Nucleotide diversity across the env gene, within each Env genotype group, is similar to that across the rest of the genome; ∼94–100 per cent (data not shown).
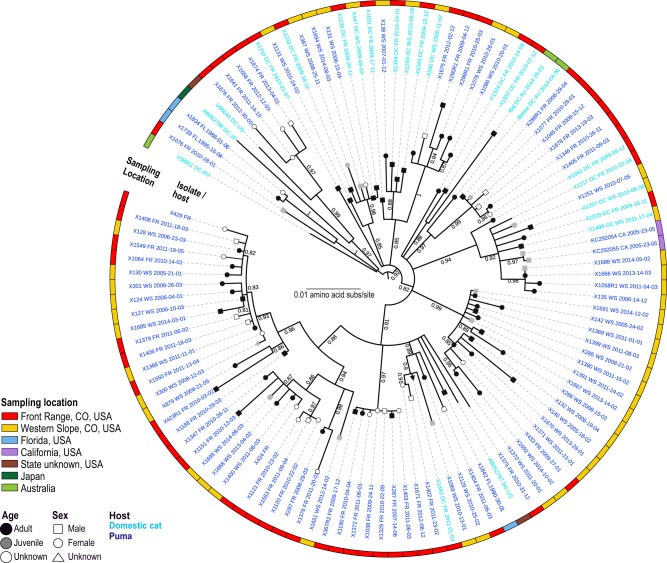
Prevalence in males and females was similar, with 60.3 per cent of males and 63.2 per cent of females identified to be qPCR positive. GLMMs analyses showed incidence of detection increased with age (43.8% for juveniles and 64.3% for adults; Supplementary Fig. S1). For a subset of tested individuals, sex and/or age information was unavailable (n = 67). Our analyses, however, showed that infection was not strongly predicted by sex or age (Supplementary Fig. S1). Phylogenetically groupings did not demonstrate distinctive specific sex or age trends; however, juvenile isolates clustered with isolates recovered from adults (Fig. 2).
Figure 2.
FFV from circulates in both domestic cats and puma. Domestic cat FFV Pol protein sequences are interspersed throughout the maximum likelihood phylogenetic tree, and cluster closely to FFV isolated from pumas. Isolate names in bold font were sequenced in this study and font color indicates host species, light blue domestic cat and dark blue puma. Location information is marked by colored squares on the periphery of the tree (red, FR, Colorado; yellow, WS, Colorado; blue, Florida; purple, California; brown, State unknown; dark green, Japan; light green, Australia) and demographic information on the branch tips (black, adult; gray, juvenile; white, unknown age and square, male; circle, female; triangle, unknown).
3.2 Phylogenetic analyses of full genome, Pol, and Env
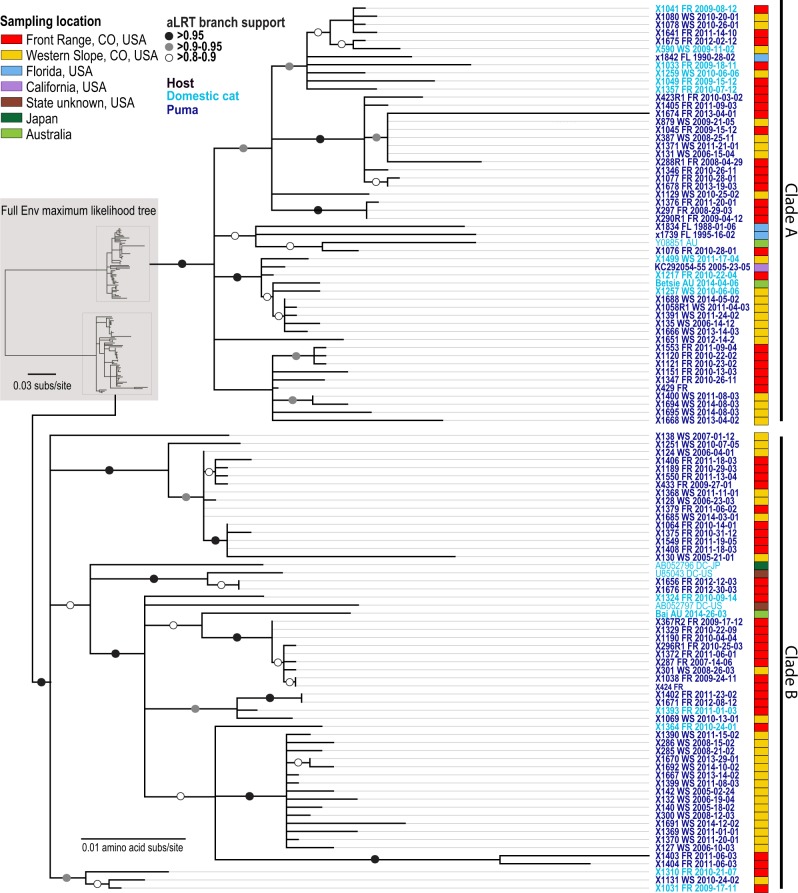
The genetic relationship of FFV in puma compared with domestic cats in maximum likelihood phylogenetic trees highlights a single virus–dual host dynamic which is clearly demonstrated in all three phylogenies representing Pol (Fig. 2), Env (Fig. 3), and the full genome (Supplementary Fig. S2). There is strong evidence for transmission of FFV between FR and WS locations, though some subclades indicate regionally specific viral populations (Figs 2 and 3). Domestic cat FFV isolates from the USA, internationally sourced isolates, and puma FFV isolates from Florida and California are interspersed throughout the trees (Figs 2 and 3). The Pol protein phylogeny documents that internationally sourced domestic cat sequences, although interspersed, sit basal to puma and domestic cat isolates from Colorado (Fig. 2). This pattern was not indicated for Env; however, two SU clades are evident in the Env protein phylogenetic tree (Fig. 3); isolates with these two highly divergent SU regions were found to circulate in both host species and across locations globally.
Figure 3.
Env protein maximum likelihood phylogenetic tree indicates FFV does not exhibit strong host or geographic structure. Clades A and B of FFV Env isolates are circulating in both host species and across locations broadly. Isolate names in bold font were sequenced in this study and font color indicates host species (light blue, domestic cat; dark blue, puma). Location information is marked by colored squares on the periphery of the tree (red, FR, Colorado; yellow, WS, Colorado; blue, Florida; purple, California; Brown, State unknown; dark green, Japan; light green, Australia). The two Env SU Clades A and B are highlighted.
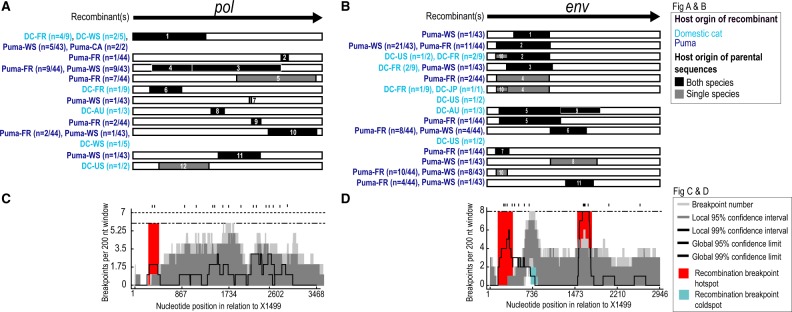
3.3 FFV recombination
Recombination analyses of pol and env gene sequences from domestic cats and puma showed high levels of recombination in both open reading frames (Fig. 4A and B). A total of twelve events in pol (Fig. 4A) and eleven in env (Fig. 4B) were strongly supported. Recombination events with inferred parental FFVs originating from both species were identified in 9/12 (75%) events in pol and 8/11 (∼72%) events in env (Fig. 4A and B and Supplementary Data S2). Recombination breakpoint hot and cold spots were identified in both genes, including one hotspot in the 5′ region of pol, a hotspot as well as a cold spot in the 5′ region of env, and a hotspot in the SU region of env (Fig. 4C and D).
Figure 4.
Recombination is common in genomes of domestic cat and puma FFV. Recombination analyses of FFV of (A) pol and (B) env sequences. Gene region is displayed at the top of each analyses for reference and recombinant regions depicted for each event below Host species origin of recombinant is indicated by color of recombinant names (light blue, domestic cat; dark blue, puma) and host species origin of minor parental sequence, single—puma or domestic cat or both—puma and domestic cat, by shading of recombinant region (black, single; gray, both). Number of individuals with recombinant sequences is shown following host and region name (C) Breakpoint frequency plot highlights recombination breakpoint hotspot in the 5′ of pol. (D) Graphic highlights recombination breakpoint hotspots, in the 5′ and SU region, and coldspot in the 5′ region of env.
3.4 Substitution rates and Bayesian ancestral reconstruction of location and host state
To evaluate substitution rates, we undertook Bayesian analyses of the pol and env recombination-free datasets. Pol displayed the slowest substitution rate [2.640 × 10−4; 95% highest posterior density (HPD) interval 1.480 × 10−4, 3.768 × 10−4], whereas env substitution rates were comparatively faster [3.872 × 10−4; 95% HPD interval 1.85 × 10−4, 6.025 × 10−4].
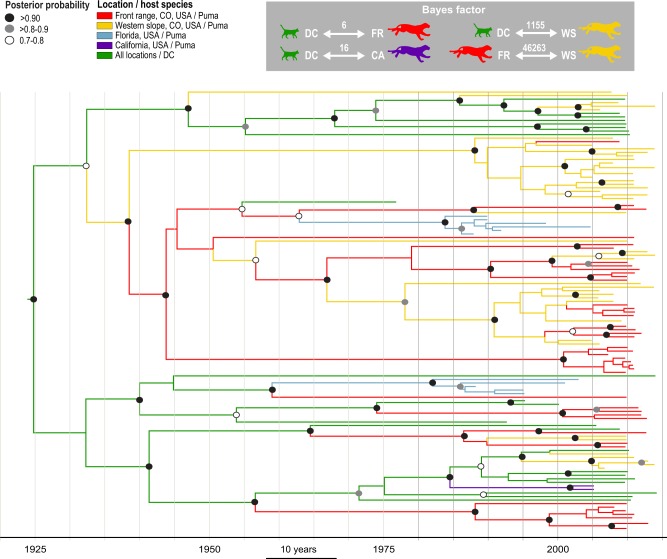
Further BEAST analyses on only the pol (the high level of diversity present in the env resulted in a lack of temporal signal), was used to investigate historical geographic movements, and host species transmission patterns. Four trends of FFV transmission were well supported (Fig. 5): 1, domestic cat—FR puma (BF = 6, posterior probability = 0.84); 2, domestic cat—California puma (BF = 16, posterior probability = 0.93); 3, domestic cat—WS puma (BF = 1,155, posterior probability = 0.99); and 4, puma—puma transmission between individuals from the FR and WS (BF = 46,263, posterior probability = 1.0). Further, analysis of host transmission using an asymmetrical model of discrete trait transition (Supplementary Fig. S3) shows two supported trends of FFV transmission: puma to domestic cat (BF = 20, posterior probability = 0.99) and domestic cat to puma (BF = 8,155, posterior probability = 1). Supported nodes indicate that FFV has likely been present in Colorado at least 70 years. Four major clades are present, one which consists primarily of domestic cats, two primarily puma and a fourth that includes both domestic cats and puma from three sites in the USA (Fig. 5).
Figure 5.
FFV phylogeny indicates puma FFV originated from cross-species transmission followed by efficient puma-to-puma spread. Bayesian maximum clade credibility tree of pol showing location and host movement of the virus over time. BF supported movements across the tree show supported movements between 1, domestic cats (green) and California puma (purple); 2, domestic cats (green) and WS puma (yellow); and 3, puma from the FR (red) and WS (yellow). Posterior probability indicated by circles as indicated in legend (white, 0.7–0.8; gray, ≥0.8–0.9, black; >0.9).
4. Discussion
4.1 High prevalence of FFV in puma
The high prevalence (∼60%) of FFV identified in Colorado puma population’s documents endemic infection in this region (Fig. 1B). This prevalence is on the high end of what has been reported in domestic cat populations (range 8–80%, (Winkler, Löchelt, and Flower 1999; Nakamura et al. 2000; Powers et al. 2018). Reported seroprevalence was even higher (77.3%) which is likely due to higher sensitivity of ELISA compared with PCR (Kechejian et al. 2019). Identification of FFV positive pumas from Florida and California indicates this virus is not limited to Colorado puma populations, and is widely spread across free-ranging populations. We found little statistical support for increased infection rate with sex (GLMM pMCMC > 0.05) or age (GLMM pMCMC > 0.05), indicating that FFV is readily transmitted between the sexes, and is not age dependent—providing strong support for social transmission mechanisms such as grooming. Although sex and age were not found to be risk factors for FFV infection, their inclusion in our GLMMs improved model fit suggesting they may still influence FFV transmission. Recent work by our research group has indicated age and/or sex to be risk factors that vary by population (Kechejian et al. 2019), so the impact of these factors on risk of infection has yet to be fully elucidated. Juvenile-originating FFV sequences were dispersed throughout the phylogeny, and commonly nested most closely to those from adults, a pattern suggesting transmission from parent to offspring (Fig. 2). Additionally, lack of phylogenetic stratification by sex indicates transmission between males and females occurs commonly (Fig. 2).
4.2 Complex phylogeny of puma FFV indicates spillover and puma to puma transmission
The phylogenetic relationship of FFV from puma and domestic cats highlights a single virus–dual host dynamic, supporting domestic cat to puma transmission and puma to puma transmission. This is displayed in both the Pol and Env protein phylogenetic trees independently (Figs 2 and 3) with the presence of 1) puma only clades and 2) domestic cat isolates cluster throughout the tree, basal to or within puma dominated clades. Viral spillover resulting from trophic interactions has been demonstrated for other retroviruses, including feline immunodeficieny virus - FIVlru, transmission from bobcat (Lynx rufus) to puma (Lee et al. 2017a), feline leukemia virus transmission from domestic cat to puma (Cunningham et al. 2008; Chiu et al. 2019), and also for other pathogens such as Mycoplasma haemominutum from bobcat and domestic cat to puma (Kellner et al. 2018). Recent reports that domestic animals are increasingly preyed upon by puma near human populated areas, including the FR study site examined here (Moss et al. 2016), is a strong indicator that increases in viral spillover from domestic cats to pumas are likely to be observed with intensified urbanization. Clades that are predominantly comprised of isolates from one geographic area in Colorado (FR or WS; Figs 2 and 3 and Supplementary Fig. S2), but include a single isolate from the other location in Colorado (FR or WS), could represent either introductions from a migrant puma, or domestic cat spillover events.
4.3 Two co-circulating genotypes of FFV are pervasive in puma and domestic cats
Phylogenetic analysis of Env proteins indicates two distinct clades denoting the two SU Env genotypes (Clades A and B; Fig. 3). Both SU Env genotypes were isolated from domestic cats and pumas, and were found in the WS and FR of Colorado, and globally among domestic cats. This indicates both genotypes are adapted to both species and frequently co-circulate in populations, thereby suggesting a synergistic relationship. The dominant presence of both genotypes circulating among both host species indicates that despite the high level of amino acid variability in this SU region, these genotypes are well adapted to both host species. This observation parallels recent studies of two unique Env SFV subtypes in non-human primates (Richard et al. 2015; Lambert et al. 2018). In domestic cats analyses clearly identified these two genotypes using serotype-specific PCR assays, sequencing and neutralizing assays (Winkler et al. 1998). A later study targeted Env-specific antibodies using a ElpSU antigen ELIZA; however, this assay was unable to distinguish-specific serotypes (Bleiholder et al. 2011). It has been suggested that variance in SU allows the virus to utilize multiple receptors in a genotype-specific manner, or perhaps they have different cell tropisms relevant for the biology of FFV. Receptor interference studies however support that all FVs, despite having a broad host range, may gain viral entry via the same cell surface receptor (Hill, Bieniasz, and McClure 1999; Berg et al. 2003). Cellular expression of a FV Env has been shown to induce superinfection resistance, thereby acting as a barrier to infection by other FV’s (Hill, Bieniasz, and McClure 1999; Berg et al. 2003). This would indicate that the two FFV genotypes also utilize the same receptor; however, additional studies of this interesting phenomenon are warranted to further explore this.
4.4 Recombination plays an important role in FFV evolution and adaptation
We identified FFV recombinant regions spanning the complete open reading frame of pol, whereas recombinant regions in env are concentrated in the 5′ and SU region. Hotspots in the 5′ region of pol (Fig. 4C), and in the 5′ and SU region of env (Fig. 4D) relate to relatively high levels of variation in the FFV genome. The env SU hotspot spans the 3′ end of the SU coding region, highlighting this site as an evolutionarily important breakpoint region in the context of two co-circulating genetically distinct Env genotypes. This env recombination breakpoint hotspot mirrors those detected in SFV’s (Galvin et al. 2013; Richard et al. 2015; Ensser et al. 2018). One recombination breakpoint coldspot was identified in env approximately at 730 nt position (Fig. 4D), a region encoding one of the receptor binding domains in env SU. The lack of recombination breakpoints detected here suggests its conservation is critical for viral function. Recombination has also been frequently observed among SFVs (Liu et al. 2008; Richard et al. 2015; Galvin et al. 2013) indicating a precedent for our observations.
4.5 Feline FV substitution rates differ from other FVs
Substitution rates in FFV pol, ∼2.5 × 10−4 substitutions per site per year (s/n/y), are considerably higher than rates reported for SFV pol (∼1.7 × 10−8 s/n/y; Switzer et al. 2005). Several factors likely contribute to this difference. First, a 425 nt region of pol was used in the SFV study (Switzer et al. 2005) compared with the full ∼3,460 nt pol gene region analyzed here. Second, time dependency, which refers to the differences between short- and long-term studies, can alter estimates of FV evolutionary rates (Aiewsakun and Katzourakis 2015; Membrebe et al. 2019). Tissue type sampled may also attribute to calculation of different substitution rates, as sampling of the virus from mucosa likely represents an active infection, whereas viral sequences sampled from the blood represent latent infection (Soliven et al. 2013). The frequent transmission events of FFV in this dual-host system may also be a factor in driving evolution rates. Nevertheless, FFV substitution rates appear to be falling between reported rates for DNA and RNA viruses, and in the range described for other retroviruses such as FIV (Peck and Lauring 2018; Krakoff et al. 2019).
4.6 Host and spatial movement patterns evident in FFV phylogeny
Puma isolates were grouped according to location, while domestic cat FFV isolates were analyzed as a single group in order to best mitigate the effects of sampling bias from puma. The BF supported transitions indicates a high probability of FFV spillover from domestic cats to puma, as well as puma to puma transmission (Fig. 5 and Supplementary Fig. S3). It’s worth noting a flexible non-parametric coalescent model was applied in these analyses, which assumes all organisms are part of the same population. Given the spatial/host dimensions of this analyses this prior although not optimal was the most appropriate currently available. Nonetheless, we found that whilst there was some support for puma to domestic cat cross-species transmission (BF = 20), overwhelmingly the model supported domestic cat to puma transmission (BF = 8,155) (Supplementary Fig. S3). This is not surprising considering that interactions between the puma predator and domestic cat prey make puma to domestic cat pathogen transmission events for directly transmitted agents biologically implausible. Spatial movement of FFV via puma to puma transmission is strongly supported between urban (FR) and rural (WS) locations (Fig. 5), which is likely propagated by dispersal of individuals. Our findings support FFV transmission between domestic cat and puma in both urban (California and Colorado FR) and rural (Colorado WS) areas, although the latter appears to be much more strongly supported. There is potentially a high propensity for cross-species transmission in an urban area due to high densities of domestic cats kept as pets (Moss et al. 2016). High risks for transmission in rural regions may be related to the higher number of outdoor cats in these areas which are at risk for predation.
5. Conclusion
This study provides the first large-scale molecular analysis of FFV infection in a free-ranging wild felid, and presents an in-depth investigation of the viral dynamics and evolution within puma populations in comparison to domestic cats. We document a remarkably high number of puma infected with FFV (including sequences from California, Colorado, and Florida), approximating or exceeding seroprevalence rates in domestic cats (Winkler, Löchelt, and Flower 1999; Phung et al. 2001). Interestingly, phylogeny indicates that FFV from domestic cats readily infects pumas suggesting limited innate or adaptive immune barriers exist to prohibit FFV cross-species transmission between these hosts. Further, the differing phylogenetic patterns between Pol and Env maximum likelihood phylogenetic trees indicate a complex and dynamic evolutionary history (Figs 2 and 3).
SFVs and FFVs share the unusual phenomenon of having two distinct and unique co-circulating genotypes which vary significantly in the receptor binding domain of Env. We show here that these two distinct FFV genotypes commonly infect both puma and domestic cats, though neither genotype exhibits species specificity. Although the biological implications and origin(s) are unknown, it appears this region of FV is highly prone to recombination in both felids and primates (Galvin et al. 2013; Richard et al. 2015; Ensser et al. 2018). Further, the near equivalence of these two genotypes in prevalence suggests that the two genotypes may be synergistic and/or dependent upon one another, as a greater fitness of one genotype versus the other would be accompanied by disproportionate occurrence of one isolate (Moya, Holmes, and González-Candelas 2004).
Unlike another retroviral infection of puma, FIVpco (Lee et al. 2014), the phylogeny of FFV sequences displays little geographic structure. Interestingly, here we see domestic cat derived FFV sequences are interspersed throughout the phylogenetic trees, and are situated either basal to, or clustered within, clades comprised of both domestic cat and puma isolates from the USA. Further, internationally derived FFV domestic cat sequences from Europe or Australia are also sporadically located across the phylogenetic tree. This pattern is most easily explained by owners relocating with their pet cats and/or historical trade movements where cats were dispersed for the purpose of rodent control, both domestically and globally (Gehrt, Riley, and Cypher 2010; Ottoni et al. 2017). Well-supported BF movements are congruent with a co-circulating history in these two host species, and strongly suggest that domestic cats may be an important factor for ‘seeding’ FFV into puma populations, followed by subsequent spread from puma to puma (Fig. 5 and Supplementary Fig. S3). Genetic and historical records indicate the introduction of domestic cats to the New World occurred with early European settlers ∼1,600 (Lipinski et al. 2008). It is thought that settlers brought domestic cats to Colorado during the Pike’s Peak gold rush of 1,858, and were well-established in the state by the 1870s (Ross 2016). In the early to mid-1900s, puma abundance throughout Colorado declined to a few hundred individuals, primarily due to unrestricted hunting and predator control. From 1965 to present, human-caused mortality in pumas was restricted (Anderson et al. 2010), and as a result the puma population in Colorado have grown from as low as a few hundred in 1965 to a few thousand adult and sub-adult pumas (Cahalane 1964; Anderson et al. 2010; Apker 2017). When puma numbers were low, interactions between pumas and domestic cats would consequently be limited. Increases in free-ranging puma populations, concurrent with increases in domestic cat ownership have likely influenced prevalence trends recorded here that are congruent with general indications of FFV phylogenetic analysis. The very high level of contemporary FFV infection/movement among pumas indicates rapid fixation in the population. A similar pattern has been seen for other pathogens, such as the ∼1980 canine parvovirus pandemic, illustrating that a very contagious pathogen that does not cause high morbidity can rapidly become endemic (Parrish and Kawaoka 2005).
Interestingly, although identified in domestic cats and puma, a homologous genotype of FFV is not known to infect bobcats or Canada Lynx (Lynx canadensis), both of which have overlapping geographic ranges with puma, and it has been shown that bobcats will predate on domestic cats (Clark 2010). This suggests that species-specific immunological or innate factors may limit FFV. FFV is present in the unique wild felid species which inhabits the island of Iriomote, Japan, an island that has been geographically isolated for 200,000 years (Masuda and Yoshida 1995). Despite harboring FFV the Iriomote wild cat population were negative for other major feline viral infections such as feline immunodeficiency virus and feline leukemia virus (Mochizuki, Akuzawa, and Nagatomo 1990), thereby supporting an ancient ancestral origin. More extensive molecular investigations into FFV infecting other wild felid species globally would shed further light on the evolutionary history of what is believed to be one of the oldest known exogenous vertebrate RNA virus families (Rethwilm and Bodem 2013).
Given its high prevalence, and the unlikelihood that FFV would be transmitted from puma to domestic cats (since the domestic cats are unlikely to survive such an encounter), FFV provides an interesting and unique viral ‘fingerprint’ that can map puma movements and augment studies of individual and population movements in this wide-ranging and secretive apex predator (Fountain-Jones et al. 2018). Although FVs evolve slowly, we document FFV substitution rates that are more rapid than predicted by previous FV mutation rate estimates. This may be due to adaptation to a new host species and pervasive inter- and intra-species transmission, and provides an opportunity to better study transmission events within and between species. The information gained here elucidating the evolution and cross-species transmission dynamics of a highly prevalent but apathogenic virus can be further applied to model more virulent disease, such as that caused by feline leukemia virus, which is a major threat to the endangered Florida panther (Cunningham et al. 2008; Chiu et al. 2019).
Supplementary Material
Acknowledgements
We thank wildlife managers, scientists, veterinarians, and support staff at Colorado Parks and Wildlife, Florida Fish and Wildlife Conservation Commission, and National Park Service including Dave Onorato, Mark Cunningham, and Roy McBride. In addition we thank the Boulder Humane Society and Community Cat Care shelter and the Ridgway Second Chance shelter for their assistance providing samples, and Dr Julia Beatty at the University of Sydney for providing domestic cat samples from Australia. We are hugely thankful to Guy Baele for his expert advice for the BEAST analyses. Thanks to Daryl Trumbo for generating map features used in Fig. 1A.
Funding
This study was supported by NSF-EEID award 1413925.
Conflict of interest: No conflicts of interest.
References
- Aiewsakun P., Katzourakis A. (2015) ‘Time Dependency of Foamy Virus Evolutionary Rate Estimates’, BMC Evolutionary Biology, 15: 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alais S. et al. (2018) ‘STLV-1 Co-Infection is Correlated with an Increased SFV Proviral Load in the Peripheral Blood of SFV/STLV-1 Naturally Infected Non-Human Primates’, PLoS Neglected Tropical Diseases, 12: e0006812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson C. et al. (2010) ‘Cougar Management in North America’, in Hornocker M., Negri S. (eds.) Cougar Ecology and Conservation, pp. 41–54. Chicago, IL: University of Chicago Press. [Google Scholar]
- Apker J. (2017). ‘Colorado Mountain Lion Status Report’, in McLaughlin, C. R., and Vieira, M. (eds.) Proceedings of the 12th Mountain Lion Workshop, pp. 74–84. Estes Park, CO: Western Association of Fish and Wildlife Agencies
- Berg A. et al. (2003) ‘Determinants of Foamy Virus Envelope Glycoprotein Mediated Resistance to Superinfection’, Virology, 314: 243–52. [DOI] [PubMed] [Google Scholar]
- Betsem E. et al. (2011) ‘Frequent and Recent Human Acquisition of Simian Foamy Viruses through Apes’ Bites in Central Africa’, PLoS Pathogens, 7: e1002306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevins S. N. et al. (2012) ‘Three Pathogens in Sympatric Populations of Pumas, Bobcats, and Domestic Cats: Implications for Infectious Disease Transmission’, PLoS One, 7: e31403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielejec F. et al. (2016) ‘SpreaD3: Interactive Visualization of Spatiotemporal History and Trait Evolutionary Processes’, Molecular Biology and Evolution, 33: 2167–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleiholder A. et al. (2011) ‘Pattern of Seroreactivity against Feline Foamy Virus Proteins in Domestic Cats from Germany’, Veterinary Immunology and Immunopathology, 143: 292–300. [DOI] [PubMed] [Google Scholar]
- Bodem J. et al. (1996) ‘Characterization of the Spliced Pol Transcript of Feline Foamy Virus: The Splice Acceptor Site of the Pol Transcript is Located in Gag of Foamy Viruses’, Journal of Virology, 70: 9024–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouillant A., Ruckerbauer G. (1984) ‘Isolation of Bovine Syncytial Virus from Lymphocytes Recovered from Fluids Used to Flush Uterus and Oviducts of Superovulated Cattle’, Canadian Journal of Comparative Medicine, 48: 332. [PMC free article] [PubMed] [Google Scholar]
- Buseyne F. et al. (2018) ‘Clinical Signs and Blood Test Results among Humans Infected with Zoonotic Simian Foamy Virus: A Case-Control Study’, The Journal of Infectious Diseases, 218: 144–51. [DOI] [PubMed] [Google Scholar]
- Cahalane V. H. 1964. A Preliminary Study of Distribution and Numbers of Cougar, Grizzly, and Wolf in North America. New York: New York Zoological Society. [Google Scholar]
- Calattini S. et al. (2004) ‘Natural Simian Foamy Virus Infection in Wild-Caught Gorillas, Mandrills and Drills from Cameroon and Gabon’, Journal of General Virology, 85: 3313–7. [DOI] [PubMed] [Google Scholar]
- Calattini S. et al. (2006) ‘Modes of Transmission and Genetic Diversity of Foamy Viruses in a Macaca tonkeana Colony’, Retrovirology, 3: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver S. et al. (2016) ‘Pathogen Exposure Varies Widely among Sympatric Populations of Wild and Domestic Felids across the United States’, Ecological Applications, 26: 367–81. [DOI] [PubMed] [Google Scholar]
- Cavalcante L. et al. (2018) ‘Clinical and Molecular Features of Feline Foamy Virus and Feline Leukemia Virus Co-Infection in Naturally-Infected Cats’, Viruses, 10: 702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary V. et al. (2018) ‘Risk of Disease Spillover from Dogs to Wild Carnivores in Kanha Tiger Reserve, India’, bioRxiv, 360271.
- Chiu E. S. et al. (2019) ‘Multiple Introductions of Domestic Cat Feline Leukemia Virus in Endangered Florida Panthers’, Emerging Infectious Diseases, 25: 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary A. et al. (2013) ‘Influence of Naturally Occurring Simian Foamy Viruses (SFVs) on SIV Disease Progression in the Rhesus Macaque (Macaca mulatta) Model’, Viruses, 5: 1414–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark H. O., Jr (2010) ‘Urban Carnivores: Ecology, Conflict, and Conservation by SD Gehrt, SPD Riley, and BL Cypher [Editors]’, Western North American Naturalist, 70: 20. [Google Scholar]
- Cunningham M. W. et al. (2008) ‘Epizootiology and Management of Feline Leukemia Virus in the Florida Puma’, Journal of Wildlife Diseases, 44: 537–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels M. et al. (1999) ‘Feline Viruses in Wildcats from Scotland’, Journal of Wildlife Diseases, 35: 121–4. [DOI] [PubMed] [Google Scholar]
- Darriba D. et al. (2011) ‘ProtTest 3: Fast Selection of Best-Fit Models of Protein Evolution’, Bioinformatics, 27: 1164–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Pietro F. et al. (2011) ‘Genomic DNA Extraction from Whole Blood Stored from 15- to 30-Years at −20°C by Rapid Phenol–Chloroform Protocol: A Useful Tool for Genetic Epidemiology Studies’, Molecular and Cellular Probes, 25: 44–8. [DOI] [PubMed] [Google Scholar]
- Drummond A. J., Rambaut A. (2007) ‘BEAST: Bayesian Evolutionary Analysis by Sampling Trees’, BMC Evolutionary Biology, 7: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ensser A. et al. (2018) ‘Isolation and Sequence Analysis of a Novel Rhesus Macaque Foamy Virus Isolate with a Serotype-1-like Env’, Archives of Virology, 163: 2507–12. [DOI] [PubMed] [Google Scholar]
- Fountain-Jones N. M. et al. (2018) ‘Towards an Eco-Phylogenetic Framework for Infectious Disease Ecology’, Biological Reviews, 93: 950–70. [DOI] [PubMed] [Google Scholar]
- Franklin S. P. et al. (2007) ‘Frequent Transmission of Immunodeficiency Viruses among Bobcats and Pumas’, Journal of Virology, 81: 10961–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvin T. A. et al. (2013) ‘Identification of Recombination in the Envelope Gene of Simian Foamy Virus Serotype 2 Isolated from Macaca cyclopis’, Journal of Virology, 87: 8792–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehrt S. D., Riley S. P., Cypher B. L. (2010) Urban Carnivores: Ecology, Conflict, and Conservation. Baltimore, MD: JHU Press. [Google Scholar]
- Guindon S. et al. (2010) ‘New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0’, Systematic Biology, 59: 307–21. [DOI] [PubMed] [Google Scholar]
- Han G.-Z., Worobey M. (2012) ‘An Endogenous Foamy-Like Viral Element in the Coelacanth Genome’, PLoS Pathogens, 8: e1002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han G.-Z., Worobey M. (2012) ‘An Endogenous Foamy Virus in the Aye-Aye (Daubentonia madagascariensis)’, Journal of Virology, 86: 7696–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatama S. et al. (2001) ‘Reactivation of Feline Foamy Virus from a Chronically Infected Feline Renal Cell Line by Trichostatin A’, Virology, 283: 315–23. [DOI] [PubMed] [Google Scholar]
- Helps C., Harbour D. (1997) ‘Comparison of the Complete Sequence of Feline Spumavirus with Those of the Primate Spumaviruses Reveals a Shorter Gag Gene’, Journal of General Virology, 78: 2549–64. [DOI] [PubMed] [Google Scholar]
- Hill C. L., Bieniasz P. D., McClure M. O. (1999) ‘Properties of Human Foamy Virus Relevant to Its Development as a Vector for Gene Therapy’, Journal of General Virology, 80: 2003–9. [DOI] [PubMed] [Google Scholar]
- Homer C. et al. (2015) ‘Completion of the 2011 National Land Cover Database for the Conterminous United States–representing a Decade of Land Cover Change Information’, Photogrammetric Engineering & Remote Sensing, 81: 345–54. [Google Scholar]
- Jarosz A. F., Wiley J. (2014) ‘What Are the Odds? A Practical Guide to Computing and Reporting Bayes Factors’, The Journal of Problem Solving, 7: 2. [Google Scholar]
- Kechejian S. R. et al. (2019) ‘Feline Foamy Virus is Highly Prevalent in Free-Ranging Puma concolor from Colorado, Florida and Southern California’, Viruses, 11: 359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehl T. et al. (2013) ‘Complete Genome Sequences of Two Novel Puma concolor Foamy Viruses from California’, Genome Announcements, 1: e00201–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehl T., Tan J., Materniak M. (2013) ‘Non-Simian Foamy Viruses: Molecular Virology, Tropism and Prevalence and Zoonotic/Interspecies Transmission’, Viruses, 5: 2169–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellner A. et al. (2018) ‘Transmission Pathways and Spillover of an Erythrocytic Bacterial Pathogen from Domestic Cats to Wild Felids’, Ecology and Evolution, 8: 9779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A. S. et al. (2018) ‘Spumaretroviruses: Updated Taxonomy and Nomenclature’, Virology, 516: 158–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krakoff E. et al. (2019) ‘Variation in Intra-Individual Lentiviral Evolution Rates: A Systematic Review of Human, Non-Human Primate and Felid Species’, Journal of Virology, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert C. et al. (2018) ‘Potent Neutralizing Antibodies in Humans Infected with Zoonotic Simian Foamy Viruses Target Conserved Epitopes Located in the Dimorphic Domain of the Surface Envelope Protein’, PLoS Pathogens, 14: e1007293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. S. et al. (2014) ‘Evolution of Puma Lentivirus in Bobcats (Lynx rufus) and Mountain Lions (Puma concolor) in North America’, Journal of Virology, 00473–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. et al. (2017a) ‘Feline Immunodeficiency Virus Cross-Species Transmission: Implications for Emergence of New Lentiviral Infections’, Journal of Virology, 91: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. S. et al. (2017b) ‘Targeted Enrichment for Pathogen Detection and Characterization in Three Felid Species’, Journal of Clinical Microbiology, 55: 1658–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefort V., Longueville J.-E., Gascuel O. (2017) ‘SMS: Smart Model Selection in PhyML’, Molecular Biology and Evolution, 34: 2422–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I., Bork P. (2016) ‘Interactive Tree of Life (iTOL) v3: An Online Tool for the Display and Annotation of Phylogenetic and Other Trees’, Nucleic Acids Research, 44: W242–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski M. J. et al. (2008) ‘The Ascent of Cat Breeds: Genetic Evaluations of Breeds and Worldwide Random-Bred Populations’, Genomics, 91: 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W. et al. (2008) ‘Molecular Ecology and Natural History of Simian Foamy Virus Infection in Wild-Living Chimpanzees’, PLoS Pathogens, 4: e1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luther P., Nuttall P., Gibbons R. (1978) ‘Isolation of Viruses from Cultures of Bovine Endometrial Cells’, Journal of Infectious Diseases, 138: 660–3. [DOI] [PubMed] [Google Scholar]
- Martin D. P. et al. (2015) ‘RDP4: Detection and Analysis of Recombination Patterns in Virus Genomes’, Virus Evolution, 1: vev003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda R., Yoshida M. C. (1995) ‘Two Japanese Wildcats, the Tsushima Cat and the Iriomote Cat, Show the Same Mitochondrial DNA Lineage as the Leopard Cat (Felis bengalensis) ’, Zoological Science, 12: 655–9. [DOI] [PubMed] [Google Scholar]
- Materniak M., Bicka L., Kuźmak J. (2006) ‘Isolation and Partial Characterization of Bovine Foamy Virus from Polish Cattle’, Polish Journal of Veterinary Sciences, 9: 207–11. [PubMed] [Google Scholar]
- Membrebe J. V. et al. (2019) ‘Bayesian Inference of Evolutionary Histories under Time-Dependent Substitution Rates’, Molecular Biology and Evolution, 36: 1793., [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minin V. N., Bloomquist E. W., Suchard M. A. (2008) ‘Smooth Skyride through a Rough Skyline: Bayesian Coalescent-Based Inference of Population Dynamics’, Molecular Biology and Evolution, 25: 1459–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki M., Akuzawa M., Nagatomo H. (1990) ‘Serological Survey of the Iriomote Cat (Felis iriomotensis) in Japan’, Journal of Wildlife Diseases, 26: 236–45. [DOI] [PubMed] [Google Scholar]
- Moss W. E. et al. (2016) ‘Human Expansion Precipitates Niche Expansion for an Opportunistic Apex Predator (Puma concolor)’, Scientific Reports, 6: 39639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouinga-Ondémé A. et al. (2012) ‘Cross-Species Transmission of Simian Foamy Virus to Humans in Rural Gabon, Central Africa’, Journal of Virology, 86: 1255–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moya A., Holmes E. C., González-Candelas F. (2004) ‘The Population Genetics and Evolutionary Epidemiology of RNA Viruses’, Nature Reviews Microbiology, 2: 279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray S. M., Linial M. (2006) ‘Foamy Virus Infection in Primates’, Journal of Medical Primatology, 35: 225–35. [DOI] [PubMed] [Google Scholar]
- Nakamura K. et al. (2000) ‘Contrastive Prevalence of Feline Retrovirus Infections between Northern and Southern Vietnam’, Journal of Veterinary Medical Science, 62: 921–3. [DOI] [PubMed] [Google Scholar]
- Ottoni C. et al. (2017) ‘The Palaeogenetics of Cat Dispersal in the Ancient World’, Nature Ecology & Evolution, 1: 0139. [Google Scholar]
- Parrish C. R., Kawaoka Y. (2005) ‘The Origins of New Pandemic Viruses: The Acquisition of New Host Ranges by Canine Parvovirus and Influenza a Viruses’, Annual Review of Microbiology, 59: 553. [DOI] [PubMed] [Google Scholar]
- Peck K. M., Lauring A. S. (2018) ‘The Complexities of Viral Mutation Rates’, Journal of Virology, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phung H. T. et al. (2001) ‘Genetic Analyses of Feline Foamy Virus Isolates from Domestic and Wild Feline Species in Geographically Distinct Areas’, Virus Research, 76: 171–81. [DOI] [PubMed] [Google Scholar]
- Posada D. (2009) ‘Selection of Models of DNA Evolution with jModelTest’ in Posada (ed.) Bioinformatics for DNA sequence analysis Methods in Molecular Biology. New York: Humana Press. [DOI] [PubMed] [Google Scholar]
- Powers J. A. et al. (2018) ‘Feline Leukemia Virus Disease Outcomes in a Domestic Cat Breeding Colony: Relationship to Endogenous FeLV and Other Chronic Viral Infections’, Journal of Virology, 00649–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A. (2012) FigTree v1. 4. Institute of Evolutionary Biology, University of Edinburgh <http://tree.bio.ed.ac.uk/software/figtree/> accessed 23 December 2019.
- Rambaut A. et al. (2016) ‘Exploring the Temporal Structure of Heterochronous Sequences Using TempEst (Formerly Path-O-Gen)’, Virus Evolution, 2: vew007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rethwilm A., Bodem J. (2013) ‘Evolution of Foamy Viruses: The Most Ancient of All Retroviruses’, Viruses, 5: 2349–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard L. et al. (2015) ‘Cocirculation of Two Env Molecular Variants, of Possible Recombinant Origin, in Gorilla and Chimpanzee Simian Foamy Virus Strains from Central Africa’, Journal of Virology, 89: 12480–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross A. (2016) Celebrity Cats of Colorado History. Denver News <https://history.denverlibrary.org/news/cats-co-not-real-title> accessed 23 December 2019.
- Soliven K. et al. (2013) ‘Simian Foamy Virus Infection of Rhesus Macaques in Bangladesh: Relationship of Latent Proviruses and Transcriptionally Active Viruses’, Journal of Virology, 01989–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stöver B. C., Müller K. F. (2010) ‘TreeGraph 2: Combining and Visualizing Evidence from Different Phylogenetic Analyses’, BMC Bioinformatics, 11: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Switzer W. M. et al. (2004) ‘Frequent Simian Foamy Virus Infection in Persons Occupationally Exposed to Nonhuman Primates’, Journal of Virology, 78: 2780–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Switzer W. M. et al. (2008) ‘Coinfection with HIV-1 and Simian Foamy Virus in West Central Africans’, The Journal of Infectious Diseases, 197: 1389–93. [DOI] [PubMed] [Google Scholar]
- Switzer W. M. et al. (2005) ‘Ancient co-Speciation of Simian Foamy Viruses and Primates’, Nature, 434: 376. [DOI] [PubMed] [Google Scholar]
- Troyer R. M. et al. (2014) ‘Novel Gammaherpesviruses in North American Domestic Cats, Bobcats and Pumas: Identification, Prevalence and Risk Factors’, Journal of Virology, 03405–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X. et al. (2019) ‘A Reptilian Endogenous Foamy Virus Sheds Light on the Early Evolution of Retroviruses’, Virus Evolution, 5: vez001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiethoelter A. K. et al. (2015) ‘Global Trends in Infectious Diseases at the Wildlife–Livestock Interface’, Proceedings of the National Academy of Sciences of the United States of America, 112: 9662–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler I. G. et al. (1998) ‘Detection and Molecular Characterisation of Feline Foamy Virus Serotypes in Naturally Infected Cats’, Virology, 247: 144–51. [DOI] [PubMed] [Google Scholar]
- Winkler I. G., Löchelt M., Flower R. L. P. (1999) ‘Epidemiology of Feline Foamy Virus and Feline Immunodeficiency Virus Infections in Domestic and Feral Cats: A Seroepidemiological Study’, Journal of Clinical Microbiology, 37: 2848–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X. et al. (2018) ‘Endogenous Retroviruses of Non-Avian/Mammalian Vertebrates Illuminate Diversity and Deep History of Retroviruses’, PLoS Pathogens, 14: e1007072. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.