HIV-1 has exceptionally high sequence diversity, much of which is found within CD8 epitopes. Therefore, the ability of CD8 T cells to recognize multiple versions of a single epitope could be important for an effective vaccine. Here, we show that two previously tested vaccines induced a similar level of CD8 cross-reactivity to that seen in acute HIV-1 infection. Although this cross-reactivity did not seem to affect viral control in vaccine recipients who became infected, we identified several ways in which CD8 cross-reactivity appeared to influence HIV-1 viral evolution. First, we saw that strains isolated from infected vaccine recipients would likely be poorly cross-recognized by the vaccine-induced response. Second, we saw that adapted CD8 epitopes were poorly cross-recognized in both vaccination and infection. Collectively, we believe these results show that CD8 cross-reactivity could be an important consideration in future HIV-1 vaccine design.
KEYWORDS: CD8 cross-reactivity, HIV-1 vaccine, HIV-1 viral evolution
ABSTRACT
Because of HIV’s vast sequence diversity, the ability of the CD8 T-cell response to recognize several variants of a single epitope is an important consideration for vaccine design. Cross-recognition of viral epitopes by CD8 T cells is associated with viral control during HIV-1 infection, but little is known about CD8 cross-reactivity in the context of HIV-1 vaccination. Here, we evaluated vaccine-induced CD8 cross-reactivity in two preventative HIV-1 vaccine efficacy trials, the MRKAd5 and DNA/rAd5 studies. Cross-reactive CD8 responses elicited by vaccination were similar in magnitude and frequency to those induced during acute HIV-1 infection. Although responses directed against variant epitopes were less avid than responses to vaccine-matched epitopes, we did not detect any difference in response polyfunctionality (the proportion of cells producing multiple effector molecules). And while depth, or the frequency of cross-reactive responses, did not correlate with viral loads in recipients who became infected, cross-reactivity did appear to influence early viral evolution. In comparing viral sequences of placebo versus vaccine recipients, we found that viral sequences from vaccinees encoded CD8 epitopes with more substitutions and greater biochemical dissimilarity. In other words, breakthrough sequences of vaccinees would be less cross-recognized by vaccine-induced responses. Additionally, vaccine-induced CD8 T cells poorly cross-recognized variant epitopes encoding HLA-I-associated adaptations, further supporting our conclusion that these responses play a role in driving early HIV-1 viral evolution.
IMPORTANCE HIV-1 has exceptionally high sequence diversity, much of which is found within CD8 epitopes. Therefore, the ability of CD8 T cells to recognize multiple versions of a single epitope could be important for an effective vaccine. Here, we show that two previously tested vaccines induced a similar level of CD8 cross-reactivity to that seen in acute HIV-1 infection. Although this cross-reactivity did not seem to affect viral control in vaccine recipients who became infected, we identified several ways in which CD8 cross-reactivity appeared to influence HIV-1 viral evolution. First, we saw that strains isolated from infected vaccine recipients would likely be poorly cross-recognized by the vaccine-induced response. Second, we saw that adapted CD8 epitopes were poorly cross-recognized in both vaccination and infection. Collectively, we believe these results show that CD8 cross-reactivity could be an important consideration in future HIV-1 vaccine design.
INTRODUCTION
Although only one vaccine trial has provided protection against HIV-1 infection (1), studies of CD8 T cells induced by inefficacious vaccines suggest that they can contribute to viral control in recipients who became infected (2) and to vaccine efficacy in a subset of recipients (3). In HIV Vaccine Trials Network (HVTN) 502, commonly known as the Step Study or MRKAd5, researchers found that the HIV-1 sequences isolated from vaccine recipients who became infected were more distant from the vaccine-encoded sequence than those from placebo recipients. These differences were only seen in proteins that were included in the vaccine and suggested that vaccine-induced CD8s exerted immune pressure on the infecting virus (4). In this same vaccine trial, the Gag-specific breadth of the vaccine-induced CD8 T-cell response (CD8 response) was found to inversely correlate with viral load in vaccine recipients who became infected (2). We recently reported a similar relationship between overall CD8 T-cell response breadth and viral load in MRKAd5 recipients who became infected (5). In another vaccine trial, HVTN 505 (DNA/rAd5), polyfunctional Env-specific CD8s, or CD8 T cells capable of producing multiple effector molecules simultaneously, were associated with a decreased risk of infection (3). Together, these studies suggest that an effective CD8 T-cell response is important for the biology of the immune response against HIV and could be an important component of an effective vaccine.
In HIV-1 infection, there is evidence that CD8 T-cell cross-reactivity could be important for natural viral control. In the context of the protective HLA-I alleles, B*27 and B*57, T-cell receptor (TCR) clonotypes from natural controllers have been found to be more effective at suppressing viruses encoding escape mutations (6, 7), and greater cross-recognition of variant epitopes has been associated with lower viral loads (8). One computational study found that a larger fraction of B*57-restricted CD8 T-cell repertoires could cross-recognize variant epitopes than other HLA-restricted CD8s (9). A major limitation of many CD8 cross-reactivity studies has been their focus on responses during chronic HIV-1 infection. In these studies, it is difficult to distinguish whether a given CD8 T-cell response is due to previous exposure to variant epitopes during the course of viral evolution or if they are truly exhibiting cross-reactivity. Vaccination presents a unique opportunity to examine CD8 T-cell cross-reactivity, since, unlike natural HIV-1 infection, the epitope priming the CD8 response is known, and cross-reactive responses can be clearly defined.
Given that HIV-1 harbors incredible sequence diversity and that much of that variability exists within epitopes targeted by CD8s, vaccine-induced CD8 cross-reactivity is likely an important aspect of the vaccine-induced immune response. However, the ability of vaccine-induced CD8s to cross-recognize variant epitopes not encoded by the vaccine has remained understudied. One past study of chronically infected participants showed that variant epitope recognition by vaccine-induced CD8s was constricted by similar factors to those found in infection, namely, the number of amino acid substitutions and the biochemical conservation of the amino acid change. However, this study did not expand upon these cross-reactive responses in vaccination (10). Another study showed that vaccine-elicited CD8s were not able to suppress variant epitope-encoding viruses as well as CD8s from chronically infected individuals. However, because this study did not examine responses at the epitope level, it is difficult to ascertain whether this relatively poor viral inhibition was due to a lack of cross-reactivity or a different aspect of the CD8 response, such as breadth or polyfunctionality (11). While variant recognition has been correlated with viral control in HIV-1-infected individuals (12), the relationship between vaccine-induced CD8 cross-reactivity and viral evolution or control remains unknown.
In this study, we examined the epitope-level cross-reactivity of CD8s induced by two previously tested adenovirus type 5 (Ad5)-vectored HIV-1 vaccines, MRKAd5 and DNA/rAd5. Using the interferon gamma (IFN-γ) enzyme-linked immunosorbent spot (ELISpot) assay, we detected a wide range of cross-recognition of variant epitopes not encoded by the vaccine. A higher number of amino acid substitutions, greater biochemical dissimilarity, and anchor mutations hindered cross-recognition of variant epitopes. Although the antigen sensitivity of these cross-reactive responses was lower when responding to variant epitopes, the polyfunctionality of the CD8s was similar after stimulation with vaccine and variant epitopes. There was no evidence that CD8 cross-reactivity contributed to viral control in MRKAd5 recipients who became infected; however, we observed differences between the breakthrough sequences of vaccine versus placebo recipients, where epitope sequences in Pol that were isolated from vaccinees were less likely to be cross-recognized by the vaccine-induced response. We also found that vaccine-induced CD8 responses were less able to cross-recognize variant epitopes encoding HLA-I-associated adaptations, further arguing that CD8 cross-reactivity shapes HIV-1 viral evolution. Collectively, our results support the idea that maximizing CD8 cross-reactivity could be an important component of future HIV-1 vaccines.
RESULTS
Cross-reactive CD8 responses elicited by vaccination are similar in magnitude and depth to those induced in acute HIV-1 infection.
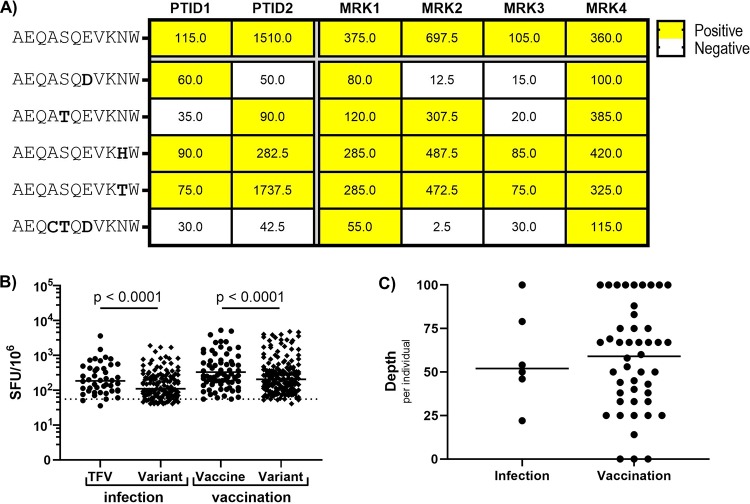
In order to assess CD8 cross-reactivity, we first mapped responses of HIV-1-infected individuals to epitopes encoded by their transmitted founder virus (TFV) and of vaccine recipients to epitopes encoded by the vaccine. Each positive TFV or vaccine-directed CD8 response was then assessed for cross-reactivity to the most common circulating epitope variants (see Materials and Methods for epitope selection details). Six HIV-1-infected individuals, described in Table 1, were tested in acute infection a median of 43 days postinfection (DPI). Of previously evaluated vaccine recipients (5), 46 who mounted vaccine-directed responses were screened for cross-reactive CD8 T-cell responses 4 weeks following the final vaccination time point (37 from MRKAd5 and 9 from DNA/Ad5). Vaccine and cross-reactive responses were assayed using IFN-γ ELISpot. An example of an epitope group tested for cross-reactivity is shown in Fig. 1A. This epitope (Pol-B*44-AEQASQEVKNW) was encoded by the TFV in two HIV-1-infected individuals as well as the MRKAd5 and DNA/Ad5 vaccines. In both vaccination and infection, we observed varied potential to recognize variants of this epitope. These results indicate that while CD8 responses in acute infection and vaccination are often cross-reactive on some level and some variants are consistently recognized, cross-reactivity is not necessarily consistent for a given epitope across individuals.
TABLE 1.
Clinical data of HIV-1-infected individuals included in the study
| PTID | DPI | CD4 count | Viral load | ART status |
|---|---|---|---|---|
| PTID1 | 47 | 1.0 × 105 | No | |
| PTID2 | 89 | 466 | 9.2 × 101 | Yes (21 days) |
| PTID3 | 34 | 862 | 4.0 × 105 | No |
| PTID4 | 92 | 1,043 | 5.3 × 105 | No |
| PTID5 | 23 | 479 | 3.0 × 106 | No |
| PTID6 | 38 | 242 | 6.7 × 105 | No |
FIG 1.
Cross-reactive CD8 responses elicited by vaccination are similar in magnitude and depth to those induced in acute HIV-1 infection. (A) Representative example of an epitope (B*44 restricted) tested in 2 infected individuals (PTID) and 4 MRKAd5 recipients (MRK). The top epitope row is the TFV and vaccine-encoded sequence (AEQASQEVKNW), with variant epitopes listed below. Net ELISpot magnitudes are shown in each cell, and positive responses are shaded in yellow. (B) Net ELISpot magnitudes of positive responses against TFV/vaccine-encoded epitopes and their variant epitope counterparts. (Dotted line indicates the positive threshold for ELISpot responses [55 SFU/106]; a few values fall below this line, as net values are shown. A mixed-effect model was used to account for intraindividual correlation to multiple epitopes.) (C) The depth, or the percentage of variants that are recognized, per individual. For B and C, Ninfection = 6 and Nvaccine = 46.
In examining the positive responses in HIV-1-infected individuals, we observed that TFV-specific responses were higher in magnitude than responses to variants. Similarly, among vaccine recipients, responses to vaccine-matched epitopes were higher than responses to variants of the same epitope (Fig. 1B). In addition to the magnitude of variant-directed responses, response depth is an important metric of cross-reactivity. The depth of the response is the number of positive responses to variant epitopes divided by the number of variant epitopes screened, or the frequency of cross-reactive recognition of variants. We found no significant difference in the depth of CD8 responses in acutely infected versus vaccinated individuals (Fig. 1C). All infected individuals mounted some level of cross-reactivity, and the majority of vaccine recipients were also able to cross-recognize at least one variant epitope (43/46, 93%). Collectively, these data show that CD8 cross-reactivity in acute HIV-1 infection and vaccination are similar in magnitude and depth.
CD8 cross-reactivity is dampened by increased amino acid substitutions, decreased biochemical similarity, and HLA anchor site mutations.
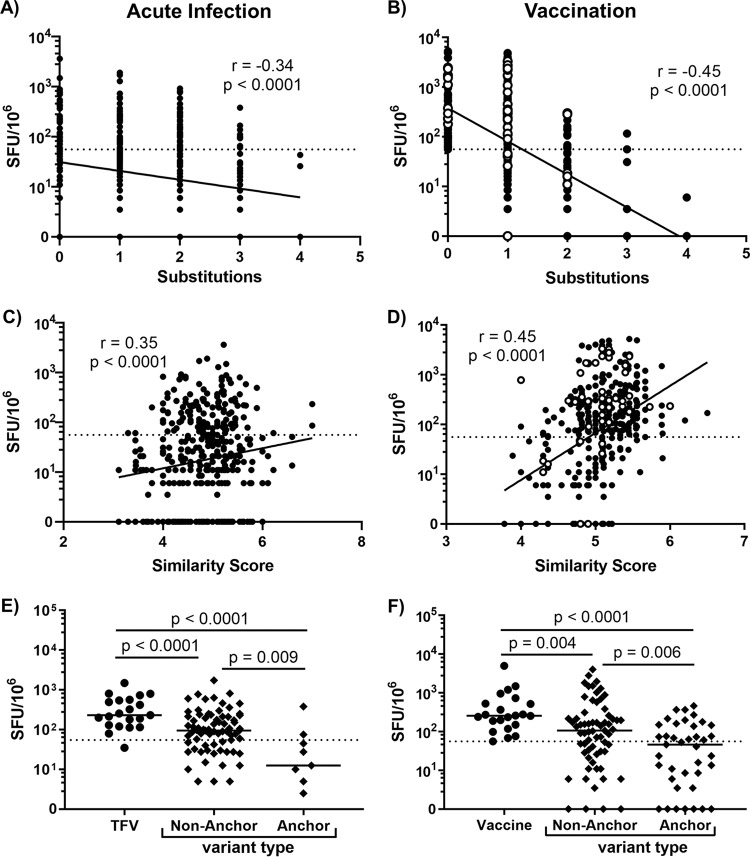
Previous investigations into vaccine-induced CD8 T-cell cross-reactivity demonstrated that cross-recognition was impaired as the number of amino acid substitutions increased and as biochemical similarity decreased (10). We also found that as the number of amino acid substitutions increased, the ELISpot responses to variant epitopes decreased in both acute infection (Fig. 2A) and vaccination (Fig. 2B). We calculated the biochemical similarity of variant epitopes compared to their TFV- or vaccine-encoded counterpart and found that as the biochemical similarity of variant epitopes increased (similarity score increased), cross-recognition also increased (Fig. 2C and D). We also examined the impact of HLA anchor mutations on cross-recognition, where the peptide’s second amino acid residue (P2) and C-terminal end were defined as HLA-I binding sites. Initially, we did not see any difference between nonanchor and anchor site mutated variants (data not shown). A past study showing decreased recognition of anchor mutated epitopes by CD8 responses in chronic HIV-1 infection limited its analysis exclusively to 9-mers (13). When we did the same, we found that anchor site mutated variants were less cross-recognized than variant epitopes with mutations outside HLA-I anchor sites, in both infection and vaccination (Fig. 2E and F). It is possible that this difference between nonanchor and anchor mutated variant epitope responses only arises when the data set is limited to 9-mers because the P2/C-terminal assumption of HLA-I binding is not an accurate assumption for peptides of all lengths. Overall, these results demonstrate that, as expected, vaccine-induced CD8 T cells most easily recognize variant epitopes that closely resemble the vaccine immunogen.
FIG 2.
CD8 cross-reactivity is dampened by increased amino acid substitutions, decreased biochemical similarity, and HLA anchor site mutations. (A and B) Net magnitude of responses to variant epitopes with one to four amino acid substitutions compared to the TFV-encoded or vaccine-encoded epitope (substitutions = 0) in acute infection and vaccination, respectively. (C and D) Net magnitude of responses to variant epitopes with various levels of biochemical similarity to the TFV-encoded or vaccine-encoded epitope in acute infection and vaccination, respectively. (E and F) Net magnitude of responses of anchor or nonanchor mutated variant 9-mers in acute infection and vaccination, respectively. In vaccine plots, closed symbols represent MRKAd5 recipient responses; open symbols represent DNA/rAd5 recipient responses. For all statistical comparisons, substitutions, similarity score, and variant type were modeled as predictors of magnitude in a mixed-effect model that accounted for intraindividual correlation. Data from 6 infected individuals and 46 vaccinees are shown here.
MRKAd5-induced CD8 cross-reactivity did not impact viral control in recipients who became infected.
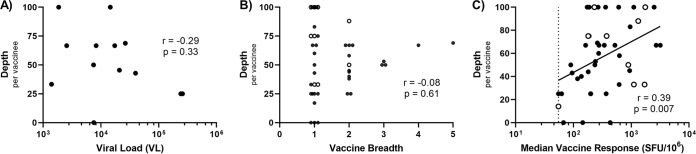
Because the breadth of the vaccine-induced response (i.e., number of vaccine-matched epitopes recognized) was inversely correlated with viral load among MRKAd5 recipients who became infected (2, 5), we asked whether CD8 cross-reactivity impacted viral control in a similar manner. Viral load for each vaccine recipient was calculated as the geometric mean of viral loads (VLs) from all visits for 1 year following the first positive Western blot or until the participant started antiretroviral therapy (ART), as we have previously done (14). We did not observe any relationship between depth and viral load in MRKAd5 recipients who became infected (Fig. 3A). In fact, the depth of an individual’s response did not correlate with that individual’s breadth (Fig. 3B). It is worth noting, however, that vaccine recipients who mounted three or more vaccine responses all cross-recognized at least 50% of the variant epitopes tested. Additionally, there is a clear relationship between response magnitude and depth, where individuals with higher-magnitude vaccine responses also exhibited higher depth, or more cross-reactivity (Fig. 3C). Although recent studies have also implicated CD8 T cells as decreasing infection risk in vaccine recipients (3), we did not observe a difference in the level of CD8 T-cell cross-reactivity between MRKAd5 recipients who became infected and those who did not (data not shown). Our results suggest that, even though MRKAd5 induced cross-reactive CD8 responses, these responses did not influence viral control or infection risk.
FIG 3.
MRKAd5-induced CD8 cross-reactivity did not impact viral control in recipients who became infected. (A) Depth per vaccinee, or frequency of cross-reactive responses, versus viral load following infection (Spearman correlation, n = 13). (B) Depth versus breadth of the vaccine-induced response per vaccinee (Spearman correlation, n = 46). (C) Depth of positive vaccine responses versus median IFN-γ ELISpot magnitude of that response (Spearman correlation, n = 46).
Cross-reactive CD8 T-cell responses are functionally similar to responses against vaccine-encoded epitopes.
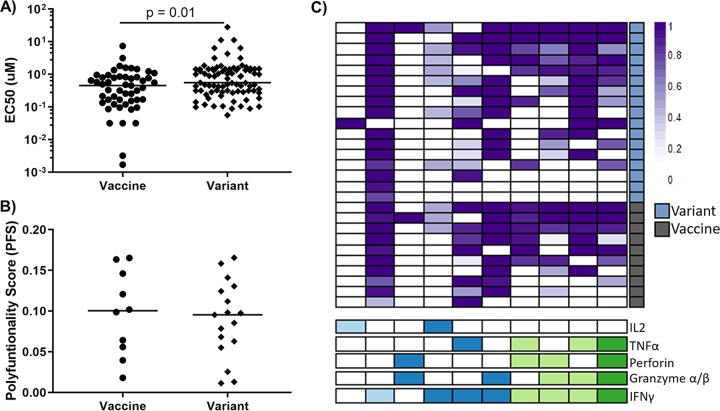
Because cross-reactivity did not appear to play a role in viral control, we next asked if this may be due to poor functionality of these CD8 T cells when stimulated with the variant epitope compared to when stimulated with the vaccine-encoded epitope. An important functional measure of the CD8 response is antigen sensitivity, or the amount of antigen required to induce a response, which has been shown to correlate with viral suppression (15). We found that vaccine-induced CD8 responses had a slightly lower antigen sensitivity toward variant epitopes than toward vaccine epitopes (Fig. 4A). However, when we assessed the polyfunctionality of a subset of the cross-reactive responses, quantified as a polyfunctionality score, or PFS, by the R package COMPASS (16), we did not find a difference between cells stimulated by the vaccine versus the variant epitope (Fig. 4B and C). Since polyfunctionality is a metric that has been linked to vaccine efficacy (3), and cross-reactive responses are similarly polyfunctional to vaccine-matched and variant epitopes, these findings suggest that cross-reactive responses can be an important parameter to be induced by preventative vaccines.
FIG 4.
Cross-reactive CD8 T-cell responses are functionally similar to responses against vaccine-encoded epitopes. (A) Antigen sensitivity of vaccine versus variant responses, quantified as an EC50 value (a mixed-effect model was used to account for intraindividual correlation; data from 37 vaccine recipients). (B) Polyfunctionality, quantified as a polyfunctionality score (PFS), for vaccine versus variant responses. (C) Heatmap of posterior probabilities of each combination of effector molecules. Each row represents a different epitope-specific response. Right-hand boxes indicate responses to variant epitopes (blue) or vaccine-matched epitopes (gray). The combination of effector molecules (IL-2, TNF-α, perforin, granzyme α or β, and/or IFN-γ) produced by each subset is indicated below the heatmap. The number of effector molecules is indicated by dark green (n = 4), light green (n = 3), dark blue (n = 2), and light blue (n = 1). (Panels B and C include data from 8 vaccine recipients: 17 responses to variant epitopes and 10 responses to vaccine-matched epitopes).
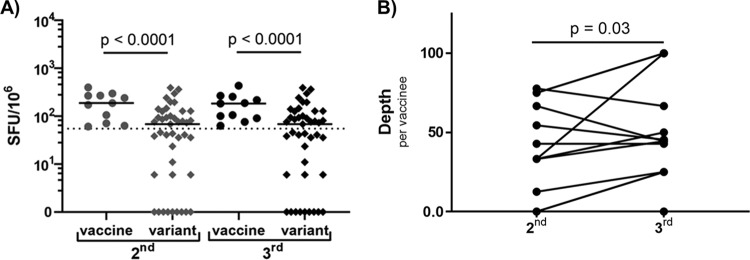
Frequency of cross-reactive responses increased over homologous sequential MRKAd5 vaccination.
In order to determine how cross-reactive responses may change over the course of a vaccine schedule, we compared the responses of 13 vaccine recipients after the second vaccination at week 8 (2nd) to their responses following the third, and final, vaccination at week 30 (3rd). We found that cross-reactive responses to variant epitopes were consistently lower in magnitude than their vaccine counterparts at both time points, but no significant difference was detected in the magnitude of responses after the 2nd vaccination and after the 3rd vaccination (Fig. 5A). However, we did observe a significant increase in the depth of the response over the sequential vaccinations (Fig. 5B), indicating that the frequency of cross-recognition increases following the 2nd vaccination to the 3rd vaccination. Immunogenicity studies of the MRKAd5 study did not show a significant increase in magnitude or frequency of vaccine-induced responses from the second to the third vaccination time point (17), so it is intriguing that cross-reactivity of these responses does increase.
FIG 5.
Frequency of cross-reactive responses increased over homologous sequential MRKAd5 vaccination (n = 13). (A) Net response magnitudes following the 2nd and 3rd vaccinations to vaccine or variant epitopes (mixed-effect model). (B) Response depth per vaccinee to variant epitopes at these time points (Wilcoxon signed-rank test).
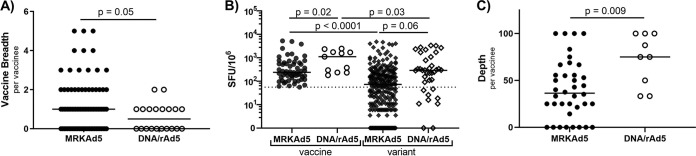
The DNA/rAd5 vaccine elicited CD8 responses with greater depth than the MRKAd5 vaccine.
While the two vaccine studies examined here, MRKAd5 and DNA/rAd5, were both designed with the goal of eliciting CD8s, the MRKAd5 vaccine insert encoded gag/pol/nef, while DNA/rAd5 encoded gag/pol and three versions of env. Additionally, the MRKAd5 vaccine was administered as a homologous prime boost, while the DNA/rAd5 vaccine was administered in a heterologous prime boost regimen. We asked if the CD8 cross-reactivity induced by the two vaccines differed. In comparing responses between the two, we observed a trend toward a lower breadth of response in DNA/rAd5 recipients (Fig. 6A), which is consistent with published immunogenicity of the efficacy study (18). We limited our comparisons of these studies to gag and pol-specific responses, as both vaccines encoded these genes. However, it is possible that the inclusion of three env genes in the DNA/rAd5 insert drew responses away from gag and pol. We found that DNA/rAd5 vaccine-elicited CD8 responses were higher in magnitude than those elicited by MRKAd5, and the cross-reactive responses in DNA/rAd5 also tended to have a higher magnitude than those in MRKAd5 (Fig. 6B). DNA/rAd5 recipients also clearly mounted more cross-reactive responses than MRKAd5 recipients (greater depth), with all of the examined DNA/rAd5 recipients responding to at least a third of the variant epitopes screened (Fig. 6C). These data suggest that the DNA/rAd5 heterologous prime boost vaccination more effectively induced cross-reactive CD8 T-cell responses.
FIG 6.
DNA/rAd5 vaccine-elicited CD8 responses with greater depth than the MRKAd5 vaccine. (A) Vaccine breadth per MRKAd5 or DNA/rAd5 recipient (Mann-Whitney U test). (B) Magnitude of vaccine and variant responses in each study (mixed-effect model). (C) Depth, or frequency of cross-reactive responses, per vaccine recipient (Mann-Whitney U test). NMRKAd5 = 37, NDNA/rAd5 = 9.
Vaccine-generated CD8 T-cell cross-reactivity influences breakthrough viral sequences in recipients who became infected.
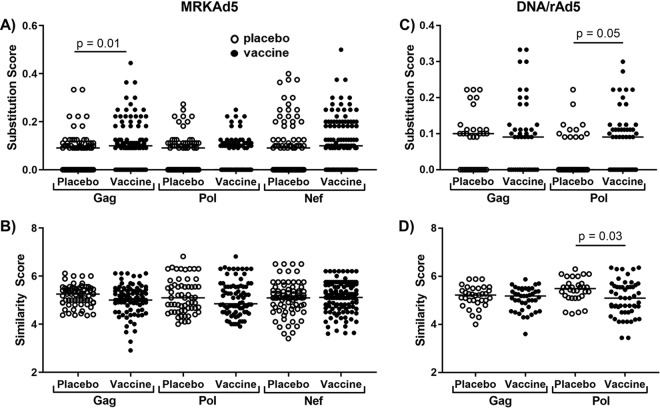
Because CD8 cross-reactivity did not appear to impact viral control in vaccine recipients who became infected, we next asked whether the cross-reactivity of the vaccine response influenced early viral evolution in these individuals. Prior publications have highlighted the distance between the vaccine immunogen and the breakthrough sequences in vaccine recipients who became infected as evidence of a vaccine “sieve effect” (4, 19–21). We examined the breakthrough viral sequences of both MRKAd5 and DNA/rAd5 recipients for evidence of immune pressure exerted by vaccine-generated CD8 cross-reactivity. Our analysis focused only on those HLA-I-restricted epitopes that were relevant for each individual and that encoded HLA-I-associated polymorphisms (14). This is the same strategy we used in our ELISpot experiments to map cross-reactivity, and we believe this analysis is more focused on responses actually targeted by vaccinees, whereas previous sieving analyses examined all NetMHC-predicted epitopes. As illustrated previously in Fig. 2, two major factors impacted the immunogenicity of cross-reactive responses, the number of amino acid substitutions and their biochemical similarity to the vaccine-matched epitope. We hypothesized that CD8 epitopes under postinfection selection pressure from vaccine-induced CD8 cross-reactivity would have more mutations and be less biochemically similar to the vaccine sequence.
In our analysis, we observed an increased number of substitutions in the Gag epitopes restricted by MRKAd5 recipients compared to their placebo counterparts (Fig. 7A). However, no differences were seen in biochemical similarity between breakthrough epitopes in MRKAd5 vaccine and placebo recipients (Fig. 7B). In the previous sieving analysis of MRKAd5 recipients, a greater distance between the breakthrough Gag sequences of vaccine recipients and the vaccine immunogen than those of placebo recipients was shown at the whole-gene level and at the predicted cytotoxic T lymphocyte (CTL) epitope level (4). Overall, our finding of increased amino acid substitutions in Gag sequences of MRKAd5 vaccine recipients is consistent with the previous study of MRKAd5 breakthrough sequences (4).
FIG 7.
Breakthrough viral sequences from vaccine recipients who became infected encode CD8 T-cell epitopes with more substitutions than and less biochemically similar to the vaccine immunogen. (A) Substitution score, or number of substitutions divided by the length of the epitope, of HLA-I restricted epitopes encoded by the breakthrough sequences in MRKAd5 recipients relative to the vaccine-encoded sequence. (B) Similarity scores, or cumulative BLOSUM score divided by length of the epitope, of breakthrough sequence epitopes in MRKAd5 recipients compared to the vaccine-encoded sequence. Substitution scores (C) and similarity scores (D) for DNA/rAd5 vaccine versus placebo recipients. Open symbols indicate placebo recipients, and closed symbols indicate vaccine recipients. All significance was assessed by a mixed-effect model accounting for repeated measures for each vaccine recipient. NMRKAd5 = 65, NDNA/rAd5 = 41.
In DNA/rAd5 recipients, Pol epitopes encoded by vaccinees’ breakthrough sequences harbored more substitutions (Fig. 7C), and we also observed that Pol epitopes in DNA/rAd5 vaccine recipients were less biochemically similar to the vaccine than those in placebo recipients (Fig. 7D). This finding indicates that the breakthrough sequence-encoded epitopes in vaccinees would be less cross-recognized by a vaccine-induced responses in those individuals. Additionally, the fact that we see this effect in DNA/rAd5 recipients for both the number of substitutions and the biochemical similarity is in line with our finding that the DNA/rAd5 vaccine induced stronger CD8 cross-reactivity than the MRKAd5 vaccine (Fig. 6). Our analysis was limited to Gag and Pol, as higher variability in Env sequences precludes accurate HLA-I-associated polymorphism predictions. Meanwhile, previous sieving analysis of DNA/rAd5 primarily focused on effects seen in Env breakthrough sequences but did identify a sieving site in Pol (19).
To then identify epitope-level responses that may be driving this effect in Gag for MRKAd5 and Pol for DNA/rAd5 sequences and to better compare these results with previous work, we iteratively reran our analysis and excluded one epitope at a time. Rolland et al. identified Gag84 as the only significant amino acid site of sieving in MRKAd5 breakthrough sequences (4). Our analysis included three epitopes that encode this site: A*01-SLYNTVATLY, A*11-ATLYCVHQK, and A*31-TLYCVHQK (Gag84 underlined). Although the P value remained significant for the MRKAd5 substitution analysis through each iteration when individual epitopes were excluded one by one, exclusion of all three Gag84-encoding epitopes resulted in a nonsignificant P value, indicating that Gag84 is the driving force in our analysis as well (data not shown). When we iteratively excluded epitopes from the Pol analysis of DNA/rAd5, we identified three DNA/rAd5-encoded epitopes, where exclusion of these epitopes resulted in a lack of statistical significance (P > 0.05) for both substitution and similarity scores: B*13:02-GQGQWTYQI, A*30-GQGQWTYQIY, B*18:01-NETPGIRYQY. At the single epitope-level, these sequences demonstrate the overall effect seen where vaccine recipient sequences encode a higher number of amino acid substitutions, greater biochemical dissimilarity, and more anchor mutations (Table 2). While Decamp et al. (19) identified Pol 238 as a signature site, the three epitopes shown here do not overlap with this site, indicating that our analysis has identified unique T-cell-mediated sieving in DNA/rAd5 recipients. Together, these data suggest that vaccine-induced CD8 cross-reactivity may have influenced the early viral evolution of transmitted HIV-1 strains in vaccine recipients.
TABLE 2.
Placebo versus vaccine sequences of epitopes driving DNA/rAd5 Pol effecta
| PubIDb | RXc | HLA | Sequence | Similarity score | No. of substitutions | Anchor mutation |
|---|---|---|---|---|---|---|
| 5050122 | Placebo | B*13:02 | GQGQWTYQI | 6.00 | 0 | |
| 5051958 | Placebo | B*13:02 | GQGQWTYQI | 6.00 | 0 | |
| 5051993 | Vaccine | B*13:02 | GL GQWS YQI | 4.78 | 2 | Yes |
| 5050122 | Placebo | A*30 | GQGQWTYQIY | 6.10 | 0 | |
| 5050695 | Placebo | A*30 | GQGQWTYQIY | 6.10 | 0 | |
| 5051958 | Placebo | A*30 | GQGQWTYQIY | 6.10 | 0 | |
| 5051144 | Vaccine | A*30 | GQGQWTYQIY | 6.10 | 0 | |
| 5051226 | Vaccine | A*30 | EL GQWTYQV Y | 4.50 | 3 | Yes |
| 5052474 | Vaccine | A*30 | E QGQWTYQIY | 5.30 | 1 | No |
| 5050102 | Placebo | B*18:01 | NEA PGIRYQY | 5.20 | 1 | No |
| 5050396 | Placebo | B*18:01 | NETPGIRYQY | 5.70 | 0 | |
| 5050821 | Placebo | B*18:01 | NETPGIRYQY | 5.70 | 0 | |
| 5051012 | Vaccine | B*18:01 | NETPGIRYQY | 5.70 | 0 | |
| 5051144 | Vaccine | B*18:01 | NA TPGIRYQY | 5.10 | 1 | Yes |
| 5051763 | Vaccine | B*18:01 | NETPGIRYQY | 5.70 | 0 |
Vaccine recipients are highlighted in gray. Amino acid substitutions compared to vaccine sequence are shown in bold.
PubID, publication identifier of vaccine recipients.
RX, treatment arm (vaccine or placebo).
Evidence of CD8 T-cell cross-reactivity impacting population-level HIV-1 sequence evolution.
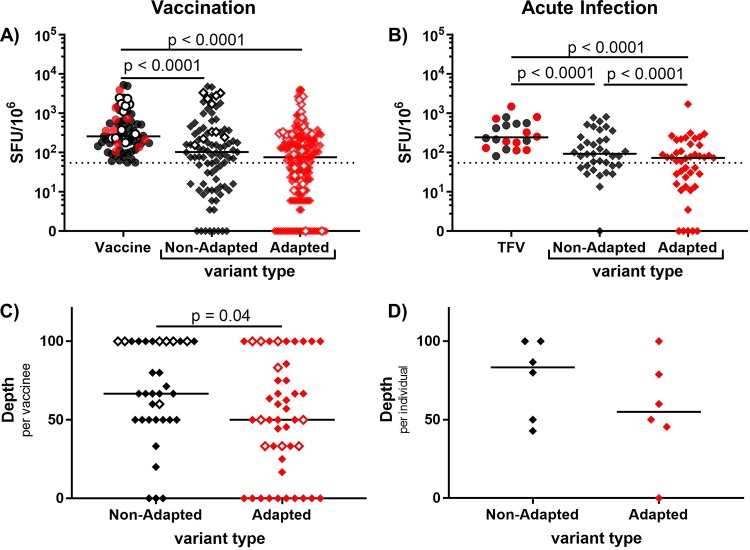
Our analysis of breakthrough sequences in MRKAd5 and DNA/rAd5 vaccine recipients indicates that CD8 T-cell cross-reactivity significantly influences viral evolution. One way to identify CD8 T-cell-mediated pressure in natural HIV-1 infection is through HLA-I-associated polymorphisms/adaptations, so we examined whether cross-reactive CD8 responses were able to equivalently recognize variants that encoded adaptations (adapted variants) versus those that did not (nonadapted variants). Individuals infected with viruses encoding a greater number of these adaptations, defined as HLA-I-associated polymorphisms, have previously been shown to have accelerated disease progression (higher viral loads and faster CD4 T-cell decline) (14). If cross-reactive CD8 responses influenced HIV-1 viral evolution at the population level, we hypothesized that vaccine and TFV-induced CD8 T-cell responses would be less able to cross-recognize adapted variants, which encoded relevant HLA-I-associated polymorphisms, compared to nonadapted variants. While we did not detect a significant difference in magnitude between cross-reactive responses to nonadapted and adapted variants in vaccine recipients (Fig. 8A), we did find that TFV-directed responses in acute HIV-1 infection were less able to cross-recognize adapted variants (Fig. 8B). Additionally, the frequency of vaccine-induced responses cross-recognizing nonadapted variants was higher than that for adapted variants (Fig. 8C). Although the number of infected individuals was small (n = 6), the response depth against nonadapted variants again appeared to be higher than it was for the adapted variants (Fig. 8D). These results suggest that CD8 cross-reactivity plays a role in shaping HIV-1 viral evolution and that HLA-I adaptations, to some extent, are a response to the pressure exerted by cross-reactive CD8s.
FIG 8.
CD8 T cells poorly cross-recognize variant epitopes encoding adapted polymorphisms. (A) Net magnitude of vaccine and cross-reactive responses against nonadapted or adapted variant epitopes (mixed-effect model). (B) Net magnitude of TFV and cross-reactive responses against nonadapted and adapted variants. (C) Depth, or proportion of variant epitopes cross-recognized, per individual of nonadapted and adapted variants in vaccination (Mann-Whitney U test). (D) Depth of acute infection-induced responses (Mann-Whitney U test). Closed symbols represent MRKAd5 recipients, and open symbols represent DNA/rAd5 recipients. All adapted epitope responses are denoted by red. Data from 46 vaccinees and 6 infected individuals are shown here.
DISCUSSION
This study represents the first comprehensive assessment of HIV-1 vaccine-induced CD8 T-cell cross-reactivity. Here, we show that two past Ad5-based HIV-1 vaccines elicited a broad range of cross-reactive responses, similar to what is seen in acute HIV-1 infection, but that CD8 cross-reactivity did not correlate with viral load in vaccine recipients who became infected. These variant-specific responses had a lower magnitude and were less antigen sensitive than vaccine-specific responses but exhibited similar polyfunctionality. CD8 responses were also less able to cross-recognize variants with an increasing number of amino acid substitutions, reduced biochemical similarity, and encoding anchor mutations, and these factors were used to assess the immune pressure exerted by vaccine-induced cross-reactivity on breakthrough sequences of trial participants. We found that epitopes encoded by HIV-1 breakthrough sequences of DNA/rAd5 vaccine recipients would probably be poorly cross-recognized by vaccine-induced CD8 T cells. We also observed that vaccine-induced responses were less able to cross-recognize variant epitopes containing HLA-I-associated adaptations, supporting the idea that CD8 cross-reactivity is a driver of HIV-1 viral evolution.
Although sieve analyses have previously been conducted on both MRKAd5 and DNA/rAd5 study recipients (4, 19), our approach differed from those studies by focusing solely on relevant HLA-I restricted epitopes, identified based on HLA-I-associated polymorphisms (14). In the breakthrough sequences of participants in the DNA/rAd5 study, we saw an increased number of amino acid substitutions and decreased biochemical similarity in breakthrough Pol epitopes in individuals who received the vaccine. While the previous sieving analysis did identify a sieving effect in Pol (19), we found specific CD8 T-cell epitopes unique from previously reported sites, and we propose CD8 cross-reactivity as the mechanism for T-cell-driven sieving in DNA/rAd5. These findings suggest that cross-recognition by vaccine-induced CD8 responses significantly impacted early viral evolution in vaccinees who became infected.
In contrast to our sieving analysis of MRKAd5 breakthrough sequences, the CD8 cross-reactivity sieving effect appeared stronger in the DNA/rAd5 recipients, where both the number of amino acid substitutions was higher and the biochemical dissimilarity was greater than placebo sequences. This is in line with the higher magnitude and greater frequency of CD8 cross-reactive responses elicited by the DNA/rAd5 vaccine compared to the MRKAd5 one (Fig. 6). In addition to the impact on breakthrough sequences, we observed decreased cross-recognition of variant epitopes containing HLA-I-associated adaptations, indicating that HIV-1 evolves away from cross-reactive CD8 T cell response during natural infection in a way that is detectable at the population level. We believe these findings demonstrate that cross-reactivity is an important component of an effective CD8 response.
Our data put forward a few strategies to boost vaccine-induced CD8 cross-reactivity. The strong relationship observed between the magnitude of a vaccine-induced CD8 response and cross-reactivity of that response suggests that strategies to boost the magnitude of the CD8 T-cell response will coincidentally boost the ability of these responses to cross-recognize variant epitopes. We should note that very high-magnitude responses, some of which are presented in Fig. 1A, still may not be able to cross-recognize certain variant epitopes, indicating that there may be elements of cross-recognition that are not solely linked to response magnitude. The heterologous prime-boost regimen of DNA/rAd5 also elicited both higher magnitude and more frequent CD8 cross-reactivity. Unfortunately, we were not able to assess how the presence of multiple variants of a given epitope within an insert, such as in the case of mosaic vaccines, would influence the cross-reactivity of the CD8 T-cell response. Preliminary studies in nonhuman primates suggest that mosaic vaccines increase the depth of the CD8 response, and we would hypothesize that the inclusion of variant epitopes would induce a more polyclonal CD8 T-cell response, thus broadening the cross-recognition of variant epitopes. However, mosaic vaccines also encode a greater proportion of adapted epitopes, which we have previously shown are poorly immunogenic (5) and which we show here are poorly cross-recognized.
Overall, we observed remarkable consistency between the CD8 cross-reactivity induced by these vaccines and the CD8 cross-reactivity elicited in early HIV-1 infection. Magnitude, depth, and factors impacting variant recognition were all similar between vaccination and infection. The HIV-infected individuals studied here do not naturally control virus, so it is unreasonable to expect that similar levels of cross-reactivity induced by a vaccine could impact viral control in recipients who became infected. We show using several approaches that CD8 cross-reactive responses influence HIV evolution, suggesting that if cross-reactivity were boosted, these responses could go from influencing sequence evolution to controlling viral load.
MATERIALS AND METHODS
Vaccine samples.
HVTN 502 participants (MRKAd5, ClinicalTrials.gov identifier NCT00095576) were randomized to receive the Ad5 vaccine with HIV-1 gene inserts (gag, pol, nef) or placebo (22). HVTN 505 participants (DNA/rAd5, ClinicalTrials.gov identifier NCT00865566) were randomized to receive the DNA/rAd5 vaccine with HIV-1 gene inserts (gag, pol, env a/b/c, nef [DNA only]) or placebo (18). Informed consent was obtained from all participants, and all relevant guidelines of the authors’ institutions were followed. A total of 46 vaccine recipients (37 from MRKAd5 and 9 from DNA/Ad5) who were previously found to have positive responses to vaccine-encoded epitopes (5) were screened for cross-reactive CD8 T-cell responses for this study. Of the MRKAd5 recipients, 13 were individuals who eventually became HIV-1 infected. Five placebo recipients from MRKAd5 and two from DNA/Ad5 were tested for vaccine responses, and no positive responses were detected in these recipients.
Infection samples.
Six HIV-1-infected individuals were tested for cross-reactive CD8 responses in acute HIV-1 infection. The median days postinfection (DPI) was 43 days for acute infection samples; Table 1 provides the full clinical details of these individuals. TFV sequences were inferred from the plasma of these patients at Fiebig stage III or earlier using a single-genome amplification method, as described previously (23).
HLA typing.
HLA-I alleles for MRKAd5 vaccine recipients were provided by the HVTN. HLA typing was performed on DNA/rAd5 samples as previously described (24). Briefly, four-digit genotyping was generated by sequencing-based typing (Abbott Molecular, Inc., Des Plaines, IL) and automated DNA hybridization with oligonucleotide probes (Innogenetics, Inc., Alpharetta, GA).
IFN-γ ELISpot.
ELISpot assays were performed as previously described (25). In brief, peripheral blood mononuclear cells (PBMCs) were thawed and rested overnight in RPMI medium supplemented with 10% human AB serum (R10 Abs) at 37°C and 5% CO2. Plates were coated with anti-IFN-γ antibody at 4°C overnight and then blocked with R10 Abs for 2 hours at 37°C and 5% CO2. PBMCs were plated at 100,000 cells/well with the peptide of interest at a 10-μM final concentration in duplicate and incubated together at 37°C and 5% CO2 for 22 hours. A negative control of medium only and a positive control of phytohemagglutinin (PHA) were included on each plate for each sample. Plates were then washed and developed with biotinylated anti-IFN-γ antibody (2 h), streptavidin (45 min), and nitroblue tetrazolium/5-bromo-4-chloro-3-indolylphosphate (NBT/BCIP) substrate solution (10 min) sequentially. Once dry, plates were scanned and counted using the ImmunoSpot analyzer and software. Results were normalized and reported as the average spot-forming units per 1 million cells (SFU/106). The positive threshold for a response was at least 55 SFU/106 and at least 3 times the medium-only wells. Antigen sensitivity was assessed using IFN-γ ELISpot by performing logfold serial dilution of peptide from 10 μM to 10−2 μM. A dose-response curve was fit for each epitope response and used to calculate the EC50 value, or the amount of peptide required to elicit 50% of a maximal response.
CD8 T-cell response mapping strategy.
Individuals were first screened for CD8 T-cell responses to TFV- or vaccine-encoded epitopes. Only those epitopes predicted to be restricted by each individual’s HLA-I alleles were screened. A range of 4 to 17 epitopes were tested per vaccine recipient (median = 10), and 1 to 4 epitope-specific responses were detected in each individual (median = 1). Then, we tested positive responses for cross-reactivity against variant epitopes. The three to seven most common variant epitopes were selected based on population frequencies found in the Los Alamos HIV-1 Sequence Database (26). Variant epitopes were designed to represent at least 90% of the circulating sequences at that epitope site, with a maximum of seven variants designed for each vaccine epitope. Cross-reactive responses were summarized as the depth of the response or frequency of responses to variant epitopes. All peptides (8- to 11-mer) were previously predicted using Microsoft Research’s Epipred software and were synthesized by New England Peptide in a 96-well array format.
Flow cytometry.
Cytokine and effector molecule production was measured by flow cytometry as previously described (27). Briefly, PBMCs were thawed and stimulated with the relevant peptide or peptide pool in the presence of anti-CD28, anti-CD49d, and anti-CD107a-FITC (BD Biosciences) for 1 hour at 37°C and 5% CO2. After the addition of monensin and brefeldin-A (BD Biosciences), cells were incubated for an additional 11 hours. Cells were then surface stained with the following antibodies for 30 min at 4°C: dead cell dye (Invitrogen), anti-CD3-Alexa780 (eBioscience), anti-CD4-Qdot655 (Invitrogen), anti-CD8-V500 (BD Pharmingen), anti-CD14-Percp/CY5.5 (BD Pharmingen), and anti-CD19-Percp/CY5.5 (BD Pharmingen). Cells were permeabilized with Cytoperm/Cytofix (BD Biosciences) and then intracellularly stained with the following for 30 min at 4°C: anti-IFN-γ-Alexa700 (BD Biosciences), anti-IL-2-APC (BD Biosciences), anti-TNF-α-PECy7 (BD Biosciences), anti-perforin-PE (eBioscience), and anti-granzymeA-PacBlue and anti-granzymeB-V450 (BD Biosciences). Cells were fixed in 5% formalin. Events were acquired on an LSRII flow cytometer (BD Immunocytometry Systems) and analyzed using FlowJo version 10 (Tree Star, Inc.).
Breakthrough sequence analysis.
Sequences and treatment arm assignments were downloaded from the publicly available SieveSifter tool (20). MRKAd5 sequences were previously submitted to GenBank with the accession numbers JF320002 to JF320643 (4). DNA/rAd5 sequences were previously submitted to GenBank with the accession numbers MG196642 to MG197219 (19). HLA-I alleles for these individuals were obtained from the HVTN. The sequences of epitopes restricted by the HLA-I alleles of vaccine and placebo recipients were compared to the vaccine-encoded sequence. Only epitopes encoding an HLA-I-associated polymorphism were included, as these were the epitopes we tested for cross-reactivity in immune assays and as we believe these epitopes elicit the CD8 T-cell responses that exert significant immune pressure. Epitopes were extracted and analyzed from 40 MRKAd5 vaccine recipients, 26 MRKAd5 placebo recipients, 24 DNA/rAd5 vaccine recipients, and 17 DNA/rAd5 placebo recipients. Across the two vaccine studies, 33 Gag, 35 Pol, and 18 Nef epitopes were analyzed. Although Env was included in the DNA/rAd5 vaccine, it was not included in our analysis since there were three Env sequences included in the insert, making cross-reactivity more difficult to accurately assess. The number of amino acid substitutions compared to the vaccine insert sequence was normalized to the length of the epitope. The biochemical similarity of the breakthrough epitope to the vaccine insert sequence was quantified by a sum of the BLOSUM-62 (blocking substitution matrix) (28) scores for each residue of the aligned breakthrough and vaccine epitopes, again normalized to the length of the epitope.
Statistical analysis.
Graphs were created in Prism version 8, and all mixed-effect models were built in R (29). In analyzing ELISpot responses, net magnitudes (SFU/106 minus background) were log(x + 1) transformed. Both individual and epitope groupings were added as random effects to account for the multiple epitopes that were tested for each individual and multiple variants that were tested for each responsive epitope. In the analysis of breakthrough sequences, vaccine versus placebo comparisons were done using a mixed effect model accounting for intraindividual correlation among epitopes. Polyfunctionality was summarized as a polyfunctionality score (PFS) using the R package COMPASS (16).
ACKNOWLEDGMENTS
Peptide design was assisted by Victor Du and Kai Qin.
This work was supported by the National Institute of Allergy and Infectious Diseases (NIAID) at the National Institutes of Health (F30AI140829 [S.B.], R01AI112566 [P.G.], UM1 AI068614 [HVTN Leadership Operations Center], UM1 AI068618 [HVTN Laboratory Center], and UM1 AI068635 [HVTN Statistical Data Management Center]).
We thank the Center for AIDS Research at UAB’s Flow Cytometry Core for assistance (P30 AI027767). We also thank NIAID and the NIAID-funded HIV-1 Vaccine Trials Network (HVTN) for providing specimens and data from the HVTN 502 and HVTN 505 studies. We thank members of the HVTN 502 and HVTN 505 Protocol Teams for their support of this project and gratefully acknowledge the study participants. We thank staff across the HVTN Leadership Operations Center, the Statistical Data Management Center, and the Laboratory Center for assistance with this project.
REFERENCES
- 1.Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, Premsri N, Namwat C, de Souza M, Adams E, Benenson M, Gurunathan S, Tartaglia J, McNeil JG, Francis DP, Stablein D, Birx DL, Chunsuttiwat S, Khamboonruang C, Thongcharoen P, Robb ML, Michael NL, Kunasol P, Kim JH, MOPH–TAVEG Investigators. 2009. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med 361:2209–2220. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- 2.Janes H, Friedrich DP, Krambrink A, Smith RJ, Kallas EG, Horton H, Casimiro DR, Carrington M, Geraghty DE, Gilbert PB, McElrath MJ, Frahm N. 2013. Vaccine-induced gag-specific T cells are associated with reduced viremia after HIV-1 infection. J Infect Dis 208:1231–1239. doi: 10.1093/infdis/jit322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Janes HE, Cohen KW, Frahm N, De Rosa SC, Sanchez B, Hural J, Magaret CA, Karuna S, Bentley C, Gottardo R, Finak G, Grove D, Shen M, Graham BS, Koup RA, Mulligan MJ, Koblin B, Buchbinder SP, Keefer MC, Adams E, Anude C, Corey L, Sobieszczyk M, Hammer SM, Gilbert PB, McElrath MJ. 2017. Higher T-cell responses induced by DNA/rAd5 HIV-1 preventive vaccine are associated with lower HIV-1 infection risk in an efficacy trial. J Infect Dis 215:1376–1385. doi: 10.1093/infdis/jix086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rolland M, Tovanabutra S, deCamp AC, Frahm N, Gilbert PB, Sanders-Buell E, Heath L, Magaret CA, Bose M, Bradfield A, O’Sullivan A, Crossler J, Jones T, Nau M, Wong K, Zhao H, Raugi DN, Sorensen S, Stoddard JN, Maust BS, Deng W, Hural J, Dubey S, Michael NL, Shiver J, Corey L, Li F, Self SG, Kim J, Buchbinder S, Casimiro DR, Robertson MN, Duerr A, McElrath MJ, McCutchan FE, Mullins JI. 2011. Genetic impact of vaccination on breakthrough HIV-1 sequences from the STEP trial. Nat Med 17:366–371. doi: 10.1038/nm.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boppana S, Sterrett S, Files J, Qin K, Fiore-Gartland A, Cohen KW, De Rosa SC, Bansal A, Goepfert PA. 2019. HLA-I associated adaptation dampens the CD8 T-cell response in HIV Ad5-vectored vaccine recipients. J Infect Dis 220:1620–1628. doi: 10.1093/infdis/jiz368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ladell K, Hashimoto M, Iglesias MC, Wilmann PG, McLaren JE, Gras S, Chikata T, Kuse N, Fastenackels S, Gostick E, Bridgeman JS, Venturi V, Arkoub ZA, Agut H, van Bockel DJ, Almeida JR, Douek DC, Meyer L, Venet A, Takiguchi M, Rossjohn J, Price DA, Appay V. 2013. A molecular basis for the control of preimmune escape variants by HIV-specific CD8+ T cells. Immunity 38:425–436. doi: 10.1016/j.immuni.2012.11.021. [DOI] [PubMed] [Google Scholar]
- 7.Chen H, Ndhlovu ZM, Liu D, Porter LC, Fang JW, Darko S, Brockman MA, Miura T, Brumme ZL, Schneidewind A, Piechocka-Trocha A, Cesa KT, Sela J, Cung TD, Toth I, Pereyra F, Yu XG, Douek DC, Kaufmann DE, Allen TM, Walker BD. 2012. TCR clonotypes modulate the protective effect of HLA class I molecules in HIV-1 infection. Nat Immunol 13:691–700. doi: 10.1038/ni.2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turnbull EL, Lopes AR, Jones NA, Cornforth D, Newton P, Aldam D, Pellegrino P, Turner J, Williams I, Wilson CM, Goepfert PA, Maini MK, Borrow P. 2006. HIV-1 epitope-specific CD8+ T cell responses strongly associated with delayed disease progression cross-recognize epitope variants efficiently. J Immunol 176:6130–6146. doi: 10.4049/jimmunol.176.10.6130. [DOI] [PubMed] [Google Scholar]
- 9.Kosmrlj A, Read EL, Qi Y, Allen TM, Altfeld M, Deeks SG, Pereyra F, Carrington M, Walker BD, Chakraborty AK. 2010. Effects of thymic selection of the T-cell repertoire on HLA class I-associated control of HIV infection. Nature 465:350–354. doi: 10.1038/nature08997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malhotra U, Nolin J, Horton H, Li F, Corey L, Mullins JI, McElrath MJ. 2009. Functional properties and epitope characteristics of T-cells recognizing natural HIV-1 variants. Vaccine 27:6678–6687. doi: 10.1016/j.vaccine.2009.08.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayes PJ, Cox JH, Coleman AR, Fernandez N, Bergin PJ, Kopycinski JT, Nitayaphan S, Pitisuttihum P, de Souza M, Duerr A, Morgan C, Gilmour JW. 2016. Adenovirus-based HIV-1 vaccine candidates tested in efficacy trials elicit CD8+ T cells with limited breadth of HIV-1 inhibition. AIDS 30:1703–1712. doi: 10.1097/QAD.0000000000001122. [DOI] [PubMed] [Google Scholar]
- 12.Sunshine J, Kim M, Carlson JM, Heckerman D, Czartoski J, Migueles SA, Maenza J, McElrath MJ, Mullins JI, Frahm N. 2014. Increased sequence coverage through combined targeting of variant and conserved epitopes correlates with control of HIV replication. J Virol 88:1354–1365. doi: 10.1128/JVI.02361-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bronke C, Almeida CA, McKinnon E, Roberts SG, Keane NM, Chopra A, Carlson JM, Heckerman D, Mallal S, John M. 2013. HIV escape mutations occur preferentially at HLA-binding sites of CD8 T-cell epitopes. AIDS 27:899–905. doi: 10.1097/QAD.0b013e32835e1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carlson JM, Du VY, Pfeifer N, Bansal A, Tan VYF, Power K, Brumme CJ, Kreimer A, DeZiel CE, Fusi N, Schaefer M, Brockman MA, Gilmour J, Price MA, Kilembe W, Haubrich R, John M, Mallal S, Shapiro R, Frater J, Harrigan PR, Ndung’u T, Allen S, Heckerman D, Sidney J, Allen TM, Goulder PJ, Brumme ZL, Hunter E, Goepfert PA. 2016. Impact of pre-adapted HIV transmission. Nat Med 22:606–613. doi: 10.1038/nm.4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Almeida JR, Sauce D, Price DA, Papagno L, Shin SY, Moris A, Larsen M, Pancino G, Douek DC, Autran B, Sáez-Cirión A, Appay V. 2009. Antigen sensitivity is a major determinant of CD8+ T-cell polyfunctionality and HIV-suppressive activity. Blood 113:6351–6360. doi: 10.1182/blood-2009-02-206557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin L, Finak G, Ushey K, Seshadri C, Hawn TR, Frahm N, Scriba TJ, Mahomed H, Hanekom W, Bart P-A, Pantaleo G, Tomaras GD, Rerks-Ngarm S, Kaewkungwal J, Nitayaphan S, Pitisuttithum P, Michael NL, Kim JH, Robb ML, O’Connell RJ, Karasavvas N, Gilbert P, De Rosa SC, McElrath MJ, Gottardo R. 2015. COMPASS identifies T-cell subsets correlated with clinical outcomes. Nat Biotechnol 33:610–616. doi: 10.1038/nbt.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McElrath MJ, De Rosa SC, Moodie Z, Dubey S, Kierstead L, Janes H, Defawe OD, Carter DK, Hural J, Akondy R, Buchbinder SP, Robertson MN, Mehrotra DV, Self SG, Corey L, Shiver JW, Casimiro DR, Step Study Protocol Team. 2008. HIV-1 vaccine-induced immunity in the test-of-concept Step Study: a case-cohort analysis. Lancet 372:1894–1905. doi: 10.1016/S0140-6736(08)61592-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hammer SM, Sobieszczyk ME, Janes H, Karuna ST, Mulligan MJ, Grove D, Koblin BA, Buchbinder SP, Keefer MC, Tomaras GD, Frahm N, Hural J, Anude C, Graham BS, Enama ME, Adams E, DeJesus E, Novak RM, Frank I, Bentley C, Ramirez S, Fu R, Koup RA, Mascola JR, Nabel GJ, Montefiori DC, Kublin J, McElrath MJ, Corey L, Gilbert PB, HVTN 505 Study Team. 2013. Efficacy trial of a DNA/rAd5 HIV-1 preventive vaccine. N Engl J Med 369:2083–2092. doi: 10.1056/NEJMoa1310566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.deCamp AC, Rolland M, Edlefsen PT, Sanders-Buell E, Hall B, Magaret CA, Fiore-Gartland AJ, Juraska M, Carpp LN, Karuna ST, Bose M, LePore S, Miller S, O’Sullivan A, Poltavee K, Bai H, Dommaraju K, Zhao H, Wong K, Chen L, Ahmed H, Goodman D, Tay MZ, Gottardo R, Koup RA, Bailer R, Mascola JR, Graham BS, Roederer M, O’Connell RJ, Michael NL, Robb ML, Adams E, D’Souza P, Kublin J, Corey L, Geraghty DE, Frahm N, Tomaras GD, McElrath MJ, Frenkel L, Styrchak S, Tovanabutra S, Sobieszczyk ME, Hammer SM, Kim JH, Mullins JI, Gilbert PB. 2017. Sieve analysis of breakthrough HIV-1 sequences in HVTN 505 identifies vaccine pressure targeting the CD4 binding site of Env-gp120. PLoS One 12:e0185959. doi: 10.1371/journal.pone.0185959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fiore-Gartland A, Kullman N, deCamp AC, Clenaghan G, Yang W, Magaret CA, Edlefsen PT, Gilbert PB. 2017. SieveSifter: a Web-based tool for visualizing the sieve analyses of HIV-1 vaccine efficacy trials. Bioinformatics 33:2386–2388. doi: 10.1093/bioinformatics/btx168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edlefsen PT, Rolland M, Hertz T, Tovanabutra S, Gartland AJ, deCamp AC, Magaret CA, Ahmed H, Gottardo R, Juraska M, McCoy C, Larsen BB, Sanders-Buell E, Carrico C, Menis S, Kijak GH, Bose M, RV144 Sequencing Team, Arroyo MA, O’Connell RJ, Nitayaphan S, Pitisuttithum P, Kaewkungwal J, Rerks-Ngarm S, Robb ML, Kirys T, Georgiev IS, Kwong PD, Scheffler K, Pond SL, Carlson JM, Michael NL, Schief WR, Mullins JI, Kim JH, Gilbert PB. 2015. Comprehensive sieve analysis of breakthrough HIV-1 sequences in the RV144 vaccine efficacy trial. PLoS Comput Biol 11:e1003973. doi: 10.1371/journal.pcbi.1003973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buchbinder SP, Mehrotra DV, Duerr A, Fitzgerald DW, Mogg R, Li D, Gilbert PB, Lama JR, Marmor M, Del Rio C, McElrath MJ, Casimiro DR, Gottesdiener KM, Chodakewitz JA, Corey L, Robertson MN, Step Study Protocol Team. 2008. Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the Step Study): a double-blind, randomised, placebo-controlled, test-of-concept trial. Lancet 372:1881–1893. doi: 10.1016/S0140-6736(08)61591-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salazar-Gonzalez JF, Salazar MG, Keele BF, Learn GH, Giorgi EE, Li H, Decker JM, Wang S, Baalwa J, Kraus MH, Parrish NF, Shaw KS, Guffey MB, Bar KJ, Davis KL, Ochsenbauer-Jambor C, Kappes JC, Saag MS, Cohen MS, Mulenga J, Derdeyn CA, Allen S, Hunter E, Markowitz M, Hraber P, Perelson AS, Bhattacharya T, Haynes BF, Korber BT, Hahn BH, Shaw GM. 2009. Genetic identity, biological phenotype, and evolutionary pathways of transmitted/founder viruses in acute and early HIV-1 infection. J Exp Med 206:1273–1289. doi: 10.1084/jem.20090378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang J, Bansal A. 2012. Protocol for analyzing human leukocyte antigen variants and sexually transmitted infections: from genotyping to immunoassays. Methods Mol Biol 903:359–380. doi: 10.1007/978-1-61779-937-2_25. [DOI] [PubMed] [Google Scholar]
- 25.Bansal A, Yue L, Conway J, Yusim K, Tang J, Kappes J, Kaslow RA, Wilson CM, Goepfert PA. 2007. Immunological control of chronic HIV-1 infection: HLA-mediated immune function and viral evolution in adolescents. AIDS 21:2387–2397. doi: 10.1097/QAD.0b013e3282f13823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Foley B, Leitner T, Apetrei C, Hahn B, Mizrachi I, Mullins J, Rambaut A, Wolinsky S, Korber B. 2018. HIV sequence compendium. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, NM: LA-UR 18–25673. [Google Scholar]
- 27.Bansal A, Jackson B, West K, Wang S, Lu S, Kennedy JS, Goepfert PA. 2008. Multifunctional T-cell characteristics induced by a polyvalent DNA prime/protein boost human immunodeficiency virus type 1 vaccine regimen given to healthy adults are dependent on the route and dose of administration. J Virol 82:6458–6469. doi: 10.1128/JVI.00068-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Henikoff S, Henikoff JG. 1992. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci U S A 89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.R Core Team. 2017. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]