ABSTRACT
Malaria is an infectious disease caused by parasitic protozoa in the Plasmodium genus. A complete understanding of the biology of these parasites is challenging in view of their need to switch between the vertebrate and insect hosts. The parasites are also capable of becoming highly motile and of remaining dormant for decades, depending on the stage of their life cycle. Malaria elimination efforts have been implemented in several endemic countries, but the parasites have proven to be resilient. One of the major obstacles for malaria elimination is the development of antimalarial drug resistance. Ineffective treatment regimens will fail to remove the circulating parasites and to prevent the local transmission of the disease. Genomic epidemiology of malaria parasites has become a powerful tool to track emerging drug-resistant parasite populations almost in real time. Population-scale genomic data are instrumental in tracking the hidden pockets of Plasmodium in nationwide elimination efforts. However, genomic surveillance data can be useful in determining the threat only when combined with a thorough understanding of the malarial resistome – the genetic repertoires responsible for causing and potentiating drug resistance evolution. Even though long-term selection has been a standard method for drug target identification in laboratories, its implementation in large-scale exploration of the druggable space in Plasmodium falciparum, along with genome-editing technologies, have enabled mapping of the genetic repertoires that drive drug resistance. This Review presents examples of practical use and describes the latest technology to show the power of real-time genomic epidemiology in achieving malaria elimination.
KEY WORDS: Drug resistance, Genomic epidemiology, Malaria
Summary: This Review discusses the challenges in malaria elimination and how implementation of national-scale genomic surveillance programmes in combination with resistome analyses could provide a powerful solution.
Introduction
Malaria is a serious mosquito-borne tropical infectious disease caused by protozoa in the Plasmodium genus. Malaria elimination (see Glossary, Box 1) has become a real possibility once again after previous attempts have proven that elimination of malaria parasites on a global scale is elusive (Li et al., 2019). The new hope springs from an unprecedented scale of investment and commitment, resulting in new drug candidates and genetic toolkits considered unattainable just a decade ago. A new paradigm of nearly real-time genomic epidemiology has provided insights into emerging populations of drug-resistant parasites almost at the same time as their impact on the treatment outcome was recognized. The availability of large-scale genomic data arising from extensive surveillance networks is a critical element in the development of an effective reconnaissance strategy to ‘search and destroy’ every pocket of parasite transmission. Failure to respond to the looming threat from malaria drug resistance with effective treatment regimens and control measures would permit residual parasites to evade the elimination efforts. Thus, elimination plans can only succeed when the genomic surveillance data are paired with information on the molecular mechanisms underlying the modes of action, and the resistance mechanisms associated with existing antimalarial drugs and the new candidates in the research and development (R&D) pipeline.
Box 1. Glossary.
Antifolate: the drug family that inhibits folate production, a key substrate in DNA synthesis, making the drugs effective against actively propagating targets such as bacteria, cancer cells and parasites. The antimalarials pyrimethamine and sulfadoxine are antifolates.
Artemisinin: a sesquiterpene lactone compound with a peroxide bridge, originally extracted from Artemisia annua. It is a fast-acting antimalarial drug currently used for treating malaria worldwide. The success of this drug is one of the reasons behind the recent reduction in malaria mortality.
Chloroquine: a 4-aminoquinoline antimalarial drug that used to be a part of the treatment regimens due to its fast-acting and safety profile. Currently, the drug is ineffective for treating drug-resistant P. falciparum malaria, but is still the drug of choice for P. vivax malaria.
Druggable space: a compendium of biomolecules, mostly proteins that can be perturbed by (small) molecules to achieve desirable pharmacological properties. Not every essential gene product is druggable, since putative targets must provide a certain degree of specificity to avoid cross-reaction and toxicity.
Kelch: a large protein family containing the Kelch motif, a blade-like structure built from beta-sheets to form a propeller. A member of this family, Kelch13, has been linked to artemisinin resistance.
Malaria elimination: the reduction of locally transmitted malaria cases to zero in a defined geographical region. Any imported case needs to be managed quickly. This term should not be confused with eradication, which is a worldwide goal of zero incidence of malaria cases. Eradication can be achieved when the regional elimination efforts have been achieved.
piggyBac: a mobile transposable element originally isolated from a moth species. In conjunction with a transposase, it is a useful tool in genetic engineering due to its strong insertional mutagenesis activity.
Resistome: the compendium of all resistance genes including the precursor genes capable of evolving to cause drug resistance.
Tajima's D: a statistical test named after Fumio Tajima. It determines and compares the degree of genetic diversity in the population scale. The test can be used to indicate an evolutionary process under selection.
Saturation screen: a screen for the genetic factors capable of functionally perturbing a phenotype of interest. The screen is supposed to reach saturation when all genes are experimentally tested.
Zinc finger: one of the DNA-binding protein motifs. Its zinc-coordinated scaffold interacts with DNA with high specificity, which could be rationally engineered to recognize a specific region. It is the key sequence-recognition subunit of zinc finger nucleases (ZFNs), which can be customized to cut DNA at a predetermined site, enabling precise genome editing.
In recent years, novel genetic toolkits in combination with the classic long-term drug selection approach and the analysis of drug resistance evolution have become powerful tools in identifying drug targets. These approaches have also shed light on the evolutionary process by which parasites overcome antimalarial drugs. A series of long-term selection experiments have proven to be useful beyond drug target identification. On the surface, drug target identification and genomic surveillance are not related, but the two fields have become intertwined, affecting global malaria control efforts. Genome-wide screens exploring potential drug targets and resistance mechanisms have also extensively investigated their putative evolutionary paths to the point at which the data can now be combined with genomic surveillance to identify emerging parasite populations that carry existential threats to malaria treatment and control. This Review discusses how these powerful molecular toolkits can work side by side with genomic surveillance to accomplish malaria elimination.
A brief history of antimalarial drugs and their resistance
Malaria has left its footprint throughout the history of human civilization. Even during the past few hundred years, eight Presidents of the United States, including George Washington, were afflicted by the disease (https://www.impatientoptimists.org/Posts/2012/01/Cherry-Trees-Log-Cabins-and-Malaria). At present, almost half of the world population still live under the threat of malaria, with over 200 million cases every year, and antimalarial drugs remain the only proven effective means to treat patients (https://www.who.int/malaria/publications/world-malaria-report-2018/en/). The medical and research communities are currently working towards malaria elimination, and have already achieved a promising global reduction in the malaria mortality rate. However, almost all effective antimalarial drugs have been lost to drug-resistant parasites (White et al., 2014). Even though more new families of antimalarial drug candidates are available in the pipeline now than ever before, the emergence of new multidrug resistance in parasites could negate years of elimination efforts (Hooft van Huijsduijnen and Wells, 2018).
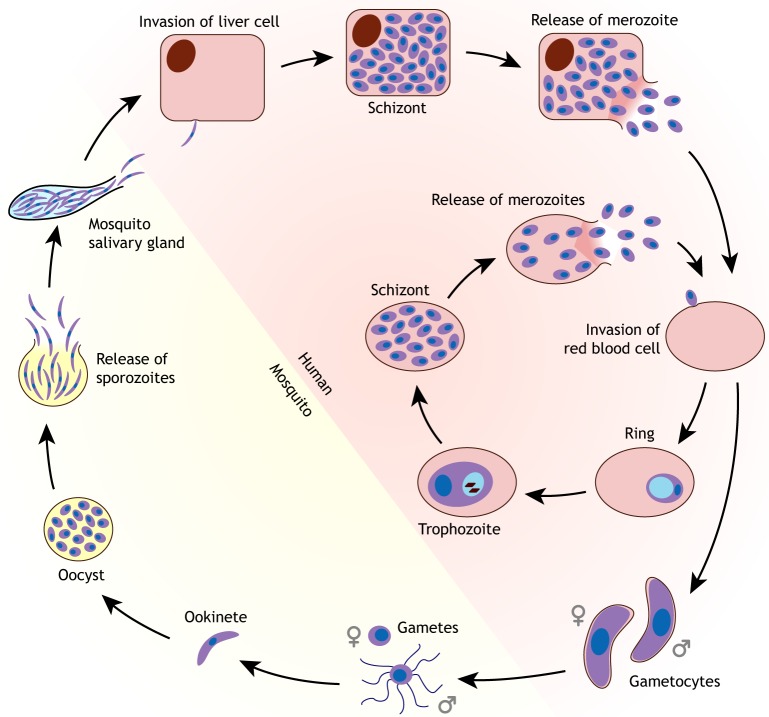
Human malaria is caused by the single-celled parasites in the genus Plasmodium; namely, Plasmodium falciparum, Plasmodium vivax, Plasmodium malariae and Plasmodium ovale (White et al., 2014). The zoonotic Plasmodium knowlesi, a monkey malaria parasite, was found to be able to infect humans living near the forest in Southeast Asia (Cox-Singh et al., 2008). Malaria parasites are transmitted between humans via blood feeding of female mosquitoes in the Anopheles species complex (Fig. 1) (Phillips et al., 2017). After entering the human host, the parasites first invade liver cells, multiply and egress to infect red blood cells (Fig. 1) (Phillips et al., 2017). The parasitic protozoa propagating and residing inside the human red blood cells are responsible for most of the malaria-related symptoms. P. falciparum malaria is the deadliest form of human malaria because patients can develop severe symptoms including coma, organ failure and anaemia (White et al., 2014).
Fig. 1.
Malaria life cycle. The parasitic protozoa switch between the vertebrate and the mosquito hosts. The sporozoite released via a mosquito bite enters the liver cell of the vertebrate host to produce progenies for invading the red blood cells. The majority of the parasites undergo the asexual cycle inside the vertebrate host. A portion of the parasites enter the sexual cycle, producing gametocytes for mating inside the mosquitoes. Most antimalarial drugs kill the blood-stage asexual parasites. Only primaquine and its derivatives (Box 1) are capable of killing the liver-stage and gametocytic stages of the parasites, thus preventing further transmission to the mosquito host.
During the 1960s, the availability of chloroquine and antifolates (Box 1), in addition to active dichlorodiphenyltrichloroethane (DDT) mosquito spraying, gave an illusion of worldwide malaria elimination (Nájera et al., 2011). However, the failure of this elimination campaign resulting from the spread of multidrug-resistant parasites was a rude awakening, demonstrating how a few mutations in the parasite population can turn the tide, with millions of human lives lost as a result (White, 2012). Half a century later, our generation is equipped with several new drugs, giving us the same opportunity to eliminate malaria and to inevitably face the same challenge from new drug-resistant Plasmodium strains. During the past 50 years, studies on malaria drug resistance have focused on chloroquine, antifolate and, more recently, artemisinin (Box 1). Chloroquine and antifolate saved millions of lives during the primes of their clinical uses. At present, artemisinin combination therapy is considered the most effective regimen for P. falciparum malaria treatment (White, 2008). The drug was found to be highly effective in clearing the parasites, but has a short half-life. It therefore requires co-administration with another antimalarial drug with a long half-life. The key to the success of artemisinin is its fast parasite clearance time. Its implementation has saved millions and has become a hallmark for the triumph of traditional medicine in modern time (Su and Miller, 2015). The first sign of artemisinin resistance has recently emerged in malaria cases that require more time to clear the parasites when exposed to artemisinin. The delayed clearance following artemisinin treatment was first observed in few cases at the border between Cambodia and Thailand, but these parasites have now spread to many Greater Mekong Subregion countries in Southeast Asia (Amato et al., 2018; Dondorp et al., 2009; Imwong et al., 2017). It is obvious that the problem is growing, but the current artemisinin regimen and dosage can still treat malaria patients (Chookajorn, 2018). However, the spread of full-blown resistance to Africa could cost millions of lives. To prevent this catastrophe, drug efficacy and genomic surveillance projects have been ongoing in Southeast Asia. Control efforts also include the implementation of new and additional partner drugs to artemisinin. Mefloquine, piperaquine and lumefantrine have been the partner drugs of choice in Cambodia, Thailand and Laos, respectively. Pyronaridine is also being explored in a limited area. These partner antimalarial drugs are vital in protecting artemisinin from drug resistance, and any sign of ineffectiveness must be detected as quickly as possible. Nevertheless, little information is available for tracking drug targets and resistance mechanisms, and this includes drug candidates in the antimalarial R&D pipeline.
Exploring the resistome for malaria elimination
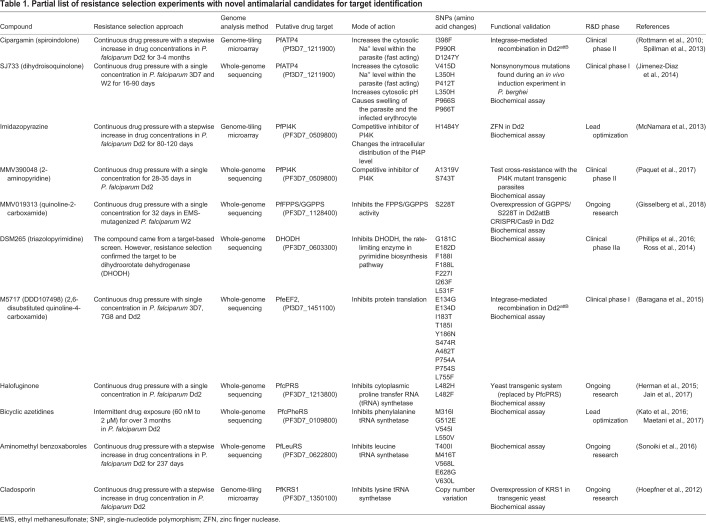
Long-term resistance selection is the experimental method of choice for target identification in malaria research (Luth et al., 2018). The selection process also reveals the resistance mechanisms for different antimalarial drugs. This drug pressure selection approach was originally employed with rodent and avian malaria parasites (Rollo, 1952), where drug-sensitive isolates were exposed to antimalarial drugs to select for resistant clones. For example, selection with antifolate pyrimethamine in Plasmodium chabaudi and Plasmodium yoelii gave rise to a mutation in the gene encoding dihydrofolate reductase, the target of pyrimethamine, and successfully selected for the S106N mutation, the equivalent of the critical pyrimethamine-resistance-associated S108N mutation in P. falciparum (Cheng and Saul, 1994; Cowman and Lew, 1990; Peterson et al., 1990). The modern standard for long-term drug selection relies on an in vitro culture system. The first in vitro resistance selection experiment in P. falciparum was reported in 1978 by continuously exposing the African FCR-3 line to chloroquine, yielding a stable phenotype capable of tolerating higher doses of chloroquine (Nguyen-Dinh and Trager, 1978). This finding forewarned the fate of chloroquine treatment in Africa. The common selection method is performed in batches and exposing the parasites to the drug either continuously or in pulses (Fig. 2). The surviving parasites are supposedly selected by acquiring mutations in the genes encoding either the drug target or a resistance mechanism. The individual resistant clones are isolated to ensure genetic homogeneity and validated by dose-response analysis. This approach reliably elucidated and confirmed the drug targets and the resistance pathways of multiple novel drug candidates in the pipeline (Table 1; the rationale behind target identification is explained in Box 2). From the examples shown in Table 1, there are several factors that could shape the selection process, including the drug concentration (incremental or fixed) and the exposure time (pulsed or prolonged) (Nzila and Mwai, 2010). There is no definite rule on the best selection protocol, and the selection workflow must be optimized for each drug.
Fig. 2.
Combination of long-term selection and genomic surveillance workflows. The selection workflow in the light-blue box represents a series of selections performed in parallel (R1, R2 and R3). The surviving parasites are cloned and tested for changes in drug sensitivity levels as determined by the half-maximal inhibitory concentration (IC50) analysis. The mutations arising from this selection process, especially those found in all the parallel selection experiments, are functionally validated by genome editing. The list of mutations can be used as a filter to flag emerging variants from genomic surveillance efforts in the field that use blood spot samples from malaria patients. These variants will be closely monitored and experimentally tested for their ability to alter drug sensitivity levels. This surveillance data will be converted into a report card summarizing the drug sensitivity profile and the parasite population structure to inform malaria treatment and elimination efforts.
Table 1.
Partial list of resistance selection experiments with novel antimalarial candidates for target identification
Box 2. Why we still need new drugs and more targets for malaria elimination.
Malaria is eliminated when there are no new cases from local transmission. During the past half-century, several countries, especially in the developed world, successfully eliminated malaria (Li et al., 2019). However, the elimination efforts at the epicentre of multidrug-resistant malaria in Southeast Asia have not been successful yet (Newby et al., 2015; Smith Gueye et al., 2014). A major limitation in malaria elimination is the failure to systemically kill parasites in two Plasmodium developmental stages; namely, the sexual gametocyte and liver stages (Fig. 1). This is one of the reasons why the modern-day standard for an ideal antimalarial drug requires multi-stage killing capacity with the emphasis on liver hypnozoites and sexual gametocytes (Burrows et al., 2017). Activities against both stages are instrumental in the global malaria elimination effort because the parasites in the liver hypnozoite form can lay hidden for years, and the unchallenged transition to sexually committed gametocytes could become an escape route to reach the mosquito host for another round of mating. The success of malaria elimination in developed countries relies on effective control measures blocking the cycle of transmission and preventing the outbreak from imported cases, a luxury unavailable to countries with limited resources and porous borders. The multiple candidate drug families currently under development are promising. As the targets for many of these drug candidates were already identified, one might ask why target identification is still needed. The answer to this question lies in the fact that many attractive drug candidates were discovered through high-throughput screening (Gamo et al., 2010; Guiguemde et al., 2010). The chemical libraries used for screening remain largely untapped resources that contain the compounds with desirable drug properties, and knowing their potential targets will allow researchers to monitor for drug resistance evolution. Another benefit of target identification is the opportunity to explore the druggable space of the precious known antimalarial drugs. For example, primaquine is the only drug capable of killing the liver-stage and gametocytic stages (Graves et al., 2018). Its 8-aminoquinoline scaffold is similar to a more classical antimalarial 4-aminoquinoline found in the widely used chloroquine, piperaquine and mefloquine (Vale et al., 2009). The mystery is why a simple switch in the position of a nitrogen atom in the quinoline scaffold could broaden its antimalarial activity. This question has a larger implication beyond an academic exercise. Even though primaquine is an ideal drug for radical treatment, it can cause a severe adverse effect in individuals with glucose-6-phosphate dehydrogenase (G6PD) deficiency (Howes et al., 2013), where the presence of metabolized primaquine was proposed to instigate oxidative damage, especially to the red blood cells with a compromised redox system from low G6PD (Ganesan et al., 2012). Conventional structure and activity relationship (SAR) analysis is a strategy for identifying the core pharmacophore conferring the drug activity and for removing undesirable chemical moieties causing adverse effects. Implementing this approach for primaquine would allow the development of its pharmacophore scaffold toward a safer drug for the population with G6PD deficiency. The question is how one can mimic the activity of 8-aminoquinoline. Unlocking the secret behind its unique killing activity requires the understanding of the mode of action and its putative targets.
The mutations found after drug selection need to be functionally validated. The malaria research field was limited by the availability of molecular tools to specifically modify the candidate genes. The application of zinc finger (Box 1) nuclease (ZFN) in P. falciparum provided a possible solution (Straimer et al., 2012). However, each ZFN needs to be custom tailored to the target DNA base pairs, making cost and reliability the major issues limiting widespread adoption (Bibikova et al., 2003; Shaw and Aroonsri, 2017). Recently, testing the functional significance of each variant has become easier and faster by the application of clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 genome editing in P. falciparum. This system can achieve genome editing in P. falciparum by introducing a point mutation and disrupting the gene of interest (Ghorbal et al., 2014; Wagner et al., 2014). The method requires three major components; namely, the Streptococcus pyogenes Cas9 (SpCas9) endonuclease, a single-guide RNA (sgRNA) and a donor DNA template. The sgRNA is designed to lead SpCas9 to the target genomic site using complementary base pairing. The donor template containing a mutation of choice is introduced into the genome at the SpCas9-induced breakage site by homology-directed repair. The benefit of CRISPR/Cas9 compared to conventional recombinant DNA technology is that the former does not leave an extra plasmid component in the chromosome. The dependency on matching nucleotides provides a cost-effective and versatile system for gene editing, surpassing ZFN. The feasibility of CRISPR/Cas9 in P. falciparum was first proven by testing the effect of kelch13 mutations (Box 1) on parasite susceptibility to artemisinin (Ghorbal et al., 2014). Subsequently, the CRISPR/Cas9 machinery was proven to be functional in P. falciparum without plasmid transformation: Crawford et al. (2017) used the Cas9-sgRNA ribonucleoprotein complex with a single-stranded oligodeoxynucleotide as a donor template to introduce mutations in the gene encoding the PfATP4 sodium transporter. As PfATP4 is the target of the new fast-acting antimalarial drug candidate SJ733, the mutations introduced by this plasmid-free approach reduced the antimalarial effect of SJ733 as expected. In addition, applying selection-linked integration (SLI) can significantly speed up the selection process of edited parasites (Birnbaum et al., 2017). SLI is a plasmid-based system that relies on the integration-linked resistance marker located at the 3′ end of the homologous template with a self-cleavage 2A peptide to separate the modified protein and the drug-resistant selection protein. This technique can improve the rate of the gene-editing process from months to just a few weeks, rendering the discovery of putative genetic markers of resistance more efficient.
Interestingly, the ease of genome editing in P. falciparum has expanded the scope of druggable space (Box 1) and resistance mechanisms. A network of researchers have systematically chosen potent antimalarial compounds and performed drug selection on a large scale (Antonova-Koch et al., 2018; Corey et al., 2016; Cowell et al., 2018). The emerging mutations were tested for their ability to shift drug sensitivity. The studies revealed new putative targets, including aminophospholipid-transporting P-type ATPase and farnesyltransferase (Cowell et al., 2018), and the approach has now been expanded to target multiple Plasmodium life-cycle stages, especially the sexual and liver stages, with a larger drug candidate collection (Antonova-Koch et al., 2018). A companion approach is genome-wide mutation screening, which has also played a prominent role in mapping drug targets and resistance mechanisms during the past 2 years. The piggyBac (Box 1) system uses a transposable element to disrupt functional genes (Balu et al., 2005). A saturation screen (Box 1) was performed by allowing piggyBac elements to randomly insert into the malarial genes and determine their dispensability. A collection of parasite clones with piggyBac insertion at various genes (∼40% of the P. falciparum genes) was exposed to artemisinin, and the abrogation of the genes related to the drug's mode of action and cellular response affected the drug sensitivity level (Zhang et al., 2018). This gene-based approach successfully identified the role of the proteasome degradation pathway, including that of Kelch13, in artemisinin sensitivity (Zhang et al., 2018). A similar large-scale knockout screen in Plasmodium berghei, a murine malaria parasite known to be more genetically malleable than its human counterparts, revealed the degree of dispensability of almost every malarial gene during the asexual blood stage (Bushell et al., 2017). These recent large-scale projects have allowed saturation screening to reach levels not possible in malaria research during the past decades and have opened up the druggable space, including the putative resistome of malaria parasites (Cowell and Winzeler, 2018). The concept of resistome (Box 1) originally arose from studies of bacterial antibiotics (Wright, 2007). The size and diversity of genomic data accelerate the discoveries of resistance genes and their precursors (Crofts et al., 2017). Structural and expression changes of the drug target are the most common evolutionary pathways to resistance. However, proto-resistance genes that have no direct role in the biochemical and physiological pathway could evolve to reduce the collateral biological damage caused by the drug (Crofts et al., 2017). Predicting all possible evolutionary pathways towards drug resistance in Plasmodium might seem farfetched, but recent reports on both long-term selection and genome-wide screening have already generated a plethora of data. The genes and variants identified in these studies could be developed into a sentinel filter, identifying the potential threats of emerging genetic variants as part of the global genomic malaria surveillance effort. For example, mutations in the genes encoding the ABC transporter PfABCI3 and amino acid transporter PfAAT1 were associated with reduced sensitivity for several antimalarial drug candidates (Cowell et al., 2018). These genes were not previously known to be linked with drug resistance, but their functions and broad biological effects warrant further functional studies.
Combining resistome research and genomic surveillance in malaria elimination
In order to appreciate the power of the malarial resistome in elimination efforts, the broader community needs to become aware of the unprecedented large-scale genomic surveillance efforts in several endemic countries. These involve mass blood spot collection from patients, making the malaria genome database one of the best genomic databases in terms of diversity and size (Box 3). Despite the initial success, implementation of genomic reconnaissance in malaria elimination needs to be precise and almost in real time to root out the last pockets of parasite populations. It is not an overstatement to say that the ‘last mile’ of malaria elimination is likely to be the hardest. Parties involved in malaria elimination can be lulled into false security if residual parasite populations eluding the efforts linger unnoticed in asymptomatic individuals and re-emerge once control measures are removed. The current phase of malaria genomic surveillance is fostered by collaborations at the national level. The GenRe-Mekong programme works with national malaria control programmes in the Greater Mekong Subregion in Southeast Asia to genotype malaria parasites nationwide in each country. The genomic data are translated into a report card summarizing drug sensitivity profile and population movement (Fig. 2). Actionable epidemiology data are purported to be a guide in malaria control and elimination for the national public health authorities based on the effective treatment regimen for each area and the origin of the circulating parasites. However, the scale of genomic epidemiology surveillance also means that a plethora of genetic variants that naturally emerge in Plasmodium populations will be reported. In the past, genetic association studies have identified mutations with strong linkage to drug resistance, revealing the causal mutations and/or putative resistance markers (Wang et al., 2016). The emerging variants are often fixed as dominating alleles due to their selective advantage from either conferring drug resistance or fitness compensation (Chookajorn and Kumpornsin, 2011). Routine genomic epidemiology surveillance will detect emerging variants instantaneously even before they become fixed in the population. In the most straightforward scenario, a specific drug-resistant allele is favoured within a population and undergoes a ‘hard’ sweep into fixation (Fig. 3) (Pritchard et al., 2010). However, several mutations could emerge in response to drug selection, and multiple evolutionary paths towards drug resistance or any advantageous traits could occur simultaneously without a clear winner. More than one single genetic variant from multiple genes or even within the same gene can accumulate over time. The ‘soft’ sweep scenario means that no clear selective signal will be detected during genomic surveillance, and it will take a relatively long time before a particular allele becomes a major feature in a parasite population (Fig. 3) (Pritchard et al., 2010). The common parameters for determining selection, such as Tajima's D (Box 1), are less useful in tracking the soft sweep (Fig. 3) (Messer and Petrov, 2013). The foreseeable challenge for large-scale genomic surveillance projects is to evaluate the rise of emerging variants in the context of antimalarial drug resistance. In fact, real success would be to identify potential threats from emerging alleles and to suppress them before a strong drug-resistance driver becomes fixed in the parasite population.
Box 3. A genetic variant call and database for the malaria research community.
At the start of malaria genomic research, the identification of P. falciparum genetic variants was achieved by polymerase chain reaction (PCR) amplification of the target region followed by Sanger sequencing. This workflow was sufficient for studying known polymorphic sites, but is not suitable for novel variant discovery. The implementation of next-generation sequencing technology has paved the way to track new variants on the genomic scale. The most popular next-generation sequencing technology for P. falciparum is the Illumina-based method. This method has satisfactorily produced the read quality suitable for the AT-rich P. falciparum genome. For the discovery of structural variants (duplications, large indels and translocations), Illumina sequencing can be combined with either nested PCR coupled to Sanger sequencing or with long-read sequencing methods such as PacBio. The long reads from PacBio can extend across the AT-rich regions, making the assembly of the whole chromosome possible (Otto et al., 2018). For genes known to be associated with drug resistance, an amplicon sequencing approach targets the regions of interest for sequencing, reducing the cost and the error by performing sequencing of indexed samples in a large batch.
Global collaborations have resulted in several Plasmodium variant collection databases. PlasmoDB (https://plasmodb.org/) provides access to the variant data from laboratory isolates and parasite collections. The P. falciparum Community Project (https://www.malariagen.net/projects/p-falciparum-community-project) and the Pf3k project (https://www.malariagen.net/projects/Pf3k) contain the variant and the geographical data from P. falciparum across the globe (Malaria Genomic Epidemiology Network, 2008). The January 2016 release of the P. falciparum Community Project contains genotype data from 3394 samples from 22 countries. The Pf3k project is a collaboration of research groups at the Broad Institute, the University of Oxford and the Wellcome Sanger Institute to comprehensively analyse the genome variations. The current release (Pf3k pilot data release 5) covers 2512 field isolates, five clonal samples of laboratory-adapted strains, 96 samples from the progenies of genetic crosses and 27 mixed samples of laboratory strains experimentally mixed together as controls. These databases are continuously updated, and additional data for further analysis can be found on their respective websites.
Fig. 3.
Hard and soft sweeps in drug resistance evolution. In the hard sweep scenario, the evolutionary process toward a drug-resistant phenotype can be driven by a few mutations, and they quickly dominate over other alleles in a population, giving a strong selective signature, as shown with a negative Tajima's D value (D). However, the evolutionary process is often less than ideal, and the accumulation of several mutations either within the same gene or in multiple genes contribute to the eventual drug resistance outcome, making the selective signature less obvious and less easy to track.
Functional validation, using the techniques discussed above, of emerging variants is instrumental in making sense of the surveillance data. Predicting the functional significance of emerging variants based on known resistome repertoires could provide a solution to this problem. This notion is not a mere abstract concept. In the early days of emerging artemisinin resistance, there was no clear genetic variant to be used as a candidate artemisinin resistance marker. It was obvious at the time that the degree of diversification suggested a strong selective pressure on the parasite populations in Cambodia, the site of emerging artemisinin resistance (Miotto et al., 2013). Genome-wide association studies identified candidates and narrowed the linkage sites to certain chromosomes (Cheeseman et al., 2012). An important consideration for this discussion is that emerging artemisinin resistance does not fit a conventional drug resistance phenotype (Chookajorn, 2018), which is defined as treatment failure at the recommended dosage and treatment regimen. Artemisinin and its derivatives can still kill parasites at the recommended dose, but the clearance time is delayed by hours (Ashley et al., 2014). The thorough and well-organized drug efficacy monitoring effort in Cambodia revealed a clear shift in clearance time, but the problem has been detected before the parasites reached full drug resistance (Ashley et al., 2014). As this ongoing artemisinin resistance evolution did not yet reach the end stage, the selection for a resistance genetic marker was less obvious than in typical full-blown drug resistance. The populations of P. falciparum in Cambodia originally emerged as multiple sympatric subpopulations (Miotto et al., 2013). They later evolved towards a shared genetic background with many genetic factors predisposing for artemisinin resistance (Miotto et al., 2015). To provide functional clues, researchers conducted a long-term selection experiment. A P. falciparum isolate from Africa was subjected to intermittent artemisinin pressure for 5 years (Ariey et al., 2014). During the selection process, parasites were periodically sampled for artemisinin sensitivity testing and genomic sequencing. The accumulation of novel mutations causing the gradual loss of artemisinin susceptibility revealed a series of evolutionary changes needed for emerging artemisinin resistance. As expected, multiple mutations arose throughout the selection period. The list of genes and mutations by itself was not informative, but, after matching them to the variants found in Cambodia, kelch13 mutations have become a notable candidate. By the degree of association alone, variants in other genes in the Cambodian population had statistical significance, but they were not found in the selection experiment. CRISPR/Cas9 genome editing experiments confirmed the effect of kelch13 mutations on improving parasite survival under artemisinin pulses (Ghorbal et al., 2014). kelch13 mutations alone are not likely to be the sole driver of artemisinin resistance, but the implementation of kelch13 mutations as a molecular marker played a pivotal role in tracking the spread of the parasites with reduced artemisinin sensitivity (Amato et al., 2018; Imwong et al., 2017; Sa et al., 2018; Wilairat et al., 2016). It is not yet possible to ignore the foreseeable threat of full-blown artemisinin resistance (Chookajorn, 2018). Nevertheless, the studies on kelch13 and its role in emerging artemisinin resistance have undoubtedly established a new paradigm for a combination of genomic surveillance in the field, with long-term drug selection and genome-editing experiments to prevent and mitigate the spread of drug-resistant pathogens.
The resistome data described above could be used to construct a filter to assess the emerging variants in different parasite populations. Mutations in the genes considered to be components of the malarial resistome need to be subsequently functionally validated, perhaps by CRISPR technology. One possible challenge is to validate a specific mutation among various genetic backgrounds. If parasites with a similar genetic background are cultivated, it will be ideal to introduce mutations in the same background to exclude any confounding effect from genetic interactions. Nevertheless, emerging variants might not be causal, and their roles in fitness compensation must be carefully considered. The story of kelch13 mutations provides an excellent example. These mutations alone have a relatively weak drug resistance selection effect, but they fit the clinical phenotype of artemisinin resistance. At this point, the significance of kelch13 as a molecular marker has already contributed significantly to the utility of the resistome in genomic surveillance and malaria elimination.
The ‘last mile’ of malaria elimination
It is undeniable that good science is vital to malaria elimination, but science alone will not translate into successful elimination. The challenge is to share the scientific data with national and regional control programmes, and to present the shared data in an actionable format beyond gigantic DNA sequence files and variant collections (Dalmat et al., 2019). Translation and distribution of big data in an actionable form such as a genetic report card would ensure that the genomic surveillance data become a part of the decision-making process in a national malaria elimination policy. It also means that the national control programmes in each malaria-endemic country have to form a local team capable of understanding the basic concepts of genomic epidemiology and implement it in an existing public health structure. This requires a capacity-building scheme to train the local workforce, either within a degree-granting system or as short-course training. The structure for any genomic surveillance programme is sustainable only when local workforce feels empowered and appreciates how the genetic data gathered benefit the health and welfare of their fellow compatriots.
Acknowledgements
We thank Drs Marcus Lee, Olivo Miotto and Prapon Wilairat for their invaluable advice. We are grateful for many excellent suggestions from the referee during the review process.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
This work was supported by a Newton Advanced Fellowship from the Thailand Research Fund/Royal Society (DBG5980010). K.K. was supported by a grant from the Bill and Melinda Gates Foundation (OPP1054480).
References
- Amato R., Pearson R. D., Almagro-Garcia J., Amaratunga C., Lim P., Suon S., Sreng S., Drury E., Stalker J., Miotto O. et al. (2018). Origins of the current outbreak of multidrug-resistant malaria in southeast Asia: a retrospective genetic study. Lancet Infect. Dis. 18, 337-345. 10.1016/S1473-3099(18)30068-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonova-Koch Y., Meister S., Abraham M., Luth M. R., Ottilie S., Lukens A. K., Sakata-Kato T., Vanaerschot M., Owen E., Jado J. C. et al. (2018). Open-source discovery of chemical leads for next-generation chemoprotective antimalarials. Science 362, eaat9446 10.1126/science.aat9446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariey F., Witkowski B., Amaratunga C., Beghain J., Langlois A. C., Khim N., Kim S., Duru V., Bouchier C., Ma L. et al. (2014). A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature 505, 50-55. 10.1038/nature12876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashley E. A., Dhorda M., Fairhurst R. M., Amaratunga C., Lim P., Suon S., Sreng S., Anderson J. M., Mao S., Sam B. et al. (2014). Spread of artemisinin resistance in Plasmodium falciparum malaria. N. Engl. J. Med. 371, 411-423. 10.1056/NEJMoa1314981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balu B., Shoue D. A., Fraser M. J. Jr and Adams J. H (2005). High-efficiency transformation of Plasmodium falciparum by the lepidopteran transposable element piggyBac. Proc. Natl. Acad. Sci. USA 102, 16391-16396. 10.1073/pnas.0504679102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baragana B., Hallyburton I., Lee M. C., Norcross N. R., Grimaldi R., Otto T. D., Proto W. R., Blagborough A. M., Meister S., Wirjanata G. et al. (2015). A novel multiple-stage antimalarial agent that inhibits protein synthesis. Nature 522, 315-320. 10.1038/nature14451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M., Beumer K., Trautman J. K. and Carroll D (2003). Enhancing gene targeting with designed zinc finger nucleases. Science 300, 764 10.1126/science.1079512 [DOI] [PubMed] [Google Scholar]
- Birnbaum J., Flemming S., Reichard N., Soares A. B., Mesén-Ramírez P., Jonscher E., Bergmann B. and Spielmann T (2017). A genetic system to study Plasmodium falciparum protein function. Nat. Methods 14, 450-456. 10.1038/nmeth.4223 [DOI] [PubMed] [Google Scholar]
- Burrows J. N., Duparc S., Gutteridge W. E., Hooft van Huijsduijnen R., Kaszubska W., Macintyre F., Mazzuri S., Möhrle J. J. and Wells T. N. C. (2017). New developments in anti-malarial target candidate and product profiles. Malar. J. 16, 26 10.1186/s12936-016-1675-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushell E., Gomes A. R., Sanderson T., Anar B., Girling G., Herd C., Metcalf T., Modrzynska K., Schwach F., Martin R. E. et al. (2017). Functional profiling of a plasmodium genome reveals an abundance of essential genes. Cell 170, 260-272.e8. 10.1016/j.cell.2017.06.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I. H., Miller B. A., Nair S., Nkhoma S., Tan A., Tan J. C., Al Saai S., Phyo A. P., Moo C. L., Lwin K. M. et al. (2012). A major genome region underlying artemisinin resistance in malaria. Science 336, 79-82. 10.1126/science.1215966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Q. and Saul A (1994). The dihydrofolate reductase domain of rodent malarias: point mutations and pyrimethamine resistance. Mol. Biochem. Parasitol. 65, 361-363. 10.1016/0166-6851(94)90087-6 [DOI] [PubMed] [Google Scholar]
- Chookajorn T. (2018). How to combat emerging artemisinin resistance: Lessons from “The Three Little Pigs”. PLoS Pathog. 14, e1006923 10.1371/journal.ppat.1006923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chookajorn T. and Kumpornsin K (2011). ‘Snakes and Ladders’ of drug resistance evolution. Virulence 2, 244-247. 10.4161/viru.2.3.16194 [DOI] [PubMed] [Google Scholar]
- Corey V. C., Lukens A. K., Istvan E. S., Lee M. C., Franco V., Magistrado P., Coburn-Flynn O., Sakata-Kato T., Fuchs O., Gnadig N. F. et al. (2016). A broad analysis of resistance development in the malaria parasite. Nat. Commun. 7, 11901 10.1038/ncomms11901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowell A. and Winzeler E (2018). Exploration of the plasmodium falciparum resistome and druggable genome reveals new mechanisms of drug resistance and antimalarial targets. Microbiol. Insights 11, 1178636118808529 10.1177/1178636118808529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowell A. N., Istvan E. S., Lukens A. K., Gomez-Lorenzo M. G., Vanaerschot M., Sakata-Kato T., Flannery E. L., Magistrado P., Owen E., Abraham M. et al. (2018). Mapping the malaria parasite druggable genome by using in vitro evolution and chemogenomics. Science 359, 191-199. 10.1126/science.aan4472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowman A. F. and Lew A. M (1990). Chromosomal rearrangements and point mutations in the DHFR-TS gene of Plasmodium chabaudi under antifolate selection. Mol. Biochem. Parasitol. 42, 21-29. 10.1016/0166-6851(90)90109-Y [DOI] [PubMed] [Google Scholar]
- Cox-Singh J., Davis T. M., Lee K. S., Shamsul S. S., Matusop A., Ratnam S., Rahman H. A., Conway D. J. and Singh B (2008). Plasmodium knowlesi malaria in humans is widely distributed and potentially life threatening. Clin. Infect. Dis. 46, 165-171. 10.1086/524888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford E. D., Quan J., Horst J. A., Ebert D., Wu W. and DeRisi J. L (2017). Plasmid-free CRISPR/Cas9 genome editing in Plasmodium falciparum confirms mutations conferring resistance to the dihydroisoquinolone clinical candidate SJ733. PLoS ONE 12, e0178163 10.1371/journal.pone.0178163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crofts T. S., Gasparrini A. J. and Dantas G (2017). Next-generation approaches to understand and combat the antibiotic resistome. Nat. Rev. Microbiol. 15, 422-434. 10.1038/nrmicro.2017.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmat R., Naughton B., Kwan-Gett T. S., Slyker J. and Stuckey E. M (2019). Use cases for genetic epidemiology in malaria elimination. Malar. J. 18, 163 10.1186/s12936-019-2784-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dondorp A. M., Nosten F., Yi P., Das D., Phyo A. P., Tarning J., Lwin K. M., Ariey F., Hanpithakpong W., Lee S. J. et al. (2009). Artemisinin resistance in Plasmodium falciparum malaria. N. Engl. J. Med. 361, 455-467. 10.1056/NEJMoa0808859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamo F. J., Sanz L. M., Vidal J., de Cozar C., Alvarez E., Lavandera J. L., Vanderwall D. E., Green D. V., Kumar V., Hasan S. et al. (2010). Thousands of chemical starting points for antimalarial lead identification. Nature 465, 305-310. 10.1038/nature09107 [DOI] [PubMed] [Google Scholar]
- Ganesan S., Chaurasiya N. D., Sahu R., Walker L. A. and Tekwani B. L (2012). Understanding the mechanisms for metabolism-linked hemolytic toxicity of primaquine against glucose 6-phosphate dehydrogenase deficient human erythrocytes: evaluation of eryptotic pathway. Toxicology 294, 54-60. 10.1016/j.tox.2012.01.015 [DOI] [PubMed] [Google Scholar]
- Ghorbal M., Gorman M., Macpherson C. R., Martins R. M., Scherf A. and Lopez-Rubio J. J (2014). Genome editing in the human malaria parasite Plasmodium falciparum using the CRISPR-Cas9 system. Nat. Biotechnol. 32, 819-821. 10.1038/nbt.2925 [DOI] [PubMed] [Google Scholar]
- Gisselberg J. E., Herrera Z., Orchard L. M., Llinas M. and Yeh E (2018). Specific inhibition of the bifunctional farnesyl/geranylgeranyl diphosphate synthase in malaria parasites via a new small-molecule binding Site. Cell Chem. Biol. 25, 185-193.e5. 10.1016/j.chembiol.2017.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves P. M., Choi L., Gelband H. and Garner P (2018). Primaquine or other 8-aminoquinolines for reducing Plasmodium falciparum transmission. Cochrane Database Syst. Rev. 2, CD008152 10.1002/14651858.CD008152.pub5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiguemde W. A., Shelat A. A., Bouck D., Duffy S., Crowther G. J., Davis P. H., Smithson D. C., Connelly M., Clark J., Zhu F. et al. (2010). Chemical genetics of Plasmodium falciparum. Nature 465, 311-315. 10.1038/nature09099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman J. D., Pepper L. R., Cortese J. F., Estiu G., Galinsky K., Zuzarte-Luis V., Derbyshire E. R., Ribacke U., Lukens A. K., Santos S. A. et al. (2015). The cytoplasmic prolyl-tRNA synthetase of the malaria parasite is a dual-stage target of febrifugine and its analogs. Sci. Transl. Med. 7, 288ra77 10.1126/scitranslmed.aaa3575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoepfner D., McNamara C. W., Lim C. S., Studer C., Riedl R., Aust T., McCormack S. L., Plouffe D. M., Meister S., Schuierer S. et al. (2012). Selective and specific inhibition of the plasmodium falciparum lysyl-tRNA synthetase by the fungal secondary metabolite cladosporin. Cell Host Microbe 11, 654-663. 10.1016/j.chom.2012.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooft van Huijsduijnen R. and Wells T. N. (2018). The antimalarial pipeline. Curr. Opin. Pharmacol. 42, 1-6. 10.1016/j.coph.2018.05.006 [DOI] [PubMed] [Google Scholar]
- Howes R. E., Battle K. E., Satyagraha A. W., Baird J. K. and Hay S. I (2013). G6PD deficiency: global distribution, genetic variants and primaquine therapy. Adv. Parasitol. 81, 133-201. 10.1016/B978-0-12-407826-0.00004-7 [DOI] [PubMed] [Google Scholar]
- Imwong M., Suwannasin K., Kunasol C., Sutawong K., Mayxay M., Rekol H., Smithuis F. M., Hlaing T. M., Tun K. M., van der Pluijm R. W. et al. (2017). The spread of artemisinin-resistant Plasmodium falciparum in the Greater Mekong subregion: a molecular epidemiology observational study. Lancet Infect. Dis. 17, 491-497. 10.1016/S1473-3099(17)30048-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain V., Yogavel M., Kikuchi H., Oshima Y., Hariguchi N., Matsumoto M., Goel P., Touquet B., Jumani R. S., Tacchini-Cottier F. et al. (2017). Targeting prolyl-tRNA synthetase to accelerate drug discovery against malaria, leishmaniasis, toxoplasmosis, cryptosporidiosis, and coccidiosis. Structure 25, 1495-1505.e6. 10.1016/j.str.2017.07.015 [DOI] [PubMed] [Google Scholar]
- Jimenez-Diaz M. B., Ebert D., Salinas Y., Pradhan A., Lehane A. M., Myrand-Lapierre M. E., O'Loughlin K. G., Shackleford D. M., Justino de Almeida M., Carrillo A. K. et al. (2014). (+)-SJ733, a clinical candidate for malaria that acts through ATP4 to induce rapid host-mediated clearance of Plasmodium. Proc. Natl. Acad. Sci. USA 111, E5455-E5462. 10.1073/pnas.1414221111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato N., Comer E., Sakata-Kato T., Sharma A., Sharma M., Maetani M., Bastien J., Brancucci N. M., Bittker J. A., Corey V. et al. (2016). Diversity-oriented synthesis yields novel multistage antimalarial inhibitors. Nature 538, 344-349. 10.1038/nature19804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. H., Kondrashin A., Greenwood B., Lindblade K., Loku Galappaththy G. and Alonso P (2019). A historical review of WHO certification of malaria elimination. Trends Parasitol. 35, 163-171. 10.1016/j.pt.2018.11.011 [DOI] [PubMed] [Google Scholar]
- Luth M. R., Gupta P., Ottilie S. and Winzeler E. A (2018). Using in vitro evolution and whole genome analysis to discover next generation targets for antimalarial drug discovery. ACS Infect Dis. 4, 301-314. 10.1021/acsinfecdis.7b00276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maetani M., Zoller J., Melillo B., Verho O., Kato N., Pu J., Comer E. and Schreiber S. L (2017). Synthesis of a bicyclic azetidine with in vivo antimalarial activity enabled by stereospecific, directed C(sp(3))-H arylation. J. Am. Chem. Soc. 139, 11300-11306. 10.1021/jacs.7b06994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malaria Genomic Epidemiology Network (2008). A global network for investigating the genomic epidemiology of malaria. Nature 456, 732-737. 10.1038/nature07632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara C. W., Lee M. C., Lim C. S., Lim S. H., Roland J., Nagle A., Simon O., Yeung B. K., Chatterjee A. K., McCormack S. L. et al. (2013). Targeting plasmodium PI(4)K to eliminate malaria. Nature 504, 248-253. 10.1038/nature12782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messer P. W. and Petrov D. A (2013). Population genomics of rapid adaptation by soft selective sweeps. Trends Ecol. Evol. 28, 659-669. 10.1016/j.tree.2013.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miotto O., Almagro-Garcia J., Manske M., Macinnis B., Campino S., Rockett K. A., Amaratunga C., Lim P., Suon S., Sreng S. et al. (2013). Multiple populations of artemisinin-resistant Plasmodium falciparum in Cambodia. Nat. Genet. 45, 648-655. 10.1038/ng.2624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miotto O., Amato R., Ashley E. A., MacInnis B., Almagro-Garcia J., Amaratunga C., Lim P., Mead D., Oyola S. O., Dhorda M. et al. (2015). Genetic architecture of artemisinin-resistant Plasmodium falciparum. Nat. Genet. 47, 226-234. 10.1038/ng.3189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nájera J. A., González-Silva M. and Alonso P. L (2011). Some lessons for the future from the global malaria eradication programme (1955-1969). PLoS Med. 8, e1000412 10.1371/journal.pmed.1000412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newby G., Hwang J., Koita K., Chen I., Greenwood B., von Seidlein L., Shanks G. D., Slutsker L., Kachur S. P., Wegbreit J. et al. (2015). Review of mass drug administration for malaria and its operational challenges. Am. J. Trop. Med. Hyg. 93, 125-134. 10.4269/ajtmh.14-0254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen-Dinh P. and Trager W (1978). Chloroquine resistance produced in vitro in an African strain of human malaria. Science 200, 1397-1398. 10.1126/science.351801 [DOI] [PubMed] [Google Scholar]
- Nzila A. and Mwai L (2010). In vitro selection of Plasmodium falciparum drug-resistant parasite lines. J. Antimicrob. Chemother. 65, 390-398. 10.1093/jac/dkp449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto T. D., Bohme U., Sanders M., Reid A., Bruske E. I., Duffy C. W., Bull P. C., Pearson R. D., Abdi A., Dimonte S. et al. (2018). Long read assemblies of geographically dispersed Plasmodium falciparum isolates reveal highly structured subtelomeres. Wellcome Open Res. 3, 52 10.12688/wellcomeopenres.14571.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquet T., Le Manach C., Cabrera D. G., Younis Y., Henrich P. P., Abraham T. S., Lee M. C. S., Basak R., Ghidelli-Disse S., Lafuente-Monasterio M. J. et al. (2017). Antimalarial efficacy of MMV390048, an inhibitor of Plasmodium phosphatidylinositol 4-kinase. Sci. Transl. Med. 9 10.1126/scitranslmed.aad9735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson D. S., Milhous W. K. and Wellems T. E (1990). Molecular basis of differential resistance to cycloguanil and pyrimethamine in Plasmodium falciparum malaria. Proc. Natl. Acad. Sci. USA 87, 3018-3022. 10.1073/pnas.87.8.3018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips M. A., White K. L., Kokkonda S., Deng X., White J., El Mazouni F., Marsh K., Tomchick D. R., Manjalanagara K., Rudra K. R. et al. (2016). A triazolopyrimidine-based dihydroorotate dehydrogenase inhibitor with improved drug-like properties for treatment and prevention of malaria. ACS Infect Dis. 2, 945-957. 10.1021/acsinfecdis.6b00144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips M. A., Burrows J. N., Manyando C., van Huijsduijnen R. H., Van Voorhis W. C. and Wells T. N. C. (2017). Malaria. Nat. Rev. Dis. Primers 3, 17050 10.1038/nrdp.2017.50 [DOI] [PubMed] [Google Scholar]
- Pritchard J. K., Pickrell J. K. and Coop G (2010). The genetics of human adaptation: hard sweeps, soft sweeps, and polygenic adaptation. Curr. Biol. 20, R208-R215. 10.1016/j.cub.2009.11.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollo I. M. (1952). ‘Daraprim’ resistance in experimental malarial infections. Nature 170, 415 10.1038/170415a0 [DOI] [PubMed] [Google Scholar]
- Ross L. S., Gamo F. J., Lafuente-Monasterio M. J., Singh O. M., Rowland P., Wiegand R. C. and Wirth D. F (2014). In vitro resistance selections for Plasmodium falciparum dihydroorotate dehydrogenase inhibitors give mutants with multiple point mutations in the drug-binding site and altered growth. J. Biol. Chem. 289, 17980-17995. 10.1074/jbc.M114.558353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottmann M., McNamara C., Yeung B. K., Lee M. C., Zou B., Russell B., Seitz P., Plouffe D. M., Dharia N. V., Tan J. et al. (2010). Spiroindolones, a potent compound class for the treatment of malaria. Science 329, 1175-1180. 10.1126/science.1193225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sa J. M., Kaslow S. R., Krause M. A., Melendez-Muniz V. A., Salzman R. E., Kite W. A., Zhang M., Moraes Barros R. R., Mu J., Han P. K. et al. (2018). Artemisinin resistance phenotypes and K13 inheritance in a Plasmodium falciparum cross and Aotus model. Proc. Natl. Acad. Sci. USA 115, 12513-12518. 10.1073/pnas.1813386115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw P. J. and Aroonsri A (2017). Tools for attenuation of gene expression in malaria parasites. Int. J. Parasitol. 47, 385-398. 10.1016/j.ijpara.2016.11.006 [DOI] [PubMed] [Google Scholar]
- Smith Gueye C., Newby G., Hwang J., Phillips A. A., Whittaker M., MacArthur J. R., Gosling R. D. and Feachem R. G. (2014). The challenge of artemisinin resistance can only be met by eliminating Plasmodium falciparum malaria across the Greater Mekong subregion. Malar. J. 13, 286 10.1186/1475-2875-13-286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoiki E., Palencia A., Guo D., Ahyong V., Dong C., Li X., Hernandez V. S., Zhang Y. K., Choi W., Gut J. et al. (2016). Antimalarial benzoxaboroles target plasmodium falciparum Leucyl-tRNA synthetase. Antimicrob. Agents Chemother. 60, 4886-4895. 10.1128/AAC.00820-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spillman N. J., Allen R. J. W., McNamara C. W., Yeung B. K. S., Winzeler E. A., Diagana T. T. and Kirk K (2013). Na(+) regulation in the malaria parasite Plasmodium falciparum involves the cation ATPase PfATP4 and is a target of the spiroindolone antimalarials. Cell Host Microbe 13, 227-237. 10.1016/j.chom.2012.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straimer J., Lee M. C. S., Lee A. H., Zeitler B., Williams A. E., Pearl J. R., Zhang L., Rebar E. J., Gregory P. D., Llinás M. et al. (2012). Site-specific genome editing in Plasmodium falciparum using engineered zinc-finger nucleases. Nat. Methods 9, 993-998. 10.1038/nmeth.2143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X.-Z. and Miller L. H (2015). The discovery of artemisinin and the Nobel Prize in Physiology or Medicine. Sci. China Life Sci. 58, 1175-1179. 10.1007/s11427-015-4948-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale N., Moreira R. and Gomes P (2009). Primaquine revisited six decades after its discovery. Eur. J. Med. Chem. 44, 937-953. 10.1016/j.ejmech.2008.08.011 [DOI] [PubMed] [Google Scholar]
- Wagner J. C., Platt R. J., Goldfless S. J., Zhang F. and Niles J. C (2014). Efficient CRISPR-Cas9-mediated genome editing in Plasmodium falciparum. Nat. Methods 11, 915-918. 10.1038/nmeth.3063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Cabrera M., Yang J., Yuan L., Gupta B., Liang X., Kemirembe K., Shrestha S., Brashear A., Li X. et al. (2016). Genome-wide association analysis identifies genetic loci associated with resistance to multiple antimalarials in Plasmodium falciparum from China-Myanmar border. Sci. Rep. 6, 33891 10.1038/srep33891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White N. J. (2008). Qinghaosu (artemisinin): the price of success. Science 320, 330-334. 10.1126/science.1155165 [DOI] [PubMed] [Google Scholar]
- White N. J. (2012). Counter perspective: artemisinin resistance: facts, fears, and fables. Am. J. Trop. Med. Hyg. 87, 785 10.4269/ajtmh.2012.12-0573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White N. J., Pukrittayakamee S., Hien T. T., Faiz M. A., Mokuolu O. A. and Dondorp A. M (2014). Malaria. Lancet 383, 723-735. 10.1016/S0140-6736(13)60024-0 [DOI] [PubMed] [Google Scholar]
- Wilairat P., Kümpornsin K. and Chookajorn T (2016). Plasmodium falciparum malaria: Convergent evolutionary trajectories towards delayed clearance following artemisinin treatment. Med. Hypotheses 90, 19-22. 10.1016/j.mehy.2016.02.022 [DOI] [PubMed] [Google Scholar]
- Wright G. D. (2007). The antibiotic resistome: the nexus of chemical and genetic diversity. Nat. Rev. Microbiol. 5, 175-186. 10.1038/nrmicro1614 [DOI] [PubMed] [Google Scholar]
- Zhang M., Wang C., Otto T. D., Oberstaller J., Liao X., Adapa S. R., Udenze K., Bronner I. F., Casandra D., Mayho M. et al. (2018). Uncovering the essential genes of the human malaria parasite Plasmodium falciparum by saturation mutagenesis. Science 360, eaap7847 10.1126/science.aap7847 [DOI] [PMC free article] [PubMed] [Google Scholar]