Monoclonal antibodies have been isolated against conserved epitopes on the HIV trimer and are being investigated for passive immunization. Some of the challenges associated with full-sized antibody proteins may be overcome by using single-chain variable fragments (scFv). These smaller forms of antibodies can be produced more efficiently, may show fewer off-target effects with increased tissue penetration, and are more adaptable to vectored-mediated expression than IgG. Here, we demonstrate that scFv of four HIV-directed bNAbs (CAP256-VRC26.25, PGT121, 3BNC117, and 10E8v4) had significant neutralizing activity against diverse global strains of HIV. Loss of potency and/or breadth was shown to be due to increased dependence of the scFv on key residues within the epitope. These smaller antibody molecules with functional activity in the therapeutic range may be suitable for further development as passive immunity for HIV prevention.
Keywords: HIV, broadly neutralizing antibodies, scFv, epitope mapping, passive immunization
ABSTRACT
Passive administration of HIV-directed broadly neutralizing antibodies (bNAbs) can prevent infection in animal models, and human efficacy trials are under way. Single-chain variable fragments (scFv), comprised of only the variable regions of antibody heavy and light chains, are smaller molecules that may offer advantages over full-length IgG. We designed and expressed scFv of HIV bNAbs prioritized for clinical testing that target the V2-apex (CAP256-VRC26.25), V3-glycan supersite (PGT121), CD4 binding site (3BNC117), and MPER (10E8v4). The use of either a 15- or 18-amino-acid glycine-serine linker between the heavy- and light-chain fragments provided adequate levels of scFv expression. When tested against a 45-multisubtype virus panel, all four scFv retained good neutralizing activity, although there was variable loss of function compared to the parental IgG antibodies. For CAP256-VRC26.25, there was a significant 138-fold loss of potency that was in part related to differential interaction with charged amino acids at positions 169 and 170 in the V2 epitope. Potency was reduced for the 3BNC117 (13-fold) and PGT121 (4-fold) scFv among viruses lacking the N276 and N332 glycans, respectively, and in viruses with a longer V1 loop for PGT121. This suggested that scFv interacted with their epitopes in subtly different ways, with variation at key residues affecting scFv neutralization more than the matched IgGs. Remarkably, the scFv of 10E8v4 maintained breadth of 100% with only a minor reduction in potency. Overall, scFv of clinically relevant bNAbs had significant neutralizing activity, indicating that they are suitable for passive immunization to prevent HIV-1 infection.
IMPORTANCE Monoclonal antibodies have been isolated against conserved epitopes on the HIV trimer and are being investigated for passive immunization. Some of the challenges associated with full-sized antibody proteins may be overcome by using single-chain variable fragments (scFv). These smaller forms of antibodies can be produced more efficiently, may show fewer off-target effects with increased tissue penetration, and are more adaptable to vectored-mediated expression than IgG. Here, we demonstrate that scFv of four HIV-directed bNAbs (CAP256-VRC26.25, PGT121, 3BNC117, and 10E8v4) had significant neutralizing activity against diverse global strains of HIV. Loss of potency and/or breadth was shown to be due to increased dependence of the scFv on key residues within the epitope. These smaller antibody molecules with functional activity in the therapeutic range may be suitable for further development as passive immunity for HIV prevention.
INTRODUCTION
Broadly neutralizing antibodies (bNAbs) block infection of diverse HIV strains by targeting relatively conserved epitopes on the HIV envelope trimer (1–3). An HIV vaccine will likely need to elicit these types of antibodies, but this has not yet been achieved. However, some HIV-infected individuals naturally generate bNAb responses, enabling the isolation of monoclonal antibodies with high potency and exceptional breadth and creating the possibility of passive immunization for HIV prevention (4, 5). Animal studies support this approach, with bNAbs such as CAP256-VRC26.25, PGT121, 3BNC117, 10-1074, PGDM1400, VRC01, and VRC07 shown to prevent simian-human immunodeficiency virus infection in nonhuman primate models (6–9). In addition, passively infused bNAbs 3BNC117, 10-1074, and VRC01 substantially reduce viral levels in viremic individuals, confirming their neutralization capacity in vivo (10–15). This has led to the antibody-mediated prevention (AMP) trial, a proof-of-concept study in Africa, Switzerland, and the Americas that is under way to assess whether the bNAb VRC01 can prevent HIV infection in humans (https://ampstudy.org.za/).
The promise of passive immunization opens up possibilities for using bNAbs in other formats. Small antibody fragments such as single-chain variable fragments (scFv) may offer advantages over IgG (16–18). scFv consist of the antibody variable heavy and variable light chains and are substantially smaller than IgG molecules (30 kDa versus approximately 147 kDa) (19). This may enhance some of their pharmacokinetic properties and improve their distribution throughout the body and absorption into various tissues. In the context of HIV, the smaller size may enhance diffusion into genital tract mucosal tissues, where most HIV infections occur (5, 16, 20–22). Moreover, as scFv do not have an Fc region, they are less likely to display nonspecific cytotoxicity through cytokine release, although this reduces their half-life significantly and removes their ability to perform Fc-mediated effector functions (16, 22). Antibody scFv have been successfully used in clinical settings particularly for cancer therapy, where their enhanced tissue penetration aids in tumor targeting, and are also being developed for neurological diseases, such as Parkinson’s disease, although some anti-drug antibody responses were noted (16, 23, 24).
Antibody scFv targeting the HIV envelope have previously been isolated through phage display, but their neutralization capacity was limited (25, 26). Initial attempts to engineer scFv, immunoadhesins, or other antibody fragments from bNAbs VRC01, PG9, and PG16 showed poor expression and loss of function compared to the IgG (18). The availability of a larger number of broad and potent bNAbs has revived interest in this area. We therefore made scFvs of bNAbs that target 4 distinct epitopes on the HIV envelope trimer, focusing on those undergoing human clinical testing. These included CAP256-VRC26.25 (which binds the V2 apex and is referred to here as CAP256.25) (28), PGT121 (V3-glycan supersite) (29), 3BNC117 (CD4 binding site [CD4bs]) (30), and 10E8v4 (MPER) (31). We found that a reduction in neutralization breadth and/or potency of the scFv compared to the IgG was due to differences in dependence on specific residues in the epitopes. However, overall, scFv largely retained breadth and potency against diverse global HIV isolates and may prove to be useful for future clinical development to prevent HIV acquisition.
RESULTS
Construction and optimization of scFv derived from HIV bNAbs.
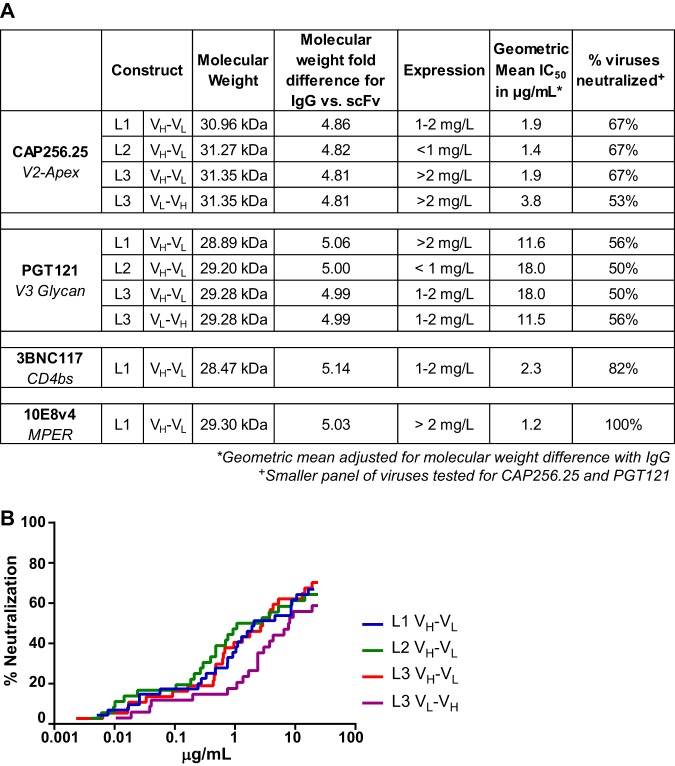
We designed scFv of bNAbs that target four major epitopes on the HIV-1 envelope (CAP256.25, PGT121, 3BNC117, and 10E8v4) to determine if they retain neutralization breadth and potency. These scFv consisted of the variable heavy chain and the variable light chain of the parent IgG connected by a glycine-serine linker with a histidine tag for purification (amino acid sequences of each construct used to generate scFv are shown in Table 1). As linker length can be a major determinant of paratope formation, we tested linkers of three different lengths. Heavy (VH)- and light (VL)-chain orientation was also tested for one of the linkers. CAP256.25 and PGT121 were used to first assess expression and function of these different constructs (Fig. 1A). For CAP256.25 scFv, the 18-amino-acid linker (termed L3) in both VH-VL and VL-VH orientations had the highest expression. The 15-amino-acid (GGGGS)3 linker (L1) showed lower expression levels, and the 20-amino-acid (GGGGS)4 linker (L2) was poorly expressed. For PGT121, the highest expression was observed for L1 in the VH-VL orientation. We next tested whether the different linkers affected neutralization breadth and potency using panels of sensitive viruses tailored to each bNAb. Since scFv are roughly 5 times smaller than IgG, the 50% inhibitory concentration (IC50) values were multiplied by the molecular weight fold difference between each scFv and its matched IgG (values shown in Fig. 1A and explained in Materials and Methods).
TABLE 1.
Amino acid sequences of four scFv constructs derived from HIV bNAbsa
The variable heavy chain (light blue) and variable light chain (yellow) are indicated, as well as the glycine-serine linker (purple) and the 8× histidine tag (red), which is preceded by an HRV3C site (italicized) and is separated from the light chain by a small GG spacer.
FIG 1.
Design, expression, and testing of scFv constructs of HIV bNAbs. (A) A total of 10 scFv constructs were made, with different glycine-serine linkers for CAP256.25 and PGT121 and L1 (GGGGS)3, L2 (GGGGS)4, and L3 (GGSSRSSSSGGGGSGGGG) for VH-VL and L3 for VL-VH. For 3BNC117 and 10E8v4, only L1 VH-VL was made. The molecular weights and protein expression levels for each scFv are shown together with the adjusted geometric mean neutralization potency. A selected panel of viruses was used to assess CAP256.25 scFv (34 viruses) and PGT121 scFv (18 viruses), whereas the full 45-virus panel is shown for 3BNC117 and 10E8v4. (B) Breadth-potency curves of CAP256.25 scFv made with 3 different linkers in the VH-VL orientation and for L3 in the VL-VH orientation. (scFv is corrected for molecular weight size difference by multiplying the IC50 by the fold difference in molecular weight between the IgG and the scFv, a value of ∼5×).
For CAP256.25, all three linkers in the VH-VL orientation showed similar neutralizing activity against a panel of 36 viruses, while the VL-VH L3 (purple) had a 2-fold lower geometric mean potency (Fig. 1B). We therefore used CAP256.25 scFv VH-VL L3, which showed the highest expression levels, to conduct further experiments. Similarly, no significant differences were observed for the different PGT121 scFv constructs against an 18-virus panel (Fig. 1A), and PGT121 VH-VL L1, which showed the highest expression levels, was used. Based on this, 3BNC117 and 10E8v4 scFv were expressed in the VH-VL conformation with the L1 linker. Both expressed relatively well and showed good neutralizing activity (Fig. 1A).
HIV bNAb scFv retain good neutralization breadth and potency.
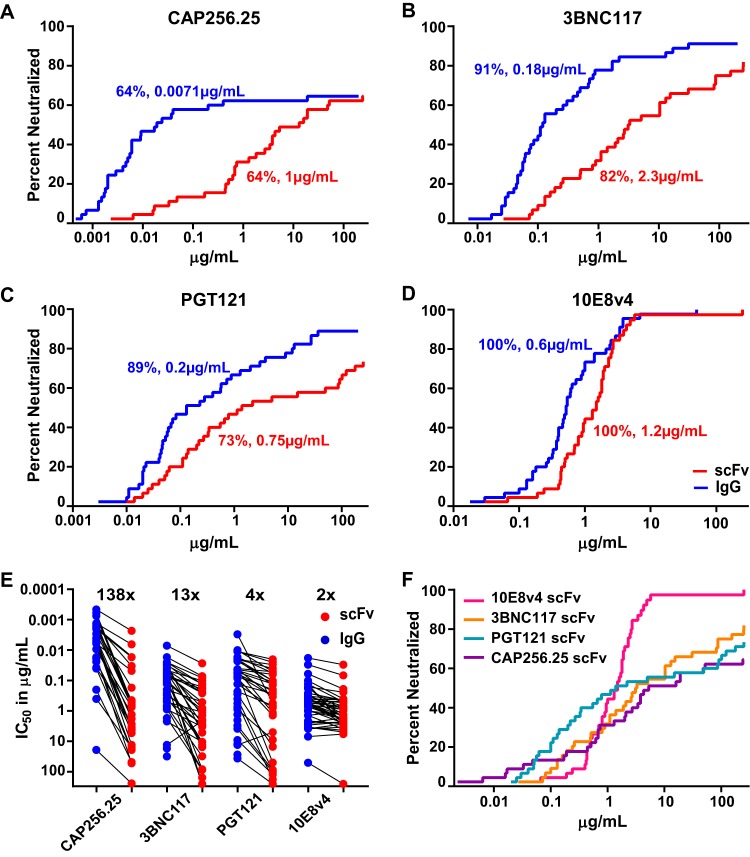
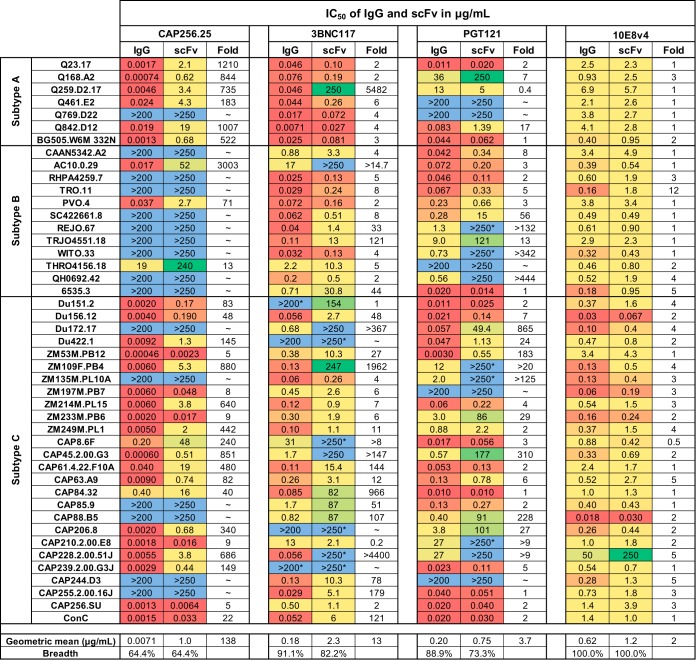
All four scFv were tested for neutralizing activity against a larger multiclade panel of 45 viruses for breadth and potency. Compared to their parental IgG proteins, all scFv retained substantial neutralizing activity, although they showed some reduction in breadth and/or potency. CAP256.25 scFv maintained breadth of 64% equivalent to the full IgG, but there was a significant (P < 0.0001) 138-fold decrease in geometric mean potency compared to that of IgG (from 0.0071 to 1 μg/ml) (Fig. 2A). While all viruses in the panel were less potently neutralized by CAP256.25 scFv, some viruses were more severely affected, with >1,000-fold loss of potency for Q23.17, Q842.D12, and AC10.0.29 (Fig. 3).
FIG 2.
Neutralization breadth and potency of scFv compared to those of IgG for 4 HIV bNAbs. (A to D) Breadth-potency curves of scFv (red) and IgG (blue) of CAP256.25 (A), 3BNC117 (B), PGT121 (C), and 10E8v4 (D) using data from a 45-virus multisubtype panel tested in the TZM-bl neutralization assay. The IC50 of viruses against the percentage of viruses neutralized is given. Percent neutralization and geometric mean IC50 potency, in micrograms per milliliter (corrected for molecular weight size difference by multiplying their IC50 by the fold difference in molecular weight between IgG and scFv, ∼5×), are shown in each plot. (E) Potency losses of scFv (red) versus IgG (blue) against the same panel. Viruses resistant to both IgG and scFv were excluded. (F) Comparison of breadth-potency curves for all 4 scFv bNAbs against the 45-virus panel.
FIG 3.
Neutralization data for IgG and scFv of HIV bNAbs tested against the 45-virus panel. The IC50 values for both IgG and scFv against 7 subtype A, 12 subtype B, and 26 subtype C viruses are shown in micrograms per milliliter. IgG was tested at a starting concentration of 50 μg/ml, whereas scFv started at 250 μg/ml. Fold changes between IgG and scFv are shown. The colors indicate the potency at which the viruses were neutralized, with the warmer colors indicating higher potency. Resistant viruses are colored in light blue. Maximum levels of neutralization for scFv and IgG of <50% but >20% are indicated with an asterisk. Geometric mean titers, fold difference from IgG, and breadth against the 45-virus panel are shown at the bottom of the table. (scFv titers were corrected for molecular weight size difference by multiplying their IC50 by the fold difference in molecular weight between the IgG and the scFv, as noted in Fig. 1A).
The scFv of 3BNC117 and PGT121 showed an overall loss in both potency and breadth of neutralization. For 3BNC117, the scFv was 13-fold less potent than the IgG (P < 0.0001) and had slightly lower breadth of 82% compared to 91% for the IgG (Fig. 2B). PGT121 scFv had 4-fold reduced potency (P < 0.0001) and breadth of 73% compared to 89% for the IgG (Fig. 2C). This was a result of the scFv losing activity against viruses such as REJO.67 and QH0692.42 (Fig. 3). However, even in these cases, the scFv showed low levels of maximum neutralization (>20% but <50%) at the highest concentration tested, indicating that the scFv still bound to resistant viruses (highlighted in Fig. 3 with an asterisk). Despite the significant potency losses for some viruses, there were a number of viruses that were neutralized equally well by the scFv and by the IgG of PGT121 and 3BNC117, a phenomenon that was not seen with CAP256.25 scFv (Fig. 3).
The MPER antibody 10E8v4, when expressed as an scFv, showed no loss of function. It maintained 100% breadth of the panel with a minor 2-fold reduction in geometric mean potency compared to the IgG version (Fig. 2D). A significant reduction in potency was noted for only one virus, TRO.11, although it was still neutralized (Fig. 3).
Comparison of the potency of all four scFv relative to their IgG clearly demonstrates the significant loss of function for CAP256.25 scFv (Fig. 2E). Nonetheless, the breadth-potency curve of CAP256.25 scFv still overlapped that of the other scFv (Fig. 2F), a consequence of the exceptional potency of the parental CAP256.25 IgG. The superior neutralization breadth of the 10E8v4 scFv was also evident in these breadth-potency curves. Overall, the median IC50 for all 4 scFv were in the range of 1 to 2 μg/ml, which is close to the therapeutic range for VRC01 that is currently undergoing human efficacy testing.
Loss of potency in CAP256.25 scFv linked to epitope charge.
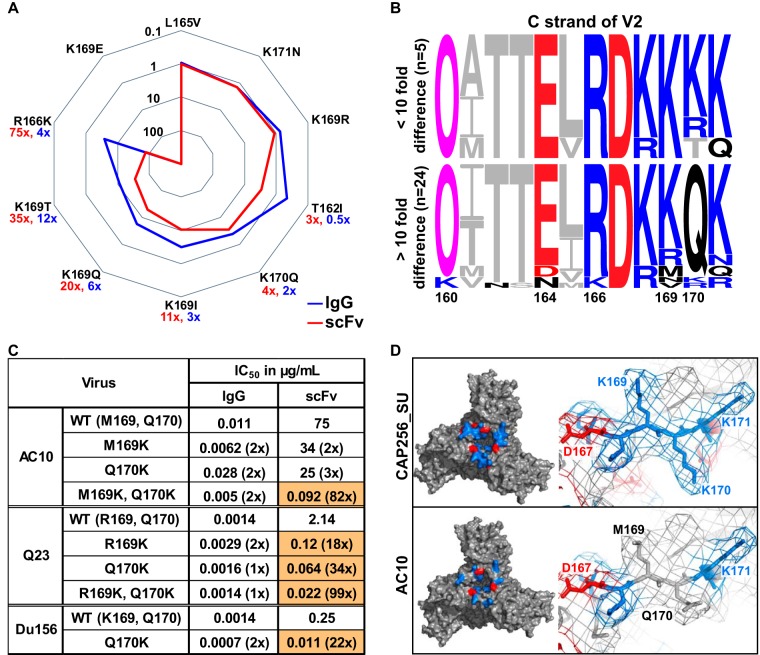
In order to probe the loss of potency for CAP256.25 scFv, we tested the effect of point mutations in the V2 epitope of the autologous virus CAP256_SU. Mutations L165V and K171N had no effect on either the scFv or IgG (Fig. 4A). Similarly, a conservative K169R mutation did not impact the neutralizing activity of either protein. The T162I mutation (which removes the conserved glycan at position 160) and the K170Q mutations had a small effect on the scFv (3- and 4-fold, respectively) but not on the IgG (below 3-fold). However, the scFv was more affected by the K169I substitution (11-fold for the scFv versus 3-fold for the IgG), the K169Q (20- versus 6-fold), the K169T (35- versus 12-fold), and the R166K (75- versus 4-fold) mutations. For both scFv and IgG, a K169E mutation resulted in complete neutralization resistance. Therefore, while neutralization by the scFv and IgG were overall similarly influenced by mutations in the epitope, the consequence of individual mutations was more pronounced for the scFv.
FIG 4.
Charged amino acids in V2 reduced CAP256.25 scFv neutralization. (A) Spiderplot showing fold change in neutralization of V2 mutants relative to CAP256_SU wild type (WT) by the CAP256.25 IgG (blue) and the scFv (red). A fold change of 1 indicates no difference from the WT virus. Values closer to the center indicate resistance, whereas those on the outer bounds indicate a sensitivity. (B) Amino acid sequence alignment of viruses neutralized by scFv and IgG with similar potency (top) or more than 10-fold decrease in potency (bottom). The N160 glycan is denoted by an O and is colored magenta. Negatively and positively charged residues are indicated by red and blue, respectively. (C) IgG and scFv neutralization (IC50) of three viruses with point mutations at positions 169 and 170. Fold changes relative to the wild type are indicated in parentheses, and those greater than 5-fold are colored orange. scFv is corrected for the molecular weight size difference (multiplied ∼5×). (D) Structural representation of the V2 apex of the HIV envelope trimer of a CAP256.25 scFv-sensitive virus (CAP256_SU) and a less sensitive virus (AC10.0.29). The apex of the trimer is shown on the left, and a close-up of positions 167 to 171 is shown on the right. Positive charges are colored blue, whereas negative charges are colored red. (Model is based on 5V8L structure of BG505 SOSIP.664 [78] and was modified using PyMOL software.)
To further explore this, we partitioned CAP256.25-sensitive viruses into those with a greater than 10-fold potency loss for the scFv compared to that of the IgG (24 viruses) versus those with less than a 10-fold change (5 viruses) (Fig. 4B; glycans, N(X)T/S, are indicated by a pink O in the logograms). Differences at residue 160 or 166 between these two groups did not explain the general loss of potency by the scFv. Some variation was noted at position 169, with 169M (2/24), 169N (1/24), 169V (1/24), and 169R (3/24) found in viruses where the scFv lost potency. However, in 21/24 viruses with a large potency loss, an uncharged glutamine (Q) was present at position 170 rather than a charged lysine (K) or arginine (R). We therefore introduced Q170K into three viruses (AC10.0.29, Q23.17, and Du156.12) along with 169K in AC10.0.29 (M169K) and Q23.17 (R169K). Individual 169K and 170K mutations significantly improved neutralization by the scFv in both Du156.12 and Q23.17 (but not AC10.0.29) but had no significant effect on the IgG (Fig. 4C). However, the combination of these mutations significantly improved scFv neutralization in both Q23.17 (70-fold) and AC10.0.29 (86-fold) compared to a minimal effect (2- and 3-fold, respectively) for the IgG. These data suggested that CAP256.25 scFv neutralization was more dependent on the charge of the epitope than the matched IgG.
To visualize the effect of charge in the V2 apex of different envelopes, models of the HIV Env of CAP256_SU and AC10.0.29 were generated (Fig. 4D). CAP256_SU, which is similarly neutralized by both scFv and IgG, has charged lysine residues at positions 169 and 170 (Fig. 4D). These 169K and 170K residues formed a cluster of positive charges at the apex of the trimer. In contrast, AC10 displayed a reduced charge in the epitope, with neutral amino acids occupying these positions (M169 and Q170), possibly explaining the observed virus-specific loss of potency.
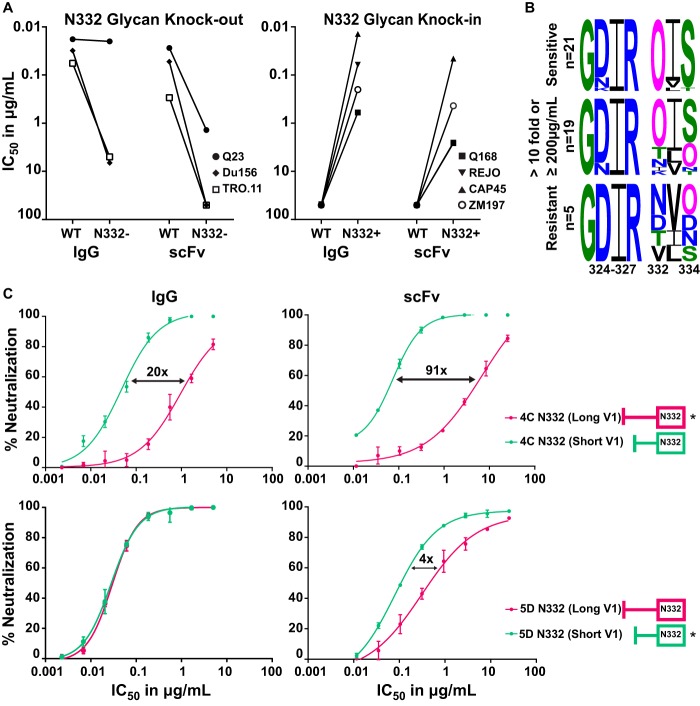
PGT121 scFv neutralization affected by both the N332 glycan and V1 length.
The glycan at position N332 is a key part of the epitope of V3-directed bNAbs, such as PGT121 (29, 32). To determine the dependence of the scFv on the N332 glycan, we deleted this glycan in three viruses (Q23.17, Du156.12, and TRO.11) through an N332A mutation. There was a significant loss of scFv potency for all 3 viruses, while for the IgG, this effect was seen in 2 of the 3 viruses (Fig. 5A, left). We also introduced this glycan into resistant viruses Q168.A2, REJO.67, CAP45.2.00.G3, and ZM197M.PB7, which conferred neutralization sensitivity to both the scFv and the IgG of PGT121 (Fig. 5A, right), although not always to the same level as the IgG.
FIG 5.
PGT121 scFv neutralization was dependent on the N332 glycan and the V1 loop length. (A) IC50 of IgG and scFv against 3 sensitive viruses (WT) with their N332 glycan knockouts (N332−) (left) and 4 resistant viruses (WT) with matched N332 glycan knockins (N332+) (right). (B) Noncontiguous amino acid sequences of the V3 region (positions 324 to 327 and 332 to 334, encompassing the GDIR motif and the N332/4 glycans). Viruses are divided into three groups: viruses neutralized equally well by IgG and scFv of PGT121 (top), those with >10-fold difference and/or ≥200 μg/ml for scFv (middle), and viruses resistant to both scFv and IgG (bottom). Glycans [N(X)T/S] are shown as a pink O. (C) Neutralization curves of IgG and scFv (in micrograms per milliliter) against two CAP177 clones, 4C, which has a long V1 (dark pink, star), and 5D, with a short V1 (green, star), and their V1 mutant swaps (no stars). Fold differences between the wild-type and mutant swaps are indicated. scFv is corrected for the molecular weight size difference (multiplied ~5 times).
We next analyzed the sequences of the 45 viruses tested, dividing them into 3 groups: viruses equally sensitive to the IgG and scFv (Fig. 5B, upper, 21 viruses), viruses with a 10-fold-reduced sensitivity to the scFv and/or IC50 of ≥200 μg/ml for the scFv (Fig. 5B, middle, 19 viruses), and viruses resistant to both the IgG and the scFv (Fig. 5B, lower, 5 viruses). There was no difference in the GDIR motif between the groups. As expected, 21/21 viruses sensitive to PGT121 had a glycan at position N332 (indicated by a pink O in the logograms). In contrast, viruses resistant to both scFv and IgG lacked the N332 glycan, with 2 viruses showing a glycan shift to N334 and 3 lacking either glycan. In viruses where the scFv lost potency (Fig. 5B, middle), only 7 out of 19 had either a glycan shift to N334 or lacked a glycan at N332/N334. This suggested that while both the PGT121 IgG and scFv were dependent on the N332 glycan, the reduced potency and breadth of the scFv was only partly due to an increased dependence on this glycan.
V3-directed bNAbs are also affected by the length of the V1 region (33, 34), so we assessed whether this contributed to the loss of scFv neutralization. We used two related viruses isolated from the same individual, donor CAP177, that showed differential sensitivity to PGT121. The CAP177-4C virus contains a long V1 loop (dark pink) (Fig. 5C, upper), while CAP177-5D has a short V1 loop (green) (Fig. 5C, lower). V1 swaps between these 2 clones were made and tested for sensitivity to both scFv and IgG. Inserting a short V1 in CAP177-4C (Fig. 5C, upper) increased the potency of PGT121 IgG by 20-fold (from 0.94 μg/ml to 0.047 μg/ml) but had a substantially greater 91-fold effect on the scFv (from 6.7 μg/ml to 0.074 μg/ml). Similarly, there was a 4-fold loss in potency for the scFv when a long V1 was introduced into CAP177-5D (from 0.080 μg/ml to 0.31 μg/ml), with no effect on the IgG (Fig. 5C, lower). The length of V1, therefore, had a greater effect on the neutralization potency of the scFv than the IgG of PGT121.
Loss of potency for the 3BNC117 scFv more pronounced for subtype C viruses.
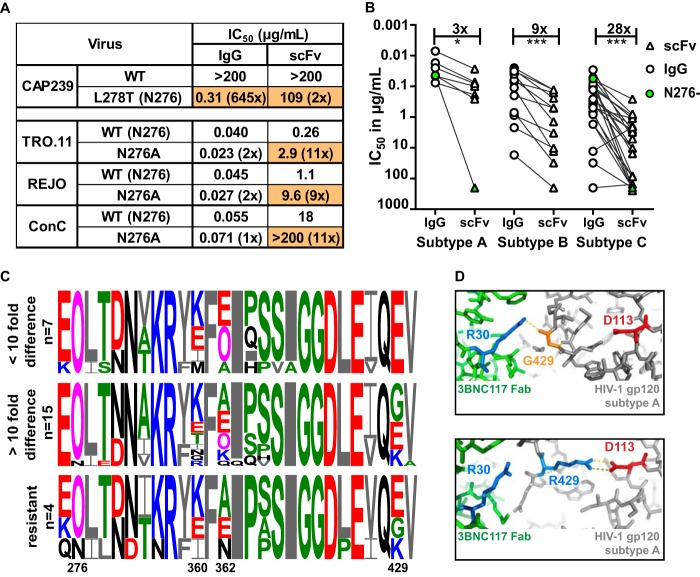
The N276 glycan is highly conserved and a major contact residue for CD4 binding site antibodies, including 3BNC117 (30, 35). To interrogate the loss of potency for the 3BNC117 scFv, we introduced this glycan into a resistant virus or removed it from three sensitive viruses (Fig. 6A). Insertion of the N276 glycan into CAP239.2.00.G3J (through an L278T mutation) rendered this resistant virus highly sensitive to IgG, although the effect on the scFv was marginal. However, removal of the N276 glycan in three viruses had a more significant effect on the scFv than the IgG, with the N276A mutation causing a 10-fold increase in resistance or a knockout effect (Fig. 6A). This indicated that the scFv was more dependent on the 276 glycan for neutralization than the IgG.
FIG 6.
Loss of potency for the 3BNC117 scFv was more pronounced for subtype C viruses. (A) IC50 of IgG and scFv neutralization of one virus that lacked the N276 glycan with a glycan knockin (CAP239) and 3 viruses in which the N276 glycan has been deleted (scFv corrected for molecular weight difference). Fold changes compared to the wild type are shown in parentheses, with changes of >5-fold colored in orange. (B) IC50 titers in micrograms per milliliter of IgG (circle) and scFv (triangle) against HIV-1 subtypes A, B, and C. Viruses lacking an N276 glycan are colored green. IgG- and scFv-resistant viruses are excluded. Fold differences are indicated above the graph and excluded the 2 viruses that lacked an N276 glycan. (C) Logogram of amino acid sequences of the CD4 binding site of HIV-1 subtype C viruses neutralized at the same level by scFv and IgG (<10-fold), those less well neutralized by scFv (>10-fold difference), or those resistant to both IgG and scFv. The N276 glycan and sites 360, 362, and 429, which exhibit the most variability, are indicated. Negatively and positively charged residues are colored red and blue, respectively. (D) Structural representation of the interaction of 3BNC117 Fab (green) and HIV Env (gray) containing an arginine (blue) or glycine (orange) at position 429. The aspartic acid at residue 113 in the HIV trimer is colored red, whereas the arginine at position 30 in the Fab is colored blue. Polar bonds are indicated by dotted lines (PDB entries 4JPV and 5V8L) (78, 79).
CD4bs antibodies have been shown to have some subtype preference, often exhibiting lower efficacy against subtype C viruses (36). To explore whether this also impacted the scFv, we compared the loss of potency, relative to that of IgG, for subtype A, B, and C viruses. There was a significant 9-fold difference in the IC50 between the scFv and IgG for subtype B viruses (Fig. 6B). However, there was a 28-fold difference for subtype C viruses, suggesting that the subtype-specific genetic differences have more of an impact on the scFv than the IgG. For subtype A, only one virus, Q259.D2.17, which lacked the N276 glycan (marked in green), showed more than a 10-fold difference (Fig. 6B). This glycan was also missing in the subtype C virus, CAP228.51, which was significantly less well neutralized by the scFv.
We next analyzed the viral amino acid sequences in the CD4 binding site of subtype C viruses to identify other residues that might contribute to the potency difference between IgG and scFv (Fig. 6C). Viruses were separated into 3 groups based on their relative sensitivity to IgG and scFv. There was significant viral variation at positions 360, 362, and 429, all of which have been previously implicated in the binding of CD4bs bNAbs (36, 37), although position 429 has not specifically been shown to impact 3BNC117.
To further explore the possibility that residue 429 impacted the scFv, we compared crystal structures of the 3BNC117 Fab in complex with gp120 (PDB entries 4JPV and 5V8L). We found that a neutral or negative charge at residue 429 interacted with a positively charged residue 30 (arginine) on the 3BNC117 Fab (Fig. 6D). In contrast, a positive charge at position 429 in the HIV-1 trimer could prevent 3BNC117 binding to this site by folding back and binding to an aspartic acid at position 113 in gp120, occluding this residue. Therefore, the 3BNC117 scFv binding may in part be more reliant on the neutral or negative charge at position 429 than its IgG counterpart. Thus, virus-specific loss of potency for the 3BNC117 scFv was likely impacted by viral subtype, the 276 glycan, and charged residues within the epitope.
DISCUSSION
We have designed and expressed scFv of HIV bNAbs that are undergoing human clinical testing to assess whether they maintained neutralization capacity in this format. These bNAbs targeted one of four well-defined epitopes on the viral envelope, namely, the V2 apex (CAP256.25), the V3 glycan (PGT121), the CD4bs (3BNC117), and the MPER (10E8v4). Remarkably, these much smaller scFv, engineered from the variable regions of the IgG, retained substantial breadth and potency, and 10E8v4 was equivalent to the full IgG molecule. While CAP256.25, 3BNC117, and PGT121 scFv lost potency, they were nonetheless in the therapeutic range of VRC01, a CD4bs antibody that is currently in clinical efficacy testing.
Newly designed scFv showed good levels of expression with different linker lengths of 15, 18, or 20 amino acids having minimal to no impact on neutralizing activity. This suggested that the loss of activity relative to IgG was due to the inherent nature rather than the design of the scFv, which at ∼30 kDa are 5 times smaller than IgG. Antibody scFv were more likely to lose neutralization potency rather than breadth, although for 3BNC117 and PGT121 the loss of potency often resulted in reduced breadth. Potency loss was most severe for CAP256.25 and was evident against all viruses tested, while for 3BNC117 and PGT121 this occurred in a virus-specific manner. Epitope mapping studies on selected viruses showed that scFv bound to their epitope in ways subtly different from those of the matched IgG. Thus, charged residues at positions 169 and 170 in V2 affected CAP256.25 scFv neutralization more than the IgG. For PGT121, interactions with the N332 glycan and the length of V1 impacted the scFv to a greater degree than the IgG, and for 3BNC117, loss of binding to the N276 glycan was more detrimental to the scFv than the IgG. Thus, while scFv retained the ability to interact with known epitopes, they were more sensitive to variations at single residues within these epitopes. Whether these potency losses are unique to each scFv or are dependent on the epitopes being targeted would need to be determined by designing and testing a larger number of scFv versions of bNAbs.
scFv have a single binding site compared to the 2 binding sites of IgG, which likely reduces their affinity for the antigen. It is known that smaller proteins tend to have higher off rates due to lower binding affinity (38–40). This was previously demonstrated for scFv-based fragments of VRC01 that had a 4-fold lower affinity than the IgG version (18). Affinity is influenced by noncovalent intermolecular interactions between proteins, such as electrostatic charge and hydrogen bonding. Our data suggest that a charge change in the epitope had a greater effect than the loss of a glycan, although both likely impacted the binding affinity of the scFv, accounting for the potency losses. Loss of affinity could also be due to the inability of scFv with a single binding site to effectively cross-link protomers on the trimer. Even though trimers are present at low density on the HIV virion, polarization of trimers at the cell contact site would allow the IgG but not the scFv to cross-link (41–45).
The constant region affects the angle at which the antibody paratope engages the viral epitope and also helps to stabilize the heavy-light chain interactions (46). As a result, scFv may have a somewhat less rigid paratope than IgG. Removal of the Fc region could, therefore, alter these interactions, contributing to the scFv being more dependent on key residues in the epitope. Structural studies in combination with affinity studies of scFv are needed to further inform the molecular interactions with the HIV trimer and provide insights into the stability of scFv paratopes. Others have shown that the lack of an Fc region also significantly reduces the half-life of scFv and removes their ability to mediate effector functions, which compromises their clinical development (16, 22). However, scFv may have lower immunogenicity and off-target effects than IgG as a result of not having an antibody Fc region to engage host Fc receptors (18, 47).
scFv may have better access to their epitopes than IgG due to their smaller size, as has been reported using other antibody constructs. For example, antigen-binding fragments (Fab) of antibodies targeting the CD4-induced (CD4i) epitope showed significant improvement in potency over the IgG versions (48–50). Moreover, scFv isolated from phage display libraries demonstrated good breadth and potency (25, 51). A number of antibody epitopes on the HIV trimer are not readily accessible by IgG molecules due to steric hindrance, including the MPER on gp41 (53–55). Remarkably, the scFv of 10E8v4, an MPER antibody that targets an epitope close to the viral membrane, largely maintained potency similar to that of the IgG. The improved access of the 10E8v4 scFv to this recessed site could offset the loss of affinity that results from having only a single paratope. Alternatively, scFv targeting the MPER may be partially buried in the lipid membrane, which could stabilize the interaction with the epitope (54–56). scFv may also be important in preventing cell-cell transmission, where spatial restrictions within the virological synapse limit antibody access (57).
scFv may be useful in the context of passive immunization; however, the caveats associated with this format will need to be addressed, in particular their reduced half-life. Adeno-associated virus (AAV) vectors, which are being developed for continuous delivery of antibody-based products as vectored immunoprophylaxis against HIV infection (58–61), may provide a solution. scFv, with their smaller gene inserts, may be more suitable for expression in vectors, such as AAV-8, that have been previously deployed for some of the IgG bNAbs described here (61). This approach could be particularly important in the context of bNAb combinations needed to optimize the coverage of global HIV strains (62–64). Since multiple scFv could be expressed from the same vector, their use in combination treatments may be a distinct advantage (47, 58, 62, 65). scFv derived from other HIV bNAbs will need to be tested to determine which combinations show optimal complementarity in their neutralization profiles given that scFv neutralization cannot be predicted.
Overall, our data indicate that scFv retain considerable breadth and potency and could perform better than IgG equivalents as combination therapeutics. In particular, 10E8v4 should be considered a leading candidate for human clinical testing, given that it retains all its functional activity as an scFv. Although there are some challenges with this format, there are also unique advantages of scFv over IgG. These smaller antibody proteins, therefore, have considerable potential in passive immunization strategies to prevent HIV infection.
MATERIALS AND METHODS
Construction of scFv.
scFv containing the variable regions of various bNAbs were cloned by overlapping PCR or ordered from GenScript (NJ, USA) (Table 1). For CAP256.25 and PGT121, 3 different glycine-serine linkers, namely, a 15-amino-acid linker [linker 1, (GGGGS)3], a 20-amino-acid linker [linker 2, (GGGGS)4], and an 18-amino-acid linker (linker 3, GGSSRSSSSGGGGSGGGG) were tested in the VH-VL orientation. L3 was also tested for the VL-VH orientation. We used an 8×-histidine tag for purification purposes preceded by an HRV-3C site in case removal of the histidine tag was needed for future experiments. Table 1 shows the scFv constructs in VH-VL orientation with L3 for CAP256.25. All other scFv (3BNC117 and 10E8v4) were expressed in the VH-VL conformation with linker 1 as specified.
Protein expression.
All constructs were cloned into the pCMVR vector (AIDS Reagent Program, Division of AIDS, NIAID, NIH). For the lambda chain of PGT121, the pBR322-based IG-lambda expression vector was used (AIDS Reagent Program, Division of AIDS, NIAID, NIH). The constructs were grown in JM109 bacterial cells and extracted using a plasmid Maxiprep kit (Qiagen, Hilden Germany). Sequences were confirmed using the Applied Biosystems 3500xL Genetic Analyzer. Constructs were expressed as previously described (66). In short, HEK293F suspension cells at 1.5 × 106 to 2 × 106 cells/ml were cotransfected with linear polyethylenimine hydrochloride (molecular weight, 40,000) at a 3:1 ratio with 1 μg of plasmid per 1 ml of culture. Supernatants were harvested after 6 days.
Purification of scFv.
scFv were purified using Ni-Sepharose beads (GE Healthcare, Massachusetts USA). Proteins were washed using a 30 mM imidazole–phosphate-buffered saline (PBS) solution and eluted using 400 mM imidazole in PBS. Glycerol was added to the elution at a final concentration of 5% to limit aggregation. scFv were applied to hiload Superdex 75 or Superdex 200 columns (GE Healthcare) equilibrated with PBS at pH 6.5 (5% glycerol with 0.02% sodium azide). The fractions corresponding to the size of the scFv were collected, pooled, and concentrated using Vivaspin concentrators or Vivapore static concentrators (GE Healthcare). The samples were dialyzed overnight at room temperature to remove sodium azide. Concentrations were measured on a NanoDrop device (Thermofisher, MA, USA), with extinction coefficients at 1% calculated using Expasy ProtParam (69) and characterized by SDS-PAGE. Molar weight was determined by using Expasy ProtParam (Fig. 1A). scFv proteins were stored at −75°C.
IgG production.
IgG constructs were expressed in HEK293F cells as described previously (66). Supernatants were harvested after 6 days and purified using a protein A affinity column. Proteins were eluted using a 0.15 M glycine buffer at pH 2.5 in 1 M Tris, pH 8, and were concentrated and dialyzed into PBS, pH 6.5, containing 5% glycerol. Concentrations were measured on a NanoDrop device using an extinction coefficient of 13.7 and a 1% solution. The molecular weight of the IgG was calculated using Expasy ProtParam (69) for CAP256.25 (150.71 kDa), 10E8v4 (147.29 kDa), 3BNC117 (146.24 kDa), and PGT121 (146.2 kDa). IgG and proteins were stored at −75°C.
Neutralization assay.
Single-round infection pseudoviruses were grown and titers determined as described previously (70–72). Briefly, plasmids were transfected with the pseudovirus backbone pSG3Δenv into 293T cells at ratios of 1:3 PEI Max. Viruses were harvested after 48 h, filtered, frozen, and titrated. A panel of 43 viruses (73) plus BG505 N332 and CAP256_SU (CAP256.3 mo.9C) (74, 75), representing HIV-1 clades A, B, and C, was used to compare neutralization titers of IgG and scFv. Neutralization assays were performed in TZM-bl cells as described previously (76, 77). Proteins were tested at 200 μg/ml for the IgG (∼146 kDa) and 50 μg/ml for the scFv (28 to 32 kDa). scFv neutralization titers were recalculated by compensating for the fold difference in molecular weight (values for each scFv are shown in Fig. 1A). (The IC50 data for scFv and IgG molar concentrations are available on request). All assays were repeated at least twice. Geometric mean potency was calculated for both IgG and scFv using sensitive viruses only. When comparing viruses mutated by site-directed mutagenesis, a fold change of more than 3 compared to the parent virus was considered significant.
Site-directed mutagenesis.
Mutations in the envelope gene of viruses were made using the QuikChange Lightning site-directed mutagenesis kit (Agilent). Sequences were confirmed using the Applied Biosystems 3500xL Genetic Analyzer. Pseudoviruses were grown as described above.
Statistics and sequence analysis.
Wilcoxon matched-pair signed-rank test was used to assess differences between IgG and scFv within a subtype.
Sequence alignments were made using the Analyze Align tool from the HIV LanL website (https://www.hiv.lanl.gov/content/sequence/ANALYZEALIGN/analyze_align.html).
Structural modeling.
We used the PyMOL Molecular Graphics System (version 2.1.1; Schrödinger, LLC) to model a BG505 SOSIP.664 trimer (PDB entry 5V8L). Residues corresponding to the sequences in the CATNAP LANL for CAP256_SU and AC10.0.29 were modeled and colored based on charge.
For 3BNC117, we used Fab structures, either in complex with a clade A gp120 from 93TH057 with G429 (4JPV) or in complex with the clade A BG505 SOSIP.664 trimer with R429 (PDB entry 5V8L). We used PyMOL to visualize the 429 residue and used the built-in software to predict polar bonds.
ACKNOWLEDGMENTS
We thank H. Marcotte (Karolinska Institute) for the donation of the CAP256 VH-VL L1 scFv and C. Anthony and C. Williamson from the University of Cape Town for the CAP177 plasmids. We thank the AIDS Reagent Program, Division of AIDS, NIAID, NIH, for the use of the pCMVR vector and the pBR322-based IG-lambda expression vector. We also thank J. Mascola, the VRC, USA, for the donation of the PGT121 heavy- and light-chain plasmids.
We acknowledge research funding from the South African Medical Research Council (SAMRC) Flagship Project, the NIH through a U01 grant (U01AI116086), the Poliomyelitis Research Fund (PRF) through a PRF research grant (17/15), and the Centre for the AIDS Program of Research (CAPRISA). CAPRISA is funded by the South African HIV/AIDS Research and Innovation Platform of the South African Department of Science and Technology and was initially supported by the U.S. NIAID, NIH, U.S. Department of Health and Human Services grant U19 AI51794. R.T.V.D. is supported by a Poliomyelitis Research Foundation Ph.D. bursary (W 16/72). P.L.M. is supported by the South African Research Chairs Initiative of the Department of Science and Technology and National Research Foundation of South Africa (grant no. 98341). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
REFERENCES
- 1.Bhiman JN, Lynch RM. 2017. Broadly neutralizing antibodies as treatment: effects on virus and immune system. Curr HIV/AIDS Rep 14:54–62. doi: 10.1007/s11904-017-0352-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burton DR, Hangartner L. 2016. Broadly neutralizing antibodies to HIV and their role in vaccine design. Annu Rev Immunol 34:635–659. doi: 10.1146/annurev-immunol-041015-055515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Julg B, Barouch DH. 2019. Neutralizing antibodies for HIV-1 prevention. Curr Opin HIV AIDS 14:318–324. doi: 10.1097/COH.0000000000000556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mascola JR, Stiegler G, VanCott TC, Katinger H, Carpenter CB, Hanson CE, Beary H, Hayes D, Frankel SS, Birx DL, Lewis MG. 2000. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat Med 6:207–210. doi: 10.1038/72318. [DOI] [PubMed] [Google Scholar]
- 5.Cheeseman HM, Olejniczak NJ, Rogers PM, Evans AB, King DFL, Ziprin P, Liao H-X, Haynes BF, Shattock RJ. 2017. Broadly neutralizing antibodies display potential for prevention of HIV-1 infection of mucosal tissue superior to that of nonneutralizing antibodies. J Virol 91:e01762-16. doi: 10.1128/JVI.01762-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Julg B, Liu P-T, Wagh K, Fischer WM, Abbink P, Mercado NB, Whitney JB, Nkolola JP, McMahan K, Tartaglia LJ, Borducchi EN, Khatiwada S, Kamath M, LeSuer JA, Seaman MS, Schmidt SD, Mascola JR, Burton DR, Korber BT, Barouch DH. 2017. Protection against a mixed SHIV challenge by a broadly neutralizing antibody cocktail. Sci Transl Med 9:eaao4235. doi: 10.1126/scitranslmed.aao4235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Julg B, Tartaglia LJ, Keele BF, Wagh K, Pegu A, Sok D, Abbink P, Schmidt SD, Wang K, Chen X, Joyce MG, Georgiev IS, Choe M, Kwong PD, Doria-Rose NA, Le K, Louder MK, Bailer RT, Moore PL, Korber B, Seaman MS, Abdool Karim SS, Morris L, Koup RA, Mascola JR, Burton DR, Barouch DH. 2017. Broadly neutralizing antibodies targeting the HIV-1 envelope V2 apex confer protection against a clade C SHIV challenge. Sci Transl Med 9:eaal1321. doi: 10.1126/scitranslmed.aal1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gautam R, Nishimura Y, Pegu A, Nason MC, Klein F, Gazumyan A, Golijanin J, Buckler-White A, Sadjadpour R, Wang K, Mankoff Z, Schmidt SD, Lifson JD, Mascola JR, Nussenzweig MC, Martin MA. 2016. A single injection of anti-HIV-1 antibodies protects against repeated SHIV challenges. Nature 533:105–109. doi: 10.1038/nature17677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pegu A, Hessell AJ, Mascola JR, Haigwood NL. 2017. Use of broadly neutralizing antibodies for HIV-1 prevention. Immunol Rev 275:296–312. doi: 10.1111/imr.12511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caskey M, Klein F, Lorenzi JCC, Seaman MS, West AP Jr, Buckley N, Kremer G, Nogueira L, Braunschweig M, Scheid JF, Horwitz JA, Shimeliovich I, Ben-Avraham S, Witmer-Pack M, Platten M, Lehmann C, Burke LA, Hawthorne T, Gorelick RJ, Walker BD, Keler T, Gulick RM, Fatkenheuer G, Schlesinger SJ, Nussenzweig MC, West AP, Fätkenheuer G. 2015. Viraemia suppressed in HIV-1-infected humans by broadly neutralizing antibody 3BNC117. Nature 522:487–491. doi: 10.1038/nature14411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caskey M, Schoofs T, Gruell H, Settler A, Karagounis T, Kreider EF, Murrell B, Pfeifer N, Nogueira L, Oliveira TY, Learn GH, Cohen YZ, Lehmann C, Gillor D, Shimeliovich I, Unson-O'Brien C, Weiland D, Robles A, Kümmerle T, Wyen C, Levin R, Witmer-Pack M, Eren K, Ignacio C, Kiss S, West AP, Mouquet H, Zingman BS, Gulick RM, Keler T, Bjorkman PJ, Seaman MS, Hahn BH, Fätkenheuer G, Schlesinger SJ, Nussenzweig MC, Klein F. 2017. Antibody 10-1074 suppresses viremia in HIV-1-infected individuals. Nat Med 23:185–191. doi: 10.1038/nm.4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lynch RM, Boritz E, Coates EE, DeZure A, Madden P, Costner P, Enama ME, Plummer S, Holman L, Hendel CS, Gordon I, Casazza J, Conan-Cibotti M, Migueles SA, Tressler R, Bailer RT, McDermott A, Narpala S, O'Dell S, Wolf G, Lifson JD, Freemire BA, Gorelick RJ, Pandey JP, Mohan S, Chomont N, Fromentin R, Chun T-W, Fauci AS, Schwartz RM, Koup RA, Douek DC, Hu Z, Capparelli E, Graham BS, Mascola JR, Ledgerwood JE. 2015. Virologic effects of broadly neutralizing antibody VRC01 administration during chronic HIV-1 infection. Sci Transl Med 7:319ra206. doi: 10.1126/scitranslmed.aad5752. [DOI] [PubMed] [Google Scholar]
- 13.Mendoza P, Gruell H, Nogueira L, Pai JA, Butler AL, Millard K, Lehmann C, Suárez I, Oliveira TY, Lorenzi J, Cohen YZ, Wyen C, Kümmerle T, Karagounis T, Lu C-L, Handl L, Unson O’Brien C, Patel R, Ruping C, Schlotz M, Witmer-Pack M, Shimeliovich I, Kremer G, Thomas E, Seaton KE, Horowitz J, West AP, Bjorkman PJ, Tomaras GD, Gulick RM, Pfeifer N, Fätkenheuer G, Seaman MS, Klein F, Caskey M, Nussenzweig MC. 2018. Combination therapy with anti-HIV-1 antibodies maintains viral suppression. Nature 561:479–484. doi: 10.1038/s41586-018-0531-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bar KJ, Sneller MC, Harrison LJ, Justement JS, Overton ET, Petrone ME, Salantes DB, Seamon CA, Scheinfeld B, Kwan RW, Learn GH, Proschan MA, Kreider EF, Blazkova J, Bardsley M, Refsland EW, Messer M, Clarridge KE, Tustin NB, Madden PJ, Oden K, O'Dell SJ, Jarocki B, Shiakolas AR, Tressler RL, Doria-Rose NA, Bailer RT, Ledgerwood JE, Capparelli EV, Lynch RM, Graham BS, Moir S, Koup RA, Mascola JR, Hoxie JA, Fauci AS, Tebas P, Chun T-W. 2016. Effect of HIV antibody VRC01 on viral rebound after treatment interruption. N Engl J Med 375:2037–2050. doi: 10.1056/NEJMoa1608243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scheid JF, Horwitz JA, Bar-On Y, Kreider EF, Lu C-L, Lorenzi JCC, Feldmann A, Braunschweig M, Nogueira L, Oliveira TY, Shimeliovich I, Patel R, Burke LA, Cohen YZ, Hadrigan S, Settler A, Witmer-Pack M, West AP Jr, Juelg B, Keler T, Hawthorne T, Zingman BS, Gulick RM, Pfeifer N, Learn GH, Seaman MS, Bjorkman PJ, Klein F, Schlesinger SJ, Walker BD, Hahn BH, Nussenzweig MC, Caskey M. 2016. HIV-1 antibody 3BNC117 suppresses viral rebound in humans during treatment interruption. Nature 535:556–560. doi: 10.1038/nature18929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahmad ZA, Yeap SK, Ali AM, Ho WY, Alitheen NBM, Hamid M. 2012. scFv antibody: principles and clinical application. Clin Dev Immunol 2012:980250–980215. doi: 10.1155/2012/980250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen W, Dimitrov DS. 2009. Human monoclonal antibodies and engineered antibody domains as HIV-1 entry inhibitors. Curr Opin HIV AIDS 4:112–117. doi: 10.1097/COH.0b013e328322f95e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.West AP, Galimidi RP, Gnanapragasam PNP, Bjorkman PJ. 2012. Single-chain Fv-based anti-HIV proteins: potential and limitations. J Virol 86:195–202. doi: 10.1128/JVI.05848-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang S, Zheng C, Liu Y, Zheng H, Wang Z. 2008. Construction of multiform scFv antibodies using linker peptide. J Genet Genomics 35:313–316. doi: 10.1016/S1673-8527(08)60045-4. [DOI] [PubMed] [Google Scholar]
- 20.Colcher D, Pavlinkova G, Beresford G, Booth BJ, Choudhury A, Batra SK. 1998. Pharmacokinetics and biodistribution of genetically-engineered antibodies. Q J Nucl Med 42:225–241. [PubMed] [Google Scholar]
- 21.Yokota T, Milenic DE, Whitlow M, Schlom J. 1992. Rapid tumor penetration of a single-chain Fv and comparison with other immunoglobulin forms. Cancer Res 52:3402–3408. [PubMed] [Google Scholar]
- 22.Unverdorben F, Richter F, Hutt M, Seifert O, Malinge P, Fischer N, Kontermann RE. 2016. Pharmacokinetic properties of IgG and various Fc fusion proteins in mice. MAbs 8:120–128. doi: 10.1080/19420862.2015.1113360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holliger P, Hudson PJ. 2005. Engineered antibody fragments and the rise of single domains. Nat Biotechnol 23:1126–1136. doi: 10.1038/nbt1142. [DOI] [PubMed] [Google Scholar]
- 24.Bivi N, Moore T, Rodgers G, Denning H, Shockley T, Swearingen CA, Gelfanova V, Calderon B, Peterson DA, Hodsdon ME, Siegel RW, Higgs RE, Konrad RJ. 2019. Investigation of pre-existing reactivity to biotherapeutics can uncover potential immunogenic epitopes and predict immunogenicity risk. MAbs 11:861–869. doi: 10.1080/19420862.2019.1612699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khan L, Kumar R, Thiruvengadam R, Parray HA, Makhdoomi MA, Kumar S, Aggarwal H, Mohata M, Hussain AW, Das R, Varadarajan R, Bhattacharya J, Vajpayee M, Murugavel KG, Solomon S, Sinha S, Luthra K. 2017. Cross-neutralizing anti-HIV-1 human single chain variable fragments (scFvs) against CD4 binding site and N332 glycan identified from a recombinant phage library. Sci Rep 7:45163. doi: 10.1038/srep45163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burton DR, Barbas CF, Persson MA, Koenig S, Chanock RM, Lerner RA. 1991. A large array of human monoclonal antibodies to type 1 human immunodeficiency virus from combinatorial libraries of asymptomatic seropositive individuals. Proc Natl Acad Sci U S A 88:10134–10137. doi: 10.1073/pnas.88.22.10134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reference deleted.
- 28.Doria-Rose NA, Bhiman JN, Roark RS, Schramm CA, Gorman J, Chuang G-Y, Pancera M, Cale EM, Ernandes MJ, Louder MK, Asokan M, Bailer RT, Druz A, Fraschilla IR, Garrett NJ, Jarosinski M, Lynch RM, McKee K, O'Dell S, Pegu A, Schmidt SD, Staupe RP, Sutton MS, Wang K, Wibmer CK, Haynes BF, Abdool-Karim S, Shapiro L, Kwong PD, Moore PL, Morris L, Mascola JR. 2016. New member of the V1V2-directed CAP256-VRC26 lineage that shows increased breadth and exceptional potency. J Virol 90:76–91. doi: 10.1128/JVI.01791-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien J-P, Wang S-KS-K, Ramos A, Chan-Hui P-Y, Moyle M, Mitcham JL, Hammond PW, Olsen OA, Phung P, Fling S, Wong C-H, Phogat SK, Wrin T, Simek MD, Principal Investigators PG, Koff WC, Wilson IA, Burton DR, Poignard P, Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Chan-Hui P-Y, Moyle M, Mitcham JL, Hammond PW, Olsen OA, Phung P, Principal Investigators PG, Koff WC, Wilson IA, Burton DR, Poignard P, Protocol G Principal Investigators. 2011. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scheid JF, Mouquet H, Ueberheide B, Diskin R, Klein F, Oliveira TYK, Pietzsch J, Fenyo D, Abadir A, Velinzon K, Hurley A, Myung S, Boulad F, Poignard P, Burton DR, Pereyra F, Ho DD, Walker BD, Seaman MS, Bjorkman PJ, Chait BT, Nussenzweig MC. 2011. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science 333:1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kwon YD, Georgiev IS, Ofek G, Zhang B, Asokan M, Bailer RT, Bao A, Caruso W, Chen X, Choe M, Druz A, Ko S-Y, Louder MK, McKee K, O'Dell S, Pegu A, Rudicell RS, Shi W, Wang K, Yang Y, Alger M, Bender MF, Carlton K, Cooper JW, Blinn J, Eudailey J, Lloyd K, Parks R, Alam SM, Haynes BF, Padte NN, Yu J, Ho DD, Huang J, Connors M, Schwartz RM, Mascola JR, Kwong PD. 2016. Optimization of the solubility of HIV-1-neutralizing antibody 10E8 through somatic variation and structure-based design. J Virol 90:5899–5914. doi: 10.1128/JVI.03246-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Julien J-P, Sok D, Khayat R, Lee JH, Doores KJ, Walker LM, Ramos A, Diwanji DC, Pejchal R, Cupo A, Katpally U, Depetris RS, Stanfield RL, McBride R, Marozsan AJ, Paulson JC, Sanders RW, Moore JP, Burton DR, Poignard P, Ward AB, Wilson IA. 2013. Broadly neutralizing antibody PGT121 allosterically modulates CD4 binding via recognition of the HIV-1 gp120 V3 base and multiple surrounding glycans. PLoS Pathog 9:e1003342. doi: 10.1371/journal.ppat.1003342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anthony C, York T, Bekker V, Matten D, Selhorst P, Ferreria R-C, Garrett NJ, Karim SSA, Morris L, Wood NT, Moore PL, Williamson C. 2017. Cooperation between strain-specific and broadly neutralizing responses limited viral escape and prolonged the exposure of the broadly neutralizing epitope. J Virol 91:e00828-17. doi: 10.1128/JVI.00828-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deshpande S, Patil S, Kumar R, Hermanus T, Murugavel KG, Srikrishnan AK, Solomon S, Morris L, Bhattacharya J. 2016. HIV-1 clade C escapes broadly neutralizing autologous antibodies with N332 glycan specificity by distinct mechanisms. Retrovirology 13:60. doi: 10.1186/s12977-016-0297-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McGuire AT, Hoot S, Dreyer AM, Lippy A, Stuart A, Cohen KW, Jardine J, Menis S, Scheid JF, West AP, Schief WR, Stamatatos L. 2013. Engineering HIV envelope protein to activate germline B cell receptors of broadly neutralizing anti-CD4 binding site antibodies. J Exp Med 210:655–663. doi: 10.1084/jem.20122824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bricault CA, Yusim K, Seaman MS, Yoon H, Theiler J, Giorgi EE, Wagh K, Theiler M, Hraber P, Macke JP, Kreider EF, Learn GH, Hahn BH, Scheid JF, Kovacs JM, Shields JL, Lavine CL, Ghantous F, Rist M, Bayne MG, Neubauer GH, McMahan K, Peng H, Chéneau C, Jones JJ, Zeng J, Ochsenbauer C, Nkolola JP, Stephenson KE, Chen B, Gnanakaran S, Bonsignori M, Williams LD, Haynes BF, Doria-Rose N, Mascola JR, Montefiori DC, Barouch DH, Korber B. 2019. HIV-1 neutralizing antibody signatures and application to epitope-targeted vaccine design. Cell Host Microbe 25:59–72. doi: 10.1016/j.chom.2018.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Q, Lai YT, Zhang P, Louder MK, Pegu A, Rawi R, Asokan M, Chen X, Shen CH, Chuang GY, Yang ES, Miao H, Wang Y, Fauci AS, Kwong PD, Mascola JR, Lusso P. 2019. Improvement of antibody functionality by structure-guided paratope engraftment. Nat Commun 10:721. doi: 10.1038/s41467-019-08658-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schreiber G, Haran G, Zhou H-X. 2009. Fundamental aspects of protein−protein association kinetics. Chem Rev 109:839–860. doi: 10.1021/cr800373w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pan AC, Borhani DW, Dror RO, Shaw DE. 2013. Molecular determinants of drug-receptor binding kinetics. Drug Discov Today 18:667–673. doi: 10.1016/j.drudis.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 40.Miller DC, Lunn G, Jones P, Sabnis Y, Davies NL, Driscoll P. 2012. Investigation of the effect of molecular properties on the binding kinetics of a ligand to its biological target. Med Chem Commun 3:449. doi: 10.1039/c2md00270a. [DOI] [Google Scholar]
- 41.Klasse PJ. 2007. Modeling how many envelope glycoprotein trimers per virion participate in human immunodeficiency virus infectivity and its neutralization by antibody. Virology 369:245–262. doi: 10.1016/j.virol.2007.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chien MP, Lin CH, Chang DK. 2009. Recruitment of HIV-1 envelope occurs subsequent to lipid mixing: a fluorescence microscopic evidence. Retrovirology 6:20. doi: 10.1186/1742-4690-6-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cohen FS, Melikyan GB. 2004. The energetics of membrane fusion from binding, through hemifusion, pore formation, and pore enlargement. J Membr Biol 199:1–14. doi: 10.1007/s00232-004-0669-8. [DOI] [PubMed] [Google Scholar]
- 44.Louder MK, Sambor A, Chertova E, Hunte T, Barrett S, Ojong F, Sanders-Buell E, Zolla-Pazner S, McCutchan FE, Roser JD, Gabuzda D, Lifson JD, Mascola JR. 2005. HIV-1 envelope pseudotyped viral vectors and infectious molecular clones expressing the same envelope glycoprotein have a similar neutralization phenotype, but culture in peripheral blood mononuclear cells is associated with decreased neutralization sensitivity. Virology 339:226–238. doi: 10.1016/j.virol.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 45.Cohen YZ, Lorenzi JCC, Seaman MS, Nogueira L, Schoofs T, Krassnig L, Butler A, Millard K, Fitzsimons T, Daniell X, Dizon JP, Shimeliovich I, Montefiori DC, Caskey M, Nussenzweig MC. 2017. Neutralizing activity of broadly neutralizing anti-HIV-1 antibodies against clade B clinical isolates produced in peripheral blood mononuclear cells. J Virol 92:e01883-17. doi: 10.1128/JVI.01883-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Janda A, Bowen A, Greenspan NS, Casadevall A. 2016. Ig constant region effects on variable region structure and function. Front Microbiol 7:22. doi: 10.3389/fmicb.2016.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Monnier P, Vigouroux R, Tassew N. 2013. In vivo applications of single chain Fv (variable domain) (scFv) fragments. Antibodies 2:193–208. doi: 10.3390/antib2020193. [DOI] [Google Scholar]
- 48.Labrijn AF, Poignard P, Raja A, Zwick MB, Delgado K, Franti M, Binley JM, Vivona V, Grundner C, Huang C-C, Venturi M, Petropoulos C, Wrin T, Dimitrov DS, Robinson JE, Kwong PD, Wyatt RT, Sodroski JG, Burton DR. 2003. Access of antibody molecules to the conserved coreceptor binding site on glycoprotein gp120 is sterically restricted on primary human immunodeficiency virus type 1. J Virol 77:10557–10565. doi: 10.1128/jvi.77.19.10557-10565.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang MY, Shu Y, Sidorov I, Dimitrov DS. 2004. Identification of a novel CD4i human monoclonal antibody Fab that neutralizes HIV-1 primary isolates from different clades. Antiviral Res 61:161–164. doi: 10.1016/j.antiviral.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 50.Tanaka K, Kuwata T, Alam M, Kaplan G, Takahama S, Valdez KPR, Roitburd-Berman A, Gershoni JM, Matsushita S. 2017. Unique binding modes for the broad neutralizing activity of single-chain variable fragments (scFv) targeting CD4-induced epitopes. Retrovirology 14:44. doi: 10.1186/s12977-017-0369-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kumar S, Kumar R, Khan L, Makhdoomi MA, Thiruvengadam R, Mohata M, Agarwal M, Lodha R, Kabra SK, Sinha S, Luthra K. 2017. CD4-binding site directed cross-neutralizing scFv monoclonals from HIV-1 subtype C infected Indian children. Front Immunol 8:1568. doi: 10.3389/fimmu.2017.01568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reference deleted.
- 53.Zwick MB, Labrijn AF, Wang M, Spenlehauer C, Saphire EO, Binley JM, Moore JP, Stiegler G, Katinger H, Burton DR, Parren P. 2001. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J Virol 75:10892–10905. doi: 10.1128/JVI.75.22.10892-10905.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Irimia A, Sarkar A, Stanfield RL, Wilson IA. 2016. Crystallographic identification of lipid as an integral component of the epitope of HIV broadly neutralizing antibody 4E10. Immunity 44:21–31. doi: 10.1016/j.immuni.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Irimia A, Serra AM, Sarkar A, Jacak R, Kalyuzhniy O, Sok D, Saye-Francisco KL, Schiffner T, Tingle R, Kubitz M, Adachi Y, Stanfield RL, Deller MC, Burton DR, Schief WR, Wilson IA. 2017. Lipid interactions and angle of approach to the HIV-1 viral membrane of broadly neutralizing antibody 10E8: insights for vaccine and therapeutic design. PLoS Pathog 13:e1006212. doi: 10.1371/journal.ppat.1006212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee JH, Ozorowski G, Ward AB. 2016. Cryo-EM structure of a native, fully glycosylated, cleaved HIV-1 envelope trimer. Science 351:1043–1048. doi: 10.1126/science.aad2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang MY, Borges AR, Ptak RG, Wang Y, Dimitrov AS, Alam SM, Wieczorek L, Bouma P, Fouts T, Jiang S , Polonis VR, Haynes BF, Quinnan GV , Montefiori DC, Dimitrov DS. 2010. Potent and broad neutralizing activity of a single chain antibody fragment against cell-free and cell-associated HIV-1. MAbs 2:266–274. doi: 10.4161/mabs.2.3.11416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Naso MF, Tomkowicz B, Perry WL, Strohl WR. 2017. Adeno-associated virus (AAV) as a vector for gene therapy. BioDrugs 31:317–334. doi: 10.1007/s40259-017-0234-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Balazs AB, Chen J, Hong CM, Rao DS, Yang L, Baltimore D. 2011. Antibody-based protection against HIV infection by vectored immunoprophylaxis. Nature 481:81–86. doi: 10.1038/nature10660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lin A, Balazs AB. 2018. Adeno-associated virus gene delivery of broadly neutralizing antibodies as prevention and therapy against HIV-1. Retrovirology 15:66. doi: 10.1186/s12977-018-0449-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van den Berg FT, Makoah NA, Ali SA, Scott TA, Mapengo RE, Mutsvunguma LZ, Mkhize NN, Lambson BE, Kgagudi PD, Crowther C, Abdool Karim SS, Balazs AB, Weinberg MS, Ely A, Arbuthnot PB, Morris L. 2019. AAV-mediated expression of broadly neutralizing and vaccine-like antibodies targeting the HIV-1 envelope V2 region. Mol Ther Methods Clin Dev 14:100–112. doi: 10.1016/j.omtm.2019.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wagh K, Bhattacharya T, Williamson C, Robles A, Bayne MG, Garrity J, Rist M, Rademeyer C, Yoon H, Lapedes A, Gao H, Greene K, Louder MK, Kong R, Karim SA, Burton DR, Barouch DH, Nussenzweig MC, Mascola JR, Morris L, Montefiori DC, Korber B, Seaman MS. 2016. Optimal combinations of broadly neutralizing antibodies for prevention and treatment of HIV-1 clade C infection. PLoS Pathog 12:e1005520. doi: 10.1371/journal.ppat.1005520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Doria-Rose NA, Louder MK, Yang Z, O'Dell S, Nason M, Schmidt SD, McKee K, Seaman MS, Bailer RT, Mascola JR. 2012. HIV-1 neutralization coverage is improved by combining monoclonal antibodies that target independent epitopes. J Virol 86:3393–3397. doi: 10.1128/JVI.06745-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kong R, Louder MK, Wagh K, Bailer RT, DeCamp A, Greene KM, Gao H, Taft JD, Gazumyan A, Liu C, Nussenzweig MC, Korber BT, Montefiori DC, Mascola JR. 2015. Improving neutralization potency and breadth by combining broadly reactive HIV-1 antibodies targeting major neutralization epitopes. J Virol 89:2659–2671. doi: 10.1128/JVI.03136-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu Z, Chen O, Wall JBJ, Zheng M, Zhou Y, Wang L, Ruth Vaseghi H, Qian L, Liu J. 2017. Systematic comparison of 2A peptides for cloning multi-genes in a polycistronic vector. Sci Rep 7:2193. doi: 10.1038/s41598-017-02460-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Portolano N, Watson PJ, Fairall L, Millard CJ, Milano CP, Song Y, Cowley SM, Schwabe JW. 2014. Recombinant protein expression for structural biology in HEK 293F suspension cells: a novel and accessible approach. J Vis Exp 92:e51897. doi: 10.3791/51897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Reference deleted.
- 68.Reference deleted.
- 69.Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. 2009. Protein identification and analysis tools on the ExPASy server, p 571–607. In Walker JM. (ed), The proteomics protocols handbook. Humana Press, Totowa, NJ. [Google Scholar]
- 70.Mascola JR, D'Souza P, Gilbert P, Hahn BH, Haigwood NL, Morris L, Petropoulos CJ, Polonis VR, Sarzotti M, Montefiori DC. 2005. Recommendations for the design and use of standard virus panels to assess neutralizing antibody responses elicited by candidate human immunodeficiency virus type 1 vaccines. J Virol 79:10103–10107. doi: 10.1128/JVI.79.16.10103-10107.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Seaman MS, Janes H, Hawkins N, Grandpre LE, Devoy C, Giri A, Coffey RT, Harris L, Wood B, Daniels MG, Bhattacharya T, Lapedes A, Polonis VR, McCutchan FE, Gilbert P, Self SG, Korber BT, Montefiori DC, Mascola JR. 2010. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J Virol 84:1439–1452. doi: 10.1128/JVI.02108-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Todd CA, Greene KM, Yu X, Ozaki DA, Gao H, Huang Y, Wang M, Li G, Brown R, Wood B, D'Souza MP, Gilbert P, Montefiori DC, Sarzotti-Kelsoe M. 2012. Development and implementation of an international proficiency testing program for a neutralizing antibody assay for HIV-1 in TZM-bl cells. J Immunol Methods 375:57–67. doi: 10.1016/j.jim.2011.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gray ES, Madiga MC, Hermanus T, Moore PL, Wibmer CK, Tumba NL, Werner L, Mlisana K, Sibeko S, Williamson C, Abdool Karim SS, Morris L. 2011. The neutralization breadth of HIV-1 develops incrementally over four years and is associated with CD4+ T cell decline and high viral load during acute infection. J Virol 85:4828–4840. doi: 10.1128/JVI.00198-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Moore PL, Sheward DJ, Nonyane M, Ranchobe N, Hermanus T, Gray ES, Abdool Karim SS, Williamson C, Morris L. 2013. Multiple pathways of escape from HIV broadly cross-neutralizing V2-dependent antibodies. J Virol 87:4882–4894. doi: 10.1128/JVI.03424-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Doria-Rose NA, Schramm CA, Gorman J, Moore PL, Bhiman JN, DeKosky BJ, Ernandes MJ, Georgiev IS, Kim HJ, Pancera M, Staupe RP, Altae-Tran HR, Bailer RT, Crooks ET, Cupo A, Druz A, Garrett NJ, Hoi KH, Kong R, Louder MK, Longo NS, McKee K, Nonyane M, O'Dell S, Roark RS, Rudicell RS, Schmidt SD, Sheward DJ, Soto C, Wibmer CK, Yang Y, Zhang Z, Mullikin JC, Binley JM, Sanders RW, Wilson IA, Moore JP, Ward AB, Georgiou G, Williamson C, Abdool Karim SS, Morris L, Kwong PD, Shapiro L, Mascola JR. 2014. Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature 509:55–62. doi: 10.1038/nature13036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Montefiori DC. 2009. Measuring HIV neutralization in a luciferase reporter gene assay, p 395–405. In Prasad VR, Kalpana GV (ed), HIV protocols. Methods in molecular biology. Humana Press, Totowa, NJ. [DOI] [PubMed] [Google Scholar]
- 77.Sarzotti-Kelsoe M, Bailer RT, Turk E, Lin C, Bilska M, Greene KM, Gao H, Todd CA, Ozaki DA, Seaman MS, Mascola JR, Montefiori DC. 2014. Optimization and validation of the TZM-bl assay for standardized assessments of neutralizing antibodies against HIV-1. J Immunol Methods 409:131–146. doi: 10.1016/j.jim.2013.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lee JH, Andrabi R, Su C-Y, Yasmeen A, Julien J-P, Kong L, Wu NC, McBride R, Sok D, Pauthner M, Cottrell CA, Nieusma T, Blattner C, Paulson JC, Klasse PJ, Wilson IA, Burton DR, Ward AB. 2017. A broadly neutralizing antibody targets the dynamic HIV envelope trimer apex via a long, rigidified, and anionic β-hairpin structure. Immunity 46:690–702. doi: 10.1016/j.immuni.2017.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Klein F, Diskin R, Scheid JF, Gaebler C, Mouquet H, Georgiev IS, Pancera M, Zhou T, Incesu RB, Fu BZ, Gnanapragasam PNP, Oliveira TY, Seaman MS, Kwong PD, Bjorkman PJ, Nussenzweig MC. 2013. Somatic mutations of the immunoglobulin framework are generally required for broad and potent HIV-1 neutralization. Cell 153:126–138. doi: 10.1016/j.cell.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]