Abstract
Objective
The inter-α-trypsin inhibitor heavy chain 4 (ITIH4) protein is involved in the development of tumors. However, the relationship between ITIH4 and ovarian cancer (OC) has not been extensively examined. This study aimed to explore the effect of ITIH4 on OC and to identify its underlying mechanism.
Methods
Expressions of ITIH4 in OC tissues and cells were determined using quantitative reverse transcription polymerase chain reaction (RT-qPCR) and western blots. The function of ITIH4 in the OC cell line HO8910pm was tested via ITIH4 knockdown. The cell growth rate was measured using MTT and colony formation assays. Flow cytometry was performed to evaluate cell cycle progression. Cell migration and invasion abilities were observed using the transwell migration assay.
Results
ITIH4 was downregulated in OC tissues and cells. ITIH4 knockdown promoted cell growth and cell cycle progression. Consistent with these results, inhibition of ITIH4 in OC cells significantly increased cell migration and invasion abilities. Cox regression analysis suggests that ITIH4 expression alone is not a good predictor of the prognosis of malignant ovarian tumors in patients.
Conclusions
ITIH4 inhibits the progression of OC, suggesting that ITIH4 may be a useful biomarker for OC. This study may provide a potential novel target for the treatment of OC.
Keywords: Ovarian cancer, inter-α-trypsin inhibitor H4 protein, quantitative reverse transcription PCR, RNAi
Introduction
Ovarian cancer (OC) has the highest mortality rate among all gynecological malignancies, with a survival rate of approximately 40% after five years (1,2). In China, the mortality rate of OC patients ranks the second in female reproductive system (3). Since OC is generally asymptomatic in the early stage and plus lack of effective screening approaches, about 75% of OC patients are not diagnosed until the disease is advanced and the best window for successful treatment has disappeared (4). Therefore, finding sensitive biomarkers with higher diagnostic accuracy in the early screening process is urgent.
Our previous study identified and validated differences in certain proteins between epithelial ovarian carcinoma cells and healthy control cells. The increased protein fragment m/z 3272 in epithelial ovarian carcinoma cells was viewed as a potential new tumor biomarker (5). Further studies identified m/z 3272 as a fragment of inter-α-trypsin inhibitor heavy chain 4 (ITIH4), a protein that combines with other four homologous heavy chain and light chain molecules to form the inter-α-trypsin inhibitor (ITI) (6,7). Multiple reports suggest that ITIH4 may be involved in the pathogenesis of various cancers. Van den Broek et al. found that ITIH4 serum concentration was significantly increased in breast cancer patients compared to control patients (8). Abdullah-Soheimi et al. confirmed that a 39 kDa fragment of ITIH4 was detected in the urine of patients with ovarian carcinoma (9). These studies demonstrate that the fragments of ITIH4 could be used as complementary biomarkers in the development of new, noninvasive methods for the diagnosis and screening of ovarian carcinoma. However, the molecular mechanism that explains how ITIH4 regulates malignant ovarian tumors is unclear. This study measured ITIH4 in OC and analyzed the relationship between ITIH4 gene expression and the clinical pathology characteristics of malignant ovarian tumors. This study used siRNA technology to explore the clinical significance of ITIH4 in the biological mechanisms of OC.
Materials and methods
Clinical data
All patients in this study visited the Affiliated Tumor Hospital of Guangxi Medical University between January 2008 and January 2014. Information collected from all patients included age, International Federation of Gynecology and Obstetrics (FIGO) stage, surgery history, adjuvant therapy history, and relapse characteristics. During the study, 80 patients with malignant ovarian tumors were identified. The median age at diagnosis was 45 (range, 19−68) years old. Tumors were classified and graded according to World Health Organization (WHO) standards. The group consisted of 62 patients with malignant epithelial tumors and 18 patients with malignant non-epithelial tumors, as well as 46 patients with poorly differentiated tumors and 34 patients with well-differentiated tumors. The surgical-pathologic stage was determined according to the FIGO 2006 standard: 28 cases at stage I−II and 52 cases at stage III−IV. All patients with malignant ovarian tumors underwent cytoreductive surgery plus platinum-based chemotherapy. All patients received the longest possible follow-up. The median patient-survival rate was 29 months. Eight cases left the study. Twenty-four samples were collected from benign ovarian lesions of patients aged 45−66 years old, with an average age of 49.2 years old. Of those samples, 10 cases were benign cystadenomas and 14 cases were teratomas. Twenty-six samples were collected from normal ovarian tissue that came from uterine fibroids or patients who underwent hysterectomies or salpingo-oophorectomies between the ages of 47 and 69 years old, with an average age of 50.3 years old. A pathologist confirmed these normal samples. All samples were taken from specimens collected during surgery. This study was approved by the Ethics Committee of Guangxi Medical University Affiliated Tumor Hospital (No. LW2019004). Informed consents were obtained from the patients or their families.
Reagents
The First Strand cDNA Synthesis Kit was purchased from Fermentas Co., Ltd (St.Leon-Rot, Germany). The RT-PCR Reaction Kit was provided by Sangon Biological Engineering Technology Co., Ltd (Shanghai, China). The gel extraction kit was purchased from Tektronix Co., Ltd (Beijing, China). The human epithelial OC cell lines (A2780, SKOV3, HO8910, and HO8910pm) were provided by the laboratory of Guangxi Medical University Affiliated Tumor Hospital. The 2x Long Taq PCR Master Mix was purchased from Tiangen Biotech Co., Ltd (Beijing, China). The pTG19-T vector plasmid and T4 DNA ligase were provided by Takala Biotech Co., Ltd (Shiga, Japan). The pGPU6/GFP/Neo carrier plasmid was acquired from Gene Pharma Biotech Co., Ltd. (Shanghai, China). Fibronectin (FN), transwell chambers, and Matrigel were purchased from Costar (New York, USA).
Cell culture
Human OC cell lines (A2780, SKOV3, HO8910pm, and HO8910) were maintained in RIPA1640 (Gibco, USA) supplemented with 1% penicillin-streptomycin and 10% fetal bovine serum (FBS) at 37 °C with 5% CO2.
Quantitative reverse transcription polymerase chain reaction (RT-qPCR)
Total RNA was extracted with TRIzol reagent (Ambino, USA) and quantified on a NanoDrop 2000. Total RNA (1 μg) was reversely transcribed into cDNA with oligo-dT primers. RT-qPCR was carried out on an ABI Prism 7500 Fast System (Applied Biosystems, USA) with SYBR Premix Ex Taq II (Takara, Dalian, China). The reaction was performed in 25 µL. The reaction conditions were: 94 °C denaturation for 3 min; 35 cycles of 94 °C for 30 s, 61 °C for 30 s, and 72 °C for 20 s; then 72 °C final extension for 10 min. Primer sets used were as follows: for ITIH4, 5’-GGTCTCATCCCGATTTGC-3’ (forward), 5’-TGATCCCTGGGTAGGTCAT-3’ (reverse); for GAPDH, 5’-GAAGGTGAAGGTCGGAGT-3’ (forward), 5’-GAAGATGGTGATGGGATTTC-3’ (reverse). Data analysis was performed using the 2−ΔΔCt method. All amplification reactions were performed in triplicate.
Lentivirus production
siRNA was designed and synthesized by Gene Pharma Biotech Co., Ltd (Shanghai, China). The siRNA sequences used were: ITIH4-546: GGGAAGCCCTAATCAAGATCCTTCAAGAGAGGATCTTGATTAGGGCTTCCCTT; ITIH4-795: GATCGAGAACGGCTACTTTGTTTCAAGAGAACAAAGTAGCCGTTCTCGATCTT; ITIH4-917: GCACCTGCAGATGGACATTCATTCAAGAGATGAATGTCCATCTGCAGGTGCTT; ITIH4-1568: GGAAGCTGCCTACACAGAACATTCAAGAGATGTTCTGTGTAGGCAGCTTCCTT; GAPDH: GTTCTCCGAACGTGTCACGTCAAGAGATTACGTGACACGTTCGGAGAATT; NC: GTTCTCCGAACGTGTCACGTCAAGAGATTACGTGACACGTTCGGAGAATT. GAPDH antisense RNA was used as a positive control. An unrelated scramble was used as a negative control (ITIH4-NC). Cells were plated in tissue culture flasks and incubated at 37 °C overnight. Cells were then transfected with Lipofectamine 2000 (Invitrogen) using 1 g of the siRNA, scrambled negative control, or positive control.
Cell growth assays
Growth of the attached OC cells was assessed using the MTT assay. Plates with cells were incubated for up to 7 d with inhibitors or vehicle supplemented every 3−4 d. MTT assay was used to assess anchorage-independent cell growth. Cells were seeded in 96-well plates pre-coated with 0.5% soft agar in 10% RPMI plus 0.3% soft agar at a density of 1,000−2,000 cells per 50 μL per well, and incubated overnight at 37 °C. The cell density in treated wells was expressed as a percent of the vehicle-treated control wells. The experiments included duplicate samples and were repeated at least three times.
Colony forming assay
A total of 100 stably transfected cells were seeded per well in a 24-well plate. Cells were incubated at 37 °C in a humidified incubator containing 5% CO2. After 5−7 d, colonies were stained with methanol and Giemsa. Colonies in each well were counted under a microscope. A group of 15−50 cells was defined as a standard colony. Colony formation rate (%) = number of colonies/seeded cells × 100%.
Flow cytometry
Cells were collected, washed twice with phosphate buffer saline (PBS), and fixed in ice-cold 70% ethanol overnight at 4 °C. The cells were then resuspended and filtered through a size 400 mesh sieve. Next, the cells were exposed to a propidium iodide solution at 37 °C for 30 min. Finally, the cells were run on a FACS Calibur Flow Cytometer (BD Biosciences, USA) for cell cycle phase analysis.
Invasion and migration assays
Cell invasion was evaluated in a 24-well Transwell plate system. The Transwell unit was loaded with PBS containing fibronectin (5 μg in 50 μL, Sigma, USA), and then Matrigel (Costar, USA) was added to the upper chamber of the Transwell unit before incubation at 37 °C for 4−5 h. Cells were seeded into the upper chamber of the Transwell unit at a density of 2.5×104 cells per well and incubated in RPMI 1640 medium for 20−24 h. The cells in the upper and lower chambers were evaluated by MTT assays and the absorbance was read at 490 nm. Invasive ability of cells (%) = upper chamber (490 nm)/lower chamber (490 nm) × 100%. Cell migration ability was assessed using the same method described for invasion assays, except that no Matrigel was added to the upper chamber. Migration ability of cells (%) = upper chamber (490 nm)/lower chamber (490 nm) × 100%.
Statistical analysis
All statistical data were calculated using SPSS 13.0 statistical software (SPSS Inc., Chicago, IL, USA). Data were compared with analysis of variance (ANOVA), Kruskal-Wallis, unpaired t-tests, Mann-Whitney, or Wilcoxon signed rank tests, as appropriate. ANOVA analyses or Chi-square tests were performed to identify differences. The results were defined as significant when P<0.05.
Results
Expression level of ITIH4 mRNA in OC
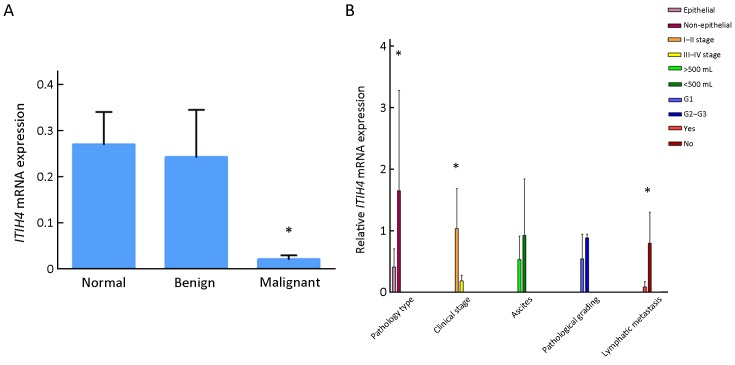
As shown in Figure 1A , the mRNA expression level of ITIH4 was the lowest in the OC (malignant group) samples, and relatively higher in the samples of patients with benign ovarian lesions, and even higher in the samples of the normal group (P<0.05).
1.
Expression of inter-α-trypsin inhibitor heavy chain 4 (ITIH4) mRNA in different tissues and comparison of it with different clinicopathologic factors. (A) Quantitative reverse transcription polymerase chain reaction (RT-qPCR) detects the expression of ITIH4 mRNA in different tissues; the mRNA expression of ITIH4 was the lowest in ovarian cancer (malignant group), compared with benign ovarian lesions and normal ovarian lesions. *, P<0.05; (B) Comparison of ITIH4 expression with different clinicopathological factors. Epithelialvs. Non-epithelial, P=0.015; Stage I−IIvs. Stage III−IV, P=0.026; Lymphatic metastasisvs. No lymphatic metastasis, P=0.042. *, P<0.05.
Expression of ITIH4 in cancerous ovarian tissue is correlated with clinicopathologic factors
OC progress is divided into four stages according to the FIGO 2006 standard. The expression of ITIH4 in stages III−IV was lower than that in stages I−II (Figure 1B ). Samples with lymph node metastasis have lower ITIH4 than those without lymph node metastasis (P<0.05). However, no differences were observed in the expression of ITIH4 based on the different pathologic grade and ascites (P>0.05) (Figure 1B ).
Correlation of ITIH4 expression in OC tissue with prognosis of patients
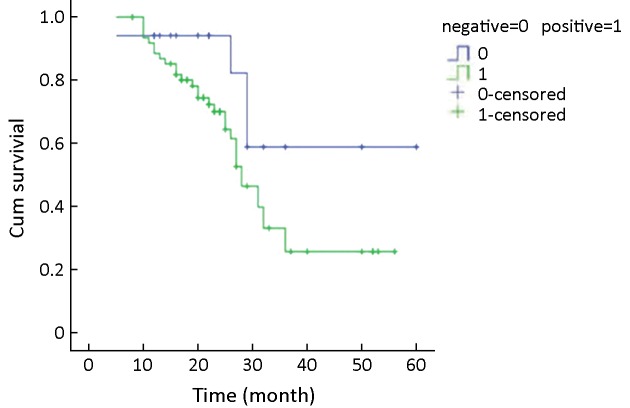
A receiver operating characteristic (ROC) curve was created to set the sum of the degree of sensitivity and specific highs (0.0159) to a positive threshold, therefore making it positive below this value and negative above this value. A Kaplan-Meier survival curve (Figure 2 ) showed the median survival time of 28 months for the positive group and 29 months for the negative group. There was no significant difference between the groups (P>0.05).
2.
Relationship between inter-α-trypsin inhibitor heavy chain 4 (ITIH4) expression and prognosis in ovarian cancer tissue. Kaplan-Meier survival curves were drawn and results showed that there was no significant difference in median survival time between positive group and negative group.
The Cox regression model was applied for the analysis of multiple prognosis factors, including age, pathologic grade, clinical stage, metastasis, ascites, residual foci, carbohydrate antigen 125 (CA125) levels, ITIH4 expression levels, and ITIH4 positive expression (Table 1 ). The results showed that the clinical stage, pathologic grades, and pelvic metastasis were the factors affecting the prognosis of patients with OC (P<0.05), but ITIH4 expression was not the determining factor (P>0.05). These results suggest thatITIH4 gene is not an independent factor affecting the prognosis of OC patients.
1.
Multivariate analysis of prognostic factors for patients with ovarian cancer
| Clinical factors | β | SE | Wald | df | Sig. | Exp (B) | 95% CI for Exp (B) | |
| Lower | Upper | |||||||
| CA, carbohydrate antigen; ITIH4, inter-α-trypsin inhibitor heavy chain 4; SE, standard error; 95% CI, 95% confidence interval. | ||||||||
| Clinical staging | −1.044 | 0.461 | 5.133 | 1 | 0.023 | 0.352 | 0.143 | 0.869 |
| Pathologic types | −1.140 | 0.593 | 3.699 | 1 | 0.054 | 0.320 | 0.100 | 1.022 |
| Tissue grade | 1.392 | 0.533 | 6.817 | 1 | 0.009 | 4.021 | 1.415 | 11.430 |
| Ascites | 0.215 | 0.400 | 0.289 | 1 | 0.591 | 1.239 | 0.566 | 2.713 |
| Pelvic metastasis | −1.192 | 0.472 | 6.381 | 1 | 0.012 | 0.304 | 0.120 | 0.766 |
| Lymphatic metastasis | −0.447 | 0.422 | 1.119 | 1 | 0.290 | 0.640 | 0.280 | 1.463 |
| Residual | −0.149 | 0.554 | 0.072 | 1 | 0.788 | 0.862 | 0.291 | 2.550 |
| CA125 | 0.336 | 0.571 | 0.346 | 1 | 0.556 | 1.099 | 0.457 | 4.286 |
| Neoadjuvant chemotherapy | 0.093 | 0.265 | 0.124 | 1 | 0.725 | 1.097 | 0.653 | 1.843 |
| Age | 0.013 | 0.015 | 0.737 | 1 | 0.391 | 1.013 | 0.983 | 1.044 |
| ITIH4 expression quantity | −0.204 | 0.365 | 0.313 | 1 | 0.576 | 0.815 | 0.399 | 1.667 |
| ITIH4 expression | 0.982 | 0.570 | 2.972 | 1 | 0.085 | 2.669 | 0.874 | 8.151 |
Construction, transfection and identification of plasmids with downregulated ITIH4
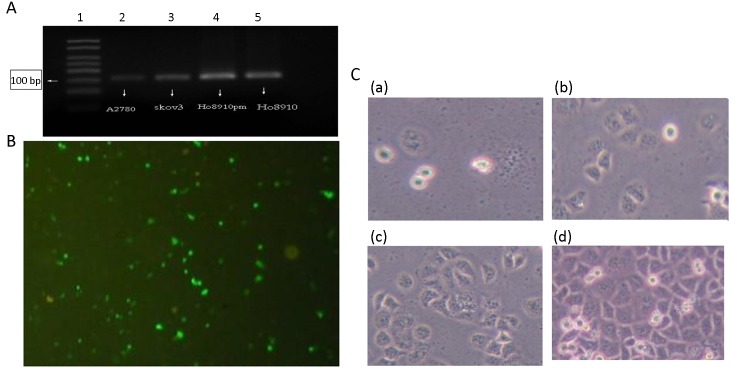
We observed the highest mRNA level of ITIH4 in HO8910pm cells (Figure 3A ) that were chosen for the knockdown experiment. Among the siRNAs designed to target ITIH4, the ITIH4-917 siRNA demonstrated the highest ITIH4-silencing rate of 87.2% [Silencing rate = (1−experimental group/control group) × 100%]. Thus, this fragment was selected to construct the ITIH4-silencing plasmid named pGPU6/GFP/Neo-shRNA-ITIH4-917. This plasmid was then transfected into HO8910pm cells with a transfection efficiency of about 60% after 48 h (Figure 3B ). After 14 d of G418 (800 μg/mL) selection, a single positive clone of pGPU6/GFP/Neo-shRNA-ITIH4-917-HO8910pm cells were harvested [Figure 3C(a) ], and as shown in Figure3C(b−d) , we can see the growth of monoclonal cells after 3 d, 7 d and 14 d of culture. At the same time, pGPU6/GFP/Neo-shRNA-HO8910pm control cells and pGPU6/GFP/Neo-shRNA-ITIH4-NC-HO8910pm control cells were obtained. As depicted in Figure 3C(d) , pGPU6/GFP/Neo-shRNA-ITIH4-917-HO8910pm cell colonies were successfully formed after G418 selection.
3.
Construction, transfection and identification of plasmids with downregulated inter-α-trypsin inhibitor heavy chain 4 (ITIH4). (A) ITIH4 gene was expressed in A2780, SKOV3, HO8910pm, HO8910 ovarian cancer cells with the highest expression in HO8910pm cells; (B) ITIH4-917 siRNA fragment had a maximal ITIH4-silencing rate of 87.2%. This fragment was selected to construct the ITIH4-silencing plasmid pGPU6/GFP/Neo-shRNA-ITIH4-917, and then transfected into HO8910pm cells with 60% transfection efficiency; (C) (a) represents the monoclonal pGPU6/GFP/Neo-shRNA-ITIH4-917-HO8910pm cells screened by G418 (800 μg/mL) selection after 14 d. (b−d) represent the state of monoclonal cells after 3 d, 7 d, and 14 d of culture, respectively. ×200.
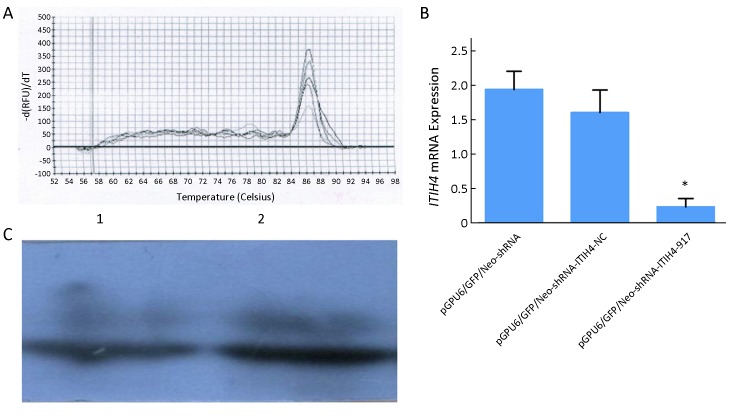
As shown in Figure 4A ,B , the ITIH4 mRNA level in pGPU6/GFP/Neo-shRNA-ITIH4-917-HO8910pm cells were lower than that in pGPU6/GFP/Neo-shRNA-ITIH4-NC-HO8910pm and pGPU6/GFP/Neo-shRNA-HO8910pm control cells (P=0.032 and P=0.008, respectively). Similar results were observed in protein level (Figure 4C ) (P1=0.008, P2=0.014).
4.
Confirmation of inter-α-trypsin inhibitor heavy chain 4 (ITIH4) knockdown. (A,B) Results of quantitative reverse transcription polymerase chain reaction (RT-qPCR) indicated that expression of ITIH4 mRNA in pGPU6/GFP/Neo-shRNA-ITIH4-917-HO8910pm cells was lower than that in pGPU6/GFP/Neo-shRNA-ITIH4-NC-HO8910pm and pGPU6/GFP/Neo-shRNA-HO8910pm cells. *, P<0.05; (C) Western blot indicated that the expression of ITIH4 protein in pGPU6/GFP/Neo-shRNA-ITIH4-917-HO8910pm cells was lower than that in pGPU6/GFP/Neo-shRNA-ITIH4-NC-HO8910pm and pGPU6/GFP/Neo-shRNA-HO8910pm cells; the difference was statistically significant (P1=0.008, P2=0.014). 1, Expression of ITIH4 protein in pGPU6/GFP/Neo-shRNA-ITIH4-917 group; 2, Expression of ITIH4 protein in pGPU6/GFP/Neo-shRNA-ITIH4-NC group.
Cell growth
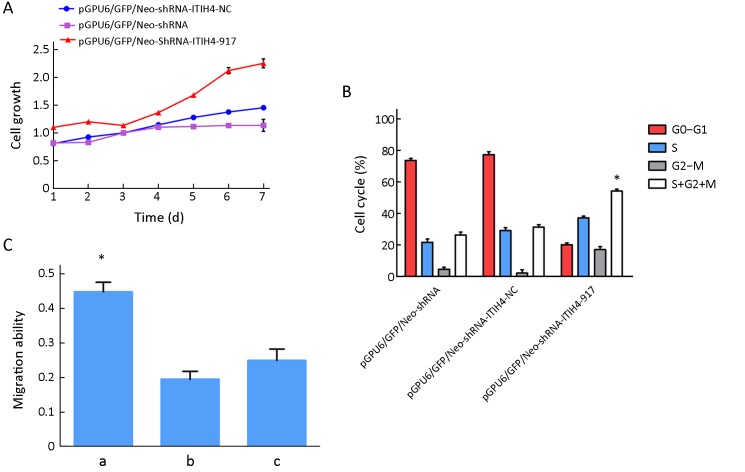
As shown in Figure 5A , MTT assay indicated increased cell viability in pGPU6/GFP/Neo-shRNA-ITIH4-917-HO8910pm cells compared to the empty vector and negative control groups.
5.
Role of inter-α-trypsin inhibitor heavy chain 4 (ITIH4) knockdown in cell growth, cell cycle, migration and invasion. (A) Cell growth was assessed by MTT. Blue line represents the growth curve of pGPU6/GFP/Neo-shRNA-ITIH4-NC; red line represents the growth curve of pGPU6/GFP/Neo-shRNA-ITIH4-917 and the purple line represents the growth curve of pGPU6/GFP/Neo-shRNA. Cell growth rates were increased in pGPU6/GFP/Neo-shRNA-ITIH4-917-HO8910pm cells compared with the other two groups; (B) Cell phase of S+G2+M was significantly increased in pGPU6/GFP/Neo-shRNA-ITIH4-917-HO8910pm cells. The rate of S+G2+M phase cells between the other two groups was no difference. *, P<0.05; (C) Cell migration abilities. a, pGPU6/GFP/Neo-shRNA-ITIH4-917-HO8910pm cells; b, pGPU6/GFP/Neo-shRNA-ITIH4-NC-HO8910pm cells; c, pGPU6/GFP/Neo-shRNA-HO8910pm cells. Compared a with b, P<0.05; Compared a with c, P<0.05; Compared b with c, P>0.05. *, P<0.05.
Colony formation
The colony formation rate of pGPU6/GFP/Neo-shRNA-ITIH4-917-HO8910pm cells (55.7±0.7)% was higher than those of pGPU6/GFP/Neo-shRNA-HO8910 cells (29.7±0.9)% and pGPU6/GFP/Neo-shRNA-ITIH4-NC-HO8910pm cells (31.4±0.2)%. The differences were statistically significant (P=0.043 and P=0.037, respectively), and there was no difference between the empty vector group and the negative control group.
Cell cycle
The percent of cells in G0/G1, S, or G2-M phase is shown in Figure 5B . The proportion of cells in the S+G2+M phases was significantly increased in pGPU6/GFP/Neo-shRNA ITIH4-917-HO8910pm cells compared to control groups (P<0.05). The number of cells in the S+G2+M phases was not significantly different between the two control groups.
Cell migration and invasion abilities
Inhibition of ITIH4 expression by shRNA significantly enhanced cells migration (0.40±0.18) compared to pGPU6/GFP/Neo-shRNA-ITIH4-NC-HO8910pm and pGPU6/GFP/Neo-shRNA-HO8910pm cells (P=0.028 and P=0.031, respectively) (Figure 5C ). There was no difference between the empty vector group and the negative control group. In addition, there was no significant difference in the invasive ability of cells among these three groups.
Discussion
ITI protein is a rich plasma protein whose physiological function has just begun to be discovered. Previous studies have shown that ITI helps maintain the stability of the extracellular matrix (ECM) (10) and functions as a potential modulator of ECM (11). Our work has verified and validated differential expression of ITI in the serum of patients with epithelial OC. We also found that the m/z 3272 fragment of ITIH4 is upregulated in OC (2) and that the C-terminal poly-proline region within ITIH4 is a conserved and ethnic-specific region that has been shown to be closely related to a variety of tumors (12). Zhang et al. found that a cleavage fragment of ITIH4 was upregulated in the early stages of OC (13). Other researchers have found that the m/z 2358 fragment of ITIH4 is significantly elevated in breast cancer (14), and that multiple ITIH4 fragments (m/z 3272, 2358, and 3156) are significantly increased in prostate cancer and other tumors (15). These studies suggest that ITIH4 plays a role in the development of malignant tumors, and that a large amount of protein may bind to the C-terminal poly-proline region in ITIH4. In addition, different ITIH4 cleavage fragments appear at different stages of tumor progression accompanied by different ITIH4 levels. However, the mechanism for explaining the relationship between cleavage fragments and biological effects remains unclear.
Hamm et al. found that ITIH genes are downregulated in multiple human tumors, including breast, colon, and lung cancer (16). Loss of ITIH2 expression was determined in 70% of breast cancer cases (n=50, CPA). Thus, ITIH genes may represent a family of putative tumor suppressors. We showed that ITIH4 gene expression in malignant OC is downregulated compared to benign ovarian tumors and healthy control samples. In patients with late clinical stage OC or abdominal cavity lymph node metastasis, ITIH4 expression was lower than patients with early clinical stage or no abdominal cavity lymph node metastasis (P<0.05). These findings suggest that ITIH4 may be cleaved during translation process to increase amounts of the m/z 3272 fragment. Interestingly, ITIH4 expression can be used to determine the features of benign and malignant ovarian tumors through the calculation of the area under the ROC curve. The lower the expression of ITIH4 is, the greater the likelihood of developing a malignant tumor will be. It would be interesting to study whether ITIH functions as tumor suppressor in tumor development.
There is evidence that the role of ITIH4 in cancer is mainly associated with its interaction with ECM. The proteolytic destruction of ECM is often the first sign of tumor invasion. During tumor metastasis, the ECM can be hydrolyzed by metalloproteinases and serine proteases secreted by tumor cells (17). Schmarier et al. found that hyaluronic acid binding protein is the serine protease that degrades the single-chain urokinase plasminogen activator (PHBP) and produces some dimers of fragments derived from ITIH proteins (18). This process activates fibrinolytic pathways and induces ECM degradation, which in turn causes ECM collapse and channel formation that allow the accumulation of a large number of tumor cells, thereby promoting the invasion of tumor cells into normal adjacent tissues (12). Therefore, it suggests that the protease inhibitory activity of ITIH4 can prevent proteolytic destruction of ECM inhibiting tumor cell growth and metastasis. We observed that the expression of ITIH4 gene in malignant ovarian tumors was lower than that in benign ovarian tumors and normal ovarian tissues, which was inconsistent with our previous findings. This may be due to the development of tumor growth and the destruction of ECM.
Song et al. reported that ITIH4 is a precursor of the plasma kallikrein activity peptide that is highly sensitive to plasma kallikrein (12). By regulating vascular structure and disease procession, plasma kallikrein participates in blood coagulation and fibrinolysis via the release of bradykinin. Upon clinical detection of peripheral ITIH4 cleavage fragments, the measured value of ITIH4 levels is relatively stable regardless of serum collection and measurement methods. It may be able to replace the endogenous proteases as a diagnostic marker of OC. In this study, although ITIH4 gene expression was not found to be an independent prognostic indicator for OC, its expression was significantly correlated with cell invasion, metastasis, histological differentiation and clinical stage. Meanwhile, these data suggest that ITIH4 plays a role in the development, invasion and metastasis of OC, and could be utilized as a potential tumor marker for diagnosis.
The mechanism of ITIH4 function as a tumor suppressor remains unclear. Given that ITIH4 is a potential regulator of ECM, this study investigated its role in migration and invasion by knockdown ITIH4. This study revealed that ITIH4 knockdown in HO8910pm cells significantly increased migration without any change in invasion. Previous studies have found that the inhibition of growth and metastasis of tumor cells is due to the stability of ECM, which is mediated by ITIH4 though covalent binding interactions with hyaluronic acid (19). Zhuo et al. found that down-conformation of hyaluronic acid may lead to angiogenesis (20). Because the stabilizing effect of hyaluronic acid on ECM largely depends on ITIH, downregulation of ITIH4 may play a role in tumor vascularization. We reported that ITIH4 fragments were significantly increased in the peripheral blood of OC patients, but ITIH4 mRNA level was significantly reduced in tumor tissues. It should be noted that ITIH4 binds to hyaluronic acid as a structural protein to maintain the stability of the ECM, thereby inhibiting the growth and metastasis of tumor cells. Thus, ITIH4 may be cleaved in the presence of carcinogens to increase the number of sheared ITIH4 fragments in the peripheral blood of OC patients, although the mRNA level of ITIH4 in tumor tissues is significantly reduced. Furthermore, the decrease in covalent binding interactions between ITIH4 and hyaluronic acid promotes the invasion and metastasis of tumor cells.
Conclusions
The decreased ITIH4 mRNA expression in OC is correlated with tumorigenesis and progression of OC. ITIH4 knockdown promoted cell growth and migration in OC cells. Meanwhile, these results suggest that ITIH4 is involved in the development, invasion and metastasis of OC and may represent a potential marker for tumor diagnosis.
Acknowledgements
This study was supported by Guangxi Scientific Research and Technology Development Project (No. 14124004-1-24).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Torre LA, Trabert B, DeSantis CE, et al Ovarian cancer statistics, 2018. CA Cancer J Clin. 2018;68:284–96. doi: 10.3322/caac.21456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wright JD, Chen L, Hou JY, et al Association of hospital volume and quality of care with survival for ovarian cancer. Obstet Gynecol. 2017;130:545–53. doi: 10.1097/AOG.0000000000002164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen W, Sun K, Zheng R, et al Cancer incidence and mortality in China, 2014. Chin J Cancer Res. 2018;30:1–12. doi: 10.21147/j.issn.1000-9604.2018.01.01>. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chien J, Poole EM Ovarian cancer prevention, screening, and early detection: report from the 11th biennial ovarian cancer research symposium. Int J Gynecol Cancer. 2017;27:S20–S2. doi: 10.1097/IGC.0000000000001118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Q, Li L, Li DR, et al Identification of serum biomarkers for ovarian cancer using protein chips and time of flight mass spectrometry technology. Zhonghua Fu Chan Ke Za Zhi. 2006;41:544–8. [PubMed] [Google Scholar]
- 6.Cai T, Yu P, Monga SP, et al Identification of mouse itih-4 encoding a glycoprotein with two EF-hand motifs from early embryonic liver. Biochim Biophys Acta. 1998;1398:32–7. doi: 10.1016/S0167-4781(98)00049-9. [DOI] [PubMed] [Google Scholar]
- 7.Piñeiro M, Alava MA, González-Ramón N, et al ITIH4 serum concentration increases during acute-phase processes in human patients and is up-regulated by interleukin-6 in hepatocarcinoma HepG2 cells. Biochem Biophys Res Commun. 1999;263:224–9. doi: 10.1006/bbrc.1999.1349. [DOI] [PubMed] [Google Scholar]
- 8.van den Broek I, Sparidans RW, van Winden AW, et al The absolute quantification of eight inter-α-trypsin inhibitor heavy chain 4(ITIH4)-derived peptides in serum from breast cancer patients. Proteomics Clin Appl. 2010;4:931–9. doi: 10.1002/prca.201000035. [DOI] [PubMed] [Google Scholar]
- 9.Abdullah-Soheimi SS, Lim BK, Hashim OH, et al Patients with ovarian carcinoma excrete different altered levels of urine CD59, kininogen-1 and fragments of inter-alpha-trypsin inhibitor heavy chain H4 and albumin. Proteome Sci. 2010;8:58. doi: 10.1186/1477-5956-8-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Venna VR, Roy-O’Reilly MA, Howe MD, et al Circulating Inter-alpha inhibitor protein levels decline after stroke in humans: exogenous supplementation is protective in multiple animal models of ischemic stroke. Stroke. 2018;49:A71. doi: 10.1161/str.49.suppl_1.71>. [DOI] [Google Scholar]
- 11.Barrios-Anderson A, Chen X, Lim YP, et al Inter-alpha inhibitor proteins reduce the presence of neutrophils and matrix metalloproteinase-9 positive neutrophils in damaged brain regions of neonates with hypoxic-ischemic (HI) brain injury. Stroke. 2017;48:AWP288. doi: 10.1161/str.48.suppl_1.60>. [DOI] [Google Scholar]
- 12.Song J, Patel M, Rosenzweig CN, et al Quantification of fragments of human serum inter-alpha-trypsin inhibitor heavy chain 4 by a surface-enhanced laser desorption/ionization-based immunoassay. Clin Chem. 2006;52:1045–53. doi: 10.1373/clinchem.2005.065722. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Z, Bast RC Jr, Yu Y, et al Three biomarkers identified from serum proteomic analysis for the detection of early stage ovarian cancer. Cancer Res. 2004;64:5882–90. doi: 10.1158/0008-5472.CAN-04-0746. [DOI] [PubMed] [Google Scholar]
- 14.van Winden AW, Gast MC, Beijnen JH, et al Validation of previously identified serum biomarkers for breast cancer with SELDI-TOF MS: a case control study. BMC Med Genomics. 2009;2:4. doi: 10.1186/1755-8794-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mohamed E, Abdul-Rahman PS, Doustjalali SR, et al Lectin-based electrophoretic analysis of the expression of the 35 kDa inter-alpha-trypsin inhibitor heavy chain H4 fragment in sera of patients with five different malignancies. Electrophoresis. 2008;29:2645–50. doi: 10.1002/elps.200700828. [DOI] [PubMed] [Google Scholar]
- 16.Hamm A, Veeck J, Bektas N, et al Frequent expression loss of Inter-alpha-trypsin inhibitor heavy chain (ITIH) genes in multiple human solid tumors: A systematic expression analysis. BMC Cancer. 2008;8:25. doi: 10.1186/1471-2407-8-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li X, Li B, Li B, et al ITIH4: effective serum marker, early warning and diagnosis, hepatocellular carcinoma. Pathol Oncol Res. 2018;24:663–70. doi: 10.1007/s12253-017-0285-4. [DOI] [PubMed] [Google Scholar]
- 18.Schmaier AH The plasma kallikrein-kinin system counterbalances the renin-angiotensin system. J Clin Invest. 2002;109:1007–9. doi: 10.1172/JCI15490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garantziotis S, Zudaire E, Trempus CS, et al Serum inter-alpha-trypsin inhibitor and matrix hyaluronan promote angiogenesis in fibrotic lung injury. Am J Resp Crit Care Med. 2008;178:939–47. doi: 10.1164/rccm.200803-386OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhuo L, Hascall VC, Kimata K Inter-alpha-trypsin inhibitor, a covalent protein-glycosaminoglycan-protein complex. J Biol Chem. 2004;279:38079–82. doi: 10.1074/jbc.R300039200. [DOI] [PubMed] [Google Scholar]