Abstract
Objective
We investigated the correlations between cyclin-dependent kinase 4/6 (CDK4/6) levels and human papillomavirus (HPV) infection state in head and neck squamous cell cancer (HNSCC). The aim was to explore the potential value of CDK4/6 inhibitors in the treatment of HNSCC.
Methods
Multiomic sequencing data for HNSCC were obtained from The Cancer Genome Atlas (TCGA), and the mRNA levels and copy number variations (CNVs) of CDK4 and CDK6 were strictly analyzed. Overall survival (OS) curves were produced using the Kaplan-Meier method, and survival differences between groups were assessed by the log-rank test. Next, gene set enrichment analysis (GSEA) was applied to interrogate CDK4/6-associated molecular pathways in HPV-positive (HPV+) and HPV-negative (HPV−) HNSCC. Last, lymphoid cell infiltrates in each type of HNSCC were explored, and the correlations between CDK4/6 expression and lymphoid infiltrates were explored by Tumor Immune Estimation Resource (TIMER) analysis.
Results
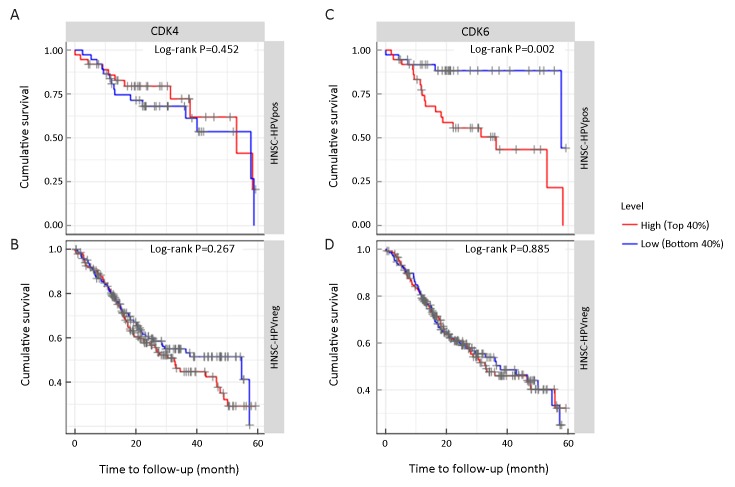
Overexpression of either CDK6 or CDK4 was not a relevant factor for OS in HPV− HNSCC (CDK6: top 40%vs. bottom 40%, P=0.885; CDK4: top 40% vs. bottom 40%, P=0.267). In HPV+ HNSCC, CDK6 but not CDK4 was a relevant factor for OS (CDK6: top 40% vs. bottom 40%, P=0.002; CDK4: top 40% vs. bottom 40%, P=0.452). GSEA found that overexpressed CDK6 in HPV+ HNSCC inhibited pathways involved in the tumor immune response, suggesting its roles in antitumor immunity. TIMER analysis results revealed that CDK6 but not CDK4 accumulation was negatively correlated with the number of tumor-infiltrating lymphocytes specific for HPV+ HNSCC, which led to tumor response suppression.
Conclusions
CDK6, but not CDK4, is a poor prognostic marker specific in HPV+ HNSCC patients. Overexpressed CDK6 might stimulate tumor progression by suppressing lymphocytes infiltration independent of its kinase activity. Only abrogating its kinase activity using current CDK4/6 inhibitors was not enough to block its tumor promotion function.
Keywords: HNSCC, HPV, CDK4/6, immune infiltration
Introduction
Every year, more than 600,000 people are diagnosed with head and neck squamous cell carcinoma (HNSCC) worldwide, but only 40%−50% of them would survive for 5 years (1,2). Currently, the most frequently used strategy remains the combination of chemotherapy and radiotherapy (3,4). Cetuximab, the only available second-line targeted therapy, has a response rate of less than 15% (5,6). In recent years, significant efforts have been expended to develop alternative therapeutics for HNSCC, including cell cycle-targeted therapies.
The development and progression of cancer are usually related to the dysregulation of normal cell cycle control due to alterations in cell cycle machinery components (7). Under normal conditions, cyclin-dependent kinases (CDKs), the core components of the cell cycle machinery, ensure orderly transitions between cell cycle phases. In the event of an abnormality, deregulated CDK activity may result in aberrant responses to cell damage and senescence, sometimes even contributing to malignant transformation. The idea to develop anticancer therapeutic strategies by targeting CDKs can be backdated to the late 1980s (8-10). Although the first generation of “pan-CDK” inhibitors failed as a result of significant toxicity and a low response rate, two CDK4/6-specific inhibitors (palbociclib and ribociclib) have been approved by the U.S. Food and Drug Administration (FDA) for the treatment of breast cancer, which offers an encouraging perspective (11).
Since loss of control of the cell cycle, especially the G1/S transition, has been proven to be the major characteristic of HNSCC (12,13), researchers have been investigating the therapeutic value of CDK4/6 inhibitors in HNSCC. For example, more than 10 relevant clinical trials have been established worldwide, including NCT03179956, NCT02429089, and NCT02499120. When used as a single agent, a CDK4/6 inhibitor showed only modest activity in a preclinical study, but combined use with chemotherapy, immunotherapy or epidermal growth factor receptor (EGFR) inhibitors may produce a better effect (14). In a phase I study, two out of nine patients with recurrent or metastatic HNSCC showed a partial response after receiving combined treatment with palbociclib and cetuximab (15), which supports the use of combination treatment. To ensure optimal therapeutic effects, it is worth determining what type of pharmaceuticals should be combined and what indications should be used for screening patients.
Herein, we analyze clinical data generated by The Cancer Genome Atlas (TCGA). All samples were first stratified into different subsets according to their human papillomavirus (HPV) status and CDK4 and CDK6 gene expression levels. The survival profiles of the individual subsets showed that the expression level of CDK6 but not CDK4 was an excellent prognostic indicator in HPV-positive (HPV+) HNSCC patients. Next, we performed gene set enrichment analysis (GSEA) and lymphocyte quantification, which linked CDK6 expression to tumor infiltration by lymphocytes and the antitumor immune response. In HPV+ HNSCC tumors, overexpressed CDK6 suppressed the lymphocyte infiltration process, attenuating CDK6 protein level might elevate the number of filtered lymphocytes in tumor site. Therefore, we speculate that using molecules that could downregulated protein level of CDK6, rather than the currently used CDK4/6 kinase inhibitors in combination with immune checkpoint inhibitor-based immunotherapy could be an effective treatment strategy for CDK6+/HPV+ subtype HNSCC patients.
Materials and methods
Data source
A digital gene expression dataset (level 3) as well as clinical information, such as sex, age, HPV infection status, and survival time, were downloaded for a total of 496 HNSCC patients from the official website of the TCGA. Normalization of the expression data was performed following the standard method summarized by the TCGA. Before statistical analysis, we performed logarithmic transformation of the expression levels.
Survival curve analysis
Tumor Immune Estimation Resource (TIMER) web server ( https://cistrome.shinyapps.io/timer/) was used to generate Kaplan-Meier plots to visualize survival differences according to the expression levels of target genes (e.g., CDK6 and CDK4). To determine optimal grouping criteria, the samples in both HPV+ and HPV-negative (HPV−) groups were ranked in descending order according to the expression level of CDK6 or CDK4. We then chose equal numbers of the samples (i.e. 10%, 20%, 30%, 40% and 50% of total, respectively) at the top and bottom of the list and compared their survival curves of up to 5 years (data not shown). Based on these results, the high- and low-level groups were ultimately defined as the top and bottom 40% to include as many samples as possible while ensuring sufficiently significant differences between the two groups.
GSEA
GSEA was carried out to investigate whether selected gene sets were negatively or positively correlated with the CDK6 expression level in HPV+ HNSCC tumors. Gene sets were selected from the MsigDB database version 6.1. In our study, GSEA was run with the default parameters (i.e., permutation number = 1,000, permutation type = “gene-sets”, and recompute time set to 1,000 times). All genes in the dataset were ranked using a signal-to-noise metric. The enrichment score (ES), normalized enrichment score (NES), and false discovery rate (FDR) were calculated on the basis of the ranked list of genes (16). Gene set with statistical significance (Q value <0.001) were considered to represent biological processes with significant expression differences.
Copy number alterations (CNAs)
Somatic CNAs (SCNAs) were defined by GISTIC 2.0 (Version 2.0.23, ftp://ftp.broadinstitute.org/pub/GISTIC2.0/GISTIC_2_0_23.tar.gz), which include deep deletion (−2), arm-level deletion (−1), diploid/normal (0), arm-level gain (1), and high amplification (2). The distributions of immune subsets at each copy number status in either HPV+ or HPV− HNSCC are given in box plots. The infiltration level for each SCNA category was compared with the normal level using a two-sided Wilcoxon rank sum test.
Results
CDK6 is a poor prognostic marker in HPV+ HNSCC patients
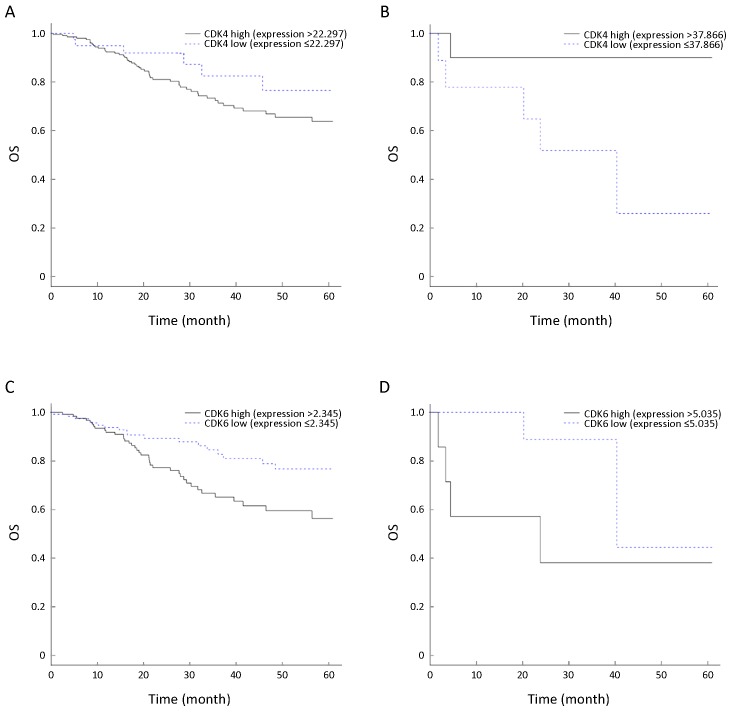
HPV infection is one of the most commonly reported carcinogenic factors involved in the development of HNSCC. Comprehensive integrative genomic analysis suggested that HPV+ and HPV− HNSCC harbored different molecular features and drove the cell cycle through different pathways (12). As the most prominently distinct HNSCC subclasses, HPV-related and HPV-unrelated tumors are often considered to be different diseases and should be analyzed separately (17-19). In total, 496 HNSCC patients were stratified into two separate groups according to their HPV infection status, i.e., HPV+ HNSCC and HPV− HNSCC. For each group, the patients were further divided into subsets based on the mRNA expression level of either CDK4 or CDK6 (Table 1). The association between the mRNA level of CDK4 and survival was statistically nonsignificant (P>0.05) regardless of the HPV infection status, as shown inFigure 1A,B. In contrast, the prognostic value of CDK6 showed distinct differences between the HPV+ and HPV− subtypes of HNSCCFigure 1C,D. More specifically, in the HPV+ HNSCC subset, the patients with a low CDK6 level showed a median survival time over 60 months, which was significantly longer than the median survival time of approximately 30 months in patients with a high CDK6 level (P=0.002). Nevertheless, no significant differences (Student’s t test, P>0.05) in age or tumor stage were observed between patients with high CDK6 expression and patients with low CDK6 expression, indicating the excellent prognostic value of CDK6 in HPV+ HNSCC.
1.
Sample information and stratified subtypes
| Variables | HPV+, CDK6 [n (%)] | HPV−, CDK6 [n (%)] | HPV+, CDK4 [n (%)] | HPV−, CDK4 [n (%)] | |||||||
| Top 40% (N=35) | Bottom 40% (N=35) | Top 40% (N=164) | Bottom 40% (N=164) | Top40% (N=35) | Bottom 40% (N=35) | Top40% (N=164) | Bottom 40% (N=164) | ||||
| CDK6, cyclin-dependent kinase 6; CDK4, cyclin-dependent kinase 4; HPV, human papillomavirus; HNSCC, head and neck squamous cell cancer. | |||||||||||
| Gender | |||||||||||
| Male | 29 (82.86) | 34 (97.14) | 118 (71.95) | 120 (73.17) | 33 (94.29) | 29 (82.86) | 125 (76.22) | 105 (64.02) | |||
| Female | 6 (17.14) | 1 (2.86) | 46 (28.05) | 44 (26.83) | 2 (5.71) | 6 (17.14) | 39 (23.78) | 59 (35.98) | |||
| Age (year) | |||||||||||
| <55 | 12 (34.29) | 14 (40.00) | 52 (31.71) | 38 (23.17) | 13 (37.14) | 15 (42.86) | 35 (21.34) | 51 (31.10) | |||
| ≥55 | 23 (65.71) | 12 (60.00) | 112 (68.29) | 125 (76.22) | 22 (62.86) | 20 (57.14) | 129 (78.66) | 112 (68.29) | |||
| Stage | |||||||||||
| I−III | 11 (31.43) | 9 (25.71) | 64 (39.02) | 51 (31.10) | 10 (28.57) | 7 (20.00) | 56 (34.15) | 68 (41.46) | |||
| IV | 17 (48.57) | 13 (37.14) | 87 (53.05) | 94 (57.32) | 12 (34.29) | 19 (54.29) | 88 (53.66) | 82 (50.00) | |||
1.
Survival profiles of head and neck squamous cell cancer (HNSCC) patient subsets according to their human papillomavirus (HPV) status and cyclin-dependent kinase 4 (CDK4) and cyclin-dependent kinase 6 (CDK6) gene expression levels. (A,B) CDK4 expression did not correlate with overall survival (OS) in HPV-positive (HPV+) (A) or HPV-negative (HPV−) HNSCC (B) patients by Kaplan-Meier survival analysis (P=0.452 and 0.267, respectively); (C,D) CDK6 expression correlated with OS in HNSCC patients with HPV infection (C) but not in HNSCC patients without HPV infection (D) by Kaplan-Meier survival analysis (P=0.002 and 0.885, respectively).
CDK6 contributes to immune response in HPV+ HNSCC
Because HPV+ HNSCC usually has a high expression level of p16 and a low expression level of retinoblastoma 1 (RB1), the CDK4/6-D-type cyclin complex is unnecessary to overcome the G1/S transition barrier established by RB1. This characteristic indicated that the overexpression of the CDK6 protein could promote the progression of HPV+ HNSCC through a cell proliferation-independent pathway. To evaluate the possibility of such a mechanism, GSEA was carried out with HPV+ HNSCC and HPV− HNSCC data to globally evaluate the molecular functions of CDK6 in HNSCC.
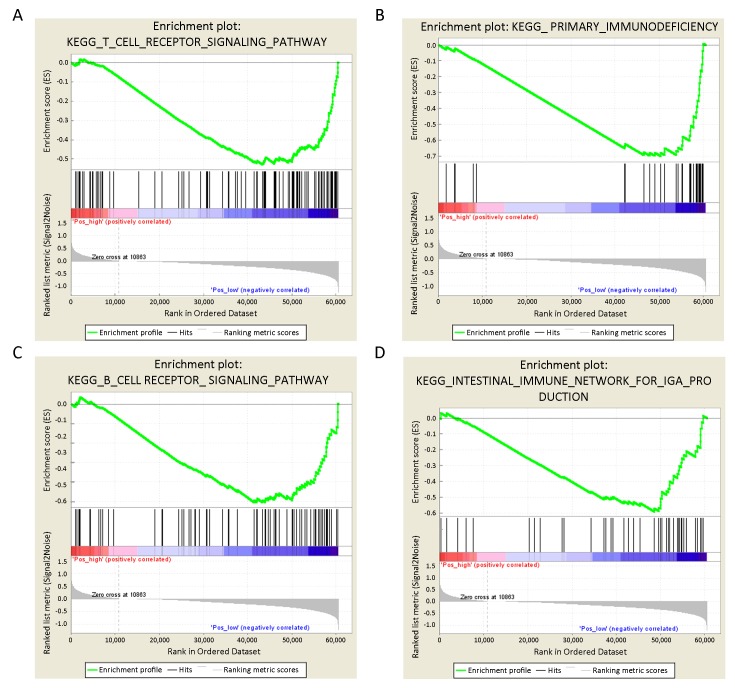
In HPV+ HNSCC, we found that a high level of CDK6 expression was most positively correlated with the cell-cell adhesion process, including an ECM receptor interaction (NES=2.03, P<0.0001) and focal adhesion (NES=1.76, P<0.0001), suggesting potential roles for CDK6 in cell migration (Supplementary Table S1). Nevertheless, a high level of CDK6 expression was most negatively correlated with immune response-associated pathways. More specifically, of the top 10 enriched KEGG pathways in the context of low CDK6 expression, 4 were relevant to the immune response, including T cell receptor signaling pathway (Figure 2A, NES=−2.90, P<0.0001), primary immunodeficiency (Figure 2B, NES=−2.33, P<0.0001), B cell receptor signaling pathway (Figure 2C, NES=−2.18, P<0.0001) and the intestinal immune network for IgA production (Figure 2D, NES=−2.56, P<0.0001) (Supplementary Table S2). Indeed, T cell receptor signaling, the most important antitumor immune pathway, was the most significantly enriched pathway in the context of low CDK6 expression.
S1.
Top 10 pathways enriched in patients expressed high level of CDK6 in HPV+ HNSCC
| Name | Size | ES | NES | Nominal
P value |
FDR
Q value |
FWER
P value |
| CDK6, cyclin-dependent kinase 6; HPV+, human papillomavirus positive; HNSCC, head and neck squamous cell cancer; ES, enrichment score; NES, normalized enrichment score; FDR, false discovery rate; FWER, family wise-error rate. | ||||||
| KEGG_ECM_RECEPTOR_INTERACTION | 84 | 0.641343 | 2.027304 | 0 | 0 | 0 |
| KEGG_FOCAL_ADHESION | 197 | 0.544853 | 1.75837 | 0 | 0 | 0 |
| KEGG_ARRHYTHMOGENIC_RIGHT_
VENTRICULAR_CARDIOMYOPATHY_ARVC |
74 | 0.41215 | 1.461838 | 0 | 0.3605968 | 0.6 |
| KEGG_GLIOMA | 65 | 0.325062 | 1.279453 | 0 | 0.61244035 | 0.8 |
| KEGG_GLYCOSAMINOGLYCAN_BIOSYNTHESIS_
CHONDROITIN_SULFATE |
22 | 0.447463 | 1.164818 | 0.25 | 0.5968058 | 1 |
| KEGG_SMALL_CELL_LUNG_CANCER | 84 | 0.31889 | 1.156354 | 0.25 | 0.54958457 | 1 |
| KEGG_PATHOGENIC_ESCHERICHIA_COLI_
INFECTION |
56 | 0.395632 | 1.125031 | 0.25 | 0.59326184 | 1 |
| KEGG_TGF_BETA_SIGNALING_PATHWAY | 85 | 0.33044 | 1.238055 | 0.33333334 | 0.65543085 | 1 |
| KEGG_REGULATION_OF_ACTIN_CYTOSKELETON | 211 | 0.355829 | 1.22164 | 0.33333334 | 0.61781913 | 1 |
| KEGG_GAP_JUNCTION | 88 | 0.313264 | 1.08623 | 0.33333334 | 0.57261807 | 1 |
2.
Gene set enrichment analysis (GSEA). GSEA results of human papillomavirus-positive (HPV+) head and neck squamous cell cancer (HNSCC) showed negative correlations between CDK6 expression and T cell receptor signaling pathway (NES=−2.90, P<0.0001) (A); primary immunodeficiency (NES=−2.33, P<0.0001) (B); B cell receptor signaling pathway (NES=−2.18, P<0.0001) (C) and the intestinal immune network for IgA production (NES=−2.56, P<0.0001) (D). NES, normalized enrichment score.
S2.
Top 10 pathways enriched in patients expressed low level of CDK6 in HPV+ HNSCC
| Name | Size | ES | NES | Nominal
P value |
FDR
Q value |
FWER
P value |
| CDK6, cyclin-dependent kinase 6; HPV+, human papillomavirus positive; HNSCC, head and neck squamous cell cancer; ES, enrichment score; NES, normalized enrichment score; FDR, false discovery rate; FWER, family wise-error rate. | ||||||
| KEGG_T_CELL_RECEPTOR_SIGNALING_PATHWAY | 106 | −0.52969 | −2.90334 | 0 | 0 | 0 |
| KEGG_DNA_REPLICATION | 36 | −0.6954 | −2.59264 | 0 | 0 | 0 |
| KEGG_INTESTINAL_IMMUNE_NETWORK_FOR_IGA_
PRODUCTION |
46 | −0.58877 | −2.56313 | 0 | 0 | 0 |
| KEGG_PRIMARY_IMMUNODEFICIENCY | 35 | −0.70077 | −2.33397 | 0 | 0 | 0 |
| KEGG_VALINE_LEUCINE_AND_ISOLEUCINE_
DEGRADATION |
44 | −0.62555 | −2.23644 | 0 | 0 | 0 |
| KEGG_PROPANOATE_METABOLISM | 32 | −0.62538 | −2.22141 | 0 | 0 | 0 |
| KEGG_REGULATION_OF_AUTOPHAGY | 34 | −0.47799 | −2.21522 | 0 | 0 | 0 |
| KEGG_B_CELL_RECEPTOR_SIGNALING_PATHWAY | 74 | −0.5054 | −2.17945 | 0 | 0 | 0 |
| KEGG_BASE_EXCISION_REPAIR | 35 | −0.7105 | −2.11501 | 0 | 0 | 0 |
| KEGG_COLORECTAL_CANCER | 62 | −0.45481 | −2.05097 | 0 | 0 | 0 |
To investigate whether the correlation between CDK6 expression and T cell suppression in the tumor site was related to HPV infection, we also performed GSEA with HPV-HNSCC tissue samples (Supplementary Table S3,4). Interestingly, we did not find any obvious correlations between the CDK6 level and T cell receptor signaling or other immune response parameters. Overall, CDK6 overexpression inhibited the immune response in HPV+ HNSCC but not in HPV− HNSCC.
S3.
Top 10 pathways enriched in patients expressed high level of CDK6 in HPV− HNSCC
| Name | Size | ES | NES | Nominal
P value |
FDR
Q value |
FWER
P value |
| CDK6, cyclin-dependent kinase 6; HPV−, human papillomavirus negative; HNSCC, head and neck squamous cell cancer; ES, enrichment score; NES, normalized enrichment score; FDR, false discovery rate; FWER, family wise-error rate. | ||||||
| KEGG_FOCAL_ADHESION | 197 | 0.69635 | 3.340728 | 0 | 0 | 0 |
| KEGG_PATHWAYS_IN_CANCER | 321 | 0.586894 | 3.025786 | 0 | 0 | 0 |
| KEGG_N_GLYCAN_BIOSYNTHESIS | 46 | 0.625422 | 2.987428 | 0 | 0 | 0 |
| KEGG_COLORECTAL_CANCER | 62 | 0.572911 | 2.869832 | 0 | 0 | 0 |
| KEGG_ADHERENS_JUNCTION | 68 | 0.708096 | 2.813825 | 0 | 0 | 0 |
| KEGG_GAP_JUNCTION | 88 | 0.557052 | 2.749696 | 0 | 0 | 0 |
| KEGG_SMALL_CELL_LUNG_CANCER | 84 | 0.70374 | 2.619811 | 0 | 0 | 0 |
| KEGG_UBIQUITIN_MEDIATED_PROTEOLYSIS | 130 | 0.580366 | 2.50688 | 0 | 0 | 0 |
| KEGG_REGULATION_OF_ACTIN_CYTOSKELETON | 211 | 0.59045 | 2.484044 | 0 | 0 | 0 |
| KEGG_DILATED_CARDIOMYOPATHY | 90 | 0.538568 | 2.438433 | 0 | 0 | 0 |
S4.
Top 10 pathways enriched in patients expressed low level of CDK6 in HPV− HNSCC
| Name | Size | ES | NES | Nominal P value | FDR Q value | FWER P value |
| CDK6, cyclin-dependent kinase 6; HPV−, human papillomavirus negative; HNSCC, head and neck squamous cell cancer; ES, enrichment score; NES, normalized enrichment score; FDR, false discovery rate; FWER, family wise-error rate. | ||||||
| KEGG_ALLOGRAFT_REJECTION | 35 | −0.79215 | −2.5456 | 0 | 0 | 0 |
| KEGG_TYPE_I_DIABETES_MELLITUS | 41 | −0.75306 | −2.43268 | 0 | 0 | 0 |
| KEGG_GRAFT_VERSUS_HOST_DISEASE | 37 | −0.80384 | −2.42367 | 0 | 0 | 0 |
| KEGG_SYSTEMIC_LUPUS_ERYTHEMATOSUS | 135 | −0.50422 | −2.39379 | 0 | 0 | 0 |
| KEGG_RIBOSOME | 88 | −0.8923 | −2.37643 | 0 | 0 | 0 |
| KEGG_PARKINSONS_DISEASE | 101 | −0.75302 | −2.13886 | 0 | 0 | 0 |
| KEGG_ALZHEIMERS_DISEASE | 145 | −0.57017 | −2.08944 | 0 | 0 | 0 |
| KEGG_HUNTINGTONS_DISEASE | 161 | −0.55314 | −2.08907 | 0 | 0 | 0 |
| KEGG_AUTOIMMUNE_THYROID_DISEASE | 50 | −0.62345 | −2.0359 | 0 | 0 | 0 |
| KEGG_OXIDATIVE_PHOSPHORYLATION | 102 | −0.78112 | −1.96961 | 0 | 0 | 0 |
CDK6 suppressed tumor-infiltrating lymphocytes
We have shown that elevated CDK6 expression could negatively regulate the antitumor immune response in HPV+ HNSCC, and we then investigated whether CDK6 is a molecular marker that can predict the response to immune checkpoint inhibitor-based therapy. Given that infiltrating lymphocytes in the tumor site are the central players in the antitumor immune response, we then quantified the amounts of different types of lymphocytes, including B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils and DCs in each HNSCC sample and analyzed the roles of CDK6 at the level of lymphoid cell infiltration ( https://cistrome.shinyapps.io/timer/).
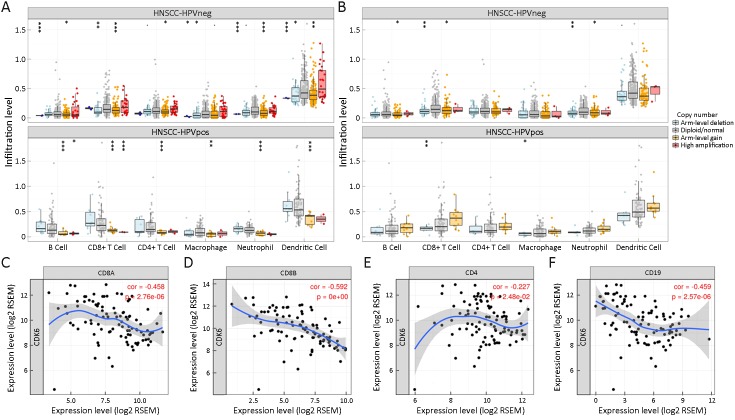
GISTIC results showed that the copy number variation (CNV) state of CDK6 was highly correlated with the abundance of each type of lymphocyte in HPV+ HNSCC. When compared to patients with a normal CDK6 copy number state, patients with high CDK6 amplification contained the lowest levels of B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils and dendritic cells (DCs), followed by patients with high CDK6 arm-level gain. In fact, the amount of CD8+ T cells, the major antitumor immunocyte, showed the most significant difference among HPV+ HNSCC samples with different CDK6 copy number states (Figure 3A). However, the CNV state of CDK4 did not show predictive value in estimating the abundance of tumor-infiltrating lymphocytes (Figure 3B).
3.
Cyclin-dependent kinase 6 (CDK6) reduces the number of tumor-infiltrating lymphocytes in human papillomavirus-positive (HPV+) head and neck squamous cell cancer (HNSCC). (A) The expression level of CDK6 negatively correlated with tumor-infiltrating lymphocytes in HPV+ HNSCC, but not in HPV− HNSCC; (B) The expression level of CDK4 does not correlated with tumor infiltrating lymphocytes in both HPV+ and HPV− HNSCC; (C−F) Negative correlations between the mRNA levels of CDK6 and CD8A (C), CD8B (D), CD4 (E) and CD19 (F). CNV, copy number variation.
To further confirm the correlation between CD8+ T cell abundance and the CDK6 CNV state, we next applied a correlation analysis using transcriptomic data. Consistent with our expectations, the mRNA level of CDK6 was highly negatively correlated with the mRNA levels of the CD8+ T cell markers CD8A and CD8B, CD4+ T cell marker CD4, and B cell marker CD19 in HPV+ HNSCC (Figure 3C−F).
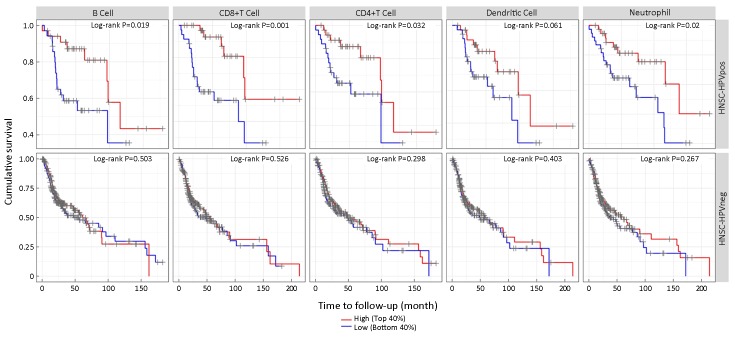
We also investigated the ability of each type of lymphocyte to predict OS outcomes in HNSCC (Figure 4). In contrast to the survival results for CDK6, patients with a higher level of B cells, CD8+ T cells, CD4+ T cells, neutrophils or DCs showed extended survival. However, in HPV− HNSCC, neither the copy number state ofCDK6 nor that of CDK4 could influence the amount of each type of tumor-infiltrating cell. Similarly, the amounts of tumor-infiltrating lymphocytes did not influence OS in HPV− HNSCC. These results indicated that CDK6 might regulate the progression of HPV+ HNSCC through tumor-infiltrating lymphoid cells and the subsequent immune response.
4.
The number of tumor-infiltrating lymphocytes correlated with overall survival in human papillomavirus-positive (HPV+) head and neck squamous cell cancer (HNSCC) but not in HPV-negative (HPV−) HNSCC.
Discussion
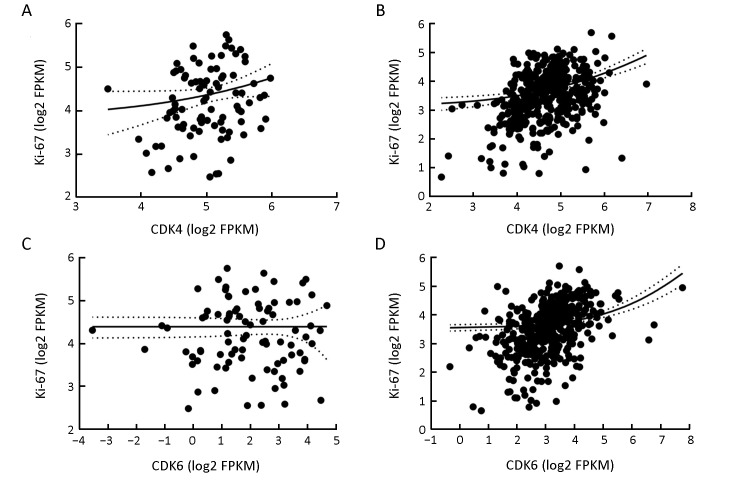
It is recognized that the tumor suppresser gene P16 is often inactivated in HPV− HNSCC (20). The loss function of P16INK4a, which is encoded by the geneP16, leads to increased activity of CDK4/6 kinases and abnormal regulation of the cell cycle. However, according to our analysis in this study, patients with different expression levels of CDK4/6 showed no differences in survival. One possible explanation is that the expression levels of CDKs are not equivalent to their kinase activities. To evaluate this assumption, we further analyzed the correlation between the expression of CDK4/6 and Ki-67 (21), which is a marker of cell proliferation, in both HPV+ and HPV− patients. In HPV− patients, the expression levels of both CDK4 and CDK6 were weakly correlated with that of Ki-67, indicating that the mRNA levels of CDK4/6 cannot precisely reflect their kinase activities. On the other hand, almost no correlation was found between the expression levels of CDK4/6 and Ki-67 in the HPV+ patients (Supplementary Figure S1). This is consistent with our knowledge that the proliferation of HPV+ HNSCC cells is not dependent on CDK4/6 kinase activity. To further investigate the correlation between CDK4/6 expression and HPV infection in cancer progression, we also conducted a similar analysis of patients with squamous carcinoma of the cervix (CESC), which also frequently harbors HPV infection. Consistent with the results observed for HNSCC, the results for CESC showed that the level of CDK4 did not show any prognostic value in CESC, regardless of the HPV state. Only in HPV+ CESC did patients with a high level of CDK6 expression have notably poorer outcomes than patients with low CDK6 expression (P=0.015). Meanwhile, the survival curves of the remaining HPV− patients did not differ statistically (P>0.05) (Supplementary Figure S2). Although the number of HPV− CESC patients was limited (total number=21), this feature would not impair the prognostic value of the CDK6 level in HPV+ CESC. CDK6 and its close homolog CDK4 share high sequence identity (>70%) and play similar roles in cell cycle regulation (22). However, only CDK6 has been reported to have kinase-independent functions thus far. On the basis of this knowledge, we hypothesized that CDK6 might play a role in the progression of HPV+ HNSCC in a kinase-independent manner. As was observed in the HPV+ patient subgroup, CDK kinase activity is not the primary driver of tumor progression. If this hypothesis holds, the phenomenon that CDK6 and CDK4 exhibit distinct prognostic values only in HPV+ HNSCC can be well explained. To evaluate this hypothesis, we performed GSEA, which linked CDK6 expression to tumor-infiltrating lymphocytes. Interestingly, the prognostic role of CD8+ T cell infiltration was previously reported in HNSCC by another research team (23). Subsequent GISTIC analysis also confirmed that the mRNA level of CDK6 was negatively correlated with the mRNA levels of a series of lymphocyte infiltration-related markers (e.g., the CD8+ T cell markers CD8A and CD8B, CD4+ T cell marker CD4, and the B cell marker CD19) in HPV+ HNSCC but not in HPV− HNSCC. However, the association between CDK6 and tumor infiltration is not sufficient to support the hypothesis that CDK6 contributes to tumor progression in a kinase-independent manner. Therefore, we further studied the correlation between CDK6 and vascular endothelial growth factor (VEGF)-A in HPV+ HNSCC patients. The reason for doing so was that CDK6 has been reported to be part of a transcriptional complex that induces the expression of VEGF-A in lymphoid malignancies (24). In addition, VEGF signaling is able to suppress T lymphocyte infiltration into the tumor microenvironment (25). Subsequently, a weak positive correlation between CDK6 and VEGF-A was found. Although our result was not statistically significant, we believe that cytology experiments might provide further evidence to support our hypothesis.
S1.
Correlation between expression of cyclin-dependent kinase 4/6 (CDK4/6) and Ki-67. (A) The expression of CDK4 and Ki-67 is not correlated in human papillomavirus-positive (HPV+) head and neck squamous cell cancer (HNSCC) (R=0.196; P=0.067); (B) The expression of CDK 4 is positively correlated with Ki-67 in HPV-negative (HPV−) HNSCC (R=0.001; P=0.998); (C) The expression of CDK6 and Ki-67 is not correlated in HPV+ HNSCC (R=0.293; P<0.0001); (D) The expression of CDK6 is positively correlated with Ki-67 in HPV- HNSCC (R=0.270; P<0.001).
S2.
Subtype-specific survival curve of cervical carcinoma patients according to their human papillomavirus (HPV) status and cyclin-dependent kinase 4/6 (CDK4/6) expression level. (A,B) CDK4 expression does not correlate with overall survival (OS) in HPV-positive (HPV+) (A) or HPV-negative (HPV−) (P=0.313) (B) squamous carcinoma of the cervix (CESC) (P=0.107); (C) CDK6 expression correlates with OS in HPV+ CESC (P=0.015); (D) CDK6 expression does not correlate with OS in HPV− CESC (P=0.067).
Conclusions
In this work, we demonstrated that CDK6 is a potential prognostic marker in HPV+ HNSCC. Although the underlying mechanism remains unclear, we presume that inhibiting CDK6 expression instead of its kinase activity in HPV+ HNSCC patients might alter the antitumor immune response, thus improving the outcomes of immune checkpoint-based therapy.
Acknowledgements
This study was supported by the Beijing Municipal Science and Technology Commission (No. Z171100001017127) and the Fund for National Natural Science Foundation of China (No. 81472186).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Contributor Information
Chuanhao Tang, Email: tangchuanhao@pkuih.edu.cn.
Jun Liang, Email: liangjun1959@aliyun.com.
References
- 1.Marur S, Forastiere AA Head and neck squamous cell carcinoma: update on epidemiology, diagnosis, and treatment. Mayo Clin Proc. 2016;91:386–96. doi: 10.1016/j.mayocp.2015.12.017. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Sun K, Zheng R, et al Cancer incidence and mortality in China, 2014. Chin J Cancer Res. 2018;30:1–12. doi: 10.21147/j.issn.1000-9604.2018.01.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Price KA, Cohen EE Current treatment options for metastatic head and neck cancer. Curr Treat Options Oncol. 2012;13:35–46. doi: 10.1007/s11864-011-0176-y. [DOI] [PubMed] [Google Scholar]
- 4.Specenier P, Vermorken JB Optimizing treatments for recurrent or metastatic head and neck squamous cell carcinoma. Expert Rev Anticancer Ther. 2018:901–15. doi: 10.1080/14737140.2018.1493925. [DOI] [PubMed] [Google Scholar]
- 5.Concu R, Cordeiro MNDS Cetuximab and the head and neck squamous cell cancer. Curr Top Med Chem. 2018;18:192–8. doi: 10.2174/1568026618666180112162412. [DOI] [PubMed] [Google Scholar]
- 6.Bonner JA, Harari PM, Giralt J, et al Radiotherapy plus cetuximab for squamous-cell carcinoma of the head and neck. N Engl J Med. 2006;354:567–78. doi: 10.1056/NEJMoa053422. [DOI] [PubMed] [Google Scholar]
- 7.Foster I Cancer: A cell cycle defect. Radiography. 2008;14:144–9. doi: 10.1016/j.radi.2006.12.001>. [DOI] [Google Scholar]
- 8.Garrett MD, Fattaey A CDK inhibition and cancer therapy. Curr Opin Genet Dev. 1999;9:104–11. doi: 10.1016/S0959-437X(99)80015-X. [DOI] [PubMed] [Google Scholar]
- 9.Fischer PM, Lane DP Inhibitors of cyclin-dependent kinases as anti-cancer therapeutics. Curr Med Chem. 2000;7:1213–45. doi: 10.2174/0929867003374048. [DOI] [PubMed] [Google Scholar]
- 10.Senderowicz AM, Sausville EA Preclinical and clinical development of cyclin-dependent kinase modulators. J Natl Cancer Inst. 2000;92:376–87. doi: 10.1093/jnci/92.5.376. [DOI] [PubMed] [Google Scholar]
- 11.Sherr CJ, Beach D, Shapiro GI Targeting CDK4 and CDK6: from discovery to therapy. Cancer Discov. 2016;6:353–67. doi: 10.1158/2159-8290.CD-15-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cancer Genome Atlas Network Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–82. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ritta M, De Andrea M, Mondini M, et al Cell cycle and viral and immunologic profiles of head and neck squamous cell carcinoma as predictable variables of tumor progression. Head Neck. 2009;31:318–27. doi: 10.1002/hed.20977. [DOI] [PubMed] [Google Scholar]
- 14.Kalu NN, Johnson FM Do CDK4/6 inhibitors have potential as targeted therapeutics for squamous cell cancers? Expert Opin Investig Drugs. 2017;26:207–17. doi: 10.1080/13543784.2017.1274731. [DOI] [PubMed] [Google Scholar]
- 15.Michel L, Ley J, Wildes TM, et al Phase I trial of palbociclib, a selective cyclin dependent kinase 4/6 inhibitor, in combination with cetuximab in patients with recurrent/metastatic head and neck squamous cell carcinoma. Oral Oncol. 2016;58:41–8. doi: 10.1016/j.oraloncology.2016.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Subramanian A, Tamayo P, Mootha VK, et al Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leemans CR, Braakhuis BJ, Brakenhoff RH The molecular biology of head and neck cancer. Nat Rev Cancer. 2011;11:9–22. doi: 10.1038/nrc2982. [DOI] [PubMed] [Google Scholar]
- 18.Kobayashi K, Hisamatsu K, Suzui N, et al A review of HPV-related head and neck cancer. J Clin Med. 2018;7:pii: E241. doi: 10.3390/jcm7090241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang W, Edwards A, Fang Z, et al Integrative genomics and transcriptomics analysis reveals potential mechanisms for favorable prognosis of patients with HPV-positive head and neck carcinomas. Sci Rep. 2016;6:24927. doi: 10.1038/srep24927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reed AL, Califano J, Cairns P, et al High frequency of p16 (CDKN2/MTS-1/INK4A) inactivation in head and neck squamous cell carcinoma. Cancer Res. 1996;56:3630–3. [PubMed] [Google Scholar]
- 21.Ikeda Y, Oda K, Ishihara H, et al Prognostic importance of CDK4/6-specific activity as a predictive marker for recurrence in patients with endometrial cancer, with or without adjuvant chemotherapy. Br J Cancer. 2015;113:1477–83. doi: 10.1038/bjc.2015.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tadesse S, Yu M, Kumarasiri M, et al Targeting CDK6 in cancer: State of the art and new insights. Cell Cycle. 2015;14:3220–30. doi: 10.1080/15384101.2015.1084445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Ruiter EJ, Ooft ML, Devriese LA, et al The prognostic role of tumor infiltrating T-lymphocytes in squamous cell carcinoma of the head and neck: A systematic review and meta-analysis. Oncoimmunology. 2017;6:e1356148. doi: 10.1080/2162402X.2017.1356148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kollmann K, Heller G, Schneckenleithner C, et al A kinase-independent function of CDK6 links the cell cycle to tumor angiogenesis. Cancer Cell. 2016;30:359–60. doi: 10.1016/j.ccell.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang H, Langenkamp E, Georganaki M, et al VEGF suppresses T-lymphocyte infiltration in the tumor microenvironment through inhibition of NF-κB-induced endothelial activation. FASEB J. 2015;29:227–38. doi: 10.1096/fj.14-250985. [DOI] [PubMed] [Google Scholar]