Abstract
Recognition of histone-modified nucleosomes by specific reader domains underlies the regulation of chromatin-associated processes. Whereas structural studies revealed how reader domains bind modified histone peptides, it is unclear how reader domains interact with modified nucleosomes. Here we report the cryo-electron microscopy (cryo-EM) structure of the PWWP reader domain of human transcriptional coactivator LEDGF in complex with a H3K36-methylated nucleosome at 3.2 Å resolution. The structure reveals multivalent binding of the reader domain to the methylated histone tail and to both gyres of nucleosomal DNA, explaining the known cooperative interactions. The observed cross-gyre binding may contribute to nucleosome integrity during transcription. The structure also explains how human PWWP domain-containing proteins are recruited to H3K36-methylated regions of the genome for transcription, histone acetylation and methylation, and for DNA methylation and repair.
Introduction
Covalent modifications of nucleosomes regulate chromatin-based processes such as DNA transcription, replication, and repair. Many modifications occur on the accessible histone tails that protrude from the nucleosome core particle. These modifications include acetylation and methylation of lysine residues and are recognized by ‘reader’ domains that recruit various proteins. The molecular basis for how reader domains recognize histone modifications has been provided by structural studies of reader domain-histone peptide complexes1.
Some reader domains not only bind modified histone tails, but can additionally bind to DNA. Out of over 20 types of reader domains, at least four (PWWP, Tudor, Chromo and Bromo) are known to bind both, the modified histone and DNA2,3. These reader domains are predicted to recognize the histone modification in the context of nucleosomal DNA, with both types of interactions contributing to the affinity of the domain for modified nucleosomes. However, such multivalent binding of a reader domain was thus far not observed directly.
A widespread type of reader domain that can engages in multivalent interactions is the PWWP (Pro-Trp-Trp-Pro motif) domain. This domain was identified in the protein product of the Wolf-Hirschhorn syndrome candidate gene 1 (WHSC1) and in proteins related to hepatoma-derived growth factor4,5. It was later found in the DNA methyltransferase DNMT3B and the DNA repair protein MSH66,7. The PWWP domain is a reader of methylated histone tails and comprises a five-stranded N-terminal β-barrel and two C-terminal α-helices8. The β-barrel harbors a conserved aromatic cage that binds a methyl-lysine residue9, as observed also for other reader domains of the ‘Royal’ superfamily such as Tudor, Chromo, Agenet and MBT domains10.
Most PWWP domains bind the histone H3 N-terminal tail that is di- or tri-methylated at lysine 36 (H3K36me2 or H3K36me3)11–16. The H3K36me3 modification occurs in gene bodies of transcribed chromatin in eukaryotic species from yeast to human17. This modification is mainly generated by SETD2 (KMT3A), a lysine methyltransferase associated with elongating RNA polymerase II (Pol II)18,19. H3K36me3 has functions in transcription elongation, alternative splicing, DNA methylation, DNA damage signaling, and repression of cryptic transcription and histone exchange20.
The PWWP domain binds methylated H3K36 histone tail peptides with much lower affinity11,15 than other methyl-lysine reader domains such as PHD fingers21. To achieve high-affinity binding, PWWP domains rely on additional interactions with DNA. Indeed, the PWWP domain was first described as a DNA-binding fold7,22, and the PWWP domain in Pdp1 was found to bind both a methylated histone peptide and DNA23. Nucleosomal DNA was shown to contribute strongly to high-affinity binding of a PWWP domain to a H3K36-methylated nucleosome24,25, and it was predicted that the domain may bind both DNA gyres25. Very recently, structures were reported of the PWWP domain of HDGF in complex with a 10-bp DNA fragment (PDB 5XSK) and of the PWWP domain of HRP3 (also called HDGFL3) with a H3K36me3 peptide and DNA26. However, there is no structure of a PWWP domain bound to a nucleosome.
To investigate how a reader domain binds a modified nucleosome, we solved the cryo-EM structure of the PWWP domain of LEDGF in complex with a H3K36me3 analog-containing nucleosome. The association of LEDGF with both H3K36me3 and DNA has been well studied14,24,25,27,28. LEDGF is also known as PSIP1, p75 or p52 and functions as a transcription coactivator29 and as an interactor of HIV-1 integrase28. LEDGF helps RNA polymerase (Pol) II to overcome the nucleosome barrier during transcription30. Our structure explains cooperative interactions of the PWWP domain with the histone tail and DNA, and recruitment of many diverse human PWWP domain-containing factors to H3K36-methylated chromatin regions.
Results
Linker DNA stabilizes LEDGF binding to a modified nucleosome
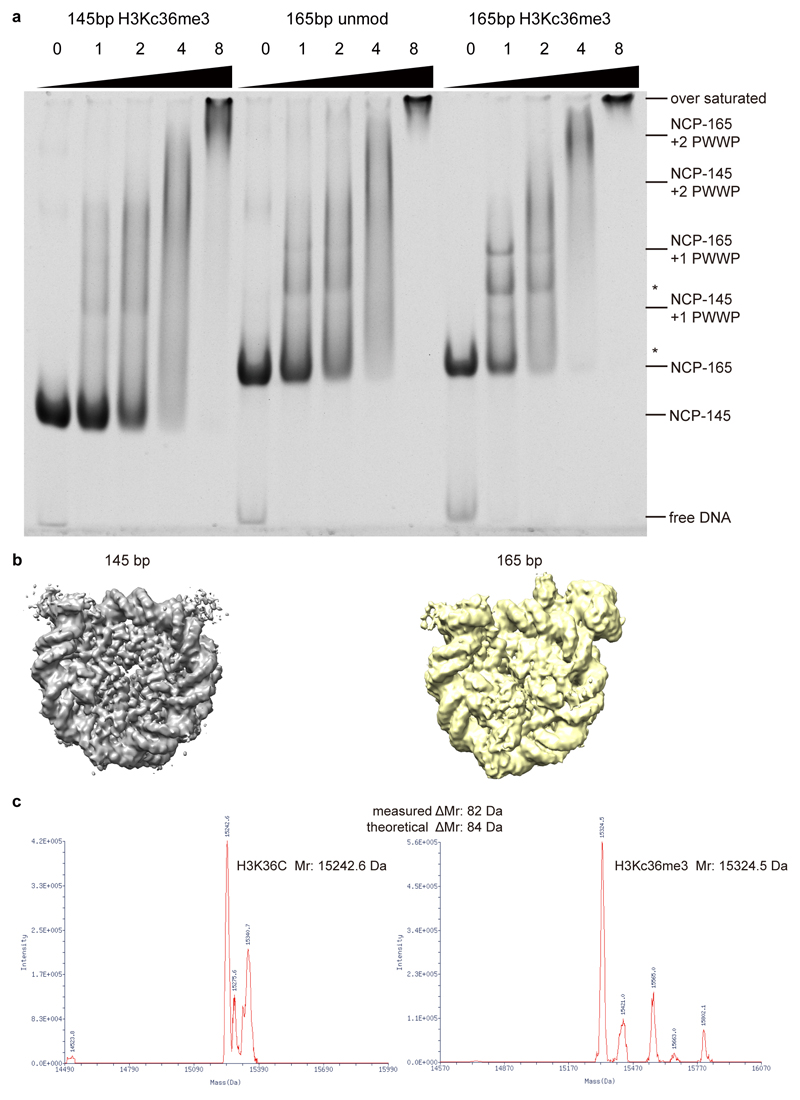
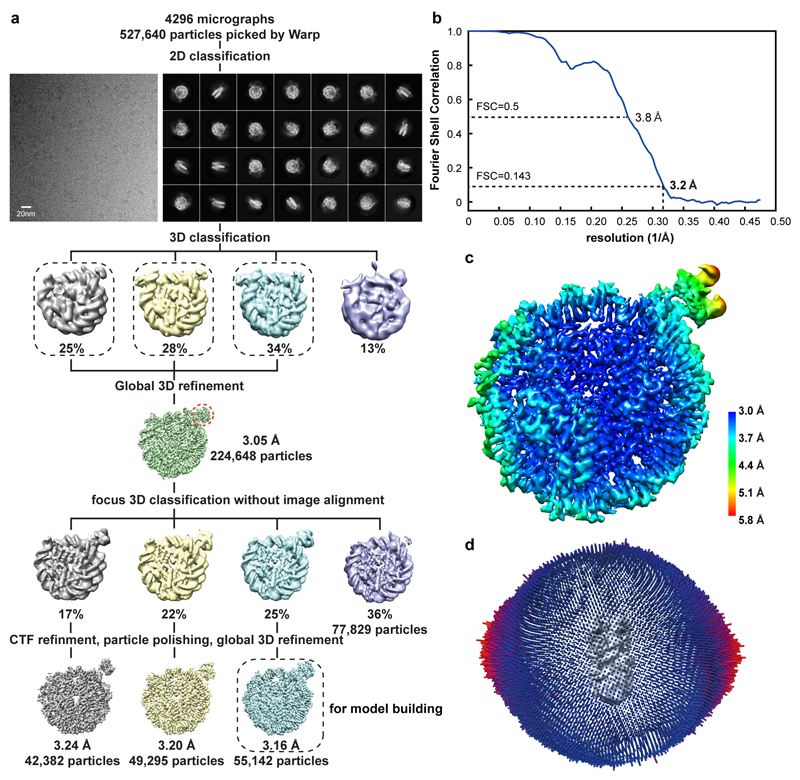
We assembled recombinant nucleosomes with H3K36me3 mimicked by a methyl-lysine analog31. The lysine is mutated to cysteine and alkylated to form a thioether mimicking methylated lysine (H3KC36me3). Purified recombinant full-length LEDGF bound to the H3KC36me3-modified nucleosome, as seen in an electrophoretic mobility shift assay (EMSA) (Extended Data Fig. 1a). The complex was assembled by mixing LEDGF and a H3KC36me3-modified nucleosome with 145 base pairs (bp) DNA with the canonical Widom 601 sequence at a molar ratio of 2:1. The complex was then isolated by size-exclusion chromatography, cross-linked with glutaraldehyde, and used for the preparation of cryo-EM grids. Cryo-EM data were collected on a Titan Krios microscope (FEI) with a K2 detector (Gatan) (Methods).
Analysis of the cryo-EM data revealed weak density for LEDGF (Extended Data Fig. 1b left). To stabilize LEDGF on the nucleosome, we prepared H3KC36me3-modified nucleosomes with an additional 20 bp of extranucleosomal linker DNA at the exit site. Indeed, LEDGF bound more tightly to the extended nucleosome containing 165 bp of DNA (Extended Data Fig. 1a). Cryo-EM analysis revealed a defined additional density for the PWWP domain of LEDGF (Extended Data Fig. 1b right). We therefore subjected the modified nucleosome-LEDGF complex with the 165 bp DNA to structure determination.
Cryo-EM structure of nucleosome-PWWP complex
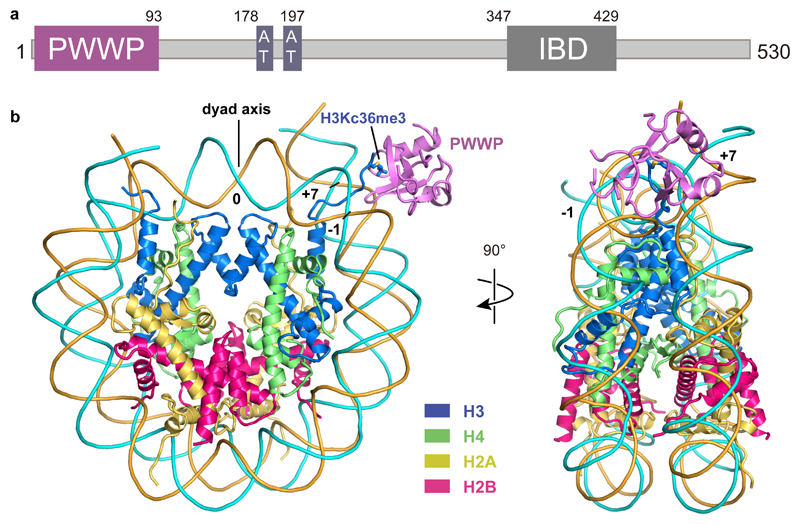
The cryo-EM structure was determined from 55,142 particles at a resolution of 3.2 Å (Methods, Extended Data Fig. 2). Whereas the nucleosome and the PWWP domain of LEDGF were well defined in the density, the two AT hook regions and a HIV integrase binding domain (IBD) of LEDGF27,32 were mobile (Fig. 1a, b). This is consistent with results that the PWWP domain mediates chromatin tethering of LEDGF33. The nucleosome core was resolved at 3.0 Å resolution, whereas the PWWP domain was resolved at local resolutions of 3.7-5.8 Å (Extended Data Fig. 2c).
Figure 1. Structure of H3KC36me3-modified nucleosome bound to the PWWP domain of LEDGF.
(a) Domain architecture of LEDGF. Only the PWWP domain is visible in the structure. (b) Overview of the structure in front view (left) and side view (right). H2A, H2B, H3, H4, forward strand, reverse strand and LEDGF are colored in yellow, red, blue, green, cyan, orange and purple respectively. The color code is used throughout. H3KC36me3 residue is depicted as a stick model. SHLs 0, -1 and +7 are indicated with numbers.
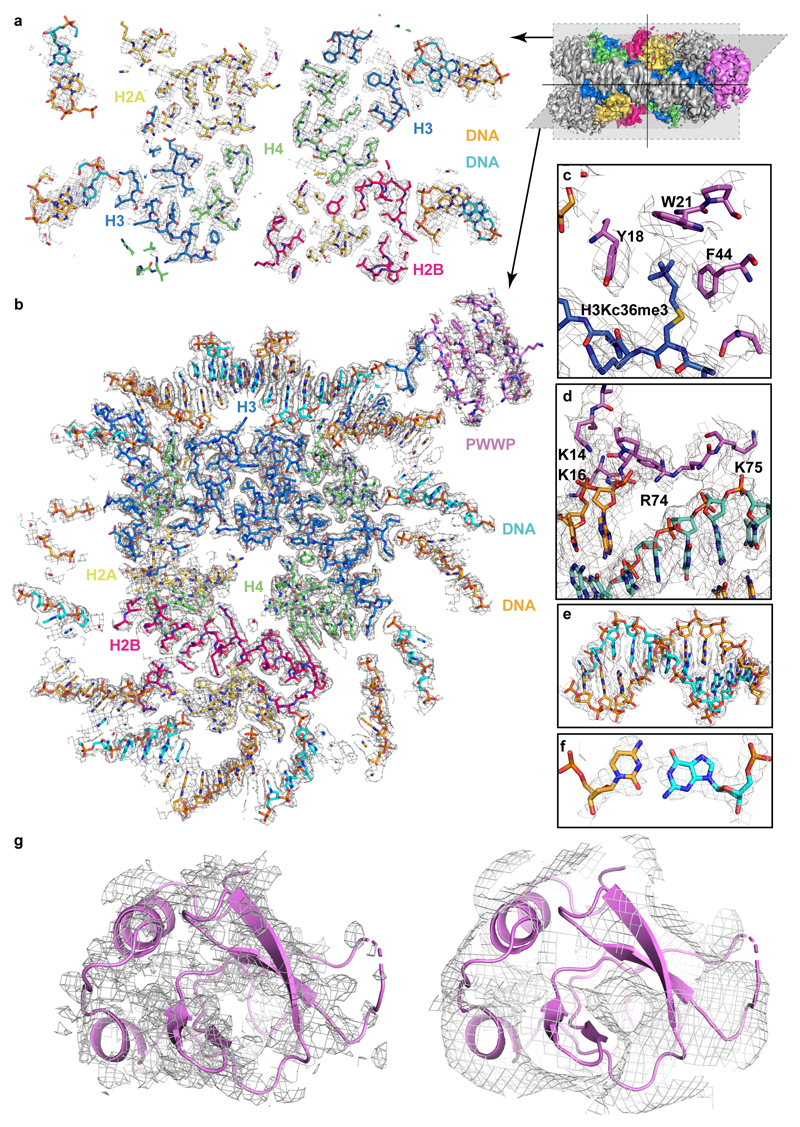
Density for purine and pyrimidine bases could be distinguished around the dyad from SHL -4 to SHL +4 and enabled tracing of the nucleosomal DNA sequence (Extended Data Fig. 3e, f). Of the 20 bp of extranucleosomal DNA, the proximal 5 bp could be traced in the final model. We also built the H3 tail from the octamer core to residue V35 (Extended Data Fig. 3). The side chain of residue H3KC36me3 was clearly visible (Extended Data Fig. 3c). Fragmented density for a second PWWP domain was observed on the other side of the nucleosome around the second methylated H3 tail, indicating that two copies of LEDGF could bind simultaneously to a methylated nucleosome (Extended Data Fig. 1b). However, the second protein copy could not be included in the model due to its poor density. The structure was subjected to real-space refinement and showed very good stereochemistry (Methods, Table 1).
Table 1. Cryo-EM data collection, refinement and validation statistics.
| 165bp H3KC36me3 nucleosome-LEDGF (EMD-10069, PDB 6S01) | |
|---|---|
| Data collection and processing | |
| Magnification | 130,000 |
| Voltage (kV) | 300 |
| Electron exposure (e–/Å2) | 43.2 |
| Defocus range (μm) | –1.25 to –2.75 |
| Pixel size (Å) | 1.05 |
| Symmetry imposed | C1 |
| Initial particle images (no.) | 527,640 |
| Final particle images (no.) | 55,142 |
| Map resolution (Å) | 3.2 |
| FSC threshold | 0.143 |
| Map resolution range2 (Å) | 3.0-5.8 |
| Refinement | |
| Initial models used (PDB code) | 3MVD, 4FU6 |
| Model resolution (Å) | 3.1 |
| FSC threshold | 0.143 |
| Model resolution range (Å) | -- |
| Map sharpening B factor (Å2) | -124 |
| Model composition | |
| Non-hydrogen atoms | 12959 |
| Protein residues | 853 |
| Nucleic acid residues | 300 |
| B factor (Å2) | |
| Protein | 88.0 |
| Nucleic acid | 142.7 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.01 |
| Bond angles (°) | 0.73 |
| Validation | |
| MolProbity score | 1.44 |
| Clashscore | 3.93 |
| Poor rotamers (%) | 0.42 |
| Ramachandran plot | |
| Favored (%) | 96.13 |
| Allowed (%) | 3.87 |
| Disallowed (%) | 0 |
Multivalent nucleosome binding by the PWWP domain
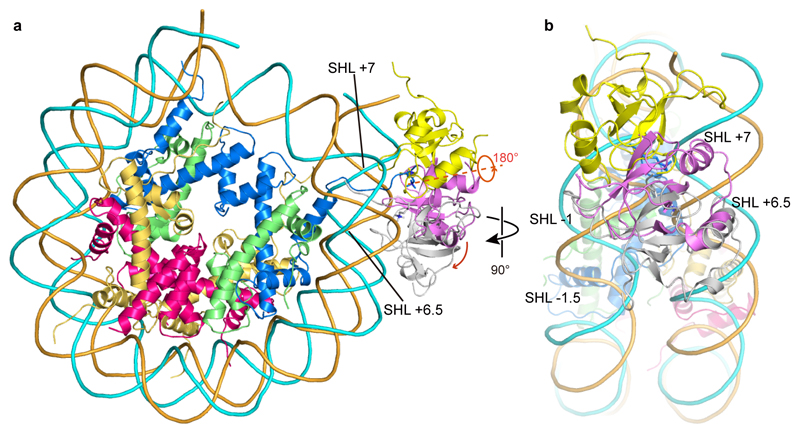
The structure shows that the PWWP domain binds the methylated H3 tail and both DNA gyres of the nucleosome (Fig. 1b). The PWWP domain contacts DNA at super-helical locations (SHLs) +7 and -1, where the H3 tail protrudes from the nucleosome core between the two DNA gyres (Fig. 1b). The same face of the PWWP domain interacts with the H3 tail and both DNA gyres (Extended Data Fig. 4a). The multivalent binding mode observed here has not been seen in previous nucleosome-protein complexes and explains why the binding affinity of the PWWP domain to a H3K36me3-modified nucleosome is ~10,000-fold higher than to a free H3K36me3-modified peptide24,25. The structures remain almost unaltered upon binding, with root-mean-square deviations (RMSDs) of 1.1 Å and 1.3 Å compared to the free nucleosome34 and PWWP structures (PDB 4FU6), respectively.
Interaction with methylated histone tail
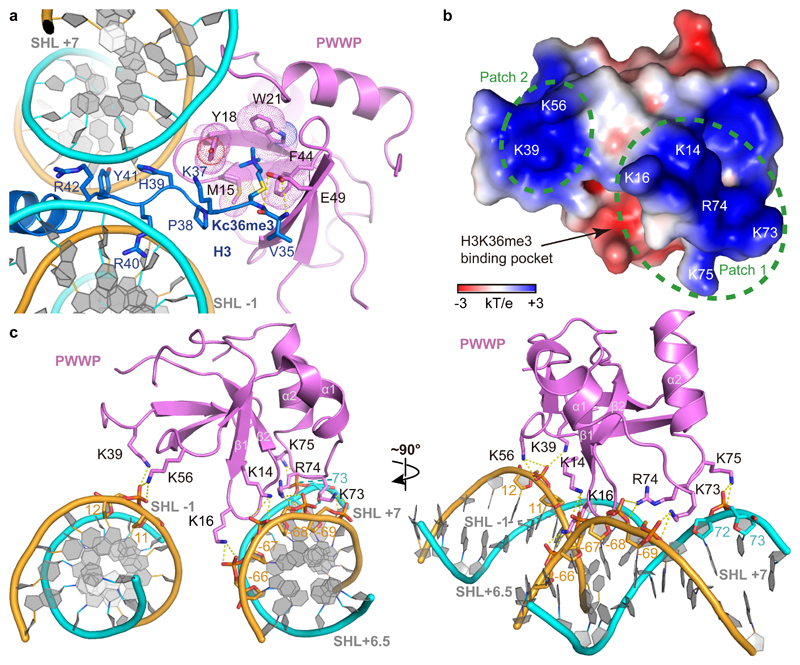
The aromatic cage of the PWWP domain binds the H3KC36me3 side chain via cation-π interaction as previously observed in H3K36me3 peptide-bound structures of the PWWP domain of BRPF1, ZMYND11 and DNMT3B11,35,36 (Fig. 2a). The aromatic cage is formed by residues M15, Y18, W21 and F44. The histone tail interacts with the PWWP domain not only via its KC36me3 side chain, but also via the main chain of residues V35 and KC36me3 with the side chain of residue E49 in the PWWP domain (Fig. 2a). Overall, these interactions are highly similar to those observed in other isolated histone H3 tail peptide-PWWP domain structures.
Figure 2. Nucleosome-PWWP interactions.
(a) Details of the interactions between PWWP and the methylated H3 tail. PWWP residues involved in H3KC36me3 recognition and H3 tail residues are shown as sticks. Selected hydrogen bonds are shown as yellow dashed lines. (b) Electrostatic surface of the PWWP domain calculated using the APBS tool in a range of -3 kT/e to +3 kT/e. Two positively charged surface patches involved in DNA interaction are indicated with green dashed circles; residues inside are denoted in white. (c) Details of DNA interactions. Electrostatic interactions and hydrogen bonds are shown as yellow dashes. SHLs are denoted. H3 tail is not shown for clarity.
Interactions with both DNA gyres
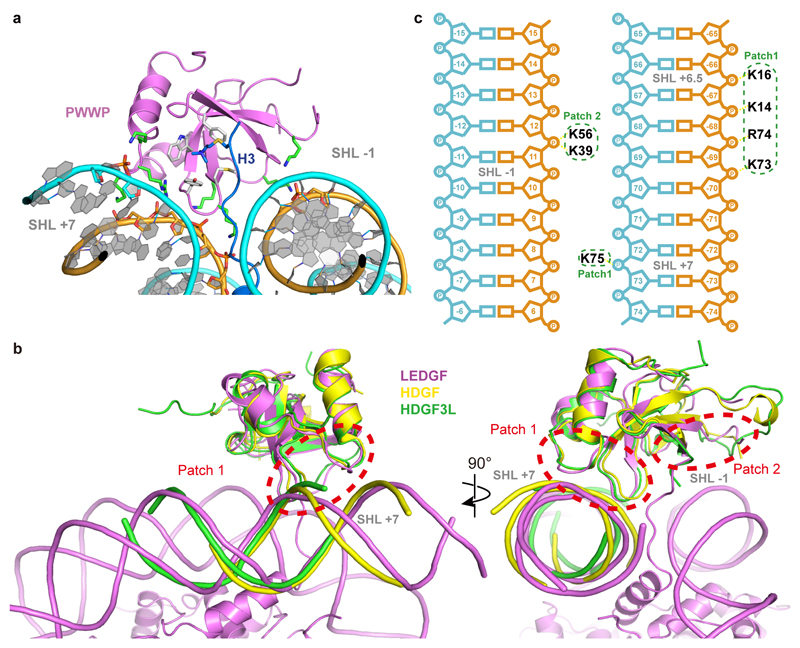
Interactions of the PWWP domain are exclusively with the phosphodiester backbone in the minor grooves of DNA, and no interactions with DNA bases are observed. This indicates that the binding between the PWWP domain and nucleosomal DNA is not sequence specific. Two positively charged patches that flank the aromatic cage of the PWWP domain contact negatively charged backbones of both DNA gyres (Fig. 2b). Whereas one patch (patch 1) binds the DNA gyre at SHL +7, the other patch (patch 2) binds the second DNA gyre at SHL -1. Whereas the former contact was recently observed in crystal structures of PWWP domains with a short DNA fragments26 (Extended Data Fig. 4b), it was not known that it reflects binding to SHL +7.
In more detail, the PWWP domain patch 1 is formed by residues from loops β1-β2 and α1-α2. Side chains of K14 and K16 from the β1-β2 loop and of K73 and R74 from loop α1-α2 form a network of electrostatic interactions and hydrogen bonds with the phosphates of nucleotides -66 to -69 on the reverse strand of DNA at SHL +7 (Fig. 2c, Extended Data Fig. 4c). In addition, the side chain of K75 in loop α1-α2 forms two hydrogen bonds with the phosphate groups of nucleotides 72 and 73 on the forward chain at SHL +7. This interaction contributes to stabilizing the LEDGF-nucleosome interaction on the exit site where linker DNA was added. The positively charged patch 2 of the PWWP domain contains residues K39 and K56 and interacts with the phosphate backbone groups of nucleotides 11 and 12 at SHL -1 (Fig. 2c, Extended Data Fig. 4c).
Conserved mode of PWWP-nucleosome interaction
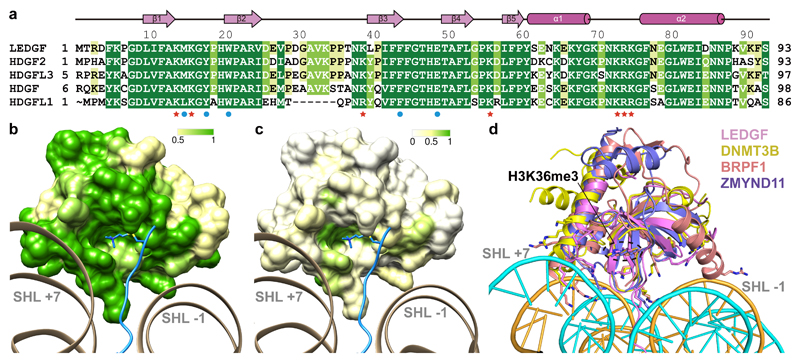
Most proteins that recognize H3K36-methylated nucleosomes employ a PWWP domain. The human genome codes for more than 20 proteins that contain a PWWP domain. PWWP domains can be categorized into six subfamilies based on variable insertion motifs8,15,37 (Supplementary Note). LEDGF belongs to the subfamily of HDGF-related proteins (HRP), and LEDGF residues involved in methylated nucleosome recognition are highly conserved in this subfamily (Fig. 3a). This predicts that our nucleosome-PWWP structure is a good model for HRP subfamily members that bind with their PWWP domains to methylated nucleosomes (Fig. 3b). Although these PWWP domains differ in the presence of specific insertions, they all share the two DNA-interacting positively charged surface patches that face the DNA gyres (Supplementary Note and Fig. 3c), suggesting that all PWWP domains interact with nucleosomes in a similar way. This is supported by a superposition of the three known structures of PWWP domains bound to an H3K36me3-containing histone H3 peptide (BRPF1, ZMYND11, and DNMT3B)11,35,36 onto our structure (Fig. 3d).
Figure 3. Conserved mode of nucleosome-PWWP interaction.
(a) Sequence alignment of PWWP domains within the HDGF-related protein (HRP) subfamily. Residues involved in H3K36me3 recognition are marked with blue circles; residues involved in DNA interaction are marked with red stars. Other conserved residues are highlighted from green to yellow with decreasing conservation. Secondary structure elements of the PWWP domain are shown above the sequences. (b) Conservation of PWWP domain surface within the HRP subfamily. Surface of the PWWP domain colored according to sequence conservation from green (identical) via yellow (conserved) to white (non-conserved). The H3 N-terminal tail is shown in blue. (c) Conservation of PWWP domain surface over all protein families. Color code as for (b). Note that both DNA patches and the methyllysine-binding region are at least partially conserved. (d) Superposition of known H3K36me3 peptide bound PWWP domain structures onto the nucleosome-PWWP structure presented here. PWWP domains of LEDGF, DNMT3B, BRPF1 and ZMYND11 are shown in cartoon and colored as indicated.
Discussion
Here we report the cryo-EM structure of a PWWP reader domain bound to a nucleosome with a trimethylation mark at residue K36 of histone H3. The structure reveals that the PWWP domain uses a single composite binding face to contact the methylated H3 tail and both DNA gyres flanking the tail. This binding mode is distinct from classical nucleosome interactions, where factors generally bind to the H2A-H2B acidic patch34,38. The observed binding mode is also distinct from recent structural studies of nucleosomes in complex with a deubiquitinase module or histone methyltransferases, which all bind to the ubiquitinated histone octamer face39,40, and not to the edge of the nucleosome, as observed for the PWWP domain here.
Our structural observations are highly consistent with previous mutagenesis results that identified key residues involved in nucleosome-PWWP interaction. In particular, van Nuland et al mutated the positively charged residues in the PWWP domain of LEDGF and identified key residues involved in DNA interactions25. Mutation of each of the positively charged residues located in both DNA-interacting patches (K14A, K16A, K73A, R74A and K75A in patch 1, and K39A and K56A in patch 2) observed in our structure had been previously shown to lead to reduced binding affinity to nucleosome25. Substitution of arginine R74 in the LEDGF PWWP domain abolished its interaction with the nucleosome in vitro25, and was shown to dramatically reduce chromatin tethering and HIV-1 infectivity in vivo33,41. These published functional data validate our structural results.
Comparison of our structure with available structures of PWWP domains with isolated H3 peptides or DNA fragments or both revealed that binding of the methylated H3 tail and of one of the two DNA gyres is similar (Extended Data Fig. 4b). This shows that structures of minimal complexes provide binding sites that are relevant for understanding domain interactions with the nucleosome. However, from these structures, the detailed contacts and the exact position of the PWWP domain on the methylated nucleosome could not be predicted. Several models were proposed for how PWWP domains bind to a methylated nucleosome24–26,36, but these deviate from our nucleosome-PWWP complex structure (Extended Data Fig. 5). Although the models placed the PWWP domain near the H3 tail as observed, and predicted cross-gyre binding25,26, they suggested distinct DNA positions and different domain orientations, apparently because DNA fragments in available structures could not be assigned to one of the two DNA gyres and the SHLs of the nucleosome.
The structure is also a good model for understanding the binding of other PWWP domain proteins to H3K36me3-modified nucleosomes. For example, the DNA methyltransferase DNMT3B is recruited to transcribed genes, which leads to their preferential methylation42. DNMT3B recruitment to transcribed genes requires SETD2-mediated methylation of H3K36 and a functional PWWP domain in DNMT3B42. Our structure thus suggests a general mechanism for recruitment of many other PWWP domain-containing proteins to H3K36-methylated chromatin regions, including proteins involved in transcription elongation35, histone acetylation11 and methylation16, and DNA methylation42 and repair43.
A very interesting finding from our work is that the PWWP reader domain can bind across both DNA gyres of the nucleosome. Such cross-gyre binding of nucleosome-interacting proteins was proposed already 25 years ago44, but only observed very recently, when a study revealed that transcription factors of the T-box family use such cross-gyre binding45. Recent structures of chromatin remodeling enzymes and a retroviral intasome bound to nucleosomes also showed cross-gyre binding through multiple domains46–50, but with different geometries and at distinct SHL positions.
Our structure also suggests a mechanism that could contribute to preventing nucleosome loss during gene transcription. The SETD2 methyltransferase associates with transcribing Pol II and introduces the H3K36me3 modification co-transcriptionally18,19. It was shown in yeast that this results in decreased nucleosome turnover and maintains intact chromatin within actively transcribed regions51,52. Stabilization of nucleosomes after Pol II passage can be achieved indirectly, by recruitment of a histone deacetylase that triggers chromatin closure and a chromatin remodeling complex that prevents histone exchange during transcription in yeast53,54. Our structure suggest that nucleosome stabilization could also be achieved directly, by cross-gyre binding of proteins with PWWP domains to H3K36me3-modified nucleosomes. Thus, binding of PWWP domains to H3K36-methylated nucleosomes that occurs in the wake of transcribing Pol II could help to prevent histone loss and spurious transcription initiation from inside transcribed genes.
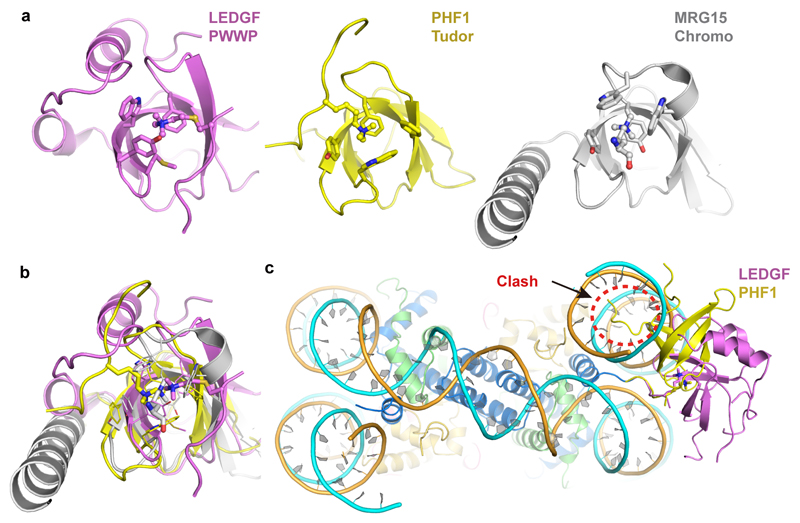
Our structure also has implications for understanding nucleosome interactions of other proteins of the ‘royal’ family that bind methylated H3K36 and DNA, such as the Tudor domain of Polycomb-like (PCL) proteins PHF1 and PHF19 and the Chromo domain of MRG1555–58. These domains share a β-barrel with the PWWP domain but differ in subsidiary motifs (Extended Data Fig. 6a). However, structural superposition shows that the H3 tail is bound differently to the domain surface and the H3K36me3 moiety inserts into the aromatic cage differently (Extended Data Fig. 6b). Assuming that the position of the H3 tail protruding between the two DNA gyres is restricted, the Tudor domain of PHF1 would clash with one end of nucleosomal DNA and would not be able to bind to the nucleosome as observed here (Extended Data Fig. 6c). This predicts that Tudor domain binding to the H3K36-methylated nucleosome destabilizes the nucleosome and leads to unwrapping of terminal DNA. This could account for the known increase in DNA accessibility upon binding of PHF1 to the nucleosome59.
There are several other reader domains that were reported to bind both a modified histone tail and DNA simultaneously. In particular, the PWWP domain of Pdp1 and the Chromo domain of MSL3 are thought to interact with nucleosomes methylated at H4K2023,60, and the Chromo domain of CBX2, CBX8 and their homologs with nucleosomes containing H3K27me32,61. Combined histone tail and DNA interactions were also proposed for a double Bromodomain in TAF162, and recently shown with binding assays for the Bromodomains of BRDT and BRG163,64. However, the structural basis for how these reader domains recognize various histone modifications in the context of the nucleosome cannot be predicted and awaits future studies.
Methods
Protein expression and purification
Full-length LEDGF was cloned in a modified pFastBac vector containing an N-terminal His6-MBP tag followed by a TEV cleavage site [a gift of Scott Gradia, UC Berkeley, vector 438-C (Addgene: 55220)] via ligation independent cloning. Protein expression in insect cells was performed as described47. Briefly, the recombinant vector was transformed into DH10EMBacY cells (Geneva Biotech, Geneva, Switzerland) by electroporation to generate bacmid. After virus amplification, 300 μL of V1 virus were added to 600 mL of Hi5 cells grown in ESF-921 media (Expression Systems, Davis, CA, United States). Cells were grown for 48-72 h at 27 °C, harvested by centrifugation (238 xg, 4°C, 30 min), and resuspended in lysis buffer (20 mM HEPES-Na pH 7.5, 300 mM NaCl, 10% glycerol (v/v), 1 mM DTT, 30 mM imidazole pH 8.0, 0.284 μg/ml leupeptin, 1.37 μg/ml pepstatin A, 0.17 mg/ml PMSF, 0.33 mg/ml benzamidine). The cell resuspension was frozen in liquid nitrogen and stored at -80 °C.
Frozen cell pellets were thawed and lysed by sonication. Lysates were cleared by centrifugation (18,000 g, 4 °C, 30 min and 235,000 g, 4°C, 60 min). The supernatant containing LEDGF was filtered using 0.8 μm syringe filters (Millipore) and applied onto a GE HisTrap HP 5 mL (GE Healthcare, Little Chalfont, United Kingdom), pre-equilibrated in lysis buffer. After sample application, the column was washed with 10 CV lysis buffer, 5 CV high salt buffer (20 mM HEPES-Na pH 7.5, 1 M NaCl, 10% glycerol (v/v), 1 mM DTT, 30 mM imidazole pH 8.0, 0.284 μg/ml leupeptin, 1.37 μg/ml pepstatin A, 0.17 mg/ml PMSF, 0.33 mg/ml benzamidine), and 5 CV lysis buffer. The protein was eluted with a gradient of 0-100% elution buffer (20 mM HEPES-Na pH 7.5, 300 mM NaCl, 10% glycerol (v/v), 1 mM DTT, 500 mM imidazole pH 8.0). Peak fractions were pooled and dialyzed for 16 hours against 600 mL dialysis buffer (20 mM HEPES-Na pH 7.5, 300 mM NaCl, 10% glycerol (v/v), 1 mM DTT, 30 mM imidazole) in the presence of 2 mg His6-TEV protease. The dialyzed sample was applied to a GE HisTrap HP 5 mL. The flow-through containing LEDGF was concentrated using an Amicon Millipore 15 mL 10,000 MWCO centrifugal concentrator and applied to a GE Superdex 200 10/300 size exclusion column, pre-equilibrated in gel filtration buffer (20 mM HEPES-Na pH 7.5, 300 mM NaCl, 10% glycerol (v/v), 1 mM DTT). Peak fractions were concentrated to ~100 μM, aliquoted, flash frozen, and stored at -80 °C.
Preparation of unmodified and H3KC36me3-modified nucleosomes
Xenopus laevis histones were expressed, purified, and assembled into nucleosomes with Widom 601 sequence as described65. To generate the H3KC36me3-modified histone, a single lysine-to-cysteine mutation (K36C) was introduced into the H3 sequence by site-directed mutagenesis. Cysteine-engineered histone H3K36C protein was alkylated as described31. Briefly, purified K36C H3 protein was reduced with DTT before addition of a 50-fold molar excess of trimethylammonium bromide (Sigma 117196–25G). The reaction mixture was incubated for 4 h at 50 °C before quenching with 5 mM β-mercaptoethanol. The modified protein was separated and desalted using a PD-10 desalting column (GE Healthcare) pre-equilibrated in water supplemented with 2 mM β-mercaptoethanol and lyophilized. The incorporation of alkylation agents was confirmed by MALDI-TOF mass spectrometry (Extended Data Fig. 1c).
Widom 601 145 bp DNA was purified as described from the pUC19 8 × 145 bp 601-sequence plasmid using EcoRV restriction enzyme to digest the DNA into fragments65. Widom 601 165 bp DNA was generated by PCR using purified 145bp Widom 601 DNA as a template and two primers (forward: ATCAGAATCCCGGTGCCG, reverse: GTCGCTGTTCAATACATGCAATCGATGTATATATCTGACAC). PCR products were pooled from two 48-well PCR plates (100 μL per well). The products were ethanol precipitated and resuspended in 1 mL TE buffer (10 mM Tris pH 8.0, 1 mM EDTA pH 8.0). The resuspended DNA was applied to a Mono Q 1 mL (GE Healthcare) and eluted with a gradient from 20-100 % TE high salt buffer (10 mM Tris pH 8.0, 1 M NaCl, 1 mM EDTA pH 8.0). Peak fractions were analyzed on a 1 % (v/v) TAE agarose gel and fractions containing the desired DNA product were pooled. The sample was ethanol precipitated, resuspended in 100 μL TE buffer, and stored at 4 °C prior to use.
To reconstitute nucleosomes, histone octamers and DNA were mixed at a 1:1 molar ratio in high salt buffer (20 mM HEPES-Na pH 7.5, 1 mM EDTA pH 8, 2 M KCl), and gradient dialyzed against low salt buffer (20 mM HEPES-Na pH 7.5, 1 mM EDTA pH 8, 30 mM KCl) over 18 hours.
Electrophoretic mobility shift assay
Nucleosome and LEDGF were incubated in EMSA buffer (20mM HEPES-Na 7.5, 50mM NaCl, 1mM TCEP, 5% Glycerol) for 1 h on ice and analyzed by native 6% 0.5 × TBE PAGE. Each reaction contained 1 pmol of nucleosome and increasing amounts of LEDGF (0, 1, 2, 4, 8 pmol). Gels were run at 120 V for 3 h and stained with SyberGold (Invitrogen).
Formation of nucleosome-LEDGF complex
LEDGF and the H3KC36me3-modified nucleosome were mixed at a molar ratio of 2:1 and incubated for 1 hour on ice. The mixture was applied to a Superose 6 Increase 3.2/300 column equilibrated in gel filtration buffer (20 mM HEPES-Na pH 7.5, 50 mM NaCl, 1 mM DTT). The peak fraction was cross-linked with 0.05 % (v/v) glutaraldehyde on ice for 10 minutes and quenched for 10 min using 10 mM Tris-HCl (pH 7.5), 2 mM lysine and 8 mM aspartate. The sample was transferred to a Slide-A-Lyzer MINI Dialysis Unit 20,000 MWCO (Thermo Scientific), and dialyzed for 6 hours against 500 mL dialysis buffer (20 mM HEPES-Na pH 7.5, 50 mM NaCl, 1 mM DTT).
Cryo-EM grid preparation and data collection
The sample was applied to glow-discharged UltrAuFoil 2/2 grids (Quantifoil, Grossloebichau, Germany) by applying 2 μL on each side of the grid. After incubation of 10 seconds, the sample was blotted for 4 seconds and vitrified by plunging into liquid ethane via a Vitrobot Mark IV (FEI Company, Hillsboro, OR, United States) operated at 4 °C and 100 % humidity. Cryo-EM data was acquired on a FEI Titan Krios transmission electron microscope (TEM) operated at 300 keV, equipped with a K2 summit direct detector and a GIF quantum energy filter (Gatan, Pleasanton, CA, United States). Automated data acquisition was carried out using FEI EPU software at a nominal magnification of 130,000x, resulting in a physical pixel size corresponding to 1.05 Å. Movies of 40 frames were collected in counting mode over 9 s at a defocus range from 1.25-2.75 μm. The dose rate was 5.29 e- per Å2 per second resulting in 1.08 e- per Å2 per frame. A total of 4296 and 1268 movies were collected for the nucleosome-LEDGF complexes with 165 bp and 145 bp DNA, respectively.
Image processing and model building
Movie stacks were motion-corrected, CTF corrected, and dose-weighted using Warp66. Particles were auto-picked by Warp, yielding 527,640 particle images. Image processing was performed with RELION 3.0.567. Particles were extracted using a box size of 2562 pixels, and normalized. Reference-free 2D classification was performed to remove poorly aligned particles. An ab initio model generated from cryoSPARC68 was used as an initial model for subsequent 3D classification. All classes containing nucleosome density were combined and used for a global 3D refinement. A reconstructed map at 3.1 Å resolution was obtained from 224,648 particles. To obtain an improved density map for the PWWP domain, the nucleosome part in the refined particles was subtracted by back-projection employing a mask. The remaining density of the particles were subjected to further 3D classification without image alignment. All classes containing PWWP density were subjected to CTF refinement, Bayesian polishing, and 3D refinement. Post-processing of refined models was performed using automatic B-factor determination in RELION and reported resolutions are based on the gold-standard Fourier shell correlation 0.143 criterion (-123.97 Å2 B factor and 3.2 Å resolution for the best class). Local resolution estimates were obtained using the built-in local resolution estimation tool of RELION and previously estimated B-factors.
The model was built into the density of the class which showed the best local resolution of the PWWP domain. A nucleosome structure with 145bp Widom 601 DNA (PDB 3MVD)34 and the crystal structure of the LEDGF PWWP domain (PDB 4FU6) were placed into the density map by rigid-body fitting in Chimera. Both structures were manually adjusted and the extra linker DNA was built using COOT. The model was subjected to alternating real-space refinement and manual adjustment using PHENIX69,70 and COOT71, resulting in very good stereochemistry as assessed by Molprobity72.
Extended Data
Extended Data Fig. 1. Binding of LEDGF to H3KC36me3-modified nucleosome.
a. EMSA reveals that full-length LEDGF preferentially binds to a H3KC36me3-modified nucleosome with longer (165 bp) DNA. Molar ratio of full-length LEDGF to indicated nucleosomes are shown on the top of each lane. Bands correspond to each component and complexes are labeled on the right. Bands of degraded LEDGF-bound nucleosomes are denoted with *. For gel source data, see Source Data Extended Data Fig. 1.
b. Reconstructed EM density maps of 145bp and 165bp H3KC36me3-modified nucleosome with LEDGF. Note that the presence of the extra DNA in the latter complex leads to a defined additional density for the PWWP domain.
c. Mass spectrometry measurement of the H3KC36me3 modified histone H3. Left: molecular weight measurement of H3K36C mutant. Right: molecular weight measurement of H3KC36me3 modified H3.
Extended Data Fig. 2. Cryo-EM data processing.
a. Data processing procedure for the complex of the 165 bp H3KC36me3-modified nucleosome with LEDGF using Warp and Relion.
b. Fourier Shell Correlation (FSC) plot for the reconstruction using 55,142 particles in the indicated class (enclosed by dashed line in the final step in a). The overall resolution is 3.2 Å as determined by the FSC 0.143 criterion.
c. Local resolution assessment of the final cryo-EM map.
d. Euler angle distribution of particles used in the final 3D reconstruction.
Extended Data Fig. 3. Cryo-EM density.
a. A vertical slice through the structure. Models of all chains are shown as sticks and the cryo-EM density is shown as a gray mesh.
b. A horizontal slice through the structure. Models of all chains are shown as sticks and the EM density is shown as a gray mesh.
c. Density of histone H3 residue KC36me3 and its interacting residues of the aromatic cage in the PWWP domain.
d. Density of DNA-interacting residues of patch 1.
e. Density of part of the nucleosomal DNA at SHL 0.
f. Density of the dyad DNA base pair.
g. Density of the B-factor sharpened (left) and unsharpened (right) PWWP domain.
Extended Data Fig. 4. Nucleosome-PWWP interface and comparison with other PWWP-DNA structures.
a. Nucleosome-PWWP interface. Residues colored in white recognize H3KC36me3 and residues colored in green interact with DNA.
b. Front and side view of DNA conformation comparison with other known PWWP-DNA structures. PDB code of the structures used are: 5XSK (HDGF) and 6IIS (HDGF3L, also known as HRP3).
c. Schematic view of DNA interactions. Electrostatic interactions and hydrogen bonds are shown as yellow dashes. SHLs are denoted.
Extended Data Fig. 5. Comparison of the location of the PWWP domain in our nucleosome-PWWP complex structure with previously proposed models.
a. Front view of the comparison with two models proposed for LEDGF (gray) 25 or its highly conserved homolog HDGFL3 (yellow)26 with our structure (pink). Whereas in one model (yellow) the domain is rotated by around 180 degrees and shifted to SHL -1 on one DNA gyre, in another model (Gray) the domain is moved to SHL +6.5 and -1.5, and placed in the major groove of the DNA gyres.
b. Side view of the comparison shown in panel a.
Extended Data Fig. 6. Comparison with other ‘royal’ family domains bound to methylated H3K36 peptides.
a. Structures of the PWWP, Tudor and Chromo domain bound with methylated H3K36 peptides. PDB codes of structures used here are: 4HCZ (PHF1), 2F5K (MRG15) and 4PLI (H3K36me3 of MRG2).
b. Superposition of all three structures shown in a.
c. Placement of the PHF1 Tudor domain structure (yellow)60 onto our nucleosome-PWWP structure based on superposition of the H3 peptides in both structures reveals a clash between the Tudor domain and the nucleosomal DNA (red dashed circle). This shows that the Tudor domain must bind differently, and may unwind the end of nucleosomal DNA or alter the conformation of the H3 tail, or both.
Supplementary Material
Acknowledgements
We thank C. Oberthür for help with LEDGF protein purification, U. Neef and S. Vos for maintaining insect cell stocks, S. Aibara for helpful discussion. H.W. was supported by an EMBO Long-Term Fellowship (ALTF 650-2017). P.C. was supported by the Deutsche Forschungsgemeinschaft (SFB860, SPP1935, SPP2191), the Advanced Grant ‘TRANSREGULON’ from the European Research Council (grant agreement No 693023), and the Volkswagen Foundation.
Footnotes
Data availability. Electron microscopy densities were deposited in the EM Data Bank with the accession codes EMD-10069. The coordinate file for the LEDGF PWWP-165bp H3KC36me3-modified nucleosome structure was deposited in the Protein Data Bank with the accession code 6S01.
Author contributions
H.W and L.F. initiated the project. H.W. designed and conducted all the experiments and data analysis unless stated otherwise. L.F. cloned and purified full-length LEDGF protein. C.D. maintained the EM facility and advised on microscope setup. P.C. supervised research. H.W. and P.C. wrote the manuscript with input from all authors.
Competing interests
The authors declare no competing interests.
References
- 1.Patel DJ, Wang Z. Readout of Epigenetic Modifications. Annual Review of Biochemistry. 2013;82:81–118. doi: 10.1146/annurev-biochem-072711-165700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weaver T, Morrison E, Musselman C. Reading more than histones: the prevalence of nucleic acid binding among reader domains. Molecules. 2018;23:2614. doi: 10.3390/molecules23102614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marabelli C, et al. A Tail-Based Mechanism Drives Nucleosome Demethylation by the LSD2/NPAC Multimeric Complex. Cell Reports. 2019;27:387–399.e7. doi: 10.1016/j.celrep.2019.03.061. [DOI] [PubMed] [Google Scholar]
- 4.Stec I, et al. WHSC1, a 90 kb SET domain-containing gene, expressed in early development and homologous to a Drosophila dysmorphy gene maps in the Wolf-Hirschhorn syndrome critical region and is fused to IgH in t (1; 14) multiple myeloma. Human molecular genetics. 1998;7:1071–1082. doi: 10.1093/hmg/7.7.1071. [DOI] [PubMed] [Google Scholar]
- 5.Izumoto Y, Kuroda T, Harada H, Kishimoto T, Nakamura H. Hepatoma-derived growth factor belongs to a gene family in mice showing significant homology in the amino terminus. Biochemical and biophysical research communications. 1997;238:26–32. doi: 10.1006/bbrc.1997.7233. [DOI] [PubMed] [Google Scholar]
- 6.Stec I, Nagl SB, van Ommen GJB, den Dunnen JT. The PWWP domain: a potential protein-protein interaction domain in nuclear proteins influencing differentiation? Febs Letters. 2000;473:1–5. doi: 10.1016/s0014-5793(00)01449-6. [DOI] [PubMed] [Google Scholar]
- 7.Qiu C, Sawada K, Zhang X, Cheng XD. The PWWP domain of mammalian DNA methyltransferase Dnmt3b defines a new family of DNA-binding folds. Nature Structural Biology. 2002;9:217–224. doi: 10.1038/nsb759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rona GB, Eleutherio EC, Pinheiro AS. PWWP domains and their modes of sensing DNA and histone methylated lysines. Biophysical reviews. 2016;8:63–74. doi: 10.1007/s12551-015-0190-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hughes RM, Wiggins KR, Khorasanizadeh S, Waters ML. Recognition of trimethyllysine by a chromodomain is not driven by the hydrophobic effect. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:11184–11188. doi: 10.1073/pnas.0610850104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maurer-Stroh S, et al. The Tudor domain 'Royal Family': Tudor, plant Agenet, Chromo, PWWP and MBT domains. Trends in Biochemical Sciences. 2003;28:69–74. doi: 10.1016/S0968-0004(03)00004-5. [DOI] [PubMed] [Google Scholar]
- 11.Vezzoli A, et al. Molecular basis of histone H3K36me3 recognition by the PWWP domain of Brpf1. Nat Struct Mol Biol. 2010;17:617–9. doi: 10.1038/nsmb.1797. [DOI] [PubMed] [Google Scholar]
- 12.Dhayalan A, et al. The Dnmt3a PWWP domain reads histone 3 lysine 36 trimethylation and guides DNA methylation. J Biol Chem. 2010;285:26114–20. doi: 10.1074/jbc.M109.089433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vermeulen M, et al. Quantitative Interaction Proteomics and Genome-wide Profiling of Epigenetic Histone Marks and Their Readers. Cell. 2010;142:967–980. doi: 10.1016/j.cell.2010.08.020. [DOI] [PubMed] [Google Scholar]
- 14.Pradeepa MM, Sutherland HG, Ule J, Grimes GR, Bickmore WA. Psip1/Ledgf p52 Binds Methylated Histone H3K36 and Splicing Factors and Contributes to the Regulation of Alternative Splicing. Plos Genetics. 2012;8 doi: 10.1371/journal.pgen.1002717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu H, et al. Structural and histone binding ability characterizations of human PWWP domains. PLoS One. 2011;6:e18919. doi: 10.1371/journal.pone.0018919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sankaran SM, Wilkinson AW, Elias JE, Gozani O. A PWWP Domain of Histone-Lysine N-Methyltransferase NSD2 Binds to Dimethylated Lys-36 of Histone H3 and Regulates NSD2 Function at Chromatin. Journal of Biological Chemistry. 2016;291:8465–8474. doi: 10.1074/jbc.M116.720748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 18.Li J, Moazed D, Gygi SP. Association of the Histone Methyltransferase Set2 with RNA Polymerase II Plays a Role in Transcription Elongation. Journal of Biological Chemistry. 2002;277:49383–49388. doi: 10.1074/jbc.M209294200. [DOI] [PubMed] [Google Scholar]
- 19.Sun X-J, et al. Identification and characterization of a novel human histone H3 lysine 36-specific methyltransferase. Journal of Biological Chemistry. 2005;280:35261–35271. doi: 10.1074/jbc.M504012200. [DOI] [PubMed] [Google Scholar]
- 20.Wagner EJ, Carpenter PB. Understanding the language of Lys36 methylation at histone H3. Nature Reviews Molecular Cell Biology. 2012;13:115–126. doi: 10.1038/nrm3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H, et al. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 2006;442:91. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ge YZ, et al. Chromatin targeting of de novo DNA methyltransferases by the PWWP domain. Journal of Biological Chemistry. 2004;279:25447–25454. doi: 10.1074/jbc.M312296200. [DOI] [PubMed] [Google Scholar]
- 23.Qiu Y, et al. Solution structure of the Pdp1 PWWP domain reveals its unique binding sites for methylated H4K20 and DNA. Biochem J. 2012;442:527–38. doi: 10.1042/BJ20111885. [DOI] [PubMed] [Google Scholar]
- 24.Eidahl JO, et al. Structural basis for high-affinity binding of LEDGF PWWP to mononucleosomes. Nucleic Acids Research. 2013;41:3924–3936. doi: 10.1093/nar/gkt074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Nuland R, et al. Nucleosomal DNA binding drives the recognition of H3K36-methylated nucleosomes by the PSIP1-PWWP domain. Epigenetics & Chromatin. 2013;6:12. doi: 10.1186/1756-8935-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tian W, et al. The HRP3 PWWP domain recognizes the minor groove of double-stranded DNA and recruits HRP3 to chromatin. Nucleic Acids Research. 2019 doi: 10.1093/nar/gkz294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turlure F, Maertens G, Rahman S, Cherepanov P, Engelman A. A tripartite DNA-binding element, comprised of the nuclear localization signal and two AT-hook motifs, mediates the association of LEDGF/p75 with chromatin in vivo. Nucleic Acids Research. 2006;34:1653–1665. doi: 10.1093/nar/gkl052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Llano M, et al. Identification and characterization of the chromatin-binding domains of the HIV-1 integrase interactor LEDGF/p75. Journal of Molecular Biology. 2006;360:760–773. doi: 10.1016/j.jmb.2006.04.073. [DOI] [PubMed] [Google Scholar]
- 29.Ge H, Si YZ, Roeder RG. Isolation of cDNAs encoding novel transcription coactivators p52 and p75 reveals an alternate regulatory mechanism of transcriptional activation. Embo Journal. 1998;17:6723–6729. doi: 10.1093/emboj/17.22.6723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.LeRoy G, et al. LEDGF and HDGF2 relieve the nucleosome-induced barrier to transcription. bioRxiv. 2018 doi: 10.1126/sciadv.aay3068. 500926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simon MD, et al. The site-specific installation of methyl-lysine analogs into recombinant histones. Cell. 2007;128:1003–1012. doi: 10.1016/j.cell.2006.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsutsui KM, Sano K, Hosoya O, Miyamoto T, Tsutsui K. Nuclear protein LEDGF/p75 recognizes supercoiled DNA by a novel DNA-binding domain. Nucleic Acids Research. 2011;39:5067–5081. doi: 10.1093/nar/gkr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hendrix J, et al. The transcriptional co-activator LEDGF/p75 displays a dynamic scan-and-lock mechanism for chromatin tethering. Nucleic Acids Research. 2011;39:1310–1325. doi: 10.1093/nar/gkq933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Makde RD, England JR, Yennawar HP, Tan S. Structure of RCC1 chromatin factor bound to the nucleosome core particle. Nature. 2010;467:562–6. doi: 10.1038/nature09321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wen H, et al. ZMYND11 links histone H3.3K36me3 to transcription elongation and tumour suppression. Nature. 2014 doi: 10.1038/nature13045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rondelet G, Dal Maso T, Willems L, Wouters J. Structural basis for recognition of histone H3K36me3 nucleosome by human de novo DNA methyltransferases 3A and 3B. Journal of Structural Biology. 2016;194:357–367. doi: 10.1016/j.jsb.2016.03.013. [DOI] [PubMed] [Google Scholar]
- 37.Qin S, Min JR. Structure and function of the nucleosome-binding PWWP domain. Trends in Biochemical Sciences. 2014;39:536–547. doi: 10.1016/j.tibs.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 38.McGinty RK, Tan S. Recognition of the nucleosome by chromatin factors and enzymes. Current Opinion in Structural Biology. 2016;37:54–61. doi: 10.1016/j.sbi.2015.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morgan MT, et al. Structural basis for histone H2B deubiquitination by the SAGA DUB module. Science. 2016;351:725–8. doi: 10.1126/science.aac5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Worden EJ, Hoffmann NA, Hicks CW, Wolberger C. Mechanism of Cross-talk between H2B Ubiquitination and H3 Methylation by Dot1L. Cell. 2019;176:1490–1501. e12. doi: 10.1016/j.cell.2019.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shun M-C, et al. Identification and Characterization of PWWP Domain Residues Critical for LEDGF/p75 Chromatin Binding and Human Immunodeficiency Virus Type 1 Infectivity. Journal of Virology. 2008;82:11555–11567. doi: 10.1128/JVI.01561-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baubec T, et al. Genomic profiling of DNA methyltransferases reveals a role for DNMT3B in genic methylation. Nature. 2015;520:243–7. doi: 10.1038/nature14176. [DOI] [PubMed] [Google Scholar]
- 43.Li F, et al. The Histone Mark H3K36me3 Regulates Human DNA Mismatch Repair through Its Interaction with MutSα. Cell. 2013;153:590–600. doi: 10.1016/j.cell.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wolffe AP. Architectural transcription factors. Science. 1994;264:1100–1. doi: 10.1126/science.8178167. [DOI] [PubMed] [Google Scholar]
- 45.Zhu F, et al. The interaction landscape between transcription factors and the nucleosome. Nature. 2018;562:76–81. doi: 10.1038/s41586-018-0549-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maskell DP, et al. Structural basis for retroviral integration into nucleosomes. Nature. 2015;523:366. doi: 10.1038/nature14495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Farnung L, Vos SM, Wigge C, Cramer P. Nucleosome–Chd1 structure and implications for chromatin remodelling. Nature. 2017;550:539. doi: 10.1038/nature24046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eustermann S, et al. Structural basis for ATP-dependent chromatin remodelling by the INO80 complex. Nature. 2018;556:386–390. doi: 10.1038/s41586-018-0029-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ayala R, et al. Structure and regulation of the human INO80–nucleosome complex. Nature. 2018;556:391–395. doi: 10.1038/s41586-018-0021-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Willhoft O, et al. Structure and dynamics of the yeast SWR1-nucleosome complex. Science. 2018;362:eaat7716. doi: 10.1126/science.aat7716. [DOI] [PubMed] [Google Scholar]
- 51.Venkatesh S, et al. Set2 methylation of histone H3 lysine 36 suppresses histone exchange on transcribed genes. Nature. 2012;489:452. doi: 10.1038/nature11326. [DOI] [PubMed] [Google Scholar]
- 52.Venkatesh S, Workman JL. Histone exchange, chromatin structure and the regulation of transcription. Nat Rev Mol Cell Biol. 2015;16:178–89. doi: 10.1038/nrm3941. [DOI] [PubMed] [Google Scholar]
- 53.Carrozza MJ, et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 54.Smolle M, et al. Chromatin remodelers Isw1 and Chd1 maintain chromatin structure during transcription by preventing histone exchange. Nature Structural & Molecular Biology. 2012;19:884. doi: 10.1038/nsmb.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang P, et al. Structure of human MRG15 chromo domain and its binding to Lys36-methylated histone H3. Nucleic Acids Research. 2006;34:6621–6628. doi: 10.1093/nar/gkl989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Musselman CA, et al. Molecular basis for H3K36me3 recognition by the Tudor domain of PHF1. Nat Struct Mol Biol. 2012;19:1266–72. doi: 10.1038/nsmb.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ballare C, et al. Phf19 links methylated Lys36 of histone H3 to regulation of Polycomb activity. Nat Struct Mol Biol. 2012;19:1257–65. doi: 10.1038/nsmb.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brien GL, et al. Polycomb PHF19 binds H3K36me3 and recruits PRC2 and demethylase NO66 to embryonic stem cell genes during differentiation. Nat Struct Mol Biol. 2012;19:1273–81. doi: 10.1038/nsmb.2449. [DOI] [PubMed] [Google Scholar]
- 59.Musselman CA, et al. Binding of PHF1 Tudor to H3K36me3 enhances nucleosome accessibility. Nat Commun. 2013;4 doi: 10.1038/ncomms3969. 2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim D, et al. Corecognition of DNA and a methylated histone tail by the MSL3 chromodomain. Nature Structural & Molecular Biology. 2010;17:1027–1029. doi: 10.1038/nsmb.1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Connelly KE, et al. Engagement of DNA and H3K27me3 by the CBX8 chromodomain drives chromatin association. Nucleic Acids Res. 2019;47:2289–2305. doi: 10.1093/nar/gky1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jacobson RH, Ladurner AG, King DS, Tjian R. Structure and Function of a Human TAFII250 Double Bromodomain Module. Science. 2000;288:1422–1425. doi: 10.1126/science.288.5470.1422. [DOI] [PubMed] [Google Scholar]
- 63.Miller TC, et al. A bromodomain-DNA interaction facilitates acetylation-dependent bivalent nucleosome recognition by the BET protein BRDT. Nat Commun. 2016;7 doi: 10.1038/ncomms13855. 13855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Morrison EA, et al. DNA binding drives the association of BRG1/hBRM bromodomains with nucleosomes. Nat Commun. 2017;8 doi: 10.1038/ncomms16080. 16080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dyer PN, et al. Reconstitution of nucleosome core particles from recombinant histones and DNA. Chromatin and Chromatin Remodeling Enzymes, Pt A. 2004;375:23–44. doi: 10.1016/s0076-6879(03)75002-2. [DOI] [PubMed] [Google Scholar]
- 66.Tegunov D, Cramer P. Real-time cryo-EM data pre-processing with Warp. bioRxiv. 2018 338558. [Google Scholar]
- 67.Zivanov J, et al. RELION-3: new tools for automated high-resolution cryo-EM structure determination. bioRxiv. 2018 doi: 10.7554/eLife.42166. 421123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nature Methods. 2017;14:290. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 69.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallographica Section D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Afonine PV, et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallographica Section D: Structural Biology. 2018;74 doi: 10.1107/S2059798318006551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallographica Section D. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallographica Section D: Biological Crystallography. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.