Rabies is a severe zoonotic disease caused by rabies virus (RABV). However, the nature of RABV entry remains unclear, which has hindered the development of therapy for rabies. It is suggested that modulations of RABV glycoprotein and multiple host factors are responsible for RABV invasion. Here, we showed that integrin β1 (ITGB1) directly interacts with RABV glycoprotein, and both proteins are internalized together into host cells. Differential expression of ITGB1 in mature muscle and cerebral cortex of mice led to A-4 (ITGB1-specific antibody), and RGD peptide (competitive inhibitor for interaction between ITGB1 and fibronectin) blocked street RABV infection via intramuscular but not intracerebral inoculation in mice, suggesting that ITGB1 plays a role in RABV peripheral entry. Our study revealed this distinct cellular factor in RABV infection, which may be an attractive target for therapeutic intervention.
KEYWORDS: rabies virus, integrin β1, protein interaction, viral entry, fibronectin
ABSTRACT
Rabies virus (RABV) is a widespread pathogen that causes fatal disease in humans and animals. It has been suggested that multiple host factors are involved in RABV host entry. Here, we showed that RABV uses integrin β1 (ITGB1) for cellular entry. RABV infection was drastically decreased after ITGB1 short interfering RNA knockdown and moderately increased after ITGB1 overexpression in cells. ITGB1 directly interacts with RABV glycoprotein. Upon infection, ITGB1 is internalized into cells and transported to late endosomes together with RABV. The infectivity of cell-adapted RABV in cells and street RABV in mice was neutralized by ITGB1 ectodomain soluble protein. The role of ITGB1 in RABV infection depends on interaction with fibronectin in cells and mice. We found that Arg-Gly-Asp (RGD) peptide and antibody to ITGB1 significantly blocked RABV infection in cells in vitro and street RABV infection in mice via intramuscular inoculation but not the intracerebral route. ITGB1 also interacts with nicotinic acetylcholine receptor, which is the proposed receptor for peripheral RABV infection. Our findings suggest that ITGB1 is a key cellular factor for RABV peripheral entry and is a potential therapeutic target for postexposure treatment against rabies.
IMPORTANCE Rabies is a severe zoonotic disease caused by rabies virus (RABV). However, the nature of RABV entry remains unclear, which has hindered the development of therapy for rabies. It is suggested that modulations of RABV glycoprotein and multiple host factors are responsible for RABV invasion. Here, we showed that integrin β1 (ITGB1) directly interacts with RABV glycoprotein, and both proteins are internalized together into host cells. Differential expression of ITGB1 in mature muscle and cerebral cortex of mice led to A-4 (ITGB1-specific antibody), and RGD peptide (competitive inhibitor for interaction between ITGB1 and fibronectin) blocked street RABV infection via intramuscular but not intracerebral inoculation in mice, suggesting that ITGB1 plays a role in RABV peripheral entry. Our study revealed this distinct cellular factor in RABV infection, which may be an attractive target for therapeutic intervention.
INTRODUCTION
Rabies, a serious zoonotic disease caused by rabies virus (RABV), is responsible for ∼59,000 human deaths and heavy economic burden annually worldwide (1). More than 99% of human deaths are caused by dog-mediated rabies. The global community aims to eliminate human deaths from dog-mediated rabies by 2030 (2). Rabies is a 100% vaccine-preventable disease. Regrettably, rural poor populations and free-roaming dogs are neglected in vaccination campaigns. Humans become infected by rabies-infected animals, and the virus finally invades the central nervous system (CNS) and then causes rabies symptoms (3). Once clinical signs are present, the mortality rate is almost 100%. Until now, only a few cases have recovered with prolonged intensive care, and currently, no therapy has been shown to prevent death (4).
RABV belongs to the genus Lyssavirus of the Rhabdoviridae family and can infect almost all warm-blooded animals. The RABV genome encodes five proteins: nucleoprotein (N), phosphoprotein (P), matrix protein (M), glycoprotein (G), and large polymerase protein (L). The viral RNA is encapsidated by N to form a helical nucleocapsid and, together with P and L, forms the ribonucleoprotein that constitutes the core of the bullet-shaped virion and the active viral replication unit. M is located beneath the viral membrane and bridges the nucleocapsid and lipid bilayer. G is an integral transmembrane protein that is thought to be of prime importance in virus-receptor binding during infection and in vaccine development (5–8). The broad tropism of RABV infection suggests that multiple cellular factors are involved in virus-host entry. So far, nicotinic acetylcholine receptor α1 (nAChRα1) (9), neural cell adhesion molecule (NCAM) (10), and metabotropic glutamate receptor 2 (mGluR2) (11) have been identified as host receptors for RABV. RABV uses different factors during progress from the periphery to the CNS. Researchers have been successfully studying the fundamental molecular mechanism of RABV infection for many years. Further explication of RABV invasion and pathogenesis is still urgently needed for the development of rabies therapy and, ultimately, elimination.
We previously used a global RNA interference (RNAi) strategy to screen potential host factors for RABV infection with a recombinant RABV Evelyn-Rokitnicki-Abelseth (ERA) strain expressing enhanced green florescence protein (ERA-eGFP) in HEK293 cells. We found that downregulation of integrin β1 (ITGB1), a type I transmembrane glycoprotein that facilitates early infection with human cytomegalovirus (12), Ebola virus (13), parvovirus (14), and reovirus (15), significantly decreased infection with ERA-eGFP.
In the present study, we demonstrated that downregulation and overexpression of ITGB1 significantly affected RABV infection, and ITGB1 directly interacted with RABV G. ITGB1 was internalized into cells and transported to late endosomes together with RABV. ITGB1 ectodomain soluble protein neutralized the infectivity of cell-adapted RABV in cells and street RABV in mice. The role of ITGB1 on RABV infection depended on interaction with fibronectin in cells and mice. Antibody to ITGB1 and RGD peptide significantly blocked cell-adapted RABV infection in cells and street RABV infection in mice via intramuscular but not intracerebral inoculation.
RESULTS
ITGB1 is required for RABV infection.
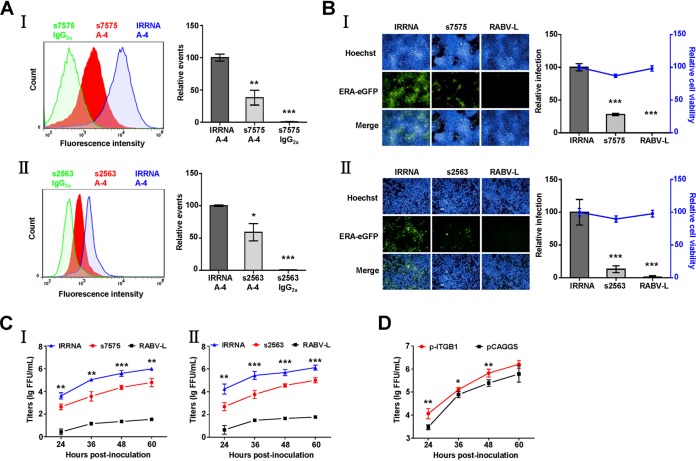
To examine whether reduced ITGB1 expression decreased RABV infection, HEK293 cells were transfected with short interfering RNA (siRNA) s7575, targeting human integrin β1 mRNA, which reduced 62% expression of ITGB1 on the cell surface according to flow cytometry analysis (Fig. 1A, I). Compared to that of irrelative siRNA (IRRNA)-transfected cells, the relative infection rate of ERA-eGFP decreased by 72% at 48 h postinoculation (h p.i.) (Fig. 1B, I), and ERA-eGFP growth titers were significantly lower at different times after RABV inoculation in ITGB1-silenced HEK293 cells (Fig. 1C, I). N2a cells were also transfected with siRNA s2563, targeting mouse integrin β1 mRNA, to confirm that ITGB1 was required for RABV replication. Expression of ITGB1 was reduced by 48% on the cell surface (Fig. 1A, II), which resulted in an 87% drop in relative infection rate of ERA-eGFP (Fig. 1B, II) at 48 h p.i. and a significant decrease in ERA-eGFP growth titers (Fig. 1C, II). We further determined whether overexpression of ITGB1 enhanced RABV infection by transient transfection with ITGB1 cDNA (p-ITGB1) into HEK293 cells. Overexpression of ITGB1 moderately increased the ERA-eGFP growth titers of RABV at 24, 36, and 48 h p.i. (Fig. 1D). All of these results demonstrate that ITGB1 is a key host factor for RABV infection.
FIG 1.
siRNA silencing and overexpression of ITGB1 affect RABV infection in cells. (A) Transfection with siRNA s7575 or s2563 resulted in downregulation of ITGB1. HEK293 (I) or N2a (II) cells were transfected with siRNA s7575 or s2563, respectively, at 37°C for 48 h. Cells were collected and stained with mouse anti-ITGB1 MAb (A-4) and fluorescein isothiocyanate (FITC)-mouse IgG and analyzed using flow cytometry. siRNA s7575- or s2563-transfected cells stained with mouse IgG2a and the irrelative siRNA (IRRNA)-transfected cells stained with A-4 were used as controls. The amount of cell surface ITGB1 was shown by comparison with that of the IRRNA-transfected cells stained with A-4 group, which was set as 100. The statistical differences were assessed using Student's t test. *, P < 0.05; ***, P < 0.001. (B) Downregulation of ITGB1 inhibited ERA-eGFP infection. HEK293 (I) and N2a (II) cells were transfected with siRNAs at 37°C for 48 h and infected with ERA-eGFP at a multiplicity of infection (MOI) of 0.1 at 37°C for 48 h. siRNA targeting the RABV L gene (RABV-L) and IRRNA were used as controls. Cells were used to detect cell viability using a CellTiter-Glo kit and stained for imaging using the PerkinElmer Operetta high-content system, and infection rate of RABV was analyzed using Columbus software. Cell viability and infection rate were shown as the relative cell viability and relative infection compared with those of the IRRNA-transfected group, which were set as 100. (C) Downregulation of ITGB1 inhibited the growth of ERA-eGFP. HEK293 (I) and N2a (II) cells were transfected with siRNAs at 37°C for 48 h and infected with ERA-eGFP at an MOI of 0.1. The supernatants were harvested at different time points for virus titration, and virus titers were determined in BSR cells and expressed as focus-forming units (FFU) per milliliter. (D) Overexpression of ITGB1 enhanced growth of ERA-eGFP. HEK293 cells were transfected with plasmid expressing ITGB1 (p-ITGB1) at 37°C for 36 h and infected with ERA-eGFP at an MOI of 5 for 1 h at 4°C, washed with prechilled DMEM three times, and incubated with new DMEM (supplemented with 2% FBS) at 37°C. The plasmid vector (pCAGGS) was used as a control. Virus titers of the supernatants were determined as FFU/ml in BSR cells. All data were considered statistically significant using Student's t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
ITGB1 directly interacts with RABV G.
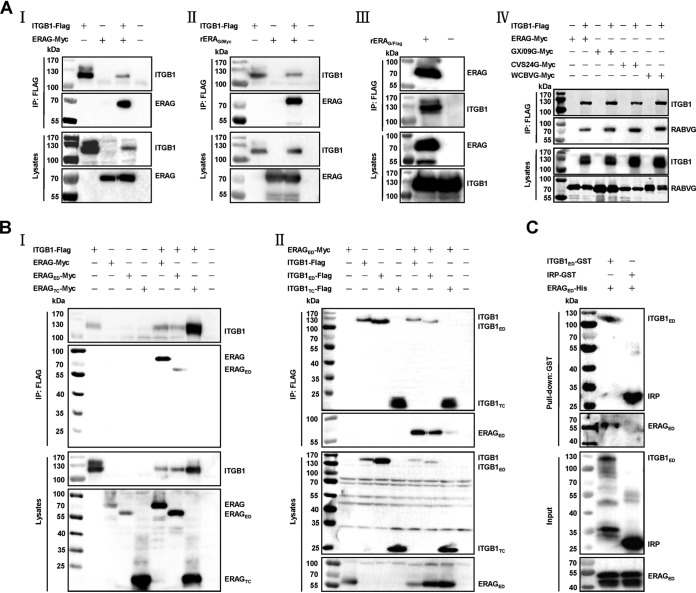
ITGB1 is proposed to facilitate early infection of human cytomegalovirus (12), Ebola virus (13), and parvovirus (14) and mediate reovirus internalization (15). RABV G is thought to be of prime importance in early entry and virus-host interaction (5). Consequently, ITGB1 may affect RABV infection by direct interaction between ITGB1 and RABV G. Coimmunoprecipitation (co-IP) assays of ITGB1 and RABV G derived from the cell-adapted strain ERA were carried out. Flag (DYKDDDDK)-tagged ITGB1 (ITGB1-Flag) interacted with Myc (EQKLISEEDL)-tagged ERA G (ERAG-Myc) in HEK293 cell lysates with plasmid coexpression (Fig. 2A, I). Plasmid-expressed ITGB1-Flag interacted with ERAG-Myc expressed from recombinant RABV rERAG/Myc that was tagged with a Myc at the C terminus of ERAG in HEK293 cell lysates (Fig. 2A, II). Endogenous ITGB1 also interacted with ERAG-Flag expressed from recombinant RABV rERAG/Flag that was tagged with a Flag at the C terminus of ERA G in HEK293 cell lysates (Fig. 2A, III). We carried out co-IP with ITGB1 and RABV G derived from the mouse-adapted strain CVS-24 (16), street strain GX/09 (isolated from the brain of a rabid dog in Guangxi Province of China in 2009), and west Caucasian bat virus (WCBV; another member of the lyssavirus family). ITGB1-Flag also interacted with Myc (EQKLISEEDL)-tagged CVS-24 G (CVS24G-Myc), GX/09 G (GX/09G-Myc), and WCBV G (WCBVG-Myc) in HEK293 cell lysates with plasmid coexpression (Fig. 2A, IV).
FIG 2.
Interactions between ITGB1 and RABV G. (A) ITGB1 interacted with RABV G. (I) Plasmid ITGB1-Flag and ERAG-Myc were cotransfected in HEK293 cells at 37°C for 48 h, and ITGB1-Flag interacted with ERAG-Myc in co-IP assays with HEK293 cell lysates. (II) HEK293 cells were infected with a recombinant ERA fused with a Myc tag at the C terminus of glycoprotein (rERAG/Myc) at an MOI of 0.1 for 36 h after transfection with plasmid ITGB1-Flag for 12 h, and ITGB1-Flag interacted with ERAG-Myc from rERAG/Myc in co-IP assays. (III) HEK293 cells were infected with a recombinant ERA fused with a Flag tag at the C terminus of glycoprotein (rERAG/Flag) at an MOI of 0.1 for 48 h, and endogenous ITGB1 interacted with ERAG-Flag from rERAG/Flag in co-IP assays. (IV) Myc-tagged RABV G plasmids from mouse-adapted strain CVS-24 (CVS24G-Myc), street virus strain GX/09 (GX/09G-Myc), or west Caucasian bat virus (WCBVG-Myc) were cotransfected with plasmid ITGB1-Flag in HEK293 cells for 48 h, and the indicated RABV G interacted with ITGB1-Flag in co-IP assays. (B) ITGB1 ectodomain (aa 1 to 728, ITGB1ED-Flag) interacted with ERA G ectodomain (aa 20 to 459, ERAGED-Myc). (I) ITGB1-Flag interacted with ERAGED-Myc but not ERA G transmembrane and cytoplasmic domain (aa 460 to 524, ERAGTC-Myc) in co-IP assays with plasmid-cotransfected HEK293 cell lysates. (II) ITGB1ED-Flag but not ITGB1 transmembrane and cytoplasmic domain (aa 729 to 798, ITGB1TC-Flag) interacted with ERAGED-Myc in co-IP assays with plasmid-cotransfected HEK293 cell lysates. (C) Purified GST-tagged ITGB1 ectodomain (aa 21 to 728, ITGB1ED), but not GST-tagged irrelative protein (IRP), pulled down purified His-tagged ERAG ectodomain (aa 41 to 450, ERAGED) using pulldown assay.
We then found that there was interaction between the Flag-tagged ITGB1 ectodomain (ITGB1ED-Flag, amino acids [aa] 1 to 728) and Myc-tagged ERA G ectodomain (ERAGED-Myc, aa 20 to 459) but no interactions between ITGB1-Flag and Myc-tagged ERA G transmembrane/cytoplasmic domain (ERAGTC-Myc, aa 460 to 524), ERAGED-Myc, and Flag-tagged ITGB1 transmembrane/cytoplasmic domain (ITGB1TC-Flag, aa 729 to 798) (Fig. 2B, I and II).
To further examine whether ITGB1 interacted directly with RABV G, N-terminal glutathione S-transferase (GST)-tagged ITGB1 ectodomain (ITGB1ED, aa 21 to 728) and N-terminal His-tagged ERAG ectodomain (ERAGED, aa 41 to 450) were purified and pooled to perform the pulldown assay. ERAGED was pulled down by ITGB1ED but not the GST-tagged irrelative protein (IRP) (Fig. 2C). These results demonstrated that ITGB1 directly interacted with ERA G.
ITGB1 and RABV are internalized into cells and transported together to endosomes.
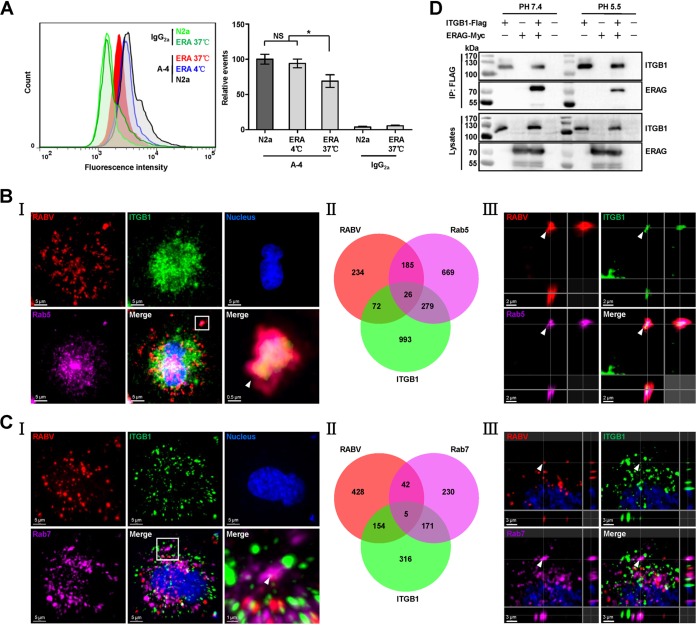
Given that ITGB1 directly interacts with RABV G, we next determined whether ITGB1 is internalized with RABV. Flow cytometry results in N2a cells first showed that the cellular ITGB1 level after RABV infection significantly decreased (Fig. 3A), indicating that ITGB1 was internalized upon infection. N2a cells were infected with recombinant RABV expressing an additional ERA nucleoprotein fused with a red fluorescent protein, mCherry (ERA-N/mCherry) (17). The cells were then stained immunofluorescently for RABV particles, ITGB1, and Rab5 or Rab7, using the tyramide signal amplification (TSA) method (11). Subcellular colocalization of RABV, ITGB1, and Rab5 was indicated as white spots in the merged image (Fig. 3B, I) and counted (Fig. 3B, II). Colocalization of RABV-ITGB1 complex with Rab5 in early endosomes is indicated by the white arrowhead in the three-dimensionally (3D) rendered image using Imaris software (Fig. 3B, III). Subcellular colocalization of RABV, ITGB1, and Rab7 is shown in Fig. 3C. Interaction between ITGB1 and ERA G under acidic conditions (pH 5.5) was also confirmed by co-IP assays (Fig. 3D), supporting our observation that the RABV-ITGB1 complex existed in late endosomes. These results demonstrate that ITGB1 is internalized into cells and transported to the cellular endosomal compartments along with RABV.
FIG 3.
RABV and ITGB1 are internalized into cells and transported to early and late endosomes together. (A) RABV infection results in downregulation of the cell surface ITGB1. N2a cells were infected with ERA at 37°C for 30 min, stained with A-4 and fluorescein isothiocyanate (FITC)-mouse IgG, and analyzed using flow cytometry. Noninfected cells (shown as N2a) stained with A-4 or mouse IgG2a, cells infected with ERA (shown as ERA) at 37°C and stained with mouse IgG2a, and cells infected with ERA at 4°C and stained with A-4 were used as controls. The amount of cell surface ITGB1 was shown as the relative events compared with that of the noninfected cells stained with A-4 group, which was set as 100. Significant differences were assessed using the Student's t test. NS, no significant difference. *, P < 0.05. (B and C) RABV and ITGB1 colocated in early and late endosomes. N2a cells were infected with recombinant RABV expressing an additional ERA nucleoprotein fused with a red fluorescent protein, mCherry (ERA-N/mCherry), at an MOI of 5 for 1 h at 4°C, washed with prechilled DMEM three times, and incubated with new DMEM (supplemented with 2% FBS) at 37°C for 30 min. Cells were stained using the tyramide signal amplification immunofluorescent method. RABV antigen (red), ITGB1 (green), Rab5 or Rab7 (purple), and the cell nuclei (blue) were examined in single-fluorescence channels. Colocalization of the RABV-ITGB1 complex with Rab5 or Rab7 was observed (I) and counted (II). The images in the lower right corner of panel I represent amplified random colocalization spots in the merged image within the small white box. (III) The 3D-rendered images were generated using Imaris software, and colocalization of the RABV-ITGB1 complex with Rab5 or Rab7 from the three-single fluorescence channels is indicated with the white arrowhead. (D) ITGB1-Flag interacted with ERAG-Myc under acidic conditions. ITGB1-Flag and ERAG-Myc plasmids were cotransfected in HEK293 cells at 37°C for 48 h, and ITGB1-Flag interacted with ERAG-Myc in co-IP assays with HEK293 cell lysates at pH 5.5 and pH 7.4.
ITGB1 ectodomain soluble protein neutralizes infectivity of RABV in cells.
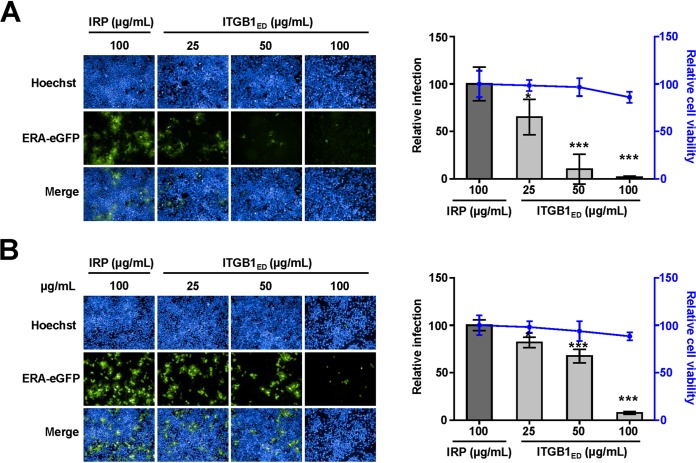
Based on the direct binding and coendocytosis of ITGB1 with RABV, we speculated that soluble ITGB1 protein should neutralize RABV infection. We performed a neutralization assay using ITGB1ED and ERA-eGFP. The infectivity of ERA-eGFP was neutralized by ITGB1ED in HEK293 (Fig. 4A) and N2a (Fig. 4B) cells, based on confirmation that cell viability was unaffected by ITGB1ED at a concentration of 100 μg/ml. The relative infection rate of ERA-eGFP decreased by 35% in HEK293 cells and 18% in N2a cells at 48 h p.i. after premixing with 25 μg/ml ITGB1ED for 1 h at 4°C. Infection rates were drastically reduced by 98% in HEK293 cells and 92% in N2a cells after treatment with 100 μg/ml ITGB1ED, which were significantly different from those after treatment with 100 μg/ml IRP in HEK293 and N2a cells.
FIG 4.
Soluble GST-tagged ITGB1 ectodomain (ITGB1ED) neutralizes infectivity of RABV. ITGB1ED neutralized ERA-eGFP infection in HEK293 (A) and N2a (B) cells in a dose-dependent manner. Different concentrations of ITGB1ED were mixed with ERA-eGFP at an MOI of 0.1 at 4°C for 1 h and added to the cells. GST-tagged irrelative protein (IRP) at high concentration was used as a control. After incubation at 37°C for 48 h, cells were used to detect cell viability, stained for imaging, and analyzed for the infection rate of RABV. The cell viability and infection rate were shown as the relative cell viability and relative infection compared with those of the IRP-treated group, which were set as 100. The statistical differences were assessed using Student's t test. *, P < 0.05; ***, P < 0.001.
Antibody against ITGB1 blocks infectivity of RABV in cells.
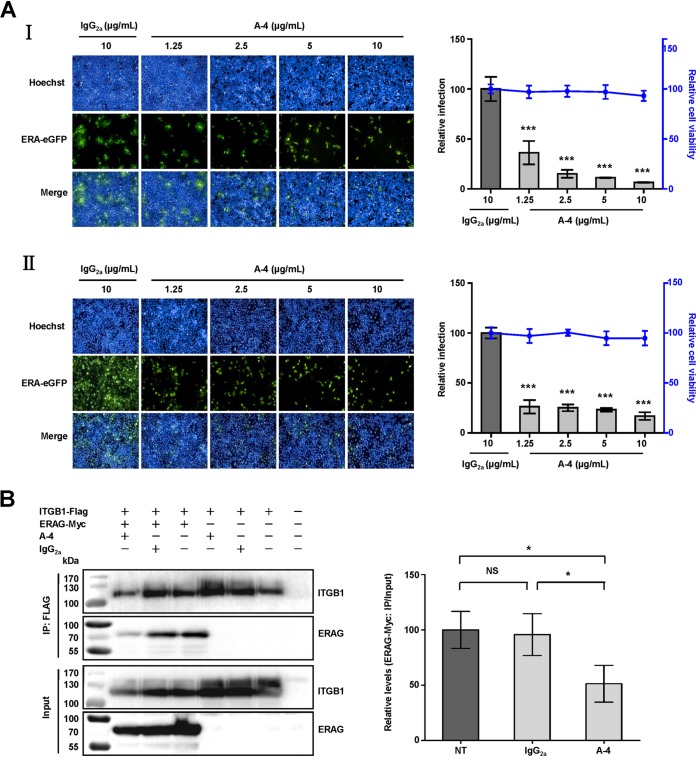
Integrin-specific antibodies have been proposed to decrease viral infection (18, 19). We speculated that RABV infection would be decreased by ITGB1-specific antibody. Antibody-blocking assays were performed in vitro using a mouse monoclonal antibody (MAb) against ITGB1 (A-4) and ERA-eGFP. Infectivity of ERA-eGFP was blocked by A-4 in HEK293 (Fig. 5A, I) and N2a (Fig. 5A, II) cells in a dose-dependent manner, based on the finding that cell viability was unaffected by A-4 at the highest concentration (10 μg/ml). The relative infection rate of ERA-eGFP decreased by 63% in HEK293 and 73% in N2a cells at 48 h p.i. after treatment with A-4 (1.25 μg/ml). The infection rate decreased with increasing doses of A-4, which differed significantly from that after treatment with mouse antibody IgG2a (10 μg/ml) in HEK293 and N2a cells.
FIG 5.
Antibody against ITGB1 blocks RABV infection. (A) Mouse monoclonal antibody against ITGB1 (A-4) blocked RABV infection in HEK293 (I) and N2a (II) cells in a dose-dependent manner. Different concentrations of A-4 were mixed with ERA-eGFP at an MOI of 0.1 at 4°C for 1 h, added to the cells, and incubated at 37°C for 48 h. Mouse IgG2a at high concentration was used as a control. The relative cell viability and infection were assessed compared with those of the IgG2a-treated group, which were set as 100. (B) A-4 weakened the interaction between ITGB1 and RABV G. Plasmid expressing ITGB1-Flag was incubated overnight with anti-Flag antibody-conjugated agarose beads, incubated with A-4 (10 μg/ml) at 4°C, and mixed with plasmid expressing ERAG-Myc at 4°C for co-IP assays. Non-antibody-treated ITGB1-Flag (NT) and IgG2a (10 μg/ml)-treated ITGB1-Flag (IgG2a) were used as controls. The levels of interaction between ITGB1 and ERAG were determined by the ratio of ERAG-Myc before and after co-IP assays (ERAG-Myc: IP/Input) using Image J software and are shown as relative levels compared with that of the NT group, which was set as 100. Significant differences were assessed using Student's t test. *, P < 0.05; ***, P < 0.001.
We clarified whether the interaction between ITGB1 and RABV G was weakened by A-4. ITGB1-Flag or ERAG-Myc was expressed in plasmid-transfected HEK293 cells, followed by coincubation after preincubation with A-4 or IgG2a for the co-IP assays. As expected, the level of ITGB1-ERA G complex was decreased by 49% after pretreatment with A-4, and there was a significant difference compared with that (4% drop) after pretreatment with IgG2a (Fig. 5B). This indicated that A-4 competitively weakened the interaction between ITGB1 and RABV G. These results demonstrate that blocking cell surface ITGB1 inhibits RABV infection in vitro.
ITGB1 affects RABV infection by depending on interaction with fibronectin.
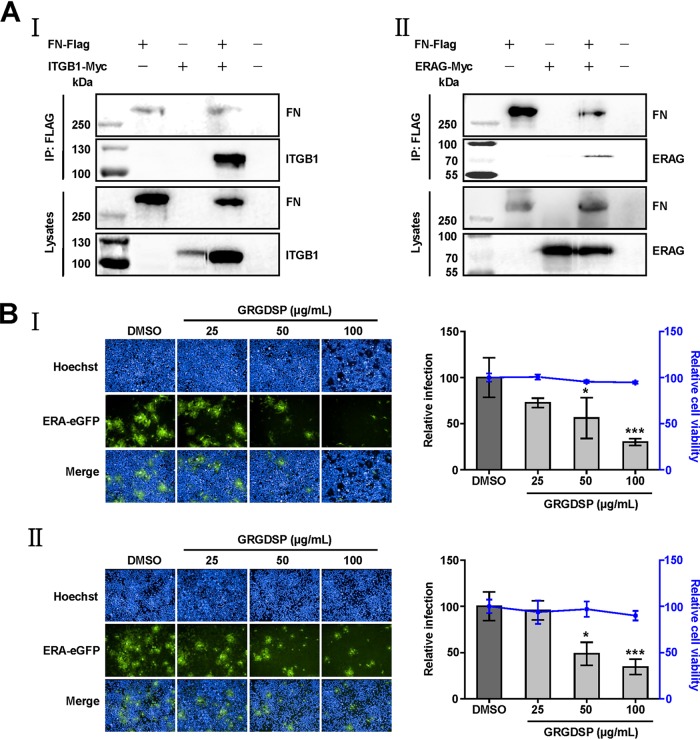
Given the importance of the ITGB1 ectodomain, we investigated whether fibronectin (FN), which is the natural ligand of ITGB1, interacted with ITGB1 through Arg-Gly-Asp (RGD) motifs (20). We performed co-IP assays to investigate the interactions between FN and ITGB1 or RABV G. As expected, Flag-tagged FN (FN-Flag) interacted with Myc-tagged ITGB1 (ITGB1-Myc) (Fig. 6A, I) and ERAG-Myc (Fig. 6A, II) in plasmid-coexpressed HEK293 cell lysates.
FIG 6.
Fibronectin is involved in RABV infection. (A) Fibronectin (FN) interacts with ITGB1 and RABV G. Plasmid FN-Flag was cotransfected with plasmid ITGB1-Myc or ERAG-Myc in HEK293 cells at 37°C for 48 h, and FN-Flag interacted with ITGB1-Myc (I) and ERAG-Myc (II) in co-IP assays with HEK293 cell lysates. (B) RGD peptide (GRGDSP) blocked RABV infection in HEK293 (I) and N2a (II) cells in a dose-dependent manner. Different concentrations of GRGDSP were mixed with ERA-eGFP at an MOI of 0.1 at 4°C for 1 h, added to the cells, and incubated at 37°C for 48 h. Dimethyl sulfoxide (DMSO) with the minimum dilution ratio of GRGDSP was used as a control. Relative cell viability and infection were assessed compared with those of the DMSO-treated group, which were set as 100. The data shown are the mean titers ± SD from four independent experiments. The statistical differences were assessed using Student's t test. *, P < 0.05; ***, P < 0.001.
To test the importance of the interaction between FN and ITGB1 for RABV infection, we used RGD peptide GRGDSP to competitively weaken the interaction between FN and ITGB1 and then detected the infectivity of ERA-eGFP. We found that cell viability was unaffected by GRGDSP (100 μg/ml), and the infectivity of ERA-eGFP was blocked by GRGDSP in HEK293 (Fig. 6B, I) and N2a (Fig. 6B, II) cells in a dose-dependent manner. The relative infection rate of ERA-eGFP decreased by 44% in HEK293 cells and 51% in N2a cells at 48 h p.i. by GRGDSP (50 μg/ml), which differed significantly compared with those after treatment with dimethyl sulfoxide (DMSO) in HEK293 and N2a cells. These results indicated that the function-blocking peptide specific for ITGB1-FN blocked RABV infection in vitro.
ITGB1 interacts with nAChRα1 and coexists with RABV particles at the inoculation site of RABV in mouse muscle.
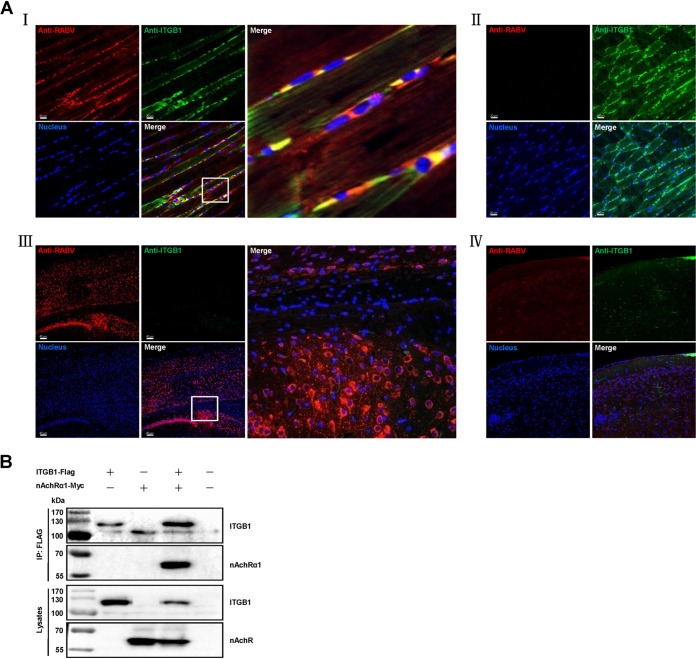
ITGB1 is expressed in muscle but not mature nerves and is important for axon generation during development of the peripheral nervous system (PNS), as well as regeneration of the vertebrate neuromuscular junction in adult PNS (21–25). To clarify whether ITGB1 facilitates RABV cell entry in vivo, 6-week-old female BALB/c mice were injected intramuscularly (i.m.) at the hind leg with phosphate-buffered saline (PBS) or 10 MLD50 (50% mouse lethal dose) GX/09 in 100 μl PBS, or intracerebrally (i.c.) with PBS or 5 MLD50 GX/09 in 30 μl PBS, and then killed 4 days postchallenge. The injected hind leg or brain was subjected to immunohistofluorescence analysis for ITGB1 and RABV antigen in the thigh muscle or cerebral cortex of mice infected with GX/09 as described previously (11). ITGB1 was stained and distributed throughout the cells of thigh muscle (Fig. 7A, II) and coexisted with RABV in GX/09-infected muscle cells (Fig. 7A, I). In contrast, ITGB1 was not stained in cerebral cortical neurons (Fig. 7A, V) and remained unstained after GX/09 infection (Fig. 7A, III). These results indicate that ITGB1 plays a role in RABV peripheral infection in mice.
FIG 7.
RABV and ITGB1 coexist in RABV-inoculated muscle of mice and ITGB1 interacts with nAChRα1. (A) RABV street virus GX/09 and ITGB1 coexist in muscle cells but not cerebral cortex at the RABV-inoculated site. Six-week-old BALB/c mice were i.m. injected with 10 MLD50 GX/09 or i.c. injected with 5 MLD50 GX/09 for 4 days, and the RABV-inoculated thigh muscle and cerebral cortex were collected and fluorescently stained. Mice i.m. or i.c. injected with PBS were used as controls and processed in parallel. RABV antigen (red), ITGB1 (green), and the cell nuclei (blue) were observed in single-fluorescence channels using a Carl Zeiss LSM700 microscope. RABV antigen was observed in thigh muscle (I) and cerebral cortex (III). ITGB1 was stained in thigh muscle (I and II) but not cerebral cortex (III and IV) and colocalized with RABV in thigh muscle (I). The images on the right (I and III) represent amplified random spots in the merged image within the small white box. (B) ITGB1 interacted with nAChRα1. Plasmid ITGB1-Flag and nAChRα1-Myc were cotransfected in HEK293 cells at 37°C for 48 h, and ITGB1-Flag interacted with ERAG-Myc in co-IP assays with HEK293 cell lysates.
nAChRα1 is located at the postsynaptic muscle membrane of neuromuscular junctions and facilitates RABV peripheral infection in vivo (9). To clarify the role of ITGB1 in RABV peripheral infection, the interaction between ITGB1 and nAChRα1 was detect by co-IP assay with plasmid-transfected HEK293 cells. ITGB1 interacts with nAChRα1 (Fig. 7B), which indicates that ITGB1 is involved in RABV peripheral infection.
ITGB1ED, A-4, and GRGDSP neutralize or block infectivity of RABV in mice.
To confirm whether ITGB1 facilitates RABV cell entry in vivo, neutralization assays of ITGB1ED and infection-blocking assays with A-4 and GRGDSP in adult mice via i.m. or i.c. challenge were performed by using RABV street virus strain GX/09. Different amounts of ITGB1ED or IRP, A-4 or mouse IgG2a, and GRGDSP or DMSO were paired and premixed with a fixed concentration of GX/09 and then inoculated into mice via the i.m. route with 10 MLD50 or the i.c. route with 5 MLD50 of GX/09.
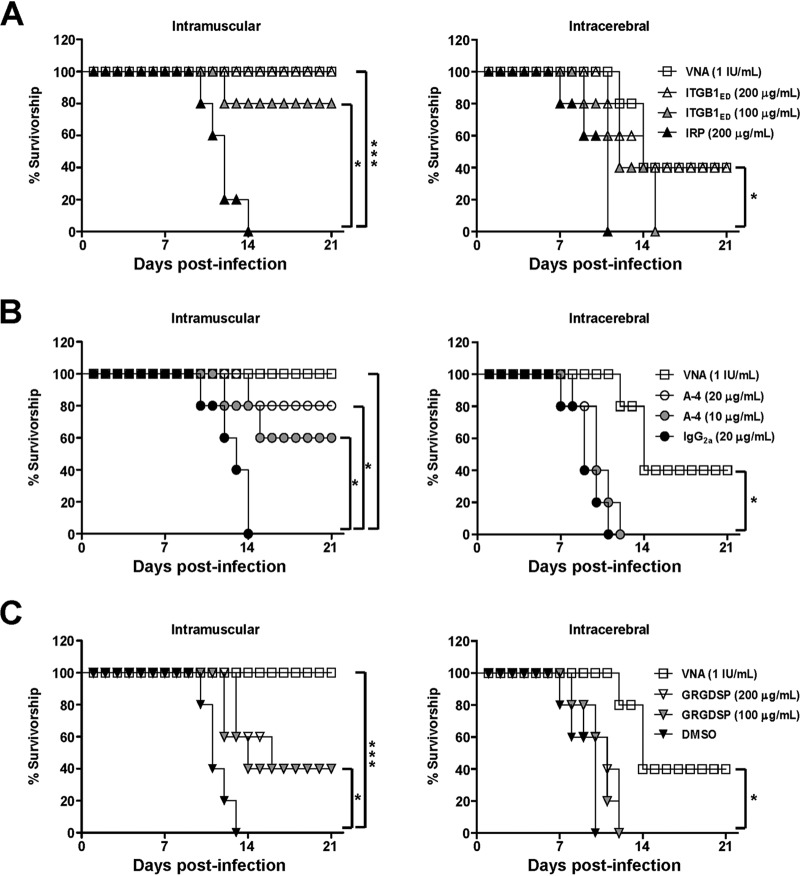
In ITGB1ED neutralization assays, infectivity of GX/09 was neutralized by ITGB1ED in mice following i.m. and i.c. inoculation after a 21-day observation period (Fig. 8A). ITGB1ED at 200 μg/ml conferred complete (i.m.) or 40% (i.c.) protection upon the treated mice, which maintained a survival pattern similar to that of mice treated with 1 IU/ml rabies virus neutralizing antibody (VNA). In contrast, IRP (200 μg/ml) showed no neutralizing effect after i.m. or i.c. challenge. These results demonstrate that the soluble ITGB1 neutralizes RABV infection in mice.
FIG 8.
ITGB1ED, A-4, and GRGDSP protect mice from lethal RABV challenge. (A) ITGB1ED neutralized the infectivity of GX/09 via both i.m. and i.c. challenge. (B and C) Both A-4 (B) and GRGDSP (C) blocked infectivity of GX/09 via i.m. but not i.c. challenge. Different concentrations of ITGB1ED, A-4, and GRGDSP were premixed with 10 MLD50 GX/09 at 4°C for 1 h and then i.m. injected into mice or premixed with 5 MLD50 GX/09 at 4°C for 1 h and then i.c. injected into mice. IRP, mouse IgG2a, DMSO, and rabies virus neutralizing antibody (VNA) were used at the indicated concentrations as controls and processed in parallel. Survival rates of mice were obtained from 21 days of observation. The log-rank (Mantel-Cox) test was used to analyze the statistical difference between the survival rates of the challenged mice. *, P < 0.05; ***, P < 0.001.
In A-4- or GRGDSP-blocking assays, A-4 conferred 80% protection upon mice treated with 20 μg/ml and 60% protection upon mice treated with 10 μg/ml via i.m. challenge after a 21-day observation period, which differed significantly from protection after treatment with 20 μg/ml IgG2a (Fig. 8B). Meanwhile, GRGDSP conferred 40% protection upon mice treated with 20 or 10 μg/ml via i.m. challenge, which also differed significantly from protection after treatment with DMSO (Fig. 8C). Surprisingly, i.c. treatment with both A-4 and GRGDSP (Fig. 8B and C) showed no blocking effect in mice. These results demonstrate that both GRGDSP and A-4 blocked RABV peripheral infection in mice.
DISCUSSION
In this study, we found that ITGB1 is an important host factor for RABV infection. The results in vitro from siRNA silencing, protein interaction, 3D-rendered imaging, soluble protein neutralization, and antibody or peptide blocking, as well as in vivo from immunohistofluorescence assays, soluble protein neutralization, and antibody or peptide blocking, strongly suggest that ITGB1 is an important host factor for RABV peripheral infection.
Integrins belong to the family of transmembrane cell surface molecules that consist of one α and one β subunit, and they are ubiquitously expressed in all metazoan cell types. To date, 18 α and 8 β subunits have been identified that noncovalently form 24 integrin proteins in vertebrates. ITGB1 links with 12 α partners to form the largest and most diverse integrin heterodimers (26). Here, we focused on ITGB1 and did not study the role of different α partners. The ability of ITGB1 to transmit signals inside/outside cells is reliant on interaction with numerous ligands (27). ITGB1 is an RGD-dependent fibronectin receptor, and RGD peptide competitively weakens the interaction between fibronectin and ITGB1 (20). Our studies show that ITGB1 and fibronectin interact with RABV G, and RGD peptide (GRGDSP) functionally blocks the RABV infection in vitro and in vivo, strongly suggesting that ITGB1 facilitates RABV infection depending on fibronectin.
Studies suggest that integrins on the surface of the cell membrane can be endocytosed to enter cells using macropinocytosis mediated by circular dorsal ruffles (28), carbohydrate-binding protein galectin-3 (29), caveolin, Ras homolog A (RhoA), or clathrin-mediated internalization (30). The degradative pathway of integrins is related to the activity and dynamics of integrin itself, and most integrins are recycled back to the cellular membrane from late endosomes (31) or tubular Arf6-containing endosomes (32). ITGB1 shares the currently established traffic mechanisms of other integrins, including clathrin-dependent endocytosis, which is consistent with the well-known endocytosis of RABV (33). Here, we found that ITGB1 and RABV are internalized into cells, which indicates that ITGB1 uses clathrin-dependent endocytosis to enter cells after RABV infection in nature. We also observed that ITGB1 colocalized with RABV in early and late endosomes, which indicates that ITGB1 is involved in RABV traffic after internalization. However, the mechanism still needs to be further explored. In addition, it is also important to detect whether ITGB1 associated with RABV is recycled back to the cellular membrane or degraded.
ITGB1 has been proposed to facilitate viral infection of multiple viruses and plays different roles in different viruses. ITGB1 mediates Kaposi’s sarcoma-associated herpesvirus (KSHV) and Epstein-Barr virus (EBV) attachment and entry into target cells via the RGD motif of viral glycoprotein (34, 35). Similarly, ITGB1 links with integrin αv-mediated avian metapneumovirus (aMPV) membrane fusion and virus infection via the similar RGD motif of viral fusion protein (19). ITGB1 functions as an entry receptor for human cytomegalovirus (HCMV) and is involved in internalization but not cellular attachment via the conserved disintegrin-like domain of HCMV glycoprotein B (12), and it also mediates internalization of mammalian reovirus via the RGD motif of viral λ2 protein (15). In addition, ITGB1 links with integrin α5 are necessary for Ebolavirus entry but not for binding or internalization via assisting the requirement for endosomal cathepsins of viral infection (13) and also are used for porcine hemagglutinating encephalomyelitis virus (PHEV) invasion via the integrin-focal adhesion kinase (FAK) signaling pathway (36). These results were obtained by several assays in vitro from different antibodies, peptides, siRNAs, and/or knocked down cell lines and suggest that ITGB1 affects different stages of early viral infection. Unfortunately, relevant experiments in vivo have rarely been performed. Here, we showed that ITGB1 is a key host factor for RABV early entry based on the results from assays in vitro as well as in vivo. To date, ITGB1 is the first important host factor investigated to inhibit RABV infection in vivo through antibody or peptide blocking. We will extend the in vivo studies with mice lacking ITGB1 and test whether antibody against ITGB1 and RGD peptide can still effectively inhibit the infection of RABV as well as other ITGB1-dependent viruses in vivo.
Previous studies have suggested that ITGB1 is essential for normal morphological development of PNS and CNS, including neuronal and nonneuronal cells (21, 37, 38). Genetic ablation of ITGB1 in mice shows severe perturbation of the PNS, which results in no development at the embryonic stage (39, 40). ITGB1 expressed in muscle is also important for the regeneration of vertebrate neuromuscular junctions in the adult PNS (22, 23, 25). In the present study, we found that antibody against ITGB1 and RGD peptide blocked RABV infection in vivo via intramuscular but not intracerebral challenge, suggesting that ITGB1 plays different roles in RABV PNS and CNS infection in adult mice. In addition, we observed that ITGB1 expression is abundant in skeletal muscle, whereas it is rare in the cerebral cortex. Actually, integrins are excluded from the axons of mature, polarized CNS neurons (41, 42). These results indicate that ITGB1 serves as a key host factor for RABV peripheral infection, and other cellular membrane proteins in the brain that can compensate for the function of ITGB1 may be exploited for RABV CNS infection.
RABV is a highly neurotropic virus that can infect different neuronal and nonneuronal cells with attachment to multiple cellular proteins in vitro and in vivo (43). Previous studies have shown that proposed receptors (nAChRα1, NCAM, and mGluR2) (9 to 11) and some molecules, including phospholipids, gangliosides, sialic acid, carbohydrates, and heparan sulfate, are involved in RABV infection (44–47). nAChRα1, the first suggested receptor for RABV infection, is located at the postsynaptic muscle membrane of neuromuscular junctions and facilitates RABV peripheral infection in muscle cells and subsequent invasion to nerve terminals (9, 48). However, the exact role of nAChRα1 during RABV uptake in vitro and in vivo, and whether it also combines other host factors for peripheral infection, has not yet been determined. Here, we found that ITGB1 interacted with nAChRα1 in co-IP assays. Given the importance of ITGB1 for RABV peripheral infection, it would be valuable to clarify how ITGB1 associated with nAChRα1 facilitates RABV peripheral infection.
Rabies is still a worldwide public health problem, and it carries a heavy economic burden worldwide (1). Currently, no therapy has been shown to prevent death (4). Although rabies vaccines are highly effective for postexposure prophylaxis (PEP) in rabies control (3), a few PEP failures have occurred in developing countries due to deviations from the WHO-recommended prophylaxis protocol, including delay in prophylaxis, poor-quality rabies vaccine, and/or lack of administration of biological products against RABV (49). Four classes of biological product are available for passive immunization, including human rabies immunoglobulin, equine rabies immunoglobulin, purified F(ab´)2 fragments of equine immunoglobulin, and antibodies against RABV (50–52). These biological products contribute to the delay of rabies development in cases of complex PEP. However, rabies immunoglobulins and specific antibodies often require more consideration, such as safety, availability, purity, and cost, which leads to short supply globally. Hence, high-quality and affordable anti-RABV agents and chemotherapy are sought for PEP prophylaxis and therapy. In this study, we showed that antibody A-4 and GRGDSP could block the street RABV infection in vivo via intramuscular inoculation. With proper administration, A-4 and GRGDSP have the potential to delay rabies development in nature and, as an alternative complement to vaccines, may avoid PEP failures. The safety and efficacy of A-4 and GRGDSP need to be further monitored before being recommended for use in public health programs.
MATERIALS AND METHODS
Ethics statement.
All animal experiments were performed by strictly following the Guide for the Care and Use of Laboratory Animals (53) and approved by the Committee on the Ethics of Animal Experiments of the Harbin Veterinary Research Institute, Chinese Academy of Agricultural Sciences. RABV experiments were performed under biosafety level 2 (BSL-2) and animal BSL-3 conditions.
Cells and viruses.
Human embryonic kidney cells (HEK293) (ATCC CRL-1573) and mouse neuroblastoma cells (N2a cells) (ATCC CCL-131) were maintained in Dulbecco’s modified Eagle’s medium (DMEM) plus 10% fetal bovine serum (FBS), and BSR cells were maintained in DMEM plus 5% FBS at 37°C with 5% CO2. RABV Evelyn-Rokitnicki-Abelseth (ERA) strain, recombinant ERA expressing eGFP (ERA-eGFP), recombinant ERA fused with a Flag (DYKDDDDK) tag (rERAG/Flag) or a Myc (EQKLISEEDL) tag (rERAG/Myc) at the C terminus of glycoprotein, and recombinant ERA expressing an additional ERA nucleoprotein fused with a red fluorescent protein, mCherry (ERA-N/mCherry), were propagated in BSR cells and maintained in our laboratory (54). RABV street virus strain GX/09, isolated from the brain of a rabid dog in Guangxi Province, China, in 2009, was propagated in mouse brain and titrated in adult mice by i.m. inoculation and expressed as MLD50 per milliliter (7). All viruses were stored at −70°C before use.
Plasmids.
Recombinant pCAGGS plasmids expressing ITGB1 (p-ITGB1; GenBank accession no. NM_002211) or different C-terminal Flag- or Myc-tagged proteins were constructed, including ITGB1 (ITGB1-Flag and ITGB1-Myc), ITGB1 ectodomain (aa 1 to 728, ITGB1ED-Flag), ITGB1 transmembrane/cytoplasmic domain (aa 729 to 798, ITGB1TC-Flag), fibronectin (FN-Flag; GenBank accession no. NM_212482), ERA G (ERAG-Myc; GenBank accession no. J02293.1), ERA G ectodomain (aa 20 to 459, ERAGED-Myc), ERA G transmembrane/cytoplasmic domain (aa 460 to 524, ERAGTC-Myc), CVS24 G (CVS24G-Myc) (16), GX/09 G (GX/09G-Myc; GenBank accession no. GQ472537.1), WCBV (WCBVG-Myc; GenBank accession no. EF614258.1), and nAChRα1-Myc (GenBank accession no. NM_001039523).
RNAi.
siRNA s7575 (Ambion), targeting human integrin β1, s2563 (Ambion), targeting mouse integrin β1, RABV L (sense, 5′-GGAAUGCACUUUCGAUAUATT-3′; antisense, 5′-UAUAUCGAAAGUGCAUUCCTT-3′), targeting the RABV L gene, and irrelative siRNA (IRRNA) (Ambion) were used. s7575, RABV L, or IRRNA (200 nM, 5 μl/well) was mixed with 35 μl Opti-MEM medium (Invitrogen) containing 0.15 μl Lipofectamine RNAiMAX transfection reagent (Invitrogen) on 96-well cell carrier plates (PerkinElmer) and incubated for 30 min, and 60 μl Opti-MEM medium containing 1.0 × 104 HEK293 cells was added. s2563, RABV L, or IRRNA at the same dose was used in N2a cells and processed in parallel. The cells were incubated for 48 h at 37°C to knock down gene expression.
In vitro RABV infection, multistep growth assay, and viral titration.
HEK293 or N2a cells were infected with ERA-eGFP at a multiplicity of infection (MOI) of 0.1. The supernatants were harvested at 24, 36, 48, and 60 h postinoculation (h p.i.) for virus titration, and virus titers were determined in BSR cells and expressed as focus-forming units (FFU) per milliliter (54). Cells were fixed with 3% paraformaldehyde at 48 h p.i. and stained with Hoechst 33342 (Invitrogen) in PBS for 30 min. The stained cells were imaged using the PerkinElmer Operetta high-content system (PerkinElmer). The infection rate, determined according to the numbers of infected cells versus the total number of cells per well with Columbus software (PerkinElmer), is shown as relative infection by comparing the infection ratio with that of the control group, which was set as 100 (11).
Overexpression assay.
HEK293 cells in 24-well plates (105 cells/well) were transfected with TransIT-293 transfection reagent (Mirus Bio) and 0.5 μg p-ITGB1 or pCAGGS. At 36 h posttransfection, cells were infected with ERA-eGFP at an MOI of 5 for 1 h at 4°C, washed with Dulbecco’s modified Eagle’s medium (DMEM) three times, and then incubated at 37°C. The supernatants were harvested at different time points for virus titration.
Flow cytometry.
To determine whether ITGB1 expression was downregulated, siRNA-silenced HEK293 or N2a cells were washed twice with PBS containing 3% fetal bovine serum (FBS) and collected after being dispersed with 0.25% trypsin, fixed with 3% paraformaldehyde, and then stained with mouse anti-ITGB1 MAb A-4 (Santa Cruz Biotechnology) as the primary antibody and fluorescein-isothiocyanate-conjugated goat anti-mouse antibody (Invitrogen) as the secondary antibody. IRRNA-treated cells stained with A-4 served as the reference population for ITGB1 fluorescence, and siRNA-silenced cells stained with the mouse IgG2a (Santa Cruz Biotechnology) served as the reference population for background fluorescence. To determine whether RABV infection results in the downregulation of cell surface ITGB1, N2a cells were infected with ERA at 37°C for 30 min and then washed, dispersed, fixed, and lastly stained with antibodies as described above. Uninfected cells and cells infected with ERA at 4°C for 30 min stained with A-4 served as the reference population for ITGB1 fluorescence, whereas uninfected cells and cells infected with ERA at 37°C for 30 min stained with IgG2a served as the reference population for background fluorescence. The levels of cellular ITGB1 were measured as the cell surface fluorescence density and determined using an FC500 flow cytometer (Beckman Coulter). Data were analyzed with FlowJo software, and the relative events of ITGB1 were determined using the comparative fluorescence density with IRRNA-treated or uninfected cells stained with A-4 as a control, which was set as 100.
Cell viability.
The CellTiter-Glo kit (Promega) was used to determine cell viability. HEK293 or N2a cells in 96-well plates with opaque walls were incubated with 100 μl CellTiter-Glo reagent (Promega) per well for 10 min on a shaker to induce cell lysis. Luminescence was then measured with a GloMax 96 microplate luminometer (Promega).
Multiplex immunofluorescence and Imaris 3D rendering.
N2a cells were incubated with ERA-N/mCherry at an MOI of 5 for 1 h at 4°C, washed with prechilled DMEM, and then cultured with new DMEM (supplemented with 2% FBS) at 37°C for 30 min. Cells were fixed in prechilled 3% paraformaldehyde in PBS at 4°C for 20 min, treated with 0.3% hydrogen peroxide in PBS at room temperature for 20 min to quench the endogenous peroxidase activity, and then permeabilized by 0.25% Triton X-100 in PBS at room temperature for 15 min. Following blocking steps (Zsbio), cells were incubated with primary antibody at 4°C overnight followed by horseradish peroxidase (HRP)-conjugated secondary antibody (Zsbio) at 4°C for 30 min and then incubated with the TSA amplification reagent (PerkinElmer) diluted 1:100 in reaction buffer at room temperature for 30 to 120 s to achieve the best signal intensity and signal-to-noise ratio. After a brief rinse with stripping buffer at 37°C for 30 min to remove the first primary/secondary antibodies and retain the TSA signal, cells were serially incubated with specific primary/secondary antibodies and spectrally different TSA reagents to detect the other antigens by following the above-described method. The antibodies used in this study were rabbit anti-integrin β1 MAb D6S1W (Cell Signaling Technology), rabbit anti-mCherry polyclonal antibody (pAb) (Abcam), rabbit anti-Rab5 MAb (Abcam), rabbit anti-Rab7 MAb (Abcam), and HRP-conjugated anti-rabbit IgG (GenScript). Data were processed using Bitplane Imaris software (Bitplane AG) to generate 3D-rendered images (11). The red channel (RABV), purple channel (Rab5 or Rab7), green channel (ITGB1), and new channels that represent the merges of two or three channels were established and analyzed for colocalization of RABV, ITGB1, and Rab5 or Rab7.
Cells were scanned for 24 layers along the z axis with a pixel dwell time of 1 μs to acquire images using a Zeiss LSM880 laser-scanning confocal microscope (Carl Zeiss AG) with Airyscan (objective, 63×, numerical aperture, 1.4; Plan-Apochromat). The 3D-rendered images then were generated using Bitplane Imaris software (Bitplane AG) in a stepwise fashion (11). The red (RABV) and purple (Rab5 and Rab7) channels were processed using the “surface module;” the surface results of the purple and red channels were then inputted as “cell” and “nuclei,” respectively, under the “cell module.” The green channel (ITGB1) next was processed using the surface module. After that, a merged channel representing the colocalization of RABV and ITGB1 was established by merging the red and green channels using the “mask data set” of the “coloc” module. Spots in the merged channel were counted by using the “spot” module. Finally, the spots results were input into “cell,” and the new spots representing the colocalization of RABV, ITGB1, and Rab5 or Rab7 (displayed as white spots in the 3D-rendered image) were counted.
Co-IP assays.
HEK293 cells cotransfected with targeted pCAGGS plasmids were washed with PBS 48 h posttransfection and lysed with 1% NP-40–PBS buffer for 1 h. Cell debris were removed by centrifugation (12,000 rpm) for 10 min, and the supernatant was collected and then mixed with protein G agarose (Roche Diagnostics GmbH) for 4 h. Following centrifugation, the supernatant was collected and incubated overnight with anti-Flag antibody-conjugated agarose beads (Sigma-Aldrich). All steps were done at 4°C on a flip shaker. Following extensive washes with prechilled 1% NP-40–PBS buffer, the beads with protein complexes were subjected to SDS-PAGE by adding SDS sample buffer and heating for 10 min. Immunoprecipitates were analyzed by Western blotting using anti-Flag or Myc MAb (Genscript) and Super ECL Star (US Everbright, Inc.).
For co-IP assay of ITGB1 and ERA G from RABV, HEK293 cells were infected with rERAG/Myc (MOI of 0.1) 12 h posttransfection of ITGB1-Flag, and then co-IP assay was carried out at 36 h p.i. using the same method as that described above. For co-IP assay of endogenous ITGB1 and ERA G from RABV, HEK293 cells were infected with rERAG/Flag (MOI of 0.1), and then co-IP assay was carried out at 48 h p.i., except that the anti-Myc MAb of Western blotting was replaced with A-4. For co-IP assay of ITGB1 and ERA G under acidic conditions, cotransfected HEK293 cells were lysed with acidic NP-40 PBS buffer (pH 5.5) and then co-IP assay was carried out. For co-IP assay of ITGB1 and ERA G under antibody blocking, HEK293 cells were transfected with ITGB1-Flag or ERAG-Myc. After the collection of protein supernatants, samples of ITGB1-Flag were incubated overnight with anti-Flag antibody-conjugated agarose beads and then mixed with samples of ERAG-Myc at 4°C for 2 h after incubation with A-4 or IgG2a (10 μg/ml) at 4°C for 1 h. Following washes, beads with protein complexes were subjected to SDS-PAGE and Western blotting.
Pulldown assays.
GST-tagged ITGB1ED and His-tagged ERAGED were expressed and purified (FriendBio Technology). ITGB1ED (10 μg) was mixed with 100 μl of glutathione Sepharose 4B beads (GE Healthcare Bioscience) for 2 h. Following washes with 1% NP-40–PBS buffer, the beads were collected and resuspended in 100 μl of PBS and then mixed with 5 μg of ERAG-His for 2 h. All steps were done at 4°C on a flip shaker, and the GST-tagged irrelative protein (IRP; GenScript) was used as a control and processed in parallel. Following extensive washes, the beads with protein complexes were subjected to SDS-PAGE and Western blotting using anti-GST or His MAb (GenScript) and Super ECL Star (US Everbright Inc.).
Infectivity neutralization and blocking assays.
HEK293 or N2a cells were plated in 96-well carrier plates. At 100 μl per well, different concentrations of ITGB1ED (25, 50, and 100 μg/ml) or IRP (100 μg/ml) for infectivity neutralization, A-4 (2.5, 5, and 10 μg/ml) or mouse IgG2a (10 μg/ml) for antibody-blocking assays, or GRGDSP (25, 50, and 100 μg/ml) (Sigma-Aldrich) or DMSO for ligand-blocking assays were paired and mixed with ERA-eGFP at an MOI of 0.1 in DMEM (supplemented with 2% FBS) at 4°C for 1 h and added to the cells. Cells were incubated at 37°C for 48 h and fixed and stained for imaging and analysis of relative infection.
Mouse challenge tests.
Groups of 10 6-week-old female BALB/c mice (Vital River Laboratories) were randomly divided. Different amounts of ITGB1ED (100 and 200 μg/ml) or IRP (200 μg/ml), A-4 (10 and 20 μg/ml) or mouse IgG2a (20 μg/ml), and GRGDSP (100 and 200 μg/ml) or DMSO (5%) were paired and mixed with 10 MLD50 RABV GX/09 in 100 μl PBS at 4°C for 1 h and injected into the thigh muscle of mice for i.m. challenge or mixed with 5 MLD50 GX/09 in 30 μl PBS at 4°C for 1 h and injected into the skull cavity of mice for i.c. challenge. Mice were housed in cages under controlled conditions of humidity and observed for 21 days for definitive clinical signs of rabies. Survival rates obtained with the different infection groups were compared.
Immunohistofluorescence.
Six-week-old BALB/c mice that had been i.m. (10 MLD50) or i.c. (5 MLD50) injected with RABV street virus GX/09 for 4 days were humanely euthanized. The GX/09-inoculated hind leg or brain were collected and fixed in 10% (vol/vol) formaldehyde–PBS buffer, dehydrated and embedded in paraffin wax, and sectioned at 1.5-μm thickness. Following antigen retrieval with a citrate acid (pH 7.4)–sodium citrate buffer solution (pH 8.0) at 121°C for 30 min, the sections were treated with a 3% H2O2 methanol solution at room temperature for 30 min to quench the endogenous peroxidase activity and blocked with 5% skim milk in PBS for 30 min at room temperature. Following thorough washes, sections were first stained with 1:50 diluted rabbit anti-integrin β1 MAb D6S1W and 1:100 diluted fluorescein goat anti-rabbit IgG (Vector Labs), next stained with 1:10 diluted mouse anti-RABV P protein MAb (prepared in our laboratory) and 1:100 diluted DyLight 680-labeled anti-mouse IgG (SeraCare), and lastly stained with Fluoroshield with 4′,6-diamidino-2-phenylindole (DAPI) (Sigma-Aldrich). Slides were imaged using a Carl Zeiss LSM700 microscope (Carl Zeiss).
Statistical analysis.
The assays described above were independently repeated four times. Data shown were the means plus standard deviations (SD) and generated using GraphPad Prism software. Statistical analyses were carried out using a paired Student's t test and log-rank (Mantel-Cox) test.
ACKNOWLEDGMENTS
This work was supported by the National Key Research and Development Program of China (2016YFD0500403) and the National Natural Science Fund of China (31800138). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
REFERENCES
- 1.Rupprecht C, Kuzmin I, Meslin F. 2017. Lyssaviruses and rabies: current conundrums, concerns, contradictions and controversies. F1000Res 6:184. doi: 10.12688/f1000research.10416.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Minghui R, Stone M, Semedo MH, Nel L. 2018. New global strategic plan to eliminate dog-mediated rabies by 2030. Lancet Glob Health 6:e828–e829. doi: 10.1016/S2214-109X(18)30302-4. [DOI] [PubMed] [Google Scholar]
- 3.Davis BM, Rall GF, Schnell MJ. 2015. Everything you always wanted to know about rabies virus (but were afraid to ask). Annu Rev Virol 2:451. doi: 10.1146/annurev-virology-100114-055157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mahadevan A, Suja MS, Mani RS, Shankar SK. 2016. Perspectives in diagnosis and treatment of rabies viral encephalitis: insights from pathogenesis. Neurotherapeutics 13:477–492. doi: 10.1007/s13311-016-0452-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Etessami R, Conzelmann KK, Fadai-Ghotbi B, Natelson B, Tsiang H, Ceccaldi PE. 2000. Spread and pathogenic characteristics of a G-deficient rabies virus recombinant: an in vitro and in vivo study. J Gen Virol 81:2147–2153. doi: 10.1099/0022-1317-81-9-2147. [DOI] [PubMed] [Google Scholar]
- 6.Wang X, Feng N, Ge J, Shuai L, Peng L, Gao Y, Yang S, Xia X, Bu Z. 2012. Recombinant canine distemper virus serves as bivalent live vaccine against rabies and canine distemper. Vaccine 30:5067–5072. doi: 10.1016/j.vaccine.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 7.Ge J, Wang X, Tao L, Wen Z, Feng N, Yang S, Xia X, Yang C, Chen H, Bu Z. 2011. Newcastle disease virus-vectored rabies vaccine is safe, highly immunogenic, and provides long-lasting protection in dogs and cats. J Virol 85:8241–8252. doi: 10.1128/JVI.00519-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Albertini AA, Schoehn G, Weissenhorn W, Ruigrok RW. 2008. Structural aspects of rabies virus replication. Cell Mol Life Sci 65:282–294. doi: 10.1007/s00018-007-7298-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lentz TL, Burrage TG, Smith AL, Crick J, Tignor GH. 1982. Is the acetylcholine receptor a rabies virus receptor? Science 215:182–184. doi: 10.1126/science.7053569. [DOI] [PubMed] [Google Scholar]
- 10.Thoulouze MI, Lafage M, Schachner M, Hartmann U, Cremer H, Lafon M. 1998. The neural cell adhesion molecule is a receptor for rabies virus. J Virol 72:7181–7190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang JL, Wang ZL, Liu RQ, Shuai L, Wang XX, Luo J, Wang C, Chen WY, Wang XJ, Ge JY, He XJ, Wen ZY, Bu ZG. 2018. Metabotropic glutamate receptor subtype 2 is a cellular receptor for rabies virus. PLoS Pathog 14:e1007189. doi: 10.1371/journal.ppat.1007189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feire AL, Koss H, Compton T. 2004. Cellular integrins function as entry receptors for human cytomegalovirus via a highly conserved disintegrin-like domain. Proc Natl Acad Sci U S A 101:15470–15475. doi: 10.1073/pnas.0406821101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schornberg KL, Shoemaker CJ, Dube D, Abshire MY, Delos SE, Bouton AH, White JM. 2009. Alpha5beta1-integrin controls ebolavirus entry by regulating endosomal cathepsins. Proc Natl Acad Sci U S A 106:8003–8008. doi: 10.1073/pnas.0807578106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weigel-Kelley KA, Yoder MC, Srivastava A. 2003. Alpha5beta1 integrin as a cellular coreceptor for human parvovirus B19: requirement of functional activation of beta1 integrin for viral entry. Blood 102:3927–3933. doi: 10.1182/blood-2003-05-1522. [DOI] [PubMed] [Google Scholar]
- 15.Maginnis MS, Forrest JC, Kopecky-Bromberg SA, Dickeson SK, Santoro SA, Zutter MM, Nemerow GR, Bergelson JM, Dermody TS. 2006. Beta1 integrin mediates internalization of mammalian reovirus. J Virol 80:2760–2770. doi: 10.1128/JVI.80.6.2760-2770.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wenqiang J, Xiangping Y, Xuerui L, Jixing L. 2013. Complete genome sequence of rabies virus CVS-24 from China. Arch Virol 158:1993–2000. doi: 10.1007/s00705-013-1657-z. [DOI] [PubMed] [Google Scholar]
- 17.Zhao DD, Shuai L, Ge JY, Wang JL, Wen ZY, Liu RQ, Wang C, Wang XJ, Bu ZG. 2019. Generation of recombinant rabies virus ERA strain applied to virus tracking in cell infection. J Integr Agric 18:2361. doi: 10.1016/S2095-3119(19)62717-6. [DOI] [Google Scholar]
- 18.Cseke G, Maginnis MS, Cox RG, Tollefson SJ, Podsiad AB, Wright DW, Dermody TS, Williams JV. 2009. Integrin alphavbeta1 promotes infection by human metapneumovirus. Proc Natl Acad Sci U S A 106:1566–1571. doi: 10.1073/pnas.0801433106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yun BL, Guan XL, Liu YZ, Zhang Y, Wang YQ, Qi XL, Cui HY, Liu CJ, Zhang YP, Gao HL, Gao L, Li K, Gao YL, Wang XM. 2016. Integrin alphavbeta1 modulation affects subtype B avian metapneumovirus fusion protein-mediated cell-cell fusion and virus infection. J Biol Chem 291:14815–14825. doi: 10.1074/jbc.M115.711382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagae M, Re S, Mihara E, Nogi T, Sugita Y, Takagi J. 2012. Crystal structure of alpha5beta1 integrin ectodomain: atomic details of the fibronectin receptor. J Cell Biol 197:131–140. doi: 10.1083/jcb.201111077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leone DP, Relvas JB, Campos LS, Hemmi S, Brakebusch C, Fassler R, Ffrench-Constant C, Suter U. 2005. Regulation of neural progenitor proliferation and survival by beta1 integrins. J Cell Sci 118:2589–2599. doi: 10.1242/jcs.02396. [DOI] [PubMed] [Google Scholar]
- 22.Schwander M, Shirasaki R, Pfaff SL, Muller U. 2004. Beta1 integrins in muscle, but not in motor neurons, are required for skeletal muscle innervation. J Neurosci 24:8181–8191. doi: 10.1523/JNEUROSCI.1345-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kashani AH, Chen BM, Grinnell AD. 2001. Hypertonic enhancement of transmitter release from frog motor nerve terminals: Ca2+ independence and role of integrins. J Physiol 530:243–252. doi: 10.1111/j.1469-7793.2001.0243l.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hawthorne AL, Hu H, Kundu B, Steinmetz MP, Wylie CJ, Deneris ES, Silver J. 2011. The unusual response of serotonergic neurons after CNS injury: lack of axonal dieback and enhanced sprouting within the inhibitory environment of the glial scar. J Neurosci 31:5605–5616. doi: 10.1523/JNEUROSCI.6663-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eva R, Andrews MR, Franssen EH, Fawcett JW. 2012. Intrinsic mechanisms regulating axon regeneration: an integrin perspective. Int Rev Neurobiol 106:75–104. doi: 10.1016/B978-0-12-407178-0.00004-1. [DOI] [PubMed] [Google Scholar]
- 26.Brower DL, Brower SM, Hayward DC, Ball EE. 1997. Molecular evolution of integrins: genes encoding integrin beta subunits from a coral and a sponge. Proc Natl Acad Sci U S A 94:9182–9187. doi: 10.1073/pnas.94.17.9182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo BH, Carman CV, Springer TA. 2007. Structural basis of integrin regulation and signaling. Annu Rev Immunol 25:619–647. doi: 10.1146/annurev.immunol.25.022106.141618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gu Z, Noss EH, Hsu VW, Brenner MB. 2011. Integrins traffic rapidly via circular dorsal ruffles and macropinocytosis during stimulated cell migration. J Cell Biol 193:61–70. doi: 10.1083/jcb.201007003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lakshminarayan R, Wunder C, Becken U, Howes MT, Benzing C, Arumugam S, Sales S, Ariotti N, Chambon V, Lamaze C, Loew D, Shevchenko A, Gaus K, Parton RG, Johannes L. 2014. Galectin-3 drives glycosphingolipid-dependent biogenesis of clathrin-independent carriers. Nat Cell Biol 16:595–606. doi: 10.1038/ncb2970. [DOI] [PubMed] [Google Scholar]
- 30.Mai A, Muharram G, Barrow-McGee R, Baghirov H, Rantala J, Kermorgant S, Ivaska J. 2014. Distinct c-Met activation mechanisms induce cell rounding or invasion through pathways involving integrins, RhoA and HIP1. J Cell Sci 127:1938–1952. doi: 10.1242/jcs.140657. [DOI] [PubMed] [Google Scholar]
- 31.Dozynkiewicz MA, Jamieson NB, Macpherson I, Grindlay J, van den Berghe PV, von Thun A, Morton JP, Gourley C, Timpson P, Nixon C, McKay CJ, Carter R, Strachan D, Anderson K, Sansom OJ, Caswell PT, Norman JC. 2012. Rab25 and CLIC3 collaborate to promote integrin recycling from late endosomes/lysosomes and drive cancer progression. Dev Cell 22:131–145. doi: 10.1016/j.devcel.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen PW, Luo R, Jian X, Randazzo PA. 2014. The Arf6 GTPase-activating proteins ARAP2 and ACAP1 define distinct endosomal compartments that regulate integrin alpha5beta1 traffic. J Biol Chem 289:30237–30248. doi: 10.1074/jbc.M114.596155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Piccinotti S, Kirchhausen T, Whelan SP. 2013. Uptake of rabies virus into epithelial cells by clathrin-mediated endocytosis depends upon actin. J Virol 87:11637–11647. doi: 10.1128/JVI.01648-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Akula SM, Pramod NP, Wang FZ, Chandran B. 2002. Integrin alpha3beta1 (CD 49c/29) is a cellular receptor for Kaposi’s sarcoma-associated herpesvirus (KSHV/HHV-8) entry into the target cells. Cell 108:407–419. doi: 10.1016/S0092-8674(02)00628-1. [DOI] [PubMed] [Google Scholar]
- 35.Tugizov SM, Berline JW, Palefsky JM. 2003. Epstein-Barr virus infection of polarized tongue and nasopharyngeal epithelial cells. Nat Med 9:307–314. doi: 10.1038/nm830. [DOI] [PubMed] [Google Scholar]
- 36.Lv X, Li Z, Guan J, Hu S, Zhang J, Lan Y, Zhao K, Lu H, Song D, He H, Gao F, He W. 2019. Porcine hemagglutinating encephalomyelitis virus activation of the integrin alpha5beta1-FAK-Cofilin pathway causes cytoskeletal rearrangement to promote its invasion of N2a cells. J Virol 93:e01736-18. doi: 10.1128/JVI.01736-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Förster E, Tielsch A, Saum B, Weiss KH, Johanssen C, Graus-Porta D, Müller U, Frotscher M. 2002. Reelin, Disabled 1, and beta 1 integrins are required for the formation of the radial glial scaffold in the hippocampus. Proc Natl Acad Sci U S A 99:13178–13183. doi: 10.1073/pnas.202035899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gleeson JG, Walsh CA. 2000. Neuronal migration disorders: from genetic diseases to developmental mechanisms. Trends Neurosci 23:352–359. doi: 10.1016/s0166-2236(00)01607-6. [DOI] [PubMed] [Google Scholar]
- 39.Brakebusch C, Hirsch E, Potocnik A, Fassler R. 1997. Genetic analysis of beta1 integrin function: confirmed, new and revised roles for a crucial family of cell adhesion molecules. J Cell Sci 110:2895–2904. [DOI] [PubMed] [Google Scholar]
- 40.Pietri T, Eder O, Breau MA, Topilko P, Blanche M, Brakebusch C, Fassler R, Thiery JP, Dufour S. 2004. Conditional beta1-integrin gene deletion in neural crest cells causes severe developmental alterations of the peripheral nervous system. Development 131:3871–3883. doi: 10.1242/dev.01264. [DOI] [PubMed] [Google Scholar]
- 41.Bi X, Lynch G, Zhou J, Gall CM. 2001. Polarized distribution of alpha5 integrin in dendrites of hippocampal and cortical neurons. J Comp Neurol 435:184–193. doi: 10.1002/cne.1201. [DOI] [PubMed] [Google Scholar]
- 42.Eva R, Fawcett J. 2014. Integrin signalling and traffic during axon growth and regeneration. Curr Opin Neurobiol 27:179–185. doi: 10.1016/j.conb.2014.03.018. [DOI] [PubMed] [Google Scholar]
- 43.Charlton KM, Nadin-Davis S, Casey GA, Wandeler AI. 1997. The long incubation period in rabies: delayed progression of infection in muscle at the site of exposure. Acta Neuropathol 94:73–77. doi: 10.1007/s004010050674. [DOI] [PubMed] [Google Scholar]
- 44.Lafon M. 2005. Rabies virus receptors. J Neurovirol 11:82–87. doi: 10.1080/13550280590900427. [DOI] [PubMed] [Google Scholar]
- 45.Superti F, Derer M, Tsiang H. 1984. Mechanism of rabies virus entry into CER cells. J Gen Virol 65:781–789. doi: 10.1099/0022-1317-65-4-781. [DOI] [PubMed] [Google Scholar]
- 46.Sasaki M, Anindita PD, Ito N, Sugiyama M, Carr M, Fukuhara H, Ose T, Maenaka K, Takada A, Hall WW, Orba Y, Sawa H. 2018. The role of heparan sulfate proteoglycans as an attachment factor for rabies virus entry and infection. J Infect Dis 217:1740–1749. doi: 10.1093/infdis/jiy081. [DOI] [PubMed] [Google Scholar]
- 47.Conti C, Superti F, Tsiang H. 1986. Membrane carbohydrate requirement for rabies virus binding to chicken embryo related cells. Intervirology 26:164–168. doi: 10.1159/000149696. [DOI] [PubMed] [Google Scholar]
- 48.Lewis P, Fu Y, Lentz TL. 2000. Rabies virus entry at the neuromuscular junction in nerve-muscle cocultures. Muscle Nerve 23:720–730. doi:. [DOI] [PubMed] [Google Scholar]
- 49.Rupprecht CE, Briggs D, Brown CM, Franka R, Katz SL, Kerr HD, Lett S, Levis R, Meltzer MI, Schaffner W, Cieslak PR. 2009. Evidence for a 4-dose vaccine schedule for human rabies post-exposure prophylaxis in previously non-vaccinated individuals. Vaccine 27:7141–7148. doi: 10.1016/j.vaccine.2009.09.029. [DOI] [PubMed] [Google Scholar]
- 50.Lang J, Attanath P, Quiambao B, Singhasivanon V, Chanthavanich P, Montalban C, Lutsch C, Pepin-Covatta S, Le Mener V, Miranda M, Sabchareon A. 1998. Evaluation of the safety, immunogenicity, and pharmacokinetic profile of a new, highly purified, heat-treated equine rabies immunoglobulin, administered either alone or in association with a purified, Vero-cell rabies vaccine. Acta Trop 70:317–333. doi: 10.1016/s0001-706x(98)00038-2. [DOI] [PubMed] [Google Scholar]
- 51.Muller T, Dietzschold B, Ertl H, Fooks AR, Freuling C, Fehlner-Gardiner C, Kliemt J, Meslin FX, Franka R, Rupprecht CE, Tordo N, Wanderler AI, Kieny MP. 2009. Development of a mouse monoclonal antibody cocktail for post-exposure rabies prophylaxis in humans. PLoS Negl Trop Dis 3:e542. doi: 10.1371/journal.pntd.0000542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chao TY, Ren S, Shen E, Moore S, Zhang SF, Chen L, Rupprecht CE, Tsao E. 2017. SYN023, a novel humanized monoclonal antibody cocktail, for post-exposure prophylaxis of rabies. PLoS Negl Trop Dis 11:e0006133. doi: 10.1371/journal.pntd.0006133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.National Research Council. 2011. Guide for the care and use of laboratory animals, p 220–225, 8th ed. National Academies Press, Washington, DC. [Google Scholar]
- 54.Shuai L, Feng N, Wang X, Ge J, Wen Z, Chen W, Qin L, Xia X, Bu Z. 2015. Genetically modified rabies virus ERA strain is safe and induces long-lasting protective immune response in dogs after oral vaccination. Antiviral Res 121:9–15. doi: 10.1016/j.antiviral.2015.06.011. [DOI] [PubMed] [Google Scholar]