Despite the use of effective live vaccines, mumps outbreaks still occur worldwide. Mumps virus (MuV) infection typically causes flu-like symptoms and parotid gland swelling but sometimes leads to orchitis, oophoritis, and neurological complications, such as meningitis, encephalitis, and deafness. MuV enters the host cell through membrane fusion mediated by two viral proteins, a receptor-binding attachment protein, and a fusion protein, but its detailed mechanism is not fully understood. In this study, we show that the tetramer (dimer of dimers) formation of the MuV attachment protein head domain is supported by an anion located at the interface between two dimers and that the dimer-dimer interaction plays an important role in triggering the activation of the fusion protein and causing membrane fusion. These results not only further our understanding of MuV entry but provide useful information about a possible target for antiviral drugs.
KEYWORDS: mumps, paramyxovirus, hemagglutinin-neuraminidase, entry, fusion
ABSTRACT
Mumps virus (MuV), an enveloped negative-strand RNA virus belonging to the family Paramyxoviridae, enters the host cell through membrane fusion mediated by two viral envelope proteins, an attachment protein hemagglutinin-neuraminidase (MuV-HN) and a fusion (F) protein. However, how the binding of MuV-HN to glycan receptors triggers membrane fusion is not well understood. The crystal structure of the MuV-HN head domain forms a tetramer (dimer of dimers) like other paramyxovirus attachment proteins. In the structure, a sulfate ion (SO42−) was found at the interface between two dimers, which may be replaced by a hydrogen phosphate ion (HPO42−) under physiological conditions. The anion is captured by the side chain of a positively charged arginine residue at position 139 of one monomer each from both dimers. Substitution of alanine or lysine for arginine at this position compromised the fusion support activity of MuV-HN without affecting its cell surface expression, glycan-receptor binding, and interaction with the F protein. Furthermore, the substitution appeared to affect the tetramer formation of the head domain as revealed by blue native-PAGE analysis. These results, together with our previous similar findings with the measles virus attachment protein head domain, suggest that the dimer-dimer interaction within the tetramer may play an important role in triggering membrane fusion during paramyxovirus entry.
IMPORTANCE Despite the use of effective live vaccines, mumps outbreaks still occur worldwide. Mumps virus (MuV) infection typically causes flu-like symptoms and parotid gland swelling but sometimes leads to orchitis, oophoritis, and neurological complications, such as meningitis, encephalitis, and deafness. MuV enters the host cell through membrane fusion mediated by two viral proteins, a receptor-binding attachment protein, and a fusion protein, but its detailed mechanism is not fully understood. In this study, we show that the tetramer (dimer of dimers) formation of the MuV attachment protein head domain is supported by an anion located at the interface between two dimers and that the dimer-dimer interaction plays an important role in triggering the activation of the fusion protein and causing membrane fusion. These results not only further our understanding of MuV entry but provide useful information about a possible target for antiviral drugs.
INTRODUCTION
Mumps virus (MuV), an enveloped nonsegmented negative-strand RNA virus in the family Paramyxoviridae, is the causative agent of mumps, which is characterized by unilateral or bilateral parotid gland swelling accompanied by flu-like symptoms (1). Despite the presence of effective live vaccines, outbreaks still occur worldwide, even in developed countries. The virus initially enters through the nose or mouth and infects various tissues and organs in addition to salivary glands, causing meningitis, pancreatitis, orchitis, oophoritis, mastitis, encephalitis, and deafness in some cases (1).
Paramyxoviruses, which include clinically important human and animal pathogens, such as MuV, measles virus (MeV), parainfluenza viruses (PIVs), and Newcastle disease virus (NDV), have two envelope glycoproteins, an attachment protein and a fusion (F) protein. The attachment proteins are referred to as the hemagglutinin (H), hemagglutinin-neuraminidase (HN), or glycoprotein (G), depending on the virus (2). The MuV attachment protein is the HN protein (because it has both hemagglutinating and neuraminidase activities) and binds glycan receptors like other paramyxovirus HN proteins. The MuV HN protein (MuV-HN) consists of the N-terminal cytoplasmic tail, transmembrane region, stalk region, and C-terminal head domain. The head domain is responsible for receptor binding, whereas the helix-bundled stalk region is likely involved in the interaction with the F protein (2, 3). Recently, we demonstrated that the trisaccharide containing α2,3-linked sialic acid, rather than the terminal sialic acid alone, acts as a receptor for MuV (4, 5).
Although the detailed molecular mechanism of paramyxovirus cell entry remains to be understood, the binding of the attachment protein to a cellular receptor is thought to trigger the activation of the F protein, causing fusion of the viral envelope with the plasma membrane and allowing the viral genome to enter the host cell (2). The crystal structures of the HN protein head domain have been determined for NDV, human PIV3 (hPIV3), PIV5, and MuV (4, 6–8). They exhibit a six-bladed β-propeller fold and form a homodimer. Two dimers in turn form a tetramer (dimer of dimers). The crystal structure of the NDV-HN ectodomain exhibits a “4-heads down” conformation in which two of the four head domains interact with the tetrameric helix-bundled stalk (3). Conversely, the crystal structure of the PIV5 HN ectodomain complexed with a glycan receptor shows a “2-heads up/2-heads down” conformation (9). One pair of disulfide-linked two-head domains is in the up position, whereas another pair of two-head domains is in the down position with one head domain interacting with the stalk. These structural arrangements of paramyxovirus HN proteins, combined with functional studies of various mutant proteins, led to the proposal that the fusion process is initialed by the rearrangements of the HN head domains upon receptor binding, and the stalk domain plays an important role in the activation of paramyxovirus F proteins (10–15). The 4-heads down conformation of the NDV-HN is thought to prevent the stalk domain from interacting with the F protein, representing the form before receptor binding. The 2-heads up/2-heads down conformation may be the intermediate form, and the “4-heads up” conformation may represent the form after receptor binding (9, 16, 17). A mutagenesis study in PIV5 and NDV-HN proteins indicated that the loop region connecting the head and stalk domains facilitates the flexible swinging motion of the head domain, which is important for the activation of the F proteins (18). It has been shown that the head domain dimer (monomer-monomer) interface is also crucial for efficiently transmitting the fusion-triggering signal, upon receptor binding, to the stalk region in NDV, hPIV3, and Nipah virus attachment proteins (19–22).
The MeV H protein (MeV-H) head domain also shows a homodimer conformation, although the relative orientation of the two monomers forming the homodimer is highly tilted compared with other paramyxoviruses (23, 24). Importantly, the MeV-H head domain in complex with its receptor CD150 exhibits two forms of tetramers (24). Mutations in the putative dimer-dimer interface of the head domain in either form inhibited the ability of MeV-H to support membrane fusion without affecting its cell surface expression, receptor binding, and interaction with the F protein (25). Based on these findings, we proposed that the dimer-dimer interaction of the MeV-H head domain contributes to triggering membrane fusion and that conformational shift of head domain tetramers may play a role in this process (24, 25).
Recently, we reported high-resolution crystal structures of the MuV-HN head domain both in the unliganded form and in complex with a glycan receptor analogue, 3′-sialyllactose (3′-SL) (4). Examination of the structures led us to notice the presence of a sulfate ion at the interface between two dimers comprising a tetramer, which is unique to the structure of the MuV-HN head domain and has not been observed in the attachment protein head domains of other paramyxoviruses. In this study, we examined whether the dimer-dimer interaction, aided by this anion at the interface, plays a role in triggering membrane fusion.
RESULTS
A sulfate ion located at the dimer-dimer interface of the MuV-HN head domain is a putative “linker” of two dimers.
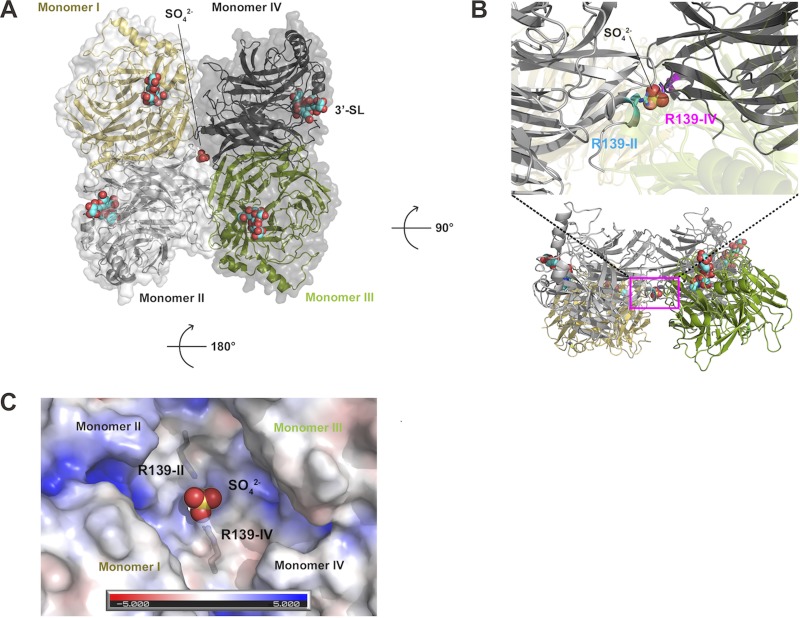
The crystal structure of the MuV-HN head domain forms a tetrameric “dimer of dimers” assembly (4). In view of our previous finding that the dimer-dimer interaction of the MeV-H head domain is involved in triggering membrane fusion (25), we closely examined the structure of the MuV-HN head domain (PDB accession no. 5B2D) and found a sulfate ion (SO42−) located at the interface between two dimers (Fig. 1A). The sulfate ion interacts with the side chain of the positively charged arginine residue at position 139 (R139) of one monomer each (monomer II and monomer IV) from both dimers (Fig. 1B and C). The ion appears to support the tetramer formation of the MuV-HN head domain by serving as a “linker” connecting two dimers.
FIG 1.
MuV-HN head domain tetramer. (A) Top view of MuV-HN head domain tetramer (dimer of dimers). Monomer I (yellow) and II (pale gray) form one dimer, whereas monomer III (green) and IV (dark gray) form another. The receptor analogue 3′-SL and anion SO42− are shown in spheres. (B) Side view of MuV-HN head domain tetramer. Magnified view of the dimer-dimer interface. The sulfate ion (SO42−) at the interface connects two dimers by interacting with the side chain of R139 from monomer II (R139-II) and monomer IV (Arg139-IV). (C) Electrostatic representation of the MuV-HN dimer-dimer interface seen from the stalk side. Red and blue surfaces indicate negatively and positively charged areas, respectively. Oxygen atom, red; sulfur atom, yellow.
Replacement of the residue interacting with the “linker” anion compromises the fusion support activity of MuV-HN.
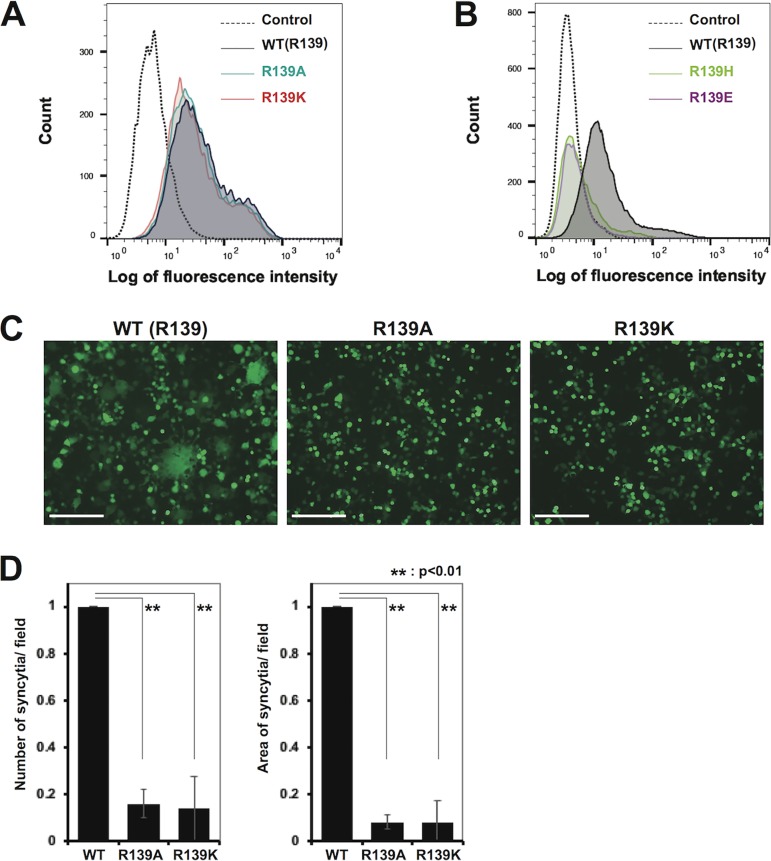
To examine whether the interaction between MuV-HN R139 and the anion at the dimer-dimer interface plays a role in triggering membrane fusion, the arginine residue was substituted by alanine (a noncharged residue), lysine (a positively charged residue), histidine (a positively charged residue), or glutamic acid (a negatively charged residue). HEK293 cells were transfected with the expression plasmid encoding the wild type (WT) or one of the mutant HN proteins (R139A, R139K, R139H, and R139E). Flow cytometric analysis showed that mutant HN proteins with R139A or R139K were expressed on the cell surface at comparable levels to the WT protein (Fig. 2A), whereas mutant HN proteins with R139H or R139E were hardly detected (Fig. 2B). Therefore, further studies were performed for the R139A and R139K mutants. HEK293 cells were transfected with the expression plasmid encoding the WT or one of the mutant HN proteins (R139A and R139K), together with expression plasmids respectively encoding the MuV F protein and enhanced green fluorescent protein (EGFP), and examined at 2 days posttransfection. The mutant HN proteins did not support syncytium formation as efficiently as the WT protein (Fig. 2B). Both the number and the size (total area) of syncytia in cells transfected with the mutant HN proteins were strongly reduced (∼10% or less of those in cells transfected with the WT protein) (Fig. 2C).
FIG 2.
Cell surface expression and fusion support activity of WT and mutant HN proteins. (A, B) Cell surface expression of the WT (black) and mutant ([A] R139A, green; R139K, pink and [B] R139H, light green; R139E, purple) HN proteins on transfected HEK293 cells was examined by flow cytometry. The empty plasmid served as the control (dotted line). (C) HEK293 cells transfected with the expression plasmids respectively encoding the HN protein (WT or mutant), F protein, and EGFP were observed for syncytia formation under a fluorescence microscope at 2 days after transfection (scale bar, 200 μm). (D) The average number (left) and area (right) of syncytia in the assay described in panel C were quantified by using the hybrid cell count system (Keyence). The value obtained with the WT HN protein is set to 1, and the data indicate the mean ± standard deviation (SD) of 5 randomly selected fields. **, P < 0.01, two-tailed Student's t test.
The binding abilities of the MuV-HN mutants to receptor and the F protein are intact.
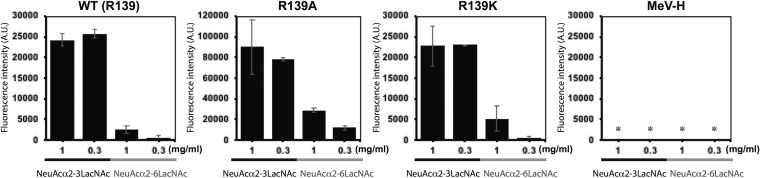
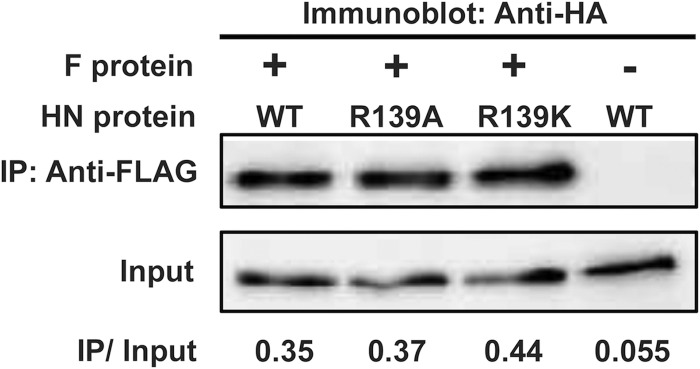
We also evaluated the abilities of the two MuV-HN mutants to bind receptor by using purified HN protein head domains (amino acid residues 96 to 582) and glycoconjugate-spotted slides. The biologically relevant glycan receptors for MuV have been reported recently (4, 5). Based on those findings, Neu5Acα2,3Galβ1,4GlcNAc and Neu5Acα2,6Galβ1,4GlcNAc were used as receptor and nonreceptor glycans on the slides, respectively. As expected from the location of position 139 in MuV-HN, which is far away from the receptor-binding pocket (Fig. 1), both mutant HN proteins could preferentially and efficiently bind Neu5Acα2,3Galβ1,4GlcNAc, like the WT protein (Fig. 3). The head domain of MeV-H, which uses proteinaceous receptors (24), bound neither of the glycans (Fig. 3, lower right). We then examined the abilities of the mutant HN proteins to interact with the F protein. HEK293 cells were transfected with the expression plasmid encoding the WT HN protein or one of the mutants, together with the expression plasmid encoding the F protein. The mutant HN proteins was coprecipitated with the F protein as efficiently as the WT protein (Fig. 4), indicating that the R139A or R139K substitution does not affect the binding ability of the HN protein with the F protein.
FIG 3.
Binding of MuV-HN head domains to sialylglycans. Binding of purified WT MuV-HN head domain, mutant MuV-HN head domains (R139A and R139K), and purified MeV-H head domain to sialylglycans (Neu5Acα2,3Galβ1,4GlcNAc or Neu5Acα2,6Galβ1,4GlcNAc) were examined. These glycans were attached to BSA as a substrate and spotted onto glass slides at a concentration of 1 or 0.3 mg/ml. The fluorescence intensity from each spot was corrected by subtracting that of BSA as the background. Data are the mean ± SD of three samples. *, Below background levels.
FIG 4.
Coimmunoprecipitation of MuV-HN with the F protein. WT and mutant HN proteins were HA tagged, whereas the F protein was FLAG tagged. HEK293 cell transiently expressing HN and F proteins were lysed and immunoprecipitated (IP) with anti-FLAG Ab. The signal intensities of IP and corresponding whole-cell lysate (Input) samples on the membrane as detected with anti-HA Ab were quantified by using the VersaDoc 3000 imager. The ratio of the intensity of the IP sample to that of Input is indicated below.
Blue native-PAGE analysis of MuV-HN.
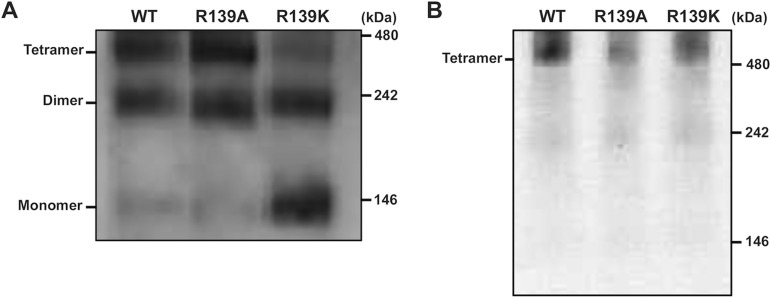
To examine whether the MuV-HN head domain alone could form tetramers (dimer of dimers) under physiological conditions, and if so, whether R139 contributes to the tetramer formation, we performed blue native-PAGE analysis by using purified head domains from the WT and mutant HN proteins. The head domain proteins as well as the anode and cathode buffers for electrophoresis were prepared in the phosphate-buffered saline (PBS) solution so that the experimental conditions would be close to physiological ones. The MuV-HN head domain exhibited three discrete bands migrating approximately at 400, 200, and 100 kDa on blue native-PAGE (Fig. 5A) like the MeV-H head domain previously studied in the same manner (25). Thus, we assumed that the three bands represent tetramers, dimers, and monomers of the MuV-HN head domain, respectively. In the WT and R139A mutant proteins, tetramers and dimers were predominant forms whereas in the R139K mutant protein, tetramers were reduced and monomers greatly increased in proportion (Fig. 5A). By contrast, when the full-length WT and mutant HN proteins transiently expressed in cells were solubilized and examined on blue native-PAGE, all of them predominantly formed tetramers (Fig. 5B) like other paramyxovirus attachment proteins (3). The results indicate that not only the full-length HN protein but the head domain alone is also capable of forming tetramers, to which R139 contributes.
FIG 5.
Blue native-PAGE and immunoblot analysis of WT and mutant HN proteins. (A) Blue native-PAGE analysis of purified WT and mutant HN protein head domains. These proteins were His tagged and subjected to immunoblot analysis with anti-His Ab. (B) Blue native-PAGE analysis of full length HN proteins. HEK293S GnTI(−) cells were transiently transfected with the expression plasmid encoding HA-tagged WT and mutant HN proteins. The cells were treated with 1% n-dodecyl-β-d-maltoside, and the lysates were subjected to blue native-PAGE and immunoblot analysis with anti-HA Ab.
DISCUSSION
In the crystal structure of the MuV-HN head domain, we found an anion SO42− at the interface between two dimers comprising a tetramer. The ion, captured by a positively charged arginine residue on one monomer each from both dimers, presumably connects the two dimers. Although SO42− is present at the dimer-dimer interface in the crystal structure (the crystallization buffer used contains ammonium sulfate), a hydrogen phosphate ion (HPO42−), a more abundant divalent anion in the body fluid, may take the place of the sulfate ion under physiological conditions. Substitution of alanine or lysine for the arginine residue capturing the anion compromised the fusion support activity of MuV-HN without reducing its cell surface expression, glycan-receptor binding, and interaction with the F protein. Thus, the dimer-dimer interaction within the head domain tetramer appears to be essential for triggering the F protein for membrane fusion. Conversely, histidine and glutamic acid substitutions compromised the cell surface expression of the MuV-HN protein (Fig. 2B). The property of the residue at position 139 may affect either intracellular trafficking of the HN protein or its stability on the cell surface. A divalent anion at the dimer-dimer interface is observed only in the MuV-HN head domain tetramer but not in the HN tetramers of hPIV3, PIV5, and NDV (3, 7, 8). Interestingly, a Ca2+ ion is found at the common site within each HN monomer of hPIV3, PIV5, and NDV but not MuV, and it is reported that Ca2+ is required for neuraminidase activity in NDV (26). An SO42− ion is present near the HN monomer-monomer interface of hPIV3 (7). The divalent cation and anion observed in HN proteins of these paramyxoviruses may also play roles in their life cycle.
This is reminiscent of our previous finding with MeV-H, in which substitutions at the dimer-dimer interface of the head domain inhibited the ability of MeV-H to support membrane fusion without affecting its other functions (25). In the MeV study, amino acid substitutions were introduced to disrupt interactions between residues on different protomers (one monomer each from both dimers comprising a tetramer), whereas in the present study, interactions of residues on MuV-HN with an anion located at the dimer-dimer interface were disrupted. In either case, the proper interaction between two dimers comprising a tetramer would be affected by substitutions. MuV and MeV belong to different genera of the family Paramyxoviridae (the genus Rubulavirus versus the genus Morbillivirus), have different attachment proteins (the HN protein versus the H protein), and utilize different types of cellular receptors (glycans versus proteins). Furthermore, their attachment proteins interact differently with receptors (via the top pocket versus the lateral surface of the head domain) (4, 24, 27, 28). The finding that the dimer-dimer interaction is important for triggering membrane fusion in these two viruses with such differences may suggest that this rule applies to paramyxoviruses in general.
In blue native-PAGE analysis of the MeV-H head domain, dimers were much more abundant than tetramers (25), consistent with the crystal structure of the NDV-HN ectodomain (the head domain plus stalk region), in which the head domain dimers do not associate with each other to form tetramers (3). The MuV-HN head domain is different from MeV and NDV attachment protein head domains in that tetramers were formed as abundantly as dimers. The anion located at the dimer-dimer interface may facilitate and/or stabilize tetramer formation of the MuV-HN head domain by acting as a linker for two dimers.
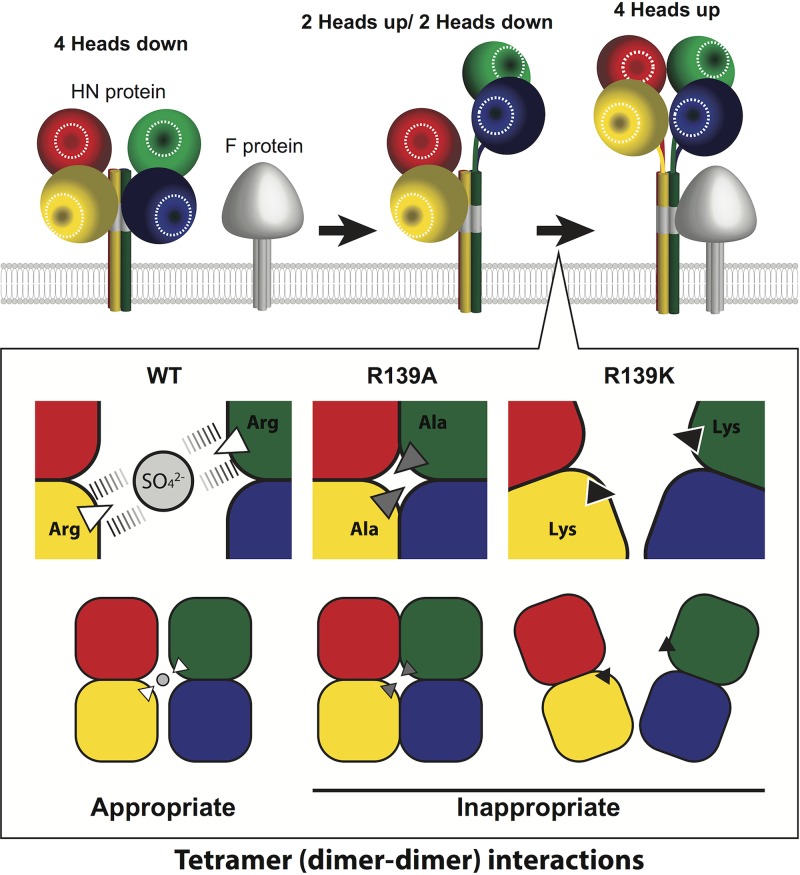
However, the R139A mutant is still capable of forming tetramers like the WT head domain, suggesting that the anion at the interface is not the only driving force for tetramer formation. It is possible that the R139A mutant makes an aberrant (non-fusion-triggering) form of tetramer through the interaction of alanine with other hydrophobic residues (Fig. 6). By contrast, the R139K mutant hardly exhibits the tetramer form. The physical and chemical properties of lysine may not be appropriate to capture the anion at the interface, unlike those of arginine. The side chains of R139 from both dimers capture the anion through quadruple electrostatic interactions in the crystal structure (Fig. 6), whereas those of lysine could, if at all, form only double electrostatic interactions with the anion (one interaction each by a single lysine residue). Additionally, the slightly longer side chain of arginine, as compared with that of lysine, may be crucial for the proper spacing and accommodation of the anion at the interface. Thus, even in the presence of mutations in the head domain, the full-length MuV-HN may exist as a tetramer because of the tetrameric helical bundle-forming stalk region (3, 29) (Fig. 5B), but the proper conformation of MuV-HN head domain tetramers, not the mere formation of tetramers, must be essential for triggering membrane fusion (Fig. 6).
FIG 6.
A model for tetrameric (dimer of dimers) conformations of WT and mutant MuV-HN head domains. (Top) A schematic illustration for rearrangements of paramyxovirus HN proteins in membrane fusion triggering. The receptor-binding sites in HN proteins are labeled with white circles. The “4-heads down” conformation is thought to be a receptor-free, inactive form in which HN protein head domains mask the F interaction site in the stalk region (gray). The “2-heads up/2 heads down” conformation is thought to be an intermediate state, and the “4-heads up” conformation is most likely an active form that can trigger the structural change of the F protein by exposing the F interaction site. (Bottom) In MuV, the dimer-dimer interaction of the MuV-HN head domain through the appropriate tetrameric conformation, driven by the proper spacing and accommodation of the anion at the interface, is essential for triggering membrane fusion.
Based on the structures of several paramyxovirus HN proteins, models have been proposed for fusion triggering (2, 16, 17, 19, 21, 30–33). Upon receptor binding, the receptor-unbound “4-heads down” conformation may change to the receptor-bound “4-heads up” conformation via the potential intermediate “2-heads up/2-heads down” conformation (Fig. 6). Tetramerization of the HN head domain presumably representing the 4-heads up conformation may allow the F protein to make contact with the HN stalk domain (12). A possible mechanism for the reduced fusion support activity of the R139A and R139K mutants is that they are unable to form or maintain the 4-heads up conformation due to the lack of appropriate connection between two dimers (Fig. 6). Since the complete ectodomain structure of MuV-HN remains unknown, we cannot exclude the possibility that the R139 mutants affect fusion triggering through other mechanisms. Nevertheless, it is certain that MuV-HN R139 plays an important role in forming the proper head domain tetramer conformation and triggering the F protein for membrane fusion. The role of the dimer (monomer-monomer) interface in fusion triggering has also been reported in NDV, hPIV3, MeV, and Nipah virus attachment proteins (19–22, 34). In these studies, fusion-triggering conformational changes were shown to be relayed from the receptor-binding domain of the attachment proteins to the stalk through the monomer-monomer interface. Thus, dynamic structural changes of the paramyxovirus attachment protein, including those at monomer-monomer and dimer-dimer interfaces, may be required for the activation of the F protein.
Since the region around R139 is conserved among different genotypes, it may serve as a good target for antiviral drugs. Interestingly, a monoclonal antibody (Ab) 1648 raised against the MuV Urabe strain (genotype B) was found to exhibit neutralizing activity against multiple genotypes (35), including genotype A possessing a unique antigenicity among MuV genotypes (36, 37). An escape mutant MuV from Ab 1648 had the L to P substitution at position 138 in MuV-HN (35), which is located at the interface between two dimers in our crystal structure of MuV-HN. Thus, it is possible that Ab 1648 exhibits its neutralizing activity by directly binding to the interface between two dimers, thereby masking R139 and inhibiting the tetramer formation. Alternatively, a local structural change caused by the L to P substitution might indirectly prevent the access of the Ab.
Although many models have been proposed (9, 12, 16, 17, 19–22, 24, 25, 30–34, 38–41), how receptor engagement by the paramyxovirus attachment protein leads to the activation of the F protein remains to be determined. Our results with MeV and MuV strongly suggest that the head domain dimer-dimer interaction within the tetramer is of critical importance for the conformational changes of the attachment protein required for the refolding of the F protein and eventual membrane fusion.
MATERIALS AND METHODS
Construction of expression plasmids.
The DNA fragments encoding the full-length HN and F proteins were amplified by PCR from the template plasmids of the MuV Hoshino strain (GenBank accession no. AB470486) and cloned into the expression plasmid pCA7 (42). The mutant DNA fragments encoding MuV-HN with alanine or lysine substitution at amino acid position 139 were generated by PCR-based mutagenesis. The WT and mutant HN fragments were amplified with the reverse primer containing the influenza virus HA tag sequence so that the expressed HN proteins contain the HA tag at their C terminus. The expression plasmids encoding the F protein and HA-tagged HN protein were used for the fusion assay. The expression plasmids encoding the HA-tagged HN protein were also used for blue native-PAGE analysis. The F protein was also amplified by PCR with the reverse primer containing the FLAG tag, and the expression plasmid encoding the FLAG-tagged F protein was used for coimmunoprecipitation. The expression plasmid encoding the MuV-HN head domain (amino acid position 96 to 582) was constructed in the vector pHLsec containing the N-terminal secretion signal sequence and the C-terminal His6 tag sequence as described previously (4). For the glycan-binding assay, the expression plasmids encoding the HN protein head domain and its mutants were produced from the MuV SBL-1 strain sequence because this strain shows stronger affinity to glycans with binding specificity similar to that of the Hoshino strain (4). The expression plasmid encoding the MeV-H head domain was described previously (4).
Fusion assay.
HEK293 cells in a 12-well dish (Corning) were replenished with Opti-MEM serum-free medium (Gibco) and then transfected with 0.4 μg, 1.0 μg, and 0.5 μg of the expression plasmids encoding the HN, F, and EGFP proteins, respectively. The culture medium was changed to Dulbecco's modified Eagle medium (DMEM) (Wako) supplemented with 10% (vol/vol) fetal bovine serum (FBS) (Sigma-Aldrich) and penicillin-streptomycin (Gibco) 5 h later. The cells were observed under fluorescence microscopy 48 h posttransfection. Syncytia were quantified by using the BZ-X analyzer (Keyence). Six fields were photographed for each sample under a microscope, and the number and the total area of syncytia in each image were quantified by setting the threshold of contiguous EGFP-positive area at 3,413.7 μm2, which defines syncytia.
Flow cytometric analysis.
To examine the cell surface expression of the WT and mutant HN proteins, HEK293 cells cultured in a 12-well dish were transfected with 2 μg of the expression plasmid encoding the HA-tagged HN protein or the empty vector by using Lipofectamine LTX and PLUS reagent (Invitrogen). At 48 h posttransfection, the cells were washed with PBS(−) (without Ca2+ and Mg2+) and detached from the dish with 2 mM EDTA/PBS(−). The cells were incubated with the first Ab (Y-11, anti-HA tag Ab [Santa Cruz Biotechnology]) in 1% FBS/PBS(−) for 90 min at 4°C, then washed with PBS(−), and incubated with the second Ab (anti-rabbit IgG fluorescein isothiocyanate conjugate; Sigma-Aldrich) in 1% FBS/PBS(−) for 45 min at 4°C. After washing with PBS(−), the cells were analyzed on a FACSCalibur flow cytometer (BD Biosciences).
Protein expression and purification.
The expression and purification of the MuV-HN head domain (amino acid position 96 to 582) was described previously (4). Briefly, the expression plasmids encoding WT and mutant HN proteins were individually transfected into HEK293S cells lacking N-acetylglucosaminyltransferase I [HEK293S GnTI(−) cells] (43). The secreted MuV-HN proteins were collected from supernatants at 6 days posttransfection. They were purified using a Ni2+-nitrilotriacetic acid (NTA) affinity column (Cosmogel His-Accept; Nacalai Tesque) and subsequent gel filtration was performed with a Superdex 200 GL 10/300 column (GE Healthcare). MuV-HN head domain proteins were adjusted to the optimum concentrations by using Amicon Ultra centrifugal filters (Merck Millipore). The MeV-H head domain protein was expressed and purified with the same procedures.
Glycan-binding assay.
Purified WT and mutant MuV-HN head domains (amino acid position 96 to 582) and the MeV-H head domain (amino acid positions 149 to 617) were diluted at a concentration of 200 μg/ml in the binding buffer (Rexxam). They were incubated with the glycoconjugate-spotted Bio-Rex epoxy-coated glass slide (Rexxam) for 120 min at room temperature with gentle shaking. Bovine serum albumin (BSA)-mounted spots were also prepared on each slide to evaluate the background binding. After removal of the protein solution, anti-His6 tag cy3-conjugated Ab (Rockland) diluted at a concentration of 2.5 ng/ml in the binding buffer reacted with the glass slide for 120 min at room temperature with gentle shaking. The fluorescence was detected with a Bio-Rex Scan 200 scanner at 1 min of the exposure time (Rexxam) and analyzed by using a BZ-X analyzer (Keyence).
Coimmunoprecipitation.
HEK293 cells prepared in a 6-well dish (Corning) were transfected using Lipofectamine LTX and PLUS reagent, with the expression plasmid encoding the FLAG-tagged F protein and that encoding the WT or mutant HN protein. In the control well, the cells were transfected with the expression plasmid encoding the WT HN protein and the empty vector. At 24 h posttransfection, the cells were lysed in the immunoprecipitation buffer (25 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 5% glycerol) containing protease inhibitors (Sigma-Aldrich) followed by centrifugation. One twentieth of each supernatant was reserved as a cell lysate (input) sample. The rest of the supernatant was incubated with anti-FLAG monoclonal Ab M2 (Sigma-Aldrich) and protein A-Sepharose (GE Healthcare) at 4°C with gentle stirring for 24 h. The immunoprecipitated samples were centrifuged and washed six times with the immunoprecipitation buffer. They were subjected, together with the input samples, to SDS-PAGE under a reducing condition and immunoblotted with anti-HA tag mouse monoclonal Ab (Wako).
Blue native-PAGE and immunoblot analysis.
Purified WT and mutant MuV-HN head domains (1 μg each) were treated with the NativePAGE sample buffer (Invitrogen) containing 1× PBS(−) solution and Coomassie brilliant blue G-250 and separated on a 4 to 16% NativePAGE bis-Tris gel in the anode and cathode buffers, which also contained 1× PBS(−) solution. The gel was transferred onto a membrane and decolored with 100% methanol. After washing with Tris-buffered saline with Tween 20 (TBST) (20 mM Tris-HCl, pH 7.4, 137.5 mM NaCl, 0.05% Tween 20), the membrane was blocked with 2.5% fat-free milk and incubated with the first Ab (anti-His rabbit polyclonal Ab [MBL]) at 4°C overnight. The membrane was washed with TBST and incubated with the second Ab (peroxidase-conjugated goat anti-rabbit Ab; Jackson ImmunoResearch Laboratories, Inc.) at room temperature for 60 min, followed by washing with TBST. The membrane was treated with Chemi-Lumi One Super (Nacalai Tesque), and the chemiluminescent signals were detected using a VersaDoc 3000 imager (Bio-Rad). HEK293S GnTI(−) cells (43), cultured in a 12-well dish, were transfected with the expression plasmid encoding the full length WT or mutant MuV-HN by using Lipofectamine LTX and PLUS reagent. HEK293S GnTI(−) cells lack GnTI(−) activity so that the glycosylation patterns of glycoproteins in them are simpler compared with those produced in normal cells. At 48 h posttransfection, the cells were harvested and lysed with NativePAGE sample buffer containing Coomassie brilliant blue G-250, 1% n-dodecyl-β-d-maltoside, and protease inhibitors. These samples were treated and detected using the same procedures described above. For immunoblotting, anti-HA mouse monoclonal Ab (Wako) and peroxidase-conjugated goat anti-mouse Ab (Santa Cruz Biotechnology) were respectively used as the 1st and the 2nd Abs.
ACKNOWLEDGMENTS
We thank N. Kurisaki for technical help.
This study was supported by a Grant-in-Aid for Scientific Research on Innovative Areas from the Ministry of Education, Culture, Science, Sports, and Technology (MEXT) of Japan (24115005 and 17H05820) (to Y.Y.), by the Research Program on Emerging and Re-emerging Infectious Diseases from AMED (JP18fk0108014h) (to T.H.), by AMED J-PRIDE (JP19fm0208022h) (to T.H.), by the Takeda Science Foundation (to T.H.), by the Terumo Foundation for Life Sciences and Arts (to T.H.), and by the Japan Society for the Promotion of Science (Postdoctoral Fellowship DC2) (to M.K.).
REFERENCES
- 1.Rubin SA, Sauder CJ, Carbone KM. 2013. Mumps virus, p 1024–1041. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed, vol I Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Lamb RA, Parks GD. 2013. Paramyxoviridae, p 957–995. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed, vol I Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 3.Yuan P, Swanson KA, Leser GP, Paterson RG, Lamb RA, Jardetzky TS. 2011. Structure of the Newcastle disease virus hemagglutinin-neuraminidase (HN) ectodomain reveals a four-helix bundle stalk. Proc Natl Acad Sci U S A 108:14920–14925. doi: 10.1073/pnas.1111691108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kubota M, Takeuchi K, Watanabe S, Ohno S, Matsuoka R, Kohda D, Nakakita SI, Hiramatsu H, Suzuki Y, Nakayama T, Terada T, Shimizu K, Shimizu N, Shiroishi M, Yanagi Y, Hashiguchi T. 2016. Trisaccharide containing alpha2,3-linked sialic acid is a receptor for mumps virus. Proc Natl Acad Sci U S A 113:11579–11584. doi: 10.1073/pnas.1608383113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kubota M, Matsuoka R, Suzuki T, Yonekura K, Yanagi Y, Hashiguchi T. 2019. Molecular mechanism of the flexible glycan receptor recognition by mumps virus. J Virol 93:e00344-19. doi: 10.1128/JVI.00344-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crennell S, Takimoto T, Portner A, Taylor G. 2000. Crystal structure of the multifunctional paramyxovirus hemagglutinin-neuraminidase. Nat Struct Biol 7:1068–1074. doi: 10.1038/81002. [DOI] [PubMed] [Google Scholar]
- 7.Lawrence MC, Borg NA, Streltsov VA, Pilling PA, Epa VC, Varghese JN, McKimm-Breschkin JL, Colman PM. 2004. Structure of the haemagglutinin-neuraminidase from human parainfluenza virus type III. J Mol Biol 335:1343–1357. doi: 10.1016/j.jmb.2003.11.032. [DOI] [PubMed] [Google Scholar]
- 8.Yuan P, Thompson TB, Wurzburg B, Paterson RG, Lamb R, Jardetzky TS. 2005. Structural studies of the parainfluenza virus 5 hemagglutinin-neuraminidase tetramer in complex with its receptor, sialyllactose. Structure 13:803–815. doi: 10.1016/j.str.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 9.Welch BD, Yuan P, Bose S, Kors CA, Lamb RA, Jardetzky TS. 2013. Structure of the parainfluenza virus 5 (PIV5) hemagglutinin-neuraminidase (HN) ectodomain. PLoS Pathog 9:e1003534. doi: 10.1371/journal.ppat.1003534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee JK, Prussia A, Paal T, White LK, Snyder JP, Plemper RK. 2008. Functional interaction between paramyxovirus fusion and attachment proteins. J Biol Chem 283:16561–16572. doi: 10.1074/jbc.M801018200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mirza AM, Aguilar HC, Zhu Q, Mahon PJ, Rota PA, Lee B, Iorio RM. 2011. Triggering of the Newcastle disease virus fusion protein by a chimeric attachment protein that binds to Nipah virus receptors. J Biol Chem 286:17851–17860. doi: 10.1074/jbc.M111.233965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bose S, Zokarkar A, Welch BD, Leser GP, Jardetzky TS, Lamb RA. 2012. Fusion activation by a headless parainfluenza virus 5 hemagglutinin-neuraminidase stalk suggests a modular mechanism for triggering. Proc Natl Acad Sci U S A 109:E2625–E2634. doi: 10.1073/pnas.1213813109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brindley MA, Suter R, Schestak I, Kiss G, Wright ER, Plemper RK. 2013. A stabilized headless measles virus attachment protein stalk efficiently triggers membrane fusion. J Virol 87:11693–11703. doi: 10.1128/JVI.01945-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Talekar A, Moscona A, Porotto M. 2013. Measles virus fusion machinery activated by sialic acid binding globular domain. J Virol 87:13619–13627. doi: 10.1128/JVI.02256-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ader-Ebert N, Khosravi M, Herren M, Avila M, Alves L, Bringolf F, Orvell C, Langedijk JP, Zurbriggen A, Plemper RK, Plattet P. 2015. Sequential conformational changes in the morbillivirus attachment protein initiate the membrane fusion process. PLoS Pathog 11:e1004880. doi: 10.1371/journal.ppat.1004880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bose S, Jardetzky TS, Lamb RA. 2015. Timing is everything: fine-tuned molecular machines orchestrate paramyxovirus entry. Virology 479–480:518–531. doi: 10.1016/j.virol.2015.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jardetzky TS, Lamb RA. 2014. Activation of paramyxovirus membrane fusion and virus entry. Curr Opin Virol 5:24–33. doi: 10.1016/j.coviro.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adu-Gyamfi E, Kim LS, Jardetzky TS, Lamb RA. 2016. Flexibility of the head-stalk linker domain of paramyxovirus HN glycoprotein is essential for triggering virus fusion. J Virol 90:9172–9181. doi: 10.1128/JVI.01187-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Porotto M, Salah Z, DeVito I, Talekar A, Palmer SG, Xu R, Wilson IA, Moscona A. 2012. The second receptor binding site of the globular head of the Newcastle disease virus hemagglutinin-neuraminidase activates the stalk of multiple paramyxovirus receptor binding proteins to trigger fusion. J Virol 86:5730–5741. doi: 10.1128/JVI.06793-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Porotto M, Palmer SG, Palermo LM, Moscona A. 2012. Mechanism of fusion triggering by human parainfluenza virus type III: communication between viral glycoproteins during entry. J Biol Chem 287:778–793. doi: 10.1074/jbc.M111.298059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu R, Palmer SG, Porotto M, Palermo LM, Niewiesk S, Wilson IA, Moscona A. 2013. Interaction between the hemagglutinin-neuraminidase and fusion glycoproteins of human parainfluenza virus type III regulates viral growth in vivo. mBio 4:e00803-13. doi: 10.1128/mBio.00803-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iketani S, Shean RC, Ferren M, Makhsous N, Aquino DB, Des Georges A, Rima B, Mathieu C, Porotto M, Moscona A, Greninger AL. 2018. Viral entry properties required for fitness in humans are lost through rapid genomic change during viral isolation. mBio 9:e00898-18. doi: 10.1128/mBio.00898-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hashiguchi T, Kajikawa M, Maita N, Takeda M, Kuroki K, Sasaki K, Kohda D, Yanagi Y, Maenaka K. 2007. Crystal structure of measles virus hemagglutinin provides insight into effective vaccines. Proc Natl Acad Sci U S A 104:19535–19540. doi: 10.1073/pnas.0707830104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hashiguchi T, Ose T, Kubota M, Maita N, Kamishikiryo J, Maenaka K, Yanagi Y. 2011. Structure of the measles virus hemagglutinin bound to its cellular receptor SLAM. Nat Struct Mol Biol 18:135–141. doi: 10.1038/nsmb.1969. [DOI] [PubMed] [Google Scholar]
- 25.Nakashima M, Shirogane Y, Hashiguchi T, Yanagi Y. 2013. Mutations in the putative dimer-dimer interfaces of the measles virus hemagglutinin head domain affect membrane fusion triggering. J Biol Chem 288:8085–8091. doi: 10.1074/jbc.M112.427609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Munoz Barroso I, Moralejo FJ, Villar E. 1994. Ionic dependence of the sialidase activity of hemagglutinin-neuraminidase glycoprotein in Newcastle disease virus membrane. Biochem Soc Trans 22:366S. doi: 10.1042/bst022366s. [DOI] [PubMed] [Google Scholar]
- 27.Zhang X, Lu G, Qi J, Li Y, He Y, Xu X, Shi J, Zhang CW, Yan J, Gao GF. 2013. Structure of measles virus hemagglutinin bound to its epithelial receptor nectin-4. Nat Struct Mol Biol 20:67–72. doi: 10.1038/nsmb.2432. [DOI] [PubMed] [Google Scholar]
- 28.Santiago C, Celma ML, Stehle T, Casasnovas JM. 2010. Structure of the measles virus hemagglutinin bound to the CD46 receptor. Nat Struct Mol Biol 17:124–129. doi: 10.1038/nsmb.1726. [DOI] [PubMed] [Google Scholar]
- 29.Yuan P, Leser GP, Demeler B, Lamb RA, Jardetzky TS. 2008. Domain architecture and oligomerization properties of the paramyxovirus PIV 5 hemagglutinin-neuraminidase (HN) protein. Virology 378:282–291. doi: 10.1016/j.virol.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iorio RM, Mahon PJ. 2008. Paramyxoviruses: different receptors - different mechanisms of fusion. Trends Microbiol 16:135–137. doi: 10.1016/j.tim.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Plattet P, Plemper RK. 2013. Envelope protein dynamics in paramyxovirus entry. mBio 4: e00413-13. doi: 10.1128/mBio.00413-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bossart KN, Fusco DL, Broder CC. 2013. Paramyxovirus entry. Adv Exp Med Biol 790:95–127. doi: 10.1007/978-1-4614-7651-1_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Plattet P, Alves L, Herren M, Aguilar HC. 2016. Measles virus fusion protein: structure, function and inhibition. Viruses 8:112. doi: 10.3390/v8040112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Navaratnarajah CK, Oezguen N, Rupp L, Kay L, Leonard VH, Braun W, Cattaneo R. 2011. The heads of the measles virus attachment protein move to transmit the fusion-triggering signal. Nat Struct Mol Biol 18:128–134. doi: 10.1038/nsmb.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yates PJ, Afzal MA, Minor PD. 1996. Antigenic and genetic variation of the HN protein of mumps virus strains. J Gen Virol 77:2491–2497. doi: 10.1099/0022-1317-77-10-2491. [DOI] [PubMed] [Google Scholar]
- 36.Inou Y, Nakayama T, Yoshida N, Uejima H, Yuri K, Kamada M, Kumagai T, Sakiyama H, Miyata A, Ochiai H, Ihara T, Okafuji T, Okafuji T, Nagai T, Suzuki E, Shimomura K, Ito Y, Miyazaki C. 2004. Molecular epidemiology of mumps virus in Japan and proposal of two new genotypes. J Med Virol 73:97–104. doi: 10.1002/jmv.20065. [DOI] [PubMed] [Google Scholar]
- 37.Örvell C, Alsheikhly AR, Kalantari M, Johansson B. 1997. Characterization of genotype-specific epitopes of the HN protein of mumps virus. J Gen Virol 78:3187–3193. doi: 10.1099/0022-1317-78-12-3187. [DOI] [PubMed] [Google Scholar]
- 38.Plemper RK, Hammond AL, Cattaneo R. 2001. Measles virus envelope glycoproteins hetero-oligomerize in the endoplasmic reticulum. J Biol Chem 276:44239–44246. doi: 10.1074/jbc.M105967200. [DOI] [PubMed] [Google Scholar]
- 39.Plemper RK, Hammond AL, Gerlier D, Fielding AK, Cattaneo R. 2002. Strength of envelope protein interaction modulates cytopathicity of measles virus. J Virol 76:5051–5061. doi: 10.1128/jvi.76.10.5051-5061.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Connolly SA, Leser GP, Jardetzky TS, Lamb RA. 2009. Bimolecular complementation of paramyxovirus fusion and hemagglutinin-neuraminidase proteins enhances fusion: implications for the mechanism of fusion triggering. J Virol 83:10857–10868. doi: 10.1128/JVI.01191-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hashiguchi T, Maenaka K, Yanagi Y. 2011. Measles virus hemagglutinin: structural insights into cell entry and measles vaccine. Front Microbiol 2:247. doi: 10.3389/fmicb.2011.00247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takeda M, Ohno S, Seki F, Nakatsu Y, Tahara M, Yanagi Y. 2005. Long untranslated regions of the measles virus M and F genes control virus replication and cytopathogenicity. J Virol 79:14346–14354. doi: 10.1128/JVI.79.22.14346-14354.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reeves PJ, Callewaert N, Contreras R, Khorana HG. 2002. Structure and function in rhodopsin: high-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc Natl Acad Sci U S A 99:13419–13424. doi: 10.1073/pnas.212519299. [DOI] [PMC free article] [PubMed] [Google Scholar]