Human bocavirus 1 (HBoV1) is one of the significant pathogens causing acute respiratory tract infections in young children worldwide. HBoV1 encodes a small nonstructural protein (NP1) that plays an important role in the maturation of viral mRNAs encoding capsid proteins as well as in viral DNA replication. Here, we identified a critical host factor, CPSF6, that directly interacts with NP1, mediates the nuclear import of NP1, and plays a role in the maturation of capsid protein-encoding mRNAs in the nucleus. The identification of the direct interaction between viral NP1 and host CPSF6 provides new insights into the mechanism by which a viral small nonstructural protein facilitates the multiple regulation of viral gene expression and replication and reveals a novel target for potent antiviral drug development.
KEYWORDS: human bocavirus 1, NP1, CPSF6, mRNA processing, DNA replication, parvovirus
ABSTRACT
Human bocavirus 1 (HBoV1), which belongs to the genus Bocaparvovirus of the Parvoviridae family, causes acute respiratory tract infections in young children. In vitro, HBoV1 infects polarized primary human airway epithelium (HAE) cultured at an air-liquid interface (HAE-ALI). HBoV1 encodes a small nonstructural protein, nuclear protein 1 (NP1), that plays an essential role in the maturation of capsid protein (VP)-encoding mRNAs and viral DNA replication. In this study, we determined the broad interactome of NP1 using the proximity-dependent biotin identification (BioID) assay combined with mass spectrometry (MS). We confirmed that two host mRNA processing factors, DEAH-box helicase 15 (DHX15) and cleavage and polyadenylation specificity factor 6 (CPSF6; also known as CFIm68), a subunit of the cleavage factor Im complex (CFIm), interact with HBoV1 NP1 independently of any DNA or mRNAs. Knockdown of CPSF6 significantly decreased the expression of capsid protein but not that of DHX15. We further demonstrated that NP1 directly interacts with CPSF6 in vitro and colocalizes within the virus replication centers. Importantly, we revealed a novel role of CPSF6 in the nuclear import of NP1, in addition to the critical role of CPSF6 in NP1-facilitated maturation of VP-encoding mRNAs. Thus, our study suggests that CPSF6 interacts with NP1 to escort NP1 imported into the nucleus for its function in the modulation of viral mRNA processing and viral DNA replication.
IMPORTANCE Human bocavirus 1 (HBoV1) is one of the significant pathogens causing acute respiratory tract infections in young children worldwide. HBoV1 encodes a small nonstructural protein (NP1) that plays an important role in the maturation of viral mRNAs encoding capsid proteins as well as in viral DNA replication. Here, we identified a critical host factor, CPSF6, that directly interacts with NP1, mediates the nuclear import of NP1, and plays a role in the maturation of capsid protein-encoding mRNAs in the nucleus. The identification of the direct interaction between viral NP1 and host CPSF6 provides new insights into the mechanism by which a viral small nonstructural protein facilitates the multiple regulation of viral gene expression and replication and reveals a novel target for potent antiviral drug development.
INTRODUCTION
Human bocavirus 1 (HBoV1), a member of the genus Bocaparvovirus in the subfamily Parvovirinae of the Parvoviridae family (1), causes respiratory tract infections in young children worldwide (2–11). The genus Bocaparvovirus also includes bovine parvovirus 1 (BPV1) and minute virus of canines (MVC), in addition to HBoV1 to HBoV4 (12). Human embryonic kidney 293 (HEK293) cells support the replication of an HBoV1 double-stranded DNA (dsDNA) genome clone (pIHBoV1) and progeny virion production but not virus infection (13, 14). In vitro, HBoV1 infects only polarized (well-differentiated) human airway epithelium (HAE) cultured at an air-liquid interface (HAE-ALI) (13, 15–17).
The HBoV1 replicative-form (RF) dsDNA genome transcribes one precursor mRNA (pre-mRNA) from a single RNA polymerase II (Pol II) promoter (P5) at the left end of the genome (10, 15, 18–20) and one long noncoding RNA (lncRNA), bocavirus-expressed small RNA (BocaSR), from an intragenic RNA Pol III promoter at the 3′-end noncoding region (NCR) (21). The viral pre-mRNA is processed through alternative splicing and alternative polyadenylation to generate 8 major species of mature viral mRNAs, which are translated to six major nonstructural (NS) proteins, NS1, NS1-70, NS2, NS3, NS4, and nuclear protein 1 (NP1), and three structural/capsid proteins (VP), VP1, VP2, and VP3 (18–20). Internal (proximal) polyadenylation [(pA)p] sites terminate the transcription/processing of the viral pre-mRNA, and thus, the resultant short viral mRNAs fail to encode VP. Previously, we discovered that HBoV1 NP1 expression is essential to the maturation of viral mRNAs encoding VP. It not only facilitates the effective splicing of viral pre-mRNA at the splice acceptor site upstream of the (pA)p sites but also assists the RNA Pol II complex to read through the (pA)p sites for the production of long viral mRNAs terminated at the distal polyadenylation [(pA)d] sites, which yields the VP-encoding mRNAs; therefore, NP1 is crucial to capsid protein expression (19, 22). A comprehensive analysis of viral mRNA transcripts during HBoV1 infection of HAE-ALI using transcriptome sequencing identified two (pA)p sites, (pA)p1 and (pA)p2, whose cleavage sites are nucleotide (nt) 3503 and nt 3341, respectively, and two (pA)d sites, (pA)d1 and (pA)dREH, which end viral mRNAs at nt 5171 and nt 5443, respectively (23). The two (pA)p sites were utilized at an equivalent efficiency in HBoV1-infected HAE-ALI as well as in HBoV1 RF dsDNA-transfected HEK293 cells (22, 23). However, the (pA)d1 site was used more frequently than the (pA)dREH site during virus infection of HAE-ALI (23), but the opposite was found in viral RF dsDNA-transfected HEK293 cells (24). In addition, HBoV1 NP1 is localized in viral DNA replication centers and is required for efficient viral DNA replication (13, 25). How NP1 regulates viral DNA replication remains unknown.
NP1 is unique among the nonstructural proteins expressed from members of the Parvoviridae family. MVC NP1 was the first nonstructural protein found in all parvoviruses to govern the production of both viral nonstructural and structural proteins (26, 27). Similar to the findings for HBoV1 NP1 described above, MVC NP1 suppresses the polyadenylation of viral pre-mRNA at the (pA)p sites, which ensures the accumulation of viral mRNAs polyadenylated at the (pA)d site (26) and which facilitates the processing of viral pre-mRNA at the splice acceptor upstream of the (pA)p sites. MVC NP1 interacts with a cellular mRNA 3′-end processing factor, cleavage and polyadenylation specificity factor 6 (CPSF6) (28), also known as CFIm68, the 68-kDa subunit of the cleavage factor Im (CFIm) complex (29). The knockout of CPSF6 significantly accumulated viral mRNAs polyadenylated at the (pA)p sites but not at the (pA)d site (28). As MVC NP1 interacts with viral mRNAs (28) and CPSF6 indirectly binds to mRNAs by interacting with the 25-kDa subunit of the CFIm complex (CFIm25) (30), which directly binds to a UGUA enhancer upstream of the hexanucleotide AAUAAA site (31), the interaction could be mediated by viral mRNAs.
HBoV1 NP1 localizes in the viral DNA replication centers in the nucleus and plays an important role in viral DNA replication (25, 32). As a small viral nonstructural protein of only 25 kDa, the dual roles of NP1 in both viral pre-mRNA processing and viral DNA replication are intriguing. In this study, we profiled the NP1 interactome using a proximity-dependent biotin identification (BioID) assay, and the following mass spectrometry (MS) identified over 300 host proteins that interacted with NP1, among which at least two mRNA processing factors, DEAH-box helicase 15 (DHX15) and CPSF6, were found to directly interact with NP1 without the involvement of DNA or RNA. Although DHX15 was not confirmed to play a role in the expression of viral capsid proteins, the interaction of CPSF6 and NP1 was essential to the production of viral capsid proteins through the accumulation of VP-encoding viral mRNAs that are polyadenylated at the (pA)d sites. Importantly, we revealed that CPSF6 mediates the import of NP1 into the nucleus, which is critical to its function in viral pre-mRNA processing and viral DNA replication.
RESULTS
Development of a biotin proximity labeling assay to identify host proteins that interact with HBoV1 NP1.
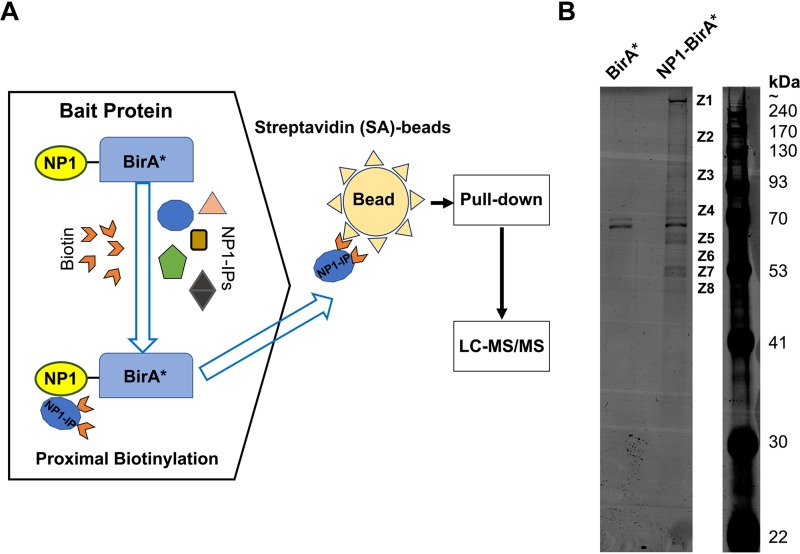
HBoV1 NP1 plays an important role in the production of capsid proteins through the regulation of viral pre-mRNA transcription and processing (19, 22) and also in viral DNA replication (25). To identify the proteins associated with NP1 during HBoV1 replication, we used a proximity-dependent BioID assay (Fig. 1A).
FIG 1.
Identification of NP1-interacting proteins using a proximity-dependent biotin identification (BioID) assay. (A) BioID assay. BirA* is a mutant of E. coli-derived biotin ligase with a catalytic site mutation (R118G) which promiscuously biotinylates proximal proteins. NP1-BirA* is a fusion protein in which the ORF of BirA* is fused at the C terminus of the HBoV1 NP1 protein, serving as the bait. When the cells expressing NP1-BirA* are incubated with excess biotins, the BirA* biotin ligase covalently attaches biotins to neighboring proteins (candidate NP1-interacting proteins [NP1-IPs]) in a close proximity of ∼10 nm. Then biotinylated proteins are captured on streptavidin (SA)-conjugated agarose beads for identification by liquid chromatography-tandem mass spectrometry (LC-MS/MS). (B) SDS-polyacrylamide gel electrophoresis (PAGE) of the biotinylated proteins. HEK293 cells were cotransfected with pIHBoV1ΔNP1 and pNP1-BirA*. At 24 h posttransfection, biotins were added to the cells at a final concentration of 50 μM to induce biotinylation. After 48 h, the cells were harvested and lysed. Biotinylated proteins in the lysates were isolated with SA-conjugated beads by biotin-affinity capture and resolved on SDS-PAGE gels (8% acrylamide). Cells cotransfected with pIHBoV1ΔNP1 and pBirA* were used as a control (BirA*). The protein bands (Z1 to Z8) pulled down from NP1-BriA*-expressing cells but not from BirA*-expressing cells were excised and subject to LC-MS/MS.
pIHBoV1ΔNP1 is a dsDNA genome clone of HBoV1, but its NP1 expression is disabled by the premature termination of translation (13). To complement the NP1 function of pIHBoV1ΔNP1 and biotinylate the neighboring NP1-interacting proteins (NP1-IPs), we constructed a plasmid (pNP1-BirA*) to express the bait protein NP1-BirA*, in which the open reading frame (ORF) of BirA*, derived from the Escherichia coli biotin ligase (BirA) with a catalytic site mutation (R118G), is fused to the C terminus of the HBoV1 NP1 protein. Cotransfection of pIHBoV1ΔNP1 and pNP1-BirA* resulted in replication of the HBoV1 dsDNA genome, but the cotransfection of pIHBoV1ΔNP1 and pBirA*, which served as the control group, did not (data not shown). These two groups of transfected HEK293 cells were subjected to the BioID assay, and the streptavidin (SA) affinity-captured biotinylated proteins were resolved on sodium dodecyl sulfate (SDS)- polyacrylamide gel electrophoresis (PAGE) gels, which revealed 8 protein bands in the pNP1-BirA*-transfected group unique from those in the control (pBirA*-transfected) group (Fig. 1B). The following mass spectrometry analyses of these eight protein bands (Z1 to Z8) identified ∼300 proteins that potentially interact with NP1 directly or indirectly during HBoV1 replication (see Table S1 in the supplemental material).
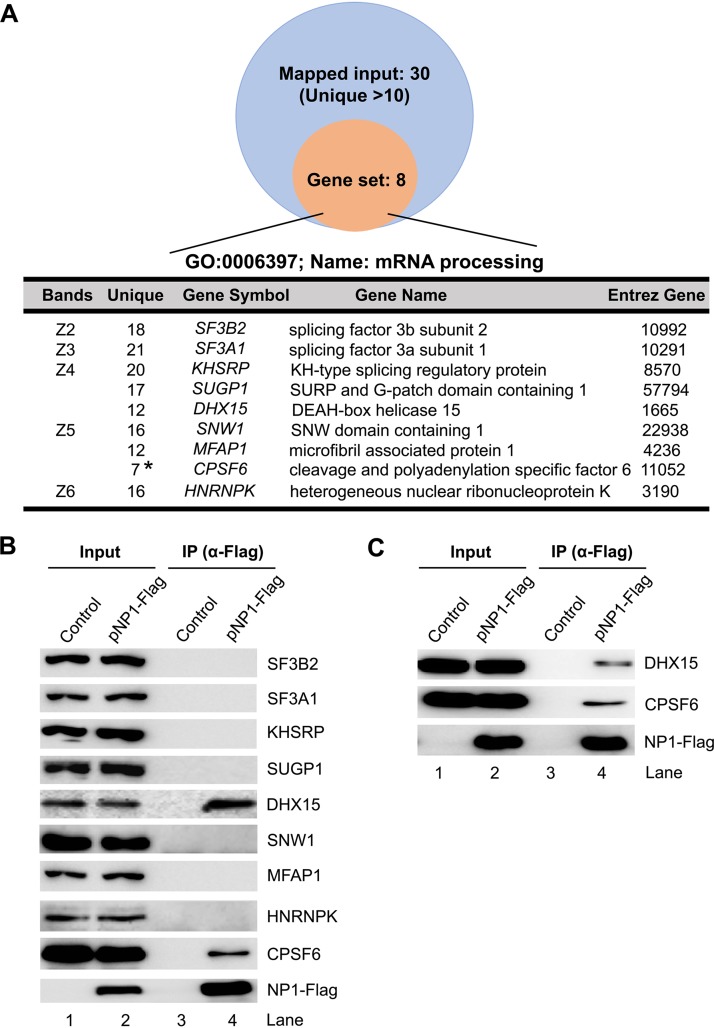
Our primary objective was to elucidate the NP1 function in viral pre-mRNA processing. To this end, we performed a Gene Ontology (GO) analysis with the BioID assay-identified proteins that have >10 unique reads in each protein band. This analysis revealed 8 proteins that are functionally involved in pre-mRNA processing (Fig. 2A). We next performed coimmunoprecipitation (co-IP) to confirm the direct binding of these proteins to NP1. To this end, HEK293 cells were transfected with pNP1-Flag or mock transfected with empty plasmid as a control. The cell lysates were incubated with anti-Flag affinity-conjugated beads for co-IP. The result showed that only DHX15 and CPSF6 were pulled down by anti-Flag-conjugated beads (Fig. 2B), which was further confirmed by the same co-IP but on which nuclease (Benzonase) was included in the co-IP buffer (Fig. 2C), suggesting that the interactions between NP1 and DHX15 or CPSF6 do not associate with any DNA or RNA.
FIG 2.
Confirmation of the interactions of the BioID-identified mRNA processing proteins with NP1 using coimmunoprecipitation (co-IP). (A) Gene Ontology (GO) analysis of NP1-interacting proteins. The identified proteins with 11 or more unique reads were analyzed in function of mRNA processing of the GO biological process category using the WebGestalt website (http://www.webgestalt.org/). (B and C) HEK293 cells were cotransfected with pNP1-Flag for 48 h. Immunoprecipitation (IP) was performed with anti-Flag-conjugated agarose beads in the absence (B) or presence (C) of nuclease (Benzonase), followed by Western blotting using antibodies against the proteins, as indicated. Lysates of untransfected cells and pIHBoV1ΔNP1- and pNP1-Flag-transfected cells were used as controls. Ten percent of the lysates used for co-IP (lanes 3 and 4) were loaded as inputs (lanes 1 and 2).
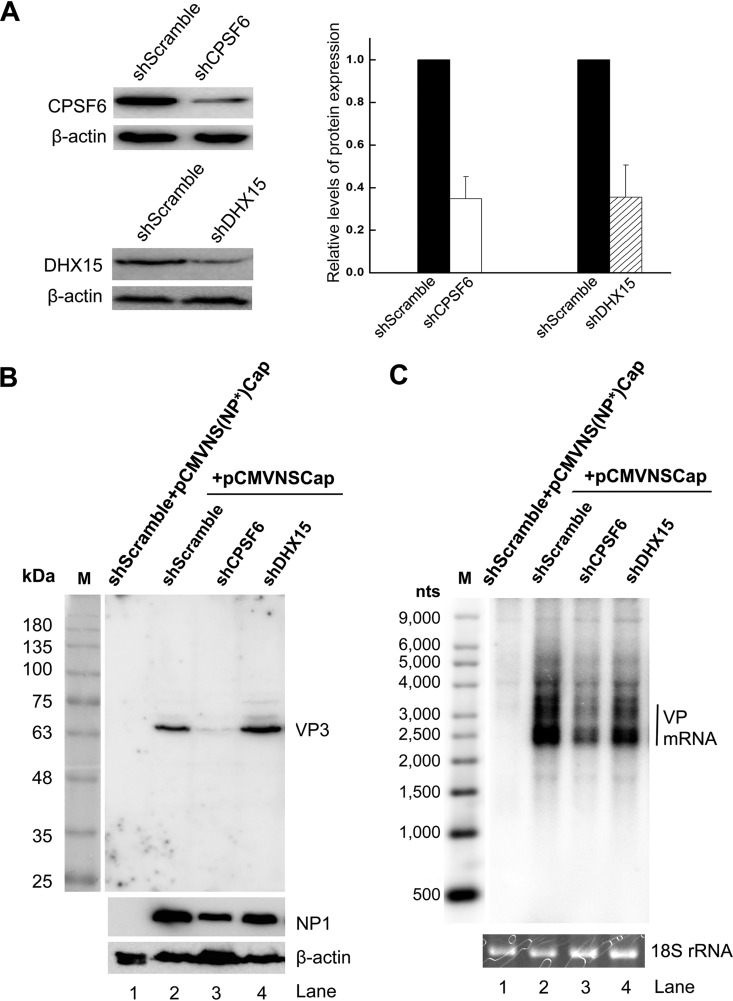
To investigate the functional requirement for DHX15 and CPSF6 in the expression of HBoV1 capsid proteins and mRNAs, we knocked down the expression of DHX15 and CPSF6 in HEK293 cells, respectively, using lentiviral vectors to introduce the stable expression of short hairpin RNAs (shRNAs) (Fig. 3A). Next, these gene-specific or scrambled shRNA-expressing cells were transfected with pCMVNSCap, which expresses HBoV1 genes under the control of a cytomegalovirus (CMV) promoter and which was previously used to investigate NP1 function in viral capsid expression (19). When the expression of DHX15 was knocked down, Western blotting did not show any decrease in VP expression, and Northern blotting also did not show a decrease in the expression of VP-encoding mRNA (Fig. 3B and C; compare lanes 4 with lanes 2). However, the knockdown of CPSF6 (Fig. 3A) obviously decreased the expression of both capsid proteins and VP-encoding mRNAs (Fig. 3B and C; compare lanes 3 with lanes 2).
FIG 3.
Knockdown of NP1-interacting protein CPSF6 but not that of DHX15 decreases the expression of HBoV1 capsid proteins. (A) Knockdown of NP1-interacting proteins in HEK293 cells. (Left) HEK293 cells were transduced with a lentiviral vector expressing gene-specific shRNA (shCPSF6 and shDHX15) or scrambled shRNA (shScramble), as indicated. Cell lysates were probed by Western blotting with the respective antibodies (top) or anti-β-actin antibody (bottom). (Right) The protein bands detected were quantified with Quantity One software (GE Healthcare), and the quantities are expressed as relative levels. (B and C) Western blotting and Northern blotting. HEK293T cells expressing the indicated shRNAs were transfected with pCMVNSCap (lanes 2 to 4). Scrambled shRNA-expressing cells were also transfected with pCMVNS(NP*)Cap to serve as a no-NP1 control (lane 1). At 48 h posttransfection, the protein extracts were analyzed by Western blotting for capsid proteins (B; lane M, protein size markers), and the total RNA was analyzed by Northern blotting for VP-encoding mRNAs (C). Western blotting used anti-VP, anti-NP1, and anti-β-actin antibodies. Northern blotting used the Cap probe, which specifically detects VP mRNAs. Ethidium bromide-stained 18S rRNA bands are shown as loading controls, and the VP-encoding mRNA bands are indicated. An Ambion Millennium RNA ladder (Thermo Fisher Scientific) was loaded, and its specific probes were used as size markers (lane M).
In summary, the BioID assay combined with mass spectrometry suggested over 300 proteins that potentially interact with NP1, and some of these are involved in a function in viral pre-mRNA processing. We identified that both DHX15 and CPSF6 interact with NP1 in cells, but only CPSF6 was confirmed to play a direct role in the expression of viral VP-encoding mRNAs and capsid proteins. In the following experiments, our study focused on the interaction of CPSF6 with NP1.
CPSF6 specifically interacts with NP1 in vitro, and the knockout of CPSF6 totally abolishes the expression of both capsid proteins and their encoding mRNAs.
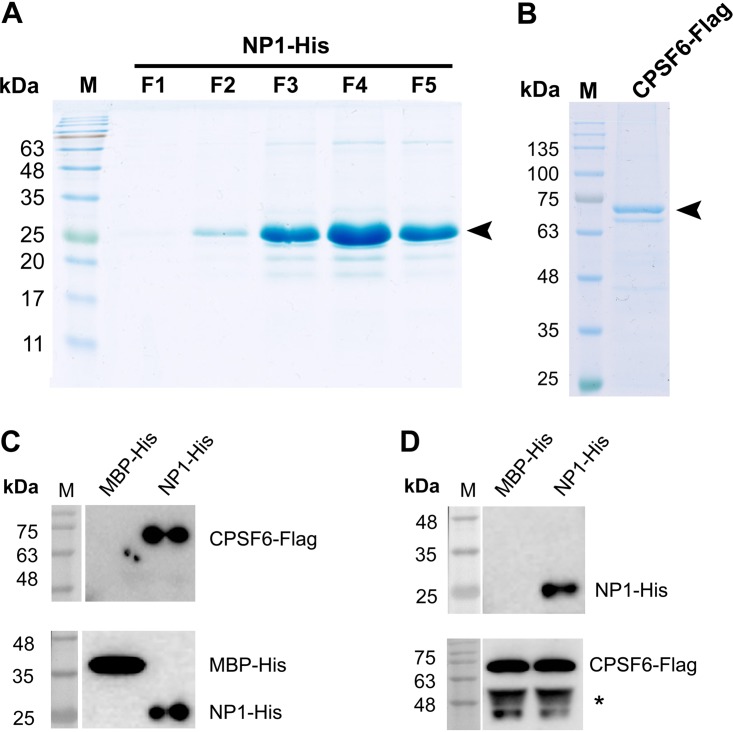
An in vitro binding assay was conducted using purified NP1 and CPSF6 proteins. Recombinant C-terminally His-tagged NP1 (NP1-His) was produced in E. coli (Fig. 4A), and C-terminally Flag-tagged CPSF6 (CPSF6-Flag) was produced in suspension-cultured 293-ES cells (Fig. 4B). The results showed that NP1-His was able to pull down CPSF6-Flag (Fig. 4C) and vice versa (Fig. 4D), confirming the direct interaction between NP1 and CPSF6.
FIG 4.
NP1 specifically interacts with CPSF6 in vitro. (A) NP1-His purification. NP1-His was purified as described in Materials and Methods. The NP1-His protein eluted from fractions 1 to 5 (lanes F1 to F5, respectively) was run on an SDS-PAGE gel (15% acrylamide) and then stained with Coomassie brilliant blue dye. (B) CPSF6-Flag purification. CPSF6-Flag protein was expressed in suspension-culture 293-ES cells by transfection and purified with anti-Flag beads. Four micrograms of the purified CPSF6-Flag protein was analyzed on an SDS-PAGE gel (10% acrylamide) and then stained with Coomassie brilliant blue dye. (C and D) In vitro pulldown assay. (C) Purified NP1-His and control MBP-His were used as the bait to pull down purified CPSF6-Flag using Ni-NTA agarose beads. The proteins pulled down by the Ni-NTA agarose beads were analyzed by Western blotting using anti-His (bottom) and anti-Flag (top). (D) Purified CPSF6-Flag was used to coat anti-Flag M2 affinity beads, which were used to pull down purified NP1-His and MBP-His (a negative control). The proteins pulled down by the anti-Flag M2 affinity beads were analyzed by Western blotting using anti-His (top) and anti-Flag (bottom). Lanes M, molecular mass markers.
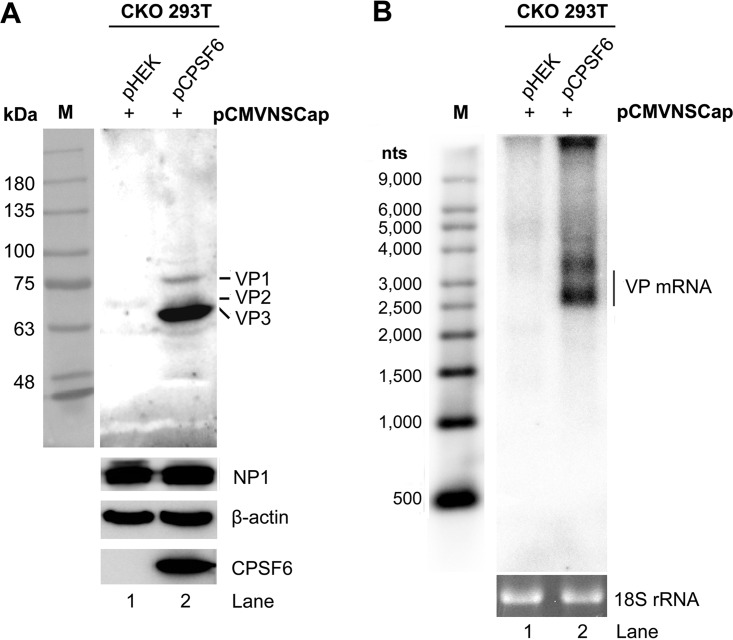
Knockdown of CPSF6 expression in HEK293 cells, evidenced by Western blotting (Fig. 3A), moderately decreased the level of HBoV1 VP expression (Fig. 3B), with a corresponding reduction in the level of VP mRNAs being observed (Fig. 3C). We further used an HEK293T cell line in which CPSF6 was knocked out (CKO 293T cells), a kind gift from Alan N. Engelman at Harvard University (33), to interrogate the function of CPSF6 in VP expression. Western blotting and Northern blotting showed that CPSF6 knockout completely abolished the expression of both VP and VP-encoding mRNAs, while the transfection of CPSF6-Flag rescued the expression in the CPSF6-knockout cells (Fig. 5A and B).
FIG 5.
Complementation of CPSF6 in CKO 293T cells rescues the expression of capsid proteins. CPSF6-knockout HEK293T cells (CKO 293T cells) were transfected with pCMVNSCap together with the pHEK empty vector or pCPSF6-Flag. Western blotting and Northern blotting were performed at 48 h posttransfection. (A) Western blot analysis of capsid proteins. Cell lysates were analyzed by Western blotting using anti-VP, anti-NP1, anti-CPSF6, and anti-β-actin antibodies. The detected protein bands are indicated. Lane M, protein size markers. (B) Northern blot analysis of VP-encoding mRNAs. Total RNAs were analyzed by Northern blotting using the Cap probe. Ethidium bromide-stained 18S rRNA bands are shown as the loading control, and the VP-encoded mRNA bands are indicated. A Millennium RNA ladder was used as a size marker (lane M).
Taken together, we confirmed the interaction of CPSF6 with NP1 in vitro and the absolute requirement of CPSF6 for capsid protein expression.
CPSF6 colocalizes with NP1 in HBoV1 dsDNA genome-transfected HEK293 cells as well as in HBoV1-infected HAE cells.
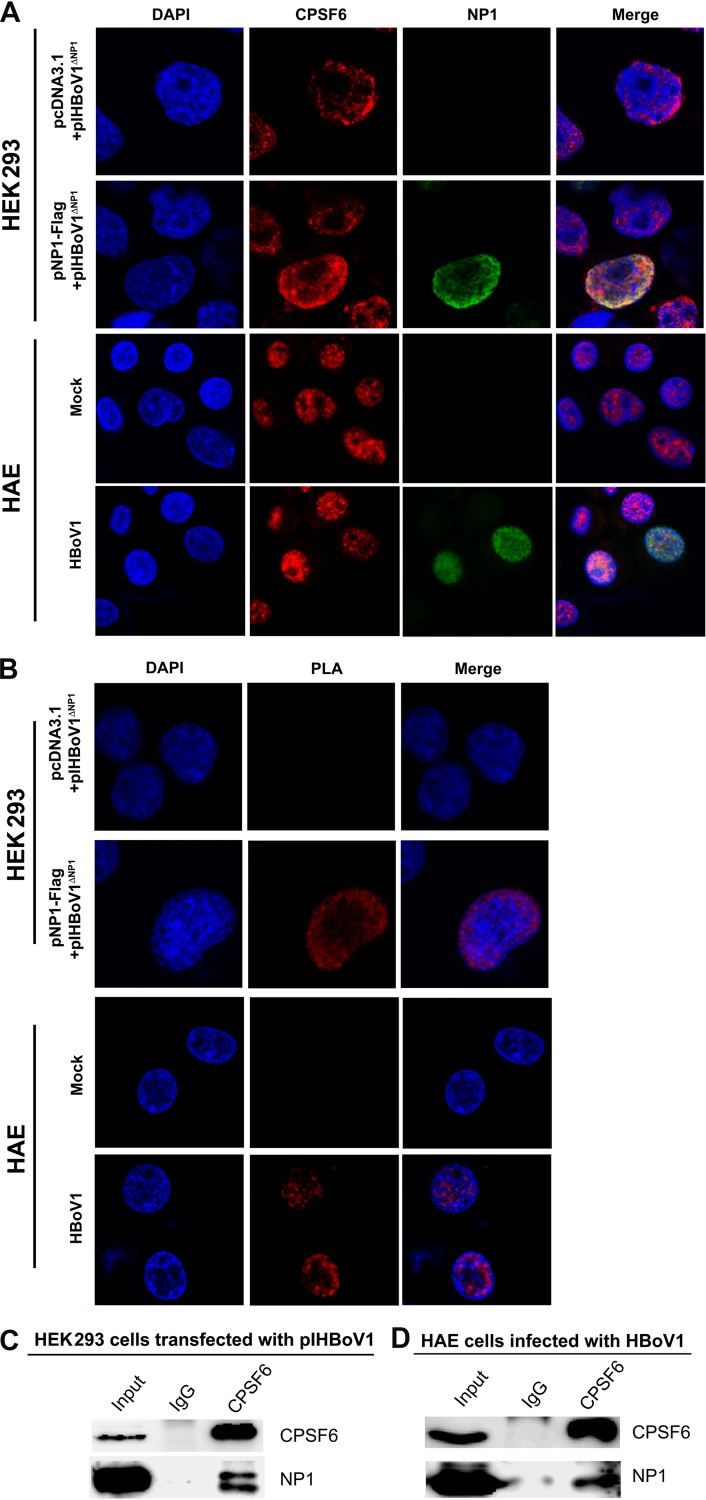
To investigate the direct interaction of NP1 and CPSF6 during viral replication and infection in vivo, we first performed an immunofluorescence assay in HEK293 cells cotransfected with pIHBoV1ΔNP1 and pNP1-Flag. Confocal microscopy revealed a clear association between anti-CPSF6 (red) and anti-NP1 (green) staining (Fig. 6A, HEK293). This association was further confirmed in a proximity ligation assay (PLA) (Fig. 6B, HEK293), which determined an interaction between two proteins in close proximity of ∼40 nm (34), and in the co-IP in the presence of nuclease (Fig. 6C). More importantly, in the HBoV1-infected HAE cells, we also observed the colocalization of CPSF6 with NP1 by confocal microscopy and PLA (Fig. 6A and B, HAE) and also their interaction by co-IP (Fig. 6D).
FIG 6.
NP1 is tightly associated with cellular CPSF6 in both HEK293 cells transfected with an HBoV1 replicative genome and HAE cells infected with HBoV1. HEK293 cells were cotransfected with pIHBoV1ΔNP1 and pNP1-Flag or pIHBoV1ΔNP1 and the pcDNA3.1 vector. HAE-ALI cultures were infected with HBoV1. At 48 h posttransfection or 7 days postinfection, the cells were analyzed as follows. For infected HAE-ALI, the HAE cells were trypsinized off the transwell inserts and cytospun onto slides. (A) Immunofluorescence assay. The cells (HEK293 or HAE cells) were permeabilized and costained for CPSF6 (red) and NP1 (green). (B) Proximity ligation assay (PLA). The cells (HEK293 or HAE cells) were cytospun onto slides and were costained with mouse anti-CPSF6 and rabbit anti-NP1 for PLA. The PLA-amplified signals are shown in red. The nuclei were stained with DAPI (blue). Images were taken using a Leica TCS SPE confocal microscope at a ×63 magnification. (C and D) Coimmunoprecipitation (co-IP) assay. pIHBoV1-transfected HEK293 cells (C) or HBoV1-infected HAE-ALI cultures (D) were lysed at 48 h posttransfection or at 7 days postinfection. The cell lysate was incubated with anti-CPSF6 and pulled down by protein G beads. Western blotting was performed to identify the pulldown yield.
Collectively, these results strongly suggest that CPSF6 interacts with NP1 in cells during HBoV1 replication and infection.
CPSF6 plays an important role in the nuclear import of NP1 and in viral DNA replication thereafter.
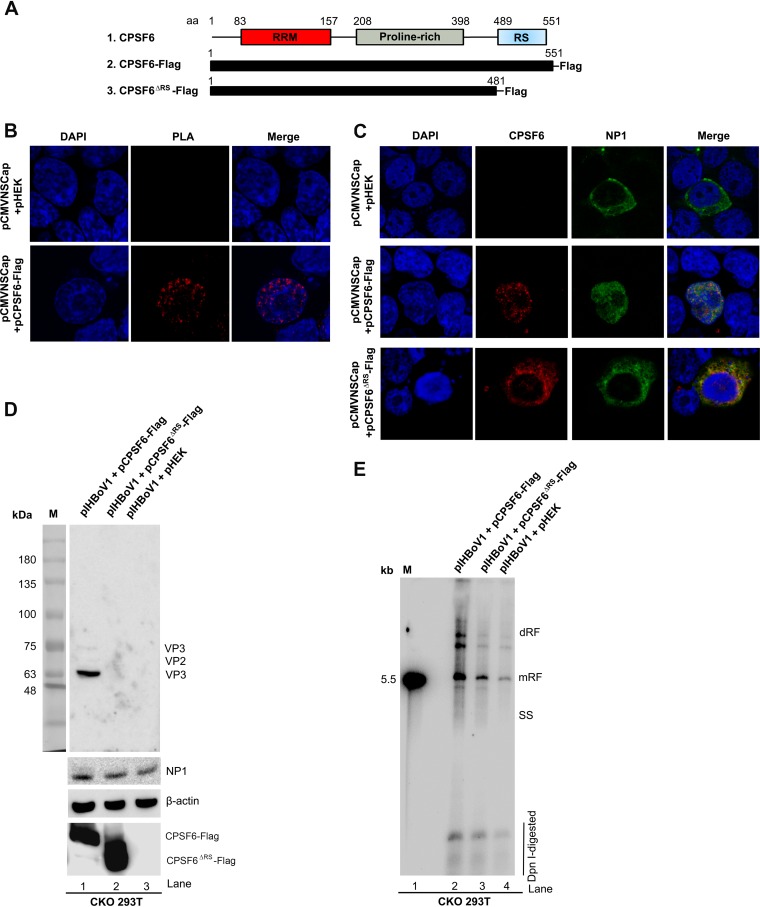
CPSF6 was identified to play an important role in the transportation of the HIV capsid into the nucleus, which is a critical step in the early phase of HIV infection (33, 35). The arginine-serine-rich (RS) domain is thought to be critical for the nuclear import of CPSF6 (36) (Fig. 7A). Thus, we wondered if CPSF6 also functions in the nuclear import of NP1. To this end, we performed a PLA in CKO 293T cells cotransfected with pCMVNSCap and pCPSF6-Flag. We observed an association of CPSF6 with NP1 (amplified signals in red) in the nucleus (Fig. 7B). Interestingly, when CKO 293T cells were transfected with pCMVNSCap and the empty plasmid control (pHEK), NP1 was expressed in the cytoplasm (Fig. 7C, top), suggesting the failure of NP1 nuclear import in the absence of CPSF6 expression, although NP1 is thought to bear a noncanonical nuclear localization signal (NLS) (37).
FIG 7.
CPSF6 knockout and expression of the RS domain-deleted mutant (CPSF6ΔRS) in CFSF6-knockout HEK293T cells (CKO 293T cells) resulted in the failure of NP1 nuclear entry. (A) Schematic representation of the protein domains of wild-type CPSF6 and the truncated mutant, CPSF6ΔRS. The RNA recognition motif (RRM), proline-rich domain, and arginine-serine-rich domain (RS) of CPSF6 are diagrammed. Flag-tagged CPSF6 and the RS domain-deleted mutant (CPSF6ΔRS) expression cassettes are diagrammed. (B) Proximity ligation assay (PLA). CKO 293T cells were cotransfected with pCMVNSCap and the pHEK empty vector or pCMVNSCap and pCPSF6-Flag. At 48 h posttransfection, the cells were cytospun onto slides and costained with mouse anti-CPSF6 antibody and rabbit anti-NP1 antibody. The PLA-amplified signals are shown in red. (C) Immunofluorescence assay. CKO 293T cells were cotransfected with pCMVNSCap and one of the following plasmids, as indicated: the pHEK empty vector, pCPSF6-Flag, or pCPSF6ΔRS-Flag. At 48 h posttransfection, the cells were cytospun onto slides and costained for CPSF6 (red) and NP1 (green). The cells were then visualized using a Leica TCS SPE confocal microscope at a magnification of × 63. The nuclei were stained with DAPI (blue). (D and E) CKO 293T cells were cotransfected with pIHBoV1 and one of the following plasmids, as indicated: pCPSF6-Flag, pCPSF6ΔRS-Flag, or the pHEK empty vector. At 48 h posttransfection, the transfected cells were harvested for analyses. (D) Western blot analysis. Cell lysates were analyzed by Western blotting using anti-VP, anti-NP1, anti-Flag, and anti-β-actin antibodies. The detected protein bands are indicated. Lane M, molecular size marker. (E) Southern blot analysis. Hirt DNA samples were extracted from the cells and examined for viral DNA replication by Southern blotting with a [α-32P]dCTP-labeled HBoV1 NSCap probe. The double-replicative-form (dRF), monomer RF, and single-stranded (SS) DNA bands and DpnI-digested DNA are indicated. The 5.5-kb HBoV1 RF DNA marker is shown on the left (lane M).
We then constructed an RS domain-deleted CPSF6 mutant (CPSF6ΔRS-Flag) (Fig. 7A). While the nuclear presence of NP1 was again observed in cells supplemented with a full-length CPSF6-Flag (Fig. 7C, middle), it was not in cells supplemented with the mutant CPSF6ΔRS-Flag (Fig. 7C, bottom). In the latter cells, NP1 and CPSF6ΔRS colocated in the cytoplasm. These results suggest that the nuclear import of NP1 relies on the nuclear transport function of CPSF6.
We next investigated capsid protein expression and viral DNA replication from the transfected infectious clone pIHBoV1 in CKO 293T cells supplemented with pCPSF6-Flag, the plasmid harboring the RS-deleted CPSF6 mutant (pCPSF6ΔRS-Flag), or an empty vector control. Western blotting showed that capsid proteins VP1 to VP3 were expressed from the transfected HBoV1 dsDNA genome (pIHBoV1) in CPSF6-Flag-expressing CKO 293T cells but not in CPSF6ΔRS-Flag-expressing or the control CKO 293T cells (Fig. 7D). Southern blotting of Hirt DNAs extracted from these transfected cells showed that CPSF6ΔRS did not rescue the level of the replicative-form (RF) DNA to that in cells supplemented with full-length CPSF6 (Fig. 7E).
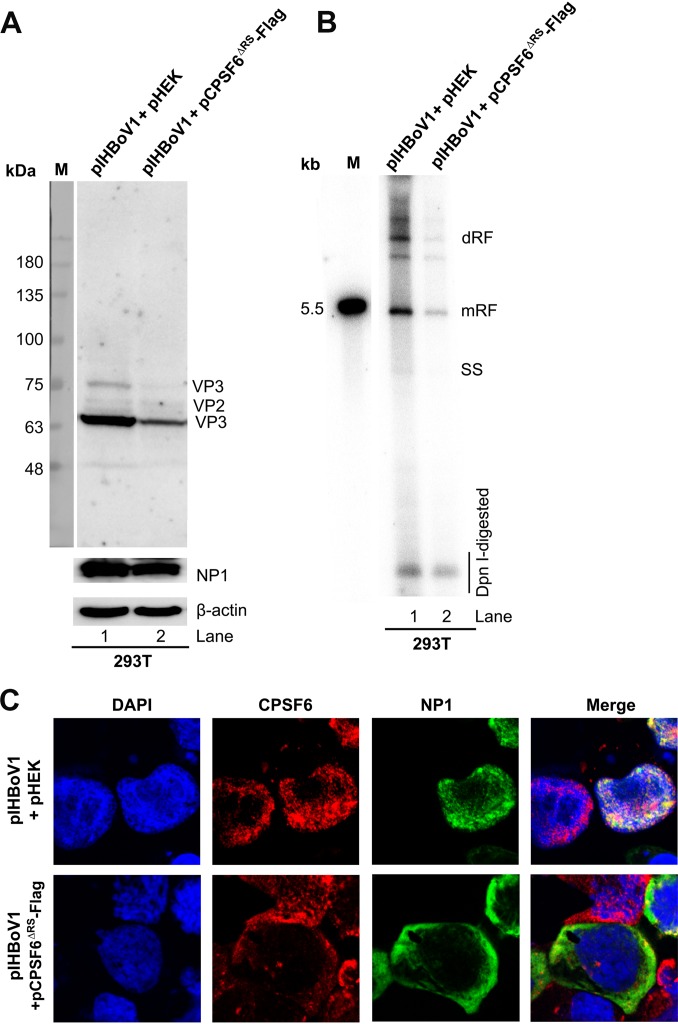
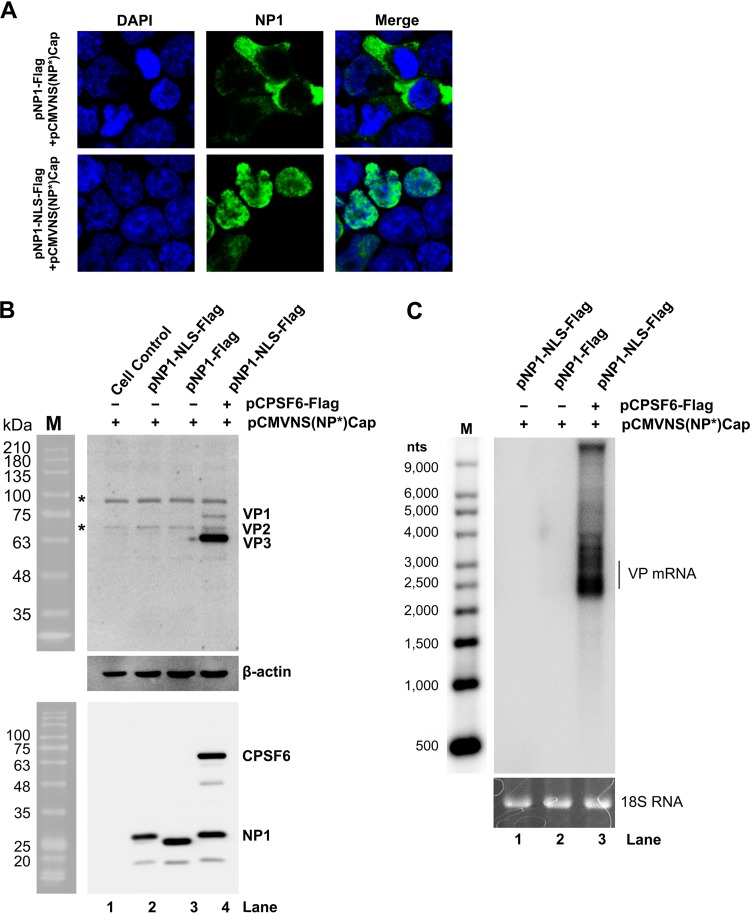
Furthermore, in regular HEK293T cells, overexpression of CPSF6ΔRS remarkably decreased the levels of capsid proteins (Fig. 8A) and viral DNA replication (Fig. 8B) compared with those in the empty vector (pHEK) control-transfected cells. Consistent with the role of CPSF6 in the nuclear import of NP1, the overexpression of CPSF6ΔRS resulted in the dominant expression of NP1 in the cytoplasm (Fig. 8C), echoing the role of NP1 in the expression of VP and VP-encoding mRNAs in the nucleus (Fig. 6A and B). Notably, fusing a strong NLS (the c-Myc NLS [38]) to the C terminus of NP1 successfully facilitated the expression of NP1 in the nucleus (Fig. 9A). However, NP1-NLS expression in CKO 293T cells transfected with pCMV(NP*)Cap still failed to confer the function of NP1 in viral capsid protein expression (Fig. 9B, lanes 2 versus 4) and the accumulation of VP-encoding mRNAs (Fig. 9C, lanes 1 versus 3).
FIG 8.
Overexpression of the RS domain-deleted mutant (CPSF6ΔRS) decreased HBoV1 replication in HEK293T cells. HEK293T cells were cotransfected with pIHBoV1 and the pHEK empty vector or pIHBoV1 and pCPSF6ΔRS-Flag. (A) Western blot analysis. At 48 h posttransfection, 10% of the transfected cells were collected, lysed, and immunoblotted with anti-HBoV1 VP, anti-HBoV1 NP1, and anti-β-actin antibodies. The proteins detected are indicated. Lane M, molecular size markers. (B) Southern blot analysis. At 48 h posttransfection, 80% of the collected cells were lysed for Hirt DNA extraction. Hirt DNA samples were digested with DpnI and analyzed by Southern blotting with a 32P-labeled HBoV1 NSCap probe. The double-replicative-form (dRF), monomer RF, and single-stranded (SS) DNA bands and DpnI-digested DNA are indicated. The 5.5-kb HBoV1 marker is shown on the left (lane M). (C) Immunofluorescence assay. At 48 h posttransfection, the transfected cells were cytospun onto slides and costained for CPSF6 (red) and NP1 (green). The cells were then visualized using a Leica TCS SPE confocal microscope at a magnification of ×63. The nuclei were stained with DAPI (blue).
FIG 9.
The nuclear expression of NP1 is not sufficient to confer its function in viral capsid expression in the absence of CPSF6 expression. (A) NLS-fused NP1 was expressed in the nucleus. CPSF6-knockout HEK293T cells (CKO 293T cells) were cotransfected with pCMVNS(NP*)Cap and pNP1-Flag (top) or pCMVNS(NP*)Cap and pNP1-NLS-Flag (bottom). At 2 days posttransfection, the cells were analyzed by an immunofluorescent assay using anti-NP1 (green). Images were taken under a Leica TCS SPE confocal microscope at a magnification of ×63. The nuclei were stained with DAPI (blue). (B and C) NP1-NLS did not rescue the expression of capsid proteins in CKO 293T cells. CKO 293T cells were cotransfected with (i) pCMVNS(NP*)Cap, pNP1-NLS-Flag, and pHEK (empty vector), (ii) pCMVNS(NP*)Cap, pNP1-Flag, and pHEK, or (iii) pCMVNS(NP*)Cap, pNP1-NLS-Flag, and pCPSF6-Flag, as indicated. (B) Western blot analysis of capsid proteins. At 48 h posttransfection, the cells were analyzed for the expression of VP, NP1, and CPSF6, as indicated, using Western blotting. β-Actin was probed as a loading control. Untransfected cells were used as a control (lane 1). Lane M, protein size markers. The bands indicated by asterisks were nonspecific bands. (C) Northern blot analysis of VP-encoding mRNAs. At 48 h posttransfection, total RNA was extracted from transfected cells and analyzed by Northern blotting using the Cap probe. Ethidium bromide-stained 18S rRNA bands are shown as loading controls, and the VP-encoding mRNAs are indicated. The Millennium RNA ladder RNA size markers are shown (lane M).
Taken all together, these results strongly suggest that CPSF6 first helps import NP1 into the nucleus and then further facilitates the function of NP1 in the expression of capsid proteins and viral DNA replication.
DISCUSSION
In the present study, we identified a critical role of CPSF6 in the nuclear import of HBoV1 NP1. Additionally, the interaction of NP1 with CPSF6 plays a direct role in viral pre-mRNA processing, because when NP1 is imported into the nucleus through a CPSF6-independent route, CPSF6 is still required for the accumulation of VP-encoding mRNAs.
Splicing of HBoV1 pre-mRNA at the A3 acceptor (3′ splice site) is essential for read-through of the VP-encoding mRNAs (15, 18, 39). Previous studies have shown that NP1 is critical to the splicing of viral pre-mRNA at the A3 acceptor (19, 28). The main role of NP1 in viral pre-mRNA processing is to accumulate VP-encoding mRNAs that are cleaved and polyadenylated at the (pA)d sites (19, 22). The U2 small nuclear ribonucleoprotein (snRNP) complex deposited on the A3 acceptor is critical for the communication between the A3 acceptor and the polyadenylation (pA) site that interacts with the 3′-end cleavage and polyadenylation specificity factor (CPSF) complex. The interaction between the U2 snRNP and CPSF complexes defines the size of the last exon of viral mRNAs (40): a short exon that encodes no proteins or a large exon that encodes VP. While the detailed mechanism of how NP1 interacts with the CPSF complex in viral pre-mRNA processing remains unclear, NP1 is absolutely required to define the large VP-encoding exon. We speculate that when NP1 is not present, the short exon is preferably defined, as the distance between the last A3 acceptor and the (pA)p signals is short, which permits a strong interaction between the U2 snRNP and CPSF complexes. When NP1 is expressed, its interaction with CPSF6 weakens the interaction of the CPSF complex on the (pA)p sites, which generates a stronger interaction of the U2 snRNP on the A3 acceptor with the CPSF complex on the (pA)d sites. How NP1 decreases the interaction between the CPSF complex and the (pA)p sites of the viral pre-mRNA warrants further investigations.
The (pA)dREH site is also unique among the four HBoV1 pA sites, as it does not have a consensus CPSF-binding (AAUAAA) site (23, 24), indicating that the building of the CPSF complex is different from that of a regular one, which may be less dependent on the binding of the CFIm complex to the UGUA signal upstream of the (pA)dREH site. Of note, we also observed an interaction of Fip1 with NP1 in a co-IP experiment. However, this interaction was abolished by addition of a nuclease in the binding buffer (data not shown), supporting the suggestion that NP1 is associated within the CPSF complex that is mediated by viral mRNA.
HBoV1 NP1 is 219 amino acids (aa) in length and shares only 46% homology (identity) with MVC NP1. Although the NP1 proteins of bocaparvovirus (BPV), HBoV1, and MVC share a low percent identity (39%), they all feature in viral DNA replication (39). Both the BPV1 and HBoV1 NP1 proteins can complement the loss of NP1 during MVC DNA replication to some extent (39). It is now clear at least that the NP1 proteins of MVC and HBoV1 share features in viral pre-mRNA processing, including activation of the splicing of VP-encoding mRNA at the A3 acceptor upstream of the (pA)p sites and facilitation of the read-through of the (pA)p site (19, 22). In terms of the function of HBoV1 NP1 in splicing of the A3 acceptor, as MVC NP1 is an RNA-binding protein (28), it is possible that HBoV1 NP1 brings CPSF6 in close proximity to the A3 acceptor and that the RS domain of CPSF6 interacts with the U2AF heterogeneous dimer or a component of the U2 snRNP complex, which would enhance spliceosome assembly. Whether HBoV1 NP1 has a preferred binding site close to the A3 acceptor requires further investigation. While the RS domain of MVC NP1 is required to facilitate splicing at the A3 acceptor, the RS domain of HBoV1 NP1 is not required (22, 26), suggesting that there are certainly differences in the regulation of viral pre-mRNA processing between the involvement of MVC NP1 and that of HBoV1 NP1, which requires further investigation.
It has been reported that a noncanonical NLS is present in the amino acid sequence (aa 7 to 50) of HBoV1 NP1, in which a canonical bipartite NLS (aa 14 to 28) is embedded (37). In this study, we identified that the NP1 interaction with CPSF6 is important for the nuclear import of NP1, which is mediated by the RS domain of CPSF6, which has been confirmed to be the NLS of CPSF6 (36). NP1 also contains a nuclear export signal (NES) and consensus binding sites for 14-3-3 and Crm1 proteins (32), but the functions of these sites have not yet been evidenced. It remains for further clarification whether the nuclear import of NP1 is regulated by a balance of multiple signals of NLS and NES.
MATERIALS AND METHODS
Ethics.
The use of primary human airway epithelial cells was approved by the Institutional Review Board (IRB) of the University of Iowa (IRB approval no. 9507432).
Cell lines and primary cultures. (i) HEK293 cells.
HEK293 cells (ATCC CRL-1573), HEK293T cells (ATCC CRL-11268), and CKO 293T cells (a CPSF6-knockout HEK293T cell line that was a gift from Alan N. Engelman at Harvard University) were cultured in Dulbecco’s modified Eagle’s medium (DMEM; catalog no. SH30022.01; HyClone; GE Healthcare Life Sciences, Logan, UT) supplemented with 10% fetal bovine serum (catalog no. F0926; Sigma, St. Louis, MO) at 37°C under a 5% CO2 atmosphere. A suspension of 293-ES cells (catalog no. 94-007S) obtained from Expression Systems, LLC (Davis, CA), was cultured in mammalian cell culture serum-free medium (SFM; catalog no. 98-001-01; Expression Systems, LLC) in shake flasks at 37°C under a 5% CO2 atmosphere.
(ii) HAE-ALI culture.
Polarized human airway epithelium (HAE) cultured at an air-liquid interface (ALI), termed HAE-ALI (Transwell inserts; catalog no. 3470; Corning, NY), was obtained from the Cell Culture Core of the Center for Gene Therapy, University of Iowa, as previously described (16, 41). They were differentiated and maintained at an air-liquid interface either in a medium supplemented with 2% Ultraser G serum substitute (Pall Life Sciences) or in PneumaCult-ALI medium (StemCell, Vancouver, BC, Canada) in 5% CO2 at 37°C. The differentiation of the polarized HAE-ALI cultures was determined based on the transepithelial electrical resistance (TEER); the cultures that had a TEER of over 1,000 Ω/cm2 were used for HBoV1 infection in this study.
Plasmid constructs. (i) pIHBoV1-based constructs.
HBoV1 infectious clone plasmid pIHBoV1 and its mutant, pIHBoV1ΔNP1, which does not express the NP1 protein, were described previously (13).
(ii) pCMVNSCap-based constructs.
The pCMVNSCap plasmid contains HBoV1 DNA from nt 282 to 5395 (GenBank accession no. JQ923422) and expresses all HBoV1 proteins under the control of the human cytomegalovirus (CMV) promoter, as described previously (19). The pCMVNS(NP*)Cap plasmid bears an early stop codon in the NP1 ORF, which causes the premature termination of translation of the NP1 protein (19).
(iii) NP1-expressing plasmids.
pNP1-Flag has been described previously (42). pNP1-NLS-Flag was constructed by inserting a c-Myc NLS (38) between the NP1 ORF and the Flag tag in pNP1-Flag. pNP1-BirA* was constructed by fusing an ORF of E. coli-derived biotin ligase (BirA*) with a catalytic site mutation (R118G) to the C terminus of the HBoV1 NP1 protein.
(iv) CPSF6-expressing plasmids.
pCPSF6-Flag, which expresses C-terminally Flag-tagged CPSF6, was constructed by inserting a coding sequence of human CPSF6 into the pHEK vector (TaKaRa Bio Inc.), using the XhoI and XbaI sites. pCPSF6ΔRS-Flag was constructed by deleting the RS domain (aa 482 to 551) in pCPSF6-Flag.
(v) pLKO constructs.
The lentiviral vector pLKO.1-mCherry (41) was used to clone shRNA sequences between the AgeI and EcoRI sites. pLKO.1-mCherry containing a scrambled shRNA sequence was used as a control (41). The following shRNAs were obtained from Sigma (St. Louis, MO) for knocking down CPSF6 (shCPSF6) and DHX15 (shDHX15): for shCPSF6, 5′-CCG GGT TGT AAC TCC ATG CAA TAA ACT CGA GTT TAT TGC ATG GAG TTA CAA CTT TTT G-3′; and for shDHX15, 5′-CCG GGT TGG TTC GAT AAT GGC CTT TCT CGA GAA AGG CCA TTA TCG AAC CAA CTT TTT G-3′.
(vi) Bacterial expression plasmids.
pET30a-NP1 was constructed by inserting an NP1 ORF into the pET30a(+) vector (EMD Millipore) through the NdeI and XhoI sites. pMBP-His was constructed by fusing 6× histidines (His) to the maltose binding protein (MBP) ORF at the C terminus in pMAL-c5X (New England Biolabs) through the SacI site, as described previously (43).
Plasmid DNA transfection.
HEK293 cells, HEK293T cells, and CKO 293T cells were transfected by using the PEImax transfection reagent (molecular weight, 40,000; catalog no. 24765-2; Polysciences, Inc.). The total amounts of plasmid DNA were kept constant (4 μg per 60-mm dish) in each group by supplementation with an empty vector to balance the transfection efficiency. Suspension-cultured 293-ES cells were transfected by using the TransIT-Pro transfection reagent (catalog no. MIR 5760; Mirus Bio LLC), according to the manufacturer’s instructions.
Viruses and infection.
HAE-ALI cultures were infected with HBoV1 at a multiplicity of infection (MOI) of 100 DNase I-resistant particles (DRP) per cell as previously described (41). Viruses were diluted in 100 μl of culture medium and incubated on the apical side for 1 h.
Lentivirus production.
We produced lentiviruses according to the instructions provided by Addgene (http://www.addgene.org/tools/protocols/plko) and purified them as described previously (44, 45). We transduced cells with lentiviral vectors at an MOI of ∼5 transduction units/cell, as described previously (44, 45).
BioID assay.
We followed a published method (46) to perform the BioID assay. HEK293 cells were transfected with pIHBoV1ΔNP1 and pNP1-BirA* or pIHBoV1ΔNP1 and pBirA*. At 24 h posttransfection, the cells were incubated with biotins (50 μM) for another 24 h. After three washes with phosphate-buffered saline (PBS), the cells were lysed at room temperature in lysis buffer (50 mM Tris-HCl, pH 7.4, 500 mM NaCl, 0.2% SDS, 1 mM dithiothreitol [DTT], protease inhibitors [catalog no. S8820; Sigma]) and sonicated. Then, Triton X-100 was added to a final concentration of 2%. After further sonication, an equal volume of prechilled 50 mM Tris-Cl (pH 7.4) buffer was added, before additional sonication (subsequent steps were performed at 4°C) and centrifugation at 16,500 × g. The collected supernatants were incubated with 100 μl of prewashed streptavidin-conjugated agarose beads overnight. The beads were collected and washed twice for 5 min each time at 25°C (all subsequent steps were performed at 25°C) in wash buffer 1 (2% SDS in double-distilled H2O). This was repeated once with 50 mM Tris-Cl (pH 7.4) buffer for 5 min. Finally, bound proteins were removed from the beads with an equal volume of 2× Laemmli buffer saturated with biotin at 98°C for 5 min and then analyzed by SDS-polyacrylamide gel electrophoresis (PAGE). Bands differentiated from the control bands were excised and subjected to liquid chromatography-tandem mass spectrometry (LC-MS/MS) at the Taplin Biological Mass Spectrometry Facility, Harvard University.
Coimmunoprecipitation (co-IP) assay.
For candidates that interacted with the NP1 protein, the pNP1-Flag-transfected cells were washed twice with cold PBS and then lysed with lysis buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% NP-40, protease inhibitors) with constant agitation for 30 min at 4°C. The cell lysates were treated without or with 250 units of nuclease (Benzonase; Thermo Fisher Scientific), followed by centrifugation at 10,000 × g for 15 min at 4°C. The supernatant was collected and incubated with 25 μl of prewashed anti-Flag G1 affinity resin (GenScript, Piscataway, NJ) with rotation at 4°C overnight. Finally, the beads were pelleted down and washed 3 times with washing buffer (25 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% NP-40, 1 mM EDTA), before mixing with loading dye for Western blotting.
For pIHBoV1-transfected HEK293 cells and HBoV1-infected HAE-ALI cells, the supernatant of the cell lysate was preincubated with either 4 μg of mouse anti-CPSF6 or an equivalent amount of normal mouse IgG (0.4 mg/ml; Santa Cruz, Dallas, TX) for 3 h at 4°C with rotation. The mixture was incubated with 25 μl of prewashed protein G beads (Thermo Fisher Scientific) for at least 2 h at 4°C with rotation. The bound proteins were removed from the protein G beads by dissolving them in 1× Laemmli sample buffer at 98°C for 5 min and were then analyzed by Western blotting.
Protein expression and purification. (i) Recombinant proteins expressed in bacteria.
MBP-His protein was expressed in E. coli BL21(DE3) cells as described previously (43) and purified using amylose resin according to the manufacturer’s instructions (New England Biolabs). NP1-His was expressed in E. coli BL21(DE3) cells and purified by using Ni-nitrilotriacetic acid (NTA) agarose (Qiagen), as we have described previously (47).
(ii) Recombinant proteins expressed in 293-ES cells in suspension.
One liter of a suspension culture of 293-ES cells (2 × 106 cells/ml) was transfected with pCPSF6-Flag. At 3 days posttransfection, the cells were lysed in lysis buffer (25 mM Tris, pH 7.4, 150 mM NaCl, and 1 mM EDTA supplemented with protease inhibitors). Crude lysate was sonicated for 3 min at a frequency of 15 s on and 25 s off at a 70% pulse with a VCX130 sonicator (Sonics & Materials Inc., Newtown, CT). The lysate was then centrifuged at 10,000 rpm for 15 min and filtered through a 0.2-μm-pore-size filter. The filtered lysate was incubated with 1 ml of phosphate-buffered saline (PBS; pH 7.4)-prewashed anti-Flag G1 affinity resin (GenScript, Piscataway, NJ) at 4°C for at least 1 h. Then, the beads were washed with washing buffer (50 mM Tris, pH 7.4, 500 mM NaCl, 0.05% Triton X-100, protease inhibitors) at a volume of 5 times the resin volume and were eluted with 3× Flag peptides (APExBIO, Houston, TX) at a concentration of 200 μg/ml. Finally, the eluted protein was dialyzed against PBS twice and against binding buffer (20 mM Tris-HCl, pH 8.0, 125 mM NaCl, 10% glycerol, 1% NP-40, 5 mM DTT, protease inhibitors) once and was concentrated 10 times using polyethylene glycol (PEG) 6000. The concentrated protein was quantified, aliquoted, and stored at −80°C.
In vitro pulldown assay.
For the pulldown using Ni-NTA beads, 1.5 μg each of purified NP1-His and MBP-His proteins was immobilized on 25 μl of prewashed Ni-NTA agarose (Qiagen) in binding buffer (25 mM Tris, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% NP-40) for 2 h at 4°C with rotation. The purified CPSF6-Flag protein (1.5 μg) was then added to the beads and the mixture was incubated for 2 h. The beads were washed with washing buffer (25 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% NP-40, 1 mM EDTA) 3 times. The bound proteins were eluted by boiling in 1× Laemmli sample buffer and visualized using Western blotting. In a similar fashion, for anti-Flag G1 affinity resin pulldown, 1.5 μg of the purified CPSF6-Flag was first bound to the beads in binding buffer and then 1.5 μg of NP1-His was incubated with the anti-Flag beads. MBP-His protein (1.5 μg) was used as a negative control. After briefly washing three times, the beads were boiled for 5 min in 1× Laemmli sample buffer for Western blotting.
Western blotting.
Western blotting was carried out as previously described (45, 48). Briefly, the cell lysates or other protein samples were loaded, along with 2.5 μl of a prestained protein ladder (catalog no. P008; GoldBio, St. Louis, MO), and separated on SDS-PAGE gels. The proteins were transferred onto a polyvinylidene difluoride (PVDF) membrane (catalog no. IPVH00010; Millipore, Bedford, MA), which was sequentially blocked and probed with primary and secondary antibodies. The signals were visualized by enhanced chemiluminescence, and the prestained protein ladder (molecular mass markers) was imaged under bright light simultaneously using a Fuji LAS4000 imaging system. The chemiluminescent image and the protein ladder image were then aligned using Multi Gauge software (Fujifilm Life Sciences), which generated all the Western blot images.
Northern blotting.
At 48 h posttransfection, total RNA was extracted from transfected cells using the TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. RNA (5 μg) was separated on a 1.4% denaturation agarose gel and visualized using ethidium bromide staining. The stained 18S rRNA bands served as loading controls. Northern blot analysis was performed as previously described (48), using a 32P-labeled HBoV1 Cap DNA (nt 3921 to 5200) probe (19). An RNA ladder (Invitrogen) was used as a size marker.
In vivo DNA replication analysis. (i) Low-molecular-weight (Hirt) DNA extraction.
HEK293T and CKO 293T cells were collected at 48 h posttransfection for low-molecular-weight (Hirt) DNA extraction (49). Cells were washed once with PBS and lysed with Hirt lysis buffer (10 mM Tris, pH 8.0, 10 mM EDTA, 0.6% SDS) for 15 min. Cell lysates were transferred into Eppendorf tubes, adjusted to a final NaCl concentration of 1.5 M, and incubated on ice overnight, before being cleared by centrifugation at 17,000 × g for 20 min. The supernatants were collected and treated with proteinase K at a final concentration of 1 mg/ml for 1 h. Hirt DNA was purified with a DNA gel extraction kit (Qiagen) according to the manufacturer’s instructions.
(ii) Southern blotting.
Southern blotting was performed according to our previously reported methods (25, 49, 50). Briefly, the Hirt DNA samples were digested with DpnI, resolved on a 1% agarose gel, blotted onto a nitrocellulose membrane, and probed with an [α-32P]dCTP-labeled probe of the HBoV1 NSCap gene (25). Hybridization signals were captured by using a storage phosphor screen and visualized on a Typhoon FLA 9000 biomolecular imager (GE Healthcare).
IFA.
Immunofluorescence analysis (IFA) was performed as previously described (45). Briefly, the cells were collected and cytospun onto slides. Then they were fixed with 3.7% paraformaldehyde (PFA) in PBS, pH 7.4, at room temperature for 15 min, washed in PBS three times for 5 min each time, and permeabilized with 0.5% Triton X-100 for 5 min. The primary antibodies were diluted in PBS with 2% fetal calf serum. After incubation for 1 h at 37°C, fluorescent dye-conjugated secondary antibodies were applied, followed by staining of the nuclei with DAPI (4′,6-diamidino-2-phenylindole). The cells were then visualized using a Leica TCS SPE confocal microscope at the Confocal Core Facility of the University of Kansas.
PLA.
A Duolink proximity ligation assay (PLA) kit (Sigma) was used to perform PLA according to the manufacturer’s instructions as previously described (41). Briefly, the cells were cytospun onto slides and were then fixed with 3.7% PFA for 15 min, permeabilized with 0.2% Triton X-100 for 5 min, and blocked with Duolink blocking buffer for 30 min. Then, the cells were incubated with anti-CPSF6 and anti-NP1 primary antibodies for 1 h at room temperature. Two diluted PLA probes were applied to the cells, and the slides were incubated for 1 h at 37°C. Hybridized oligonucleotides were then ligated in the ligation solution for 30 min at 37°C and amplified in the amplification solution for 100 min. The cells were then washed and mounted with Duolink in situ mounting medium with DAPI. The slides were observed under a Leica TCS SPE confocal microscope.
Antibodies used in the study. (i) First antibodies.
Antibodies against the HBoV1 NP1 and VP proteins were previously described (18). A rabbit anti-HBoV1 NP1 antibody (32), which was a gift from Peter Tattersall from Yale University, was used in immunofluorescent assays and PLA. The following first antibodies were purchased: mouse anti-Flag (catalog no. 200-301-B13) from Rockland, Limerick, PA; anti-SF3A1 (catalog no. A301-601A), anti-CPSF6 (catalog no. A301-356A), anti-DHX15 (catalog no. A300-390A), anti-KHSRP (catalog no. A302-021A), anti-SUGP1 (catalog no. A304-675A), anti-MFAP1 (catalog no. A304-647), and anti-hnRNPK (catalog no. A300-674A) from Bethyl, Montgomery, TX; anti-SF3B2 (catalog no. AP16199c) from Abgent, San Diego, CA; anti-SNW1 (catalog no. A14580) from Abclone, Woburn, MA; anti-β-actin (catalog no. A5441) from Sigma-Aldrich, St. Louis, MO; monoclonal anti-CPSF6 (catalog no. sc-100692) from Santa Cruz Biotechnology, Dallas, TX; and anti-His tag (catalog no. 12698S) from Cell Signaling Technology, Danvers, MA.
(ii) Secondary antibodies.
Horseradish peroxidase (HRP)-conjugated anti-mouse IgG and HRP-conjugated anti-rabbit IgG were purchased from Sigma, and fluorescein isothiocyanate (FITC)- and rhodamine-conjugated anti-mouse IgG and Alex Fluor 594-conjugated anti-rabbit IgG were purchased from Jackson ImmunoResearch Inc. (West Grove, PA).
Supplementary Material
ACKNOWLEDGMENTS
We thank the members of the J. Qiu lab for technical support and valuable discussions. We are indebted to Alan N. Engelman at Harvard University for the CPSF6-knockout HEK293T (CKO 293T) cells and Peter Tattersall at Yale University for the rabbit anti-HBoV1 antibody. We thank David Pintel for information prior to publication. We acknowledge the Confocal Microscopy Core Laboratory, The University of Kansas Medical Center, for help with confocal microscopy.
This study was supported by PHS grants R01AI070723 and R21AI139572 from the National Institute of Allergy and Infectious Diseases, P30DK054759 from the National Institute of Diabetes and Digestive and Kidney Diseases, and P01HL051670 from the National Heart, Lung, and Blood Institute. This study was also supported by grant YAN19XX0 from the Cystic Fibrosis Foundation and grants 31470268 and 31700873 from the National Natural Science Foundation of China. The Confocal Microscopy Core Laboratory, The University of Kansas Medical Center, is supported, in part, by NIH/NIGMS COBRE grant P30GM122731.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Cotmore SF, Agbandje-McKenna M, Canuti M, Chiorini JA, Eis-Hubinger AM, Hughes J, Mietzsch M, Modha S, Ogliastro M, Penzes JJ, Pintel DJ, Qiu J, Soderlund-Venermo M, Tattersall P, Tijssen P, ICTV Report Consortium. 2019. ICTV virus taxonomy profile: Parvoviridae. J Gen Virol 100:367–368. doi: 10.1099/jgv.0.001212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allander T, Jartti T, Gupta S, Niesters HG, Lehtinen P, Osterback R, Vuorinen T, Waris M, Bjerkner A, Tiveljung-Lindell A, van den Hoogen BG, Hyypiä T, Ruuskanen O. 2007. Human bocavirus and acute wheezing in children. Clin Infect Dis 44:904–910. doi: 10.1086/512196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin F, Zeng A, Yang N, Lin H, Yang E, Wang S, Pintel D, Qiu J. 2007. Quantification of human bocavirus in lower respiratory tract infections in China. Infect Agents Cancer 2:3. doi: 10.1186/1750-9378-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christensen A, Nordbø SA, Krokstad S, Rognlien AG, Døllner H. 2010. Human bocavirus in children: mono-detection, high viral load and viraemia are associated with respiratory tract infection. J Clin Virol 49:158–162. doi: 10.1016/j.jcv.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deng Y, Gu X, Zhao X, Luo J, Luo Z, Wang L, Fu Z, Yang X, Liu E. 2012. High viral load of human bocavirus correlates with duration of wheezing in children with severe lower respiratory tract infection. PLoS One 7:e34353. doi: 10.1371/journal.pone.0034353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Don M, Söderlund-Venermo M, Valent F, Lahtinen A, Hedman L, Canciani M, Hedman K, Korppi M. 2010. Serologically verified human bocavirus pneumonia in children. Pediatr Pulmonol 45:120–126. doi: 10.1002/ppul.21151. [DOI] [PubMed] [Google Scholar]
- 7.Edner N, Castillo-Rodas P, Falk L, Hedman K, Söderlund-Venermo M, Allander T. 2012. Life-threatening respiratory tract disease with human bocavirus-1 infection in a four-year-old child. J Clin Microbiol 50:531–532. doi: 10.1128/JCM.05706-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kantola K, Hedman L, Allander T, Jartti T, Lehtinen P, Ruuskanen O, Hedman K, Söderlund-Venermo M. 2008. Serodiagnosis of human bocavirus infection. Clin Infect Dis 46:540–546. doi: 10.1086/526532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin ET, Kuypers J, McRoberts JP, Englund JA, Zerr DM. 2015. Human bocavirus-1 primary infection and shedding in infants. J Infect Dis 212:516–524. doi: 10.1093/infdis/jiv044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qiu J, Söderlund-Venermo M, Young NS. 2017. Human parvoviruses. Clin Microbiol Rev 30:43–113. doi: 10.1128/CMR.00040-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christensen A, Kesti O, Elenius V, Eskola AL, Dollner H, Altunbulakli C, Akdis CA, Soderlund-Venermo M, Jartti T. 2019. Human bocaviruses and paediatric infections. Lancet Child Adolesc Health 3:418–426. doi: 10.1016/S2352-4642(19)30057-4. [DOI] [PubMed] [Google Scholar]
- 12.Johnson FB, Qiu J. 2011. Bocavirus, Parvoviridae, Parvovirinae, p 1209–1215. In Tidona C, Darai G (ed), The Springer index of viruses, 2nd ed Springer, New York, NY. [Google Scholar]
- 13.Huang Q, Deng X, Yan Z, Cheng F, Luo Y, Shen W, Lei-Butters DC, Chen AY, Li Y, Tang L, Söderlund-Venermo M, Engelhardt JF, Qiu J. 2012. Establishment of a reverse genetics system for studying human bocavirus in human airway epithelia. PLoS Pathog 8:e1002899. doi: 10.1371/journal.ppat.1002899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deng X, Xu P, Zou W, Shen W, Peng J, Liu K, Engelhardt JF, Yan Z, Qiu J. 2016. DNA damage signaling is required for replication of human bocavirus 1 DNA in dividing HEK293 cells. J Virol 91:e01831-16. doi: 10.1128/JVI.01831-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dijkman R, Koekkoek SM, Molenkamp R, Schildgen O, van der Hoek L. 2009. Human bocavirus can be cultured in differentiated human airway epithelial cells. J Virol 83:7739–7748. doi: 10.1128/JVI.00614-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deng X, Yan Z, Luo Y, Xu J, Cheng Y, Li Y, Engelhardt J, Qiu J. 2013. In vitro modeling of human bocavirus 1 infection of polarized primary human airway epithelia. J Virol 87:4097–4102. doi: 10.1128/JVI.03132-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deng X, Li Y, Qiu J. 2014. Human bocavirus 1 infects commercially available primary human airway epithelium cultures productively. J Virol Methods 195:112–119. doi: 10.1016/j.jviromet.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen AY, Cheng F, Lou S, Luo Y, Liu Z, Delwart E, Pintel D, Qiu J. 2010. Characterization of the gene expression profile of human bocavirus. Virology 403:145–154. doi: 10.1016/j.virol.2010.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zou W, Cheng F, Shen W, Engelhardt JF, Yan Z, Qiu J. 2016. Nonstructural protein NP1 of human bocavirus 1 plays a critical role in the expression of viral capsid proteins. J Virol 90:4658–4669. doi: 10.1128/JVI.02964-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen W, Deng X, Zou W, Cheng F, Engelhardt JF, Yan Z, Qiu J. 2015. Identification and functional analysis of novel non-structural proteins of human bocavirus 1. J Virol 89:10097–10109. doi: 10.1128/JVI.01374-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Z, Shen W, Cheng F, Deng X, Engelhardt JF, Yan Z, Qiu J. 2017. Parvovirus expresses a small noncoding RNA that plays an essential role in virus replication. J Virol 91:e02375-16. doi: 10.1128/JVI.02375-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dong Y, Fasina OO, Pintel DJ. 2018. The human bocavirus 1 NP1 protein is a multifunctional regulator of viral RNA processing. J Virol 92:e01187-18. doi: 10.1128/JVI.01187-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zou W, Xiong M, Deng X, Engelhardt JF, Yan Z, Qiu J. 2019. A comprehensive RNA-seq analysis of human bocavirus 1 transcripts in infected human airway epithelium. Viruses 11:E33. doi: 10.3390/v11010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hao S, Zhang J, Chen Z, Xu H, Wang H, Guan W. 2017. Alternative polyadenylation of human bocavirus at its 3′ end is regulated by multiple elements and affects capsid expression. J Virol 91:e02026-16. doi: 10.1128/JVI.02026-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen W, Deng X, Zou W, Engelhardt JF, Yan Z, Qiu J. 2016. Analysis of the cis and trans requirements for DNA replication at the right end hairpin of the human bocavirus 1 genome. J Virol 90:7761–7777. doi: 10.1128/JVI.00708-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fasina OO, Dong Y, Pintel DJ. 2016. NP1 Protein of the bocaparvovirus minute virus of canines controls access to the viral capsid genes via its role in RNA processing. J Virol 90:1718–1728. doi: 10.1128/JVI.02618-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fasina OO, Stupps S, Figueroa-Cuilan W, Pintel DJ. 2017. Minute virus of canines NP1 protein governs the expression of a subset of essential nonstructural proteins via its role in RNA processing. J Virol 91:e00260-17. doi: 10.1128/JVI.00260-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dong Y, Fasina OO, Pintel DJ. 2018. Minute virus of canines NP1 protein interacts with the cellular factor CPSF6 to regulate viral alternative RNA processing. J Virol 93:e01530-18. doi: 10.1128/JVI.01530-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hardy JG, Norbury CJ. 2016. Cleavage factor Im (CFIm) as a regulator of alternative polyadenylation. Biochem Soc Trans 44:1051–1057. doi: 10.1042/BST20160078. [DOI] [PubMed] [Google Scholar]
- 30.Yang Q, Gilmartin GM, Doublie S. 2010. Structural basis of UGUA recognition by the Nudix protein CFI(m)25 and implications for a regulatory role in mRNA 3′ processing. Proc Natl Acad Sci U S A 107:10062–10067. doi: 10.1073/pnas.1000848107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu Y, Wang X, Forouzmand E, Jeong J, Qiao F, Sowd GA, Engelman AN, Xie X, Hertel KJ, Shi Y. 2018. Molecular mechanisms for CFIm-mediated regulation of mRNA alternative polyadenylation. Mol Cell 69:62–74. doi: 10.1016/j.molcel.2017.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mihaylov IS, Cotmore SF, Tattersall P. 2014. Complementation for an essential ancillary non-structural protein function across parvovirus genera. Virology 468–470:226–237. doi: 10.1016/j.virol.2014.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sowd GA, Serrao E, Wang H, Wang W, Fadel HJ, Poeschla EM, Engelman AN. 2016. A critical role for alternative polyadenylation factor CPSF6 in targeting HIV-1 integration to transcriptionally active chromatin. Proc Natl Acad Sci U S A 113:E1054–E1063. doi: 10.1073/pnas.1524213113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soderberg O, Gullberg M, Jarvius M, Ridderstrale K, Leuchowius KJ, Jarvius J, Wester K, Hydbring P, Bahram F, Larsson LG, Landegren U. 2006. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Methods 3:995–1000. doi: 10.1038/nmeth947. [DOI] [PubMed] [Google Scholar]
- 35.Lee K, Ambrose Z, Martin TD, Oztop I, Mulky A, Julias JG, Vandegraaff N, Baumann JG, Wang R, Yuen W, Takemura T, Shelton K, Taniuchi I, Li Y, Sodroski J, Littman DR, Coffin JM, Hughes SH, Unutmaz D, Engelman A, KewalRamani VN. 2010. Flexible use of nuclear import pathways by HIV-1. Cell Host Microbe 7:221–233. doi: 10.1016/j.chom.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jang S, Cook NJ, Pye VE, Bedwell GJ, Dudek AM, Singh PK, Cherepanov P, Engelman AN. 2019. Differential role for phosphorylation in alternative polyadenylation function versus nuclear import of SR-like protein CPSF6. Nucleic Acids Res 47:4663–4683. doi: 10.1093/nar/gkz206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Q, Zhang Z, Zheng Z, Ke X, Luo H, Hu Q, Wang H. 2013. Identification and characterization of complex dual nuclear localization signals in human bocavirus NP1: identification and characterization of complex dual nuclear localization signals in human bocavirus NP1. J Gen Virol 94:1335–1342. doi: 10.1099/vir.0.047530-0. [DOI] [PubMed] [Google Scholar]
- 38.Ray M, Tang R, Jiang Z, Rotello VM. 2015. Quantitative tracking of protein trafficking to the nucleus using cytosolic protein delivery by nanoparticle-stabilized nanocapsules. Bioconjug Chem 26:1004–1007. doi: 10.1021/acs.bioconjchem.5b00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun Y, Chen AY, Cheng F, Guan W, Johnson FB, Qiu J. 2009. Molecular characterization of infectious clones of the minute virus of canines reveals unique features of bocaviruses. J Virol 83:3956–3967. doi: 10.1128/JVI.02569-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kyburz A, Friedlein A, Langen H, Keller W. 2006. Direct interactions between subunits of CPSF and the U2 snRNP contribute to the coupling of pre-mRNA 3′ end processing and splicing. Mol Cell 23:195–205. doi: 10.1016/j.molcel.2006.05.037. [DOI] [PubMed] [Google Scholar]
- 41.Deng X, Yan Z, Cheng F, Engelhardt JF, Qiu J. 2016. Replication of an autonomous human parvovirus in non-dividing human airway epithelium is facilitated through the DNA damage and repair pathways. PLoS Pathog 12:e1005399. doi: 10.1371/journal.ppat.1005399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Z, Deng X, Zou W, Engelhardt JF, Yan Z, Qiu J. 2017. Human bocavirus 1 is a novel helper for adeno-associated virus replication. J Virol 91:e00710-17. doi: 10.1128/JVI.00710-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu P, Chen AY, Ganaie SS, Cheng F, Shen W, Wang X, Kleiboeker S, Li Y, Qiu J. 2018. The nonstructural protein 11-kDa of human parvovirus B19 facilitates viral DNA replication by interacting with Grb2 through its proline-rich motifs. J Virol 92:e01464-18. doi: 10.1128/JVI.01464-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen AY, Kleiboeker S, Qiu J. 2011. Productive parvovirus B19 infection of primary human erythroid progenitor cells at hypoxia is regulated by STAT5A and MEK signaling but not HIF alpha. PLoS Pathog 7:e1002088. doi: 10.1371/journal.ppat.1002088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lou S, Luo Y, Cheng F, Huang Q, Shen W, Kleiboeker S, Tisdale JF, Liu Z, Qiu J. 2012. Human parvovirus B19 DNA replication induces a DNA damage response that is dispensable for cell cycle arrest at G2/M phase. J Virol 86:10748–10758. doi: 10.1128/JVI.01007-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roux KJ, Kim DI, Burke B, G May D. 2018. BioID: a screen for protein-protein interactions. Curr Protoc Protein Sci 91:19.23.1–19.23.15. doi: 10.1002/cpps.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu P, Ganaie SS, Wang X, Wang Z, Kleiboeker S, Horton NC, Heier RF, Meyers MJ, Tavis JE, Qiu J. 2018. Endonuclease activity inhibition of the NS1 protein of parvovirus B19 as a novel target for antiviral drug development. Antimicrob Agents Chemother 63:e01879-18. doi: 10.1128/AAC.01879-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu Z, Qiu J, Cheng F, Chu Y, Yoto Y, O'Sullivan MG, Brown KE, Pintel DJ. 2004. Comparison of the transcription profile of simian parvovirus with that of the human erythrovirus B19 reveals a number of unique features. J Virol 78:12929–12939. doi: 10.1128/JVI.78.23.12929-12939.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guan W, Cheng F, Yoto Y, Kleiboeker S, Wong S, Zhi N, Pintel DJ, Qiu J. 2008. Block to the production of full-length B19 virus transcripts by internal polyadenylation is overcome by replication of the viral genome. J Virol 82:9951–9963. doi: 10.1128/JVI.01162-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen Z, Chen AY, Cheng F, Qiu J. 2010. Chipmunk parvovirus is distinct from members in the genus Erythrovirus of the family Parvoviridae. PLoS One 5:e15113. doi: 10.1371/journal.pone.0015113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.