Kaposi’s sarcoma-associated herpesvirus (KSHV) is causally linked to Kaposi’s sarcoma and several lymphoproliferative diseases. KSHV, like other herpesviruses, intermittently reactivates from latency and enters a lytic cycle in which numerous lytic mRNAs and proteins are produced, culminating in infectious virion production. These lytic proteins may also contribute to tumorigenesis. Reactivation from latency is controlled by processes that restrict or activate the transcription of KSHV lytic genes. KSHV gene expression is modulated by binding of the host cell proteins CTCF and cohesin complex to the KSHV genome. These proteins bind to and modulate the conformation of chromatin, thereby regulating transcription. We have analyzed the interdependence of binding of CTCF and cohesin and demonstrate that while CTCF is required for cohesin binding to KSHV, they have very distinct effects, with cohesin primarily restricting KSHV lytic transcription. Furthermore, we show that cohesin and CTCF also exert effects on the host cell that promote antiviral defenses.
KEYWORDS: Kaposi's sarcoma-associated herpesvirus, chromatin, cohesin, CTCF, transcriptional regulation, viral replication
ABSTRACT
CTCF and the cohesin complex modify chromatin by binding to DNA and interacting with each other and with other cellular proteins. Both proteins regulate transcription by a variety of local effects on transcription and by long-range topological effects. CTCF and cohesin also bind to herpesvirus genomes at specific sites and regulate viral transcription during latent and lytic cycles of replication. Kaposi’s sarcoma-associated herpesvirus (KSHV) transcription is regulated by CTCF and cohesin, with both proteins previously reported to act as restrictive factors for lytic cycle transcription and virion production. In this study, we examined the interdependence of CTCF and cohesin binding to the KSHV genome. Chromatin immunoprecipitation sequencing (ChIP-seq) analyses revealed that cohesin binding to the KSHV genome is highly CTCF dependent, whereas CTCF binding does not require cohesin. Furthermore, depletion of CTCF leads to the almost complete dissociation of cohesin from sites at which they colocalize. Thus, previous studies that examined the effects of CTCF depletion actually represent the concomitant depletion of both CTCF and cohesin components. Analysis of the effects of single and combined depletion indicates that CTCF primarily activates KSHV lytic transcription, whereas cohesin has primarily inhibitory effects. Furthermore, CTCF or cohesin depletion was found to have regulatory effects on cellular gene expression relevant for the control of viral infection, with both proteins potentially facilitating the expression of multiple genes important in the innate immune response to viruses. Thus, CTCF and cohesin have both positive and negative effects on KSHV lytic replication as well as effects on the host cell that enhance antiviral defenses.
IMPORTANCE Kaposi’s sarcoma-associated herpesvirus (KSHV) is causally linked to Kaposi’s sarcoma and several lymphoproliferative diseases. KSHV, like other herpesviruses, intermittently reactivates from latency and enters a lytic cycle in which numerous lytic mRNAs and proteins are produced, culminating in infectious virion production. These lytic proteins may also contribute to tumorigenesis. Reactivation from latency is controlled by processes that restrict or activate the transcription of KSHV lytic genes. KSHV gene expression is modulated by binding of the host cell proteins CTCF and cohesin complex to the KSHV genome. These proteins bind to and modulate the conformation of chromatin, thereby regulating transcription. We have analyzed the interdependence of binding of CTCF and cohesin and demonstrate that while CTCF is required for cohesin binding to KSHV, they have very distinct effects, with cohesin primarily restricting KSHV lytic transcription. Furthermore, we show that cohesin and CTCF also exert effects on the host cell that promote antiviral defenses.
INTRODUCTION
Kaposi’s sarcoma-associated herpesvirus (KSHV) (human herpesvirus 8 [HHV8]) is a gammaherpesvirus causally related to Kaposi’s sarcoma, primary effusion lymphoma, and multicentric Castleman’s disease (for a review see reference 1). KSHV maintains latent infection as an episome but intermittently may reactivate from latency and express a regulated cascade of lytic cycle genes, which culminates in infectious virion production. Expression of lytic cycle proteins may also exert pleiotropic effects on the host cell and modulate the innate immune response, inflammation, cell proliferation, and survival (2).
CTCF and the cohesin complex, cellular proteins important in regulating transcription and chromatin conformation, have been shown to bind to specific sites on herpesvirus genomes and regulate both latent and lytic gene expression (3–15). CTCF binds to approximately 25 major sites on the KSHV genome (6). Most of the CTCF sites also exhibit colocalized cohesin binding, although the amount of cohesin binding is overall smaller (6). Cohesin, a complex of three core proteins, SMC1, SMC3, and SCC1/Rad21, essential for chromatid segregation (16), has also been recognized as a global regulator of transcription (17). Its importance in transcriptional regulation, in addition to its role in mitosis, is emphasized by the fact that several mutations affecting cohesin function are associated with Cornelia de Lange syndrome, a human disease associated with physical malformations and abnormalities in mental development (18). The cohesin proteins form a ring-shaped structure that encloses sister chromatids (16, 19). Several other proteins are associated with cohesin and regulate the dynamic association of cohesin with chromatin as it is sequentially loaded and dissociated from chromosomes during mitosis and segregation (20). The specificity of cohesin localization is complex and likely mediated by multiple proteins, including NIPBL, mediator, transcription factors, and CTCF (21, 22). Thus, while cohesin binds to many human chromosomal CTCF sites, it also binds to some sites independently of CTCF.
Cohesin and CTCF may have both positive and negative effects on transcription. Many of the effects of cohesin and CTCF are thought to be mediated by facilitating and stabilizing long-range interactions between promoters and enhancers (23). The most likely mechanism is that CTCF binds to specific chromosomal DNA sites and that cohesin facilitates the topological linking of DNA sequences in cis, similar to its role in chromatid linkage in trans. Conversely, CTCF may perform insulator functions, preventing promoter-enhancer interactions and spreading of transcription into silenced gene loci (24). Cohesin also is involved in the regulation of RNA polymerase II (RNAP II) pausing at promoters and relieves pausing, promoting RNA elongation (25).
Recently, we demonstrated that both cohesin and CTCF appear to act as restriction factors for KSHV lytic gene expression and virion production (6). Depletion of either protein limits lytic gene transcription, such that KSHV RNA accumulation and viral replication are enhanced. Depletion of either protein results in markedly increased production of infectious virions. Rad21 appears to exert a greater effect, as the knockdown (KD) of Rad21 resulted in >100-fold increases in virus yields, approximately five times more than the knockdown of CTCF. Both proteins dissociate from the viral genome as lytic viral replication proceeds, but Rad21 dissociates from viral genomes earlier and more completely than CTCF. The loss of cohesin from the KSHV genome is site specific, as it remains bound to some latent gene regions and to the terminal repeats. CTCF also exhibits site-specific changes in KSHV genome occupancy during lytic replication, with decreased binding at most but not all sites during lytic replication (6).
Our findings that cohesin and CTCF may play distinct roles in regulating KSHV reactivation and virion production are mirrored by the differing effects of their knockdown on KSHV lytic gene transcription (6). Consistent with the more profound effects of Rad21 KD on virion production, Rad21 depletion consistently leads to greater increases in KSHV lytic gene expression overall than does CTCF KD (3, 6). Furthermore, the individual depletion of each of the two proteins leads to distinguishable effects on the lytic transcriptional profile. Whereas most KSHV lytic genes are repressed by both CTCF and cohesin, some are increased upon CTCF or cohesin KD, suggesting that the transcription of a subset of genes is directly or indirectly stimulated by CTCF and/or cohesin binding (6). In addition, some genes show decreased expression during CTCF KD but increased expression upon cohesin KD, suggesting that CTCF and cohesin exert opposite effects on specific KSHV genes, with CTCF being required for efficient transcription and cohesin inhibiting transcription (6).
Interpretation of these findings, however, is complicated by a lack of knowledge regarding the interactions between cohesin and CTCF occupancy of the KSHV genome. Previous findings on the interaction between cohesin components, such as Rad21, and CTCF, when bound to the human genome, indicated that cohesin binding and localization may require CTCF (26–28), but a similar analysis of the KSHV genome has not been carried out. We first performed knockdowns of either CTCF, cohesin, or both proteins simultaneously and measured virion production and lytic gene transcription. To investigate the mutual interdependence of cohesin and CTCF binding, we performed experiments in which either CTCF or Rad21 was knocked down individually, followed by chromatin immunoprecipitation (ChIP) for both proteins, allowing us to ask whether KSHV genome occupancy of either protein was affected by the other. The results of these experiments demonstrated that CTCF and cohesin have distinct but interrelated effects on KSHV lytic replication. We have determined that the effects of Rad21 and CTCF on each other’s binding to the KSHV genome are not reciprocal and that the pattern of interdependence varies significantly compared to binding to the human genome. Since depletions of CTCF and cohesin have been reported to differentially affect cellular chromatin structure and gene expression (29), we asked whether we could detect changes in cellular gene expression relevant to viral replication in this study. We found that both CTCF and Rad21 depletions have effects on cellular gene expression, including the innate immune system, that have the potential to affect viral replication indirectly.
RESULTS
Distinct effects of CTCF and Rad21 depletion on KSHV virion production.
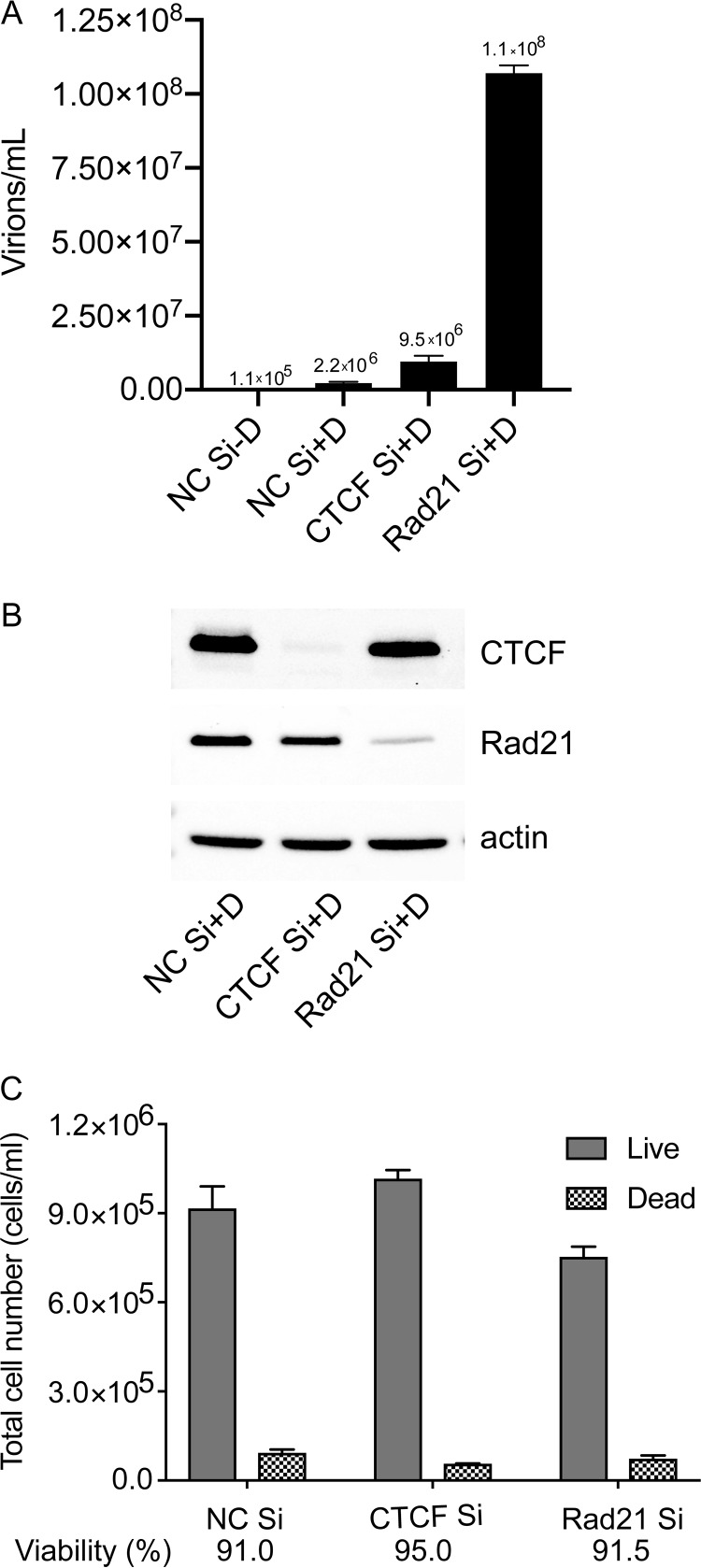
We have previously shown that CTCF depletion prior to the induction of lytic replication led to increased KSHV virion production and lytic gene expression (6). In addition, depletion of Rad21, an essential component of cohesin (30), consistently led to greater increases in KSHV transcription and virion production than CTCF depletion. The more potent effects of Rad21 depletion on infectious virus production are shown in Fig. 1A. The iSLK cell line, a renal epithelial cell-derived line infected with the KSHV Bac16 bacmid, transduced with a doxycycline-inducible Rta-expressing lentivirus, and robustly and synchronously induced to permit KSHV lytic replication by doxycycline treatment, was used for these experiments (31, 32). iSLK cells were transfected with either control small interfering RNAs (siRNAs), CTCF siRNAs, or Rad21 siRNAs, and after 48 h to allow for complete depletion of the targeted proteins, KSHV lytic replication was induced by doxycycline treatment. After 5 days to permit maximal infectious virion production, serial dilutions of filtered cell supernatants were incubated with 293T cells for 48 h. Infected 293T cells that became green fluorescent protein (GFP) positive due to infection by recombinant GFP-expressing KSHV were counted by flow cytometry, and infectious virion titers were calculated as previously described (33). The results confirm that Rad21 has a greater inhibitory effect on KSHV lytic replication and are consistent with our previous report that lytic gene transcription is enhanced by either CTCF or Rad21 knockdown, with Rad21 depletion having a greater enhancing effect on lytic gene expression (6). The efficacy of CTCF and Rad21 depletion is shown in Fig. 1B. Viabilities and numbers of cells after each knockdown were similar, as shown in Fig. 1C.
FIG 1.
Effect of CTCF or Rad21 depletion on KSHV virion production. (A) KSHV-infected iSLK cells were transfected with either control siRNA (NC Si), CTCF-specific siRNA (CTCF Si) or Rad21-specific siRNA (Rad21 Si). KSHV replication was induced by doxycycline treatment (+D) or mock induced (−D). Supernatants from induced cells were used to infect 293T cells. Virus passage was quantitated by flow cytometry of GFP-positive 293T cells. Each transfection/induction was performed in triplicate, and three replicate infections were performed with each supernatant. Error bars denote standard errors of the means (SEM) for all experiments. (B) Western blot showing the efficacy of CTCF and Rad21 KD with actin as a loading control. (C) Cells were counted after each KD with vital dye staining. A minimum of 200 cells per sample were counted.
CTCF-cohesin interdependence when binding to the KSHV genome.
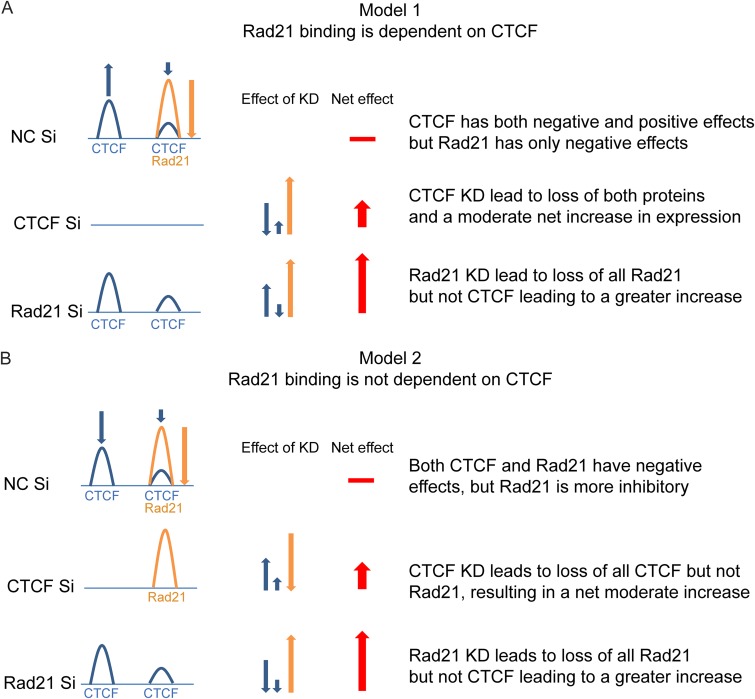
CTCF binding is thought to recruit cohesin components to chromosomal sites where they colocalize, and the depletion of CTCF leads to a variable loss of cohesin occupancy (26). If a similar mechanism operated in KSHV, it would be expected that CTCF depletion would lead to a concomitant loss of Rad21 and other cohesin components. Yet the specific depletion of Rad21 alone caused greater increases in KSHV lytic transcription than the depletion of CTCF (6), suggesting that the loss of CTCF may limit maximal transcriptional activation. Such an outcome could be explained if CTCF generally has net positive effects on KSHV transcription, but cohesin has larger negative or restrictive effects on transcription (model 1) (Fig. 2A). The depletion of CTCF thus would lead to a loss of both CTCF and Rad21, a loss of CTCF’s positive effects, and a concomitant loss of Rad21’s negative effects. Since CTCF KD is postulated to have a net inhibitory effect on transcription, the enhancing effects of Rad21 KD would be partially offset compared to KD of Rad21 alone, which is hypothesized to have no effect on CTCF occupancy. Variations of this model are also possible, in which Rad21 KD causes a loss of CTCF at colocalized sites but does not affect sites at which CTCF alone is bound.
FIG 2.
Models for functional interaction of CTCF and Rad21 on the KSHV genome. (A) Model 1: Rad21 binding is dependent on CTCF binding. Individual or colocalized CTCF (blue) and Rad21 (orange) binding is shown at a regulated gene locus in the KSHV genome. At baseline, CTCF binding can have either positive (blue up arrow) or negative (blue down arrow) effects on transcription, whereas Rad21 has only a negative effect (orange down arrow). Red arrows represent the additive effects of CTCF and/or Rad21 KD on expression at the gene locus. The horizontal red bar depicts the baseline level of gene expression without KD. The predicted effect of CTCF KD is that the loss of CTCF leads to a net decrease in transcription, whereas the concomitant loss of Rad21 leads to a greater net increase in transcription, cumulatively resulting in a moderate increase. KD of Rad21 leads to a loss of Rad21 alone, producing a large transcriptional increase, without affecting the net positive effects of bound CTCF, resulting in a larger net increase than with CTCF KD. (B) Model 2: Rad21 does not depend on CTCF binding. CTCF and Rad21 binding sites are depicted as described above for panel A. CTCF binding, however, is postulated to have only negative effects, similar to Rad21, albeit not as great as those of Rad21. CTCF KD therefore leads to a moderate increase in transcription, and Rad21 KD leads to a larger increase. Neither KD affects the effect of the other protein on transcription.
Conversely, it is possible that Rad21 and CTCF binding are completely independent and that the depletion of either one does not affect the binding of the other (model 2) (Fig. 2B). In this case, the different effects of CTCF and Rad21 KD on the magnitude of transcriptional activation could still be explained but would require that the net effects of both proteins are inhibitory, with CTCF having a smaller restrictive potential than Rad21. In this case, the depletion of either protein would independently have an enhancing effect on transcription, with Rad21 KD having a greater enhancing effect.
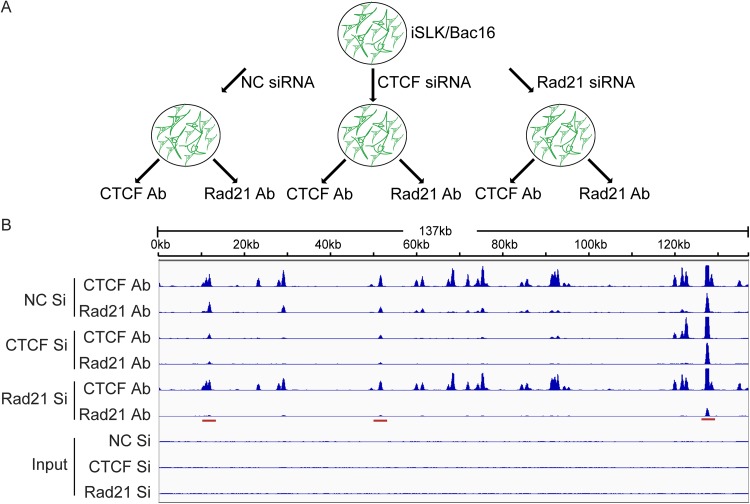
In order to distinguish between these two models, we performed a reciprocal ChIP experiment to directly examine the effect of depleting either protein on KSHV genome occupancy by the other protein. After depletion of either Rad21 or CTCF with specific siRNAs, or treatment with nontargeting control siRNAs, DNA was chemically cross-linked, sonicated, and immunoprecipitated with an antibody (Ab) to either CTCF or Rad21 (Fig. 3A). Deep sequencing of the immunoprecipitated DNA then allowed the determination of the effect of CTCF or Rad21 depletion on the binding of both proteins. The results clearly demonstrated that model 1 more closely describes the interdependence of CTCF and Rad21 binding than model 2. CTCF KD led to an almost complete loss of Rad21 at sites where they bind together and where CTCF is lost. At sites where CTCF remained bound despite KD (avid sites), Rad21 also remained bound (Fig. 3B). The removal effect of CTCF KD on Rad21 occupancy was almost as efficient as that of Rad21 KD itself. Conversely, Rad21 KD had no detectable effect on CTCF occupancy, despite the efficient removal of Rad21 at most sites.
FIG 3.
Interdependence of CTCF and Rad21 binding to the KSHV genome. (A) Strategy for CTCF and Rad21 reciprocal ChIP. iSLK/Bac16 cells were transfected with negative-control siRNA (NC Si), CTCF Si, or Rad21 Si. At 2 days posttransfection, each group was divided into two, and ChIP-seq was performed with Rad21 or CTCF to determine the effect of the depletion of either protein on the binding of the other protein. (B) Graphical representation of reciprocal ChIP-seq. Relative read numbers are plotted on the vertical axis versus the reference KSHV genome map on the horizontal axis. CTCF and Rad21 ChIP results for each sample are shown in the top six panels, and the corresponding input samples (IN) are shown in the bottom three panels. As shown in tracks 3 and 4, CTCF KD leads to a loss of Rad21 binding, whereas Rad21 depletion does not affect CTCF binding. Rad21 KD is more effective than CTCF KD in causing a loss of Rad21 binding at some sites (cf. tracks 6 and 4, highlighted with a red bar). The tracks depict coverage per base, scaled per million mapped KSHV reads.
Complex effects of CTCF and Rad21 depletion on KSHV gene expression and virion production.
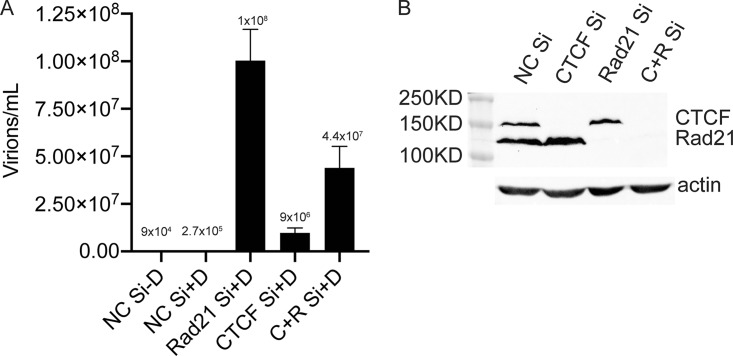
Based on these findings, one would predict that KD of both CTCF and Rad21 simultaneously would have essentially the same effect as CTCF KD on both virus production and lytic transcription. We therefore examined KSHV infectious virion production under these conditions of individual and double KD. KSHV-infected iSLK cells were transfected with either Rad21 siRNA, CTCF siRNA, both siRNAs together, or control siRNA. Forty-eight hours later, KSHV replication was induced with doxycycline as described above, and infectious virus titers in the cell supernatants were determined by infection of 293T cells and flow cytometry as described above (Fig. 4A). As expected, Rad21 KD yielded a 368-fold increase over control siRNA KD, and CTCF KD resulted in a lower 36-fold increase in the virus titer. Interestingly, double KD of both CTCF and Rad21 yielded a larger amount of virus than KD of CTCF alone. Western blotting of cell lysates from each KD confirmed that KD of each protein did not affect levels of the other and that depletions of soluble CTCF and Rad21 were complete (Fig. 4B). These results suggested that subtle differences in residual Rad21 or CTCF that remained bound to the viral genomes could explain the differences between CTCF KD and double KD of both Rad21 and CTCF. Indeed, a close examination of the magnitude of the residual ChIP binding after each KD suggested that specific Rad21 KD led to the more complete removal of Rad21 from the genome than CTCF1 KD (Fig. 3B, compare CTCF Si/Rad21 Ab to Rad21 Si/Rad21 Ab). Thus, more complete removal of Rad21 would occur upon double KD. Therefore, if Rad21 primarily has restrictive effects and CTCF has net positive effects on KSHV transcription, double KD with more complete Rad21 removal would be expected to increase virus production more than CTCF KD with incomplete removal of Rad21.
FIG 4.
KSHV production in cells depleted of CTCF, Rad21, or both CTCF and Rad21. CTCF and/or Rad21 was depleted in iSLK/Bac16 cells prior to the induction of lytic replication. Cells were transfected with either negative-control siRNA (NC Si), siRNA specific for CTCF (CTCF Si), siRNA specific for Rad21 (Rad21 Si), or both CTCF Si and Rad21 siRNAs (C+R Si). (A) KSHV replication was induced by treatment with doxycycline. Five days later, supernatants from induced cells were used to infect 293T cells. Virus passage was quantitated by flow cytometry of GFP-positive 293T cells. Each transfection/induction was performed in triplicate, and three replicate infections were performed with each supernatant. (B, top) Cell lysates were prepared 48 h after transfection and analyzed by immunoblotting with an anti-CTCF antibody or anti-Rad21 antibody. (Bottom) Blots were stripped and reprobed with an anti-actin antibody as a loading control.
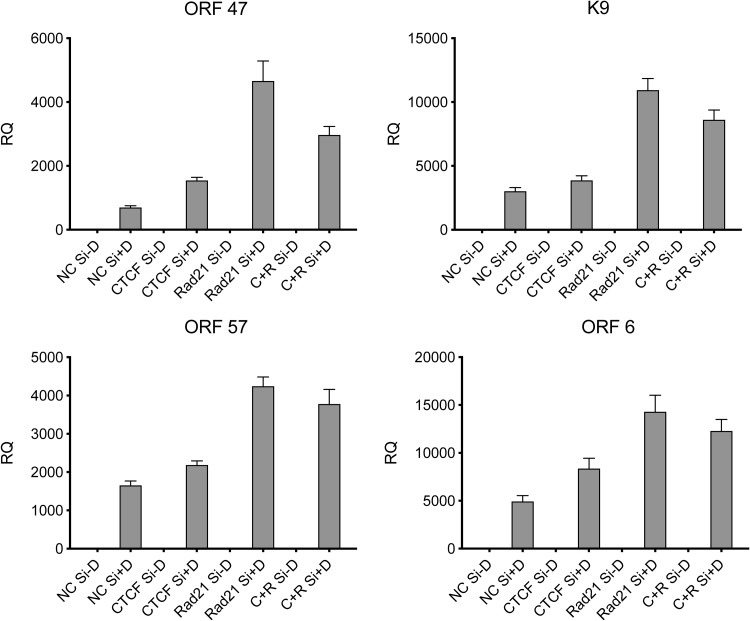
In order to ask whether the transcription of individual KSHV genes was also affected similarly by the depletion of CTCF and Rad21, we performed quantitative PCR (qPCR) on samples from cells treated in the same manner as those in which virus production was measured, as described above. Knockdown of either CTCF, Rad21, or both proteins in iSLK/Bac16 cells was performed. KSHV replication was induced by the addition of doxycycline or mock induced, and RNA was harvested 48 h after the induction of replication. The expression of KSHV lytic RNAs was measured by qPCR under each condition of depletion and induction. As shown in Fig. 5, the pattern of gene expression enhancement due to the knockdown of either CTCF or Rad21 was consistent with our previous findings, showing that Rad21 KD led to consistently greater increases in gene expression. Similar to the effects on virion production, double KD resulted in a level of RNA expression intermediate between those of individual CTCF KD and Rad21 KD in each case. These data indicate that the interrelated effects of CTCF and Rad21 on transcription mediate the observed effects on virus production. Furthermore, these results imply that the net effect of CTCF on KSHV gene transcription is positive but that KD of CTCF, due to the concomitant loss of cohesin binding, leads to the derepression of KSHV gene expression. It should also be noted that we previously demonstrated that CTCF and/or Rad21 KD does not affect the amounts of the KSHV lytic transactivator Rta (open reading frame 50 [ORF50]) expressed in this ORF50-inducible cell line (6).
FIG 5.
Effect of individual or combined depletion of CTCF and Rad21 on KSHV lytic cycle gene expression. iSLK/Bac16 cells were depleted of CTCF (CTCF Si) and/or Rad21 (Rad21 Si) or mock depleted (NC Si) by siRNA transfection followed by treatment with doxycycline (−D, mock treatment; +D, doxycycline treatment) to induce lytic replication. RNA was prepared 48 h after the induction of replication, and relative quantification of mRNA expression (RQ) for each lytic gene was performed by qPCR. Results for the early genes K9, ORF6, and ORF57 and the late gene ORF47 are shown. Each transfection was performed in triplicate, and qPCR was performed with three technical replicates per sample. The level of expression of each RNA was normalized to the level of expression in uninduced cells.
Interactions of CTCF and Rad21 during binding to the human genome.
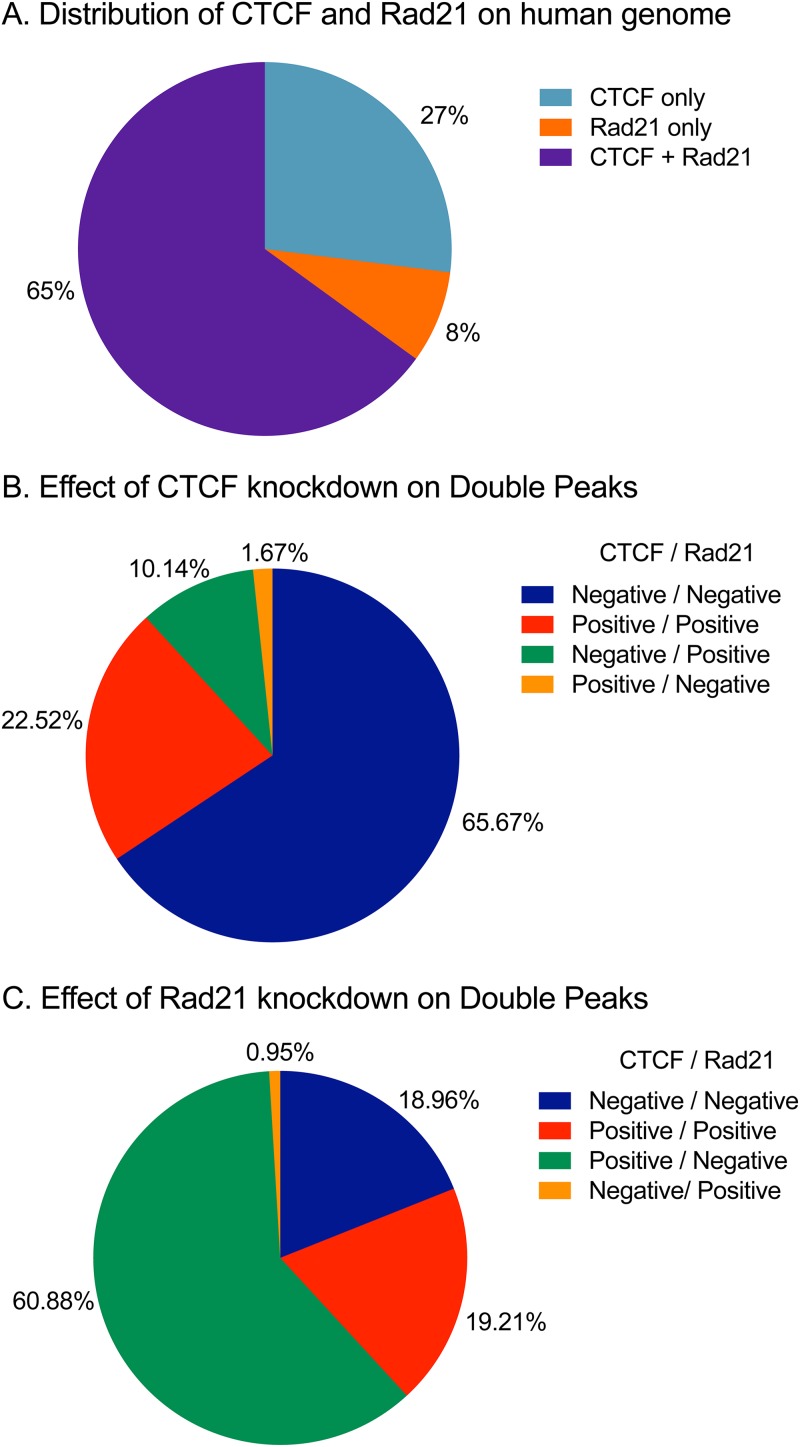
Similar interactions between CTCF and Rad21 binding at sites where they colocalize on the human genome have been reported, showing that Rad21 binding is mostly dependent on CTCF binding but not vice versa (26). In order to more closely examine the interdependence of CTCF and Rad21 binding to the human genome, we analyzed the binding patterns of CTCF and Rad21 by ChIP sequencing (ChIP-seq) at baseline and after the depletion of either protein. In contrast to the pattern observed with KSHV, there were 29,363 (65%) sites that were CTCF and Rad21 double positive, 12,164 (27%) that were CTCF-only sites, and 3,426 (8%) that were Rad21-only sites (Fig. 6A). Thus, unlike the situation with KSHV, only ∼70% of the CTCF binding sites exhibited colocalization of Rad21, and a small number of Rad21 peaks did not coincide with CTCF. The interdependence of CTCF and Rad21 was also distinct from that observed with binding to the KSHV genome. While the depletion of CTCF also led to a loss of Rad21 at most doubly bound sites, there was a significant percentage of sites where Rad21 binding was largely unaffected despite the loss of CTCF (10%) (Fig. 6B). In addition, CTCF remained bound at 22% of the sites despite KD; the percentage of such avid CTCF sites was thus higher than that observed with KSHV. The effect of Rad21 KD on CTCF occupancy was also different from that observed with KSHV (Fig. 6C). At the majority of sites where Rad21 was removed (61%), there was no effect on CTCF occupancy, similar to KSHV. However, at 19% of the sites, Rad21 KD led to a concomitant loss of CTCF binding. Similar to CTCF, ∼19% of the sites displayed tight Rad21 binding, with no change in Rad21 occupancy after KD. It therefore appears that overall, Rad21 binding to the KSHV genome is much more dependent on CTCF than binding to the human genome. Binding of both proteins to the host cell genome also does not appear to be as labile as binding to the viral genome, with a larger percentage of bound sites being resistant to either CTCF or Rad21 depletion. Furthermore, the interdependence of binding also appears to be more complex, with a significant percentage of sites where the depletion of one protein does not affect the maintained binding of the other.
FIG 6.
Interdependence of CTCF and Rad21 binding to the human genome. (A) Distribution of CTCF and Rad21 binding sites on the human genome. Peaks were called using MACS2 as described in Materials and Methods. (B) Effect of CTCF depletion on the binding of each protein at colocalized sites. Where CTCF depletion leads to a loss of CTCF at a site, it is denoted CTCF negative; where it leads to a loss of Rad21, it is denoted Rad21 negative. (C) Effects of Rad21 depletion on Rad21 and CTCF occupancy. Sites where Rad21 depletion led to a loss of both Rad21 and CTCF, sites where neither protein is lost, and sites at which Rad21 depletion causes a loss of Rad21 but does not affect CTCF occupancy are shown (61%).
Effects of CTCF and Rad21 depletion on host cell gene expression.
The global effects of CTCF and Rad21 depletion on the human transcriptome have not been extensively characterized. Given the complex patterns of effects on binding observed upon the depletion of CTCF and Rad21, it was possible that these proteins could have unique effects on cell gene expression with relevance to viral replication. We therefore examined the effect of either CTCF or Rad21 KD on cellular transcription by high-throughput RNA sequencing (RNA-seq). RNA from iSLK cells in which either CTCF or Rad21 KD had been performed was analyzed by deep sequencing. We separated the genes that were most highly affected by either KD using a threshold of a log2-fold change of 2. Using these criteria, we classified all human transcripts based on the pattern of the response to either CTCF or Rad21 KD (Table 1). Since the level of an individual transcript could either increase, decrease, or remain unchanged with either KD, there are theoretically nine categories of gene responses, as shown in Table 1. Again, unlike the effect on KSHV transcript levels, where virtually all lytic genes are activated by CTCF and even more so by Rad21 depletion, the level of expression of the vast majority of detectably expressed cellular genes (96%) was not highly affected by either KD. Of the remaining eight categories, there were between 100 and 300 genes in each category, except for one (there were no genes whose expression increased in response to Rad21 KD but decreased in response to CTCF KD). We performed gene ontology (GO) analyses using the PANTHER database to investigate whether there were any distinguishable pathways or functional groups of gene expression relevant to virus replication that were affected by CTCF or Rad21 KD. Interestingly, in the category of genes whose expression decreased by >2-log2-fold upon KD of either CTCF or Rad21, several genes classified as being important in the interferon (IFN) response were enriched, including all three 2′,5′-oligoadenylate synthase isoforms (OAS1, OAS2, and OAS3). Analysis of the subset of genes whose expression was not affected by Rad21 KD but was decreased by CTCF, i.e., genes whose expression is dependent on CTCF, revealed a strong enrichment of genes involved in homophilic cell adhesion via plasma membrane adhesion molecules (Table 2). Interestingly, 27 members of the protocadherin gene cluster were found to be downregulated by CTCF KD. CTCF was previously identified as a master regulator of protocadherin expression (34, 35), confirming the validity of our analyses.
TABLE 1.
Cellular gene transcripts categorized using a threshold of a 4-fold change based on the pattern of response to either CTCF or Rad21 KDa
| Category | Response |
No. of genes | |
|---|---|---|---|
| CTCF KD | Rad21 KD | ||
| 1 | ↑↑↑ | ↑↑↑ | 145 |
| 2 | ↑↑↑ | − | 287 |
| 3 | ↑↑↑ | ↓↓↓ | 2 |
| 4 | − | ↑↑↑ | 208 |
| 5 | − | − | 23,921 |
| 6 | − | ↓↓↓ | 124 |
| 7 | ↓↓↓ | ↑↑↑ | 0 |
| 8 | ↓↓↓ | − | 209 |
| 9 | ↓↓↓ | ↓↓↓ | 99 |
| Total | 24,995 | ||
↑↑↑, gene expression was upregulated; −, gene expression did not change; ↓↓↓, gene expression was downregulated.
TABLE 2.
Cellular genes involved in homophilic cell adhesion downregulated with CTCF KD using a threshold of a 4-fold change
| Gene IDa | Mapped ID | Gene name | PANTHER protein class (PANTHER ID) |
|---|---|---|---|
| HUMAN|HGNC=8669|UniProtKB=Q9Y5H8 | PCDHA3 | Protocadherin alpha-3 | Cadherin (PC00069) |
| HUMAN|HGNC=8667|UniProtKB=Q9Y5I0 | PCDHA13 | Protocadherin alpha-13 | Cadherin (PC00069) |
| HUMAN|HGNC=8664|UniProtKB=Q9Y5I2 | PCDHA10 | Protocadherin alpha-10 | Cadherin (PC00069) |
| HUMAN|HGNC=8663|UniProtKB=Q9Y5I3 | PCDHA1 | Protocadherin alpha-1 | Cadherin (PC00069) |
| HUMAN|HGNC=8694|UniProtKB=Q9Y5E1 | PCDHB9 | Protocadherin beta-9 | Cadherin (PC00069) |
| HUMAN|HGNC=8671|UniProtKB=Q9Y5H7 | PCDHA5 | Protocadherin alpha-5 | Cadherin (PC00069) |
| HUMAN|HGNC=8665|UniProtKB=Q9Y5I1 | PCDHA11 | Protocadherin alpha-11 | Cadherin (PC00069) |
| HUMAN|HGNC=1756|UniProtKB=Q12864 | CDH17 | Cadherin-17 | Cadherin (PC00069) |
| HUMAN|HGNC=8681|UniProtKB=Q9UN67 | PCDHB10 | Protocadherin beta-10 | Cadherin (PC00069) |
| HUMAN|HGNC=8670|UniProtKB=Q9UN74 | PCDHA4 | Protocadherin alpha-4 | Cadherin (PC00069) |
| HUMAN|HGNC=8682|UniProtKB=Q9Y5F2 | PCDHB11 | Protocadherin beta-11 | Cadherin (PC00069) |
| HUMAN|HGNC=8677|UniProtKB=Q9Y5I4 | PCDHAC2 | Protocadherin alpha-C2 | Cadherin (PC00069) |
| HUMAN|HGNC=8672|UniProtKB=Q9UN73 | PCDHA6 | Protocadherin alpha-6 | Cadherin (PC00069) |
| HUMAN|HGNC=8673|UniProtKB=Q9UN72 | PCDHA7 | Protocadherin alpha-7 | Cadherin (PC00069) |
| HUMAN|HGNC=8692|UniProtKB=Q9Y5E2 | PCDHB7 | Protocadherin beta-7 | Cadherin (PC00069) |
| HUMAN|HGNC=8668|UniProtKB=Q9Y5H9 | PCDHA2 | Protocadherin alpha-2 | Cadherin (PC00069) |
| HUMAN|HGNC=8675|UniProtKB=Q9Y5H5 | PCDHA9 | Protocadherin alpha-9 | Cadherin (PC00069) |
| HUMAN|HGNC=8676|UniProtKB=Q9H158 | PCDHAC1 | Protocadherin alpha-C1 | Cadherin (PC00069) |
| HUMAN|HGNC=8666|UniProtKB=Q9UN75 | PCDHA12 | Protocadherin alpha-12 | Cadherin (PC00069) |
| HUMAN|HGNC=8674|UniProtKB=Q9Y5H6 | PCDHA8 | Protocadherin alpha-8 | Cadherin (PC00069) |
| HUMAN|HGNC=8685|UniProtKB=Q9Y5E9 | PCDHB14 | Protocadherin beta-14 | Cadherin (PC00069) |
Species and HGNC and UniProt accession numbers for each gene are as listed.
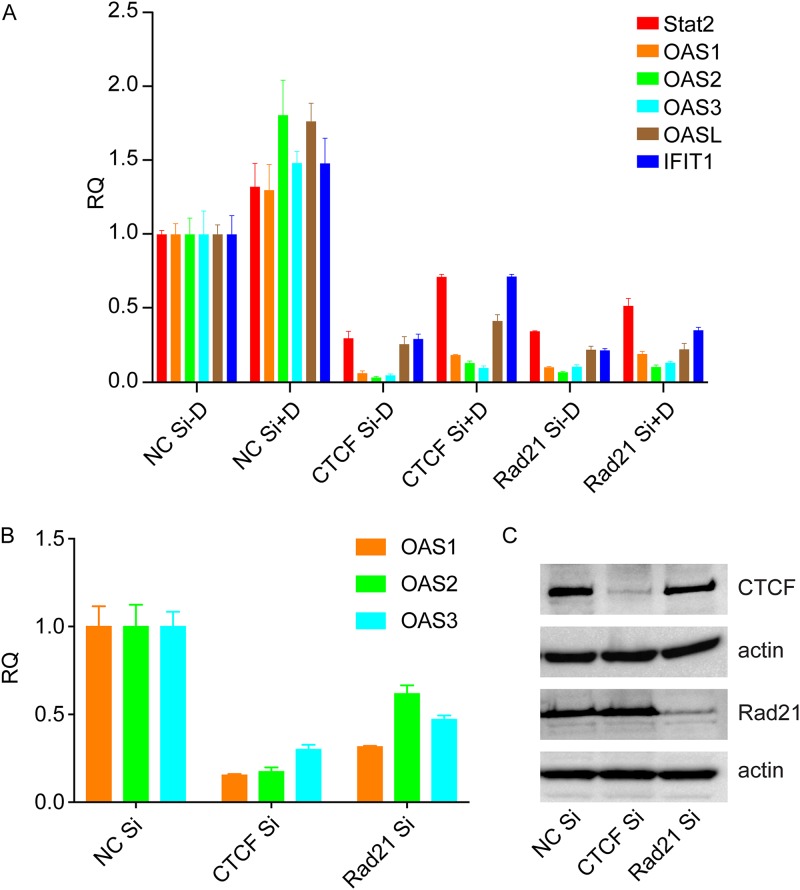
We then performed additional analyses where the threshold for fold change was set at ≥2. The number of genes in each category then increased, as expected (Table 3). The number of genes whose expression decreased with either CTCF KD or Rad21 KD then increased to 658. A gene ontology analysis of these genes also revealed highly significant enrichment for genes involved in the cellular response to viral infection, particularly those induced by interferons (Table 4). A list of the genes most highly enriched in the type I IFN signaling pathway that are downregulated by either CTCF or Rad21 KD is shown in Table 5. In order to confirm the effect of CTCF or Rad21 on these transcripts, we performed qPCR to measure the levels of five interferon-stimulated genes (ISGs). As shown in Fig. 7, the expression of ISGs increased with the induction of KSHV replication, as might be expected. Furthermore, ISG expression was inhibited by KD of either CTCF or Rad21, confirming the RNA-seq findings. These data therefore suggest not only that CTCF and Rad21 have direct effects on KSHV transcription but also that both proteins may play a positive role in the innate immune response and interferon-mediated antiviral defenses.
TABLE 3.
Cellular gene transcripts categorized using a threshold of a 2-fold change based on the pattern of the response to either CTCF or Rad21 KDa
| Category | Response |
No. of genes | |
|---|---|---|---|
| CTCF KD | Rad21 KD | ||
| 1 | ↑↑↑ | ↑↑↑ | 938 |
| 2 | ↑↑↑ | − | 950 |
| 3 | ↑↑↑ | ↓↓↓ | 49 |
| 4 | − | ↑↑↑ | 709 |
| 5 | − | − | 20,172 |
| 6 | − | ↓↓↓ | 584 |
| 7 | ↓↓↓ | ↑↑↑ | 20 |
| 8 | ↓↓↓ | − | 915 |
| 9 | ↓↓↓ | ↓↓↓ | 658 |
| Total | 24,995 | ||
↑↑↑, gene expression was upregulated; −, gene expression did not change; ↓↓↓, gene expression was downregulated.
TABLE 4.
GO enrichment analysis of cellular genes whose levels decreased 2-fold with either CTCF or Rad21 KD
| GO biological process | Expecteda | Fold enrichment | P value |
|---|---|---|---|
| Type I interferon signaling pathway | 1.6 | 11.87 | 8.62E−11 |
| Cellular response to type I interferon | 1.6 | 11.87 | 8.62E−11 |
| Response to type I interferon | 1.7 | 11.16 | 2.53E−10 |
| Negative regulation of viral genome replication | 1.27 | 11.02 | 7.16E−07 |
Number of genes expected to occur by chance in each gene set for the listed GO pathways.
TABLE 5.
Cellular genes in the type I interferon pathway downregulated with either CTCF or Rad21 KD
| Gene IDa | Mapped ID | Gene name | PANTHER protein class(es) (PANTHER ID[s]) |
|---|---|---|---|
| HUMAN|HGNC=11363|UniProtKB=P52630 | STAT2 | Signal transducer and activator of transcription 2 | Nucleic acid binding (PC00171), transcription factor (PC00218) |
| HUMAN|HGNC=5413|UniProtKB=Q01629 | IFITM2 | Interferon-induced transmembrane protein 2 | |
| HUMAN|HGNC=7533|UniProtKB=P20592 | MX2 | Interferon-induced GTP-binding protein Mx2 | Hydrolase (PC00121), microtubule family cytoskeletal protein (PC00085), small GTPase (PC00157) |
| HUMAN|HGNC=1119|UniProtKB=Q10589 | BST2 | Bone marrow stromal antigen 2 | |
| HUMAN|HGNC=30932|UniProtKB=Q6GPH4 | XAF | XIAP-associated factor 1 | |
| HUMAN|HGNC=6121|UniProtKB=O14896 | IRF6 | Interferon regulatory factor 6 | Nucleic acid binding (PC00171), transcription factor (PC00218) |
| HUMAN|HGNC=4963|UniProtKB=P30511 | HLA-F | HLA class I histocompatibility antigen, alpha chain F | Immunoglobulin receptor superfamily (PC00090), major histocompatibility complex antigen (PC00124) |
| HUMAN|HGNC=5411|UniProtKB=O14879 | IFIT3 | Interferon-induced protein with tetratricopeptide repeats 3 | |
| HUMAN|HGNC=5399|UniProtKB=P80217 | IFI35 | Interferon-induced 35-kDa protein | |
| HUMAN|HGNC=7532|UniProtKB=P20591 | MX1 | Interferon-induced GTP binding protein Mx1 | Hydrolase (PC00121), microtubule family cytoskeletal protein (PC00085), small GTPase (PC00157) |
| HUMAN|HGNC=8086|UniProtKB=P00973 | OAS1 | 2′,5′-Oligoadenylate synthase 1 | Defense/immunity protein, (PC00090), nucleic acid binding (PC00171), nucleotidyltransferase (PC00220) |
| HUMAN|HGNC=8090|UniProtKB=Q15646 | OASL | 2′,5′-Oligoadenylate synthase-like protein | Defense/immunity protein (PC00090), nucleic acid binding (PC00171), nucleotidyltransferase (PC00220) |
| HUMAN|HGNC=30908|UniProtKB=Q8WXG1 | RSAD2 | Radical S-adenosylmethionine domain-containing protein 2 | |
| HUMAN|HGNC=6130|UniProtKB=Q96AZ6 | ISG20 | Interferon-stimulated gene 20-kDa protein | Exoribonuclease (PC00171) |
| HUMAN|HGNC=5407|UniProtKB=P09914 | IFIT1 | Interferon-induced protein with tetratricopeptide repeats 1 | |
| HUMAN|HGNC=4054|UniProtKB=P09912 | IFI6 | Interferon alpha-inducible protein 6 | |
| HUMAN|HGNC=4053|UniProtKB=P05161 | ISG15 | Ubiquitin-like protein ISG15 | Ribosomal protein (PC00171) |
| HUMAN|HGNC=8088|UniProtKB=Q9Y6K5 | OAS3 | 2′,5′-Oligoadenylate synthase 3; OAS3; ortholog | Defense/immunity protein (PC00090), nucleic acid binding (PC00171), nucleotidyltransferase (PC00220) |
| HUMAN|HGNC=8087|UniProtKB=P29728 | OAS2 | 2′,5′-Oligoadenylate synthase 2 | Defense/immunity protein (PC00090), nucleic acid binding (PC00171), nucleotidyltransferase (PC00220) |
Species and HGNC and UniProt accession numbers for each gene are as listed.
FIG 7.
Effect of depletion of CTCF or Rad21 on cellular interferon-regulated gene expression. (A) Depletion of CTCF and/or Rad21 in iSLK/Bac16 cells followed by KSHV lytic replication induction was performed. RNA was prepared 48 h after the induction of replication. Relative quantification of mRNA expression (RQ) for each lytic gene was performed by qPCR. Results for Stat2, OAS1, OAS1, OAS2, OAS3, OASL, and IFIT1 are shown. Each transfection was performed in triplicate, and qPCR was performed with three technical replicates per sample. The level of expression of each RNA was normalized to the level of expression in uninduced cells (NC Si, negative-control siRNAs). (B) Uninfected iSLK cells were transfected with either negative-control siRNA, CTCF siRNA, or Rad21 siRNA. RNA was harvested, and qPCR was performed for ISGs as described above for panel A. (C) Efficiency of CTCF and Rad21 KD in panel B measured by Western blotting, with actin as a loading control.
Since both CTCF and Rad21 KD led to increased KSHV replication, it was possible that the effects of CTCF and Rad21 KD on the innate immune response were indirectly mediated by viral gene products. Although possible, such effects are unlikely given that the effect on ISGs was also observed in iSLK/Bac16 cells that were not induced to permit lytic replication, in which the expression of lytic KSHV genes and virion production is negligible. Nevertheless, we also examined ISG expression in iSLK cells that were not infected by KSHV. As shown in Fig. 7B, the expression of several ISGs was also decreased upon CTCF or Rad21 depletion in uninfected iSLK cells.
DISCUSSION
Modification of chromatin conformation by CTCF and cohesin plays a major role in mammalian transcriptional regulation. Although most of the focus has been on the topological organization of chromatin via loop formation and its effects on gene activation and repression, local and direct effects of CTCF and cohesin on promoter activity are also important (36). CTCF may act as a barrier insulator, preventing the spread of transcription from regions of active to inactive chromatin (37, 38). CTCF also binds to RNAP II and can facilitate RNAP II recruitment and elongation, while cohesin may regulate pausing of RNAP II (25, 39, 40). The relative contributions of these mechanisms to gammaherpesvirus transcription remain to be fully defined. Work from the Lieberman laboratory has demonstrated the formation of intragenomic CTCF-dependent loops in KSHV (5, 7, 41). Using 3C and other chromatin conformation assays, they showed that a major CTCF and cohesin binding site in the LANA intron formed multiple loops with other KSHV genomic locations. Two such interactions were characterized in detail, with the ORF50 promoter region and the 3′ end of the latency region. Mutation of the CTCF binding sites in the latency region, or depletion of CTCF, led to decreased loop formation between the latency region and ORF50 as well as decreased ORF50 transcription. In addition, local effects of CTCF binding were also reported, with mutation of CTCF binding sites in the LANA intron affecting RNAP II binding at K12 and LANA promoter regions. The loss of CTCF binding decreased K12 transcription but increased LANA transcription and altered histone modification patterns and nucleosome occupancy, indicating that CTCF may have complex local effects independent of its ability to mediate long-range DNA interactions. The depletion of cohesin leading to a loss of KSHV genome binding acts to activate lytic gene transcription and virion production (3, 6). Because CTCF depletion also had similar effects, it initially appeared that both CTCF and cohesin are negative regulators of lytic transcription. However, our finding that CTCF depletion cannot be achieved without a concomitant loss of cohesin from the KSHV genome complicates the interpretation of findings from experiments involving CTCF depletion or mutation of CTCF binding sites. It should also be noted that during lytic replication, newly produced linear genomes are not chromatinized when packaged. The extent to which CTCF and cohesin may affect potential lytic transcription from such templates prior to encapsidation remains to be determined.
Our data are consistent with multifactorial mechanisms for CTCF regulation of KSHV lytic gene transcription. The depletion of CTCF led to the loss of Rad21 at all KSHV sites where CTCF binding was lost. In addition, CTCF KD led to the loss of CTCF at several other sites that were not cooccupied by cohesin. As shown by the reciprocal ChIP experiments, the interdependence of CTCF and cohesin binding to the KSHV genome is unilateral; i.e., cohesin binding is almost entirely CTCF dependent but not vice versa. This leads to a situation where CTCF removal leads to the removal of cohesin as well. The effects of CTCF depletion are therefore actually the effects of the removal of both cohesin and CTCF. The overall effects of CTCF on KSHV lytic cycle transcription and virion production are thus likely to be a combination of its local effects, its effect on long-range interactions, and local effects of cohesin occupancy that require CTCF. The clearly more potent effects of cohesin removal in activating lytic transcription than those of CTCF (and concomitantly cohesin) removal suggest that CTCF has generally activating effects on KSHV lytic cycle transcription. While loop formation between the LANA and ORF50 sites involving both CTCF and cohesin may enhance lytic cycle transcription under some circumstances (5), the depletion of cohesin alone leads to lytic cycle activation and virion production (3, 6), suggesting that repressive effects of cohesin play a greater role in regulating KSHV lytic transcription. In addition, in the experimental system used here, initial Rta protein is derived from an inducible transgene, demonstrating that cohesin and CTCF have effects on lytic cycle transcription independent of the regulation of the ORF50 promoter. In summary, CTCF appears to be coopted by KSHV to act primarily as a positive activator of lytic cycle transcription. The stimulatory effects of CTCF depletion, however, cannot be uncoupled from its effects on cohesin binding, which is primarily inhibitory to KSHV lytic cycle transcription.
Interestingly, the wholly one-sided dependence of cohesin on CTCF binding to the KSHV genome was different from that observed in the cellular genome. While the general pattern of cohesin dependence on CTCF was maintained, there was a significant number of sites where Rad21 occupancy was maintained despite CTCF depletion and even sites where Rad21 depletion appeared to decrease CTCF binding, suggesting that local effects of cohesin and CTCF may play complex roles in regulating gene expression in addition to their effects on chromatin conformation. While we cannot rule out a small amount of residual CTCF after KD at such sites, cohesin has also been shown to bind to the human genome together with transcription factors, independently of CTCF, and to have a functional role in estrogen-regulated transcription (42). Such a model of individual CTCF and cohesin effects is consistent with the intriguing finding that either cohesin or CTCF depletion affected the expression of several key components of the interferon response. These genes are present on several different chromosomes, unlike the protocadherin cluster, which is coordinately regulated by CTCF (34, 43, 44). While long-range topological effects of CTCF/cohesin on each individual interferon-induced gene cannot be ruled out, it leaves open the possibility that binding of CTCF and/or cohesin may independently and locally activate the transcription of these genes. We previously demonstrated that IFIT1, IFIT2, and IFIT3, as well as OAS2 proteins, inhibit KSHV lytic replication and virion production (45). Experiments using depletion of CTCF or cohesin or mutation of CTCF binding sites therefore have the potential to alter the cellular phenotype and viral biology by mechanisms unrelated to their direct effects on viral DNA genomes, which should be considered in the interpretation of findings from such studies. The finding that CTCF and cohesin may broadly regulate the interferon response also raises the possibility that cohesinopathies, human diseases in which various genes in the cohesin pathway are mutated, may result in subtle immunodeficiency and affect resistance to viral infections.
MATERIALS AND METHODS
Cells and plasmids.
293T cells were grown at 37°C in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and glutamine. iSLK cells (31) (gift of Don Ganem, UCSF) were maintained in DMEM containing 10% charcoal-stripped FBS (Sigma) and 1% glutamine with 250 μg/ml G-418 and 1 μg/ml puromycin. iSLK cells were infected with wild-type (WT) KSHV derived from the bacmid Bac16, expressing enhanced green fluorescent protein (eGFP) and hygromycin resistance (32). Bac16 KSHV-infected iSLK cells (iSLK/Bac16 cells) were maintained in a solution containing 1.2 mg/ml hygromycin, 250 μg/ml G-418, and 1 μg/ml puromycin.
CTCF and Rad21 knockdown.
On-target plus smart pool siRNAs for CTCF (catalog no. L-020165-00-0005), Rad21 (catalog no. L-006832-00-0005), and the negative control (catalog no. D-001210-03-20) were purchased from Thermo Scientific. Each siRNA at a 10 nM final concentration was transfected into iSLK/Bac16 cells using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer’s protocol. Immunoblotting was performed to verify the knockdown of the relevant protein.
Induction of lytic gene expression and virus replication and quantification of infectious virus release.
To induce KSHV lytic gene expression or virus replication, iSLK/Bac16 cells were depleted of CTCF, Rad21, or both CTCF and Rad21 simultaneously. Two days later, cells were treated with 1 μg/ml doxycycline to induce lytic replication. Cells were harvested at 48 h postinduction for RNA preparation. At 48 h postinduction, cell viability was greater than 94% by vital dye staining. For virus production, supernatants of the cells were harvested at 5 days postinduction, cleared by centrifugation twice, and filtered through a 0.80-μm-pore-size cellulose acetate filter. Serial dilutions of supernatants were used to infect 293T cells. Forty-eight hours after infection, flow cytometry was performed to measure the number of GFP-positive cells, each representing a cell infected by a GFP-expressing KSHV virion (33). Each infection was done in triplicate, and each infected cell sample was assayed by flow cytometry. Based on the dilution factor, infectious virus titers in the iSLK cell supernatant were calculated.
Immunoblot analysis.
Protein samples from lysed cells were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotted with rabbit polyclonal anti-CTCF (Millipore), anti-Rad21 (Bethyl), or anti-actin monoclonal antibody (Sigma) and horseradish peroxidase-conjugated secondary antibody (GE Healthcare), followed by visualization with the Clarity Western ECL substrate.
RNA isolation and analysis.
Total cellular RNA was isolated from washed cell pellets using Qiazol and Qiagen miRNeasy columns according to the manufacturer’s protocols. Reverse transcription-PCR (RT-PCR) was performed with Power SYBR green RNA-to-CT master mix (Applied Biosystems) according to the manufacturer’s protocol. Each sample was analyzed in triplicate with gene-specific primers, and β-actin was used as the endogenous control. The gene-specific primers were as follows: K9/vIRF1 Q1F (5′-CGGCATAGCTGTGCTTACCA-3′), K9/vIRF1 Q1R (5′-CATTGTCCCGCAACCAGACT-3′), OAS1 Q1F (5′-GCGCCCCACCAAGCTCAAGA-3′), OAS1 Q1R (5′-GCTCCCTCGCTCCCAAGCAT-3′), OAS2 Q1F (5′-ACCCGAACAGTTCCCCCTGGT-3′), OAS2 Q1R (5′-ACAAGGGTACCATCGGAGTTGCC-3′), OAS3 Q1F (5′-TGCTGCCAGCCTTTGACGCC-3′), OAS3 Q1R (5′-TCGCCCGCATTGCTGTAGCTG-3′), OASL Q1F (5′-GCGGAGCCCATCACGGTCAC-3′), OASL Q1R (5′-AGCACCACCGCAGGCCTTGA-3′), ORF6 Q1F (5′-CTGCCATAGGAGGGATGTTTG-3′), ORF6 Q1R (5′-CCATGAGCATTGCTCTGGCT-3′), ORF47 Q1F (5′-AGCCTCTACCCTGCCGTTGTTCT-3′), ORF47 Q1R (5′-ACGACCGCGACTAAAAATGACCT-3′), ORF57 Q1-5 (5′-GCAGAACAACACGGGGCGGA-3′), ORF57 Q2-3 (5′-GTCGTCGAAGCGGGGGCTCT-3′), Stat2 Q1F (5′-ATCATCCGCCATTACCAGTTGC-3′), Stat2 Q1R (5′-CGGGGGATTCGGGGATAGA-3′), β-actin Q1F (5′-TCAAGATCATTGCTCCTCCTGAG-3′), β-actin Q1R (5′-ACATCTGCTGGAAGGTGGACA-3′), IFIT1 Q1F (5′-GGAATACACAACCTACTAGCC-3′), and IFIT1 Q1R (5′-CCAGGTCACCAGACTCCTCA-3′).
ChIP assays.
A chromatin immunoprecipitation (ChIP) assay was performed as previously described (6), with some modifications. Briefly, iSLK/Bac16 cells were transfected with CTCF siRNA and/or Rad21 siRNA or negative-control siRNA at a final concentration of 10 nM. A total of 50 million iSLK cells were harvested 48 h after transfection for reciprocal ChIP. After sonication, each sample was split into two parts and immunoprecipitated with anti-CTCF antibody or anti-Rad21 antibody. Libraries were constructed from the chromatin-immunoprecipitated DNA and input samples. Single-end reads of 50 cycles were sequenced on an Illumina HiSeq2000 platform. Sequence reads were mapped to the KSHV genome (GenBank accession no. GQ994935.1). Library preparations, Illumina sequencing, and sequencing data analysis were performed by the University of Utah Huntsman Cancer Institute microarray facility.
High-throughput deep sequencing of RNA and data analysis.
RNA samples from iSLK/Bac16 cells were prepared and analyzed as previously described (6). Briefly, iSLK/Bac16 cells were transfected with a 10 nM final concentration of CTCF siRNA and/or Rad21 siRNA or negative-control siRNA (10 nM final concentration). Two days later, cells were treated with 1 μg/ml doxycycline. Cells were collected at 48 h postinduction. RNA samples were prepared using Qiagen miRNeasy kits. A total of 1.5 mg of each RNA was poly(A) selected, and libraries were prepared using the Illumina TruSeq RNA sample preparation protocol (catalog no. RS-930-2001) and validated using an Agilent bioanalyzer. RNA sequencing libraries were sequenced (50-cycle single-end reads) using an Illumina HiSeq2000 instrument.
Reference fasta files were generated by combining the standard chromosome sequences from hg19 and the KSHV sequence from NCBI accession no. NC_009333.1. Ensembl transcript annotations for hg19 were downloaded from the UCSC table browser and combined with the KSHV gene annotations listed under NCBI accession no. NC_009333.1. Gene annotations were created by merging transcripts with the same gene identifier. All possible splice junction sequences from each gene’s transcripts were generated using USeq’s MakeTranscriptome application using a radius of 46. These splice junction sequences were added to the combined hg19 and KSHV sequences and run through Novoindex (v2.8) to create the RNA-seq reference index. Reads were aligned to the transcriptome reference index described above using Novoalign (v2.08.01), allowing up to 50 alignments for each read. USeq’s SamTranscriptomeParser application was used to select the best alignment for each read and convert the coordinates of reads aligning to splices back to genomic space. Differential gene expression was measured using USeq’sDefined Region Differential Seq application. Briefly, the numbers of reads aligned to each gene annotation were calculated. The counts were then used in DESeq, which normalizes the signal and determines differential expression (46).
Bioinformatic analysis of ChIP-seq data.
(i) Peak calling. Reads were aligned to the combined human (hg19) and KSHV (GenBank accession no. GQ994935.1) genomic sequences using Novoalign (v2.08.01). Peaks were called between each pair of ChIP and input samples using MACS2. The results were then filtered to retain peaks with at least a 5-fold signal over the background.
(ii) Peak counts. Read counts for each peak were generated for the six ChIP samples using bedtools (v2.27.0) multicov. The control Si/CTCF Ab sample peak definitions were used for all CTCF ChIP samples, and the control Si/Rad21 Ab sample peak definitions were used for all Rad21 ChIP samples.
(iii) Count normalization. ChIP samples were grouped into four pairs: control Si/CTCF Ab plus CTCF Si/CTCF Ab, control Si/CTCF Ab plus Rad21 Si/CTCF Ab, control Si/Rad21 Ab plus CTCF Si/Rad21 Ab, and control Si/Rad21 Ab plus Rad21 Si/Rad21Ab. The number of mapped reads in the top shared peaks was calculated for each sample in the pair. First, shared peaks for each pair were identified using the “bedtools intersect,” requiring that at least 50% of each peak overlapped (–f 0.50 –r). Next, read counts were summed for the 10% of shared peaks with the highest combined count across the pair. Reads were normalized by dividing the raw counts by the number of mapped reads in the top shared peaks and then multiplying by 1 million.
(iv) CTCF/Rad21 intersection. Shared CTCF and Rad21 peaks were identified using the bedtools intersect command on the control Si/CTCF Ab and control Si/Rad21 Ab peak definitions, requiring that at least 50% of each peak overlapped.
(v) KD peak calling. A normalized peak count ratio was calculated for each pair: control Si/CTCF Ab plus CTCF Si/CTCF Ab, control Si/CTCF Ab plus Rad21 Si/CTCF Ab, and control Si/Rad21 Ab plus CTCF Si Rad21 Ab. If the CTCF Si/Rad21 Si normalized peak count was less than 50% of the control Si peak count, the peak was considered absent in the knockdown. Each peak was then categorized based on the presence or absence of peaks after knockdown in the four pairs.
Data availability.
The data discussed in this publication have been deposited in the NCBI Gene Expression Omnibus (GEO) (47) and are accessible through GEO accession no. GSE138105 and GSE138937.
ACKNOWLEDGMENTS
S.S. was supported by grants 1I01BX002262 from HS R&D, Veterans Health Affairs (Merit Review), and R01CA81133 from the National Cancer Institute, National Institutes of Health. Flow Cytometry was supported by the HSC core, and bioinformatics support was provided by the Huntsman Cancer Center Bioinformatics Core at the University of Utah School of Medicine.
REFERENCES
- 1.Ganem D. 2007. Kaposi’s sarcoma-associated herpesvirus, p 2847–2888. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (ed), Fields virology, 5th ed, vol 2 Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Cesarman E, Damania B, Krown SE, Martin J, Bower M, Whitby D. 2019. Kaposi sarcoma. Nat Rev Dis Primers 5:9. doi: 10.1038/s41572-019-0060-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen HS, Wikramasinghe P, Showe L, Lieberman PM. 2012. Cohesins repress Kaposi’s sarcoma-associated herpesvirus immediate early gene transcription during latency. J Virol 86:9454–9464. doi: 10.1128/JVI.00787-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kang H, Lieberman PM. 2009. Cell cycle control of Kaposi’s sarcoma-associated herpesvirus latency transcription by CTCF-cohesin interactions. J Virol 83:6199–6210. doi: 10.1128/JVI.00052-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kang H, Wiedmer A, Yuan Y, Robertson E, Lieberman PM. 2011. Coordination of KSHV latent and lytic gene control by CTCF-cohesin mediated chromosome conformation. PLoS Pathog 7:e1002140. doi: 10.1371/journal.ppat.1002140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li DJ, Verma D, Mosbruger T, Swaminathan S. 2014. CTCF and Rad21 act as host cell restriction factors for Kaposi’s sarcoma-associated herpesvirus (KSHV) lytic replication by modulating viral gene transcription. PLoS Pathog 10:e1003880. doi: 10.1371/journal.ppat.1003880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stedman W, Kang H, Lin S, Kissil JL, Bartolomei MS, Lieberman PM. 2008. Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. EMBO J 27:654–666. doi: 10.1038/emboj.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Toth Z, Smindak RJ, Papp B. 2017. Inhibition of the lytic cycle of Kaposi’s sarcoma-associated herpesvirus by cohesin factors following de novo infection. Virology 512:25–33. doi: 10.1016/j.virol.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kang H, Lieberman PM. 2011. Mechanism of glycyrrhizic acid inhibition of Kaposi’s sarcoma-associated herpesvirus: disruption of CTCF-cohesin-mediated RNA polymerase II pausing and sister chromatid cohesion. J Virol 85:11159–11169. doi: 10.1128/JVI.00720-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martinez FP, Cruz R, Lu F, Plasschaert R, Deng Z, Rivera-Molina YA, Bartolomei MS, Lieberman PM, Tang Q. 2014. CTCF binding to the first intron of the major immediate early (MIE) gene of human cytomegalovirus (HCMV) negatively regulates MIE gene expression and HCMV replication. J Virol 88:7389–7401. doi: 10.1128/JVI.00845-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lang F, Li X, Vladimirova O, Hu B, Chen G, Xiao Y, Singh V, Lu D, Li L, Han H, Wickramasinghe JM, Smith ST, Zheng C, Li Q, Lieberman PM, Fraser NW, Zhou J. 2017. CTCF interacts with the lytic HSV-1 genome to promote viral transcription. Sci Rep 7:39861. doi: 10.1038/srep39861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee JS, Raja P, Pan D, Pesola JM, Coen DM, Knipe DM. 2018. CCCTC-binding factor acts as a heterochromatin barrier on herpes simplex viral latent chromatin and contributes to poised latent infection. mBio 9:e02372-17. doi: 10.1128/mBio.02372-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Washington SD, Musarrat F, Ertel MK, Backes GL, Neumann DM. 2018. CTCF binding sites in the herpes simplex virus 1 genome display site-specific CTCF occupation, protein recruitment, and insulator function. J Virol 92:e00156-18. doi: 10.1128/JVI.00156-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tempera I, Wiedmer A, Dheekollu J, Lieberman PM. 2010. CTCF prevents the epigenetic drift of EBV latency promoter Qp. PLoS Pathog 6:e1001048. doi: 10.1371/journal.ppat.1001048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chau CM, Zhang XY, McMahon SB, Lieberman PM. 2006. Regulation of Epstein-Barr virus latency type by the chromatin boundary factor CTCF. J Virol 80:5723–5732. doi: 10.1128/JVI.00025-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sumara I, Vorlaufer E, Gieffers C, Peters BH, Peters J-M. 2000. Characterization of vertebrate cohesin complexes and their regulation in prophase. J Cell Biol 151:749–762. doi: 10.1083/jcb.151.4.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Merkenschlager M, Nora EP. 2016. CTCF and cohesin in genome folding and transcriptional gene regulation. Annu Rev Genomics Hum Genet 17:17–43. doi: 10.1146/annurev-genom-083115-022339. [DOI] [PubMed] [Google Scholar]
- 18.Deardorff MA, Noon SE, Krantz ID. 1993. Cornelia de Lange syndrome, p 1993–2019. In Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Stephens K, Amemiya A (ed), GeneReviews. University of Washington, Seattle, WA. [Google Scholar]
- 19.Gligoris T, Lowe J. 2016. Structural insights into ring formation of cohesin and related Smc complexes. Trends Cell Biol 26:680–693. doi: 10.1016/j.tcb.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Makrantoni V, Marston AL. 2018. Cohesin and chromosome segregation. Curr Biol 28:R688–R693. doi: 10.1016/j.cub.2018.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wendt KS, Peters JM. 2009. How cohesin and CTCF cooperate in regulating gene expression. Chromosome Res 17:201–214. doi: 10.1007/s10577-008-9017-7. [DOI] [PubMed] [Google Scholar]
- 22.Wutz G, Varnai C, Nagasaka K, Cisneros DA, Stocsits RR, Tang W, Schoenfelder S, Jessberger G, Muhar M, Hossain MJ, Walther N, Koch B, Kueblbeck M, Ellenberg J, Zuber J, Fraser P, Peters JM. 2017. Topologically associating domains and chromatin loops depend on cohesin and are regulated by CTCF, WAPL, and PDS5 proteins. EMBO J 36:3573–3599. doi: 10.15252/embj.201798004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dorsett D, Merkenschlager M. 2013. Cohesin at active genes: a unifying theme for cohesin and gene expression from model organisms to humans. Curr Opin Cell Biol 25:327–333. doi: 10.1016/j.ceb.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, Yahata K, Imamoto F, Aburatani H, Nakao M, Imamoto N, Maeshima K, Shirahige K, Peters JM. 2008. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature 451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 25.Fay A, Misulovin Z, Li J, Schaaf CA, Gause M, Gilmour DS, Dorsett D. 2011. Cohesin selectively binds and regulates genes with paused RNA polymerase. Curr Biol 21:1624–1634. doi: 10.1016/j.cub.2011.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, Jarmuz A, Canzonetta C, Webster Z, Nesterova T, Cobb BS, Yokomori K, Dillon N, Aragon L, Fisher AG, Merkenschlager M. 2008. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell 132:422–433. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 27.Busslinger GA, Stocsits RR, van der Lelij P, Axelsson E, Tedeschi A, Galjart N, Peters JM. 2017. Cohesin is positioned in mammalian genomes by transcription, CTCF and Wapl. Nature 544:503–507. doi: 10.1038/nature22063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rubio ED, Reiss DJ, Welcsh PL, Disteche CM, Filippova GN, Baliga NS, Aebersold R, Ranish JA, Krumm A. 2008. CTCF physically links cohesin to chromatin. Proc Natl Acad Sci U S A 105:8309–8314. doi: 10.1073/pnas.0801273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zuin J, Dixon JR, van der Reijden MI, Ye Z, Kolovos P, Brouwer RW, van de Corput MP, van de Werken HJ, Knoch TA, van IJcken WF, Grosveld FG, Ren B, Wendt KS. 2014. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc Natl Acad Sci U S A 111:996–1001. doi: 10.1073/pnas.1317788111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nasmyth K, Haering CH. 2009. Cohesin: its roles and mechanisms. Annu Rev Genet 43:525–558. doi: 10.1146/annurev-genet-102108-134233. [DOI] [PubMed] [Google Scholar]
- 31.Myoung J, Ganem D. 2011. Generation of a doxycycline-inducible KSHV producer cell line of endothelial origin: maintenance of tight latency with efficient reactivation upon induction. J Virol Methods 174:12–21. doi: 10.1016/j.jviromet.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brulois KF, Chang H, Lee AS, Ensser A, Wong LY, Toth Z, Lee SH, Lee HR, Myoung J, Ganem D, Oh TK, Kim JF, Gao SJ, Jung JU. 2012. Construction and manipulation of a new Kaposi’s sarcoma-associated herpesvirus bacterial artificial chromosome clone. J Virol 86:9708–9720. doi: 10.1128/JVI.01019-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Verma D, Li DJ, Krueger B, Renne R, Swaminathan S. 2015. Identification of the physiological gene targets of the essential lytic replicative Kaposi’s sarcoma-associated herpesvirus ORF57 protein. J Virol 89:1688–1702. doi: 10.1128/JVI.02663-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Golan-Mashiach M, Grunspan M, Emmanuel R, Gibbs-Bar L, Dikstein R, Shapiro E. 2012. Identification of CTCF as a master regulator of the clustered protocadherin genes. Nucleic Acids Res 40:3378–3391. doi: 10.1093/nar/gkr1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo Y, Monahan K, Wu H, Gertz J, Varley KE, Li W, Myers RM, Maniatis T, Wu Q. 2012. CTCF/cohesin-mediated DNA looping is required for protocadherin alpha promoter choice. Proc Natl Acad Sci U S A 109:21081–21086. doi: 10.1073/pnas.1219280110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Phillips JE, Corces VG. 2009. CTCF: master weaver of the genome. Cell 137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cho DH, Thienes CP, Mahoney SE, Analau E, Filippova GN, Tapscott SJ. 2005. Antisense transcription and heterochromatin at the DM1 CTG repeats are constrained by CTCF. Mol Cell 20:483–489. doi: 10.1016/j.molcel.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 38.Filippova GN, Thienes CP, Penn BH, Cho DH, Hu YJ, Moore JM, Klesert TR, Lobanenkov VV, Tapscott SJ. 2001. CTCF-binding sites flank CTG/CAG repeats and form a methylation-sensitive insulator at the DM1 locus. Nat Genet 28:335–343. doi: 10.1038/ng570. [DOI] [PubMed] [Google Scholar]
- 39.Pena-Hernandez R, Marques M, Hilmi K, Zhao T, Saad A, Alaoui-Jamali MA, del Rincon SV, Ashworth T, Roy AL, Emerson BM, Witcher M. 2015. Genome-wide targeting of the epigenetic regulatory protein CTCF to gene promoters by the transcription factor TFII-I. Proc Natl Acad Sci U S A 112:E677–E686. doi: 10.1073/pnas.1416674112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chernukhin I, Shamsuddin S, Kang SY, Bergstrom R, Kwon YW, Yu W, Whitehead J, Mukhopadhyay R, Docquier F, Farrar D, Morrison I, Vigneron M, Wu SY, Chiang CM, Loukinov D, Lobanenkov V, Ohlsson R, Klenova E. 2007. CTCF interacts with and recruits the largest subunit of RNA polymerase II to CTCF target sites genome-wide. Mol Cell Biol 27:1631–1648. doi: 10.1128/MCB.01993-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kang H, Cho H, Sung GH, Lieberman PM. 2013. CTCF regulates Kaposi’s sarcoma-associated herpesvirus latency transcription by nucleosome displacement and RNA polymerase programming. J Virol 87:1789–1799. doi: 10.1128/JVI.02283-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmidt D, Schwalie PC, Ross-Innes CS, Hurtado A, Brown GD, Carroll JS, Flicek P, Odom DT. 2010. A CTCF-independent role for cohesin in tissue-specific transcription. Genome Res 20:578–588. doi: 10.1101/gr.100479.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yokota S, Hirayama T, Hirano K, Kaneko R, Toyoda S, Kawamura Y, Hirabayashi M, Hirabayashi T, Yagi T. 2011. Identification of the cluster control region for the protocadherin-beta genes located beyond the protocadherin-gamma cluster. J Biol Chem 286:31885–31895. doi: 10.1074/jbc.M111.245605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hirayama T, Tarusawa E, Yoshimura Y, Galjart N, Yagi T. 2012. CTCF is required for neural development and stochastic expression of clustered Pcdh genes in neurons. Cell Rep 2:345–357. doi: 10.1016/j.celrep.2012.06.014. [DOI] [PubMed] [Google Scholar]
- 45.Li D, Swaminathan S. 2019. Human IFIT proteins inhibit lytic replication of KSHV: a new feed-forward loop in the innate immune system. PLoS Pathog 15:e1007609. doi: 10.1371/journal.ppat.1007609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol 11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Edgar R, Domrachev M, Lash AE. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data discussed in this publication have been deposited in the NCBI Gene Expression Omnibus (GEO) (47) and are accessible through GEO accession no. GSE138105 and GSE138937.