Human cytomegalovirus (HCMV) is a herpesvirus present in up to 85% of some populations. Like all herpesviruses, HCMV infection is for life. No vaccine is currently available, neutralizing antibody therapies are ineffective, and current antivirals have limited long-term efficacy due to side effects and potential for viral mutation and resistance. The significance of this research is in understanding how HCMV manipulates the host mitochondria to support bioenergetic and biosynthetic requirements for replication. Despite a large genome, HCMV relies exclusively on host cells for metabolic functions. By understanding the dependency of HCMV on the mitochondria, we could exploit these requirements and develop novel antivirals.
KEYWORDS: CMV, OXPHOS, cytomegalovirus, electron transport chain, membrane potential, mitochondria, mitochondrial biogenesis, mtDNA, oxidative stress, reactive oxygen species
ABSTRACT
Human cytomegalovirus (HCMV) is a large DNA herpesvirus that is highly prevalent in the human population. HCMV can result in severe direct and indirect pathologies under immunosuppressed conditions and is the leading cause of birth defects related to infectious disease. Currently, the effect of HCMV infection on host cell metabolism as an increase in glycolysis during infection has been defined. We have observed that oxidative phosphorylation is also increased. We have identified morphological and functional changes to host mitochondria during HCMV infection. The mitochondrial network undergoes fission events after HCMV infection. Interestingly, the network does not undergo fusion. At the same time, mitochondrial mass and membrane potential increase. The electron transport chain (ETC) functions at an elevated rate, resulting in the release of increased reactive oxygen species. Surprisingly, despite the stress applied to the host mitochondria, the network is capable of responding to and meeting the increased bioenergetic and biosynthetic demands placed on it. When mitochondrial DNA is depleted from the cells, we observed severe impairment of viral replication. Mitochondrial DNA encodes many of the ETC components. These findings suggest that the host cell ETC is essential to HCMV replication. Our studies suggest the host cell mitochondria may be a therapeutic target.
IMPORTANCE Human cytomegalovirus (HCMV) is a herpesvirus present in up to 85% of some populations. Like all herpesviruses, HCMV infection is for life. No vaccine is currently available, neutralizing antibody therapies are ineffective, and current antivirals have limited long-term efficacy due to side effects and potential for viral mutation and resistance. The significance of this research is in understanding how HCMV manipulates the host mitochondria to support bioenergetic and biosynthetic requirements for replication. Despite a large genome, HCMV relies exclusively on host cells for metabolic functions. By understanding the dependency of HCMV on the mitochondria, we could exploit these requirements and develop novel antivirals.
INTRODUCTION
Human cytomegalovirus (HCMV) is a ubiquitous herpesvirus found in the majority of the population (1). Usually asymptomatic, HCMV infection can lead to serious morbidity and mortality in immunocompromised settings, including transplantation and HIV-positive individuals (2). HCMV is the leading cause of viral associated congenital infections resulting in microcephaly, cognitive disabilities, and hearing loss and has been linked to increased fragility and mortality in elderly populations (3–6). HCMV cellular tropism is diverse and includes epithelial, endothelial, fibroblast, and most immune cell types (7). CD34+ progenitor cells are the site of latency in humans and, like all herpesviruses, HCMV can reactivate and produce new viral progeny (8). HCMV is a large, enveloped DNA virus (∼230 kbp) that encodes over 190 open reading frames, enabling manipulation and control over many host cell pathways (9). These viral properties likely contribute to the slow viral replication kinetics observed with HCMV infection. As a result, there are large energetic and biosynthetic demands on infected host cells during HCMV replication and production of viral progeny.
In eukaryotic cells, mitochondria are the central source of energy production and contribute to cellular proliferation, differentiation, signaling, cell cycle, and cell death pathways (10, 11). Cytosolic glycolysis and mitochondrial oxidative phosphorylation (OXPHOS) pathways converge at the outer and inner membrane mitochondrial membranes (OMM and IMM, respectively) driving energy metabolism and maintaining ATP/ADP ratios (12). Mitochondria are dynamic organelles that undergo fission and fusion events, involving both the OMM and the IMM (13, 14). Changes to mitochondrial shape can result in altered mitochondrial function and homeostasis, while maintaining mitochondrial quality control (15). Mitochondrial fission is mediated by dynamin-related protein 1 (Drp1), a cytosolic dynamin-related GTPase, and results in smaller, individual mitochondria (16). After recruitment to the mitochondria, Drp1 interacts with mitochondrial fission 1 protein (Fis1) located on the OMM. This interaction initiates a scission event, resulting in a parent and daughter organelle. Fusion of mitochondria is the result of a multistep process that includes organelle transport, followed by OMM and IMM fusion events. These events are mediated by optic atrophy 1 (Opa1) protein and mitofusins 1 and 2 (Mfn1 and Mfn2) dynamin-related GTPases (17, 18). In healthy cells, fusion is followed by fission to segregate healthy mitochondria from membranes displaying reduced potential. Ideally, fission is balanced by fusion, ensuring energy demands are met and mitochondrial distribution is maintained. Oxidative stress can lead to mtDNA damage by disrupting mitochondrial proteins, nucleic acids and lipids (19). Damaged or dysfunctional mitochondria are normally removed by mitophagy. This process is balanced by mitochondrial biogenesis that requires the synthesis, import and use of proteins and lipids, and mitochondrial DNA (mtDNA) replication. The master regulator of mitochondrial biogenesis is peroxisome proliferator-activated receptor-gamma coactivators 1α (PGC-1α). PGC-1α activation is associated with upregulation of transcription factors related to mitochondrial metabolism, mitochondrial proteins, energy usage, and import of metabolites (20, 21). Disruption of any of these events results in dysfunctional mitochondria and impaired mitochondrial homeostasis that has been associated with developmental defects, metabolic disease, cancer, neurodegeneration, and numerous other pathologies (14, 22, 23).
As obligate intracellular parasites, viruses rely on host cell metabolic pathways for successful replication. Defining the metabolic requirements during viral infections has been a growing field of interest over the last decade. Viruses have been shown to influence mitochondrial function directly and indirectly through viral proteins, dysregulated calcium homeostasis, and oxidative stress (24–32). Interestingly, HCMV-infected cells exhibit many similarities to the Warburg Effect (aerobic glycolysis), first described in cancer cells (33, 34). HCMV-infected fibroblasts display increased glycolysis and glutaminolysis (35–37). Citrate derived from extracellular glucose is anaplerotically shuttled out of the tricarboxylic acid (TCA) cycle and used for fatty acid synthesis and glutamine is used to feed the TCA cycle (35, 38). Mitochondrial function plays a role in these altered metabolic pathways and is essential to host cell metabolism, and yet the importance of mitochondrial function during HCMV infection is poorly defined. Previous studies have observed that HCMV infection induced mitochondrial fragmentation and biogenesis (25, 39–41). Also, mitochondrial transcription and translation machineries have been reported to be highly upregulated using transcriptome sequencing approaches (25, 42–44). However, it is unclear if host mitochondria are directly involved or even required for HCMV replication. Specifically, the role of the electron transport chain (ETC) during HCMV infection is understudied. We hypothesize that HCMV reprograms mitochondrial function to enhance its replication. To test our hypothesis, we evaluated an acute HCMV infection and changes to mitochondrial morphology and function in the classic fibroblast tissue-culture model. We provide evidence that HCMV infection alters host cell mitochondrial morphology and mitochondrial function, particularly OXPHOS, as a strategy to respond to the stress of viral replication and that correct functioning of mitochondrial metabolism is essential for proper HCMV replication. These data suggest that therapies targeting mitochondria may effectively impair the ability of HCMV to replicate.
RESULTS
HCMV infection of fibroblasts results in a hybrid glycolysis/OXPHOS metabolic phenotype.
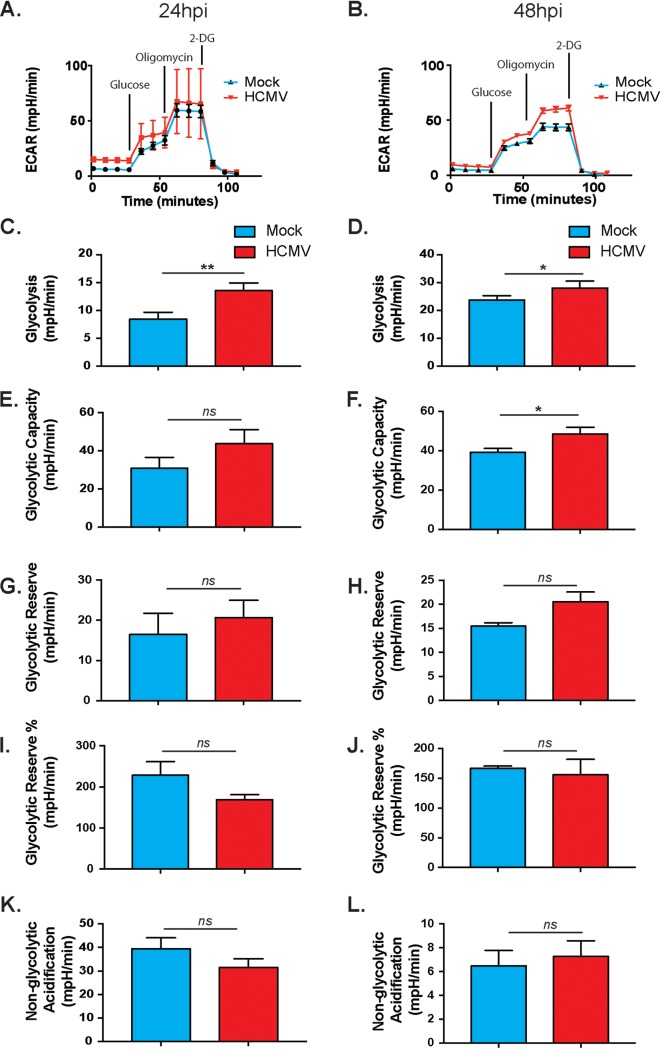
To develop a comprehensive understanding of the metabolic changes occurring during HCMV infection, we compared the two major metabolic pathways: glycolysis and OXPHOS. Using the Seahorse XFe24 Bioanalyzer, we first examined changes to glycolysis following HCMV infection of fibroblasts as measured by extracellular acidification rate (ECAR) (Fig. 1). Briefly, basal levels of glycolysis were measured (time points 1 to 4), followed by a saturating concentration of glucose to determine glycolytic flux (time points 5 to 7). Oligomycin is then added to inhibit mitochondrial ATP production and switch energy production to glycolysis (time points 8 to 10). Lastly, 2-deoxy-d-glucose, a glucose analog, is added to inhibit glycolysis (time points 11 to 13). Consistent with previous studies using targeted metabolomics (36), glycolysis was increased significantly in HCMV-infected cells at 24 and 48 h postinfection (hpi) (Fig. 1C and D). HCMV-infected cells utilized glycolysis for energy production during inhibition of mitochondrial function, as shown by an increase in glycolytic capacity with infection (Fig. 1E and F). The glycolytic reserve, or the potential to increase glycolytic output to maximum capability, was increased in infected cells, but the change did not reach significance (Fig. 1G and H). There were also nonsignificant decreases in glycolytic reserve percentage and nonglycolytic acidification (Fig. 1I to L). These data confirm that HCMV increases glycolysis during HCMV infection.
FIG 1.
HCMV-infected cells display increased glycolysis. The extracellular acidification rate was measured at 24 h (A) or 48 h (B) after HFFs were mock or HCMV infected using the Seahorse XFe24 glycolytic stress assay. Glycolysis (C and D), glycolytic capacity (E and F), glycolytic reserve (G and H), glycolytic reserve % (I and J), and nonglycolytic acidification (K and L) were derived from these measurements. All samples were normalized to protein. The data are representative of pooled data from at least three independent experiments. Error bars indicate ± the standard errors of the mean (SEM) of at least triplicates. *, P < 0.05; **, P < 0.01; ns, not significant.
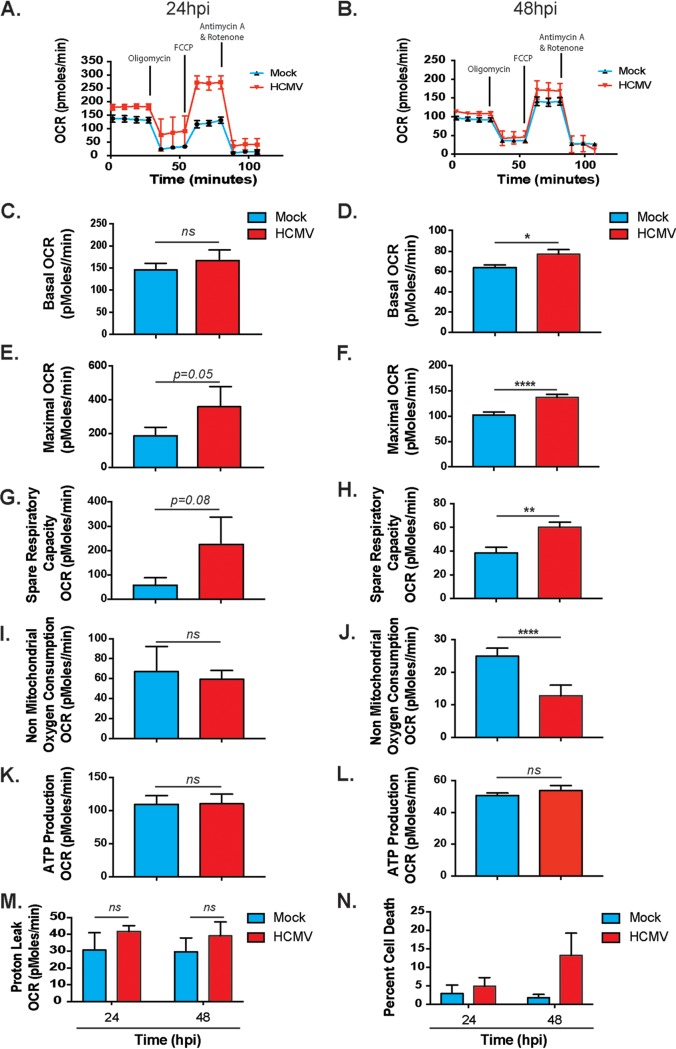
A significant increase in glycolysis may suggest less reliance on or a decrease in mitochondrial respiration. Previous studies have indicated that mitochondria are altered during HCMV infection (25, 39, 42). We observed that OXPHOS activity was increased in HCMV-infected cells, as measured by the oxygen consumption rate (OCR) using the Seahorse XFe24 platform (Fig. 2A to L). Briefly, basal respiration is measured (time points 1 to 4) prior to addition of an ATP synthase inhibitor, oligomycin (time points 5 to 7). This allows the determination of mitochondrial respiration associated cellular ATP production. Carbonyl cyanide-4 (trifluoromethoxy) phenylhydrazone (FCCP) is then added to disrupt the proton gradient and membrane potential (time points 8 to 10). Lastly, ETC complex inhibitors rotenone and antimycin A are added to assess nonmitochondrial respiration (time points 11 to 13). Basal respiration and maximal respiration are increased with HCMV infection at 24 hpi (Fig. 2C and Fig. 2E), reaching significance at 48 hpi (Fig. 2D and Fig. 2F). The spare respiratory capacity, or mitochondrial bioenergetic potential, in HCMV-infected cells increased significantly at 48 hpi, suggesting that HCMV enhances mitochondrial function (Fig. 2H). The increase in OCR observed does not appear to be a result of an increase in the activity of other oxygen-dependent systems, as indicated by the decrease in nonmitochondrial oxygen consumption during HCMV infection (Fig. 2I and J). Consistent with this, proton leak is not significantly increased during infection (Fig. 2M), suggesting that the mitochondria are functional and not simply consuming oxygen futilely. Using indirect Seahorse-derived measurements, we observed no change to ATP levels in infected cells at our measured time points (Fig. 2K and L).
FIG 2.
HCMV-infected cells display increased OXPHOS. The OCR was measured at 24 h (A) or 48 h (B) after HFFs were mock or HCMV infected using the Seahorse XFe24 MitoStress kit. Basal OCR (C and D), maximal OCR (E and F), spare respiratory capacity (G and H), nonmitochondrial oxygen consumption (I and J), ATP production (K and L), and proton leak (M) were derived from these measurements. All samples were normalized to protein. (N) Flow cytometry viability assays were used to measure cell death in mock- and HCMV-infected cells. Data pooled from at least three independent experiments. Error bars indicate ± the SEM of at least triplicates. *, P < 0.05; **, P < 0.01; ****, P < 0.0001; ns, not significant.
To confirm that HCMV-induced cell death was not a confounding factor in our observed metabolic phenotypes, we also analyzed cell viability by fluorescence-activated cell sorting using a live/dead stain. We observed less than 15% cell death at 24 and 48 hpi (Fig. 2N). Thus, the HCMV-induced changes to metabolic function are unlikely related to cell death, although this cannot be completely eliminated as a factor. Collectively, these data suggest glycolysis is not a dominant metabolic phenotype during HCMV infection. Rather, glycolysis and mitochondrial oxidation energy pathways are both required for bioenergetic and biosynthetic purposes during HCMV replication.
HCMV increases mitochondrial function of host cells.
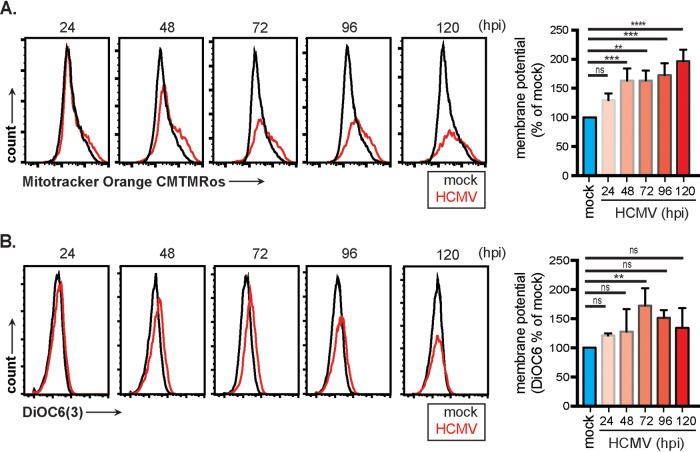
To evaluate the oxidative activity of mitochondria under mock- or HCMV-infected conditions, cells were labeled with MitoTracker Orange CM-H2TMRos (CMTMRos), a nonfluorescent reduced form of tetramethylrosamine that is oxidized to a fluorescent state by molecular oxygen in actively respiring cells. CMTMRos values gradually increased in infected cells between 24 and 120 hpi (Fig. 3A). Respiration is dependent on mitochondrial membrane potential. To assess mitochondrial membrane potential, cells were labeled with the cationic lipophilic fluorochrome 3,3′-dihexyloxacarbocyanine iodide (DiOC6), which accumulates in mitochondria. DiOC6 levels peaked significantly at 72 hpi before declining (Fig. 3B) but remaining above mock-treated levels. Both approaches show a trend of increased respiration in HCMV-infected cells. Together, these data suggest that HCMV infection is driving and sustaining increased mitochondrial function.
FIG 3.
Mitochondrial respiration and membrane potential are increased in HCMV-infected fibroblasts. HFFs infected with HCMV were loaded with MitoTracker CMTMRos dye (A) or DiOC6 dye (B) at the indicated times and analyzed by flow cytometry. Samples were gated on live cells only. Graphs represent pooled data from three independent experiments. Mean ± the SEM of at least triplicates. **, P < 0.01; **, P < 0.001; ****, P < 0.0001; ns, not significant.
Markers of mitochondrial stress are increased after HCMV infection.
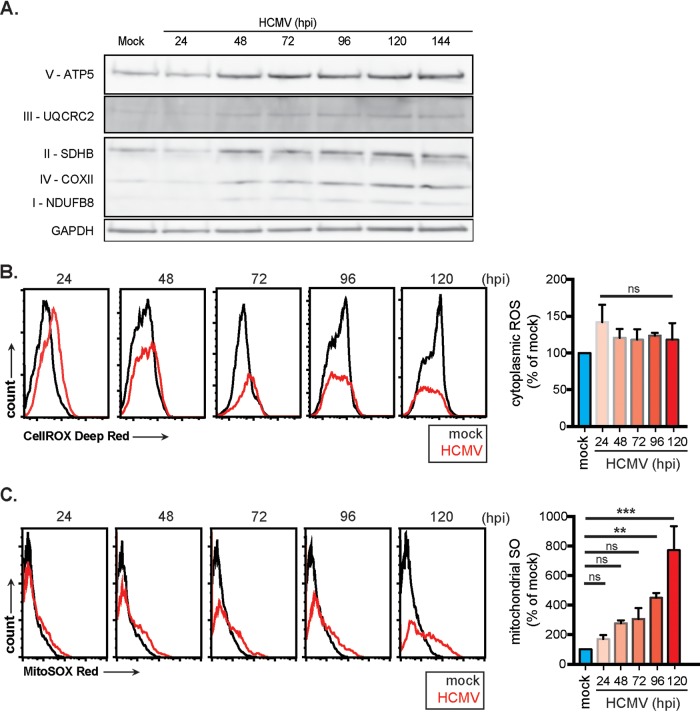
Next, we investigated whether the molecular components of OXPHOS are increased, as would be predicted with increased mitochondrial respiration. Mock- and HCMV-infected fibroblast protein lysates were collected over a 6-day period. Western blot analysis was used to measure protein levels of the four electron transport chain (ETC) complexes (I to IV) and the FO/F1 ATPase. NADH dehydrogenase (ubiquinone) 1 beta subcomplex subunit 8 (NDUFB8)—a subunit of ETC complex I—protein levels increased minimally during HCMV infection (Fig. 4A). Succinate dehydrogenase complex, subunit D (SDHD) (complex II), cytochrome bc1 complex subunit 2 (UQCRC2) (complex III), and cytochrome c oxidase subunit 2 (COXII) (complex IV) all increase markedly after 24 hpi. A by-product of increased mitochondrial function is reactive oxygen species (ROS). Therefore, we used flow cytometric analysis of mock- and HCMV-infected cells loaded with CellROX Deep Red dye to detect total ROS or MitoSOX red dye to detect superoxide (SO). Total ROS levels increased within 24 hpi and persisted over 120 hpi (Fig. 4B). SO levels gradually increased over time beginning at 24 hpi before sharply increasing at 120 hpi (Fig. 4C). These data are consistent with a model in which HCMV acts as a metabolic stressor on the mitochondria that induces a subsequent increase in mitochondrial respiration.
FIG 4.
HCMV upregulates ETC complex proteins and increases reactive oxygen species levels. (A) Whole-cell extracts were prepared from mock- and HCMV-infected cells. ETC complexes I to V and GAPDH protein levels were determined by Western blotting. Total ROS (detected by CellROX Deep Red) (B) and superoxide (detected using MitoSOX Red) (C) levels were obtained by flow cytometric analysis. The data of three independent experiments were pooled. Means ± the SEM are shown.
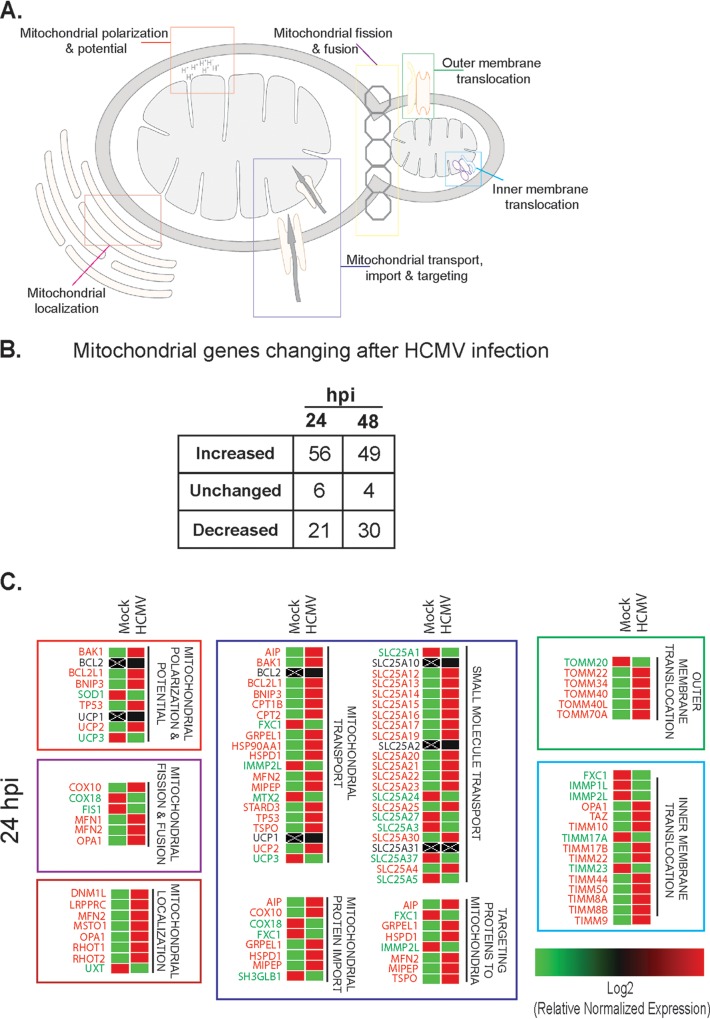
We reasoned that an increase in mitochondrial function during HCMV infection should lead to an increase in the molecular components necessary in controlling mitochondrial function. Using a transcriptomic PCR-based array, we identified changes to mitochondrion-related pathways during HCMV infection (Fig. 5A to C). At 24 hpi, ∼67% of the mitochondrial genes targeted were upregulated compared to mock-infected cells (Fig. 5B). This includes genes responsible for function, morphology, and localization or movement of proteins and small molecules (Fig. 5A and C). Many of these changes continued through 48 hpi as ∼59% of queried genes remained upregulated (Fig. 5B). An example is dynamin-1-like protein (DNM1L). This gene is upregulated at 24 h and remains increased at 48 hpi. The protein product (Drp1) can be seen increasing at 48 hpi and remains elevated through 96 hpi (Fig. 6B).
FIG 5.
Changes in mitochondrial gene expression after HCMV infection of HFFs. (A) Schematic of mitochondrial functions controlled by genes altered in HCMV-infected HFFs. (B) Table displaying the total numbers of mitochondrion-associated genes increased, unchanged, or decreased in expression at 24 and 48 h after HCMV infection. (C) Heat map organized by mitochondrial function showing differential gene expression at 24 h postinfection (n = 3 biological replicates).
FIG 6.
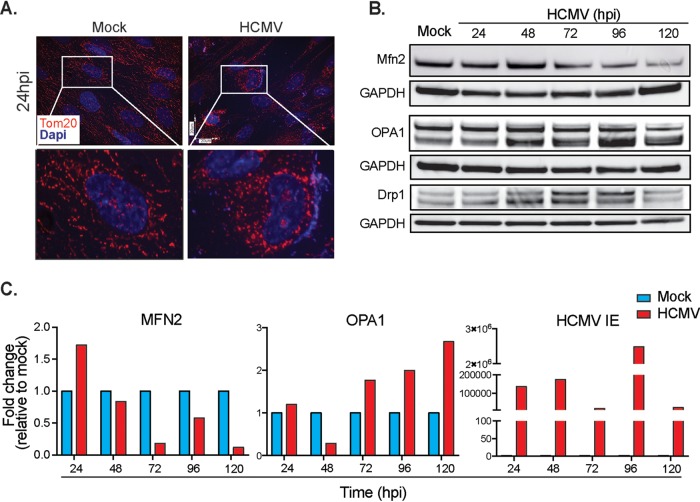
HCMV infection induces mitochondrial fission. (A) Representative microscopy images of mock- and HCMV-infected HFFs stained with an antibody to Tom20 located on the outer mitochondrial membrane. (B) The time course of fission/fusion protein expression was analyzed by Western blotting. (C) Expression of genes related to fusion (MFN2) and fission (OPA1), as well as HCMV IE, was determined by qRT-PCR. The data are shown as the means ± the SEM of three independent experiments.
Mitochondrial fission is upregulated during HCMV infection.
Changes to the mitochondrial network and individual structure of mitochondria correlate with altered function (45). Immunofluorescent imaging using the mitochondrion-specific Tom20 antibody revealed long tubular mitochondrial networks in mock-infected cells at 24 hpi (Fig. 6A). However, in HCMV-infected cells, mitochondrial networks begin to exhibit a fragmented appearance. Data from our PCR array showed altered expression of mitochondrial fission and fusion related genes in HCMV-infected cells supporting this hypothesis (Fig. 5). We further validated these results by quantitative reverse transcription-PCR (qRT-PCR). An initial increase of transcriptional activity is apparent for OMM MFN2, but after 24 hpi the transcriptional levels decline (Fig. 6C). Consistent with these data, protein levels of the OMM MFN2 were increased at 48 hpi and then decreased below control levels (Fig. 6B). Interestingly, mitochondrial dynamin-like GTPase (OPA1) mRNA and protein levels also increased during the course of infection (Fig. 6B and C). Although OPA1 is involved in mitochondrial fusion of the inner mitochondrial membrane, it also plays an important role in cristae structure regulation (46). Importantly, we see an increase in Drp1 protein levels between 48 and 96 hpi (Fig. 6B), verifying our previous observations that HCMV induces mitochondrial fission.
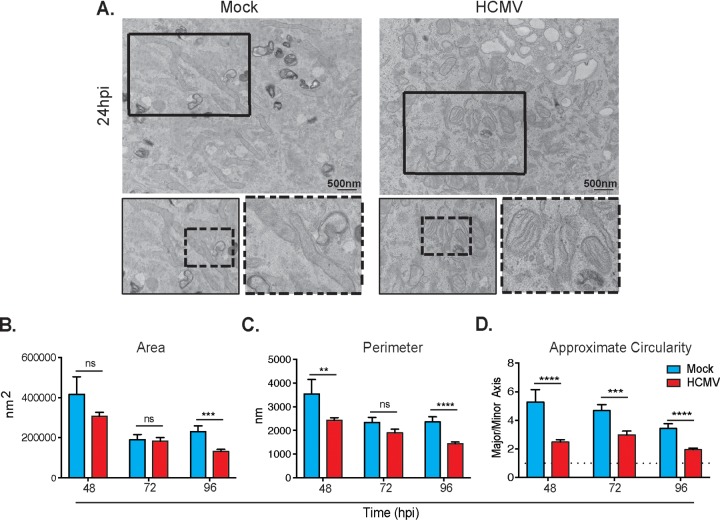
Using transmission electron microscopy (TEM), we imaged mock- and HCMV-infected cells between 48 and 96 hpi (Fig. 7A). The mitochondrial size became smaller and, structurally, the mitochondria adopted a rounder, less tubular phenotype after infection by HCMV compared to mock-infected cells. Quantifying mitochondrial physical characteristics between the mock- and HCMV-infected groups reveals that the average mitochondrial area in HCMV-infected cells decreases significantly at 96 hpi compared to mock-infected cells (Fig. 7B), and the average mitochondrial perimeter is significantly decreased at 48 and 96 hpi (Fig. 7C). The approximate circularity of the mitochondria was determined as a ratio of the longer (major axis) and the shorter (minor axis) of individual mitochondria. During the time course of 48 to 96 hpi, HCMV-infected mitochondria are significantly more circular (Fig. 7D). Surprisingly, the cristae structure appears unchanged between mock- and HCMV-infected cells. Collectively, these observations suggest that mitochondria in infected cells are undergoing morphological changes and fragmentation through mitochondrial fission processes while maintaining normal cristae structure.
FIG 7.
Ultrastructure of mitochondria after HCMV infection. (A) Representative electron micrographs of mock-infected (left) and HCMV-infected (right) HFFs. Mock-infected cells display tubular mitochondria. HCMV-infected cells show circular mitochondria with intact cristae. (A to D) The areas (B), perimeters (C), and circularities (D) of individual mitochondria decrease with time in infected cells. The image is representative of n = 3 biological controls. Scale bar, 500 nm. Magnification, ×6,000. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant.
HCMV induces mitochondrial biogenesis.
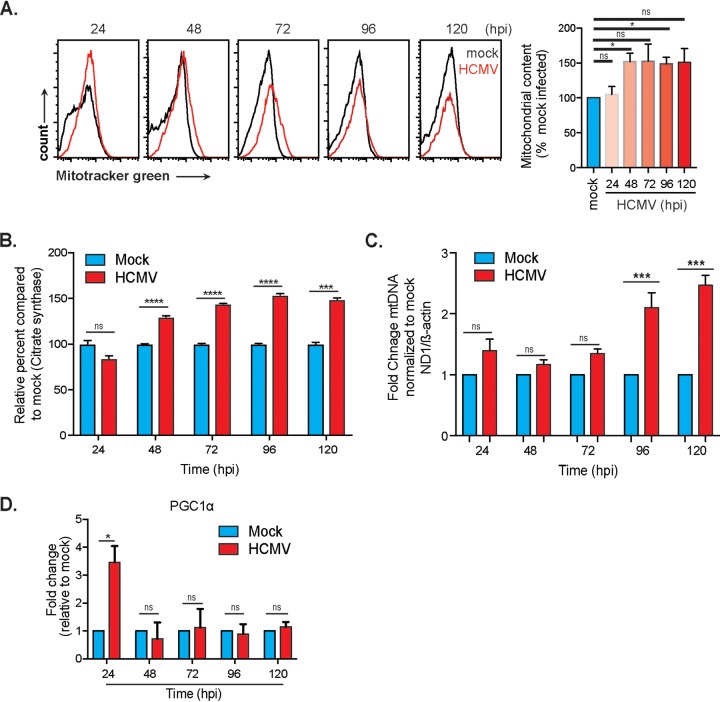
An increase in mitochondrial respiration is typically supported by an increase in mitochondrial mass. To determine whether this is true during HCMV infection, cells were labeled with MitoTracker Green FM, a cell-permeable dye retained specifically in the mitochondria, independent of the membrane potential. Using flow cytometry analysis, we observed an increase in mitochondrial mass in HCMV-infected cells with significance at 48 and 96 hpi (Fig. 8A). Citrate synthase activity, a biomarker of mitochondrial density, was also increased in HCMV-infected cells (Fig. 8B). To further validate these results, mitochondrial genome copy number was determined by qPCR as the ratio of a mitochondrial gene copy number (ND1) to a single-copy nuclear gene (β-actin). Infection with HCMV increased mitochondrial genome copy number over the course of infection, becoming significant at 96 hpi (Fig. 8C). We also analyzed the RNA expression level of a master regulator of mitochondrial biogenesis, PGC1α. A significant increase was detected at 24 hpi, decreasing through 48 hpi before an increase in expression occurs at 72 hpi (Fig. 8D). RNA levels increase again at 120 hpi. Together, these data suggest that HCMV induces mitochondrial biogenesis.
FIG 8.
HCMV infection induces mitochondrial biogenesis. (A) Flow cytometry analysis of mitochondrial mass (content) using MitoTracker green accumulation. (B) Citrate synthase activity in lysates from mock- and HCMV-infected HFFs. (C) qPCR analysis of fold change mtDNA copy number from mock- and HCMV-infected HFFs. (D) RNA expression of PGC1α in HCMV-infected HFFs was determined using qRT-PCR. Citrate synthase data are representative of a single experiment. All other data represent means ± the SEM from three independent experiments. *, P = 0.05; ***, P = 0.001; ****, P = 0.0001.
Host cell mitochondrial function, specifically the ETC is essential for HCMV replication.
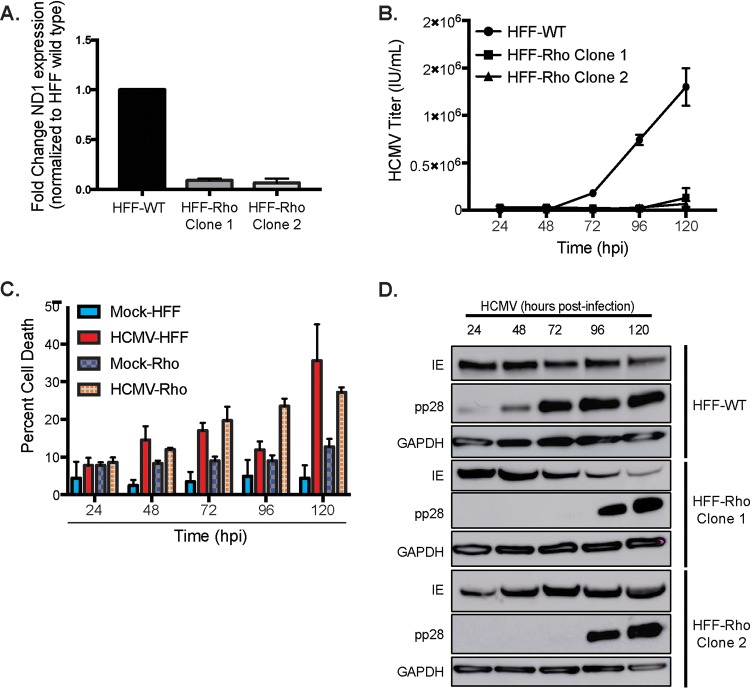
To fully appreciate the importance of mitochondrial respiration during HCMV infection, we created a human foreskin fibroblast (HFF) cell line devoid of mtDNA. These cell lines, termed Rho cells (ρ0 cells), have previously been used to examine the relationship between mtDNA-dependent proteins and mitochondrial function (47, 48). Using established protocols, we depleted the mtDNA in HFF cells using low doses of ethidium bromide (EtBr). Clones were isolated and propagated in media supplemented with uridine (for pyrimidine biosynthesis), pyruvate (as a buffer and glycolysis supplement), and high glucose (ρ0 cells are dependent on glucose fermentation for ATP synthesis). Significant depletion of mtDNA was confirmed by qPCR by measuring the mitochondrion-specific ND1 gene (Fig. 9A). ND1 expression was decreased in each of the ρ0 clones we derived. We next examined the effect of mtDNA depletion on viral replication. Wild-type and ρ0 HFF cells were infected with HCMV (MOI = 3). Culture medium was collected every 24 h for 120 h. Standard HCMV IE protein immunofluorescent viral titration assays showed almost complete inhibition of HCMV replication in ρ0 cells (Fig. 9B). To ensure this effect was due to mtDNA knockdown, flow cytometry cell viability assays were used to compare cell viability rates between different conditions. Although apoptosis was increased in infected cells, there was no significant difference between HCMV-infected wild-type HFF and ρ0 HFF cells (Fig. 9C). Interestingly, we did not observe changes to viral protein production in the ρ0 cells (Fig. 9D). Together, this suggests that mtDNA stability, and specifically mtDNA transcription and translation, is required for the production and/or release of infectious HCMV progeny.
FIG 9.
Host cell mtDNA integrity is required for HCMV replication. (A) RNA expression of mitochondrial gene ND1 in wild-type (WT) and ρ0 clone HFFs. (B) HFF wild-type and HFF ρ0 cells were infected, and the titers of released HCMV progeny were determined. (C) The levels of apoptosis were compared by using eFluor780 fluorescent flow cytometry detection assays. The data are representative of one experimental replicate. (D) HCMV viral protein expression from HFF wild-type (WT) or ρ0 (Rho) cell lysate was determined by using Western blot analysis. The data are representative of at least three independent experiments unless otherwise indicated. Error bars indicate ± the SEM of at least triplicates.
DISCUSSION
The goal of the present study was to define the broad morphological and functional changes to the host mitochondria during HCMV infection. Previous literature has reported altered mitochondrial function, but these observations were typically associated with apoptosis. Indeed, apoptosis and the mitochondria are intimately linked, but we sought to define the dependency of HCMV on mitochondria in relation to metabolism. Successful viral replication depends upon the coordination and availability of host biosynthesis and energy production. Building upon previous research indicating the need for increased glycolysis and fatty acid synthesis to provide macromolecules for the construction of new mature virions, we show that HCMV also requires functional mitochondrial respiration to drive this process. HCMV infection increases mitochondrial respiration during replication and enables metabolic plasticity to adequately meet replication mediated biosynthetic and bioenergetic stress. During replication of HCMV, we highlight an increase in gene expression related to mitochondrial function, network fragmentation, and biogenesis. This provides increased mitochondrial capacity to the infected cell, further supporting the increased oxidative requirements of HCMV replication. We have also shown that a functional ETC may be a critical component required for efficient replication of HCMV. Viruses encode no metabolic transcripts of their own, and yet it is clear that host cell metabolism is vital to viral replication.
Many viruses have been reported to damage host cell mtDNA (49, 50). Targeting host cell mtDNA is a strategy that allows viruses to control energy production, metabolism, cell signaling, immune responses, and cell cycle. While many viruses have been reported to induce apoptosis and reduce mitochondrial function, HCMV prevents apoptosis and maintains mitochondrial function. This may be a related to the physical characteristics of HCMV. HCMV is a large virus with a slow replication cycle that induces cytopathology within the host cell. This would suggest a large demand for biosynthetic building blocks, bioenergetic pathways, and avoidance of apoptosis. In fact, our work and other studies showing increased metabolic function in HCMV-infected cells suggest that host cell apoptosis is not due to energy deficits. Early studies on the effects of HCMV replication on host cell metabolism show a need for increased glycolysis primarily to produce the macromolecular building blocks the virus requires for new progeny (36, 37). When glycolysis or related pathways are inhibited or stripped of their carbon source, HCMV fails to properly replicate despite minimal changes to viral gene expression (35, 37). Earlier studies on HCMV metabolism suggest a state that mimicked the Warburg effect in cancer, namely, an increased reliance upon aerobic glycolysis. Our data (Fig. 1) suggest that HCMV establishes a more complex metabolic state with the ability to rely upon both glycolysis and OXPHOS to meet metabolic demand and respond to environmental stress (Fig. 1 and 2), a phenotype similar to that of cancer stem cells.
At the cellular level, mitochondrial morphology is unchanged after initial HCMV infection. Beginning at 24 hpi, mitochondrial fission events are observed (Fig. 6 and 7) (25). Early work by Karbowski et al. showed that fission events during HCMV infection were driven by expression of vMIA (UL37.1) and the viral proteins interaction with Bax and Bak (40, 41). These morphological changes are maintained during the complete replication cycle and do not disrupt normal mitochondrial function. Fusion events facilitate the exchange of mtDNA, proteins, and metabolites. Fusion also directly impacts OXPHOS, membrane potential, and mtDNA replication/repair. The long-term impact of prolonged fission events on the microenvironment is unknown, but dysregulated fusion events usually result in decreased respiratory capacity (51). Notably, in other work, the fragmentation of mitochondrial network through fission leads to diminished OXPHOS (52). It is also interesting that at 24 hpi, MFN2 transcript levels were increased, and the resultant protein levels were also elevated at 48 h. The role of HCMV in changing or suppressing expression of fusion mediators is unknown but intriguing. In the case of HCMV infection, the initial stress induced by entry of the virion and initiation of the lytic program may induce fragmentation of the mitochondrial network as an antiapoptotic response (25). It should be noted that mitochondrial fission is not essential for HCMV replication since vMIA (UL37.1)-deficient HCMV strains replicate under tissue culture conditions (53). Whether these events are mediated by host cellular responses or viral factors is an important question that is not addressed here.
In accordance with our metabolic and morphological data, we observed increased mitochondrial biogenesis (Fig. 8). The induction of mitochondrion-associated genes important in maintaining mitochondrial function occurs within 24 hpi with a steady increase in mitochondrial genome copy number and an overall upregulation of the master controller of mitochondrial biogenesis, PGC-1α (Fig. 8). These data are consistent with previous reports showing increased mitochondrial biogenesis and upregulation of mitochondrial transcription and translation pathways (39, 42, 54).
HCMV-induced morphological changes result in functional changes by altering membrane potential. HCMV-infected fibroblasts displayed an increase in membrane potential (Fig. 3) and mitochondrial matrix protein expression (Fig. 4), in agreement with previous reports (42, 55). Alternatively, HCMV infection has been reported to depolarize mitochondria after infection (56). Regardless, ATP levels are unchanged initially after HCMV infection and remain steady for the duration of replication (Fig. 2), as observed using a Seahorse bioanalyzer and reported previously using alternative approaches (28, 36, 56). Despite minimal changes to ATP levels, ETC complexes, specifically complex IV proteins, have been reported to increase (42, 44). Indeed, we observe an increase in ETC-associated gene products after HCMV infection (Fig. 4). To complicate matters, a HCMV protein, viral mitochondrion-localized inhibitor of apoptosis (vMIA or pUL37.1) has been shown to interact with the host mitochondria (24, 57–59). The importance of vMIA has been obscured by conflicting reports. vMIA has been reported to be essential for HCMV replication under some conditions and nonessential under others. vMIA-expressing cells induce fragmentation of the mitochondrial network with rounder, smaller mitochondria that display reduced cristae (25, 60). Other attributes of vMIA are reduced ATP levels and unchanged mitochondrial mass (52, 60). Our data support fragmentation of mitochondria, but we do not observe changes to cristae structure. Based on our TEM images, the cristae in HCMV-infected cells appear similar to mock-infected cells. We also did not observe reduced ATP levels and recorded an increase in mitochondrial mass. These discrepancies are currently being explored further in our laboratory. Further clarification of the role for HCMV interactions with host cell mitochondria would be of benefit.
Under normal conditions, mitochondrial function metabolizes oxygen and, through this process, ROS is generated. At low concentrations, ROS has a known role in redox signaling (61). It is hypothesized that increased ROS levels may promote increased stress resistance, resulting in reduced oxidative stress over time (62). Excess production of ROS, specifically SO, allows for the formation of secondary hydroxyl radicals that can damage DNA, proteins, and lipids. HCMV seems to modulate ROS levels until late in the viral replication cycle when SO formation becomes unregulated (Fig. 4). Whether HCMV utilizes low ROS concentrations early in the replication cycle to alter immune function, differentiation, or repair pathways is unexplored. The role of ROS in HCMV replication requires further investigation.
How HCMV induces such robust changes to metabolic pathways in host cells so quickly after viral entry is poorly defined. One possibility is the release of calcium by vMIA (52). Mitochondrial fragmentation induced soon after HCMV infection may be utilized to inhibit apoptosis and sequester calcium by signaling through 5′ AMP-activated protein kinase (AMPK) and calcium/calmodulin-dependent protein kinase kinase 2 (CaMKK2) (63). This may be in conflict with prior research that showed decreases in ATP content and indicators of mitochondrial function upon expression of vMIA (64). As our data show, the metabolic state of viral replication is still acutely responsive to metabolic environmental conditions and the use of different cell lines, variations in growth medium of cells, purification of viral stock, and timing of observations of viral replication may all affect observations of mitochondrial function. Alternatively, other HCMV-derived products known to interact with the host mitochondria, including noncoding Beta2.7 RNA (β2.7lncRNA) and others, may play roles (24, 25, 28, 57, 65, 66). In addition to being highly expressed at 24 hpi (β2.7lncRNA) and 48 hpi (vMIA or pUL37.1), these viral products have been shown to alter mitochondrial morphology or function in the context of apoptosis-related pathways (25, 28), specifically β2.7lncRNA, which has been shown to directly interact with mitochondrial respiratory complex I (28). This interaction was shown to fortify complex I ensuring unaltered ETC activity and thus ATP production. A β2.7lncRNA knockout HCMV strain was still able to replicate, but decreases in metabolic output could be detected, and the degree of change was dependent on cell type.
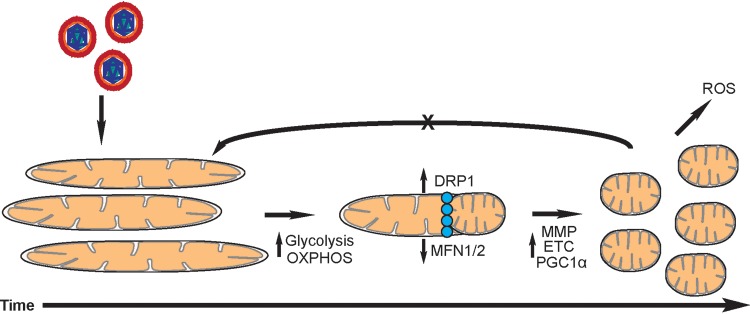
Given the cumulative data supporting increased OXPHOS during HCMV infection presented here, it is evident that HCMV requires mitochondrial respiration for proper and efficient replication. Our current working model is illustrated in Fig. 10. HCMV infection of fibroblasts induces increased biosynthetic and bioenergetic demands on the host mitochondria. Metabolic plasticity is required to meet these virally induced demands creating a glycolytic-OPXHOS hybrid. The host cell mitochondria undergo fission in response to increased metabolic demands. Mitochondrial activity remains elevated for the entirety of the infection and fusion events are inhibited through unknown mechanisms. We were surprised that HCMV was unable to replicate in ρ0 HFF cells. It is possible that the biosynthetic and bioenergetics demands of HCMV replication are too large for the restricted pathways in ρ0 HFF cells. However, this places further significance on the importance of host mitochondria during HCMV replication. Inhibiting ETC complexes using pharmacological inhibitors has been reported to reduce HCMV titers (28). Despite the importance of the ETC, there is a precedent for this approach as a therapeutic. Drugs such as metformin (ETC complex I target), a first-line diabetic drug, are being pursued as cancer therapeutics by specifically targeting metabolism (67). The energy required by viral replication should be substantial, requiring a high rate of ATP use and regeneration to fuel the process. A better understanding of the changes in host cell metabolism induced by HCMV infection is useful in identifying the broader context in which HCMV replicates. It also necessitates future research in the in vivo context of viral infection and in vitro tests in variable contexts, such as CD34+ progenitor, epithelial, or endothelial cell infections to understand whether metabolic dysregulation is a feature of HCMV infection in fibroblasts alone or a general feature of lytic replication. Understanding how HCMV affects mitochondrial respiration is critical in understanding how HCMV may shape and affect disparate tissues in the host body that may give rise to pathologies correlated with HCMV infection. Changes in mitochondrial function may be especially important in the context of aging. Mitochondrial dysfunction is a hallmark of aging and, as we show here, HCMV induces major changes to host mitochondria during an acute infection. How reactivation or reinfection alters host mitochondria over the lifetime of a host remains unknown. Understanding the role of mitochondria may also offer alternative prevention or treatment strategies for HCMV disease.
FIG 10.
Model depicting changes to host cell mitochondria after HCMV infection. Changes to mitochondrial morphology are induced due to increasing glycolytic and OXPHOS demands. Mitochondrial fission occurs enabling increased mitochondrial membrane potential, ETC output, and mitochondrial biogenesis. This results in increased ROS production. HCMV prevents fusion events through unknown mechanisms.
MATERIALS AND METHODS
Cells, virus, and reagents.
HFFs (American Type Culture Collection) were cultured in Dulbecco modified Eagle medium (DMEM) containing 4.5 g/liter glucose and l-glutamine (Gibco) supplemented with 10% fetal bovine serum (FBS; Atlanta Biologicals) at 37°C and 5% CO2, unless otherwise specified. HFF-Rho (ρ0) cells were generated by adding 50 ng/ml of ethidium bromide (EtBr) to DMEM plus 10% FBS, 2 mM l-glutamine, 100 μg/ml sodium pyruvate (Invitrogen), and 50 μg/ml uridine (Sigma). The media and EtBr were replaced twice weekly. This process was continued for 4 weeks total. Cells that remained after 4 weeks were collected and serially plated to obtain single cell suspensions in 24-well culture dishes. At this point, EtBr was no longer added to the culture media. Cells were collected, and PCR was used to determine the presence of mitochondrial DNA. Cells with large decreases in mitochondrial DNA levels were expanded and used in experiments.
Virus stocks were propagated in confluent HFFs at a multiplicity of infection (MOI) of 0.05 and cultured as described above until reaching nearly complete cell lysis. Virus was purified using standard techniques. Briefly, supernatant was collected and centrifuged to remove cellular debris. The remaining supernatant was transferred to polyallomer tubes (Seton Scientific), underlaid with a 20% sucrose solution in Tris-sodium (TN) buffer, and ultracentrifuged for 1.5 h at 22,000 rpm. Pellets were resuspended in TN buffer prior to titering. Viral titers were determined as previously described (68). Briefly, HFFs were infected with serial dilutions of purified virus (in triplicate) in 96-well plates. After 48 h, the cells were washed with phosphate-buffered saline (PBS) and fixed in ice-cold ethanol for 10 min prior to staining with primary mouse anti-HCMV IE1 antibody (kindly provided by William Britt) and anti-mouse Alexa Fluor 555-conjugated secondary antibody (Molecular Probes). Stained cells were visualized using a Nikon Eclipse TE300 microscope and counted manually at a ×10 magnification.
Viral infection was completed as follows. Confluent HFFs were serum starved overnight in base media (DMEM only) prior to infection at an MOI of 3 with HCMV TR, unless otherwise specified. Cells were infected with virus for 1.5 h in DMEM plus 2% FBS to allow viral entry. After 1.5 h, the medium was removed and replaced with fresh DMEM plus 10% FBS unless otherwise noted. Drug treatments or vehicle controls were added to infected cells at 1.5 h postinfection during the medium change.
Seahorse glycolysis and respiration measurements.
Glycolysis and mitochondrial respiration activity were determined using the Seahorse flux analyzer XFe24 according to the manufacturer’s instructions. Briefly, HFFs were seeded on Seahorse 24-well plates and incubated overnight in complete DMEM. Cells were mock or HCMV infected with HCMV TR strain (MOI = 3) in fresh medium for 48 h. The ECAR and OCR rates were an average of at least three independent experiments and normalized by protein. Samples were mixed (3 min), incubated (2 min), and measured (3 min). For glycolytic stress, glucose (10 mM), oligomycin (1 μM) and 2-deoxyglucose (100 mM) were injected at the indicated time points. MitoStress reagents, oligomycin (1 μM), carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP; 1 μM), and rotenone (0.5 μM) were injected at the indicated time points. All wells were normalized to the protein immediately after the Seahorse bioanalyzer measurements were completed using a BCA kit (Pierce).
Gene arrays.
HFF cells (5 × 105) were seeded to 80% confluence and serum starved (0% FBS) overnight. Cells were mock or HCMV infected (TR, MOI = 3) in 2% FBS for 24, 48, or 72 h. Samples were collected using a Qiagen RNeasy kit according to the manufacturer’s instructions. Samples were subjected to an ethanol precipitation step after RNA isolation. Samples were precipitated at – 20°C for 1 h, pelleted, and washed twice with ice-cold 75% ethanol before being air dried. Synthesis of cDNA from total RNA was prepared by using an iScript reverse transcription kit (Bio-Rad). Real-time PCRs were conducted using SsoAdvanced universal supermix (Bio-Rad) and run on PrimePCR Mitochondria 96-well gene arrays (Bio-Rad) using a three-step amplification repeated 40 times on a CFX96 thermocycler (Bio-Rad). Data were normalized to at least three internal controls and quantified using CFX Manager software.
PCR.
RNA was purified by using a RNeasy minikit (Qiagen) according to the manufacturer’s instructions. The RNA template was used to synthesize cDNA by using an iScript cDNA synthesis kit (Bio-Rad). qRT-PCR was performed using cDNA template, gene-specific primers, and SsoAdvanced universal supermix (Bio-Rad). The gene specific primers are listed in Table 1 . For DNA PCR analysis, DNA was extracted using a RNA/DNA extract kit (Qiagen) according to the manufacturer’s instructions. Gene-specific primers are listed in Table 1.
TABLE 1.
Primers used in PCR assays
| Primer | Description | Primer (5′–3′) |
|---|---|---|
| HCMV intermediate early gene | Forward | CAAGTGACCGAGGATTGCAA |
| Reverse | CACCATGTCCACTCGAACCTT | |
| Mitofusin 2 (MFN2) | PrimePCR probe | Bio-Rad probe |
| Mitochondrial dynamin like GTPase (OPA1) | PrimePCR probe | Bio-Rad probe |
| DNA polymerase gamma (POLG) | Forward | AGCGACGGGCAGCGGCGGCGGCA |
| Reverse | CCCTCCGAGGATAGCACTTGCGGC | |
| Mitochondrially encoded NADH: ubiquinone oxidoreductase core subunit 1 (ND1) | PrimePCR SYBR Green | Bio-Rad |
| Peroxisome proliferator-activated receptor gamma coactivator 1α (PGC-1α) | PrimePCR SYBR Green | Bio-Rad |
Transmission electron microscopy.
HFFs were grown on Aclar coverslips and serum starved overnight (0% FBS). Cells were mock or HCMV infected with the TR strain (MOI = 3) in 2% FBS media. Coverslips were washed in 0.15 M cacodylate buffer (pH 7.4) and then fixed in electron microscopy (EM)-grade 2.5% glutaraldehyde, 2% paraformaldehyde, 0.15 M cacodylate buffer (pH 7.4), and 2 mM calcium chloride for 5 min at 37°C, followed by 1 h at room temperature. Fixative was replaced with 1% glutaraldehyde–0.15 M cacodylate buffer and stored at 4°C until shipping. Images were generated at the Center for Cellular Imaging at Washington University in St. Louis Center for Cellular Imaging using a JEOL JEM-1400 120-kV transmission electron microscope. Mitochondrial parameters were measured using ImageJ software. The length/pixel was set for each image, and the longest axis (major), the short axis perpendicular to the major axis (minor), and the perimeter of each mitochondrion was measured for length, perimeter, and bounded area values. The major axis/minor axis ratio was taken for each as an approximation of circularity with a value of 1 equaling a perfect circle.
Immunofluorescence microscopy.
Cells were plated on 8-well glass chamber slides and mock or HCMV infected as described above. At completion of infection, slides were fixed in 4% EM-grade paraformaldehyde solution for 15 min at room temperature. After a wash step with PBS, the cells were permeabilized using 0.1% Triton-X solution for 5 min. Unspecific binding was blocked using 5% bovine serum albumin (BSA) plus 0.05% Triton-X in PBS for 1 h. Tom20 antibody (Cell Signaling) diluted in PBS/BSA (1%) at a concentration of 1:250, and the cells were incubated for 1 h at room temperature. Alexa Fluor 555-conjugated anti-mouse antibody (Thermo Fisher) was added at a concentration of 1:500 for 1 h at room temperature. The cells were washed again in PBS, and DAPI (4′,6′-diamidino-2-phenylindole; 1:500) was added for 5 min. Slides were cured in antifade reagent (Prolong Gold; Thermo Fisher) prior to sealing. Images were acquired using a Zeiss Axioplan II microscope with Slidebook software (v6; Intelligent Imaging Innovations). Images were deconvolved using Velocity software (Perkin-Elmer).
Mitochondrial DNA quantification.
HFFs cells (2 × 105) were seeded at 80% confluence and serum starved (0% FBS) overnight. Cells were mock or HCMV infected (TR, MOI = 3) in 2% FBS-DMEM. Samples were collected every 24 h postinfection for 120 h using a QIAamp DNA minikit (Qiagen) according to the manufacturer’s instructions. NanoDrop quantification was used to control for DNA content in each reaction. Real-time PCRs were conducted using SsoAdvanced universal supermix (Bio-Rad). A mitochondrion-specific gene, NADH dehydrogenase subunit 1 (ND1) and a nuclear specific gene, β-actin, were amplified using a three-step amplification repeated 40 times on a CFX96 thermocycler (Bio-Rad). The mtND1/β-actin ratio of was determined.
Citrate synthase activity assay.
HFFs were plated to near confluence in 60-mm plates, serum starved (0% FBS) overnight, and infected at an MOI of 3 with TR or mock infected over a 5-day period. At the completion of all infection time points, the cells were washed with ice-cold PBS and lysed using radioimmunoprecipitation assay (RIPA) buffer and protease inhibitor cocktail (Sigma). Protein concentrations were quantified by using a BCA assay. A MitoCheck citrate synthase activity assay kit was used to quantify citrate synthase activity in lysates according to the manufacturer’s suggested protocol. Mock- and HCMV-infected lysates, along with a positive control and a RIPA-only negative control, were diluted 1:100 and loaded in triplicate at the recommended amount onto a 96-well flat-bottom plate. Acetyl coenzyme A and developer solution were added to each well, followed by oxaloacetate solution to start the reaction. The plate was analyzed on a BioTek Synergy H1 plate reader set at 25°C for kinetic reads utilizing sweep speed for 30-s intervals over a period of 20 min, reading the absorbance at 412 nm. The slope (reaction rate) was calculated from the linear portion of the kinetic read, and the activity was determined by the following equation: [(reaction rate/5.712 mm–1) × (0.1 ml/0.03 ml)] × sample dilution = activity in μmol/min/ml. This value was then normalized to the protein concentration (mg/ml) to determine the specificity activity for each sample (expressed as μmol/min/mg).
Viability assays.
The viability dye eFluor 780 was added to cells for 30 min according to the manufacturer’s protocol. Fluorescence was analyzed using a BD Biosciences LSRFortessa. Viability was quantified using FlowJo software.
Mitochondrial mass, membrane potential, and reactive oxygen species.
HFFs were seeded (5 × 105) and mock or HCMV infected (TR at MOI = 3) for the time points indicated. Cells were trypsinized, pelleted by centrifugation at 400 × g for 5 min, and washed in PBS. For the mitochondrial mass, cells were stained with 100 nM MitoTracker Green (Thermo Scientific) for 30 min. Data were acquired using a BD Biosciences LSRFortessa using the FITC channel, and data were quantified by FlowJo software. To measure the membrane potential, the cells were stained with 20 nM DiOC6 or 100 nM MitoTracker Orange CMTMRos (Thermo Scientific) for 30 min. Data were acquired using a BD Biosciences LSRFortessa with fluorescein isothiocyanate or phycoerythrin-Texas Red (PE-Tx-Red) YG channels, respectively. To measure the ROS and SO, the cells were incubated in 5 μM CellROX Deep Red (oxidative stress indicator) or 5 μM MitoSox Red (mitochondrial superoxide indicator) (Thermo Scientific) for 30 min at 37°C. The cells were analyzed by flow cytometry, and data were analyzed using the APC and PE-Tx-Red YG channels, respectively.
Western blotting.
Cells treated as indicated were washed with ice-cold PBS and lysed using RIPA buffer with protease and phosphatase inhibitor cocktails (Sigma). Protein concentrations were quantified using a BCA assay. Proteins were separated on 4 to 12% or 10% polyacrylamide gels (Invitrogen) and transferred onto nitrocellulose membranes using an iBlot device (Invitrogen). Membranes were incubated in TBS plus 0.1% Tween (TBST) and blocked using 5% BSA (wt/vol) or 5% dried nonfat milk (wt/vol) in TBST to inhibit nonspecific antibody binding. Membranes were incubated overnight in 1% blocking solution in TBST and primary antibodies. Membranes were washed in TBST and incubated with secondary antibodies at room temperature for 1 h. Immunoblots were visualized by ECL using a GE Amersham imaging system.
Statistical analysis.
All data were analyzed using Prism 7 (GraphPad Software) or Bio-Rad software. Unpaired t tests were used to compare data between mock- and HCMV-infected samples. One-way analysis of variance Dunnett’s multiple-comparison test was used to compare assays over time. A P value of <0.05 was considered significant.
ACKNOWLEDGMENTS
This study was supported by National Institute of General Medical Sciences of the National Institutes of Health (NIH) pilot grant P20GM103629 (to K.J.Z), Tulane University Faculty Research Enhancement Funds (to K.J.Z), National Institute of General Medical Sciences of the NIH pilot grant P20GM103629 (to D.E.S), and National Institute of Neurological Disorders and Stroke and National Institute of General Medical Sciences of the NIH grant NS094834 (to P.V.K.).
The funders had no role in any content of this work, including study design, data analysis, or the publication process, which is solely the responsibility of the authors.
REFERENCES
- 1.Cannon MJ, Schmid DS, Hyde TB. 2010. Review of cytomegalovirus seroprevalence and demographic characteristics associated with infection. Rev Med Virol 20:202–213. doi: 10.1002/rmv.655. [DOI] [PubMed] [Google Scholar]
- 2.Azevedo LS, Pierrotti LC, Abdala E, Costa SF, Strabelli TMV, Campos SV, Ramos JF, Latif AZA, Litvinov N, Maluf NZ, Caiaffa Filho HH, Pannuti CS, Lopes MH, Santos V. A d, Linardi C. d C G, Yasuda MAS, Marques H. H d S. 2015. Cytomegalovirus infection in transplant recipients. Clinics (Sao Paulo) 70:515–523. doi: 10.6061/clinics/2015(07)09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fowler KB, Boppana SB. 2018. Congenital cytomegalovirus infection. Semin Perinatol 42:149–154. doi: 10.1053/j.semperi.2018.02.002. [DOI] [PubMed] [Google Scholar]
- 4.Lim Y, Lyall H. 2017. Congenital cytomegalovirus: who, when, what-with, and why to treat? J Infect 74(Suppl 1):S89–S94. doi: 10.1016/S0163-4453(17)30197-4. [DOI] [PubMed] [Google Scholar]
- 5.Pawelec G, McElhaney JE, Aiello AE, Derhovanessian E. 2012. The impact of CMV infection on survival in older humans. Curr Opin Immunol 24:507–511. doi: 10.1016/j.coi.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 6.Aiello AE, Chiu YL, Frasca D. 2017. How does cytomegalovirus factor into diseases of aging and vaccine responses, and by what mechanisms? Geroscience 39:261–271. doi: 10.1007/s11357-017-9983-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sinzger C, Digel M, Jahn G. 2008. Cytomegalovirus cell tropism. Curr Top Microbiol Immunol 325:63–83. doi: 10.1007/978-3-540-77349-8_4. [DOI] [PubMed] [Google Scholar]
- 8.Sinclair J. 2008. Human cytomegalovirus: latency and reactivation in the myeloid lineage. J Clin Virol 41:180–185. doi: 10.1016/j.jcv.2007.11.014. [DOI] [PubMed] [Google Scholar]
- 9.Murphy E, Rigoutsos I, Shibuya T, Shenk TE. 2003. Reevaluation of human cytomegalovirus coding potential. Proc Natl Acad Sci U S A 100:13585–13590. doi: 10.1073/pnas.1735466100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Antico Arciuch VG, Elguero ME, Poderoso JJ, Carreras MC. 2012. Mitochondrial regulation of cell cycle and proliferation. Antioxid Redox Signal 16:1150–1180. doi: 10.1089/ars.2011.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chandel NS. 2014. Mitochondria as signaling organelles. BMC Biol 12:34. doi: 10.1186/1741-7007-12-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roberts DJ, Miyamoto S. 2015. Hexokinase II integrates energy metabolism and cellular protection: Akting on mitochondria and TORCing to autophagy. Cell Death Differ 22:364. doi: 10.1038/cdd.2014.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farmer T, Naslavsky N, Caplan S. 2018. Tying trafficking to fusion and fission at the mighty mitochondria. Traffic 19:569. doi: 10.1111/tra.12573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan DC. 2006. Mitochondria: dynamic organelles in disease, aging, and development. Cell 125:1241–1252. doi: 10.1016/j.cell.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 15.Picard M, Taivassalo T, Gouspillou G, Hepple RT. 2011. Mitochondria: isolation, structure, and function. J Physiol 589:4413–4421. doi: 10.1113/jphysiol.2011.212712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Westermann B. 2010. Mitochondrial fusion and fission in cell life and death. Nat Rev Mol Cell Biol 11:872–884. doi: 10.1038/nrm3013. [DOI] [PubMed] [Google Scholar]
- 17.Koshiba T, Detmer SA, Kaiser JT, Chen H, McCaffery JM, Chan DC. 2004. Structural basis of mitochondrial tethering by mitofusin complexes. Science 305:858–862. doi: 10.1126/science.1099793. [DOI] [PubMed] [Google Scholar]
- 18.Song Z, Chen H, Fiket M, Alexander C, Chan DC. 2007. OPA1 processing controls mitochondrial fusion and is regulated by mRNA splicing, membrane potential, and Yme1L. J Cell Biol 178:749–755. doi: 10.1083/jcb.200704110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wallace DC. 2005. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet 39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scarpulla RC. 2008. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol Rev 88:611–638. doi: 10.1152/physrev.00025.2007. [DOI] [PubMed] [Google Scholar]
- 21.Handschin C, Spiegelman BM. 2008. The role of exercise and PGC1α in inflammation and chronic disease. Nature 454:463–469. doi: 10.1038/nature07206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chan DC. 2012. Fusion and fission: interlinked processes critical for mitochondrial health. Annu Rev Genet 46:265–287. doi: 10.1146/annurev-genet-110410-132529. [DOI] [PubMed] [Google Scholar]
- 23.Ishihara N, Nomura M, Jofuku A, Kato H, Suzuki SO, Masuda K, Otera H, Nakanishi Y, Nonaka I, Goto Y, Taguchi N, Morinaga H, Maeda M, Takayanagi R, Yokota S, Mihara K. 2009. Mitochondrial fission factor Drp1 is essential for embryonic development and synapse formation in mice. Nat Cell Biol 11:958–966. doi: 10.1038/nc907. [DOI] [PubMed] [Google Scholar]
- 24.Goldmacher VS. 2002. vMIA, a viral inhibitor of apoptosis targeting mitochondria. Biochimie 84:177–185. doi: 10.1016/s0300-9084(02)01367-6. [DOI] [PubMed] [Google Scholar]
- 25.McCormick AL, Smith VL, Chow D, Mocarski ES. 2003. Disruption of mitochondrial networks by the human cytomegalovirus UL37 gene product viral mitochondrion-localized inhibitor of apoptosis. J Virol 77:631–641. doi: 10.1128/jvi.77.1.631-641.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharon-Friling R, Goodhouse J, Colberg-Poley AM, Shenk T. 2006. Human cytomegalovirus pUL37x1 induces the release of endoplasmic reticulum calcium stores. Proc Natl Acad Sci U S A 103:19117–19122. doi: 10.1073/pnas.0609353103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharon-Friling R, Shenk T. 2014. Human cytomegalovirus pUL37x1-induced calcium flux activates PKCalpha, inducing altered cell shape and accumulation of cytoplasmic vesicles. Proc Natl Acad Sci U S A 111:E1140–E1148. doi: 10.1073/pnas.1402515111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reeves MB, Davies AA, McSharry BP, Wilkinson GW, Sinclair JH. 2007. Complex I binding by a virally encoded RNA regulates mitochondria-induced cell death. Science 316:1345–1348. doi: 10.1126/science.1142984. [DOI] [PubMed] [Google Scholar]
- 29.Luo X, Hong L, Cheng C, Li N, Zhao X, Shi F, Liu J, Fan J, Zhou J, Bode AM, Cao Y. 2018. DNMT1 mediates metabolic reprogramming induced by Epstein-Barr virus latent membrane protein 1 and reversed by grifolin in nasopharyngeal carcinoma. Cell Death Dis 9:619. doi: 10.1038/s41419-018-0662-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Corcoran JA, Saffran HA, Duguay BA, Smiley JR. 2009. Herpes simplex virus UL12.5 targets mitochondria through a mitochondrial localization sequence proximal to the N terminus. J Virol 83:2601–2610. doi: 10.1128/JVI.02087-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pal AD, Basak NP, Banerjee AS, Banerjee S. 2014. Epstein-Barr virus latent membrane protein-2A alters mitochondrial dynamics promoting cellular migration mediated by Notch signaling pathway. Carcinogenesis 35:1592–1601. doi: 10.1093/carcin/bgu069. [DOI] [PubMed] [Google Scholar]
- 32.Shi CS, Qi HY, Boularan C, Huang NN, Abu-Asab M, Shelhamer JH, Kehrl JH. 2014. SARS-coronavirus open reading frame-9b suppresses innate immunity by targeting mitochondria and the MAVS/TRAF3/TRAF6 signalosome. J Immunol 193:3080–3089. doi: 10.4049/jimmunol.1303196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Warburg O, Wind F, Negelein E. 1927. The metabolism of tumors in the body. J Gen Physiol 8:519–530. doi: 10.1085/jgp.8.6.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Warburg O. 1956. On the origin of cancer cells. Science 123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 35.Chambers JW, Maguire TG, Alwine JC. 2010. Glutamine metabolism is essential for human cytomegalovirus infection. J Virol 84:1867–1873. doi: 10.1128/JVI.02123-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Munger J, Bajad SU, Coller HA, Shenk T, Rabinowitz JD. 2006. Dynamics of the cellular metabolome during human cytomegalovirus infection. PLoS Pathog 2:e132. doi: 10.1371/journal.ppat.0020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu Y, Maguire TG, Alwine JC. 2011. Human cytomegalovirus activates glucose transporter 4 expression to increase glucose uptake during infection. J Virol 85:1573–1580. doi: 10.1128/JVI.01967-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Munger J, Bennett BD, Parikh A, Feng XJ, McArdle J, Rabitz HA, Shenk T, Rabinowitz JD. 2008. Systems-level metabolic flux profiling identifies fatty acid synthesis as a target for antiviral therapy. Nat Biotechnol 26:1179–1186. doi: 10.1038/nbt.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaarbø M, Ager-Wick E, Osenbroch PØ, Kilander A, Skinnes R, Müller F, Eide L. 2011. Human cytomegalovirus infection increases mitochondrial biogenesis. Mitochondrion 11:935–945. doi: 10.1016/j.mito.2011.08.008. [DOI] [PubMed] [Google Scholar]
- 40.Karbowski M, Norris KL, Cleland MM, Jeong SY, Youle RJ. 2006. Role of Bax and Bak in mitochondrial morphogenesis. Nature 443:658–662. doi: 10.1038/nature05111. [DOI] [PubMed] [Google Scholar]
- 41.Norris KL, Youle RJ. 2008. Cytomegalovirus proteins vMIA and m38.5 link mitochondrial morphogenesis to Bcl-2 family proteins. J Virol 82:6232–6243. doi: 10.1128/JVI.02710-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karniely S, Weekes MP, Antrobus R, Rorbach J, van Haute L, Umrania Y, Smith DL, Stanton RJ, Minczuk M, Lehner PJ, Sinclair JH. 2016. Human cytomegalovirus infection upregulates the mitochondrial transcription and translation machineries. mBio 7:e00029. doi: 10.1128/mBio.00029-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Galinato M, Shimoda K, Aguiar A, Hennig F, Boffelli D, McVoy MA, Hertel L. 2019. Single-cell transcriptome analysis of CD34+ stem cell-derived myeloid cells infected with human cytomegalovirus. Front Microbiol 10:577. doi: 10.3389/fmicb.2019.00577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hertel L, Mocarski ES. 2004. Global analysis of host cell gene expression late during cytomegalovirus infection reveals extensive dysregulation of cell cycle gene expression and induction of pseudomitosis independent of US28 function. J Virol 78:11988–12011. doi: 10.1128/JVI.78.21.11988-12011.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liesa M, Palacin M, Zorzano A. 2009. Mitochondrial dynamics in mammalian health and disease. Physiol Rev 89:799–845. doi: 10.1152/physrev.00030.2008. [DOI] [PubMed] [Google Scholar]
- 46.Patten DA, Wong J, Khacho M, Soubannier V, Mailloux RJ, Pilon-Larose K, MacLaurin JG, Park DS, McBride HM, Trinkle-Mulcahy L, Harper ME, Germain M, Slack RS. 2014. OPA1-dependent cristae modulation is essential for cellular adaptation to metabolic demand. EMBO J 33:2676–2691. doi: 10.15252/embj.201488349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Slonimski PP, Perrodin G, Croft JH. 1968. Ethidium bromide induced mutation of yeast mitochondria: complete transformation of cells into respiratory deficient non-chromosomal “petites. Biochem Biophys Res Commun 30:232–239. doi: 10.1016/0006-291x(68)90440-3. [DOI] [PubMed] [Google Scholar]
- 48.Wilkins HM, Carl SM, Swerdlow RH. 2014. Cytoplasmic hybrid (cybrid) cell lines as a practical model for mitochondriopathies. Redox Biol 2:619–631. doi: 10.1016/j.redox.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martinez R, Shao L, Bronstein JC, Weber PC, Weller SK. 1996. The product of a 1.9-kb mRNA which overlaps the HSV-1 alkaline nuclease gene (UL12) cannot relieve the growth defects of a null mutant. Virology 215:152–164. doi: 10.1006/viro.1996.0018. [DOI] [PubMed] [Google Scholar]
- 50.Wiedmer A, Wang P, Zhou J, Rennekamp AJ, Tiranti V, Zeviani M, Lieberman PM. 2008. Epstein-Barr virus immediate-early protein Zta co-opts mitochondrial single-stranded DNA binding protein to promote viral and inhibit mitochondrial DNA replication. J Virol 82:4647–4655. doi: 10.1128/JVI.02198-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen H, Chomyn A, Chan DC. 2005. Disruption of fusion results in mitochondrial heterogeneity and dysfunction. J Biol Chem 280:26185–26192. doi: 10.1074/jbc.M503062200. [DOI] [PubMed] [Google Scholar]
- 52.Seo JY, Yaneva R, Hinson ER, Cresswell P. 2011. Human cytomegalovirus directly induces the antiviral protein viperin to enhance infectivity. Science 332:1093–1097. doi: 10.1126/science.1202007. [DOI] [PubMed] [Google Scholar]
- 53.Reboredo M, Greaves RF, Hahn G. 2004. Human cytomegalovirus proteins encoded by UL37 exon 1 protect infected fibroblasts against virus-induced apoptosis and are required for efficient virus replication. J Gen Virol 85:3555–3567. doi: 10.1099/vir.0.80379-0. [DOI] [PubMed] [Google Scholar]
- 54.Furukawa T, Sakuma S, Plotkin SA. 1976. Human cytomegalovirus infection of WI-38 cells stimulates mitochondrial DNA synthesis. Nature 262:414–416. doi: 10.1038/262414a0. [DOI] [PubMed] [Google Scholar]
- 55.Landini MP, Rugolo M. 1984. Increased accumulation of a lipophilic cation (tetraphenylphosphonium) in human embryo fibroblasts after infection with cytomegalovirus. J Gen Virol 65:2269–2272. doi: 10.1099/0022-1317-65-12-2269. [DOI] [PubMed] [Google Scholar]
- 56.Crowe WE, Maglova LM, Ponka P, Russell JM. 2004. Human cytomegalovirus-induced host cell enlargement is iron dependent. Am J Physiol Cell Physiol 287:C1023–C1030. doi: 10.1152/ajpcell.00511.2003. [DOI] [PubMed] [Google Scholar]
- 57.Bozidis P, Williamson CD, Colberg-Poley AM. 2008. Mitochondrial and secretory human cytomegalovirus UL37 proteins traffic into mitochondrion-associated membranes of human cells. J Virol 82:2715–2726. doi: 10.1128/JVI.02456-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Colberg-Poley AM, Patel MB, Erezo DP, Slater JE. 2000. Human cytomegalovirus UL37 immediate-early regulatory proteins traffic through the secretory apparatus and to mitochondria. J Gen Virol 81:1779–1789. doi: 10.1099/0022-1317-81-7-1779. [DOI] [PubMed] [Google Scholar]
- 59.Goldmacher VS, Bartle LM, Skaletskaya A, Dionne CA, Kedersha NL, Vater CA, Han JW, Lutz RJ, Watanabe S, Cahir McFarland ED, Kieff ED, Mocarski ES, Chittenden T. 1999. A cytomegalovirus-encoded mitochondria-localized inhibitor of apoptosis structurally unrelated to Bcl-2. Proc Natl Acad Sci U S A 96:12536–12541. doi: 10.1073/pnas.96.22.12536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Poncet D, Pauleau AL, Szabadkai G, Vozza A, Scholz SR, Le Bras M, Briere JJ, Jalil A, Le Moigne R, Brenner C, Hahn G, Wittig I, Schagger H, Lemaire C, Bianchi K, Souquere S, Pierron G, Rustin P, Goldmacher VS, Rizzuto R, Palmieri F, Kroemer G. 2006. Cytopathic effects of the cytomegalovirus-encoded apoptosis inhibitory protein vMIA. J Cell Biol 174:985–996. doi: 10.1083/jcb.200604069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thannickal VJ, Fanburg BL. 2000. Reactive oxygen species in cell signaling. Am J Physiol Lung Cell Mol Physiol 279:L1005–L1028. doi: 10.1152/ajplung.2000.279.6.L1005. [DOI] [PubMed] [Google Scholar]
- 62.Ristow M, Zarse K. 2010. How increased oxidative stress promotes longevity and metabolic health: the concept of mitochondrial hormesis (mitohormesis). Exp Gerontol 45:410–418. doi: 10.1016/j.exger.2010.03.014. [DOI] [PubMed] [Google Scholar]
- 63.McArdle J, Schafer XL, Munger J. 2011. Inhibition of calmodulin-dependent kinase kinase blocks human cytomegalovirus-induced glycolytic activation and severely attenuates production of viral progeny. J Virol 85:705–714. doi: 10.1128/JVI.01557-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Seo JY, Cresswell P. 2013. Viperin regulates cellular lipid metabolism during human cytomegalovirus infection. PLoS Pathog 9:e1003497. doi: 10.1371/journal.ppat.1003497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McCormick AL, Meiering CD, Smith GB, Mocarski ES. 2005. Mitochondrial cell death suppressors carried by human and murine cytomegalovirus confer resistance to proteasome inhibitor-induced apoptosis. J Virol 79:12205–12217. doi: 10.1128/JVI.79.19.12205-12217.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Greenaway PJ, Wilkinson GW. 1987. Nucleotide sequence of the most abundantly transcribed early gene of human cytomegalovirus strain AD169. Virus Res 7:17–31. doi: 10.1016/0168-1702(87)90055-4. [DOI] [PubMed] [Google Scholar]
- 67.Wheaton WW, Weinberg SE, Hamanaka RB, Soberanes S, Sullivan LB, Anso E, Glasauer A, Dufour E, Mutlu GM, Budigner GS, Chandel NS. 2014. Metformin inhibits mitochondrial complex I of cancer cells to reduce tumorigenesis. Elife 3:e02242. doi: 10.7554/eLife.02242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Britt WJ. 2010. Human cytomegalovirus: propagation, quantification, and storage. Curr Protoc Microbiol Chapter 14:Unit 14E3. [DOI] [PubMed] [Google Scholar]