Significance
Cross-linking mass spectrometry (XLMS) has become an established approach to complement the more common techniques for protein structure elucidation for those molecular machines where the traditional techniques are unable to obtain sufficient resolution. The success of XLMS relies on the ability to obtain high coverage of cross-linked peptides which are then used as restraints to guide modeling efforts. Here we describe advances in chemical and computational methods to obtain density of 1 cross-link per 7 amino acids of protein sequence. We show that this level of coverage is sufficient to produce a medium-resolution model a large protein subcomplex without using structural information for the subunits, as well as to define specific interfaces at higher resolution.
Keywords: cross-linking, mass spectrometry, integrative modeling, protein docking
Abstract
Detailed mechanistic understanding of protein complex function is greatly enhanced by insights from its 3-dimensional structure. Traditional methods of protein structure elucidation remain expensive and labor-intensive and require highly purified starting material. Chemical cross-linking coupled with mass spectrometry offers an alternative that has seen increased use, especially in combination with other experimental approaches like cryo-electron microscopy. Here we report advances in method development, combining several orthogonal cross-linking chemistries as well as improvements in search algorithms, statistical analysis, and computational cost to achieve coverage of 1 unique cross-linked position pair for every 7 amino acids at a 1% false discovery rate. This is accomplished without any peptide-level fractionation or enrichment. We apply our methods to model the complex between a carbonic anhydrase (CA) and its protein inhibitor, showing that the cross-links are self-consistent and define the interaction interface at high resolution. The resulting model suggests a scaffold for development of a class of protein-based inhibitors of the CA family of enzymes. We next cross-link the yeast proteasome, identifying 3,893 unique cross-linked peptides in 3 mass spectrometry runs. The dataset includes 1,704 unique cross-linked position pairs for the proteasome subunits, more than half of them intersubunit. Using multiple recently solved cryo-EM structures, we show that observed cross-links reflect the conformational dynamics and disorder of some proteasome subunits. We further demonstrate that this level of cross-linking density is sufficient to model the architecture of the 19-subunit regulatory particle de novo.
Recent advances in elucidating the network of protein interactions in the cell (1) promise to improve our understanding of cellular protein machinery. However, a higher-resolution view of the structure of proteins and complexes involved will help provide more detailed insights (2). Traditional methods for structure determination remain expensive and labor-intensive. Cross-linking mass spectrometry (XLMS) allows identification of proximal structural regions on amino acid level (3). Protein samples are combined with reagents that form covalent bonds in solution, and upon protein digestion, resulting peptide pairs can be identified by tandem mass spectrometry. Cross-linked position pairs can then be used as geometric constraints for structural modeling of proteins (4), docking (5), and modeling of large protein assemblies (6–8).
Recent improvements in XLMS coincided with the resolution revolution in cryogenic electron microscopy (Cryo-EM) as well as development of integrative modeling software. Combining these technologies with other data sources resulted in impressive models of previously intractable protein complexes (9–13). For modeling purposes, the key factor in the utility of a dataset is the cross-linking density or sequence coverage of the component proteins, and attaining high coverage has been a major challenge. Multiple chemistries have been proposed, including isotopic labels (14), cleavable cross-linkers (15), and photoactivatable cross-linkers (16). Various approaches to improve coverage have been reported, such as affinity enrichment via additional chemical handles (17), peptide fractionation, and use of multiple proteases (18). These and other methodological developments have been recently thoroughly reviewed (3, 15, 19).
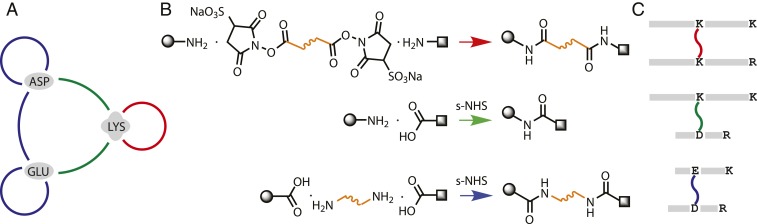
Lysine side chains are highly reactive and abundant on protein surfaces, which is why the field has traditionally focused on cross-linkers targeting them, particularly using homobifunctional NHS-ester reagents. However, orthogonal chemistries targeting other residue pairings have also been used (16, 20, 21). Carbodiimide chemistry has been used for decades for coupling carboxylic acids and primary amines in proteins using 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide—a water-soluble 0-length cross-linker (EDC). EDC reaction proceeds via an unstable intermediate and preferentially at pH < 6; however, activating agents, such as sulfo-NHS (22, 23), have been used to increase reaction efficiency with proteins in conditions closer to native. In addition to carboxyl–primary amine pairs, EDC has been extended with dihydrazides to link carboxyl groups in a proof-of-principle study to further improve cross-linking coverage (24). In a similar approach, (4-(4,6-dimethoxy-1,3,5-triazin-2-yl)-4-methyl-morpholinium chloride) (DMTMM) 0-length chemistry, also extended with dihydrazides, targeting the same residues pairs under physiological conditions was applied to several large protein complexes (25). Very recently, an N-hydroxybenzotriazole activating agent was proposed for use with EDC and extended with a diamine linker (26). Despite these developments, 0-length and related chemistries targeting orthogonal residue pairs have not been shown to consistently outperform traditional Lys–Lys cross-linking approaches and remain underutilized (27).
Building on preceding work, we use a combination of orthogonal chemistries allowing all possible cross-links between Lys, Glu, and Asp residues (Fig. 1 A and B). EDC/sNHS chemistry with and without a diamine linker allows cross-links between 5 amino acid pairs and should theoretically result in better density than the single Lys–Lys pair. We refer to it below as Extended-EDC/sNHS (E-EDC/sNHS). Note that lysines modified by the cross-linker are not susceptible to commonly used tryptic digestion, leading to larger, more difficult to identify peptides (Fig. 1C).
Fig. 1.
Overview of cross-linking chemistry and search/FDR strategy. (A) Targeting 3 of the most abundant residues—Lysine (LYS), Aspartate (ASP), and Glutamate (GLU)—on protein surfaces and all 6 possible cross-links among them with 3 reactions. (B) Bifunctional NHS-ester—the most common type of reagent—cross-links primary amines (red). EDC/sNHS chemistry is used to link carboxyl groups to primary amines, resulting in 0-length cross-links (green). Addition of a diamine linker to the EDC/sNHS reaction (E-EDC/sNHS) results in cross-links between carboxyl groups (blue). (C) Following tryptic digestion, the 3 cross-link types lead to peptides that theoretically should be 4, 3, and 2 times the size of a typical tryptic peptide, due to missed cleavage at the cross-link site.
Many of the recent chemical and analytical developments help apply XLMS at proteome scale to discover and monitor protein interactions, but here we focus on improving coverage of affinity purified proteins and protein complexes for purposes of structural modeling. We use a simple XLMS pipeline that relies on very cheap and readily available reagents without isotopic labels, cleavable backbones, or affinity handles and acquire MS/MS data in single-shot analyses. We develop computational methods to streamline the search and statistical analysis of resulting datasets and show that our approach scales from small proteins and complexes to one of the largest molecular machines produced by the cell.
Results
Simplified Calculation of False Discovery Rate at Reduced Computational Cost.
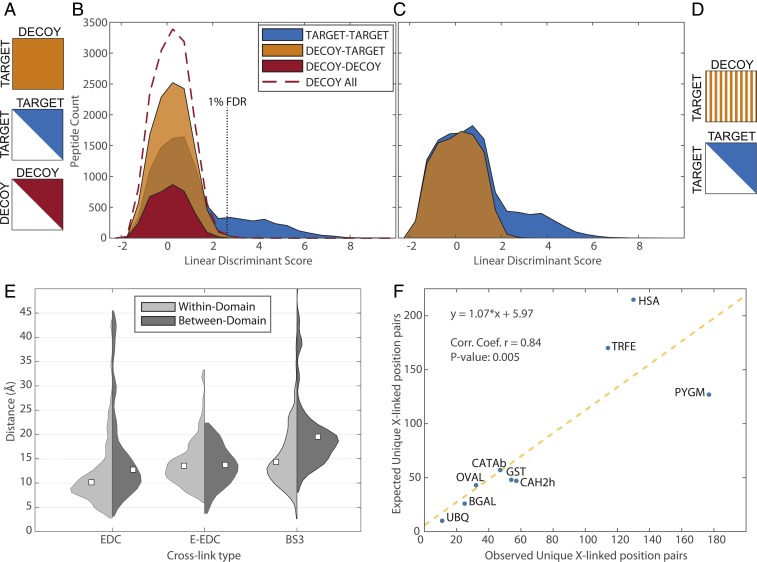
Identification of cross-linked peptide spectra is more challenging than a traditional peptide search (SI Appendix, Fig. S1A) due to search space size when considering all possible combinations of peptide pairs, as well as due to difficulties in estimating the false discovery rate (FDR). The standard in bottom-up proteomics has been to use the target–decoy approach (28). While a similar approach has been used for identification of cross-linked peptides, the statistical analysis is more complicated (29, 30). Cross-linked peptide search space consists of target–target (TT), decoy–target (DT), and decoy–decoy (DD) subsets. Since the DT subset can arise from any combination of decoy and target peptides, it is twice the size of TT and DD (Fig. 1D). This skewed ratio of target–decoy search space makes it more difficult to estimate FDR and requires making assumptions that the 1:2:1 ratio of TT:DT:DD cross-linked peptide hits holds across the full range of matches, independently of their likelihood. DT matches are common, requiring 1 or a few spurious peak matches to the decoy peptide, with most of the score value contributed by the target peptide. They contribute heavily to the relevant part of the decoy distribution (29). DD matches require multiple spurious matches from both peptides. While these types of matches are observed in the middle of the distribution, they are exceedingly rare in the range of interest of 1 to 5% FDR (Fig. 1E). Since DD corresponds to a quarter of the computational expense of the search while providing little discriminative value, we propose to eliminate that subset of the search space.
We further note that the size of DT could be reduced by half, thus matching the size of TT, while still providing sufficient discriminative value. We therefore propose to uniformly sample half of DT (Fig. 2 C and D and SI Appendix). These adjustments cut the computational cost of a search in half while simultaneously simplifying the FDR estimation procedure to that analogous to a standard peptide search. Scores and other features calculated upon completion of the search can be used as input features into a standard statistical analysis method to estimate FDR without cross-link–specific adjustments.
Fig. 2.
Summary of search/FDR strategy and model protein dataset. (A) A full-scale cross-linked search needs to consider decoy–decoy (DD), target–target (TT), and decoy–target (DT) subspaces, with DT being twice the size of the other 2. (B) Scores and features of the resulting matches are combined into a discriminant score. Distribution of these scores can be used to estimate FDR, but the imbalance between target and decoy search spaces leads to complications. (C and D) Removing DD and reducing DT in half simplifies FDR estimation while cutting computational costs. Striped pattern of DT subspace represents uniform sampling of half the space. Distributions in B and C are based on E-EDC/sNHS sample mix of model proteins. Cross-linked distance distributions. (E) The expected progression from 0-length (EDC) to extended-EDC to BS3. White squares denote medians of respective distributions. (F) Observed cross-links are in good agreement with those expected from known structure. Sum of all 3 cross-link types is used for both observed (unique position pairs) and predicted values for each protein.
Model Proteins.
Nine diverse proteins were chosen to evaluate the search algorithm and FDR estimation. Each protein was cross-linked with EDC/sNHS, E-EDC/sNHS, and BS3 reagents individually and digested, and peptides were mixed by cross-link type. Peptides were then subjected to liquid chromatography-mass spectrometry (LC-MS) in a total of 3 runs without enrichment or fractionation. Data were searched with our PIXL (Protein Interactions from Cross-Linking) algorithm and stringently filtered to 1% FDR (SI Appendix), resulting in the identification of 3,761 cross-linked peptides, which reduced to 610 unique cross-linked position pairs (SI Appendix, Table S1). This corresponds to 1 unique cross-linked position for every 7.5 amino acids of sequence. Note that our E-EDC/sNHS approach outperforms the Lys–Lys experiment by 50%, with BS3 spanning a longer cross-link distance, both theoretical and observed. A previous study comparing acidic and 0-length cross-links with BS3 for a large number of proteins showed them to be, at best, equal (25).
Since each of the model proteins was cross-linked individually, the dataset can be used to evaluate our FDR estimation procedure by computing the rate of interprotein cross-links (29) among the 9 proteins (expected to be 1%; Methods). This turned out to be 0.94% combined for the 3 runs, thus validating the quality of the dataset and FDR calculation methods. Mapping cross-links onto the available structures shows the expected increase in median linked distance from 0-length EDC/sNHS to E-EDC/sNHS to BS3 (Fig. 2A). Stratifying the data based on PFAM (31) domains suggests greater conformational flexibility between domains based on 0-length and amine-reactive cross-linkers, although the difference was not detected for carboxyl–carboxyl cross-linkers. This could be explained by propensity of lysine residues to appear in mobile loops, perhaps near domain interfaces (32).
Despite the recent progress in XLMS, cross-linking coverage has been quite variable. Here we use our model dataset to try to understand the source of this variability and find a sequence-based predictor of success for a cross-linking experiment. First, we show that the numbers of observed unique cross-linked positions correlate well with those computed from structures (R = 0.84; Fig. 2F). However, sequence length turns out to be a poor predictor of coverage (mean R2 = 0.24). We then considered a linear model based on the sum of arginine and lysine residues, which performed much better (mean R2 = 0.57). An even better predictor turned out to be the number of theoretical tryptic peptides containing a cross-linkable amino acid. This value is straightforward to calculate and on average explains 80% of the variance in the resulting number of cross-links (SI Appendix, Fig. S2). While this model does not explicitly consider cross-links due to intersubunit interactions and may underestimate their prediction, it is nevertheless useful to point out sequences with poor distributions of cleavage sites and cross-linkable residues for larger systems where doing so manually would be time-consuming.
Inhibitor of Carbonic Anhydrase.
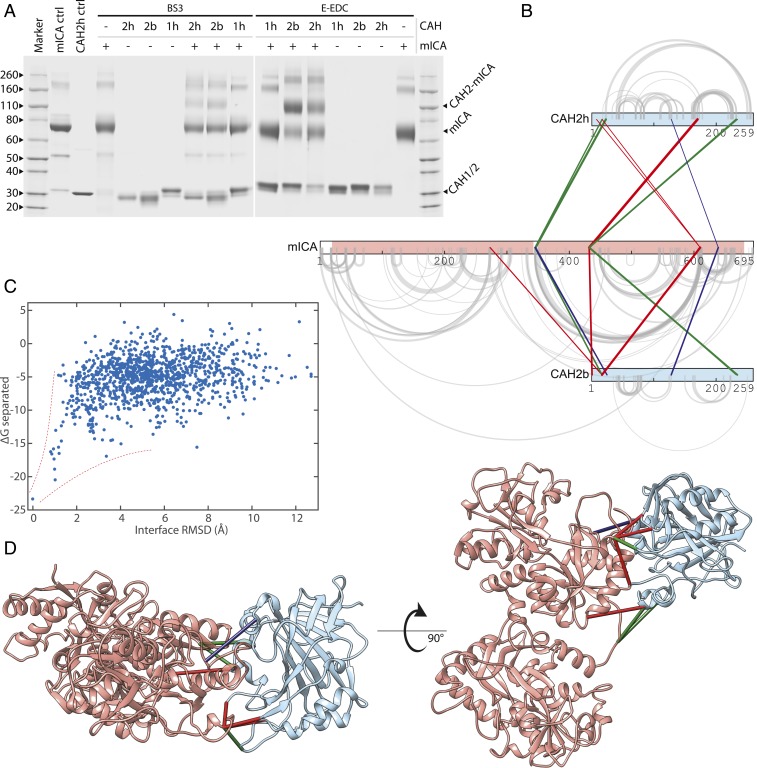
Members of the Carbonic Anhydrase (CA) family of zinc-containing enzymes catalyze the conversion of CO2 to bicarbonate and protons. They are involved in respiration, transport and regulation of CO2, pH homeostasis, and other processes (33). Many of the CA isozymes have been identified as drug targets to treat diseases such as glaucoma, obesity, and cancer (34, 35). Several sulfonamide-based inhibitors of CA are used in the clinic, but they show poor selectivity among the isozymes (33). Interestingly, a protein inhibitor of members of the family was identified in some mammals. The murine inhibitor of CA (mICA) was shown to be a nanomolar inhibitor of several isoforms, including the ubiquitous CAII enzyme (35). The structures of CAII in different species (36, 37) have been well studied, and the structure of mICA was recently solved (38); however, the structure of the enzyme-inhibitor complex remains elusive (35). Identifying the interface would be the first step to the design of a new class of protein-based CA family inhibitors. Here mICA inhibitor was cross-linked using the BS3 and E-EDC/sNHS reagents with both human and bovine versions of CAHII as well as human CAHI. Human as well as bovine CAII with both cross-linkers showed strong Coomassie-stained bands corresponding to a 1:1 complex (Fig. 3A). We did not detect a complex with CAI, in agreement with previous studies. CAII samples were digested in solution and analyzed by mass spectrometry with methods analogous to those used for the model dataset above. Among hundreds of cross-linked peptides, we identified 10 unique interprotein position pairs for human and 9 for bovine CAII. These data, mapped onto protein sequences, show a self-consistent pattern, pointing to the C-terminal domain of mICA as primarily responsible for CAII binding (Fig. 3B), in agreement with previous observations (39). Despite 80% sequence identity between the human and bovine CAII isoforms, every cross-linked peptide differs in at least 1 amino acid from its equivalent in the other species (SI Appendix, Table S2) and therefore serves as a biological replicate, validating our results. The matching set of identifications was filtered to select 5 representative high-confidence cross-linked position pairs for use in modeling the CAII–mICA complex with Rosetta (40, 41) (SI Appendix). Unbiased docking simulations typically require the generation of hundreds of thousands of decoys to make sure every relative orientation of the 2 molecules is well sampled. Here the cross-links provided sufficient geometric constraint that all solutions converged on essentially the same orientation (SI Appendix, Fig. S3) with model binding energies forming a characteristic funnel shape leading toward the lowest-energy structure (Fig. 3C). Note that all 3 cross-link types are identified in the interface and contribute to constrain modeling. The identified cross-links mapped onto our model of the enzyme/inhibitor complex (Fig. 3D) are consistent between human/bovine replicates and point to the same interface region across cross-link types and corresponding conditions. The model is also in agreement with much of the existing literature, confirming that the inhibitor C-terminal domain binds near the entrance to the active site, thus blocking it without directly interacting with key residues conserved among CAs. The interface buries 1,340 Å2 of solvent accessible surface area, providing an opportunity to use the inhibitor for tuning isoform selectivity.
Fig. 3.
Modeling CA–mICA complex structure from cross-link data. (A) Both human (h) and bovine (b) Carbonic Anhydrase (CAH) II (2h and 2b) cross-link with the murine Inhibitor of Carbonic Anhydrase (mICA), while human Carbonic Anhydrase I (1h) does not. (B) Cross-links exhibit self-consistent patterns in a 2D representation [produced with xiNET (75)]. (C) Using cross-links as geometric constraints in Rosetta produces a characteristic energy funnel with respect to the lowest-energy model. (D) Best-scoring model with cross-links mapped on them illustrates how different cross-link types help define the interface and reinforce each other. Cross-link colors are as follows: red, BS3; green, EDC; blue, E-EDC.
Yeast Proteasome.
Overview and initial analysis of cross-links.
To apply our methods to a larger system, we chose the proteasome complex, the focus of many structural studies over the years, including using XLMS (25, 42). The proteasome is a 2.5-MDa symmetric assembly made up of the core particle (CP) and 2 regulatory particles (RP). The CP is a cylinder composed of 2 layers of α and β rings carrying out the proteolytic function. RP contains a hexameric ring of AAA-ATPases, which deubiquitinate the substrate and promote its translocation to the proteolytic core of the CP, as well as subunits responsible for substrate recognition and binding. The RP is made up of the Base and Lid subcomplexes, which assemble independently before associating into the functional complex. Recent cryo-EM studies have identified 3 conformational states of the proteasome with the s1 ground state preferred in the presence of ATP and absence of substrate (43).
Yeast proteasomes, affinity purified via RPN11 tag, were cross-linked in the presence of ATP under 3 conditions (EDC/sNHS, E-EDC/sNHS, and BS3) and processed using methods analogous to those described above with some differences described in Methods. Three LC-MS runs with no offline peptide enrichment or fractionation were searched and filtered to 1% FDR on unique peptide level, resulting in a dataset with 8,296 total identified cross-linked peptide-spectral matches, reducing to 3,893 unique cross-linked peptides after combining the data to remove redundant 0-length cross-links. Further collapsing the data and removing cross-links between positions close in sequence results in 2,549 unique positions pairs, of which 1,704 are among the 33 proteasome subunits, with combined density of 6.8 amino acids per position pair (similar density to what was observed for the model proteins above; SI Appendix, Table S3). This is 4 times the size of a recently published cross-linking analysis of human proteasome, which utilized pulldowns of multiple subunits and cleavable reagents (42); 57% of our cross-links are intersubunit, and the median distances between cross-linked positions were similar to those observed in the model dataset.
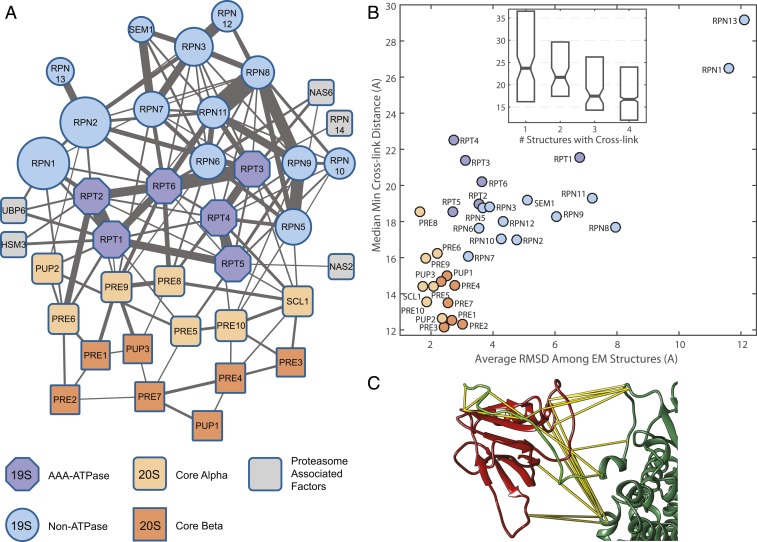
One way to evaluate overall quality of the dataset is by considering the 2-dimensional (2D) topology of a network of subunits, with edges between them weighted by the summed peptide spectral counts. We filtered the dataset to a 1% FDR on the protein interaction level, resulting in a network of 121 protein nodes with 219 edges (Fig. 4A and SI Appendix, Fig. S5B). Cytoscape’s (44) “organic” layout algorithm readily recapitulated the topology of 26S proteasome with layout of the core, base, and lid subunits congruent with cryo-EM data. Most of the cross-links outside of the proteasome are among subunits of the ribosome, with 68 of 79 known subunits represented (SI Appendix, Fig. S5B).
Fig. 4.
Cross-linking the yeast proteasome. (A) Cytoscape’s (44) layout algorithm recapitulates subunit architecture of the complex in 2D based on XLMS data. Tightly intertwined subunits of the AAA-ATPase ring and the MPN heterodimer (Mpr1, Pad1 N-terminal domain-containing RPN8 and RPN11) exhibit the strongest edges. Known chaperones controlling RP assembly are shown connected to the AAA-ATPase subunits. See SI Appendix, Fig. S5B, for the rest of the network. (B) Cross-links that map to longer distances in static published structures (Protein Data Bank IDs 4CR2, 5MPD, 5WVI, and 3JCP) correspond to observed conformational dynamics of the s1 state. Correlation between average cryo-EM structure RMSD and median of minimum cross-link distance is 0.76 (P < 1e-7). (Inset) The more likely both cross-linked residues are to be resolved in published structures, the smaller the minimum observed distance between them. (C) Cross-links confirm proposed orientation of RPN13 (red)–RPN2 (dark green) interaction and allow accurate modeling with unresolved RPN2 C-terminal tail (light green model); see SI Appendix, Fig. S7, for more details.
Conformational flexibility and structure-based FDR.
Typically, to assess dataset quality, previous studies have defined a cutoff distance based on theoretical length of the cross-linker with an additional adjustment for side-chain length and protein conformational dynamics, but aside from some simulations (45), there has not been a way to evaluate the effect of flexibility and partial disorder on cross-linked distance measured on static structures. Cross-links bridging distances in available structures that fall beyond the cutoff are considered violating the theoretical assumptions and presumed to stem from a combination of conformational flexibility, heterogeneity, and false positives (25, 42, 46, 47). Our initial data are conservatively filtered to 1% FDR on the unique peptide level and, upon collapsing to unique cross-linked positions, reach a combined 1.5% FDR. However, we reasoned the size of the dataset allows us to estimate a structure-based FDR, using a statistical approach. False positive cross-link identifications are expected to map to a random distance distribution arising from all possible cross-linkable residue distances. On the other hand, residues cross-linked as a result of conformational flexibility or local structural disorder are likely to follow a distribution closely resembling that expected from theoretical cross-links but potentially extending somewhat beyond theoretical distances. Fitting the distance distributions of observed and theoretical cross-linked distances to a bimodal mixture model allows us to estimate a value of 3.2% structure-based FDR for the combined dataset with a 95% confidence interval (2.0 and 4.5%) (SI Appendix, Fig. S4B). While this is higher than our peptide and position-level estimates, it should be sufficient for subsequent modeling. Some reasons for the higher apparent FDR are discussed below.
XLMS technology has been successfully applied to investigate protein conformational switching between states upon perturbation (48, 49) in vitro and in vivo (50); however, sparse structural data have limited the possibility of fully exploring the effect of protein steady-state flexibility on cross-linking data. Two aspects of the proteasome complex enable us to directly evaluate this relationship: 1) the recent surge in published high-resolution cryo-EM structures and 2) the fact that the CP and RP subcomplex conformational dynamics are known to be very different, allowing for contrasting measurements within the same complex in the same experiment. We aligned and compared the 4 available cryo-EM structures of the s1 state (43, 51–54) against each other and then related those differences to the closest cross-link distances observed in our data, averaged by subunit. The resulting correlation of 0.76 is highly significant, with the more rigid CP subunit cross-links closely clustered at lower distances, while the more conformationally flexible regulatory particle subunits show more variation. The most dynamic subunits—RPN1 and RPN13—exhibit the largest differences, with over 4-fold higher RMSD among cryo-EM structures, corresponding to cross-linked distances double that of CP subunits (Fig. 4B). Furthermore, the more confidently resolved residues correspond to shorter cross-linked distances in static structures (Fig. 4 B, Inset). These data explicitly demonstrate on a large scale that XLMS reflects steady-state conformational dynamics of protein structures and suggest caution when applying strict theoretical distance cutoffs.
Outlier cross-links.
Focusing on parts of the proteasome with the largest number of cross-links that appeared to be in violation of the available structures, we observed that many of them involved RPN11 intralinks, as well as RPN11 interlinks with RPN5, RPN8, and RPN9. Since these lid subunits have been shown to undergo extensive conformational changes upon assembly of the full proteasome, we mapped these interactions onto the recently solved cryo-EM structure of the isolated lid subcomplex (55). Forty-three cross-links (2.4% of total) appear to be specific to the unbound lid (SI Appendix, Fig. S6), possibly resulting from affinity enrichment retaining a fraction of unassembled lid subcomplex or partial dissociation (56) during postpurification sample handling and cross-linking reaction. Cross-links like this could potentially be used to quantify stoichiometry of proteasome conformational states.
Cross-links involving RPN13 ubiquitin receptor appeared inconsistent with some available structures (SI Appendix, Fig. S7 A and B). Its human ortholog ADRM1 was not resolved in the latest human proteasome structures. However, RPN13 is important for proteasome function and has recently been identified as a target for cancer therapy (57). In yeast, RPN13 was initially placed in the structure manually (43, 51), and subsequent structures show a somewhat similar orientation (52–54). Wehmer et al. were able to achieve higher local resolution in one of the states and suggested a new orientation, confirmed by 2 cross-links (54). These 2 are among 22 unique cross-linked positions found in our dataset, and 16 of them could be mapped onto resolved residues of the 4 structures, supporting the Wehmer et al. orientation (SI Appendix, Fig. S7B). In addition, we observed several cross-links between RPN13 and the unresolved C terminus of RPN2, which is necessary and sufficient for binding (58). Using the high-confidence cross-link between RPN13 Glu72 and RPN2 C terminus, together with RPN2 chain continuity as the only constraints and starting with extended conformation of the C-terminal peptide, we used Rosetta (41, 59) to model the interaction interface (SI Appendix, Fig. S7C). The best-scoring model (Fig. 4C) shows the C-terminal RPN2 tail binding against 2 loops and the beta-sheet of RPN13. The model is 3.4 Å RMSD from the recently published human homolog structure (60), illustrating the power of cross-linking data, in combination with modern modeling software, to enhance cryo-EM models and provide medium- to high-resolution details for some important protein interfaces.
Detailed analysis of the 4 available structures of the s1 state also identified 2 regions of disagreement among them: C-terminal helix of RPN8, recently shown to be crucial for lid subcomplex assembly and part of the lid helical bundle (61) (SI Appendix, Fig. S8), and the N-terminal helix of RPT1 (SI Appendix, Fig. S9). High-confidence cross-links available for these helical regions suggest potential register errors, where cross-linking data could be used to validate candidate cryo-EM models.
Of the remaining cross-links that are not in agreement with s1 state cryo-EM structures, most prominent are the ones involving the RPT subunits. Given the presence in the network of chaperones known to govern assembly of the Base subunits, we hypothesize that these cross-links represent partially assembled states of the regulatory particle, lacking CP.
De Novo Modeling of the Yeast Proteasome Regulatory Particle.
In recent years, development of the Integrative Modeling Platform (IMP) (62) has facilitated the elucidation of the molecular architecture for large protein assemblies. Typically, such modeling would take advantage of all available structural information for the subunits. However, given the high density of our cross-linking dataset, we wanted to ask what kind of model quality is possible if relying on it as the sole source. Starting with the crystal structure of the proteasome core particle (63), our cross-linking dataset described above, and no other structural information, we used IMP to model the entire 19-subunit RP. Each subunit was modeled as a series of connected beads representing at least 25 amino acids (SI Appendix, Fig. S10). Candidate models produced by IMP were each scored, combining contributions from cross-linking, excluded volume, and sequence connectivity restraints. Starting from random initial orientations, 40,000 models were computed, optimizing the position of the beads, representing RP subunits, with respect to each other and to the CP, modeled as rigid body.
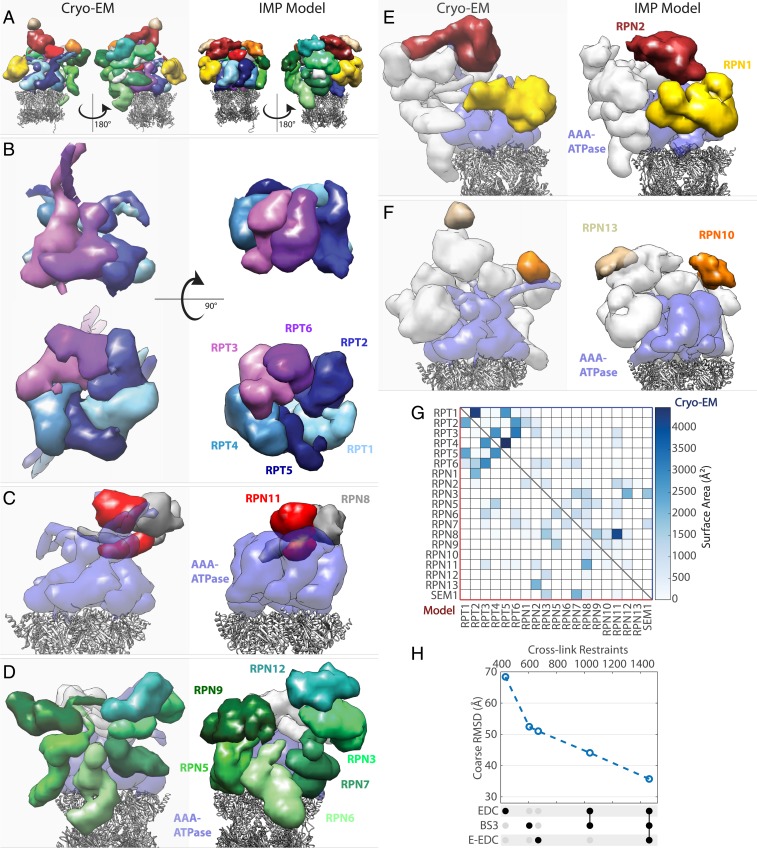
We used recently developed protocols (64) to show the sampling precision of this cluster converges around 27 Å RMSD, allowing us to compute a model at 21 Å (RMSF). Splitting the ensemble into 2 samples results in very similar models, suggesting that the sampling is exhaustive (SI Appendix, Fig. S11). The resulting density map shows that cross-linking data alone can capture enough information to reproduce the general architecture of the RP (Fig. 5 A–G). Compared with a map derived from an atomic model (54), we compute an average density correlation (weighted by volume) of 0.79. To assess the accuracy of our model, we computed 35.7 Å average Cα RMSD (with respect to Wehmer et al.), where the coordinates of every Cα atom in the dominant cluster were set to the center position of the bead representing the corresponding residue. While a crude upper-bound estimate due to coarseness of representation, it nevertheless shows that a cross-linking dataset derived from just 3 runs can lead to models with subdomain level precision and accuracy.
Fig. 5.
Comparison of proteasome RP density maps based on atomic model (5MP9, 5MPD, and Cryo-EM) and IMP model computed from cross-linking data alone, with no other structural data or homology modeling. (A) Overall structure with RP colored by subunit, where CP is represented by gray ribbons. IMP model represents full-length sequences, while cryo-EM–based model includes only sequence-assigned portions. (B) AAA-ATPase subunits. (Top) Side view. (Bottom) View from below. (C) MPN heterodimer of RPN11/RPN8. (D) Proteasome component (PCI) domain-containing subunits. (E) PC-repeat containing subunits. (F) Ubiquitin receptor subunits. See main text for additional information on RPN13 localization and modeling. (G) Comparison of intersubunit surface areas within a 7-Å cutoff shows that the integrative model captures most of the interaction patterns among subunits. (H) Models derived from dataset subsets result in higher RMSD with respect to cryo-EM structure, illustrating the value of increasing cross-link density.
To further evaluate model quality, we compute and compare contacting surface areas using Chimera (65), to show that our model captures the main contacts among all RP and relevant CP alpha subunits (Fig. 5G). Note that some contacts are missing in the cryo-EM–based matrix due to poorly resolved regions. For example, RPN2–RPN13 interaction is missing because of the unresolved RPN2 tail but is observed in our model. Similarly, more extensive contacts between RPN1 and RPN2 are observed in our model, likely due to unassigned cryo-EM density. The volume of RPN1 subunit in the IMP model (Fig. 5 A and E) is ∼10% greater than the corresponding volume mapped from 5MPD. A more extensive RPN1–RPN2 interface was recently resolved in human proteasomes (66).
Despite a good agreement of the overall subunit architecture, the cryo-EM structure of RP contains a few very prominent features that are not very well reproduced in the model. One such feature is the RPN13 subunit, whose poor localization is discussed above. Another set of features consists of 3 pairs of unusually long, intertwined N-terminal helices of AAA-ATPase subunits (with additional, unresolved 40- to 50-amino acid-long arms) that protrude radially outward from the center of the heterohexamer (Fig. 5B). Unfortunately, these cannot be well modeled using 25-amino acid beads. Perhaps future iterations of our approach could take advantage of secondary structure prediction to better model nonglobular regions of structure.
To confirm the effect of combining orthogonal cross-linkers as well as evaluate increased constraint density in general, we repeated the above modeling procedure on subsets of the full dataset, using the same representation and quality control criteria. The results, summarized in Fig. 5H, show a near-linear improvement in model quality with increasing cross-link density, suggesting a path toward higher-resolution de novo models.
Discussion
The field of XLMS has been diversifying into using more complex reagents with affinity handles, isotopic labels, and cleavable spacers, and these are promising for larger-scale analysis of whole-cell lysates, organelles, and tissues (67–69). However, for the purposes of structural modeling of even large affinity-purified protein complexes, we show that simple, cheap, and readily available reagents can produce detailed, high-density datasets without offline peptide enrichment or fractionation.
With E-EDC/sNHS, we show that at pH 6.5 and with higher reagent concentrations, it tends to produce better coverage than a traditional Lysine–lysine cross-linker, the current gold standard for XLMS studies. With the diamine linker used here, E-EDC/sNHS cross-links also result in shorter constraints, both in theory and in practice. Searching EDC-based data is more computationally expensive due to the abundance of acidic residues and the resulting number of possible monolinked peptide variants. However, the PIXL algorithm makes improvements in handling the search space, reducing that computational complexity in half, with the added benefit of also streamlining the FDR calculation. Even for the E-EDC/sNHS experiments, PIXL can handle up to ∼200 sequences for its cross-linked search component, which should be sufficient for any affinity purification experiment.
Like many methods aimed at studying protein structure, XLMS has limitations. In particular, the EDC-based methods used here require that the protein or complex be stable at pH 6.5. Furthermore, because both EDC and the diamine need to be introduced into the reaction as acid salts to maintain proper pH, the system under study may need to be tolerant to higher ionic strength. Here we adjusted the buffer conditions to maintain similar ionic strength, using NaCl for BS3 reactions to make up for contributions of EDC and diamine linker in EDC-based reactions. For the proteasome, we chose to lower the linker concentration significantly out of caution, to ensure that CP and RP particles remain bound, and still identified 10% more unique cross-linked position pairs than with BS3. Aside from buffer conditions, there is concern that modifying many charged residues on protein surface may cause conformational changes. Data presented here, especially for the proteasome, suggest that cross-linking data reflect natural protein conformational dynamics. Perhaps the stabilization of protein structure by covalent cross-links serves to offset the potential destabilization of modified surface charges.
Interpreting cross-linking data for proteins and complexes subject to conformational flexibility and partially disordered regions is a challenge because the results appear to conflict with theoretical limits of cross-linker reach. We use the proteasome cross-linking data in conjunction with multiple recently published cryo-EM structures to examine the effect of such flexibility on the measured cross-links. To validate our methods, we further propose a structure-based method to estimate FDRs and independently confirm the quality of the data.
Despite the recent successes of cryo-EM, a significant fraction of obtained maps fall beyond 4-Å cutoff where structures can be solved de novo (70). This is especially true for the more dynamic or disordered regions of structure. High-resolution structural data may not always be available and become a limiting factor when working with less studied, orphan protein complexes. Here we test the limits of constraints derived from high-density cross-linking data to build a model of the proteasome regulatory particle while not taking advantage of the available structures. Even with good constraint coverage, modeling of more conformationally flexible complexes is challenging as current modeling methods do not account for void space and force models to be more compact, closer to average protein density. This limitation is evident in Fig. 5A. Although ours is a coarse model, it captures many salient interaction details and can serve as a valuable starting point for further refinement.
While our data show the E-EDC/sNHS approach is superior on average (SI Appendix, Tables S1A and S3A), the key to attaining high cross-link density is the combination of experiments to identify all 6 possible cross-link pairs among 3 residues. These 3 chemistries can be analyzed in just 2 or 3 LC-MS runs. By increasing the probability of cross-linking events, we achieve a dataset-average density of 1 cross-link for every 7 residues. With state-of-the-art modeling software, the cross-linking coverage achieved here appears promising both for higher-level modeling of protein complex architectures, as well as producing more detailed models of protein interfaces or refining lower-resolution regions of structures solved by other methods.
Methods
Cross-Linking Reactions.
Zero-length reactions were carried out for 1 h at room temperature in 50 mM Pipes Buffer, pH 6.5, with 100 mM EDC and 15 to 20 mM sulfo-NHS.
E-EDC reactions were performed analogously to the 0-length reactions but with the addition of 2,2′-(Ethylenedioxy)diethylamine (EDDA, NH2CH2CH2OCH2CH2OCH2CH2NH2) (SIGMA-Aldrich) linker and also included 15 to 20 mM sulfo-NHS. The linker stock solution (275 mM) was prepared fresh with Pipes buffer and pH-adjusted by adding HCl at approximately double the concentration of linker. Sulfo-NHS and EDC stock solutions were also prepared fresh in Pipes at 0.5 and 1 M, respectively. For model proteins, we used 100 mM EDC with 100 mM linker. For mICA experiments, we used 150 mM EDC with 100 mM linker. For the proteasome, we used 100 mM EDC but lowered the linker concentration to ∼27 mM to ensure the RP and CP particles remain associated.
BS3 reactions at 2 mM concentration were carried out for 1 h at room temperature in 50 mM Hepes Buffer, pH 7.8. All 9 model protein reactions contained 100 mM NaCl, and the β-Galactosidase reaction additionally contained 10 mM MgCl2. mICA/CAH reactions omitted NaCl for the E-EDC reactions but used 25 mM NaCl for the BS3 reactions. The proteasome reactions omitted the NaCl but added 5 mM MgCl2.
All cross-linking reactions were quenched with hydroxylamine to final concentration of 100 mM for 20 to 30 min, which resulted in raising the pH above ∼8, making the samples ready for subsequent tryptic digestion.
Model Proteins.
Bovine Ubiquitin, Ovalbumin, Human Carbonic Anhydrase II, Human Serum Albumin, Bovine Liver Catalase, and Rabbit Phosphorylase a were all obtained from Sigma-Aldrich. GST and β-Galactosidase were obtained from Rockwell, Inc. Human Holo-Transferrin was obtained from EMD Millipore. Model proteins were cross-linked at concentrations of ∼25 to 50 µM.
mICA/CAH.
Carbonic Anhydrase isoforms were all obtained from Sigma-Aldrich. N-His–tagged nonglycosylated version of mICA containing a signal peptide substituted from human Transferrin was purified from BHK cells obtained from Anne B. Mason, University of Vermont, Burlington, VT following published protocols with minor modifications (71). Briefly, cells were initially grown in T175 flasks in DMEM and 10% FBS. Subsequently, the media was changed to Pro293S (Lonza) with addition of 2.5 mM l-Glutamine and 1 mM butyric acid (Sigma-Aldrich). Cell culture media was collected and saved every 2 to 4 d and subsequently used to purify mICA with HisPur resin (Qiagen). For cross-linking reactions, the concentration of the carbonic anhydrase variants was 0.2 mg/mL with mICA at 0.4 mg/mL
26S Proteasome.
Saccharomyces cerevisiae proteasome samples were generously provided by the Finley laboratory, where they were produced following established affinity purification protocols (72) from Ecm29-deletion strains (73). Concentration of proteasome was 0.5 to 1 mg/mL during the cross-linking reactions.
Mass Spectrometry and Data Analysis.
Sample preparation and mass spectrometry methods as well as data processing, search, filtering, and analysis are described in SI Appendix.
Data Availability.
The mass spectrometry data have been deposited to the ProteomeXchange Consortium via the PRIDE (74) partner repository with the dataset identifier PXD011296.
Supplementary Material
Acknowledgments
We are very grateful to G. Tian and D. Finley for the proteasome sample and critical reading of the manuscript. We are also grateful to J. Paolo and R. Kunz for instrument help. We thank A.B. Mason and M.P. Weekes for help and advice with cell culture, as well as M. Liao, D.K. Schweppe, and W. Haas for critical reading of the manuscript. This work was funded by NIH Grant (GM067945) to S.P.G.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The mass spectrometry data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD011296.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1902931116/-/DCSupplemental.
References
- 1.Huttlin E. L., et al. , The bioplex network: A systematic exploration of the human interactome. Cell 162, 425–440 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Robinson C. V., Sali A., Baumeister W., The molecular sociology of the cell. Nature 450, 973–982 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Yu C., Huang L., Cross-linking mass spectrometry: An emerging technology for interactomics and structural biology. Anal. Chem. 90, 144–165 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brodie N. I., Popov K. I., Petrotchenko E. V., Dokholyan N. V., Borchers C. H., Solving protein structures using short-distance cross-linking constraints as a guide for discrete molecular dynamics simulations. Sci. Adv. 3, e1700479 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vreven T., et al. , Integrating cross-linking experiments with ab initio protein-protein docking. J. Mol. Biol. 430, 1814–1828 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ward A. B., Sali A., Wilson I. A., Biochemistry. Integrative structural biology. Science 339, 913–915 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferber M., et al. , Automated structure modeling of large protein assemblies using crosslinks as distance restraints. Nat. Methods 13, 515–520 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Karaca E., Rodrigues J. P. G. L. M., Graziadei A., Bonvin A. M. J. J., Carlomagno T., M3: An integrative framework for structure determination of molecular machines. Nat. Methods 14, 897–902 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Lasker K., et al. , Molecular architecture of the 26S proteasome holocomplex determined by an integrative approach. Proc. Natl. Acad. Sci. U.S.A. 109, 1380–1387 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erzberger J. P., et al. , Molecular architecture of the 40S⋅eIF1⋅eIF3 translation initiation complex. Cell 158, 1123–1135 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robinson P. J., et al. , Molecular architecture of the yeast Mediator complex. elife 4, e08719 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim S. J., et al. , Integrative structure and functional anatomy of a nuclear pore complex. Nature 555, 475–482 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zelter A., et al. , The molecular architecture of the Dam1 kinetochore complex is defined by cross-linking based structural modelling. Nat. Commun. 6, 8673 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seebacher J., et al. , Protein cross-linking analysis using mass spectrometry, isotope-coded cross-linkers, and integrated computational data processing. J. Proteome Res. 5, 2270–2282 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Sinz A., Divide and conquer: Cleavable cross-linkers to study protein conformation and protein-protein interactions. Anal. Bioanal. Chem. 409, 33–44 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Belsom A., Schneider M., Fischer L., Brock O., Rappsilber J., Serum albumin domain structures in human blood serum by mass spectrometry and computational biology. Mol. Cell. Proteomics 15, 1105–1116 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trester-Zedlitz M., et al. , A modular cross-linking approach for exploring protein interactions. J. Am. Chem. Soc. 125, 2416–2425 (2003). [DOI] [PubMed] [Google Scholar]
- 18.Leitner A., et al. , Expanding the chemical cross-linking toolbox by the use of multiple proteases and enrichment by size exclusion chromatography. Mol. Cell. Proteomics 11, M111.014126 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chavez J. D., Bruce J. E., Chemical cross-linking with mass spectrometry: A tool for systems structural biology. Curr. Opin. Chem. Biol. 48, 8–18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kalkhof S., Ihling C., Mechtler K., Sinz A., Chemical cross-linking and high-performance Fourier transform ion cyclotron resonance mass spectrometry for protein interaction analysis: Application to a calmodulin/target peptide complex. Anal. Chem. 77, 495–503 (2005). [DOI] [PubMed] [Google Scholar]
- 21.Helgeson L. A., et al. , Human Ska complex and Ndc80 complex interact to form a load-bearing assembly that strengthens kinetochore-microtubule attachments. Proc. Natl. Acad. Sci. U.S.A. 115, 2740–2745 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Staros J. V., Wright R. W., Swingle D. M., Enhancement by N-hydroxysulfosuccinimide of water-soluble carbodiimide-mediated coupling reactions. Anal. Biochem. 156, 220–222 (1986). [DOI] [PubMed] [Google Scholar]
- 23.Grabarek Z., Gergely J., Zero-length crosslinking procedure with the use of active esters. Anal. Biochem. 185, 131–135 (1990). [DOI] [PubMed] [Google Scholar]
- 24.Novak P., Kruppa G. H., Intra-molecular cross-linking of acidic residues for protein structure studies. Eur. J. Mass Spectrom. (Chichester) 14, 355–365 (2008). [DOI] [PubMed] [Google Scholar]
- 25.Leitner A., et al. , Chemical cross-linking/mass spectrometry targeting acidic residues in proteins and protein complexes. Proc. Natl. Acad. Sci. U.S.A. 111, 9455–9460 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fioramonte M., et al. , XPlex: An effective, multiplex cross-linking chemistry for acidic residues. Anal. Chem. 90, 6043–6050 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Rivera-Santiago R. F., Sriswasdi S., Harper S. L., Speicher D. W., Probing structures of large protein complexes using zero-length cross-linking. Methods 89, 99–111 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elias J. E., Gygi S. P., Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Walzthoeni T., et al. , False discovery rate estimation for cross-linked peptides identified by mass spectrometry. Nat. Methods 9, 901–903 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Yang B., et al. , Identification of cross-linked peptides from complex samples. Nat. Methods 9, 904–906 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Finn R. D., et al. , The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 44, D279–D285 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Linding R., et al. , Protein disorder prediction: Implications for structural proteomics. Structure 11, 1453–1459 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Supuran C. T., Carbonic anhydrases: Novel therapeutic applications for inhibitors and activators. Nat. Rev. Drug Discov. 7, 168–181 (2008). [DOI] [PubMed] [Google Scholar]
- 34.Neri D., Supuran C. T., Interfering with pH regulation in tumours as a therapeutic strategy. Nat. Rev. Drug Discov. 10, 767–777 (2011). [DOI] [PubMed] [Google Scholar]
- 35.Durdagi S., et al. , Protein-protein interactions: Inhibition of mammalian carbonic anhydrases I-XV by the murine inhibitor of carbonic anhydrase and other members of the transferrin family. J. Med. Chem. 55, 5529–5535 (2012). [DOI] [PubMed] [Google Scholar]
- 36.Saito R., Sato T., Ikai A., Tanaka N., Structure of bovine carbonic anhydrase II at 1.95 A resolution. Acta Crystallogr. D Biol. Crystallogr. 60, 792–795 (2004). [DOI] [PubMed] [Google Scholar]
- 37.Fisher Z., et al. , Structural and kinetic characterization of active-site histidine as a proton shuttle in catalysis by human carbonic anhydrase II. Biochemistry 44, 1097–1105 (2005). [DOI] [PubMed] [Google Scholar]
- 38.Eckenroth B. E., Mason A. B., McDevitt M. E., Lambert L. A., Everse S. J., The structure and evolution of the murine inhibitor of carbonic anhydrase: A member of the transferrin superfamily. Protein Sci. 19, 1616–1626 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mason A. B., et al. , Evolution reversed: The ability to bind iron restored to the N-lobe of the murine inhibitor of carbonic anhydrase by strategic mutagenesis. Biochemistry 47, 9847–9855 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kahraman A., et al. , Cross-link guided molecular modeling with ROSETTA. PLoS One 8, e73411 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Das R., Baker D., Macromolecular modeling with rosetta. Annu. Rev. Biochem. 77, 363–382 (2008). [DOI] [PubMed] [Google Scholar]
- 42.Wang X., et al. , Molecular details underlying dynamic structures and regulation of the human 26S proteasome. Mol. Cell. Proteomics 16, 840–854 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Unverdorben P., et al. , Deep classification of a large cryo-EM dataset defines the conformational landscape of the 26S proteasome. Proc. Natl. Acad. Sci. U.S.A. 111, 5544–5549 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shannon P., et al. , Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Merkley E. D., et al. , Distance restraints from crosslinking mass spectrometry: Mining a molecular dynamics simulation database to evaluate lysine-lysine distances. Protein Sci. 23, 747–759 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rappsilber J., The beginning of a beautiful friendship: Cross-linking/mass spectrometry and modelling of proteins and multi-protein complexes. J. Struct. Biol. 173, 530–540 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ding Y.-H., et al. , Modeling protein excited-state structures from “over-length” chemical cross-links. J. Biol. Chem. 292, 1187–1196 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walzthoeni T., et al. , xTract: Software for characterizing conformational changes of protein complexes by quantitative cross-linking mass spectrometry. Nat. Methods 12, 1185–1190 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen Z., et al. , Quantitative cross-linking/mass spectrometry reveals subtle protein conformational changes. Wellcome Open Res. 1, 5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chavez J. D., Schweppe D. K., Eng J. K., Bruce J. E., In vivo conformational dynamics of hsp90 and its interactors. Cell Chem. Biol. 23, 716–726 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Beck F., et al. , Near-atomic resolution structural model of the yeast 26S proteasome. Proc. Natl. Acad. Sci. U.S.A. 109, 14870–14875 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ding Z., et al. , High-resolution cryo-EM structure of the proteasome in complex with ADP-AlFx. Cell Res. 27, 373–385 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Luan B., et al. , Structure of an endogenous yeast 26S proteasome reveals two major conformational states. Proc. Natl. Acad. Sci. U.S.A. 113, 2642–2647 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wehmer M., et al. , Structural insights into the functional cycle of the ATPase module of the 26S proteasome. Proc. Natl. Acad. Sci. U.S.A. 114, 1305–1310 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dambacher C. M., Worden E. J., Herzik M. A., Martin A., Lander G. C., Atomic structure of the 26S proteasome lid reveals the mechanism of deubiquitinase inhibition. elife 5, e13027 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Leggett D. S., et al. , Multiple associated proteins regulate proteasome structure and function. Mol. Cell 10, 495–507 (2002). [DOI] [PubMed] [Google Scholar]
- 57.Anchoori R. K., et al. , A bis-benzylidine piperidone targeting proteasome ubiquitin receptor RPN13/ADRM1 as a therapy for cancer. Cancer Cell 24, 791–805 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.He J., et al. , The structure of the 26S proteasome subunit Rpn2 reveals its PC repeat domain as a closed toroid of two concentric α-helical rings. Structure 20, 513–521 (2012). [DOI] [PubMed] [Google Scholar]
- 59.Raveh B., London N., Zimmerman L., Schueler-Furman O., Rosetta FlexPepDock ab-initio: Simultaneous folding, docking and refinement of peptides onto their receptors. PLoS One 6, e18934 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.VanderLinden R. T., Hemmis C. W., Yao T., Robinson H., Hill C. P., Structure and energetics of pairwise interactions between proteasome subunits RPN2, RPN13, and ubiquitin clarify a substrate recruitment mechanism. J. Biol. Chem. 292, 9493–9504 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Estrin E., Lopez-Blanco J. R., Chacón P., Martin A., Formation of an intricate helical bundle dictates the assembly of the 26S proteasome lid. Structure 21, 1624–1635 (2013). [DOI] [PubMed] [Google Scholar]
- 62.Shi Y., et al. , A strategy for dissecting the architectures of native macromolecular assemblies. Nat. Methods 12, 1135–1138 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Groll M., et al. , Structure of 20S proteasome from yeast at 2.4 A resolution. Nature 386, 463–471 (1997). [DOI] [PubMed] [Google Scholar]
- 64.Viswanath S., Chemmama I. E., Cimermancic P., Sali A., Assessing exhaustiveness of stochastic sampling for integrative modeling of macromolecular structures. Biophys. J. 113, 2344–2353 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pettersen E. F., et al. , UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 66.Schweitzer A., et al. , Structure of the human 26S proteasome at a resolution of 3.9 Å. Proc. Natl. Acad. Sci. U.S.A. 113, 7816–7821 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu F., Rijkers D. T. S., Post H., Heck A. J. R., Proteome-wide profiling of protein assemblies by cross-linking mass spectrometry. Nat. Methods 12, 1179–1184 (2015). [DOI] [PubMed] [Google Scholar]
- 68.Schweppe D. K., et al. , Mitochondrial protein interactome elucidated by chemical cross-linking mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 114, 1732–1737 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chavez J. D., et al. , Chemical crosslinking mass spectrometry analysis of protein conformations and supercomplexes in heart tissue. Cell Syst. 6, 136–141.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schmidt C., Urlaub H., Combining cryo-electron microscopy (cryo-EM) and cross-linking mass spectrometry (CX-MS) for structural elucidation of large protein assemblies. Curr. Opin. Struct. Biol. 46, 157–168 (2017). [DOI] [PubMed] [Google Scholar]
- 71.Wang F., et al. , A novel murine protein with no effect on iron homoeostasis is homologous with transferrin and is the putative inhibitor of carbonic anhydrase. Biochem. J. 406, 85–95 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Leggett D. S., Glickman M. H., Finley D., Purification of proteasomes, proteasome subcomplexes, and proteasome-associated proteins from budding yeast. Methods Mol. Biol. 301, 57–70 (2005). [DOI] [PubMed] [Google Scholar]
- 73.Kleijnen M. F., et al. , Stability of the proteasome can be regulated allosterically through engagement of its proteolytic active sites. Nat. Struct. Mol. Biol. 14, 1180–1188 (2007). [DOI] [PubMed] [Google Scholar]
- 74.Vizcaíno J. A., et al. , 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44, D447–D456 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Combe C. W., Fischer L., Rappsilber J., xiNET: Cross-link network maps with residue resolution. Mol. Cell. Proteomics 14, 1137–1147 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry data have been deposited to the ProteomeXchange Consortium via the PRIDE (74) partner repository with the dataset identifier PXD011296.